Abstract
The amino acids at the V - J rearrangement junction of TCR are encoded by the D region, and by N or P nucleotides. Together they comprise the NDN region, the specific pMHC selection surface of the TCR β-chain. As an extension of our earlier work on the recall response to influenza M158-66 in HLA-A2 individuals, we have been analyzing the circulating BV19 CD8 T cell repertoires. We observed that NDN regions of the CDR3 often start at positions that are V-region encoded. Here we examine NDN encoded amino acid motifs of BV19 rearrangements in circulating CD8 T cells based on the CDR3 length, the CDR3 start position of the NDN, and the motif length. Motifs that start at V region-encoded positions could be expected to be CDR3 length independent as indeed is the case. Motifs that included sequential proline and glycine showed a CDR3 length independent distribution and examining codon usage indicates that a large proportion of these can be explained by P-nucleotide addition from the 5’ end of the D region. Other examples of skewed codon usage were observed indicating possible additional rearrangement mechanisms. Another pattern of motif distributions was a shift of position along the CDR3 as a function of the CDR3 length. As these data were collected from an older healthy individual they can be used to model successful repertoire selection and to further define characteristics associated with a positive history of responses to pathogen exposures.
Keywords: TCR beta, CDR3 amino acid motifs, NDN rearrangement patterns, P nucleotide addition
INTRODUCTION
The T cell response is based on the recognition of a peptide-MHC complex (pMHC) by the clonotypic T cell receptor under the appropriate ancillary sensing conditions. T cells that have been proven as part of a response can be promoted to memory status, especially if the pathogen is recurring. The latter would include possible cross-reactive recognition. Responses to a pathogen tend to be oligo- or polyclonal and each individual builds up a memory T cell repertoire to the pathogens encountered while the thymus is still active. At any point along this history we refer to the ensemble of available T cells as the repertoire. After thymic involution the repertoire in an individual is considered established and mature.
Advances in DNA sequencing have provided the possibility of examining a large number of members of a repertoire (Robbins et al, 2009, Qi et al, 2014, Wang et al, 2010) and override the need for any special and possibly limiting selection/isolation steps. Indeed some of the first such analyses pointed out the inclusion of the same CDR3 sequence in different CD45-defined repertoires (Wang et al, 2010). In this light we performed an analysis of circulating CD8 T cells expressing the TCRBV19-1 gene from a healthy individual 68 years old at the time of enrollment in our study. A mature repertoire in a healthy individual represents the culmination of the selection events that took place earlier in that individual's life. Examination of such repertoires should provide informative characteristics of CDR3 motifs associated with the antigen exposure history of the individual. The repertoire data are generated from PBMC collected at six times over almost a two year period to minimize short term fluctuations. TCRBV19-1 will be referred to as BV19 in the manuscript.
We chose BV19 because of our extensive history of examining recall repertoire to the matrix-derived influenza A epitope, M158-66. HLA-A2 individuals who have been exposed to influenza generate memory repertoires that predominantly utilize CD8 T cells expressing the BV19 β-chain gene (Moss et al, 1991, Lehner et al, 1995) and a restricted number of α-chain genes (Moss et al, 1991, Naumov et al 2008). One BV19 CDR3 length, L11, predominates and clonotypes of this length encode a conserved RS motif at CDR3 positions 5 and 6. In spite of these restrictions, the memory repertoire is polyclonal (Naumov et al 1998, Zhou et al 2013), and the clonotype distribution can be described as power law-like (Naumov et al, 2003). The polyclonality in part reflects the complexity of a pathogen encounter, and different clonotypes may be invoked at different stages of pathogen density (Naumov et al, 2006). The repertoire is also relatively cross-reactive with ~ 50% of clonotypes capable of recognizing epitopes with structurally similar substitutions in TCR contact residues (Petrova et al, 2011). As substitutions at TCR contact residues become more pronounced, BV19 stops being used in favor of other BV families (Petrova and Gorski, 2012). The individual analyzed here has a strong recall response to M158-66.
The sequence data for all six time points were cleaned and the nucleotide sequence analysis including mathematical description of the clonotypic repertoire distribution is presented elsewhere (Yassai et al, 2016). These analyses showed that the BV19 circulating repertoire has self-similar power law-like characteristics that we had already described for the M158-66 recall response (Naumov et al 2003). We also observed that a large proportion of the NDN regions started within the portion of the CDR3 that is V-encoded. We would expect that if the NDN started at the second or third codon of the V-encoded region, the amino acids observed would be located at a fixed position within the CDR3 region irrespective of CDR3 length. Here we focus on the amino acid defined CDR3 motifs of this dataset, in terms of the same parameters as were established in our analysis of the M1-specific recall repertoire: CDR3 length, position along the CDR3 at which the motif is observed, and add a third parameter, the start position of the NDN region in the CDR3. The NDN start represents the rearrangement point which can be an important parameter in understanding motif geometry. We discuss the data in terms of rearrangement mechanisms and selection.
RESULTS
We use a genetic definition of the CDR3 as the amino acids starting immediately after the conserved cysteine in the V region and extending to the amino acid immediately before the conserved phenylalanine-glycine in the J region. The CDR3 has three genetic components, the 3’ end of V region, the D region, and the 5’ end of the J region. Flanking the D region are untemplated nucleotides added by terminal transferase (N-nucleotides), and presumably less frequently by hairpin loop resolution (P nucleotides). For any V - J pair, the amino acids from the V and J are fixed, with the diversity being a function of the sequence and position of the NDN component. Thus, the portion of the CDR3 derived from the NDN generates the most diversity in the TCR. While the rearrangement site is identified at the nucleotide level, in terms of amino acid motif analysis we describe the NDN start site by the CDR3 amino acid position at which it occurs. Thus, an amino acid is considered to be NDN encoded even if the rearrangement is at the third codon position and result in a synonymous substitution. Our CDR3 definitions and nomenclature have been described previously (Yassai et al 2009). BV19 encodes four amino acids after the cysteine and the first two bases of either Asp or Glu (CASSID/E). Thus, the NDN can start from CDR3 amino acid position 1 to 5. Our final dataset included 12,690 clonotypes represented by 203,660 sequences. None of the clonotypes had an NDN start at CDR3 position 1. Slightly more than half of NDN regions start at CDR3 position 4. The description of the diversity of the repertoire at the clonotype (nucleotide) level as well as the clonotype distribution of the overall repertoire and its subsets is presented elsewhere (Yassai et al, 2015).
NDN-encoded two amino acid motifs
We start by considering the simplest motifs, those composed of two amino acids. The analysis of the response to M158-66 has shown that such motifs are relevant and sufficient for defining responding T cells. The analysis presented is restricted to clonotypes of CDR3 length 10 to 15, which account for 93.2% of the data. Of the 400 possible doublet motifs, 382 (95.5%) were observed. It should be pointed out that longer NDN regions can encode multiple doublet motifs. This form of analysis makes no assumptions about the register of a motif with respect to the NDN start site, and assumes the possibility that the same TCR CDR3 may use doublets motifs in different frames for different responses.
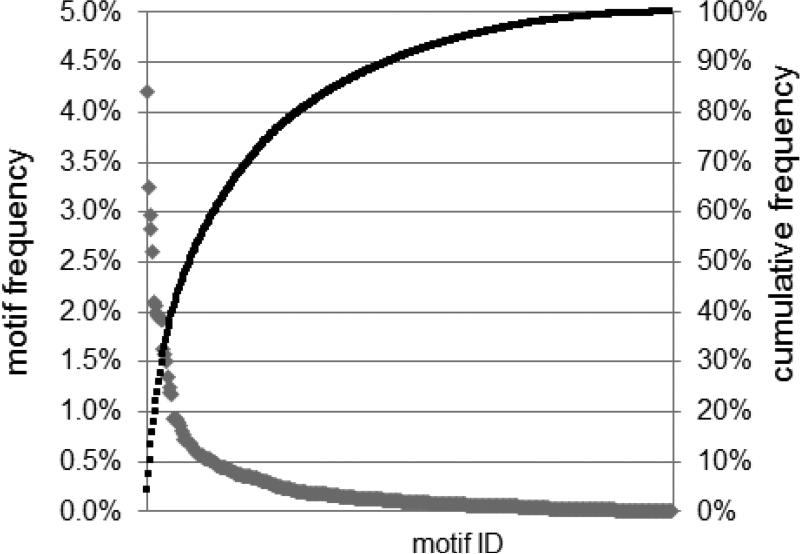
The frequency of the various motifs is shown in Figure 1 as the number of clonotypes which encode the motif as percent of the total number. There are a few motifs present at higher frequencies and a large number present at lower frequencies. A quantitative/mathematical description of motif distributions is presented elsewhere (Yassai et al, 2016). The cumulative distribution of the motifs is shown on the secondary axis. The first 32 motifs represent 50% of the observations. Table 1 enumerates the 30 most frequent motifs, which correspond to 48.6% of the observations in Figure 1. For motifs in which at least one amino acid is D-region encoded, the D-encoded contribution is also shown. The last column shows the percentage of the motif count that is D-encoded. The bottom row shows the number of total doublet motifs and D-encoded motifs for the entire dataset. Just over 25% of the doublet motifs are D-encoded.
Figure 1. Doublet motif frequency.
The frequency of doublet motifs is plotted with respect to the primary Y-axis. The motif ID has been omitted for sake of readability as there are 382 motifs. The cumulative frequency is plotted on the secondary axis.
TABLE 1.
The total count represents the number of clonotypes that encode the doublet motif. The 30 most frequent motifs are shown. The number of motifs whose codon usage is compatible with D-region encoding is also shown. A minimum of five continuous bases were required. Percentages are relative to the entire dataset.
| Total | Dencoded | |||
|---|---|---|---|---|
| motif | count | percent | count | percent |
| GG | 1981 | 4.20% | 1126 | 2.39% |
| SG | 1529 | 3.25% | 1062 | 2.25% |
| AG | 1400 | 2.97% | 1069 | 2.27% |
| TG | 1332 | 2.83% | 890 | 1.89% |
| GT | 1229 | 2.61% | 1023 | 2.17% |
| GL | 992 | 2.11% | 508 | 1.08% |
| RG | 972 | 2.06% | 851 | 1.81% |
| PG | 939 | 1.99% | 0 | 0.00% |
| GS | 919 | 1.95% | 0 | 0.00% |
| QG | 914 | 1.94% | 844 | 1.79% |
| GA | 910 | 1.93% | 555 | 1.18% |
| LA | 904 | 1.92% | 766 | 1.63% |
| GR | 764 | 1.62% | 292 | 0.62% |
| TS | 739 | 1.57% | 451 | 0.96% |
| GQ | 708 | 1.50% | 553 | 1.17% |
| GP | 640 | 1.36% | 0 | 0.00% |
| IG | 585 | 1.24% | 0 | 0.00% |
| LG | 562 | 1.19% | 0 | 0.00% |
| GV | 552 | 1.17% | 0 | 0.00% |
| SS | 441 | 0.94% | 50 | 0.11% |
| GD | 440 | 0.93% | 0 | 0.00% |
| DR | 437 | 0.93% | 326 | 0.69% |
| SP | 434 | 0.92% | 0 | 0.00% |
| VG | 426 | 0.90% | 0 | 0.00% |
| RD | 405 | 0.86% | 0 | 0.00% |
| SL | 380 | 0.81% | 0 | 0.00% |
| RA | 367 | 0.78% | 0 | 0.00% |
| SR | 343 | 0.73% | 0 | 0.00% |
| AS | 340 | 0.72% | 110 | 0.23% |
| MG | 330 | 0.70% | 0 | 0.00% |
| TOTAL | 47117 | 11859 | 25.17% |
Two amino acid motif distributions
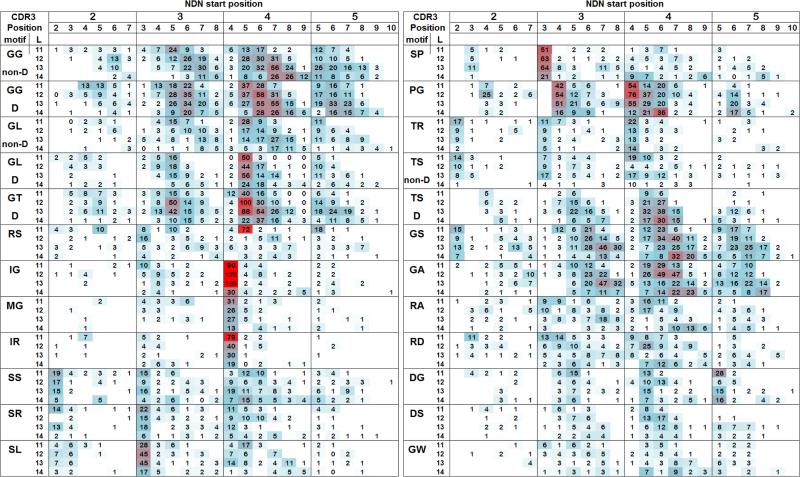
We generated a fine grained analysis of the doublet motifs by counting the number of clonotypes encoding each motif in terms of CDR3 length, NDN start and CDR3 position. To simplify the presentation of the data we focus on a few motifs that are indicative of some general characteristics of the dataset (Fig. 2). The number of clonotypes observed at each CDR3 length is shaded white (zero or low) to blue (medium) to red (high) to reflect relative frequency at that CDR3 length. For each motif, the data are separated by the CDR3 position of the NDN start site and by the actual CDR3 position at which the motif starts. For each motif the data are grouped by CDR3 length to facilitate comparison of length effects. Thus, a GG doublet motif observed at CDR3 position 5 when the NDN start site was at CDR3 position 3, would belong to an xxGG NDN sequence. It should be pointed out that the number of clonotypes at each length differs. Thus, 17 observations of the motif in the L14 data set (N=7539) is approximately equivalent to 31 observations in the L13 dataset (N=14670).
Figure 2. Doublet motif distributions.
The number of observations of clonotypes encoding the identified NDN doublet motif is shown. The motifs are divided on the basis of 1) the NDN start site as defined by the CDR3 amino acid position, and 2) the CDR3 position at which the motif starts. The doublet motif and CDR3 lengths in amino acids (L) are identified in the left column. Possible D-encoded motifs are divided into those where D encoding is identified (D) and those where it is not (non-D). The data for the different CDR3 lengths are stacked for each motif. The shading represents the relative frequency with respect to other motifs at that length from white, none or low, blue, medium, and red highest.
Change in motif distribution as a function of CDR3 length
The highest frequency doublet motif, GG, occurs at any CDR3 length, NDN start, and most CDR3 positions. GG can be encoded by both D1 and D2 using three of the possible sixteen codon combinations. Guanines are also preferentially added by terminal transferase (Basu et al, 1983, Lieber et al, 1998). The distribution of the GG motif moves to higher CDR3 positions at longer CDR3 lengths, as if maintaining the relative position of the motif in the CDR3 (staircase). This is observed at all four NDN starts, and for both D non-encoded and D encoded motifs. For the GL motif, the non-D-encoded distribution also shifts right. However, the D-encoded motifs at NDN start positions 3, 4, and 5 have a maximum at position 5, irrespective of CDR3 length. However, there is also a rightward trend as L increases. For the GT motif, the non-D data are too sparse to define a trend (not shown), whereas the D-encoded data show a position 5 maximum with some rightward movement as CDR3 length increases. It should be pointed out that both GL and GT are encoded by the 5’ end of the D regions, whereas GG is encoded by the 3’ end of the D regions. This staircase effect represents an interesting novel observation.
The influenza-response associated motif, RS
The RS-motif occurs at highest frequency at CDR3 position 5 at L11, NDN start site 4. This would yield the CDR3 sequence, CASSxRS. The higher frequency occurrence at NDN start site 5 yields the CASSIRS sequence, with the Ile being V-encoded. The RS doublet also occurs with higher frequency at start 3, position 3, at L11 and 12. RS doublets occurring at CDR3 positions other than 5 or CDR3 lengths other than 11 are not associated with the recall response to the M158-66 peptide and thus can represent selection for another epitope. Interestingly RS was the only exemplar of a high frequency motif with a clear CDR3 length-dependent distribution.
Motifs occurring at the same position independent of CDR3 length
On the basis of the high frequency of rearrangements within the V region-encoded portion of the CDR3, we expected to observe motifs that occur predominantly at one CDR3 position, that are NDN start site dependent, and length independent. Doublets where the NDN starts at position 4 and in which the first amino acid is Ile (e.g. IG) or Met (e.g. MG) are examples of rearrangements in which the first two bases from the V-region are kept as part of the CDR3. This pattern is maintained for other doublets starting with I or M. If the motifs starting with I or M at CDR3 position 4 are treated as subset of the repertoire and are examined in the same manner as above, the doublets C-terminal to the initial I or M show the same general rightward shift as described above (not shown).
The SS doublet at position 2 start 2 or position 3 start 3 is another example of a V-gene encoded NDN start site, as ~90% of these have the first amino acid encoded as AGC, with the NDN starting at the third base of the codon. All six codons are used relative evenly for encoding the second Ser. However, not all doublet motifs starting with S follow the same pattern. SR shows a stronger preference for position3 start3 and a CDR3 length preference at position 2 start 2 for L11 and L13. SL is also preferentially observed at CDR3 position 3, NDN start site3, but has an even preference for CDR3 position 2 or 3 at NDN start site 2, all in a length-independent manner. SP shows a very strong preference for position3 start3 and is absent at start position 2. SI (not shown) has a pattern similar to that of SP. Sx doublet motifs at CDR3 positions 4 or 5 cannot be V-dependent. They are still best represented at motifs associated with an NDN start at position 4, but the data distribution shows no discernable pattern.
The PG motif is present at high frequencies at CDR3 position 4 when the NDN starts at positions 3 or 4 in CDR3 of length 11, 12, and 13. Of interest is the selection of PG at position 6 for NDN start 4 and 5, predominantly at L14. In general, other motifs starting with P behaved in a similar manner.
Doublet motifs starting with Thr are frequent at the NDN start site for all CDR3 lengths with TR being an example. Interestingly the TS motif that is not encoded by the D region shows a similar pattern as TR with highest frequency at each NDN start site. However, the D-encoded TS motif shows a different and more complicated pattern. Similar to SP, TP is predominantly associated with position 3, start 3 (not shown).
Preferential observation of DG at position 5, start 5, is also based on the first two bases being encoded by the V gene. DG is also observed at position 5 at earlier NDN start positions indicating selection for the particular motif at this position. Dx doublet motifs are more common than Ex motifs, although both could be encoded from the V region. Most other Dx motifs show a similar pattern. Of interest is the DS motif, which does not show a strong position 5 preference but rather shows the rightward shift in frequency at increasing length.
Other distributions
The GS and GA motifs showed a tendency for a rightward shift with increasing CDR3 length, however they both showed a length and start-based variation in the motif frequencies. The GS motif also showed a length-independent increased frequency at NDN start site 2.
The GW motif is present at low frequencies at most CDR3 positions where the NDN start is at CDR3 position 3 and 4. This relatively flat distribution is uncommon.
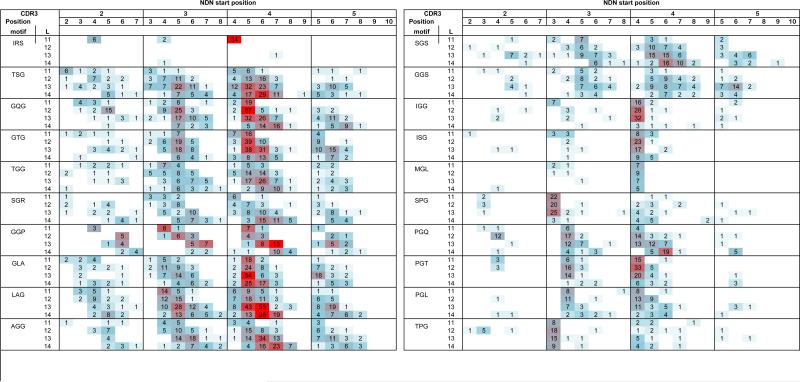
Three amino acid motifs
Three amino acid motifs indicated a similar pattern to that of the two amino acid motifs (Fig 3). The analysis shown is not separated on the basis of D-contribution as was done for the doublet motifs. Examination of longer NDN motifs did not change the generalizations reached from the doublet data. The length restriction of the IRS motif (Fig 3.top panel) is more striking than that of the RS motif. The pattern in which motif frequencies shift to higher CDR3 positions with increasing CDR3 length is present and most obvious with multiple Gly containing motifs. Start position restriction of motifs with Ile or Met in the initial position is maintained. The SP doublet frequency increase at NDN start site 3 continues to be observed at start site 3 in the form of SPx triplets (SPG shown). The PG doublet motif associated with position 4 is now extended to PGx, with PGQ, PGT and PGL being shown. The TP motif association with position 3, start 3, is carried over to TPx motifs with TPG being shown.
Figure 3. Triplet motif distributions.
The number of observations of clonotypes encoding the identified NDN doublet motif is shown. The motif is identified in the left column. The data for the different CDR3 lengths are stacked for each motif. The shading represents the relative frequency with respect to other motifs at that length.
Analysis of motifs 4 and 5 amino acids in length (not shown) generate the same patterns as established with doublet and triplet motifs but represented sparser datasets.
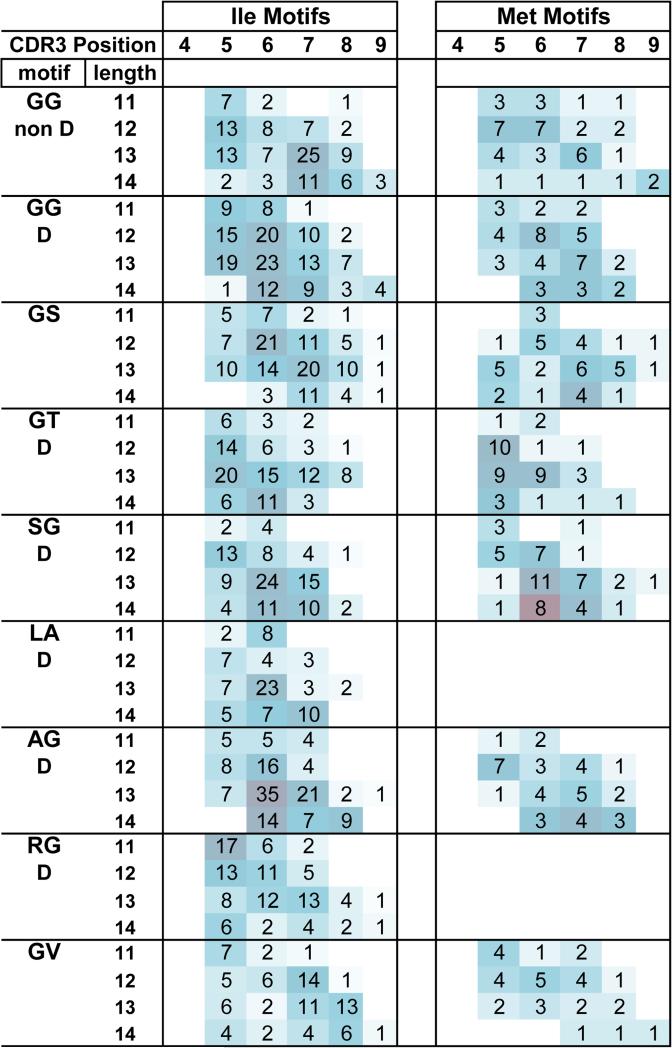
NDN motifs starting with Ile or Met
Of interest was to examine longer motifs that were observed starting at CDR3 position 4 and that were CDR3 length independent to see if they could be broken down into components. We focused on motifs that started with Ile or Met as these were the most common exemplars of motifs that start at position 4 at all CDR3 lengths. We show the frequency distribution of a number of doublet motifs that are all part of an extended motif that starts with these two amino acids in Figure 4. It is evident that while all these motifs start at position 4, the doublets examined, show the same staircase distribution as a function of CDR3 length as was observed for some of the motifs in Figures 2 and 3.
Figure 4. Motifs starting with Ile and Met.
The 1774 clonotypes whose NDN region starts with Ile at CDR3 position 4, and the 676 clonotypes whose NDN region starts with Met at CDR3 position 4 were analyzed for the distribution of doublet motifs in the same manner at presented in Figure 2. Results are shown for doublets present at higher frequencies. The shading represents the relative frequency with respect to other motifs at that length.
A large proportion of motifs starting with PG and associated with D regions can result from P nucleotide addition from the 5’ D-region hairpin loop
Frequent CDR3 motifs may not be exclusively due to selective expansion based on sequence fit for a pathogen peptide. We have already proposed that the more frequent exemplars of the RS motif examined in CD8 single positive thymocytes may partially result from increased initial frequencies, in this particular case due to long P nucleotide addition from J2.7 (Yassai et al, 2011).
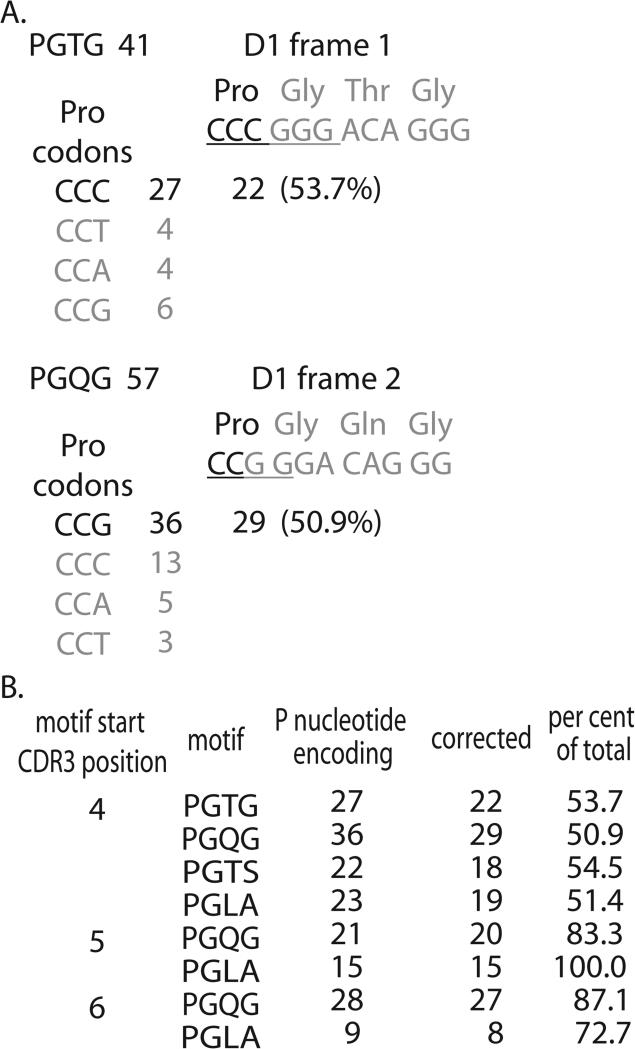
The PG doublet is striking in its high frequency at position 4 (Fig 2) irrespective of CDR3 length. Furthermore the PGQ, PGT, and PGL triplet motifs, all three of which are associated with the start of a D region reading frame, are also high frequency (Fig 3). This indicates a strong possibility that the Pro could be encoded from the D region via P nucleotides. Therefore we examined the codon usage of a number of four amino acid motifs that each start with Pro and also contain the first three amino acids from a D region. Figure 5A shows starts for the PGTG motif of which there are 41 examples in the dataset. The GTG portion represents the first reading frame of D1. Since selection is on the amino acid, and since encoding by N region diversity is considered random, all four codons that encode proline should be equally represented ahead of the D-region encoded glycine. However, if the Pro was the result of resolution of the hairpin loop (p-loop) during rearrangement, the palindromic addition would be encoded by CCC (dark font) and this codon should be in higher frequency. Indeed, this codon was observed 27 times. The three Proline codons that would not be associated with P nucleotide addition were observed an average of five times (4, 4, 6). Thus, N addition would account for ~5 exemplars of each codon if it was the only mechanism functioning. Subtracting this average from the number of CCC codons observed should provide an estimate of the p-loop contribution. The 22 putative examples of the p-loop derived proline (27 total – 5 ascribed to N-addition) represent ~50% (22/41) of the occurrences of proline as part of the motif.
Figure 5. Amino acid encoding data for D-region containing motifs starting with a proline.
A. The analysis of the two motifs derived from D1 for motifs starting at CDR3 position 4 is shown in detail. The PTGT motif was observed 41 times. The encoding expected of a P-loop mechanism (dark font) and the D region is shown with the palindromic nucleotides underlined. The Pro codon usage is shown with the data for the codon expected from the P-loop mechanism in the darker font. The average of the three non-p-loop examples is used to estimate a background, and this is subtracted from the candidate encoding to provide a corrected estimation of the p-loop contribution. The corrected number is shown and used to calculate the percentage of PGTG motifs generated by the mechanism. A similar example is shown for the PGQG motif. B. Summary of data for motifs appearing at three CDR3 positions. PGTG and PGTS frequencies at CDR3 positions 5 and 6 were too low for analysis.
The PGQG motif uses the second D1 reading frame, so the first nucleotide of the D region would be the last base of the Pro codon (second panel of Fig 5A). The palindromic additions (two C in dark font) would result in a Pro encoded by CCG. CCG was encoded 36 times in the 57 cases of Pro adjacent to D1 encoded GQG. The other three encodings of Pro were represented seven times on average. Correcting for the average N addition gives 29 putative p-loop generated exemplars, represents ~50% of the motif observed.
The data are summarized in Figure 5B for the three most frequent CDR3 positions at which the PGL, PGT and PGQ motifs are observed; positions 4, 5, and 6 (see Fig 3). Only the PGQG and PGLA motifs that start at CDR3 positions 5 or 6 are at sufficient frequencies to analyze. Approximately half of the PGxx motifs examined that started at CDR3 position 4 could be accounted for by a P-loop mechanism. For motif starts at CDR3 position 5, a corrected 20 of 21 of PGQG motifs could be accounted for by the p-loop mechanism. For PGLA, all 15 exemplars of 5’-complete D2 regions were associated with a Pro that could arise from the p-loop. For motifs starting at CDR3 position 6, 87% of the PGQG motifs, and ~73% of PGLA motifs could be generated by the p-loop mechanism.
Analysis of PG doublets shows CDR3 length and NDN start position effects on P nucleotide addition
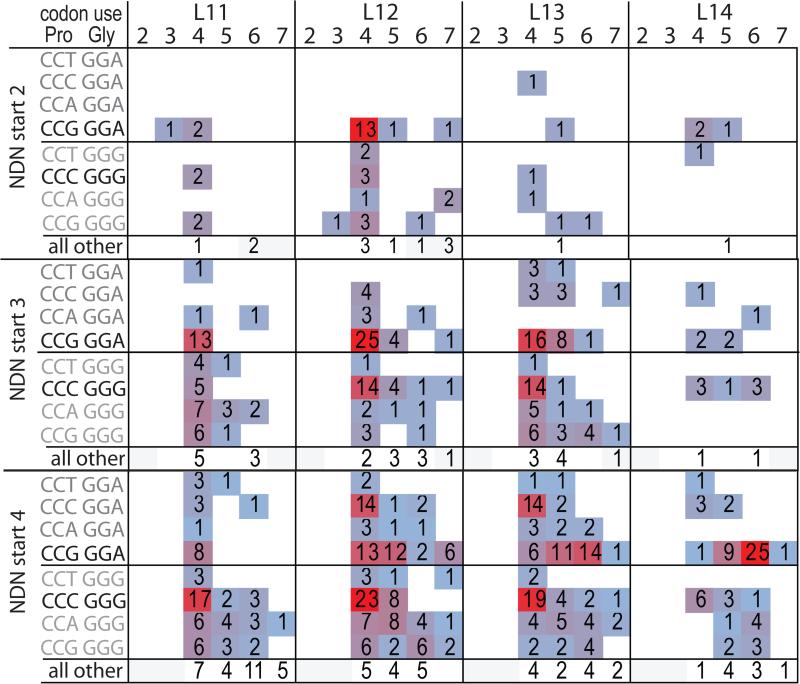
The above analysis of the four amino acid motifs that start with PG led us to analyze the codon use of the PG doublet motifs. Analysis of doublet motifs increases the numbers of observations at the expense of decreasing the confidence that the glycine is D-region derived. Analysis of the PG encoding indicates that in ~85% of cases the glycine was encoded by GGA or GGG, in almost equal proportions. Both of these codons are compatible with possible origin from the 5’ end of either D-region, with GGA representing reading frame 2, and GGG the representing reading frame 1. The codon usage for the PG doublet is shown in more detail in Figure 5 by including the effect of CDR3 length and CDR3 position as identified across the top, and NDN start position as identified on the left. For each NDN start position, the upper panel shows the glycine codon that is D-derived from the 2nd frame, GGA, and the middle panel shows the D-derived 1st frame codon, GGG. It should be noted that in the upper panel, the D region provides the final base in the proline codon as underlined, CCG GGA. The sum of all the other PG codons used at each length and CDR3 position is shown in the bottom panel.
When the NDN starts at CDR3 position 2, there is only strong evidence for increased frequency of the codon associated p-loop generation when the D-derived Gly is GGA-encoded (upper NDN2 start panel) and for CDR3 of L12 and when the doublet occurs at CDR3 position 4. All of the GGA-encoded glycine motifs are associated with the CCG-encoded proline expected from the p-loop mechanism. When the glycine is encoded by GGG, the proline codon usage does not show any preference. Thus, the p-loop insertion is restricted in D-region alignment (only frame 2), for CDR3 length (L12) and CDR3 position (position 4) when the NDN starts deep in the V-encoded region.
For NDN starts at CDR3 position 3, both possible alignments are observed, at multiple CDR3 lengths and for one of these lengths at two CDR3 positions. When the glycine is GGA encoded (upper panel, D-reading frame 2) there is a frequency skew in favor of the CCG encoding at position 4 for L11 to L13. At L13 there is also an increased CCG-encoded proline at position 5. For the GGG proline encoding (bottom panel), at L12 and L13 there is an increase in CCC encoding at CDR3 position 4. After correction for possible N-addition, ~60% and ~40% of the GGG doublets at the two CDR3 lengths respectively can be attributable to the p-loop mechanism. Thus as the p-loop insertion site occurs closer to the end of the V-encoded portion of the CDR3, it occurs more frequently at with less restriction.
Codon use skewing in PG motifs that cannot be explained by current mechanisms
There is an important effect of CDR3 length on observed codon usage when the NDN starts at CDR3 position 4 and the glycine is GGA encoded (top panel). At L12, the Pro codon, CCG (p-loop compatible), is equally represented at positions 4 and 5. Interestingly, at CDR3 position 4 there is an equivalent frequency skewing for the CCC codon which is not compatible with the p-loop mechanism. At L13, CDR3 position 4, the CCC codon skew (non-p loop) remains whereas the CCG skew (p-loop) is not strong. However, the CCG codon skew is high at positions 5 and 6. At L14, the doublet predominantly starts at position 6 and the skew for the CCG (p-loop) encoding is very pronounced. For the GGG encoding of proline, the PG doublets data is similar to that seen for NDN starts at position 3 except that there is also a skewed appearance of the CCCGGG-encoded doublet compatible with the p-loop mechanism at position 4, length 11. Thus, the increased frequency of the CCCGGG palindromic PG codon pair is always associated with CDR3 position 4, and observed at more CDR3 lengths as the NDN start position occurs further away from the conserved V region cysteine. The same is true for the PG codon pair CCGGGA, for NDN starts at CDR3 position 2 and 3. When the NDN starts at CDR3 position 4, the increased frequency of the CCGGGA codon pair moves as a function of CDR3 length, from CDR3 position 4 (L11), to 4 and 5 (L12), to 5 and 6 (L13), to 6 alone at L14. In addition there is an increased frequency of the CCCGGA codon pair at CDR3 position 4, at L12 and L13. This codon pair is not compatible with a simple p-loop mechanism.
We also examined triplet motifs starting with PG that could be D region encoded. The dataset was smaller, but the same patterns were observed, including the CDR3 length and start position effects that show a CCCGGA-codon frequency skew that is not explained by p-loop resolution.
DISCUSSION
T cell repertoires that are the culmination of a long period of adaptive responses can be source of novel information about the mechanisms that generate functional diversity. The decision to analyze the BV19 repertoire was taken because of our existing understanding of the role of BV19 clonotypes encoding the RS motif in response to the HLA-A2-restricted influenza A M158-66 epitope. The subject analyzed has a strong recall response to this epitope. Over 80% of the clonotypes identified in the recall response were observed in this data set. The details of comparisons between the recall and circulating repertoires in the M158-66 response are in preparation. The analysis of the circulating BV19 CDR3 amino acid sequence motifs indicated that the CDR3 length restriction observed for the M158-66 epitope is not a general rule. The IR doublet of the IRS motif can be observed at all lengths examined, with a noted increase at L11. It is the RS doublet that shows the L11 restriction (Fig 2) and this length restriction is obvious for the triplet motif IRS (Fig 3). Other IRx triplet motifs (IRT, IRG, IRQ), while not observed at the same high frequency as IRS, were observed at all lengths (not shown) and therefore the IRS length restriction appears to be a function of negative selection on the RS at lengths other than L11.
We had previously observed that NDN start sites most frequently occur within the V encoded portion of the CDR3. Just over 50% of the NDN starts occur at CDR3 position 4. Depending on the actual codon position of the NDN start will encode the same amino acid as occurs at that CDR3 position (3rd base synonymous substitutions) or the more restricted possibilities associated with second base substitutions (Yassai et al 2016). We expected that motifs that start at V-encoded positions will be observed irrespective of CDR3 length. The data indicate that this is indeed the case. However in addition to the amino acids that would be expected on the basis of the V region sequence two others occurred at relatively high frequency in a CDR3 length independent manner. Proline occurs frequently at position 4, and this is often in conjunction with the beginning of the first two reading frames of the D-regions. Examining the codon frequencies of the proline observed in PG motifs that incorporate the D-region 5’ end, indicated that a minimum of 50% of the proline initiated PG motifs are compatible with P-nucleotide addition. P-nucleotides arise from unsymmetrical resolution of the hairpin loop (p-loop) that closes the coding ends of the DNA after RAG-mediated double strand cleavage. Palindromic CDR3 sequences in human TCR have also been reported in recent high-throughput sequencing data (Srivastava and Robins, 2012).
It should be pointed out that both the evidence presented here with the 5’ end of D-regions, and our previous publication on longer P nucleotide addition from the 5’ end of the J-region (Yassai et al, 2011) involve the same directionality to the process. We searched for CDR3 sequences that would be compatible with P nucleotides from the V- or 3’ portion of the D-region, and found some examples attributable to P-nucleotides being added, but these were observed at a much lower frequency than those from 5’ ends of D or J regions. A similar bias was observed by Srivastava and Robins (2012). This may indicate an underlying asymmetry in the rearrangement process.
Our data indicate a relationship between the extent of trimming of the V-encoded portion of the CDR3 and the extent to which the p-loop insertion can take place. This can be due to the inherent characteristic of the mechanisms, which are still incompletely understood. Alternatively, the initial rearrangements may be unrestricted, but selection at the β-selection stage or later may be responsible for our observations.
There was also PG motif codon utilization skewing observed under certain CDR3 length and NDN start conditions that could not be explained by the p-loop mechanism as we currently understand it. Their presence and significance was evidenced because of the detailed examination of the codon usage based on the different CDR3 parameters. Continuing analysis of highly selected CDR3 sequences in a more detailed manner may bring to light additional mechanisms involved in the rearrangement process.
In this light we observed another high frequency doublet motif, SP, strongly associated with CDR3 position 3 when the NDN started at position 3 (Fig 2). Approximately half of these doublets were part of the extended SPG motif (Fig 3). The encoding of the SP motifs showed that 88% are encoded as AGC, indicative of a rearrangement at the last base of this position.
Threonine also appears at higher frequency at positions 2, 3, and 4 (Yassai et al 2016), and many Tx doublet motifs at these positions occur independent of CDR3 length. TP motif occurs at high frequency at position 3, over a third of the doublets forming the extended TPG motif (Fig 3). In this case the 77% of the Thr are ACC encoded. These observations could either indicate very intense selection or an effect of incoming D-regions on rearrangement site choice.
An interesting motif frequency distribution is represented examples where the highest frequency shifted towards later CDR3 positions as CDR3 length increased (staircase effect). As this motif may be in a different position and thus recognize a novel pMHC component. Alternatively, the shift may recenter the motif in the same relative position in the CDR3 loop. The latter possibility would be dependent on the nature of the sidechain of the N-terminal residue(s) causing the shift. Often these will be glycines as these are the most common amino acids in the CDR3. Glycine stretches in CDR3 regions are proposed to increase TCR structural plasticity (Naumov et al, 2008) in part to explain TCR cross-reactivity (Selin et al 2006). This rightward shift of motifs containing glycines was also observed upon more detailed analysis of NDN-encoded motifs that start with Ile or Met at position 4 (Fig 4). It will be interesting to fit such motif shifts into molecular models.
The data analyzed has been restricted to the motifs observed frequently in BV19 clonotypes. Arguing on the basis of the self-similar characteristic of the BV19 repertoire (Naumov et al, 2003), we hypothesize that increasing the number of observations will reveal similar patterns in those motifs that were below the current analysis threshold. We also hypothesize that the observations made here for BV19 will generalize to the other BV genes in the CD8 repertoire. However, whether the observations made here are applicable to CD4 T cells is an open question.
MATERIALS AND METHODS
The human research conducted here was authorized by Institutional Review Board of BloodCenter of Wisconsin under BC 05-11, “Generation and Decay of Memory T Cells in Older Populations,” which is still open for data analysis. Written consent was obtained. UPN204 was 68 at time of enrollment.
T cell sequence analysis are described in more detail elsewhere (Yassai et al 2016) including error estimation, and steps taken in cleaning the nucleotide sequence data. In brief, PBMC corresponding to six different time points collected over ~ 1.5 years were used. PCR amplification was done using our standard BV19 and BC primers (Maślanka et al 1995). Amplicons were analyzed using high throughput sequencing on a Roche GS-FLX Genome Sequencer at the Human and Molecular Genomic Center Sequencing Facility (www.hmgc.mcw.edu) of the Medical College of Wisconsin. Sequences derived from each sample were downloaded in fasta format and analyzed using our proprietary “CDR3Reader” software, which assigns clonotype names according to the naming convention described by Yassai et al (2009). Data were analyzed using Microsoft Excel. “Clonotype” is used here to refer to the unique CDR3 nucleotide sequence of the TCR β-chain gene, which owing to allelic exclusion identifies a lineage, but because of the likeli5hood of different α-chain rearrangements can refer to multiple distinct T cells.
Figure 6. Amino acid encoding data for PG doublet motifs.
Codon usage for all four Pro possibilities is shown for the two Gly encodings that can be D region-encoded, GGA and GGG. The number of clonotypes using the encoding listed in the left column is divided based on CDR3 length and CDR3 position. Codon encoding for possible P-nucleotide encoding is darker. All other PG encodings are summarized on the bottom row of each panel. The codon usage data is further divided by NDN start. Frequency is displayed by both number and shading.
ACKNOWLEDGEMENTS
This work was supported by NIH Contract NO1 AI-50032 (JG). We thank Dr Liz Worthy and Mike Tschannen at the Human and Molecular Genetics Center of the Medical College of Wisconsin for 454 sequencing. JG has overall responsibility for the study, performed data analysis and wrote the manuscript. MBY and WD prepared PBMC for analysis, oversaw the sequencing, and prepared the sequences for more detailed analyses. The samples were collected and stored by the Center for Human Immunology of the Blood Research Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no commercial or financial conflict of interest.
REFERENCES
- Basu M, Hegde MV, Modak MJ. Synthesis of compositionally unique DNA by terminal deoxynucleotidyl transferase. Biochem Biophys Res Commun. 1983;111:1105–1112. doi: 10.1016/0006-291x(83)91413-4. [DOI] [PubMed] [Google Scholar]
- Lehner PJ, Wang EC, Moss PA, Williams S, Platt K, Friedman SM, Bell JI, Borysiewicz LK. Human HLA-A0201-restricted cytotoxic T lymphocyte recognition of influenza A is dominated by T cells bearing the Vβ17 gene segment. J. Exp. Med. 1995;181:79–91. doi: 10.1084/jem.181.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR, Hesse JE, Mizuuchi K, Gellert M. Lymphoid V(D)J recombination: nucleotide insertion at signal joints as well as coding joints. Proc Natl Acad Sci USA. 1988;85:8588–8592. doi: 10.1073/pnas.85.22.8588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maślanka K, Piatek T, Gorski J, Yassai M, Gorski J. Molecular analysis of T cell repertoires. Spectratypes generated by multiplex polymerase chain reaction and evaluated by radioactivity or fluorescence. Hum Immunol. 1995;44:28–34. doi: 10.1016/0198-8859(95)00056-a. [DOI] [PubMed] [Google Scholar]
- Moss PA, Moots RJ, Rosenberg WM,WM, Rowland-Jones SJ, Bodmer HC, McMichael AJ, Bell JI. Extensive conservation of α-β-chains of the human T cell antigen receptor recognizing HLA-A2 influenza A matrix peptide. Proc. Natl. Acad. Sci. USA. 1991;88:8987–8990. doi: 10.1073/pnas.88.20.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumov YN, Hogan KT, Naumova EN, Pagel JT, Gorski J. A class I MHC-restricted recall response to a viral peptide is highly polyclonal despite stringent CDR3 selection: implications for establishing memory T cell repertoires in “real-world” conditions. J. Immunol. 1998;160:2842–2852. [PubMed] [Google Scholar]
- Naumov YN, Naumova EN, Hogan KT, Selin LK, Gorski J. A fractal clonotype distribution in the CD8+ memory T cell repertoire could optimize potential for immune responses. J. Immunol. 2003;170:3994–4001. doi: 10.4049/jimmunol.170.8.3994. [DOI] [PubMed] [Google Scholar]
- Naumov YN, Naumova EN, Clute SC, Watkin LB, Kota K, Gorski J, Selin LK. Complex T cell memory repertoires participate in recall responses at extremes of antigenic load. J Immunol. 2006;177:2006–2014. doi: 10.4049/jimmunol.177.3.2006. [DOI] [PubMed] [Google Scholar]
- Naumov YN, Naumova EN, Yassai MB, Kota K, Welsh RM, Selin LK. Multiple glycines in TCR alpha-chains determine clonally diverse nature of human T cell memory to influenza A virus. J Immunol. 2008;181:7407–7419. doi: 10.4049/jimmunol.181.10.7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova G, Naumova EN, Gorski J. The polyclonal CD8 T cell response to influenza M158-66 generates a fully connected network of cross-reactive clonotypes to structurally related peptides: a paradigm for memory repertoire coverage of novel epitopes or escape mutants. J Immunol. 2011;186:6390–6397. doi: 10.4049/jimmunol.1004031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova GV, Gorski J. Cross-reactive responses to modified M158-66 peptides by CD8+ T cells that use noncanonical BV genes can describe unknown repertoires. Eur J Immunol. 2012;42:3001–3008. doi: 10.1002/eji.201242596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Q, Liu Y, Cheng Y, Glanville J, Zhang D, Lee JY, Olshen RA, Weyand CM, Boyd SD, Goronzy JJ. Diversity and clonal selection in the human T-cell repertoire. Proc Natl Acad Sci U S A. 2014;111:13139–13144. doi: 10.1073/pnas.1409155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins HS, Campreghe PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, Riddell SR, Warren EH, Carlson CS. Comprehensive assessment of T cell receptor β chain diversity in αβ T cells. Blood. 2009;114:4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava SK, Robins HS. Palindromic nucleotide analysis in human T cell receptor rearrangements. PLoS One. 2012;7:e52250. doi: 10.1371/journal.pone.0052250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selin LK, Brehm MA, Naumov YN, Cornberg M, Kim SK, Clute SC, Welsh RM. Memory of mice and men: CD8+ T-cell cross-reactivity and heterologous immunity. Immunol. Rev. 2006;11:164–181. doi: 10.1111/j.0105-2896.2006.00394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Sanders CM, Yang Q, Schroeder HW, Jr, Wang E, Babrzadeh F, Gharizadeh B, Myers RM, Hudson JR, Jr, Davis RW, Han J. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc Natl Acad Sci USA. 2010;107:1518–1523. doi: 10.1073/pnas.0913939107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassai MB, Naumov YN, Naumova EN, Gorski J. A clonotype nomenclature for T cell receptors. Immunogenetics. 2009;61:493–502. doi: 10.1007/s00251-009-0383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassai M, Bosenko D, Unruh M, Zacharias G, Reed E, Demos W, Ferrante A, Gorski J. Naive T cell repertoire skewing in HLA-A2 individuals by a specialized rearrangement mechanism results in public memory clonotypes. J Immunol. 2011;186:2970–2977. doi: 10.4049/jimmunol.1002764. [DOI] [PubMed] [Google Scholar]
- Zhou V, Yassai MB, Regunathan J, Box J, Bosenko D, Vashishath Y, Demos W, Lee F, Gorski J. The functional CD8 T cell memory recall repertoire responding to the influenza A M1(58-66) epitope is polyclonal and shows a complex clonotype distribution. Hum Immunol. 2013;74:809–817. doi: 10.1016/j.humimm.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]