Abstract
This study determined the anti-neoplastic activity and nephrotoxicity of epigenetic inhibitors in vitro. The therapeutic efficacy of epigenetic inhibitors was determined in human prostate cancer cells (PC-3 and LNCaP) using the DNA methyltransferase inhibitor 5-azacytidine (5-Aza) and the histone deacetylase inhibitor trichostatin A (TSA). Cells were also treated with carbamazepine (CBZ), an anti-convulsant with histone deacetylase inhibitor-like properties. 5-Aza, TSA or CBZ alone did not decrease MTT staining in PC-3 or LNCaP cells after 48 hr. In contrast, docetaxel, a frontline chemotherapeutic induced concentration-dependent decreases in MTT staining. Pretreatment with 5-Aza or TSA increased docetaxel-induced cytotoxicity in LNCaP cells, but not PC-3 cells. TSA pretreatment also increased cisplatin-induced toxicity in LNCaP cells. Carfilzomib (CFZ), a protease inhibitor approved for the treatment of multiple myeloma had minimal effect on LNCaP cell viability, but reduced MTT staining 50% in PC-3 cells compared to control, and pretreatment with 5-Aza further enhanced toxicity. Treatment of normal rat kidney (NRK) and human embryonic kidney 293 (HEK293) cells with the same concentrations of epigenetic inhibitors used in prostate cancer cells significantly decreased MTT staining in all cell lines after 48 hr. Interestingly, we found that the toxicity of epigenetic inhibitors to kidney cells was dependent on both the compound and the stage of cell growth. The effect of 5-Aza and TSA on DNA methyltransferase and histone deacetylase activity, respectively, was confirmed by assessing the methylation and acetylation of the CDK inhibitor p21. Collectively, these data show that combinatorial treatment with epigenetic inhibitors alters the efficacy of chemotherapeutics in cancer cells in a compound- and cell-specific manner; however, this treatment also has the potential to induce nephrotoxic cell injury.
Keywords: epigenetic inhibitors, prostate cancer, kidney cells, nephrotoxicity
1. Introduction
DNA methylation is an epigenetic event that affects cell function by altering gene expression [1]. Essentially, it is the covalent addition of a methyl group, catalyzed by DNA methyltransferase, to the 5-carbon of a cytosine in a CpG dinucleotide [2]. This methyl group impedes the binding of transcription proteins, thereby decreasing transcription and expression. DNA methylation plays a crucial role in carcinogenesis through methylation of cytosines at promoter regions of various genes, silencing their transcription [3]. A number of tumor suppressor genes can also be silenced via methylation [4].
Histone modifications can also affect gene expression. Histone acetylation generally induces gene activation through post-transcriptional acetylation of lysine residues at the N-terminal tail of each of four histones: H2A, H2B, H3, and H4 [5, 6]. Histone acetyltransferase catalyzes the transfer of an acetyl group from acetyl-coenzyme A to the histone. The addition of an acetyl group decreases the interaction of the histone with the negatively charged phosphate groups of DNA. This results in less condensed chromatin, referred to as euchromatin, which is more readily and easily transcribed. Histone deacetylases remove these acetyl groups. Alterations in promoter-specific histone modifications can result in improper silencing or transcription, which could lead to cellular transformation, carcinogenesis, or cancer progression [7, 8].
Growing evidence indicates that epigenetic abnormalities contribute to the dysregulation in gene expression associated with cancer formation and progression [9]. Prostate cancer is the second leading cause of cancer death among men in the United States. Although the mechanisms underlying prostate cancer formation are not well understood, irregular epigenetic events including alterations in DNA methylation and histone modifications appear to contribute to its development [10–14]. This is one reason why epigenetic inhibitors are being proposed as potential treatments for prostate cancer, among other cancer types, both alone and in combination with established chemotherapeutics [15–18].
Several studies have investigated the role of epigenetic alterations on the expression of specific genes and how these genes contribute to prostate cancer growth [12–14]. In contrast, fewer studies have examined the effect of concomitant treatments of epigenetic inhibitors and known chemotherapeutics on prostate cancer cell growth. One such study assessed the effect of pretreatment of prostate cancer cells with the DNA methyltransferase inhibitor 5-azacytidine followed by the chemotherapeutic docetaxel, and found that this co-treatment increased docetaxel-induced apoptosis in DU-145 cells [15]. Another study found that combinatorial treatment with a histone deacetylase inhibitor and androgen receptor antagonist synergistically reduced prostate cancer cell proliferation and increased cell death [19].
A limitation of the above studies, as well as other studies, is that the anti-cancer activities of these treatments were not compared directly to their non-target organ toxicity. The kidney is often subject to adverse effects by chemotherapeutic treatments [20, 21]. Further, epigenetic inhibitors are known to induce cell- and compound-specific nephrotoxicity. For example, 5-azacytidine induces renal tubular dysfunction in patients with advanced acute leukemia [20, 22]. Dong et al. [23] found that histone deacetylase inhibitors induce apoptosis in renal tubular cells. DNA methyltransferase inhibitors and histone deacetylase inhibitors have also been shown to induce nephrotoxicity via increased oxidative stress [24]. These studies support the need to examine the effect of concomitant treatment schedules of epigenetic inhibitors with established chemotherapeutics on nephrotoxicity. This study determined the cytotoxicity of DNA methylation and histone deacetylation inhibition alone and in combination with various chemotherapeutics on prostate cancer and kidney cells.
2. Materials and Methods
2.1. Materials
Normal rat kidney (NRK) cells, human embryonic kidney (HEK-293) cells, human prostate cancer (PC-3, LNCaP) cells, DMEM media, F-12K media, RPMI 1640 media and fetal bovine serum were purchased from American Type Culture Collection (Mannasas, VA). Antibiotic antimycotic solution was purchased from Sigma-Aldrich (St. Louis, MO). Docetaxel, cisplatin, carfilzomib (CFZ), 5-azacytidine (5-Aza), carbamezapine (CBZ), trichostatin A (TSA) and 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) were purchased from Sigma-Aldrich (St. Louis, MO). All compounds were dissolved in dimethyl sulfoxide (DMSO) from Fisher Scientific (Pittsburg, PA). DNeasy blood and tissue kit was purchased from Qiagen (Valencia, CA), EZ DNA methylation-lightening kit from Zymo Research (Irvine CA), NucleoSpin gel and PCR clean-up kit from Macherey-Nagel (Düren, Germany), PCR master mix from Promega (Madison, WI), EpiQuick Acetyl-Histone H3 ChIP kit from Epigentek (Farmingdale, NY) and MiSeq reagent v3 kit from Illumina Inc (San Diego, CA)
2.2. Cell Lines and Treatments
The human PC-3 cell line was initiated from bone metastasis of a prostatic adenocarcinoma. These cells are p53-null, androgen insensitive and malignant. LNCaP cells are derived from a human lymph node metastasis of prostate carcinoma and are p53-positive, androgen sensitive and less aggressive. All cells were grown at 37°C in 5% CO2. Docetaxel is a frontline chemotherapeutic for prostate cancer and acts through tubulin disturbance [25]. Cisplatin is a common chemotherapeutic that acts through DNA alkylation. Carfilzomib is a selective proteasome inhibitor approved for the treatment of myeloma [26]. 5-Aza is a DNA methyltransferase (DNMT) inhibitor, TSA is a histone deacetylase (HDAC) inhibitor and CBZ is a clinically used anticonvulsant with HDAC inhibitor-like properties [27–29].
2.3. MTT Staining
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) staining was used as an indicator of toxicity and was corroborated with examination of cellular morphology using phase contrast microscopy. All cell lines were seeded in 48-well plates at 100,000 cells/mL. The prostate cancer cells were incubated 24 hr to reach log phase growth. The kidney cells were grown for 48 hr to obtain confluence. These conditions were selected to more accurately reflect the biological state of both in the body, as cancer cells are generally actively dividing, while, under normal circumstances, the kidney is not. After this time, kidney cells and prostate cancer cells were pretreated with varying concentrations of epigenetic inhibitors (5-Aza, CBZ, TSA) or vehicle control for 30 min, followed by addition of chemotherapeutics or vehicle control for a total of 48 hr. The total amount of DMSO was never above 0.1% of the total volume per well. After 48 hr, 0.25 mg/mL of MTT was added to each well and the plates were incubated for 2 hr. The media was then aspirated and replaced with DMSO. The plates were shaken at 100 rpm for 15 min to dissolve all precipitates and the absorbance of each well at 544 nm, including control and blank wells, was measured with a FLUOstar OPTIMA plate reader (BMG Lab Technologies, Inc., Durham, NC).
2.4. Confirmation of the effect of 5-Aza
The effect of 5-Aza on DNA methylation was investigated using next-generation sequencing (NGS) of bisulfite converted DNA and verified via decreases in methylation of a specific target gene (CDKN1a or p21). HEK293 cells were seeded and incubated for 24 hr, then treated with 5-Aza or control for 6 days. DNA was extracted from 5 x 106 cells using Qiagen’s DNeasy blood and tissue kit, followed by bisulfite conversion using Zymo Research’s EZ DNA methylation-lightening kit following manufacturer’s protocol. PCR of the bisulfite converted DNA was performed to amplify the p21 promoter region (sense: 5’-TTTTTTGAGTTTTAGTTTTT TTAGTAGTG-3’ and anti-sense: 5’-AACCAAAATAATTTTTCAATCCC-3’, designed using Methprimer [30]). The amplicons were further processed for NGS on the Illumina MiSeq platform using custom adapters [31] and 600 cycle v3 kits to obtain paired-end 300 base reads. Comparison of sequences of the bisulfite-treated DNA from 5-Aza-treated and control cells with a reference genome allowed the identification of sites with 5-methylcytosine. More than 10,000 sequence reads per sample were aligned and analyzed using Bismark bisulfite mapper to obtain percent DNA methylation data.
2.5. Confirmation of the effect of TSA
The effect of TSA on histone acetylation was investigated using Epigentek’s EpiQuick Acetyl-Histone H3 chromatin immunoprecipitation (ChIP) kit following the manufacturer’s protocol and using p21 as a reference gene. For this, NRK cells were incubated for 24 hr and then treated with or TSA or control for 72 hr. 3 x 106 cells were used for ChIP. PCR of the eluted DNA was performed to amplify the rat p21 promoter region with primers as used by Yuan H et al. [32], and to amplify the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH, sense: 5’-CACGGCAAGTTCAACGGCACAGTCA-3’ and anti-sense: 5’-GTGAAGACGCCAGTAG ACTCCAGGAC-3’). The amplicons were analyzed by densitometry using Flourchem HD2 system (ProteinSimple, San Jose, CA).
2.6. Statistical Analysis
Cells isolated from a distinct passage represented one experiment (n = 1). Data are represented as the mean ± standard deviation (SD) of 6–12 separate experiments (n = 6–12). A one-way analysis of variance (ANOVA) was performed for each data set followed by Bonferonni post-hoc test using GraphPad Prism software with P < 0.05 being considered indicative of a statistically significant difference between mean values. NGS generates thousands of read sequences for each sample, unlike traditional bisulfite sequencing with only 10–20 sequences. The greater the read number, the higher the statistical power of analysis for detecting even subtle differences in methylation between samples [33]. ChIP data are represented as the mean ± SD of 6 different treatments (n=6) and analyzed by two-tailed paired t-test with significance level P < 0.05 as above.
3. Results
3.1. Effect of epigenetic inhibitors on prostate cancer and kidney cytotoxicity
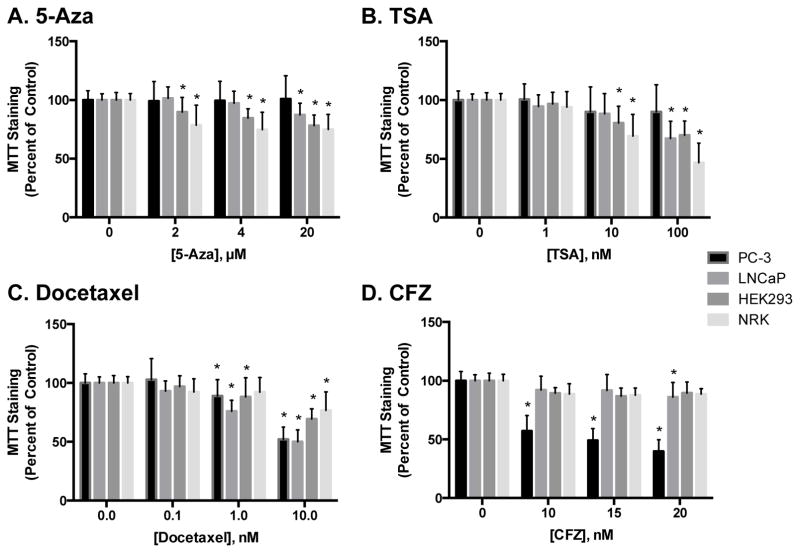
Both kidney and prostate cancer cells were treated with epigenetic inhibitors for 48 hr. The DNA methyltransferase inhibitor 5-azacytidine (5-Aza) was used at concentrations ranging from 0–20 μM, the HDAC inhibitor trichostatin A (TSA) was used at concentrations of 0–100 nM and carbamezapine (CBZ) was used at 0–100 μM. These concentrations were chosen based on literature studies investigating DNMT and HDAC activity [29, 34, 35]. 5-Aza had minimal effects on MTT staining in PC-3 or LNCaP cells as compared to controls. In contrast, 5-Aza treatment decreased MTT staining in NRK and HEK293 cells at concentrations as low as 2 μM (Figure 1A). TSA treatment decreased MTT staining in kidney cells at concentrations of 10 nM and higher, and decreased MTT staining in LNCaP cells at a concentration of 100 nM. Similar to 5-Aza, TSA had no effect on MTT staining in PC-3 cells at any concentration (Figure 1B). On the other hand, CBZ treatment only altered MTT staining at concentrations above 100 μM, where it significantly decreased MTT staining in LNCaP, HEK293 and NRK cells compared to control (data not shown).
Figure 1.
Effect of epigenetic inhibitors and chemotherapeutics on prostate cancer and kidney cytotoxicity. Cells were treated with 5-Aza (A), TSA (B), docetaxel (C) or CFZ (D) for 48 hr and cytotoxicity was assessed using MTT staining. Data are indicative of results from at least 6 separate passages per cell line (n ≥ 6) and are expressed as mean ± SD. (* p < 0.05 compared to respective control by One-way ANOVA followed by Bonferonni post-hoc test)
3.2. Effect of chemotherapeutics on prostate cancer and kidney cytotoxicity
Docetaxel is a frontline chemotherapeutic for the treatment of prostate cancer and acts via tubulin disruption [25]. Unlike 5-Aza and TSA, docetaxel decreased MTT staining in all cell lines tested, with initial decreases being detected at concentrations as low as 1 nM (Figure 1C). As anticipated, docetaxel had a greater effect in prostate cancer cells than in kidney cells.
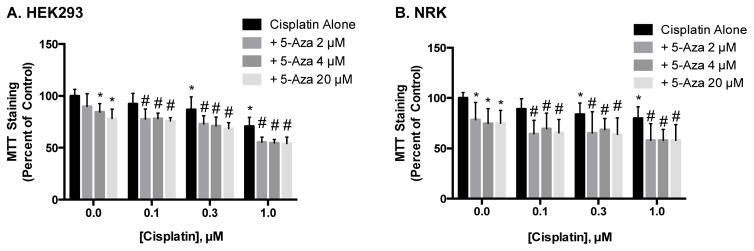
We also examined the effects of carfilzomib (CFZ), a selective proteasome inhibitor approved for the treatment of myeloma, on kidney and prostate cancer cytotoxicity at concentrations from 0–20 nM. Interestingly, treatment of cells with only 10 nM, the lowest concentration used for this study, caused an approximate 50% decrease in MTT staining in PC–3 cells, but had minimal effects on MTT staining in LNCaP cells. CFZ also had a minimal effect on MTT staining in kidney cells (Figure 1D). Cisplatin, another prominent chemotherapeutic and well-known nephrotoxicant, was also studied at concentrations ranging from 0–10 μM, and induced approximately a 25% decrease in MTT staining in each cell line at the highest concentration used (Supplemental Figure 1 ).
3.3. Effect of epigenetic inhibitors on chemotherapeutic-induced cytotoxicity in prostate cancer cells
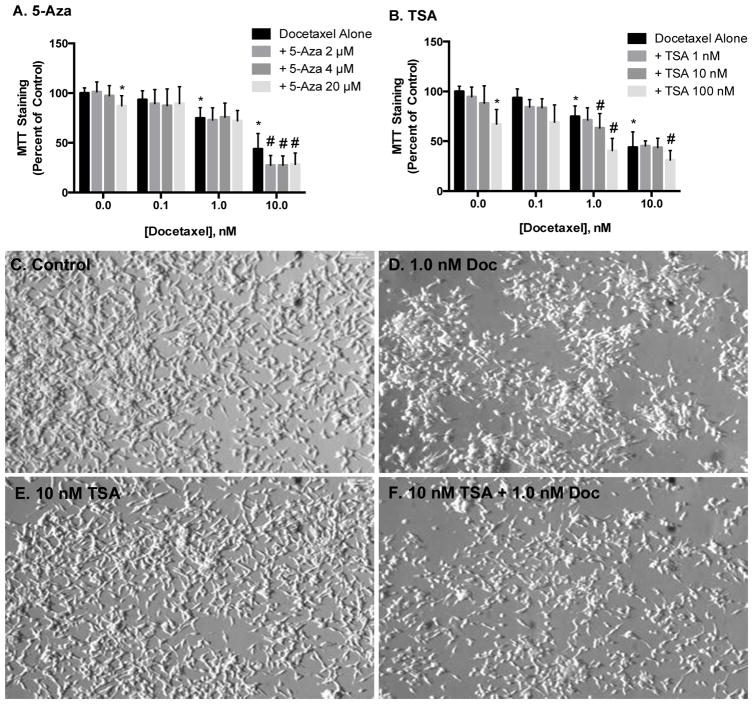
Prostate cancer cells were treated with epigenetic inhibitors for 30 min prior to 48 hr exposure to chemotherapeutics. Pretreatment with 5-Aza, TSA or CBZ had no effect on docetaxel-induced cytotoxicity in PC-3 cells (data not shown). 5-Aza and TSA treatment alone had little to no effect on MTT staining or cell number and morphology in LNCaP cells, while docetaxel induced a concentration-dependent decrease. Pretreatment with any concentration of 5-Aza tested further decreased MTT staining by approximately 20% compared to 10 nM docetaxel alone (Figure 2A). Similar effects were observed with TSA, as combinatorial treatments with docetaxel and TSA at 1 nM and 10 nM, respectively, decreased MTT staining to levels comparable to 10 nM docetaxel (Figure 2B ). These data were confirmed using phase contrast microscopy (Figure 2B–F).
FIGURE 2.
Effect of epigenetic inhibitors on docetaxel-induced cytotoxicity in LNCaP cells. Cells were pretreated with 5-Aza or TSA for 30 min prior to 48 hr exposure to docetaxel and cytotoxicity was measured via changes in MTT staining (A, B) and phase contrast microscopy (C-F). Data in A and B are indicative of results from at least 6 separate passages per cell line (n ≥ 6) and are expressed as mean ± SD. (* p < 0.05 compared to control, # p < 0.05 compared to respective docetaxel treatment alone by One-way ANOVA followed by Bonferonni post-hoc test)
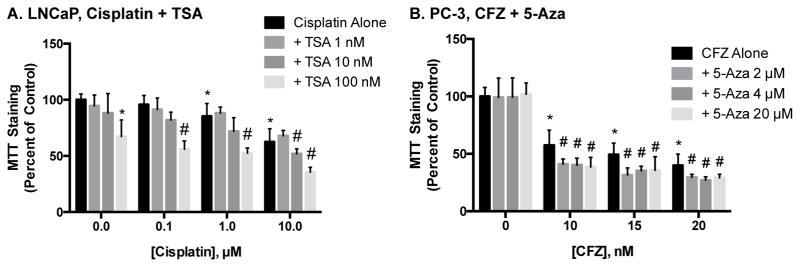
We further explored the ability of epigenetic inhibitors to augment chemotherapeutic efficacy in prostate cancer cells by studying the effect of TSA and 5-Aza on cisplatin- and CFZ- induced toxicity. Similar to the results reported above, pretreatment of LNCaP cells with TSA decreased MTT staining compared to cells treated with cisplatin alone (Figure 3A). However, these same results were not seen with 5-Aza or CBZ (data not shown). While CFZ induced a concentration-dependent decrease in MTT staining in PC-3 cells, neither TSA nor CBZ altered these decreases (data not shown). In contrast, pretreatment of PC-3 cells with 5-Aza resulted in modest, but significant, decreases in MTT staining, compared to cells exposed to only CFZ (Figure 3B). The results of these combinatorial treatments are summarized in Table 1.
Figure 3.
Effect of epigenetic inhibitors on cisplatin- and CFZ-induced cytotoxicity in prostate cancer cells. LNCaP (A) cells were combinatorial treated with TSA and cisplatin and PC-3 (B) cells with 5-Aza and CFZ. The effect on cytotoxicity was measured using MTT staining and data are indicative of results from 6 separate passages per cell line (n ≥ 6) and are expressed as mean ± SD. (* p < 0.05 compared to control, # p < 0.05 compared to respective chemotherapeutic treatment alone by One-way ANOVA followed by Bonferonni post-hoc test)
Table 1.
Effect of epigenetic inhibitors on chemotherapeutic-induced toxicity in prostate cancer cellsa
| + 5-Azab | + CBZ | + TSA | ||||||
|---|---|---|---|---|---|---|---|---|
| LNCaP | PC-3 | LNCaP | PC-3 | LNCaP | PC-3 | LNCaP | PC-3 | |
| Docetaxel | ✓c | ✓ | + | - | - | - | + | - |
| Cisplatin | ✓ | ✓ | - | - | - | - | + | - |
| CFZ | - | ✓ | - | + | - | - | - | - |
Cells were exposed to docetaxel, cisplatin or carfilzomib (CFZ) for 48 hr, after which toxicity was assessed using MTT staining.
Cells were pretreated for 30 min with 5-Aza = 5-azacytidine, CBZ = carbamezapine or TSA = trichostatin A prior to chemotherapeutic exposure.
✓ indicates that the given chemotherapeutic was toxic, while - indicates no significant effect and + indicates increased toxicity in the presence of the specified epigenetic inhibitor.
3.4. Effect of epigenetic inhibitors on chemotherapeutic-induced cytotoxicity in kidney cells
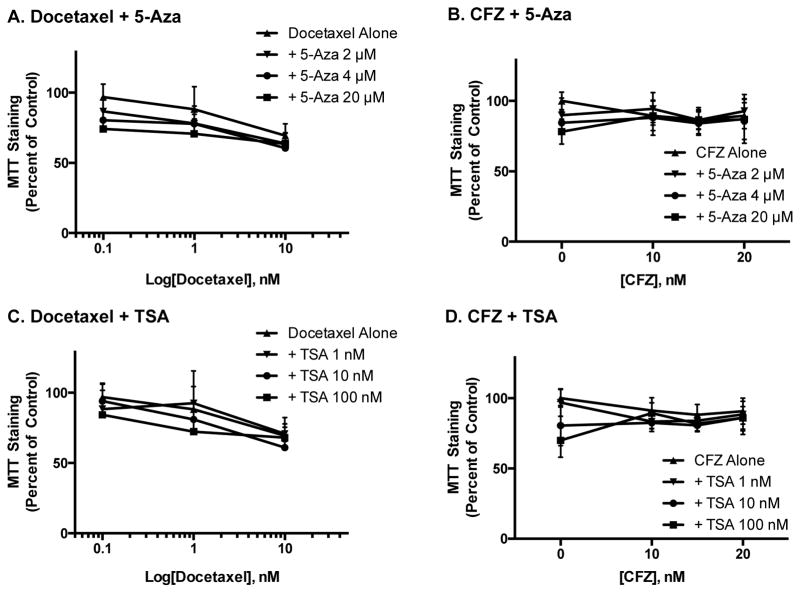
While epigenetic inhibitors may increase cancer cell death on their own, or even augment the effect of other chemotherapeutics, these beneficial clinical results may be for naught if they also induce nephrotoxicity. To address this possibility, we exposed both rat and human renal cell lines to the same treatment protocols used for prostate cancer cells. As reported above, the epigenetic inhibitors alone did prove to be more toxic to kidney cells than to the prostate cancer cells (Figure 1). Further, treatment of HEK293 cells with either 5-Aza or TSA prior to exposure to docetaxel or CFZ did not further decrease MTT staining, compared to cells treated with either chemotherapeutic alone (Figure 4). Similar results were seen in NRK cells exposed to TSA and 5-Aza in combination with docetaxel and CFZ (Supplemental Figure 2) and in kidney cells treated with CBZ and either chemotherapeutic (Supplemental Figure 3). In contrast, treatment of HEK293 and NRK cells with 5-Aza prior to exposure to cisplatin significantly decreased MTT staining, compared to cells treated with cisplatin alone (Figure 5). Treatment with TSA or CBZ prior to cisplatin had no effect on MTT staining in HEK293 or NRK cells (Supplemental Figure 4).
Figure 4.
Effect of epigenetic inhibitors on docetaxel and CFZ-induced toxicity in HEK293 cells. Cells were exposed to the same treatments as prostate cancer cells and the effects of epigenetic inhibitors on docetaxel- (A, C) and CFZ-induced (B, D) toxicity were examined using MTT staining. Data are indicative of results from 6 separate passages per cell line (n≥ 6) and are expressed as mean ± SD.
Figure 5.
Effect of 5-Aza pretreatment on cisplatin-induced toxicity in kidney cells. HEK293 (A) and NRK (B) cells were pretreated with 5-Aza prior to 48 hr exposure to cisplatin and analysis via MTT staining. Data are indicative of results from 6 separate passages per cell line (n ≥ 6) and are expressed as mean ± SD. (* p < 0.05 compared to control, # p < 0.05 compared to respective chemotherapeutic treatment alone by One-way ANOVA followed by Bonferonni post-hoc test)
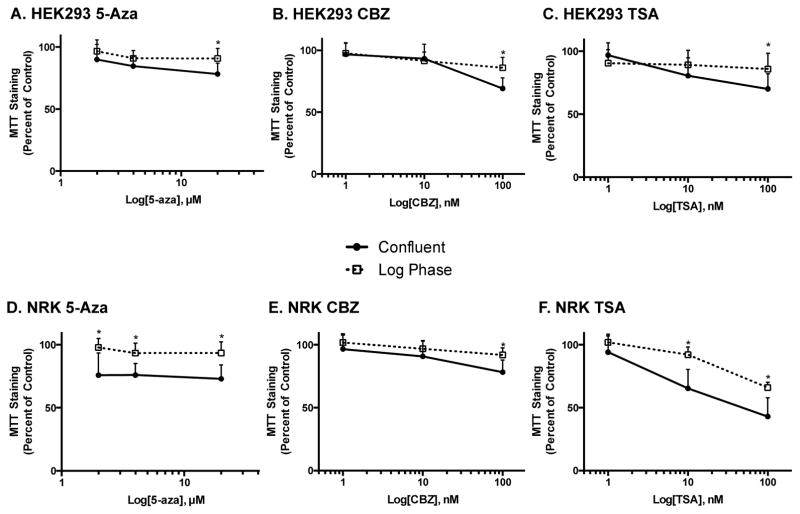
3.5. Effect of stage of cell growth on chemotherapeutic- and epigenetic inhibitor-induced toxicity in kidney cells
For the experiments discussed above, kidney cells were exposed to treatment at confluence to more closely mimic the biological state of the kidney. We also exposed kidney cells to epigenetic inhibitors and chemotherapeutics during log phase growth to maintain consistency with the experiments performed on prostate cancer cells. As shown in Figure 6A–C, decreases in MTT staining in HEK293 cells were similar at almost all concentrations of epigenetic inhibitors, with slight differences being seen at the highest concentrations tested. As expected, the toxicity of the chemotherapeutics was dependent on the stage of cell growth (data not shown). In contrast to HEK293 cells, the stage of cell growth had a more significant effect on epigenetic inhibitor-induced decreases in MTT staining in NRK cells. Of even greater interest was the fact that decreases in MTT staining were greatest in confluent cells. For example, 5-Aza exposure resulted in a 20% greater decrease in MTT staining in confluent NRK cell cultures, as compared to cells exposed during log phase growth (Figure 6D). This same trend was seen with TSA, but not with CBZ (Figure 6E and F).
Figure 6.
Effect of the stage of cell growth on epigenetic inhibitor-induced toxicity in kidney cells. HEK293 (A–C) and NRK cells (D–F) were treated with epigenetic inhibitors after 24 hr (Log Phase; open squares) or 48 hr of growth (Confluent; closed circles) and the difference observed using MTT staining. Data are indicative of results from 6 separate passages per cell line (n ≥ 6) and are expressed as mean ± SD. (* p < 0.05 compared to Confluent by One-way ANOVA followed by Bonferonni post-hoc test)
3.6. Validation of the activity of epigenetic inhibitors
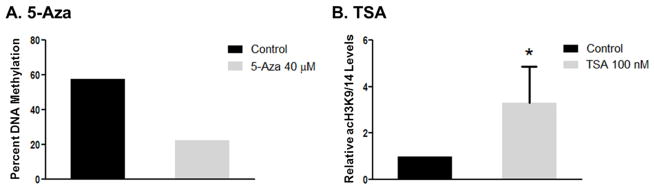
As mentioned above, the concentrations chosen for the epigenetic inhibitors used in this study were based on previous studies demonstrating DNMT and HDAC inhibitory activity [34, 35]. In addition, there have been numerous studies performed and published with these compounds at similar concentrations using prostate cancer cell lines [36–39]. While not wanting to repeat the above studies, we still wished to validate the activity of these inhibitors in these cell lines. Thus, we assessed changes in a specific gene whose expression we recently demonstrated to be mediated by both methylation and histone acetylation, namely p21 [40, 41]. Further, we, along with others, have also shown p21 to be an important mediator of renal toxicity [42–44]. To verify the effect of 5-Aza on p21 methylation, DNA was isolated from HEK293 cells treated with 5-Aza or control and the methylation of p21 was determined using NGS. The data showed that 5-Aza decreased DNA methylation by 35% compared to control (Figure 7A). These data represent the methylation status in over 10,000 sequence reads, extensively minimizing any error, and verifying the activity of 5-Aza.
Figure 7.
Effect of 5-Aza and TSA on the methylation and histone acetylation of p21. The effect of 5-Aza on the methylation of p21 in HEK293 cells was determined using next-generation sequencing (A) and p21 H3 acetylation in NRK cells following TSA was determined by ChIP and is represented as the relative histone-H3-lysine9/14-acetylation (acH3K9/14) levels normalized to GAPDH (B). Data in A represent the overall percent methylation of the p21 promoter region obtained from 10,000 reads. Data in B represent the relative acetylation indicative of 6 different treatments. Data are expressed as mean ± SD. (*p < 0.05 compared with control)
To confirm the effect of TSA on histone acetylation, cells were treated with TSA or control and the acetylation of p21 was determined using ChIP. TSA treatment induced a 2-fold increase in acH3K9/14 of the rat p21 promoter region as compared to control (Figure 7B). These data indicate that, similar to 5-Aza, TSA is functioning as expected at the doses used.
4. Discussion
Multiple studies have shown that epigenetic inhibitors, including DNA methyltransferase and histone deacetylase inhibitors, induce nephrotoxicity [22, 24]. Even so, these compounds are continuously being investigated as potential chemotherapeutics. In support of this research, data in this study showed that pretreatment of prostate cancer cells with these inhibitors enhanced the toxicity of several diverse chemotherapeutics. Unfortunately, data in this study also showed that epigenetic inhibitors alone were more toxic to kidney cells than to prostate cancer cells. Further, combinatorial treatment of kidney cells with certain epigenetic inhibitors increased the toxicity of select chemotherapeutics. Fortunately, these increases were only seen for the combination of 5-Aza and cisplatin.
While data in this study showed that epigenetic inhibitors in general did not increase chemotherapeutic-induced nephrotoxicity, they also demonstrated that these inhibitors did not protect against this toxicity. This is in contrast to data from Tikoo et al. [45] who reported that 5-Aza decreased the nephrotoxicity of cisplatin in vivo while enhancing its anti-neoplastic activity against colon and breast cancer. It should be noted, however, that the concentrations of 5-Aza used in the aforementioned study did not induce nephrotoxicity alone, unlike those used for our study. Thus, these differences could be due in part to the different concentrations used. In support of this hypothesis, Dong et al. [46] observed that TSA also lessened cisplatin-induced nephrotoxicity; however, again, concentrations of TSA shown to be nontoxic alone were used for this study. Additionally, TSA undergoes extensive hepatic metabolism [47], so while we observe toxicity in vitro, this effect may be limited in vivo. Conversely, it is also possible that differences between in vitro and in vivo studies may be related to differences in kinetics. For example, renal excretion is the primary route of 5-Aza clearance [48], which, in combination with the data presented here, suggests the need for a DNA methyltransferase inhibitor with alternate pharmacokinetic properties for use as a therapeutic treatment.
The finding that kidney cells were more susceptible to epigenetic inhibitor-induced toxicity when exposed at confluence compared to during log phase growth was initially surprising. In general, chemotherapeutics target dividing cells, more of which are present during log phase growth. The mechanism of action of these epigenetic inhibitors, however, is different from that of chemotherapeutics, and these differences may explain our findings. The hypomethylating activity of 5-Aza has been studied and proven in various systems, both in vivo and in vitro, using various methods of analyses [49–51]. 5-Aza is one of the prominent epigenetic drugs approved by the FDA for the treatment of acute myeloid leukemia and MDS [52, 53]. 5-Aza incorporates into DNA where it irreversibly binds to DNMTs rendering them inactive resulting in depletion of cellular DNMT levels [27, 54]. Increased DNMT expression and activity, such as that present in actively dividing cells, could decrease the effectiveness of 5-Aza. In support of this hypothesis, DNMT activity has been shown to be downregulated during the G0 phase of the cell cycle [55, 56]. Therefore, less 5-Aza could be necessary to saturate DNMT activity in confluent cells than in cells at log phase growth, shifting the dose response. Similarly, TSA binds directly to HDACs, so the same principal could be behind the toxicity pattern observed with this compound. The histone deacetylase inhibitory activity of TSA has also been extensively studied and proven [32, 57, 58].
Although NRK cells are not of human origin like the other cell lines used for this study, the use of this cell line for nephrotoxicity studies is not uncommon. Our laboratory has published multiple studies using NRK cells as a model for renal toxicity induced by the environmental oxidant bromate, and the data obtained have aligned well with in vivo results [44, 59–61]. Furthermore, similar bromate-induced nephrotoxicity studies have been performed in HEK293 cells, with the results being comparable to that observed in NRK cells [40, 44]. Nevertheless, there are known limitations with these cell lines and future experiments using other cell lines or primary cultures may be needed to solidify and confirm the results obtained from this study.
In general, epigenetic inhibitors had little to no effect on chemotherapeutic-induced toxicity in the kidney cells tested; however, pretreatment with epigenetic inhibitors did enhance chemotherapeutic-induced toxicity in prostate cancer cells in a cell- and compound-specific manner, as outlined in Table 1. This is an exciting and potentially clinically beneficial finding, but the mechanisms behind these enhancements remain unknown. Furthermore, it should be mentioned that while epigenetic events are integral to cancer formation and progression, studies suggest that these combinatorial effects may actually be independent of epigenetic alterations. For example, studies by Abbruzzese and Frost [62] indicate that DNA methyltransferase inhibitor incorporation into DNA, not the resulting hypomethylation, may increase cisplatin binding, resulting in synergistic cytotoxicity. Therefore, further studies are needed to discern the mechanisms mediating alteration in toxicity induced by combinatorial treatment of epigenetic inhibitors and chemotherapeutics. Specifically, studies determining if cell death is mediated by apoptosis or necrosis are needed.
The differences in chemotherapeutic-induced toxicity observed between LNCaP and PC-3 cells, while interesting, were not fully unexpected. In fact, these two prostate cancer cell lines were largely chosen because of their different phenotypes. Cang et al. [63] observed elevated HDAC activity in both prostate cancer cell lines when compared to non-malignant prostate cells; however, the HDAC activity in PC-3 cells was 20–30% higher than that in LNCaP cells. In the same study, LNCaP and PC-3 cells were found to have distinctly different patterns of histone acetylation [63]. Another potentially relevant dissimilarity is the lack of p53 in the PC-3 cell line. Fortson et al. [64] found that TSA induces acetylation of p53, which has been shown to increase DNA binding and transcriptional activation of p21, a well-known tumor suppressor gene [65, 66]. Therefore, the presence of p53 in the LNCaP cells could enhance oncogenic potential of the HDAC inhibitor; an effect which is not observed in the p53-null PC-3 cells. Any combination of the aforementioned variations could account for the differences in chemotherapeutic-induced toxicity observed between the two cell lines in the presence of TSA. Additionally, Yegnasubramanian et al. observed that DNA hypomethylation, although the most prominent in late metastatic stages of prostate cancer, was heterogeneous across different metastatic sites, even within a single individual [67]. This heterogeneity could contribute to the differences observed between PC-3 and LNCaP cells following combinatorial treatment with 5-Aza, as PC-3 cells are obtained from bone metastases and LNCaP from the lymph node. Heterogeneity remains a significant challenge for cancer treatment, as different cell types are known to respond to therapeutics differently; the data presented here further corroborate this fact. Moreover, studies have shown that the population of cells which initiate prostate cancer have distinct phenotypes from those which drive cancer progression [68]. Such findings indicate the importance of investigating chemotherapeutic efficacy in multiple, if not varied, cell types for a given disease.
The innovative design of this study investigated the anti-cancer activity of combinatorial treatments with various epigenetic inhibitors and chemotherapeutics in tandem with non-target organ toxicity. The data presented here corroborate the potential therapeutic use of epigenetic inhibitors, as indicated by increased chemotherapeutic-induced toxicity in prostate cancer cells with select co-treatments. However, the DNA methyltransferase and histone deacetylase inhibitors used proved more toxic to kidney cells than prostate cancer cells. Further, we report the novel finding that epigenetic-inhibitor induced toxicity in kidney cells is dependent on the stage of cell growth at the time of exposure, with confluent cells being more susceptible than those in log phase growth. Taken together, these data further validate the need to thoroughly examine non-target toxicity when developing potential chemotherapeutic treatments.
Supplementary Material
The anti-cancer activity and nephrotoxicity of epigenetic inhibitors was determined
Combinatorial therapy altered the efficacy of select chemotherapeutics
This treatment also has the potential to induce nephrotoxic cell injury
Epigenetic inhibitor nephrotoxicity was dependent on the stage of cell growth
Acknowledgments
Funding: This work was supported by the National Institutes of Health (NIH); National Institute of Biomedical Imaging and Bioengineering (NIBIB) (EB016100 to B.S.C., R.D.A.); Sloan Graduate Minority Fellowship (to N.E.S.); Interdisciplinary Toxicology Program, University of Georgia (to N.E.S., R.T.K.).
Abbreviations
- 5-Aza
5-Azacytidine
- CBZ
carbamezapine
- CDK
cyclin-dependent kinase
- CDKN1a
cyclin-dependent kinase inhibitor 1a
- CFZ
carfilzomib
- ChIP
chromatin immunoprecipitation
- DMSO
dimethyl sulfoxide
- DNMT
DNA methyltransferase
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- HEK293
human embryonic kidney 293
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NGS
next-generation sequencing
- NRK
normal rat kidney
- TSA
trichostatin A
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Phillips T. The role of DNA methylation in gene expression. Nature Education. 2008;1 [Google Scholar]
- 2.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature genetics. 2003;33(Suppl):245–54. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 3.Yu N, Wang M. Anticancer drug discovery targeting DNA hypermethylation. Current medicinal chemistry. 2008;15:1350–75. doi: 10.2174/092986708784567653. [DOI] [PubMed] [Google Scholar]
- 4.Mathews LA, Crea F, Farrar WL. Epigenetic gene regulation in stem cells and correlation to cancer. Differentiation; research in biological diversity. 2009;78:1–17. doi: 10.1016/j.diff.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaquero A, Loyola A, Reinberg D. The constantly changing face of chromatin. Science of aging knowledge environment : SAGE KE. 2003;2003:Re4. doi: 10.1126/sageke.2003.14.re4. [DOI] [PubMed] [Google Scholar]
- 6.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 7.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nature genetics. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 8.Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:13225–30. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones PA, Baylin SB. The Epigenomics of Cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L-C, Carroll PR, Dahiya R. Epigenetic Changes in Prostate Cancer: Implication for Diagnosis and Treatment. Journal of the National Cancer Institute. 2005;97:103–15. doi: 10.1093/jnci/dji010. [DOI] [PubMed] [Google Scholar]
- 11.Dobosy JR, Roberts JLW, Fu VX, Jarrard DF. The Expanding Role of Epigenetics in the Development, Diagnosis and Treatment of Prostate Cancer and Benign Prostatic Hyperplasia. The Journal of Urology. 2007;177:822–31. doi: 10.1016/j.juro.2006.10.063. [DOI] [PubMed] [Google Scholar]
- 12.Bhatia N, Agarwal R. Detrimental effect of cancer preventive phytochemicals silymarin, genistein and epigallocatechin 3-gallate on epigenetic events in human prostate carcinoma DU145 cells. The Prostate. 2001;46:98–107. doi: 10.1002/1097-0045(20010201)46:2<98::aid-pros1013>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 13.Yu S, Khor TO, Cheung KL, Li W, Wu TY, Huang Y, et al. Nrf2 expression is regulated by epigenetic mechanisms in prostate cancer of TRAMP mice. PloS one. 2010;5:e8579. doi: 10.1371/journal.pone.0008579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majid S, Kikuno N, Nelles J, Noonan E, Tanaka Y, Kawamoto K, et al. Genistein induces the p21WAF1/CIP1 and p16INK4a tumor suppressor genes in prostate cancer cells by epigenetic mechanisms involving active chromatin modification. Cancer research. 2008;68:2736–44. doi: 10.1158/0008-5472.CAN-07-2290. [DOI] [PubMed] [Google Scholar]
- 15.Ramachandran K, Gopisetty G, Gordian E, Navarro L, Hader C, Reis IM, et al. Methylation-mediated repression of GADD45α in prostate cancer and its role as a potential therapeutic target. Cancer research. 2009;69:1527–35. doi: 10.1158/0008-5472.CAN-08-3609. [DOI] [PubMed] [Google Scholar]
- 16.Li SY, Sun R, Wang HX, Shen S, Liu Y, Du XJ, et al. Combination therapy with epigenetic-targeted and chemotherapeutic drugs delivered by nanoparticles to enhance the chemotherapy response and overcome resistance by breast cancer stem cells. Journal of controlled release : official journal of the Controlled Release Society. 2014 doi: 10.1016/j.jconrel.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 17.Juergens RA, Wrangle J, Vendetti FP, Murphy SC, Zhao M, Coleman B, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer discovery. 2011;1:598–607. doi: 10.1158/2159-8290.CD-11-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silverman LR, Demakos EP, Peterson BL, Kornblith AB, Holland JC, Odchimar-Reissig R, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J Clin Oncol. 2002;20:2429–40. doi: 10.1200/JCO.2002.04.117. [DOI] [PubMed] [Google Scholar]
- 19.Marrocco DL, Tilley WD, Bianco-Miotto T, Evdokiou A, Scher HI, Rifkind RA, et al. Suberoylanilide hydroxamic acid (vorinostat) represses androgen receptor expression and acts synergistically with an androgen receptor antagonist to inhibit prostate cancer cell proliferation. Molecular cancer therapeutics. 2007;6:51–60. doi: 10.1158/1535-7163.MCT-06-0144. [DOI] [PubMed] [Google Scholar]
- 20.Kintzel P. Anticancer Drug—Induced Kidney Disorders. Drug-Safety. 2001;24:19–38. doi: 10.2165/00002018-200124010-00003. [DOI] [PubMed] [Google Scholar]
- 21.Ries F, Klastersky J. Nephrotoxicity induced by cancer chemotherapy with special emphasis on cisplatin toxicity. American journal of kidney diseases : the official journal of the National Kidney Foundation. 1986;8:368–79. doi: 10.1016/s0272-6386(86)80112-3. [DOI] [PubMed] [Google Scholar]
- 22.Peterson BA, Collins AJ, Vogelzang NJ, Bloomfield CD. 5-Azacytidine and renal tubular dysfunction. Blood. 1981;57:182–5. [PubMed] [Google Scholar]
- 23.Dong G, Wang L, Wang CY, Yang T, Kumar MV, Dong Z. Induction of apoptosis in renal tubular cells by histone deacetylase inhibitors, a family of anticancer agents. J Pharmacol Exp Ther. 2008;325:978–84. doi: 10.1124/jpet.108.137398. [DOI] [PubMed] [Google Scholar]
- 24.Nadasi E, Clark JS, Szanyi I, Varjas T, Ember I, Baliga R, et al. Epigenetic modifiers exacerbate oxidative stress in renal proximal tubule cells. Anticancer research. 2009;29:2295–9. [PubMed] [Google Scholar]
- 25.Omlin A, Pezaro C, Gillessen Sommer S. Sequential use of novel therapeutics in advanced prostate cancer following docetaxel chemotherapy. Therapeutic advances in urology. 2014;6:3–14. doi: 10.1177/1756287213509677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siegel DS, Martin T, Wang M, Vij R, Jakubowiak AJ, Lonial S, et al. A phase 2 study of single-agent carfilzomib (PX-171-003-A1) in patients with relapsed and refractory multiple myeloma. Blood. 2012;120:2817–25. doi: 10.1182/blood-2012-05-425934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christman JK. 5-Azacytidine and 5-aza-2'-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–95. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 28.Yoshida M, Horinouchi S, Beppu T. Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. BioEssays : news and reviews in molecular, cellular and developmental biology. 1995;17:423–30. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]
- 29.Beutler AS, Li S, Nicol R, Walsh MJ. Carbamazepine is an inhibitor of histone deacetylases. Life sciences. 2005;76:3107–15. doi: 10.1016/j.lfs.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 30.Li L-C, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–31. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 31.Glenn TC, Nilsen R, Kieran TJ, Finger JW, Pierson TW, Bentley KE, et al. Adapterama I: Universal stubs and primers for thousands of dual-indexed Illumina libraries (iTru & iNext) 2016 doi: 10.7717/peerj.7755. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan H, Reddy MA, Sun GD, Lanting L, Wang M, Kato M, et al. Involvement of p300/CBP and epigenetic histone acetylation in TGF-beta 1-mediated gene transcription in mesangial cells. Am J Physiol-Renal. 2013;304:F601–F13. doi: 10.1152/ajprenal.00523.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morrill BH, Cox L, Ward A, Heywood S, Prather RS, Isom SC. Targeted DNA Methylation Analysis by High Throughput Sequencing in Porcine Peri-attachment Embryos. J Reprod Develop. 2013;59:314–20. doi: 10.1262/jrd.2012-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vigushin DM, Ali S, Pace PE, Mirsaidi N, Ito K, Adcock I, et al. Trichostatin A is a histone deacetylase inhibitor with potent antitumor activity against breast cancer in vivo. Clinical cancer research : an official journal of the American Association for Cancer Research. 2001;7:971–6. [PubMed] [Google Scholar]
- 35.Creusot F, Acs G, Christman JK. Inhibition of DNA methyltransferase and induction of Friend erythroleukemia cell differentiation by 5-azacytidine and 5-aza-2'-deoxycytidine. The Journal of biological chemistry. 1982;257:2041–8. [PubMed] [Google Scholar]
- 36.Festuccia C, Gravina GL, D'Alessandro AM, Muzi P, Millimaggi D, Dolo V, et al. Azacitidine improves antitumor effects of docetaxel and cisplatin in aggressive prostate cancer models. Endocrine-related cancer. 2009;16:401–13. doi: 10.1677/ERC-08-0130. [DOI] [PubMed] [Google Scholar]
- 37.Gravina GL, Marampon F, Sanita P, Mancini A, Colapietro A, Scarsella L, et al. Increased expression and activity of p75NTR are crucial events in azacitidine-induced cell death in prostate cancer. Oncology reports. 2016 doi: 10.3892/or.2016.4832. [DOI] [PubMed] [Google Scholar]
- 38.Liu Z, Marquez M, Nilsson S, Holmberg AR. Incubation with somatostatin, 5-aza decitabine and trichostatin up-regulates somatostatin receptor expression in prostate cancer cells. Oncology reports. 2008;20:151–4. [PubMed] [Google Scholar]
- 39.Rokhlin OW, Glover RB, Guseva NV, Taghiyev AF, Kohlgraf KG, Cohen MB. Mechanisms of cell death induced by histone deacetylase inhibitors in androgen receptor-positive prostate cancer cells. Molecular cancer research : MCR. 2006;4:113–23. doi: 10.1158/1541-7786.MCR-05-0085. [DOI] [PubMed] [Google Scholar]
- 40.Scholpa NE, Zhang X, Kolli RT, Cummings BS. Epigenetic Changes in p21 Expression in Renal Cells after Exposure to Bromate. Toxicological sciences : an official journal of the Society of Toxicology. 2014;141:432–40. doi: 10.1093/toxsci/kfu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scholpa NE, Briggs SB, Wagner JJ, Cummings BS. Cyclin-Dependent Kinase Inhibitor 1a (p21) Modulates Response to Cocaine and Motivated Behaviors. J Pharmacol Exp Ther. 2016;357:56–65. doi: 10.1124/jpet.115.230888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price PM, Safirstein RL, Megyesi J. The cell cycle and acute kidney injury. Kidney Int. 2009;76:604–13. doi: 10.1038/ki.2009.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X, Bull RJ, Fisher J, Cotruvo JA, Cummings BS. The synergistic effect of sodium chlorite and bromochloroacetic acid on BrO3--induced renal cell death. Toxicology. 2011;289:151–9. doi: 10.1016/j.tox.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X, De Silva D, Sun B, Fisher J, Bull RJ, Cotruvo JA, et al. Cellular and molecular mechanisms of bromate-induced cytotoxicity in human and rat kidney cells. Toxicology. 2010;269:13–23. doi: 10.1016/j.tox.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 45.Tikoo K, Ali IY, Gupta J, Gupta C. 5-Azacytidine prevents cisplatin induced nephrotoxicity and potentiates anticancer activity of cisplatin by involving inhibition of metallothionein, pAKT and DNMT1 expression in chemical induced cancer rats. Toxicology letters. 2009;191:158–66. doi: 10.1016/j.toxlet.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 46.Dong G, Luo J, Kumar V, Dong Z. Inhibitors of histone deacetylases suppress cisplatin-induced p53 activation and apoptosis in renal tubular cells. American journal of physiology Renal physiology. 2010;298:F293–300. doi: 10.1152/ajprenal.00410.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanderson L, Taylor GW, Aboagye EO, Alao JP, Latigo JR, Coombes RC, et al. Plasma pharmacokinetics and metabolism of the histone deacetylase inhibitor trichostatin a after intraperitoneal administration to mice. Drug metabolism and disposition: the biological fate of chemicals. 2004;32:1132–8. doi: 10.1124/dmd.104.000638. [DOI] [PubMed] [Google Scholar]
- 48.Troetel WM, Weiss AJ, Stambaugh JE, Laucius JF, Manthei RW. Absorption, distribution, and excretion of 5-azacytidine (NSC-102816) in man. Cancer chemotherapy reports Part 1. 1972;56:405–11. [PubMed] [Google Scholar]
- 49.Broday L, Lee YW, Costa M. 5-azacytidine induces transgene silencing by DNA methylation in Chinese hamster cells. Mol Cell Biol. 1999;19:3198–204. doi: 10.1128/mcb.19.4.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bott SRJ, Arya M, Kirby RS, Williamson M. p21(WAF1/CIP1) gene is inactivated in metastatic prostatic cancer cell lines by promoter methylation. Prostate Cancer P D. 2005;8:321–6. doi: 10.1038/sj.pcan.4500822. [DOI] [PubMed] [Google Scholar]
- 51.Shin JY, Kim HS, Park J, Park JB, Lee JY. Mechanism for inactivation of the KIP family cyclin-dependent kinase inhibitor genes in gastric cancer cells. Cancer Research. 2000;60:262–5. [PubMed] [Google Scholar]
- 52.Estey EH. Epigenetics in clinical practice: the examples of azacitidine and decitabine in myelodysplasia and acute myeloid leukemia. Leukemia. 2013;27:1803–12. doi: 10.1038/leu.2013.173. [DOI] [PubMed] [Google Scholar]
- 53.Follo MY, Finelli C, Mongiorgi S, Clissa C, Bosi C, Testoni N, et al. Reduction of phosphoinositide-phospholipase C beta1 methylation predicts the responsiveness to azacitidine in high-risk MDS. P Natl Acad Sci USA. 2009;106:16811–6. doi: 10.1073/pnas.0907109106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stresemann C, Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. International journal of cancer Journal international du cancer. 2008;123:8–13. doi: 10.1002/ijc.23607. [DOI] [PubMed] [Google Scholar]
- 55.Hayes O, Ramos B, Rodríguez LL, Aguilar A, Badía T, Castro FO. Cell confluency is as efficient as serum starvation for inducing arrest in the G0/G1 phase of the cell cycle in granulosa and fibroblast cells of cattle. Animal Reproduction Science. 2005;87:181–92. doi: 10.1016/j.anireprosci.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 56.Robertson KD, Keyomarsi K, Gonzales FA, Velicescu M, Jones PA. Differential mRNA expression of the human DNA methyltransferases (DNMTs) 1, 3a and 3b during the G(0)/G(1) to S phase transition in normal and tumor cells. Nucleic Acids Research. 2000;28:2108–13. doi: 10.1093/nar/28.10.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mateen S, Raina K, Jain AK, Agarwal C, Chan D, Agarwal R. Epigenetic modifications and p21-cyclin B1 nexus in anticancer effect of histone deacetylase inhibitors in combination with silibinin on non-small cell lung cancer cells. Epigenetics-Us. 2012;7:1161–72. doi: 10.4161/epi.22070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sowa Y, Orita T, Minamikawa S, Nakano K, Mizuno T, Nomura H, et al. Histone deacetylase inhibitor activates the WAF1/Cip1 gene promoter through the Sp1 sites. Biochem Bioph Res Co. 1997;241:142–50. doi: 10.1006/bbrc.1997.7786. [DOI] [PubMed] [Google Scholar]
- 59.Kolisetty N, Bull RJ, Muralidhara S, Costyn LJ, Delker DA, Guo Z, et al. Association of brominated proteins and changes in protein expression in the rat kidney with subcarcinogenic to carcinogenic doses of bromate. Toxicology and applied pharmacology. 2013;272:391–8. doi: 10.1016/j.taap.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 60.Kolisetty N, Delker DA, Muralidhara S, Bull RJ, Cotruvo JA, Fisher JW, et al. Changes in mRNA and protein expression in the renal cortex of male and female F344 rats treated with bromate. Archives of toxicology. 2013;87:1911–25. doi: 10.1007/s00204-013-1052-2. [DOI] [PubMed] [Google Scholar]
- 61.Zhang X, Bull RJ, Fisher J, Cotruvo JA, Cummings BS. The synergistic effect of sodium chlorite and bromochloroacetic acid on BrO3(-)-induced renal cell death. Toxicology. 2011;289:151–9. doi: 10.1016/j.tox.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 62.Abbruzzese JL, Frost P. Studies on the mechanism of the synergistic interaction between 2'-deoxy-5-azacytidine and cisplatin. Cancer chemotherapy and pharmacology. 1992;30:31–6. doi: 10.1007/BF00686482. [DOI] [PubMed] [Google Scholar]
- 63.Cang S, Feng J, Konno S, Han L, Liu K, Sharma SC, et al. Deficient histone acetylation and excessive deacetylase activity as epigenomic marks of prostate cancer cells. International journal of oncology. 2009;35:1417–22. doi: 10.3892/ijo_00000459. [DOI] [PubMed] [Google Scholar]
- 64.Fortson WS, Kayarthodi S, Fujimura Y, Xu H, Matthews R, Grizzle WE, et al. Histone deacetylase inhibitors, valproic acid and trichostatin-A induce apoptosis and affect acetylation status of p53 in ERG-positive prostate cancer cells. International journal of oncology. 2011;39:111–9. doi: 10.3892/ijo.2011.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nature reviews Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 66.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 67.Yegnasubramanian S, Haffner MC, Zhang Y, Gurel B, Cornish TC, Wu Z, et al. DNA hypomethylation arises later in prostate cancer progression than CpG island hypermethylation and contributes to metastatic tumor heterogeneity. Cancer research. 2008;68:8954–67. doi: 10.1158/0008-5472.CAN-07-6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stoyanova T, Goldstein AS. Distinct phases of human prostate cancer initiation and progression can be driven by different cell-types. Cancer cell & microenvironment. 2014;1:e90. doi: 10.14800/ccm.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.