Abstract
Purpose
The optic nerve (ON) represents the sole pathway between the eyes and brain; consequently, diseases of the ON can have dramatic effects on vision. However, qMT applications in the ON have been limited to ex vivo studies, in part due to the fatty connective tissue, which surrounds the ON, confounding the MT experiment. Therefore, the aim of this study was to implement a multi-echo Dixon fat-water separation approach to remove the fat component from MT images.
Methods
MT measurements were taken in a single slice of the ON and frontal lobe using a three-echo Dixon readout, and the water and out-of-phase images were applied to a two-pool model in ON tissue and brain white matter to evaluate the effectiveness of using Dixon fat-water separation to remove fatty tissue from MT images.
Results
White matter data showed no significant differences between image types, however, there was a significant increase (p < 0.05) in variation in the out-of-phase images in the ON relative to the water images.
Conclusions
The results of this study demonstrate that Dixon fat-water separation can be robustly utilized for accurate MT quantification of anatomies susceptible to partial volume effects due to fat.
Keywords: Dixon Method, Fat-Water Separation, qMT, quantitative magnetization transfer, optic nerve
Introduction
The optic nerve (ON) is responsible for mediating visual information from the eyes to the optic chiasm and represents the sole pathway between the eyes and brain. The integrity of these nerves is imperative in maintaining visual function; consequently, diseases of the ON can have dramatic effects on daily function. One significant challenge in the management of patients with optic neuropathies (such as glaucoma, optic neuritis) has been the lack of robust, quantitative tools capable of characterizing the underlying pathophysiology. For example, conventional MRI scan contrasts, e.g. spin-density, T1- and T2-weighted, currently do not provide significant diagnostic (3) or prognostic value over conventional ophthalmologic exams in optic neuropathies.
Magnetization transfer (MT) imaging has emerged as an MRI technique capable of quantifying myelin density changes (4-6) brought about by neurodegenerative diseases affecting the brain, spinal cord, and peripheral nerves (7-16); it may therefore provide similar quantitative information in ON diseases. MT MRI exploits the fact that there are protons residing on immobile macromolecules in tissue in addition to the free water protons observed with conventional MRI (17). Conventional MRI cannot image these protons directly because their T2 relaxation times are too short (≈10 μs) to be captured by typical readout schemes. However, these macromolecular protons communicate with the surrounding water and can be indirectly imaged by exploiting this exchange, which is referred to as the MT effect.
The contrast in an MT experiment is generated via application of a radiofrequency (RF) irradiation pulse at an offset frequency with respect to water (Δω) to selectively saturate the spectrally broad macromolecular proton resonance. This saturation is transferred to the free water pool via MT, resulting in an observed signal attenuation. The MT effect is semi-quantitatively characterized via the magnetization transfer ratio (MTR), which has been shown to correlate with myelin content (6). Unfortunately, the MTR is also sensitive to pulse sequence design, RF transmit field (B1), and magnetic static field (B0) inhomogeneity (18), as well as by tissue relaxation times and other non-MT-specific NMR parameters (19,20), limiting researchers and clinicians from creating a standard MTR metric to define pathology. To overcome some of these limitations and derive indices that are directly reflective of MT phenomena, quantitative MT (qMT) has been developed and implemented in the brain and spinal cord (20-24). qMT typically requires images to be acquired at multiple RF irradiation powers and/or frequency offsets, generating a so-called MT z-spectrum for each voxel (25). The resulting z-spectrum can be fit to a two (or more)-pool model to estimate quantitative indices, such as the pool size ratio (PSR), defined as the macromolecular pool size divided by the free pool size, the MT exchange rate from the macromolecular pool to the free pool (kmf), and the transverse and longitudinal relaxation times for each pool (20,26). Often, the focus is on the PSR, as it has been shown to correlate well with white matter (WM) myelin density (6,27-29), and may offer a biomarker of demyelination and axonal loss in WM pathologies.
MT imaging of the ON, therefore, has the potential to report on microstructural changes preceding atrophy, potentially offering greater insight into changes that may persist after transient inflammation has subsided. Despite this promise, qMT applications in the ON have been limited to ex vivo studies (20,30), largely due to the technical challenges associated with quantitative imaging of the ON. First, qMT methods often require long scan times for accurate data fitting (31). The ON is approximately 3 mm in diameter (32), necessitating longer, higher-resolution scans for whole-nerve quantification. Short scan times are, however, often required for quantitative ON imaging, as eye fixation is advisable to reduce to impact of constant eye motion. Secondly, fatty connective tissue surrounds the ON and exhibits markedly different MT properties than nerve, requiring fat suppression techniques to accurately separate the nerve from the surrounding anatomy. For conventional MRI, spectral fat suppression techniques, such as spectral presaturation with inversion recovery (SPIR) (33) or spectral attenuated inversion recovery (SPAIR) (34), are typically employed. Unfortunately, these approaches are often incompatible with qMT imaging because they are performed with an off-resonance pulse that can affect MT quantification. Short TI inversion recovery (STIR) methods have also been proposed (35), however, STIR results in significant water signal attenuation and a reduction in SNR. Lastly on-resonant fat suppression techniques such as binomial spatial-spectral excitation pulses can be applied, however, they require a large, 3-D field of view and increased gradient demands, which may not always be achievable in vivo.
We, therefore, propose a multi-echo Dixon fat-water separation approach (36-39) to remove the fat component from the MT images. The Dixon method takes advantage of the differences in precession rates between water and fat by acquiring data at multiple echo times to separate the fat and water components of the signal (40). While whole-brain fat and water images have been successfully implemented in slice locations that cover the ON (41), to the authors’ knowledge, Dixon-separated qMT has not been studied. The importance of the work presented here is to evaluate the stability of qMT under multi-echo signal combinations and utilize this information to derive qMT indices reflective of myelination at resolutions that are typically prohibitive for qMT methods. Toward this end, we performed in vivo qMT in the brain and ON of healthy volunteers using a three-echo Dixon method, and evaluated this technique against traditional single-echo out-of-phase imaging data.
Methods
All numerical simulations and data analyses were performed with scripts written in MATLAB 2014a & 2015a (The MathWorks, Inc; Natick, MA).
Simulations
To predict the effect from fitting a two-pool (water + MT) qMT model to a tissue that contains three effective pools (water + MT + fat), data were numerically generated from a three-pool model based on the equations from Portnoy and Stanisz (42). A set of typical parameters for WM and fat (WM: R1obs = 0.9 s−1, T2f = 30 ms, T2m = 11 μs, kmf = 10 s−1, PSR = 0.15; Fat: R1obs,fat = 0.365 s−1, T2,fat = 133 ms) (43) were used to model the tissue, and the sampling scheme included two saturation powers at nominal flip angles (αMT) of 900° and 1200°, with frequency offsets (Δω) of 1, 1.5, 2, 2.5, 8, 16, 32, and 100 kHz. The fat fractions were varied between 0.0 and 0.95, stepped by 0.05. Gaussian noise was added to the data to simulate an SNR of 100 at thermal equilibrium (to match experimental parameters), and the two-pool model described by Yarnykh and Yuan (44) & Yarnykh (31), which does not account for the effect of fat on the observed signal fraction, was used to fit the noisy signal data at each fat fraction over 10,000 noise realizations per fat fraction. The mean and standard deviation of the fitted qMT parameters were plotted as a function of fat fraction and used to predict the effect of an un-modeled fat component on the qMT analysis.
Experimental Data
Subjects
Eight healthy controls were recruited and imaged for this study (mean age 29.4 ± 5.4, 4 female). The local Institutional Review Board approved this study, and signed informed consent was obtained prior to the examination.
Data Acquisition
All data was acquired on a 3.0 tesla Philips Achieva scanner (Philips Healthcare, Best, The Netherlands). A 2-channel, multi-transmit body coil was used for excitation and an 8-channel head coil was used for signal reception. The MT acquisition consisted of a single-slice with the field-of-view (FOV) centered along the ethmoid bone (using high-resolution T2 images (32)), perpendicular to both ONs, and spanned, at minimum, the width of the head in all subjects. Parallel imaging with sensitivity encoding (SENSE) and second-order shimming over both ONs was used to minimize image artifacts arising from susceptibility differences between bone, tissue, and air. All images had an FOV of 150 × 150 mm2, a slice thickness of 3 mm, and included both the ON and the brain (see Fig. 1a,b). MT-weighted images were acquired using a 2D MT-prepared, three-echo spoiled gradient echo sequence (SPGR) (22). Nominal in-plane resolution was 1 × 1 mm2, (reconstructed to 0.3 × 0.3 mm2) with 2 signal averages, and a SENSE factor of 2. MT weighting was achieved using a 20 ms, single-lobed sinc-Gauss pulse, and the parameters listed in Table 1. The water (W), in-phase, out-of-phase (OP), and fat images were reconstructed using the seven peak fat spectrum, multi-echo mDixon option in the Philips scanner software (37,38). Optic nerve motion artifacts were minimized by employing fixation every other dynamic using the Psychophysics Toolbox MATLAB extensions (45-48), resulting in 32 dynamics with two full sets of (fixated and non-fixated) qMT data. This consisted of an “on” dynamic, where subjects were asked to fixate on a white cross against a black background, and an “off” dynamic, where there was a black screen. To help address eye fatigue, the subjects were encouraged to not focus during the “off” dynamics.
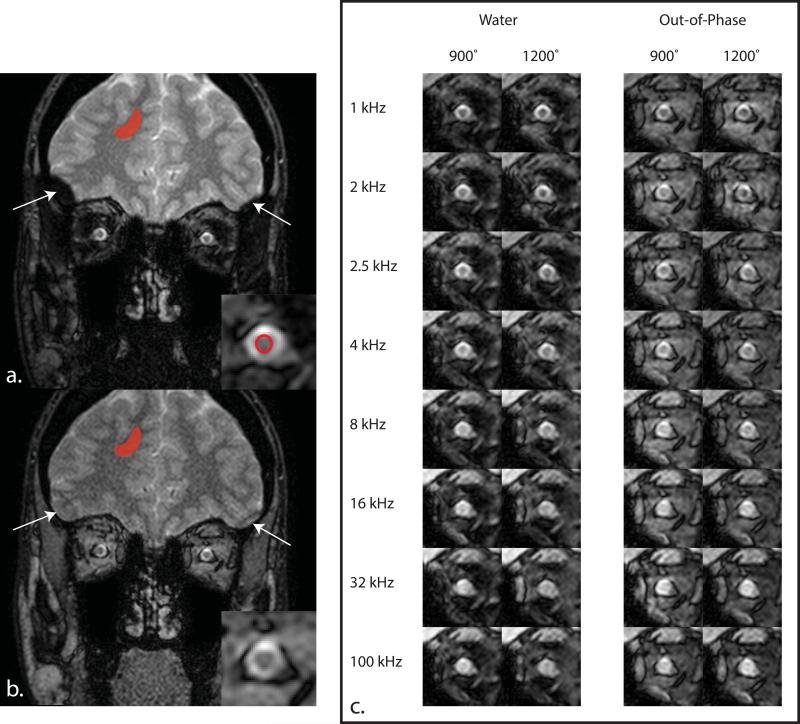
Figure 1.
Panels (a.) and (b.) illustrate the MT effect at 1000 kHz with a saturation power of 900° for the (a.) water and (b.) out-of-phase images for a slice immediately posterior to the globe (proximal retrobulbar). The cutoff area of the brain on the outer edges of the volume in the water image is indicative of poor shimming in these regions, and thus, a mislabeling effect of the Dixon method (see arrows). The ROIs in (a.) and (b.) display the area used to compare the water and out-of-phase qMT parameters in the brain and optic nerve. The optic nerve ROI in (a.) is propagated to (b.) for analysis in the out-of-phase images. (c) Example MT-weighted data in the optic nerve for the water and out-of-phase images. Notice the increased conspicuity of the optic nerve against the surrounding tissue in the water images compared to the out-of-phase images.
Table 1.
Scan parameters and MT prepulse parameters for the MT, B1, B0, and T1 scans.
| Scan: | Scan Parameters: | MT Prepulse Parameters: | Scan Time (m:ss) | |
|---|---|---|---|---|
| Δω (kHz) | Powers | |||
| MT | TR/TE1/ΔTE/α: 78/5.8/3.1 ms/10° | 1, 2, 2.5, 4, 8, 16, 32, 100 | 900°, 1200° | 6:32 |
| B1 | TR1/TR2/TE/α: 30/130/5.7 ms/60° | — | — | 0:21 |
| B0 | TR/TE1/TE2/α: 50/5.8/8.1 ms/25° | — | — | 0:34 |
| T1 | TR/TE: 20/5.7 ms α: 5, 15, 20, 25, 30° | — | — | 2:00 |
| Total Time: | 9:27 | |||
To correct for B1 and B0 inhomogeneities present in the imaging volume, B1 (using the dual-TR actual flip angle GRE method (49)) and B0 (using dual-TE GRE) maps were acquired using fast 3D techniques over 3 slices, centered on the MT acquisition. T1 mapping was performed using a multiple flip angle (MFA) acquisition over 3 slices, centered on the MT acquisition. Fixation was not employed for the B1, and B0 acquisitions because it was assumed that B1 and B0 inhomogeneities will be slowly varying over the volume, while the T1 acquisition did not utilize fixation in order to save time on the acquisition due to eye fatigue. Detailed scan parameters and acquisition times are listed in Table 1.
Data Postprocessing
Brain Data
To determine the effects of the Dixon method on MT data, we first considered the W and OP data in the brain regions for all subjects. Using the Brain Extraction Tool (BET (50,51)) and FLIRT (52,53) packages from FSL (54), the skull and ONs were removed, and the data was co-registered to the first MT offset for each image type, respectively. Large ROIs were drawn in the WM of the right hemisphere of the brain using the Δω = 1000 kHz offset (see Figure 1a,b), and these were propagated to the MT-weighted, T1, B1, and B0 images. OP data was used to approximate a normal out-of-phase acquisition, and to ensure user error during ROI selection did not contribute to differences in parameter values.
Optic Nerve Data
ROIs were drawn in the W only images for each offset of the MT-weighted data of the ON (see Figure 1a) in each eye, as well as for the T1, B1, and B0 data. As the W and OP images were taken from the same acquisition, the ROIs in the W images were also propagated to the OP images. Following the ROI selection, W and OP qMT parameters were generated in both ONs for each volunteer using the methods described above.
Parameter Generation
The resulting data was used to generate qMT parameters for each volunteer using the qMT model described in Yarnykh (24) & Yarnykh and Yuan (44). This model contains six independent parameters: R1m, R1f, T2m, T2f, PSR = M0m/M0f, and kmf = kfm/PSR. The R1obs (1/T1obs) maps were independently reconstructed by regressing MFA data to the SPGR signal equation in the steady-state (55); these maps were used during MT parameter estimation (below) to estimate the parameter R1f (24,44). Henkelman, et al. (19) & Morrison and Henkelman (30) showed that the signal dependence on R1m is weak; therefore, it was set equal to the R1f as well (31). The remaining MT parameters (PSR, kmf, T2f, and T2m) were estimated for the W and OP brain data by fitting the qMT data to the two-pool model (24,44). For all fitting, the nominal offset frequency and RF amplitudes were corrected using B0 and B1 maps, respectively (21).
Statistical Analysis
The mean and standard deviation for each image type (W and OP) were calculated for each qMT-derived index in the brain and ONs. Statistical variations between the left and right ON (for both W and OP image types) and between the W and OP brain data were evaluated using the non-parametric Wilcoxon signed rank test, with a threshold for significance of p < 0.05, for each qMT parameter (PSR, kmf, T2f, and T2m).
MAD Analysis
While the statistical analysis above is straightforward in the brain, due to the relatively homogeneous ROIs taken in each image type, the Wilcoxon signed rank test in the ON is more complicated (see Results), because this approach does not capture the potential for nested variance structures or an asymmetric number of voxels between eyes, and thus may potentially underestimate the true variance between each image type. Therefore, we also computed the mean absolute difference (MAD) of all possible left and right ON voxel pairs drawn from an empirically determined distribution (below). If there are n voxels in the right and m voxels in the left ON for a single subject, then the MAD is given by
| [1] |
where hk(Left) denotes a measure (e.g., PSR) on the kth voxel in the left ON, and similarly, hl(Right) denotes the same measure (e.g., PSR) on the lth voxel of the right ON. The distribution of data for each qMT parameter was empirically constructed via the bootstrap method with 3000 iterations (56). For each iteration, the subject-level MT data were resampled with replacement to generate a Z-spectrum, and qMT parameter maps were determined from this sampled MT data. The ROI-derived values for the T1, B1, and B0 data from each subject was used in each case to avoid biases due to the variance that could occur from these parameters. The data for each subject was then combined, and a total MAD score was calculated. The empirical 95% confidence interval of the MAD for each parameter of interest was computed and examined for statistical significance.
We assumed no differences would exist in healthy subjects for each qMT parameter between the left and right ON, and that a low MAD indicates a greater symmetry between the right and left ON for a given qMT parameter and image type. Comparing across image types, we calculated the ΔMAD% as the difference in MAD values for each technique relative to the MAD for the OP acquisition as summarized in the following equation:
| [2] |
A negative ΔMAD% indicates a lower W MAD score compared with the OP MAD, and the ΔMAD% is an estimate of the relative improvement (or worsening) when using the Dixon method relative to using an OP image.
Results
Simulations
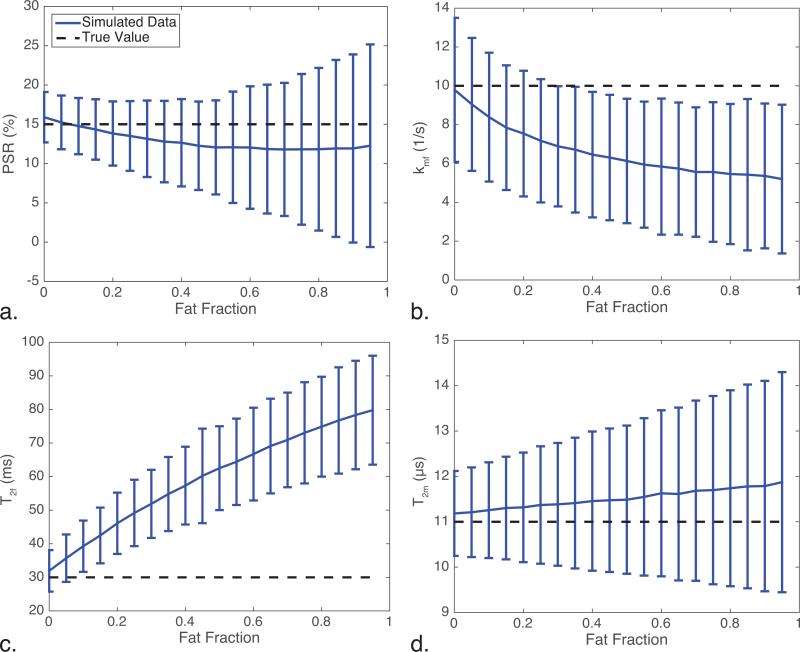
Figure 2 illustrates the effect of fat on the qMT parameters where the qMT-derived indices (from simulation) are shown against the fat fraction. Note the dotted black line is the true, expected value and the blue line shows the fit results for each parameter, with the error bars representing one standard deviation over 10,000 noise iterations at each point. The PSR never significantly diverges from the true PSR value (larger than one standard deviation from the true value); however, the kmf and T2f parameters significantly diverge from the true value for fat fractions greater than 0.3 and 0.1, respectively. Furthermore, the variance in the PSR and T2m increase as a function of fat fraction, which can generally be assumed as instability in the fitting algorithm at higher fat fractions.
Figure 2.
Results of the numerical simulations of the qMT parameters as a function of fat fraction, focusing on (a.) PSR, (b.) kmf, (c.) T2f, and (d.) T2m. Notice that (a.), (b.), and (d.) all show increased variance with increasing fat fraction, while (b.) and (c.) show significantly different deviations from the true value at relatively low fat fractions (20% and 30% fat, respectively).
Experimental Data
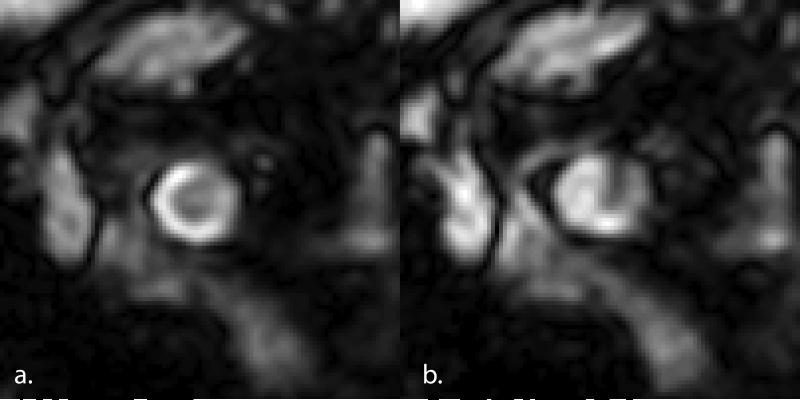
Representative MT data for the W and OP image types are displayed in Fig. 1. The conspicuity between the ON and surrounding tissue increases dramatically in the W images compared to the OP images (Fig. 1c). Furthermore, fixation provides a marked increase in ON visualization, as can be seen in Fig. 3a.
Figure 3.
Example MT data with (a.) and without (b.) fixation. The nerve is much more clearly delineated when using fixation. An example ROI is also presented in (a.) to illustrate the area used to quantify the MT data.
Brain Data
The results comparing the qMT-derived indices between the W and OP image types in the brain are shown in Table 2. The mean values across methods are indistinguishable indicating no effect from combining multiple echoes for all qMT-derived indices. Further, all parameter p-values are well above the threshold for significance (p-values: PSR = 0.64, kmf = 0.95, T2f = 0.38, T2m = 0.25), indicating there is no statistical significance between the W and OP images when no fat is expected to be present. Therefore, the Dixon method does not alter the MT-weighted observations and resulting fits during the multi-echo reconstruction.
Table 2.
Mean and Standard Deviation of qMT Parameters in the Brain.
| PSR (%) | kmf (1/s) | T2f (10−2) | T2m (μs) | R1obs (1/s) | |
|---|---|---|---|---|---|
| Water | 17.0±1.7 | 17.2±5.2 | 1.8±0.2 | 11.1±0.8 | 1.15±0.17 |
| Out-of-Phase | 17.6±3.7 | 17.6±7.1 | 1.9±0.3 | 11.3±1.3 |
Optic Nerve: W vs OP Data
qMT-derived indices for the ON are presented in Table 3, with an example of the fits displayed in Fig. 4. Compared to the brain, the qMT-derived indices from the W ON data are different in the ON (lower for PSR, kmf, and T2m, higher for T2f), which may be driven by differences in the neurological tissue between the brain and ON (as is seen in the spinal cord (23)), or minor partial volume effects from cerebrospinal fluid (CSF).
Table 3.
Mean and Standard Deviation of qMT Parameters for Left and Right Optic Nerves.
| PSR (%) | kmf (1/s) | T2f (10−2) | T2m (μs) | R1obs (1/s) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Left | Right | Left | Right | Left | Right | Left | Right | Left | Right | |
| Water | 15.8±4.4 | 14.7±2.7 | 11.1±3.6 | 10.2±3.2 | 2.9±1.8 | 2.3±1.0 | 8.5±2.7 | 8.3±1.1 | 0.94±0.19 | 0.82±0.12 |
| Out-of-Phase | 34.0±31.6 | 18.2±11.2 | 28.5±44.3 | 41.5±41.6 | 2.9±1.8 | 2.0±0.6 | 9.9±2.8 | 7.9±3.4 | ||
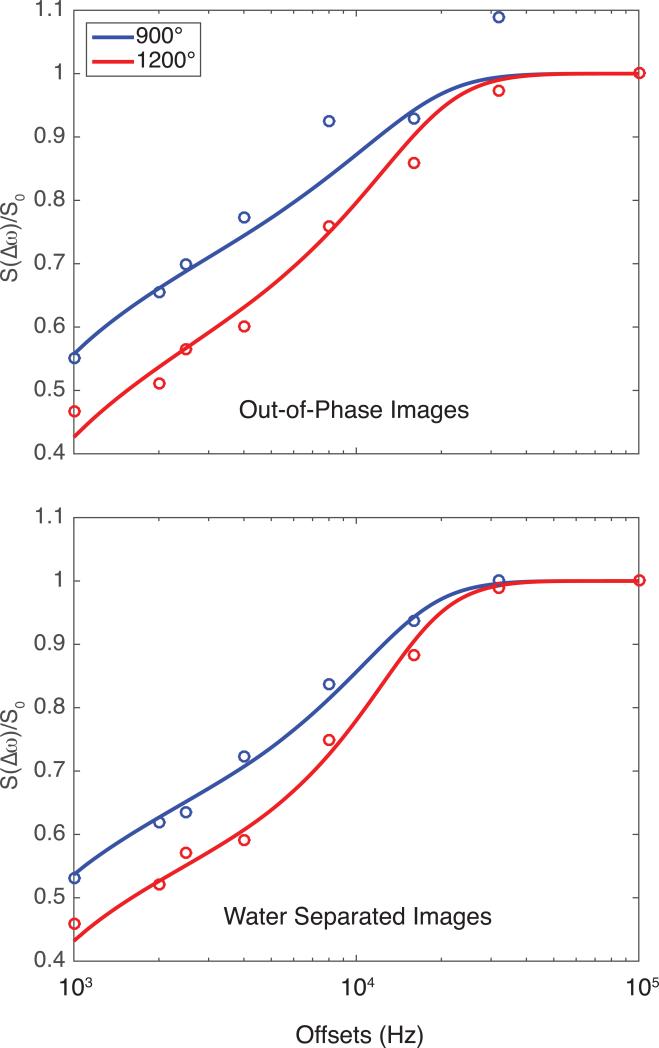
Figure 4.
Example optic nerve Z-spectra for the out-of-phase and water images for a single subject. The water images (normalized residual = 0.0035) show better goodness of fit, and visually performed better compared to the out-of-phase images (normalized residual = 0.0228).
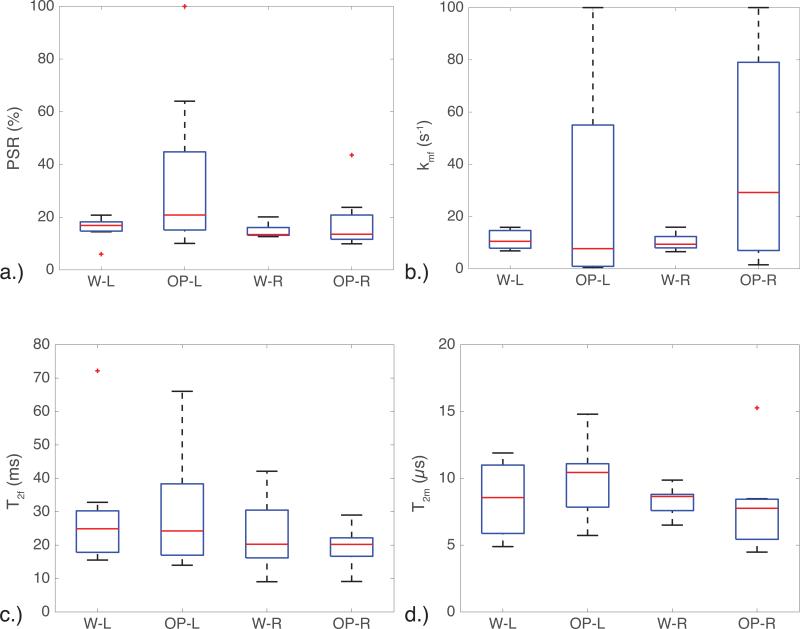
However, the kmf in the OP data has a much higher mean and standard deviation than the W data (Table 3). Although the OP images have approximately more noise than the W images, this does not fully account for the larger standard deviation seen in the OP images relative to the W images, as can be seen in Fig. 5, which displays a box plot of the qMT parameters. This is important, as the PSR, T2f, and T2m seem to be relatively consistent across image types, indicating that the kmf may be sensitive to the unmodeled fat components in the tissue. When considering the MT-weighted data and the resulting fits (Figs. 4 and 5), the fits for the OP data were worse than those for the water separated data, indicating that some fat may be influencing the OP image data (see Fig. 4 for representative residual values).
Figure 5.
Box plots illustrating the variance in the a.) PSR, b.) kmf, c.) T2f, and d.) T2m for the left (L) and right (R) optic nerves in both the water (W) and out-of-phase (OP) images. The kmf shows a large amount of variation in the OP images, and small variation in the W images. A similar trend is seen in the PSR, indicating that the two-pool qMT model used here did not fit the OP data well.
Optic Nerve: Left vs. Right Data
Comparing the left and right ON data for each method, no significant differences can be seen for either method (p-values range from 0.11-0.95). This is expected, as the ONs in healthy subjects should not be significantly different from one another. We used this result to increase the power of the statistical test between image types (W versus OP) by combining the left and right ON data. Evaluating the mean qMT-derived indices in the W and OP data, there were no significant differences between any of the qMT parameters when employing the Wilcoxon signed rank test (p-values: PSR = 0.13, kmf = 0.44, T2f = 0.64, T2m = 0.92). However, this is most likely due to the large variances found in the OP parameters (as seen in Fig. 5), which ensures a high likelihood that each image type will be determined to be from the same distribution. The kmf in particular displayed a large variance in the fitting derived from the OP images (L/R = 28.5±44.3/41.5±41.6 s−1), implying that the OP images did not always yield high-quality fits to the data (see Table 3).
MAD Analysis
Table 4 provides the results of the MAD calculation (mean absolute difference between the left and right ONs drawn from an empirically determined distribution). We report the initial MAD value, the 95% CI for MAD from 3000 bootstrap iterations, and the ΔMAD%, which is the relative difference between the W and OP parameter values (note a negative value indicates improved MAD in the W images relative to the OP images). Recall that the MAD is a reflection of the variation across eyes rather than a variation of the mean qMT-derived index of interest. The variation across eyes for each qMT parameter are approximately the same between image types, and indeed, most of the 95% CI overlap one another (PSR, T2f, T2m). However, the kmf 95% CI for each image type (W and OP) do not overlap, indicating that they may be drawn from separate distributions, and thus, these image types may show significantly different fits.
Table 4.
MAD Between Left and Right Optic Nerves at the Initial Point and 95% CI (MAD) and relative improvement (−) or Worsening (+) of Dixon Water Images Versus Out-of-Phase Images.
| PSR (%) | kmf (s−1) | T2f (ms) | T2m (μs) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAD | 95% CI | ΔMAD% | MAD | 95% CI | ΔMAD% | MAD | 95% CI | ΔMAD% | MAD | 95% CI | ΔMAD% | |
| Water | 7.69 | [7.11, 7.83] | −4.46 | 9.16 | [8.92, 9.57] | −6.76 | 6.50 | [5.99, 7.08] | −5.63 | 1.24 | [1.13, 1.27] | −11.32 |
| Out-of-Phase | 8.05 | [7.40, 8.04] | 9.83 | [9.80, 10.19] | 6.88 | [6.24, 7.35] | 1.40 | [1.22, 1.40] | ||||
We next assessed the ΔMAD%, which is a reflection of the relative improvement (a negative ΔMAD%) or worsening (a positive ΔMAD%) of the variability across ONs (assumed to be small) when considering two different techniques. The ΔMAD% is negative for each qMT-derived index, indicating a general reduction in the variation across eyes derived from the W images compared to the OP images. Therefore, the W imaging data from which we derive our qMT-indices outperform the same data considered from only OP images.
Discussion
The goal of this study was to demonstrate that qMT techniques will not be adversely affected by the use of the Dixon method. Furthermore, when considering regions, like the ON, where fat may influence the image, Dixon qMT imaging may derive higher fidelity fits and more accurate quantitatively derived indices. We compared the impact of fat on qMT-derived PSR, kmf, T2f, and T2m via simulations (indicating the importance of accurate fat removal), and across healthy volunteers in the brain, where there is no fat, and in the ON, which is surrounded by a fatty sheath. Lastly, we evaluated the statistical impact of removing fat contamination from qMT data in difficult anatomies. We show that the OP images do not provide robust estimations of qMT-derived indices in the presence of fat, and that the Dixon method can be employed to reliably separate fat and water without altering the ability to characterize the MT effect using a two-pool qMT model. The ability to generate accurate qMT parameter maps while utilizing the Dixon method may increase the clinical applicability of qMT, as it provides a method to apply MT saturation in anatomies where fat may be in abundance, such as in the muscle (60), breast (61), or even the peripheral nervous system where fat is interlaced within the nerves (8).
Strength of Dixon Method
While on-resonant fat separation methods exist, they do not have the same advantages as the Dixon method. Binomial pulses may be used for fat suppression, however, clinically available gradient strengths/slew rates require implementation of large 3D volumes, which may not be suitable in anatomies prone to significant amounts of motion, such as the ON or abdomen, and may also significantly increase scan time. While echo planar imaging trains can be used to reduce scan time, employing these sequences in anatomies with large B0 inhomogeneities, such as the ON and parts of the abdomen, may cause undesirable image distortion artifacts (62). The Dixon method, however, can be employed over any chosen (i.e. smaller) volume, reducing scan time, to accurately separate fat and water images. It should also be noted that the Dixon method is relatively robust to B0 inhomogeneities, which can be problematic across different anatomies (36).
Statistical Implications
While the Wilcoxon signed rank test failed to statistically quantify the differences between the W and OP images in the ON, several conclusions can be drawn from the data itself. The kmf parameter has a much higher variance in the OP images compared with the W images (Fig. 5), suggesting that kmf is sensitive to the underlying distribution. Therefore, if there are tissue components in a voxel that are not accounted for in the model, kmf will be difficult to characterize, thus producing inaccuracies in the other qMT parameters. This is further confirmed utilizing the MAD data (Table 4). The ΔMAD% illustrates an improvement in the consistency of each qMT-derived index between eyes when the W only images are considered relative to the OP images. Furthermore, although there is a factor of difference in SNR between the OP and W images, the MAD utilizes empirical data, which reduces the influence of nested variance and differences in noise between image types. Particularly important to this argument is the fact that our results from the brain indicate these differences are not due to variations driven by the different reconstruction methods (Dixon vs. non-Dixon) (see Table 2), suggesting the kmf in tissues where voxels have a combination of tissue types is poorly characterized.
Simulations
Considering the simulations further (Fig. 2), T2f and kmf deviate significantly as the fat fraction increases. Additionally, the PSR and T2m show larger variance in their fit as the fat fraction increases, indicating greater instability in the fitting algorithm due to the presence of fat. This indicates that the errors from the kmf and T2f may be influencing the stability of the PSR and T2m. This may also be the case in the OP ON images: although the PSR, T2f, and T2m are not significantly different from the W images, the kmf is significantly different, indicating there may be errors present within the other OP qMT parameters as well. Therefore, ensuring all parameters in the qMT model are well determined is important to accurately model the MT effect, particularly in tissues where there may be significant partial volume effects present.
Data Reduction Strategies
While this study utilized a single slice acquisition to explore the Dixon method for fat suppression, an expanded 3D volume may be considered. However, scan time cannot be disregarded with any qMT method due to the number of powers and/or offsets that are required for appropriate fitting. A single-point qMT model (1 MT-weighted volume with the addition of T1, B1, and B0 maps) has been introduced to reduce the scan time to a clinically appropriate length, and has been applied to the brain (31), spinal cord (23) and muscle (63). However, the assumption for single-point qMT is that the parameters kmf, T2f, and T2m are well-determined from prior scans utilizing a full qMT acquisition. Therefore, without adequate fat suppression, variance in the assumed model parameters may result in a poorly constrained model and will be hampered in tissues where fat cannot be excluded such as the ON, peripheral nerves, and muscle. Importantly, these experiments utilized out-of-phase images to perform the T1 mapping. However, this may introduce fat signal into the T1 data, confounding the results. While the observed T1 does not seem to be adversely affected by the presence of fat here, this will potentially limit the scan if appropriate fat suppression techniques are not employed.
This study used a fixation cross to help minimize motion across dynamics, due to the large saturation and readout times for each MT dynamic (approximately 10 seconds), which resulted in large amounts of motion blur (Fig. 3). To minimize tiring during fixation, we utilized a 1-on and 1-off acquisition scheme (one dynamic under fixation and the next dynamic without), which effectively doubles the scan time. Requiring fewer dynamics would significantly alleviate these challenges.
Limitations
While fat was the target of this study, the CSF surrounding the ON may also influence the signal from nerve tissue due to partial volume effects. CSF will reduce the MT effect within the nerve and thus partial volume effects will result in an underestimation of the PSR, kmf, and T2m and overestimation of T2f (Tables 2 and 3). Researchers have investigated the use of inversion recovery pulses in diffusion imaging for CSF suppression (65,66) however, this would add significantly to the scan time and may introduce extraneous off-resonance effects for nearby slices. Additionally, while techniques such as DANTE (67) may provide advantages over inversion recovery methods, they would still introduce extra off-resonance saturation effects, which would alter the observed MT effect and would need to be incorporated into the model.
Therefore, to preserve the fidelity of the observed MT effect in tissue, alternative techniques can be considered to accurately suppress CSF. A three-pool model (2 MT pools, and 1 non-exchanging pool) was recently introduced by Mossahebi, et al. (68) to fit the CSF pool (non-exchanging) in the brain. Their results suggest that the PSR can be accurately quantified even in the presence of significant amounts of CSF contamination. Research has also been performed to separate the CSF sheath from the ON using a model derived using T2-weighted 3-D spin echo sequences with a turbo spin echo readout (32). These sequences have similar contrast to the small offset MT scans, and thus may be applicable to the MT-weighted data. Thus, further research is warranted to address the removal of the CSF sheath from the ON.
Conclusions
The results of this study demonstrate the ability to successfully remove the fat component from MT images using the Dixon fat-water separation method. The development of this technique provides a method to perform qMT while in the presence of significant B0 inhomogeneities, and over small imaging volumes. Future work includes further investigation of this technique in the presence of disease, and application of this method in other anatomies with large fat components.
Acknowledgements
The authors would like to thank Ms. Leslie Mcintosh, Ms. Kristen George-Durrett, Ms. Clair Kurtenbach, and Mr. Christopher Thompson, who provided invaluable assistance with data collection. This work was supported by NIH/NIBIB K01 EB009120, NIH/NIBIB K25 EB013659, NIH/NCI R25 CA136440, NIH/NIBIB R01 EY023240 01A1, NIH/NIBIB R21 NS087465-01, DOD W81XWH-13-0073, and NIH/NCATS UL1 TR000445.
References
- 1.Pane A, Burdon MA, Miller NR. The Neuro-ophthalmology Survival Guide. Mosby/Elsevier; London: 2007. [Google Scholar]
- 2.Toosy AT, Mason DF, Miller DH. Optic neuritis. Lancet neurology. 2014;13(1):83–99. doi: 10.1016/S1474-4422(13)70259-X. [DOI] [PubMed] [Google Scholar]
- 3.Shams PN, Plant GT. Optic neuritis: a review. International MS journal / MS Forum. 2009;16(3):82–89. [PubMed] [Google Scholar]
- 4.Koenig SH. Cholesterol of myelin is the determinant of gray-white contrast in MRI of brain. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 1991;20(2):285–291. doi: 10.1002/mrm.1910200210. [DOI] [PubMed] [Google Scholar]
- 5.Kucharczyk W, Macdonald PM, Stanisz GJ, Henkelman RM. Relaxivity and magnetization transfer of white matter lipids at MR imaging: importance of cerebrosides and pH. Radiology. 1994;192(2):521–529. doi: 10.1148/radiology.192.2.8029426. [DOI] [PubMed] [Google Scholar]
- 6.Schmierer K, Tozer DJ, Scaravilli F, Altmann DR, Barker GJ, Tofts PS, Miller DH. Quantitative magnetization transfer imaging in postmortem multiple sclerosis brain. Journal of magnetic resonance imaging : JMRI. 2007;26(1):41– 51. doi: 10.1002/jmri.20984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen-Adad J, El Mendili MM, Lehericy S, Pradat PF, Blancho S, Rossignol S, Benali H. Demyelination and degeneration in the injured human spinal cord detected with diffusion and magnetization transfer MRI. NeuroImage. 2011;55(3):1024–1033. doi: 10.1016/j.neuroimage.2010.11.089. [DOI] [PubMed] [Google Scholar]
- 8.Dortch RD, Dethrage LM, Gore JC, Smith SA, Li J. Proximal nerve magnetization transfer MRI relates to disability in Charcot-Marie-Tooth diseases. Neurology. 2014;83(17):1545–1553. doi: 10.1212/WNL.0000000000000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellingson BM, Ulmer JL, Kurpad SN, Schmit BD. Diffusion tensor MR imaging in chronic spinal cord injury. AJNR American journal of neuroradiology. 2008;29(10):1976–1982. doi: 10.3174/ajnr.A1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filippi M, Agosta F. Imaging biomarkers in multiple sclerosis. Journal of magnetic resonance imaging : JMRI. 2010;31(4):770–788. doi: 10.1002/jmri.22102. [DOI] [PubMed] [Google Scholar]
- 11.Freund P, Wheeler-Kingshott C, Jackson J, Miller D, Thompson A, Ciccarelli O. Recovery after spinal cord relapse in multiple sclerosis is predicted by radial diffusivity. Multiple sclerosis. 2010;16(10):1193–1202. doi: 10.1177/1352458510376180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nair G, Carew JD, Usher S, Lu D, Hu XP, Benatar M. Diffusion tensor imaging reveals regional differences in the cervical spinal cord in amyotrophic lateral sclerosis. NeuroImage. 2010;53(2):576–583. doi: 10.1016/j.neuroimage.2010.06.060. [DOI] [PubMed] [Google Scholar]
- 13.Naismith RT, Xu J, Klawiter EC, Lancia S, Tutlam NT, Wagner JM, Qian P, Trinkaus K, Song SK, Cross AH. Spinal cord tract diffusion tensor imaging reveals disability substrate in demyelinating disease. Neurology. 2013;80(24):2201–2209. doi: 10.1212/WNL.0b013e318296e8f1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poloni G, Minagar A, Haacke EM, Zivadinov R. Recent developments in imaging of multiple sclerosis. The neurologist. 2011;17(4):185–204. doi: 10.1097/NRL.0b013e31821a2643. [DOI] [PubMed] [Google Scholar]
- 15.Smith SA, Golay X, Fatemi A, Mahmood A, Raymond GV, Moser HW, van Zijl PC, Stanisz GJ. Quantitative magnetization transfer characteristics of the human cervical spinal cord in vivo: application to adrenomyeloneuropathy. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2009;61(1):22–27. doi: 10.1002/mrm.21827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zackowski KM, Smith SA, Reich DS, Gordon-Lipkin E, Chodkowski BA, Sambandan DR, Shteyman M, Bastian AJ, van Zijl PC, Calabresi PA. Sensorimotor dysfunction in multiple sclerosis and column-specific magnetization transfer-imaging abnormalities in the spinal cord. Brain : a journal of neurology. 2009;132(Pt 5):1200–1209. doi: 10.1093/brain/awp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolff SD, Balaban RS. Magnetization transfer contrast (MTC) and tissue water proton relaxation in vivo. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 1989;10(1):135–144. doi: 10.1002/mrm.1910100113. [DOI] [PubMed] [Google Scholar]
- 18.Berry I, Barker GJ, Barkhof F, Campi A, Dousset V, Franconi JM, Gass A, Schreiber W, Miller DH, Tofts PS. A multicenter measurement of magnetization transfer ratio in normal white matter. Journal of magnetic resonance imaging : JMRI. 1999;9(3):441–446. doi: 10.1002/(sici)1522-2586(199903)9:3<441::aid-jmri12>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 19.Henkelman RM, Huang X, Xiang QS, Stanisz GJ, Swanson SD, Bronskill MJ. Quantitative interpretation of magnetization transfer. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 1993;29(6):759–766. doi: 10.1002/mrm.1910290607. [DOI] [PubMed] [Google Scholar]
- 20.Stanisz GJ, Odrobina EE, Pun J, Escaravage M, Graham SJ, Bronskill MJ, Henkelman RM. T1, T2 relaxation and magnetization transfer in tissue at 3T. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2005;54(3):507–512. doi: 10.1002/mrm.20605. [DOI] [PubMed] [Google Scholar]
- 21.Levesque IR, Sled JG, Narayanan S, Giacomini PS, Ribeiro LT, Arnold DL, Pike GB. Reproducibility of quantitative magnetization-transfer imaging parameters from repeated measurements. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2010;64(2):391–400. doi: 10.1002/mrm.22350. [DOI] [PubMed] [Google Scholar]
- 22.Sled JG, Pike GB. Quantitative imaging of magnetization transfer exchange and relaxation properties in vivo using MRI. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2001;46(5):923–931. doi: 10.1002/mrm.1278. [DOI] [PubMed] [Google Scholar]
- 23.Smith AK, Dortch RD, Dethrage LM, Smith SA. Rapid, high-resolution quantitative magnetization transfer MRI of the human spinal cord. NeuroImage. 2014;95:106–116. doi: 10.1016/j.neuroimage.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yarnykh VL. Pulsed Z-spectroscopic imaging of cross-relaxation parameters in tissues for human MRI: theory and clinical applications. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2002;47(5):929– 939. doi: 10.1002/mrm.10120. [DOI] [PubMed] [Google Scholar]
- 25.Hinton DP, Bryant RG. 1H magnetic cross-relaxation between multiple solvent components and rotationally immobilized protein. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonancein Medicine / Society of Magnetic Resonance in Medicine. 1996;35(4):497–505. doi: 10.1002/mrm.1910350408. [DOI] [PubMed] [Google Scholar]
- 26.Gochberg DF, Kennan RP, Robson MD, Gore JC. Quantitative imaging of magnetization transfer using multiple selective pulses. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 1999;41(5):1065–1072. doi: 10.1002/(sici)1522-2594(199905)41:5<1065::aid-mrm27>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 27.Odrobina EE, Lam TY, Pun T, Midha R, Stanisz GJ. MR properties of excised neural tissue following experimentally induced demyelination. NMR in biomedicine. 2005;18(5):277–284. doi: 10.1002/nbm.951. [DOI] [PubMed] [Google Scholar]
- 28.Ou X, Sun SW, Liang HF, Song SK, Gochberg DF. The MT pool size ratio and the DTI radial diffusivity may reflect the myelination in shiverer and control mice. NMR in biomedicine. 2009;22(5):480–487. doi: 10.1002/nbm.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Underhill HR, Rostomily RC, Mikheev AM, Yuan C, Yarnykh VL. Fast bound pool fraction imaging of the in vivo rat brain: association with myelin content and validation in the C6 glioma model. NeuroImage. 2011;54(3):2052–2065. doi: 10.1016/j.neuroimage.2010.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrison C, Henkelman RM. A model for magnetization transfer in tissues. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 1995;33(4):475–482. doi: 10.1002/mrm.1910330404. [DOI] [PubMed] [Google Scholar]
- 31.Yarnykh VL. Fast macromolecular proton fraction mapping from a single offresonance magnetization transfer measurement. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2012;68(1):166–178. doi: 10.1002/mrm.23224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrigan RL, Plassard AJ, Mawn LA, Galloway RL, Smith SA, Landman BA. Constructing a statistical atlas of the radii of the optic nerve and cerebrospinal fluid sheath in young healthy adults. Proc Soc Photo Opt Instrum Eng. 2015:9413. doi: 10.1117/12.2081887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaldoudi E, Williams SC, Barker GJ, Tofts PS. A chemical shift selective inversion recovery sequence for fat-suppressed MRI: theory and experimental validation. Magnetic resonance imaging. 1993;11(3):341–355. doi: 10.1016/0730-725x(93)90067-n. [DOI] [PubMed] [Google Scholar]
- 34.Zhou XJ, Bernstein MA, King KF. Handbook of MRI Pulse Sequences. Academic Press; Burlington: 2004. Chapter 6 - Adiabatic Radiofrequency Pulses. pp. 177– 212. [Google Scholar]
- 35.Dwyer AJ, Frank JA, Sank VJ, Reinig JW, Hickey AM, Doppman JL. Short-Ti inversion-recovery pulse sequence: analysis and initial experience in cancer imaging. Radiology. 1988;168(3):827–836. doi: 10.1148/radiology.168.3.3406412. [DOI] [PubMed] [Google Scholar]
- 36.Berglund J, Kullberg J. Three-dimensional water/fat separation and T2* estimation based on whole-image optimization--application in breathhold liver imaging at 1.5 T. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2012;67(6):1684–1693. doi: 10.1002/mrm.23185. [DOI] [PubMed] [Google Scholar]
- 37.Dimitrov I. mDIXON Quant delivers robust, high quality fat quantification. 2015 http://clinical.netforum.healthcare.philips.com/global/Explore/White-Papers/MRI/mDIXON-Quant-delivers-robust-high-quality-fat-quantification: Philips Healthcare; 2014. [Google Scholar]
- 38.Eggers H, Brendel B, Duijndam A, Herigault G. Dual-echo Dixon imaging with flexible choice of echo times. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2011;65(1):96–107. doi: 10.1002/mrm.22578. [DOI] [PubMed] [Google Scholar]
- 39.Glover GH, Schneider E. Three-point Dixon technique for true water/fat decomposition with B0 inhomogeneity correction. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 1991;18(2):371–383. doi: 10.1002/mrm.1910180211. [DOI] [PubMed] [Google Scholar]
- 40.Dixon WT. Simple proton spectroscopic imaging. Radiology. 1984;153(1):189–194. doi: 10.1148/radiology.153.1.6089263. [DOI] [PubMed] [Google Scholar]
- 41.Hardy PA, Hinks RS, Tkach JA. Separation of fat and water in fast spin-echo MR imaging with the three-point Dixon technique. Journal of magnetic resonance imaging : JMRI. 1995;5(2):181–185. doi: 10.1002/jmri.1880050213. [DOI] [PubMed] [Google Scholar]
- 42.Portnoy S, Stanisz GJ. Modeling pulsed magnetization transfer. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2007;58(1):144– 155. doi: 10.1002/mrm.21244. [DOI] [PubMed] [Google Scholar]
- 43.Gold GE, Han E, Stainsby J, Wright G, Brittain J, Beaulieu C. Musculoskeletal MRI at 3.0 T: relaxation times and image contrast. AJR American journal of roentgenology. 2004;183(2):343–351. doi: 10.2214/ajr.183.2.1830343. [DOI] [PubMed] [Google Scholar]
- 44.Yarnykh VL, Yuan C. Cross-relaxation imaging reveals detailed anatomy of white matter fiber tracts in the human brain. NeuroImage. 2004;23(1):409–424. doi: 10.1016/j.neuroimage.2004.04.029. [DOI] [PubMed] [Google Scholar]
- 45.Pelli DG. The VideoToolbox software for visual psychophysics: transforming numbers into movies. Spat Vis. 1997;10(4):437–442. [PubMed] [Google Scholar]
- 46.Zhang Y, Nateras OS, Peng Q, Kuranov RV, Harrison JM, Milner TE, Duong TQ. Lamina-specific anatomic magnetic resonance imaging of the human retina. Investigative ophthalmology & visual science. 2011;52(10):7232–7237. doi: 10.1167/iovs.11-7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beenakker JW, van Rijn GA, Luyten GP, Webb AG. High-resolution MRI of uveal melanoma using a microcoil phased array at 7 T. NMR in biomedicine. 2013;26(12):1864–1869. doi: 10.1002/nbm.3041. [DOI] [PubMed] [Google Scholar]
- 48.Brainard DH. The Psychophysics Toolbox. Spat Vis. 1997;10(4):433–436. [PubMed] [Google Scholar]
- 49.Yarnykh VL. Actual flip-angle imaging in the pulsed steady state: a method for rapid three-dimensional mapping of the transmitted radiofrequency field. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2007;57(1):192–200. doi: 10.1002/mrm.21120. [DOI] [PubMed] [Google Scholar]
- 50.Smith SM. Fast robust automated brain extraction. Human brain mapping. 2002;17(3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jenkinson M, Pechaud M, Smith S. BET2: MR-based estimation of brain, skull and scalp surfaces. 2005 [Google Scholar]
- 52.Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Medical image analysis. 2001;5(2):143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- 53.Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17(2):825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 54.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. Fsl. NeuroImage. 2012;62(2):782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 55.Fram EK, Herfkens RJ, Johnson GA, Glover GH, Karis JP, Shimakawa A, Perkins TG, Pelc NJ. Rapid calculation of T1 using variable flip angle gradient refocused imaging. Magnetic resonance imaging. 1987;5(3):201–208. doi: 10.1016/0730-725x(87)90021-x. [DOI] [PubMed] [Google Scholar]
- 56.Jeong HK, Dewey BE, Hirtle JA, Lavin P, Sriram S, Pawate S, Gore JC, Anderson AW, Kang H, Smith SA. Improved diffusion tensor imaging of the optic nerve using multishot two-dimensional navigated acquisitions. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2015;74(4):953–963. doi: 10.1002/mrm.25469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wansapura JP, Holland SK, Dunn RS, Ball WS., Jr. NMR relaxation times in the human brain at 3.0 tesla. Journal of magnetic resonance imaging : JMRI. 1999;9(4):531–538. doi: 10.1002/(sici)1522-2586(199904)9:4<531::aid-jmri4>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 58.Lu H, Nagae-Poetscher LM, Golay X, Lin D, Pomper M, van Zijl PC. Routine clinical brain MRI sequences for use at 3.0 Tesla. Journal of magnetic resonance imaging : JMRI. 2005;22(1):13–22. doi: 10.1002/jmri.20356. [DOI] [PubMed] [Google Scholar]
- 59.Smith SA, Edden RA, Farrell JA, Barker PB, Van Zijl PC. Measurement of T1 and T2 in the cervical spinal cord at 3 tesla. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2008;60(1):213–219. doi: 10.1002/mrm.21596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li K, Dortch RD, Welch EB, Bryant ND, Buck AK, Towse TF, Gochberg DF, Does MD, Damon BM, Park JH. Multi-parametric MRI characterization of healthy human thigh muscles at 3.0 T - relaxation, magnetization transfer, fat/water, and diffusion tensor imaging. NMR in biomedicine. 2014;27(9):1070–1084. doi: 10.1002/nbm.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bonini RH, Zeotti D, Saraiva LA, Trad CS, Filho JM, Carrara HH, de Andrade JM, Santos AC, Muglia VF. Magnetization transfer ratio as a predictor of malignancy in breast lesions: preliminary results. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2008;59(5):1030–1034. doi: 10.1002/mrm.21555. [DOI] [PubMed] [Google Scholar]
- 62.Schmitt F, Stehling MK, Turner R. Echo-Planar Imaging. Springer; Berlin, Germany: 1998. [Google Scholar]
- 63.Li K, Dortch RD, Kroop SF, Huston JW, Gochberg DF, Park JH, Damon BM. A Rapid Approach for Quantitative Magnetization Transfer Imaging in Thigh Muscles using the Pulsed Saturation Method. Magnetic resonance imaging. 2015 doi: 10.1016/j.mri.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramani A, Dalton C, Miller DH, Tofts PS, Barker GJ. Precise estimate of fundamental in-vivo MT parameters in human brain in clinically feasible times. Magnetic resonance imaging. 2002;20(10):721–731. doi: 10.1016/s0730-725x(02)00598-2. [DOI] [PubMed] [Google Scholar]
- 65.Falconer JC, Narayana PA. Cerebrospinal fluid-suppressed high-resolution diffusion imaging of human brain. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 1997;37(1):119–123. doi: 10.1002/mrm.1910370117. [DOI] [PubMed] [Google Scholar]
- 66.Bhagat YA, Beaulieu C. Diffusion anisotropy in subcortical white matter and cortical gray matter: changes with aging and the role of CSF-suppression. Journal of magnetic resonance imaging : JMRI. 2004;20(2):216–227. doi: 10.1002/jmri.20102. [DOI] [PubMed] [Google Scholar]
- 67.Li L, Miller KL, Jezzard P. DANTE-prepared pulse trains: a novel approach to motion-sensitized and motion-suppressed quantitative magnetic resonance imaging. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2012;68(5):1423–1438. doi: 10.1002/mrm.24142. [DOI] [PubMed] [Google Scholar]
- 68.Mossahebi P, Alexander AL, Field AS, Samsonov AA. Removal of cerebrospinal fluid partial volume effects in quantitative magnetization transfer imaging using a three-pool model with nonexchanging water component. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2014 doi: 10.1002/mrm.25516. [DOI] [PMC free article] [PubMed] [Google Scholar]