Abstract
For more than a century, bacteriophage (or phage) research has enabled some of the most important discoveries in biological sciences and has equipped scientists with many of the molecular biology tools that have advanced our understanding of replication, maintenance, and expression of genetic material. Phages have also been recognized and exploited as natural antimicrobial agents and nanovectors for gene therapy, but their potential as therapeutics has not been fully exploited in Western medicine because of challenges such as narrow host range, bacterial resistance, and unique pharmacokinetics. However, increasing concern related to the emergence of bacteria resistant to multiple antibiotics has heightened interest in phage therapy and the development of strategies to overcome hurdles associated with bacteriophage therapeutics. Recent progress in sequencing technologies, DNA manipulation, and synthetic biology allowed scientists to refactor the entire bacterial genome of Mycoplasma mycoides, thereby creating the first synthetic cell. These new strategies for engineering genomes may have the potential to accelerate the construction of designer phage genomes with superior therapeutic potential. Here, we discuss the use of phage as therapeutics, as well as how synthetic biology can create bacteriophage with desirable attributes.
The threat posed by antibiotic-resistant bacteria has triggered interest in the development of phage therapies. But several challenges (e.g., narrow host range and unique pharmacokinetics of phage therapies) must be addressed.
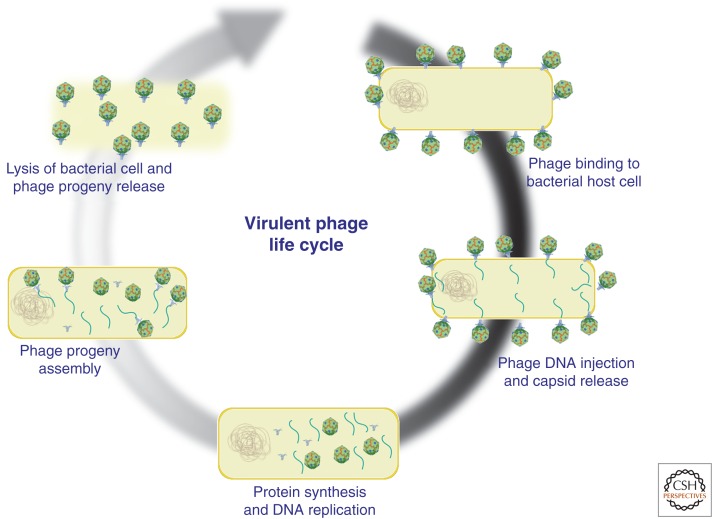
Lysogenic (temperate) and lytic bacteriophages are viruses that specifically infect bacteria, and have long been considered as natural antimicrobial agents for treatment of bacterial infections (d’Herelle 1931; Twort 1936). On binding to a bacterial surface moiety, phages inject their genetic material into the host cell. In the case of temperate phage, the lysogenic cycle is initiated by viral genome integration into the bacterial chromosome (prophage) where it remains until specific lytic genes are activated and phage virions are produced, eventually lysing the host cell. Before the activation of the lytic cycle, the phage genome remains repressed and can be transferred to daughter cells during bacterial replication. Temperate phages are largely not considered for therapeutic use, as many have been shown to encode bacterial virulence factors or horizontally transfer virulence genes in a process known as transduction (Boyd and Brussow 2002; Ptashne 2004; Tinsley et al. 2006). In contrast, virulent phages immediately begin replication and quickly subvert host-cell metabolism, which results in the destruction of the host cell within minutes to hours (Fig. 1) (Sulakvelidze et al. 2001; Hanlon 2007). Virulent phages do not enter a prophage state and have not shown the ability to transfer virulence factors between bacterial cells, making them suitable for phage therapy (Hendrix 2003; Hatfull 2008).
Figure 1.
Lytic cycle of bacteriophage. Schematic representation of virulent phage infection, replication, and lysis of bacterial host cells.
The clinical potential of virulent bacteriophages as antibacterial agents was first recognized by Felix d’Herelle, one of the pioneers of the phage therapy field (d’Herelle et al. 1931). Although early successes of personalized clinical applications with tailored formulations were reported, the use of phage therapy against bacterial pathogens in large patient populations was never achieved in part because of its displacement by antibiotics (Merril et al. 2003, 2006). The growing threat posed by highly antibiotic-resistant bacteria combined with the decreasing numbers of newly licensed drugs has recently triggered interest in the development of phage antimicrobials. However, several challenges associated with phage therapy limit their therapeutic potential.
OVERCOMING PHAGE THERAPY CHALLENGES IN THE ERA OF SYNTHETIC BIOLOGY
Host Specificity
Viral attachment and genome injection are regimented and stepwise processes that are tightly controlled by specific receptor–ligand interactions (Furukawa et al. 1983). Phage–host selectivity permits phage therapy to be directed at specific bacterial populations, preventing secondary infections by leaving nontarget bacteria unaffected (Sulakvelidze et al. 2001). Host discernment prevents phage therapy from enriching for resistance, a major problem associated with antibiotic use. Historically, the diversity and malleability of bacterial surface ligands, even at the species level, makes it difficult to identify phage capable of treating a clinical infection without prescreening against phage libraries (Keen 2012). The specificity of viral–host interactions has made phage therapy attractive, but has also required either the targeting of pathogens that are largely phylogenetically constrained (clonal) or the use of large phage cocktails capable of covering the diversity of a bacterial population (O’Flaherty et al. 2005; McVay et al. 2007).
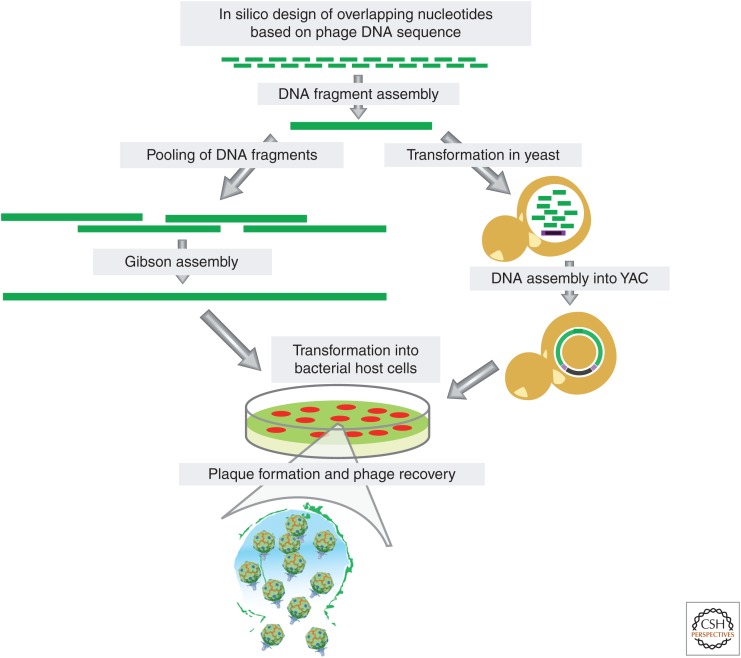
The plummeting cost of DNA sequencing and new molecular biology techniques may allow scientists to leverage phenotypic screening, comparative genomics, and viral engineering to overcome the challenges associated with host specificity. As bacterial phylogeny correlates with phage host range, the ability to rapidly sequence clinical isolates will increase both the accuracy and the speed at which lytic phage can be paired with potentially susceptible bacterial infections (DeLappe et al. 2003). Last, newly developed viral engineering and DNA manipulation methods could be used to expand the host range of bacteriophages through rational design, potentially decreasing the number of phages needed to cover bacterial diversity. For example, ϕX174 genome (5.4 kb) was assembled from a pool of synthetic nucleotides and successfully launched in bacteria (Smith et al. 2003). Furthermore, the entire bacterial genome of Mycoplasma mycoides was chemically synthesized from DNA fragments (Gibson et al. 2009). Other technologies, such as bacteriophage recombineering of electroporated DNA (BRED) and genome editing via targetrons and recombinases (GETR), have been developed to edit phage genomes (Marinelli et al. 2008; Enyeart et al. 2013). Assembly of genomes in yeast artificial chromosome and plasmids and clustered regularly interspaced short palindromic repeats (CRISPR)/Cas-editing systems in bacteria have also been recently used to manipulate phage DNA (Lu and Koeris 2011; Jaschke et al. 2012; Kiro et al. 2014; Martel and Moineau 2014; Nobrega et al. 2015). Together, these methods can be integrated to construct synthetic phages with desired characteristics and precise genomic content (Fig. 2).
Figure 2.
Synthetic assembly of bacteriophage genomes. Step-by-step construction of phage genomes starting from overlapping nucleotides and recovery of functional phage particles from host cells following transformation. (Left) Chemical assembly of DNA fragments synthesized from overlapping nucleotides using Gibson assembly. (Right) Assembly of DNA fragments synthesized from overlapping nucleotides in yeast artificial chromosome (YAC, in purple).
Development of Resistance
The ability of bacteria to rapidly acquire resistance to a single selective pressure hinders the success of both standard small molecule and nonstandard antimicrobials, such as phages (Neu 1992; Labrie et al. 2010). Phage resistance most commonly arises through the down-regulation, mutation, or shielding of the viral receptor (Labrie et al. 2010). This simple fact has encouraged the use of phages that bind to receptors that are highly conserved or required for in vivo virulence (O’Flaherty et al. 2005; Ojala et al. 2013). However, bacteria can also acquire resistance to phages through a diverse array of restriction modification, toxin–antitoxin, and CRISPR/Cas systems (Labrie et al. 2010). Unlike purified small molecules, phages respond to selective pressure through evolution, which has supplied scientists with a plethora of genetically encoded counter-resistance systems capable of overcoming bacterial resistance mechanisms (Samson et al. 2013). In a clinical setting, it is important to remember that phage and antibiotic resistance mechanisms do not overlap; therefore, loss of sensitivity to phage is unlikely to affect antibiotic susceptibility (Labrie et al. 2010; Blair et al. 2015). In fact, CRISPR/Cas-mediated resistance to phage is associated with decreased antibiotic resistance in enterococci (Palmer and Gilmore 2010).
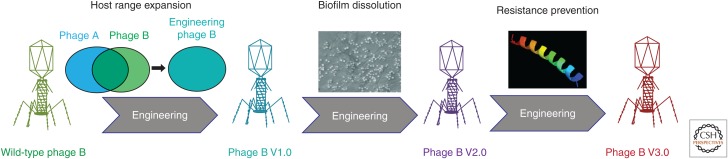
The success of chemical cocktails, phage cocktails, and multivalent vaccines has shown that targeting multiple conserved but independent ligands drastically reduces the incidence of pathogen evasion. Thus, endowing single bacteriophage with an independent bactericidal payload could prevent bacterial resistance to phages (Fig. 3).
Figure 3.
Iterative engineering of phage genomes to create phages with superior therapeutic potential. Phage B V1.0 represents a phage with enhanced host range (includes the host range specific to phage A). Phage B V2.0 delineates iterative engineering of phage B V1.0 to include moieties aiding in biofilm disruption. Similarly, phage B V3.0 captures the properties of phage B V2.0 and is engineered to express antimicrobial payloads that may aid in elimination of bacterial subpopulation that lost susceptibility to phage.
Unique Pharmacological Properties of Viral Therapeutics
Biologics are large molecules with desirable therapeutic and prophylactic uses, which are very complex and are often 200 to more than 1000 times the size of small molecule drugs (Kingham et al. 2014). Similar to biologics, the large size of phage poses distinctive pharmacological challenges when compared with chemical products. Additionally, therapeutic viruses, such as oncolytic virions or bacteriophage, are capable of replicating in vivo, further differentiating them from static biologics or chemicals (Chiocca 2002).
The large size of biologics prevents them from diffusing into cells or biofilms, crossing the blood–brain barrier, and makes administration of large doses challenging (Baumann 2006). Fortunately, bacteriophages are capable of density-dependent productive amplification within a target pathogen at the site of infection. Whereas autoamplification may require a certain concentration of phage particles to accumulate at the site of infection, it is believed that amplification can substantially lower the dose required to treat an infection (Abedon 2011). As replication absolutely requires the presence of the target bacteria, phages are self-limiting and quickly cleared following pathogen eradication (Abedon 2011). Phage replication results in the lysis of target cells and potential release of bacterial toxins, an important concern especially for endotoxin encoding Gram-negative pathogens (Hagens et al. 2004). However, no toxic effects have been reported in bacteremic patients successfully treated with phages (Kutter et al. 2014).
Using molecular biology, scientists have already created bacteriophages coated in cell-permeable peptides, allowing them to diffuse into human cells or through the blood–brain barrier (Staquicini et al. 2011; Rangel et al. 2012). Additionally, phages are known to encode and can be engineered to produce enzymes, allowing them to degrade bacterial biofilms (Lu and Collins 2007). Researchers have also created nonreplicating or lysis-deficient phages to deliver bacterial static toxins, enabling inhibition of bacterial growth without release of toxins through lysis (Hagens et al. 2004; Matsuda et al. 2005; Fairhead 2009). Whereas viral size is unlikely to be meaningfully altered, genetically programing phages with the ability to produce small peptides with useful properties could offset many of the size constraints inherent to classic biologics (Fig. 3).
Human Immune Response and Phage Clearance
The mammalian innate immune system recognizes and clears viral particles, creating a major hurdle for any viral-based therapeutic (Geier et al. 1973). Phagocytic cells located in the reticular connective tissue play a key role in rapidly clearing circulating phage particles through the innate immune mononuclear phagocyte system (MPS) (Merril et al. 1996). Whereas the MPS may form the first major barrier to phage circulation, the adaptive immune response provides the second. The adaptive response mediated by B/T cells takes 5–7 d to produce robust antibody titers against phage in naïve humans, providing an ample window for phage therapeutic activity (Ochs et al. 1971; Wedgwood et al. 1975). However, it is hypothesized that the adaptive memory response may prevent the reuse of phage therapy vectors within the same patient, as protective antibodies will already have been raised (Górski et al. 2012).
Long-circulating phage harboring single amino acid mutations in the major capsid protein have already been selected in vivo and shown to provide an increased evasion of MPS by more than 10,000-fold, drastically improving phage therapy efficacy (Merril et al. 1996; Vitiello et al. 2005). Thus, engineering of mutations that enhance pharmacokinetics properties of phage capsids may improve the therapeutic potential of multiple phage vectors for antimicrobial and phage-based therapeutic nanovector applications.
Regulatory and Production Challenges
Bacteriophages are large and complex therapeutics more similar to other genetically engineered biologics than small molecule antibiotics. Like other biologics, phage are difficult to fully characterize, and are sensitive to manufacturing and handling conditions, making their production much more complex than chemical drugs (Kingham et al. 2014). However, a handful of phage products have been approved by American regulatory agencies, including the U.S. Food and Drug Administration (FDA), granting multiple bacteriophages generally recognized as safe (GRAS) status when used as antimicrobial agents in human food (Soni and Nannapaneni 2010; Carter et al. 2012; Woolston et al. 2013). Further, phage cocktails for human therapeutic use have been produced for decades within the former Soviet Republics of Eastern Europe, indicating some level of safety and efficacy (Sulakvelidze et al. 2001). Scalable oncolytic virus current good manufacturing practice (cGMP)-compliant processes provide significant insight into the methods and regulatory oversight required for viral therapeutic production. Interestingly, the substantially higher cost of producing biologics than chemical therapeutics has not prevented their rapid ascension into the ranks of the best selling drugs in the United States. Unfortunately, phage therapeutic production costs relative to multidrug-resistant (MDR) infection rates may still provide the largest single barrier to phage therapy success. Many phage therapy products currently being tested in clinical trials or used in Eastern Europe constitute a cocktail of multiple independent types of phages (Wright et al. 2009; Abedon et al. 2011). Current Western medical standards would require each independent phage to be produced and purified under cGMP-compliant processes, multiplying production costs to prohibitive levels (Kingham et al. 2014).
The ability to generate whole bacteriophage genomes directly from synthetic oligonucleotides will be attractive to regulators, as it decreases contamination with adventitious agents. Additionally, launching and amplifying bacteriophage in cell-free reactions could significantly lower production costs and contamination (Kerr and Sadowski 1974; Gunther et al. 1993; Shin et al. 2012). These synthetic biology advances coupled with our growing ability to engineer bacteriophage with expanded host range, antiresistance mechanisms, and immune evasion capabilities may lower the number of phages and cost of production to a level attractive to regulators and commercial entities (Merril et al. 1996; Tetart et al. 1996, 1998; Samson et al. 2013). Last, the decreasing cost of sequencing will greatly aid the speed and accuracy of matching a particular therapeutic phage with a susceptible strain, further limiting the known challenge of phage host range.
PHAGES AS ANTI-INFECTIVES
Numerous strategies have been used to show that virulent phages are realistic alternatives to standard antibiotics. However, insight into the potential clinical and therapeutic relevance of preclinical efficacy models of phage treatment is often obscured by a lack of standardized techniques and protocols and, as a consequence, vast differences in therapeutic outcomes. Whereas some studies report superior therapeutic value (McVay et al. 2007; Oliveira et al. 2010; Gu et al. 2012; Hall et al. 2012; Jaiswal et al. 2013), others note marginal reduction in bacterial loads (Rozema et al. 2009). Nonetheless, results stemming from recent attempts to show efficacy of phage therapy exemplify how delivery routes, single or multiple treatments, monophage versus polyphage therapy, and dosage/timing of administration contribute to therapeutic success in preclinical models of infection.
Lessons Learned from Preclinical Phage Therapy
Oral Administration of Phages
Per os administration of phage has been found suitable for treatment of gastrointestinal infections. For example, a cocktail of three bacteriophages administered orally in a single dose (108 PFU/mL) resulted in significant reduction of Escherichia coli O157:H7 in the feces of mice (99%). Further examination revealed that the most successful administration, as determined by reduction in E. coli load, was daily oral ingestion of high amounts of phage (1010 PFU/mL) (Tanji et al. 2005). Similar studies found that a threshold of an orally administered phage cocktail was required to reduce the colonization of mice infected with pediatric diarrhea E. coli clinical isolates (Chibani-Chennoufi et al. 2004). The effect of phage treatment on fecal counts was negligible, which led the investigators to hypothesize that either phages lose infectivity because of exposure to acid during gastric passage or that once in the gut E. coli becomes resistant to phage infection. Although subsequent investigations showed that intestinal transit has no impact on phage infectivity, protection from gastric acidity by microencapsulation or coadministration with antacid may enhance their efficacy (Stanford et al. 2010). Thus, acid sensitivity must be characterized for individual phage before oral administration.
The preventative and therapeutic efficacy of oral administration of phage was also examined in a chicken model of Campylobacter jejuni decolonization (Wagenaar et al. 2005). To show therapeutic effect, poultry received an oral dose of 105 CFU C. jejuni, followed by inoculation with a single phage (1010 PFU) for six successive days starting 5 d after the bacterial challenge. Initially, a 30-fold decrease in C. jejuni viable counts was observed in the therapeutic group. However, after 5 d, phage and bacteria reached equilibrium most likely because of the presence of a resistant subpopulation of microbes. Hence, using a cocktail or an engineered phage with a wide host range rather than a single phage may overcome bacterial resistance.
The timing of treatment also affects the success of phage therapy. Prophylaxis with phage KPP1 in a model of Pseudomonas aeruginosa–induced gut-derived septicemia had little to no effect on mice survival, whereas concomitant administration of phage and bacteria induced significant protection against infection.
Parenteral Administration of Phages
Intramuscular injection (i.m.) of different concentrations of phage R in chickens and calves suffering from E. coli–derived septicemia showed that significant protection is conferred by a high concentration of phages (Barrow et al. 1998). Similarly, intraperitoneal (i.p.) injection of ENB6 phage in a vancomycin-resistant Enterococcus faecium mouse bacteremia model within an hour of bacterial challenge rescued all experimental animals (Biswas et al. 2002). Several other reports highlighted the rapid distribution and bioavailability of phage on i.m. or i.p. injection. For instance, phage ΦMR11 appeared in the blood shortly after i.p. injection into a Staphylococcus aureus mouse bacteremia model, and high levels of circulating phage could be detected until bacteria were eliminated (Matsuzaki et al. 2003). Most importantly, McVay et al. (2007) showed that, in the case of a P. aeruginosa–specific phage cocktail, i.p. injection of phage has enhanced their therapeutic potential. In the absence of phage administration, almost all wounded mice (94%) succumbed to the infection within 3 d. In contrast, the rate of mortality was reduced to 12% when the phage cocktail was delivered by i.p. injection. A low percentage of animals (28% or 22%, respectively) were rescued by i.m. or subcutaneous phage delivery. In summary, these results suggest that the route of parenteral administration must be carefully chosen based on empirical phage pharmacokinetics data and tailored to the type of infection requiring treatment. At the same time, the dosage levels appear to directly correlate with the success of phage therapy.
Local Application of Phage
Successful topical administration of phage has been widely reported. Although early studies focused almost solely on wound healing, more recent investigations also include otic and ocular applications (Kumari et al. 2009; Hawkins et al. 2010; Fukuda et al. 2012). For certain microbes, skin and mucosal colonization represents a prerequisite for infection (Casadevall and Pirofski 2000). Therefore, effective decontamination with an antimicrobial that has no effect on normal microbiota significantly decreases the risk of disease. Early attempts to use phage for decolonization of chicken skin indicated that Salmonella enterica, C. jejuni, and Campylobacter spp. could be effectively eliminated when the multiplicity of infection (MOI) of viral particles to bacteria was above 100. However, the minimum levels of phage particles necessary to significantly reduce colonization were specific for each organism within a species (Goode et al. 2003).
Recent reports highlighted the successful treatment of Klebsiella pneumoniae–induced burn wound infection in mice with the Klebsiella-specific phage Kpn5 incorporated in a 3% hydroxypropylmethylcellulose hydrogel (Kumari et al. 2010). On bacterial challenge, mice were treated once with low and high doses of phage hydrogel (MOI of 1 or 100, respectively). Significant levels of protection against invasive K. pneumonia infection were achieved only by high titers of phage. These results indicate that the dosage of phage is important for effective phage therapy. However, whether application of high titers of phage triggers bacterial death because of a productive infection or lysis from without remains to be determined. It is possible that the need for excessive MOIs may be overcome by multiple applications of phage following an empirically determined treatment schedule.
Otic administration of a phage cocktail in dogs suffering from P. aeruginosa otitis has shown great promise. Two days after instillation of a single dose of phage into the auditory canal of one ear, bacterial counts decreased significantly. Concomitantly, phage titers increased and no adverse effects were registered. Although the cocktail formulation was not disclosed, otic phage therapy may be a suitable alternative to antibiotics. Moreover, amplification of phage at the site of infection indicates that the infectious agent was susceptible to at least one phage administered and the dose applied was amenable to a productive viral infection (Hawkins et al. 2010).
Inhalation of Phage
In vivo efficacy of phage therapy against Burkholderia cepacia (isolated from the sputum of patients with cystic fibrosis) respiratory tract infections was evaluated in a mouse model of acute lung infection (Carmody et al. 2010). Twenty-four hours postintratracheal bacterial challenge, mice were treated by i.p. injection or intranasal inhalation with a single phage at an MOI of 100. No significant differences in lung bacterial load or inflammatory cytokine levels were found between untreated mice and rodents treated intranasally with phage. In contrast to inhalation, systemic administration of phages was moderately effective, suggesting that the systemic application of phages may facilitate accessibility to bacteria present in the lungs.
Debarbieux et al. (2010) reported multiple studies of successful intranasal phage treatment of P. aeruginosa pulmonary infection in a mouse model. In a first study, phage were administered either 24 h before or 2 h after bacterial challenge with a bioluminescent strain of P. aeruginosa. No significant difference in survival of untreated animals and mice treated with the lowest dose of phage (MOI of 0.1) was observed. A substantially higher dose of phage (MOI of 10) facilitated complete recovery of infected mice. Further analysis revealed that phage propagated within lungs, underscoring the importance of dosage and timing in phage therapy. Given that treatment was delivered shortly after bacterial infection, it is difficult to predict whether inhalation of phage would be successful for treating lower pulmonary tract or chronic infections. These data indicate that inhalation of phage may confer therapeutic and prophylactic protection against pulmonary infection.
Prevention of Biofilm Formation and Medical Device–Associated Infections
Indwelling medical devices are often used to restore function or regenerate tissue. However, the propensity to become colonized by bacteria and serve as abiotic support for biofilm formation leading to medical device–associated infections undermines their use in medical practice. Over the past couple of decades, therapeutic management of implant-associated infections has been further complicated by the emergence of antibiotic-resistant bacteria (Arias and Murray 2009; Boucher et al. 2013).
Mitigation of Staphylococcus epidermidis biofilm formation on hydrogel-coated catheters has been achieved by pretreatment with bacteriophage (Curtin and Donlan 2006). In similar studies, Fu et al. (2010) investigated the effect of pre-, post-, and recurring treatment of Foley catheters with a cocktail of phage on P. aeruginosa biofilm formation. Significant reduction of bacterial density was observed regardless of the timing of treatment (Fu et al. 2010). However, regrowth of biofilm occurred between 24 and 48 h. Importantly, sequential treatments had triggered mature biofilm dissolution. Another study reported prevention of biofilm formation on Foley catheters following impregnation of neutral hydrogel (Lubri-Sil, Bard, Covington, GA) loaded with lytic bacteriophages (Carson et al. 2010). The densities of E. coli and Proteus mirabilis on the silicone tubing were reduced by 90% after 24 h of exposure to the gel. These results highlight the potential use of phage cocktails for mitigation of biofilm formation by clinically relevant bacteria on the surfaces of indwelling medical devices. Moreover, wide host range phage engineered to express biofilm disruptive enzymes may become the next-generation coating materials for indwelling devices.
Anti-Infective Phage Therapy in Humans
Before the late 1930s, phage therapy was used worldwide to treat infections. In the United States, phage therapy was of interest mainly between the 1920s and the 1930s. However, the advent of antibiotics and a couple of controversial opinions regarding the preparation and efficacy of phage lysates led to decreased interest in phage therapy (Eaton and Bayne-Jones 1934). Nonetheless, seven phage products for human use were produced by Eli Lilly (Indianapolis, IN). Another product, Staph Phage Lysate, was developed by Delmont Laboratories (Swarthmore, PA) and licensed for human use in 1959 and veterinary use in 1987 for treatment of canine pyoderma. Phage therapy continued to be used in France until the 1990s when phage preparations were discontinued. Ever since then, phage therapists have continued their practice with products obtained from Poland and the United States, where phage therapy is included in the current health care standard (Abedon et al. 2011).
Many studies have been conducted to evaluate the efficacy and safety of phage therapy and much clinical experience has been accumulated, especially in Eastern Europe (reviewed in Alisky et al. 1998; Summers 2001; Sulakvelidze et al. 2001; Górski et al. 2009; Kutter et al. 2010; Abedon et al. 2011; Burrowes et al. 2011; Harper et al. 2011; Chanishvili 2012). However, these trials did not follow current Western rigorous standards. The first double-blind, randomized, placebo-controlled phase I trial to show the safety of phage treatment was performed by Nestle Research Center (Lausanne, Switzerland) (Bruttin and Brussow 2005). Investigators showed no significant side effects following administration of phage and also showed that oral administration of T4 did not disturb the natural gut E. coli population. In subsequent studies, the group performed a detailed metagenomics analysis of their entire T4-related antidiarrheagenic phage collection and assessed the clinical risk of a subset of phages following oral administration in healthy adults (Sarker et al. 2012).
The first reported randomized controlled trial evaluated both safety and efficacy of phage therapy against chronic otitis caused by antibiotic-resistant P. aeruginosa (Wright et al. 2009). No treatment-related adverse events were reported, and the results showed that topical administration of a therapeutic cocktail (Biophage-PA, Biocontrol, UK) in the ear results in an improvement of clinical manifestation of the infection. However, the patients were selected based on being infected with a P. aeruginosa strain susceptible to one or more of the phage included in the cocktail, which shows that, similar to antibiotics, a phage antibiogram is necessary before treatment.
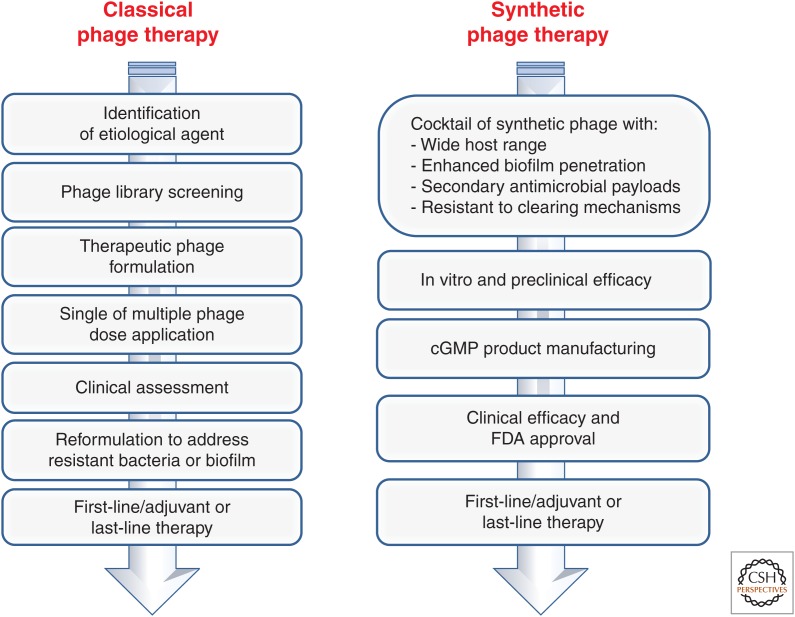
The first FDA-approved phase I clinical trial of phage therapy was performed in 2007 at the Southwest Regional Wound Care Center in Lubbock, Texas to evaluate local administration of a small set of well-characterized phage in patients with chronic venous leg ulcers (Rhoads et al. 2009). The study showed that topical phage administration neither had any adverse effects nor affects wound healing. Although outside of the scope of the clinical trial, efficacy could not be established. Currently, several other clinical studies are being conducted, which highlights the interest of the scientific community in developing bacteriophage as antimicrobials (Kutter et al. 2014). However, the development of phage for contemporary clinical use will likely rely on the synthesis of phage with built-in characteristics (such as wide host range, biofilm-degrading enzymes, and secondary antimicrobial payloads) that can be evaluated in rigorous clinical trials (Fig. 4).
Figure 4.
Phage therapy approaches. Classical (or Eastern European approach) relies on extensive banks of phages that can be used for treatment on a case-by-case basis by a physician specializing in phage therapy and adjusted based on clinical assessment. The synthetic phage therapy approach aims to develop phage with drug-like properties that can be used, much like antibiotics, in standardized Western treatment approaches. cGMP, Current good manufacturing practice.
PHAGE IN THE TREATMENT OF CANCER AND DEGENERATIVE DISEASES
Natural phages have been long considered poor gene delivery vehicles because they only infect bacteria (Ivanenkov et al. 1999). Recognizing the potential of phages for gene therapy, Hajitou et al. (2006) engineered a new generation of hybrid prokaryotic–eukaryotic nanovectors as a chimera between eukaryotic adeno-associated virus (AAV) and the filamentous M13 bacteriophage, referred to as adeno-associated virus/phage (AAVP). This phage particle expresses the cyclic peptide RGD4C (CDCRGDCFC) ligand on the phage pIII minor coat protein, allowing systemic and specific targeting to the αvβ3-integrin receptor, which is present primarily on tumor vasculature and tumor cells, and is expressed at barely detectable levels in normal endothelium and tissues (Folkman 1997). The hybrid nanovector genome was also modified by inserting an engineered AAV (recombinant adeno-associated virus [rAAV]) transgene cassette into an intergenomic region of the phage genome, under the regulation of the cytomegalovirus (CMV) promoter and flanked by full-length inverted terminal repeats (ITRs) from AAV serotype 2. The ITRs improve transduction efficiency and enhance transgene expression by maintaining and forming concatemers of the eukaryotic transgene cassette (Hajitou et al. 2006; Trepel et al. 2009). Further, this vector has been engineered to carry the herpes simplex thymidine kinase gene cassette suitable for tumor treatment with ganciclovir and positron emission tomography imaging for theranostic approaches (Soghomonyan et al. 2007; Hajitou et al. 2007, 2008). The RGD4C-AAVP has been used to deliver tumor necrosis factor α (TNF-α) to the angiogenic vasculature of human melanoma xenografts in nude mice (Tandle et al. 2009). Following systemic administration of phage, the TNF-α expression was shown to be specifically localized in tumors, leading to apoptosis in tumor blood vessels and significant inhibition of tumor growth, while remaining virtually undetectable in all other tissues, notably the liver and spleen. The efficacy of targeted RGD4C-AAVP expressing the TNF-α has been also shown in dogs with soft tissue sarcoma (Paoloni et al. 2009). This vector was modified even further to replace the CMV promoter with the tumor-specific promoter Grp78, which drives expression only in the targeted tumor vasculature (Kia et al. 2012).
Recent studies have shown that the linear structure of M13 bacteriophage permits binding to β-amyloid and α-synuclein proteins, leading to plaque disaggregation in models of Alzheimer’s and Parkinson’s diseases. Convection-enhanced delivery of M13 phage to the brain of nonhuman primates confirmed distribution into both white and gray matter, which makes filamentous phage very attractive theranostics for direct plaque dissolution, as well as delivery of therapeutic and imaging agents into the brain (Frenkel and Solomon 2002; Ksendzovsky et al. 2012).
PHAGE-BASED VACCINOLOGY
Recombinant DNA technology allows synthesis of vaccine candidates that are subunits of pathogens. However, such vaccines have limited immunogenic features and require adjuvants or effective delivery systems for proper activation of the immune system (Petrovsky and Aguilar 2004; Barrett and Stanberry 2008). Thus, phages are now being evaluated as vaccine delivery agents because of their inherited ability to stimulate humoral and cell-mediated immunity (Burdin et al. 2004). Two strategies are often combined to produce phage vaccines: (1) phage display, when virions are decorated with peptides selected for their ability to bind antigen-presenting cells; and (2) phage DNA vaccines, when viral DNA is engineered to carry a foreign antigen gene under the control of a strong eukaryotic promoter and has the ability to deliver this element to mammalian cells (Clark and March 2004a; Zanghi et al. 2007).
Filamentous phage M13 has been the first virus manipulated to express a melanoma-specific tumor antigen fragment and has been successfully used to raise an immune response capable of reducing tumor growth in animal models (Benhar 2001; Fang et al. 2005). Currently, several vaccines for infectious diseases are prepared by using the T4 phage display system, which has shown promising results in animal models (Jiang et al. 1997). Similarly, phage T7 has been engineered to display vascular endothelial growth factor (VEGF) and has been successfully used to break immunologic tolerance and produce a strong immune response against Lewis lung cell carcinoma (Li et al. 2006).
Alternatively, phage can be exploited to transfer genes into mammalian cells. In these vectors, the antigens, under the control of a eukaryotic promoter, are cloned inside of a nonessential region of a phage genome. When injected in a mammalian system, these phage particles, acting as a DNA vaccine, can induce potent immune response by expressing foreign antigen inside of anaphase-promoting complexes (APCs) or other cells (Clark and March 2006). Several λ-based DNA vaccines for infectious diseases have been prepared that have shown promising results in animal models (Clark and March 2004b; March et al. 2004). It should be noted that these viruses are manipulated to penetrate mammalian cells and, in the absence of engineering, phages are only capable of infecting bacteria.
CONCLUDING REMARKS
The global escalation of bacterial resistance to antibiotics raises the possibility of returning to the clinical equivalent of the preantibiotic era and requires the development of new classes of antimicrobials that could strengthen the effectiveness of current use drugs. Phage therapy is a promising, yet challenging, strategy for the development of new antimicrobial therapies that can enable more strategic treatment approaches that do not affect beneficial normal microbiota. Another major benefit of phage therapy is that the mode of action of bacteriophage is distinct from those of antibiotics; thus, bacterial strains that are resistant to antibiotics are nevertheless susceptible to phage infection. However, limitations such as narrow host range, bacterial resistance to phage, cost of manufacturing, and challenging dose finding/delivery methods need to be addressed. Recent advances in synthetic genome assembly and viral genome engineering can be used to create phage with superior therapeutic properties. Designer synthetic phage may enable new intellectual property and attract interest from large pharmaceutical companies in these products. Moreover, these techniques can also be applied to engineering novel theranostic nanovectors with improved tissue penetration and payload carrying potential for treatment of cancer and other diseases.
Footnotes
Editors: Daniel G. Gibson, Clyde A. Hutchison III, Hamilton O. Smith, and J. Craig Venter
Additional Perspectives on Synthetic Biology available at www.cshperspectives.org
REFERENCES
- Abedon S. 2011. Phage therapy pharmacology: Calculating phage dosing. Adv Appl Microbiol 77: 1–40. [DOI] [PubMed] [Google Scholar]
- Alisky J, Iczkowski K, Rapoport A, Troitsky N. 1998. Bacteriophages show promise as antimicrobial agents. J Infect 36: 5–15. [DOI] [PubMed] [Google Scholar]
- Arias CA, Murray BE. 2009. Antibiotic-resistant bugs in the 21st century—A clinical super-challenge. N Engl J Med 360: 439–443. [DOI] [PubMed] [Google Scholar]
- Barrett ADT, Stanberry LR. 2008. Vaccines for biodefence and emerging and neglected diseases. Academic, Waltham, MA. [Google Scholar]
- Barrow P, Lovell M, Berchieri A Jr. 1998. Use of lytic bacteriophage for control of experimental Escherichia coli septicemia and meningitis in chickens and calves. Clin Diag Lab Immun 5: 294–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann A. 2006. Early development of therapeutic biologics—Pharmacokinetics. Curr Drug Metab 7: 15–21. [DOI] [PubMed] [Google Scholar]
- Benhar I. 2001. Biotechnological applications of phage and cell display. Biotech Adv 19: 1–33. [DOI] [PubMed] [Google Scholar]
- Biswas B, Adhya S, Washart P, Paul B, Trostel AN, Powell B, Carlton R, Merril CR. 2002. Bacteriophage therapy rescues mice bacteremic from a clinical isolate of vancomycin-resistant Enterococcus faecium. Infect Immun 70: 204–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher HW, Talbot GH, Benjamin DK Jr, Bradley J, Guidos RJ, Jones RN, Murray BE, Bonomo RA, Gilbert D, Infectious Diseases Society of America. 2013. 10 × ’20 Progress—Development of new drugs active against Gram-negative bacilli: An update from the Infectious Diseases Society of America. Clin Infect Dis 56: 1685–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd EF, Brussow H. 2002. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol 10: 521–529. [DOI] [PubMed] [Google Scholar]
- Bruttin A, Brussow H. 2005. Human volunteers receiving Escherichia coli phage T4 orally: A safety test of phage therapy. Antimicrob Agents Chemother 49: 2874–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdin N, Guy B, Moingeon P. 2004. Immunological foundations to the quest for new vaccine adjuvants. BioDrugs 18: 79–93. [DOI] [PubMed] [Google Scholar]
- Burrowes B, Harper DR, Anderson J, McConville M, Enright MC. 2011. Bacteriophage therapy: Potential uses in the control of antibiotic-resistant pathogens. Expert Rev Anti Infect Ther 9: 775–785. [DOI] [PubMed] [Google Scholar]
- Carmody LA, Gill JJ, Summer EJ, Sajjan US, Gonzalez CF, Young RF, LiPuma JJ. 2010. Efficacy of bacteriophage therapy in a model of Burkholderia cenocepacia pulmonary infection. J Infect Dis 201: 264–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson L, Gorman SP, Gilmore BF. 2010. The use of lytic bacteriophages in the prevention and eradication of biofilms of Proteus mirabilis and Escherichia coli. FEMS Immunol Med Microbiol 59: 447–455. [DOI] [PubMed] [Google Scholar]
- Carter CD, Parks A, Abuladze T, Li M, Woolston J, Magnone J, Senecal A, Kropinski AM, Sulakvelidze A. 2012. Bacteriophage cocktail significantly reduces Escherichia coli O157: H7 contamination of lettuce and beef, but does not protect against recontamination. Bacteriophage 2: 178–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, Pirofski LA. 2000. Host–pathogen interactions: Basic concepts of microbial commensalism, colonization, infection, and disease. Infect Immun 68: 6511–6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanishvili N. 2012. Phage therapy—History from Twort and d’Herelle through Soviet experience to current approaches. Adv Vir Res 83: 3–40. [DOI] [PubMed] [Google Scholar]
- Chibani-Chennoufi S, Sidoti J, Bruttin A, Kutter E, Sarker S, Brussow H. 2004. In vitro and in vivo bacteriolytic activities of Escherichia coli phages: Implications for phage therapy. Antimicrob Agents Chemother 48: 2558–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiocca EA. 2002. Oncolytic viruses. Nat Rev Cancer 2: 938–950. [DOI] [PubMed] [Google Scholar]
- Clark JR, March JB. 2004a. Bacterial viruses as human vaccines? Exp Rev Vaccines 3: 463–476. [DOI] [PubMed] [Google Scholar]
- Clark JR, March JB. 2004b. Bacteriophage-mediated nucleic acid immunization. FEMS Immunol Med Microbiol 40: 21–26. [DOI] [PubMed] [Google Scholar]
- Clark JR, March JB. 2006. Bacteriophages and biotechnology: Vaccines, gene therapy and antibacterials. Trends Biotechnol 24: 212–218. [DOI] [PubMed] [Google Scholar]
- Curtin JJ, Donlan RM. 2006. Using bacteriophages to reduce formation of catheter-associated biofilms by Staphylococcus epidermidis. Antimicrob Agents Chemother 50: 1268–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debarbieux L, Leduc D, Maura D, Morello E, Criscuolo A, Grossi O, Balloy V, Touqui L. 2010. Bacteriophages can treat and prevent Pseudomonas aeruginosa lung infections. J Infect Dis 201: 1096–1104. [DOI] [PubMed] [Google Scholar]
- DeLappe N, O’Halloran F, Fanning S, Corbett-Feeney G, Cheasty T, Cormican M. 2003. Antimicrobial resistance and genetic diversity of Shigella sonnei isolates from western Ireland, an area of low incidence of infection. J Clin Microbiol 41: 1919–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Herelle F. 1931. An address on bacteriophagy and recovery from infectious diseases. Can Med Assoc J 24: 619–628. [PMC free article] [PubMed] [Google Scholar]
- Eaton MD, Bayne-Jones S. 1934. Bacteriophage therapy: Review of the principles and results of the use of bacteriophage in the treatment of infections. JAMA 103: 1769–1776. [Google Scholar]
- Enyeart PJ, Chirieleison SM, Dao MN, Perutka J, Quandt EM, Yao J, Whitt JT, Keatinge-Clay AT, Lambowitz AM, Ellington AD. 2013. Generalized bacterial genome editing using mobile group II introns and Cre-lox. Mol Syst Biol 9: 685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhead H. 2009. SASP gene delivery: A novel antibacterial approach. Drug News Perspect 22: 197–203. [DOI] [PubMed] [Google Scholar]
- Fang J, Wang G, Yang Q, Song J, Wang Y, Wang L. 2005. The potential of phage display virions expressing malignant tumor specific antigen MAGE-A1 epitope in murine model. Vaccine 23: 4860–4866. [DOI] [PubMed] [Google Scholar]
- Folkman J. 1997. Addressing tumor blood vessels. Nat Biotech 15: 510. [DOI] [PubMed] [Google Scholar]
- Frenkel D, Solomon B. 2002. Filamentous phage as vector-mediated antibody delivery to the brain. Proc Natl Acad Sci 99: 5675–5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Forster T, Mayer O, Curtin JJ, Lehman SM, Donlan RM. 2010. Bacteriophage cocktail for the prevention of biofilm formation by Pseudomonas aeruginosa on catheters in an in vitro model system. Antimicrob Agents Chemother 54: 397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda K, Ishida W, Uchiyama J, Rashel M, Kato S, Morita T, Muraoka A, Sumi T, Matsuzaki S, Daibata M, et al. 2012. Pseudomonas aeruginosa keratitis in mice: Effects of topical bacteriophage KPP12 administration. PloS ONE 7: e47742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H, Kuroiwa T, Mizushima S. 1983. DNA injection during bacteriophage T4 infection of Escherichia coli. J Bacteriol 154: 938–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geier MR, Trigg ME, Merril CR. 1973. Fate of bacteriophage λ in non-immune germ-free mice. Nature 246: 221–223. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Goode D, Allen VM, Barrow PA. 2003. Reduction of experimental Salmonella and Campylobacter contamination of chicken skin by application of lytic bacteriophages. Appl Env Microbiol 69: 5032–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Górski A, Miedzybrodzki R, Borysowski J, Weber-Dabrowska B, Lobocka M, Fortuna W, Letkiewicz S, Zimecki M, Filby G. 2009. Bacteriophage therapy for the treatment of infections. Curr Opin Investig Drugs 10: 766–774. [PubMed] [Google Scholar]
- Górski A, Miedzybrodzki R, Borysowski J, Dabrowska K, Wierzbicki P, Ohams M, Korczak-Kowalska G, Olszowska-Zaremba N, Lusiak-Szelachowska M, Klak M, et al. 2012. Phage as a modulator of immune responses: Practical implications for phage therapy. Adv Virus Res 83: 41–71. [DOI] [PubMed] [Google Scholar]
- Gu J, Liu X, Li Y, Han W, Lei L, Yang Y, Zhao H, Gao Y, Song J, Lu R, et al. 2012. A method for generation phage cocktail with great therapeutic potential. PloS ONE 7: e31698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther EJ, Murray NE, Glazer PM. 1993. High efficiency, restriction-deficient in vitro packaging extracts for bacteriophage λ DNA using a new E. coli lysogen. Nucleic Acids Res 21: 3903–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagens S, Habel A, von Ahsen U, von Gabain A, Blasi U. 2004. Therapy of experimental pseudomonas infections with a nonreplicating genetically modified phage. Antimicrob Agents Chemother 48: 3817–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajitou A, Trepel M, Lilley CE, Soghomonyan S, Alauddin MM, Marini FC 3rd, Restel BH, Ozawa MG, Moya CA, Rangel R, et al. 2006. A hybrid vector for ligand-directed tumor targeting and molecular imaging. Cell 125: 385–398. [DOI] [PubMed] [Google Scholar]
- Hajitou A, Rangel R, Trepel M, Soghomonyan S, Gelovani JG, Alauddin MM, Pasqualini R, Arap W. 2007. Design and construction of targeted AAVP vectors for mammalian cell transduction. Nat Protocols 2: 523–531. [DOI] [PubMed] [Google Scholar]
- Hajitou A, Lev DC, Hannay JA, Korchin B, Staquicini FI, Soghomonyan S, Alauddin MM, Benjamin RS, Pollock RE, Gelovani JG, et al. 2008. A preclinical model for predicting drug response in soft-tissue sarcoma with targeted AAVP molecular imaging. Proc Natl Acad Sci 105: 4471–4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AR, De Vos D, Friman VP, Pirnay JP, Buckling A. 2012. Effects of sequential and simultaneous applications of bacteriophages on populations of Pseudomonas aeruginosa in vitro and in wax moth larvae. Appl Env Microbiol 78: 5646–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon GW. 2007. Bacteriophages: An appraisal of their role in the treatment of bacterial infections. Int J Antimicrob Agents 30: 118–128. [DOI] [PubMed] [Google Scholar]
- Harper DR, Anderson J, Enright MC. 2011. Phage therapy: Delivering on the promise. Ther Deliv 2: 935–947. [DOI] [PubMed] [Google Scholar]
- Hatfull GF. 2008. Bacteriophage genomics. Curr Opin Microbiol 11: 447–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins C, Harper D, Burch D, Anggard E, Soothill J. 2010. Topical treatment of Pseudomonas aeruginosa otitis of dogs with a bacteriophage mixture: A before/after clinical trial. Vet Microbiol 146: 309–313. [DOI] [PubMed] [Google Scholar]
- Hendrix RW. 2003. Bacteriophage genomics. Curr Opin Microbiol 6: 506–511. [DOI] [PubMed] [Google Scholar]
- Ivanenkov V, Felici F, Menon AG. 1999. Uptake and intracellular fate of phage display vectors in mammalian cells. Biochem Biophys Acta 1448: 450–462. [DOI] [PubMed] [Google Scholar]
- Jaiswal A, Koley H, Ghosh A, Palit A, Sarkar B. 2013. Efficacy of cocktail phage therapy in treating Vibrio cholerae infection in rabbit model. Microbes Infect 15: 152–156. [DOI] [PubMed] [Google Scholar]
- Jaschke PR, Lieberman EK, Rodriguez J, Sierra A, Endy D. 2012. A fully decompressed synthetic bacteriophage ϕX174 genome assembled and archived in yeast. Virology 434: 278–284. [DOI] [PubMed] [Google Scholar]
- Jiang J, Abu-Shilbayeh L, Rao VB. 1997. Display of a PorA peptide from Neisseria meningitidis on the bacteriophage T4 capsid surface. Infect Immun 65: 4770–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen EC. 2012. Phage therapy: Concept to cure. Frontiers Microbiol 3: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr C, Sadowski PD. 1974. Packaging and maturation of DNA of bacteriophage T7 in vitro. Proc Natl Acad Sci 71: 3545–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kia A, Przystal JM, Nianiaris N, Mazarakis ND, Mintz PJ, Hajitou A. 2012. Dual systemic tumor targeting with ligand-directed phage and Grp78 promoter induces tumor regression. Mol Cancer Ther 11: 2566–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingham R, Klasa G, Carver KH. 2014. Key regulatory guidelines for the development of biologics in the United States and Europe. In Biological drug products: Development and strategies, 1st ed. (ed. Wang W and Singh M), pp. 75–109. Wiley, New York. [Google Scholar]
- Kiro R, Shitrit D, Qimron U. 2014. Efficient engineering of a bacteriophage genome using the type I-E CRISPR-Cas system. RNA Biol 11: 42–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksendzovsky A, Walbridge S, Saunders RC, Asthagiri AR, Heiss JD, Lonser RR. 2012. Convection-enhanced delivery of M13 bacteriophage to the brain. J Neurosurgery 117: 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S, Harjai K, Chhibber S. 2009. Efficacy of bacteriophage treatment in murine burn wound infection induced by Klebsiella pneumoniae. J Microbiol Biotechnol 19: 622–628. [DOI] [PubMed] [Google Scholar]
- Kumari S, Harjai K, Chhibber S. 2010. Topical treatment of Klebsiella pneumoniae B5055 induced burn wound infection in mice using natural products. J Infect Dev Ctries 4: 367–377. [PubMed] [Google Scholar]
- Kutter E, De Vos D, Gvasalia G, Alavidze Z, Gogokhia L, Kuhl S, Abedon ST. 2010. Phage therapy in clinical practice: Treatment of human infections. Curr Pharmacol Biotechnol 11: 69–86. [DOI] [PubMed] [Google Scholar]
- Kutter E, Borysowski J, Międzybrodzki R, Górski A, Weber-Dąbrowska B, Kutateladze M, Alavidze Z, Goderdzishvili M, Adamia R. 2014. Clinical phage therapy. In Phage therapy: Current research and applications (ed. Borysowski J, et al. ), pp. 257–288. Caister Academic, Norfolk, UK. [Google Scholar]
- Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat Rev Microbiol 8: 317–327. [DOI] [PubMed] [Google Scholar]
- Li XH, Tang L, Liu D, Sun HM, Zhou CC, Tan LS, Wang LP, Zhang PD, Zhang SQ. 2006. Antitumor effect of recombinant T7 phage vaccine expressing xenogenic vascular endothelial growth factor on Lewis lung cancer in mice. Chinese J Cancer 25: 1221–1226. [PubMed] [Google Scholar]
- Lu TK, Collins JJ. 2007. Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci 104: 11197–11202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TK, Koeris MS. 2011. The next generation of bacteriophage therapy. Curr Opin Microbiol 14: 524–531. [DOI] [PubMed] [Google Scholar]
- March JB, Clark JR, Jepson CD. 2004. Genetic immunisation against hepatitis B using whole bacteriophage λ particles. Vaccine 22: 1666–1671. [DOI] [PubMed] [Google Scholar]
- Marinelli LJ, Piuri M, Swigonova Z, Balachandran A, Oldfield LM, van Kessel JC, Hatfull GF. 2008. BRED: A simple and powerful tool for constructing mutant and recombinant bacteriophage genomes. PloS ONE 3: e3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel B, Moineau S. 2014. CRISPR-Cas: An efficient tool for genome engineering of virulent bacteriophages. Nucleic Acids Res 42: 9504–9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Freeman TA, Hilbert DW, Duff M, Fuortes M, Stapleton PP, Daly JM. 2005. Lysis-deficient bacteriophage therapy decreases endotoxin and inflammatory mediator release and improves survival in a murine peritonitis model. Surgery 137: 639–646. [DOI] [PubMed] [Google Scholar]
- Matsuzaki S, Yasuda M, Nishikawa H, Kuroda M, Ujihara T, Shuin T, Shen Y, Jin Z, Fujimoto S, Nasimuzzaman MD, et al. 2003. Experimental protection of mice against lethal Staphylococcus aureus infection by novel bacteriophage phi MR11. J Infect Dis 187: 613–624. [DOI] [PubMed] [Google Scholar]
- McVay CS, Velasquez M, Fralick JA. 2007. Phage therapy of Pseudomonas aeruginosa infection in a mouse burn wound model. Antimicrob Agents Chemother 51: 1934–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merril CR, Biswas B, Carlton R, Jensen NC, Creed GJ, Zullo S, Adhya S. 1996. Long-circulating bacteriophage as antibacterial agents. Proc Natl Acad Sci 93: 3188–3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merril CR, Scholl D, Adhya SL. 2003. The prospect for bacteriophage therapy in Western medicine. Nat Rev Drug Discov 2: 489–497. [DOI] [PubMed] [Google Scholar]
- Merril CR, Scholl D, Adhya S. 2006. Phage therapy. In The bacteriophages, 2nd ed. (ed. Calendar RL, Abedon S), pp. 725–741. Oxford University Press, London. [Google Scholar]
- Neu HC. 1992. The crisis in antibiotic resistance. Science 257: 1064–1073. [DOI] [PubMed] [Google Scholar]
- Nobrega FL, Costa AR, Kluskens LD, Azeredo J. 2015. Revisiting phage therapy: New applications for old resources. Trends Microbiol 23: 185–191. [DOI] [PubMed] [Google Scholar]
- Ochs HD, Davis SD, Wedgwood RJ. 1971. Immunologic responses to bacteriophage ϕX 174 in immunodeficiency diseases. J Clin Invest 50: 2559–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Flaherty S, Ross RP, Meaney W, Fitzgerald GF, Elbreki MF, Coffey A. 2005. Potential of the polyvalent anti-Staphylococcus bacteriophage K for control of antibiotic-resistant staphylococci from hospitals. Appl Env Mocrobiol 71: 1836–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala V, Laitalainen J, Jalasvuori M. 2013. Fight evolution with evolution: Plasmid-dependent phages with a wide host range prevent the spread of antibiotic resistance. Evol Appl 6: 925–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira A, Sereno R, Azeredo J. 2010. In vivo efficiency evaluation of a phage cocktail in controlling severe colibacillosis in confined conditions and experimental poultry houses. Vet Microbiol 146: 303–308. [DOI] [PubMed] [Google Scholar]
- Palmer KL, Gilmore MS. 2010. Multidrug-resistant enterococci lack CRISPR-cas. mBio 1: e00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoloni MC, Tandle A, Mazcko C, Hanna E, Kachala S, Leblanc A, Newman S, Vail D, Henry C, Thamm D, et al. 2009. Launching a novel preclinical infrastructure: Comparative oncology trials consortium directed therapeutic targeting of TNF α to cancer vasculature. PloS ONE 4: e4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovsky N, Aguilar JC. 2004. Vaccine adjuvants: Current state and future trends. Immunol Cell Biol 82: 488–496. [DOI] [PubMed] [Google Scholar]
- Ptashne M. 2004. A genetic switch: Phage λ revisited. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Rangel R, Guzman-Rojas L, le Roux LG, Staquicini FI, Hosoya H, Barbu EM, Ozawa MG, Nie KD Jr, Langley RR, et al. 2012. Combinatorial targeting and discovery of ligand-receptors in organelles of mammalian cells. Nat Commun 3: 788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads DD, Wolcott RD, Kuskowski MA, Wolcott BM, Ward LS, Sulakvelidze A. 2009. Bacteriophage therapy of venous leg ulcers in humans: Results of a phase I safety trial. J Wound Care 18: 237–238, 240–233. [DOI] [PubMed] [Google Scholar]
- Rozema EA, Stephens TP, Bach SJ, Okine EK, Johnson RP, Stanford K, McAllister TA. 2009. Oral and rectal administration of bacteriophages for control of Escherichia coli O157:H7 in feedlot cattle. J Food Prot 72: 241–250. [DOI] [PubMed] [Google Scholar]
- Samson JE, Magadan AH, Sabri M, Moineau S. 2013. Revenge of the phages: Defeating bacterial defences. Nat Rev Microbiol 11: 675–687. [DOI] [PubMed] [Google Scholar]
- Sarker SA, McCallin S, Barretto C, Berger B, Pittet AC, Sultana S, Krause L, Huq S, Bibiloni R, Bruttin A, et al. 2012. Oral T4-like phage cocktail application to healthy adult volunteers from Bangladesh. Virology 434: 222–232. [DOI] [PubMed] [Google Scholar]
- Shin J, Jardine P, Noireaux V. 2012. Genome replication, synthesis, and assembly of the bacteriophage T7 in a single cell-free reaction. ACS Synth Biol 1: 408–413. [DOI] [PubMed] [Google Scholar]
- Smith HO, Hutchison CA 3rd, Pfannkoch C, Venter JC. 2003. Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides. Proc Natl Acad Sci 100: 15440–15445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soghomonyan S, Hajitou A, Rangel R, Trepel M, Pasqualini R, Arap W, Gelovani JG, Alauddin MM. 2007. Molecular PET imaging of HSV1-tk reporter gene expression using [18F]FEAU. Nat Protoc 2: 416–423. [DOI] [PubMed] [Google Scholar]
- Soni KA, Nannapaneni R. 2010. Bacteriophage significantly reduces Listeria monocytogenes on raw salmon fillet tissue. J Food Prot 73: 32–38. [DOI] [PubMed] [Google Scholar]
- Srinivasan S, Alexander JF, Driessen WH, Leonard F, Ye H, Liu X, Arap W, Pasqualini R, Ferrari M, Godin B. 2013. Bacteriophage associated silicon particles: Design and characterization of a novel theranostic vector with improved payload carrying potential. J Mater Chem B Mater Biol Med 1: 5218–5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford K, McAllister TA, Niu YD, Stephens TP, Mazzocco A, Waddell TE, Johnson RP. 2010. Oral delivery systems for encapsulated bacteriophages targeted at Escherichia coli O157:H7 in feedlot cattle. J Food Prot 73: 1304–1312. [DOI] [PubMed] [Google Scholar]
- Staquicini FI, Ozawa MG, Moya CA, Driessen WH, Barbu EM, Nishimori H, Soghomonyan S, Flores LG II, Liang X, Paolillo V, et al. 2011. Systemic combinatorial peptide selection yields a non-canonical iron-mimicry mechanism for targeting tumors in a mouse model of human glioblastoma. J Clin Invest 121: 161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulakvelidze A, Alavidze Z, Morris JG Jr. 2001. Bacteriophage therapy. Antimicrob Agents Chemother 45: 649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers WC. 2001. Bacteriophage therapy. Ann Rev Microbiol 55: 437–451. [DOI] [PubMed] [Google Scholar]
- Tandle A, Hanna E, Lorang D, Hajitou A, Moya CA, Pasqualini R, Arap W, Adem A, Starker E, Hewitt S, et al. 2009. Tumor vasculature-targeted delivery of tumor necrosis factor-α. Cancer 115: 128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji Y, Shimada T, Fukudomi H, Miyanaga K, Nakai Y, Unno H. 2005. Therapeutic use of phage cocktail for controlling Escherichia coli O157:H7 in gastrointestinal tract of mice. J Biosci Bioeng 100: 280–287. [DOI] [PubMed] [Google Scholar]
- Tetart F, Repoila F, Monod C, Krisch HM. 1996. Bacteriophage T4 host range is expanded by duplications of a small domain of the tail fiber adhesin. J Mol Biol 258: 726–731. [DOI] [PubMed] [Google Scholar]
- Tetart F, Desplats C, Krisch HM. 1998. Genome plasticity in the distal tail fiber locus of the T-even bacteriophage: Recombination between conserved motifs swaps adhesin specificity. J Mol Biol 282: 543–556. [DOI] [PubMed] [Google Scholar]
- Tinsley CR, Bille E, Nassif X. 2006. Bacteriophages and pathogenicity: More than just providing a toxin? Microbes Infect 8: 1365–1371. [DOI] [PubMed] [Google Scholar]
- Trepel M, Stoneham CA, Eleftherohorinou H, Mazarakis ND, Pasqualini R, Arap W, Hajitou A. 2009. A heterotypic bystander effect for tumor cell killing after adeno-associated virus/phage-mediated, vascular-targeted suicide gene transfer. Mol Cancer Ther 8: 2383–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twort FW. 1936. Further investigations on the nature of ultra-microscopic viruses and their cultivation. J Hygiene 36: 204–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitiello CL, Merril CR, Adhya S. 2005. An amino acid substitution in a capsid protein enhances phage survival in mouse circulatory system more than a 1000-fold. Virus Res 114: 101–103. [DOI] [PubMed] [Google Scholar]
- Wagenaar JA, Van Bergen MA, Mueller MA, Wassenaar TM, Carlton RM. 2005. Phage therapy reduces Campylobacter jejuni colonization in broilers. Vet Microbiol 109: 275–283. [DOI] [PubMed] [Google Scholar]
- Wedgwood RJ, Ochs HD, Davis SD. 1975. The recognition and classification of immunodeficiency diseases with bacteriophage ϕΧ 174. Birth Defects Orig Artic Ser 11: 331–338. [PubMed] [Google Scholar]
- Woolston J, Parks AR, Abuladze T, Anderson B, Li M, Carter C, Hanna LF, Heyse S, Charbonneau D, Sulakvelidze A. 2013. Bacteriophages lytic for rapidly reduce contamination on glass and stainless steel surfaces. Bacteriophage 3: e25697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A, Hawkins CH, Anggard EE, Harper DR. 2009. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin Otolaryngol 34: 349–357. [DOI] [PubMed] [Google Scholar]
- Zanghi CN, Sapinoro R, Bradel-Tretheway B, Dewhurst S. 2007. A tractable method for simultaneous modifications to the head and tail of bacteriophage λ and its application to enhancing phage-mediated gene delivery. Nucleic Acids Res 35: e59. [DOI] [PMC free article] [PubMed] [Google Scholar]