Abstract
Background
Semaphorin 4D (Sema4D) is highly expressed in certain types of tumors and functions in the regulation of tumor angiogenesis and growth. However, it is still not clear regarding the roles of Sema4D in breast cancer. This study was designed to explore the effects of Sema4D on proliferation, cell cycle progression, apoptosis, invasion, migration, tumor growth, and angiogenesis in breast cancer.
Materials and methods
The expression level of Sema4D was investigated in MCF10A, 184A1, HCC1937, MDA-MB-468, MDA-MB-231, Hs578T, BT474, MCF-7, and T47D breast cancer cell lines by Western blotting analysis. Sema4D downregulation or overexpression was established by infection with lentiviruses-encoding Sema4D short hairpin RNA (shRNA) or Sema4D. To evaluate the effects of Sema4D on cell proliferation, cell cycle progression, apoptosis, invasion, and migration of MDA-MB-231 and MDA-MB-468 cells, methods including MTT assay, flow cytometry, wound healing assay, and transwell experiments were applied. BALB/c nude mice were injected with MDA-MB-231 cells, which were respectively infected with lentiviruses-encoding Sema4D, Sema4D shRNA, and GFP, followed by tumor angiogenesis assay.
Results
Sema4D was expressed at higher levels in breast cancer cell lines compared with the normal human breast epithelial cell lines, especially in MDA-MB-231 and MDA-MB-468 cells. Cell proliferation ability was remarkably inhibited in Sema4D downregulated condition, whereas the proportions of cells in the G0/G1 phase and apoptosis increased in MDA-MB-231 and MDA-MB-468 cells. In addition, the invasion and migration abilities of these cells were obviously reduced. Xenograft growth as well as angiogenesis was inhibited when infected with lentiviruses-encoding Sema4D shRNA in vivo.
Conclusion
Downregulation of Sema4D had notable influence on cell proliferation ability, invasion, migration, and apoptosis of both MDA-MB-231 and MDA-MB-468 cells. Furthermore, infection with lentiviruses-encoding Sema4D shRNA obviously inhibited tumor growth and angiogenesis in BALB/c nude mice. Our results showed that Sema4D may represent a novel therapeutic target for human breast cancer.
Keywords: semaphorin 4D, breast cancer, apoptosis, angiogenesis, proliferation
Introduction
Human breast cancer is a highly malignant tumor, making it urgent and important to develop new biomarkers and therapeutic targets to treat this type of cancer. Recent studies have shown that semaphorin 4D (Sema4D) is overexpressed in several carcinomas, including head and neck squamous cell carcinoma, colorectal cancer (CRC), lung cancer, ovarian cancer, oral squamous cell carcinoma, pancreatic cancer, and prostate cancer. It functions in the regulation of immune function, epithelial morphogenesis, invasion, growth, and the vascularity of tumor xenografts in vivo.1–11 It was reported that tumors that overexpress Sema4D are highly aggressive with poor prognosis and not easy to control during treatment.1,7,12,13 Emerging evidence indicates that Sema4D might promote tumor growth and angiogenesis.8,9,14,15 On the other hand, small GTPase Ras homolog (Rho) proteins, including RhoA, are highly expressed in human cancers.16,17 Rho kinases (also termed ROCKs) are involved in cancer invasion and metastasis through their involvement in the Rho/ROCK pathway.18,19 The results reported by several research groups support the hypothesis that the inhibition of RhoA phosphorylation might be effective in preventing invasion and metastasis.20,21 This study investigated the role of Sema4D on the proliferation, cell cycle progression, apoptosis, invasion, and migration of breast cancer cells in vitro, and investigated the role of Sema4D in tumor growth and angiogenesis in vivo to determine the role of Sema4D on tumor development and angiogenesis in human breast cancer; the study further explored the effect of Sema4D on the phosphorylation of RhoA in breast cancer.
Materials and methods
Cell culture
All breast cell lines and packaging 293T cells were gifts from the Kunming Institute of Zoology, Chinese Academy of Sciences (Kunming, People’s Republic of China). All cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM)/F12 supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, MA, USA). Cells were grown under a humidified atmosphere containing 5% CO2 at 37°C. Cell transfection was carried out using Lipofectamine2000™ (Thermo Fisher Scientific).
Lentiviral vector system
The lentiviral vector system used was a gift from Dr John R Basile (University of Maryland Dental School, Baltimore, MD, USA). The following lentiviral vectors were included: Sema4D, Sema4D short hairpin RNA (shRNA), and control lentivirus. The oligonucleotide used for Sema4D shRNA was same as the previous report.6 Reference gene accession number for Sema4D was NM_006378.3. Sema4D expression constructs applied in the study encoded for full-length Sema4D. The lentiviral viruses were prepared and used as previously reported.22–26
Western blotting analysis
The expression level of Sema4D was detected in the following breast cancer cell lines by Western blotting: HCC1937, MDA-MB-231, MDA-MB-468, Hs578T, BT474, MCF-7, and T47D. Briefly, samples were harvested using RIPA lysis buffer (Solarbio, Beijing, People’s Republic of China) containing protease inhibitor cocktail (Hoffman-La Roche Ltd., Basel, Switzerland); then, protein concentrations were measured by the BCA Protein Assay Kit (TIANGEN Biotech, Beijing, People’s Republic of China). Then, the samples were loaded for sodium dodecyl sulfate-polyacrylamide gel electrophoresis electrophoreses and then transferred onto a polyvinylidene difluoride membrane (EMD Millipore, Billerica, MA, USA). β-Actin was used as a loading control. The membrane was then blocked with 5% fat-free milk and incubated with the following primary antibodies separately: mouse anti-Sema4D antibody (BD Biosciences, San Jose, CA, USA), mouse anti-RhoA monoclonal antibody (Santa Cruz Biotechnology Inc., Dallas, TX, USA), mouse anti-phosphorylated-RhoA (Santa Cruz Biotechnology Inc.), and mouse anti-β-actin antibody (Santa Cruz Biotechnology Inc.). The membranes were then incubated with the secondary antibody (goat anti-mouse IgG; KPL, Gaithersburg, MD, USA). Protein bands were visualized using Super Signal reagents (Thermo Fisher Scientific) and then imaged using Image Quant LAS 4000 (GE Healthcare Bio-Sciences Corp., Piscataway, NJ, USA).
Cell proliferation assay
The number of viable cells was estimated using the MTT assay. Twenty-four hours after infection, MDA-MB-231 and MDA-MB-468 cells were plated in 96-well plates in a medium containing 10% FBS at a density of 2,000 cells/well. To quantitate cell viability, the cultures were stained after 1, 2, 3, 4, and 5 days; 100 μL MTT (Sigma-Aldrich Co., St Louis, MO, USA) working solution was added into the wells for 4 hours at 37°C, after which the culture medium was removed and 150 μL dimethyl sulfoxide was added. The absorbance was measured at 450 nm using an Epoch Multi-Volume Spectrophotometer System (BioTek, Winooski, VT, USA).
Wound healing assay
Wound healing assays were generally performed as per the previous report.1 Briefly, the cells were cultured to 90% confluence and starved in a serum-free medium overnight. The cell monolayer was then scratched using a 1 mL pipette tip across the diameter of the wells. After aspiration of non-adherent cells, the plates were rinsed twice with phosphate-buffered saline (PBS). Fresh medium containing 2% FBS was added to the cells every 24 hours. The wound edges were observed under microscope within 48 hours; the images were then analyzed by NIH Image software. A minimum of five randomly chosen areas was measured.
Cell migration assay
Boyden chamber (EMD Millipore) was used to determine the ability of cell migration; 1×105 lentivirus-infected cells were suspended in DMEM/F12 (1:1) containing 0.1% bovine serum albumin (BSA) and then placed onto the top of each chamber. Medium containing 10% FBS was added to the bottom of the chamber. The cells were incubated for 24 hours, and then the cells on the upside of membrane were wiped off for removing the non-migrated cells. Cells that migrated to the underside of the membrane were stained with crystal violet and visualized under a microscope; four random fields (magnification ×100) were scanned and analyzed using NIH Image software. The results are expressed as a pixel intensity bar graph, and the averages and standard deviations were calculated.
Cell invasion assay
A fixed number of cells (1×105 cells), suspended in DMEM/F12 (1:1) containing 0.1% BSA, was added to the top of the Boyden chamber of the Boyden chamber (EMD Millipore), which was coated with 50 μL Matrigel (1 mg/mL final concentration); the lower chamber contained 10% serum-containing medium. After incubation for 24 hours, the cells on the upside of membrane were wiped off to remove the non-invaded cells. The invaded cells were detected by staining with crystal violet and visualized under a microscope (magnification ×100); four random fields were scanned and analyzed using NIH Image software. The results are expressed as a pixel intensity bar graph, and the averages and standard deviations were calculated.
Cell apoptosis assay
Cell apoptosis was performed using an Annexin V-Fluorescein Isothiocyanate Apoptosis Detection Kit (BioVision, Milpitas, CA, USA) and analyzed using a Becton-Dickinson Accuri C6 flow cytometer and CellQuest (version 3.3) software (BD Biosciences). After 48 hours of lentivirus infection, the cells were washed with PBS, fixed in 2% paraformaldehyde for 30 minutes, and permeabilized using 0.1% Triton-X 100 in PBS for 30 minutes. The cells were centrifuged for 5 minutes at 1000× g, and then the cells were resuspended at a density of 1×106 cells/mL in 1× binding buffer and stained with annexin V-fluorescein isothiocyanate and propidium iodide for 5 minutes, followed by flow cytometry detection. The rate of apoptosis was analyzed using CellQuest software (version 3.3) (BD Biosciences).
Cell cycle analysis
Lentivirus-infected cells were cultured in 6 cm dishes to 70%–80% confluence. After 48 hours of incubation, both floating and adherent cells were collected after trypsinization and washed with ice-cold PBS; the cells were then fixed overnight using 70% ethanol in PBS at −20°C. Fixed cells were then treated with 20 μg/mL RNaseA and 50 μg/mL propidium iodide (Sigma-Aldrich Co.) for 30 minutes at room temperature in the dark; the stained cells were then immediately analyzed using an Accuri C6 flow cytometer (BD Biosciences). The data were analyzed using the accompanying software (CellQuest), and the averages and standard deviations were calculated. Ten thousand events per sample were counted, and the experiments were repeated three times.
Tumor cell injections and animal studies in BALB/C nude mice
Twenty nude mice were randomly divided into four groups: 1) lentivirus coding for Sema4D shRNA, 2) lentivirus coding for Sema4D, 3) lentivirus coding for GFP, and 4) 0.9% normal saline. MDA-MB-231 cells (2×106) that had been infected with different lentiviruses were inoculated into the back of BALB/C nude mice. MDA-MB-231 cells were resuspended in 100 μL of serum-free DMEM with the same volume of liquid Matrigel (BD Biosciences) and injected subcutaneously into immunocompromised nude mice. The mice were euthanized on day 24 following the injections, and the tumors were removed. Tumor sizes were measured during the experiment. The long-diameter (a) and short-diameter (b) of the tumors were measured, and then the volume (V) was calculated using the formula V =1/2a × b2. After the tumor had been photographed, the animals were euthanized, and the tumors were removed, weighed, and processed for immunohistochemistry and immunofluorescence. Procedures involving animals and their care were conducted in conformity with NIH guidelines (NIH Pub. No. 85-23, revised 2010) and was approved by Animal Ethical and Welfare Committee of the Affiliated Children’s Hospital of Kunming Medical University.
Immunohistochemistry
The immunohistochemistry analysis was performed as previously described.9 Briefly, tumor tissues were fixed, and then the slides were rinsed with PBS and blocked in 1% BSA at 4°C overnight. The primary antibody used was mouse anti-Sema4D (BD Biosciences). The slides were then washed in PBS followed by incubation with secondary antibody (goat anti-mouse IgG antibody; KPL) for 30 minutes at 37°C. The slides were developed using an HRP-DAB (TIANGEN Biotech) and then stained with hematoxylin, dehydrated, and mounted. Images were observed using a Nikon Eclipse TE2000-U microscope.
Immunofluorescence
The immunofluorescence analysis of tumor tissues was performed as previously described.9 The slides were rehydrated and blocked in 1% BSA and then incubated with primary antibodies (rat anti-mouse CD31; BD Biosciences Pharmingen, San Diego, CA, USA). The slides were then washed thrice with PBS followed by incubation with secondary antibody (goat anti-rat IgG; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA). The slides were mounted and observed under a Nikon Eclipse TE2000-U microscope. The vascular content of the sections was determined by the average vessel density in five stained slides from each group. Vessel density was analyzed using ImageJ software.
Statistical analysis
Independent Student’s t-tests were used for statistical analysis between different groups. Experimental values were described as mean ± standard deviation. P-values <0.05 indicated significant difference between two groups.
Results
Sema4D is highly expressed in breast cancer cell lines
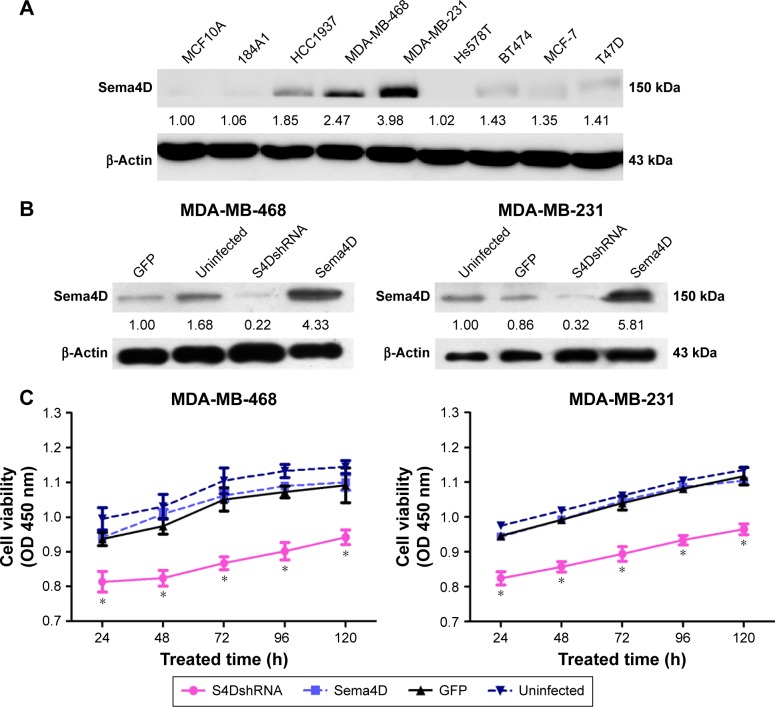
The expression level of Sema4D was investigated in the breast cancer cell lines HCC1937, MDA-MB-231, MDA-MB-468, Hs578T, BT474, MCF-7, and T47D using Western blotting analysis. The results showed that all cell lines expressed Sema4D at higher levels than those found in the normal human breast epithelial cell lines MCF10A and 184A1, with the exception of Hs578T (Figure 1A). In addition, our results showed that Sema4D expression was significantly higher in the MDA-MB-231 and MDA-MB-468 cell lines compared with the others (Figure 1A).
Figure 1.
The expression of Sema4D in seven breast cancer cell lines and in normal human breast epithelial cell lines.
Notes: (A) Sema4D protein levels were detected by Western blotting; β-actin was used as a loading control (lower panels). The following cell lines were analyzed: HCC1937, MDA-MB-231, MDA-MB-468, Hs578T, BT474, MCF-7, T47D, and the normal human breast epithelial cell lines MCF10A and 184A1. Protein levels are quantified below each blot as the fold increase relative to MCF10A cells. Bars indicate the means of three individual experiments ± standard errors. (B) Western blotting analysis of Sema4D in the indicated cell lines; β-actin was used as the loading control. Protein levels are quantified below each blot as the fold increase relative to loading controls. Each cell line was separately infected by control lentivirus-expressing enhanced GFP, lentivirus coding for Sema4D shRNA, and lentivirus coding for Sema4D. The following cell lines were included in the analysis: MDA-MB-231 and MDA-MB-468 (*P<0.05, relative to both the uninfected and GFP groups). (C) The proliferation of Sema4D in MDA-MB-231 and MDA-MB-468 cells was detected using the MTT assay.
Abbreviations: Sema4D, semaphorin 4D; shRNA, short hairpin RNA.
Proliferation of MDA-MB-231 and MDA-MB-468 cells was significantly inhibited by Sema4D interference
Western blotting analysis was used to determine the expression level of Sema4D in MDA-MB-231 and MDA-MB-468 cells; according to the results, Sema4D was significantly overexpressed or downregulated in MDA-MB-231 and MDA-MB-468 cells after infection with lentivirus-expressing Sema4D or Sema4D shRNA when compared with the uninfected cell group (both P<0.05) (Figure 1B).
Proliferation of MDA-MB-231 and MDA-MB-468 cells was determined using the MTT assay. The OD values of Sema4D shRNA-infected MDA-MB-231 cells at 24, 48, 48, 96, and 120 hours were significantly lower than those in the uninfected group (P<0.05), indicating that cell growth was significantly inhibited. Cell proliferation was not significantly different in the GFP group and the Sema4D group, compared with the uninfected group (P>0.05). Similarly, the OD values of Sema4D shRNA-infected MDA-MB-468 cells at 24, 48, 72, 96, and 120 hours were significantly lower than those recorded for the uninfected group (P<0.05), indicating that cell growth was significantly inhibited. Cell proliferations of the GFP group and Sema4D group were not significantly changed, compared with the uninfected group (P>0.05). The results demonstrate that interference of Sema4D significantly affects the proliferation of MDA-MB-231 and MDA-MB-468 cells (Figure 1C).
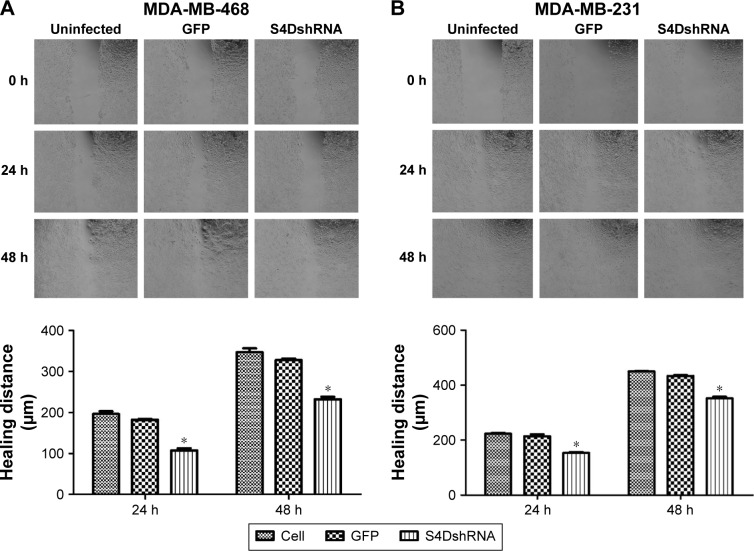
The lateral migration ability of the cells was significantly decreased in response to the downregulation of Sema4D expression
According to the wound healing assay, the wound healing distances of the uninfected MDA-MB-468 cell group at 24 and 48 hours were 196.3±11.5 and 346.3±16.9 μm, respectively, whereas those of the Sema4D shRNA group were 107.6±8.7 and 232.3±10.3, respectively (Figure 2A). The lateral migration ability of the cells was significantly decreased (P<0.05) in response to the downregulation of Sema4D expression. Similarly, the wound healing distances of the uninfected MDA-MB-231 cell group at 24 and 48 hours were 223±6.3 and 450.3±2.7 μm, respectively. The corresponding values for the Sema4D shRNA group were 153.6±5.5 and 352.3±10.3 μm, respectively (Figure 2B). Based on these results, the interference of Sema4D significantly decreased the lateral migration ability of MDA-MB-231 and MDA-MB-468 cells (P<0.05).
Figure 2.
Wound healing assay in MDA-MB-231 and MDA-MB-468 cells.
Notes: Breast cancer cells (uninfected or infected by the indicated lentivirus) were examined in six-well plates. Two breast cancer cell lines were included: MDA-MB-231 and MDA-MB-468. Three groups of each cell line were tested: uninfected, infected with control lentivirus-expressing GFP, and lentivirus coding for Sema4D shRNA. The results for MDA-MB-468 (A) and MDA-MB-231 (B) are shown (×40). Bar graphs show the healing distances, as analyzed using NIH Image software. Error bars represent the standard deviation of the results for three wells (*P<0.05, relative to both the uninfected and GFP groups).
Abbreviations: Sema4D, semaphorin 4D; shRNA, short hairpin RNA.
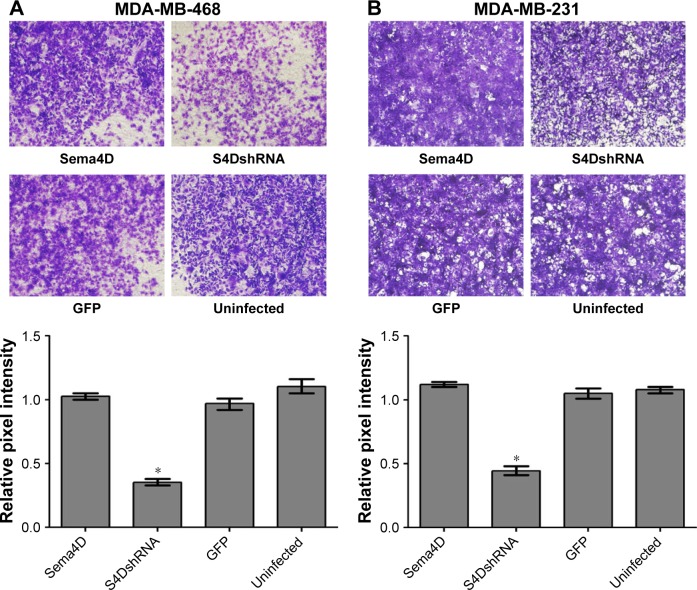
The migration ability of the MDA-MB-468 and MDA-MB-231 cells was obviously reduced by the downregulation of Sema4D expression
To evaluate the contributions of Sema4D to migration in the MDA-MB-231 and MDA-MB-468 cells, the cells were infected with lentiviruses-expressing Sema4D, Sema4D shRNA, and GFP and analyzed using transwell experiments. The results showed that the migration abilities of the MDA-MB-231 and MDA-MB-468 cells were obviously reduced (P<0.05) by the downregulation of Sema4D expression (Figure 3A and B). By contrast, the group infected with Sema4D-expressing lentivirus showed no significant difference between the control groups, perhaps due to the high Sema4D background in the MDA-MB-231 and MDA-MB-468 cells.
Figure 3.
In vitro migration assay of MDA-MB-231 and MDA-MB-468 cells.
Notes: Breast cancer cells, uninfected or infected by the indicated lentivirus, were examined in a transwell plate. Two breast cancer cell lines were included: MDA-MB-231 and MDA-MB-468. Four groups of each cell line were tested, including cells that were uninfected or infected with the indicated lentivirus: a control lentivirus-expressing GFP, a Sema4D-coding lentivirus, or a lentivirus coding for Sema4D shRNA. MDA-MB-468 (A) and MDA-MB-231 (B) cells on the membranes were stained with crystal violet (×100). Bar graphs below the stained migration membranes show the results of the migration assay, as analyzed using NIH Image software and expressed as the pixel intensity of scanned stained migration membranes relative to negative controls. Error bars represent the standard deviation of the results obtained from six wells (*P<0.05, relative to both the uninfected and GFP groups).
Abbreviations: Sema4D, semaphorin 4D; shRNA, short hairpin RNA.
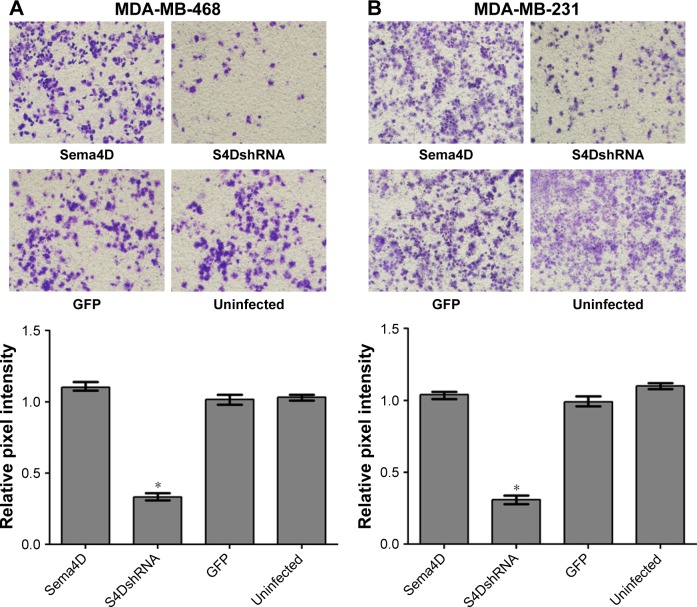
The invasive ability of the MDA-MB-468 and MDA-MB-231 cells was obviously reduced by the downregulation of Sema4D expression
To evaluate the contributions of Sema4D to invasion in the MDA-MB-468 and MDA-MB-231 cell lines, the cells were infected with lentiviruses-expressing Sema4D, Sema4D shRNA, and GFP and then analyzed using transwell experiments. The invasive abilities of the MDA-MB-468 and MDA-MB-231 cells were obviously inhibited (P<0.05) after infection with lentivirus-encoding Sema4D shRNA (Figure 4A and B). By contrast, the group that was infected with Sema4D-expressing lentivirus showed no obvious difference between the control groups, perhaps due to the high Sema4D background in the MDA-MB-468 and MDA-MB-231 cells.
Figure 4.
In vitro invasion assay of MDA-MB-231 and MDA-MB-468 cells.
Notes: Breast cancer cells, uninfected or infected by the indicated lentivirus, were examined in a transwell plate assay. Two breast cancer cell lines were included: MDA-MB-231 and MDA-MB-468. Four groups of each cell line were tested, including cells that were uninfected or infected with the indicated lentivirus: control lentivirus-expressing GFP, a lentivirus coding for Sema4D, or a lentivirus coding for Sema4D shRNA. MDA-MB-468 (A) and MDA-MB-231 (B); cells on the membranes were stained with crystal violet (×100). Bar graphs below the stained membranes show the results of the invasion assay, as analyzed using NIH Image software and expressed as the pixel intensity of scanned stained membranes relative to negative controls. Error bars represent the standard deviation of the results obtained from six wells (*P<0.05, relative to both the uninfected and GFP groups).
Abbreviations: Sema4D, semaphorin 4D; shRNA, short hairpin RNA.
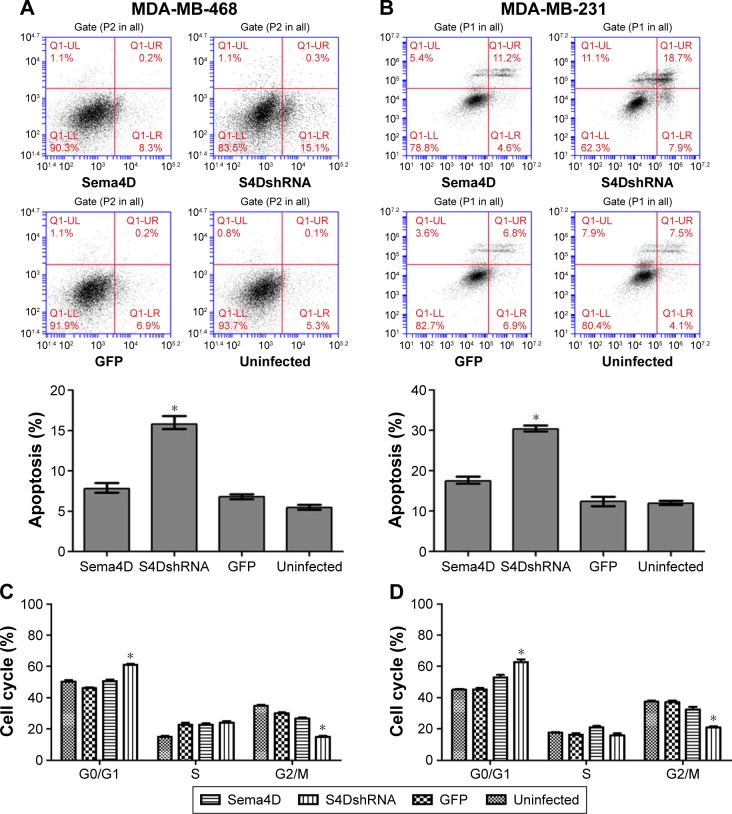
Downregulation of Sema4D promoted the apoptosis of the MDA-MB-231 cells and MDA-MB-468 cells
The percentage of apoptotic cells (15.4%) in the MDA-MB-468 cells infected with Sema4D shRNA was significantly higher than that in the uninfected group (5.4%) (P<0.05); however, no significant difference was observed between the other groups (P>0.05). Similarly, the percentage of apoptotic cells (18.7%) in the MDA-MB-231 cells infected with Sema4D shRNA was significantly higher than that in the uninfected group (4.1%) (P<0.05); however, no significant difference was observed between the other groups (P>0.05). These results indicate that the downregulation of Sema4D promoted apoptosis in the MDA-MB-231 and MDA-MB-468 cells (Figure 5A and B). By contrast, the group that was infected with Sema4D-expressing lentivirus did not differ from the control group, perhaps due to the high Sema4D background in the MDA-MB-468 and MDA-MB-231 cells.
Figure 5.
Effects of Sema4D interference on apoptosis and cell cycle progression in MDA-MB-231 and MDA-MB-468 cell lines.
Notes: Cells were treated in four groups, uninfected or infected with the following indicated lentiviruses: a control lentivirus-expressing GFP, a Sema4D-coding lentivirus, or a lentivirus coding for Sema4D shRNA. Apoptosis and cell cycle stage were analyzed by flow cytometry in MDA-MB-468 (A and C) and MDA-MB-231 (B and D) breast cancer cells (*P<0.05, relative to both the uninfected and GFP groups). The bar graph summarizes the data from results of Sema4D interference on apoptosis and cell cycle progression in MDA-MB-231 and MDA-MB-468 cell lines. Bars indicate mean of three individual experiments ± standard error.
Abbreviations: Sema4D, semaphorin 4D; shRNA, short hairpin RNA.
Sema4D downregulation affected the MDA-MB-231 and MDA-MB-468 cell cycle distribution
The cell cycle distribution of the MDA-MB-231 and MDA-MB-468 cells at 48 hours after infection with lentivirus was monitored by flow cytometry. The results revealed that the cells that had been infected with Sema4D shRNA exhibited more abundant cells at G1 phase and less abundant cells at G2 phase compared with the GFP and uninfected cells; by contrast, there was no distinguishable difference between the GFP and uninfected cells. The proportion of cells at G0/G1 phase significantly increased (P<0.05), indicating that Sema4D downregulation affected the MDA-MB-231 and MDA-MB-468 cell cycle distribution. By contrast, the group that had been infected with Sema4D-expressing lentivirus exhibited no significant difference compared with the control group (Figure 5C and D); this result was perhaps due to the high Sema4D background in the MDA-MB-468 and MDA-MB-231 cells.
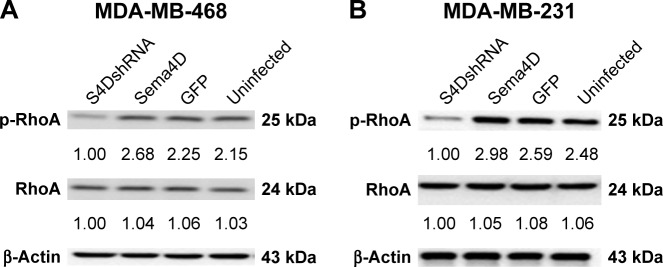
Sema4D inhibition can affect the phosphorylation of RhoA in breast cancer
The expressions of RhoA and phosphorylated RhoA (p-RhoA) were analyzed in MDA-MB-231 and MDA-MB-468 cells that had been infected with lentiviruses-expressing Sema4D, Sema4D shRNA, and GFP. Cell lysates were harvested and then analyzed using Western blotting. The total expression levels of RhoA were not affected in the MDA-MB-231 and MDA-MB-468 cells; by contrast, the p-RhoA expression levels measured for the Sema4D shRNA group were lower in both the cell lines than those found in the uninfected group (P<0.05). By contrast, the group that had been infected with Sema4D-expressing lentivirus showed no obvious difference with respect to the control group (Figure 6A and B), perhaps due to high Sema4D background in the MDA-MB-468 and MDA-MB-231 cells.
Figure 6.
RhoA and p-RhoA expression in MDA-MB-468 and MDA-MB-231 cell lines.
Notes: Cells were treated in four different groups, uninfected or infected with the indicated lentivirus: a control lentivirus-expressing GFP, a Sema4D-coding lentivirus, or a lentivirus coding for Sema4D shRNA. Western blotting analysis for RhoA and p-RhoA in MDA-MB-468 (A) and MDA-MB-231 (B) breast cancer cell lines; β-actin was used as a loading control (lower panels). Protein levels are quantified below each blot as the fold increase relative to the loading controls (P<0.05, relative to both the uninfected and GFP groups).
Abbreviations: RhoA, Ras homolog A; p-RhoA, phosphorylated RhoA; Sema4D, semaphorin 4D; shRNA, short hairpin RNA.
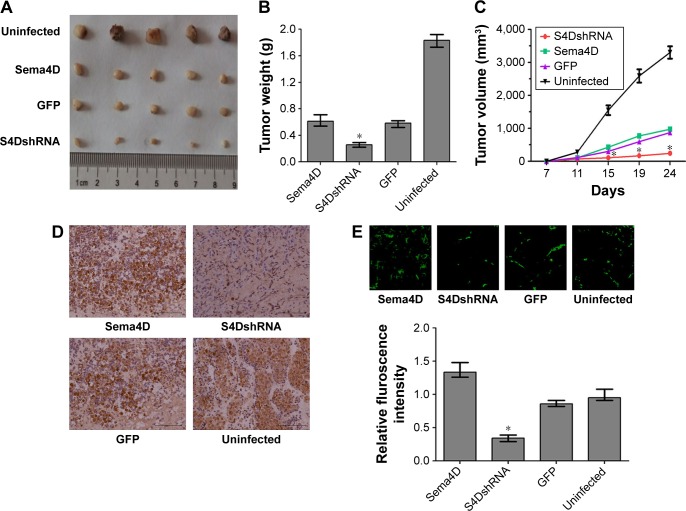
Sema4D interference contributes to the inhibition of tumor growth and vascularity in MDA-MB-231 cells
To examine the effect of Sema4D in tumor growth and vascularity in MDA-MB-231 cells, the cells were infected with lentiviruses-expressing Sema4D, Sema4D shRNA, and GFP, and inoculated into the back of BALB/C nude mice. The tumors resulting from cells that had been infected by Sema4D shRNA lentiviruses weighed approximately ten times less than the corresponding controls. Tumors from uninfected MDA-MB-231 cells were the largest (Figure 7A–D), and tumors that overexpressed Sema4D were heavier. Vascularity was the most inhibited in tumors resulting from cells that had been infected by Sema4D shRNA lentiviruses and consisted of scattered small vessels in the tumors; this finding indicates that the downregulation of Sema4D clearly reduced tumor growth and vascularity (Figure 7E). Although the tumors resulting from cells that had been infected with lentiviruses coding for Sema4D were larger, these tumors showed much higher vessel density than the control group. Some similarities in vessel distribution were observed between the uninfected group and the control group. Tumors from the uninfected group were the heaviest and largest among all the groups, while the vascular density only exceeded in the Sema4D-overexpressing groups by ~20%. Tumor growth and angiogenesis were inhibited in BALB/c nude mice after the injection of MDA-MB-231 cells that had been infected with lentivirus coding for Sema4D-shRNA. Lentivirus itself probably influences the in vivo growth rate of MDA-MB-231 cells, thus causing the uninfected cells to exhibit different results than the other three lentivirus-infected groups.27,28
Figure 7.
Results of an in vivo tumorigenesis assay for MDA-MB-231 cells.
Notes: Cells were treated in four different groups, uninfected or infected with the indicated lentivirus: a control lentivirus-expressing GFP, a Sema4D-coding lentivirus, or a lentivirus coding for Sema4D shRNA. Cells from the four groups were injected subcutaneously into nude mice along with basement membrane matrix. (A) MDA-MB-231 xenografts were infected by the indicated lentiviruses. (B) Tumor weights in the four groups of MDA-MB-231 cells are shown as a bar graph (n=5; *P<0.05, relative to both the uninfected and GFP groups). Bars indicate the means of five tumor weights ± standard errors. (C) Tumor growth curve for the four groups of MDA-MB-231 cells and tumor volume measurements (mm3) are shown, (n=5; *P<0.05, relative to both the uninfected and GFP groups). (D) Immunohistochemical measurements of Sema4D in tumor tissues harvested from the four indicated groups. Clear differences in Sema4D expression were found among the four indicated groups. (E) CD31 immunofluorescence staining for the evaluation of vascular density; CD31 immunofluorescence staining of frozen section of tumors derived from the four indicated groups is shown here. The bar graphs show the vascular contents of these tumors, which were determined based on the average vessel density in five CD31-stained sections from each group (n=5; *P<0.05, relative to both the uninfected and GFP groups). Bars indicate means of vessel density in ten CD31-stained sections ± standard errors. Scale bars =100 μm.
Abbreviations: Sema4D, semaphorin 4D; shRNA, short hairpin RNA.
Discussion
Human breast cancer is a highly malignant tumor; thus, the development of new biomarkers and therapeutic targets has become critical. The main cause of poor prognosis in breast cancer patients is the high invasive and metastatic potential of this cancer. Sema4D is a ligand that is known to be expressed in head and neck squamous cell carcinoma, CRC, lung cancer, ovarian cancer, oral squamous cell carcinoma, pancreatic cancer, and prostate cancer. It is reportedly involved in the regulation of immune functions, epithelial morphogenesis, tumor growth, and metastasis.6,10–12,26 Cell–cell signaling of semaphorin ligands through interaction with their plexin receptors is important for the homeostasis and morphogenesis of many tissues and has been widely studied in terms of its role in neural connectivity, cell migration, apoptosis, and immune responses.29–31 The overexpression of Sema4D is significantly correlated with clinical factors, such as lymph node metastasis, distant metastasis, and poor prognosis in patients with CRC.2 In the present study, we report that Sema4D mediates cell invasion, apoptosis, and tumor growth in breast cancer. We found that Sema4D was highly expressed among seven human breast cancer cells with different metastatic potentials (with the exception of the Hs578T cell line), and significantly higher Sema4D expression was confirmed in MDA-MB-231 and MDA-MB-468 cells. Using in vitro analysis, we found that the proliferation of MDA-MB-231 and MDA-MB-468 cells was significantly inhibited in the Sema4D shRNA interference group (P<0.05), and the proportions of cells in the G0/G1 phase and undergoing apoptosis significantly increased (P<0.05). In addition, the invasion and migration of breast cancer cells were obviously reduced (P<0.05), suggesting that a blockade of Sema4D might diminish the invasive potential and development of breast cancer cells. Twenty-four hours after infection was chosen as the time point in cell invasion assay in order to avoid the influence of cell viability. As to wound healing assay, a medium containing 2% FBS was used other than 10% FBS, which is used in the MTT assay, in order to avoid the influencing of viability. The experiments to evaluate the effect of Sema4D on cell migration, invasion, and wound healing were also performed in the low Sema4D background cell line (Hs578T) by overexpression of Sema4D. Our results showed that overexpressed Sema4D might increase the invasive potential and migration of Hs578T, a breast cancer cell line with low Sema4D background (Figures S1–S3). Inhibition of tumor growth and angiogenesis was observed in nude mice after the injection of MDA-MB-231 cells that had been infected with lentivirus coding for Sema4D shRNA for 24 days; this result provides evidence that Sema4D down-regulation might play a significant role in tumor growth and angiogenesis in breast cancer. Our findings provide a new insight into possible prognostic biomarkers and therapeutic targets in breast cancer patients.
It has been reported that the binding of Sema4D to plexinB1 induces the activation of members of the small GTPase Rho gene family, resulting in the phosphorylation of MAPK and Akt.29,32–34 Upregulation or overexpression of the Rho protein correlates with poor prognosis in gastric cancer patients.35,36 Rho GTPases including RhoA are highly expressed in human cancers.17,18 ROCKs are downstream effector molecules of Rho that are involved in cancer invasion and metastasis through their role in the Rho/ROCK pathway.21 The results of a recent study support the hypothesis that the inhibition of RhoA phosphorylation might provide an effective way for the reduction of invasion and metastasis.37,38 We also found that p-RhoA could be down-regulated when the Sema4D level was reduced by infecting with lentivirus-encoding Sema4D shRNA. However, the role of Sema4D in human breast cancer malignancy and the associated signaling pathway warrant further study.39
In conclusion, Sema4D could play a significant role in tumor development and angiogenesis in breast cancer. Sema4D might represent a valuable target for antiangiogenic therapies in breast cancer.
Supplementary materials
Wound healing assay in Hs578T cells.
Notes: Breast cancer Hs578T cells (infected by the indicated lentivirus) were examined in six-well plates. Three groups of cells were tested: Sema4D-coding lentivirus, control lentivirus-expressing GFP, and lentivirus coding for Sema4D shRNA. The results are shown (×40). Bar graphs show the healing distances, as analyzed using NIH Image software. Error bars represent the standard deviation of the results for three wells (*P<0.05, relative to the GFP group).
Abbreviations: Sema4D, semaphorin 4D; shRNA, short hairpin RNA.
In vitro migration assay of Hs578T cells.
Notes: Breast cancer Hs578T cells, uninfected or infected by the indicated lentivirus, were examined in a transwell plate. Four groups of each cell line were tested, including cells that were uninfected or infected with the indicated lentivirus: control lentivirus-expressing GFP, Sema4D-coding lentivirus, or lentivirus coding for Sema4D shRNA. Cells migrated to the lower side of the membranes were stained with crystal violet (×100). Bar graphs below the stained migration membranes show the results of the migration assay, as analyzed using NIH Image software and expressed as the pixel intensity of stained migration membranes relative to negative control. Error bars represent the standard deviation of the results obtained from six wells (*P<0.05, relative to the uninfected group).
Abbreviations: Sema4D, semaphorin 4D; shRNA, short hairpin RNA.
In vitro invasion assay of Hs578T cells.
Notes: Breast cancer Hs578T cells, uninfected or infected by the indicated lentivirus, were examined in a transwell plate assay. Four groups of each cell line were tested, including cells that were uninfected or infected with the indicated lentivirus: control lentivirus-expressing GFP, lentivirus coding for Sema4D, or lentivirus coding for Sema4D shRNA; cells on the lower side of membranes were stained with crystal violet (×100). Bar graphs below the stained membranes show the results of the invasion assay, as analyzed using NIH Image software and expressed as the pixel intensity of stained membranes relative to negative control. Error bars represent the standard deviation of the results obtained from six wells (*P<0.05, relative to the uninfected group).
Abbreviations: Sema4D, semaphorin 4D; shRNA, short hairpin RNA.
Acknowledgments
The authors would like to thank Dr John R Basile from the University of Maryland Dental School for technical assistance. This research was supported by the National Natural Science Foundation of China (grant number 81171946), the Natural Science Foundation of Yunnan Province (grant numbers 2011CA016 and 2012FB188), the Association Foundation Program of Yunnan Science and Technology Department and Kunming Medical University (grant number 2014FZ058), the Innovation Foundation of Kunming Medical University for PhD candidates (grant number 2014D07), the Training Project for Medical Science and Technology Experts in Kunming City (grant number SW-75), and the Foundation of Yunnan Medical Science and Technology (grant number 2016NS124).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Ruan SS, Li RC, Han Q, et al. Expression and clinical significance of semaphorin4D in non-small cell lung cancer and its impact on malignant behaviors of A549 lung cancer cells. J Huazhong Univ Sci Technolog Med Sci. 2014;34(4):491–496. doi: 10.1007/s11596-014-1304-2. [DOI] [PubMed] [Google Scholar]
- 2.Mu L, Wang J, Chen Y, et al. Hypoxia-inducible factor-1alpha and semaphorin4D genes involved with tumor-associated macrophage-induced metastatic behavior and clinical significance in colon cancer. Chinese Med J (Engl) 2014;127(20):3568–3575. [PubMed] [Google Scholar]
- 3.Bismuth G, Boumsell L. Controlling the immune system through semaphorins. Sci STKE. 2002;2002(128):re4. doi: 10.1126/stke.1282002re4. [DOI] [PubMed] [Google Scholar]
- 4.Neufeld G, Shraga-Heled N, Lange T, Guttmann-Raviv N, Herzog Y, Kessler O. Semaphorins in cancer. Front Biosci. 2005;10:751–760. doi: 10.2741/1569. [DOI] [PubMed] [Google Scholar]
- 5.Neufeld G, Lange T, Varshavsky A, Kessler O. Semaphorin signaling in vascular and tumor biology. Adv Exp Med Biol. 2007;600:118–131. doi: 10.1007/978-0-387-70956-7_10. [DOI] [PubMed] [Google Scholar]
- 6.Sun Q, Zhou H, Binmadi NO, Basile JR. Hypoxia-inducible factor-1-mediated regulation of semaphorin 4D affects tumor growth and vascularity. J Biol Chem. 2009;284(46):32066–32074. doi: 10.1074/jbc.M109.057166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Zhang L, Pan Y, Ren X, Hao Q. Over-expression of sema-phorin4D, hypoxia-inducible factor-1α and vascular endothelial growth factor is related to poor prognosis in ovarian epithelial cancer. Int J Mol Sci. 2012;13(10):13264–13274. doi: 10.3390/ijms131013264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou H, Yang YH, Binmadi NO, Proia P, Basile JR. The hypoxia-inducible factor-responsive proteins semaphorin 4D and vascular endothelial growth factor promote tumor growth and angiogenesis in oral squamous cell carcinoma. Exp Cell Res. 2012;318(14):1685–1698. doi: 10.1016/j.yexcr.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding X, Qiu L, Zhang L, et al. The role of semaphorin 4D as a potential biomarker for antiangiogenic therapy in colorectal cancer. Onco Targets Ther. 2016;9:1189–1204. doi: 10.2147/OTT.S98906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujioka S, Masuda K, Toguchi M, et al. Neurotrophic effect of semaphorin 4D in PC12 cells. Biochem Biophys Res Commun. 2003;301(2):304–310. doi: 10.1016/s0006-291x(02)03023-1. [DOI] [PubMed] [Google Scholar]
- 11.Kato S, Kubota K, Shimamura T, et al. Semaphorin 4D, a lymphocyte semaphorin, enhances tumor cell motility through binding its receptor, plexinB1, in pancreatic cancer. Cancer Sci. 2011;102(11):2029–2037. doi: 10.1111/j.1349-7006.2011.02053.x. [DOI] [PubMed] [Google Scholar]
- 12.Liu H, Yang Y, Xiao J, et al. Semaphorin 4D expression is associated with a poor clinical outcome in cervical cancer patients. Microvasc Res. 2014;93:1–8. doi: 10.1016/j.mvr.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Zhang L, Lv R, Zhang WQ. Overexpression of Semaphorin4D indicates poor prognosis and prompts monocyte differentiation toward M2 macrophages in epithelial ovarian cancer. Asian Pac J Cancer Prev. 2013;14(10):5883–5890. doi: 10.7314/apjcp.2013.14.10.5883. [DOI] [PubMed] [Google Scholar]
- 14.Basile JR, Holmbeck K, Bugge TH, Gutkind JS. MT1-MMP controls tumor-induced angiogenesis through the release of semaphorin 4D. J Biol Chem. 2007;282(9):6899–6905. doi: 10.1074/jbc.M609570200. [DOI] [PubMed] [Google Scholar]
- 15.Zhou H, Binmadi NO, Yang YH, Proia P, Basile JR. Semaphorin 4D cooperates with VEGF to promote angiogenesis and tumor progression. Angiogenesis. 2012;15(3):391–407. doi: 10.1007/s10456-012-9268-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Zhang Y, Zhang B. D4-GDI, a Rho GTPase regulator, promotes breast cancer cell invasiveness. Cancer Res. 2006;66(11):5592–5598. doi: 10.1158/0008-5472.CAN-05-4004. [DOI] [PubMed] [Google Scholar]
- 17.Malliri A, Collard JG. Role of Rho-family proteins in cell adhesion and cancer. Curr Opin Cell Biol. 2003;15(5):583–589. doi: 10.1016/s0955-0674(03)00098-x. [DOI] [PubMed] [Google Scholar]
- 18.Horiuchi A, Imai T, Wang C, et al. Up-regulation of small GTPases, RhoA and RhoC, is associated with tumor progression in ovarian carcinoma. Lab Invest. 2003;83(6):861–870. doi: 10.1097/01.lab.0000073128.16098.31. [DOI] [PubMed] [Google Scholar]
- 19.Walker K, Olson MF. Targeting Ras and Rho GTPases as opportunities for cancer therapeutics. Curr Opin Genet Dev. 2005;15(1):62–68. doi: 10.1016/j.gde.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Shinto E, Tsuda H, Matsubara O, Mochizuki H. Significance of RhoC expression in terms of invasion and metastasis of colorectal cancer. Nihon Rinsho. 2003;61(suppl 7):215–219. [PubMed] [Google Scholar]
- 21.van Golen KL, Bao L, DiVito MM, Wu Z, Prendergast GC, Merajver SD. Reversion of RhoC GTPase-induced inflammatory breast cancer phenotype by treatment with a farnesyl transferase inhibitor. Mol Cancer Ther. 2002;1(8):575–583. [PubMed] [Google Scholar]
- 22.Paddison PJ, Silva JM, Conklin DS, et al. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428(6981):427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- 23.Paddison PJ, Caudy AA, Sachidanandam R, Hannon GJ. Short hairpin activated gene silencing in mammalian cells. Methods Mol Biol. 2004;265:85–100. doi: 10.1385/1-59259-775-0:085. [DOI] [PubMed] [Google Scholar]
- 24.Naldini L, Blomer U, Gage FH, Trono D, Verma IM. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci U S A. 1996;93(21):11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naldini L, Blomer U, Gallay P, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272(5259):263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 26.Basile JR, Castilho RM, Williams VP, Gutkind JS. Semaphorin 4D provides a link between axon guidance processes and tumor-induced angiogenesis. Proc Natl Acad Sci U S A. 2006;103(24):9017–9022. doi: 10.1073/pnas.0508825103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao F, Xie X, Gollan T, et al. Comparison of gene-transfer efficiency in human embryonic stem cells. Mol Imaging Biol. 2010;12(1):15–24. doi: 10.1007/s11307-009-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lafitte M, Rousseau B, Moranvillier I, et al. In vivo gene transfer targeting in pancreatic adenocarcinoma with cell surface antigens. Mol Cancer. 2012;11:81. doi: 10.1186/1476-4598-11-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Basile JR, Barac A, Zhu T, Guan KL, Gutkind JS. Class IV semaphorins promote angiogenesis by stimulating Rho-initiated pathways through plexin-B. Cancer Res. 2004;64(15):5212–5224. doi: 10.1158/0008-5472.CAN-04-0126. [DOI] [PubMed] [Google Scholar]
- 30.Basile JR, Afkhami T, Gutkind JS. Semaphorin 4D/plexin-B1 induces endothelial cell migration through the activation of PYK2, Src, and the phosphatidylinositol 3-kinase-Akt pathway. Mol Cell Biol. 2005;25(16):6889–6898. doi: 10.1128/MCB.25.16.6889-6898.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito Y, Oinuma I, Katoh H, Kaibuchi K, Negishi M. Sema4D/plexin-B1 activates GSK-3beta through R-Ras GAP activity, inducing growth cone collapse. EMBO Rep. 2006;7(7):704–709. doi: 10.1038/sj.embor.7400737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vikis HG, Li W, Guan KL. The plexin-B1/Rac interaction inhibits PAK activation and enhances Sema4D ligand binding. Genes Dev. 2002;16(7):836–845. doi: 10.1101/gad.966402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soong J, Chen Y, Shustef EM, Scott GA. Sema4D, the ligand for Plexin B1, suppresses c-Met activation and migration and promotes melanocyte survival and growth. J Invest Dermatol. 2012;132(4):1230–1238. doi: 10.1038/jid.2011.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aurandt J, Li W, Guan KL. Semaphorin 4D activates the MAPK pathway downstream of plexin-B1. Biochem J. 2006;394(Pt 2):459–464. doi: 10.1042/BJ20051123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meili R, Sasaki AT, Firtel RA. Rho Rocks PTEN. Nat Cell Biol. 2005;7(4):334–335. doi: 10.1038/ncb0405-334. [DOI] [PubMed] [Google Scholar]
- 36.Kandpal RP. Rho GTPase activating proteins in cancer phenotypes. Curr Protein Pept Sci. 2006;7(4):355–365. doi: 10.2174/138920306778018025. [DOI] [PubMed] [Google Scholar]
- 37.Jin J, Ryu HS, Lee KB, Jang JJ. High expression of protein tyrosine kinase 7 significantly associates with invasiveness and poor prognosis in intrahepatic cholangiocarcinoma. PloS One. 2014;9(2):e90247. doi: 10.1371/journal.pone.0090247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cardone RA, Bagorda A, Bellizzi A, et al. Protein kinase A gating of a pseudopodial-located RhoA/ROCK/p38/NHE1 signal module regulates invasion in breast cancer cell lines. Mol Biol Cell. 2005;16(7):3117–3127. doi: 10.1091/mbc.E04-10-0945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fisher TL, Seils J, Reilly C, et al. Saturation monitoring of VX15/2503, a novel semaphorin 4D-specific antibody, in clinical trials. Cytometry B Clin Cytom. 2016;90(2):199–208. doi: 10.1002/cyto.b.21338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Wound healing assay in Hs578T cells.
Notes: Breast cancer Hs578T cells (infected by the indicated lentivirus) were examined in six-well plates. Three groups of cells were tested: Sema4D-coding lentivirus, control lentivirus-expressing GFP, and lentivirus coding for Sema4D shRNA. The results are shown (×40). Bar graphs show the healing distances, as analyzed using NIH Image software. Error bars represent the standard deviation of the results for three wells (*P<0.05, relative to the GFP group).
Abbreviations: Sema4D, semaphorin 4D; shRNA, short hairpin RNA.
In vitro migration assay of Hs578T cells.
Notes: Breast cancer Hs578T cells, uninfected or infected by the indicated lentivirus, were examined in a transwell plate. Four groups of each cell line were tested, including cells that were uninfected or infected with the indicated lentivirus: control lentivirus-expressing GFP, Sema4D-coding lentivirus, or lentivirus coding for Sema4D shRNA. Cells migrated to the lower side of the membranes were stained with crystal violet (×100). Bar graphs below the stained migration membranes show the results of the migration assay, as analyzed using NIH Image software and expressed as the pixel intensity of stained migration membranes relative to negative control. Error bars represent the standard deviation of the results obtained from six wells (*P<0.05, relative to the uninfected group).
Abbreviations: Sema4D, semaphorin 4D; shRNA, short hairpin RNA.
In vitro invasion assay of Hs578T cells.
Notes: Breast cancer Hs578T cells, uninfected or infected by the indicated lentivirus, were examined in a transwell plate assay. Four groups of each cell line were tested, including cells that were uninfected or infected with the indicated lentivirus: control lentivirus-expressing GFP, lentivirus coding for Sema4D, or lentivirus coding for Sema4D shRNA; cells on the lower side of membranes were stained with crystal violet (×100). Bar graphs below the stained membranes show the results of the invasion assay, as analyzed using NIH Image software and expressed as the pixel intensity of stained membranes relative to negative control. Error bars represent the standard deviation of the results obtained from six wells (*P<0.05, relative to the uninfected group).
Abbreviations: Sema4D, semaphorin 4D; shRNA, short hairpin RNA.