Abstract
MicroRNAs (miRNAs) are short, single-stranded RNA that regulate post-transcriptional control of mRNA translation. Knowledge on the role of these critical regulators in toxicological responses in increasing, but is still limited. Atrazine is a herbicide used throughout the Midwestern US that is reported to frequently contaminate potable water supplies above the maximum contaminant level of 3 parts per billion. Atrazine is a suspected endocrine disrupting chemical and studies have begun to investigate the genetic mechanisms of toxicity; however, studies investigating epigenetic mechanisms are limited. In this study both zebrafish and human miRNAs were significantly altered in response to an embryonic atrazine exposure of 0.3, 3, or 30 ppb in zebrafish. Altered miRNAs are known to play a role in angiogenesis, cancer, or neuronal development, differentiation, and maturation. Targeted analysis of altered human miRNAs with genes previously identified to be altered by atrazine exposure revealed several targets linked to cell cycle and cell signaling. Further analysis of hsa-miRNA-126-3p, which had altered expression in all three atrazine treatments at 72 hpf, revealed alterations also occurred at 60 hpf in the 30 ppb treatment group. Results from this study indicate miRNA deregulation in zebrafish and human miRNAs following an embryonic atrazine exposure in zebrafish.
Keywords: atrazine, development, miRNA, targeting analysis, transcriptomics, zebrafish
1. Introduction
Particular interest in recent years has shifted towards the study of the underlying epigenetic mechanisms of gene regulation and the role it plays in developmental reprogramming of the genome and disease susceptibility. Epigenetics can be defined as the study of the molecular mechanisms that regulate gene expression often leading to permanent, yet reversible, changes that can be stable throughout the lifespan of an organism and potentially be heritable (Goldberg et al., 2007; Portela and Esteller, 2010). The term was initially coined by Conrad Waddington in the 1940s while describing the outward appearance, or phenotype, that an organism displays based upon the genetic material and functions of the genes in an organism (Hanson and Gluckman, 2014). Three types of known molecular epigenetic mechanisms include DNA methylation, histone modifications, and small non-coding RNAs that regulate gene expression (Jirtle and Skinner, 2007). Of these, the alteration of DNA methylation patterns and subsequent reprogramming of developmental processes by changing transcriptional gene expression has received most attention, especially as it pertains to the heritability of these epigenetic alterations (Anway et al., 2005, 2006; Guerrero-Bosagna et al., 2010; Inawaka et al., 2009; Stouder et al., 2010; Vandegehuchte et al., 2009). More recently, studies have implicated the importance of post-transcriptional regulation by non-coding RNAs in regulating gene expression, namely through short sequences of RNA referred to as microRNAs (miRNAs).
miRNAs are short (~22 nucleotides in length), single-stranded RNA genes that regulate post-transcriptional gene expression by targeting messenger RNAs (mRNAs) through their complementary sequences in the 3’ untranslated region and repressing their translation. MicroRNAs are associated with a broad spectrum of cellular and developmental processes including responses to xenobiotic stresses. Thus, miRNAs are being widely studied for their mechanistic role in toxicological outcomes and have been implicated in cardiovascular, developmental, liver, and neurotoxicity pathways (Tal and Tanguay, 2012; Yokoi and Nakajima, 2001). miRNAs are found in diverse organisms suggesting evolutionary conservation of mechanisms related to miRNA regulation (Li et al., 2010; Lim et al., 2003). However, the majority of miRNA functions are unknown and different animal models are being used to identify miRNA functions and differences in miRNA expression in laboratory studies (Ason et al., 2006). Moreover, few studies have investigated the role of miRNAs in toxicological responses thereby limiting the knowledge of relevance of these critical regulators in mechanisms of toxicity.
Endocrine disrupting chemicals (EDCs) are exogenous agents that alter the endocrine system through multiple pathways. EDCs are diverse in structure and are present in many products such as plasticizers, pharmaceuticals, and pesticides (Roy et al., 2009; Swedenborg et al., 2009). Rapid increases in industrialization and in the production of these chemicals have increased the risk of human exposure, therefore, heightening public concern and the need for investigation into their mechanisms of action. Studies have reported that exposure to EDCs can cause irreversible changes in tissue formation, decreased reproductive potential, obesity, and cancer (Cooper et al., 2000; Hatch et al., 2011; Roy et al., 2009; Swedenborg et al., 2009; Wetzel et al., 1994). Moreover, evidence suggests that exposure to EDCs can cause adverse effects not only in organisms that come into contact with them, but also to future progeny of exposed individuals (Anway et al., 2005, 2006).
Atrazine (2-chloro-4-ethylamino-6-isopropylamino-1,3,5-triazine) is a pre-emergent herbicide used to prevent the growth of broadleaf and grassy weeds on crops such as corn, sorghum grass, sugar cane, and wheat and is reported to have endocrine disrupting effects (Barr et al., 2007; Eldridge et al., 1999; Ochoa-Acuña et al., 2009; Solomon et al., 2008). The United States Environmental Protection Agency (U.S. EPA) estimates approximately 76.5 million pounds of atrazine are applied annually in the United States, making it one of the most widely used herbicides (Rinsky et al., 2012). The U.S. EPA has set the maximum contaminant level (MCL) for atrazine at 3 parts per billion (ppb; μg/L) in drinking water supplies; however, during spring and summer months, this level is often exceeded (Barr et al., 2007; Ochoa-Acuña et al., 2009; Rohr and McCoy, 2010; U.S. EPA, 2002). Numerous studies have investigated the adverse health effects of atrazine on the neuroendocrine system. Results report that atrazine decreases gonadotropin releasing hormone (GnRH) release, the pre-ovulatory surge of luteinizing hormone (LH), follicle stimulating hormone (FSH), and prolactin (PRL) (Cooper et al., 2000; Foradori et al., 2009, 2013). The genetic and cellular mechanisms behind the observed hormonal alterations are also under investigation both in vitro and in vivo (Fa et al., 2013; Kucka et al., 2012; Pogrmic-Majkic et al., 2010, 2014; Quignot et al., 2012). Furthermore, epidemiological studies highlight the need for understanding the developmental and reproductive effects of atrazine exposure and its impact on rural communities as exposure is evident in areas where this herbicide is used (Munger et al., 1997; Ochoa-Acuña et al., 2009; Winchester et al., 2009).
We have previously reported that developmental atrazine exposure results in morphological alterations and expression alterations in genes associated with reproductive system function and development, cell cycle regulation, and cancer in zebrafish larvae (Weber et al., 2013). The zebrafish provides a strong complementary vertebrate model when investigating epigenetic mechanisms of toxicity and developmental toxicity including ex utero fertilization and development, short developmental periods, a relatively short life span, and high genetic homology to humans. In addition, the zebrafish has structural and functional homology of the central nervous system (CNS) (de Esch et al., 2012; Howe et al., 2013).
In the present study we exposed zebrafish embryos to 0.3, 3 or 30 ppb atrazine from 1-72 hours post fertilization (hpf; the end of embryogenesis) and identified zebrafish and human miRNAs that were significantly altered in response to atrazine. Targeted analysis was then performed to determine the regulation of gene expression by miRNAs on altered gene targets from our previous transcriptome analysis (Weber et al., 2013). Furthermore, developmental characterization and atrazine toxicity of hsa-miR-126-3p, a miRNA reported to be altered in all atrazine treatments, was established.
2. Materials and Methods
2.1 Zebrafish husbandry and experimental design
Zebrafish (wild-type AB strain) were housed in a Z-Mod System (Aquatic Habitats, Apopka, FL) on a 14:10 hour light:dark cycle and maintained at 28°C (±1°C) with a pH of 7.0 - 7.2 and conductivity range of 470-520 μ S. Adult zebrafish were bred in cages and embryos were collected, staged, and rinsed with system water as described previously (Peterson et al., 2011 ). Embryos were dosed with 0, 0.3, 3, or 30 ppb atrazine (CAS #1912-24-9; Chem Service, 98% purity) from 1-72 hours post fertilization (hpf) as previously described (Weber et al., 2013; Wirbisky et al., 2015). Four biological replicates (n=4), each from a different clutch, were included in each of the experiments. All animal protocols were approved and performed in accordance with Purdue University’s Institutional Animal Care and Use Committee guidelines.
2.2 miRNA microarray analysis of 72 hpf zebrafish larvae
Upon completion of the exposure period, 50 embryos from each treatment group of each replicate were collected, pooled, and homogenized in Trizol reagent and flash frozen in liquid nitrogen. Total RNA was isolated from the pooled embryo samples using a miRNeasy kit (Qiagen). The Agilent miRNA Complete Labeling and Hyb Kit was used to fluorescently label samples along with the MicroRNA Spike-In Kit to measure the efficiency of the labeling process. A total of 100 ng total RNA from each sample was used for labeling. Samples were dephosphorylated using Calf Intestinal Phosphatase and then denatured with dimethyl sulfoxide. Following denaturation, samples were cooled and subsequently labeled with Cyanine 3-pCp using a T4 RNA Ligase. Labeled samples were purified using MicroBioSpin 6 columns (Bio-Rad, Hercules, CA) and dried using a Savant Speed Vac on medium-high heat for 1 hour. Dried samples were resuspended in nuclease-free water and prepared for hybridization using GE Blocking Agent and Hi-RPM Hybridization Buffer. Samples were loaded onto a custom designed 8X60K Agilent miRNA array in which all human and zebrafish miRNAs were included (based on miRBase release 18.0). Arrays were loaded into SureHyb chambers and hybridized at 55°C for 20 hours in an Agilent Microarray Hybridization Oven. The following day, arrays were removed from the oven, washed with Agilent Gene Expression Wash Buffers and scanned on an Agilent SureScan Microarray scanner. Probe information was extracted from generated tif images using Feature Extraction image analysis software 9.5.3 using QC Metric Set to evaluate the quality of labeling and hybridization. A single replicate was removed from the 30 ppb treatment group as it did not meet QC standards. Data were background-subtracted, normalized to the 90th percentile, and analyzed for differential expression in GeneSpring 12.5 software using a one-way analysis of variance (ANOVA) with a Tukey’s post hoc test (p<0.05) to determine which atrazine treatment groups were different from the control.
2.3 miRNA targeted analysis of gene expression microarrays
miRNAs found to be differentially expressed were uploaded in Ingenuity Pathway Analysis (IPA) software program for targeting analysis of previously obtained 72 hpf gene expression data (Weber et al., 2013). The microRNA Target Filter in IPA provides insights into the biological effects of microRNAs and filters microRNA-mRNA data relationships using experimentally validated interactions from TarBase, miRecords, and peer-reviewed biomedical literature. This software currently does not support this analysis for zebrafish miRNAs.
2.4 Deposition of data
Array data has been deposited at the gene expression data base GEO (GSE78805).
2.5 Quantitative polymerase chain reaction (qPCR) developmental characterization of mir-126-3p
Zebrafish embryos were exposed to control aquaria water for completion of the developmental characterization of hsa-miR-126-3p. Embryos were collected at 12, 24, 36, 48, 60, and 72 hpf (n=4). In addition, to determine the effects of atrazine on hsa-miR-126-3p throughout development, zebrafish embryos were exposed to control aquaria water or an atrazine treatment of 0.3, 3, or 30 ppb throughout embryogenesis and collected at 12, 24, 36, 48, and 60 hpf (n=3). Following the specified developmental period (for both developmental characterization and atrazine exposed embryos), 50 embryos from each treatment group in each replicate were collected, pooled, and homogenized in Trizol reagent and flash frozen in liquid nitrogen. miRNA was isolated from the pooled embryo samples using a miRNeasy kit (Qiagen). miRNA was synthesized to cDNA using the Universal RT cDNA synthesis kit from Exiqon following manufacturer’s recommendations (Exiqon, Woburn, MA). The probe specific to hsa-miR-126-3p was designed using locked nucleic acid technology by Exiqon. qPCR was performed using Exiqon SYBR Green kit according to manufacturer’s recommendation (Exiqon, Woburn, MA). qPCR was performed following similar methods as previously described (Weber et al., 2013; Wirbisky et al., 2015) following the minimum information for publication of quantitative real-time PCR experiments (MIQE) guidelines (Bustin et al., 2009). Several genes were assessed to determine the best reference to be used for this data set (data not shown). U6 was found to be most consistent and least variable for this analysis. Experimental samples were run in triplicate (technical replicates) and gene expression was normalized to U6. Efficiency and specificity were checked with melting and dilution curve analysis and no-template controls. The developmental time course and atrazine-treated samples were analyzed for statistical significance with an ANOVA and a post-hoc least significant difference test when a significant ANOVA was observed (p<0.05).
3. Results
3.1 Developmental atrazine exposure alters zebrafish and human miRNAs
Microarray analysis using a platform containing all known sequences for zebrafish and human miRNAs revealed only one human miRNA (hsa-miR-126-3p) with altered expression in the 0.3 ppb atrazine treatment group following an embryonic exposure. In the 3 ppb atrazine treatment group, expression of two zebrafish miRNAs and three human miRNAs were significantly decreased. The 30 ppb atrazine treatment group had 16 zebrafish miRNAs and 6 human miRNAs with a significant decrease in expression. Overall a dose response increase in the number of miRNAs affected was observed (Table 1).
Table 1.
List of significantly altered zebrafish and human miRNAs following embryonic atrazine exposure.
| Atrazine Treatment |
Systematic Name | Log2 | p-value |
|---|---|---|---|
| 0.3 ppb | hsa-miR-126-3p | −2.9067 | 0.0028 |
| 3 ppb | dre-miR-10a-5p | −0.1808 | 0.0087 |
| dre-miR-10c | −0.2451 | 0.0139 | |
| hsa-miR-126-3p | −2.9201 | 0.0028 | |
| hsa-miR-3907 | −0.3769 | 0.0021 | |
| hsa-miR-455-3p | −0.1223 | 0.0037 | |
| 30 ppb | dre-miR-10a-3p | −0.2872 | 0.0277 |
| dre-miR-10a-5p | −0.1961 | 0.0087 | |
| dre-miR-10d-5p | −0.1709 | 0.0161 | |
| dre-miR-143 | −0.2389 | 0.0168 | |
| dre-miR-153c | −0.1718 | 0.0260 | |
| dre-miR-16a | −0.2870 | 0.0188 | |
| dre-miR-16b | −0.1513 | 0.0290 | |
| dre-miR-18a | −0.1510 | 0.0046 | |
| dre-miR-18b-5p | −0.1335 | 0.0426 | |
| dre-miR-216b | −0.1909 | 0.0301 | |
| dre-miR-217 | −0.2326 | 0.0295 | |
| dre-miR-218b | −0.2144 | 0.0162 | |
| dre-miR-23a | −0.1612 | 0.0489 | |
| dre-miR-24 | −0.1186 | 0.0242 | |
| dre-miR-26a | −0.2229 | 0.0213 | |
| dre-miR-30e-5p | −0.1302 | 0.0353 | |
| hsa-miR-101-3p | −0.4652 | 0.0432 | |
| hsa-miR-10b-3p | −4.0142 | 0.0011 | |
| hsa-miR-124-3p | −0.1533 | 0.0220 | |
| hsa-miR-126-3p | −2.9791 | 0.0028 | |
| hsa-miR-3907 | −0.4577 | 0.0021 | |
| hsa-miR-455-3p | −0.1079 | 0.0037 | |
3.2 Targeting analysis with altered miRNAs and past transcriptomic results
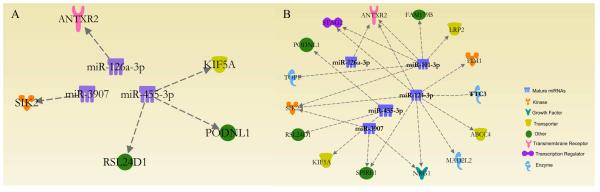
Targeting analysis between the microarray miRNA results with genes previously identified to be altered at the same atrazine exposures was completed for the human miRNAs using Ingenuity Pathway Analysis (IPA). This analysis revealed no targeted genes in the 0.3 ppb treatment by hsa-miR-126-3p. The 3 ppb atrazine treatment showed three human miRNAs (hsa-126-3p, hsa-miR-3907, and hsa-miR-455-3p) targeting 5 genes (ANTXR2, SIK2, KIF5A, PODNL1, and RSL24D1) (Table 2; Figure 1A). The most robust response occurred in the 30 ppb treatment with 5 miRNAs (hsa-miR-101-3p; hsa-miR-124-3p, hsa-126-3p, hsa-miR-3907, and hsa-miR-455-3p) targeting 15 unique genes (Table 3; Figure 1B).
Table 2.
Targeting analysis of human miRNAs and genes altered at 72 hpf in the 3 ppb atrazine treatmenta.
| Systematic Name |
miRNA p-value | Gene Symbol |
mRNA Log2 | Biological Processes |
|---|---|---|---|---|
| hsa-miR-126-3p | 0.00285 | ANTXR2 | 0.9949 | Unknown |
| hsa-miR-3907 | 0.00213 | SIK2 | 1.5266 | Cellular Processes |
| hsa-miR-455-3p | 0.00374 | KIF5A | 1.2455 | Cellular Processes |
| PODNL1 | 1.5216 | Unknown | ||
| RSL24D1 | 0.5926 | Translation | ||
Gene data is previously published in Weber et al. [35].
Figure 1. Schematic diagram of the targeted analysis of the 3 and 30 ppb atrazine treatments.
Targeted analysis of the altered miRNAs in the 3 ppb atrazine treatment to the previously altered mRNAs identified 3 miRNAs targeting 5 mRNAs through the Ingenuity Pathway Analysis (IPA) program (A). Targeted analysis of the altered miRNAs in the 30 ppb atrazine treatment to the previously altered mRNAs identified 5 miRNAs targeting 15 mRNAs through the IPA program (B). Overall a dose response was observed with similar miRNAs and more miRNAs changed as you increase in dose. In addition, as expected the down regulation of miRNA expression resulted in an increase in mRNA expression for all targets.
Table 3.
Targeting analysis of human miRNAs and genes altered at 72 hpf in the 30 ppb atrazine treatmenta
| Systematic Name |
miRNA p-value | Gene Symbol |
mRNA Log2 | Biological Processes |
|---|---|---|---|---|
| hsa-miR-101-3p | 0.0432 | ANTXR2 | 1.2624 | Unknown |
| FAM179B | 0.8655 | Unknown | ||
| LRP2 | 0.9030 | Cellular Signaling | ||
| PIM1 | 0.8883 | Cellular Processes | ||
| SIK2 | 1.6906 | Cellular Processes | ||
| STAG2 | 0.6248 | Cell Division | ||
| TOP1 | 0.6645 | Cell Cycle | ||
| hsa-miR-124-3p | 0.0220 | ABCC4 | 0.6933 | Metabolic Processes |
| ANTXR2 | 1.2624 | Unknown | ||
| MAD2L2 | 1.4547 | Cell Cycle | ||
| NRG1 | 0.7991 | Cell Signaling | ||
| PIM1 | 0.8883 | Cellular Processes | ||
| SIK2 | 1.6906 | Cellular Processes | ||
| SPIRE1 | 0.7190 | Cellular Processes | ||
| STAG2 | 0.6248 | Cell Division | ||
| TTC3 | 1.0902 | Differentiation | ||
| hsa-miR-126-3p | 0.0029 | ANTXR2 | 1.2624 | Unknown |
| hsa-miR-3907 | 0.0021 | NRG1 | 0.7991 | Cell Signaling |
| SIK2 | 1.6906 | Cellular Processes | ||
| SPIRE1 | 0.7190 | Cellular Organization | ||
| hsa-miR-455-3p | 0.0037 | KIF5A | 0.8041 | Cellular Processes |
| PODNL1 | 1.6064 | Unknown | ||
| RSL24D1 | 0.6708 | Translation |
Gene data is previously published in Weber et al. [35].
3.3 Quantitative polymerase chain reaction (qPCR) developmental characterization of hsa-miR-126-3p
As hsa-miR-126-3p was identified to be altered in all three atrazine treatment groups, analysis on the developmental time course expression was first conducted to further characterize this miRNA. Expression at 12 hpf was significantly different from all other time points with minimal expression observed. Expression at 24 and 48 hpf showed statistically similar medial expression, while 36, 60, and 72 hpf displayed higher expression levels (p<0.0001) (Figure 2).
Figure 2. Developmental expression characterization of hsa-miR-126-3p in the zebrafish.
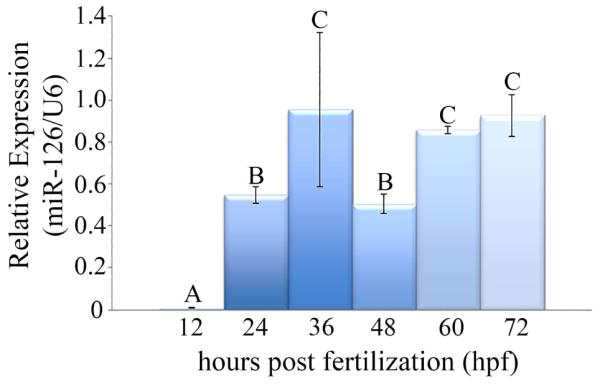
Zebrafish embryos were exposed to control aquaria water from 1-72 hpf and collected at 12, 24, 36, 48, 60, and 72 hpf for developmental characterization of miR-126. Expression of miR-126 was time point specific (p<0.0001). A dramatic increase in expression occurred after 12 hpf with peak expression at 36 hpf. After a decrease in expression at 48 hpf, expression increased again at 60 and 72 hpf. (n=4; different letters indicate that time points are significantly different from each other; miR-126 expression was normalized to U6 as a reference. Error bars indicate standard deviation).
3.4 qPCR analysis of hsa-miR-126-3p expression following embryonic atrazine exposure at additional developmental time points
To determine if atrazine exposure would alter expression of hsa-miR-126-3p at additional developmental time points, qPCR analysis was conducted at 12, 24, 36, 48, and 60 hpf. Expression at 12 hpf was minimal and did not meet expression threshold for analysis. No significant alterations in hsa-miR-126-3p was found at 24, 36, or 48 hpf in any of the atrazine concentrations (p=0.1497; p=0.2557; p=0.0998, respectively) (Figure 3A-C). However, a significant increase was observed at 60 hpf in the 30 ppb atrazine treatment (p=0.0281) (Figure 3D).
Figure 3. Expression analysis of hsa-miR-126-3p at multiple developmental time points following atrazine exposure in zebrafish.

miR-126 expression was examined at five developmental time points (12, 24, 36, 48, and 60 hpf) following an atrazine exposure of 0.3, 3, or 30 ppb or a control treatment. Expression at 12 hpf was minimal and below the threshold for analysis. No significant differences were observed at 24 (A), 36 (B), or 48 hpf (C). Expression levels were significantly increased in the 30 ppb atrazine treatment at 60 hpf (D). (n=3; *p<0.05; miR-126 expression was normalized to U6 as a reference. Error bars indicate standard deviation.)
4. Discussion
A limited number of studies to date have investigated miRNA expression alterations in response to EDC exposure in zebrafish and humans (Avissar-Whiting et al., 2010; Hsu et al., 2009; Jenny et al., 2012; Tilghman et al., 2012; Veiga-Lopez et al., 2013). Furthermore, there are limited to no published reports on the disruption of epigenetic regulators in response to atrazine exposure in human or zebrafish studies. The investigation into the adverse effects of an embryonic atrazine exposure on miRNAs is important as miRNAs are essential for embryogenesis, tissue morphogenesis, and cellular processes. In addition, miRNAs are thought to contribute to disease progression and genetic disorders (Bhattacharya et al., 2016; Esteller 2011; Tal and Tanguay, 2012). Thus, our goal in this study was to identify miRNAs that were altered in response to atrazine exposure using miRNA expression arrays interrogating both human and zebrafish miRNAs. This approach was chosen based on conservation of miRNA sequences and the fact that zebrafish miRNAs are still being identified. We identified a total of 17 zebrafish and 6 human unique miRNAs significantly altered in response to atrazine exposure, albeit, robustness in change was not high. A dose response in the number of miRNAs altered was also observed with additional miRNAs changed as treatment concentration increased.
An interesting finding was the alteration of several members of the zebrafish miR-10 family in the 3 and 30 ppb atrazine treatments. The miR-10 family is located in an evolutionary conserved region of the genome which contains the Hox cluster of genes. The Hox genes are critical regulators of the anterior-posterior body axis and the miR-10 family regulates the expression of HoxB1a and HoxB3a and seemingly works in conjunction with the HoxB4 gene (Woltering and Durston, 2008). Alterations of the miR-10 family could be a contributing factor in the previously reported increase in head length following embryonic atrazine exposure (Weber et al., 2013). Furthermore, as epigenetic alterations can be inheritable, we have also previously shown offspring of adult zebrafish that were only exposed to 0.3 or 3 ppb atrazine during embryogenesis displayed a significant increase in their head width to total body length ratio, while offspring of adult zebrafish exposed to 30 ppb atrazine during embryogenesis had a significant decrease in their head length to total body (Wirbisky et al., 2016). In addition, a recent study employing both the zebrafish model and human umbilical vein endothelial cells, found that miR-10 members regulate angiogenic signaling, making it a potential marker for modulation of angiogenesis (Hassel et al., 2012). Other significantly altered miRNAs involved in the regulation of angiogenesis include miR-23a and miR-24. These miRNAs are members of the miR-23/27/24 cluster which is highly conserved through vertebrate animals (Bang et al., 2011).
miRNA dysregulation has also been reported as a key regulator in the development of various cancers (Heneghan et al., 2010; Hu et al., 2010; Lawrie et al., 2008). In this study we observed alterations within the miR-16 family (miR-16a and miR-16b). miRNAs of the miR-16 family have been evaluated as a tool in the identification of prostate, gastric, and breast cancer (Mobarra et al., 2015). The miR-16 family has also been identified to inhibit cell proliferation, regulate cell cycle, and promote apoptosis, further supporting its role in tumorigenesis (Aluru et al., 2013; Mobarra et al., 2015). Embryonic atrazine exposure also elicited a decrease in miR-18a and miR-18b-5p. These miRNAs are members of the miR-17-92 cluster, and are highly expressed in several cancers (Komatsu et al., 2014). Other miRNAs significantly altered include miR-143, miR-216b, and miR-217. These miRNAs are also primary tools in cancer development as they function as tumor suppressors (Faraji et al., 2015; Shen et al., 2014).
miRNAs are also implicated to play a role in brain development and neurogenesis with dysregulation contributing to various neurological diseases (Sun and Shi, 2015). We observed zebrafish miR-26a altered in the 30 ppb atrazine treatment group. miR-26 functions in neuronal development and is shown to target the ctdsp2 gene in zebrafish which is important in the differentiation process of neural stem cells to neurons. Intriguingly, this miRNA is encoded in an intron of the ctdsp2 gene and is expressed along with the gene rather than independently (Dill et al., 2012).
In addition to the altered zebrafish miRNAs, we examined the effects of atrazine on human miRNAs and report 6 unique significantly altered miRNAs. The human miRNA miR-126-3p was found to be significantly and robustly down-regulated in all three atrazine treatments with just over a 7-fold decrease in expression. miR-126 has been investigated in a number of cancer-based studies with several reports pointing towards a potential tumor-suppressive role in non-small cell lung cancer, breast cancer, pancreatic and gastric cancers (Feng et al., 2010; Hamada et al., 2012; Yang et al., 2012; Zhang et al., 2013). In addition, a significantly reduced human miRNA in the 30 ppb atrazine treatment belongs to the miR-124 family (miR-124-3p). This miRNA is specifically expressed in the central nervous system in post-mitotic neurons and promotes neuronal differentiation and maturation (Sun and Shi, 2015). The role of miR-124 in development and neurogenesis has also been studied in rodent and zebrafish models (Aluru et al., 2013; Cheng et al., 2009). Results also showed human miR-10b to be significantly altered in the 30 ppb atrazine treatment which affirms the high homology between the zebrafish and human miR-10 family.
Human miRNAs that were found to be significantly altered were further analyzed using IPA to determine if these miRNAs targeted genes previously altered by a developmental atrazine exposure (Weber et al., 2013). In our previous study, alterations in larval morphology, mRNA, and corresponding protein levels of selected mRNA targets were altered with mRNA and proteins linked to neuroendocrine function, cell cycle, and carcinogenesis (Weber et al., 2013). Only human miRNAs could be included in this analysis, as the software doesn’t currently allow zebrafish miRNAs. Targeting analysis of the previously identified 23 altered genes revealed no altered targeted mRNAs in the 0.3 ppb treatment. Targeting analysis of the 62 previously altered genes in the 3 ppb treatment group revealed three miRNAs (hsa-miR126-3p, hsa-miR-3907, and hsa-miR-455-3p) which targeted 5 genes (ANTXR2, SIK2, KIF5A, PODNL1, and RSL24D1). Of key interest is the targeted regulation of SIK2. SIK2 is a kinase of the AMPK family that plays a role in CREB1 mediated gene transcription and numerous cell signaling processes including insulin signaling and gluconeogenesis (Henriksson et al., 2012; Walkinshaw et al., 2013). In addition, as we have previously discussed, numerous miRNAs altered by atrazine play a role in tumorigenesis; therefore, alterations in SIK2, which is currently under investigation as a target in cancer therapy further supports miRNA regulation of cancer targets (Ahmed et al., 2010; Bon et al., 2015).
Targeting analysis of the 30 ppb gene set showed the highest number of genes targeted by miRNAs with 5 miRNAs targeting 15 unique genes. Alterations in cell cycle progression are reported to occur following atrazine exposure resulting in an accumulation of cells in S-phase in vitro (Freeman et al., 2006; Powell et al., 2011). Our previous gene expression analysis revealed alterations in numerous genes associated with cell cycle progression and proliferation (AVP, BRCA2, MAD2L2, MCM7, PIM1, and TPD52L1) (Weber et al., 2013). Here, we report that targeting analysis revealed 2 miRNAs which targeted PIM1; miR-101-3p and miR-124-3p. PIMI has a known function of regulating cell cycle progression from late G1 through S-phase. In addition, STAG2, TOP1, and MAD2L2 were also miRNA targets that are involved in cell cycle processes including formation of the cohesion complex which is necessary for the separation of sister chromatids, alterations of topologic states, and accurate mitosis (Li et al., 2015; Listovsky et al., 2013; Xu et al., 2015). One limitation of the targeting analysis is that the samples analyzed in the current study and in the past transcriptomic analysis was from different biological samples. Although both sets of samples were analyzed at the same developmental time point (72 hpf) the rate of development is known to differ slightly among clutches with data providing a snapshot of expression. As such, this may create some difficulties in a direct comparison of the direction of change between the miRNA and mRNA, but observations in this study were in agreement with expected results in that decreases in miRNA expression were associated with increased mRNA expression signifying that there would have been less miRNA to repress mRNA expression.
As previously stated, we reported a significant and robust alteration in the human miRNA hsa-126-3p in all three atrazine concentrations. miR-126 is conserved among species and is found within intron 7 of the EGF-like domain-containing protein 7 (EGFL7) gene in vertebrates (Meister and Schmidt, 2010). miR-126 is found in highly vascularized tissues such as the heart, liver, and lung, and is the only miRNA known to be specifically expressed in the endothelial cell lineage, hematopoietic progenitor cells, and endothelial cell lines (Meister and Schmidt, 2010). Studies indicate that miR-126 is involved in regulating multiple cellular processes including blood cell development, inflammation, and angiogenesis (Meister and Schmidt, 2010).
Due to the consistent and robust alteration in miR-126 across all atrazine treatments, we sought to characterize the basal levels of miR-126 expression throughout zebrafish development. Results indicated that the expression of miR-126 is developmental time point specific. At 12 hpf, miR-126 had the lowest expression level with rapid increases at 36, 60, and 72 hpf. Due to miR-126’s known involvement with angiogenesis and vascular integrity, the variation in expression observed in this study is hypothesized to be linked to the rapid vascular development occurring during zebrafish embryogenesis. In early zebrafish development, angiogenesis occurs in two surges to form the intersegmental vessels (ISVs). The first wave begins at 20– 24 hpf, with the formation of the first ISVs from the dorsal aorta (Ellertsdóttir et al., 2010). Around 30 hpf, each new sprout formed from the dorsal aorta is made up of 3 to 4 cells with distinct positional fates (Baldessari et al., 2008). The differentiated functions of these cells allow each ISV to grow dorsally to form segmental arteries and eventually connect with neighboring ISVs to form the dorsal longitudinal anastomotic vessel (Ellertsdóttir et al., 2010). In the current study, miR-126 was found to have minimal expression at 12 hpf and then increase in expression at 24 hpf corresponding to the first wave of angiogenesis. At 32–34 hpf, the second wave of angiogenesis begins with the formation of small branches from the posterior cardinal vein. These new vessels merge with existing segmental arteries to form segmental veins or grow separately to contribute to the lymphatic vasculature (Ellertsdóttir et al., 2010). Our observed increase of miR-126 at 36 hpf, suggests a correlation to the second wave of angiogenesis. Furthermore, a study by Nicoli and colleagues (Nicoli et al., 2010) analyzed angiogenesis of the accessory fifth aortic arch (AA5x), which occurs around 60 hpf, and its dependence on blood flow. Results revealed that the flow-dependent transcription factor, krueppel-like factor 2 (klf2a), was responsible for activating miR-126 to induce vegf signaling and allow for the formation of a patent circulatory connection (Nicoli et al., 2010). The increase in miR-126 expression later in zebrafish development at 60 and 72 hpf suggests a relationship to this mechanism.
A previous study conducted by Zou and colleagues (2011) utilized the Tg(fli1:EGFP)y1 transgenic zebrafish line and also analyzed miR-126 expression throughout zebrafish embryogenesis (Zou et al., 2011). This study examined the expression of miR-126 at similar developmental time points (12, 24, 36, 48, and 72 hpf) and revealed increasing expression as development progressed; however, statistical analysis was not completed in order to determine statistical significance between developmental time points. While findings from Zou and colleagues (2011) are in agreement that miR-126 expression increases after 12 hpf, they differ in their analysis at 48 hpf. This difference may be attributed to the use of differing zebrafish lines. The current study used wild type AB zebrafish, while Zou and colleagues (2011) used the Tg(fli1:EGFP)y1 transgenic line (Zou et al., 2011). Thus, genetic profiles of the zebrafish are different and may also result in a slightly different maturation progression.
Investigating the effects of EDCs on epigenetics and in particular, miRNAs, is beginning to come into view in order to define mechanisms of toxic action (Casati et al., 2015; Hsu et al., 2009; Tilghman et al., 2012). To date, no studies have been completed examining the effects of atrazine on miRNA expression. Therefore, in the present study, we also demonstrated that miR-126 deregulation was not observed in the majority of the developmental time points following atrazine exposure. However, zebrafish embryos treated with 30 ppb atrazine through 60 hpf also showed a significant increase in expression. While atrazine does not appear to have an effect on ISV formation due to the lack of deregulation at 24 and 36 hpf, the up regulation at 60 hpf may have an effect on the mechanism governing angiogenesis in the aortic arches, specifically AA5x. As mentioned above, angiogenesis during AA5x development is induced by blood flow, which activates klf2a to induce miR-126, which results in enhanced vegf expression (Nicoli et al., 2010).. Although vascular integrity was not addressed in this study, results warrant further investigation.
While the objectives of this study surround the developmental toxicity of atrazine exposure, previous studies from our laboratory have focused on defining the later-in-life effects of an embryonic atrazine exposure (Wirbisky et al., 2015, 2016). Our previous studies show that an embryonic atrazine exposure alters 5-hydroxyindoleacetic acid (5-HIAA) and serotonin turnover in adult female brain tissue in addition to numerous gene alterations throughout the serotonergic system (Wirbisky et al., 2015). In addition, we have identified reproductive dysfunction through a decrease in spawning events, increases in follicular atresia and ovarian progesterone, and alterations in gene expression throughout the hypothalamus-pituitary-gonadal (HPG) axis in adult female zebrafish (Wirbisky et al., 2016). Studies investigating epigenetic alterations in various adult tissues following an embryonic atrazine exposure are needed as epigenetics are thought to contribute to the developmental origins hypothesis of disease onset.
5. Conclusions
In this study, we identified alterations in miRNA expression associated with a developmental atrazine exposure. We assessed both zebrafish and human miRNAs in this approach and found changes in miRNA expression for both species which further corroborates the high homology of miRNAs across species. Members of the miR-10 family of miRNAs were found to be altered in both zebrafish and human targets analyzed suggesting that this family of miRNAs may be a specific target of atrazine toxicity. Multiple zebrafish miRNAs associated with cancer were also identified, further supporting previous transcriptome analysis identifying numerous genes associated with carcinogenesis following an embryonic atrazine exposure. Targeting analysis of human miRNAs with previously identified altered gene sets from a developmental exposure to atrazine was carried out to identify downstream functional pathways that are affected from the exposure. These results suggest epigenetic regulators of cell cycle and cell signaling may be targeted by atrazine. Lastly, our characterization and atrazine profile of miR-126 shows support that atrazine could affect cellular processes associated with angiogenesis and warrants future studies into the potential lifespan effects of these alterations
Supplementary Material
Highlights.
Developmental atrazine exposure alters zebrafish and human miRNAs
Targeted analysis links miRNAs to mRNA involved in cell signaling and cell cycle
hsa-miR-126-3p was characterized through development and following developmental atrazine exposure
Acknowledgements
Grants from the National Institutes of Health, National Institute of Environmental Health Sciences to JLF and MSS [R15 ES019137], from the Purdue Center for Cancer Research Innovative Research Pilot Project to JLF and MSS, from the Purdue Research Foundation to JLF and GJW, and from the Cancer Prevention Internship Program administered by the Oncological Sciences Center and the Discovery Learning Research Center at Purdue University to JLF and KES provided support for this study.
Abbreviations
- ANOVA
analysis of variance
- CNS
central nervous system
- EDCs
endocrine disrupting chemicals
- FSH
follicle stimulating hormone
- GnRH
gonadotropin releasing hormone
- hpf
hours post fertilization
- IPA
Ingenuity Pathway Analysis
- ISVs
intersegmental vessels
- LH
luteininzing hormone
- miRNA
microRNA
- ppb
parts per billion
- PRL
prolactin
- US EPA
United States Environmental Protection Agency
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting Data: Array data has been deposited at the gene expression data basebase GEO (GSE78805).
Competing Interests: The authors declare that they have no competing interests.
References
- 1.Ahmed AA, Lu Z, Jennings NB, Etemadmoghadam D, Capalbo L, Jacamo RO, Barbosa-Morais N, Le XF, Australian Ovarian Cancer Study Group. Vivas-Mejia P, Lopez-Berestein G, Grandjean G, Bartholomeusz G, Liao W, Andreeff M, Bowtell D, Glover DM, Sood AK, Bast RC., Jr. SIK2 is a centrosome kinase required for bipolar mitotic spindle formation that provides a potential target for therapy in ovarian cancer. Cancer. Cell. 2010;18:109–121. doi: 10.1016/j.ccr.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aluru N, Deak KL, Jenny MJ, Hahn ME. Developmental exposure to valproic acid alters the expression of miRNAs involved in neurodevelopment in zebrafish. Neurotoxicol. Teratol. 2013;40:46–58. doi: 10.1016/j.ntt.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anway MD, Memon MA, Uzumcu M, Skinner MK. Transgenerational effect of the endocrine disruptor vinclozolin on male spermatogenesis. J. Androl. 2006;27:868–879. doi: 10.2164/jandrol.106.000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ason B, Darnell DK, Wittbrodt B, Berezikov E, Kloosterman WP, Wittbrodt J, Antin PB, Plasterk RH. Differences in vertebrate microRNA expression. Proc. Natl. Acad. Sci. U. S. A. 2006;103:14385–14389. doi: 10.1073/pnas.0603529103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avissar-Whiting M, Veiga KR, Uhl KM, Maccani MA, Gagne LA, Moen EL, Marsit CJ. Bisphenol A exposure leads to specific microRNA alterations in placental cells. Reprod. Toxicol. 2010;29:401–406. doi: 10.1016/j.reprotox.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baldessari D, Mione M. How to create the vascular tree? (Latest) help from the zebrafish. Pharmacol. Ther. 2008;118:206–230. doi: 10.1016/j.pharmthera.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 8.Bang C, Fiedler J, Thum T. Cardiovascular importance of the microRNA-23/27/24 family. Microcirculation. 2011;19:208–214. doi: 10.1111/j.1549-8719.2011.00153.x. [DOI] [PubMed] [Google Scholar]
- 9.Barr DB, Panuwet P, Nguyen JV, Udunka S, Needham LL. Assessing exposure to atrazine and its metabolites using biomonitoring. Environ. Health. Perspect. 2007;115:1474–1478. doi: 10.1289/ehp.10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhattacharya M, Sharma AR, Sharma G, Patra BC, Nam J-S, Chakraborty C, Lee S-S. The crucial role and regulations of miRNAs in zebrafish development. Protoplasma. 2016 doi: 10.1007/s00709-015-0931-1. doi: 10.1007/s00709-015-0931-1. [DOI] [PubMed] [Google Scholar]
- 11.Bon H, Wadhwa K, Schreiner A, Osborne M, Carroll T, Ramos-Montoya A, Ross-Adams H, Visser M, Hoffmann R, Ahmed AA, Neal DE, Mills IG. Salt-inducible kinase 2 regulates mitotic progression and transcription in prostate cancer. Mol. Cancer. Res. 2015;13:620–635. doi: 10.1158/1541-7786.MCR-13-0182-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE Guidelines: Minimum information of publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 13.Casati L, Sendra R, Sibilia V, Celotti F. Endocrine disrupters: the new players able to affect the epigenome. Front. Cell. Dev. Biol. 2015;3:37. doi: 10.3389/fcell.2015.00037. doi: 10.3389/fcell.2015.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper RL, Stoker TE, Tyrey L, Goldman JM, McElroy WK. Atrazine disrupts the hypothalamic control of pituitary-ovarian function. Toxicol. Sci. 2000;53:297–307. doi: 10.1093/toxsci/53.2.297. [DOI] [PubMed] [Google Scholar]
- 16.de Esch C, Slieker R, Wolterbeek A, Woutersen R, De Groot D. Zebrafish as potential model for developmental neurotoxicity testing: A mini review. Neurotoxicol. Teratol. 2012;34:545–553. doi: 10.1016/j.ntt.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Dill H, Linder B, Fehr A, Fischer U. Intronic miR-26b controls neuronal differentiation by repressing its host transcript, ctdsp2. Genes. Dev. 2012;26:25–30. doi: 10.1101/gad.177774.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eldridge JC, Wetzel LT, Stevens JT, Simpkins JW. The mammary tumor response in triazine-treated female rats: a threshold-mediated interaction with strain and species-specific reproductive senescence. Steroids. 1999;64:672–678. doi: 10.1016/s0039-128x(99)00051-3. [DOI] [PubMed] [Google Scholar]
- 19.Ellertsdóttir E, Lenard A, Blum Y, Krudewig A, Herwig L, Affolter M, Belting H. Vascular morphogenesis in the zebrafish embryo. Dev. Biol. 2010;341:56–65. doi: 10.1016/j.ydbio.2009.10.035. [DOI] [PubMed] [Google Scholar]
- 20.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 21.Fa S, Pogrmic-Majkic K, Samardzija D, Glisic B, Kaisarevic S, Kovacevic R, Andric N. Involvement of ERK1/2 signaling pathway in atrazine action on FSH-stimulated LHR and CYP19A1 expression in rat granulosa cells. Toxicol. Appl. Pharmacol. 2013;270:1–8. doi: 10.1016/j.taap.2013.03.031. [DOI] [PubMed] [Google Scholar]
- 22.Faraji F, Hu Y, Wu G, Goldberger NE, Walker RC, Zhang J, Hunter KW. An integrated systems genetics screen reveals the transcriptional structure of inherited predisposition to metastatic disease. Genome. Res. 2015;24:227–240. doi: 10.1101/gr.166223.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng R, Chen X, Yu Y, Su L, Yu B, Li J, Cai Q, Yan M, Liu B, Zhu Z. miR-126 functions as a tumour suppressor in human gastric cancer. Cancer. Lett. 2010;298:50–63. doi: 10.1016/j.canlet.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Foradori CD, Hinds LR, Hanneman WH, Legare ME, Clay CM, Handa RJ. Atrazine inhibits pulsatile luteinizing hormone release without altering pituitary sensitivity to a gonadotropin-releasing hormone receptor agonist in female Wistar rats. Biol. Reprod. 2009;81:40–45. doi: 10.1095/biolreprod.108.075713. [DOI] [PubMed] [Google Scholar]
- 25.Foradori CD, Zimmerman AD, Hinds LR, Zuloaga KL, Breckenridge CB, Handa RJ. Atrazine inhibits pulsatile gonadotropin-releasing hormone (GnRH) release without altering GnRH messenger RNA or protein levels in the female rat. Biol. Reprod. 2013;88:1–7. doi: 10.1095/biolreprod.112.102277. [DOI] [PubMed] [Google Scholar]
- 26.Freeman JL, Raybrun AL. Aquatic herbicides and herbicide contaminants: In vitro cytotoxicity and cell-cycle analysis. Environ. Toxicol. 2006;21:256–263. doi: 10.1002/tox.20179. [DOI] [PubMed] [Google Scholar]
- 27.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 28.Guerrero-Bosagna C, Settles M, Lucker B, Skinner MK. Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS. One. 2010;5:Pii. doi: 10.1371/journal.pone.0013100. e13100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamada S, Satoh K, Fujibuchi W, Hirota M, Kanno A, Unno J, Masamune A, Kikuta K, Kume K, Shimosegawa T. MiR-126 acts as a tumor suppressor in pancreatic cancer cells via the regulation of ADAM9. Mol. Cancer. Res. 2012;10:3–10. doi: 10.1158/1541-7786.MCR-11-0272. [DOI] [PubMed] [Google Scholar]
- 30.Hanson MA, Gluckman PD. Early developmental conditioning of later health and disease: physiology or pathophysiology? Phyisol. Rev. 2014;94:1027–1076. doi: 10.1152/physrev.00029.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hassel D, Cheng P, White MP, Ivey KN, Kroll J, Augustin HG, Katus HA, Stainier DY, Srivastava D. MicroRNA-10 regulates the angiogenic behavior of zebrafish and human endothelial cells by promoting vascular endothelial growth factor signaling. Circ. Res. 2012;111:1421–1433. doi: 10.1161/CIRCRESAHA.112.279711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hatch EE, Troisi R, Wise LA, Titus-Ernstoff L, Hyer M, Palmer JR, Strohsnitter WC, Robboy SJ, Anderson D, Kaufman R, Adam E, Hoover RN. Preterm birth, fetal growth, and age at menarche among women exposed prenatally to diethylstilbestrol (DES) Reprod. Toxicol. 2011;31:151–157. doi: 10.1016/j.reprotox.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann. Surg. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- 34.Henriksson E, Jones HA, Patel K, Peggie M, Morrice N, Sakamoto K, Göransson O. The AMPK-related kinase SIK2 is regulated by cAMP via phosphorylation at Ser358 in adipocytes. J. Biochem. 2012;444:503–514. doi: 10.1042/BJ20111932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsu PY, Deatherage DE, Rodriguez BA, Liyanarachchi S, Weng YI, Zuo T, Liu J, Cheng AS, Huang TH. Xenoestrogen-induced epigenetic repression of microRNA-9-3 in breast epithelial cells. Cancer. Res. 2009;15:5936–5945. doi: 10.1158/0008-5472.CAN-08-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, Chen Y, Xu L, Zen K, Zhang C, Shen H. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small cell lung cancer. J. Clin. Oncol. 2010;28:1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 38.Inawaka K, Kawabe M, Takahashi S, Doi Y, Tomigahara Y, Tarui H, Abe J, Kawamura S, Shirai T. Maternal exposure to anti-androgenic compounds, vinclozolin, flutamide and procymidone, has no effects on spermatogenesis and DNA methylation in male rats of subsequent generations. Toxicol. Appl. Pharmacol. 2009;237:178–187. doi: 10.1016/j.taap.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 39.Jenny MJ, Aluru N, Hahn ME. Effects of short-term exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin on microRNA expression in zebrafish embryos. Toxicol. Appl. Pharmacol. 2012;264:262–273. doi: 10.1016/j.taap.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komatsu S, Ichikawa D, Takeshita H, Morimura R, Hirajima S, Tsujiura M, Kawaguchi T, Miyamae M, Nagata H, Konishi H, Shiozaki A, Otsuji E. Circulating miR-18a: a sensitive cancer screening biomarker in human cancer. In Vivo. 2014;28:239–297. [PubMed] [Google Scholar]
- 42.Kucka M, Pogrmic-Majkic K, Fa S, Stojilkovic SS, Kovacevic R. Atrazine acts as an endocrine disrupter by inhibiting cAMP-specific phosphodiesterase-4. Toxicol. Appl. Pharmacol. 2012;265:19–26. doi: 10.1016/j.taap.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, Hatton CS, Harris AL. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 44.Li SC, Chan WC, Hu LY, Lai CH, Hsu CN, Lin WC. Identification of homologous microRNAs in 56 animal genomes. Genomics. 2010;96:1–9. doi: 10.1016/j.ygeno.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 45.Li X, Zhang TW, Tang JL, Fa PP, Lu JX, Qi FM, Cai ZM, Liu CX, Sun XJ. Loss of STAG2 causes aneuploidy in normal human bladder cells. Genet. Mol. Res. 2015;14:2638–2646. doi: 10.4238/2015.March.30.24. [DOI] [PubMed] [Google Scholar]
- 46.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 47.Listovsky T, Sale JE. Sequestration of CDH1 by MAD2L2 prevents premature APC/C activation prior to anaphase onset. J. Cell. Biol. 2013;203:87–100. doi: 10.1083/jcb.201302060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meister J, Schmidt MH. miR-126 and miR-126*: New players in cancer. Scientific. World. Journal. 2010;10:2090–2100. doi: 10.1100/tsw.2010.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mobarra N, Shafiee A, Rad SM, Tasharrofi N, Soufi-Zomorod M, Hafizi M, Movahed M, Kouhkan F, Soleimani M. Overexpression of microRNA-16 declines cellular growth, proliferation and induces apoptosis in human breast cancer cells. In Vitro. Cell. Dev. Biol. Anim. 2015;51:604–611. doi: 10.1007/s11626-015-9872-4. [DOI] [PubMed] [Google Scholar]
- 50.Nicoli S, Standley C, Walker P, Hurlstone A, Fogarty KE, Lawson ND. MicroRNA-mediated integration of haemodynamics and vegf signaling during angiogenesis. Nature. 2010;464:1196–2000. doi: 10.1038/nature08889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ochoa-Acuña H, Frankenberger J, Hahn L, Carbajo C. Drinking-water herbicide exposure in Indiana and prevalence of small-for-gestational-age and preterm delivery. Environ. Health. Perspect. 2009;117:1619–1624. doi: 10.1289/ehp.0900784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peterson SM, Zhang J, Weber G, Freeman JL. Global gene expression analysis reveals dynamic and developmental stage-dependent enrichment of lead-induced neurological gene alterations. Envrion. Health. Perspect. 2011;119:615–621. doi: 10.1289/ehp.1002590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pogrmic-Majkic K, Fa S, Dakic V, Kaisarevic S, Kovacevic R. Upregulation of peripubertal rat Leydig cell steroidogenesis following 24 h in vitro and in vivo exposure to atrazine. Toxicol. Sci. 2010;118:52–60. doi: 10.1093/toxsci/kfq227. [DOI] [PubMed] [Google Scholar]
- 54.Pogrmic-Majkic K, Samardzija D, Fa S, Hrubik J, Glisic B, Kaisarevic S, Andric N. Atrazine enhances progesterone production through activation of multiple signaling pathways in FSH-stimulated rat granulosa cells: evidence for premature luteinization. Biol. Reprod. 2014;91:1–10. doi: 10.1095/biolreprod.114.122606. [DOI] [PubMed] [Google Scholar]
- 55.Portela A, Esteller M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 56.Powell ER, Faldladdin N, Rand AD, Pelzer D, Schrunk EM, Dhanwada KR. Atrazine exposure leads to altered growth of HepG2 cells. Toxicol. In Vitro. 2011;25:644–651. doi: 10.1016/j.tiv.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 57.Quignot N, Arnaud M, Robidel F, Lecomte A, Tournier M, Cren-Olivé C, Barouki R, Lemazurier E. Characterization of endocrine-disrupting chemicals based on hormonal balance disruption in male and female adult rats. Reprod. Toxicol. 2012;33:339–352. doi: 10.1016/j.reprotox.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 58.Rinsky JL, Hopenhayn C, Golla V, Browning S, Bush HM. Atrazine exposure in public drinking water and preterm birth. Public. Health. Rep. 2012;127:72–80. doi: 10.1177/003335491212700108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rohr JR, McCoy KA. A qualitative meta-analysis reveals consistent effects of atrazine on freshwater fish and amphibians. Environ. Health. Perspect. 2010;118:20–32. doi: 10.1289/ehp.0901164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roy JR, Chakraborty S, Chakraborty TR. Estrogen-like endocrine disrupting chemicals affecting puberty in humans--a review. Med. Sci. Monit. 2009;15:137–145. [PubMed] [Google Scholar]
- 61.Shen JZ, Zhang YY, Fu HY, Wu DS, Zhou HR. Overexpression of microRNA-143 inhibits growth and induces apoptosis in human leukemia cells. Oncol. Rep. 2014;31:2035–2042. doi: 10.3892/or.2014.3078. [DOI] [PubMed] [Google Scholar]
- 62.Solomon KR, Carr JA, Du Preez LH, Giesy JP, Kendall RJ, Smith EE, Van Der Kraak GJ. Effects of atrazine on fish, amphibians, and aquatic reptiles: a critical review. Crit. Rev. Toxicol. 2008;38:721–772. doi: 10.1080/10408440802116496. [DOI] [PubMed] [Google Scholar]
- 63.Stouder C, Paoloni-Giacobino A. Transgenerational effects of the endocrine disruptor vinclozolin on the methylation pattern of imprinted genes in the mouse sperm. Reproduction. 2010;139:373–379. doi: 10.1530/REP-09-0340. [DOI] [PubMed] [Google Scholar]
- 64.Sun E, Shi Y. MicroRNAs: Small molecules with big roles in neurodevelopment and diseases. Exp. Neurol. 2015;268:46–53. doi: 10.1016/j.expneurol.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 65.Swedenborg E, Rüegg J, Mäkelä S, Pongratz I. Endocrine disruptive chemicals: mechanisms of action and involvement in metabolic disorders. J. Mol. Endocrinol. 2009;43:1–10. doi: 10.1677/JME-08-0132. [DOI] [PubMed] [Google Scholar]
- 66.Tal TL, Tanguay RL. Non-coding RNAs--novel targets in neurotoxicity. Neurotoxicology. 2012;33:530–544. doi: 10.1016/j.neuro.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tilghman SL, Bratton MR, Segar HC, Martin EC, Rhodes LV, Li M, McLachlan JA, Wiese TE, Nephew KP, Burow ME. Endocrine disruptor regulation of microRNA expression in breast carcinoma cells. PLoS. One. 2012;7:e32754. doi: 10.1371/journal.pone.0032754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.U.S. EPA . List of Contaminants and Their MCLs. EPA 816-F-02-013. U.S. Environmental Protection Agency; Washington, DC: 2002. [Google Scholar]
- 69.Vandegehuchte MB, Lemière F, Vanhaecke L, Vanden Berghe W, Janssen CR. Direct and transgenerational impact on Daphnia magna of chemicals with a known effect on DNA methylation. Comp. Biochem. Physiol. C. Toxicol. Pharmacol. 2009;151:278–285. doi: 10.1016/j.cbpc.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 70.Veiga-Lopez A, Luense LJ, Christenson LK, Padmanabhan V. Developmental programming: gestational bisphenol-A treatment alters trajectory of fetal ovarian gene expression. Endocrinology. 2013;154:1873–1884. doi: 10.1210/en.2012-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Walkinshaw DR, Weist R, Kim GW, You L, Xiao L, Nie J, Li CS, Zhao S, Xu M, Yang XJ. The tumor suppressor kinase LKB1 activates the downstream kinases SIK2 and SIK3 to stimulate nuclear export of class IIa histone deacetylases. J. Biol. Chem. 2013;288:9345–9362. doi: 10.1074/jbc.M113.456996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weber GJ, Sepúlveda MS, Peterson SM, Lewis SS, Freeman JL. Transcriptome alterations following developmental atrazine exposure in zebrafish are associated with disruption of neuroendocrine and reproductive system function, cell cycle, and carcinogenesis. Toxicol. Sci. 2013;132:458–466. doi: 10.1093/toxsci/kft017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wetzel LT, Luempert LG, 3rd., Breckenridge CB, Tisdel MO, Stevens JT, Thakur AK, Extrom PJ, Eldridge JC. Chronic effects of atrazine on estrus and mammary tumor formation in female Sprague-Dawley and Fischer 344 rats. J. Toxicol. Environ. Health. 1994;43:169–182. doi: 10.1080/15287399409531913. [DOI] [PubMed] [Google Scholar]
- 74.Wirbisky SE, Weber GJ, Sepulveda M,S, Lin TL, Jannasch AS, Freeman JL. An embryonic atrazine exposure results in reproductive dysfunction in adult zebrafish and morphological alterations in their offspring. Sci. Rep. 2016;6:21337. doi: 10.1038/srep21337. doi: 10.1038/srep21337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wirbisky SE, Weber GJ, Sepúlveda MS, Xiao C, Cannon JR, Freeman JL. Developmental origins of neurotransmitter and transcriptome alterations in adult female zebrafish exposed to atrazine during embryogenesis. Toxicology. 2015;333:156–167. doi: 10.1016/j.tox.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Woltering JM, Durston AJ. MiR-10 represses HoxB1a and HoxB3a in zebrafish. PLoS. One. 2008;3:e1396. doi: 10.1371/journal.pone.0001396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu Y, Her C. Inhibition of Topoisomerase (DNA) I (TOP1): DNA Damage Repair and Anticancer Therapy. Biomolecules. 2015;5:1652–1670. doi: 10.3390/biom5031652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang J, Lan H, Huang X, Liu B, Tong Y. MicroRNA-126 inhibits tumor cell growth and its expression level correlates with poor survival in non-small cell lung cancer patients. PLoS. One. 2012;7:e42978. doi: 10.1371/journal.pone.0042978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yokoi T, Nakajima M. Toxicological implications of modulation of gene expression by microRNAs. Toxicol. Sci. 2001;123:1–14. doi: 10.1093/toxsci/kfr168. [DOI] [PubMed] [Google Scholar]
- 80.Zhang Y, Yang P, Sun T, Li D, Xu X, Rui Y, Li C, Chong M, Ibrahim T, Mercatali L, Amadori D, Lu X, Xie D, Li QJ, Wang XF. miR-126 and miR-126* repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat. Cell. Biol. 2013;15:284–294. doi: 10.1038/ncb2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zou J, Li W, Li Q, Li X, Zhang J, Liu G, et al. Two functional microRNA-126s repress a novel target gene p21-activated kinase 1 to regulate vascular integrity in zebrafish. Circ. Res. 2011;108:201–209. doi: 10.1161/CIRCRESAHA.110.225045. [DOI] [PubMed] [Google Scholar]
- 82.Winchester PD, Huskins J, Ying J. Agrichemicals in surface water and birth defects in the United States. Acta Paediatr. 2009;98:664–669. doi: 10.1111/j.1651-2227.2008.01207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.