Abstract
Detection of bacterial urease activity has been utilized successfully to diagnose Helicobacter pylori (H. pylori). While Mycobacterium tuberculosis (M. tb) also possesses an active urease, it is unknown whether detection of mycobacterial urease activity by oral urease breath test (UBT) can be exploited as a rapid point of care biomarker for tuberculosis (TB) in humans. We enrolled 34 individuals newly diagnosed with pulmonary TB and 46 healthy subjects in Bamako, Mali and performed oral UBT, mycobacterial sputum culture and H. pylori testing. Oral UBT had a sensitivity and specificity (95% CI) of 70% (46–88%) and 11% (3–26%), respectively, to diagnose culture-confirmed M. tb disease among patients without H. pylori, and 100% sensitivity (69–100%) and 11% specificity (3–26%) to diagnose H. pylori among patients without pulmonary TB. Stool microbiome analysis of controls without TB or H. pylori but with positive oral UBT detected high levels of non-H. pylori urease producing organisms, which likely explains the low specificity of oral UBT in this setting and in other reports of oral UBT studies in Africa.
Keywords: oral urease breath test, urease, TB, H. pylori
INTRODUCTION
Despite the availability of inexpensive and effective treatment, tuberculosis (TB) still accounts for millions of deaths worldwide [1, 2]. The highest burden of disease is borne by developing countries where disease prevalence correlates with malnutrition, densely-populated communities, lack of access to affordable health care, and synergy with HIV co-infection. Sputum smear microscopy, which is more than 100 years old and lacks sensitivity, remains the most widely used diagnostic tool. Other tests with improved sensitivity, such as sputum culture and molecular based testing are often either technically difficult or too expensive for low-income countries with endemic TB. There is therefore a pressing need for the development of inexpensive and sensitive tools for the diagnosis of active TB in a point of care setting. Indeed, a recent report by UNITAID and the WHO has emphasized the need for a rapid, point-of-care screening tool for TB in resource-poor settings [3,4].
Ureases are found in numerous microbial species, including bacteria, but are not present in humans [5,6]. Assays that detect bacterial urease activity have an essential role in the diagnosis and clinical management of Helicobacter pylori (H. pylori) gastritis [7]. In H. pylori infection, the bacterial urease catalyzes the hydrolysis of urea into ammonia and carbon dioxide (CO2), which increases stomach pH and allows local survival of H. pylori. Carbon dioxide, a byproduct of this chemical reaction, is exhaled, where it can be detected in breath samples [7,8]. This phenomenon has been exploited for use in diagnostic tests for H. pylori, including the oral urease breath test (UBT) and rapid urease test (RUT) [9,10].
The oral UBT administers a dose of labeled 13C-urea by mouth; if urease is present, it hydrolyzes the stable isotope labeled urea, and 13CO2 is produced [11]. The 13CO2 to 12CO2 ratio in exhaled air is readily measured with portable infrared spectrometers, and the entire procedure can be completed within 30 minutes [12]. The test was approved by the Food and Drug Administration (FDA) for the diagnosis of H. pylori infection in 1997 [13], and it is also used to confirm eradication after antibiotic therapy [14]
Like H. pylori, Mycobacterium tuberculosis (M. tb) also possesses an active urease [15]. In vitro urease testing has long been used for mycobacterial species identification [16], but whether in situ detection of mycobacterial urease activity can be exploited for clinical diagnosis or monitoring of M. tb treatment in humans has not been explored. As part of our efforts to develop a breath test for TB, in preclinical studies, we found that rabbits with pulmonary TB rapidly convert [13C]-labeled urea into 13CO2, and that signal can be detected by UBT in a matter of minutes [17–19]. Furthermore, the UBT signal diminished as the rabbits were exposed to antimicrobial therapy and then increased again after therapy was halted, which suggested that the UBT may have a promising role in monitoring response to therapy [18]. Extrapolating from those data, lack of UBT signal inhibition during antibiotic treatment may be a rapid indicator of treatment failure among individuals with drug-resistant TB who are being treated with inadequate drug regimens. A study on TB and H. pylori co-infection in Iran showed the disappearance of UBT signal when patients were treated with standard TB therapy (without concomitant H. pylori treatment), which suggests that M. tb infection may contribute to the positivity of UBT [20].
Both H. pylori and M. tb are widely endemic infections in Mali with a prevalence of 92 per 100,000 for TB and an estimated percentage of H. pylori infection ranging from 41 to 97% [2, 21]. As co-infection with H. pylori and M. tb may confound results of the oral UBT, we evaluated the sensitivity and specificity of oral UBT to diagnose active TB and H. pylori in patients who were co-infected with H. pylori and M. tb, mono-infected with M. tb, and mono-infected with H. pylori. Patients who were double negative for both TB and H. pylori infections were used as negative controls.
METHODS
Study Population
From June of 2011 to December 2012, individuals who were 18 years of age or older were consecutively recruited from six referring health centers of Bamako and Point-G Hospital of Bamako, Mali. A total of 34 individuals who were newly diagnosed with active pulmonary TB (with or without H. pylori infection) and 46 healthy volunteers (with or without H. pylori infection) were enrolled and completed the study protocol. Participation was voluntary, written informed consent was obtained prior to enrollment, and participation did not alter provision of standard medical care to patients. This study was approved by the Institutional Review Board of the US National Institutes of Allergic and Infectious Diseases (NIAID) and the Ethics Committee of the Faculty of Medicine, Pharmacy and Dentistry (FMPOS) of the University of Sciences, Techniques and Technology of Bamako (USTTB). It was registered with ClinicalTrials.gov, identifier NCT01301144.
Study Procedures
All participants underwent a physical exam and a baseline evaluation of his or her medical history. Newly diagnosed patients with pulmonary TB were enrolled on the basis of two positive acid-fast bacilli (AFB) sputa assessments and prior to initiation of any TB drugs. Sputum microscopy assessments were performed by both Ziehl-Neelsen staining at local referral centers and by auramine rhodamine staining using fluorescence microscopy performed at the central SEREFO BSL-3 laboratory. Active pulmonary TB was later confirmed by mycobacterial culture in both MGIT and on solid agar 7H11 plates. To meet enrollment criteria, the apparently healthy volunteers had to be without any signs or symptoms of active TB. This included lack of documented fever, cough or weight loss, and a microbiological assessment that included two sputum specimens that were negative for AFB. The healthy controls were all ultimately confirmed to be TB culture negative. H. pylori stool antigen testing was performed using ImmunoCard HPSA® (Meridian Diagnostics). Additional study testing included HIV-1/2 serology (Determine® HIV-1/2, Abbott) and pregnancy testing for female participants. Key exclusion criteria included: use of any antibiotic (including TB drugs), proton pump inhibitors or bismuth containing preparations in the 15 days prior to study enrollment, HIV positivity and pregnancy.
TB and H. pylori Case Definitions
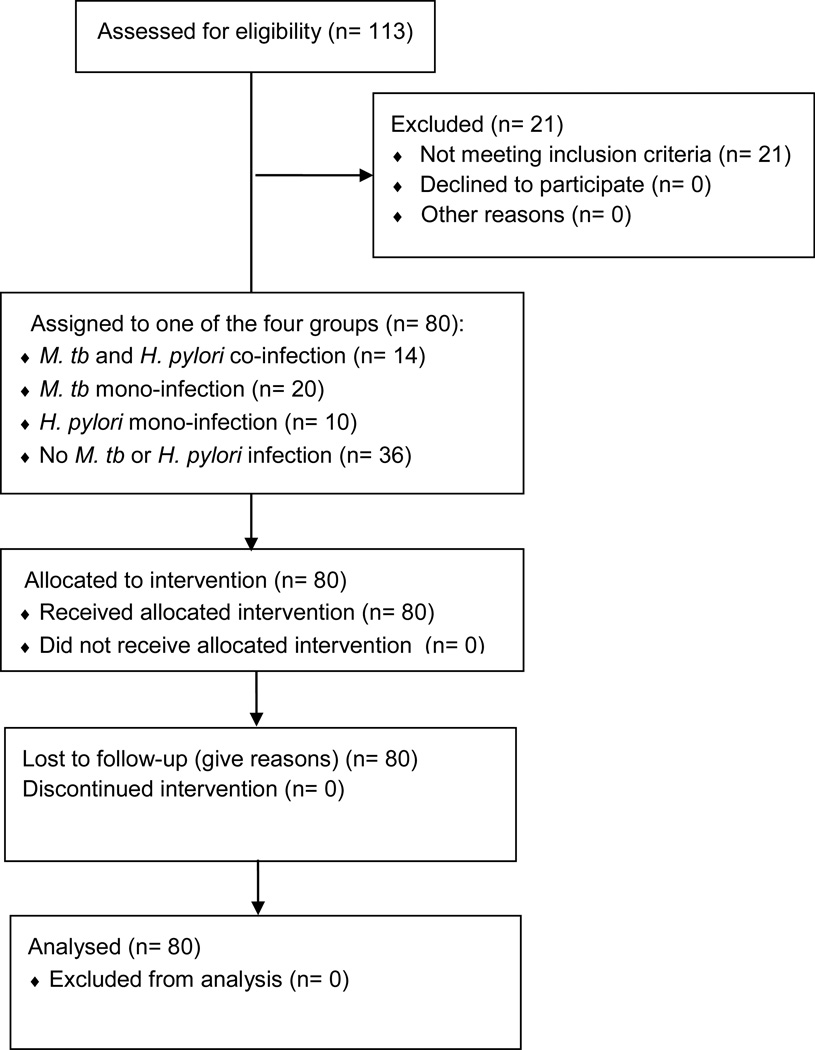
Pulmonary TB cases were defined by positive mycobacterial culture by MGIT and/or 7H11 culture. H. pylori stool antigen testing was used to define the patient’s H. pylori status. Patients were enrolled on the basis of initial TB and H. pylori testing and were later assigned to one of the four following diagnostic categories on the basis of their study mycobacterial culture and H. pylori stool antigen results: (1) active pulmonary TB with H. pylori co-infection (TB+/HP+, Group 1); (2) active pulmonary TB without H. pylori co-infection (TB+/HP−, Group 2); (3) no apparent TB but H. pylori infection (TB−/HP+, Group 3); and (4) no apparent TB and no H. pylori infection (TB1/HP−, Group 4)(Figure 1). This study included a total of 80 study subjects distributed in the four cited groups (Table 1).
Figure 1.
CONSORT Flow Diagram shows study’s groups and number of participants.
Table 1.
Demographic characteristics at time of enrollment
| Group 1: M. tb and H. pylori co- infection (n = 14) |
Group 2: M. tb mono- infection (n = 20) |
Group 3: H. pylori mono- infection (n = 10) |
Group 4: No M. tb or H. pylori infection (n = 36) |
|
|---|---|---|---|---|
| Male, n (%) | 13 (93) | 18 (90) | 9 (90) | 26 (72) |
| Mean age, yr (range) | 36 (23–63) | 29 (19–51) | 30 (19–50) | 28 (20–70) |
Urea Breath Testing
BreathTek™ UBT Collection Kit (Otsuka America Pharmaceutical Co., Ltd.) was utilized for oral UBT in this study. Subjects ingested 75 mg of a synthetic 13C-urea (Pranactin®-Citric) reconstituted with water. Measurement of the ratio of 13CO2 to 12CO2 in a post-dose breath sample was performed at baseline (before the Pranactin®-Citric) and at three different time points (10, 20, and 45 minutes) following oral urea administration. The measured difference between the ratios of 13CO2 to 12CO2 values before and after administration of Pranactin-Citric solution is reported as Delta over Baseline (DOB), with a value greater than or equal to 2.4-fold above baseline considered a positive test. The study time points were selected as optimal based on a pilot study. For patients with presumed pulmonary TB (AFB positive at the time of enrollment), UBT sampling was performed on two consecutive days prior to initiation of any TB antimicrobials and mean DOB values were used. Healthy controls were assessed only once.
Stool Microbiome Assessment
Stool samples (approximately 5g) were collected from 10 study participants who had a positive UBT, but no clinical or laboratory evidence of either pulmonary TB or H. pylori. These participants tested negative for H. pylori stool antigens (ImmunoCard HPSA®, Meridian Diagnostics) as well as negative for H. pylori serum antibodies (ImmunoCard H. pylori IgG®, Meridian Diagnostics). Whole metagenome shotgun sequencing was performed at the University of Maryland Institute for Genome Sciences using the Illumina HiSeq2000 platform to generate 101bp paired-end reads for analysis. Microbiome analysis was performed at The Broad Institute, Boston, MA.
Data analysis
The sensitivity and specificity along with 95% confidence intervals of oral UBT for diagnosis of TB and H. pylori infection was calculated using Stata version 12.1, with a UBT threshold for positivity being a DOB value of 2.4-fold above baseline. The organismal composition of microbial communities from stool was determined using alignment-based approaches that employed either a database of unique microbial genomes [22], or marker genes representing organisms at various taxonomic levels [23]. Functional metabolic pathways were predicted using the HUMAnN pipeline [24], and the relative abundance of predicted ureases was compared to the abundance found within stool samples from the Human Microbiome Project focused on healthy individuals from the United States [25].
RESULTS
Patient Population
Of the 80 volunteers enrolled in this study, 34 (42.5%) were confirmed to have pulmonary TB on the basis of a positive sputum culture (TB+), whereas 46 (57.5%) were mycobacterial culture negative (TB−) and were used as healthy comparators. Ultimately, 24 (30%) patients were determined to have a positive stool antigen test for H. pylori, and were considered actively infected with H. pylori (HP+), whereas, 56 (70%) had negative stool antigen testing and presumed H. pylori uninfected (HP−). When patients were distributed into groups on the basis of both their M. tb and H. pylori testing results, there was a relatively similar distribution of gender and ages among the four groups; however, the healthy controls were somewhat younger and more likely to be female (Table 1). The majority of the participants, 66 (82.5%), were men. As per the enrollment criteria, all participants were determined to be HIV seronegative.
Sensitivity and specificity of oral UBT for Pulmonary TB and H. pylori
Among TB mono-infected patients (Group 2, TB+/HP−) the highest oral UBT signal was detected at the 20-minute time point (data not shown); however, the signal levels were similar over the course of the three time points tested (10, 20 and 45 minutes) (data not shown). Hence we used the 20 minute time point to determine UBT positivity or negativity, and patients with DOB20 ≥ 2.4 were considered to be UBT positive.
Of the 80 patients enrolled in the study 14 (17.5%) were TB+/HP+ (Group 1), 20 (25%) were TB+/HP− (Group 2), 10 (12.5%) were TB−/HP+ (Group 3), and 36 (45%) were TB−/HP− (Group 4). More clinically relevant was that 32 of 36 (89%) TB−/HP− (Group 4) patients had significant urease activity with DOB20 ≥ 2.4. Also 14 of 20 (70%) TB+/HP− (Group 2) patients had DOB20 ≥ 2.4 (Table 2).
Table 2.
Group definitions and urea breath test (UBT) results using the delta-over-baseline at 20 minutes (DOB20) ≥ 2.4 as the breakpoint for UBT positivity.
| Patients | Infection | DOB20 < 2.4 (% within group) |
DOB20 ≥ 2.4 (% within group) |
|---|---|---|---|
| Group 1 | TB+/HP+ | 0 | 14 (100%) |
| Group 2 | TB+/HP− | 6 (30%) | 14 (70%) |
| Group 3 | TB−/HP+ | 0 | 10 (100%) |
| Group 4 | TB−/HP− | 4 (11%) | 32 (89%) |
Using DOB20 ≥ 2.4 as the breakpoint for UBT positivity, oral UBT was calculated to have a sensitivity of 70% (95% CI: 46–88%) to diagnose TB among H. pylori stool antigen negative patients (TB+/HP−, Group 2). Among H. pylori mono-infected patients (TB−/HP+, Group 3), oral UBT had a calculated sensitivity of 100% (95% CI: 69–100%) to diagnose H. pylori. Despite a high or relatively high sensitivity, the calculated specificity was surprisingly low for both groups, only 11% (95% CI: 3–26%) (Table 3). Both groups shared the same negative control group composed of participants who were not actively infected with either M. tb or H. pylori (TB−/HP−, Group 4) which is why the specificity was identical. Specificity for H. pylori was far below the reported specificity of 96% using BreathTek UBT, suggesting that background urease contribution to UBT could be high in the Malian population.
Table 3.
Oral UBT sensitivity and specificity for M. tb and H. pylori detection in comparison to mycobacterial culture or H. pylori stool antigen. Oral UBT was considered positive if signal 20 minutes following test initiation was greater than or equal to 2.4 fold delta-over-baseline (DOB20 ≥ 2.4).
| TP | FP | FN | TN | Sensitivity (95% CI) |
Specificity (95% CI) |
|
|---|---|---|---|---|---|---|
| M. tuberculosis | 14 | 32 | 6 | 4 | 0.70 (0.46–0.88) | 0.11 (0.03–0.26) |
| H. pylori | 10 | 32 | 0 | 4 | 1.00 (0.69–1.00) | 0.11 (0.03–0.26) |
TP: true positive; FP: false positive; FN: false negative; TN: true negative.
Stool Microbiome Assessment
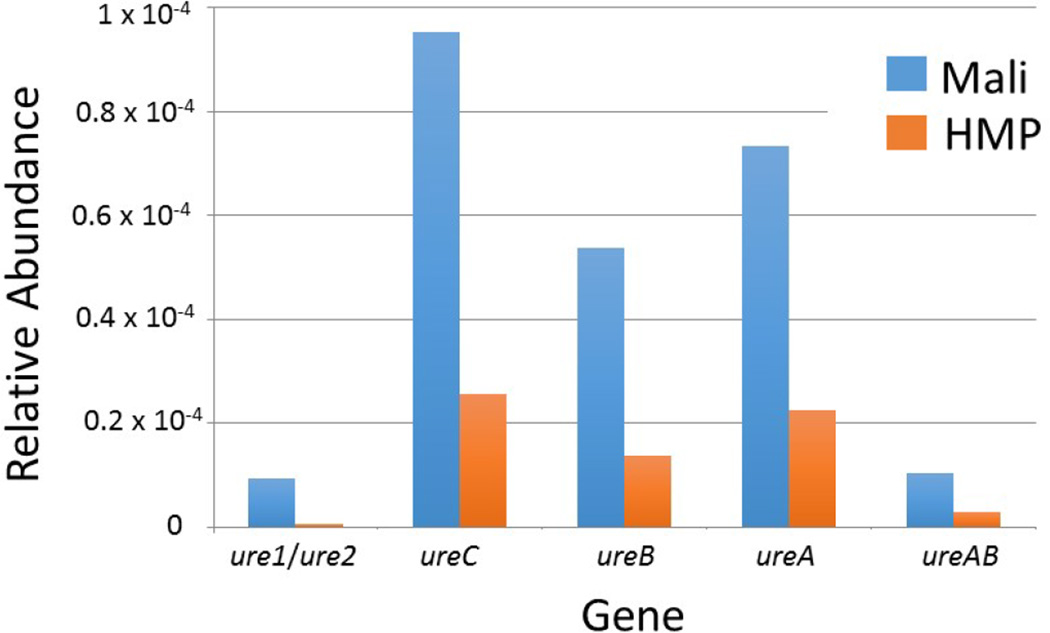
Given the low calculated specificity of oral UBT for both H. pylori and M. tb in this patient population, we analyzed DNA isolated from stool to assess for the presence of other microbes that might be contributing to a positive oral UBT test. Analysis of whole genome shotgun metagenomic data was performed on 10 healthy control participants with no clinical or laboratory evidence of either M. tb or H. pylori infection (TB−/HP−, Group 4) but with a positive oral-UBT. Neither M. tb nor H. pylori DNA was detected in any sample confirming the absence of M. tb and H. pylori infection in these patients as measured by clinical and laboratory testing. Thus, the negative sputum cultures and stool antigen results for mycobacteria and H. pylori, respectively, were not due to false negative test results. Of note, the sequencing did reveal the presence of multiple non-H. pylori ureases including bacterial encoded enzymes, ureC, ureB, ureA, and ureAB and invertebrate encoded enzymes, ure1/ure2. When compared to 136 stool samples from healthy, U.S. volunteers who participated in the Human Microbiome Project (HMP), the relative abundance of urease genes was markedly higher among the Malian samples (Figure 2). This increased relative abundance of urease genes in the Malian samples was notable for both total numbers of urease genes and for all subsets of urease genes. Of the 279 distinct ureases detected across all samples, nearly 20% of reads aligning to ureases were from 10 bacterial genera including Helicobacter (Table 4). The Helicobacter-related ureases we identified in these stool samples matched ureases from the genomes of H. hepaticus, H. mustelae, H. acinonychis and H. pylori. Though we confirmed the absence of H. pylori based on the lack of other key H. pylori genomic signatures in these samples using a whole genome-based alignment approach, these data indicate that other species, likely including close relatives of H. pylori, are present and possibly contributing to positive UBT results among healthy Malians, and low specificity for H. pylori and M. tb.
Figure 2.
Stool microbiome analysis of uninfected Malians (without M. tb or H. pylori infection but with positive oral UBT) revealed elevated levels of non-Helicobacter pylori ureases. The median relative abundances of urease gene content in the Malian samples versus those from 136 stool samples of healthy volunteers enrolled in the Human Microbiome Project (HMP) are displayed. When the relative abundance of ureases in the Malian data was compared to the HMP samples, the following fold-increases were noted: ure1 and ure2: 19.1x; ureC: 3.7x, ureB: 3.8x, urea: 3.3x, ureAB: 3.4.
Table 4.
Top 10 bacterial genera for which urease genes were identified among stool samples of M. tb culture-negative and H. pylori stool-antigen negative Malians who were nevertheless positive for the oral UBT. A total of 279 distinct ureases were identified in these stool samples. The table shows the rank order of bacterial genera that contributed the highest average relative abundance of urease genes among all detected ureases.
| Rank | Genus | Relative Abundance (Percent ± SD) |
|---|---|---|
| 1 | Bacillus | 3.56 ± 0.34 |
| 2 | Helicobacter | 2.22 ± 0.49 |
| 3 | Synechococcus | 2.14 ± 0.13 |
| 4 | Corynebacterium | 1.92 ± 0.31 |
| 5 | Clostridium | 1.90 ± 0.37 |
| 6 | Prochlorococcus | 1.84 ± 0.04 |
| 7 | Pseudomonas | 1.74 ± 0.09 |
| 8 | Paenibacillus | 1.52 ± 0.14 |
| 9 | Staphylococcus | 1.43 ± 0.28 |
| 10 | Cyanothece | 1.34 ± 0.08 |
DISCUSSION
In this study, we assessed the sensitivity and specificity of oral UBT as a diagnostic test for pulmonary TB in both H. pylori infected and uninfected populations in Mali. Our findings demonstrate only a moderate sensitivity of oral UBT (70%) for the diagnosis of active pulmonary TB among AFB-positive patients. As expected, oral UBT was 100% sensitive for the diagnosis of H. pylori infection. Surprisingly, our study revealed a remarkably low specificity of the UBT for the diagnosis of both TB and H. pylori (11%).
While the poor specificity for pulmonary TB could be partially explained by co-infections with H. pylori, the 11% specificity for H. pylori is far below the reported specificity of 96% (BreathTek). This could either be due to a higher degree of false positives in our study or a true difference between the patient populations utilized to determine the specificity of BreathTek compared to the Malian population in our study. The clinical trials used to evaluate the specificity and sensitivity of BreathTek were carried out mostly in developed countries. Those populations differ greatly from our patient sample in Mali with regards to diet, general health, genetics, and co-infections. Most importantly, H. pylori infection is endemic in Mali, which is not the case in developed countries. Consequently, the clinical trial results of BreathTek may not be representative of what we find in Mali. In addition to this, it has been recently reported that the stool antigen test is a poor diagnostic test for the detection of H. pylori in Mali, which was not known when this study was conducted. In that report, Austarheim et al. found a sensitivity of 21% only for H. pylori stool antigen test in Mali in patients with gastric ulcer, although previous studies had reported 95% prevalence of H. pylori infection in Mali [21]. This lack of an appropriate gold standard for the diagnosis of H. pylori in Mali is therefore a limitation of this study.
A further explanation for the low specificity is that the presence of other organisms gave rise to a non-specific urease signal. To explore this hypothesis, we carried out a stool microbiome analysis of individuals who lacked either clinical or laboratory evidence of either M. tb or H. pylori infection. Stool whole genome shotgun sequencing did confirm that DNA from neither M. tb nor H. pylori was present among these samples. Instead, sequencing revealed that many urease-producing organisms other than H. pylori or M. tb were present in the stool, supporting our hypothesis. Additionally, this study revealed that asymptomatic participants from Bamako had at least three times more urease genetic material than those recruited for the Human Microbiome Project (HMP), further supporting our suspicion of false positivity due to other urease producers. However, we should note the limited number and lack of controls for this microbiome analysis as only 10 samples from TB−/HP− (Group 4) participants were tested.
Oral administration of urea for the detection of pulmonary TB may not be ideal for several reasons. First, after oral administration the concentrations of 13C-urea reaching the lung are lower than those in the stomach and hence the amount of tracer available for bacterial conversion in the lung is limited. 13C-urea uptake occurs rapidly once the tracer enters the jejunum, and earlier studies suggest that serum Cmax concentrations are achieved by 30–45 minutes after ingestion [26, 27]. Although our previous study in the rabbit model of TB produced a positive signal by both intra-tracheal administration [18] and oral administration (unpublished data), the anatomical and physiological parameters of oral administration may render it a poor model for studies such as this. Second, the results are further confounded by the presence of H. pylori in the gut, particularly in an area where H. pylori is also endemic. We also considered the possibility of occult gastrointestinal tuberculosis as a potential confounder as an explanation for the high level of UBT positivity in TB−/HP− (Group 4) subjects; however, we consider this unlikely because gastrointestinal TB is rare, none of our subjects reported abdominal pain, night sweats, or fevers and our stool microbiota analysis of ten TB−/HP− (Group 4) subjects failed to identify M. tb. DNA.
In light of our findings of low specificity of oral UBT for identifying TB patients, we believe that alternative routes of 13C-urea administration such as the intravenous or inhalation routes may circumvent issues of non-specificity due to gut urease producers. In this regard we note that a phase-1 trial of inhaled 13C-urea as a pneumonia breath test is currently underway (NCT01303068) and that such a test may be more likely to render the encouraging results that were seen in M. tb.-infected animals receiving direct administration of 13C-urea into the lungs [18]. Additionally, there may be a potential role of UBT either by the oral or breath route as a biomarker of TB therapy response, although further trials addressing this question are necessary.
In summary, our data suggest that oral UBT has significant limitations as a point of care diagnostic tool for pulmonary TB in a setting with endemic H. pylori infection. Aerosolized UBT, however, may hold potential for detection of TB or monitoring patients on antitubercular therapy. Oral UBT also had a poor specificity for the diagnosis of H. pylori in this setting, which may be due to the presence of non-H. pylori ureases that we detected by stool microbiome analysis. Beyond confounding of oral UBT testing results, the clinical significance of these non-H. pylori ureases remains to be determined.
Acknowledgments
We are grateful to Otsuka America Pharmaceutical, Inc., Rockville, MD for generously providing us 650 blue and pink bags of BreathTek® UBT kits. The support of the NIAID Division of Clinical Research, NIH intramural funding to SEREFO, as well as NIH extramural grants AI36973, AI37856, AI097138 and AI069471, the Howard Hughes Medical Institute, the University of Sciences, Techniques and Technologies of Bamako, and the volunteers who participated in the study is gratefully acknowledged. This project has also been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN272200900018C and Grant Number U19AI110818 to the Broad Institute. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
REFERENCES
- 1.Tuberculosis reaches new milestones, good and bad. Lancet Infect Dis. 2015;15:1361. doi: 10.1016/S1473-3099(15)00431-4. Editors. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. TB Report. 2014 http://www.who.int/tb/publications/global_report/en/
- 3.Pai M, Schito M. Tuberculosis diagnostics in 2015: landscape, priorities, needs, and prospects. J Infect Dis. 2015;211:S21–S28. doi: 10.1093/infdis/jiu803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.UnitAID: Landscape and Technical Reports. http://wwwunitaideu/en/resources/publications/technical-reports. [Google Scholar]
- 5.Collins CM, D'Orazio SE. Bacterial ureases: structure, regulation of expression and role in pathogenesis. Mol Microbiol. 1993;9:907–913. doi: 10.1111/j.1365-2958.1993.tb01220.x. [DOI] [PubMed] [Google Scholar]
- 6.du Toit PJ, et al. In vivo effects of urease-producing bacteria involved with the pathogenesis of infection-induced urolithiasis on renal urokinase and sialidase activity. Urol Res. 1995;23:335–338. doi: 10.1007/BF00300023. [DOI] [PubMed] [Google Scholar]
- 7.Bell GD, et al. 14C-urea breath analysis, a non-invasive test for Campylobacter pylori in the stomach. Lancet. 1987;1:1367–1368. doi: 10.1016/s0140-6736(87)90664-7. [DOI] [PubMed] [Google Scholar]
- 8.Graham DY, et al. Campylobacter pylori detected noninvasively by the 13C-urea breath test. Lancet. 1987;1:1174–1177. doi: 10.1016/s0140-6736(87)92145-3. [DOI] [PubMed] [Google Scholar]
- 9.Lopes AI, Vale FF, Oleastro M. Helicobacter pylori infection - recent developments in diagnosis. World J Gastroenterol. 2014;20:9299–9313. doi: 10.3748/wjg.v20.i28.9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pourakbari B, et al. Diagnosis of Helicobacter pylori infection by invasive and noninvasive tests. Braz J Microbiol. 2013;44:795–798. doi: 10.1590/S1517-83822013005000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Rienzo TA, et al. 13C-Urea breath test for the diagnosis of Helicobacter pylori infection. Eur Rev Med Pharmacol Sci. 2013;17:51–58. [PubMed] [Google Scholar]
- 12.Ohara S, et al. Studies of 13C-urea breath test for diagnosis of Helicobacter pylori infection in Japan. J Gastroenterol. 1998;33:6–13. doi: 10.1007/pl00009968. [DOI] [PubMed] [Google Scholar]
- 13.Balon H, et al. Procedure guideline for carbon-14-urea breath test Society of Nuclear Medicine. J Nucl Med. 1998;39:2012–2014. [PubMed] [Google Scholar]
- 14.Slomianski A, Schubert T, Cutler AF. [13C] urea breath test to confirm eradication of Helicobacter pylori. Am J Gastroenterol. 1995;90:224–226. [PubMed] [Google Scholar]
- 15.Habel JE, et al. Structure of Rv1848 (UreA), the Mycobacterium tuberculosis urease gamma subunit. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:781–786. doi: 10.1107/S1744309110019536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steadham JE. Reliable urease test for identification of mycobacteria. J Clin Microbiol. 1979;10:134–137. doi: 10.1128/jcm.10.2.134-137.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jain P, et al. Reporter phage and breath tests: emerging phenotypic assays for diagnosing active tuberculosis, antibiotic resistance, and treatment efficacy. J Infect Dis. 2011;204:S1142–S1150. doi: 10.1093/infdis/jir454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jassal MS, et al. 13[C]-urea breath test as a novel point-of-care biomarker for tuberculosis treatment and diagnosis. PLoS One. 2010;5:e12451. doi: 10.1371/journal.pone.0012451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maiga M, Abaza A, Bishai WR. Current tuberculosis diagnostic tools & role of urease breath test Indian. J Med Res. 2012;135:731–736. [PMC free article] [PubMed] [Google Scholar]
- 20.Mirbagheri SA, et al. 14C-urea breath test in patients undergoing anti-tuberculosis therapy. World J Gastroenterol. 2005;11:1712–1714. doi: 10.3748/wjg.v11.i11.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Austarheim I, et al. Chromatographic immunoassays for Helicobacter pylori detection--are they reliable in Mali. West Africa Pan Afr Med J. 2013;14:72. doi: 10.11604/pamj.2013.14.72.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giannoukos G, et al. Efficient and robust RNA-seq process for cultured bacteria and complex community transcriptomes. Genome Biol. 2012;13:r23. doi: 10.1186/gb-2012-13-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segata N, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abubucker S, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oosterveld MJ, et al. Minimal sampling protocol for accurate estimation of urea production: a study with oral [13C]urea in fed and fasted piglets. Clin Nutr. 2005;24:97–104. doi: 10.1016/j.clnu.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 27.Schellekens RC, et al. Proof-of-concept study on the suitability of 13C-urea as a marker substance for assessment of in vivo behaviour of oral colon-targeted dosage forms. Br J Pharmacol. 2009;158:532–540. doi: 10.1111/j.1476-5381.2009.00302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]