Abstract
In this review we focus on new insights that challenge our understanding of homologous recombination (HR) and Rad51 regulation. Recent advances using high resolution microscopy and single molecule techniques have broadened our knowledge of Rad51 filament formation and strand invasion at double-strand break (DSB) sites and at replication forks, which are one of most physiologically relevant forms of HR from yeast to humans. Rad51 filament formation and strand invasion is regulated by many mediator proteins such as the Rad51 paralogues and the Shu complex, consisting of a Shu2/SWS1 family member and additional Rad51 paralogues. Importantly, a novel RAD-51 paralogue was discovered in C. elegans and its in vitro characterization has demonstrated a new function for the worm RAD-51 paralogues during HR. Conservation of the human RAD51 paralogues function during HR and repair of replicative damage demonstrate how the RAD51 mediators play a critical role in human health and genomic integrity. Together, these new findings provide a framework for understanding RAD51 and its mediators in DNA repair during multiple cellular contexts.
Double-strand break repair by homologous recombination
DNA double-strand breaks (DSBs) are one of the most cytotoxic DNA lesions. One mechanism to repair DSBs is homologous recombination (HR), which uses a homologous template for repair and is therefore generally considered an error-free mechanism. Underscoring the importance of HR, mutations in HR genes are found in many cancer-associated diseases including Bloom syndrome, Werner syndrome, Fanconi anemia, and ataxia telangiectasia (Bernstein et al. 2010; Ellis et al. 1995; Kitao et al. 1998; Puranam and Blackshear 1994; Savitsky et al. 1995; Seki et al. 1994; Shiloh 1997; Yu et al. 1996). After a DSB occurs and HR is engaged, the DSB ends are resected giving rise to 3′ single-stranded DNA overhangs (ssDNA; Figure 1). DNA end resection is first initiated by the Mre11-Rad50-Xrs2 (MRX) complex in the budding yeast Saccharomyces cerevisiae or the MRE11-RAD50-NBS1 (MRN) complex in human cells (Bernstein and Rothstein 2009; Cejka 2015; Mimitou and Symington 2011; Paull 2010; Stracker and Petrini 2011; Takeda et al. 2007; Williams et al. 2010). After the MRX/MRN (yeast proteins are prefaced with sc while human proteins are indicated with an h herein) complex binds to the broken DSB ends, the endonuclease scSae2/hCtIP further stimulates resection in the 5′ to 3′ direction to generate 3′ ssDNA overhangs [Figure 1 and 2; (Clerici et al. 2005; Huertas et al. 2008; Kim et al. 2008; Lengsfeld et al. 2007)]. The short-range resection by scSae2/hCtIP and the MRX/MRN complex is expanded by the redundant activities of the 5′-3′ exonuclease scExo1/hEXO1 and the scSgs1/hBLM helicase in conjunction with the endonuclease scDna2/hDNA2 (Fiorentini et al. 1997; Huang and Symington 1993; Mimitou and Symington 2008; Zhu et al. 2008). The ssDNA that is generated is immediately coated by the ssDNA binding protein complex replication protein A (RPA). RPA-coated ssDNA protects the DSB ends from further degradation and signals to the cell the presence of unrepaired DNA damage (Ghospurkar et al. 2015; Manfrini et al. 2015; Sung and Klein 2006; Wold 1997).
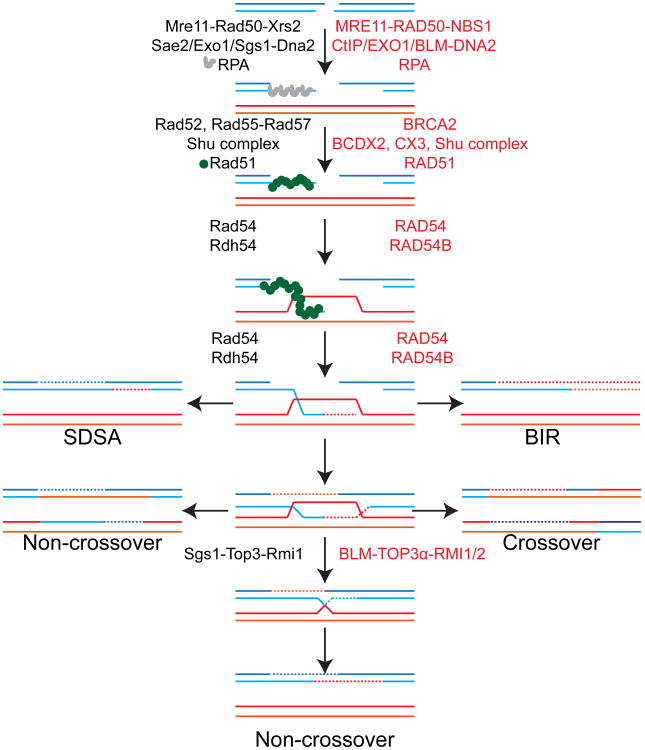
Figure 1.
The different pathways utilized during homologous recombination. Protein names in black refer to budding yeast and are used in the legend, while red names refer to human proteins. All pathways begin with recognition of the DSB, followed by resection mediated by the MRX complex in conjunction with Sae2, Exo1, and Sgs1-Dna2 to generated extended 3′ ssDNA overhangs. These overhangs are immediately coated by the ssDNA binding protein complex RPA. In order for HR to proceed, Rad51 filaments are induced to form on the ssDNA overhangs by the activity of Rad52, the Rad51 paralogues, and the Shu complex. Rad51 filaments, in conjunction with Rad54 and Rdh54, carry out the critical homology search and strand invasion steps of HR. The strand invasion step is the last common step for all the HR pathways modeled here. During SDSA, the strand invasion product is extended past the break-site before being disassembled and reannealed to the other side of the break, generating a non-crossover repair product. In BIR, the strand invasion product becomes a full replication fork and can progress to the end of the chromosome, creating widespread loss-of-heterozygosity. Alternatively, the second end of the DSB can be captured to generate a double Holliday junction (Center). This double Holliday junction can be cleaved by various nucleases to generate both non-crossover and crossover events as depicted on the left and right respectively. The double Holliday junction can also be dissolved by Sgs1-Top3-Rmi1 to yield non-crossover events. Dashed lines refer to DNA synthesized during HR.
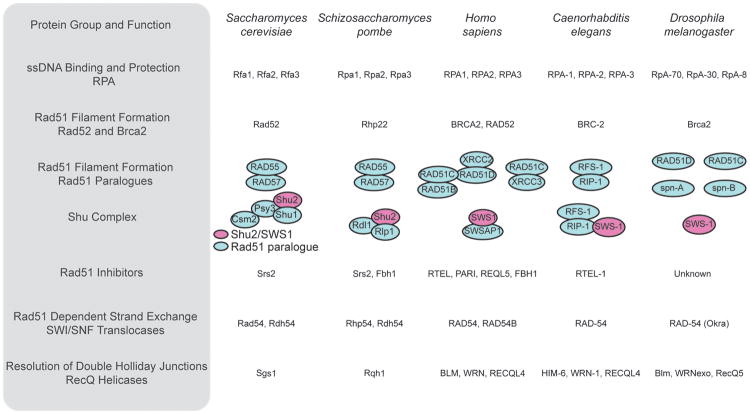
Figure 2.
A schematic of the major HR proteins discussed. Homologues in five eukaryotic lineages are shown, and the Rad51 paralogues and the Shu complex, which are discussed in detail in this review, are drawn to show the known complex members in each species.
Following ssDNA formation, RPA is displaced by formation of a Rad51 filament on the ssDNA, which is essential for the homology search and strand exchange steps that define HR in all eukaryotes [(Heyer et al. 2010); Figure 1]. Rad51 filament formation, also referred to as a presynaptic filament, is tightly regulated to prevent excessive recombination and genomic instability. The first barrier to Rad51 filament formation is the presence of RPA, which blocks Rad51 binding to ssDNA (Heyer et al. 2010). The inhibition of RPA is overcome by the activity of Rad51's regulators. In budding yeast, these include the Rad51 mediators, scRad52 and two Rad51 paralogues, scRad55 and scRad57, which form an obligate heterodimer in vitro (Krogh and Symington 2004; Lisby et al. 2004; Sung 1997; Sung and Klein 2006; West 2003). Rad51 paralogues are proteins that structurally resemble Rad51's ATPase core, and are conserved throughout eukaryotes (Figure 2). Classically, Rad51 paralogues physically associate with each other (such as yeast scRad55 with scRad57) to regulate scRad51's function (Miller et al. 2002; Sung 1997; Wiese et al. 2002). In humans, the RAD51 paralogues form three discrete complexes- including the BCDX2 complex comprised of hRAD51B, hRAD51C, hRAD51D and hXRCC2, the CX3 complex containing hRAD51C and hXRCC3 [(Liu et al. 2011b; Miller et al. 2002; Wiese et al. 2002); Figure 2]. Additionally, the human Shu complex is comprised of the RAD51 paralogue, hSWSAP1, and the SWIM domain containing protein hSWS1, which is discussed in detail below [(Liu et al. 2011b; Miller et al. 2002; Wiese et al. 2002); Figure 2]. The human RAD51 paralogues are predicted to promote HR and regulate hRAD51, but their exact mechanism of action is still under investigation.
In yeast where the Rad51 mediator proteins are the most extensively characterized, they are thought to function by forming nucleation sites for Rad51 to bind to RPA-coated ssDNA. Subsequently, the Rad51 mediators promote the elongation of Rad51 filaments by directing incoming Rad51 monomers to bind to RPA-coated ssDNA (Gibb et al. 2014). The mechanism of how RPA is removed from the ssDNA ends is not well understood but occurs, at least in part, by continuous microscopic dissociation (Gibb et al. 2014). scRad52 is functionally analogous to human BRCA2 during Rad51 filament formation (Chen et al. 1999; Davies et al. 2001; Jensen et al. 2010; Jeyasekharan et al. 2013; Liu et al. 2010; Schlacher et al. 2012; Thorslund et al. 2010). A surprising new study has revealed that scRad52 binds to RPA-coated ssDNA and suppresses its turnover where these RPA-ssDNA clusters remain interspersed along the presynaptic filament (Gibb et al. 2014). scRad51 then assembles onto the ssDNA where scRad51 filament sites are nucleated and extended. Why scRad52-RPA-ssDNA clusters remain along the pre-synaptic filament is puzzling. It has been proposed that they are important for mediating second end capture and/or stabilization of the displaced strand during strand exchange (Gibb et al. 2014). To prevent unnecessary HR, the activities of scRad52 and scRad55-Rad57 are controlled by post-translational modifications that are regulated by DNA damage checkpoint signaling (Bashkirov et al. 2000; Herzberg et al. 2006; Sacher et al. 2006; Torres-Rosell et al. 2007).
In addition to the scRad51 mediators, there are additional proteins, such as the anti-recombinase scSrs2 in budding yeast, that prevent illegitimate HR (Krejci et al. 2003; Veaute et al. 2003). scRad51, which contains Walker A and B ATP binding motifs, will only bind to ssDNA when it is ATP bound. scSrs2 functions by stimulating scRad51 to hydrolyze ATP to ADP, which releases scRad51 from ssDNA (Krejci et al. 2003; Veaute et al. 2003). In humans, numerous proteins carry out anti-recombinase functions to negatively regulate hRAD51 at both pre- and post-synaptic recombination steps, such as hRTEL, hPARI, hFBH1, and hRECQL5, although their mechanisms of action may differ from scSrs2 (Branzei and Foiani 2007; Bugreev et al. 2007; Chu et al. 2015; Hu et al. 2007; Karpenshif and Bernstein 2012; Mankouri et al. 2012; Moldovan et al. 2012). These redundant regulatory systems ensure that functional Rad51 filaments form only at sites of damage and not at other, undamaged portions of the genome.
In human cells, analogous to scRad52, hBRCA2 mediates hRAD51 binding to DSB sites (Chen et al. 1999; Davies et al. 2001; Jensen et al. 2010; Jeyasekharan et al. 2013; Liu et al. 2010; Schlacher et al. 2012; Thorslund et al. 2010). Using live cell imaging of fluorescently-tagged hBRCA2, it was observed that hBRCA2 forms oligomeric clusters (2-5 monomers) and has decreased mobility upon DNA damage [ionizing radiation (IR), hydroxyurea (HU), and mitomycin C (MMC)] suggesting that hBRCA2 is being recruited to damage sites (Reuter et al. 2014). Despite the size difference between hRAD51 and hBRCA2, hRAD51 displays a similar diffusive behavior to hBRCA2 (Reuter et al. 2014). Therefore, the interaction between hBRCA2 and hRAD51 is not limited to the initial hRAD51 nucleation steps of HR (Reuter et al. 2014). These results are consistent with EM studies finding hBRCA2 forming dimmers (Shahid et al. 2014). It is possible that positioning hBRCA2 and hRAD51 in a similar orientation will help to mediate second end capture or to attach to ssDNA independent of the initial orientation that the DNA is bound (Shahid et al. 2014).
Once a Rad51 filament forms, it is stimulated to invade duplex DNA by the Swi/Snf translocase scRad54/hRAD54 where it searches for a homologous stretch of DNA (Heyer et al. 2006; Petukhova et al. 1998). Pre-synapsis, the yeast scRad51-ssDNA samples dsDNA for 8 nucleotide tracts of homology (Qi et al. 2015). Although 8 nucleotides of contiguous homology tracts are the minimum requirement of scRad51 for dsDNA sampling, increasing the homology tract length leads to reduced dissociation rates with a minimal threshold length of 15 nucleotides (Qi et al. 2015). The 15 nucleotide threshold suggests that this is the point when the scRad51 nucleoprotein is no longer capable of sampling another dsDNA for homology and likely represents a commitment step to HR. New insights using a single molecule approach show that the strand exchange activity of scRad51 during the homology search on a complementary dsDNA occurs in three-nucleotide steps (Lee et al. 2015; Qi et al. 2015). Interestingly, a single mismatch within the three-nucleotide sequence can abolish the triplet but scRad51 has the potential to step over mismatches (Lee et al. 2015). After scRad51-mediated strand exchange, scRad51-coated ssDNA filaments are disassembled by scRad54. This allows DNA polymerases to access the heteroduplex DNA and extend the invading ssDNA by an average of 1,700 base pairs (Lo et al. 2006; Solinger et al. 2002). Using total internal reflection fluorescence (TIRF) and scanning force microscopy, the Wyman group has shown that human hRAD54 can integrate into the hRAD51 filament, both at the ends and internally under native conditions (Sanchez et al. 2013). These results support a model in which hRAD54 can both direct RAD51 strand invasion as well as dissemble hRAD51 from the invading ssDNA end to enable polymerases to access the ssDNA (Sanchez et al. 2013).
Once the strand invasion product is extended, it is disassembled through a multi-step process that can result in either a non-crossover or crossover product (Figure 1). Recent in vitro experiments using yeast and human purified proteins has revealed a novel role for scTop3/hTopoIIIα in dissolving scRad51/hRAD51-scRad54/hRAD54 D-loops in a topoisomerase-dependent manner (Fasching et al. 2015). This scTop3/hTopoIIIα activity is only observed on protein coated D-loops and is stimulated by scRmi1/hRMI1-RMI2 (Fasching et al. 2015). No direct interactions between scTop3-Rmi1 and scRad51-Rad54 were observed (Fasching et al. 2015). However, one can imagine the need for topoisomerase-mediated decatenation in D-loop resolution, especially in the context of chromatin-bound DNA. In the simplest form of repair is called synthesis dependent strand annealing (SDSA). During SDSA, the heteroduplex is unwound, the extended ssDNA end invades the other side of the DSB, and the remaining gaps are then filled in and ligated, forming a fully intact DNA helix without a crossover [(Heyer et al. 2010), Figure 1]. Alternatively, the second end of a DSB can be captured, forming a double Holliday junction (dHJ). HJs are resolved through the activity of the scSgs1-Top3-Rmi1/hBLM-TOP3α-RMI1/2 resolvases or through the activities of various nucleases, such as scMus81-Mms4/hMUS81-EME1 and scYen1/hGEN1. These proteins act to cleave the dHJ and generate either crossover or non-crossover HR products (Boddy et al. 2001; Fricke and Brill 2003; Heyer et al. 2010; Ip et al. 2008; Ira et al. 2003; Schwartz and Heyer 2011). If the second end of a DSB is not captured, an alternative form of HR called break-induced replication (BIR) can occur (Figure 1). During BIR, the strand invasion product becomes a full-fledged replication fork and can replicate the template DNA for hundreds of kilobases, potentially to the end of a chromosome (Anand et al. 2013; Costantino et al. 2014). This form of HR can lead to widespread loss of heterozygosity if the donor template is not a sister chromatid.
Novel insights into the function of homologous recombination and RAD51 at damaged replication forks
The HR pathway has been extensively studied at direct clean-ended DSBs, such as those induced by endonucleases. A primary example is the HO cut-site at the mating-type locus in budding yeastor in meiosis at scSpo11-induced DSBs (Haber 1995; Keeney et al. 1997; Weiffenbach and Haber 1985). However, in a normal replicating cell, stalled or broken replication forks are likely the major substrates acted on by the HR machinery (Li and Heyer 2008). Numerous lesions can initiate HR at a replication fork by stalling or blocking replication fork progression. For example, abasic sites produced during processing of base damage as well as bulky DNA adducts such as photoproducts can block replication fork progression. ssDNA breaks can cause replication forks to collapse into one-ended DSBs. Additionally, inter-strand crosslinks (ICLs) which prevent DNA unwinding and are repaired and bypassed, in part, by the HR pathway (Li and Heyer 2008). Each of these lesions is repaired by different mechanisms, but our primary focus here will be on HR at a stalled replication fork (Figure 3).
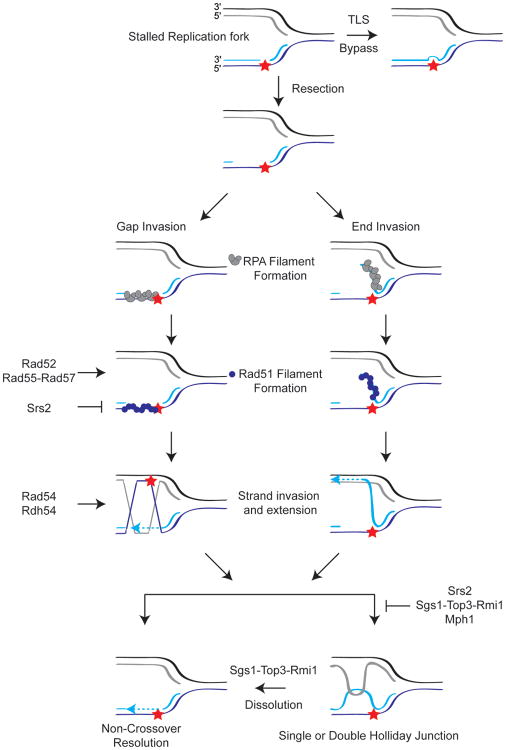
Figure 3.
A schematic highlighting three potential methods to restart a stalled replication fork in budding yeast. In this model, a blocking lesion, indicated by a red star, stalls replication behind the replication fork. This lesion can be bypassed by error-prone translesion polymerases (Top right; TLS), or it can be bypassed using high-fidelity HR (Bottom). Two potential models of how HR can progress at a damaged replication fork are shown. In the model on the left, the ssDNA gap at the replication fork serves as the template for HR. In the right-hand model, the newly synthesized, blocked DNA strand serves as the template. In both pathways, ssDNA is RPA-coated before Rad51 filaments are formed by the activities of Rad52, Rad55-Rad57, and the Shu complex. At the same time, illegitimate HR is inhibited by the anti-recombinase Srs2. Rad51-coated DNA is able to invade the undamaged, newly-synthesized sister chromatid by the activity of Rad54 and Rdh54. This allows extension by different DNA polymerases past the blocking lesion. Finally, both the gap invasion and end invasion pathways can either be dissolved into a non-crossover product (Bottom left) or result in the formation of a single or double Holliday junction (Bottom right), which is then dissolved by Sgs1-Top3-Rmi1.
Visualization of replication fork intermediates at specific replicating origins has been best characterized using two-dimensional (2D) gel electrophoresis (Brewer and Fangman 1987). It has long been suspected that stalled replication forks lead to the formation of recombination intermediates. These intermediates are observed as cruciform structures by 2D gel electrophoresis after exposure to the DNA alkylating agent methylmethane sulfonate (MMS) (Liberi et al. 2005). If the replication fork encounters a MMS-induced DNA lesion, the HR machinery can be used to repair the replication fork damage. In support of HR occurring at damaged replication forks, X-structure formation is scRAD51 dependent and X-structures accumulate in the absence of scSGS1 (Liberi et al. 2005). This genetic evidence strongly supports a model in which stalled replication forks are able to bypass the blocking lesions by HR (Figure 3). New evidence has shown that the X-structures observed by electron microscopy (EM) at MMS-damaged replication forks are indeed HR intermediates (Giannattasio et al. 2014). By directly purifying the X-structures at MMS-damaged replication forks, the Branzei group visualized these structures by EM. Importantly, these damaged forks contained numerous HR intermediates, including dHJs and the hemicatenanes proposed to form during scSgs1-mediated dissolution of dHJs (Giannattasio et al. 2014). Interestingly, in the context of this MMS damage, HR appears to occur at gapped ssDNA (Gap invasion, Figure 3), and not free ssDNA ends (Strand invasion, Figure 3). This work represents some of the strongest evidence to date in support of HR occurring at replicative damage and being initiated by the ssDNA that is produced during blocked replication (Giannattasio et al. 2014; Hashimoto et al. 2010).
Beyond the direct role HR plays in initiating bypass of lesions at stalled replication forks, two groups have independently proposed entirely novel roles for human hRAD51 in protecting these replication forks (Chen et al. 2015; Wang et al. 2015). In a study directed by the Smogorzewska group, a patient who is heterozygous for a single point mutation in the Walker A motif of hRAD51 (hRAD51-T131P) was identified to have Fanconi anemia-like symptoms and a general sensitivity to crosslinking agents (Wang et al. 2015). These findings have resulted in hRAD51 being classified as hFANCR (Wang et al. 2015). Interestingly, in vitro, purified hRAD51-T131P mutant proteins have reduced hRAD51 filament formation when co-incubated with wild-type hRAD51, suggesting that hRAD51-T131P is a co-dominant allele (Wang et al. 2015). However, when integrated into cells by CRISPR, this threshold activity of hRAD51-T131P is sufficient for HR to occur at IR-induced DNA damage sites but not at ICLs (Wang et al. 2015). These findings raise the possibility that hRAD51's role in repair of ICLs, and perhaps replication forks in general, may be distinct or differentially regulated than at IR-induced DSBs. Consistent with an alternative role for hRAD51 during replication-associated repair, an exciting new study by the Kupfer group has reported an HR-independent mechanism for hRAD51 at HU-stalled replication forks (Chen et al. 2015). They find that hRAD51 physically associates with hRAD18 and hFANCD2 specifically following HU, but not MMC treatment.hRAD18 is an E3 Ub-ligase that monoubiquitinates PCNA during replicative stress to initiate bypass of stalled lesions by translesion synthesis (TLS; Figure 3). hFANCD2 is a central protein in the ICL repair pathway that is required to initiate HR at an ICL. Furthermore, the complex of hRAD51 with hRAD18 and hFANCD2 is critical for the monoubiquitination of PCNA and subsequent recruitment of hPolη, which promotes survival in response to HU. This role for hRAD51 in translesion synthesis (TLS) at a stalled replication fork is independent of HR as it occurs in hbrca2-/- cells and when hRAD51's activity is pharmacologically inhibited with B02, a small molecule that inhibits hRAD51's nucleoprotein filament formation. Together, these studies suggest that hRAD51 has a novel role in replication-associated repair which is likely distinct from its canonical role in HR.
Links between mutations in the RAD51 regulators, cancer predisposition and Fanconi anemia
Mutation in hRAD51 itself has recently been associated with a Fanconi anemia-like disorder (hFANCR) (Ameziane et al. 2015; Wang et al. 2015). Consistently, the RAD51 mediators, such as hBRCA2 (hFANCD1) and the hBRCA2 regulator hPALB2 (hFANCN), have long been associated with Fanconi anemia (D'Andrea and Grompe 2003; Xia et al. 2007). In addition to elevated cancer risk, bi-allelic mutations in hRAD51C have been found to cause a Fanconi anemia-like disorder, leading to the classification of hRAD51C as hFANCO (Vaz et al. 2010). The multiple phenotypic consequences of hRAD51C mutations, where some defects lead to cancer predisposition and others lead to Fanconi anemia, suggest that unique functions of hRAD51C may be impaired leading to different disease outcomes. In addition to hRAD51C and hRAD51 being classified as unique Fanconi anemia complementation groups, a patient with a Fanconi anemia-like disorder was recently uncovered to have a truncation in hXRCC2 which is now classified as a Fanconi anemia-associated gene (Shamseldin et al. 2012).
In addition to mutations in hRAD51 metabolism being linked to Fanconi anemia-like disorders, heritable point mutations in hRAD51D, hRAD51C, hXRCC2, and hXRCC3 have become strongly implicated in non-hBRCA2 associated breast, ovarian, and colorectal cancer families (Blanco et al. 2014; Clague et al. 2011; Meindl et al. 2010; Michalska et al. 2015; Namazi et al. 2015; Osorio et al. 2012; Pelttari et al. 2015; Thompson et al. 2012). Understanding how these mutant alleles negatively impact hRAD51 paralogue function and protein interactions is essential for both determining how the hRAD51 paralogues normally function and how alterations in their normal biology can lead to cancer development.
Function of the human RAD51 paralogues in DSB repair and replication
Significant technical barriers have limited our understanding of the human hRAD51 paralogues function both in vitro and in vivo. For example, the low abundance and insolubility of individual hRAD51 paralogues has created a technical barrier for in vitro characterization of their function in human cells (Liu et al. 2011b; Masson et al. 2001). In addition, siRNA depletion of individual hRAD51 paralogues leads to instability of its interacting partners (Chun et al. 2013; Liu et al. 2011b). However, using yeast-2-hybrid and yeast-3-hybrid approaches, two hRAD51 paralogue containing complexes have been described including the BCDX2 (hRAD51B, hRAD51C, hRAD51D, hXRCC2) and CX3 (hRAD51C, hXRCC3) complexes (Liu et al. 2011b; Miller et al. 2002; Wiese et al. 2002). Formation of the BCDX2 and CX3 complexes was later confirmed using proteins purified from E. coli and insect cells (Compton et al. 2010; Yokoyama et al. 2004). Unlike human cells, where the hRAD51 paralogues are essential, in other model systems such as hamster (CHO) and chicken (DT40) cells the RAD51 paralogues could be deleted by homologous recombination (Johnson et al. 1999; Pierce et al. 1999; Takata et al. 2000; Takata et al. 2001). Unfortunately, mouse knockout models have resulted in embryonic lethality making our understanding of mammalian RAD51 paralogues restricted to specific cell lines (Deans et al. 2000; Kuznetsov et al. 2009; Pittman and Schimenti 2000; Prakash et al. 2015; Shu et al. 1999).
Consistent with a role for the RAD51 paralogues in HR, DT40 T lymphocytes with RAD51 paralogue disruption exhibit decreases in integration of DNA fragments, spontaneous and MMC-induced SCEs rates, and IR-induced RAD51 foci (Takata et al. 2000; Takata et al. 2001). Furthermore, RAD51 paralogue disruption results in decreased cell viability, chromosomal instability, and sensitivity to MMC and cisplatin (Takata et al. 2000; Takata et al. 2001). These data strongly suggest a role for the RAD51 paralogues during HR but what the specific functions of the BCDX2 and CX3 complexes are is an ongoing area of investigation.
Recent work in human cell lines (breast cancer cell line MCF7 and osteosarcoma cell line U2OS) has suggested separation-of-function between the two human hRAD51 paralogue sub-complexes (Chun et al. 2013). For example, transient knockdown of the BCDX2 complex by siRNA (specifically hXRCC2 or hRAD51D) reduces hRAD51 foci formation, but knockdown of hXRCC3, a unique component of the CX3 complex, does not (Chun et al. 2013). These results suggest that the BCDX2 complex is important for early steps during hRAD51 filament formation. Simultaneously depleting members from both hRAD51 paralogue complexes have no additive effect on hRAD51 foci levels (Chun et al. 2013). These findings support the notion that only BCDX2 facilitates early hRAD51 foci formation in response to IR (Chun et al. 2013). Despite the unique roles the hRAD51 paralogues may play in RAD51 regulation, depletion of either hRAD51 paralogue complex results in decreased HR at an inducible I-SceI endonuclease cut-site (Chun et al. 2013).
Where do the human RAD51 paralogue complexes function in relation to the other known hRAD51 mediator proteins such as hBRCA2? Current evidence from the Powell group suggest that recruitment of hBRCA2 to DNA damage sites following IR is independent of both the BCDX2 and CX3 complexes (Chun et al. 2013). Furthermore, hBRCA2 is epistatic to the hRAD51 paralogues as co-depletion of hBRCA2 with individual hRAD51 paralogues resembles the HR frequency observed with siBRCA2 alone (Chun et al. 2013). These results are consistent with hBRCA2 functioning as a nucleator of hRAD51 filament formation on ssDNA as has been shown for the human protein as well as other species (Carreira et al. 2009; Shahid et al. 2014; Yang et al. 2005). Intriguingly, the hRAD51 paralogues are not epistatic with human RAD52, which has a distinct function from hBRCA2 (Chun et al. 2013). Consistently, co-depletion of hRAD52 and hXRCC3 in DT40 and U2OS cells causes synthetic lethality and decreased HR rates, suggesting that hRAD52 and the hRAD51 paralogues function in discrete pathways (Chun et al. 2013; Fujimori et al. 2001).
Similar to what has been shown with hRAD51, the hRAD51 paralogues likely have dual roles in canonical HR as well as in repair of damaged replication forks. While the BCDX2 complex is important for repair of DSBs induced by the I-SceI endonuclease or IR (Chun et al. 2013), other complex lesions, such as ICLs or damaged replication forks, may require the activity of the CX3 complex. The Nagaraju group, using a DNA fiber spreading technique in chinese hamster ovary (CHO) cells, found that at HU-stalled replication forks, the CX3 complex promotes replication restart by disassembling RAD51 filaments (Somyajit et al. 2015b). They found that the hamster RAD51 paralogues localize to nascent DNA synthesized during replication and at fragile sites upon HU-induced fork stalling, where the RAD51 paralogues protect replication forks from degradation (Somyajit et al. 2015b). For example, knocking out RAD51C, XRCC2, or XRCC3 in CHO cells results in significantly shorter nascent DNA tract lengths and more parental ssDNA (Somyajit et al. 2015b). These findings are consistent with the nascent DNA around the fork being resected and thus exposing the parental ssDNA. Furthermore, disruption of the RAD51 paralogues results in elevated 53BP1 foci suggesting replication fork collapse. This work provides evidence that the RAD51 paralogues actively respond to replication fork damage where they 1) protect nascent ssDNA at stalled forks from resection, 2) prevent replication fork collapse, and 3) facilitate replication restart. Further investigation of lesion specificity of the RAD51 paralogues will shed light on their mechanistic roles in diverse repair contexts.
The Shu complex interacts with scRad52 and scRad55-Rad57 to stimulate scRad51 filament formation
Recently the budding yeast Shu complex has been uncovered to be a new scRad51 regulator and evidence suggests that this complex is conserved throughout eukaryotes (Gaines et al. 2015; Godin et al. 2015; Liu et al. 2011b; Mankouri et al. 2007; Martin et al. 2006; Shor et al. 2005). The Shu complex is an obligate heterotetramer comprised of scShu2, a SWIM domain-containing protein, in complex with scShu1, scCsm2, and scPsy3 (Ito et al. 2001; Shor et al. 2005). The Shu complex was first characterized a decade ago in budding yeast where it was found to generate the complex HR intermediates acted on by scSgs1-Top3-Rmi1 (Shor et al. 2005). Disruption of the Shu complex members results in sensitivity to replication fork stalling caused by base damage, such as by MMS, but not to direct damage induced by IR (Ball et al. 2009; Shor et al. 2005). Puzzlingly, Shu complex disruption causes no sensitivity to HU, which reduces nucleotide pools and causes replication fork stalling. However, disruption of the Shu complex strongly suppresses an sgs1Δ mutant's HU sensitivity, suggesting a nuanced role for the Shu complex at stalled replication forks. Therefore, the Shu complex likely performs a specialized function to promote HR in the context of replication-associated DNA damage (Ball et al. 2009; Xu et al. 2013). Recent work found that the Shu complex likely acts upstream of scRad54, indicating a potential role for the Shu complex in promoting scRad51 filament formation (Mankouri et al. 2007). However, how the Shu complex acts to promote scRad51 filament formation remains an unanswered question.
Several groups have crystalized two of the Shu complex subunits, scCsm2 and scPsy3, and found that these proteins mediate the DNA binding activity of the Shu complex and are structural scRad51 paralogues, despite sharing little sequence homology with scRad51 (Sasanuma et al. 2013; She et al. 2012; Tao et al. 2012). Like budding yeast, fission and humans contain a SWIM-domain protein called spSws1/hSWS1 that is also associated with the RAD51 paralogues [(Godin et al. 2013; Godin et al. 2015; Liu et al. 2011b; Martin et al. 2006) Figure 2]. Recently, scShu2/hSWS1 orthologues have been identified in every eukaryotic lineage by PSI-BLAST (Godin et al. 2015). This includes the early, divergent eukaryotic species Giardia lamblia, plant lineages such as Arabidopsis thaliana, and in all major model systems such as D. melanogaster and C. elegans (Godin et al. 2015). In all of these species the canonical SWIM domain of scShu2/hSWS1, a zinc-finger-like motif defined by CXC…Xn…CXHXXA, is invariant (Godin et al. 2015). Importantly, mutations in the SWIM domain impair budding yeast and human scShu2/hSWS1 interactions with their divergent Rad51 paralogue interacting partners (Godin et al. 2015). For example, in fission yeast Sws1 forms a complex with the spRad51 paralogues spRlp1 and spRdl1 while in human cells hSWS1 forms an obligate heterodimer with the newly identified SWS associated protein 1, hSWSAP1, which is a novel hRAD51 paralogue (Liu et al. 2011b; Martin et al. 2006). Both hSWS1 and hSWSAP1 are important for hRAD51 foci formation, HR at an inducible I-SceI cut-site, and for MMS resistance (Liu et al. 2011b; Martin et al. 2006). These results suggest that the Shu complex function is likely conserved from yeast to humans.
While it has long been known that the yeast Rad51 paralogues, scRad55-Rad57, stimulate scRad51 filament formation (Sung 1997), mechanistic insight into the function of the Shu complex has only recently been uncovered (Gaines et al. 2015). For example, we and others have shown that the budding yeast Shu complex physically associates with scRad51 and the other Rad51 mediators including scRad52 and scRad55-Rad57 by yeast-two-hybrid (Y2H) and in vitro pull downs (Gaines et al. 2015; Godin et al. 2013; Xu et al. 2013). Critically, the interaction between the Shu complex and scRad55-Rad57 is mediated by the Shu complex member, scCsm2. Furthermore scRad55-Rad57 bridges an interaction between the Shu complex and the scRad51 filament machinery (Gaines et al. 2015; Godin et al. 2013; Xu et al. 2013). New in vitro data demonstrates that the Shu complex stimulates scRad51 filament formation two- to three-fold, but only if scRad52 and scRad55-Rad57 are present (Gaines et al. 2015; Godin et al. 2013; Shor et al. 2005; Sung 1997; Xu et al. 2013). Excitingly, these results show a direct role for the Shu complex as a stimulatory co-factor for scRad51 filament formation (Gaines et al. 2015).
Rad51 paralogues may suppress inhibitors of Rad51 filament formation
In addition to the Rad51 paralogues' role during Rad51 filament formation, the Rad51 paralogues also inhibit proteins that antagonize Rad51 [Reviewed in(Karpenshif and Bernstein 2012)]. The most studied inhibitor of Rad51 filaments is the budding yeast helicase scSrs2 (Krejci et al. 2003; Veaute et al. 2003). scSrs2 interacts with scRad51's ATPase domain, causing scRad51 to hydrolyze ATP and lose its DNA binding affinity (Krejci et al. 2003; Veaute et al. 2003). For many years after its discovery, how scSrs2 would interact with other regulators of scRad51 filament formation was of particular interest, especially since scSrs2 interacts with Shu2 in both budding and fission yeast by Y2H (Ito et al. 2001; Martin et al. 2006). In addition to a physical interaction, deletion of fission yeast spSWS1 suppresses many of srs2Δ's defects such as its camptothecin sensitivity. Additionally, disruption of the budding yeast Shu complex increases scSrs2 focus formation at fluorescently labeled DSB sites by fluorescent microscopy (Bernstein et al. 2011; Martin et al. 2006). However, the most striking example of the interplay between scSrs2 and the scRad51 paralogues is the finding that scRad55-Rad57 can directly inhibit scSrs2's ability to destabilize Rad51 filaments in vitro (Liu et al. 2011a). This work complements genetic observations that budding yeast rad55Δ's hypersensitivity to IR is rescued by further deletion of scSRS2 (Liu et al. 2011a). While these findings are clear with IR damage, deletion of scSRS2 does not rescue the MMS sensitivity of either a scRAD55 or a Shu complex-disrupted cell. Indeed, for MMS-treated cells, an srs2Δ, rad55Δ, and srs2Δ rad55Δ cells exhibit the same MMS sensitivity, suggesting that these genes all act in the same pathway to promote repair in budding yeast (Xu et al. 2013). As of now, the functional significance of the physical interaction between Srs2 and the budding and fission yeast Shu complexes remains unknown. Moreover, further studies are needed to understand the interplay between scRad55 and scSrs2 at different lesions, e.g. IR-induced DSBs and MMS-induced replicative stress.
The worm RAD-51 paralogues, RIP-1 and RFS-1, reveal a novel mechanism in promoting RAD-51-dependent HR
Recently, the ceRAD-51 paralogue family in C. elegans grew to incorporate a novel, divergent ceRAD-51 paralogue, ceRIP-1 (Taylor et al. 2015). This novel ceRAD-51 paralogue physically associates with the only known ceRAD-51 paralogue in C. elegans, ceRFS-1, forming the first known ceRAD-51 paralogue complex in nematodes (Taylor et al. 2015; Ward et al. 2010). Recently, we identified ceRIP-1 to directly interact with the worm scShu2 orthologue, ceSWS-1 by Y2H and to bridge an interaction between ce-SWS-1 with ceRFS-1 by Y3H (McClendon et al. 2016). Our results show that ceSWS-1 interaction with ceRIP-1 is mediated by the SWIM domain of ce-SWS-1 and the Walker B motif of ceRIP-1 (McClendon et al. 2016). Using CRISPR to create a null sws-1 allele, sws-1 worms exhibit defects consistent with a role in HR such as DNA damage sensitivity, increased males, increased mutation rates, and fewer mitotic ceRAD-51 foci (McClendon et al. 2016). Together these results suggest that the Shu complex is conserved in higher eukaryotes.
The C. elegans ceRAD-51 paralogues share several features with the yeast and human proteins, in that they bind to ceRAD-51-coated ssDNA in vitro and their disruption in vivo inhibits normal HR (Taylor et al. 2015). However, unlike the budding yeast Rad51 paralogues, the ceRIP-1/RFS-1 heterodimer does not stimulate ceRAD-51 filament formation in vitro. Instead, the C. elegans ceRAD-51 paralogues promote ceRAD-51-dependent strand exchange (Taylor et al. 2015). The Krejci and Boulton groups furnish evidence that this stimulation is due to ceRIP-1/RFS-1 remodeling the ceRAD-51 filament into a more “open” configuration that is more accessible by various proteins, as shown by the ability of nucleases to access the DNA. This “open” configuration is hypothesized to be more amenable to the strand exchange and homology search steps that define HR (Taylor et al. 2015). These exciting findings open many new possibilities for how ceRAD-51 paralogues may function outside of C. elegans. For instance, it would be important to determine if the other eukaryotic Rad51 paralogues also stimulate Rad51 filaments to adopt a more open configuration as shown for the C. elegans proteins.
Future Directions
Clinical implications
Recently hRAD51C and hXRCC3 were found to assemble into an additional complex that is scaffolded by hPALB2, termed the “HR complex” comprised of hPALB2, hRAD51, hRAD51C, hBRCA2 and hXRCC3 (Park et al. 2014a). Mechanistically, this complex may promote hRAD51 filament formation although further investigation of this unique complex will be necessary to define its function. Clinically, this new complex is extremely intriguing for further study as heterozygous mutations in hPALB2, hBRCA2, and hRAD51C are associated with hereditary breast and ovarian cancers that arise when the WT copy is lost (Reviewed in Prakash et al. 2015). Furthermore, inherited homozygous mutations in hPALB2 (FANCN), hBRCA2 (FANCD1), and hRAD51C (FANCO) can cause FA [reviewed in (Levy-Lahad 2010; Park et al. 2014b)]. Moving forward, it will be essential to determine how different mutations in the same genes give rise to unique diseases.
Due to its clinical importance, many studies on the human RAD51 paralogues have focused on hRAD51C. hRAD51C was recently shown to promote HR as disruption of hRAD51C caused a compensatory increase in NHEJ after IR-induced DSBs (Somyajit et al. 2015a). Based on this finding, the Nagaraju group proposed a model to selectively target hRAD51C mutant cells with PARP inhibitors. Similar to the sensitivity of tumors with hBRCA1/2 mutations to PARP inhibitors, tumors with mutations in hRAD51 paralogues could likewise be targeted by PARP inhibition (Somyajit et al. 2015a). Combining hRAD51 mutations with PARP inhibitors leads to replication fork collapse and activation of the error-prone NHEJ pathway resulting in increased cell death (Somyajit et al. 2015a). Future studies will reveal whether this is a clinically viable treatment strategy.
Models of Rad51 paralogue function during HR
Although less is known about the human RAD51 mediators, new insights into the yeast proteins provide a framework for understanding how the Rad51 paralogues and the Shu complex may hypothetically contribute to repair by HR (Figure 4). After the DSB is resected and coated by RPA, then scRad52 is recruited where it stabilizes RPA-coated ssDNA in patches along the DNA (Gibb et al. 2014). Subsequently, scRad55-Rad57 and the Shu complex are assembled and enable scRad51 filament nucleation, extension and stabilization. The precise mechanism of how the scRad51 paralogues perform these functions is currently unknown. Here we propose three hypothetical models of how these mediators may function. In the first model, scRad52-RPA co-filaments are formed and scRad55-Rad57 and the Shu complex aid in the initial steps of scRad51 presynaptic filament assembly but are released from the filament (Figure 4, left model- Release). In the second model, scRad52-RPA co-filaments form and scRad55-Rad57 and the Shu complex directly bind to the ssDNA and are integrated into the filament but exhibit no further interactions with scRad52-RPA (Figure 4, middle model- Integration). Lastly, it is possible that scRad55-Rad57 and the Shu complex interact with scRad52-RPA to coat the scRad51 filament as a co-filament (Figure 4, right model- Co-filament). Alternatively, the Shu complex could act to “cap” a scRad51 filament, and direct Rad51 filaments to expand unidirectionally, as was suggested for its meiotic function (Sasanuma et al. 2013). If the Shu complex remains associated with the scRad51 filament, it will be important to determine its impact in downstream HR steps, such as the recruitment of strand invasion factors, e.g. scRad54, scRdh54, or the inhibition of scSrs2 during scRad51 filament formation. Moving forward, the models proposed in Figure 4 will need to be tested by DNA curtain, TIRF microscopy, scanning force microscopy, and other single molecule approaches.
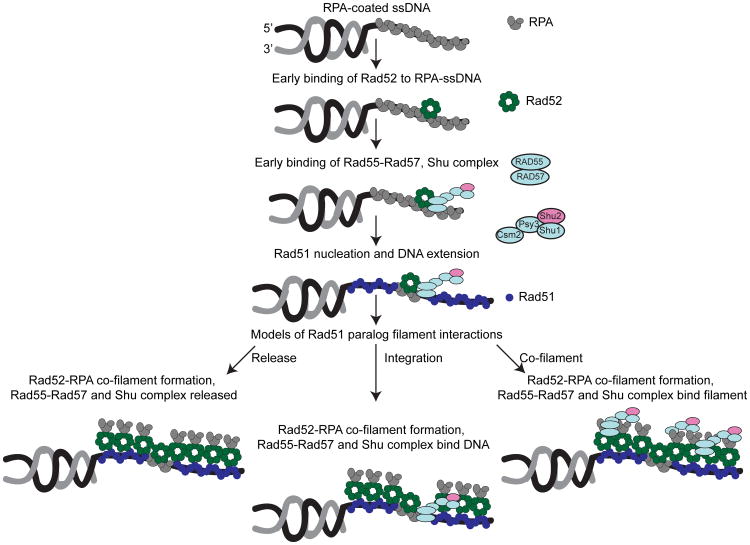
Figure 4.
Hypothetical models for how the yeast Rad51 paralogues could associate with the Rad51-Rad52-RPA filament during HR [adapted from (Gibb et al. 2014)]. A DSB is recognized and resected to generate 3′ ssDNA overhangs, which are immediately coated by the ssDNA binding complex RPA. Rad52 then binds the RPA-coated ssDNA and stabilizes a patch of RPA on the ssDNA. Next, the Rad51 paralogues, Rad55-Rad57, form a higher order ensemble with Rad52 and the Shu complex to stimulate Rad51 filament formation. Although filaments of Rad52-RPA coat the Rad51 nucleoprotein filament, whether Rad55-Rad57 and the Shu complex exhibit similar interactions with the Rad51 nucleoprotein filament remains unknown. Three hypothetical models for how the Rad51 paralogues and the Shu complex may interact with the Rad52-RPA-Rad51 nucleoprotein filament are shown. On the left, Rad55-Rad57 and the Shu complex are released after Rad51 filaments form and are not associated with the Rad52-RPA-Rad51 filament. In the middle model, Rad55-Rad57 and the Shu complex are integrated into the Rad51 nucleoprotein filament but have no additional interactions with Rad52-RPA. On the right, Rad55-Rad57 and the Shu complex interact with Rad52-RPA and together coat the Rad51 filament as a co-filament. Importantly, the middle and right-hand models are not mutually exclusive, and Rad55-Rad57 and the Shu complex could both integrate into the Rad51 filament as well as form a co-filament with Rad52-RPA.
Alternative roles for the RAD51 mediators during DNA replication
Collectively the evidence for a role for hRAD51 and the hRAD51 paralogues during repair of replicative damage raises important areas of future investigation. For example, if hRAD51's function in repair of HU-stalled replication forks is independently of HR, then will the hRAD51 mediators also exhibit HR-independent roles? Since mutations in the human hRAD51 paralogues are implicated in Fanconi anemia, do they contribute to hRAD51's HR-independent functions by regulating its activity? Different DNA intermediates may arise at replication forks depending upon DNA damaging agent that may require a unique set of RAD51 mediators for repair. Using the EM techniques pioneered by the Branzei group, it will be important to further evaluate the HR-dependent and independent roles of hRAD51 in repair of these lesions.
Concluding remarks
Here we discuss recent advances in our understanding of Rad51 regulation during HR and how repair of replicative damage has opened many new areas of investigation and experimentation. For example, although the Rad51 paralogues were identified more than twenty years ago, our understanding of their mechanistic function of the Rad51 paralogues is still limited. In humans, the BCDX2 and HR complex (hPALB2, hBRCA2, hRAD51C, hXRCC3) regulate hRAD51 filaments, but the function of the CX3 and Shu complex (hSWS1-hSWSAP1) remains enigmatic. Perhaps, like C. elegans, certain human RAD51 paralogue complexes will regulate hRAD51 presynaptic filament assembly, while other hRAD51 paralogue complexes will regulate the activity of the pre- and post-synaptic filament. Moreover, since the budding yeast Rad51 paralogues, scRad55-Rad57, and the Shu complex interact with scRad52 to stimulate scRad51 filament formation, it remains unanswered whether the human RAD51 paralogues similarly function with hRAD52 or hBRCA2 to regulate hRAD51. As new findings emerge in one model system, it is imperative that we apply those paradigms to other models such as mammalian cells. By determining how well conserved the mechanistic function of the Rad51 paralogues are from yeast to humans, we will develop novel ways of investigating the human paralogues and their role in human disease.
Acknowledgments
We thank Dana Branzei, Eric Greene, and Julieta Martino for helpful feedback and comments. This work was supported by the V Scholar Award from the V Foundation for Cancer Research and the National Institutes of Health grant ES024872 to K.A.B.
Footnotes
In this review, homologous recombination in multiple species is discussed. Where proteins from a specific species are referenced, the following prefixes are used: Saccharomyces cerevisiae (sc), Homo sapiens (h), Drosophila melanogaster (dm), Caenorhabditis elegans (ce), Schizosaccharomyces pombe (sp). When general concepts or models from multiple species are referenced, such as Rad51 filament formation, we default to using the Saccharomyces cerevisiae nomenclature with no prefix.
References
- Ameziane N, May P, Haitjema A, van de Vrugt HJ, van Rossum-Fikkert SE, Ristic D, Williams GJ, Balk J, Rockx D, Li H, Rooimans MA, Oostra AB, Velleuer E, Dietrich R, Bleijerveld OB, Maarten Altelaar AF, Meijers-Heijboer H, Joenje H, Glusman G, Roach J, Hood L, Galas D, Wyman C, Balling R, den Dunnen J, de Winter JP, Kanaar R, Gelinas R, Dorsman JC. A novel Fanconi anaemia subtype associated with a dominant-negative mutation in RAD51. Nat Commun. 2015;6:8829. doi: 10.1038/ncomms9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand RP, Lovett ST, Haber JE. Break-induced DNA replication. Cold Spring Harb Perspect Biol. 2013;5(12):a010397. doi: 10.1101/cshperspect.a010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball LG, Zhang K, Cobb JA, Boone C, Xiao W. The yeast Shu complex couples error-free post-replication repair to homologous recombination. Molecular microbiology. 2009;73(1):89–102. doi: 10.1111/j.1365-2958.2009.06748.x. [DOI] [PubMed] [Google Scholar]
- Bashkirov VI, King JS, Bashkirova EV, Schmuckli-Maurer J, Heyer WD. DNA repair protein Rad55 is a terminal substrate of the DNA damage checkpoints. Mol Cell Biol. 2000;20(12):4393–4404. doi: 10.1128/mcb.20.12.4393-4404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA helicases in DNA repair. Annu Rev Genet. 2010;44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein KA, Reid RJ, Sunjevaric I, Demuth K, Burgess RC, Rothstein R. The Shu complex, which contains Rad51 paralogues, promotes DNA repair through inhibition of the Srs2 anti-recombinase. Mol Biol Cell. 2011;22(9):1599–1607. doi: 10.1091/mbc.E10-08-0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein KA, Rothstein R. At loose ends: resecting a double-strand break. Cell. 2009;137(5):807–810. doi: 10.1016/j.cell.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco A, Gutierrez-Enriquez S, Santamarina M, Montalban G, Bonache S, Balmana J, Carracedo A, Diez O, Vega A. RAD51C germline mutations found in Spanish site-specific breast cancer and breast-ovarian cancer families. Breast Cancer Res Treat. 2014;147(1):133–143. doi: 10.1007/s10549-014-3078-4. [DOI] [PubMed] [Google Scholar]
- Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR, 3rd, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107(4):537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. RecQ helicases queuing with Srs2 to disrupt Rad51 filaments and suppress recombination. Genes Dev. 2007;21(23):3019–3026. doi: 10.1101/gad.1624707. [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51(3):463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev. 2007;21(23):3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira A, Hilario J, Amitani I, Baskin RJ, Shivji MK, Venkitaraman AR, Kowalczykowski SC. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell. 2009;136(6):1032–1043. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P. DNA End Resection: Nucleases Team Up with the Right Partners to Initiate Homologous Recombination. J Biol Chem. 2015;290(38):22931–22938. doi: 10.1074/jbc.R115.675942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CF, Chen PL, Zhong Q, Sharp ZD, Lee WH. Expression of BRC repeats in breast cancer cells disrupts the BRCA2-Rad51 complex and leads to radiation hypersensitivity and loss of G(2)/M checkpoint control. J Biol Chem. 1999;274(46):32931–32935. doi: 10.1074/jbc.274.46.32931. [DOI] [PubMed] [Google Scholar]
- Chen X, Bosques L, Sung P, Kupfer GM. A novel role for non-ubiquitinated FANCD2 in response to hydroxyurea-induced DNA damage. Oncogene. 2015;35(1):22–34. doi: 10.1038/onc.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu WK, Payne MJ, Beli P, Hanada K, Choudhary C, Hickson ID. FBH1 influences DNA replication fork stability and homologous recombination through ubiquitylation of RAD51. Nat Commun. 2015;6:5931. doi: 10.1038/ncomms6931. [DOI] [PubMed] [Google Scholar]
- Chun J, Buechelmaier ES, Powell SN. Rad51 paralog complexes BCDX2 and CX3 act at different stages in the BRCA1-BRCA2-dependent homologous recombination pathway. Mol Cell Biol. 2013;33(2):387–395. doi: 10.1128/MCB.00465-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clague J, Wilhoite G, Adamson A, Bailis A, Weitzel JN, Neuhausen SL. RAD51C germline mutations in breast and ovarian cancer cases from high-risk families. PLoS One. 2011;6(9):e25632. doi: 10.1371/journal.pone.0025632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Mantiero D, Lucchini G, Longhese MP. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J Biol Chem. 2005;280(46):38631–38638. doi: 10.1074/jbc.M508339200. [DOI] [PubMed] [Google Scholar]
- Compton SA, Ozgur S, Griffith JD. Ring-shaped Rad51 paralog protein complexes bind Holliday junctions and replication forks as visualized by electron microscopy. The Journal of biological chemistry. 2010;285(18):13349–13356. doi: 10.1074/jbc.M109.074286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, Helleday T, Haber JE, Iliakis G, Kallioniemi OP, Halazonetis TD. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343(6166):88–91. doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer. 2003;3(1):23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- Davies AA, Masson JY, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR, West SC. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001;7(2):273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- Deans B, Griffin CS, Maconochie M, Thacker J. Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. EMBO J. 2000;19(24):6675–6685. doi: 10.1093/emboj/19.24.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83(4):655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- Fasching CL, Cejka P, Kowalczykowski SC, Heyer WD. Top3-Rmi1 dissolve Rad51-mediated D loops by a topoisomerase-based mechanism. Mol Cell. 2015;57(4):595–606. doi: 10.1016/j.molcel.2015.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorentini P, Huang KN, Tishkoff DX, Kolodner RD, Symington LS. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol Cell Biol. 1997;17(5):2764–2773. doi: 10.1128/mcb.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke WM, Brill SJ. Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev. 2003;17(14):1768–1778. doi: 10.1101/gad.1105203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori A, Tachiiri S, Sonoda E, Thompson LH, Dhar PK, Hiraoka M, Takeda S, Zhang Y, Reth M, Takata M. Rad52 partially substitutes for the Rad51 paralog XRCC3 in maintaining chromosomal integrity in vertebrate cells. EMBO J. 2001;20(19):5513–5520. doi: 10.1093/emboj/20.19.5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaines WA, Godin SK, Kabbinavar FF, Rao T, VanDemark AP, Sung P, Bernstein KA. Promotion of presynaptic filament assembly by the ensemble of S. cerevisiae Rad51 paralogues with Rad52. Nat Commun. 2015;6:7834. doi: 10.1038/ncomms8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghospurkar PL, Wilson TM, Severson AL, Klein SJ, Khaku SK, Walther AP, Haring SJ. The DNA damage response and checkpoint adaptation in Saccharomyces cerevisiae: distinct roles for the replication protein A2 (Rfa2) N-terminus. Genetics. 2015;199(3):711–727. doi: 10.1534/genetics.114.173211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M, Zwicky K, Follonier C, Foiani M, Lopes M, Branzei D. Visualization of recombination-mediated damage bypass by template switching. Nat Struct Mol Biol. 2014;21(10):884–892. doi: 10.1038/nsmb.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb B, Ye LF, Kwon Y, Niu H, Sung P, Greene EC. Protein dynamics during presynaptic-complex assembly on individual single-stranded DNA molecules. Nat Struct Mol Biol. 2014;21(10):893–900. doi: 10.1038/nsmb.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin S, Wier A, Kabbinavar F, Bratton-Palmer DS, Ghodke H, Van Houten B, VanDemark AP, Bernstein KA. The Shu complex interacts with Rad51 through the Rad51 paralogues Rad55-Rad57 to mediate error-free recombination. Nucleic Acids Res. 2013;41(8):4525–4534. doi: 10.1093/nar/gkt138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin SK, Meslin C, Kabbinavar F, Bratton-Palmer DS, Hornack C, Mihalevic MJ, Yoshida K, Sullivan M, Clark NL, Bernstein KA. Evolutionary and functional analysis of the invariant SWIM domain in the conserved Shu2/SWS1 protein family from Saccharomyces cerevisiae to Homo sapiens. Genetics. 2015;199(4):1023–1033. doi: 10.1534/genetics.114.173518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE. In vivo biochemistry: physical monitoring of recombination induced by site-specific endonucleases. Bioessays. 1995;17(7):609–620. doi: 10.1002/bies.950170707. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Ray Chaudhuri A, Lopes M, Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol. 2010;17(11):1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzberg K, Bashkirov VI, Rolfsmeier M, Haghnazari E, McDonald WH, Anderson S, Bashkirova EV, Yates JR, 3rd, Heyer WD. Phosphorylation of Rad55 on serines 2, 8, and 14 is required for efficient homologous recombination in the recovery of stalled replication forks. Mol Cell Biol. 2006;26(22):8396–8409. doi: 10.1128/MCB.01317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annual review of genetics. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer WD, Li X, Rolfsmeier M, Zhang XP. Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 2006;34(15):4115–4125. doi: 10.1093/nar/gkl481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, Jasin M, Vogel H, Sung P, Luo G. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21(23):3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KN, Symington LS. A 5′-3′ exonuclease from Saccharomyces cerevisiae is required for in vitro recombination between linear DNA molecules with overlapping homology. Mol Cell Biol. 1993;13(6):3125–3134. doi: 10.1128/mcb.13.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455(7213):689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456(7220):357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115(4):401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(8):4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467(7316):678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyasekharan AD, Liu Y, Hattori H, Pisupati V, Jonsdottir AB, Rajendra E, Lee M, Sundaramoorthy E, Schlachter S, Kaminski CF, Ofir-Rosenfeld Y, Sato K, Savill J, Ayoub N, Venkitaraman AR. A cancer-associated BRCA2 mutation reveals masked nuclear export signals controlling localization. Nat Struct Mol Biol. 2013;20(10):1191–1198. doi: 10.1038/nsmb.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RD, Liu N, Jasin M. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature. 1999;401(6751):397–399. doi: 10.1038/43932. [DOI] [PubMed] [Google Scholar]
- Karpenshif Y, Bernstein KA. From yeast to mammals: recent advances in genetic control of homologous recombination. DNA Repair. 2012;11(10):781–788. doi: 10.1016/j.dnarep.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88(3):375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- Kim HS, Vijayakumar S, Reger M, Harrison JC, Haber JE, Weil C, Petrini JH. Functional interactions between Sae2 and the Mre11 complex. Genetics. 2008;178(2):711–723. doi: 10.1534/genetics.107.081331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitao S, Ohsugi I, Ichikawa K, Goto M, Furuichi Y, Shimamoto A. Cloning of two new human helicase genes of the RecQ family: biological significance of multiple species in higher eukaryotes. Genomics. 1998;54(3):443–452. doi: 10.1006/geno.1998.5595. [DOI] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423(6937):305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Kuznetsov SG, Haines DC, Martin BK, Sharan SK. Loss of Rad51c leads to embryonic lethality and modulation of Trp53-dependent tumorigenesis in mice. Cancer Res. 2009;69(3):863–872. doi: 10.1158/0008-5472.CAN-08-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Terakawa T, Qi Z, Steinfeld JB, Redding S, Kwon Y, Gaines WA, Zhao W, Sung P, Greene EC. DNA RECOMBINATION. Base triplet stepping by the Rad51/RecA family of recombinases. Science. 2015;349(6251):977–981. doi: 10.1126/science.aab2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell. 2007;28(4):638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Lahad E. Fanconi anemia and breast cancer susceptibility meet again. Nat Genet. 2010;42(5):368–369. doi: 10.1038/ng0510-368. [DOI] [PubMed] [Google Scholar]
- Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18(1):99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 2005;19(3):339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118(6):699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol. 2010;17(10):1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Renault L, Veaute X, Fabre F, Stahlberg H, Heyer WD. Rad51 paralogues Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature. 2011a;479(7372):245–248. doi: 10.1038/nature10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Wan L, Wu Y, Chen J, Huang J. hSWS1.SWSAP1 is an evolutionarily conserved complex required for efficient homologous recombination repair. The Journal of biological chemistry. 2011b;286(48):41758–41766. doi: 10.1074/jbc.M111.271080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YC, Paffett KS, Amit O, Clikeman JA, Sterk R, Brenneman MA, Nickoloff JA. Sgs1 regulates gene conversion tract lengths and crossovers independently of its helicase activity. Mol Cell Biol. 2006;26(11):4086–4094. doi: 10.1128/MCB.00136-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfrini N, Trovesi C, Wery M, Martina M, Cesena D, Descrimes M, Morillon A, d'Adda di Fagagna F, Longhese MP. RNA-processing proteins regulate Mec1/ATR activation by promoting generation of RPA-coated ssDNA. EMBO Rep. 2015;16(2):221–231. doi: 10.15252/embr.201439458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri HW, Chu WK, Hickson ID. A novel antirecombinase gains PARIty. Mol Cell. 2012;45(1):6–7. doi: 10.1016/j.molcel.2011.12.015. [DOI] [PubMed] [Google Scholar]
- Mankouri HW, Ngo HP, Hickson ID. Shu proteins promote the formation of homologous recombination intermediates that are processed by Sgs1-Rmi1-Top3. Molecular biology of the cell. 2007;18(10):4062–4073. doi: 10.1091/mbc.E07-05-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin V, Chahwan C, Gao H, Blais V, Wohlschlegel J, Yates JR, 3rd, McGowan CH, Russell P. Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. The EMBO journal. 2006;25(11):2564–2574. doi: 10.1038/sj.emboj.7601141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson JY, Tarsounas MC, Stasiak AZ, Stasiak A, Shah R, McIlwraith MJ, Benson FE, West SC. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 2001;15(24):3296–3307. doi: 10.1101/gad.947001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClendon TB, Sullivan MR, Bernstein KA, Yanowitz JL. Promotion of homologous recombination by SWS-1 in complex with the RAD-51 paralogs in Caenorhabditis elegans. Genetics. 2016 doi: 10.1534/genetics.115.185827. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meindl A, Hellebrand H, Wiek C, Erven V, Wappenschmidt B, Niederacher D, Freund M, Lichtner P, Hartmann L, Schaal H, Ramser J, Honisch E, Kubisch C, Wichmann HE, Kast K, Deissler H, Engel C, Muller-Myhsok B, Neveling K, Kiechle M, Mathew CG, Schindler D, Schmutzler RK, Hanenberg H. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet. 2010;42(5):410–414. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
- Michalska MM, Samulak D, Romanowicz H, Smolarz B. Single Nucleotide Polymorphisms (SNPs) of RAD51-G172T and XRCC2-41657C/T Homologous Recombination Repair Genes and the Risk of Triple- Negative Breast Cancer in Polish Women. Pathol Oncol Res. 2015;21(4):935–940. doi: 10.1007/s12253-015-9922-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KA, Yoshikawa DM, McConnell IR, Clark R, Schild D, Albala JS. RAD51C interacts with RAD51B and is central to a larger protein complex in vivo exclusive of RAD51. J Biol Chem. 2002;277(10):8406–8411. doi: 10.1074/jbc.M108306200. [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455(7214):770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. DNA end resection--unraveling the tail. DNA Repair (Amst) 2011;10(3):344–348. doi: 10.1016/j.dnarep.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, Dejsuphong D, Petalcorin MI, Hofmann K, Takeda S, Boulton SJ, D'Andrea AD. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol Cell. 2012;45(1):75–86. doi: 10.1016/j.molcel.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namazi A, Abedinzadeh M, Nourbaksh P, Neamatzadeh H. Association between the XRCC3 Thr241Met polymorphism and risk of colorectal cancer: a meta analysis of 5,193 cases and 6,645 controls. Asian Pac J Cancer Prev. 2015;16(6):2263–2268. doi: 10.7314/apjcp.2015.16.6.2263. [DOI] [PubMed] [Google Scholar]
- Osorio A, Endt D, Fernandez F, Eirich K, de la Hoya M, Schmutzler R, Caldes T, Meindl A, Schindler D, Benitez J. Predominance of pathogenic missense variants in the RAD51C gene occurring in breast and ovarian cancer families. Hum Mol Genet. 2012;21(13):2889–2898. doi: 10.1093/hmg/dds115. [DOI] [PubMed] [Google Scholar]
- Park JY, Singh TR, Nassar N, Zhang F, Freund M, Hanenberg H, Meetei AR, Andreassen PR. Breast cancer-associated missense mutants of the PALB2 WD40 domain, which directly binds RAD51C, RAD51 and BRCA2, disrupt DNA repair. Oncogene. 2014a;33(40):4803–4812. doi: 10.1038/onc.2013.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Zhang F, Andreassen PR. PALB2: the hub of a network of tumor suppressors involved in DNA damage responses. Biochim Biophys Acta. 2014b;1846(1):263–275. doi: 10.1016/j.bbcan.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT. Making the best of the loose ends: Mre11/Rad50 complexes and Sae2 promote DNA double-strand break resection. DNA Repair (Amst) 2010;9(12):1283–1291. doi: 10.1016/j.dnarep.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelttari LM, Kiiski JI, Ranta S, Vilske S, Blomqvist C, Aittomaki K, Nevanlinna H. RAD51, XRCC3, and XRCC2 mutation screening in Finnish breast cancer families. Springerplus. 2015;4:92. doi: 10.1186/s40064-015-0880-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393(6680):91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13(20):2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman DL, Schimenti JC. Midgestation lethality in mice deficient for the RecA-related gene, Rad51d/Rad51l3. Genesis. 2000;26(3):167–173. doi: 10.1002/(sici)1526-968x(200003)26:3<167::aid-gene1>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Prakash R, Zhang Y, Feng W, Jasin M. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol. 2015;7(4):a016600. doi: 10.1101/cshperspect.a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranam KL, Blackshear PJ. Cloning and characterization of RECQL, a potential human homologue of the Escherichia coli DNA helicase RecQ. J Biol Chem. 1994;269(47):29838–29845. [PubMed] [Google Scholar]
- Qi Z, Redding S, Lee JY, Gibb B, Kwon Y, Niu H, Gaines WA, Sung P, Greene EC. DNA sequence alignment by microhomology sampling during homologous recombination. Cell. 2015;160(5):856–869. doi: 10.1016/j.cell.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Zelensky A, Smal I, Meijering E, van Cappellen WA, de Gruiter HM, van Belle GJ, van Royen ME, Houtsmuller AB, Essers J, Kanaar R, Wyman C. BRCA2 diffuses as oligomeric clusters with RAD51 and changes mobility after DNA damage in live cells. J Cell Biol. 2014;207(5):599–613. doi: 10.1083/jcb.201405014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M, Pfander B, Hoege C, Jentsch S. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat Cell Biol. 2006;8(11):1284–1290. doi: 10.1038/ncb1488. [DOI] [PubMed] [Google Scholar]
- Sanchez H, Kertokalio A, van Rossum-Fikkert S, Kanaar R, Wyman C. Combined optical and topographic imaging reveals different arrangements of human RAD54 with presynaptic and postsynaptic RAD51-DNA filaments. Proc Natl Acad Sci U S A. 2013;110(28):11385–11390. doi: 10.1073/pnas.1306467110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasanuma H, Tawaramoto MS, Lao JP, Hosaka H, Sanda E, Suzuki M, Yamashita E, Hunter N, Shinohara M, Nakagawa A, Shinohara A. A new protein complex promoting the assembly of Rad51 filaments. Nature communications. 2013;4:1676. doi: 10.1038/ncomms2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S, Ashkenazi M, Pecker I, Frydman M, Harnik R, Patanjali SR, Simmons A, Clines GA, Sartiel A, Gatti RA, Chessa L, Sanal O, Lavin MF, Jaspers NG, Taylor AM, Arlett CF, Miki T, Weissman SM, Lovett M, Collins FS, Shiloh Y. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268(5218):1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22(1):106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz EK, Heyer WD. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma. 2011;120(2):109–127. doi: 10.1007/s00412-010-0304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Yanagisawa J, Kohda T, Sonoyama T, Ui M, Enomoto T. Purification of two DNA-dependent adenosinetriphosphatases having DNA helicase activity from HeLa cells and comparison of the properties of the two enzymes. J Biochem. 1994;115(3):523–531. doi: 10.1093/oxfordjournals.jbchem.a124369. [DOI] [PubMed] [Google Scholar]
- Shahid T, Soroka J, Kong EH, Malivert L, McIlwraith MJ, Pape T, West SC, Zhang X. Structure and mechanism of action of the BRCA2 breast cancer tumor suppressor. Nat Struct Mol Biol. 2014;21(11):962–968. doi: 10.1038/nsmb.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamseldin HE, Elfaki M, Alkuraya FS. Exome sequencing reveals a novel Fanconi group defined by XRCC2 mutation. J Med Genet. 2012;49(3):184–186. doi: 10.1136/jmedgenet-2011-100585. [DOI] [PubMed] [Google Scholar]
- She Z, Gao ZQ, Liu Y, Wang WJ, Liu GF, Shtykova EV, Xu JH, Dong YH. Structural and SAXS analysis of the budding yeast SHU-complex proteins. FEBS letters. 2012;586(16):2306–2312. doi: 10.1016/j.febslet.2012.06.024. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. Ataxia-telangiectasia and the Nijmegen breakage syndrome: related disorders but genes apart. Annu Rev Genet. 1997;31:635–662. doi: 10.1146/annurev.genet.31.1.635. [DOI] [PubMed] [Google Scholar]
- Shor E, Weinstein J, Rothstein R. A genetic screen for top3 suppressors in Saccharomyces cerevisiae identifies SHU1, SHU2, PSY3 and CSM2: four genes involved in error-free DNA repair. Genetics. 2005;169(3):1275–1289. doi: 10.1534/genetics.104.036764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Z, Smith S, Wang L, Rice MC, Kmiec EB. Disruption of muREC2/RAD51L1 in mice results in early embryonic lethality which can Be partially rescued in a p53(-/-) background. Mol Cell Biol. 1999;19(12):8686–8693. doi: 10.1128/mcb.19.12.8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinger JA, Kiianitsa K, Heyer WD. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol Cell. 2002;10(5):1175–1188. doi: 10.1016/s1097-2765(02)00743-8. [DOI] [PubMed] [Google Scholar]
- Somyajit K, Mishra A, Jameei A, Nagaraju G. Enhanced non-homologous end joining contributes toward synthetic lethality of pathological RAD51C mutants with poly (ADP-ribose) polymerase. Carcinogenesis. 2015a;36(1):13–24. doi: 10.1093/carcin/bgu211. [DOI] [PubMed] [Google Scholar]
- Somyajit K, Saxena S, Babu S, Mishra A, Nagaraju G. Mammalian RAD51 paralogs protect nascent DNA at stalled forks and mediate replication restart. Nucleic Acids Res. 2015b;43(20):9835–9855. doi: 10.1093/nar/gkv880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol. 2011;12(2):90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997;11(9):1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7(10):739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Fukushima T, Morrison C, Albala JS, Swagemakers SM, Kanaar R, Thompson LH, Takeda S. The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol Cell Biol. 2000;20(17):6476–6482. doi: 10.1128/mcb.20.17.6476-6482.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol Cell Biol. 2001;21(8):2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Nakamura K, Taniguchi Y, Paull TT. Ctp1/CtIP and the MRN complex collaborate in the initial steps of homologous recombination. Mol Cell. 2007;28(3):351–352. doi: 10.1016/j.molcel.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Tao Y, Li X, Liu Y, Ruan J, Qi S, Niu L, Teng M. Structural analysis of Shu proteins reveals a DNA binding role essential for resisting damage. The Journal of biological chemistry. 2012;287(24):20231–20239. doi: 10.1074/jbc.M111.334698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MR, Spirek M, Chaurasiya KR, Ward JD, Carzaniga R, Yu X, Egelman EH, Collinson LM, Rueda D, Krejci L, Boulton SJ. Rad51 Paralogs Remodel Pre-synaptic Rad51 Filaments to Stimulate Homologous Recombination. Cell. 2015;162(2):271–286. doi: 10.1016/j.cell.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson ER, Boyle SE, Johnson J, Ryland GL, Sawyer S, Choong DY, kConFab. Chenevix-Trench G, Trainer AH, Lindeman GJ, Mitchell G, James PA, Campbell IG. Analysis of RAD51C germline mutations in high-risk breast and ovarian cancer families and ovarian cancer patients. Hum Mutat. 2012;33(1):95–99. doi: 10.1002/humu.21625. [DOI] [PubMed] [Google Scholar]
- Thorslund T, McIlwraith MJ, Compton SA, Lekomtsev S, Petronczki M, Griffith JD, West SC. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat Struct Mol Biol. 2010;17(10):1263–1265. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, Jentsch S, Rothstein R, Aragon L, Lisby M. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol. 2007;9(8):923–931. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]
- Vaz F, Hanenberg H, Schuster B, Barker K, Wiek C, Erven V, Neveling K, Endt D, Kesterton I, Autore F, Fraternali F, Freund M, Hartmann L, Grimwade D, Roberts RG, Schaal H, Mohammed S, Rahman N, Schindler D, Mathew CG. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010;42(5):406–409. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423(6937):309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- Wang AT, Kim T, Wagner JE, Conti BA, Lach FP, Huang AL, Molina H, Sanborn EM, Zierhut H, Cornes BK, Abhyankar A, Sougnez C, Gabriel SB, Auerbach AD, Kowalczykowski SC, Smogorzewska A. A Dominant Mutation in Human RAD51 Reveals Its Function in DNA Interstrand Crosslink Repair Independent of Homologous Recombination. Mol Cell. 2015;59(3):478–490. doi: 10.1016/j.molcel.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JD, Muzzini DM, Petalcorin MI, Martinez-Perez E, Martin JS, Plevani P, Cassata G, Marini F, Boulton SJ. Overlapping mechanisms promote postsynaptic RAD-51 filament disassembly during meiotic double-strand break repair. Mol Cell. 2010;37(2):259–272. doi: 10.1016/j.molcel.2009.12.026. [DOI] [PubMed] [Google Scholar]
- Weiffenbach B, Haber JE. Homothallic switching of Saccharomyces cerevisiae mating type genes by using a donor containing a large internal deletion. Mol Cell Biol. 1985;5(8):2154–2158. doi: 10.1128/mcb.5.8.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4(6):435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- Wiese C, Collins DW, Albala JS, Thompson LH, Kronenberg A, Schild D. Interactions involving the Rad51 paralogs Rad51C and XRCC3 in human cells. Nucleic Acids Res. 2002;30(4):1001–1008. doi: 10.1093/nar/30.4.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GJ, Lees-Miller SP, Tainer JA. Mre11-Rad50-Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks. DNA Repair (Amst) 2010;9(12):1299–1306. doi: 10.1016/j.dnarep.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q, Pals G, Errami A, Gluckman E, Llera J, Wang W, Livingston DM, Joenje H, de Winter JP. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39(2):159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- Xu X, Ball L, Chen W, Tian X, Lambrecht A, Hanna M, Xiao W. The yeast Shu complex utilizes homologous recombination machinery for error-free lesion bypass via physical interaction with a Rad51 paralogue. PLoS One. 2013;8(12):e81371. doi: 10.1371/journal.pone.0081371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433(7026):653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- Yokoyama H, Sarai N, Kagawa W, Enomoto R, Shibata T, Kurumizaka H, Yokoyama S. Preferential binding to branched DNA strands and strand-annealing activity of the human Rad51B, Rad51C, Rad51D and Xrcc2 protein complex. Nucleic Acids Res. 2004;32(8):2556–2565. doi: 10.1093/nar/gkh578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin GM, Mulligan J, Schellenberg GD. Positional cloning of the Werner's syndrome gene. Science. 1996;272(5259):258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134(6):981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]