Abstract
Polypharmacology, the discovery or design of drug molecules that can simultaneously interact with multiple targets, is gaining interest in contemporary drug discovery. Serine/threonine kinases are attractive targets for therapeutic intervention in oncology due to their role in cellular phosphorylation and altered expression in cancer. Quercetin, a naturally occurring flavonoid, inhibits multiple cancer cell lines and is used as an anticancer drug in Phase I clinical trial. Quercetin glycosides have also received some attention due to their high bioavailability and activity against various diseases including cancer. However, these have been studied to a lesser extent. In this study, the structural basis of the multitarget inhibitory activity of quercetin and isoquercitrin, a glycoside derivative, on serine/threonine kinases using molecular modeling was explored. Structural analysis showed that both quercetin and isoquercitrin exhibited good binding energies and interacted with aspartate in the highly conserved Asp–Phe–Gly motif. The results indicate that isoquercitrin could be a more potent inhibitor of several members of the serine/threonine kinase family. In summary, the current structural evaluation highlights the multitarget inhibitory property of quercetin and its potential to be a chemical platform for oncological polypharmacology.
Keywords: serine/threonine kinases, quercetin, isoquercitrin, docking, polypharmacology
Introduction
The protein kinase family consists of >500 members that are involved in many cellular processes. The serine/threonine protein kinases form a diverse subfamily that phosphorylates the hydroxyl group of amino acids serine and threonine. These protein kinases interact with a diverse range of substrates such as enzymes, transcription factors, receptors, and other regulatory proteins. Perturbation of the normal activity of these kinases is associated with tumor growth and metastasis.1 Thus, these proteins are attractive targets for therapeutic intervention in cancer.
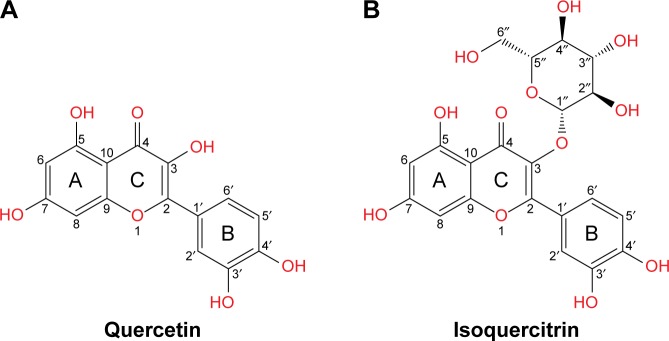
Epidemiological studies and experimental data have shown that consumption of diets rich in fruits and vegetables reduces the risk of cancer.2 Much of the protective effect can be attributed to flavonoids, a large group of polyphenolic compounds found ubiquitously in fruits and vegetables.3,4 Quercetin is one of the most abundant dietary flavonoids. It is found in onions, apples, green tea, grapes, and berries and occurs mainly as glycosides with sugar groups such as glucose, galactose, rhamnose, rutinose, and xylose bound to one of the hydroxyl groups of the flavonol.5 Quercetin and its glycosylated forms represent 60%–75% of flavonoid intake.6 The structure of quercetin (3,5,7,3′,4′-pentahydroxyflavone) is composed of three rings (A, B, and C) and five hydroxyl groups (Figure 1A). The most common quercetin glycosylation site is the hydroxyl group at the C3 carbon. Quercetin-3-glucoside (Q3G) or isoquercitrin is the major glycosidic form of quercetin (Figure 1B).
Figure 1.
Structure of (A) quercetin and (B) isoquercitrin.
Quercetin and its derivatives have been extensively studied for their role in cardiovascular disease, central nervous system disorder, and cancer.7–11 Quercetin has been shown to regulate several signal transduction pathways and is used as an anticancer drug in Phase I clinical trial.12 Several in vitro and in vivo studies have revealed the antiproliferative activity of quercetin on malignant growth and metastasis in various cancer cell lines including breast, ovarian, leukemia, hepatocellular carcinoma, and stomach cancer.13–17 Quercetin has been shown to inhibit a number of protein kinases including ABL1, Aurora-A, Aurora-B, Aurora-C, CLK1, EGFR, FLT3, JAK3, MEK1, MET, NEK4, NEK9, PAK3, PIM1, Raf1, RET, FGFR2, and PDGF-Rα.18,19 However, the biological activity of quercetin glycosides has been studied to a lesser extent. Recently, isoquercitrin has received some attention due to its high bioavailability and biological activity against cancer, cardiovascular disorders, diabetes, and allergic reactions.20 Glycosylation at C3 of quercetin has been shown to have an antiproliferative effect on breast, colon, hepatocellular, and lung cancer cell lines.21,22
Based on domain homology, serine/threonine kinases have been classified into six major groups: the AGC group (protein kinase A, G, and C), the CaMK group (calcium/calmodulin-dependent), the CMGC group (cyclin-dependent kinase [CDK], mitogen-activated protein kinase [MAPK], glycogen synthase kinase, and CDK like), the STE group (homologs of yeast sterile 7, sterile 11, and sterile 20), the CK1 group (casein kinase 1), and the tyrosine-kinase-like (TKL) group.23 Like several other kinases, the active site of these kinases is situated between a small N-terminal lobe (N-lobe), composed mainly of β-sheets, and a large C-terminal lobe (C-lobe), composed of six helices, which contains the activation loop segment that positions the Mg2+ ion and peptide substrates for catalysis (Figure 2). The catalytic active site of these proteins contains residues from the N-lobe including the glycine-rich loop, which provides flexibility for anchoring ATP. The base of the active site cleft is lined by residues from the catalytic loop and the front of the cleft is made up of residues from the activation loop. The linker region (hinge region) connects the two lobes and forms the back of the binding pocket.
Figure 2.
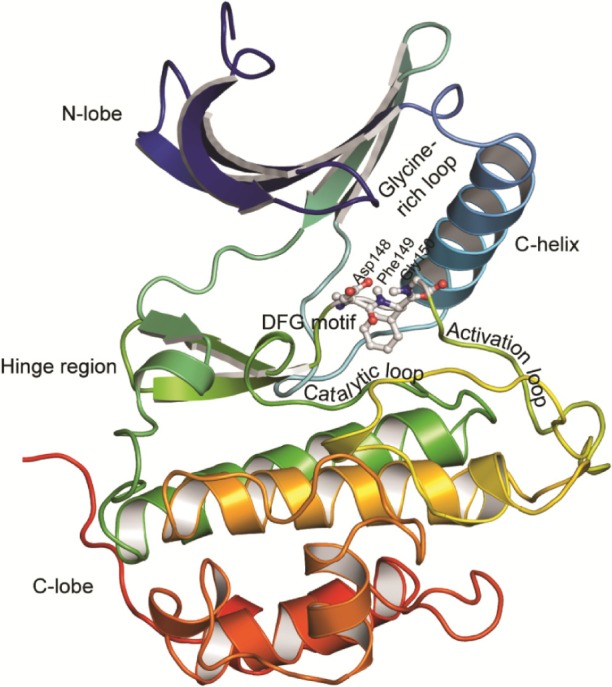
Structure of serine/threonine kinases.
Notes: The protein is shown in cartoon representation and colored in rainbow colors with violet at the N-terminus and red at the C-terminus of the structure. The N- and C-lobes with the connecting hinge region are indicated. The catalytic loop, activation loop, glycine-rich loop, C-helix, and the DFG motif are labeled. The Chk1 protein structure (PDB ID: 1ZYS) belonging to the CaMK family of serine/threonine protein kinases was used to generate this image.
Abbreviations: C-lobe, C-terminal lobe; DFG, Asp–Phe–Gly; N-lobe, N-terminal lobe; PDB, Protein Data Bank.
Computational techniques facilitate the design of novel, more potent inhibitors by closely evaluating the atomic-level receptor–drug interactions.24 Molecular docking approaches are routinely used in modern drug design to identify lead compounds by docking and scoring small molecules in appropriate binding sites of a target protein. Discovering or designing drug molecules that can interact with multiple targets is gaining interest in modern drug discovery.25 The current study provides a comprehensive overview and discusses the structural binding features of quercetin and its derivative, isoquercitrin, on multiple serine/threonine protein kinases based on molecular docking and binding energy calculations.
Materials and methods
Protein structure preparation
A total of 15 serine/threonine protein kinases were selected to represent all major subgroups of serine/threonine kinases that are involved in tumorigenesis. Three-dimensional (3D) coordinates of the X-ray crystal structure of these proteins with a resolution of <3 Å were retrieved from the RCSB Protein Data Bank (PDB)26 (Table 1). Structures with catalytically active conformation (Asp–Phe–Gly [DFG]-in), where the side chain of aspartate of the highly conserved DFG motif was positioned into the active site of the binding cleft, were used in the current study. The protein structures were processed using Schrödinger Maestro’s protein preparation wizard.27–29 This preprocessing protocol included simplification of multimeric structures, addition and optimization of hydrogen bonds, location and deletion of unnecessary water molecules, creation of disulfide bonds, proper assignment of bond orders and ionization states, conversion of selenomethionines to methionine, addition of missing atoms and side-chain residues, aligning and capping of terminal amides, and assignment of partial charges. To obtain a geometrically stable structure, minimization was performed.30
Table 1.
Proteins and PDB IDs of structures used in this study
| Groups | Protein | PDB ID | Resolution (Å) |
|---|---|---|---|
| AGC | Akt1 | 3MVH | 2.01 |
| Akt2 | 2UW9 | 2.10 | |
| Aurora kinase | 4J8M | 1.85 | |
| RSK2 | 4NW5 | 1.94 | |
| CaMK | Chk1 | 1ZYS | 1.70 |
| CMGC | CDK2 | 4EK4 | 1.55 |
| ERK2 | 4ZZN | 1.33 | |
| p38α | 4R3C | 2.06 | |
| JNK1 | 4QTD | 1.50 | |
| GSK3B | 1J1B | 1.80 | |
| CLK1 | 1Z57 | 1.70 | |
| STE | MEK1 | 3VVH | 2.00 |
| PAK4 | 2Q0N | 1.75 | |
| Plk1 | 2RKU | 1.95 | |
| TKL | B-Raf | 5CSW | 2.66 |
Abbreviation: PDB, Protein Data Bank.
Active site identification and grid generation
Schrödinger Sitemap was used to identify the binding cavity in the ATP-binding pocket of the selected structures. The highly conserved binding cleft of serine/threonine protein kinases is sandwiched between a small N-lobe, which comprises five β sheets and a single α-helix (C-helix), and a large C-lobe, which comprises six helices, and connected by a linker (Figure 2). A receptor grid was generated for each structure by incorporating all the functional residues in these regions.
Preprocessing of ligands
The two-dimensional (2D) structures of quercetin and isoquercitrin were retrieved from PubChem.31 For a comparative study, where available, 2D structure of a known inhibitor for each target protein was also retrieved from PubChem. These structures were preprocessed and conformers were generated using Schrödinger Ligprep.32 Ligand preprocessing included the conversion of 2D structures to 3D format, generation of tautomers and ionization states, addition of hydrogen atoms, neutralization of charged groups, and geometry optimization of the molecule.30
Molecular docking using extra precision glide
For predicting the binding pose and docking score of the compounds, grid-based docking was carried out using Schrödinger Glide in extra precision (XP) mode.33 No constraints were used in the docking process. Quercetin, isoquercitrin, and inhibitors were flexibly docked to the prepared protein structures. The geometry of the docked poses was improved by post-docking minimization, and other parameters of XP docking were set to the software’s default values. The protein–ligand interactions including hydrogen bonds, hydrophobic interactions, and π–π stacking were analyzed using XP visualizer.
Binding energy calculations
The ligand binding and strain energies were calculated using molecular mechanics-generalized Born surface area (MM-GBSA) method using Schrödinger Prime.29 The VSGB 2.0 solvation model was employed in these calculations.34
Results
Molecular docking was used to produce complexes of serine/threonine kinases with quercetin and isoquercitrin to elucidate the binding mode and to gain structural insights into the inhibitory mechanism. All serine/threonine kinase structures used here adopt an active conformation (DFG-in). The interactions and binding energies of quercetin, isoquercitrin, and inhibitors investigated in this study are summarized in Table 2.
Table 2.
Interactions of quercetin, isoquercitrin, and selected inhibitors with serine/threonine kinases
| Group | Target | PDB ID | Ligand | Hydrogen bonds | Hydrophobic interactions | π–π or cation–π interactions | XP GlideScore (kcal/mol) | MM–GBSA binding energy (kcal/mol) |
|---|---|---|---|---|---|---|---|---|
| AGC | Akt1 | 3MVH | Quercetin | Glu228, Ala230, Asp292 | Leu156, Val164, Ala177, Met227, Tyr229, Met281, Phe438, Phe442 | −9.79 | −64.71 | |
| Isoquercitrin | Glu228, Asp292 | Leu156, Val164, Ala177, Met227, Tyr229, Ala230, Met281, Phe438, Phe442 | −9.86 | −69.79 | ||||
| GSK690693 | Asp439 | Leu156, Phe161, Val164, Ala177, Leu181, Phe236, Phe237, Met281, Tyr437, Phe438 | Phe442 | −5.42 | −64.09 | |||
| Akt2 | 2UW9 | Quercetin | Lys160, Glu230, Ala232, Thr292, Asp293 | Leu158, Val166, Ala179, Met229, Tyr231, Met282, Phe439, Phe443 | −9.97 | −63.45 | ||
| Isoquercitrin | Lys160, Glu236, Lys277, Asp293, Asp440 | Leu158, Val166, Ala179, Met229, Phe238, Phe239, Met282, Phe439, Phe443 | −10.91 | −68.70 | ||||
| GSK690693 | Leu158, Thr213, Asn280, Asp440 | Val166, Ala179, Met229, Tyr231, Ala232, Phe239, Met282, Tyr438, Phe439, Phe443 | −7.10 | −73.94 | ||||
| Aurora | 4J8M | Quercetin | Leu139, Glu211, Ala213, Asp274 | Val147, Ala160, Leu194, Leu210, Tyr212, Leu263, Ala273 | −8.16 | −62.39 | ||
| Isoquercitrin | Glu211, Ala213, Asp274 | Leu139, Val147, Ala160, Leu194, Leu210, Tyr212, Leu263, Ala273, Ala281, Pro282 | −10.22 | −71.01 | ||||
| AT9283 | Ala213, Thr217 | Leu139, Val147, Ala160, Leu194, Leu210, Tyr212, Pro214, Tyr219, Leu263, Ala273, Ala281 | −6.71 | −70.34 | ||||
| RSK2 | 4NW5 | Quercetin | Leu74, Leu150, Asp211 | Val82, Leu147, Ala98, Phe149, Leu200 | −10.14 | −69.17 | ||
| Isoquercitrin | Gln76, Asp148, Leu150, Asn198 | Leu74, Val82, Ala98, Phe149, Leu200, Val131, Leu147 | −12.44 | −88.84 | ||||
| BI-D1870 | Lys72, Leu150 | Leu74, Val82, Ala98, Leu200, Val131, Leu147, Phe149 | −8.41 | −78.42 | ||||
| CaMK | Chk1 | 1ZYS | Quercetin | Lys38, Glu85, Cys87, Asp148 | Leu15, Tyr20, Val23, Ala36, Val68, Leu84, Tyr86, Leu137 | −8.38 | −63.43 | |
| Isoquercitrin | Glu17, Cys87, Asn135, Asp148 | Leu15, Tyr20, Val23, Ala36, Val68, Leu84, Tyr86, Leu137, Phe149 | −11.67 | −79.53 | ||||
| AZD7762 | Glu85, Cys87, Glu91 | Leu15, Tyr20, Val23, Ala36, Leu84, Val68, Tyr86, Leu137 | −7.15 | −83.52 | ||||
| CMGC | CDK2 | 4EK4 | Quercetin | Leu83, Gln131, Asp145 | Ile10, Val18, Ala31, Phe82, Leu134, Ala144 | −9.33 | −64.14 | |
| Isoquercitrin | Thr14, Lys33, Leu83, Gln131, Asp145 | Ile10, Val18, Ala31, Leu134 | −10.49 | −70.09 | ||||
| Dinaciclib | Lys33, Asp86, Lys89 | Ile10, Val18, Ala31, Val64, Phe80, Phe82, Leu83, Leu134, Ala144 | −5.57 | −75.63 | ||||
| ERK2 | 4ZZN | Quercetin | Lys52, Met106, Asp109, Asp165 | Ile29, Ala33, Tyr34, Val37, Ala50, Ile82, Leu105, Leu154, Cys164 | −6.69 | −59.57 | ||
| Isoquercitrin | Gln103, Met106, Asp109, Asp165 | Ile29, Ala33, Tyr34, Val37, Ala50, Ile82, Leu105, Leu154, Lys164 | Lys52 | −8.61 | −68.49 | |||
| SCH772984 | Lys52, Asp104, Lys112 | Ile29, Ala33, Val37, Ala50, Ile82, Leu105, Tyr111, Met106, Leu154, Lys164 | −6.61 | −74.69 | ||||
| p38α | 4R3C | Quercetin | Lys53, Met109, Asp112, Asp168 | Val30, Val38, Ala51, Ile84, Leu108, Ala111, Leu167, Met179, Tyr182 | −6.69 | −44.85 | ||
| Isoquercitrin | His107, Met109, Asp112, Asp168 | Val30, Val38, Ala51, Ile84, Leu108, Leu167, Met179, Tyr182 | −9.31 | −69.86 | ||||
| Doramapimod | Asp168 | Val30, Ala34, Tyr35, Val38, Ala51, Leu75, Ile84, Leu108, Met109, Ala111, Ala157, Leu167, Phe169 | −4.87 | −79.74 | ||||
| JNK1 | 4QTD | Quercetin | Ala36, Lys55, Glu73, Asp151, Asn156, Leu168 | Val40, Ile86, Met108, Leu172, Val187 | −8.20 | −45.53 | ||
| Isoquercitrin | Ala36, Arg69, Asp151, Ser155, Asn156, Leu168 | Val40, Ile86, Met108, Phe170, Leu172, Val186, Val187 | Arg69 | −10.60 | −50.23 | |||
| AS601245 | Lys55, Met111 | Ile32, Ala36, Val40, Ala53, Ile86, Met108, Leu110, Val158, Leu168 | −9.03 | −81.19 | ||||
| GSK3B | 1J1B | Quercetin | Lys85, Val135, Gln185, Asp200 | Ile62, Val70, Ala83, Val110, Leu132, Tyr134, Leu188, Cys199 | −9.29 | −55.35 | ||
| Isoquercitrin | Ile62, Asn64, Lys85, Val135, Arg141, Gln185, Asp200 | Val70, Val110, Leu132, Tyr134, Pro136, Tyr140, Leu188, Cys199, Phe201 | Arg141 | −10.44 | −62.59 | |||
| CHIR-99021 | Ile62, Asn64 | Val70, Ala83, Leu108, Val110, Leu132, Tyr134, Val135, Pro136, Leu188, Cys199 | Lys85, Arg141 | −5.94 | −67.09 | |||
| CLK1 | 1Z57 | Quercetin | Leu167, Lys191, Asp250, Asp325 | Phe172, Val175, Ala189, Val225, Phe241, Leu243, Leu244, Leu295, Val324 | −8.99 | −62.96 | ||
| Isoquercitrin | Gly245, Asp250, Asp325 | Leu167, Phe172, Val175, Ala189, Val225, Phe241, Leu243, Leu244, Leu246, Tyr249 Leu295, Val324 | −10.53 | −73.19 | ||||
| STE | MEK1 | 3VVH | Quercetin | Leu74, Glu144, Met146, Asn195, Asp208 | Ala76, Val82, Ala95, Val127, Met143, Leu197, Cys207 | −9.97 | −64.99 | |
| Isoquercitrin | Met146, Ser194, Asp152, Asp208 | Leu74, Ala76, Val82, Ala95, Met143, Leu197, Cys207 | −9.65 | −68.12 | ||||
| PAK4 | 2Q0N | Quercetin | Glu329, Glu396, Leu398, Asp458 | Ile327, Val335, Ala348, Val379, Met395, Phe397, Leu447 | −10.03 | −58.22 | ||
| Isoquercitrin | Glu396, Leu398, Ala402, Asp458 | Ile327, Val335, Ala348, Val379, Met395, Phe397, Leu447 | −9.86 | −60.47 | ||||
| Plk1 | 2RKU | Quercetin | Leu59, Lys82, Glu131, Cys133, Asp194 | Cys67, Ala80, Val114, Leu130, Leu132, Phe183, Phe195 | −10.89 | −62.84 | ||
| Isoquercitrin | Leu59, Lys82, Glu140, Cys133, Asp194 | Cys67, Ala80, Val114, Leu130, Leu132, Leu139 | Phe183 | −11.26 | −74.86 | |||
| TKL | B-Raf | 5CSW | Quercetin | Cys532, Asp594, Phe595 | Ile463, Phe468, Val471, Ala481, Leu505, Leu514, Ile527, Trp531, Phe583 | −9.48 | −60.46 | |
| Isoquercitrin | Cys532, Gly534, Ser536, Asn580, Phe583, Asp594 | Leu514, Ile463, Val471, Ala481, Trp531, Tyr538 | −11.07 | −67.72 |
Abbreviations: MM–GBSA, molecular mechanics–generalized Born surface area; PDB, Protein Data Bank; XP, extra precision.
AGC group of serine/threonine kinases
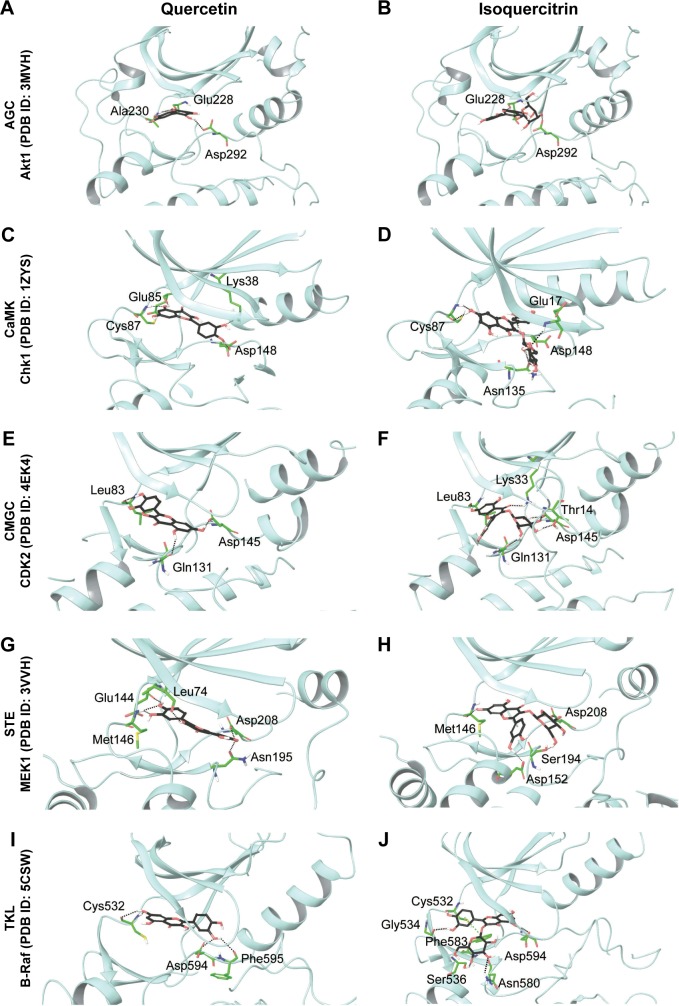
The structure of quercetin docked to Akt1 (PDB ID: 3MVH) showed an extensive network of hydrogen bonds and hydrophobic interactions in the ATP-binding pocket (Table 2). The hydroxyl group attached to C7 (A ring) of quercetin bound to the hinge region of the active site through direct hydrogen bonds with the main chain of Glu228 and Ala230 (Figure 3). Furthermore, the hydroxyl group attached to C3 (C ring) formed a hydrogen bond with the side chain of Asp292 of the DFG motif (Asp292–Phe293–Gly294). Similarly, Akt2 (PDB ID: 2UW9)–quercetin complex also produced strong hydrogen bond interactions in the ATP-binding pocket. First, the hydroxyl group on the B ring of quercetin formed hydrogen bonds with the backbone of Glu230 and Ala232 located in the hinge region. Second, the hydroxyl group on C5 (A ring) formed a hydrogen bond with Lys160 of the glycine-rich loop. Significantly, similar to the Akt1–quercetin complex, the hydroxyl group attached to C3 (C ring) formed a hydrogen bond with the side chain of Asp293 of the DFG motif (Figure S1). This supports evidence indicating that mice treated with quercetin resulted in significant downregulation of phosphorylated Akt in the myelomonocytic leukemia cell line P39.35
Figure 3.
Docked pose and hydrogen bond interactions of quercetin and isoquercitrin with serine/threonine protein kinases.
Notes: One protein has been chosen from each group. (A) Quercetin in Akt1 (AGC group), (B) isoquercitrin in Akt1, (C) quercetin in Chk1 (CaMK group), (D) isoquercitrin in Chk1, (E) quercetin in CDK2 (CMGC group), (F) isoquercitrin in CDK2, (G) quercetin in MEK1 (STE group), (H) isoquercitrin in MEK1, (I) quercetin in B-Raf (TKL group), (J) isoquercitrin in B-Raf. The protein is shown in light-blue cartoon representation, interacting residues are shown in stick representation, and the docked ligand is represented as black sticks.
Abbreviation: PDB, Protein Data Bank.
Structural analysis of aurora kinase A (PDB ID: 4J8M) with quercetin revealed that the binding mode is characterized by the quercetin-forming interactions with the hinge region in aurora kinase. The interaction mode of quercetin in aurora kinase A revealed that the hydroxyl groups of A and C rings of quercetin formed three hydrogen bonds with residues Glu211 and Ala213 in the hinge region. The hydroxyl groups of the B ring positioned at C3′ and C4′ formed hydrogen bonds with Leu139 of the glycine-rich loop. The hydroxyl group on C5 formed a hydrogen bond with Asp274 of the DFG motif (Figure S1). This could form the structural basis of quercetin’s ability to inhibit the activity of aurora kinases in several cancer cell lines.19
Similar to the other AGC members, quercetin showed favorable interactions with ribosomal protein S6 kinase alpha-3 (RSK2; PDB ID: 4NW5). A hydrogen bond was formed between the hydroxyl group on the B ring and the side chain of Asp211 of the DFG motif. In addition, it formed hydrogen bonds with Leu74 located in the N-lobe and the backbone amino and carbonyl groups of Leu150 from the hinge region (Figure S1). Thus, quercetin exhibited a conserved binding mode in the AGC family proteins with a crucial interaction with the aspartate of the DFG motif.
Isoquercitrin produced similar or better binding energies with all the selected AGC group members compared to quercetin (Table 2). Isoquercitrin bound in the ATP-binding cavity of AGC kinases with several hydrogen bonds and hydrophobic interactions (Table 2). In Akt1 (PDB ID: 3MVH), the quercetin core formed a hydrogen bond with the backbone carbonyl of Glu228 in the hinge region and the glycosyl moiety formed a hydrogen bond with the side chain of Asp292 of the DFG motif (Figure 3). In Akt2, the hydroxyl group of C3 (C ring) formed a hydrogen bond with the backbone of Lys160 of the glycine-rich loop. Moreover, the glycosyl part formed hydrogen bonds with the side chain of Glu236 in the hinge region, Lys277 in the catalytic loop, and the side chain of Asp293 of the DFG motif (Figure S1).
Isoquercitrin showed similar interactions in aurora kinase (PDB ID: 4J8M) as well. The sugar moiety of isoquercitrin formed a hydrogen bond with the side chain of Asp274 of the DFG motif, and the B ring produced three hydrogen bonds with Glu211 and Ala213 in the hinge region (Figure S1).
In the case of ribosomal protein S6 kinase alpha-3 (PDB ID: 4NW5), the hydroxyl groups attached to C3′ and C4′ (B ring) of isoquercitrin formed hydrogen bonds with the backbone of Asp148 and Leu150 in the hinge region and the glycosyl part formed three hydrogen bonds with Gln76 of the glycine-rich loop and the side chain of Asn198 located at the apex of the catalytic loop (Figure S1).
For a comparative analysis, these targets were docked with currently known inhibitors. Results showed that quercetin and isoquercitrin produced similar or better GlideScore and binding energy when compared to inhibitors of Akt (GSK690693),36 aurora kinase (AT9283),37 and ribosomal S6 kinase (BI-D1870)38 (Table 2). The binding interaction of quercetin and isoquercitrin and the associated residues are highly conserved among the AGC kinases. Both compounds showed one or two hydrogen bonds with residues located in the hinge region (Glu228 and Ala230 in Akt1, Glu230 and Ala232 in Akt2, Glu211 and Ala213 in aurora kinase, and Asp148 and Leu150 in ribosomal S6 kinase), and thereby mimic the normal hydrogen bonds formed by the adenine ring of ATP.39 Significantly, the docking results showed that the flavonoids quercetin and isoquercitrin bind deeply in the ATP-binding site, interacting with the aspartate residue of the DFG motif. This could explain how these molecules could competitively inhibit the activity of these kinases. In addition, the data indicate that isoquercitrin produces a higher binding score and binding energy than quercetin and other known inhibitors of the AGC family of kinases (Table 2).
CaMK family of serine/threonine kinases
Quercetin docked deeply in the ATP-binding pocket of checkpoint kinase (Chk1; PDB ID: 1ZYS) and formed hydrogen bonds and hydrophobic interactions (Table 2). Hydrogen bonds were formed with the backbone of residues Glu85 and Cys87 in the hinge region and also with Lys38 in the N-lobe (Figure 3). In addition, the docking pose showed the expected hydrogen bond between the hydroxyl group of C3′ (B ring) and the side chain of Asp148 of the DFG motif.
For isoquercitrin, the A ring of the quercetin core formed two hydrogen bonds with the backbone of Cys87 in the hinge region and the B ring formed a hydrogen bond with Asp148 of the DFG motif. In addition, the glycosyl part of isoquercitrin formed a hydrogen bond with Glu17 of the glycine-rich loop and also with Asn135 of the catalytic loop (Figure 3).
AZD7762, an inhibitor of Chk1,40 was docked for comparative analysis. Both quercetin and isoquercitrin showed more interactions and better binding score than the inhibitor (Table 2). Similar to the AGC family, the CaMK family protein considered in this study also produced one or two hydrogen bond interactions with the backbone residues in the hinge region and interacted with the aspartate residue of the DFG motif. Furthermore, isoquercitrin showed better GlideScore and binding energy when compared to quercetin and the control.
CMGC group of serine/threonine kinases
Table 2 summarizes the structural analysis of the docking of quercetin and isoquercitrin to CMGC kinases. When quercetin was docked to cyclin-dependent kinase 2 (CDK2; PDB ID: 4EK4), the hydroxyl groups of the B ring formed two hydrogen bonds with the backbone carbonyl of Leu83 in the hinge region (Figure 3). The hydroxyl group attached to C7 of quercetin formed hydrogen bond interactions with the side-chain carbonyl of Asp145 of the DFG motif and C5 hydroxyl group with Gln131 of the catalytic loop. Several hydrophobic interactions were also involved in stabilizing quercetin in the binding site (Table 2). This may explain the ability of quercetin to induce cell cycle arrest in G1 phase by inhibiting the activity of CDK2.41
Quercetin has been reported to induce apoptosis in human gastric adenocarcinoma cells through interactions with MAPKs.42 Docked quercetin participated in several hydrogen bonds and hydrophobic interactions with the active confirmation of several MAPKs such as mitogen-activated protein kinase 1 (MAPK1/ERK2), mitogen-activated protein kinase 14 (MAPK14/p38α MAPK), and c-Jun N-terminal kinases (MAPK 8/JNK1; Table 2). The hydroxyl groups on the A ring formed hydrogen bonds with the side chain of Met106 and Asp109 in the hinge region of ERK2 (PDB ID: 4ZZN; Figure S2. Both hydroxyl groups on the B ring formed hydrogen bonds with the side chain of Lys52 in the N-lobe and with the side chain of Asp165 of the DFG motif. Similarly, quercetin formed hydrogen bonds and hydrophobic interactions with the structure of p38α kinase (PDB ID: 4R3C). The hydroxyl groups of the B ring formed hydrogen bond interactions with the side chain of Asp168 of the DFG motif and C5 hydroxyl group formed hydrogen bond interactions with the backbone carbonyl of Met109 in the hinge region. Lys53 in the N-lobe and Asp112 in the C-lobe produced additional hydrogen bonds with A and B rings Figure S2).
Quercetin bound in the active cleft of JNK1 (PDB ID: 4QTD) as well. The 4-oxo group (C ring) of quercetin formed a hydrogen bond with Ala36 of the glycine-rich loop, and the A and C rings formed hydrogen bonds with Asp151 and Asn156 of the catalytic loop. Three hydrogen bonds were formed between the hydroxyl groups on the B ring and the side chain of Lys55 in the N-lobe, Glu73 in the C-helix, and the backbone carbonyl of Leu168 of the activation loop (Figure S2).
Quercetin also produced interactions in the ATP-binding pocket of GSK3B (PDB ID: 1J1B). The 4-oxo group of the C ring formed a hydrogen bond with the side chain of Lys85 in the N-lobe and the C5 hydroxyl group in the A ring formed a hydrogen bond with Asp200 of the DFG motif and C7 hydroxyl group formed a hydrogen bond with Gln185 of the catalytic loop. Two hydroxyl groups on the B ring also formed hydrogen bonds with the backbone carbonyl of Val135 in the hinge region (Figure S2).
CLK1 (PDB ID: 1Z57) also produced a similar pattern of interactions as the other CMGC group of proteins. The keto group of the C ring formed a hydrogen bond with Lys191 from the N-lobe and the side chain of Asp325 of the DFG motif. Hydroxyl groups of the B ring formed hydrogen bonds with Leu167 from the N-lobe and Asp250 in the C-lobe (Figure S2). Interactions of CLK1 with quercetin were reported in a myelomonocytic leukemia cell line in agreement with these results.35
The CDK2–isoquercitrin complex (PDB ID: 4EK4) showed hydrogen bond interactions with the residues from N- and C-lobes (Figure 3). The isoquercitrin core formed a hydrogen bond between the keto group of the C ring and Lys33 in the N-lobe and another between C7 hydroxyl group and the backbone carbonyl of Leu83 in the hinge region. The glycosyl moiety formed several hydrogen bond interactions including interactions with the side-chain hydroxyl group of Thr14 of the glycine-rich loop, the side-chain of residue Gln131, and the side-chain of Asp145 of the DFG loop.
Isoquercitrin docked to the ATP-binding site of ERK2 (PDB ID: 4ZZN) showed hydrogen bonds, hydrophobic interactions, and a cation–π interaction as well (Table 2). The hydroxyl groups of the glycosyl moiety formed hydrogen bonds with residues Met106 and Asp109 in the hinge region. The C7 hydroxyl group of the A ring formed hydrogen bond interactions with the side chain of Asp165 of the DFG motif. The hydroxyl group on the B ring formed a hydrogen bond with Gln103 in the N-lobe in addition to a cation–π interaction with the side chain of Lys52 in the N-lobe (Figure S2).
When isoquercitrin was docked to p38α kinase (PDB ID: 4R3C), the C7 hydroxyl group of isoquercitrin formed hydrogen bonds with the backbone of His107 and Met109 in the hinge region and the C3′ hydroxyl group (B ring) formed hydrogen bonds with the side chain of Asp112 in the C-lobe. The glycosyl moiety also formed the crucial hydrogen bond with the side chain of Asp168 of the DFG motif (Figure S2).
In the case of JNK1 (PDB ID: 4QTD), the 4-oxo group of isoquercitrin formed hydrogen bond interaction with Ala36 of the glycine-rich loop and the A ring formed a hydrogen bond with Asp151 and a cation–π interaction with Arg69. The hydroxyl groups of isoquercitrin formed a hydrogen bond with the backbone amide of Leu168 of the activation loop and with Ser155 and Asn156 of the catalytic loop (Figure S2).
Isoquercitrin docked to GSK3B (PDB ID: 1J1B) also produced several hydrogen bonds, hydrophobic interactions, and a π–π interaction (Table 2). Hydroxyl groups on the B ring formed hydrogen bonds with the backbone carbonyl of Val135 from the hinge region and Ile62 in the N-lobe. The A ring formed a hydrogen bond with the backbone amide group of Asp200 of the DFG motif. In addition, the glycosyl moiety of isoquercitrin formed hydrogen bonds with Asn64 located at the apex of the glycine-rich loop, Arg141 in the C-lobe, and with the side chain of Gln185 in the catalytic loop. Besides these hydrogen bonds, the B ring of isoquercitrin also produced a cation–π interaction with the side chain of Arg141 in the C-lobe (Figure S2).
In CLK1–isoquercitrin complex (PDB ID: 1Z57), the quercetin core of isoquercitrin formed a hydrogen bond with the side-chain carbonyl of Asp250 in the C-lobe and the carbonyl of Gly245 of the catalytic loop. The sugar moiety of isoquercitrin formed a hydrogen bond with the side chain of Asp325 of the DFG motif (Figure S2).
For all the CMGC family proteins considered in this study, quercetin and isoquercitrin showed similar or better binding scores and binding energies than inhibitors docked to these structures: dinaciclib (CDK2),43 SCH772984 (ERK2),44 doramapimod (p38α),45 AS601245 (JNK1),46 and CHIR-99021 (GSK3B)47 (Table 2). Quercetin and isoquercitrin interacted with the ATP-binding pocket of CMGC protein kinases by forming several hydrogen bonds with the backbone residues from the hinge region, the side chain of residues of the catalytic loop, as expected, and the aspartate residue of the conserved DFG motif. In addition, isoquercitrin ranked better than quercetin and inhibitors considered in this study.
STE group of serine/threonine kinases
Quercetin formed several hydrogen bonds and hydrophobic interactions with the ATP-binding pocket of the STE group of kinases (Table 2). Like other serine/threonine kinases, in MAPK1 (MEK1; PDB ID: 3VVH), the hydroxyl groups of the B ring of quercetin formed hydrogen bonds with the hinge region residues Glu144 and Met146 (Figure 3). The C3 hydroxyl group of quercetin formed a hydrogen bond with Lys74 in the N-lobe. Following the pattern observed, the C7 hydroxyl group of quercetin formed a hydrogen bond with the side chain of Asp208 of the DFG motif and Asn195 of the catalytic loop. In vitro and ex vivo analysis showed that the inhibition of MEK1 by quercetin resulted in the downregulation of its activity.48
Quercetin docked to p21-activated kinase 4 (PAK4; PDB ID: 2Q0N) also showed similar hydrogen-bonding pattern in the ATP-binding pocket (Table 2). Hydroxyl groups at C5 and C7 (A ring) formed hydrogen bonds with Glu396 and Leu398 in the hinge region. It also formed a hydrogen bond with Glu329 of the glycine-rich loop and the side chain of Asp458 of the DFG motif (Figure S3).
The third structural representative of the STE group, polo-like kinase (Plk1; PDB ID: 2RKU) showed similar pattern of interactions with quercetin. A and C rings of quercetin formed four hydrogen bonds that included interactions with Glu131 and Cys133 in the hinge region and Leu59 in the N-lobe. Hydroxyl group of the B ring interacted with Asp194 of the DFG motif and Lys82 in the N-lobe through hydrogen bonds (Figure S3).
In MEK1–isoquercitrin complex, the glycosyl moiety formed hydrogen bonds with the side chain of Asp208 of the DFG motif, Ser194 of the catalytic loop, and Asp152 in the C-lobe. C7 hydroxyl group in the A ring formed hydrogen bond with Met146 in the hinge region (Figure 3).
In the PAK4–isoquercitrin complex, a hydroxyl group on the B ring produced two hydrogen bond interactions with the backbone of Glu396 and Leu398 in the hinge region and the A ring produced hydrogen bond with Ala402 in the C-lobe. The glycosyl moiety showed hydrogen bond interaction with the side chain of Asp458 of the DFG motif (Figure S3).
Isoquercitrin formed hydrogen bonds, hydrophobic interactions, and π–π interactions with Plk1. Hydroxyl group of B ring at the C3′ position formed a hydrogen bond with Cys133 in the hinge region (Figure S3). As expected, the hydroxyl group of the A ring formed a hydrogen bond with the side chain of Asp194 of the DFG motif. The glycosyl part of isoquercitrin formed hydrogen bonds with Leu59 and Lys82 in the N-lobe and Glu140 in the C-lobe. In addition to these hydrogen bonds, the A and C rings showed π–π interactions with Phe183 of the catalytic loop.
Thus, quercetin and isoquercitrin exhibited a conserved interaction pattern in the STE group of kinases as well. This included interactions with residues in the hinge region and with the critical aspartate residue of the DFG motif.
TKL group of serine/threonine kinases
Quercetin bound in the ATP-binding site of B-Raf (PDB ID: 5CSW) with hydrogen bonds and hydrophobic interactions (Table 2). The C3′ hydroxyl group formed hydrogen bonds with the backbone amide group of Asp594 and with Phe595 of the DFG motif (Figure 3). The hydroxyl groups of the A ring formed two hydrogen bond interactions with Cys532 in the hinge region. This supports the finding that quercetin imparts chemopreventive activity to Raf1 by directly binding to the protein.48
Similar to other proteins considered, isoquercitrin produced several interactions with B-Raf (Table 2). The B ring of the isoquercitrin core formed hydrogen bonds with Cys532 and Gly534 in the hinge region. The C5 hydroxyl group of the A ring formed a hydrogen bond with the side chain of Asp594 of the DFG motif (Figure 3). The glycosyl moiety of isoquercitrin provided additional interactions for complex formation including hydrogen bonds with Ser536 in the C-lobe and Asn580 of the catalytic loop. Interactions and binding energy of isoquercitrin were more favorable than quercetin in B-Raf (Table 2).
Discussion
Docking analysis of a series of serine/threonine kinases with the flavonoids quercetin and isoquercitrin revealed that both could be potent multitarget anticancer agents. The highly conserved DFG motif plays an important role in the regulation of protein kinase activity, and targeting the DFG motif in kinases is gaining prominence.49,50 Structural studies involving phosphoinositide 3-kinase (PDB ID: 3R7R),51 aurora kinase (PDB ID: 3UP7),52 interleukin-2 inducible T-cell kinase (PDB ID: 4HCT),53 and Mapk13 (PDB ID: 4EYJ)54 have revealed that inhibitors interact through hydrogen bonds with the aspartate residue of the DFG motif. The interaction of quercetin with the DFG motif of serine/threonine kinases has also been observed in structures such as death-associated protein kinase 1 (DAPK1; PDB ID: 5AUW) and serine/threonine protein kinase 17B (PDB ID: 3LM5). The B ring of quercetin interacts with Asp161, and the C4 hydroxyl group (B ring) interacts with the side chain of Phe162 of the DFG motif of DAPK1 (PDB ID: 5AUW), a target for the treatment of endometrial adenocarcinomas.55 In serine/threonine protein kinase 17B, the C3 hydroxyl group (A ring) of quercetin forms hydrogen bond interaction with the side chain of Asp179 of the DFG motif (PDB ID: 3LM5). Both quercetin and isoquercitrin interacted with the aspartate residue of the highly conserved DFG motif in almost all structures considered in this study. Quercetin and isoquercitrin were observed to interact with one or more hydrogen bonds with the hinge region residues mimicking the interaction of the adenine ring of ATP with the hinge region.39 Besides interacting with the DFG motif, the docked poses and binding energies calculated for the glycosidic derivative of quercetin revealed that isoquercitrin could be a more potent inhibitor. Analysis of the antiproliferative activities of quercetin derivatives on various cancer cell lines including breast, colon, hepatocellular, and lung has shown that isoquercitrin exerts a more potent effect than quercetin.21 In addition to more favorable interactions, isoquercitrin is also more bioavailable and rapidly absorbed than quercetin. Compared to quercetin, isoquercitrin produced high levels of quercetin metabolites in tissues (twofold to fivefold) and in plasma (twofold to threefold) when orally administered.56 The sugar moiety enables isoquercitrin to be absorbed more efficiently than aglycones since membrane transporters have a higher affinity and pump isoquercitrin more effectively across the membrane due to the presence of the glucose moiety.57 Extending this, the derivatives of quercetin could be explored to develop novel, polypharmacological or specific, serine/threonine inhibitors.
Conclusion
Quercetin, the most abundantly found dietary flavonoid, is a promising anticancer agent. In this study, a structure-based analysis of the interaction mechanism of quercetin and, its glycosidic derivative, isoquercitrin with serine/threonine kinases was performed. Both compounds produced a conserved interaction pattern with residues in the hinge region. More importantly, these molecules specifically interacted with the aspartate of the highly conserved DFG motif that plays an important role in the regulation of kinase activity. Binding energy analysis suggested that isoquercitrin had a higher potential to inhibit several of these proteins compared to quercetin. This study illustrates the structural basis of the polypharmacological inhibitory mechanism of quercetin and isoquercitrin. Quercetin could thus be a promising platform to generate more potent derivatives targeting serine/threonine kinases.
Supplementary materials
Docked pose and hydrogen bond interactions of quercetin and isoquercitrin with AGC family serine/threonine kinases.
Notes: (A) Quercetin in Akt2, (B) isoquercitrin in Akt2, (C) quercetin in aurora kinase, (D) isoquercitrin in aurora kinase, (E) quercetin in RSK2, (F) isoquercitrin in RSK2. The protein is shown in light-blue cartoon representation, interacting residues are shown in stick representation, and the docked ligand is represented as black sticks.
Abbreviation: PDB, Protein Data Bank.
Docked pose and hydrogen bond interactions of quercetin and isoquercitrin with CMGC family serine/threonine kinases.
Notes: (A) Quercetin in ERK2, (B) isoquercitrin in ERK2, (C) quercetin in p38α, (D) isoquercitrin in p38α, (E) quercetin in JNK1, (F) isoquercitrin in JNK1, (G) quercetin in GSK3B, (H) isoquercitrin in GSK3B, (I) quercetin in CLK1, (J) isoquercitrin in CLK1. The protein is shown in light-blue cartoon representation, interacting residues are shown in stick representation, and the docked ligand is represented as black sticks.
Abbreviation: PDB, Protein Data Bank.
Docked pose and hydrogen bond interactions of quercetin and isoquercitrin with STE family serine/threonine kinases.
Notes: (A) Quercetin in PAK4, (B) isoquercitrin in PAK4, (C) quercetin in Plk1, (D) isoquercitrin in Plk1. The protein is shown in light-blue cartoon representation, interacting residues are shown in stick representation, and the docked ligand is represented as black sticks.
Abbreviation: PDB, Protein Data Bank.
Acknowledgments
This study was funded by a center-based grant (31R015) from the Zayed Center for Health Sciences, United Arab Emirates University, and a startup grant (31S156) from the United Arab Emirates University to RV.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411(6835):355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 2.Tantamango-Bartley Y, Jaceldo-Siegl K, Fan J, Fraser G. Vegetarian diets and the incidence of cancer in a low-risk population. Cancer Epidemiol Biomarkers Prev. 2013;22(2):286–294. doi: 10.1158/1055-9965.EPI-12-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross JA, Kasum CM. Dietary flavonoids: bioavailability, metabolic effects, and safety. Annu Rev Nutr. 2002;22(1):19–34. doi: 10.1146/annurev.nutr.22.111401.144957. [DOI] [PubMed] [Google Scholar]
- 4.Pace G, Lima P, Vianello F, et al. Polyphenols in fruits and vegetables and its effect on human health. Food Nutr Sci. 2014;5(5):1065–1082. [Google Scholar]
- 5.Boots AW, Haenen GR, Bast A. Health effects of quercetin: from antioxidant to nutraceutical. Eur J Pharmacol. 2008;585(2–3):325–337. doi: 10.1016/j.ejphar.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Bouktaib M, Atmani A, Rolando C. Regio- and stereoselective synthesis of the major metabolite of quercetin, quercetin-3-O-β-D-glucuronide. Tetrahedron Lett. 2002;43(35):6263–6266. [Google Scholar]
- 7.Dajas F. Life or death: neuroprotective and anticancer effects of quercetin. J Ethnopharmacol. 2012;143(2):383–396. doi: 10.1016/j.jep.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Cossarizza A, Gibellini L, Pinti M, et al. Quercetin and cancer chemo-prevention. Evid Based Complementary Altern Med. 2011;2011:591356. doi: 10.1093/ecam/neq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russo M, Spagnuolo C, Tedesco I, Bilotto S, Russo GL. The flavonoid quercetin in disease prevention and therapy: facts and fancies. Biochem Pharmacol. 2012;83(1):6–15. doi: 10.1016/j.bcp.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 10.Shen X, Si Y, Wang Z, Wang J, Guo Y, Zhang X. Quercetin inhibits the growth of human gastric cancer stem cells by inducing mitochondrial-dependent apoptosis through the inhibition of PI3K/Akt signaling. Int J Mol Med. 2016;38(2):619–626. doi: 10.3892/ijmm.2016.2625. [DOI] [PubMed] [Google Scholar]
- 11.Devipriya S, Ganapathy V, Shyamaladevi CS. Suppression of tumor growth and invasion in 9,10 dimethyl benz(a) anthracene induced mammary carcinoma by the plant bioflavonoid quercetin. Chem Biol Interact. 2006;162(2):106–113. doi: 10.1016/j.cbi.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Ferry DR, Smith A, Malkhandi J, et al. Phase I clinical trial of the flavonoid quercetin: pharmacokinetics and evidence for in vivo tyrosine kinase inhibition. Clin Cancer Res. 1996;2(4):659–668. [PubMed] [Google Scholar]
- 13.Choi J, Kim J, Lee J, et al. Induction of cell cycle arrest and apoptosis in human breast cancer cells by quercetin. Int J Oncol. 2001;19(4):837–844. doi: 10.3892/ijo.19.4.837. [DOI] [PubMed] [Google Scholar]
- 14.Pawlikowska-Pawlega B, Jakubowicz-Gil J, Rzymowska J, Gawron A. The effect of quercetin on apoptosis and necrosis induction in human colon adenocarcinoma cell line LS180. Folia Histochem Cytobiol. 2001;39(2):217–218. [PubMed] [Google Scholar]
- 15.Asaum J, Matsuzaki H, Kawasak S, et al. Effects of quercetin on the cell growth and the intracellular accumulation and retention of adriamycin. Anticancer Res. 2000;20(4):2477–2483. [PubMed] [Google Scholar]
- 16.Damianaki A, Bakogeorgou E, Kampa M, et al. Potent inhibitory action of red wine polyphenols on human breast cancer cells. J Cell Biochem. 2000;78(3):429–441. doi: 10.1002/1097-4644(20000901)78:3<429::aid-jcb8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 17.Lamson DW, Brignall MS. Antioxidants and cancer, part 3: quercetin. Altern Med Rev. 2000;5(3):196–208. [PubMed] [Google Scholar]
- 18.Boly R, Gras T, Lamkami T, et al. Quercetin inhibits a large panel of kinases implicated in cancer cell biology. Int J Oncol. 2011;38(3):833–842. doi: 10.3892/ijo.2010.890. [DOI] [PubMed] [Google Scholar]
- 19.Lee LT, Huang YT, Hwang JJ, et al. Blockade of the epidermal growth factor receptor tyrosine kinase activity by quercetin and luteolin leads to growth inhibition and apoptosis of pancreatic tumor cells. Anticancer Res. 2002;22(3):1615–1627. [PubMed] [Google Scholar]
- 20.Valentová K, Vrba J, Bancířová M, et al. Isoquercitrin: pharmacology, toxicology, and metabolism. Food Chem Toxicol. 2014;68:267–282. doi: 10.1016/j.fct.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 21.You HJ, Ahn HJ, Ji GE. Transformation of rutin to antiproliferative quercetin-3-glucoside by Aspergillus niger. J Agric Food Chem. 2010;58(20):10886–10892. doi: 10.1021/jf102871g. [DOI] [PubMed] [Google Scholar]
- 22.Lee J, Han S-I, Yun J-H, et al. Quercetin 3-O-glucoside suppresses epidermal growth factor–induced migration by inhibiting EGFR signaling in pancreatic cancer cells. Tumor Biol. 2015;36(12):9385–9393. doi: 10.1007/s13277-015-3682-x. [DOI] [PubMed] [Google Scholar]
- 23.Goldsmith EJ, Akella R, Min X, et al. Substrate and docking interactions in serine/threonine protein kinases. Chem Rev. 2007;107(11):5065–5081. doi: 10.1021/cr068221w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sliwoski G, Kothiwale S, Meiler J, Lowe EW., Jr Computational methods in drug discovery. Pharmacol Rev. 2014;66(1):334–395. doi: 10.1124/pr.112.007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anighoro A, Bajorath J, Rastelli G. Polypharmacology: challenges and opportunities in drug discovery. J Med Chem. 2014;57(19):7874–7887. doi: 10.1021/jm5006463. [DOI] [PubMed] [Google Scholar]
- 26.Sussman JL, Lin D, Jiang J, et al. Protein data bank (PDB): database of three-dimensional structural information of biological macromolecules. Acta Crystallogr D Biol Crystallogr. 1998;54(6 I):1078–1084. doi: 10.1107/s0907444998009378. [DOI] [PubMed] [Google Scholar]
- 27.Schrödinger, LLC . Epik, version 3.3. New York, NY: Schrödinger, LLC; 2015. Schrödinger Suite 2015-3 Protein Preparation Wizard. [Google Scholar]
- 28.Schrödinger, LLC . Impact, version 6.8. New York, NY: Schrödinger, LLC; 2015. [Google Scholar]
- 29.Schrödinger, LLC . Prime, version 4.1. New York, NY: Schrödinger, LLC; 2015. [Google Scholar]
- 30.Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27(3):221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37(Web Server issue):W623–W633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schrödinger, LLC . LigPrep, version 3.5. New York, NY: Schrödinger, LLC; 2015. [Google Scholar]
- 33.Friesner RA, Murphy RB, Repasky MP, et al. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49(21):6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Abel R, Zhu K, Cao Y, Zhao S, Friesner RA. The VSGB 2.0 model: a next generation energy model for high resolution protein structure modeling. Proteins. 2011;79(10):2794–2812. doi: 10.1002/prot.23106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maso V, Calgarotto AK, Franchi GC, Jr, et al. Multitarget effects of quercetin in leukemia. Cancer Prev Res (Phila) 2014;7(12):1240–1250. doi: 10.1158/1940-6207.CAPR-13-0383. [DOI] [PubMed] [Google Scholar]
- 36.Rhodes N, Heerding DA, Duckett DR, et al. Characterization of an Akt kinase inhibitor with potent pharmacodynamic and antitumor activity. Cancer Res. 2008;68(7):2366–2374. doi: 10.1158/0008-5472.CAN-07-5783. [DOI] [PubMed] [Google Scholar]
- 37.Moreno L, Marshall LV, Pearson ADJ, et al. A phase I trial of AT9283 (a selective inhibitor of aurora kinases) in children and adolescents with solid tumors: a cancer research UK study. Clin Cancer Res. 2015;21(2):267–273. doi: 10.1158/1078-0432.CCR-14-1592. [DOI] [PubMed] [Google Scholar]
- 38.Sapkota GP, Cummings L, Newell FS, et al. BI-D1870 is a specific inhibitor of the p90 RSK (ribosomal S6 kinase) isoforms in vitro and in vivo. Biochem J. 2007;401(1):29–38. doi: 10.1042/BJ20061088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Traxler P, Furet P. Strategies toward the design of novel and selective protein tyrosine kinase inhibitors. Pharmacol Ther. 1999;82(2–3):195–206. doi: 10.1016/s0163-7258(98)00044-8. [DOI] [PubMed] [Google Scholar]
- 40.Zabludoff SD, Deng C, Grondine MR, et al. AZD7762, a novel checkpoint kinase inhibitor, drives checkpoint abrogation and potentiates DNA-targeted therapies. Mol Cancer Ther. 2008;7(9):2955–2966. doi: 10.1158/1535-7163.MCT-08-0492. [DOI] [PubMed] [Google Scholar]
- 41.Casagrande F, Darbon J-M. Effects of structurally related flavonoids on cell cycle progression of human melanoma cells: regulation of cyclin-dependent kinases CDK2 and CDK11. Biochem Pharmacol. 2001;61(10):1205–1215. doi: 10.1016/s0006-2952(01)00583-4. [DOI] [PubMed] [Google Scholar]
- 42.Kim MC, Lee HJ, Lim B, et al. Quercetin induces apoptosis by inhibiting MAPKs and TRPM7 channels in AGS cells. Int J Mol Med. 2014;33(6):1657–1663. doi: 10.3892/ijmm.2014.1704. [DOI] [PubMed] [Google Scholar]
- 43.Kumar SK, LaPlant B, Chng WJ, et al. Dinaciclib, a novel CDK inhibitor, demonstrates encouraging single agent activity in patients with relapsed multiple myeloma. Blood. 2014;125(3):443–448. doi: 10.1182/blood-2014-05-573741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong DJL, Robert L, Atefi MS, et al. Antitumor activity of the ERK inhibitor SCH772984 [corrected] against BRAF mutant, NRAS mutant and wild-type melanoma. Mol Cancer. 2014;13:194. doi: 10.1186/1476-4598-13-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pargellis C, Tong L, Churchill L, et al. Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site. Nat Struct Biol. 2002;9(4):268–272. doi: 10.1038/nsb770. [DOI] [PubMed] [Google Scholar]
- 46.Cerbone A, Toaldo C, Pizzimenti S, et al. AS601245, an anti-inflammatory JNK inhibitor, and clofibrate have a synergistic effect in inducing cell responses and in affecting the gene expression profile in CaCo-2 colon cancer cells. PPAR Res. 2012;2012:269751. doi: 10.1155/2012/269751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naujok O, Lentes J, Diekmann U, Davenport C, Lenzen S. Cytotoxicity and activation of the Wnt/beta-catenin pathway in mouse embryonic stem cells treated with four GSK3 inhibitors. BMC Res Notes. 2014;7(1):273. doi: 10.1186/1756-0500-7-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee KW, Kang NJ, Heo Y-S, et al. Raf and MEK protein kinases are direct molecular targets for the chemopreventive effect of quercetin, a major flavonol in red wine. Cancer Res. 2008;68(3):946–955. doi: 10.1158/0008-5472.CAN-07-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Angiolini M. Targeting the DFG-in kinase conformation: a new trend emerging from a patent analysis. Future Med Chem. 2011;3(3):309–337. doi: 10.4155/fmc.10.294. [DOI] [PubMed] [Google Scholar]
- 50.Peng Y-H, Shiao H-Y, Tu C-H, et al. Protein kinase inhibitor design by targeting the Asp-Phe-Gly (DFG) motif: the role of the DFG motif in the design of epidermal growth factor receptor inhibitors. J Med Chem. 2013;56(10):3889–3903. doi: 10.1021/jm400072p. [DOI] [PubMed] [Google Scholar]
- 51.Staben ST, Siu M, Goldsmith R, et al. Structure-based design of thienobenzoxepin inhibitors of PI3-kinase. Bioorg Med Chem Lett. 2011;21(13):4054–4058. doi: 10.1016/j.bmcl.2011.04.124. [DOI] [PubMed] [Google Scholar]
- 52.Lawrence HR, Martin MP, Luo Y, et al. Development of o-chlorophenyl substituted pyrimidines as exceptionally potent aurora kinase inhibitors. J Med Chem. 2012;55(17):7392–7416. doi: 10.1021/jm300334d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zapf CW, Gerstenberger BS, Xing L, et al. Covalent inhibitors of interleukin-2 inducible T cell kinase (itk) with nanomolar potency in a whole-blood assay. J Med Chem. 2012;55(22):10047–10063. doi: 10.1021/jm301190s. [DOI] [PubMed] [Google Scholar]
- 54.Alevy YG, Patel AC, Romero AG, et al. IL-13–induced airway mucus production is attenuated by MAPK13 inhibition. J Clin Invest. 2012;122(12):4555–4568. doi: 10.1172/JCI64896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yokoyama T, Kosaka Y, Mizuguchi M. Structural insight into the interactions between death-associated protein kinase 1 and natural flavonoids. J Med Chem. 2015;58(18):7400–7408. doi: 10.1021/acs.jmedchem.5b00893. [DOI] [PubMed] [Google Scholar]
- 56.Paulke A, Eckert GP, Schubert-Zsilavecz M, Wurglics M. Isoquercitrin provides better bioavailability than quercetin: comparison of quercetin metabolites in body tissue and brain sections after six days administration of isoquercitrin and quercetin. Pharmazie. 2012;67(12):991–996. [PubMed] [Google Scholar]
- 57.Gee JM, DuPont MS, Day AJ, Plumb GW, Williamson G, Johnson IT. Intestinal transport of quercetin glycosides in rats involves both deglycosylation and interaction with the hexose transport pathway. J Nutr. 2000;130:2765–2771. doi: 10.1093/jn/130.11.2765. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Docked pose and hydrogen bond interactions of quercetin and isoquercitrin with AGC family serine/threonine kinases.
Notes: (A) Quercetin in Akt2, (B) isoquercitrin in Akt2, (C) quercetin in aurora kinase, (D) isoquercitrin in aurora kinase, (E) quercetin in RSK2, (F) isoquercitrin in RSK2. The protein is shown in light-blue cartoon representation, interacting residues are shown in stick representation, and the docked ligand is represented as black sticks.
Abbreviation: PDB, Protein Data Bank.
Docked pose and hydrogen bond interactions of quercetin and isoquercitrin with CMGC family serine/threonine kinases.
Notes: (A) Quercetin in ERK2, (B) isoquercitrin in ERK2, (C) quercetin in p38α, (D) isoquercitrin in p38α, (E) quercetin in JNK1, (F) isoquercitrin in JNK1, (G) quercetin in GSK3B, (H) isoquercitrin in GSK3B, (I) quercetin in CLK1, (J) isoquercitrin in CLK1. The protein is shown in light-blue cartoon representation, interacting residues are shown in stick representation, and the docked ligand is represented as black sticks.
Abbreviation: PDB, Protein Data Bank.
Docked pose and hydrogen bond interactions of quercetin and isoquercitrin with STE family serine/threonine kinases.
Notes: (A) Quercetin in PAK4, (B) isoquercitrin in PAK4, (C) quercetin in Plk1, (D) isoquercitrin in Plk1. The protein is shown in light-blue cartoon representation, interacting residues are shown in stick representation, and the docked ligand is represented as black sticks.
Abbreviation: PDB, Protein Data Bank.