Abstract
The eukaryotic translation initiation factor 4F (eIF4F) has become essentially synonymous with 5’ cap-dependent mRNA translation. Recent studies demonstrate that cells assemble variants of eIF4F to produce adaptive, cap-dependent translatomes during physiological conditions that inhibit eIF4F. These findings challenge us to reassess classical perceptions of cellular translational pathways.
Keywords: eIF4F, eIF4E, eIF4E2, eIF4G3, hypoxia, translation
Forum
eIF4F: the first of its kind
In eukaryotic cells, protein synthesis is typically initiated by the binding of eIF4F to the 7-methylguanylate (m7G) cap found on the 5’ end of the majority of mRNAs [1]. eIF4F is a heterotrimeric complex consisting of eIF4E1 (cap-binding), eIF4G1 (scaffold), and eIF4A1 (RNA helicase). An overwhelming number of studies have led to the predominant belief that productive, cap-dependent translation is driven exclusively through eIF4F, with the two terms having become all but synonymous. As the driver of the most energy-consuming cellular process, extensive research has yielded significant insight into the regulation of eIF4F. Under standard growth conditions, eIF4E-binding proteins (4E-BPs) are phosphorylated in a mechanistic target of rapamycin (mTOR)-dependent manner [2]. This prevents 4E-BP assembly with eIF4E1. mTOR activity is suppressed by a multitude of stresses, including deficiencies of growth factors, energy, amino acids, and oxygen [1]. This results in the accumulation of hypo-phosphorylated 4E-BP, which binds to eIF4E1 to prevent eIF4F cap-dependent protein synthesis. Inhibition of eIF4F activity results in a marked reduction in global rate of protein synthesis as an adaptive response to stimuli. Although the precise intensity of translation varies, overall translational output under eIF4F-inhibiting stimuli remains robust, decreasing by only 30-50% in hypoxia, for example [3]. How cells sustain such high levels of protein production during periods of eIF4F inactivation has been an area of great interest. To this end, several alternative mechanisms have been proposed to explain cellular translation during eIF4F inhibition. These include the internal ribosome entry site (IRES)-mediated, cap-independent mRNA translation [4], as well as the recently reported m6A mRNA modification, whose effects on cap-dependent and cap-independent translation remain to be resolved [5]. Despite the putative biological relevance of endogenous cap-independent translation, additional concepts are needed to explain how cells maintain robust translation intensity during episodes of eIF4F inhibition.
Condition-specific eIF4Fs: expanding the family
The existence of eIF4F component homologues has been observed across the eukaryotic lineage [6]. For instance, three variants have been identified in mammals to date, eight in Drosophila, five in C. elegans, two in S. pombe, and four in trypanosomatids [6]. In addition, eIF4E and eIF4G variants co-exist in virtually all cell types and tissues examined to date [6]. Together, the high degree of evolutionary conservation and ubiquitous expression of these initiation factor homologues underscore the biological significance of alternative eIF4F complexes, a concept that we are just starting to understand. For instance, we have begun to appreciate that previous assumptions about the roles of eIF4F component homologues were inaccurate. This has motivated the pursuit of new concepts to explain how translation is maintained during eIF4F inhibition. It is now clear that homologues of eIF4E1 are not obligatory translational inhibitors and/or non-functional in in vivo settings, as previously suggested [7]. Through early in vitro and overexpression studies, the eIF4E1 homologue eIF4E2 was reported as a competitive inhibitor of eIF4E1, whereas eIF4E3 was believed to be unable to bind 5’ (m7G) caps. These homologues do not drive translation in cells maintained under standard growth conditions, fueling the conclusion that eIF4E2 and eIF4E3 are inhibitors of protein synthesis. However, these in vitro biochemical assays and cellular studies were performed under standard conditions, which do not consider potential activity in response to stress or stimuli. For example, eIF4E2 was recently shown to drive the bulk of cap-dependent translation in hypoxic cells [8], whereas eIF4E3 mediates cap-dependent protein synthesis in a mitogen-activated protein kinase interacting kinase (MNK)-dependent manner [9]. The alleged redundancy of eIF4G1 and eIF4G3 is also not entirely accurate. While it is true that eIF4E1 can interact with both variants under basal conditions, it has not been formally shown that the translational profile and kinetics are maintained when one variant is inactive or depleted. The picture is more complete for eIF4E2 and eIF4E3, which interact with eIF4G3 and eIF4G1, respectively [9, 10]. While eIF4F complex member homologues may operate as inhibitors of translation under certain settings, they can also act as functional surrogates to the traditional eIF4F on stimuli.
These findings establish the existence of eIF4F variants that become activated in response to environmental or physiological stimuli that inactivate eIF4F. Together with eIF4A, eIF4G3 and eIF4E2 form the hypoxic eIF4F (eIF4FH) that mediates cap-dependent translation in cells exposed to low oxygen conditions [10] (Figure 1). Silencing elements of eIF4FH has no effect on normoxic cells, but effectively abolishes hypoxic translation. As for eIF4E3, this eIF4E1 homologue mediates cap-dependent translation by forming a complex with eIF4G1 and eIF4A (eIF4FM) (Figure 1). Thus, cap-dependent protein synthesis should no longer be equated solely with the canonical eIF4F, but instead be envisioned as a cellular process mediated by distinct eIF4Fs (e.g. eIF4FH) depending on the cell’s environment and physiological conditions. The existence of alternative eIF4Fs that drive cap-dependent translation provides a reasonable explanation as to how cells maintain protein production intensity in the absence of the canonical eIF4F.
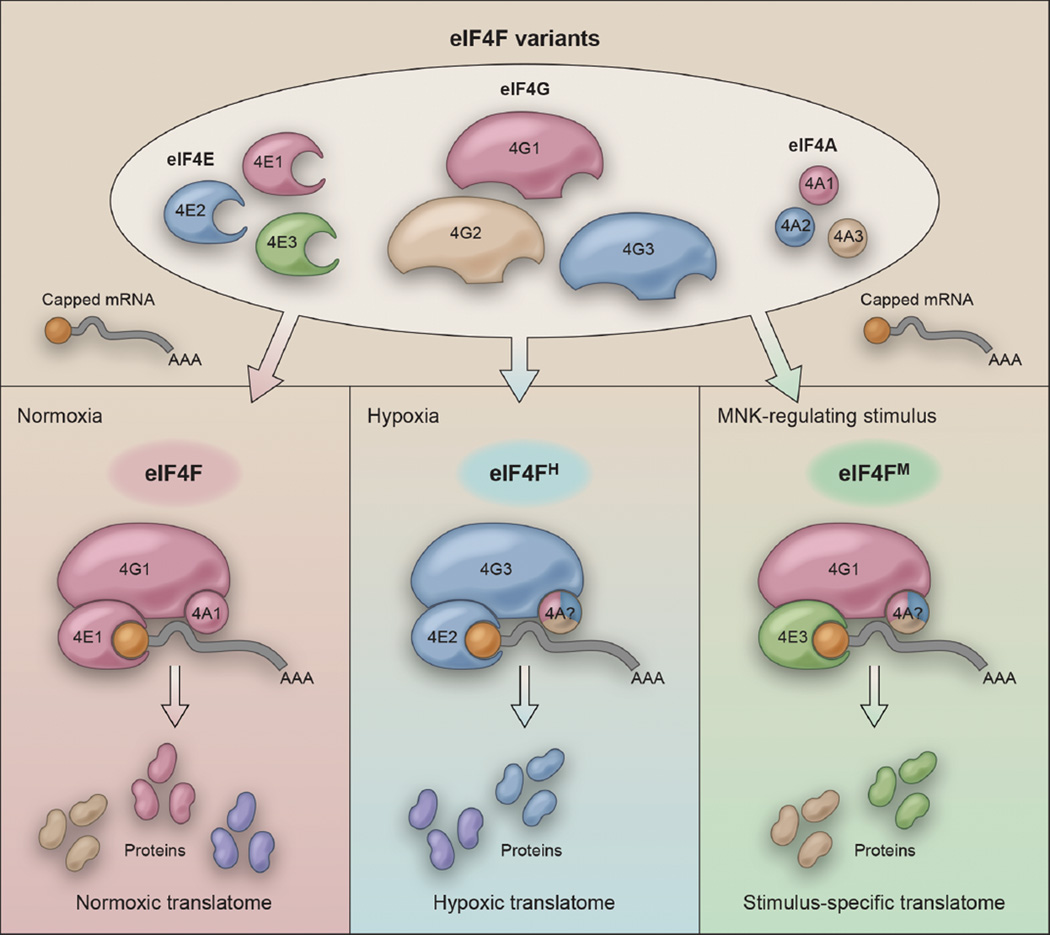
Figure 1. eIF4F variants reprogram the cellular translatome in response to distinct stimuli.
(Top panel) eIF4F-mediated cap-dependent translation in humans involves three major components, eIF4E (cap-binding protein), eIF4G (scaffold protein), and eIF4A (ATP-dependent RNA helicase), each of which has three known homologues (numbered 1, 2, and 3, respectively). Permutations of homologue-specific interactions facilitate adaptation to various stimuli through translatome reprogramming. (Bottom right panel) The basal eIF4F variant contains eIF4G1 and eIF4E1, and produces the normoxic translatome. (Bottom middle panel) Under hypoxic conditions, when eIF4F is inhibited, cells activate the hypoxic eIF4F, eIF4FH, to produce an adaptive, cap-dependent hypoxic translatome. (Bottom right panel) Other stimuli, such as those regulating MNK activity, can potentially activate eIF4E3-containing eIF4F variants (eIF4FM) to produce a stimulus-specific, cap-dependent translatome. The differential utilization of eIF4A variants in distinct eIF4F complexes remains to be tested.
Cap-dependent translation: a new world (of proteins)
The biological rationale behind the deployment of alternative eIF4Fs is an obvious and logical question. So far, the evidence indicates that switching between eIF4Fs enables cells to reprogram their translational output (i.e. translatome), such that proteins that confer adaptive benefits are preferentially synthesized [9–11]. Such translatome remodeling requires the interaction of eIF4Fs with RNA-binding proteins (RBPs), including RBM4 in the case of eIF4FH [8]. These interactions represent a critical regulatory nexus that determines the translational priorities of mRNAs, especially given the multitude of RBPs and their complex relationships with other post-transcriptional regulators such as microRNAs. The identification of eIF4F variants that mediate the production of adaptive translatomes agrees with recent findings that changes in translation efficiency, rather than mRNA concentration, is primarily responsible for stimulus-induced remodeling of the cellular proteome [12].
These observations reshape our current concept of cap-dependent translation. For instance, the traditional model states that hypoxia inhibits global cap-dependent protein synthesis, as well evidenced by mTOR-mediated inhibition of eIF4E1/eIF4F, among others [3]. This model assumes that cap-dependent translation equals eIF4F-driven translation. Instead, we propose the following model: cap-dependent protein synthesis occurs across oxygen concentrations. Hypoxia inhibits eIF4F-mediated translation, but activates eIF4FH-driven protein synthesis. This switch in cap-dependent protein synthesis machineries enables the critical remodeling of the cellular translatome for adaptation to hypoxia.
Future perspectives
As this new paradigm of eIF4F variant-mediated translatome remodeling emerges, a number of outstanding questions remain. For instance, eIF4E2 and eIF4E3 do not bind to 4E-BPs, and thus are presumably resistant to 4E-BP-dependent regulation [6]. Future studies will be required to uncover the pathways that regulate the activities of these homologues and the mechanisms that control target mRNA specificity of different eIF4F variants. These include interactions between target mRNAs and post-transcriptional regulators, such as RNA-binding proteins and microRNAs. For instance, 5’ terminal oligopyrimidine (TOP) elements, which occur in the transcripts of most ribosomal proteins, presumably confer specificity to eIF4E/4E-BP-dependent translation [2], while RBM4 contributes to the eIF4FH-dependence of its target mRNAs [8]. In summary, we have proposed a model of cap-dependent translation whereby cells assemble distinct eIF4F variants to produce adaptive translatomes in response to environmental and physiological stimuli. The findings discussed above encourage us to reexamine our traditional view of eIF4F as the primary driver of cap-dependent protein synthesis.
Acknowledgments
S.L. is funded by the Sylvester Comprehensive Cancer Center and grants from the National Institutes of General Medical Sciences (1R01GM115342) and the National Cancer Institute (1R01CA200676). J.J.D.H is a recipient of a Canadian Institutes of Health Research (CIHR) Postdoctoral Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thoreen CC, et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu LL, et al. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Molecular cell. 2006;21:521–531. doi: 10.1016/j.molcel.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell SF, Parker R. Modifications on Translation Initiation. Cell. 2015;163:796–798. doi: 10.1016/j.cell.2015.10.056. [DOI] [PubMed] [Google Scholar]
- 6.Hernandez G, Vazquez-Pianzola P. Functional diversity of the eukaryotic translation initiation factors belonging to eIF4 families. Mech Dev. 2005;122:865–876. doi: 10.1016/j.mod.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Cho PF, et al. A new paradigm for translational control: inhibition via 5'–3' mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell. 2005;121:411–423. doi: 10.1016/j.cell.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 8.Uniacke J, et al. An oxygen-regulated switch in the protein synthesis machinery. Nature. 2012;486:126–129. doi: 10.1038/nature11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landon AL, et al. MNKs act as a regulatory switch for eIF4E1 and eIF4E3 driven mRNA translation in DLBCL. Nature communications. 2014;5(5413) doi: 10.1038/ncomms6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho JJ, et al. Systemic Reprogramming of Translation Efficiencies on Oxygen Stimulus. Cell reports. 2016;14:1293–1300. doi: 10.1016/j.celrep.2016.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Truitt ML, et al. Differential Requirements for eIF4E Dose in Normal Development and Cancer. Cell. 2015;162:59–71. doi: 10.1016/j.cell.2015.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwanhausser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]