Abstract
Following the two rounds of whole-genome duplication (WGD) during deuterosome evolution, a third genome duplication occurred in the ray-fined fish lineage and is considered to be responsible for the teleost-specific lineage diversification and regulation mechanisms. As a receptor-regulated SMAD (R-SMAD), the function of SMAD3 was widely studied in mammals. However, limited information of its role or putative paralogs is available in ray-finned fishes. In this study, two SMAD3 paralogs were first identified in the transcriptome and genome of Japanese flounder (Paralichthys olivaceus). We also explored SMAD3 duplication in other selected species. Following identification, genomic structure, phylogenetic reconstruction, and synteny analyses performed by MrBayes and online bioinformatic tools confirmed that smad3a/3b most likely originated from the teleost-specific WGD. Additionally, selection pressure analysis and expression pattern of the two genes performed by PAML and quantitative real-time PCR (qRT-PCR) revealed evidence of subfunctionalization of the two SMAD3 paralogs in teleost. Our results indicate that two SMAD3 genes originate from teleost-specific WGD, remain transcriptionally active, and may have likely undergone subfunctionalization. This study provides novel insights to the evolution fates of smad3a/3b and draws attentions to future function analysis of SMAD3 gene family.
Keywords: Subfunctionalization, Smad3, Genome duplication, Selective pressure
Introduction
SMAD transcription factors are considered as the core of the TGF-β pathway, which are activated by membrane receptors and regulate target genes by transcriptional complexes (Massagué, Seoane & Wotton, 2005). According to the functional variety, SMADs can be divided into three subfamilies: receptor-activated SMADs (R-SMADs: SMAD1, SMAD2, SMAD3, SMAD5, and SMAD8), common mediator SMADs (Co-SMADs: SMAD4), and inhibitory SMADs (I-SMADs: SMAD6, SMAD7) (Miyazono, Ten Dijke & Heldin, 2000; Moustakas, Souchelnytskyi & Heldin, 2001). SMADs contain two conserved structural domains—the N-terminal MH1 domain and the C-terminal MH2 domain—and a linker region with multiple phosphorylation sites between the two domains (Shi & Massagué, 2003). As an R-SMAD, SMAD3 has various functions including regulating the pathogenesis of diseases and even cancer progression (Bonniaud et al., 2004; Ge et al., 2011; Roberts et al., 2006).
To date, SMAD genes have been found only in eumetazoan animals. Four SMAD genes have been identified in fruit fly Drosophila melanogaster, seven in nematode Caenorhabditis elegans and eight in human Homo sapiens (Newfeld & Wisotzkey, 2006). Nevertheless, much more novel SMADs exist in teleost, such as SMAD2, SMAD3, and SMAD6. As described in previous studies, these novel isoforms are highly similar and evidently caused by an additional round of whole-genome duplication (WGD) in teleost fishes rather than fragment duplication or alternative splicing (Huminiecki et al., 2009; Pang et al., 2011; Sato & Nishida, 2010).
The increased complexity and genome size of vertebrates have been derived from two verified rounds of WGD, which are thought to play major roles in promoting diversification and evolutionary innovation within vertebrates (Cañestro et al., 2013; Dehal & Boore, 2005; Hoegg & Meyer, 2005; Hoffmann, Opazo & Storz, 2011). Moreover, a fish-specific genome duplication (FSGD or 3R) occurred ∼350 million years ago which resulted in new copies of genes and provided genetic basis for evolutionary innovation (Meyer & Schartl, 1999; Meyer & Van de Peer, 2005; Taylor et al., 2001; Vandepoele et al., 2004). According to the study of an additional lineage-specific WGD in salmonids and some cyprinids, the climatic cooling and subsequent evolution of anadromy are major catalyst for salmonid speciation rather than the WGD itself (Berthelot et al., 2014; Glasauer & Neuhauss, 2014; Macqueen & Johnston, 2014). It is indicated that 3R is not directly associated with species diversity but 3R-derived duplicate genes may have subsequently undergone dosage effects regulation, lineage-specific evolution and been divergent in regulatory mechanisms, expression pattern and evolutionary rates after lineage diversification (Braasch, Salzburger & Meyer, 2006; Mulley, Chiu & Holland, 2006; Sato & Nishida, 2010; Siegel et al., 2007). According to the duplication-degeneration-complementation (DDC) model, duplicated genes will undergo three main fates, namely, nonfunctionalization (duplicates dying out as pseudogenes), subfunctionalization (partitioning of ancestral gene functions on the duplicates) and neofunctionalization (assigning a novel function to one of the duplicates) (Force et al., 1999).
In teleost zebrafish, it has been reported that SMAD3 is required for regenerative capacity of heart and mesendoderm induction (Chablais & Jaźwińska, 2012; Jia et al., 2008). However, researches investigating the origination and evolution fates of teleost SMAD3 paralogs are deficient. To better understand the origin and functional diversification of SMAD3 paralogs in teleost, we identified whole set of SMAD gene family sequences from the transcriptome and genome of Japanese flounder Paralichthys olivaceus and other teleosts. Next, gene structure, phylogenetic reconstruction, and chromosomal synteny analyses of vertebrate SMAD3 genes were performed to study the origin and evolution of two SMAD3 genes in teleosts. The analyses of motif scan, positive selection, and expression profiles of the two SMAD3 genes in Japanese flounder were performed to identify potential functional changes for the duplicated SMAD3 genes within the lineage of teleost. The results provide evidences to the duplication of teleost SMAD gene family derived from the WGD and possible subfunctionalization of teleost-specific duplicated SMAD3 genes. Moreover, this study lays the foundation for evolutionary and functionary studies of SMAD3 gene family in teleosts.
Materials and Methods
Ethics statement
Japanese flounder samples were collected from local aquatic farms. This research was conducted in accordance with the Institutional Animal Care and Use Committee of the Ocean University of China and the China Government Principles for the Utilization and Care of Vertebrate Animals Used in Testing, Research, and Training (State science and technology commission of the People’s Republic of China for No. 2, October 31, 1988. http://www.gov.cn/gongbao/content/2011/content_1860757.htm).
Fish
Healthy two-year-old Japanese flounder (three females and three males) were selected from a larger cohort population. The flounders were anesthetized and killed by severing spinal cord. Organs, including heart, liver, spleen, kidney, brain, gill, muscle, intestine, and gonad, were collected in triplicate from each fish. Samples were immediately frozen by liquid nitrogen and stored at −80 °C for extraction of total RNA.
Identification of SMAD genes in Japanese flounder and other species
The SMAD coding sequences of Amazon molly (Poecilia formosa), Japanese medaka (Oryzias latipes), and Nile tilapia (Oreochromis niloticus) were used as queries for local TBLASTX searches with an E-value of 1e-5 against the genome (Q. Zhang, 2016, unpublished data) and transcriptome (SRA, accession number: SRX500343) of Japanese flounder to identify the DNA sequences of SMAD. The coding sequences of other species (green spotted puffer Tetraodon nigroviridis, tongue sole Cynoglossus semilaevis, bicolor damselfish Stegastes partitus, gubby Poecilia reticulate, platyfish Xiphophorus maculatus) were obtained from NCBI or Ensembl database, and the accession numbers were shown in Table S1. Different abbreviations for SMAD gene orthologs exist in NCBI. For convenience reasons, SMAD was used for all vertebrate orthologs and smad3a and smad3b for the variants identified in teleost in this study.
Phylogenetic analysis of SMAD genes
In order to study the phylogenetic relations and evolution fates of SMAD genes, a phylogenetic reconstruction including all SMAD isoforms of vertebrates was performed. The whole coding sequences of SMAD isoforms were aligned by Clustal X with the default parameters (Chenna et al., 2003). Sequences used to construct phylogenetic trees were retrieved from NCBI and Ensembl (species names, gene names and accession numbers are available in Table S1). Appropriate substitution model of molecular evolution, GTR+I+G, was determined by JModelTest v2.1.4 (Darriba et al., 2012). Phylogenetic tree was constructed by Bayesian method which was implemented in MrBayes v3.2.2 (Huelsenbeck & Ronquist, 2001; Ronquist et al., 2012).
A second phylogenetic reconstruction including only SMAD3 isoforms was performed to confirm phylogenetic relations between smad3a and smad3b. The whole coding sequences of all SMAD3 isoforms were aligned by Clustal X with default parameters. Phylogenetic trees were constructed by Bayesian method and maximum likelihood method with GTR+I+G substitution model, respectively. Maximum likelihood phylogeny was constructed by phyML v3.1, and the branching reliability was tested by bootstrap resampling with 1,000 replicates (Guindon et al., 2010).
Genomic structure, motif, and synteny analysis of teleost SMAD3 paralogs
The exon-intron information of teleost SMAD3 genes was obtained by BLASTn with coding sequences against the corresponding genomic sequences. Figures of teleost SMAD3 genomic structures were obtained using an online tool Gene Structure Display Server 2.0 (GSDS: http://gsds.cbi.pku.edu.cn) with size and position information of each exon and intron (Hu et al., 2015). Alignments of the teleost smad3a/3b amino acids sequences were constructed by Clustal X (Chenna et al., 2003). MEME was applied to identify motifs of the SMAD3 coding sequences to test the possible functional divergence between teleost smad3a and smad3b (Bailey et al., 2009). Synteny comparisons of the fragments harboring SMAD3 and flanking genes were performed to test the genes’ syntenic conservation. Flanking genes of smad3a/3b used in the synteny analysis were extracted from online genome databases, such as Ensembl or NCBI. The genes were mapped according to their relative locations in the chromosome for the synteny analysis. In order to test the chromosomal synteny conservation between human and green spotted puffer, an online synteny database was used to analyze the syntenic conservation between human chromosome Hsa15, where human SMAD3 gene was located, and genome of green spotted puffer (Catchen, Conery & Postlethwait, 2009).
Positive selection test of teleost smad3a and smad3b genes
As described in the manual of PAML v4.7, the species used in the analysis should have a close genetic relationship, absolute minimum species used in the analysis is four or five, and the dS summed over all branches on the tree should be larger than 0.5. In accordance with the criteria, nine teleost species were selected to explore differences in selective pressure between smad3a and smad3b. Phylogenetic tree used for positive selection analysis was constructed by MrBayes with GTR+I+G model. Various site models (M0, M1a, M2a, M3, M7, M8, and M8a) in CODEML were applied to estimate the ratio of nonsynonymous to synonymous substitutions (dN/dS = ω) and likelihood ratio tests (LRTs) to confirm the sites that were under positive selection (Yang, 2007). In the nested site models, M0 and M3 were compared to detect whether the dN/dS was changing. Moreover, the comparisons of M2a/M1a, M8/M7, and M8a/M8 were used to estimate the positively selected sites.
RNA extraction, cDNA synthesis and distribution pattern of SMAD3 genes in Japanese flounder
Total RNA was extracted from organ samples with Trizol reagent, according to the manufacturer’ protocol (Invitrogen, Carlsbad, CA, USA). Then, DNase I (TaKaRa, Dalian, China) was applied to remove genomic DNA; protein was removed by RNAclean RNA kit (Biomed, Beijing, China). Agarose gel electrophoresis and NanoPhotometer Pearl (Implen GmbH, Munich, Germany) were used to evaluate the quality and quantity of RNA. cDNA was synthesized by M-MLV kit (TaKaRa) in accordance with the manufacturer’s instructions.
Two specific primer pairs (Table S2) for Japanese flounder smad3a and smad3b were designed by an online tool IDT (http://www.idtdna.com/Primerquest/Home/Index), in the untranslated region of both genes. Each sample was pooled from three male or female individuals, which was performed in triplicate. Pre-experiment was conducted to test product specificity. quantitative real-time PCR (qRT-PCR) was performed with SYBR Premix Ex Taq II (TaKaRa) by LightCycler 480 (Roche, Forrentrasse, Switzerland) with thermocycling consisted of 5 min at 94 °C for pre-incubation, followed by 40 cycles at 94 °C (15 s) and 60 °C (45 s). As described, 18S rRNA was used as the reference gene to determine the relative expression (Zhong et al., 2008). The target gene’s expression was analyzed by 2−ΔΔCt method. qRT-PCR data were statistically analyzed by one-way ANOVA with SPSS 20.0. P < 0.05 was considered to indicate statistical significance. All data were expressed as mean ± standard error of the mean (SEM).
Results
Identification of Japanese flounder SMAD gene family
To study the evolution history of SMAD gene family, a whole set of SMAD gene family (SMAD1, two SMAD2, two SMAD3, four SMAD4, SMAD5, two SMAD6, SMAD7, and SMAD8) was identified from Japanese flounder transcriptome and genome by TBLASTX with 1e-5. Integrated SMAD coding sequences in other specific species, such as human, house mouse, chicken, Nile tilapia, Japanese medaka, and zebrafish were also found from multiple databases. In this gene family, SMAD2, SMAD3, and SMAD6 duplicates were identified only in teleosts, except for spotted gar, while other species had only single copy. It could be speculated that these novel SMAD isoforms might derive from a teleost-specific WGD.
Genomic structures of teleost SMAD3
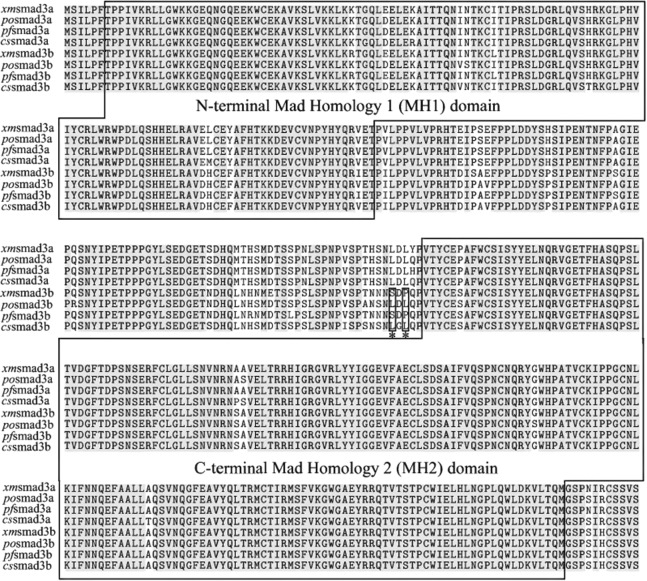
The differences between teleost smad3a and smad3b genes were shown in gene structure graphic. smad3b gene had one more exon but shorter gene full-length than smad3a because of one long intron in smad3a. The lengths of each corresponding exon were highly conserved between these two genes. Besides, the fourth exon in smad3a was divided into two exons in smad3b by an additional intron resulting in the different exon numbers (Fig. S1). Multiple sequence alignment of deduced full-length SMAD3a and SMAD3b protein sequence was constructed with selected teleost species (i.e., Japanese medaka, Japanese flounder, Amazon molly, and tongue sole) to test the similarity between the paralogs. The sequences of SMAD3a and SMAD3b were highly similar and Mad homology domains were conserved in both N-terminal and C-terminal of SMAD3a/3b. The specific mutations at MH1 and MH2 domains between the two paralogs such as tyrosine to phenylalanine, proline to serine, and in linker region such as serine to alanine, histidine to proline might lead to functional disparities between the two isoforms (Fig. 1). Taken together, it could be implied that the differences between the two SMAD3 isoforms might lead to a functional diversification.
Figure 1. Sequence alignment of the deduced SMAD3a and SMAD3b protein sequences.
Identical amino acids are in gray background. Two conserved domains, namely, MH1 and MH2 domains are marked in the figure. Two significantly positively selected sites are indicated by asterisk.
Phylogenetic reconstruction of SMAD
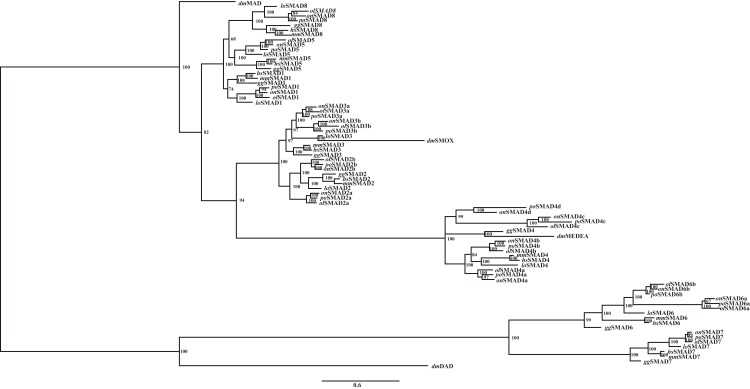
A phylogenetic tree constructed by Bayesian method with GTR+I+G model of all vertebrate SMAD isoforms was applied to test the evolution fates of SMAD gene family. Results indicated the relationships of SMAD gene family members. SMAD genes distinctly divided into four subfamilies, namely, SMAD1/5/8, SMAD2/3, SMAD4, and SMAD6/7. In addition, four fruit fly genes (Mad, Smox, Medea, and Dad) were clustered to homologous subfamily. Teleost-specific SMAD2, SMAD3, and SMAD6 paralogs were clustered into one clade and then clustered with other orthologs implying that SMAD gene family had duplicated and were most likely resulted from the WGD (Fig. 2).
Figure 2. Phylogenetic analyses of SMAD gene family.
Phylogenetic tree constructed by Bayesian method with GTR+I+G, MCMC = 800,000. Numbers at the tree nodes are posterior probabilities. Po, Paralichthys olivaceus; Hs, Homo sapiens; Mm, Mus musculus; Gg, Gallus gallus; On, Oreochromis niloticus; Lo, Lepisosteus oculatus; Dm, Drosophila melanogaster; Ol, Oryzias latipes.
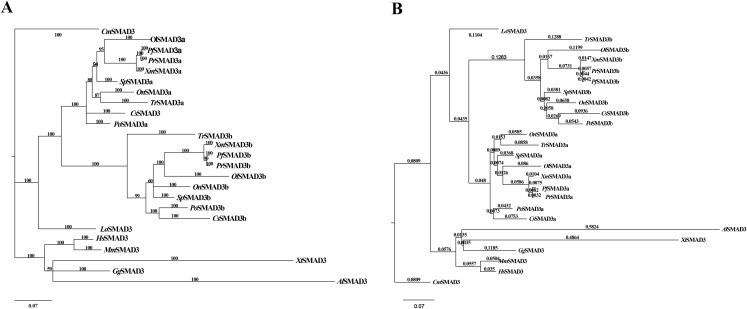
To study the duplication of SMAD3 in the lineage of teleost, a second phylogenetic reconstruction including only SMAD3 isoforms was performed by Bayesian and maximum likelihood method, respectively with GTR+I+G model and the elephant shark SMAD3 sequence was set as an outgroup. Similar topologies were inferred by the two programs (Fig. 3). Results indicated that these species gathered into two main clades, i.e., teleost clade and non-teleost clade. Teleost SMAD3 genes could be clearly divided into two well-conserved clusters, i.e., smad3a and smad3b, whereas spotted gar SMAD3 occupied a separate clade. Moreover, the branch length of smad3b cluster was longer than smad3a, and the branch lengths of frog SMAD3 and anole lizard SMAD3 were also longer than those in the other classes. These results implied that duplication of SMAD3 was widespread in teleost except for spotted gar, a species never experienced the teleost-specific WGD (Braasch et al., 2016). Combining these two phylogenetic trees, it could be deduced that teleost SMAD3 paralogs might originate from the teleost-specific WGD.
Figure 3. Phylogenetic analyses of SMAD3.
(A) Phylogenetic tree constructed by Bayesian method with GTR+I+G, MCMC = 300,000. Elephant shark SMAD3 was used as the outgroup. Numbers at the nodes are posterior probabilities. (B) Phylogenetic tree constructed by phyML with GTR+I+G. Elephant shark SMAD3 was used as the outgroup. Numbers at the nodes are bootstrap values with 1,000 replicates. Cm, Callorhinchus milii; Lo, Lepisosteus oculatus; Tr, Takifugu rubripes; Ol, Oryzias latipes; Xm, Xiphophorus maculatus; Pr, Poecilia reticulata; Pf, Poecilia formosa; Sp, Stegastes partitus; On, Oreochromis niloticus; Cs, Cynoglossus semilaevis; Po, Paralichthys olivaceus; Al, Anolis carolinensis; Xt, Xenopus tropicalis; Gg, Gallus; Mm, Mus musculus; Hs, Homo sapiens.
Synteny analysis of SMAD3 genes
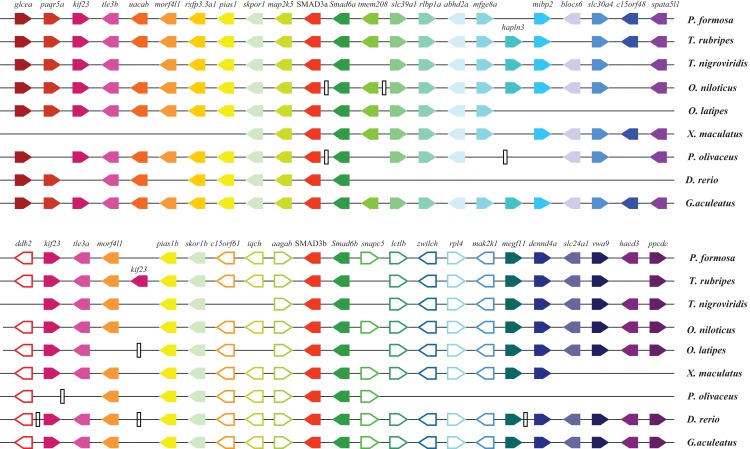
Synteny analyses were applied to testify the speculation that teleost SMAD3 paralogs originated from teleost-specific WGD rather than fragment duplication or alternative splicing. As shown in Fig. 4, the SMAD3 genes and adjoining genes were placed according to their relative locations on the scaffold or chromosome. The genes near teleost smad3a were highly conserved in teleost and shared the same direction. Similar results could be obtained from synteny analysis of smad3b. Comparison between smad3a and smad3b revealed that only SMAD3, SMAD6, and other five upstream genes (kif23, tle3, morf41l, pias1, and skor1) were conserved, and no paralogous genes were retained in the downstream of the two SMAD3 paralogs. Long fragments consisting of several genes were lost in the upstream and downstream regions of smad3a in Japanese medaka and platyfish.
Figure 4. Chromosomal segments showing the synteny of smad3a and smad3b in teleost.
Different genes are represented by different colored pentagons and gene order is determined according to their relative positions in the chromosome or scaffold; the gene names are placed on top of the pentagons. The direction of pentagons indicate the gene direction, the vertical lines represent noncontiguous regions on the scaffold or chromosome.
After comparing teleost smad3a and smad3b neighborhood gene sequences with other species, such as elephant shark, spotted gar, frog, anole lizard, chicken, and eucherian species, we found that adjoining genes of teleost smad3b shared highly conserved synteny with these species (Fig. S2). However, conserved synteny between teleost smad3a and SMAD3 sequences in these species existed only in the upstream region. Thus, the teleost smad3b gene was more likely to be the ancestor SMAD3 gene and smad3a derived from genome duplication. Moreover, long fragments were lost in the upstream regions of smad3b in spotted gar, elephant shark and anole lizard, and an inversion existed in the upstream region of teleost and other species such as chicken and ectherian. To some extent, the synteny results suggested that two teleost SMAD3 paralogs originated from WGD.
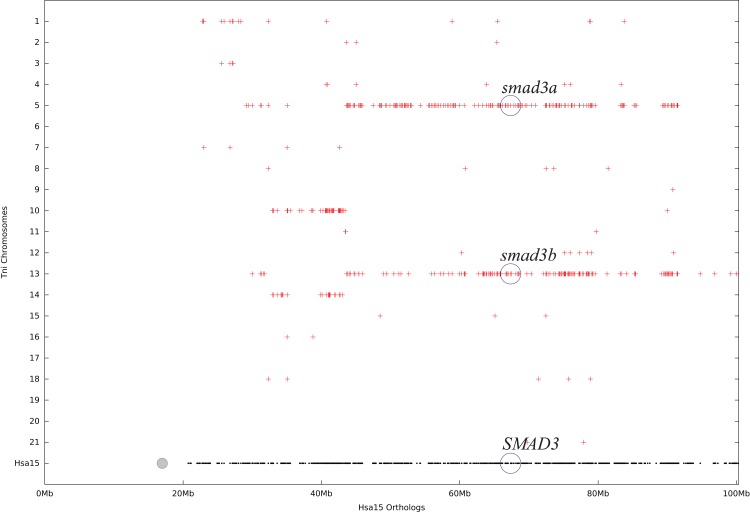
To further verify the speculation, a chromosomal synteny test was performed between the genome of human and green spotted puffer using online synteny databases to determine whether SMAD3 paralogs originated from WGD. The human SMAD3 gene was located at chromosome Hsa15, whereas green spotted puffer smad3a and smad3b were located at chromosomes Tni5 and Tni13, respectively. According to the chromosomal synteny dot plot (Fig. 5), the human SMAD3 region showed double conserved synteny with green spotted puffer chromosomes Tni5 and Tni13. In addition, highly conserved synteny could also be detected between green spotted puffer chromosomes Tni5 and Tni13. Combining the gene neighborhood analysis and chromosomal synteny analysis, it could be speculated that these two SMAD3 paralogs originated from a common ancestral gene during the teleost-specific WGD.
Figure 5. Chromosome synteny of teleost SMAD3 paralogs.
The dot plot between human SMAD3 region and green spotted puffer genome indicates that human SMAD3 gene region in chromosome Hsa15 shares double conservation with green spotted puffer smad3a gene region in chromosome Tni5 and smad3b gene region in chromosome Tni13. The black dots represent segments in human chromosome Hsa15, and the red dots represent the conserved segments in green spotted puffer genome which mostly located in chromosome Tni5 and Tni13.
Molecular evolution of teleost smad3a and smad3b
Multiple single nucleotide polymorphisms and random mutagenesis are found in protein sequence, and each substitution may have the potential to affect protein function (Ng & Henikoff, 2003). To test this potential in SMAD3 genes, we examined the protein sequence evolution in smad3a/3b by codon-based models in PAML with three model pairs M0/M3, M1a/M2a, and M7/M8 (Table 1). The phylogenetic tree used for positiveselection analysis is shown in Fig. S3.
Table 1. Results of sites model analyses on the teleost SMAD3 Bayesian gene tree.
| Tree | Model | lnL | κ | Null | LRT | df | P-value | Site | BEB |
|---|---|---|---|---|---|---|---|---|---|
| SMAD3a | M0 | −4,084.415 | 3.41131 | NA | |||||
| M1a | −4,066.241 | 3.11193 | NA | ||||||
| M2a | −4,066.241 | 3.11192 | M1a | 0 | 2 | 1.0000 | |||
| M3 | −4,063.075 | 3.06893 | M0 | 42.68 | 4 | 0.0000 | |||
| M7 | −4,067.745 | 3.02656 | NA | ||||||
| M8a | −4,065.670 | 3.08595 | NA | ||||||
| M8 | −4,065.563 | 3.08136 | M7 | 4.364 | 2 | 0.1128 | |||
| M8a | 0.214 | 1 | 0.6437 | ||||||
| SMAD3b | M0 | −4,822.425 | 2.24532 | NA | |||||
| M1a | −4,735.378 | 2.37286 | NA | ||||||
| M2a | −4,734.233 | 2.34986 | M1a | 2.29 | 2 | 0.3182 | |||
| M3 | −4,715.059 | 2.29131 | M0 | 214.732 | 4 | 0.0000 | |||
| M7 | −4,734.999 | 2.32501 | NA | ||||||
| M8a | −4,718.436 | 2.30812 | NA | ||||||
| M8 | −4,714.852 | 2.29008 | M7 | 40.294 | 2 | 0.0000 | 219 (L) | 0.991** | |
| 221 (L) | 0.994** | ||||||||
| M8a | 7.168 | 1 | 0.0074 |
According to the comparison between M3 and M0, it could be reflected that M3 was significantly better than M0 (P < 0.05) and M0 was rejected. Thus, both smad3a and smad3b were under variable alternative pressure. Comparison groups of M1a/M2a, M7/M8 and M8/M8a were then used to test the likelihood ratio. According to the results of chi2 program in PAML, we found that LRT significantly differed in M7/M8 pairs (P < 0.05) of smad3b and M8 showed better fitness to the data of smad3b, which was confirmed by an additional test between M8 and M8a. Bayes Empirical Bayes (BEB) method of M8 was applied to calculate the post probabilities of sites to identify the positive selected amino acid sites when LRT differed significantly. Five candidate positive selected sites were identified, two of which were significantly positively selected (219L**, 221L**, posterior probability > 0.99) in smad3b. However, no significantly positive selected sites were found in smad3a by comparisons (M1a/M2a, M7/M8 and M8/M8a). Positive selected sites were located in the linker region (Fig. 1). As a proline rich region, linker region determined the properties and functions of SMAD proteins by phosphorylation (Kamato et al., 2013). Mutations in this region may affect function of smad3b and result in functional diversification between smad3a and smad3b. These results indicated that there might be a functional diversification between teleost smad3a and smad3b.
Expression pattern of Japanese flounder smad3a and smad3b genes
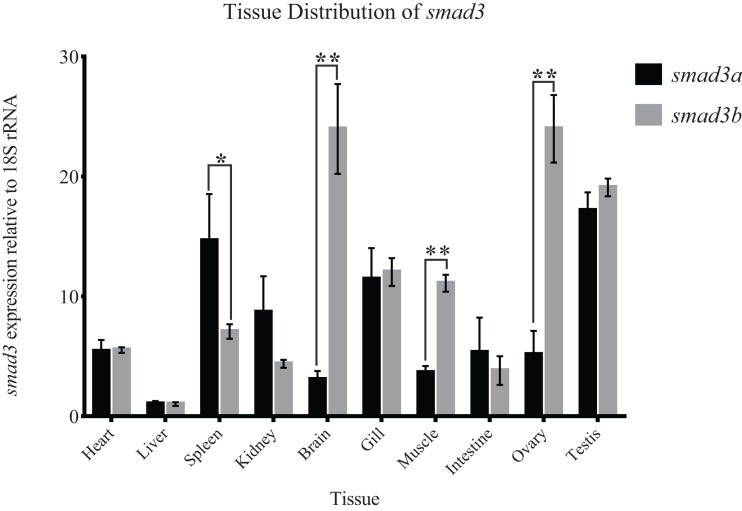
Different organ-specific expression patterns of Japanese flounder smad3a and smad3b could be deduced from the qRT-PCR results (Fig. 6). The smad3a and smad3b genes were expressed in all organs with different expression levels in specific organs. The expression levels of smad3a in spleen, kidney, and intestine were higher than smad3b, which were contrary to those in brain, gill, muscle, ovary, and testis. In addition, tissue distributions between smad3a and smad3b were significantly different in spleen, brain, muscle and ovary. The different expression patterns of Japanese flounder smad3a and smad3b might reflect the function divergence between the two genes.
Figure 6. Expression patterns of smad3a and smad3b in Japanese flounder relative to 18S rRNA.
Data are shown as mean ± SEM (n = 3). Asterisks indicate statistical significance (P < 0.05).
Discussion
Vertebrate SMAD gene family expanded by WGD
As a family of intracellular mediators, SMAD proteins are activated by serine/threonine kinase receptors and translocate signals from cytoplasm into nucleus to regulate the expression of target genes together with several kinds of transcription factors (Attisano & Lee-Hoeflich, 2001). With the development of next generation sequencing (NGS), we can easily identify SMAD gene sequences in genomes of related species. Eight SMADs were widely identified in vertebrate, namely, SMAD1 to SMAD8, and divided into three subgroups by particular function (Miyazono, Ten Dijke & Heldin, 2000; Moustakas, Souchelnytskyi & Heldin, 2001). In addition, much more lineage-specific paralogs are identified. For example, frog has two SMAD4s, SMAD4α, and SMAD4β (Masuyama et al., 1999), and additional SMAD2, SMAD3, SMAD6 isoforms exist in extant teleost fishes. It could be speculated that these novel isoforms are derived from WGD.
Phylogenetic analysis suggested that the SMAD gene family had undergone an expansion and was divided into SMAD1/5/8, SMAD2/3, SMAD4, and SMAD6/7 subfamilies. As described in previous studies, the variation of SMAD gene family is a result of WGD (Huminiecki et al., 2009). 2R-WGD affected the majority of signaling genes with strongest effect on developmental pathways, such as receptor tyrosine kinases, Wnt, and TGF-β. The genes retained after 2R were enriched in protein interaction domains and multifunctional signaling modules of Ras and MAP-kinase cascades (Huminiecki & Conant, 2012). The phylogenetic analysis indicated that teleost-specific duplication occurred because more than one SMAD2, SMAD3, SMAD4 and SMAD6 paralogs existed in the teleost. In addition, different SMAD orthologs of each species shared conserved phylogenetic relationships. These findings suggested that SMAD gene family had undergone an expansion through WGD and resulted in many teleost-specific paralogs, such as smad3a and smad3b.
Two teleost SMAD3 isoforms are generated from 3R
New genes can arise from plenty of events, such as exon shuffling, gene duplication, retroposition, mobile elements, lateral gene transfer, gene fusion/fission and de novo origination (Long et al., 2003). Duplication is a pivotal means of generating genetic material for innovation. The two rounds of WGD have resulted in complex innovations in cellular networks. During the WGD period, many classes of genes such as transcription factors, kinases, ribosomal protein, and cyclins, are duplicated more frequently (Aury et al., 2006; Seoighe & Wolfe, 1999; Wolfe & Shields, 1997). As a core member of the TGF-β family, SMAD has undergone duplication in 2R with an additional duplication in teleosts. Many paralogs have been degraded in the evolution history, which was consistent with dosage hypothesis (Edger & Pires, 2009; Papp, Pál & Hurst, 2003; Qian & Zhang, 2008), while additional SMADs, such as SMAD2s, SMAD3s, SMAD4s, and SMAD6s, were retained in teleost implying that teleost smad3a and smad3b are generated and retained from 3R.
According to the neighborhood gene synteny results, we could find that parts of adjoining genes were conserved around smad3a/3b, such as smad6, skor1, pias, morf41, tle3, and kif23 (Fig. 4). The teleost smad3b flanking genes shared more conservation with chondrichthyes, amphibians, reptiles, birds, mammals and spotted gar which had only one SMAD3 gene and did not undergo 3R (Braasch et al., 2016). The results support that smad3a and smad3b originate by WGD and that smad3b is more likely be the ancestral one.
To further verify the hypothesis, two SMAD3s genes were identified in green spotted puffer genome. Chromosomal synteny results demonstrated that chromosome Tni5 and Tni13 in green spotted puffer shared high conservation with human chromosome Hsa15 (Fig. 5). As described in an aforementioned study, Tni13 matched Tni5 and Tni19 because chromosome Tni5 was derived from ancestral chromosome E, and chromosome Tni19 was derived from ancestral chromosome F by 3R. The other copy of chromosome E and chromosome F developed into chromosome Tni13 by fusion or fragmentation (Jaillon et al., 2004). Combining with neighborhood gene synteny result, it supports that two teleost SMAD3 isoforms are generated from 3R.
Teleost smad3a and smad3b genes differ in phylogenesis and gene structure
In order to confirm the relations and differences between teleost smad3a and smad3b, phylogenetic reconstruction and genomic structure analyses were performed. As shown in Fig. 3, the phylogenetic relationships were clearly displayed. Under the teleost clade, smad3a was clearly separated from smad3b, and spotted gar SMAD3 occupied a separate clade. Thus, it could be speculated that smad3a and smad3b diverged from a common ancestral gene. According to the branch length, we could speculate that the evolution speed of teleost smad3b was faster than that of smad3a. Amphibians and reptiles occupy a special status in the evolution history: their genes may have suffered extreme selective pressure during this period, which might be reflected by the branch lengths of African clawed frog and anole lizard clades.
We also explored the conservation of gene structures of smad3a and smad3b in teleosts. Results showed that the genomic structures of smad3a and smad3b were highly conserved with minor differences: the fourth exon in smad3a was divided into two exons in smad3b by an additional intron and introns in smad3a were much longer than those in smad3b. The different exons were located in the linker region in SMAD3 protein sequence which is supposed to regulate gene function. Introns that interrupt eukaryotic protein-coding sequences are important indicators in eukaryotic evolution, and intron gain or loss rate has a significant correlation with the coding sequence evolution rate (Carmel et al., 2007). Genes can be regulated by various intronic properties, such as sequence, length, position and splicing, in the aspects of transcription initiation, transcription termination, genome organization and transcription regulation (Chorev & Carmel, 2012). Duplicate genes tend to diverge in regulatory and coding regions by amino acid-altering substitutions and/or alterations in exon-intron structure. Besides, the structural divergences have played a more important role in the evolution of duplicate genes than nonduplicate genes (Xu et al., 2012). Taken together, it could be inferred that the sequences of smad3a and smad3b had been changed under selection pressure and the different introns may regulate the expression and function of these two paralogs.
Subfunctionalization of the Japanese flounder smad3a and smad3b
Gene duplication is believed to be the primary source of new genes and plays significant roles in the evolution of genomes and genetic systems (Gu et al., 2003; Ohno, 1970). The sequences and structures of gene pairs that originated from duplication will undergo rapid changes and have different evolutionary rates (Zhang, Zhang & Rosenberg, 2002). Generally, accumulation of detrimental mutations is probably the most common fate of one of the duplicates while the other copy maintains initial function (Cañestro et al., 2013). As described in the DDC model, degenerative mutations increased the probability of a duplicated gene’s preservation, and the preservation of duplicated genes is related to the partitioning of ancestral functions but not to the evolution of new functions. Therefore, duplicated genes underwent three fates, namely, nonfunctionalization, subfunctionalization and neofunctionalization (Force et al., 1999). In our study, we hypothesized that smad3a and smad3b had undergone subfunctionalization in teleost after 3R according to plenty of analyses.
The gene structure of smad3a and smad3b differed in the lengths of introns and the fourth exon which was interrupted by an additional intron in smad3b. Beyond that, sequences of smad3a and smad3b shared high similarity. Motif scan results by MEME showed no significant differences (Fig. S4). These findings suggested that the two duplicates did not acquire novel functions.
As to the domains of SMAD3, the conserved exons between smad3a and smad3b were located in MH1 and MH2 domains, respectively, whereas the different exons were located in the linker region of SMAD3. MH1 and MH2 domains contributed to the DNA binding and protein association function of SMAD3, whereas linker region was rich of phosphorylation sites and acted as a negative region of SMAD3 function (Attisano & Lee-Hoeflich, 2001). In accordance with the selection pressure analysis, both SMAD3 paralogs were under purifying selection, and two significant mutated amino acid sites (219L, 221L) were predicted to have undergone strong positive selection among linker region in smad3b relative to smad3a. These positive selected sites may affect the function of linker region to regulate the interaction between MH1 and MH2 domains, resulting in the functional divergence between smad3a and smad3b.
The roles of SMAD3, such as in regulation of fibronectin, wound healing process, renal disease, and cell proliferation, were widely discussed in previous studies (Isono et al., 2002; Schiller, Javelaud & Mauviel, 2004; Ten Dijke et al., 2002; Wang, Koka & Lan, 2005). In the present study, we found that the expression patterns of smad3a and smad3b were different. Their expression levels were significantly different in brain, muscle, ovary, and spleen. The expression level of smad3b was higher than that of smad3a in brain, muscle, and ovary, contrary to those in spleen. Thus, a functional divergence exists between smad3a and smad3b in some biological processes in Japanese flounder. Further comprehensive in vitro and in vivo studies should be conducted to elucidate the functions of smad3a and smad3b in suitable model teleosts.
Overall, we conclude that the functions of teleost smad3a and smad3b shared conserved domain function, while functional divergence occurred because of their different evolution fates. These results provided sufficient evidence to conclude that the duplicated SMAD3 genes had undergone subfunctionalization after 3R.
Conclusion
In summary, we explored the origin of teleost smad3a/3b genes in this study and reported the general duplication of SMAD3 resulting from 3R in teleosts. This study is the first to investigate the duplication of teleost SMAD3 genes by selection pressure analysis. The results suggested probable subfunctionalization of duplicated teleost SMAD3 genes. This study provided adequate information and new insights into the teleost SMAD3 genes for further functional research in teleost.
Supplemental Information
Boxes indicate the exons and the lines indicate the introns. The length of boxes and lines are based on gene length.
Different genes are represented by different colored pentagons and gene order is determined according to their relative positions in the chromosome or scaffold; the gene names are placed on top of the pentagons. The direction of pentagons indicates the gene direction, and vertical lines represent noncontiguous regions on the scaffold or chromosome.
Tr, Takifugu rubripes; Ol, Oryzias latipes; Xm, Xiphophorus maculatus; Po, Paralichthys olivaceus; On, Oreochromis niloticus; Pf, Poecilia formosa; Cs, Cynoglossus semilaevis; Pr, Poecilia reticulata; Sp, Stegastes partitus.
Funding Statement
This work was supported by the National High-Tech Research and Development Program of China (2012AA10A402) and the National Natural Science Foundation of China (No. 31272646). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Xinxin Du performed the experiments, analyzed the data, wrote the paper, prepared figures and/or tables, reviewed drafts of the paper.
Yuezhong Liu prepared figures and/or tables, reviewed drafts of the paper.
Jinxiang Liu contributed reagents/materials/analysis tools, reviewed drafts of the paper.
Quanqi Zhang contributed reagents/materials/analysis tools, reviewed drafts of the paper.
Xubo Wang conceived and designed the experiments, reviewed drafts of the paper.
Animal Ethics
The following information was supplied relating to ethical approvals (i.e., approving body and any reference numbers):
This research was conducted in accordance with the Institutional Animal Care and Use Committee of the Ocean University of China and the China Government Principles for the Utilization and Care of Vertebrate Animals Used in Testing, Research, and Training (State science and technology commission of the People’s Republic of China for No. 2, October 31, 1988: http://www.gov.cn/gongbao/content/2011/content_1860757.htm).
Data Deposition
The following information was supplied regarding data availability:
The raw data has been supplied as Supplemental Dataset Files.
References
- Attisano & Lee-Hoeflich (2001).Attisano L, Lee-Hoeflich ST. The smads. Genome Biology. 2001;2(8):1–8. doi: 10.1186/gb-2001-2-8-reviews3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aury et al. (2006).Aury J-M, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Ségurens B, Daubin V, Anthouard V, Aiach N, Arnaiz O, Billaut A, Beisson J, Blanc I, Bouhouche K, Câmara F, Duharcourt S, Guigo R, Gogendeau D, Katinka M, Keller A-M, Kissmehl R, Klotz C, Koll F, Le Mouël A, Lepère G, Malinsky S, Nowacki M, Nowak JK, Plattner H, Poulain J, Ruiz F, Serrano V, Zagulski M, Dessen P, Bétermier M, Weissenbach J, Scarpelli C, Schächter V, Sperling L, Meyer E, Cohen J, Wincker P. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444(7116):171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- Bailey et al. (2009).Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research. 2009;37(Suppl 2):W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelot et al. (2014).Berthelot C, Brunet F, Chalopin D, Juanchich A, Bernard M, Noël B, Bento P, Da Silva C, Labadie K, Alberti A, Aury J-M, Louis A, Dehais P, Bardou P, Montfort J, Klopp C, Cabau C, Gaspin C, Thorgaard GH, Boussaha M, Quillet E, Guyomard R, Galiana D, Bobe J, Volff J-N, Genêt C, Wincker P, Jaillon O, Crollius HR, Guiguen Y. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nature Communications. 2014;5:3657. doi: 10.1038/ncomms4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonniaud et al. (2004).Bonniaud P, Kolb M, Galt T, Robertson J, Robbins C, Stampfli M, Lavery C, Margetts PJ, Roberts AB, Gauldie J. Smad3 null mice develop airspace enlargement and are resistant to TGF-β-mediated pulmonary fibrosis. Journal of Immunology. 2004;173(3):2099–2108. doi: 10.4049/jimmunol.173.3.2099. [DOI] [PubMed] [Google Scholar]
- Braasch et al. (2016).Braasch I, Gehrke AR, Smith JJ, Kawasaki K, Manousaki T, Pasquier J, Amores A, Desvignes T, Batzel P, Catchen J, Berlin AM, Campbell MS, Barrell D, Martin KJ, Mulley JF, Ravi V, Lee AP, Nakamura T, Chalopin D, Fan S, Wcisel D, Cañestro C, Sydes J, Beaudry FEG, Sun Y, Hertel J, Beam MJ, Fasold M, Ishiyama M, Johnson J, Kehr S, Lara M, Letaw JH, Litman GW, Litman RT, Mikami M, Ota T, Saha NR, Williams L, Stadler PF, Wang H, Taylor JS, Fontenot Q, Ferrara A, Searle SM, Aken B, Yandell M, Schneider I, Yoder JA, Volff J-N, Meyer A, Amemiya CT, Venkatesh B, Holland PW, Guiguen Y, Bobe J, Shubin NH, Di Palma F, Alfoldi J, Lindblad-Toh K, Postlethwait JH. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nature Genetics. 2016;48(4):427–437. doi: 10.1038/ng.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch, Salzburger & Meyer (2006).Braasch I, Salzburger W, Meyer A. Asymmetric evolution in two fish-specifically duplicated receptor tyrosine kinase paralogons involved in teleost coloration. Molecular Biology and Evolution. 2006;23(6):1192–1202. doi: 10.1093/molbev/msk003. [DOI] [PubMed] [Google Scholar]
- Cañestro et al. (2013).Cañestro C, Albalat R, Irimia M, Garcia-Fernàndez J. Impact of gene gains, losses and duplication modes on the origin and diversification of vertebrates. Seminars in Cell & Developmental Biology. 2013;24(2):83–94. doi: 10.1016/j.semcdb.2012.12.008. [DOI] [PubMed] [Google Scholar]
- Carmel et al. (2007).Carmel L, Rogozin IB, Wolf YI, Koonin EV. Evolutionarily conserved genes preferentially accumulate introns. Genome Research. 2007;17(7):1045–1050. doi: 10.1101/gr.5978207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen, Conery & Postlethwait (2009).Catchen JM, Conery JS, Postlethwait JH. Automated identification of conserved synteny after whole-genome duplication. Genome Research. 2009;19(8):1497–1505. doi: 10.1101/gr.090480.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chablais & Jaźwińska (2012).Chablais F, Jaźwińska A. The regenerative capacity of the zebrafish heart is dependent on TGFβ signaling. Development. 2012;139(11):1921–1930. doi: 10.1242/dev.078543. [DOI] [PubMed] [Google Scholar]
- Chenna et al. (2003).Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Research. 2003;31(13):3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorev & Carmel (2012).Chorev M, Carmel L. The function of introns. Frontiers in Genetics. 2012;3:55. doi: 10.3389/fgene.2012.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba et al. (2012).Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods. 2012;9(8):772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal & Boore (2005).Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biology. 2005;3(10):e2500. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger & Pires (2009).Edger PP, Pires JC. Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Research. 2009;17(5):699–717. doi: 10.1007/s10577-009-9055-9. [DOI] [PubMed] [Google Scholar]
- Force et al. (1999).Force A, Lynch M, Pickett FB, Amores A, Yan Y-L, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151(4):1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge et al. (2011).Ge X, McFarlane C, Vajjala A, Lokireddy S, Ng ZH, Tan CK, Tan NS, Wahli W, Sharma M, Kambadur R. Smad3 signaling is required for satellite cell function and myogenic differentiation of myoblasts. Cell Research. 2011;21(11):1591–1604. doi: 10.1038/cr.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasauer & Neuhauss (2014).Glasauer SMK, Neuhauss SCF. Whole-genome duplication in teleost fishes and its evolutionary consequences. Molecular Genetics and Genomics. 2014;289(6):1045–1060. doi: 10.1007/s00438-014-0889-2. [DOI] [PubMed] [Google Scholar]
- Gu et al. (2003).Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li W-H. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421(6918):63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- Guindon et al. (2010).Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hoegg & Meyer (2005).Hoegg S, Meyer A. Hox clusters as models for vertebrate genome evolution. Trends in Genetics. 2005;21(8):421–424. doi: 10.1016/j.tig.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Hoffmann, Opazo & Storz (2011).Hoffmann FG, Opazo JC, Storz JF. Whole-genome duplications spurred the functional diversification of the globin gene superfamily in vertebrates. Molecular Biology and Evolution. 2011;29(1):303–312. doi: 10.1093/molbev/msr207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu et al. (2015).Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck & Ronquist (2001).Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Huminiecki & Conant (2012).Huminiecki L, Conant GC. Polyploidy and the evolution of complex traits. International Journal of Evolutionary Biology. 2012;2012(4):292068. doi: 10.1155/2012/292068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huminiecki et al. (2009).Huminiecki L, Goldovsky L, Freilich S, Moustakas A, Ouzounis C, Heldin C-H. Emergence, development and diversification of the TGF-βsignalling pathway within the animal kingdom. BMC Evolutionary Biology. 2009;9(1):28. doi: 10.1186/1471-2148-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono et al. (2002).Isono M, Chen S, Hong SW, Iglesias-de la Cruz MC, Ziyadeh FN. Smad pathway is activated in the diabetic mouse kidney and Smad3 mediates TGF-β-induced fibronectin in mesangial cells. Biochemical and Biophysical Research Communications. 2002;296(5):1356–1365. doi: 10.1016/S0006-291X(02)02084-3. [DOI] [PubMed] [Google Scholar]
- Jaillon et al. (2004).Jaillon O, Aury J-M, Brunet F, Petit J-L, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, Nicaud S, Jaffe D, Fisher S, Lutfalla G, Dossat C, Segurens B, Dasilva C, Salanoubat M, Levy M, Boudet N, Castellano S, Anthouard V, Jubin C, Castelli V, Katinka M, Vacherie B, Biémont C, Skalli Z, Cattolico L, Poulain J, de Berardinis V, Cruaud C, Duprat S, Brottier P, Coutanceau J-P, Gouzy J, Parra G, Lardier G, Chapple C, McKernan KJ, McEwan P, Bosak S, Kellis M, Volff J-N, Guigó R, Zody MC, Mesirov J, Lindblad-Toh K, Birren B, Nusbaum C, Kahn D, Robinson-Rechavi M, Laudet V, Schachter V, Quétier F, Saurin W, Scarpelli C, Wincker P, Lander ES, Weissenbach J, Crollius HR. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431(7011):946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Jia et al. (2008).Jia S, Ren Z, Li X, Zheng Y, Meng A. smad2 and smad3 are required for mesendoderm induction by transforming growth factor-β/nodal signals in zebrafish. Journal of Biological Chemistry. 2008;283(4):2418–2426. doi: 10.1074/jbc.M707578200. [DOI] [PubMed] [Google Scholar]
- Kamato et al. (2013).Kamato D, Burch ML, Piva TJ, Rezaei HB, Rostam MA, Xu S, Zheng W, Little PJ, Osman N. Transforming growth factor-β signalling: role and consequences of Smad linker region phosphorylation. Cellular Signalling. 2013;25(10):2017–2024. doi: 10.1016/j.cellsig.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Long et al. (2003).Long M, Betrán E, Thornton K, Wang W. The origin of new genes: glimpses from the young and old. Nature Reviews Genetics. 2003;4(11):865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- Macqueen & Johnston (2014).Macqueen DJ, Johnston IA. A well-constrained estimate for the timing of the salmonid whole genome duplication reveals major decoupling from species diversification. Proceedings of the Royal Society B: Biological Sciences. 2014;281(1778):20132881. doi: 10.1098/rspb.2013.2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué, Seoane & Wotton (2005).Massagué J, Seoane J, Wotton D. Smad transcription factors. Genes & Development. 2005;19(23):2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Masuyama et al. (1999).Masuyama N, Hanafusa H, Kusakabe M, Shibuya H, Nishida E. Identification of two Smad4 proteins in Xenopus. Journal of Biological Chemistry. 1999;274(17):12163–12170. doi: 10.1074/jbc.274.17.12163. [DOI] [PubMed] [Google Scholar]
- Meyer & Schartl (1999).Meyer A, Schartl M. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Current Opinion in Cell Biology. 1999;11(6):699–704. doi: 10.1016/S0955-0674(99)00039-3. [DOI] [PubMed] [Google Scholar]
- Meyer & Van de Peer (2005).Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD) Bioessays. 2005;27(9):937–945. doi: 10.1002/bies.20293. [DOI] [PubMed] [Google Scholar]
- Miyazono, Ten Dijke & Heldin (2000).Miyazono K, Ten Dijke P, Heldin C-H. TGF-β signaling by Smad proteins. Advances in Immunology. 2000;75:115–157. doi: 10.1016/S0065-2776(00)75003-6. [DOI] [PubMed] [Google Scholar]
- Moustakas, Souchelnytskyi & Heldin (2001).Moustakas A, Souchelnytskyi S, Heldin C-H. Smad regulation in TGF-β signal transduction. Journal of Cell Science. 2001;114(24):4359–4369. doi: 10.1242/jcs.114.24.4359. [DOI] [PubMed] [Google Scholar]
- Mulley, Chiu & Holland (2006).Mulley JF, Chiu C-H, Holland PWH. Breakup of a homeobox cluster after genome duplication in teleosts. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(27):10369–10372. doi: 10.1073/pnas.0600341103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newfeld & Wisotzkey (2006).Newfeld SJ, Wisotzkey RG. Smad Signal Transduction. Dordrecht: Springer; 2006. Molecular evolution of Smad proteins; pp. 15–35. [Google Scholar]
- Ng & Henikoff (2003).Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Research. 2003;31(13):3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno (1970).Ohno S. Evolution by Gene Duplication. Berlin, Heidelberg: Springer; 1970. [Google Scholar]
- Pang et al. (2011).Pang K, Ryan JF, Baxevanis AD, Martindale MQ. Evolution of the TGF-beta signaling pathway and its potential role in the ctenophore, Mnemiopsis leidyi. PLoS ONE. 2011;6(9):e2500. doi: 10.1371/journal.pone.0024152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp, Pál & Hurst (2003).Papp B, Pál C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424(6945):194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- Qian & Zhang (2008).Qian W, Zhang J. Gene dosage and gene duplicability. Genetics. 2008;179(4):2319–2324. doi: 10.1534/genetics.108.090936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts et al. (2006).Roberts AB, Tian F, Byfield SD, Stuelten C, Ooshima A, Saika S, Flanders KC. Smad3 is key to TGF-β-mediated epithelial-to-mesenchymal transition, fibrosis, tumor suppression and metastasis. Cytokine & Growth Factor Reviews. 2006;17(1–2):19–27. doi: 10.1016/j.cytogfr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Ronquist et al. (2012).Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato & Nishida (2010).Sato Y, Nishida M. Teleost fish with specific genome duplication as unique models of vertebrate evolution. Environmental Biology of Fishes. 2010;88(2):169–188. doi: 10.1007/s10641-010-9628-7. [DOI] [Google Scholar]
- Schiller, Javelaud & Mauviel (2004).Schiller M, Javelaud D, Mauviel A. TGF-β-induced SMAD signaling and gene regulation: consequences for extracellular matrix remodeling and wound healing. Journal of Dermatological Science. 2004;35(2):83–92. doi: 10.1016/j.jdermsci.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Seoighe & Wolfe (1999).Seoighe C, Wolfe KH. Yeast genome evolution in the post-genome era. Current Opinion in Microbiology. 1999;2(5):548–554. doi: 10.1016/S1369-5274(99)00015-6. [DOI] [PubMed] [Google Scholar]
- Shi & Massagué (2003).Shi Y, Massagué J. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell. 2003;113(6):685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Siegel et al. (2007).Siegel N, Hoegg S, Salzburger W, Braasch I, Meyer A. Comparative genomics of ParaHox clusters of teleost fishes: gene cluster breakup and the retention of gene sets following whole genome duplications. BMC Genomics. 2007;8(1):312. doi: 10.1186/1471-2164-8-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor et al. (2001).Taylor JS, Van de Peer Y, Braasch I, Meyer A. Comparative genomics provides evidence for an ancient genome duplication event in fish. Philosophical Transactions of the Royal Society B: Biological Sciences. 2001;356(1414):1661–1679. doi: 10.1098/rstb.2001.0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Dijke et al. (2002).Ten Dijke P, Goumans M-J, Itoh F, Itoh S. Regulation of cell proliferation by Smad proteins. Journal of Cellular Physiology. 2002;191(1):1–16. doi: 10.1002/jcp.10066. [DOI] [PubMed] [Google Scholar]
- Vandepoele et al. (2004).Vandepoele K, De Vos W, Taylor JS, Meyer A, Van de Peer Y. Major events in the genome evolution of vertebrates: paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(6):1638–1643. doi: 10.1073/pnas.0307968100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Koka & Lan (2005).Wang W, Koka V, Lan HY. Transforming growth factor-β and Smad signalling in kidney diseases. Nephrology. 2005;10(1):48–56. doi: 10.1111/j.1440-1797.2005.00334.x. [DOI] [PubMed] [Google Scholar]
- Wolfe & Shields (1997).Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387(6634):708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Xu et al. (2012).Xu G, Guo C, Shan H, Kong H. Divergence of duplicate genes in exon–intron structure. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(4):1187–1192. doi: 10.1073/pnas.1109047109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang (2007).Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhang, Zhang & Rosenberg (2002).Zhang J, Zhang Y, Rosenberg HF. Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nature Genetics. 2002;30(4):411–415. doi: 10.1038/ng852. [DOI] [PubMed] [Google Scholar]
- Zhong et al. (2008).Zhong Q, Zhang Q, Wang Z, Qi J, Chen Y, Li S, Sun Y, Li C, Lan X. Expression profiling and validation of potential reference genes during Paralichthys olivaceus embryogenesis. Marine Biotechnology. 2008;10(3):310–318. doi: 10.1007/s10126-007-9064-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Boxes indicate the exons and the lines indicate the introns. The length of boxes and lines are based on gene length.
Different genes are represented by different colored pentagons and gene order is determined according to their relative positions in the chromosome or scaffold; the gene names are placed on top of the pentagons. The direction of pentagons indicates the gene direction, and vertical lines represent noncontiguous regions on the scaffold or chromosome.
Tr, Takifugu rubripes; Ol, Oryzias latipes; Xm, Xiphophorus maculatus; Po, Paralichthys olivaceus; On, Oreochromis niloticus; Pf, Poecilia formosa; Cs, Cynoglossus semilaevis; Pr, Poecilia reticulata; Sp, Stegastes partitus.