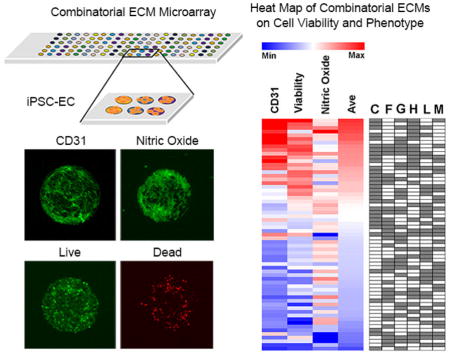
Abstract
Recent developments in cell therapy using human induced pluripotent stem cell-derived endothelial cells (iPSC-ECs) hold great promise for treating ischemic cardiovascular tissues. However, poor post-transplantation viability largely limits the potential of stem cell therapy. Although the extracellular matrix (ECM) has become increasingly recognized as an important cell survival factor, conventional approaches primarily rely on single ECMs for in vivo co-delivery with cells, even though the endothelial basement membrane is comprised of a milieu of different ECMs. To address this limitation, we developed a combinatorial ECM microarray platform to simultaneously interrogate hundreds of micro-scale multi-component chemical compositions of ECMs on iPSC-EC response. After seeding iPSC-ECs onto ECM microarrays, we performed high-throughput analysis of the effects of combinatorial ECMs on iPSC-EC survival, endothelial phenotype, and nitric oxide production under conditions of hypoxia (1% O2) and reduced nutrients (1% fetal bovine serum), as is present in ischemic injury sites. Using automated image acquisition and analysis, we identified combinatorial ECMs such as collagen IV+gelatin+heparan sulfate+laminin and collagen IV+fibronectin+gelatin+heparan sulfate+laminin that significantly improved cell survival, nitric oxide production, and CD31 phenotypic expression, in comparison to single-component ECMs. These results were further validated in conventional cell culture platforms and within three-dimensional scaffolds. Furthermore, this approach revealed complex ECM interactions and non-intuitive cell behavior that otherwise could not be easily determined using conventional cell culture platforms. Together these data suggested that iPSC-EC delivery within optimal combinatorial ECMs may improve their survival and function under the condition of hypoxia with reduced nutrients.
Keywords: pluripotent stem cell, endothelial cell, extracellular matrix, hypoxia, angiogenesis, cardiovascular
Graphical abstract
1. Introduction
Cardiovascular disease is the leading cause of death in United States, affecting more than eighty-five million people [1]. Stem cell-based therapies using autologous endothelial progenitor cells or mononuclear cells to treat myocardial infarction, critical limb ischemia, or other ischemic insults have shown only moderate benefits in clinical trials [2–6]. More recently, we and others have demonstrated that endothelial cells (ECs) differentiated from embryonic stem cells or induced pluripotent stem cells (iPSC-ECs) are another potent class of therapeutic cells for treatment of critical limb ischemia in pre-clinical animal models [7–11]. Given the infinite expansion capability of iPSCs, iPSC-ECs represent a promising therapeutic cell type for cell therapy and regenerative medicine.
Although stem cell delivery is a promising approach to treat cardiovascular diseases, numerous recent studies demonstrate the critical limitation of low post-implantation cell survival, leading to reduced therapeutic potential of the cells [12, 13]. Genetic approaches to enhance cell survival by induction of pro-survival factors such as Akt or hemoxygenase-1 have shown positive benefit [14–19]. However, due to concerns surrounding the safety of genetic modification, non-genetic approaches to improve cell survival are an attractive alternative.
In recent years, naturally derived extracellular matrix (ECM) proteins have been employed as cell delivery vehicles that provide pro-survival cues to the transplanted cells. Such ECMs include alginate [20], collagen [21], fibrin [22], hyaluronan [23], and chitosan [24]. However, a limitation of these ECMs is that they consist of single-factor ECMs, which overly simplifies the complex ECM milieu within biological tissues. In particular, ECs physiologically reside within an endothelial basement membrane that is comprised of numerous ECMs including laminin, collagen type IV, fibronectin, and heparan sulfate proteoglycans [25]. It is well-recognized that the complex ECM composition of the endothelial basement membrane provides important signaling cues for physiological maintenance of endothelial behavior and function [26]. However, so far there has been no study systematically quantifying the effects of chemically complex multi-component ECMs on endothelial survival and function under the condition of hypoxia (1% O2) and reduced nutrients (1% fetal bovine serum), as is present in ischemic injury sites.. This dearth of knowledge undermines the therapeutic potential of stem cell therapy for treatment of tissue ischemia and other cardiovascular diseases.
Under both physiological as well as pathological conditions, ECs interact with a milieu of combinatorial ECMs. Accordingly, we tested the hypothesis that combinatorial ECM interactions may be more effective than single-factor ECMs in improving iPSC-EC viability and function under hypoxia with reduced nutrient condition. Using a micro-scale high-throughput platform for simultaneous screening of iPSC-EC function and survival when cultured on hundreds of combinatorial ECMs, we demonstrate that multi-component ECM combinations such as collagen IV+gelatin+heparan sulfate+laminin (CGHL) and collagen IV+fibronectin+gelatin+heparan sulfate+laminin (CFGHL) show significant improvement over single-factor ECMs in maintaining phenotypic marker expression, as well as augmenting cell survival and function under the condition of hypoxia with reduced serum. Furthermore, this approach enables full-factorial analysis of interaction effects between ECMs, which may be critical for understanding how cells respond to complex microenvironmental cues.
2. Methods
2.1 Fabrication of ECM microarray slides
ECM microarray was generated using single ECMs or multi-component mixtures of the following ECM proteins: collagen IV (C, mouse, Southern Biotech; Cat. No. 1250–04S ), fibronectin (F, bovine, Sigma; Cat No. F1141), laminin (L, mouse, Life Technologies; Cat No. 23017–015), gelatin (G, bovine, Sigma; Cat No. G1393), heparan sulfate (H, mouse, Sigma; H4777), and Matrigel (M, mouse, BD Biosciences; Cat No. 356231) (Supp Figure 1). Stock solutions (0.5 mg/ml) of each ECM combination were loaded into micro-well reservoirs. Using the OmniGrid Accent Microarrayer (Gene Machines), combinatorial ECMs were drawn from a reservoir using circular pins and spotted onto protein binding Schott H slides (Nexterion), forming individual circular “islands” that were 0.3 mm in diameter and 0.3 mm apart from neighboring islands. Each of the 63 single or multi-component (2-factor, 3-factor, 4-factor, 5-factor, and 6-factor) ECM compositions were printed onto slides with 6 replicates, which resulted in a total of 378 ECM islands. The total ECM concentration for each ECM combination was held constant at 0.5 mg/ml. For multi-component compositions, each individual ECM component was loaded in equal mass ratios as other ECMs (ie, 1:1, 1:1:1, etc). After covalent attachment of the ECMs, the microarray slides were air dried and transferred into vacuum sealed boxes and stored in the dark at 4 °C until used. Although Matrigel is composed of non-defined ECMs derived from Engelbreth-Holm-Swarm sarcoma, it was included for comparison to defined combinatorial ECMs because it has been frequently used for in vivo cell delivery applications [13].
Green 540 Reactive Fluorescence Dye (Arrayit) was used to reveal the amount of proteins attached to the slides after fabrication based on the intensity of fluorescence. Microarray slides were incubated in Green 540 Dye (1x) for 1 hour, followed by washes with phosphate-buffered saline (PBS). Similar procedures were performed to quantify the amount of specific ECMs (laminin and fibronectin) using anti-laminin (Abcam) and anti-fibronectin (EMD Millipore) antibodies. Images were obtained using fluorescence microscope (Keyence, BZ-X710) at 4X objective. Quantification of fluorescence intensity was performed using Image J.
2.2 Generation and characterization of iPSC-ECs
Human iPSCs (HUF5 strain) were previously derived by retroviral-mediated transduction of Oct-4, Sox-2, Klf-4 and c-Myc in adult human dermal fibroblasts [27]. To generate iPSC-ECs, iPSCs were differentiated in the presence of vascular endothelial growth factor and bone morphogenetic protein-4 for two weeks as previously described [28]. Fluorescent activated cell sorting (FACS) for CD31 expression previously indicated >90% of the human iPSC-ECs expressed CD31 (Supp Figure 2A–B) [7, 28, 29]. Immunofluorescence staining demonstrated that the cells express known endothelial markers such as von Willebrand Factor and could functionally take up acetylated low density lipoprotein (Supp Figure 2C–D). Genetic, protein, and functional characterization of this strain of iPSC-ECs have been previously reported by us and others to confirm endothelial identity [28, 30].
2.3 Cell seeding on ECM microarray slides
Prior to in vitro studies, ECM microarray slides were sterilized in 1X anti-mycotic solution (Life Technologies) for 30 minutes at 37°C, followed by 3 washes in PBS. The iPSC-ECs were dissociated with Tryple Express (Life Technologies) and seeded on top of the slides at a density of 5×105 cells per slide in 5 ml EGM-2MV growth medium (Lonza) which contains growth factors and 5% fetal bovine serum (FBS). The cells were redistributed through gently shaking the slides every 1 hour to avoid cell aggregation. After 6 hours, unbound cells were removed and the medium was replaced with fresh medium. Cells seeded on the slides were incubated overnight at 37°C with 5% CO2 prior to hypoxia studies. Initial cell attachment was relatively uniform throughout the slide based on the quantification of total nuclei using Hoechst 33342 staining after 8 hour of cell seeding (Supp Figure 3).
2.4 Endothelial phenotypic marker expression of CD31 on ECM microarrays under hypoxia with reduced serum conditions
After overnight cell attachment, the cells on the ECM microarrays were subjected to conditionsfrequently found at sites of tissue ischemia, namely reduced nutrients and hypoxia. Specifically, the media was replaced with endothelial basal media (EBM, Lonza), which lacks growth factors, supplemented by 1% FBS. The cell-seeded ECM microarray slides were transferred into hypoxia chambers filled with hypoxic gas (1% O2, 5% CO2, 94% N2) and maintained at 37 °C for 48 hours. After the 48-hour incubation in hypoxia with reduced serum condition, the ECM microarray slides were fixed in 4% paraformaldehyde, permeabilized in 0.1% Triton-X100, blocked in 1% bovine serum albumin, and then incubated with an antibody targeting the endothelial phenotypic marker, CD31 (Dako). After primary antibody incubation, the samples were incubated with Alexa Fluor-488-conjugated goat anti-mouse secondary antibody (Life Technologies), followed by incubation in Hoechst 33342 nuclear dye (Life Technologies). Each ECM microarray was imaged by the ImageXpress Micro high-content imaging system (Molecular Device). Automated images were acquired for each individual ECM island in the channels of 488 (CD31) and Hoechst 33342 using 10X objectives at a focal plane that gave the maximum fluorescent signal for each channel.
The acquired images were analyzed using MetaXpress software (version 5.0) to measure the integrated fluorescence intensity of CD31 staining in each ECM island, after thresholding above the background fluorescence (n=10 independent ECM microarray slides). CD31 data for each ECM island was normalized to total cell nuclei based on the corresponding Hoechst 33342 image. For comparison of data between independent ECM microarray slide, the data was standardized by the Z-score method as described by Brafman et al. [31] in which Zi = (Xi – μ)/σ, where Xi is the log2 transformed data for ECM composition i, μ is the averaged log2 transformed data through the entire ECM slide, and σ is the standard deviation of the log2 transformed data among all spots on each array. Data from replicate spots (n = 6 per ECM composition) were averaged for each microarray slide. Heat maps were generated using Multiple Experiment Viewer (MeV, The Perl Foundation) by plotting the Z-scores from each ECM slide using a color code of red and blue representing higher and lower intensities, respectively, relative to the global average. For the ease of describing the ranked ECM combinations derived from heat maps the ECM compositions were clustered into four tiers, where Tier I compounds were associated with the highest expression of CD31.
2.5 Validation of combinatorial ECM effects on CD31 expression using a conventional tissue culture format
To verify the CD31 data from ECM microarrays, similar experiments were also performed using conventional cell culture settings.
In particular, iPSC-ECs were cultured on 4-well chamber slides that were pre-coated for 3 hours with selected ECM compositions spanning the four tiers of the heat map: C+F+G+H+L (CFGHL, Tier I), C+G+H+L (CGHL, Tier I), F (Tier III), and C (Tier IV). Cells were seeded at a density of 5 × 104 cells/well. After overnight incubation, the cells were incubated in hypoxia, followed by fixation and immunofluorescence staining against CD31 antibody and Hoechst 33342. Imaging was performed on a Zeiss LSM710 confocal microscope under 10X objective (n=6 samples). Using ImageJ software, images of CD31 staining were converted into grey scale, and then the integrated intensity was measured after thresholding above the background fluorescence. To normalize the data, total nuclei in images stained for Hoechst 33342 images were quantified by first applying thresholding to reduce the background florescence and then using the particle counting function to count total nuclei.
2.6 Cell viability on ECM microarrays under hypoxia with reduced serum condition
After overnight cell seeding on the ECM microarrays, the growth medium was replaced with EBM basal media with 1% FBS in the absence of growth factors. The ECM microarray slides were transferred into hypoxia chambers filled with hypoxic gas (1% O2, 5% CO2, 94% N2) and maintained at 37 °C for 48 hours. The Live/Dead Cytotoxicity Kit (Life Technologies) was used to determine cell viability on the slides (n=5 microarray slides). Calcein-AM (2 μM) and ethidium homodimer-1 (4 μM) vital dyes were added to the medium for 30 minutes prior to imaging to detect live cells (green) and dead cells (red), respectively. Images for each ECM island were acquired using the ImageXpress Micro high-content imaging system under 10X objectives. Automated image analysis for cell viability on each ECM island was performed using MetaXpress software by separately quantifying the number of green and red cells based on cell size and fluorescence intensity after thresholding. The percentage of viable cells was calculated by number of green cells divided by the total (green plus red) cells on each ECM island. The average of 6 replicates for each ECM slide was calculated, log2 transformed, and then used for Z-score calculation and heat map display as described in section 2.4. For the ease of describing the ranked ECM combinations derived from heat maps, the ECM compositions were clustered into four tiers, where Tier I compounds were associated with the highest cell viability.
2.7 Validation of combinatorial ECM effects on cell viability using a conventional tissue culture format
To verify the results from the ECM microarray platform, similar experiments were performed using 4-well chamber slides pre-coated with representative combinatorial and single-factor ECMs spanning the four tiers of the heat map: CFGHL, (Tier I), CGHL (Tier I), F (Tier III), and C (Tier IV) (n=3 samples). The cells were cultured for 48 hours under hypoxia in EBM media with 1% FBS prior to the addition of vital dyes. Images were taken by Zeiss confocal microscope using 10X objectives and analyzed by ImageJ software. Fluorescent images of each vital dye were acquired and converted into grey scale, and thresholding was applied to reduce the background. The numbers of green and red cells were separately counted using the particle counting function. The percentage of live cells was calculated by the number of green cells divided by the number of total (green plus red) cells.
2.8 Nitric oxide (NO) production on ECM microarrays under hypoxia with reduced serum condition
After overnight cell seeding, ECM microarray slides were incubated in EBM basal media with 1% FBS in the absence of growth factors in hypoxic gas (1% O2, 5% CO2, 94% N2). After 24 hours under hypoxia with reduced serum conditions, ECM microarray slides were then incubated with the fluorescent NO probe, 4-amino-5-methylamino-2′,7′-difluororescein diacetate (DAF-FM, 5 μM, Thermo Fisher) for 30 minutes. Afterwards, the slides were replaced with fresh medium and incubated for additional 20 minutes before high-throughput imaging using the ImageXpress Micro system under 10X objectives (n=5 microarray slides). Using the MetaXpress software, integrated fluorescence intensity of the NO probe was measured and normalized by total cell number for each ECM island in a similar manner as described in section 2.4. Cell numbers were measured using the cell counting function in MetaXpress software based on the size of the cells and the fluorescence intensity above the background. For the ease of describing the ranked ECM combinations derived from heat maps, the ECM compositions were clustered into four tiers, where Tier I compounds were associated with the highest NO production.
2.9 Validation of combinatorial ECM effects on NO production using conventional tissue culture settings
Verification of NO results from ECM microarrays was performed under a similar condition at a cell density of 5 × 104 cells/well in 4-well chamber slides pre-coated with selected combinatorial or single-factor ECMs spanning the tiers of the heat map: CFGHL, (Tier I), CGHL (Tier I), F (Tier III), and C (Tier IV) (n=4 samples). After incubation in hypoxia and reduced nutrient conditions for 24 hours, the cells were incubated with the NO probe. The NO fluorescence intensity for each treatment group was imaged by a fluorescence microscope (Keyence, BZ-X710). The integrated fluorescence intensity was quantified after thresholding and normalized to total cell number by counting green particles in each image using ImageJ software as described earlier.
2.10 Validation of combinatorial ECM effect on cell viability in three-dimensional scaffold
Bioluminescent iPSC-ECs were generated as previously described [28]. Briefly, human iPSC-ECs were transduced with a lentiviral vector (LV-pUb-Fluc-GFP) carrying an ubiquitin promoter driving firefly luciferase (Fluc) and enhanced green fluorescence protein (GFP). These cells were previously purified by FACS for dual expression of green fluorescence protein (GFP) and CD31 positive expression. The purified bioluminescent iPSC-ECs were then seeded into 3D porous polystyrene scaffolds (Biotek, 5 mm diameter, 0.6 mm thickness, 200 μm pore size) at 1×106 cells/scaffold for 24 hours under normoxic conditions in EGM2-MV, before transferring to hypoxia for 48 hours in EBM media containing 1% FBS (n=5 samples). Prior to cell seeding, the scaffolds were pre-coated by submerging in 500 μl of CGHL or C for three hours at 4 °C before washing the scaffolds with PBS. Bioluminescence imaging (BLI) was performed by the addition of reporter probe D-luciferin (150 μg/ml) to the cells, followed by imaging with the In Vivo Imaging System Spectrum (IVIS Spectrum; Caliper Life Sciences). The BLI intensity was expressed in units of radiance (p/cm2/s/sr).
2.11 Statistical analysis
All data are expressed as mean ± standard deviation. The microarray data was log2-transformed and standardized by Z-scores as previously described [31]. For ECM microarray analysis, multi-factorial Analysis of Variance (ANOVA) decomposition (main effects, 2-factor, 3-factor, 4-factor, 5-factor and 6-factor interactions) was computed using software in R with Bonferroni adjustment (63 tests) using standard factorial analysis formulae [32]. A null set of baseline values (ie, no ECM) was created by using the lowest yield from any of the experiments in order to balance the full set of data. While this is arguably artificial, it is conservative, since it overestimates the values in the true null set. For validation of ECM microarray data using chamber slides, ANOVA with Bonferroni adjustment was employed. For comparison between two groups, a Student’s t-test was used. Statistical significance was accepted at p<0.05.
3. Results
3.1 Combinatorial ECMs modulate CD31 expression in iPSC-ECs under hypoxia with reduced serum condition
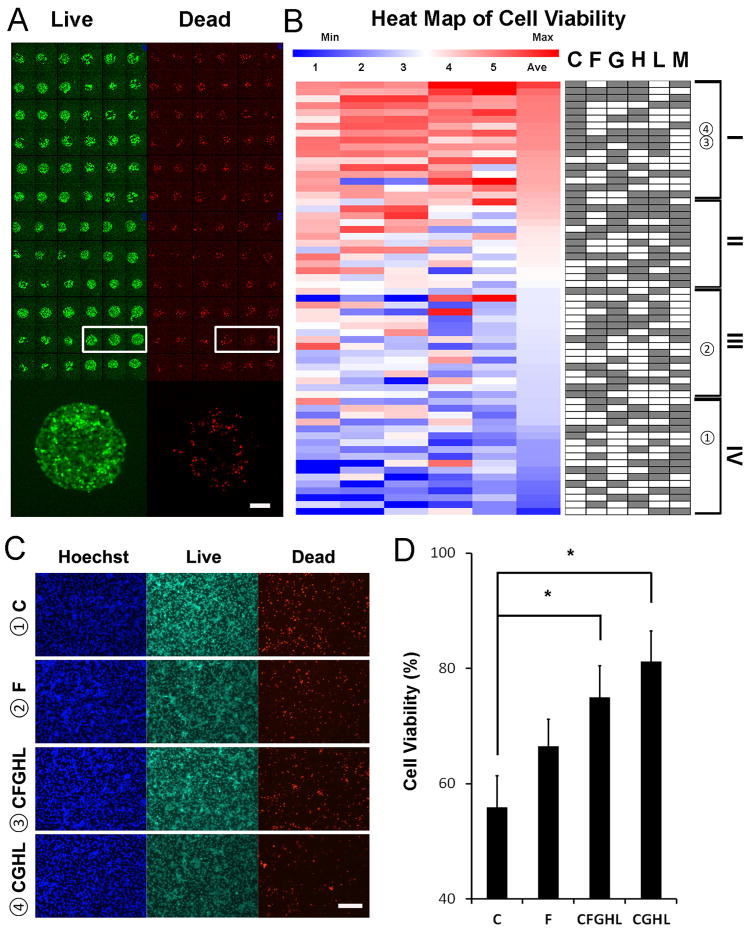
Combinatorial ECM microarrays consisting of ECMs found in the endothelial basement membrane were fabricated as previously described with some modifications [33]. Each ECM microarray slide included 63 unique ECM compositions derived from the 6 ECM proteins at equal mass ratios (1:1, 1:1:1, etc.). Each of the 63 ECM compositions were printed onto the microarray in replicates of 6, resulting in a total of 378 ECM individual islands that were 0.3 mm in diameter. Although there were slight differences in iPSC-EC attachment on different chemical compositions of ECMs, the iPSC-ECs attached reproducibly among replicates of the same combinatorial ECM. An example of consistent cell attachment among three replicates of the same ECM composition is highlighted by the white box in Figure 1A. In addition, the intensity of protein-binding fluorescence dye showed relatively consistent amount of ECMs attached to the slides, as shown in Supp Figure 4A. Further quantification was performed using specific antibodies against laminin and fibronectin (Supp Figure 4B–C). As expected, the intensity of laminin and fibronectin staining decreased from one-factor to three-factor to five-factor ECM compositions. Throughout the three in vitro assays, we observed relatively uniform cell attachment across different combinatorial ECMs (Supp Figure 5A–C). To assess the role of combinatorial ECMs in modulating iPSC-EC phenotype and function, we first quantitatively compared the expression of CD31, which is a characteristic phenotypic marker of ECs that has been shown to be differentially regulated under disease-like conditions [34]. We first compared the expression levels of CD31 in iPSC-ECs after culture on the ECM microarrays in hypoxia for 48 hours (Figure 1A & B). We used well-established image analysis software and Z-score standardization algorithms [31, 35] to compare the intensity of CD31 among combinatorial ECMs after normalization by cell number. The results were then displayed in a heat map that ranked the combinatorial ECMs compositions in four tiers. The ECMs that maintained the highest CD31 expression were located in Tier I, and they consisted predominantly of combinatorial ECMs such as C+H (CH) and C+H+L (CHL). Among single-factor ECMs, G and H were ranked within Tier I, whereas F and L were ranked in Tier III, and C and M were ranked in the bottom-most tier. These results strongly indicated that combinatorial ECMs in Tier I promoted greater expression of CD31, when compared to 4 out of 6 of the single-factor ECMs.
Figure 1. Differential expression of CD31 phenotypic marker in human iPSC-ECs cultured on ECM microarray in hypoxia with reduced serum condition.
(A) CD31 immunostaining after 48 hours of hypoxia treatment in reduced serum conditions. CD31 expression among representative replicates of the same ECM composition is shown in the white box. Enlarged image shows CD31 expression on iPSC-ECs cultured on a representative ECM island. (B) Heat map of normalized CD31 expression derived from ECM microarray data is shown for five independent slides in columns. Integrated fluorescence data was measured from each combinatorial ECM and averaged among the 6 replicates on each microarray slide. The normalized data for each dataset was then standardized by Z-score calculation to generate a heat map. The identity of all 63 combinatorial ECMs is shown on the right in which white or grey boxes denotes the absence or present, respectively, of an ECM component. Combinatorial ECMs were grouped into four tiers from high (Tier I) to low (Tier IV). (C) Human iPSC-EC expression of CD31 when cultured on 4-well chamber slides pre-coated with representative combinatorial or single-factor ECMs: ➀C (Tier IV), ➁F (Tier III), ➂CFGHL (Tier I), and ➃CGHL (Tier I). (D) Quantification of relative fold increase of CD31 expression, normalized to total cell number (*p<0.05, n=3 slides). Scale bars: 100 μm (A); 200 μm (C).
To validate these results from an ECM microarray in a conventional cell culture format, iPSC-ECs were cultured on ECM-coated chamber slides in hypoxia to compare the normalized expression of CD31 among high scoring (Tier I) combinatorial ECMs to representative single-factor ECMs: CGHL (Tier I), CFGHL (Tier I), F (Tier III), and C (Tier IV) (Figure 1C–D). After 48 hours of culture under hypoxia with reduced serum condition, the normalized relative expression levels followed a similar expression pattern to that on the ECM microarray: CGHL (189.6±13.0%), CFGHL (164.7±16.7%), F (130.1±8.3%), and C (100±0%). These results demonstrated a significant relative increase in CD31 expression in Tier I combinatorial ECM CGHL, when compared to single-factor ECMs C or F. (p<0.05). These results concurred with that of the microarray analysis of CD31 expression, suggesting that combinatorial ECMs in the first tier could augment CD31 expression, when compared to single-factor ECMs from lower tiers.
3.2 Combinatorial ECMs modulate cell viability of iPSC-ECs under hypoxia with reduced serum condition
Next, to compare the effect of combinatorial ECMs to single-factor ECMs in modulating cell survival under hypoxia with reduced serum condition, the cells were cultured on ECM microarray under 1% O2 and 1% FBS conditions for 48 hours. To quantify cell viability, the cells on the arrays were then incubated with vital dyes calcein-AM (green) for live cells and ethidium homodimer-1 (red) for dead cells (Figure 2A). Cell viability was consistent among replicates of the same combinatorial ECM (Figure 2A, white box). The heat map showing normalized cell viability across all combinatorial ECMs among 5 independent slides indicates that combinatorial ECMs promoted greater cell viability, compared to single-factor ECMs (Figure 2B). For example, Tier I combinatorial ECMs such as C+G+H+M (CGHM) and C+F+G+H (CFGH), supported higher cell viability when compared to single-factor ECMs such as F (Tier III) and C (Tier IV). The only single-factor ECM that was ranked among the Tier I ECMs was G, whereas the remaining single-factor ECMs resided in Tiers II through IV. These results suggest that combinatorial ECMs in Tier I promoted greater cell survival than 5 out of 6 single-factor ECMs.
Figure 2. Cell viability on ECM microarray in hypoxia with reduced serum condition.
(A) Viability of iPSC-ECs on ECM microarray after 48 hours of hypoxia treatment in reduced serum conditions as assessed by fluorescent vital dyes. Viability among representative replicates of the same ECM composition is shown in the white box. Enlarged image shows the proportion of live vs. dead cells within a representative ECM island. (B) Quantification of normalized cell viability from ECM microarray data arranged into a heat map from high (Tier I) to low (Tier IV). The percentage of live cells was calculated by dividing the number of green cell by total cell number (green plus red cells). Viability data between samples were then standardized by Z-score transformation. Each column represents an independent ECM microarray sample (n=5). (C) Human iPSC-EC viability after 48 hours of hypoxia when cultured on 4-well chamber slides pre-coated with selected combinatorial or single-factor ECMs: ➀C (Tier IV), ➁F (Tier III), ➂CFGHL (Tier I), and ➃CGHL (Tier I). (D) Quantification of cell viability on chamber slides (*p<0.05, n=6 slides). Scale bars: 100 μm (A); 200 μm (C).
To validate the accuracy of results from a micro-scale ECM array format, we performed a similar assay using ECM-coated chamber slides. To demonstrate consistency of results from the ECM microarray format, we compared iPSC-EC viability among representative combinatorial and single-factor ECMs: CGHL (Tier I), CFGHL (Tier I); F (Tier III); and C (Tier IV). As shown in Figure 2C–D, quantification of viability showed that CGHL and CFGHL promoted significantly higher percentage of viable cells (CGHL: 81.2±5.3%; CFGHL 75.0±5.41%), compared to C (55.84±5.5%) (p<0.05). These results confirmed that our microarray-based analysis of cell viability could be reproduced under a conventional tissue culture format.
3.3 Combinatorial ECMs modulate nitric oxide production in iPSC-ECs under hypoxia with reduced serum condition
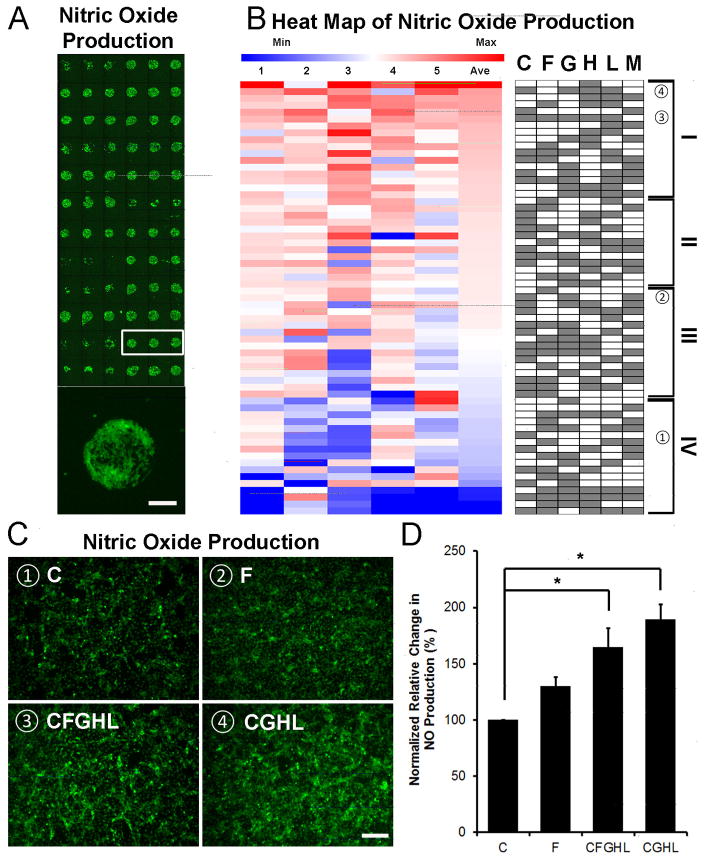
As a vasodilator and anti-atherogenic molecule that is released by ECs [36, 37], nitric oxide (NO) is an important soluble gas that is produced by ECs to maintain cardiovascular homeostasis. To assess for functional differences among iPSC-ECs on combinatorial ECM microenvironments in hypoxia with reduced serum condition, we compared NO production using DAF-FM [38], which is a fluorescent reagent that detects NO. The microarray slides seeded with iPSC-ECs were cultured for 24 hour in hypoxia and 1% FBS, followed by incubation with DAF-FM. The fluorescence intensity of DAF-FM was quantified and normalized by total cell number. As shown in the heat map of 5 independent ECM microarray slides (Figure 3A), cells produced NO at varying levels among combinatorial ECMs. The intensity of fluorescence was quantified, and the normalized values were plotted in a heap map in Figure 3B. The tier I ECM compositions included combinatorial ECMs such as CGHL and H+L+M (HLM). Among single-factor ECMs, only H was present in Tier I, whereas the other remaining 5 single-factor ECMs were ranked in the bottom two tiers. These results suggest that combinatorial ECMs in Tier I promoted greater NO production than 5 out of 6 single-factor ECMs.
Figure 3. Nitric oxide (NO) production on ECM microarrays under hypoxia with reduced serum condition.
(A) NO production was detected by fluorescence detection of the NO probe, DAF-FM diacetate. NO production among representative replicates of the same ECM composition is shown in the white box. Enlarged image shows NO production of iPSC-ECs on a representative island. (B) Heat map of NO assay showing normalized fluorescent intensity. Integrated fluorescence was measured and normalized to generate the heat map. Each column represents an independent ECM microarray sample (n=5). (C) NO production in iPSC-ECs after 48 hours of hypoxia when cultured on 4-well chamber slides pre-coated with selected combinatorial or single-factor ECMs: ➀C (Tier IV), ➁F (Tier III), ➂CFGHL (Tier I), and ➃CGHL (Tier I). (D) Quantification of NO production, normalized to total cell count (*p<0.05, n=4). Scale bars: 100 μm (A); 200 μm (C).
In order to verify our findings of NO production using conventional tissue culture format, cells were cultured on chamber slides in the presence of representative combinatorial and single-factor ECMs, namely CGHL (Tier I), CFGHL (Tier I), F (Tier III), and C (Tier IV) (Figure 3C–D). After 24 hours under hypoxia with reduced serum condition, the normalized relative NO accumulation levels in the CGHL (189.6±12.9%) and CFGHL (164.7±16.7%) treatment groups were significantly higher than that of C (100±0%) (p<0.05). These results confirmed that microscale analysis of NO production was consistent using a conventional tissue culture format.
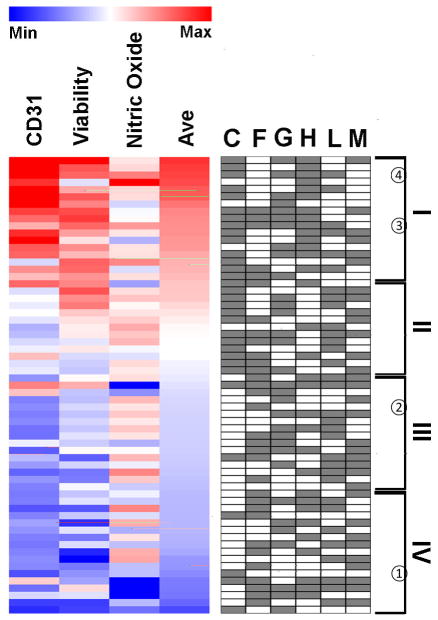
Based on the averaged levels of CD31 expression, cell viability, and NO production, combinatorial ECM compositions were ranked and plotted in a heat map (Figure 4). Although no ECM composition could produce maximum benefit in all three assays, the overall top 3 ECM compositions were identified in Tier I to be CGHM, CH, and CGHL. Within the top tier, all of the ranked ECMs were combinatorial with the exception of H. Together, the results from the three assays suggest that combinatorial ECMs in Tier I showed greater improvement in iPSC-EC viability, phenotype, and function when compared to 5 out of 6 of the single-factor ECMs.
Figure 4. Heat map of CD31 expression, viability, and NO production among combinatorial ECMs.
Combinatorial ECMs were ranked based on the average scores of the three assays.
3.4 Analysis of multi-factorial interactions and non-intuitive cellular response
Besides examining the empirical effects of combinatorial ECMs on iPSC-EC behavior and phenotype, we further applied this combinatorial approach to explore the main ECM effects. This analysis revealed fundamental insights into endothelial response to complex ECM microenvironments. A fully orthogonal basis was used for analysis of main effects and multi-factorial interactions. This analysis allows for the main and multi-factorial interactions to be tested irrespective of the others. As shown in Supp Figure 6, the bars represent the main effects measured by the change in yield in having the listed ECM component present vs. absent. The dotted line represents two-sided p-value cut-offs with Bonferroni corrections. Based on full-factorial analysis, C, G and H had overall positive effects in augmenting CD31 expression, whereas the other three ECMs (F, L, and M) had significant negative effects (Supp Figure 6A) (p<0.05). With respect to cell viability, C, G, and H had positive main effects in increasing the percentage of viable cells (p<0.05); whereas F, L or M alone did not affect cell viability significantly (Supp Figure 6B). As for the NO assay, main effect analysis showed that F was the only single ECM that significantly affected NO production, and the effect magnitude was negative (Supp Figure 6C) (p<0.05). Besides main effects, our results also suggest many significant multi-factorial interactions (up to the highest order) that also impact yield. Interactions are difficult to interpret, and more so the higher the order.
Based on iPSC-EC response to combinatorial ECMs under hypoxia with reduced serum condition, non-intuitive interaction effects among ECM components were observed. In comparing the CD31 expression among ECM combinations (Figure 5A), H or C single-factor ECMs maintained CD31 expression at levels of 1.18±0.18 and −0.95±0.13, respectively. However, the two-factor combination of C+H produced a synergistic increase in the normalized yield of 2.15±0.16. In addition, some interactions identified were redundant in nature. For example, in comparison to single-factor M (−0.68±0.04), the addition of single-factor G (1.94±0.21) showed no marked change to normalized CD31 expression yield in the two-factor combination of G+M (−0.68±0.05), suggesting that M played a dominant role over G in modulating CD31 expression. With respect to cell viability (Figure 5B), L or M single-factor ECMs could maintain cell viability at normalized yields of −0.09±0.11 and −0.01±0.17, respectively. However, the two-factor combination of L+ M reduced the normalized yields to −1.00±0.48 (in red), suggesting an inhibitory interaction of the two individual ECMs. Similarly, an inhibitory interaction was observed in NO production when iPSC-ECs were cultured on the three-factor combination of F+G+H (−1.82±0.48), in comparison to that of single-factors F (0.20±0.16), G (−0.14±0.51), or H (2.10±0.65) (Figure 5C). On the other hand, synergistic effects were also identified. For example, C as a single-factor ECM maintained cell viability at normalized yields of −0.49±0.25 (Figure 5B). However, when C was combined with L+M (−1.00±0.0.48), the normalized yield of the three-factor combination C+L+M increased synergistically to 0.85±0.09 (in green). These examples highlight the power of this combinatorial ECM microarray platform to provide new insights into iPSC-EC response to complex ECM environments, which would otherwise be difficult to achieve in conventional tissue culture plate formats.
Figure 5. Non-additive interaction effects among ECM combinations.
(A) CD31 expression was grouped by the number of ECM components and ranked among combinatorial ECMs. (B) Cell viability was grouped by the number of ECM components and ranked among combinatorial ECMs. (C) NO production was grouped by the number of ECM components and ranked among combinatorial ECMs.
3.5 Combinatorial ECMs that Enhance Cell Survival in 3D
To demonstrate whether combinatorial ECMs show consistent benefit even within a 3D system, we pre-coated 3D porous polystyrene scaffolds (Supp Figure 7) with ECM combinations CGHL (Tier I) or C (Tier IV) (Figure 4). Although the porous polystyrene scaffolds differ from the ECM microarrays in substrate stiffness and attachment chemistry, the primary purpose of the experiment was to assess whether the chemical composition of top tier combinatorial ECMs could produce a marked effect on iPSC-EC survival in a three-dimensional environment. The iPSC-ECs were cultured under hypoxia with reduced serum condition for 48 hours within the scaffolds. To quantify cell viability in 3D environments, bioluminescence imaging was performed as a measure of cell viability, where bioluminescence intensity correlates linearly with viable cell number (Supp Figure 8). Consistent to the microarray and chamber slide culture experiments, scaffolds coated with Tier I combinatorial ECM CGHL was able to significantly augment cell survival after 48 hours of hypoxia, compared to treatment with single-factor C (CGHL: 2.88±0.09×105 p/s/cm2/sr, C:1.75±0.24×105 p/s/cm2/sr, Figure 6, p<0.05). These results demonstrate the translatability of the micro-scale combinatorial ECM microarray findings to a 3D platform, which has the potential for clinical application.
Figure 6. Combinatorial ECMs modulate iPSC-EC viability in three-dimensional (3D) scaffolds.

(A) 3D porous polystyrene scaffolds were pre-coated with combinatorial ECM CGHL (Tier I) or single-factor C (Tier IV). Human iPSC-ECs were cultured under hypoxia for 48 hours in pre-coated scaffolds. Shown are bioluminescence images depicting cell survival between the treatment groups. (B) Quantification of cell survival based on bioluminescent imaging (*p<0.05, n=5).
4. Discussion
In this work, we successfully developed a high-throughput ECM microarray to assess the role of combinatorial ECMs under hypoxia with reduced serum condition. The salient findings of this work are that: 1) the Tier I combinatorial ECMs (ie CGHL & CFGHL) were capable of maintaining phenotypic marker expression and improving cell survival and function (Figures 1–4), when compared to some Tier II or Tier III ECM compositions such as C or F; 2) the ECM microarray findings could be validated in conventional tissue culture format such as chamber slides (Figures 1D, 2D, & 3D); 3) main and multi-factorial interactions analysis provides insight into how cells respond to complex cues (Supp Figure 6), although the basic signaling mechanisms that influence cellular response to combinatorial ECMs remain unknown; 4) the ECM microarray platform could identify in a facile manner the combinatorial ECMs that give rise to nonintuitive cellular responses, which would otherwise be difficult to identify in conventional cell culture platforms (Figure 5); and 5) the 2D micro-scale findings could be recapitulated in 3D microenvironments (Figure 6). For therapeutic translatability, we chose ECM combinations containing chemically defined ECM components for verifying the microarray data in conventional 2D tissue culture formats as well in a 3D scaffold environment. Based on these findings, human iPSC-EC delivery within combinatorial ECMs in Tier I (Figure 4) may improve the survival and function of these therapeutic cells under ischemic conditions such as heart failure or critical limb ischemia.
The ECM microarray platform enabled us to examine cell survival, phenotype and function in a high-throughput and systematic manner. Previous studies with ECM microarray have focused on ECM-mediated induction of stem cell differentiation and renewal. Flaim et al. employed ECM microarrays to study the effect of combinatorial ECMs on the maintenance of primary rat hepatocyte and differentiation of mouse embryonic stem cells (ESCs) toward hepatic fate [39]. This microarray system was later used by Brafman et al. to study the effects of combinatorial ECMs on modulating hepatic stellate cells phenotype [40]. In addition, the microarray platform was utilized to identify synthetic polymers that promote attachment, proliferation and self-renewal among human embryonic stem cell lines [33]. More recently, Brafman et al. reported that this platform could be used to examine ECMs that regulate endodermal differentiation in human ESCs [35], as well as identify synthetic polymers that support neuronal differentiation of neural progenitor cells [41]. These aforementioned works primarily employed ECM microarrays to identify the ideal microenvironment niche for cell maintenance and differentiation. In contrast, we believe this platform can serve as a powerful tool to advance the clinical application of stem cell therapy by improving cell survival and function under disease mimetic conditions. More importantly, our results could be validated in 3D settings to enhance the survival and function of human stem cells for therapeutic applications.
Although our ECM microarray utilized ~400 ECM islands, the ECM microarray platform is capable of testing thousands of proteins in microliter volumes at the same time under identical conditions. In addition, it utilizes a small number of cells (5×105) and small media volumes (5 ml), which is more cost-effective compared to larger cell culture dishes. Furthermore, ECM microarrays enable the assessment of interaction effects between ECMs in a systematic way. Our data clearly suggest that when multiple ECM components are present, synergistic and redundant effects were exhibited in cell survival, phenotype marker expression, and NO production (Figure 5, Supp Figure 3). In some cases, the multi-factorial interactions were significant through up to 6-factor interactions. This indicates that ECM interactions play a major role in influencing function of cells. However, a limitation of this high-throughput screening approach is that it enables identification of a general subset of possible ECMs that elicit differential responses from the iPSC-ECs, but it is not strong enough for cross-comparison of individual combinations due to the variances in response. Nevertheless, to the best of our knowledge, this is the first systematic study of multi-factorial interactions among different ECMs on cell behavior under hypoxia and reduced nutrient condition.
Similar to primary ECs, the human iPSC-ECs used in this study have shown endothelial identity by expressing multiple EC markers such as CD31, CD144 and Willebrand Factor [28]. Functionally, they form tube-like structures in matrigel, take up acetylated low density lipoprotein, undergo chemotaxis in response to vascular endothelial growth factor stimulation, exhibit tumor necrosis factor-induced upregulation of cell adhesion molecules, and polarize along the direction of laminar wall shear stress [42]. However, at the transcriptome level, there are differences in gene expression between iPSC-ECs and primary ECs [30]. The findings from this study using iPSC-ECs should be validated for consistency using primary ECs as a gold standard, since iPSC-ECs are not phenotypically identical to ECs. This limitation will be further addressed in the future investigations.
The ECMs in this study were selected based on their abundance in the basement membrane of blood vessels during physiological or pathological conditions, or due to their common usage for promoting adhesion of ECs to tissue culture Petri dishes. Laminin, collagen IV, and heparan sulfate are abundant ECMs in the basement membrane [25] of blood vessels, whereas fibronectin is secreted by ECs into the provisional matrix during angiogenesis [43, 44]. In addition, gelatin [45] and Matrigel [46] are common ECMs used in the in vitro culture of ECs, so they serve as a basis for comparison to the other ECMs. Accordingly, a limitation of this study is in the assessment of only these specific ECMs, which neglects other important ECMs that interact with ECs. In addition, drying of the ECM microarrays prior to cell seeding introduces the potential protein denaturation, which may also influence the findings.
It has been long recognized that ECMs regulate cell function, viability, and differentiation. In particular, EC function and protein expression has been shown to be modulated by ECMs. For example, endothelial NO synthase (eNOS) which catalyzes the production of NO has been shown to be down-regulated through integrin β1 interaction when primary ECs were cultured on fibronectin, collagen I, and collagen IV substrates [47]. More specifically, fibronectin has been shown to negatively modulate eNOS through a α5β1-p38 MAPK dependent pathway [48]. In addition, the glycation of ECM also contributes to NO release. It has been reported that glycated collagen reduced shear stress-mediated NO release by 50%, when comparing to that of cells on native collagen [49]. This reduction in NO release was correlated with decreased eNOS phosphorylation. In contrast to other studies, Gloe et al. showed that cells seeded on laminin substrates induced a 2-fold increase of eNOS expression in the presence of shear stress, while cells on non-coated or fibronectin-coated glass plates showed no significant effects [50]. Our microarray data also revealed that certain ECM combinations could reduce the production of NO without noticeable effects on cell viability (Figure 4). Together, our results support previously published findings that ECMs are potent modulators of endothelial behavior.
Non-intuitive interactions derived from combinatorial ECM cues have been reported not only by us, but also by others as well. For example, rat hepatocytes cultured on combinatorial ECM microarrays revealed that fibronectin or collagen IV single-component ECMs produced positive effects on albumin expression, whereas when combined fibronectin+collagen IV led to negative effects on albumin expression [39]. Conversely, single-factor ECMs of collagen III or laminin alone had negative interactions on albumin expression, whereas the two-component compositions of collagen III+laminin resulted in a positive interaction. Using the ECM microarray platform, Brafman et al. reported a number of synergistic and antagonistic interactions between ECMs. For example, in the two-factor interaction analysis, fibronectin and laminin alone had a positive effect on the activation of hepatic stellate cells (increase in proliferation, production of abnormal ECMs and inflammatory mediators), but when combined, fibronection+laminin strongly decreased the activation of these cells [40].
Although the mechanism by which combinatorial ECMs modulate cell behavior or lead to non-intuitive interactions is largely unknown, the likely contributing factors include including cell shape [51], ECM stiffness [52, 53], mechanical stimuli [54], and activation of integrin-mediated signal transduction pathways [55]. In addition, the actin cytoskeleton has been suggested to be an important component of the mechanosignaling cascade, as well as a number of transcription factors and chromatin remodeling enzymes. Manipulation of RhoA and its downstream effector, Rho kinase, appears to be important for cell differentiation [56]. The adhesion molecules also contribute to asymmetric stem cell division, including β1 integrin, CD146, and E-cadherin [57, 58]. The inhibition and synergistic effects reported in this study may likely result from crosstalk between integrin and growth factor signaling among matrix molecules [59]. For example, Kim et al. reported that fibronectin binding to integrin α5β1 enhanced endothelial cell migration on vitronectin through integrin αVβ3, without influencing cell adhesion [60]. A recent study also reported cell adhesion can be suppressed by specific integrin crosstalk using a number of integrin binding peptides, including FIB1 (integrin αVβ3), EF1zz (integrin α2β1) and 531 (integrin α3β1) [61]. It is plausible that combinatorial ECMs fine-tune cellular responses by differential activation of integrins. Together, these studies suggest that numerous signaling pathways are activated in response to ECM signaling cues, which may contribute to the non-intuitive interactions associated with combinatorial ECMs
Since cells can remodel their microenvironment by producing endogenous ECMs, it is likely that the ECM composition may be modified during the course of our studies. Although the ECM microarray platform can only control initial ECM composition, it has been shown by Brafman et al. that the starting ECM composition directs cell fate, despite endogenous production and/or remodeling of ECMs over time [35]. For this reason, the initial ECM microenvironment plays an important role in modulating cell function. It is also acknowledged that the endothelial basement ECM milieu is highly complex in not only chemical composition, but also in other factors such as secondary structure and mechanical properties. Although simple mixing of ECMs is an oversimplification of the physiological ECM microenvironment, it is an important first step towards understanding the role of combinatorial ECM chemical composition on cellular response.
When designing the ECM microarray slides, Matrigel was included as one of the components, owing to its importance in stem cell culture, maintenance, and transplantation [62–64]. However, the non-defined ECM composition of Matrigel, presence of growth factors, and batch-to-batch variability largely limits its clinical translatability [65]. Our results demonstrate that Matrigel was ranked overall in Tier III based on the three in vitro assays (Figure 4), suggesting that other combinatorial ECMs may be more suitable for iPSC-ECs than Matrigel. Since our ECM microarray is the 2D platform, we also performed cell viability assay using a 3D porous scaffold pre-coated with combinatorial ECMs to better mimic the 3D niche under hypoxia. The 3D results are promising, and future in vivo assessment of combinatorial ECMs as pro-survival carriers of stem cells is warranted for the treatment of ischemic cardiovascular diseases.
5. Conclusions
In conclusion, we demonstrate that combinatorial ECMs microenvironments such as CGHL and CFGHL could enhance iPSC-EC function and cell survival in hypoxia with reduced serum condition. These results derived from micro-scale platforms were reproducible in conventional tissue culture settings as well as in 3D scaffolds. Furthermore, main and multi-factorial analysis revealed overall positive benefits of G and H in sustaining cell viability and phenotype. Evidence of non-intuitive interaction effects highlighted the complexity of cellular response to combinatorial ECM environments. The fundamental insights gained from this micro-scale high-throughput ECM microarray platform would be precluded in conventional tissue culture formats. The findings from this study have important implications in the co-delivery of iPSC-ECs with combinatorial ECMs to improve the efficiency of stem cell therapy for treatment of ischemic cardiovascular diseases.
Supplementary Material
Statement of Significance.
Human endothelial cells (ECs) derived from induced pluripotent stem cells (iPSC-ECs) are promising for treating diseases associated with reduced nutrient and oxygen supply like heart failure. However, diminished iPSC-EC survival after implantation into diseased environments limits their therapeutic potential. Since native ECs interact with numerous extracellular matrix (ECM) proteins for functional maintenance, we hypothesized that combinatorial ECMs may improve cell survival and function under conditions of reduced oxygen and nutrients. We developed a high-throughput system for simultaneous screening of iPSC-ECs cultured on multi-component ECM combinations under the condition of hypoxia and reduced serum. Using automated image acquisition and analytical algorithms, we identified combinatorial ECMs that significantly improved cell survival and function, in comparison to single ECMs. Furthermore, this approach revealed complex ECM interactions and non-intuitive cell behavior that otherwise could not be easily determined.
Acknowledgments
This study was supported in part by grants to NFH from the US National Institutes of Health (R00HL098688, R01HL127113, and R21EB020235-01), Merit Review Award (1I01BX002310) from the Department of Veterans Affairs Biomedical Laboratory Research and Development, the Stanford Chemistry Engineering & Medicine for Human Health, and the Stanford Cardiovascular Institute. We thank Zachary Strassberg for technical assistance in bioluminescence imaging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, de Ferranti S, Despres JP, Fullerton HJ, Howard VJ, Huffman MD, Judd SE, Kissela BM, Lackland DT, Lichtman JH, Lisabeth LD, Liu S, Mackey RH, Matchar DB, McGuire DK, Mohler ER, 3rd, Moy CS, Muntner P, Mussolino ME, Nasir K, Neumar RW, Nichol G, Palaniappan L, Pandey DK, Reeves MJ, Rodriguez CJ, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Willey JZ, Woo D, Yeh RW, Turner MBC American Heart Association Statistics, S. Stroke Statistics, Heart disease and stroke statistics--2015 update: a report from the American Heart Association. Circulation. 2015;131:e29–322. doi: 10.1161/CIR.0000000000000152. [DOI] [PubMed] [Google Scholar]
- 2.Asahara T, Murohara T, Sullivan A, Silver M, van der Zee R, Li T, Witzenbichler B, Schatteman G, Isner JM. Isolation of putative progenitor endothelial cells for angiogenesis. Science. 1997;275:964–967. doi: 10.1126/science.275.5302.964. [DOI] [PubMed] [Google Scholar]
- 3.Tateishi-Yuyama E, Matsubara H, Murohara T, Ikeda U, Shintani S, Masaki H, Amano K, Kishimoto Y, Yoshimoto K, Akashi H, Shimada K, Iwasaka T, Imaizumi T I. Therapeutic Angiogenesis using Cell Transplantation Study. Therapeutic angiogenesis for patients with limb ischaemia by autologous transplantation of bone-marrow cells: a pilot study and a randomised controlled trial. Lancet. 2002;360:427–435. doi: 10.1016/S0140-6736(02)09670-8. [DOI] [PubMed] [Google Scholar]
- 4.Van Tongeren RB, Hamming JF, Fibbe WE, Van Weel V, Frerichs SJ, Stiggelbout AM, Van Bockel JH, Lindeman JH. Intramuscular or combined intramuscular/intra-arterial administration of bone marrow mononuclear cells: a clinical trial in patients with advanced limb ischemia. J Cardiovasc Surg (Torino) 2008;49:51–58. [PubMed] [Google Scholar]
- 5.Traverse JH, Henry TD, Pepine CJ, Willerson JT, Zhao DX, Ellis SG, Forder JR, Anderson RD, Hatzopoulos AK, Penn MS, Perin EC, Chambers J, Baran KW, Raveendran G, Lambert C, Lerman A, Simon DI, Vaughan DE, Lai D, Gee AP, Taylor DA, Cogle CR, Thomas JD, Olson RE, Bowman S, Francescon J, Geither C, Handberg E, Kappenman C, Westbrook L, Piller LB, Simpson LM, Baraniuk S, Loghin C, Aguilar D, Richman S, Zierold C, Spoon DB, Bettencourt J, Sayre SL, Vojvodic RW, Skarlatos SI, Gordon DJ, Ebert RF, Kwak M, Moye LA, Simari RD N. Cardiovascular Cell Therapy Research. Effect of the use and timing of bone marrow mononuclear cell delivery on left ventricular function after acute myocardial infarction: the TIME randomized trial. JAMA. 2012;308:2380–2389. doi: 10.1001/jama.2012.28726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hare JM, Fishman JE, Gerstenblith G, DiFede Velazquez DL, Zambrano JP, Suncion VY, Tracy M, Ghersin E, Johnston PV, Brinker JA, Breton E, Davis-Sproul J, Schulman IH, Byrnes J, Mendizabal AM, Lowery MH, Rouy D, Altman P, Wong Po Foo C, Ruiz P, Amador A, Da Silva J, McNiece IK, Heldman AW, George R, Lardo A. Comparison of allogeneic vs autologous bone marrow-derived mesenchymal stem cells delivered by transendocardial injection in patients with ischemic cardiomyopathy: the POSEIDON randomized trial. JAMA. 2012;308:2369–2379. doi: 10.1001/jama.2012.25321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rufaihah AJ, Huang NF, Kim J, Herold J, Volz KS, Park TS, Lee JC, Zambidis ET, Reijo-Pera R, Cooke JP. Human induced pluripotent stem cell-derived endothelial cells exhibit functional heterogeneity. Am J Transl Res. 2013;5:21–35. [PMC free article] [PubMed] [Google Scholar]
- 8.Huang NF, Niiyama H, Peter C, De A, Natkunam Y, Fleissner F, Li Z, Rollins MD, Wu JC, Gambhir SS, Cooke JP. Embryonic stem cell-derived endothelial cells engraft into the ischemic hindlimb and restore perfusion. Arteriosclerosis, Thrombosis & Vascular Biology. 2010;30:984–991. doi: 10.1161/ATVBAHA.110.202796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho SW, Moon SH, Lee SH, Kang SW, Kim J, Lim JM, Kim HS, Kim BS, Chung HM. Improvement of postnatal neovascularization by human embryonic stem cell derived endothelial-like cell transplantation in a mouse model of hindlimb ischemia. Circulation. 2007;116:2409–2419. doi: 10.1161/CIRCULATIONAHA.106.687038. [DOI] [PubMed] [Google Scholar]
- 10.Sone M, Itoh H, Yamahara K, Yamashita JK, Yurugi-Kobayashi T, Nonoguchi A, Suzuki Y, Chao TH, Sawada N, Fukunaga Y, Miyashita K, Park K, Oyamada N, Sawada N, Taura D, Tamura N, Kondo Y, Nito S, Suemori H, Nakatsuji N, Nishikawa S, Nakao K. Pathway for differentiation of human embryonic stem cells to vascular cell components and their potential for vascular regeneration. Arterioscler Thromb Vasc Biol. 2007;27:2127–2134. doi: 10.1161/ATVBAHA.107.143149. [DOI] [PubMed] [Google Scholar]
- 11.Lu SJ, Feng Q, Caballero S, Chen Y, Moore MA, Grant MB, Lanza R. Generation of functional hemangioblasts from human embryonic stem cells. Nat Methods. 2007;4:501–509. doi: 10.1038/nmeth1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Terrovitis JV, Smith RR, Marban E. Assessment and optimization of cell engraftment after transplantation into the heart. Circ Res. 2010;106:479–494. doi: 10.1161/CIRCRESAHA.109.208991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O’Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J, Murry CE. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 14.Israely E, Ginsberg M, Nolan D, Ding BS, James D, Elemento O, Rafii S, Rabbany SY. Akt suppression of TGFbeta signaling contributes to the maintenance of vascular identity in embryonic stem cell-derived endothelial cells. Stem Cells. 2014;32:177–190. doi: 10.1002/stem.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim I, Kim JH, Moon SO, Kwak HJ, Kim NG, Koh GY. Angiopoietin-2 at high concentration can enhance endothelial cell survival through the phosphatidylinositol 3′-kinase/Akt signal transduction pathway. Oncogene. 2000;19:4549–4552. doi: 10.1038/sj.onc.1203800. [DOI] [PubMed] [Google Scholar]
- 16.Matsui T, Tao J, del Monte F, Lee KH, Li L, Picard M, Force TL, Franke TF, Hajjar RJ, Rosenzweig A. Akt activation preserves cardiac function and prevents injury after transient cardiac ischemia in vivo. Circulation. 2001;104:330–335. doi: 10.1161/01.cir.104.3.330. [DOI] [PubMed] [Google Scholar]
- 17.Fujio Y, Nguyen T, Wencker D, Kitsis RN, Walsh K. Akt promotes survival of cardiomyocytes in vitro and protects against ischemia-reperfusion injury in mouse heart. Circulation. 2000;101:660–667. doi: 10.1161/01.cir.101.6.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loor G, Schumacker PT. Role of hypoxia-inducible factor in cell survival during myocardial ischemia-reperfusion. Cell Death Differ. 2008;15:686–690. doi: 10.1038/cdd.2008.13. [DOI] [PubMed] [Google Scholar]
- 19.Yet SF, Tian R, Layne MD, Wang ZY, Maemura K, Solovyeva M, Ith B, Melo LG, Zhang L, Ingwall JS, Dzau VJ, Lee ME, Perrella MA. Cardiac-specific expression of heme oxygenase-1 protects against ischemia and reperfusion injury in transgenic mice. Circ Res. 2001;89:168–173. doi: 10.1161/hh1401.093314. [DOI] [PubMed] [Google Scholar]
- 20.Aguado BA, Mulyasasmita W, Su J, Lampe KJ, Heilshorn SC. Improving viability of stem cells during syringe needle flow through the design of hydrogel cell carriers. Tissue Eng Part A. 2012;18:806–815. doi: 10.1089/ten.tea.2011.0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kutschka I, Chen IY, Kofidis T, Arai T, von Degenfeld G, Sheikh AY, Hendry SL, Pearl J, Hoyt G, Sista R, Yang PC, Blau HM, Gambhir SS, Robbins RC. Collagen matrices enhance survival of transplanted cardiomyoblasts and contribute to functional improvement of ischemic rat hearts. Circulation. 2006;114:I167–173. doi: 10.1161/CIRCULATIONAHA.105.001297. [DOI] [PubMed] [Google Scholar]
- 22.Christman KL, Vardanian AJ, Fang Q, Sievers RE, Fok HH, Lee RJ. Injectable fibrin scaffold improves cell transplant survival, reduces infarct expansion, and induces neovasculature formation in ischemic myocardium. J Am Coll Cardiol. 2004;44:654–660. doi: 10.1016/j.jacc.2004.04.040. [DOI] [PubMed] [Google Scholar]
- 23.Liang Y, Walczak P, Bulte JW. The survival of engrafted neural stem cells within hyaluronic acid hydrogels. Biomaterials. 2013;34:5521–5529. doi: 10.1016/j.biomaterials.2013.03.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suh JK, Matthew HW. Application of chitosan-based polysaccharide biomaterials in cartilage tissue engineering: a review. Biomaterials. 2000;21:2589–2598. doi: 10.1016/s0142-9612(00)00126-5. [DOI] [PubMed] [Google Scholar]
- 25.Davis GE, Senger DR. Endothelial extracellular matrix: biosynthesis, remodeling, and functions during vascular morphogenesis and neovessel stabilization. Circ Res. 2005;97:1093–1107. doi: 10.1161/01.RES.0000191547.64391.e3. [DOI] [PubMed] [Google Scholar]
- 26.Watt FM, Huck WT. Role of the extracellular matrix in regulating stem cell fate. Nat Rev Mol Cell Biol. 2013;14:467–473. doi: 10.1038/nrm3620. [DOI] [PubMed] [Google Scholar]
- 27.Byrne JA, Nguyen HN, Reijo Pera RA. Enhanced generation of induced pluripotent stem cells from a subpopulation of human fibroblasts. PLoS One. 2009;4:e7118. doi: 10.1371/journal.pone.0007118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rufaihah AJ, Huang NF, Jame S, Lee JC, Nguyen HN, Byers B, De A, Okogbaa J, Rollins M, Reijo-Pera R, Gambhir SS, Cooke JP. Endothelial cells derived from human iPSCS increase capillary density and improve perfusion in a mouse model of peripheral arterial disease. Arterioscler Thromb Vasc Biol. 2011;31:e72–79. doi: 10.1161/ATVBAHA.111.230938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang NF, Dewi RE, Okogbaa J, Lee JC, Jalilrufaihah A, Heilshorn SC, Cooke JP. Chemotaxis of human induced pluripotent stem cell-derived endothelial cells. Am J Transl Res. 2013;5:510–520. [PMC free article] [PubMed] [Google Scholar]
- 30.White MP, Rufaihah AJ, Liu L, Ghebremariam YT, Ivey KN, Cooke JP, Srivastava D. Limited gene expression variation in human embryonic stem cell and induced pluripotent stem cell-derived endothelial cells. Stem Cells. 2013;31:92–103. doi: 10.1002/stem.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brafman DA, Chien S, Willert K. Arrayed cellular microenvironments for identifying culture and differentiation conditions for stem, primary and rare cell populations. Nat Protoc. 2012;7:703–717. doi: 10.1038/nprot.2012.017. [DOI] [PubMed] [Google Scholar]
- 32.Box GE, Hunter WG, Hunter JS. Statistics for experimenters. John Wiley & Sons; New Jersey: 1978. [Google Scholar]
- 33.Brafman DA, Chang CW, Fernandez A, Willert K, Varghese S, Chien S. Long-term human pluripotent stem cell self-renewal on synthetic polymer surfaces. Biomaterials. 2010;31:9135–9144. doi: 10.1016/j.biomaterials.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rival Y, Del Maschio A, Rabiet MJ, Dejana E, Duperray A. Inhibition of platelet endothelial cell adhesion molecule-1 synthesis and leukocyte transmigration in endothelial cells by the combined action of TNF-alpha and IFN-gamma. J Immunol. 1996;157:1233–1241. [PubMed] [Google Scholar]
- 35.Brafman DA, Phung C, Kumar N, Willert K. Regulation of endodermal differentiation of human embryonic stem cells through integrin-ECM interactions. Cell Death Differ. 2013;20:369–381. doi: 10.1038/cdd.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Furchgott RF, Zawadzki JV. The obligatory role of endothelial cells in the relaxation of arterial smooth muscle by acetylcholine. Nature. 1980;288:373–376. doi: 10.1038/288373a0. [DOI] [PubMed] [Google Scholar]
- 37.Moncada S, Higgs A. The L-arginine-nitric oxide pathway. N Engl J Med. 1993;329:2002–2012. doi: 10.1056/NEJM199312303292706. [DOI] [PubMed] [Google Scholar]
- 38.Ku CJ, Karunarathne W, Kenyon S, Root P, Spence D. Fluorescence determination of nitric oxide production in stimulated and activated platelets. Anal Chem. 2007;79:2421–2426. doi: 10.1021/ac061572q. [DOI] [PubMed] [Google Scholar]
- 39.Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nat Methods. 2005;2:119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 40.Brafman DA, de Minicis S, Seki E, Shah KD, Teng D, Brenner D, Willert K, Chien S. Investigating the role of the extracellular environment in modulating hepatic stellate cell biology with arrayed combinatorial microenvironments. Integr Biol (Camb) 2009;1:513–524. doi: 10.1039/b912926j. [DOI] [PubMed] [Google Scholar]
- 41.Tsai Y, Cutts J, Kimura A, Varun D, Brafman DA. A chemically defined substrate for the expansion and neuronal differentiation of human pluripotent stem cell-derived neural progenitor cells. Stem Cell Res. 2015;15:75–87. doi: 10.1016/j.scr.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 42.Belair DG, Whisler JA, Valdez J, Velazquez J, Molenda JA, Vickerman V, Lewis R, Daigh C, Hansen TD, Mann DA, Thomson JA, Griffith LG, Kamm RD, Schwartz MP, Murphy WL. Human vascular tissue models formed from human induced pluripotent stem cell derived endothelial cells. Stem Cell Rev. 2015;11:511–525. doi: 10.1007/s12015-014-9549-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Senger DR, Davis GE. Angiogenesis. Cold Spring Harb Perspect Biol. 2011;3:a005090. doi: 10.1101/cshperspect.a005090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rowe RG, Weiss SJ. Breaching the basement membrane: who, when and how? Trends Cell Biol. 2008;18:560–574. doi: 10.1016/j.tcb.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 45.Folkman J, Haudenschild CC, Zetter BR. Long-term culture of capillary endothelial cells. Proc Natl Acad Sci U S A. 1979;76:5217–5221. doi: 10.1073/pnas.76.10.5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ades EW, Candal FJ, Swerlick RA, George VG, Summers S, Bosse DC, Lawley TJ. HMEC-1: establishment of an immortalized human microvascular endothelial cell line. J Invest Dermatol. 1992;99:683–690. doi: 10.1111/1523-1747.ep12613748. [DOI] [PubMed] [Google Scholar]
- 47.Yuan Y, Courtman D. Extracellular Matrix Differentially Regulates Endothelial Nitric Oxide Synthase Production in HUVECs and Human Blood Outgrowth Endothelial Progenitor Cells. The FASEB Journal. 2015;29:143.141. [Google Scholar]
- 48.Viji RI, Kumar VB, Kiran MS, Sudhakaran PR. Modulation of endothelial nitric oxide synthase by fibronectin. Mol Cell Biochem. 2009;323:91–100. doi: 10.1007/s11010-008-9967-2. [DOI] [PubMed] [Google Scholar]
- 49.Kemeny SF, Figueroa DS, Andrews AM, Barbee KA, Clyne AM. Glycated collagen alters endothelial cell actin alignment and nitric oxide release in response to fluid shear stress. J Biomech. 2011;44:1927–1935. doi: 10.1016/j.jbiomech.2011.04.026. [DOI] [PubMed] [Google Scholar]
- 50.Gloe T, Riedmayr S, Sohn HY, Pohl U. The 67-kDa laminin-binding protein is involved in shear stress-dependent endothelial nitric-oxide synthase expression. J Biol Chem. 1999;274:15996–16002. doi: 10.1074/jbc.274.23.15996. [DOI] [PubMed] [Google Scholar]
- 51.Ingber D. Extracellular matrix and cell shape: potential control points for inhibition of angiogenesis. J Cell Biochem. 1991;47:236–241. doi: 10.1002/jcb.240470309. [DOI] [PubMed] [Google Scholar]
- 52.Deroanne CF, Lapiere CM, Nusgens BV. In vitro tubulogenesis of endothelial cells by relaxation of the coupling extracellular matrix-cytoskeleton. Cardiovasc Res. 2001;49:647–658. doi: 10.1016/s0008-6363(00)00233-9. [DOI] [PubMed] [Google Scholar]
- 53.Engler AJ, Griffin MA, Sen S, Bonnemann CG, Sweeney HL, Discher DE. Myotubes differentiate optimally on substrates with tissue-like stiffness: pathological implications for soft or stiff microenvironments. J Cell Biol. 2004;166:877–887. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ingber DE. Mechanobiology and diseases of mechanotransduction. Ann Med. 2003;35:564–577. doi: 10.1080/07853890310016333. [DOI] [PubMed] [Google Scholar]
- 55.Cohen DM, Chen CS. Mechanical control of stem cell differentiation. StemBook; Cambridge (MA): 2008. [PubMed] [Google Scholar]
- 56.Ren XD, Kiosses WB, Schwartz MA. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 1999;18:578–585. doi: 10.1093/emboj/18.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scadden DT. The stem-cell niche as an entity of action. Nature. 2006;441:1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 58.Watt FM, Hogan BL. Out of Eden: stem cells and their niches. Science. 2000;287:1427–1430. doi: 10.1126/science.287.5457.1427. [DOI] [PubMed] [Google Scholar]
- 59.Schwartz MA, Ginsberg MH. Networks and crosstalk: integrin signalling spreads. Nat Cell Biol. 2002;4:E65–68. doi: 10.1038/ncb0402-e65. [DOI] [PubMed] [Google Scholar]
- 60.Kim S, Harris M, Varner JA. Regulation of integrin alpha vbeta 3-mediated endothelial cell migration and angiogenesis by integrin alpha5beta1 and protein kinase A. J Biol Chem. 2000;275:33920–33928. doi: 10.1074/jbc.M003668200. [DOI] [PubMed] [Google Scholar]
- 61.Hozumi K, Fujimori C, Katagiri F, Kikkawa Y, Nomizu M. Suppression of cell adhesion through specific integrin crosstalk on mixed peptide-polysaccharide matrices. Biomaterials. 2015;37:73–81. doi: 10.1016/j.biomaterials.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 62.Hughes CS, Postovit LM, Lajoie GA. Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics. 2010;10:1886–1890. doi: 10.1002/pmic.200900758. [DOI] [PubMed] [Google Scholar]
- 63.Li F, Li W, Johnson S, Ingram D, Yoder M, Badylak S. Low-molecular-weight peptides derived from extracellular matrix as chemoattractants for primary endothelial cells. Endothelium. 2004;11:199–206. doi: 10.1080/10623320490512390. [DOI] [PubMed] [Google Scholar]
- 64.Xu C, Inokuma MS, Denham J, Golds K, Kundu P, Gold JD, Carpenter MK. Feeder-free growth of undifferentiated human embryonic stem cells. Nat Biotechnol. 2001;19:971–974. doi: 10.1038/nbt1001-971. [DOI] [PubMed] [Google Scholar]
- 65.Vukicevic S, Kleinman HK, Luyten FP, Roberts AB, Roche NS, Reddi AH. Identification of multiple active growth factors in basement membrane Matrigel suggests caution in interpretation of cellular activity related to extracellular matrix components. Exp Cell Res. 1992;202:1–8. doi: 10.1016/0014-4827(92)90397-q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.