Summary
The glucocorticoid receptor (GR) binds the human genome at >10,000 sites, but only regulates the expression of hundreds of genes. To determine the functional effect of each site, we measured the glucocorticoid (GC) responsive activity of nearly all GR binding sites (GBSs) captured using chromatin immunoprecipitation (ChIP) in A549 cells. 13% of GBSs assayed had GC-induced activity. The responsive sites were defined by direct GR binding via a GC response element (GRE) and exclusively increased reporter-gene expression. Meanwhile, most GBSs lacked GC-induced reporter activity. The non-responsive sites had epigenetic features of steady state enhancers and clustered around direct GBSs. Together, our data support a model in which clusters of GBSs observed with ChIP-seq reflect interactions between direct and tethered GBSs over tens of kilobases. We further show that those interactions can synergistically modulate the activity of direct GBSs, and may therefore play a major role in driving gene activation in response to GCs.
Introduction
Regulation of transcription plays a major role in human health and disease (Olansky et al. 1992; Maurano et al. 2012; Stadhouders et al. 2014; Vockley et al. 2015). The basic mechanism of human transcriptional regulation involves transcription factors (TFs) binding to specific genomic regulatory elements. Once bound, TFs recruit transcriptional machinery to the promoter of one or more target genes. Several studies have now mapped the location of binding sites across the human genome for many TFs and in many cell types [e.g. (ENCODE 2012)]. Those studies revealed a complex landscape of TF occupancy in which a TF typically binds thousands of locations across the human genome, but only directly regulates hundreds of genes (Reddy et al. 2009; Gao et al. 2013). The discrepancy between TF binding and gene regulation can be explained in part by findings that most TF binding sites have weak regulatory activity (Melnikov et al. 2012; Kheradpour et al. 2013) and that a TF often binds multiple sites near the same target gene (Gotea et al. 2010). The multiplicity of binding may be the result of functional redundancy between sites (Somma et al. 1991), cooperative assembly of TF complexes (Hertel et al. 1997), or local diffusion of bound factors along the genome (Coleman and Pugh 1995). Furthermore, numerous studies have shown that the number and diversity of TF binding sites contributes to synergistic rather than additive regulatory activity (Smith et al. 2013; Staller et al. 2015), suggesting a relationship between clusters of TF binding sites and the activity of those sites.
Ligand inducible TFs such as the GR are a representative model system for investigating the relationship between TF binding and activity. Once bound by GCs such as the cortisol mimic dexamethasone (DEX), the GR binds thousands of locations across the genome and regulates the expression of hundreds of genes (Wang et al. 2004; So et al. 2007; Reddy et al. 2009). The GR binds the genome either directly via DNA-sequence-specific interactions with a GRE or, more often, indirectly via tethering to other proteins such as the AP-1 family of TFs (Chandler et al. 1983; Gertz et al. 2013). Direct binding sites are more often shared across cell types and more likely to occur in genomic regions with less accessible chromatin prior to induction. Conversely, AP-1 co-occupied sites are more likely to occur at regions of more accessible chromatin, are more likely to be cell type specific, and may be the basis for differences in the GC responses between tissues (Biddie et al. 2011; John et al. 2011; Gertz et al. 2013).
Here, we propose that the organization of GR binding across the human genome conforms to a model in which GREs recruit GR directly to the DNA, and then those direct sites nucleate clusters of tethered binding nearby. We quantified the activity of GR-bound DNA elements on the genome-scale, assaying 2.9 million unique reporter vectors covering 10,963 GBSs. We found that direct GBSs confer inducible enhancer function, while tethered sites do not. We further provide evidence that tethered GR binding depends on the proximity of the tethered sites to direct sites. The resulting clusters of GBSs modulate the regulatory activity of direct GBSs, potentially contributing to expression levels of cell type specific GC responsive transcription. Together, these results demonstrate that patterns of genomic GR occupancy observed with ChIP-seq reflect locally coordinated and functionally synergistic GR binding events, rather than independent and additive events. We also provide evidence that our enhancer cluster model is general to the estrogen receptor (ER), suggesting that other TFs act similarly.
Results
Quantifying DEX-induced regulatory element activity
To assess the functional diversity of GBSs, we quantified the regulatory activity of 10,963 GBSs in the A549 human lung epithelial cell line (Figure 1A). To do so, we first used ChIP to isolate GBSs from A549 cells after treating the cells for 3 h with 100 nM DEX or with equal-volume ethanol (EtOH) as a vehicle control. High-throughput sequencing of the GR ChIP DNA followed by peak calling identified 27,432 GBSs (Table S1). We then cloned the ChIP-seq library into the STARR-seq reporter assay (Arnold et al. 2013). We estimated that the resulting plasmid library contained 2.9 million unique GR ChIP fragments (Figure S1A). The fragments had a similar genomic distribution and size as the ChIP-seq library, indicating that cloning into the reporter assay was unbiased (Figure S1B and S1C). Of the cloned fragments, 6% mapped to the 27,432 called GBSs. We transfected the reporter library back into A549 cells and treated the cells with either 100 nM DEX or EtOH to assay DEX-responsive regulatory activity. After filtering to only retain sites with sufficient statistical power to make a determination of regulatory activity (Love et al. 2014b), we quantified the activity and DEX-responsiveness of 10,963 GBSs (Figure S1D). The number of GBSs assayed was similar to that identified in previous GR ChIP-seq studies (Reddy et al. 2009; Biddie et al. 2011; Reddy et al. 2012a) and included 82% of the sites reported in the same cell line previously (Reddy et al. 2009). We assayed the reporter library in three replicates per condition, and found the approach to be highly reproducible (Figure 1B). Together, these results indicate that we reproducibly assayed the activity of nearly all GBSs in A549 cells.
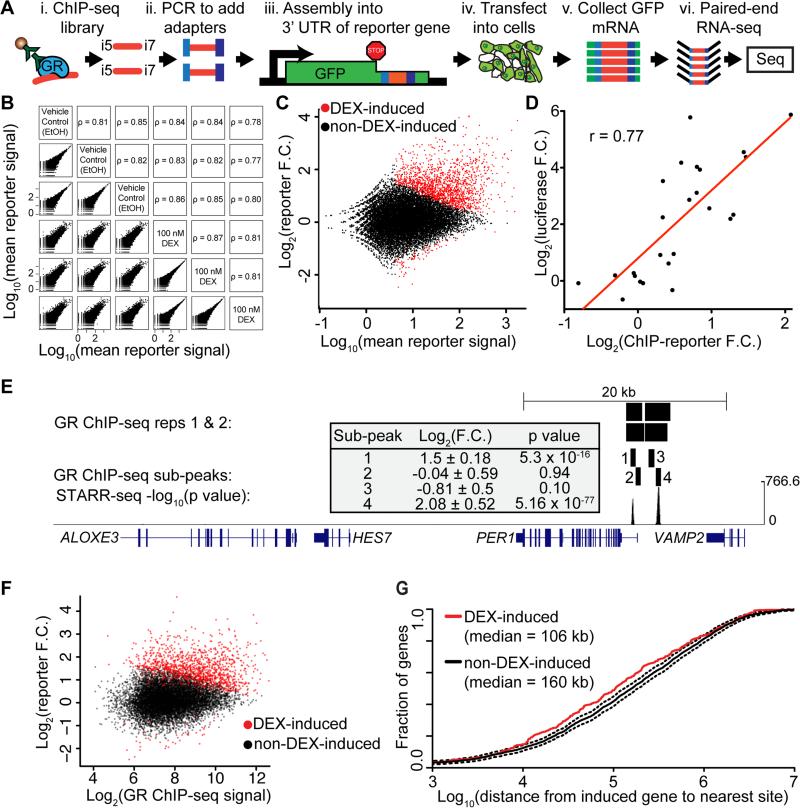
Figure 1. Chromatin immunoenriched reporter assays.
(A) For the ChIP-reporter assays, we first used PCR to add 15-bp adapters that are complementary to the reporter vector to GR ChIP-seq libraries. Adapted DNA was then ligated into the 3’ UTR of the GFP reporter gene of the STARR-seq vector. The resulting plasmid library was transfected into A549 cells. Cells were treated with 100 nM DEX or 0.02% EtOH for 3 h and RNA was collected. cDNA of the reporter GFP was amplified using gene-specific primers. The relative regulatory activity of each fragment was then estimated by counting aligned reads generated by high-throughput paired-end sequencing. (B) A comparison of the normalized ChIP-reporter signal for all 27,432 called GBS sub-peaks between all replicates and treatments. Each point represents one GBS, and all sites are included in each plot. (C) For each GBS assayed, the fold change in regulatory element activity with DEX treatment is plotted as a function of the mean sequencing coverage at that element. Red points indicate regulatory elements with a statistically significant response (FDR < 5%). (D) Correlation of dual luciferase assays with ChIP-reporter results (r = 0.77) (E) DEX-induced regulatory features of the PER1 locus on human chromosome 17. GR ChIP-seq peaks called by MACS (first two rows), GR-ChIP sub-peaks assayed by ChIP-reporters (3rd and 4th rows), −log10 (p value) from BAC-based DEX-induced STARR-seq (5th row). Inset contains ChIP-reporter log2 (fold change) in response to DEX and associated p value for each sub-peak assayed. (F) GR reporter signal is plotted against GR ChIP-seq signal for all GBSs assayed with our ChIP-reporter approach. The two datasets correlated with a Spearman's ρ = 0.22. (G) Fraction of induced genes as a function of the distance from the TSS to the nearest DEX–induced GBS (red line, median distance 106 kb) or an equivalent number of non-DEX-induced GBSs (black line, median distance 160 kb). The non-DEX-induced reporter sites were randomly subsampled to have an equivalent number as for the DEX-induced reporter sites 100 times, and the mean and standard deviation are shown in black solid and black dashed lines, respectively.
Of the assayed GBSs, 84% had a less than 2-fold difference in activity between treatments (Figure S1E), indicating that most GBSs have modest regulatory activity in our ChIP-reporter assays. That result was consistent with the results from other high-throughput reporter assay studies (Melnikov et al. 2012; Arnold et al. 2013). Of the assayed GBSs, 1,376 (13%) had statistically significant regulatory activity at a false discovery rate (FDR) < 5% (Love et al. 2014b) (Figure 1C, Figure S1F, Table S2). Larger fragments were more frequently GC-inducible (Figure S1G). While GCs are known to activate and repress gene expression (Slater et al. 1985; Sakai et al. 1988) and while STARR-seq can report activation and repression (Arnold et al. 2013) (Figure S1H), 95% (N = 1,330) of the GC-regulated GBSs assayed had increased reporter gene expression in response to DEX. We validated the results with dual luciferase reporter assays (Figure 1D, Figure S1I) and with STARR-seq performed on BACs covering 1 Mb of the human genome that was selected to contain GBSs near GC-responsive genes (Figure 1E, Table S3, Supplemental Data File 1). We observed no enrichment of plasmids containing elements that encode GC-responsive enhancers in the nuclei of cells treated with DEX, suggesting that our results are not due to biased nuclear localization of the reporter vectors (Figure S2). To test if the non-DEX-responsive sites are maximally pre-induced prior to DEX treatment, we performed additional luciferase reporter assays to evaluate if adding GREs to non-DEX-responsive sites increased enhancer activity in response to DEX. The synthetic composite sites had up to 24-fold increased DEX-responsive activity that was lost with mutation of the GRE, suggesting that non-responsive sites are capable of greater activity with direct GR binding (Figure S3A). Together, our results indicate that 13% of GBSs have GC-inducible regulatory activity in a reporter assay, and that those sites overwhelmingly act to increase gene expression. DEX-responsive activity in ChIP-reporters was largely distinct from occupancy as measured by ChIP-seq (ρ = 0.22, Figure 1F), indicating that reporters provide complementary information about mechanisms of gene regulation. We used RNA-seq to determine the set of DEX-responsive genes after the same treatment (Table S4), and found that the transcription start site (TSS) of DEX-responsive genes were closer to DEX-responsive GBSs than to an equal number of randomly sampled non-responsive sites (p = 0.005; Figure 1G).
DEX-induced GBSs are direct binding sites
The GR is known to bind DNA directly at GREs or by tethering to other TFs such as AP-1 (Ratman et al. 2013). We hypothesized that the distinct modes of binding have different regulatory activities. To evaluate that hypothesis, we investigated whether the GRE or the DNA binding motifs of known co-binding TFs [AP-1 (Herrlich 2001), FOXA1 (Belikov et al. 2009), NFκB (Wang et al. 1997) and CREB1 (Sheppard et al. 1998)] predict DEX-responsive regulatory element activity in reporter assays. Of the binding motifs evaluated, the GRE was the only one that predicted changes in regulatory element activity after DEX treatment better than random (Figure 2A and 2B, Figure S3B and S3C). Using a Gaussian mixture model, we estimated that 80% of DEX-responsive elements and 35% of non-responsive elements had a GRE (Figure S3D). A de novo motif enrichment analysis revealed AP-1 motifs in the DEX-responsive elements with GREs (MEME E-value 3 × 10−101), but not in the corresponding non-responsive elements. In contrast, binding motifs for co-binding TFs were predictive of baseline regulatory activity (Figure S3E), but were not predictive of DEX-induced regulatory activity (Figure 2A, Figure S3F, Table S5). To distinguish between tethering and GR binding directly to a weak GRE, we mapped the genomic footprints of GR using ChIP-exo. GBSs with significant reporter activity showed a footprint concordant with the known dimer structure of GR (Figure 2C, Figure S3G-H) (Starick et al. 2015). In contrast, GBSs matched for ChIP-seq signal but lacking DEX-inducible activity did not have a ChIP-exo footprint consistent with GR:DNA interactions (Figure 2D, Figure S3I). From these results, we conclude that direct GR binding to a GRE is predictive of DEX-inducible reporter activity, whereas tethered binding via other TFs is not.
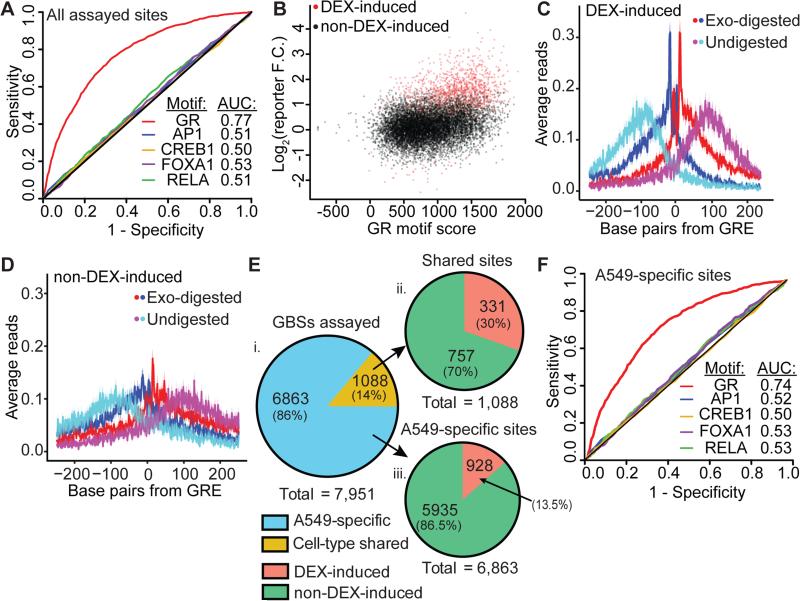
Figure 2. GRE-encoded GBSs confer DEX-inducible enhancer function.
(A) ROC analysis of DEX-induced enhancers evaluating DNA sequence motif significance for the GRE and known GR interacting proteins as predictors of enhancer function. (B) Scatter plot of GR motif score vs. ChIP-reporter activity. Significant DEX-induced elements are shown in red (r = 0.35). (C) Average GR ChIP-exo signal across DEX-inducible enhancer GBSs centered on the strongest GRE. Red and blue are the 5’ bp of reads aligned to the positive or negative strand, respectively. Cyan and magenta are the 5’ bp of reads at non-exonuclease treated fragment end aligned to the positive or negative strand, respectively. Ribbons indicate standard error. (D) GR ChIP-exo signal as above, aggregated across an equal number of sub-sampled GBSs that are not induced by DEX in reporter assays. (E) i. Fraction of GBSs in a previous study assayed using ChIP-reporters that were specific to A549 cells or common between A549 and Ishikawa cells (N = 7,771). ii. Fraction of A549-Ishikawa shared GBSs with significant DEX-induced reporter activity (N = 1088). iii. Fraction of A549-specific sites with significant DEX-induced reporter activity (N = 6,863). GBSs are categorized as specific to A549 cells or shared between A549 cells and endometrium-derived Ishikawa cells (Gertz et al. 2013). (F) ROC analysis of A549-specific DEX-inducible enhancers.
See also Figure S3.
Cell type-specific DEX-responsive enhancers are encoded by direct GBSs
GR binding sites can vary dramatically between cell types. For example, in a recent study comparing GR binding between A549 and Ishikawa cells, 15,220 (75%) of A549 GBSs were specific to A549 cells (Gertz et al. 2013). Most of the differences occur at sites that lack a GRE, suggesting that changes in the expression or binding of other TFs drive the differences in GR binding between cell types. To investigate, we asked whether DEX-responsive sites are enriched in cell-shared GBSs but depleted in cell-specific GBSs. We assayed 7,951 of the A549 GBSs identified in Gertz et al. Of those 1,088 (14%) were cell-shared and 6,863 (86%) were A549-specific. Of the cell-shared GBSs, 331 (30%) were DEX-responsive, an enrichment of 2.3-fold over the overall positive rate of 13%. Meanwhile, of A549-specific GBSs, 928 (13.5%) were DEX-responsive, similar to the overall positive rate (Figure 2E). Those results demonstrate that although cell-shared sites but not cell-specific sites are enriched for DEX-responsiveness, most (928 vs. 331) DEX-responsive enhancers are cell-specific. As was observed for all DEX-responsive GBSs, the presence of a GRE and not the motifs of tethering factors explains the DEX-response of A549-specific GBSs (Figure 2F). Together, those results demonstrate that both tethered sites and a small fraction of direct sites vary between cell types. Further, A549-specific direct GBSs act as cell type-specific DEX-responsive enhancers.
Remodeling of DEX-induced sites in the endogenous epigenome
If ChIP-reporters recapitulate endogenous gene regulation, we expect there to be differences in the endogenous epigenome between GBSs with and without DEX-responsive reporter activity. We first used DNase-seq to test for differences in chromatin accessibility between the two classes of GBSs (Song and Crawford 2010). After controlling for differences in GR occupancy between classes of GBSs, the DEX-responsive GBSs had less accessibility than non-DEX-responsive GBSs before and after DEX treatment (T-test, p < 1 × 10−100 and p = 6 × 10−55, respectively), but a significantly greater increase in accessibility after DEX (T-test p < 1 × 10−100) (Figure 3A). The increase in accessibility was distributed across many DEX-responsive GBSs, rather than due to few GBSs with large increases (Figure S4A). These data indicate that the DEX-responsive GBSs undergo more substantial chromatin remodeling after DEX treatment than non-responsive GBSs.
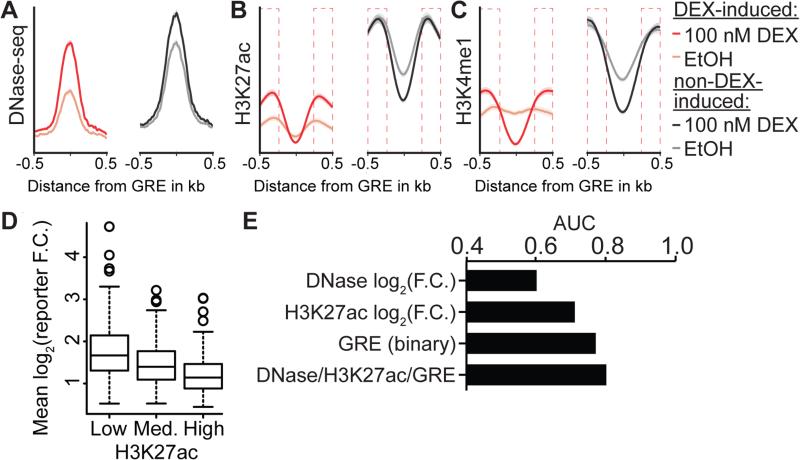
Figure 3. Distinct epigenetic remodeling DEX-inducible reporter sites.
(A) Aggregate profile plot of DNAse hypersensitivity (B) H3K27ac and (C) H3K4me1 at DEX-inducible and non-DEX-inducible sites before and after 1 h exposure to 100 nM DEX. Flanking 250 bp indicated by red dashed boxes. (D) Box plots showing the relative reporter activity of DEX-induced enhancers (FDR < 5%) binned by pre-induction H3K27ac status, where each bin contains an equal number of sites. (E) Area under ROC curves generated by using the presence of enhancer associated epigenetic marks in the endogenous genome independently or with the presence of GR motif as a predictor of enhancer activity.
See also Figure S4.
We next evaluated whether changes in covalent histone modifications at DEX-responsive and non-DEX-responsive GBSs were also consistent with the sites having distinct regulatory activities. Specifically, we evaluated whether there were distinct changes in the enrichment of two modifications that are associated with enhancer activity: histone 3 lysine 27 acetylation (H3K27ac), and H3K4 mono-methylation (H3K4me1) (ENCODE 2012) (Figure 3B and C, Table S1). In control-treated cells, non-DEX-responsive GBSs had greater H3K27ac and H3K4me1 signal than DEX-responsive GBSs before and after DEX induction. Those differences were observed both in the 500 bp window centered on the best match to the GRE, which we term the core of the GBS, (T-test, before DEX: p = 5 × 10−99, p = 3 × 10−45, respectively; after DEX: p = 1 × 10−38, p = 3 × 10−46, respectively), and in the 250 bp flanking that core on either side (T-test, before DEX: p = 3 × 10−95, p = 10 × 10−70, respectively; after DEX: p = 5 × 10−21, p = 5 × 10−32, respectively). DEX-responsive GBSs, however, had greater changes in histone modifications, especially in the flanking regions. H3K27ac and H3K4me1 increased significantly more in the flanks of the DEX-responsive GBSs than in flanks of the non-responsive GBSs (T-test, p < 1 × 10−100). At the core of the GBSs, we observed a greater DEX-dependent decrease in H3K27ac signal in non-DEX-responsive GBSs than in DEX-responsive GBSs (p < 1 × 10−100), but not for H3K4me1 (p = 0.02). We further found that, across DEX-induced GBSs, the sites with the least H3K27ac prior to DEX treatment (Figure 3D) and with the greatest increase in H3K27ac with DEX treatment (Figure S4B) also had the greatest DEX-responsive reporter activity. The histone acetyltransferase P300 is responsible for establishing H3K27ac across the human genome (Liu et al. 2008). Consistent with our observations of H3K27ac, we observed more ChIP-seq signal for P300 (ENCODE 2012) at non-DEX-responsive sites prior to hormone treatment than at DEX-responsive sites. Specifically, in control treated cells, there was P300 occupancy at 82% of non-responsive GBSs that were also bound by the AP-1 family member FOSL2. Meanwhile, P300 was bound at 6% of DEX-responsive GBSs that lack evidence of FOSL2 binding in the control treatment (Figure S4C). As anegative control, we examined H3K27 trimethylation (H3K27me3), a histone modification associated with gene repression. We found no significant correlation between endogenous H3K27me3 and reporter assay activity (Figure S4E). Integrating the endogenous epigenetic state into our ROC analysis resulted in a statistically significant increase in the area under the curve (DeLong's test, GRE motif and H3K27ac log2(fold change [F.C.]) ROC vs. GRE motif ROC, p = 9.8 × 10−3; GRE motif, DNase log2(F.C.), and H3K27ac log2(F.C.) ROC vs. GRE motif, DNase log2(F.C.), and H3K27ac ROC, p = 3.36 × 10−2, Figure 3E). The epigenetic state of both DEX-induced and non-DEX-induced GBSs reflected local chromatin remodeling around the GBS. These results show that functionally distinct classes of GBSs have distinct epigenetic states in the genome before DEX treatment, and distinct changes to epigenetic state after DEX treatment. Further, those epigenetic changes in the endogenous genome are consistent with the observed changes in regulatory activity in the ChIP-reporter assays.
GBSs cluster in the genome
TF binding sites cluster in the human genome (Gotea et al. 2010; Rye et al. 2011). Those clusters may reflect numerous independent binding sites or alternatively, dependencies between sites that arise, for example, via protein-protein interactions between TFs bound at different sites. One way to differentiate between those two possibilities is to induce the GR to bind a subset of sites, and then to ask whether those sites are a random selection of all GR binding sites, as would be expected if binding sites are independent, or instead enriched within clusters, as would be expected if there are local dependencies between GR binding sites. To do so, we analyzed data from a previous study in the same cell type in which lower concentrations of DEX (0.5 nM and 5 nM) were used to induce the GR to bind a subset of the sites that are bound at 50 nM DEX (Reddy et al. 2012a). We then asked whether the subset of sites bound at lower concentrations were randomly distributed across all GR binding sites occupied at 50 nM treatment, indicating independent binding events; or were clustered together, indicating local dependencies between sites.
Twenty-four percent of the sites bound by GR in A549 cells after a 1 h treatment with 50 nM DEX are also bound after treating cells for the same period with 5 nM DEX (Reddy et al. 2012a). The median minimal distance between sites bound by the GR at 5 nM DEX was 11 kb. In contrast, randomly distributing the number of sites that are bound at 5 nM DEX across the larger set of sites bound at 50 nM DEX and recalculating the median minimal distance between bound sites produced a distribution of distances with a median of 23 kb (Figure 4A). The sites bound by the GR at 5 nM DEX were significantly closer to each other than expected according to that permutation analysis (z = −20.1, p = 1 × 10−90). Clustered low dose GR sites should also have fewer stretches of unbound potential GR sites among the possible 50 nM DEX GR sites than expected by chance. We calculated the number of adjacent sites unbound by the GR at 5 nM DEX and compared that to the number of contiguous unbound stretches in a shuffled background model. Consistent with our hypothesis, there were significantly fewer GR unbound stretches than expected by chance (Z-test, Z = −12.9, p = 1.25 × 10−38, Figure S5A). The same clustering was tested and confirmed for 0.5 nM DEX GR sites in terms of distance to nearest adjacent GR binding site and the number of contiguous unbound regions (Z-test, Z = −8.8, p = 8.95 × 10−19; Z = −11.6, p = 1.16 × 10−31, respectively). Finally, after controlling for GR ChIP-seq counts, we did not find that GBSs bound at low doses were enriched for DEX-responsive sites (0.5 nM, p = 0.95; 5 nM, p = 0.30). These results show that GR binds genomic loci in a dose-specific coordinated manner and not as independent binding events.
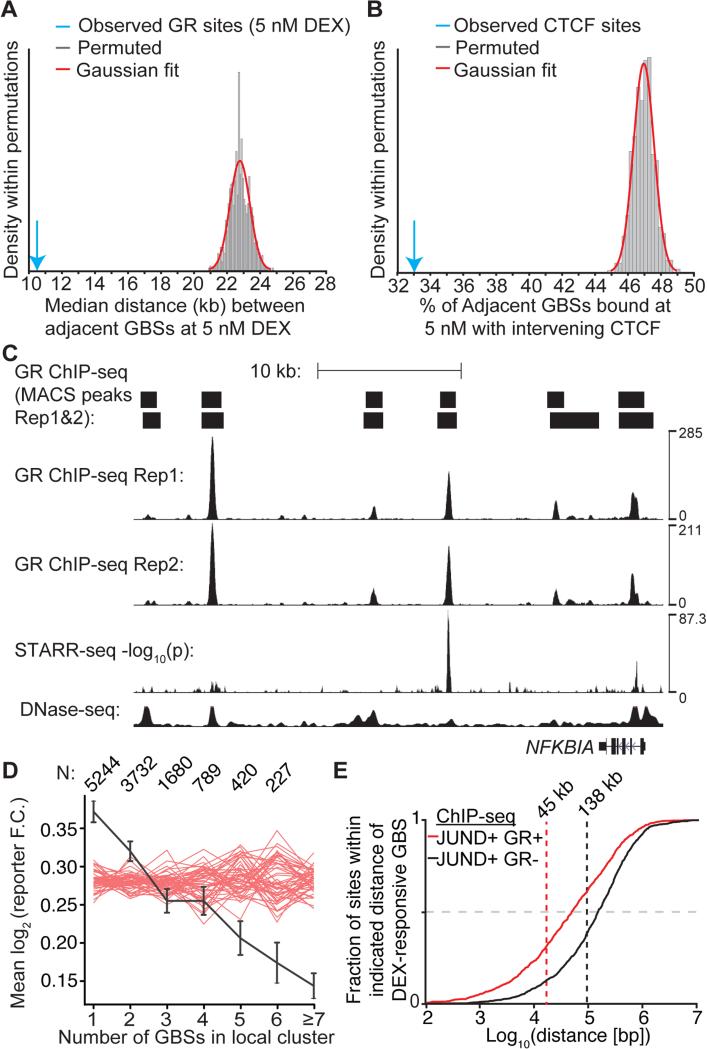
Figure 4. DEX-inducible GR bound enhancers nucleate clusters of GR binding.
(A) The median distance between sites bound by GR in A549 cells after 1 h treatment with 5 nM DEX was calculated (blue arrow). As a null model the locations GBSs bound at 5 nM DEX in A549 cells were permuted between the sites bound by the GR at 50 nM DEX in A549 cells. Sites were permuted 1,000 times and the median distance between sites was calculated after each permutation. (B) Percent of GBSs at 5 nM DEX dose with intervening CTCF sites (blue arrow) and the distribution of a null model in which the location of sites were permuted across possible binding sites occupied across doses and then assayed for the presence of an intervening CTCF binding site. (C) An example of a cluster of GBSs in A549 cells near the NFKBIA gene that is nucleated by a GC inducible enhancer. A549 DNase-seq under control conditions. (D) Each GR binding site was assigned to a cluster by grouping together sites within 5 kb of each other. The GBSs were then divided into bins based on the cardinality of their cluster. The mean and standard error of the per-cluster DEX-induced reporter activity was then plotted as a function of cluster cardinality. Red lines are 50 permutations of the reporter activity across GR binding sites. Numbers of clusters of each size indicated at top. (E) The cumulative distribution of JUND binding sites that are either bound or unbound by GR (black and red lines, respectively) in A549 cells treated with 100 nM DEX for 1 h as a function of proximity to the nearest motif encoded GR binding site.
See also Figure S5.
CTCF is depleted within GBS clusters
The three-dimensional chromatin structure of the genome is organized into topological domains that can functionally separate clusters of regulatory elements from surrounding genomic regions. CTCF is known to demarcate the boundaries of topological domains (Botta et al. 2010; Dixon et al. 2012) and can insulate the activity of adjacent regulatory elements in the genome. If clustered GR binding results from physically interacting sites, we expect CTCF to be depleted between adjacent pairs of GBSs bound at 5 nM DEX. Indeed, 33% percent of the pairs of GBSs that were both occupied at 5 nM DEX had an intervening CTCF binding site. As a control, we permuted which pairs of all possible 50 nM GBSs had an intervening CTCF site. That model resulted in a greater percentage of sites (47%) with an intervening CTCF (Z-test, Z = −22.4, p < 1 × 10−100, Figure 4B). The same result was replicated with 0.5 nM DEX GBSs (Z-test, Z = −2.4, p = 8 × 10−3). Those results show that CTCF is depleted between coordinately bound GBSs.
Tethered GBSs cluster around direct GBSs in the genome
We frequently observed that few GBSs in a cluster had reporter activity, suggesting that GBSs in a cluster may serve different functions. For example, there are six GBSs near the NFKBIA gene, but only two were DEX-responsive in ChIP-reporter assays (Figure 4C). To determine if DEX-responsive GBSs are generally depleted within clusters, we first defined GR binding clusters by identifying groups of GBSs with less than 5 kb between each site. We then quantified the average DEX-responsive activity of the GBSs in each cluster. If DEX-responsive reporter activity was evenly distributed across all GBSs in the genome, we would expect there to be no relationship between cluster cardinality and average DEX-responsive reporter activity. Instead, there was an inverse relationship such that singular clusters had greater DEX-responsive reporter activity, and the most-populated clusters had the least average reporter activity (Figure 4D). Permuting reporter activity across GBSs and repeating the analysis shows that such a trend was unexpected to occur by random. That result was general to the window size used to define clusters (Figure S5B, S5C). We observed the same trend when evaluating the average number of DEX-responsive GBSs per cluster rather than the average activity of those GBSs (Figure S5D). The fraction of clusters that harbor at least one DEX responsive GBS was enriched for small clusters, another indication that GBSs that are isolated in the genome are more likely to function autonomously in a reporter assay (Figure S3F-I). We also found that physically isolated GBSs were more likely to have stronger DEX-responsive activity (Figure S5E). Based on these results, we conclude that clusters of GBSs consist of a mixture of DEX-induced and non-DEX-induced GBSs, and that large clusters are depleted for DEX-induced GBSs while small clusters or isolated GBSs are enriched for DEX-responsiveness.
Based on those observations, we hypothesized that direct GBSs nucleate cluster formation by binding to the genome and then interacting with other TFs bound beyond the range of immediately proximal TF-TF composite activity, resulting in the appearance of non-GRE driven tethered GBSs. To evaluate that model, we focused on interactions between direct GBSs and sites where GR is tethered to AP-1. As reported previously, we found substantial co-occupancy of GR and the AP-1 subunit JUND after DEX treatment (Reddy et al. 2012a). In our analysis, 2,982 (39%) of the JUND binding sites present prior to DEX treatment were bound by GR after DEX exposure. Conversely, 1,992 (64%) of the GBSs occurred at sites also bound by JUND in the presence of DEX. If GR binding directly to the genome via a GRE coordinates GR binding at nearby AP-1 sites, we expect that JUND sites that also bind GR after DEX treatment would be closer to direct GBSs. Indeed, we found that JUND binding sites bound by the GR after DEX treatment were substantially closer to direct GBSs than expected by the genomic distribution of JUND binding (median distance 45 kb vs. 138 kb, Mann Whitney U-test p < 7.46 × 10−41, Figure 4E, Table S5), suggesting that direct GR binding coordinates tethered binding nearby. That finding agrees with a study that used a dominant negative AP-1 to show that GR binding at AP-1 co-bound sites was AP-1 dependent, but that GR binding at direct sites was not (Biddie et al. 2011). A recent study demonstrated that sites enriched for distal P300 interactions are enriched for AP-1 binding motifs and that P300 interactions gained after GC-exposure are enriched for the GRE (Kuznetsova et al. 2015). These results suggest that GR binding directly to the genome explains local clustering of AP-1 tethered GR binding sites across the genome.
Direct GBSs recruit AP-1 binding to genomic sites that lack AP-1 recognition motifs
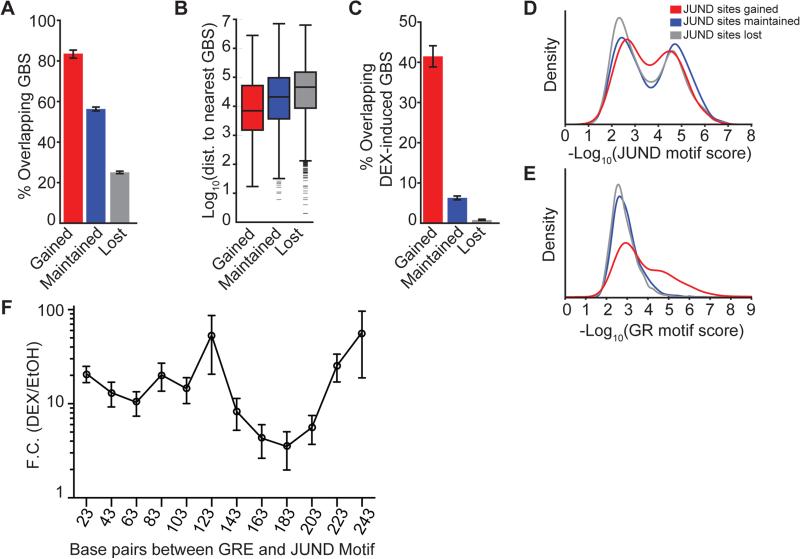
Just as we observe GR occupancy enriched at AP-1 sites that are close to direct GBSs, we also observe enriched gains of AP-1 occupancy at direct GBSs. After DEX treatment, JUND binding was gained at 352 sites, maintained at 2,629 sites and lost at 4,982 sites (Table S6). The majority of gained JUND binding sites (83%) overlapped GBSs. Meanwhile, 56% of maintained JUND binding sites and 25% of lost JUND sites overlapped GBSs (gained vs. maintained, Fisher's exact test p = 1.12 × 10−24, gained vs. lost, Fisher's exact test p < 1 × 10−100, Figure 5A). DEX-induced JUND binding sites were substantially closer to GBSs than maintained JUND binding sites (median distance 0.37 kb vs. 5.23 kb; Mann Whitney U-test p = 4.83 × 10−15) and JUND sites lost after DEX treatment (0.37 kb vs. 21.81 kb; Mann Whitney U-test p = 5.28 × 10−37; Figure 5B). The gains and losses of AP-1 were largely determined by whether the GBS had DEX-responsive reporter activity. Of the 352 AP-1 sites that were induced by DEX, 41% overlapped GBSs that were DEX-responsive in reporter assays. Meanwhile, only 6% of the 2,629 sites that maintained JUND occupancy overlapped with reporter-responsive GBSs (Fisher's exact test p = 2.65 × 10−62, Figure 5C).
Figure 5. Cluster-associated tethered GBSs amplify the expression response of DEX-inducible enhancers.
Comparisons of various and sequence genomic features among JUND binding sites that are gained, maintained, or lost with DEX treatment: (A) Percent that overlap with GBSs. (B) Distance between to the nearest non-overlapping GBS. (C) Percent that overlap GBSs. (D) Distribution of AP-1 motif scores at JUND binding sites that overlap DEX-induced GBSs. (E) GRE motif scores of JUND binding sites that overlap DEX-induced GBSs. (F) Dual luciferase assays in A549 cells treated with 100 nM DEX or vehicle control using plasmids with increasing stretches of non-DEX-responsive DNA between the TSC22D3 DEX-inducible enhancer and a canonical AP-1 binding motif.
See also Figure S6.
We investigated whether GR and AP-1 DNA binding motifs explain the gains and losses of AP-1 with DEX treatment. We computed the distribution of motif scores for the AP-1 binding motif within gained, maintained, and lost JUND sites. The distribution of motif scores in each class was bimodal, suggesting distinct classes of AP-1 motif driven and non-AP-1 motif driven JUND binding (Figure 5D, Figure S6A). Next, we computed GRE motif scores at each class of JUND binding site. While the majority of JUND binding sites had GREs consistent with a background model in which sequences were shuffled prior to motif finding, many novel JUND binding sites contained strong matches to the GRE (Figure 5E). When two-component Gaussian mixture models were fit separately to the GR motif scores of JUND gained, JUND maintained, and JUND lost sites, only JUND gained sites had a mixture component with a significantly greater GR motif score than that of matched dinucleotide shuffled sequences (Z-test, p = 3 × 10−3) (Figure S6B). We estimate that 47% of the gained JUND sites belonged to the class of sites with greater GR motif score. These results suggest that, just as GR binding to the genome at tethered sites corresponds with enrichment for AP-1 motifs, AP-1 can be observed binding at strong GREs after DEX exposure.
Epistatic interactions between GR and AP-1 modulate DEX-induced regulatory activity
GR and AP-1 act together to regulate gene expression [e.g. (Diamond et al. 1990; Herrlich 2001; Ratman et al. 2013)], and AP-1 plays a major role in determining where GR binds in different cell types (Biddie et al. 2011; Gertz et al. 2013). Our GR ChIP-reporter experiments and subsequent analyses have shown that AP-1 sites are not sufficient for DEX-responsive regulatory activity in a reporter assay, yet are clustered around direct GR binding sites in the genome. Previous studies have demonstrated that GR and AP-1 binding immediately adjacent to each other can synergistically amplify the transcriptional response to glucocorticoids, which we confirm (Figure S6C-G, Table S3) (Mittal et al. 1994; Pearce et al. 1998). Our clustering analysis also suggests that GR and AP-1 interactions occur over tens of kilobases (Figure 4). One possible explanation is that direct GBSs interact with AP-1 binding sites via DNA looping, and that those interactions also alter the GC response. To distinguish between immediately adjacent binding and looping, we varied the spacing between a DEX-induced GBS and a canonical AP-1 response element, and again tested for synergy between the sites (Figure 5F, Figure S6H-J). Consistent with previous studies (Pearce et al. 1998), AP-1 amplified the effects of GC-mediated gene activation ~20-fold when the AP-1 binding motif was between 23 and 123 nt from the GRE. The extent of activation was decreased to a minimum of 3.5 fold induction when the AP-1 response element was between 143 and 183 nt from the GRE, consistent with the axial stiffness of DNA inhibiting interactions between GR and AP-1 (Lee and Schleif 1989). Finally, reporter gene induction increased to a maximum of 58-fold when the interval between the GRE and the AP-1 binding site was between 203 and 243 nt (Figure 5F, Figure S6I). That interval is substantially more than the estimated persistence length of DNA (Lee and Schleif 1989), and is therefore consistent with DNA looping between the GR and AP-1 site. These data provide evidence that AP-1 binding sites could interact distally with DEX-inducible GR occupied enhancers to increase the potency of GC-responsive regulatory elements.
Distal binding cluster interactions are a general mechanism of gene regulation
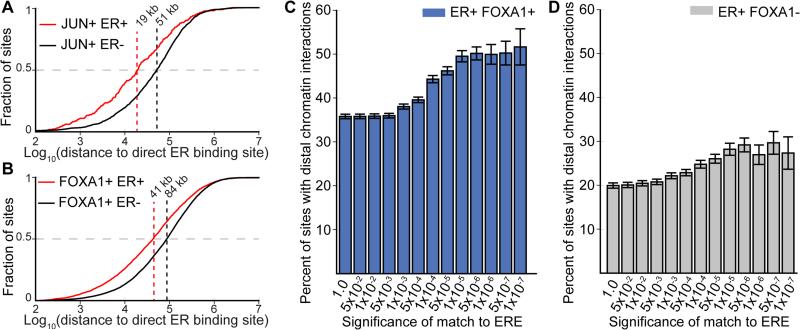
Many TFs bind to the genome in clusters, raising the possibility that our model of TF clustering via interactions between direct and tethering sites may be a general phenomenon. To address this possibility we analyzed published genomic datasets generated to study the estrogen receptor (ER) (Joseph et al. 2010; Hurtado et al. 2011). ER cooperatively interacts with both AP-1 and FOXA1. In those studies, AP-1 and FOXA1 have been described as either pioneer factors that increase chromatin accessibility prior to ER binding at composite sites or, as tethering factors that indirectly bind ER to the genome. Alternatively, we hypothesized that like the GR, the ER binds to the genome in clusters that reflect interactions between direct ER binding sites and nearby AP-1 or FOXA1 binding sites. As we observed in the case of the GR, direct ER binding sites were substantially closer to JUN binding sites that become ER bound than the genomic distribution of JUN sites (Figure 6A, 19 kb vs. 51 kb, Mann Whitney U-test p = 3.97 × 10−14). Likewise, we found that direct ER binding sites were substantially closer to FOXA1 binding sites that became bound by the ER after estrogen treatment than the genomic distribution of FOXA1 binding sites (Figure 6B, 41 kb vs. 84 kb, Mann Whitney U-test p < 2.06 × 10−71). Analysis of published ER ChIA-PET data revealed that at sites bound by the ER and FOXA1 or sites bound by the ER alone, the percentage of sites with distal chromatin interactions was directly correlated with ERE strength (Figure 6C, 6D) (Fullwood et al. 2009). These analyses demonstrate that analogous to the GR, the direct ER binding sites also likely interact with distal tethering proteins in a locally dependent manner
Figure 6. Coordinated clusters of ER binding relies distal chromatin interactions.
(A) The cumulative distribution of JUN binding sites that are either bound or unbound by ER (black and red lines, respectively) as a function of proximity to the nearest ERE motif encoded ER binding site. (B) The cumulative distribution FOXA1 binding sites that are either bound or unbound by ER (black and red lines, respectively) as a function of proximity to the nearest ERE motif encoded ER binding site. (C) Prevalence of distal chromatin interactions at ER binding sites co-bound by FOXA1 binned by the significance of the best scoring match to the ERE contained in the binding site. (D) Distal ER interactions as in panel C, for sites with no evidence of FOXA1 binding.
Discussion
Genome-wide mapping of TF-DNA interactions has revealed a vast excess of TF binding sites relative to the number of regulated genes (Reddy et al. 2009; Gao et al. 2013). Using a comprehensive empirical analysis of the regulatory activity of each GBS, we have developed an enhancer-cluster model in which direct GBSs interact with co-binding TFs kilobases away via protein-protein interactions (Figure 7). We predominantly focused on the well-known interactions between GR and AP-1. Several models have been proposed to explain those interactions such as AP-1 recruiting GR to sites that lack a GRE (Teurich and Angel 1995) or AP-1 enabling GR to bind weak versions of a GRE (Biddie et al. 2011; John et al. 2011). Our model expands on those possibilities by suggesting that GR also binds AP-1 sites via chromatin loops between AP-1 sites and distinct direct GBSs. We also show that such interactions are necessary for the DEX-responsiveness of some GBSs observed with ChIP-seq. Our model predicts the reciprocal effect of AP-1 binding at direct GREs upon DEX treatment, which we observe (Figure 5E). Our interaction model helps to explain the following observations: (i) the discrepancy between the number of TF binding sites and the number of regulated genes, (ii) the fact that only a small fraction of the binding sites for a given TF have a DNA binding motif for that TF (Gertz et al. 2013), and (iii) the distinct patterns of epigenetic remodeling at direct and tethered GBSs. Genetic studies have also shown that it is common for single nucleotide variants to disrupt TF binding without direct binding motifs for that TF (Reddy et al. 2012b; Soccio et al. 2015). According to our model, an explanation is that the variant disrupts interactions between TFs, thus leading to distal regulatory perturbations.
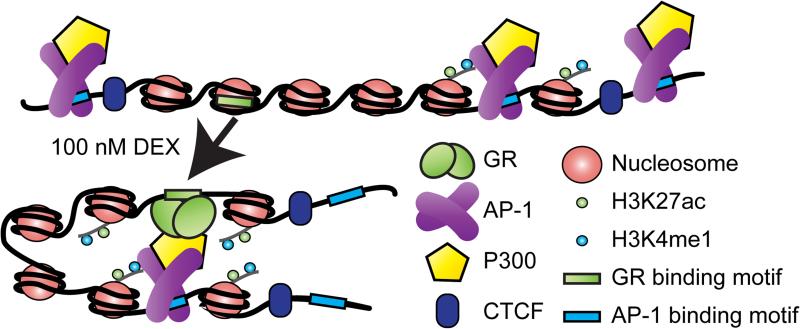
Figure 7. A model of enhancer-cluster coordinated GC-inducible enhancer activation.
Prior to hormone exposure, AP-1 binds to the genome. AP-1 bound sites are in domains of increased chromatin accessibility that are enriched for H3K27ac, H3K4me1 and P300. After hormone exposure, GR binds directly to the genome in regions of less accessible chromatin. GR interacts with distal AP-1 bound sites within the same CTCF-defined topological domain. The epigenome of the direct GBSs is remodeled, becoming enriched for H3K27ac, H3K4me1 and increased in chromatin accessibility. Direct GBSs confer GC-inducible enhancer function, while AP-1 bound interacting sites modulate the expression output of direct GBSs.
Previous studies have demonstrated that AP-1 tethered GBSs vastly outnumber direct GBSs and that tethered sites are enriched in cell-type-specific GR binding. Based on those two findings, tethered sites are thought to play a major role in cell type-specific transcriptional GC responses (Gertz et al. 2013). Meanwhile, our results show that while many tethered GBSs act as steady-state enhancers, only direct GBSs initiate GC-responsive enhancer function. A potential resolution to that discrepancy is that distal interactions between direct GBSs and tethered sites tune the activity of both cell-type-specific and cell-type-shared direct GBSs. Each direct GBS can interact with multiple nearby tethering sites. Thus, cell-type-specific tethered binding sites outnumber the cell-type-specific direct binding sites that nucleate GR binding clusters. Given the extent of amplification of reporter activity that we observe, looping interactions between direct GBSs and tethering TFs may be a major contributor to the transcriptional response to GCs. It is unclear if clusters of interacting GBSs are the result of multiple tethered sites interacting with a single direct GBS, or the result of ChIP-seq signal aggregated from a population of cells with binary GR/AP-1 interactions. Consistent with our evidence, separating direct and tethered GBSs minimally improves the ability to predict DEX-induced genes (Figure S6K). It remains to be determined which specific aspects of the interactions between direct and tethered GBSs will best predict GC-mediated expression responses.
While the results of this study demonstrate an enhancer-cluster model of GC-mediated gene induction, they do not preclude the occurrence of additional regulatory mechanisms. The specific configuration of TFs at direct and tethered sites, interactions with other TFs, GR and AP-1 phosphorylation, DNA methylation, heterochromatin formation, and the amount of GR in a cell are all likely to contribute to the transcriptional response to GCs. Thus, additional mechanisms may contribute to the lack of repression observed in our results, and to the cell-type specificity of some direct GBSs. For example, while including GR and AP-1 sites on the same plasmid amplified an activating DEX response in our experiments, others have demonstrated that tethering can also result in repression of gene expression (Luecke and Yamamoto 2005). Further clarifying the determinants and effects of interactions between direct and tethered sites will therefore likely be informative in understanding the complexities of GC-mediated gene regulation.
Finally, while the focus of this study is on GR and ER binding sites, many other TFs have similar distributions of genomic binding and binding mechanisms. It is therefore likely that our model applies to numerous additional eukaryotic TFs, and may broadly contribute to the coordination of TF binding observed across the human genome.
Methods and Resources
CONTACT FOR REAGENT AND RESOURCE SHARING
Email contact for reagent and resource sharing: Tim.Reddy@Duke.edu
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All experiments were performed in A549 cells. Cells were obtained from the Duke Cancer Center Cell Culture Facility.
METHOD DETAILS
Cell Culture
A549 cells were grown at 37 °C and 5% CO2 in F-12K medium with 10% fetal bovine serum and 1% penicillin-streptomycin. Cells were treated by adding 0.02% by volume of 5 mM DEX directly to the cell culture media. As a vehicle control, cells were treated in parallel with 0.02% by volume EtOH.
Chromatin Immunoprecipitation (ChIP) and ChIP-seq
Chromatin immunoprecipitation was performed as previously described (Reddy et al. 2009) using 2 × 107 A549 cells per replicate. Cells were sonicated using a Bioruptor XL (Diagenode) on the high setting until the resulting chromatin was fragmented to a median fragment size of ~250 nt as assayed by agarose gel electrophoresis. ChIP was performed using the 5 μg rabbit polyclonal GR antibody (Santa Cruz Biotechnology sc-1003), and 200 μl of magnetic sheep anti-rabbit beads (Life Technologies M-280). After reversal of formaldehyde crosslinks at 65 °C overnight, DNA was purified using MinElute DNA purification columns (Qiagen). Illumina sequencing libraries were then generated using the Apollo 324 liquid handling platform according to manufacturer's specifications (Wafergen).
Cell transfection and library harvesting
ChIP STARR-seq input libraries were combined in equimolar pools and transfected into T-75 (BAC STARR-seq) or T-150 flasks (ChIP STARR-seq) of A549 cells with Fugene HD (Promega) at a 4:1 ratio of Fugene:DNA. Three replicate transfections were performed per experimental condition. Cells were treated for 3 h with either 100 nM DEX or 0.02% by volume EtOH. Cells were rinsed with PBS pH 7.4 and incubated for 3 min at 37 °C with DNase I (5 mg DNAse I in 1 ml of buffer containing 10 mM Tris-HCl pH 7.5, 150 mM NaCl and 1 mM MgCl in DEPC treated water diluted to a total volume of 24 ml in PBS). Cells were rinsed again with PBS and then dissociated with Trypsin-EDTA 0.25% (Life Technologies). Trypsin was neutralized with A549 tissue culture medium and cells were pelleted via centrifugation. Cell pellets were rinsed once with PBS and then lysed in 2 ml of RLT buffer (Qiagen) with 2-mercaptoethanol (Sigma).
ChIP-reporter Input Library Construction
ChIP-seq libraries were adapted to STARR-seq by amplifying with 8 cycles of PCR (New England Biolabs Q5, GC buffer) using primers P1F and P1R to enable cloning into the STARR-seq human screening reporter backbone as previously described (Arnold et al. 2013). After cloning, the reaction was purified using 1.5X Agentcourt Ampure XP beads (SPRI beads; Beckman Coulter) and eluted in 10 μl water. Purified constructs were split into four 2 μl aliquots and electroporated into 60 μl of MegaX DH10B competent cells (Life Technologies) per aliquot. Cultures were recovered in 3 ml SOC medium for 1 h and then grown in suspension in 400 ml of Luria Broth medium for 14 h. Library plasmids were purified using the Promega Pure Yield maxiprep system.
ChIP-reporter output library construction
Total RNA was prepared using the Qiagen RNeasy kit. Poly-A RNA was isolated from 70 μg of total RNA by double selecting with Dynabead Oligo-dT(25) beads (Life Technologies). RNA was then treated with turboDNase (4 U) for 30 min at 37 °C (Invitrogen). DNase treated poly-A RNA was purified using the RNeasy Mini kit. Plasmid specific cDNA was synthesized using Superscript III (Life Technologies) incubated for 2.5 h at 55 °C and inactivated at 70 °C for 15 min. Following synthesis, cDNA was treated with RNaseA (Sigma) at 37 °C for 30 min. cDNA was purified using SPRI beads and then amplified and indexed for sequencing using a two stage PCR as described previously (Arnold et al. 2013).
Quantification of regulatory activity in ChIP-reporter assayed sites
The ChIP-reporter output libraries were sequenced using paired-end 25 bp reads on an Illumina MiSeq and analyzed as described in supplemental methods.
BAC STARR-seq Input Library Construction
Six BACs (RP11-806F7, RP11-435L21, RP11-139K17, RP11-788A16, CTD-2340K24, RP11-769H22) that contain previously identified DEX-responsive genes (Reddy et al. 2009) were amplified in E. coli and the resulting DNA was prepared using the NucleoBond BAC 100 kit (Macherey-Nagel). PCR was used to validate that the prepared DNA was from the expected genomic regions. The DNA from each BAC was pooled in equimolar ratios for tagmentation (Illumina) (Adey et al. 2010; Gertz et al. 2012). The tagmentation reactions were performed in six reactions, each with 50 ng of pooled BAC DNA. Three reactions used 1 μl of transposase, and three used 5 μl of transposase. The reactions were incubated at 55 °C for 5 min and then moved to ice. Reactions were neutralized using 30 μl QG buffer (Qiagen) and purified using SPRI beads at a 1.125X SPRI:reaction ratio. Purified reactions were eluted in 22 μl of nuclease free water (Sigma). The resulting tagged fragments were then PCR amplified to add 15 bp of sequence matching the STARR-seq backbone as for ChIP-reporter assays. Specifically, the PCR reaction was primers with primers SSV-nxt-F and SSV-nxt-R, and the DNA was amplified with Phusion High fidelity polymerase with GC buffer (New England Biolabs). The reactions were incubated at 72 °C for 3 min; 98 °C for 30 s; followed by 10 cycles of (98 °C for 10 s, 63 °C for 30s and 72 °C for 3 min). The resulting products were purified using SPRI beads at a 1.8X SPRI:reaction ratio.
The STARR-seq screening vector was digested overnight with SalI and AgeI and linearized backbone was purified with the Wizard SV Gel and PCR Clean-Up kit (Promega). 100 ng of backbone and 17 ng pooled insert were cloned in three 10 μl Infusion HD reactions (Clontech). Infusion reactions were then pooled and electroporated into MegaX DH10B electrocompetent cells at a ratio of 4 μl reaction to 20 μl of competent cells for a total of six electroporations. Transformations were recovered for 1 h in SOC medium while shaking (225 rpm, 37 °C) and then grown for 14 h in 250 ml of Luria Broth while shaking (225 rpm, 37°C). The resulting STARR-seq input libraries were then purified using the Promega Pure Yield Maxiprep kit.
To assess fragment diversity in the STARR-seq input libraries, the fragments inserted into each was sequenced on an Illumina MiSeq. 10 ng of each input library was PCR amplified using indexed Nextera primers and Phusion DNA polymerase in GC buffer (New England Biolabs). The following thermal cycling protocol was used: 98 °C for 30 s, followed by 10 cycles of (98 °C for 10 s, 65 °C for 30 s, 72 °C for 2 min), with a final extension at 72 °C for 7 min. PCR products were purified using SPRI beads (1.8X SPRI:DNA ratio) and sequenced on an Illumina MiSeq Instrument using 25 bp paired end reads.
STARR-seq output library construction
RNA from transfected A549 cells was prepared as for ChIP STARR-seq output libraries. Reverse transcription was performed in 50 μl reactions using SuperScript III scaled up from the manufacturer's protocol. Reactions were primed using Oligo(dT)12-18 primers (Life Technologies) and incubated at 42 °C for 1 h, 50 °C for 90 min and then inactivated at 70 °C for 15 min. cDNA samples were purified using SPRI beads (1.8X SPRI:DNA ratio). Purified cDNA reactions were then PCR amplified using Phusion polymerase with GC buffer under the following conditions: 98 °C for 30 s, 25 cycles of (98 °C for 10 s, 65 °C for 30 s, 72 °C for 2 min) with a final extension at 72 °C for 7 min.
Nuclear localization of plasmids
A549 cells were transfected using Fugene HD in 10 cm dishes as per manufacturer's protocol. Six transfections were performed. After 45 h, 3 plates were treated with 100 nM DEX and 3 plates were treated with 0.02% EtOH for vehicle control. Cells were harvested 3 h after treatment. Purified nuclei were harvested using the Nuclei EZ Prep Kit (Sigma-Aldrich) according to manufacturer's instructions. Purified nuclei were suspended in Qiagen Plasmid DNA mini-prep buffer P1 rather than nuclei storage buffer and processed according to standard bacterial plasmid mini-prep protocol. Sequencing libraries were generated via PCR using the same protocol used to assay input libraries. Plasmid diversity sequencing was performed using a single lane on an Illumina MiSeq with 25 bp paired end reads.
Luciferase reporter assay validations
GR binding sites were selected from regions of the genome near DEX-responsive genes containing GR binding sites assayed by both ChIP STARR-seq and BAC STARR-seq. Elements were selected to be ~400 bp in length. The elements were PCR amplified from pooled BACs (see “STARR-seq Input Library Construction”) using Q5 polymerase and the following cycling conditions: 98 °C for 30 s, 35 rounds of (98 °C for 10 s, 58 °C for 30 s, 72°C for 2 min), and a final extension at 72 °C for 7 min. Primers were designed to anneal to the target region and add 15 nucleotides that match either the HindIII or EcoRV side of the multiple cloning site of the pGL4.24 minimal promoter firefly luciferase expression vector (Promega) to enable Infusion cloning. Appropriate PCR band size was determined by electrophoresis and amplicons were purified using SPRI beads (1.5X SPRI:reaction ratio) prior to cloning.
pGL4.24 was linearized at the multiple cloning site by incubating overnight with HindIII and EcoRV. Test elements were cloned using Infusion HD (CloneTech) and transformed into Stellar competent cells (CloneTech). Colonies were picked, grown overnight and DNA was purified using PureYield Plasmid Miniprep kits (Promega), and screened for the expected insert with Sanger sequencing.
A549 cells were plated in 96-well plates (10,000 cells/well) and transfected using Fugene HD two days after plating at a 4:1 Fugene:DNA ratio. 100 ng of each test construct and 10 ng pRL-TK Renilla luciferase normalization vector (Promega) were transfected per well with 12 replicates. After 24 h, six wells of cells per vector were treated with medium containing either 100 nm DEX or 0.02% EtOH control. After 4.5 h — 3 h for response and 1.5 h for protein folding (Tyedmers, Brunke et al. 1996) — cells were harvested and assayed using the Dual-glo luciferase assay system (Promega) and plates were read using a Victor3 1420 Multilabel Counter (PerkinElmer).
The ratio of firefly luciferase activity to Renilla luciferase activity was determined for each well. The six replicates of each condition were averaged and then induction was determined by calculating the log2(average Renilla-normalized luciferase activity in the DEX condition / average Renilla-normalized luciferase activity in the EtOH condition).
AP-1 GRE combinatory experiments
For GRE/AP-1 spacing experiments, gene blocks that encode the TSC22D3 DEX-induced enhancer (Figure S6E) followed by 250 bp of non-DEX-inducible DNA (as assayed by STARR-seq; Figure S6I) and a canonical JUND binding element (Mathelier et al. 2014) (MA0489.1) were synthesized (Integrated DNA Technologies). The spacing of the JUN binding motif was varied in 20 bp increments (Figure S6H). DNA fragments were analyzed using the JASPAR database to confirm that no de novo JUN or GR binding motifs were generated while designing the constructs.
For GRE/AP-1 combination experiments, DEX responsive enhancers were synthesized in combination with JUN consensus binding motifs (Figure S6C-G) (Mathelier et al. 2014). non-DEX-induced DNA was used as intervening sequence between sites (Figure S6I). Motifs were mutated by altering the top scoring three nucleotides of the position weight matrix of each motif.
Twenty bp of DNA from the 3’ and 5’ cloning sites were added during gene block synthesis. The resulting fragments were cloned directly into the EcoRV site of the Pgl4.24 backbone (Promega) using Gibson Assembly Master Mix (NEB) according to manufacturer's instructions. DEX-induced Luciferase assays were performed as above, using 4ng pRL-sv40 per well as internal control and assayed using the GloMax multiwall luminometer (Promega). In each experiment 12 replicates were treated with 100 nM DEX and 12 replicates were treated with 0.02% EtOH. Data from these experiments are displayed in Figure S6F and Figure S6I.
GRE addition experiments
Two non-DEX-induced GBSs (~450 nt) were identified from the TSC22D3 and NFKBIA loci (Table S3). Sites were selected based on increased chromatin accessibility in control condition. The highest scoring match to the AP-1 motif was identified (Mathelier et al. 2014) (MA0489.1) in each site and constructs were designed and cloned (as described above; AP-1 GRE combinatory experiments) that contained t a minimal GRE identified from the PER1 locus cloned 20 nt 5’, 3’ or both 5’ and 3’ of the non-induced GBSs. A congruent set of vectors containing the mutated forms of the GRE (as described; AP-1 GRE combinatory experiments) were generated in each cloning position.
Dual luciferase assays (as described above; AP-1 GRE combinatory experiments) were used to determine the effects of adding GREs to non-DEX-induced elements.
DEX washout reporter assay experiments
The upstream PER1 DEX-induced enhancer was synthesized (Integrated DNA technologies) and cloned into STARR-seq as described above (STARR-seq Input Library Construction). The resulting vectors were transfected into A549 cells and treated with 100 nM DEX, or .02% EtOH vehicle control for 30 minutes. Six replicates of each condition were treated for 3 h, and 6 replicates of DEX treated cells were washed twice with medium containing EtOH vehicle and treated for an additional 3 h.
RNA was harvested (Qiagen RNeasy miniprep kit), with the addition of ERCC external control RNA cocktail added to each RNA preparation at the cell lysis step. RT-qPCR was performed using primers designed to span the splice junction of the STARR-seq transcript or ERCC external control primers (Devonshire et al. 2010) and the RNA-2-CT cDNA synthesis and SYBR green qPCR kit as per manufacturer instructions. qPCR data was analyzed using the delta-delta-cT method
RNA-seq
Total RNA was harvested from three replicates of A549 cells treated with DEX or EtOH as previously described in main methods using the Qiagen RNease Mini kit including the on-column DNase digestion. Poly-A RNA was isolated from 1 μg of total RNA as described above, and Illumina RNA-seq libraries were prepared using the Apollo 324 library prep station according to the manufacturer's protocol (Wafergen).
ChIP-exo Library Construction
Human lung adenocarcinoma cells (A549s) were grown to confluence in 15 cm plates (~20 million cells) and treated with 100 nM dexamethasone for 1 h, in triplicate. Cells were processed as described in the ChIP-seq library construction described above, through to the LiCl washes. Chromatin-bound IgG beads were then washed with 10 mM Tris-HCl (pH 7.5) and resuspended in 30 μl of end repair mix: 1X T4 Ligase Buffer, 0.4 mM dNTPs, 1.8 U T4 DNA Polymerase, 6 U T4 PNK, and 3 U DNA Polymerase I, Large (Klenow) Fragment. Beads were incubated at room temperature for 30 minutes, and then washed with 10 mM Tris-HCl (pH 8). Beads were then resuspended in 30 μl of A-tailing mix: 1X NEB Buffer 2, 0.2 mM dATP, and 3 U Klenow Fragment (3’->5’ exo-). Beads were incubated at 37°C for 30 minutes. Next, beads were washed with 10 mM Tris-HCl (pH 7.5) and resuspended in 30 μl adapter ligation mix: 1X Quick Ligase Buffer, 4 μl Quick Ligase, and 1 pmol of Illumina Adapter A. Beads were incubated at room temperature for 30 minutes. Next, the beads were washed with 10 mM Tris-HCl (pH 9.5), and resuspended in 20 μL of λ-exonuclease mix: 1X λ-exonuclease Buffer and 5 U λ-exonuclease. The beads were incubated at 37°C for 30 minutes. Next, the beads were washed with 10 mM Tris-HCl (pH 8), and resuspended in 20 μL RecJf mix: 1X NEB Buffer 2 and 15 U RecJf. The beads were incubated at 37°C for 30 minutes, and then washed with TE Buffer. The beads were resuspended in 150 μL IP Elution Buffer (1% SDS, 0.1 M NaHCO3), and incubated for 1 hr at 65°C, with vortexing every 15 minutes. The beads were centrifuged at 20,000xg for 3 minutes, and the supernatant was transferred to a new tube and incubated at 65°C overnight. DNA was purified using a MinElute PCR Purification kit (Qiagen), using 11 μl of EB Buffer for the elution. Purified DNA was mixed with 8 μL of phi29 DNA polymerase mix: 1X phi29 Buffer, 10 pmol primer P2, 375 nM dNTPs, and 4 μg BSA. The mix was heated to 95°C for 2 minutes, 63°C for 5 minutes, 30°C for 2 minutes, and held at 30°C. 10 U of phi29 DNA polymerase was added to the mix, which was then heated at 30°C for 20 minutes, 65°C for 10 minutes, and held at 4°C. DNA fragments were purified using AxyPrep Mag PCR Clean-Up beads at a 2:1 bead volume:sample volume ratio. DNA was eluted off beads using 30 μl A-tailing mix, and incubated at 37°C for 30 minutes. The DNA was then purified using AxyPrep Mag PCR Clean-Up beads as described above. DNA was eluted from beads using 30 μL of adapter ligation mix (with Illumina Adapter B instead of A), and incubated at room temperature for 30 minutes. The DNA was then purified using AxyPrep Mag PCR Clean-Up beads as described above, and eluted in 50 μl of PCR amplification mix: 1X Q5 Reaction Buffer, 200 nM dNTPs, 10 pmol Primer A, 10 pmol Primer B, and 1 U Q5 polymerase. PCR was carried out according to the manufacturer's specifications, using an annealing temperature of 65°C, and an extension time of 30 seconds, for 20 cycles. The PCR products were purified with AxyPrep Mag PCR Clean-Up beads with a 1:1 ratio, and eluted in 30 μL of EB Buffer. DNA concentration was measured on a Qubit Fluorometer (Life Technologies), and fragment size distribution was assessed on a TapeStation 2200 (Agilent Technologies). Libraries were sequenced on an Illumina HiSeq 2000 with 50 base-pair paired end reads.
DNase-seq
DNase-seq libraries were made from fresh cell cultures of the control cell line and treated cell line, with three replicates for each cell line. Library preparation and analysis was performed as described (Song and Crawford 2010) with the modification that oligo 1b was synthesized with a 5’ phosphate to increase the efficiency of ligation.
QUANTIFICATION AND STATISTICAL ANALYSIS
GR ChIP-seq analysis
GR ChIP-seq was performed in biological duplicates after 3 h treatment with either 100 nM DEX or 0.02% by volume EtOH. The libraries were sequenced on an Illumina MiSeq using single-end 50 bp reads. The sequencing reads were aligned to the hg19 version of the reference genome using Bowtie (Langmead et al. 2009), and binding sites were called in the DEX-treated samples relative to the EtOH-treated samples using the MACS peak calling software (Zhang et al. 2008; Feng et al. 2012). Peaks were then split into subpeaks using PeakSplitter (Salmon-Divon et al. 2010), and set to a fixed size of 500 bp by extending 250 bp to either side of the sub-peak summit. To combine peaks between replicates in an inclusive manner, subpeaks between replicates were merged using mergeBed from BEDTools (Quinlan 2014), requiring a 250 bp overlap to combine peaks. The resulting peaks were again fixed to a size of 500 bp by extending 250 bp upstream and downstream from the peak midpoint.
Estimating library diversity in reporter libraries
The number of unique fragments in the ChIP STARR-seq input library was estimated by sequencing on an Illumina MiSeq instrument and estimating the point of saturation. Plasmids were amplified using standard full-length Illumina sequencing adapters and sequenced using 25 bp paired end sequencing. Colony PCR was performed on individual clones of the vector plated from the initial transformation to confirm cloning efficiency. ChIP STARR-seq insert sequence data was aligned to the hg19 version of the human genome using Bowtie (Langmead and Salzberg 2012), and converted to fragments using SAMtools (Li et al. 2009) and an in-house Perl script. Insert diversity was analyzed using a custom perl script, and the number of fragments per library was estimated by Bmax in a model that accounts for saturation and non-specific sequencing errors (Y = Bmax X/(Kd active X) active NS X active Background). The model was fit using the Prism software package.
Quantification of regulatory activity in ChIP-reporter assayed sites
Reads were aligned to the hg19 build of the human genome using Bowtie (Langmead and Salzberg 2012), and the number of aligned reads per GR binding site was determined. The DEX-responsive regulatory activity of each site was evaluated using DESeq2 where read counts at each site were normalized by the total number of aligned reads in each library, and statistically significant changes in regulatory activity after DEX treatment were evaluated using a negative binomial model of the normalized read counts (Love et al. 2014a).
Estimation of Regulatory Activity in STARR-seq Assayed Sites
BAC STARR-seq output libraries were sequenced on an Illumina MiSeq Instrument using paired-end 25 bp reads. Reads were aligned to the hg19 reference genome sequence for the target BACs using Bowtie (Langmead and Salzberg 2012), paired end reads were converted to corresponding fragments, and the number of reads per fragment was counted for each replicate. The number of reads in each experiment was then normalized to the median of the number of reads per fragment divided by the geometric mean read count of that fragment. To estimate the significance of differences between EtOH and DEX treatment, a Wilcoxon signed-rank test was performed using a sliding 1 bp window across the target BACs. For each test, the normalized read counts for the fragments overlapping the window were compared between DEX and EtOH treatment conditions, and a p-value was reported.
RNA-seq Expression Analysis
RNA-seq reads were sequenced on an Illumina HiSeq using 50 bp single end reads. The resulting reads were aligned to version 19 of the GENCODE reference set of transcripts using Bowtie (Harrow et al. 2006; Langmead and Salzberg 2012). Differential expression was called using DESeq2 (Love et al. 2014) at a false discovery rate (FDR) < 5% (Benjamini and Hochberg 1995).
Estimating Proximity Between GR Binding Sites and the TSSs
Transcriptional start site coordinates used in ChIP STARR-seq—gene proximity analyses were obtained from version 19 of the GENCODE annotation for genes that were differentially expressed after treatment for 3 h with 100 nM DEX (FDR < 5%) (Harrow et al. 2006). The ChIP STARR-seq-TSS distances used for empirical cumulative density functions were found using the closestBed function from BEDTools (Quinlan 2014). GC–induced ChIP STARR-seq sites were defined as having an FDR < 5% and non-GC–induced ChIP STARR-seq sites were defined as having an FDR > 30%. Differing sizes of GC–induced ChIP STARR-seq and non-DEX-induced ChIP STARR-seq groups could confound the distribution of distances of the closest ChIP STARR-seq element to a given TSS. To account for these sample size effects, elements from the non-DEX-induced ChIP STARR-seq group were randomly sampled and distance calculations were performed 100 times to match the sample size of the DEX-induced ChIP STARR-seq group. The mean and standard deviation of these sampled calculations are shown.
Regression Model to Predict Expression Response from ChIP-seq Signal
The association between GR occupancy and proximal gene expression in response to DEX treatment was examined as follows. First, ChIP-seq signal (log fold-change in ChIP-seq in response to DEX) was summed across sites within a maximum distance D from the TSS of each DEX-responsive gene, where values of D = 50 kb, D = 100 kb, and D = 200 kb were examined; ChIP-seq signal was summed separately for reporter-positive and reporter-negative GR binding sites. Correlations between summed ChIP-seq signal and gene expression were computed via Spearman's rho and plotted separately for each value of D, for reporter-positive and reporter-negative sites (Figure S6K).
DNA Binding Motif Identification and Scoring
Match scores and p-values for motif matches in ChIP STARR-seq tested elements were found using the Motif Alignment and Search Tool (MAST) from the MEME Suite (Bailey and Gribskov 1998). The top scoring motif occurrence was computed for each ChIP STARR-seq element and used for subsequent correlations and ROC analyses.
Mixture Model and Sequence Analysis to Distinguish Sites With and Without GREs
The set of all tested GBSs was assumed to consist in a mixture of sequences with and without GRE motifs. We fit a two-component Gaussian mixture model to the negative log10 p-values for the best GRE motif match for all tested GBSs. The mean match scores corresponding to GRE motif and no GRE motif from the model that was fit with all GBSs were then fixed for reporter positive and reporter negative GBSs. Variances and mixture weights were estimated separately for each class of GBSs. We used the mixture weights to estimate of the proportion of each class of sequences that contained a GRE.
To identify motifs enriched in responsive and non-responsive GR binding sites with GREs, we constructed a matched set of 1,040 sites in each class that do or do not have a GRE according to our mixture model. We then performed a de novo motif search using MEME to find twelve enriched motifs in each set under a zero-or-one motif per sequence (ZOOPS) model. We then used Tomtom to compare the identified motifs to known TF binding motifs.
Receiver Operating Characteristic (ROC) Curve Analysis
ROC analysis was performed in R using the pROC software package (Robin et al. 2011). Putative regulatory elements were classified as enhancers if they had an FDR < 0.05 in ChIP-reporter and increased in reporter gene expression. Motif p values for tested motifs were assigned to each of the tested GR sub-peak as determined by MAST and those p-values were evaluated as predictors of enhancer status. Predictor baseline was established by repeating ROC analysis following motif prediction in randomly shuffled sequences. Shuffling was performed while preserving dinucleotide frequencies, using software uShuffle (Jiang et al. 2008) version Feb21/2008.
ChIP-exo Analysis
Read pairs were aligned to the hg19 reference genome using Bowtie2 (Langmead and Salzberg 2012) with default settings. Fragments mapping to adapter sequence or ENCODE blacklisted regions (ENCODE 2012) were removed. PCR duplicates were removed using SAMtools (Li et al. 2009), and reads across triplicates were pooled. The position of the 5’-most base pair of the first read in each read pair was determined. This position should be indicative of where λ-exonuclease was stopped by a DNA-protein interaction. These positions were split according to sense or anti-sense strand, and positions mapping to GR ChIP-seq peaks (as described in ChIP-seq analysis methods) were used for further analysis.
Aggregate profile plot of DNase-seq, histone ChIP-seq in GR binding sites
We merged aligned reads across the three replicates of DNase-seq in EtOH and DEX conditions separately, from which we created a bigwig signal file. We binned signal from selected ENCODE histone modification ChIP-seq bigwig signal files and from our DNase-seq data in 10 bp bins 500 bp upstream to 500 bp downstream of the center of each GR binding site. To produce the aggregate profile plot, we normalized by total mapped read counts (in the case of DNase-seq data) and the mean depth per base-pair covered (in the case of ENCODE data) and computed the mean and standard error of the mean in each 10 bp bin for reporter positive and reporter negative GR binding sites.
Comparison of epigenetic signal reporter positive and reporter negative GR binding sites
We merged aligned reads across the two replicates of selected ENCODE histone modification ChIP-seq datasets in EtOH and DEX conditions separately. We counted the number of reads from the above merged DNase-seq and ChIP-seq alignment files for which at least half of the read overlapped a 500 bp window centered on the best match to the GR DNA binding motif within each called sub-peak. We then normalized counts for number of mapped reads and tested for differences in GR binding. To do so, we used regression models in which the log of GR ChIP-seq summed read counts along with binary reporter activity predicted log{DEX read counts}, log{EtOH read counts}, or log{DEX read counts /EtOH read counts}. When testing specifically in the core or flanking regions, we defined the core as the −250 bp to +250 bp region centered on the best match to the GR DNA binding motif, and the flanks as the union of the −500 to −250 bp region and the +250 bp to +500 bp region.
Clustering analysis of low DEX dose GR binding sites
We merged all ENCODE ChIP-seq binding peaks for GR from 0.5 nM, 5 nM, and 50 nM DEX treatments and retained only those union peaks active at 50 nM. We then considered the clustering behavior of sets of peaks among the union that overlapped with lower DEX dose peaks (0.5 nM, 5 nM). We tested whether low dose peaks were closer to one another than expected by chance by computing the median distance for all peaks to the nearest neighboring peak. As a background model, we randomly shuffled low dose peaks among all possible peaks in the cross-DEX-dose union 1000 times and computed the median distance from each active peak to its nearest neighboring active peak for each permutation. We tested whether the set of low dose peaks had fewer stretches of contiguous unbound sites than expected by chance. Again, we randomly shuffled low dose peaks among all possible peaks in the cross-DEX-dose union 1000 times and computed the number of stretches of contiguous unbound sites. We also tested whether pairs of adjacent low dose peaks were less likely to have an intervening ENCODE DEX CTCF peak than by chance. Using the same permutation strategy, we counted the number of times a CTCF peak intervened between adjacent permuted low dose peaks. A pair of peaks was defined as adjacent at a lower dose (e.g. 5 nM) if there were no intervening peaks between the two peaks among the cross-DEX-dose union of peaks and both peaks were bound by GR at the lower dose. For all three analysis questions above, we fit Gaussian distributions to the distribution of the metrics for the permuted data and tested whether the observed data fit the permuted distribution with a Z-test.
Distance analysis of tethering factors and nuclear receptors
We tested whether certain tethering factors (JUND, JUN; FOXA1) were closer to direct motif-encoded nuclear receptor sites (GR; ER) when co-bound by nuclear receptor. We searched for the nuclear receptor motif (JASPAR,(Mathelier et al. 2014); GR: MA0113.2; ER: MA0112.2) among all nuclear receptor genomic binding sites using MAST (Bailey et al. 2009) motif search tool and designated those sites with p-value < 1 × 10−4 as direct motif-encoded. We then compared the set of tethering factor sites that intersect with nuclear receptor sites (e.g. JUND+,GR+) and the set of tethering factor sites that do not intersect with nuclear receptor sites (e.g. JUND+,GR−) in distance to nearest direct motif-encoded nuclear receptor site that does not intersect with a tethering factor (e.g. JUND−,GR+ with GR motif).
Motif analysis of JUND sites gained, maintained, or lost upon DEX exposure
We searched the central 200 bp of JUND DEX binding sites and GR binding sites for the JUND motif (MA0491.1) and the GR motif (MA0113.2) from JASPAR (Mathelier et al. 2014) using MAST motif search tool (Bailey et al. 2009). We also shuffled the dinucleotides of GR binding sites using ushuffle (Jiang et al. 2008) and searched this shuffled set of sequences for the GR motif. For unshuffled sequences, two-component Gaussian mixture models were fit to the −log10{motif p value}.
Analysis of overlap in ER and FOXA1 binding sites with established ER interactions
We took the union of all ER ChIA-PET interactions in the MCF7 cell line with q-value < 0.05 from “IHH015F” and “IHM001F” from Fullwood et al. (Fullwood et al. 2009). Using FOXA1 and ER binding peaks obtained from ChIP-seq in the MCF7 cell line, we distinguished those FOXA1 peaks that intersected ER peaks from those FOXA1 peaks that did not intersect ER peaks (Hurtado et al. 2011). For both FOXA1+,ER+ and FOXA1+,ER− peaks, we computed the best scoring p-value to the ER motif (MA0112.2; JASPAR) (Mathelier et al. 2014) using MAST motif search tool (Bailey et al. 2009). We further subsetted peaks by the strength of motif and for each subset we computed the percentage of peaks that overlapped with sites that participate in interactions as identified through ER ChIA-PET.
DATA AND SOFTWARE AVAILABILITY
Software: Software from this study has been previously published as detailed under “QUANTIFICATION AND STATISTICAL ANALYSIS”.
Data Resources: Raw and aligned sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE79432 and GSE77869.
| KEY RESOURCES TABLE | ||
|---|---|---|
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
| Antibodies | ||
| Anti-GR antibody, rabbit polyclonal | Santa Cruz Biotechnology | sc-1003 |
| Sheep anti-rabbit antibody beads | Life Technologies | M-280 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Dexamethasone | Sigma-Aldrich | D4902 |
| Critical Commercial Assays | ||
| PrepX mRNA seq kit | Wafergen | 400039 |
| PrepX ILM 32i DNA Library | Wafergen | 400044 |
| Dual-Glo Luciferase Assay | Promega | E2940 |
| Fugene HD | Promega | E2312 |
| Oligo-dT Dynabeads | ThermoFisher Sci. | 61005 |
| Superscript III reverse transcriptase | ThermoFisher Sci. | 18080044 |
| Q5 DNA Polymerase | New England Biolabs | M0491L |
| Turbo DNase | ThermoFisher Sci. | AM2238 |
| In-Fusion HD Cloning Plus | Clonetech | 638911 |
| Deposited Data | ||
| Raw and analyzed data | This study | GEO: GSE79432 and GSE77869 |
| Experimental Models: Cell Lines | ||
| A549 cells | ATCC/Duke Cancer Center Cell Culture Facility | CCL-185 |
| Experimental Models: Organisms/Strains | ||
| E.coli MegaX DH10B Electrocomp cells | ThermoFisher Sci. | C640003 |
| Recombinant DNA | ||
| pGL4.24 Lucferase Vector | Promega | E8421 |
| pRL-TK | Promega | E2241 |
| pRL-SV40 | Promega | E2231 |
| pSTARR-seq_human | Addgene | Plasmid #71509 |
| Sequence-Based Reagents | ||
| PCR primers: Luciferase assay validation sequences, See Table S3 | This study (IDT) | N/A |
| Gene block sequences: Luciferase assay AP-1-GRE spacing expts. See Table S3 | This study (IDT) | N/A |
| Gene block sequences: Luciferase assay AP-1-GRE combination expts. See Table S3 | This study (IDT) | N/A |
| PCR primer sequences: Sequencing library prep. See Table S3 | This study (IDT) | N/A |
| STARR-seq adapter primers | Arnold et al., 2013 | N/A |
| Nextera STARR-seq adapter primers, See Table S3 | This study (IDT) | N/A |
| Software and Algorithms | ||
| MACS | Zhang et al. 2008 | http://liulab.dfci.harvard.edu/MACS/ |
| PeakSplitter | Salmon-Divon et al. 2010 | http://www.bioinformatics.org/peakanalyzer/ |
| BEDTools | Quinlan 2014 | http://code.google.com/p/bedtools/ |
| Bowtie/Bowtie2 | Langmead et al. 2009; Langmead and Salzberg 2012 | http://bowtie-bio.sourceforge.net/ |
| SAMtools | Li et al. 2009 | http://samtools.sourceforge.net |
| GraphPad Prism | GraphPad Inc. | http://www.graphpad.com/scientific-software/prism/ |
| DEseq2 | Love et al. 2014 | http://www.bioconductor.org/packages/release/bioc/html/DESeq2 |
| R | R Development Core Team, 2016 | https://www.r-project.org/ |
| MEME Suite | Bailey and Gribskov 1998 | http://meme-suite.org/ |
| pROC | Robin et al. 2011 | http://web.expasy.org/pROC/ |
| uShuffle | Jiang et al. 2008 | http://digital.cs.usu.edu/~mjiang/ushuffle/ |
Supplementary Material
Acknowledgements
This work was funded by the following grants: NIH U01 HG007900 (all authors), NIH F31 HL129743 (to C.M.V.), NIH T32s GM007754 (A.M.D.) and GM071340 (I.C.M and W.H.M.). C.M.V is an NHLBI LRRC Young Investigator and was also supported by NIH U01 HL111018 through Brigid Hogan, who we thank for her helpful comments and support. We thank Alexander Stark for plasmid materials.
Footnotes
Author Contributions
C.M.V. and T.E.R. designed the study. C.M.V., A.M.D. and A.S. performed the experiments. C.M.V., A.M.D., I.C.M, W.H.M., L.S., G.E.C. and T.E.R. designed and performed the analyses. T.E.R. supervised and funded the research. C.M.V., A.M.D., I.C.M., and T.E.R. wrote the manuscript. All authors reviewed the manuscript prior to submission.
Author Information
The authors have no competing financial interests with this work.
References
- Adey A, Morrison HG, Asan, Xun X, Kitzman JO, Turner EH, Stackhouse B, MacKenzie AP, Caruccio NC, Zhang X, et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11(12):R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold CD, Gerlach D, Stelzer C, Boryn LM, Rath M, Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339(6123):1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14(1):48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- Belikov S, Astrand C, Wrange O. FoxA1 binding directs chromatin structure and the functional response of a glucocorticoid receptor-regulated promoter. Mol Cell Biol. 2009;29(20):5413–5425. doi: 10.1128/MCB.00368-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57(1):289–300. [Google Scholar]
- Biddie SC, John S, Sabo PJ, Thurman RE, Johnson TA, Schiltz RL, Miranda TB, Sung MH, Trump S, Lightman SL, et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol Cell. 2011;43(1):145–155. doi: 10.1016/j.molcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botta M, Haider S, Leung IX, Lio P, Mozziconacci J. Intra- and inter-chromosomal interactions correlate with CTCF binding genome wide. Mol Syst Biol. 2010;6:426. doi: 10.1038/msb.2010.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler VL, Maler BA, Yamamoto KR. DNA sequences bound specifically by glucocorticoid receptor in vitro render a heterologous promoter hormone responsive in vivo. Cell. 1983;33(2):489–499. doi: 10.1016/0092-8674(83)90430-0. [DOI] [PubMed] [Google Scholar]
- Coleman RA, Pugh BF. Evidence for functional binding and stable sliding of the TATA binding protein on nonspecific DNA. J Biol Chem. 1995;270(23):13850–13859. doi: 10.1074/jbc.270.23.13850. [DOI] [PubMed] [Google Scholar]
- Devonshire AS, Elaswarapu R, Foy CA. Evaluation of external RNA controls for the standardisation of gene expression biomarker measurements. BMC genomics. 2010;11:662. doi: 10.1186/1471-2164-11-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MI, Miner JN, Yoshinaga SK, Yamamoto KR. Transcription factor interactions: selectors of positive or negative regulation from a single DNA element. Science. 1990;249(4974):1266–1272. doi: 10.1126/science.2119054. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nature protocols. 2012;7(9):1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462(7269):58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Vockley CM, Pauli F, Newberry KM, Xue Y, Randell SH, Reddy TE, Hogan BL. Evidence for multiple roles for grainyhead-like 2 in the establishment and maintenance of human mucociliary airway epithelium.[corrected]. Proc Natl Acad Sci U S A. 2013;110(23):9356–9361. doi: 10.1073/pnas.1307589110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz J, Savic D, Varley KE, Partridge EC, Safi A, Jain P, Cooper GM, Reddy TE, Crawford GE, Myers RM. Distinct properties of cell-type-specific and shared transcription factor binding sites. Mol Cell. 2013;52(1):25–36. doi: 10.1016/j.molcel.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz J, Varley KE, Davis NS, Baas BJ, Goryshin IY, Vaidyanathan R, Kuersten S, Myers RM. Transposase mediated construction of RNA-seq libraries. Genome Res. 2012;22(1):134–141. doi: 10.1101/gr.127373.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotea V, Visel A, Westlund JM, Nobrega MA, Pennacchio LA, Ovcharenko I. Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome Res. 2010;20(5):565–577. doi: 10.1101/gr.104471.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl 1):S4, 1–9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrlich P. Cross-talk between glucocorticoid receptor and AP-1. Oncogene. 2001;20(19):2465–2475. doi: 10.1038/sj.onc.1204388. [DOI] [PubMed] [Google Scholar]
- Hertel KJ, Lynch KW, Maniatis T. Common themes in the function of transcription and splicing enhancers. Curr Opin Cell Biol. 1997;9(3):350–357. doi: 10.1016/s0955-0674(97)80007-5. [DOI] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43(1):27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Anderson J, Gillespie J, Mayne M. uShuffle: a useful tool for shuffling biological sequences while preserving the k-let counts. BMC bioinformatics. 2008;9:192. doi: 10.1186/1471-2105-9-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43(3):264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph R, Orlov YL, Huss M, Sun W, Kong SL, Ukil L, Pan YF, Li G, Lim M, Thomsen JS, et al. Integrative model of genomic factors for determining binding site selection by estrogen receptor-alpha. Mol Syst Biol. 2010;6:456. doi: 10.1038/msb.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheradpour P, Ernst J, Melnikov A, Rogov P, Wang L, Zhang X, Alston J, Mikkelsen TS, Kellis M. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Res. 2013;23(5):800–811. doi: 10.1101/gr.144899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova T, Wang SY, Rao NA, Mandoli A, Martens JH, Rother N, Aartse A, Groh L, Janssen-Megens EM, Li G, et al. Glucocorticoid receptor and nuclear factor kappa-b affect three-dimensional chromatin organization. Genome Biol. 2015;16:264. doi: 10.1186/s13059-015-0832-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Schleif RF. In vivo DNA loops in araCBAD: size limits and helical repeat. Proc Natl Acad Sci U S A. 1989;86(2):476–480. doi: 10.1073/pnas.86.2.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang L, Zhao K, Thompson PR, Hwang Y, Marmorstein R, Cole PA. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature. 2008;451(7180):846–850. doi: 10.1038/nature06546. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. 2014a. [DOI] [PMC free article] [PubMed]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014b;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecke HF, Yamamoto KR. The glucocorticoid receptor blocks P-TEFb recruitment by NFkappaB to effect promoter-specific transcriptional repression. Genes Dev. 2005;19(9):1116–1127. doi: 10.1101/gad.1297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, Buchman S, Chen CY, Chou A, Ienasescu H, et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42(Database issue):D142–147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov A, Murugan A, Zhang X, Tesileanu T, Wang L, Rogov P, Feizi S, Gnirke A, Callan CG, Jr., Kinney JB, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol. 2012;30(3):271–277. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal R, Kumar KU, Pater A, Pater MM. Differential regulation by c-jun and c-fos protooncogenes of hormone response from composite glucocorticoid response element in human papilloma virus type 16 regulatory region. Molecular endocrinology. 1994;8(12):1701–1708. doi: 10.1210/mend.8.12.7708058. [DOI] [PubMed] [Google Scholar]
- Olansky L, Welling C, Giddings S, Adler S, Bourey R, Dowse G, Serjeantson S, Zimmet P, Permutt MA. A variant insulin promoter in non-insulin-dependent diabetes mellitus. J Clin Invest. 1992;89(5):1596–1602. doi: 10.1172/JCI115754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce D, Matsui W, Miner JN, Yamamoto KR. Glucocorticoid receptor transcriptional activity determined by spacing of receptor and nonreceptor DNA sites. J Biol Chem. 1998;273(46):30081–30085. doi: 10.1074/jbc.273.46.30081. [DOI] [PubMed] [Google Scholar]
- Quinlan AR. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Current protocols in bioinformatics / editoral board, Andreas D Baxevanis [et al] 2014;47:11 12 11–11 12 34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2016. [Google Scholar]
- Ratman D, Vanden Berghe W, Dejager L, Libert C, Tavernier J, Beck IM, De Bosscher K. How glucocorticoid receptors modulate the activity of other transcription factors: a scope beyond tethering. Mol Cell Endocrinol. 2013;380(1-2):41–54. doi: 10.1016/j.mce.2012.12.014. [DOI] [PubMed] [Google Scholar]
- Reddy TE, Gertz J, Crawford GE, Garabedian MJ, Myers RM. The hypersensitive glucocorticoid response specifically regulates period 1 and expression of circadian genes. Mol Cell Biol. 2012a;32(18):3756–3767. doi: 10.1128/MCB.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy TE, Gertz J, Pauli F, Kucera KS, Varley KE, Newberry KM, Marinov GK, Mortazavi A, Williams BA, Song L, et al. Effects of sequence variation on differential allelic transcription factor occupancy and gene expression. Genome Res. 2012b;22(5):860–869. doi: 10.1101/gr.131201.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy TE, Pauli F, Sprouse RO, Neff NF, Newberry KM, Garabedian MJ, Myers RM. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19(12):2163–2171. doi: 10.1101/gr.097022.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Muller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rye M, Saetrom P, Handstad T, Drablos F. Clustered ChIP-Seq-defined transcription factor binding sites and histone modifications map distinct classes of regulatory elements. BMC Biol. 2011;9:80. doi: 10.1186/1741-7007-9-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai DD, Helms S, Carlstedt-Duke J, Gustafsson JA, Rottman FM, Yamamoto KR. Hormone-mediated repression: a negative glucocorticoid response element from the bovine prolactin gene. Genes Dev. 1988;2(9):1144–1154. doi: 10.1101/gad.2.9.1144. [DOI] [PubMed] [Google Scholar]
- Salmon-Divon M, Dvinge H, Tammoja K, Bertone P. PeakAnalyzer: genome-wide annotation of chromatin binding and modification loci. BMC bioinformatics. 2010;11:415. doi: 10.1186/1471-2105-11-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard KA, Phelps KM, Williams AJ, Thanos D, Glass CK, Rosenfeld MG, Gerritsen ME, Collins T. Nuclear integration of glucocorticoid receptor and nuclear factor-kappaB signaling by CREB-binding protein and steroid receptor coactivator-1. J Biol Chem. 1998;273(45):29291–29294. doi: 10.1074/jbc.273.45.29291. [DOI] [PubMed] [Google Scholar]
- Slater EP, Rabenau O, Karin M, Baxter JD, Beato M. Glucocorticoid receptor binding and activation of a heterologous promoter by dexamethasone by the first intron of the human growth hormone gene. Mol Cell Biol. 1985;5(11):2984–2992. doi: 10.1128/mcb.5.11.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RP, Taher L, Patwardhan RP, Kim MJ, Inoue F, Shendure J, Ovcharenko I, Ahituv N. Massively parallel decoding of mammalian regulatory sequences supports a flexible organizational model. Nat Genet. 2013;45(9):1021–1028. doi: 10.1038/ng.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So AY, Chaivorapol C, Bolton EC, Li H, Yamamoto KR. Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet. 2007;3(6):e94. doi: 10.1371/journal.pgen.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soccio RE, Chen ER, Rajapurkar SR, Safabakhsh P, Marinis JM, Dispirito JR, Emmett MJ, Briggs ER, Fang B, Everett LJ, et al. Genetic Variation Determines PPARgamma Function and Anti-diabetic Drug Response In Vivo. Cell. 2015;162(1):33–44. doi: 10.1016/j.cell.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somma MP, Pisano C, Lavia P. The housekeeping promoter from the mouse CpG island HTF9 contains multiple protein-binding elements that are functionally redundant. Nucleic Acids Res. 1991;19(11):2817–2824. doi: 10.1093/nar/19.11.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Crawford GE. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harbor protocols. 2010;2010(2) doi: 10.1101/pdb.prot5384. pdb prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadhouders R, Aktuna S, Thongjuea S, Aghajanirefah A, Pourfarzad F, van Ijcken W, Lenhard B, Rooks H, Best S, Menzel S, et al. HBS1L-MYB intergenic variants modulate fetal hemoglobin via long-range MYB enhancers. J Clin Invest. 2014;124(4):1699–1710. doi: 10.1172/JCI71520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staller MV, Vincent BJ, Bragdon MD, Lydiard-Martin T, Wunderlich Z, Estrada J, DePace AH. Shadow enhancers enable Hunchback bifunctionality in the Drosophila embryo. Proc Natl Acad Sci U S A. 2015;112(3):785–790. doi: 10.1073/pnas.1413877112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starick SR, Ibn-Salem J, Jurk M, Hernandez C, Love MI, Chung HR, Vingron M, Thomas-Chollier M, Meijsing SH. ChIP-exo signal associated with DNA-binding motifs provides insight into the genomic binding of the glucocorticoid receptor and cooperating transcription factors. Genome Res. 2015;25(6):825–835. doi: 10.1101/gr.185157.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teurich S, Angel P. The glucocorticoid receptor synergizes with Jun homodimers to activate AP-1-regulated promoters lacking GR binding sites. Chemical senses. 1995;20(2):251–255. doi: 10.1093/chemse/20.2.251. [DOI] [PubMed] [Google Scholar]
- Vockley CM, Guo C, Majoros WH, Nodzenski M, Scholtens DM, Hayes MG, Lowe WL, Jr., Reddy TE. Massively parallel quantification of the regulatory effects of noncoding genetic variation in a human cohort. Genome Res. 2015;25(8):1206–1214. doi: 10.1101/gr.190090.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Derynck MK, Nonaka DF, Khodabakhsh DB, Haqq C, Yamamoto KR. Chromatin immunoprecipitation (ChIP) scanning identifies primary glucocorticoid receptor target genes. Proc Natl Acad Sci U S A. 2004;101(44):15603–15608. doi: 10.1073/pnas.0407008101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang JJ, Dai W, Lei KY, Pike JW. Dexamethasone potently enhances phorbol ester-induced IL-1beta gene expression and nuclear factor NF-kappaB activation. J Immunol. 1997;159(2):534–537. [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.