Abstract
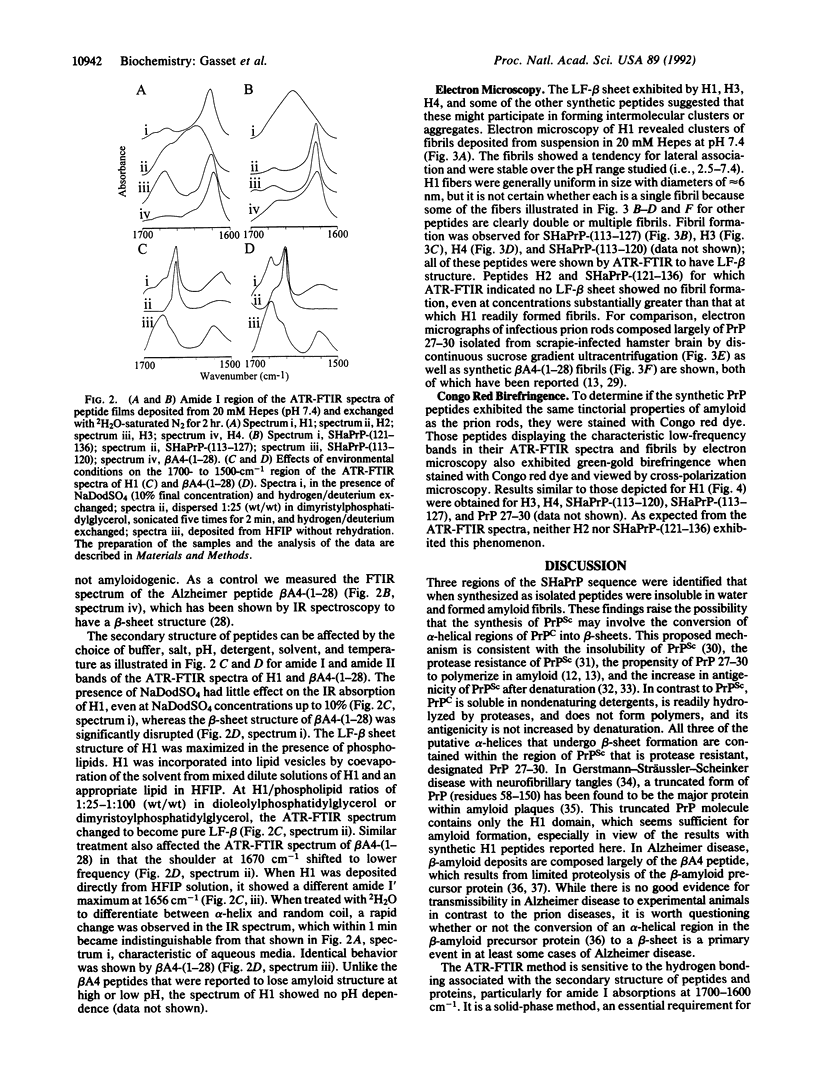
By comparing the amino acid sequences of 11 mammalian and 1 avian prion proteins (PrP), structural analyses predicted four alpha-helical regions. Peptides corresponding to these regions of Syrian hamster PrP were synthesized, and, contrary to predictions, three of the four spontaneously formed amyloids as shown by electron microscopy and Congo red staining. By IR spectroscopy, these amyloid peptides exhibited secondary structures composed largely of beta-sheets. The first of the predicted helices is the 14-amino acid peptide corresponding to residues 109-122; this peptide and the overlapping 15-residue sequence 113-127 both form amyloid. The most highly amyloidogenic peptide is AGAAAAGA, which corresponds to Syrian hamster PrP residues 113-120 and is conserved across all species for which the PrP sequence has been determined. Two other predicted alpha-helices corresponding to residues 178-191 and 202-218 form amyloids and exhibit considerable beta-sheet structure when synthesized as peptides. These findings suggest the possibility that the conversion of the cellular isoform of PrP to the scrapie isoform of PrP involves the transition of one or more putative PrP alpha-helices into beta-sheets and that prion diseases are disorders of protein conformation.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bazan J. F., Fletterick R. J., McKinley M. P., Prusiner S. B. Predicted secondary structure and membrane topology of the scrapie prion protein. Protein Eng. 1987 Feb-Mar;1(2):125–135. doi: 10.1093/protein/1.2.125. [DOI] [PubMed] [Google Scholar]
- Bendheim P. E., Barry R. A., DeArmond S. J., Stites D. P., Prusiner S. B. Antibodies to a scrapie prion protein. Nature. 1984 Aug 2;310(5976):418–421. doi: 10.1038/310418a0. [DOI] [PubMed] [Google Scholar]
- Borchelt D. R., Scott M., Taraboulos A., Stahl N., Prusiner S. B. Scrapie and cellular prion proteins differ in their kinetics of synthesis and topology in cultured cells. J Cell Biol. 1990 Mar;110(3):743–752. doi: 10.1083/jcb.110.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputo C. B., Fraser P. E., Sobel I. E., Kirschner D. A. Amyloid-like properties of a synthetic peptide corresponding to the carboxy terminus of beta-amyloid protein precursor. Arch Biochem Biophys. 1992 Jan;292(1):199–205. doi: 10.1016/0003-9861(92)90068-8. [DOI] [PubMed] [Google Scholar]
- Caughey B. W., Dong A., Bhat K. S., Ernst D., Hayes S. F., Caughey W. S. Secondary structure analysis of the scrapie-associated protein PrP 27-30 in water by infrared spectroscopy. Biochemistry. 1991 Aug 6;30(31):7672–7680. doi: 10.1021/bi00245a003. [DOI] [PubMed] [Google Scholar]
- Caughey B., Raymond G. J., Ernst D., Race R. E. N-terminal truncation of the scrapie-associated form of PrP by lysosomal protease(s): implications regarding the site of conversion of PrP to the protease-resistant state. J Virol. 1991 Dec;65(12):6597–6603. doi: 10.1128/jvi.65.12.6597-6603.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughey B., Raymond G. J. The scrapie-associated form of PrP is made from a cell surface precursor that is both protease- and phospholipase-sensitive. J Biol Chem. 1991 Sep 25;266(27):18217–18223. [PubMed] [Google Scholar]
- Cohen F. E., Abarbanel R. M., Kuntz I. D., Fletterick R. J. Turn prediction in proteins using a pattern-matching approach. Biochemistry. 1986 Jan 14;25(1):266–275. doi: 10.1021/bi00349a037. [DOI] [PubMed] [Google Scholar]
- Colonna-Cesari F., Perahia D., Karplus M., Eklund H., Brädén C. I., Tapia O. Interdomain motion in liver alcohol dehydrogenase. Structural and energetic analysis of the hinge bending mode. J Biol Chem. 1986 Nov 15;261(32):15273–15280. [PubMed] [Google Scholar]
- DeArmond S. J., McKinley M. P., Barry R. A., Braunfeld M. B., McColloch J. R., Prusiner S. B. Identification of prion amyloid filaments in scrapie-infected brain. Cell. 1985 May;41(1):221–235. doi: 10.1016/0092-8674(85)90076-5. [DOI] [PubMed] [Google Scholar]
- Doh-ura K., Tateishi J., Sasaki H., Kitamoto T., Sakaki Y. Pro----leu change at position 102 of prion protein is the most common but not the sole mutation related to Gerstmann-Sträussler syndrome. Biochem Biophys Res Commun. 1989 Sep 15;163(2):974–979. doi: 10.1016/0006-291x(89)92317-6. [DOI] [PubMed] [Google Scholar]
- Fernando Bazan J., Fletterick R. J., Prusiner S. B. AIDS virus and scrapie protein genes. Nature. 1987 Feb 12;325(6105):581–581. doi: 10.1038/325581a0. [DOI] [PubMed] [Google Scholar]
- Fraser P. E., Nguyen J. T., Surewicz W. K., Kirschner D. A. pH-dependent structural transitions of Alzheimer amyloid peptides. Biophys J. 1991 Nov;60(5):1190–1201. doi: 10.1016/S0006-3495(91)82154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabizon R., McKinley M. P., Prusiner S. B. Purified prion proteins and scrapie infectivity copartition into liposomes. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4017–4021. doi: 10.1073/pnas.84.12.4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Glenner G. G. Alzheimer's disease: its proteins and genes. Cell. 1988 Feb 12;52(3):307–308. doi: 10.1016/s0092-8674(88)80021-7. [DOI] [PubMed] [Google Scholar]
- Glenner G. G., Eanes E. D., Bladen H. A., Linke R. P., Termine J. D. Beta-pleated sheet fibrils. A comparison of native amyloid with synthetic protein fibrils. J Histochem Cytochem. 1974 Dec;22(12):1141–1158. doi: 10.1177/22.12.1141. [DOI] [PubMed] [Google Scholar]
- Glenner G. G., Eanes E. D., Page D. L. The relation of the properties of Congo red-stained amyloid fibrils to the -conformation. J Histochem Cytochem. 1972 Oct;20(10):821–826. doi: 10.1177/20.10.821. [DOI] [PubMed] [Google Scholar]
- Goormaghtigh E., Cabiaux V., Ruysschaert J. M. Secondary structure and dosage of soluble and membrane proteins by attenuated total reflection Fourier-transform infrared spectroscopy on hydrated films. Eur J Biochem. 1990 Oct 24;193(2):409–420. doi: 10.1111/j.1432-1033.1990.tb19354.x. [DOI] [PubMed] [Google Scholar]
- Hemström C., Virtanen A., Bridge E., Ketner G., Pettersson U. Adenovirus E4-dependent activation of the early E2 promoter is insufficient to promote the early-to-late-phase transition. J Virol. 1991 Mar;65(3):1440–1449. doi: 10.1128/jvi.65.3.1440-1449.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao K., Dlouhy S. R., Farlow M. R., Cass C., Da Costa M., Conneally P. M., Hodes M. E., Ghetti B., Prusiner S. B. Mutant prion proteins in Gerstmann-Sträussler-Scheinker disease with neurofibrillary tangles. Nat Genet. 1992 Apr;1(1):68–71. doi: 10.1038/ng0492-68. [DOI] [PubMed] [Google Scholar]
- Kang J., Lemaire H. G., Unterbeck A., Salbaum J. M., Masters C. L., Grzeschik K. H., Multhaup G., Beyreuther K., Müller-Hill B. The precursor of Alzheimer's disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987 Feb 19;325(6106):733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- Kascsak R. J., Rubenstein R., Merz P. A., Tonna-DeMasi M., Fersko R., Carp R. I., Wisniewski H. M., Diringer H. Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J Virol. 1987 Dec;61(12):3688–3693. doi: 10.1128/jvi.61.12.3688-3693.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamoto T., Tateishi J., Tashima T., Takeshita I., Barry R. A., DeArmond S. J., Prusiner S. B. Amyloid plaques in Creutzfeldt-Jakob disease stain with prion protein antibodies. Ann Neurol. 1986 Aug;20(2):204–208. doi: 10.1002/ana.410200205. [DOI] [PubMed] [Google Scholar]
- Laszlo L., Lowe J., Self T., Kenward N., Landon M., McBride T., Farquhar C., McConnell I., Brown J., Hope J. Lysosomes as key organelles in the pathogenesis of prion encephalopathies. J Pathol. 1992 Apr;166(4):333–341. doi: 10.1002/path.1711660404. [DOI] [PubMed] [Google Scholar]
- McKinley M. P., Braunfeld M. B., Bellinger C. G., Prusiner S. B. Molecular characteristics of prion rods purified from scrapie-infected hamster brains. J Infect Dis. 1986 Jul;154(1):110–120. doi: 10.1093/infdis/154.1.110. [DOI] [PubMed] [Google Scholar]
- McKinley M. P., Taraboulos A., Kenaga L., Serban D., Stieber A., DeArmond S. J., Prusiner S. B., Gonatas N. Ultrastructural localization of scrapie prion proteins in cytoplasmic vesicles of infected cultured cells. Lab Invest. 1991 Dec;65(6):622–630. [PubMed] [Google Scholar]
- McPhalen C. A., James M. N. Structural comparison of two serine proteinase-protein inhibitor complexes: eglin-c-subtilisin Carlsberg and CI-2-subtilisin Novo. Biochemistry. 1988 Aug 23;27(17):6582–6598. [PubMed] [Google Scholar]
- Meyer R. K., McKinley M. P., Bowman K. A., Braunfeld M. B., Barry R. A., Prusiner S. B. Separation and properties of cellular and scrapie prion proteins. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2310–2314. doi: 10.1073/pnas.83.8.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita K., Ichimura S., Zama M., James T. C. Specific codon usage pattern and its implications on the secondary structure of silk fibroin mRNA. J Mol Biol. 1988 Oct 20;203(4):917–925. doi: 10.1016/0022-2836(88)90117-9. [DOI] [PubMed] [Google Scholar]
- Oesch B., Westaway D., Wälchli M., McKinley M. P., Kent S. B., Aebersold R., Barry R. A., Tempst P., Teplow D. B., Hood L. E. A cellular gene encodes scrapie PrP 27-30 protein. Cell. 1985 Apr;40(4):735–746. doi: 10.1016/0092-8674(85)90333-2. [DOI] [PubMed] [Google Scholar]
- Prusiner S. B., Groth D. F., Bolton D. C., Kent S. B., Hood L. E. Purification and structural studies of a major scrapie prion protein. Cell. 1984 Aug;38(1):127–134. doi: 10.1016/0092-8674(84)90533-6. [DOI] [PubMed] [Google Scholar]
- Prusiner S. B., McKinley M. P., Bowman K. A., Bolton D. C., Bendheim P. E., Groth D. F., Glenner G. G. Scrapie prions aggregate to form amyloid-like birefringent rods. Cell. 1983 Dec;35(2 Pt 1):349–358. doi: 10.1016/0092-8674(83)90168-x. [DOI] [PubMed] [Google Scholar]
- Prusiner S. B. Molecular biology of prion diseases. Science. 1991 Jun 14;252(5012):1515–1522. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- Saitta B., Timpl R., Chu M. L. Human alpha 2(VI) collagen gene. Heterogeneity at the 5'-untranslated region generated by an alternate exon. J Biol Chem. 1992 Mar 25;267(9):6188–6196. [PubMed] [Google Scholar]
- Serban D., Taraboulos A., DeArmond S. J., Prusiner S. B. Rapid detection of Creutzfeldt-Jakob disease and scrapie prion proteins. Neurology. 1990 Jan;40(1):110–117. doi: 10.1212/wnl.40.1.110. [DOI] [PubMed] [Google Scholar]
- Stahl N., Baldwin M. A., Burlingame A. L., Prusiner S. B. Identification of glycoinositol phospholipid linked and truncated forms of the scrapie prion protein. Biochemistry. 1990 Sep 25;29(38):8879–8884. doi: 10.1021/bi00490a001. [DOI] [PubMed] [Google Scholar]
- Tagliavini F., Prelli F., Ghiso J., Bugiani O., Serban D., Prusiner S. B., Farlow M. R., Ghetti B., Frangione B. Amyloid protein of Gerstmann-Sträussler-Scheinker disease (Indiana kindred) is an 11 kd fragment of prion protein with an N-terminal glycine at codon 58. EMBO J. 1991 Mar;10(3):513–519. doi: 10.1002/j.1460-2075.1991.tb07977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraboulos A., Raeber A. J., Borchelt D. R., Serban D., Prusiner S. B. Synthesis and trafficking of prion proteins in cultured cells. Mol Biol Cell. 1992 Aug;3(8):851–863. doi: 10.1091/mbc.3.8.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraboulos A., Serban D., Prusiner S. B. Scrapie prion proteins accumulate in the cytoplasm of persistently infected cultured cells. J Cell Biol. 1990 Jun;110(6):2117–2132. doi: 10.1083/jcb.110.6.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronrud D. E., Schmid M. F., Matthews B. W. Structure and X-ray amino acid sequence of a bacteriochlorophyll A protein from Prosthecochloris aestuarii refined at 1.9 A resolution. J Mol Biol. 1986 Apr 5;188(3):443–454. doi: 10.1016/0022-2836(86)90167-1. [DOI] [PubMed] [Google Scholar]
- Williams R. C. Use of polylysine for adsorption of nuclei acids and enzymes to electron microscope specimen films. Proc Natl Acad Sci U S A. 1977 Jun;74(6):2311–2315. doi: 10.1073/pnas.74.6.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Lewis R. V. Structure of a protein superfiber: spider dragline silk. Proc Natl Acad Sci U S A. 1990 Sep;87(18):7120–7124. doi: 10.1073/pnas.87.18.7120. [DOI] [PMC free article] [PubMed] [Google Scholar]