Abstract
Introduction
With the emergence of the “big data” era, the biomedical research community has great interest in exploiting publicly available chemical information for drug discovery. PubChem is an example of public databases that provide a large amount of chemical information free of charge.
Areas covered
This article provides an overview of how PubChem’s data, tools, and services can be used for virtual screening and reviews recent publications that discuss important aspects of exploiting PubChem for drug discovery.
Expert opinion
PubChem offers comprehensive chemical information useful for drug discovery. It also provides multiple programmatic access routes, which are essential to build automated virtual screening pipelines that exploit PubChem data. In addition, PubChemRDF allows users to download PubChem data and load them into a local computing facility, facilitating data integration between PubChem and other resources. PubChem resources have been used in many studies for developing bioactivity and toxicity prediction models, discovering polypharmacologic (multi-target) ligands, and identifying new macromolecule targets of compounds (for drug-repurposing or off-target side effect prediction). These studies demonstrate the usefulness of PubChem as a key resource for computer-aided drug discovery and related area.
Keywords: PubChem, cheminformatics, virtual screening, computer-aided drug discovery, computational toxicology, polypharmacology, biological assay, data mining, database, quantitative structure-activity relationship (QSAR)
1. Introduction
Thanks to high-throughput screening (HTS)1,2 technology and combinational chemistry2,3, small research labs in academic institutions now can generate bioactivity data for a large number of molecules at a low cost. Through data mining and manual curation, many groups can also collect a substantial amount of chemical information from various sources, including scientific articles and patent documents. In addition, funding agencies have introduced data sharing policies for studies that they support, and journal publishers require the authors of papers to make underlying data publicly accessible. All these recent trends have led to a rapid growth in chemical information available in the public domain. With the emergence of the big data era, there is a great interest from the biomedical research community in exploiting this public information for virtual screening (VS),4,5 which uses computational techniques to explore a large compound library to select a small subset of potentially bioactive molecules that are tested in subsequent in vitro and in vivo experiments. VS is an essential part of modern drug discovery and has been reviewed in many articles.4,5
PubChem 6-8 is a public chemical information archive developed and maintained by the U.S. National Institutes of Health (NIH). PubChem collects chemical substance descriptions and their biological activities from hundreds of data sources and provides them to the public free of charge. With receiving millions of requests from tens of thousands users per day, PubChem serves as a key resource for biomedical science communities in many areas, including cheminformatics, chemical biology, medicinal chemistry, and drug discovery. Detailed information on PubChem is given elsewhere.6,7
There have been great interest in using PubChem for VS. In some studies,9,10 3-D structures of compounds downloaded from PubChem were used for molecular docking. In other studies,11,12 PubChem was searched for molecules structurally similar to known active compounds using similarity search11 or for compounds with a particular scaffold through substructure search.12 PubChem was also screened using various predictive models to identify compounds with desired bioactivity.13-20 Importantly, many studies21-35 used bioactivity data archived in PubChem to develop bioactivity or toxicity prediction models.24-35 In addition, PubChem data were used to build computational models to predict adverse drug reactions.36,37 Recently, “dark chemical matter” (DCM)38 in PubChem, defined as compounds that have never shown bioactivity after being tested repeatedly in many HTS experiments, has attracted much attention as a promising starting point for discovering lead molecules.
The present paper reviews important aspects of PubChem in the context of its application for VS. A brief overview of PubChem is given, including data organization as well as data contents relevant to VS. Information on chemical vendors and patents for compounds in PubChem is also discussed, which helps prioritize hit compounds for subsequent in vitro or in vivo screenings. In addition, this paper describes programmatic access to PubChem data, which is critical for building automated VS pipelines, and PubChemRDF, which facilitates integration of PubChem data with other resources useful for VS. A review of some important publications is given to explain how PubChem resources are used for developing bioactivity and toxicity prediction models, discovering polypharmacologic (multi-target) ligands, and identifying new macromolecule targets of chemicals (for drug-repurposing or off-target side effect prediction). Other relevant topics, such as dealing with active/inactive compound imbalances in PubChem’s bioactivity data and developing benchmarking data sets for VS from PubChem data, are also discussed.
2. An overview of PubChem as a resource for virtual screening
2.1. Data organization and chemical space coverage in PubChem
PubChem6,7 contains chemical substance descriptions and biological activity information, contributed by more than 400 data contributors.39 While PubChem’s data are primarily about small molecules, they also include other molecular entities, such as small-interfering and micro-RNAs (siRNAs and miRNAs), peptides, lipids, carbohydrates, chemically modified macromolecules and many others.
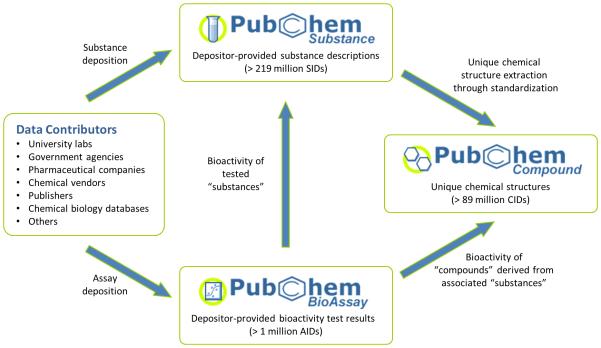
PubChem organizes these data into three primary databases: Substance, Compound, and BioAssay (Figure 1).6,7 The Substance database6,40 archives chemical substance descriptions submitted by individual data contributors. The Compound database6,41 stores unique chemical structures extracted from the Substance database through the PubChem standardization process. The BioAssay database7,42 contains the descriptions of biological assay experiments and bioactivity data for substances tested in the assays. The records in the Substance, Compound, and BioAssay databases are called substances, compounds, and assays, respectively. Similarly, SID (Substance ID), CID (Compound ID), and AID (Assay ID) are used as the record identifiers for the Substance, Compound, and BioAssay databases, respectively. As of May 2016, PubChem contains more than 219 million substances, 89 million compounds, and 230 million bioactivity outcomes from more than one million assays covering around ten thousand unique protein sequences.
Figure 1.
Data organization in PubChem. Chemical information deposited by more than 400 data contributors is organized into three primary databases: Substance, Compound, and BioAssay.
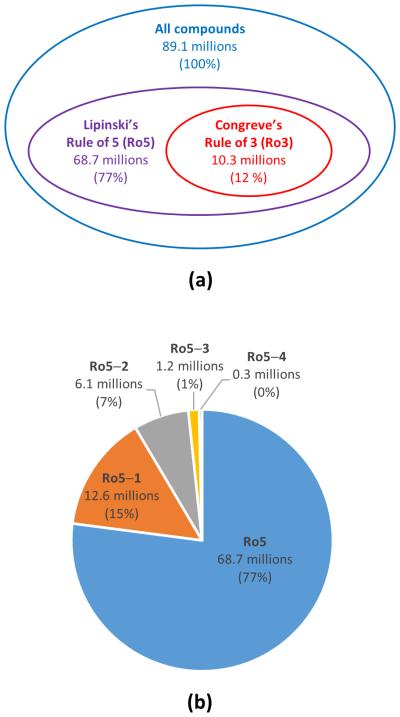
There has been much interest in analyzing the chemical space covered by molecules in PubChem. Especially, many studies43-47 have compared PubChem’s chemical space with those of other public databases of known molecules, such as DrugBank48 and ChEMBL,49 as well as those of databases of “virtual” molecules, such as chemical universe databases GDB-11 (26.4 million molecules with up to 11 atoms of C, N, O, F),43 GDB-13 (977 million molecules with up to 13 atoms of C, N, O, S, Cl),44 and GDB-17 (166.4 billion molecules up to 17 atoms of C, N, O, S, and halogens).45 As shown in Figure 2, 68.7 million compound records in PubChem (77% of the total) are drug-like compounds that satisfy Lipinski’s rule of 5.50 Among them, 10.3 millions (12% of the total) are fragment-like ones, which satisfy Congreve’s rule of 3.51
Figure 2.
Chemical Space covered by PubChem. Panel (a) shows the proportion of compounds that satisfies Lipinski’s rule of 5 (Ref. 50) and Congreve’s rule of 3 (Ref. 51). Panel (b) shows the proportion of compounds that satisfy all criteria of Lipinski’s rule of five (Ro5), and those that violate one, two, three, and four criteria of the rule of five (Ro5−1, Ro5−2, Ro5−3, and Ro5−4), respectively.
2.2. Bioactivity data in PubChem
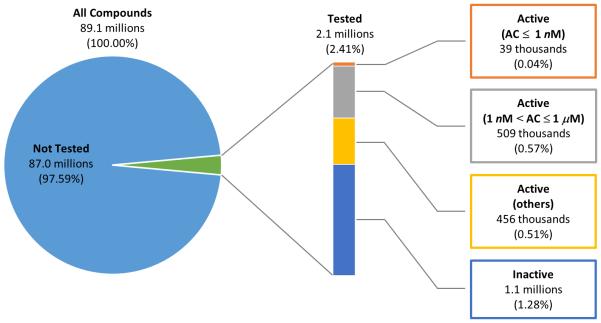
Figure 3 shows the distribution of PubChem compounds according to the availability of bioactivity data. Currently, 2.1 million compounds in PubChem have been tested in any assay in PubChem, corresponding to 2.4% of all 89.1 million compound records. About half of these tested compounds (1.0 million compounds, 1.1% of all compounds) have been declared to be active in at least one assay. 509 thousand compounds had an activity concentration between 1 nM and 1 μM, and 39 thousand compounds had an activity concentration of 1 nM or below.
Figure 3.
Distribution of tested, active, and inactive compounds in PubChem. Tested compounds are those tested in at least one assay experiment archived in PubChem. Active compounds are those which are declared as active in at least one assay in PubChem. Inactive compounds are those which are not declared as active in any assay in PubChem.
The majority of bioactivity data contained in PubChem were generated from HTS. Because HTS campaigns aim to identify hit molecules from a large compound library, HTS data typically contain a large number of inactive compounds with only a handful of active compounds, which are tested further in low-throughput experiments. Although advanced HTS technologies like quantitative HTS (qHTS)52 allow for getting bioactivity data at multiple compound concentrations in a single experiment, HTS is often run at a single concentration and therefore there is no guarantee that hit molecules from such HTS experiments would perturb the biological system in a dose-response way. In addition, HTS data may contain false hits, for example, due to aggregators,53 which non-specifically bind to multiple unrelated proteins, or autofluorescent compounds,54 which can emit light in the absence of artificial fluorescent markers used in fluorescence-based HTS assays. For these reasons, HTS data are considered to have low qualities in general.
However, PubChem also contains a substantial amount of high-quality bioactivity data extracted from scientific articles through manual curation or data mining,55 which complement the HTS data contained in PubChem. These data are contributed by various PubChem depositors, including ChEMBL,49 PDBbind,56 BindingDB,57 and IUPHAR/BPS Guide to Pharmacology.58 Data from these contributors cover different chemical domains from each other.55 For example, ChEMBL49 manually extracts bioactivity data from peer-reviewed papers published in journals in the medicinal chemistry and natural product domains. PDBbind56 collects experimentally measured binding affinity data for biomolecular complexes in the Protein Data Bank (PDB).59 BindingDB57 provides binding affinities, focusing chiefly on the interactions of protein considered to be drug targets with drug-like small molecules. The Guide to Pharmacology,58 which collects a wide range of information on important drug targets [e.g., G-protein-coupled receptors (GPCRs), ion channels, and nuclear hormone receptors (NHRs)], provides information on these proteins and their ligands.
2.3. Annotations available in PubChem
In addition to bioactivity data, PubChem contains a great deal of compound information that is useful for VS. For example, thanks to data integration with DrugBank,48 PubChem provides users with comprehensive information on FDA-approved and investigational drugs, including their drug indications, mechanisms of action, target macromolecules, interactions with proteins and genes, ADMET (absorption, distribution, excretion, metabolism, and toxicity) properties and many others. PubChem also contains toxicological information on chemicals that are of interest in environmental and human health, contributed by the Hazardous Substances Data Bank (HSDB).60
Experimentally determined 3-D structures of small molecules are also available in PubChem. The Molecular Modeling Database (MMDB)61 contributes to PubChem experimentally determined protein-bound ligand structures, derived from PDB.59 In addition, PubChem provides links to crystal structures available at the Cambridge Structural Database (CSD).62
PubChem also collects chemical information from important regulatory agencies, such as the U.S. Food and Drug Administration (FDA) and the U.S. Environmental Protection Agency (EPA). For example, information on drug products and ingredients from the FDA Orange Book63 is integrated in PubChem. PubChem also contains FDA’s Unique Ingredient Identifiers (UNIIs)64 and Pharmacologic Classes65 for drug ingredients. In addition, drug labeling information is available in PubChem, via NLM’s DailyMed.66 EPA Substance Registry Services provides PubChem with information on chemical substances tracked or regulated by EPA. Chemical data collected under the Toxic Substance Control Act and the Clean Air Act are also available in PubChem.
2.4. Availability of compounds for subsequent experiments
Because the primary goal of VS is to select a list of compounds to test in subsequent experiments, the availability of compounds is an important consideration. That is, they should be either synthesizable or purchasable. It is noteworthy that PubChem does not include “virtual” molecules. For each compound in PubChem, there are one or more data contributors who claim that they have the compound and/or information about it. Importantly, some of these contributors are chemical vendors from which one can purchase the compound.
Two important characteristics of PubChem records are worth mentioning, with respect to the availability of compounds in PubChem. First, PubChem records may become non-live, meaning that the records are not searchable although they do exist in the database. Data contributors to PubChem can revoke their substance information in PubChem for various reasons, for example, when they realize that they mistakenly submitted substances that they do not have, when they find incorrect information about the substance, or when they choose not to share their information with others. As an archive, PubChem does not remove the revoked substance information, but make it non-live (that is, not searchable). When a compound record does not have any live substance records associated, it becomes non-live. The compound can become live again if a live substance record associated with it appears in PubChem.
The second issue concerning the compound availability is that some information in PubChem is no longer maintained by data contributors (for example, because they do not have continued funding). Especially, some chemical vendors are out of business and compounds that were purchasable from them in the past are not available any more. To address this issue, PubChem introduced a “legacy” designation for collections that are not regularly updated. This legacy designation applies to projects/contributors that appear to no longer be active, as well as to their individual substance records. Legacy records will not be shown in the “Chemical Vendors” section of the Compound Summary page. Instead, they will only be found under “Legacy Depositors” in the “Substances by Category” section of the Compound Summary page. This designation will help PubChem users quickly identify records that may have out-of-date information and/or hyperlinks.
2.5. Patentability of compounds for intellectual property protection
In drug discovery programs, it is critical to make sure that identified drug candidates are patentable. PubChem currently offers links between about 6 million patent documents and more than 16 million unique chemical structures, with over 336 million chemical substance-patent links covering U.S., European, and World Intellectual Property Organization (WIPO) patent documents published since 1800. This information is contributed by various organizations, including IBM,67 SureChEMBL (formerly known as SureChem),68,69 NextMove Software,70 SCRIPDB,71 and BindingDB.57
When a compound has patent information, its Compound Summary page displays the patents associated with it in a tabular format. In addition, the compound record is annotated with WIPO International Patent Classification (IPC)72 information for the associated patents. IPC is a hierarchical classification system used to classify patent documents according to the technical fields they pertain. The IPC information is displayed under the Classification section of the Compound Summary page.
Users can search the Compound database for those associated with a particular patent document or retrieve all compounds that have patent information. Programmatic access to patent information is also possible through PUG-REST,73 which will be discussed later in this paper.
2.6. PubChem 2-D and 3-D neighbors
A disparity of available information exists among compounds contained in PubChem. Some compounds like approved drug molecules have a great deal of information, including bioactivity data, therapeutic use, mechanism of action, metabolism, literature and patents associated, and so on. However, many other structures (e.g., synthesized for HTS purposes) do not have much information other than their chemical structures. When a compound does not have desired information, it can be inferred from information available to similar compounds. PubChem assists users in finding similar chemical structures, by providing a pre-computed list of structurally similar molecules, called “neighbors,” for each compound in PubChem.74,75
PubChem neighbors come in two flavors: 2-D neighbors and 3-D neighbors (also known as “Similar Compounds” and “Similar Conformers”, respectively). Two compounds are defined as 2-D neighbors of each other when they have a 2-D molecular similarity score of 0.9 or greater, which is computed using the Tanimoto coefficient76 with the PubChem subgraph binary fingerprints.77 Computation of 3-D neighbors uses two Rapid Overlay of Chemical Structures (ROCS)78-based 3-D similarity measures: the shape-Tanimoto (ST), which quantifies the 3-D steric shape overlap between molecules, and the color-Tanimoto (CT), which evaluates the similarity in 3-D orientation of feature atoms between molecules. When one or more pairs of conformers of two compounds have a ST score of ≥ 0.80 and a CT score of ≥ 0.50, the two compounds are defined as 3-D neighbors of each other. For practical reasons, PubChem 3-D neighboring currently uses up to nine conformers per compound, although compounds in PubChem may have up to 500 conformers.74 In addition, 3-D neighboring only considers compounds with computationally generated 3-D conformer models, covering ~90% of all compounds in PubChem.74 Whereas much slower than 2-D neighboring, 3-D neighboring often identifies structural similarity that traditional 2-D graph-based structural similarity methods fail to recognize.74 Therefore, 3-D neighboring may offer complementary views on structural similarity between molecules with similar biological activities.
One may consider that PubChem neighboring is ligand-based VS against the entire PubChem Compound database with each compound as a query. The 2-D and 3-D neighbors of a compound can be accessed either through its Compound Summary page or programmatically through PUG-REST.73 They are also available in PubChemRDF,79 allowing users to import them into local computing resources and to take advantage of semantic web technologies (to be discussed later).
3. Automation of virtual screening pipelines
3.1. Programmatic access to PubChem for automated virtual screening pipelines
PubChem provides multiple programmatic access routes to its data,73 which allows one to build an automated virtual screening pipeline that exploit PubChem data. These access routes include: Entrez Utilities (also called E-Utilities or E-Utils),80 Power User Gateway (PUG),81 PUG-SOAP,82 and PUG-REST.83 The characteristics of these methods are summarized in Table 1, and more detailed information is given in our recent paper.73
Table 1.
Programmatic access routes to PubChem data. See Ref. 73 for more detail.
Entrez Utilities (E-Utilities or E-Utils) (Ref. 80)
|
Power User Gateway (PUG) (Ref. 81)
|
PUG-SOAP (Ref. 82)
|
PUG-REST (Ref. 83)
|
Among the four access routes in Table 1, PUG-REST is the simplest to use and learn because almost all information necessary to make a PUG-REST request can be encoded into a one-line Uniform Resource Locator (URL). In addition, it provides convenient access to information on PubChem records that are not accessible through the other programmatic interfaces. Importantly, PUG-REST supports various chemical structure searches commonly used in ligand-based VS, such as 2-D and 3-D similarity searches, substructure search, superstructure search, and identity search.
It should be noted that PubChem has a standard time limit of 30 seconds per web service requests. In addition, users should limit their web-requests to no more than three per second and violation of usage policies may result in the user being temporarily blocked from accessing PubChem (or NCBI) resources. See the NCBI policies and disclaimers84 for more information.
3.2. PubChemRDF for data exchange and integration
One may want to use PubChem data for building a new in-house virtual screening library or annotating an existing one. PubChemRDF,79 which is Resource Description Framework (RDF)-formatted PubChem data, can be used for this purpose. RDF85 is a World Wide Web Consortium (W3C) standard model for data interchange on the web. RDF breaks knowledge into so-called triples, each of which consists of the subject, object, and predicate. In essence, RDF expresses knowledge into a directed, labelled graph.
PubChemRDF is downloadable via the File Transfer Protocol (FTP). The RDF data on the PubChem FTP site is arranged in such a way that one can download only the desired type of information, instead of getting all RDF data. The downloaded data can be imported into a triplestore, such as Apache Jena TDB and OpenLink Virtuoso, and searched using a SPARQL query interface. Alternatively, one can load them and use the graph traversal algorithms to query the RDF graphs. In addition, PubChem provides a REST-ful interface for programmatic access to PubChemRDF data (not to be confused with PUG-REST). The PubChemRDF REST interface supports simple SPARQL-like query capabilities for grouping and filtering relevant resources.
PubChemRDF harnesses ontological frameworks to help facilitate PubChem data sharing, analysis, and integration with resources external to the National Center for Biotechnology Information (NCBI) and across scientific domains. Importantly, PubChemRDF enhances cross-integration by providing direct links to available authoritative RDF resources within applicable subdomains, including: reference, synonym, and InChIKey86 to MeSH RDF87; protein to UniProt RDF88; protein and substance to PDB RDF89; Biosystem to Reactome RDF88; substance to ChEMBL RDF88; and compound to WikiData RDF.90
4. Dealing with data imbalance issues in PubChem data
4.1. Imbalance in high-throughput screening data
Bioactivity data from HTS typically contain only a handful to a few hundred hits (active compounds) with many folds of inactive compounds. This imbalanced nature of HTS data presents a great challenge for developing an accurate prediction model from them.91-94 This issue may be addressed by generating a balanced data set through resampling of the original HTS data set. Several studies33,91-94 have applied different resampling techniques for analysis of HTS data in PubChem. They are broadly categorized into two classes: undersampling of the majority class (inactive compounds) and oversampling of the minority class (active compounds).
Li et al.91 applied the granular support vector machines (SVMs) with repetitive undersampling (GSVM-RUS)95 to develop an SVM from a highly imbalanced HTS data (with an active-to-inactive compound ratio of 1/377 and 1/379 for the training and blind test sets, respectively). The underlying idea of this method is that, because only support vectors (SVs) are important for SVM model classification, removal of non-SV samples does not substantially affect the model performance. In essence, this method enables one to extract important compounds from the data set and to eliminate unimportant ones. The best SVM model constructed in this study showed a sensitivity of 86.60% and a specificity of 88.89% for the blind test set. In some studies,33 inactive compounds were selected into the modeling set only if it had a relatively high similarity to active compounds, leading to a data set that is more challenging to establish robust prediction models.
In a study of Chang et al.,92 the simple oversampling technique was used to develop SVM models that classify compounds according to predicted cytotoxicity against the Jurkat cell line. It was demonstrated that oversampling of the minority class (toxic compounds) leads to SVM models with better predictive ability for both the training and external test sets, compared to results reported in previous studies. More recently, Hao et al.93 applied the synthetic minority oversampling technique (SMOTE)96 to tackle the HTS data set imbalance issue. Unlike the traditional oversampling method, SMOTE oversamples the minority class by creating “synthetic” samples along the line segments connecting the original minority-class samples with their k-nearest neighbors (kNN). Predictive models developed from the oversampled data set through the SMOTE algorithm was found to have better accuracies than those from simple oversampling.
Based on analysis of several common strategies for imbalanced data modeling with PubChem’s HTS data, Nicklaus and coworkers94 proposed a hybrid method that combines undersampling approach with cost-sensitive learning,97 which takes the misclassification costs into account by imposing penalties for misclassifications. The proposed method were shown to provide more accurate prediction results than other methods considered in their study.94
4.2. Imbalance in literature-extracted data
As mentioned previously, PubChem contains not only HTS data but also high-quality bioactivity data extracted from literature through data mining and/or manual curation.55 Most of scientific articles typically contain data for active compounds, but do not report much information on inactive compounds. As a result, use of literature-derived bioactivity data for virtual screening presents a data imbalance issue, which may be considered to be opposite to the imbalance in HTS data. Whereas HTS data are predominated by inactive compounds, literature-derived data have little to no inactive compounds.
In theory, resampling techniques may be used to balance literature-derived data set. However, this is not a viable option if the data set does not have any inactive compounds. In such cases, “putative” inactive compounds (also called putative negatives) may be generated to balance the data set, as proposed in a study by Han et al.98 This approach involves grouping all compounds in PubChem into clusters according to their molecular descriptors, followed by randomly selecting compounds from clusters that do not contain any known active molecules against the target. Because this method does not require known inactive compounds, it enables more expanded coverage of the inactive chemical space in case of little or no knowledge of inactive compounds. Of course, undiscovered active compounds may be included in the inactive space, leading to a reduced ability of computational models to identify novel active compounds. However, such an adverse effect is expected to be relatively small, as demonstrated in the study by Han et al.98 Many studies15,98-101 have shown that computational models derived from putative negatives can perform reasonably well in VS.
5. Computational toxicity prediction models from PubChem bioactivity data
Because experimental determination of toxicity of a large number of compounds is expensive and time-consuming, use of computational models is considered as an alternative or complement approach that can reduce the cost of experimental toxicity assessment in the early stage of drug discovery. This section summarizes computational toxicity prediction models that use PubChem’s bioactivity data.
5.1. hERG-related cardiotoxicity prediction
The human Ether-a-go-go Related Gene (hERG) protein102 is a tetrameric potassium ion channel that plays an important role in cardiac action potential. Its blockage by drug molecules is believed to be a major cause of drug-induced acquired long QT syndrome and cardiac arrhythmia called Torsades de Pointes, which are considered as electrocardiac symptoms of cardiotoxicity. Because undesirable hERG-related cardiotoxicity is a major problem in clinical studies of drug candidates and often results in withdrawal of approved drugs from the market, it is important to identify potential hERG blockers early in the drug discovery process.
Many computational predictive models for hERG blockers have been proposed, as summarized in a recent review by Villoutreix and Taboureau.102 Several studies24-27 used PubChem BioAssay data as a test or training set for developing classification models that distinguish hERG blockers from hERG nonblockers. An early example is a study by Li et al.,24 in which a SVM-based hERG classification model was developed using a training set of 495 compounds obtained from literature. The model was tested on a set of 1948 compounds whose hERG activities were available in the PubChem BioAssay database (i.e., 248 actives and 1700 inactives in AID 376), resulting in a 73% accuracy (sensitivity = 57% and specificity = 75%).
In a study of Su et al.,25 the compound set from AID 376 was reduced into a set of 876 compounds that are smaller, more condensed, and more applicable for lead optimization against the hERG receptor, by removing compounds that violate Lipinski’s rule of five50 and discarding actives with logP values of <4.1 and inactives with logP values of >2.8. These hydrophobicity constraints employed were based on the observation that the hydrophobicity of drugs tends to increase the hERG blocking effect, while hydrophilic molecules tend to decrease the hERG blocking effect. When this test set was used to evaluate binary hERG classification models derived from a continuous partial least-squares (PLS) hERG binding model, the best model showed an improved accuracy of 83% (sensitivity = 97% and specificity = 82%).
Wang et al.26 developed binary hERG classification models by employing Naïve Bayesian (NB) classification and recursive partitioning (RP) techniques in conjunction with several sets of molecular descriptors. It was found that the NB classifier outperformed the RP-based model. When applied on a test set derived from AID 376, the best Bayesian classifier at a threshold of 40 μM resulted in an accuracy of 76% (with 37% sensitivity and 82% specificity).
Whereas the PubChem hERG assay data from AID 376 were used as an external test set in all three studies24-26 mentioned above, these data have also been used as a training set for model building. For example, Shen et al.27 derived a training set of 1668 compounds from AID 376 to build SVM-based binary hERG classification models, the best of which had accuracies of 95% (with 90% sensitivity and 96% specificity) for the training set and 87% (90% sensitivity and 74% specificity) for the external set of 356 compounds.
5.2. Prediction of cytochrome P450s inhibition
The cytochrome P450s (CYPs) are a superfamily of heme-containing enzymes that catalyze the metabolism of a variety of endogenous and xenobiotic compounds. They are major enzymes involved in drug metabolism, which affects the bioavailabity of drug molecules. In addition, the broad substrate specificity of CYPs often leads to unexpected drug-drug interactions, which is an important issue in drug discovery and development as well as in their clinical applications.
The PubChem Compound database provides manually curated information on the metabolism for more than five thousand compounds, collected from data contributors such as HSDB60 and DrugBank.48 Moreover, more than nine thousand compounds have links to the corresponding records in the Human Metabolome Database (HMDB),103 which offers comprehensive information on metabolites.
The PubChem BioAssay database also contains a large amount of experimental bioactivity data for compounds tested against CYPs. While some of them were extracted from scientific articles, others were determined through HTS. Using these bioactivity data, several groups28-32 have developed computational prediction models for CYP inhibition of small molecules. For example, Cheng et al.28 constructed inhibitor prediction models for five major CYP isoforms, namely 1A2, 2C9, 2C19, 2D6, and 3A4, which account for more than 90% of drug metabolism. Their model used an algorithm that combines a back-propagation artificial neural network (BP-ANN) with other machine learning methods including kNN, SVM, NB, and C4.5 decision tree. Using a rule-based C5.0 decision tree algorithm with several molecular descriptors, Su et al.29 developed an improved prediction model, which can classify CYP inhibitors and non-inhibitors with an 81.4-93.0% accuracy.
5.3. Toxicity prediction models from cellular toxicity
When toxicity of a chemical arises from its interaction with a particular target protein, gene, or pathway, one can build a computational model that predicts whether a compound is toxic by virtue of its interaction with the target. However, chemical toxicity often comes from much more complex processes that involve many different proteins and genes in multiple pathways. In this case, a computational toxicity prediction model can be developed from cellular toxicity data generated in cell proliferation assays that do not address any specific target or underlying mechanism.
PubChem’s toxicity data have been used to develop computational toxicity prediction models. In a study of Zhu et al.,33 HTS data for cell viability of 1,408 compounds tested against six cell lines were used to construct a kNN quantitative structure-activity relationship (QSAR) model that predicts rodent carcinogenicity of chemicals. This study demonstrated that, when cell viability data were used together with chemical descriptors, the resulting kNN QSAR model had a better accuracy than those developed using chemical descriptors only.
Guha and Schürer34 built computational models to predict cell toxicity based on cell proliferation HTS data contained in PubChem. To reduce the impact of the imbalanced nature of the data set employed, their prediction models were developed using an ensemble of 30 random forest (RF) models, each of which was constructed from a training set with equal distributions of toxic and non-toxic compounds sampled from the original set. These models resulted in correct classification rates between 70% and 85% against the test sets, depending on the nature of the data sets and the descriptors employed. However, when applied to predict in vivo animal toxicity, they showed a significantly reduced accuracy although there were cases where cell toxicity strongly relates to in vivo animal toxicity.
Zhang et al.35 proposed a method to predict acute animal toxicity of compounds (represented by the LD50 values of rats), using bioactivity profiles of compounds extracted from bioassay data in the PubChem BioAssay database. Sedykh et al.104 demonstrated that the use of dose-response data from qHTS assays as biological descriptors can improve the accuracy of QSAR models for in vivo toxicity prediction when combined with chemical descriptors. More recently, a prediction model for oxidative stress-induced hepatotoxicity of chemicals105 was generated from HTS data archived in PubChem. The use of HTS data for chemical toxicity prediction is well reviewed in a recent article by Zhu et al.106
6. Application of PubChem data for polypharmacology
The term “polypharmacology”107 is used to describe a new drug development paradigm, which aims to develop a drug or a combination of drugs that simultaneously act on multiple drug targets. This multi-target approach is considered as an alternative to the traditional single-target paradigm, particularly in the treatment of complex diseases like cancer and central nervous system disorders. Polypharmacology is also very closely related to drug-repurposing, which identifies new indication for existing drugs, as well as predicting off-target adverse drug reactions (side effects), which are caused by interaction of drug molecules with unintended proteins.
PubChem data have been used in several studies that developed computational methods for identifying multi-target ligands.101,107-110 Some of these studies108 employed a combinatorial approach in which predictive models were separately constructed for each target and subsequently used for parallel screening against each target to find compounds that simultaneously bind to multiple targets. This approach may also be used for identifying selective ligands for structurally related protein targets.
Alternatively, several studies used network-based approaches for finding multi-target compounds. Chen and coworkers107 performed cross-assay analyses to investigate the polypharmacological nature of bioactivity data contained in PubChem. With 602 bioassays that had information on target proteins at that time, they constructed a network of assays, by representing each assay with a node and connecting nodes with an edge if the assays corresponding to the nodes have one or more common active compounds. Through bipartite mapping, this assay network was merged with other networks, such as drug-target network, protein-protein interaction network, and pathway. The resulting bipartite networks helped identifying compounds that are active against multiple targets, as well as interesting protein pairs that can be targeted simultaneously under the polypharmacological drug development paradigm.
Because the bipartite mapping approach used in the study by Chen et al.107 requires knowledge of the assay targets, it was not applicable to assays that have no target information (such as phenotypic assays). An alternative network-based approach has also been developed which allows for analysis of both target-based and phenotypic assays. In a study by Swamidass et al.,109 a network of 1,581 assays with at least 5,000 tested compounds was constructed based on similarity between two assays in terms of correlation between bioactivity scores of the compounds tested in both assays. The bioactivity score correlation was quantified with the promiscuity-adjusted correlation (PAC), which downweighs promiscuous compounds that were tested active in many assays. The underlying assumption in this study is that if many molecules have similar bioactivities in two assays, there is likely a strong relationship between the assays (e.g., having similar protein targets, or closely related biological pathways). This approach allows one to deduce the target and underlying biology of a phenotypic assay from information available for target-based assays connected to that assay.
7. Benchmark data sets for virtual screening derived from PubChem data
Because many VS methods have been developed, it is not easy to decide which method will be best for a particular drug discovery project. Therefore, the objective evaluation of these VS methods is an important issue. This evaluation involves a retrospective validation of VS methods using benchmark data sets that consist of known active compounds against a target protein as well as inactive compounds or untested decoys. Examples of such data sets are the Directory of Useful Decoys (DUD) set,111 and its enhanced version (DUD-E),112 Virtual Decoy Sets (VDS),113 the Demanding Evaluation Kits for Objective in Silico Screening (DEKOIS),114,115 the G-protein-coupled receptor (GPCR) ligand library (GLL)116 and GPCR decoy database (GDD)116, the Unbiased Ligand Set (ULS)117 and Unbiased Decoy Set (UDS)117.
In several studies,118-121 HTS data in the PubChem BioAssay database were used to construct benchmark data sets for VS validation. For example, Rohrer and Baumann119 developed the Maximum Unbiased Validation (MUV) benchmarking data sets from the HTS data for 17 protein targets. The MUV sets were designed to minimize the “benchmark data set bias,”119 which is caused by two critical issues in many benchmark data sets: artificial enrichment122 and analogue bias123. As a result, the MUV sets enable a more accurate and impartial evaluation of virtual screening methods.
Although some of the 17 targets covered in the MUV sets have experimentally determined 3-D structures in PDB,59 the design focus of the MUV sets were primarily on validation of ligand-based VS methods, not structure-based ones. Lindh et al.121 developed validation data sets suitable for validation of both structure-based and ligand-based VS methods, based on PubChem’s HTS data for seven protein targets whose crystal structure has been reported in PDB. Importantly, these data sets were designed to have a higher ratio of the number of inactive to active compounds than other benchmark data sets in order to reflect typical drug discovery scenarios in which hit compounds from VS are subsequently tested in an HTS experiment. Therefore, these data sets would give more realistic measures of the performance of different VS methods.
8. Conclusion
PubChem provides comprehensive chemical information collected from more than four hundred data sources. It contains experimental bioactivity data as well as other valuable information relevant to drug discovery, including pharmacology, toxicology, mechanisms of action, ADMET properties, 3-D structures, and so on. Especially, information on chemical vendors and patents helps prioritize hit compounds from VS for further screening. In addition, the pre-computed PubChem 2-D and 3-D neighboring relationships enable quick access to structurally similar compounds for a given compound.
Because information contained in PubChem can be programmatically accessed (through several methods including E-Utilities, PUG, PUG-SOAP, and PUG-REST), it is possible to build an automated VS pipeline that exploits information contained in PubChem. In addition, through PubChemRDF, users can integrate PubChem’s data into their own in-house data on a local computing machine.
PubChem data have been used in many drug discovery studies. For example, PubChem’s bioactivity data were used to build computational models for bioactivity or toxicity prediction or to discover polypharmacologic multi-target ligands. In some studies, they were used to develop benchmark data set, which allows for objective evaluation of different VS methods.
When using PubChem’s bioactivity data to construct a prediction model, one should keep in mind that they are highly imbalanced. HTS data are predominated by inactive compounds with only a few active compounds and literature-derived data often contains only active compounds without any inactive compounds. This data imbalance issue should be addressed to develop an accurate prediction model.
9. Expert opinion
PubChem is the largest source of publicly available chemical information, with more than 219 million substances, 89 million compounds, and 230 million bioactivity outcomes from more than one million assays covering around ten thousand unique protein target sequences. Therefore, the biomedical research community has great interest in exploiting PubChem’s data for drug discovery.
PubChem contains a large amount of chemical information that is useful for VS. In addition to HTS data generated by NIH’s Molecular Libraries Program and other HTS projects, PubChem contains a substantial amount of literature-extracted bioactivity information contributed by ChEMBL,49 Guide to Pharmacology,58 BindingDB,57 PDBbind,56 and so on. Moreover, through data integration with other databases such as DrugBank,48 HSDB,60 and HMDB,103 PubChem provides a broad range of annotated information on small molecules, including pharmacology, toxicology, drug target, metabolism, safety and handling and many others. PubChem also hosts data from important regulatory agencies, such as the FDA and EPA.
PubChem provides information on chemical vendors and patents for compounds. Currently it offers links between about 6 million patent documents and more than 16 million unique chemical structures, with over 329 million chemical substance-patent links covering U.S., European and WIPO patent documents published since 1800. Chemical vendor and patent information for compounds in PubChem would be useful for prioritizing hit compounds for further screening.
A large variation in the amount of available information exists among compounds contained in PubChem. For example, as shown in Figure 3, about 98% of PubChem compounds have never been tested in any assays archived in the BioAssay database. Inevitably, biological activities of these molecules need to be inferred from their structurally similar molecules that have biological activity data. PubChem helps users quickly identify similar chemical structures, by providing a pre-computed list of 2-D and 3-D neighbors74,75 for each compound.
To assist users in automating VS pipelines, PubChem provides multiple programmatic access routes, including E-Utilities, PUG, PUG-SOAP, and PUG-REST. In addition, PubChemRDF79 allows users to download PubChem data on a local computing facility and integrate them with their in-house data, facilitating data sharing and integration with other information resources. Importantly, PubChemRDF enhances cross-integration by providing direct links to available authoritative RDF resources within applicable subdomains.
When bioactivity data in PubChem are used to develop a prediction model, the data imbalance issue needs to be taken care of. Typically, HTS data in PubChem are strongly imbalanced, containing a small number of active compounds with a very large number of inactive compounds. When such imbalanced data sets are used to build computational models that predict bioactivity of molecules, they need to be balanced by undersampling inactive compounds or oversampling active compounds. Several studies proposed various sampling techniques to address the issue of data set imbalance. On the other hand, because scientific articles primarily report data for active compounds, literature-extracted bioactivity data in PubChem often lack information on inactive compounds, creating another type of data set imbalance. To use these literature-extracted data for model building, putative inactive compounds may be generated to balance the data set.
PubChem contains a large amount of toxicity data generated from HTS assays, as well as those extracted from literature through manual curation or data mining. These toxicity data have been used in many studies to construct computational models that predict toxicity of molecules. Some of these studies aimed to predict target-specific toxicities, such as cardiotoxicity due to hERG inhibition, and drug-induced liver damage due to CYP inhibition. Other studies developed prediction models for cellular toxicity, carcinogenicity, in vivo animal toxicity, which arise from much more complex mechanism involving multiple genes, targets, and pathways. These prediction models can be used for structure alerts for potentially toxic molecules during VS.
A large amount of target information for compounds in PubChem can be used to find multi-target ligands for polypharmacologic drug development. In addition, it can be used to find new targets for a compound, which allows one to predict off-target side effects of drug molecules that cause adverse drug reaction as well as to repurpose existing drug molecules for a new indication. These areas can be harnessed by using PubChemRDF, which presents a promising opportunity to exploit public chemical information not only in PubChem and but also in other chemical and biological databases.
Article Highlights.
PubChem is the largest source of publicly available chemical information, collected from more than 400 data sources.
In addition to bioactivity data generated through high-throughput screenings, PubChem contains a substantial amount of bioactivity information extracted from scientific articles.
Chemical vendor and patent information for compounds in PubChem helps prioritize hit compounds for further screening.
PubChem supports programmatic access to its data, allowing for building an automated virtual screening pipeline.
PubChemRDF allows users to download PubChem data on a local computing facility and integrate them with their own data.
PubChem data can be used for developing computational prediction models for bioactivity or toxicity of molecules.
This box summarizes key points contained in the article.
Acknowledgements
The author thanks the entire PubChem team and the NCBI staff as well as the hundreds of data contributors for making their data openly accessible within PubChem. He also thanks Evan Bolton at PubChem and Bradley Otterson at the NIH Library Editing Service for critical reading of this manuscript.
Funding:
This work was supported by the Intramural Research Program of the National Library of Medicine, NIH.
Footnotes
Declaration of Interest:
S Kim is an employee of the National Library of Medicine. He has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
- 1.Inglese J, Johnson RL, Simeonov A, et al. High-throughput screening assays for the identification of chemical probes. Nat Chem Biol. 2007;3(8):466–79. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 2.Diller DJ. The synergy between combinatorial chemistry and high-throughput screening. Curr Opin Drug Discov Dev. 2008;11(3):346–55. [PubMed] [Google Scholar]
- 3.Moos WH, Hurt CR, Morales GA. Combinatorial chemistry: oh what a decade or two can do. Mol Divers. 2009;13(2):241–45. doi: 10.1007/s11030-009-9127-y. [DOI] [PubMed] [Google Scholar]
- 4•.Scior T, Bender A, Tresadern G, et al. Recognizing Pitfalls in Virtual Screening: A Critical Review. J Chem Inf Model. 2012;52(4):867–81. doi: 10.1021/ci200528d. A comprehensive review on important issues for successful virtual screening. [DOI] [PubMed] [Google Scholar]
- 5.McInnes C. Virtual screening strategies in drug discovery. Curr Opin Chem Biol. 2007;11(5):494–502. doi: 10.1016/j.cbpa.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 6••.Kim S, Thiessen PA, Bolton EE, et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016;44(D1):D1202–D13. doi: 10.1093/nar/gkv951. The most recent overview of the PubChem Substand and Compound databases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7••.Wang YL, Suzek T, Zhang J, et al. PubChem BioAssay: 2014 update. Nucleic Acids Res. 2014;42(D1):D1075–D82. doi: 10.1093/nar/gkt978. The most recent overview of the PubChem BioAssay database. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.PubChem. National Center for Biotechnology Information; Bethesda, MD: 2004. Available at: https://pubchem.ncbi.nlm.nih.gov [Last accessed 19 July 2016] [Google Scholar]
- 9.Mahasenan KV, Li CL. Novel Inhibitor Discovery through Virtual Screening against Multiple Protein Conformations Generated via Ligand-Directed Modeling: A Maternal Embryonic Leucine Zipper Kinase Example. J Chem Inf Model. 2012;52(5):1345–55. doi: 10.1021/ci300040c. [DOI] [PubMed] [Google Scholar]
- 10.Cheng CS, Jia KF, Chen T, et al. Experimentally Validated Novel Inhibitors of Helicobacter pylori Phosphopantetheine Adenylyltransferase Discovered by Virtual High-Throughput Screening. PLoS One. 2013;8(9):11. doi: 10.1371/journal.pone.0074271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunna NR, Bandaru S, Akare UR, et al. Multiclass Comparative Virtual Screening to Identify Novel Hsp90 Inhibitors: A Therapeutic Breast Cancer Drug Target. Curr Top Med Chem. 2015;15(1):57–64. doi: 10.2174/1568026615666150112113627. [DOI] [PubMed] [Google Scholar]
- 12.Fang JS, Huang D, Zhao WX, et al. A New Protocol for Predicting Novel GSK-3 beta ATP Competitive Inhibitors. J Chem Inf Model. 2011;51(6):1431–38. doi: 10.1021/ci2001154. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh JH, Wang XS, Teotico D, et al. Differentiation of AmpC beta-lactamase binders vs. decoys using classification kNN QSAR modeling and application of the QSAR classifier to virtual screening. J Comput Aided Mol Des. 2008;22(9):593–609. doi: 10.1007/s10822-008-9199-2. [DOI] [PubMed] [Google Scholar]
- 14.Sapre NS, Gupta S, Pancholi N, et al. A Group Center Overlap Based Approach for "3D QSAR" Studies on TIBO Derivatives. J Comput Chem. 2009;30(6):922–33. doi: 10.1002/jcc.21114. [DOI] [PubMed] [Google Scholar]
- 15.Han BC, Ma XH, Zhao RY, et al. Development and experimental test of support vector machines virtual screening method for searching Src inhibitors from large compound libraries. Chem Cent J. 2012;6:14. doi: 10.1186/1752-153X-6-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kothapalli R, Khan AM, Basappa, et al. Cheminformatics-Based Drug Design Approach for Identification of Inhibitors Targeting the Characteristic Residues of MMP-13 Hemopexin Domain. PLoS One. 2010;5(8):7. doi: 10.1371/journal.pone.0012494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nolan TL, Geffert LM, Kolber BJ, et al. Discovery of Novel-Scaffold Monoamine Transporter Ligands via in Silico Screening with the S1 Pocket of the Serotonin Transporter. ACS Chem Neurosci. 2014;5(9):784–92. doi: 10.1021/cn500133b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banavath HN, Sharma OP, Kumar MS, et al. Identification of novel tyrosine kinase inhibitors for drug resistant T315I mutant BCR-ABL: a virtual screening and molecular dynamics simulations study. Sci Rep. 2014;4:11. doi: 10.1038/srep06948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bak A, Magdziarz T, Polanski J. Pharmacophore-based database mining for probing fragmental drug-likeness of diketo acid analogues. SAR QSAR Environ Res. 2012;23(1-2):185–204. doi: 10.1080/1062936X.2011.645875. [DOI] [PubMed] [Google Scholar]
- 20.Jalali-Heravi M, Mani-Varnosfaderani A, Valadkhani A. Integrated One-Against-One Classifiers as Tools for Virtual Screening of Compound Databases: A Case Study with CNS Inhibitors. Mol Inform. 2013;32(8):742–53. doi: 10.1002/minf.201200126. [DOI] [PubMed] [Google Scholar]
- 21.Khanna V, Ranganathan S. In silico approach to screen compounds active against parasitic nematodes of major socio-economic importance. BMC Bioinformatics. 2011;12:12. doi: 10.1186/1471-2105-12-S13-S25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pacureanu L, Crisan L, Bora A, et al. In silico classification and virtual screening of maleimide derivatives using projection to latent structures discriminant analysis (PLS-DA) and hybrid docking. Monatsh Chem. 2012;143(11):1559–73. [Google Scholar]
- 23.Wicht KJ, Combrinck JM, Smith PJ, et al. Bayesian models trained with HTS data for predicting beta-haematin inhibition and in vitro antimalarial activity. Bioorg Med Chem. 2015;23(16):5210–17. doi: 10.1016/j.bmc.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li QY, Jorgensen FS, Oprea T, et al. hERG classification model based on a combination of support vector machine method and GRIND descriptors. Mol Pharm. 2008;5(1):117–27. doi: 10.1021/mp700124e. [DOI] [PubMed] [Google Scholar]
- 25.Su BH, Shen MY, Esposito EX, et al. In Silico Binary Classification QSAR Models Based on 4D-Fingerprints and MOE Descriptors for Prediction of hERG Blockage. J Chem Inf Model. 2010;50(7):1304–18. doi: 10.1021/ci100081j. [DOI] [PubMed] [Google Scholar]
- 26.Wang SC, Li YY, Wang JM, et al. ADMET Evaluation in Drug Discovery. 12. Development of Binary Classification Models for Prediction of hERG Potassium Channel Blockage. Mol Pharm. 2012;9(4):996–1010. doi: 10.1021/mp300023x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen MY, Su BH, Esposito EX, et al. A Comprehensive Support Vector Machine Binary hERG Classification Model Based on Extensive but Biased End Point hERG Data Sets. Chem Res Toxicol. 2011;24(6):934–49. doi: 10.1021/tx200099j. [DOI] [PubMed] [Google Scholar]
- 28.Cheng FX, Yu Y, Shen J, et al. Classification of Cytochrome P450 Inhibitors and Noninhibitors Using Combined Classifiers. J Chem Inf Model. 2011;51(5):996–1011. doi: 10.1021/ci200028n. [DOI] [PubMed] [Google Scholar]
- 29.Su BH, Tu YS, Lin C, et al. Rule-Based Prediction Models of Cytochrome P450 Inhibition. J Chem Inf Model. 2015;55(7):1426–34. doi: 10.1021/acs.jcim.5b00130. [DOI] [PubMed] [Google Scholar]
- 30.Didziapetris R, Dapkunas J, Sazonovas A, et al. Trainable structure-activity relationship model for virtual screening of CYP3A4 inhibition. J Comput Aided Mol Des. 2010;24(11):891–906. doi: 10.1007/s10822-010-9381-1. [DOI] [PubMed] [Google Scholar]
- 31.Novotarskyi S, Sushko I, Korner R, et al. A comparison of different QSAR approaches to modeling CYP450 1A2 inhibition. J Chem Inf Model. 2011;51(6):1271–80. doi: 10.1021/ci200091h. [DOI] [PubMed] [Google Scholar]
- 32.Buchwald P. Activity-Limiting Role of Molecular Size: Size-Dependency of Maximum Activity for P450 Inhibition as Revealed by qHTS Data. Drug Metab Dispos. 2014;42(11):1785–90. doi: 10.1124/dmd.114.059717. [DOI] [PubMed] [Google Scholar]
- 33.Zhu H, Rusyn I, Richard A, et al. Use of cell viability assay data improves the prediction accuracy of conventional quantitative structure-activity relationship models of animal carcinogenicity. Environ Health Perspect. 2008;116(4):506–13. doi: 10.1289/ehp.10573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guha R, Schurer SC. Utilizing high throughput screening data for predictive toxicology models: protocols and application to MLSCN assays. J Comput Aided Mol Des. 2008;22(6-7):367–84. doi: 10.1007/s10822-008-9192-9. [DOI] [PubMed] [Google Scholar]
- 35.Zhang J, Hsieh JH, Zhu H. Profiling Animal Toxicants by Automatically Mining Public Bioassay Data: A Big Data Approach for Computational Toxicology. PLoS One. 2014;9(6):11. doi: 10.1371/journal.pone.0099863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36•.Pouliot Y, Chiang AP, Butte AJ. Predicting Adverse Drug Reactions Using Publicly Available PubChem BioAssay Data. Clinical Pharmacology & Therapeutics. 2011;90(1):90–99. doi: 10.1038/clpt.2011.81. PubChem's bioactivity data were used to build a predictive model for adverse drug reactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Cami A, Arnold A, Manzi S, et al. Predicting Adverse Drug Events Using Pharmacological Network Models. Science Translational Medicine. 2011;3(114):10. doi: 10.1126/scitranslmed.3002774. A predictive model for adverse drug events was developed using known drug-ADE relationships. [DOI] [PubMed] [Google Scholar]
- 38•.Wassermann AM, Lounkine E, Hoepfner D, et al. Dark chemical matter as a promising starting point for drug lead discovery. Nat Chem Biol. 2015;11(12):958–66. doi: 10.1038/nchembio.1936. A research article in which a new lead molecule was identified from dark chemical matter (DCM). [DOI] [PubMed] [Google Scholar]
- 39.PubChem Data Sources. National Center for Biotechnology Information; Bethesda, MD: 2016. Available at: https://pubchem.ncbi.nlm.nih.gov/sources/ [Last accessed 19 July 2016] [Google Scholar]
- 40.PubChem Substance. National Center for Biotechnology Information; Bethesda, MD: 2004. Available at: https://www.ncbi.nlm.nih.gov/pcsubstance/ [Last accessed 19 July 2016] [Google Scholar]
- 41.PubChem Compound. National Center for Biotechnology Information; Bethesda, MD: 2004. Available at: https://www.ncbi.nlm.nih.gov/pccompound/ [Last accessed 19 July 2016] [Google Scholar]
- 42.PubChem BioAssay. National Center for Biotechnology Information; Bethesda, MD: 2004. Available at: https://www.ncbi.nlm.nih.gov/pcassay/ [Last accessed 19 July 2016] [Google Scholar]
- 43.Fink T, Reymond JL. Virtual exploration of the chemical universe up to 11 atoms of C, N, O, F: Assembly of 26.4 million structures (110.9 million stereoisomers) and analysis for new ring systems, stereochemistry, physicochemical properties, compound classes, and drug discovery. J Chem Inf Model. 2007;47(2):342–53. doi: 10.1021/ci600423u. [DOI] [PubMed] [Google Scholar]
- 44.Blum LC, van Deursen R, Reymond JL. Visualisation and subsets of the chemical universe database GDB-13 for virtual screening. J Comput Aided Mol Des. 2011;25(7):637–47. doi: 10.1007/s10822-011-9436-y. [DOI] [PubMed] [Google Scholar]
- 45.Ruddigkeit L, van Deursen R, Blum LC, et al. Enumeration of 166 Billion Organic Small Molecules in the Chemical Universe Database GDB-17. J Chem Inf Model. 2012;52(11):2864–75. doi: 10.1021/ci300415d. [DOI] [PubMed] [Google Scholar]
- 46.Reymond JL, Awale M. Exploring Chemical Space for Drug Discovery Using the Chemical Universe Database. ACS Chem Neurosci. 2012;3(9):649–57. doi: 10.1021/cn3000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oprea TI, Allu TK, Fara DC, et al. Lead-like, drug-like or "Pub-like": How different are they? J Comput Aided Mol Des. 2007;21(1-3):113–19. doi: 10.1007/s10822-007-9105-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Law V, Knox C, Djoumbou Y, et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014;42(D1):D1091–D97. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bento AP, Gaulton A, Hersey A, et al. The ChEMBL bioactivity database: an update. Nucleic Acids Res. 2014;42(D1):D1083–D90. doi: 10.1093/nar/gkt1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50>•.Lipinski CA, Lombardo F, Dominy BW, et al. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2012;64:4–17. doi: 10.1016/s0169-409x(00)00129-0. A review article that describes Lipinski's rule of 5. [DOI] [PubMed] [Google Scholar]
- 51•.Congreve M, Carr R, Murray C, et al. A rule of three for fragment-based lead discovery? Drug Discovery Today. 2003;8(19):876–77. doi: 10.1016/s1359-6446(03)02831-9. A paper that describes congreve's rule of 3. [DOI] [PubMed] [Google Scholar]
- 52.Inglese J, Auld DS, Jadhav A, et al. Quantitative high-throughput screening: A titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci U S A. 2006;103(31):11473–78. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rao HB, Li ZR, Li XY, et al. Identification of Small Molecule Aggregators From Large Compound Libraries by Support Vector Machines. J Comput Chem. 2010;31(4):752–63. doi: 10.1002/jcc.21347. [DOI] [PubMed] [Google Scholar]
- 54.Su BH, Tu YS, Lin OA, et al. Rule-Based Classification Models of Molecular Autofluorescence. J Chem Inf Model. 2015;55(2):434–45. doi: 10.1021/ci5007432. [DOI] [PubMed] [Google Scholar]
- 55.Kim S, Thiessen PA, Cheng T, et al. Literature information in PubChem: associations between PubChem records and scientific articles. J Cheminform. 2016;8:32. doi: 10.1186/s13321-016-0142-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu ZH, Li Y, Han L, et al. PDB-wide collection of binding data: current status of the PDBbind database. Bioinformatics. 2015;31(3):405–12. doi: 10.1093/bioinformatics/btu626. [DOI] [PubMed] [Google Scholar]
- 57.Gilson MK, Liu T, Baitaluk M, et al. BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016;44(D1):D1045–D53. doi: 10.1093/nar/gkv1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Southan C, Sharman JL, Benson HE, et al. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. Nucleic Acids Res. 2016;44(D1):D1054–D68. doi: 10.1093/nar/gkv1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berman HM, Westbrook J, Feng Z, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fonger GC, Hakkinen P, Jordan S, et al. The National Library of Medicine's (NLM) Hazardous Substances Data Bank (HSDB): Background, recent enhancements and future plans. Toxicology. 2014;325:209–16. doi: 10.1016/j.tox.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Madej T, Lanczycki CJ, Zhang DC, et al. MMDB and VAST+: tracking structural similarities between macromolecular complexes. Nucleic Acids Res. 2014;42(D1):D297–D303. doi: 10.1093/nar/gkt1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Groom CR, Bruno IJ, Lightfoot MP, et al. The Cambridge Structural Database. Acta Crystallogr B Struct Sci Cryst Eng Mater. 2016;72:171–79. doi: 10.1107/S2052520616003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Orange Book: Approved Drug Products with Therapeutic Equivalence Evaluations. U.S. Food and Drug Administration; Silver Spring, MD: 2013. Available at: http://www.accessdata.fda.gov/scripts/cder/ob/ [Last accessed 14 July 2016] [Google Scholar]
- 64.Substance Registration System - Unique Ingredient Identifier (UNII) U.S. Food and Drug Administration; Silver Spring, MD: 2016. Available at: http://www.fda.gov/ForIndustry/DataStandards/SubstanceRegistrationSystem-UniqueIngredientIdentifierUNII/default.htm [Last accessed 14 July 2016] [Google Scholar]
- 65.Pharmacologic Class. U.S. Food and Drug Administration; Silver Spring, MD: 2015. Available at: http://www.fda.gov/ForIndustry/DataStandards/StructuredProductLabeling/ucm162549.htm [Last accessed 14 July 2016] [Google Scholar]
- 66.DailyMed. U.S. National Library of Medicine; Bethesda, MD: 2016. Available at: http://dailymed.nlm.nih.gov [Last accessed 14 July 2016] [Google Scholar]
- 67.IBM Research-Almaden . IBM; San Jose, CA: 2016. Available at: http://www.almaden.ibm.com [Last accessed 14 July 2016] [Google Scholar]
- 68.Papadatos G, Davies M, Dedman N, et al. SureChEMBL: a large-scale, chemically annotated patent document database. Nucleic Acids Res. 2016;44(D1):D1220–D28. doi: 10.1093/nar/gkv1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.SureChEMBL. EMBL European Bioinformatics Institute; Cambridgeshire, UK: 2014. Available at: https://www.surechembl.org [Last accessed 19 July 2016] [Google Scholar]
- 70.NextMove Software . NextMove Software; Cambridge, UK: 2016. Available at: https://www.nextmovesoftware.com/ [Last accessed 14 July 2016] [Google Scholar]
- 71.Heifets A, Jurisica I. SCRIPDB: a portal for easy access to syntheses, chemicals and reactions in patents. Nucleic Acids Res. 2012;40(D1):D428–D33. doi: 10.1093/nar/gkr919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.International Patent Classification (IPC) World Intellectual Property Organization; Geneva, Switzerland: 2016. Available at: http://www.wipo.int/classifications/ipc/en/ [Last accessed 14 July 2016] [Google Scholar]
- 73••.Kim S, Thiessen PA, Bolton EE, et al. PUG-SOAP and PUG-REST: web services for programmatic access to chemical information in PubChem. Nucleic Acids Res. 2015;43(W1):W605–11. doi: 10.1093/nar/gkv396. An overview of programmatic access to PubChem data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74••.Bolton EE, Chen J, Kim S, et al. PubChem3D: a new resource for scientists. J Cheminform. 2011;3:32. doi: 10.1186/1758-2946-3-32. A comprehensive overview of the PubChem3D project. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75••.Bolton EE, Kim S, Bryant SH. PubChem3D: similar conformers. J Cheminform. 2011;3:13. doi: 10.1186/1758-2946-3-13. A comprehensive overview of PubChem 2-D and 3-D neighboring computations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Holliday JD, Hu CY, Willett P. Grouping of coefficients for the calculation of inter-molecular similarity and dissimilarity using 2D fragment bit-strings. Comb Chem High Throughput Screen. 2002;5(2):155–66. doi: 10.2174/1386207024607338. [DOI] [PubMed] [Google Scholar]
- 77.PubChem substructure fingerprint description. National Center for Biotechnology Information; Bethesda, MD: 2009. Available at: ftp://ftp.ncbi.nlm.nih.gov/pubchem/specifications/pubchem_fingerprints.pdf [Last accessed 14 July 2016] [Google Scholar]
- 78.Hawkins PCD, Skillman AG, Nicholls A. Comparison of shape-matching and docking as virtual screening tools. J Med Chem. 2007;50(1):74–82. doi: 10.1021/jm0603365. [DOI] [PubMed] [Google Scholar]
- 79••.Fu G, Batchelor C, Dumontier M, et al. PubChemRDF: towards the semantic annotation of PubChem compound and substance databases. J Cheminform. 2015;7:34. doi: 10.1186/s13321-015-0084-4. A comprehensive overview of the PubChem RDF project. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Entrez Programming Utilities Help. National Center for Biotechnology Information; Bethesda, MD: 2010. Available at: http://www.ncbi.nlm.nih.gov/books/NBK25501/ [Last accessed 19 July 2016] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.PubChem Power User Gateway (PUG) Help. National Center for Biotechnology Information; Bethesda, MD: 2007. Available at: https://pubchem.ncbi.nlm.nih.gov/pug/pughelp.html [Last accessed 19 July 2016] [Google Scholar]
- 82.PUG SOAP. National Center for Biotechnology Information; Bethesda, MD: 2008. Available at: https://pubchem.ncbi.nlm.nih.gov/pug_soap/pug_soap_help.html [Last accessed 19 July 2016] [Google Scholar]
- 83.PUG REST. National Center for Biotechnology Information; Bethesda, MD: 2012. Available at: https://pubchem.ncbi.nlm.nih.gov/pug_rest/PUG_REST.html [Last accessed 19 July 2016] [Google Scholar]
- 84.NCBI Website and Data Usage Policies and Disclaimers. National Center for Biotechnology Information; Bethesda, MD: 2016. Available at: https://www.ncbi.nlm.nih.gov/home/about/policies.shtml [Last accessed 14 July 2016] [Google Scholar]
- 85.Resource Description Framework (RDF) The World Wide Web Consortium (W3C) 2014 Available at: http://www.w3.org/RDF/ [Last accessed 14 July 2016]
- 86.Heller S, McNaught A, Pletnev I, et al. InChI, the IUPAC International Chemical Identifier. J Cheminform. 2015;7:23. doi: 10.1186/s13321-015-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bushman B, Anderson D, Fu G. Transforming the Medical Subject Headings into Linked Data: Creating the Authorized Version of MeSH in RDF. J Libr Metadata. 2015;15(3-4):157–76. doi: 10.1080/19386389.2015.1099967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jupp S, Malone J, Bolleman J, et al. The EBI RDF platform: linked open data for the life sciences. Bioinformatics. 2014;30(9):1338–39. doi: 10.1093/bioinformatics/btt765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kinjo AR, Suzuki H, Yamashita R, et al. Protein Data Bank Japan (PDBj): maintaining a structural data archive and resource description framework format. Nucleic Acids Res. 2012;40(D1):D453–D60. doi: 10.1093/nar/gkr811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Erxleben F, Günther M, Krötzsch M, et al. Introducing Wikidata to the Linked Data Web. In: Mika P, Tudorache T, Bernstein A, Welty C, Knoblock C, Vrandečić D, et al., editors. The Semantic Web – ISWC 2014. Springer International Publishing; 2014. pp. 50–65. [Google Scholar]
- 91.Li QL, Wang YL, Bryant SH. A novel method for mining highly imbalanced high-throughput screening data in PubChem. Bioinformatics. 2009;25(24):3310–16. doi: 10.1093/bioinformatics/btp589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chang CY, Hsu MT, Esposito EX, et al. Oversampling to Overcome Overfitting: Exploring the Relationship between Data Set Composition, Molecular Descriptors, and Predictive Modeling Methods. J Chem Inf Model. 2013;53(4):958–71. doi: 10.1021/ci4000536. [DOI] [PubMed] [Google Scholar]
- 93.Hao M, Wang YL, Bryant SH. An efficient algorithm coupled with synthetic minority over-sampling technique to classify imbalanced PubChem BioAssay data. Anal Chim Acta. 2014;806:117–27. doi: 10.1016/j.aca.2013.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94•.Zakharov AV, Peach ML, Sitzmann M, et al. QSAR Modeling of Imbalanced High-Throughput Screening Data in PubChem. J Chem Inf Model. 2014;54(3):705–12. doi: 10.1021/ci400737s. A research article that compares different sampling strategies to address the HTS data imbalance issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tang YC, Zhang YQ, Chawla NV, et al. SVMs Modeling for Highly Imbalanced Classification. IEEE Trans Syst Man Cybern B Cybern. 2009;39(1):281–88. doi: 10.1109/TSMCB.2008.2002909. [DOI] [PubMed] [Google Scholar]
- 96.Chawla NV, Bowyer KW, Hall LO, et al. SMOTE: Synthetic minority over-sampling technique. J Artif Intell Res. 2002;16:321–57. [Google Scholar]
- 97.Ling C, Sheng V. Cost-Sensitive Learning. In: Sammut C, Webb G, editors. Encyclopedia of Machine Learning. Springer US: 2010. pp. 231–35. [Google Scholar]
- 98.Han LY, Ma XH, Lin HH, et al. A support vector machines approach for virtual screening of active compounds of single and multiple mechanisms from large libraries at an improved hit-rate and enrichment factor. J Mol Graph Model. 2008;26(8):1276–86. doi: 10.1016/j.jmgm.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 99.Ren JX, Li LL, Zheng RL, et al. Discovery of Novel Pim-1 Kinase Inhibitors by a Hierarchical Multistage Virtual Screening Approach Based on SVM Model, Pharmacophore, and Molecular Docking. J Chem Inf Model. 2011;51(6):1364–75. doi: 10.1021/ci100464b. [DOI] [PubMed] [Google Scholar]
- 100.Liu XH, Song HY, Zhang JX, et al. Identifying Novel Type ZBGs and Nonhydroxamate HDAC Inhibitors Through a SVM Based Virtual Screening Approach. Mol Inform. 2010;29(5):407–20. doi: 10.1002/minf.200900014. [DOI] [PubMed] [Google Scholar]
- 101.Ma XH, Wang R, Tan CY, et al. Virtual Screening of Selective Multitarget Kinase Inhibitors by Combinatorial Support Vector Machines. Mol Pharm. 2010;7(5):1545–60. doi: 10.1021/mp100179t. [DOI] [PubMed] [Google Scholar]
- 102.Villoutreix BO, Taboureau O. Computational investigations of hERG channel blockers: New insights and current predictive models. Adv Drug Deliv Rev. 2015;86:72–82. doi: 10.1016/j.addr.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 103.Wishart DS, Jewison T, Guo AC, et al. HMDB 3.0-The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41(D1):D801–D07. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sedykh A, Zhu H, Tang H, et al. Use of in Vitro HTS-Derived Concentration-Response Data as Biological Descriptors Improves the Accuracy of QSAR Models of in Vivo Toxicity. Environ Health Perspect. 2011;119(3):364–70. doi: 10.1289/ehp.1002476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim MT, Huang R, Sedykh A, et al. Mechanism Profiling of Hepatotoxicity Caused by Oxidative Stress Using Antioxidant Response Element Reporter Gene Assay Models and Big Data. Environ Health Perspect. 2016;124(5):634–41. doi: 10.1289/ehp.1509763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106•.Zhu H, Zhang J, Kim MT, et al. Big Data in Chemical Toxicity Research: The Use of High-Throughput Screening Assays To Identify Potential Toxicants. Chem Res Toxicol. 2014;27(10):1643–51. doi: 10.1021/tx500145h. A review article about the use of HTS data for development of computational toxicity models. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen B, Wild D, Guha R. PubChem as a Source of Polypharmacology. J Chem Inf Model. 2009;49(9):2044–55. doi: 10.1021/ci9001876. [DOI] [PubMed] [Google Scholar]
- 108.Zhang JX, Han BC, Wei XN, et al. A Two-Step Target Binding and Selectivity Support Vector Machines Approach for Virtual Screening of Dopamine Receptor Subtype-Selective Ligands. PLoS One. 2012;7(6):12. doi: 10.1371/journal.pone.0039076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Swamidass SJ, Schillebeeckx CN, Matlock M, et al. Combined Analysis of Phenotypic and Target-Based Screening in Assay Networks. J Biomol Screen. 2014;19(5):782–90. doi: 10.1177/1087057114523068. [DOI] [PubMed] [Google Scholar]
- 110.Lounkine E, Nigsch F, Jenkins JL, et al. Activity-Aware Clustering of High Throughput Screening Data and Elucidation of Orthogonal Structure-Activity Relationships. J Chem Inf Model. 2011;51(12):3158–68. doi: 10.1021/ci2004994. [DOI] [PubMed] [Google Scholar]
- 111.Huang N, Shoichet BK, Irwin JJ. Benchmarking sets for molecular docking. J Med Chem. 2006;49(23):6789–801. doi: 10.1021/jm0608356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mysinger MM, Carchia M, Irwin JJ, et al. Directory of Useful Decoys, Enhanced (DUD-E): Better Ligands and Decoys for Better Benchmarking. J Med Chem. 2012;55(14):6582–94. doi: 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wallach I, Lilien R. Virtual Decoy Sets for Molecular Docking Benchmarks. J Chem Inf Model. 2011;51(2):196–202. doi: 10.1021/ci100374f. [DOI] [PubMed] [Google Scholar]
- 114.Vogel SM, Bauer MR, Boeckler FM. DEKOIS: Demanding Evaluation Kits for Objective in Silico Screening - A Versatile Tool for Benchmarking Docking Programs and Scoring Functions. J Chem Inf Model. 2011;51(10):2650–65. doi: 10.1021/ci2001549. [DOI] [PubMed] [Google Scholar]
- 115.Bauer MR, Ibrahim TM, Vogel SM, et al. Evaluation and Optimization of Virtual Screening Workflows with DEKOIS 2.0-A Public Library of Challenging Docking Benchmark Sets. J Chem Inf Model. 2013;53(6):1447–62. doi: 10.1021/ci400115b. [DOI] [PubMed] [Google Scholar]
- 116.Gatica EA, Cavasotto CN. Ligand and Decoy Sets for Docking to G Protein-Coupled Receptors. J Chem Inf Model. 2012;52(1):1–6. doi: 10.1021/ci200412p. [DOI] [PubMed] [Google Scholar]
- 117.Xia J, Jin HW, Liu ZM, et al. An Unbiased Method To Build Benchmarking Sets for Ligand-Based Virtual Screening and its Application To GPCRs. J Chem Inf Model. 2014;54(5):1433–50. doi: 10.1021/ci500062f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schierz AC. Virtual screening of bioassay data. J Cheminform. 2009;1:21. doi: 10.1186/1758-2946-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119•.Rohrer SG, Baumann K. Maximum Unbiased Validation (MUV) Data Sets for Virtual Screening Based on PubChem Bioactivity Data. J Chem Inf Model. 2009;49(2):169–84. doi: 10.1021/ci8002649. A research article in which the Maximum Unbiased Validation (MUV) data sets were developed from PubChem's bioactivity data. [DOI] [PubMed] [Google Scholar]
- 120.Butkiewicz M, Lowe EW, Mueller R, et al. Benchmarking Ligand-Based Virtual High-Throughput Screening with the PubChem Database. Molecules. 2013;18(1):735–56. doi: 10.3390/molecules18010735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lindh M, Svensson F, Schaal W, et al. Toward a Benchmarking Data Set Able to Evaluate Ligand- and Structure-based Virtual Screening Using Public HTS Data. J Chem Inf Model. 2015;55(2):343–53. doi: 10.1021/ci5005465. [DOI] [PubMed] [Google Scholar]
- 122.Verdonk ML, Berdini V, Hartshorn MJ, et al. Virtual screening using protein-ligand docking: Avoiding artificial enrichment. J Chem Inf Comput Sci. 2004;44(3):793–806. doi: 10.1021/ci034289q. [DOI] [PubMed] [Google Scholar]
- 123.Good AC, Hermsmeier MA, Hindle SA. Measuring CAMD technique performance: A virtual screening case study in the design of validation experiments. J Comput Aided Mol Des. 2004;18(7-9):529–36. doi: 10.1007/s10822-004-4067-1. [DOI] [PubMed] [Google Scholar]