Abstract
The transforming growth factor-β (TGF-β) family of ligands elicit their biological effects by initiating new programs of gene expression. The best understood signal transducers for these ligands are the SMADs, which essentially act as transcription factors that are activated in the cytoplasm and then accumulate in the nucleus in response to ligand induction where they bind to enhancer/promoter sequences in the regulatory regions of target genes to either activate or repress transcription. This review focuses on the mechanisms whereby the SMADs achieve this and the functional implications. The SMAD complexes have weak affinity for DNA and limited specificity and, thus, they cooperate with other site-specific transcription factors that act either to actively recruit the SMAD complexes or to stabilize their DNA binding. In some situations, these cooperating transcription factors function to integrate the signals from TGF-β family ligands with environmental cues or with information about cell lineage. Activated SMAD complexes regulate transcription via remodeling of the chromatin template. Consistent with this, they recruit a variety of coactivators and corepressors to the chromatin, which either directly or indirectly modify histones and/or modulate chromatin structure.
SMADs are proteins that transduce extracellular signals from TGF-β ligands to the nucleus. They cooperate with a remarkable array of transcription factors, coactivators, and corepressors to control gene expression.
The transforming growth factor-β (TGF-β) family of ligands, which include the TGF-βs, the activins, NODAL, bone morphogenetic proteins (BMPs), and growth and differentiation factors (GDFs), elicit their pleiotropic effects on cell behavior by signaling to the nucleus and initiating new programs of gene expression. Ligand binding activates pairs of type I and type II serine/threonine kinase receptors, with specific combinations of receptors recognizing different ligands (Feng and Derynck 2005; Wakefield and Hill 2013). The type II receptor phosphorylates and activates the type I receptor, which subsequently activates the primary signal transducers of the pathway, the receptor-regulated SMADs (R-SMADs) (Massagué 2012). Although the SMADs are not the only signal transducers downstream of the receptors, they will be the only transducers discussed here, as they are the subject of this review. The R-SMADs are divided into those activated predominantly in response to TGF-β, activin, and NODAL, which are SMAD2 and SMAD3, and those primarily activated by BMPs and GDFs, which are SMAD1, SMAD5, and SMAD8 (Miyazawa et al. 2002). Receptor-mediated phosphorylation of R-SMADs occurs on two serines at the extreme carboxyl terminus in a Ser–X–Ser motif, in which X is either Val or Met. Once phosphorylated, the R-SMADs form homomeric complexes, and heteromeric complexes with the common SMAD, SMAD4. These complexes are thought to be predominantly trimers, although some evidence for dimers also exists (Chacko et al. 2001; Wu et al. 2001; Inman and Hill 2002). Activated SMAD complexes accumulate in the nucleus, where they bind DNA directly or indirectly via other transcription factors and regulate gene expression, both positively and negatively (Massagué 2012).
The SMADs behave as transcription factors that become activated in the cytoplasm as a result of extracellular signals and are directly responsible for executing programs of gene expression. Indeed, the pathway is wired so that both the level and duration of receptor activation are correlated with the level and persistence of activated SMAD complexes in the nucleus (Schmierer et al. 2008). This is achieved through constant nucleocytoplasmic shuttling of the SMADs. In the two decades since the SMADs were discovered, we have learned a substantial amount about how SMAD activity is regulated, how SMAD complexes are recruited to DNA, and which corepressors and coactivators they subsequently cooperate with. We are now just beginning to decipher the sequence of events that occur from activated SMAD complexes appearing in the nucleus to the transcription of target genes being up- or down-regulated. After a brief summary of the mechanics of transcription and the general role of transcription factors, this review will discuss current ideas concerning how SMAD complexes function in the regulation of transcription.
THE MECHANICS OF TRANSCRIPTION AND THE ROLE OF TRANSCRIPTION FACTORS
Protein-coding gene transcription is catalyzed by RNA polymerase II (Pol II) and involves positioning Pol II at the gene’s promoter, transcription initiation at the transcription start site (TSS), transcript elongation, termination, RNA transcript processing, and export from the nucleus (Fuda et al. 2009). For transcriptional regulation of a given gene, three types of DNA sequence are important—the promoter sequence just upstream of the TSS, the proximal promoter region, and enhancer sequences, which may be many kilobases (kb) away from the TSS, or even positioned in introns. In addition, the chromatin state is crucial, as this determines the availability of DNA-binding sites for transcriptional regulators and the general transcription machinery. Generally, active promoters are characterized by unmethylated CpG islands, high histone acetylation, trimethylation of Lys4 of histone H3 (H3K4), relative nucleosome depletion, and the presence of histone variants such as H2A.Z and H3.3 (Zhou et al. 2011). Active enhancers, in contrast, are marked by acetylation of H3K27, monomethylation of H3K4, and the presence of the histone acetyltransferase (HAT) p300 (now called EP300) (Rada-Iglesias et al. 2011). Repressed chromatin is characterized by trimethylation of H3K27 and H3K9, which are bound by polycomb group proteins and heterochromatin protein 1 (HP1) family members, respectively, causing chromatin remodeling and compaction, which limits the accessibility of the DNA (Beisel and Paro 2011; Zhou et al. 2011).
Initiation of transcription involves the assembly of a preinitiation complex (PIC) comprising so-called general transcription factors and Pol II. This is regulated by site-specific transcription factors bound at the proximal promoter regions or at enhancers, and chromatin looping is presumed to be involved so that enhancer-bound transcription factors can directly contact the promoter (Calo and Wysocka 2013). Enhancers act as transcription factor–binding platforms and usually contain clusters of binding sites for different transcription factors. These may be signal-activated transcription factors or cell-type-specific transcription factors, and, thus, enhancers act to integrate a number of different threads of information about the cell lineage and environmental cues (Calo and Wysocka 2013). Binding of most transcription factors at enhancers is thought to be preceded by the creation of a favorable chromatin environment. This may involve opening up of the chromatin by chromatin remodelers, or via binding of pioneer factors, such as FOXA1, which is capable of binding to condensed chromatin or nucleosomal DNA (Zaret and Carroll 2011).
Transcription factors can play different roles in transcriptional regulation, by either directly affecting PIC assembly, or by functioning indirectly through recruitment of coregulators that reorganize nucleosomes, modify histones, or alter the chromatin architecture of the gene. Chromatin remodeling can occur locally at the enhancer, perhaps allowing other transcription factors to bind, or can occur at the proximal promoter to allow the general transcription factors and Pol II access to DNA. Many transcription factors also recruit the Mediator complex, which interacts with the general transcription apparatus and promotes transcription (Fuda et al. 2009). Although much active transcriptional regulation is at the level of transcription initiation, it can also occur at the level of Pol II pause–release. In this case, Pol II is engaged at the promoter and transcribes a short nascent transcript and is then stalled. This creates an open chromatin structure that facilitates fast, synchronous gene expression in response to extracellular signals, which occurs by release of the paused Pol II (Levine 2011).
DNA BINDING OF SMAD COMPLEXES AND ITS REGULATION BY POSTTRANSLATIONAL MODIFICATIONS
As stated above, activated SMAD complexes act directly on DNA as transcription factors. All R-SMADs, with the exception of the most common spliced isoform of SMAD2, bind DNA directly, as does SMAD4 (Massagué 2012). The R-SMADs and SMAD4 have two highly conserved domains, the Mad homology 1 (MH1) domain in the amino terminus and the Mad homology 2 (MH2) domain in the carboxyl terminus, which are separated by a proline-rich linker. The domain responsible for DNA binding is the MH1 domain (Shi et al. 1998), whereas the MH2 domain is required for SMAD–receptor, SMAD–SMAD, and many SMAD–transcription factor interactions (Feng and Derynck 2005).
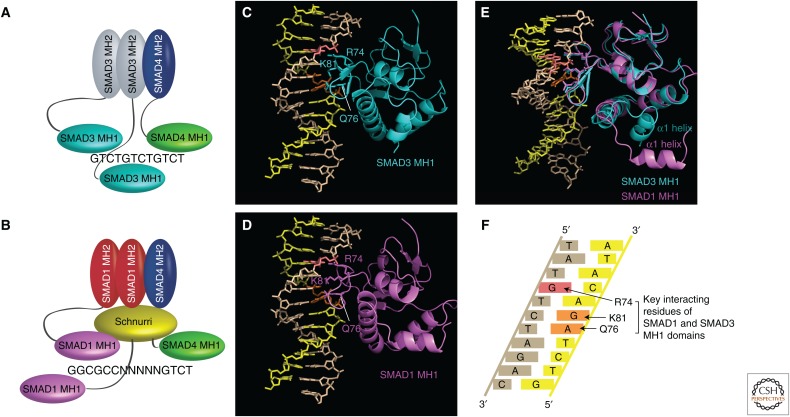
The sequence recognized by the SMAD3 and SMAD4 MH1 domains is GTCT or its reverse complement, AGAC. This sequence, which is commonly referred to as a SMAD-binding element (SBE), was first identified by DNA-binding site selection (Zawel et al. 1998). A single SBE is not sufficient to bind an activated SMAD complex, and, in vivo, SBEs have been shown to occur as direct or inverted (palindromic) repeats in the regulatory regions of TGF-β/NODAL/activin target genes by promoter analysis, or by whole-genome approaches, such as chromatin immunoprecipitation (ChIP)-sequencing or ChIP-on-chip analysis (Fig. 1A) (Dennler et al. 1998; Wong et al. 1999; Koinuma et al. 2009a; Liu et al. 2011; Mullen et al. 2011; Morikawa et al. 2013).
Figure 1.
DNA binding by SMAD1 and SMAD3. (A) Schematic showing how a trimer consisting of two SMAD3s and one SMAD4 could bind to direct repeats of SMAD-binding elements (SBEs), such as that found in the JUN enhancer (Wong et al. 1999). (B) Schematic showing how a trimer consisting of two SMAD1s and one SMAD4 could bind to the GGCGCCN5GTCT-binding site together with Schnurri. (Panel based on Gao et al. 2005.) (C) The crystal structure of the SMAD3 MH1 domain interacting with a palindromic GTCTAGAC sequence (PDB code, 1mhd) (Shi et al. 1998). For clarity, only one MH1 domain is shown. The DNA strands are colored in yellow and brown. The key interacting A and G on the yellow strand are colored orange and the key interacting G on the brown strand is colored pink. Amino acids R74, Q76, and K81 are indicated. (D) The crystal structure of the SMAD1 MH1 domain interacting with a palindromic GTCTAGAC sequence (PDB code, 3KMP) (Baburajendran et al. 2010). For clarity, only one MH1 domain is shown. The DNA strands are as in C and amino acids R74, Q76, and K81 are indicated. (E) An overlay of the two structures shown in A and B. To visualize the different positions of the α1 helices, the structures have been rotated ∼20° out of the plane of the paper. (F) A schematic showing the key amino acid–base interactions. The colors are as in C and D. (Panel based on Shi et al. 1998.) (Panels C–E were prepared using PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC.)
SMAD1/5–SMAD4 complexes, in contrast, bind distinct sites in vivo, defined as GRCGNC-N5-GTCT or GGCGCC-AN4-GNCV where N is any base, R is A or G, and V is A, C, or G (Pyrowolakis et al. 2004; Gao et al. 2005; Weiss et al. 2010). The GRCGNC sequence binds two SMAD1/5 MH1 domains, while the GTCT or GNCV binds the SMAD4 MH1 domain (Fig. 1B). The best-defined SMAD1/5-binding site is GGCGCC, which is present in the enhancers of well-characterized BMP-responsive genes, such as the ID and VENTX genes (Korchynskyi and ten Dijke 2002; Blitz and Cho 2009; Nakahiro et al. 2010; Morikawa et al. 2011). A lower affinity variant of this sequence, GGAGCC, was also identified in a ChIP-seq experiment investigating BMP-responsive elements in endothelial cells and pulmonary arterial smooth muscle cells (Morikawa et al. 2011).
Crystal structures have been solved for the MH1 domains of SMAD1, SMAD3, and SMAD4 bound to the palindromic GTCTAGAC sequence, and they revealed a novel protein fold (reviewed in Macias et al. 2015). In all cases, an 11-residue β-hairpin was shown to be responsible for DNA sequence recognition, and is embedded in the major groove of DNA (Fig. 1C–E) (Shi et al. 1998; Baburajendran et al. 2010, 2011). The amino acids responsible for specific contacts with the bases are conserved in all three MH1 domains (Fig. 1C–F). SMAD2 does not bind DNA because of an insert encoded by exon 3 just upstream of the DNA-binding β-hairpin (Shi et al. 1998). An isoform lacking exon 3 (SMAD2Δexon3) binds DNA equivalently to SMAD3 (Yagi et al. 1999).
The crystal structure of the SMAD1 MH1 domain revealed that the DNA contact interface of SMAD1 is rearranged compared with that of SMAD3, and, as a consequence, several key DNA backbone interactions are inhibited (Baburajendran et al. 2010). In the SMAD1 MH1 domain, the amino-terminal α1 helix is dislodged from the intramolecular binding site seen in the SMAD3 MH1 domain (Fig. 1E). This results in a decreased affinity for DNA, but additional cooperativity when binding to palindromic DNA motifs (Baburajendran et al. 2010). Interestingly, isolated MH1 domains of SMAD1 can bind to GC-rich compressed palindromes, such as GGCGCC, as efficiently as they do to GTCT palindromes, suggesting that the preferential binding of SMAD1/5–SMAD4 complexes to the GC-rich motifs in vivo is not determined by DNA selectivity of the MH1 domains per se (Baburajendran et al. 2010). What exactly determines the DNA-binding specificity of different types of SMAD complexes (SMAD1/5–SMAD4 versus SMAD3–SMAD4 complexes) in vivo is still not fully understood, but the cooperative nature of the SMAD1 MH1 DNA binding may be crucial.
Several different posttranslational modifications of the SMADs have been shown to regulate DNA-binding activity. Acetylation of SMAD3 and SMAD2Δexon 3 on Lys19 by p300 and the highly related HAT CREB-binding protein (CREBBP) promoted DNA binding both in vitro and in vivo as well as subsequent transcriptional activation (Simonsson et al. 2006). Monomeric SMADs are thought to exist in an inhibitory conformation, with the MH1 domain forming an intramolecular interaction with the carboxy-terminal MH2 domain (Hata et al. 1997). It was proposed that acetylation of Lys19 in SMAD2Δexon3 promotes DNA binding by releasing the MH1 domain from this inhibitory interaction. Another posttranslational modification that regulates DNA binding of SMAD3–SMAD4 complexes is poly-ADP-ribosylation (PARylation), which is induced by poly(ADP-ribose) polymerase-1 (PARP1) (Lönn et al. 2010). Both SMAD3 and SMAD4 were found to be PARylated, and in SMAD3, the sites of PARylation were mapped to Glu50 and Glu52 in the MH1 domain. SMAD3 and SMAD4 PARylation causes dissociation of SMAD3–SMAD4 complexes from DNA, leading to reduced transcriptional activity. How SMAD PARylation is regulated is not yet known. A third posttranslational modification that negatively regulates DNA binding is monoubiquitylation, which again induces disruption of DNA-bound SMAD complexes. Monoubiquitylation at Lys519 and, to a lesser extent, Lys507 in the MH2 domain of SMAD4 by the E3 ubiquitin ligase TIF1γ (transcriptional intermediary factor 1γ, also known as TRIM33 or ectodermin) disrupts the formation of SMAD2/3–SMAD4 complexes and as a result their ability to bind DNA (Dupont et al. 2009). This mechanism will be discussed in more detail below.
SMAD-INTERACTING TRANSCRIPTION FACTORS
The affinity of SMADs for DNA is weak (Kd ≈ 1 × 10−7 M) and, in the case of the long form of SMAD2, negligible (Shi et al. 1998). Thus, high-affinity and high-specificity recruitment of SMADs to DNA usually requires additional transcription factors. As discussed in this section, these cooperating transcription factors play a number of different biological roles, and can be either activators or repressors (for tables summarizing SMAD-interacting transcription factors, see Feng and Derynck 2005; Ross and Hill 2008). In some cases, they are absolutely required for recruitment of activated SMAD complexes to DNA, whereas, in others, they may bind cooperatively with the SMADs at enhancers. Moreover, they may be themselves targets of other signaling pathways or cell-type-specific factors and, thus, their cooperativity with the SMADs as enhancers functions to integrate information about extracellular signals or cell lineage with the signals from TGF-β family ligands. Other transcription factors act as pioneer factors to open up the chromatin allowing the activated SMAD complexes access to their binding sites. Finally, examples of the so-called self-enabling mechanism will be discussed. This is a mechanism whereby activated SMAD complexes induce expression of a transcriptional regulator that subsequently interacts with the SMAD complexes at the enhancers of other target genes, resulting in their transcriptional activation/repression.
Transcription Factors Interacting with SMAD2/3
The first SMAD-interacting transcription factor identified was Xenopus Foxh1 (originally called FAST-1), which was shown to be absolutely required in Xenopus embryos for recruitment of an activated Smad2–Smad4 complex to an activin-responsive element in the Xenopus mix.2 gene (Chen et al. 1996, 1997). Binding sites for Foxh1–Smad complexes have subsequently been found in numerous mouse genes during embryonic development and in NODAL-responsive genes in human and fish (Silvestri et al. 2008; Yoon et al. 2011; Lenhart et al. 2013). The concept that different SMAD-interacting transcription factors could dictate the cell-type specificity of signals from TGF-β family ligands then emerged when members of the AP1 family of transcription factors (FOS and JUN) were shown to cooperate with SMAD3–SMAD4 complexes to activate transcription from 12-O-tetradecanoyl-13-acetate (TPA)-responsive gene promoter elements (TREs) (Derynck et al. 1998; Zhang et al. 1998). This concept was subsequently validated with the discovery of a number of other transcription factors that can recruit SMAD complexes to DNA including homeodomain-containing transcription factors of the Xenopus Mix family (Mixer, Bix2, and Bix3). These were shown to interact with Smad2 via a SMAD interaction motif that is also present in Foxh1 (Germain et al. 2000; Randall et al. 2002). Importantly, in zebrafish, loss of both Foxh1 and Mixer resulted in a phenotype consistent with a substantial reduction of Nodal activity (Kunwar et al. 2003). Complete loss of Nodal signaling in zebrafish, however, results in a more severe phenotype, suggesting that additional Smad2-recruiting transcription factors exist (Kunwar et al. 2003; Slagle et al. 2011).
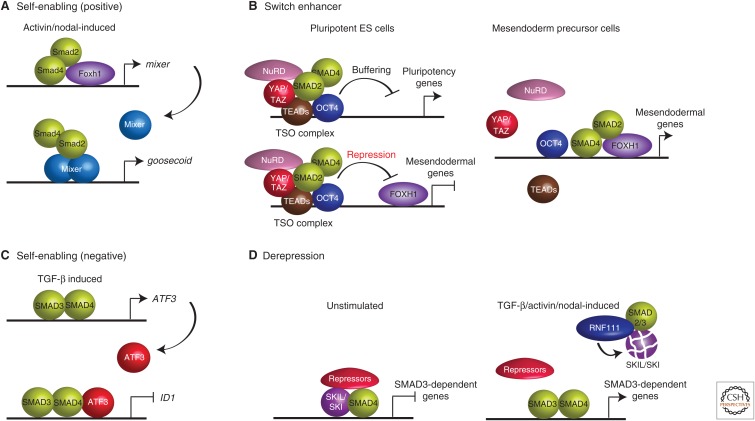
Interestingly, the Mixer–Smad interaction in Xenopus and zebrafish is an example of what was subsequently termed the self-enabling phenomenon (see below) (Kang et al. 2003). The mixer gene itself is induced by Nodal via Foxh1–Smad2–Smad4 complexes and then Mixer subsequently recruits the Smad complexes to the enhancers of target genes, examples of which are goosecoid and casanova (Fig. 2A) (Germain et al. 2000; Kunwar et al. 2003; Ober et al. 2003).
Figure 2.
Examples of different modes of SMAD-mediated transcriptional regulation. (A) Self-enabling (positive). In Xenopus, mixer is a NODAL/activin-induced gene that is induced via a Foxh1–SMAD2–SMAD4 complex. Mixer itself then recruits SMAD2–SMAD4 complexes to the goosecoid enhancer (Germain et al. 2000). (B) Switch enhancer. In pluripotent human embryonic stem cells (ESCs), the TSO complex together with the NuRD complex acts to inhibit transcription of mesendodermal genes and to buffer pluripotency genes. On differentiation to mesendoderm precursor cells, the TSO complex is disrupted and the SMADs bind FOXH1 to activate transcription of mesendodermal genes. (Panel based on Beyer et al. 2013.) (C) Self-enabling (negative). ATF3 is a TGF-β-induced gene that is induced via SMAD3–SMAD4 complexes. ATF3 then cooperates with SMAD3 and SMAD4 to repress transcription of ID1 (Kang et al. 2003). (D) Derepression. SKI and SKIL bind with SMAD4 together with corepressors to SMAD-binding elements (SBEs) to keep SMAD3-dependent genes repressed. On TGF-β/activin/NODAL signaling, RNF111 (Arkadia) is recruited to SKI/SKIL via activated SMAD2/3 to induce rapid degradation of SKI/SKIL. This exposes the SBEs to allow binding of activated SMAD3–SMAD4 complexes for transcriptional activation (Levy et al. 2007).
The idea of lineage-specific SMAD-interacting transcription factors determining cell-type responses was elaborated when whole-genome ChIP-seq showed that SMAD3 co-occupies the genome with master transcription factors, such as OCT4 (now called POU5F1), SOX2, and NANOG in embryonic stem cells (ESCs), MYOD in myoblasts, and PU.1 in pro-B cells (Mullen et al. 2011). However, it is still not clear whether these transcription factors actively recruit the SMAD complexes, or whether they act as pioneer factors to create a favorable chromatin environment that allows SMAD complexes to bind (Lee et al. 2013; Leichsenring et al. 2013).
In ESCs, NODAL/activin signaling via SMAD2/3 is required for both pluripotency and for differentiation into the mesendoderm. SMAD-interacting transcription factors have recently been shown to underlie the signaling specificity that determines these different cell fates (Beyer et al. 2013). Consistent with the study described above, these investigators showed that, in pluripotent ESCs, NODAL/activin signaling regulates the expression of pluripotency genes through OCT4–SMAD2/3 complexes. In addition, though, they found that transcriptional effectors of the Hippo pathway (TAZ/YAP/TEADs) were also present (Beyer et al. 2013), and they named the resulting complexes TSO for TEAD, SMAD, and OCT4. Because loss of TSO resulted in an increase in the levels of pluripotency genes, the investigators concluded that this complex had an inhibitory effect, which they refer to as “buffering” (Fig. 2B). Interestingly, TSO complexes were also present on mesendodermal genes, close to occupied FOXH1-binding sites. However, in this case, these genes were kept repressed (Fig. 2B). In both cases, the corepressor nucleosome remodeling deacetylase (NuRD) complex was thought to be responsible for the transcriptional repression. When cells were switched to differentiation conditions, the TSO complex was disrupted and the activated SMAD complexes were able to interact with FOXH1 and activate the mesendodermal genes (Fig. 2B). This “switch enhancer” mechanism is an attractive one to explain how one pathway can result in multiple outputs, although it still begs a number of questions, the most important of which is the mechanism underlying TSO disruption on cell differentiation (Beyer et al. 2013).
The discussion above focused on transcription factors that actively recruit the SMADs to DNA. In other cases, transcription factors bind adjacent to repeated SBEs and synergize with the SMADs to activate transcription, in some instances acting to integrate TGF-β signals with those from other signaling pathways. Examples of the former are FOXO1/3/4, HMGA2 (high-mobility group A protein 2, now called HMGB3) and C/EBPβ (CCAAT/enhancer-binding protein β, now called CEBPB) (Seoane et al. 2004; Gomis et al. 2006; Thuault et al. 2008). FOXO1/3/4 is required for TGF-β-induced activation of CDKN1A (which encodes p21CIP1/WAF1), stress-response genes, and genes involved in adaptive cell signaling (Seoane et al. 2004; Gomis et al. 2006). HMGA2 associates with TGF-β-induced SMAD3–SMAD4 to activate SNAI1 (Thuault et al. 2008), and C/EBPβ, which is required for TGF-β-induced activation of CDKN2B (which encodes p15INK4b) and repression of MYC (Gomis et al. 2006). AP1 family members and SP1 have also been shown to cooperate with SMAD2/3 (Feng and Derynck 2005; Ross and Hill 2008), and this list has expanded with the generation of whole-genome ChIP-on-chip or ChIP-seq datasets for SMAD2/3 and SMAD4 to include ETS factors, TFAP2A, OCT1 (now called POU2F1), GATA6, HNF4A, and E2F (Koinuma et al. 2009a,b; Qin et al. 2009; Liu et al. 2011; Mizutani et al. 2011).
Cooperating transcription factors can also function to integrate another signaling pathway with TGF-β signaling. An example of this phenomenon is the interaction between p53 (now called TP53) and SMAD2/3, which only occurs when p53 is phosphorylated in response to receptor tyrosine kinase (RTK)/Ras/MAP kinase signaling (Cordenonsi et al. 2007). Phosphorylation occurs on Ser6 and Ser9 of p53 and is catalyzed by the δ and ɛ isoforms of casein kinase 1 (CK1). p53 is thought to bind to its cognate site and to interact with SMAD2/3 via their MH1 domains; the SMAD complex is anchored through SMAD4 at an adjacent SBE, enabling SMAD2/3 to act as a bridge between p53 and the SBE (Elston and Inman 2012). p53 and SMAD3 cooperation has also been reported to integrate redox signals with TGF-β signaling (Overstreet et al. 2014). Another example of integration between signaling pathways is between TGF-β/NODAL/activin signaling and Wnt signaling through SMAD–β-catenin cooperation at the enhancers of siamois and xtwn genes during Xenopus gastrulation and at MSX2, MYC, and CTGF enhancers in mammalian cells (Attisano and Wrana 2013). TGF-β signaling also cooperates with the Hippo pathway through interactions between the SMADs and TAZ/YAP (Varelas et al. 2008). Finally, SMADs have been shown to actively direct the binding of other transcriptional regulators, as in the context of the TGF-β-induced liver fibrotic response in which TGF-β-induced SMAD3 directs the binding of ligand-bound nuclear vitamin D receptor (Ding et al. 2013).
The examples of SMAD2/3-interacting transcription factors given above all promote transcriptional activation. The opposite has also been shown to occur, with transcription regulators cooperating with SMAD2/3 to induce transcriptional repression. One of the first examples of this involved RUNX2 (formerly known as CBFA1). RUNX2 was shown to cooperate with SMAD3 to repress transcription of the BGLAP (formerly called osteocalcin) gene (Alliston et al. 2001; Kang et al. 2005). In this case, phosphorylated SMAD3 recruits two class IIa histone deacetylases (HDAC) 4 and 5 to deacetylate histone H4, thus conferring transcriptional repression (Kang et al. 2005) (see also discussion below concerning chromatin-remodeling enzymes). ATF3 (activating transcription factor 3) is another example of a transcription factor that recruits activated SMAD3-containing complexes to repress transcription. ATF3 is itself a TGF-β target gene and was shown to cooperate with SMAD3 to repress ID1 expression in a delayed response to TGF-β. This was defined as a self-enabling response because of its feedforward nature (Fig. 2C) (Kang et al. 2003). Additional examples of SMAD-mediated repression are mediated by the SNAI1–SMAD3–SMAD4 complex that has been shown to repress CAR/CXADR and CDH1, which encode tight junction and adherens junction proteins, respectively, during TGF-β-driven epithelial-to-mesenchymal transition (EMT) in breast epithelial cells (Vincent et al. 2009), and the SMAD3–E2F4/5–DP1–p107 complex that represses MYC transcription in response to TGF-β (Chen et al. 2002). Finally, an alternative mode of SMAD-mediated repression has been shown to occur in the context of TGF-β-mediated inhibition of skeletal muscle differentiation. In this case, instead of active SMAD complexes directly mediating transcriptional repression, activated SMAD3 interferes with an active transcription factor complex (Liu et al. 2001). In particular, SMAD3 was shown to interact with the helix–loop–helix domains of MYOD or MYOG (myogenin), which interferes with the heterodimerization of these factors with their obligatory partner E12/47. This decreases their DNA-binding affinity and results in repression of transcription from muscle-specific enhancers.
Transcription Factors Interacting with SMAD1/5
The best understood SMAD1/5-interacting transcription factor is Schnurri. Originally described in Drosophila (Arora et al. 1995; Grieder et al. 1995; Staehling-Hampton et al. 1995), Schnurri binding to the SMAD1/5–SMAD4- binding sites GRCGNC-N5-GTCT described above is thought explain the requirement for the N5 spacing (Fig. 1B) (Pyrowolakis et al. 2004). In Drosophila, Schnurri is predominantly found to be a repressor, and, importantly, it was shown that the other class of SMAD1/5–SMAD4-binding sites GGCGCC-AN4-GNCV cannot bind Schnurri and, hence, function as activating sites (Weiss et al. 2010). The best-known target of the Schnurri–SMAD complex is the gene encoding the transcriptional repressor Brinker (Pyrowolakis et al. 2004; Affolter et al. 2008). Mad–Medea–Schnurri complexes (in which Mad and Medea are Drosophila SMAD1 and SMAD4, respectively) repress Brinker expression in response to the Drosophila BMP ligand Decapentaplegic (Dpp). Brinker itself is responsible for repressing many other Dpp-responsive genes as its binding site (GGCGYY) completely overlaps with a subset of Mad (SMAD1/5)-binding sites (GRCGNC). Thus, repression of Brinker expression by Dpp results in activation of these target genes (Affolter et al. 2008). Although, for the most part, Schnurri is thought to be a repressor in Drosophila, there is one example where it acts as an activator, in the Dpp-induced transcription of the Ultrabithorax (Ubx) gene (Dai et al. 2000). The GRCGNC-N5-GTCT-binding sites for SMAD1/5–SMAD4 complexes are conserved in vertebrates and there are three vertebrate homologs of Schnurri. However, in vertebrates, the Schnurri homologs appear to act as transcriptional activators (Yao et al. 2006; Blitz and Cho 2009).
Another SMAD1/5-interacting transcription factor is RUNX2, mentioned above, which is essential for bone formation. It has been shown to be responsible, together with SMAD1, for BMP-induced transcription of a number of genes involved in osteoblast formation, such as BGLAP (Osteocalcin), SPP1 (Osteopontin), COL10A1 (Colx), and SMAD6 (Zhang et al. 2000; Drissi et al. 2003; Wang et al. 2007; Blitz and Cho 2009). In another example of signal integration, the collaboration between RUNX2 and SMAD1 on the COL10A1 promoter in chick chondrocytes integrates signals from BMP2 and retinoic acid (Drissi et al. 2003). SMAD1 has also been shown to interact with ZNF423 (OAZ), HOXC8, NKX3-2, YY1, β-catenin/Lef1 complex, and GATA4/5/6 (Blitz and Cho 2009). Furthermore, consistent with the findings described above for SMAD3, SMAD1 also appears to bind on a genome-wide scale with lineage-specific transcription factors, such as the myeloid lineage regulator CEBPA (C/EBPα) and the erythroid regulators GATA1/2 (Trompouki et al. 2011).
SKI and SKIL Are Transcriptional Repressors that Interact with R-SMADs and SMAD4
SKI and SKIL (formerly called SnoN) are important transcriptional repressors that function in the TGF-β family pathways, but their mechanism of action is distinctly different from those described above for other repressors. SKI and SKIL bind SMAD4 and SMAD2/3 (Luo 2004), and, strikingly, both SKI and SKIL are rapidly degraded on TGF-β stimulation (Stroschein et al. 1999; Sun et al. 1999). SKI and SKIL were shown by DNA-binding site selection to bind to GTCTAGAC elements (Nicol and Stavnezer 1998), which were subsequently identified as the SBEs described above. The relevance of this became clear when SMAD4 and SKIL were shown to form a complex on repeated SBEs in the absence of signal, which functions to keep target genes repressed (Stroschein et al. 1999), as a result of recruitment of corepressors such as NCOR (formerly N-CoR) or SIN3A (Luo 2004). After TGF-β induction, SKIL is rapidly degraded, exposing the SBEs and allowing activated SMAD3–SMAD4 complexes to bind (Stroschein et al. 1999). It is likely that SKI also functions via this mechanism (Le Scolan et al. 2008; Deheuninck and Luo 2009). Moreover, SKIL is a TGF-β-induced target gene and, thus, its levels recover gradually after the TGF-β signal fades, and it can once more repress TGF-β target genes (Fig. 2D). The E3 ubiquitin ligase responsible for ligand-induced degradation of SKI and SKIL is RNF111 (formerly called Arkadia) (Levy et al. 2007; Nagano et al. 2007; Le Scolan et al. 2008). Consistent with the binding specificity of SKI and SKIL for SBEs, only SMAD3- or SMAD2Δexon3-dependent TGF-β responses were found to be dependent on RNF111 (Levy et al. 2007). Whereas the role of the SKIL/SKI–SMAD4 interaction appears to be DNA binding, the activated SMAD2/3–SKIL/SKI interaction is important for RNF111-induced SKIL/SKI degradation (Levy et al. 2007). In addition, an alternative mechanism for SKIL/SKI-mediated repression has been suggested and involves these repressors directly binding DNA with transcriptionally inactive complexes of phosphorylated SMAD2/3 and SMAD4 (Wu et al. 2002). This may be relevant at longer times after ligand induction, because in most cell types SKI and SKIL are degraded within 30 min of TGF-β/NODAL/activin stimulation, and levels of SKIL in particular recover quickly as they are induced in response to ligand (Stroschein et al. 1999; Levy et al. 2007; Le Scolan et al. 2008). SKIL binding with activated SMAD2/3-containing complexes has been proposed to explain repression of mesendodermal genes in human ESCs (Tsuneyoshi et al. 2012).
SMAD INTERACTIONS WITH COACTIVATORS AND COREPRESSORS LEAD TO CHROMATIN REMODELING AND DEFINE TRANSCRIPTIONAL OUTPUT
Coactivators
As mentioned above, transcription factors may either function by promoting assembly of the PIC, by recruiting the Mediator and/or by recruiting coregulators that modify chromatin structure. The first hints as to how the SMADs may be acting came with the discovery that activated R-SMADs and SMAD4 could bind the HAT, p300, and this was required for TGF-β-induced gene expression (Feng et al. 1998; Janknecht et al. 1998; Pouponnot et al. 1998; de Caestecker et al. 2000). Subsequently, another HAT, GCN5, was identified as a direct binding partner of SMAD2/3 and a coactivator of TGF-β signaling (Kahata et al. 2004). SMAD recruitment of HATs seems to be a general phenomenon as GCN5 also potentiates BMP signaling (Kahata et al. 2004). A close relative of GCN5 is the HAT P/CAF (p300/CBP-associated factor), which has been shown to interact with SMAD2/3 and potentiate TGF-β signaling, although not BMP signaling (Itoh et al. 2000). As well as being able to recruit HATs to chromatin, the SMADs can bind Mediator components. A direct interaction between SMAD2 and the Mediator component ARC105 (activator-recruited cofactor 105), and ARC105 was shown to be important for SMAD2-mediated transcription in Xenopus embryos (Kato et al. 2002).
Several years after these initial observations, more insights as to how the SMADs regulate transcription were gleaned from in vitro transcription studies. Using recombinant phosphorylated SMAD2–SMAD4 (pSMAD2–SMAD4) complexes, it was shown that despite binding efficiently, the SMADs showed no transcriptional activity on naked DNA templates, unlike, for instance, p53, suggesting that the SMADs were unable to promote assembly of the PIC (Ross et al. 2006). In contrast, the same SMAD complexes efficiently activated transcription from chromatin templates, leading to the proposal that the SMADs function predominantly via chromatin remodeling (Ross et al. 2006). This fitted with the requirement for p300 and other HATs and it was shown that the HAT activity of p300 was indeed required for SMAD2-dependent transcription (Ross et al. 2006). Intriguingly, binding of p300 to pSMAD2–SMAD4 complexes appeared to affect its specificity, as in this context it preferentially acetylated nucleosomal histone H3, but did not acetylate histone H4. In contrast, when recruited by p53, p300 acetylated both H3 and H4 (Ross et al. 2006). How the SMADs influence the specificity of p300 is still not known.
As well as recruiting HATs, the pSMAD2–SMAD4 complex also interacts with the ATPase SMARCA4 (formerly called BRG1), which is a component of the nucleosome-remodeling complex, SWI/SNF. SMARCA4 interacts with SMAD2 in a TGF-β-dependent manner (Ross et al. 2006). It was shown to bind to SMAD2-dependent enhancers and is specifically required for TGF-β-induced expression of endogenous SMAD2 target genes, such as Lefty1 in the mouse embryonic carcinoma cell line P19. Another study extended these findings by showing on a genome-wide scale that SMARCA4 is required for most TGF-β gene responses in human keratinocytes, although to different extents (Xi et al. 2008). Interestingly, TGF-β-induced transcription of SKIL and SMAD7, which encodes key negative regulators in the pathway, was not dependent on SMARCA4. The precise role that SWI/SNF plays at the enhancers of TGF-β-induced target genes has yet to be defined. Intriguingly, its knockdown had a stronger effect on acute TGF-β-induced transcription compared with that elicited by chronic signaling, suggesting that it is required for initial transcriptional activation, but not its maintenance (Ross et al. 2006).
Another key coregulator of SMAD complexes is TIF1γ, for which two different modes of action have been proposed. As discussed above, TIF1γ was shown to be an E3 ubiquitin ligase that monoubiquitylates SMAD4 on its MH2 domain, causing disruption of active SMAD complexes. Its carboxy-terminal tandem PHD finger-bromodomain was shown to specifically recognize histone H3 tails unmodified at Arg2 and Lys4 and acetylated on two lysines, preferentially Lys18 and Lys23 (Agricola et al. 2011). In its inactive state, TIF1γ is autoinhibited with its amino-terminal RING domain forming intramolecular interactions with the PHD finger-bromodomain. Recruitment to chromatin releases the RING domain and activates the E3 ubiquitin ligase activity. The proposed functional model involves SMAD complexes binding to SBEs and recruiting HATs, such as p300 to acetylate H3 tails. These tails then bind TIF1γ, which ubiquitylates SMAD4, disrupting the SMAD complexes. In this model, TIF1γ regulates the residence time of SMAD complexes on the chromatin (Agricola et al. 2011). It is likely that this sequence of events is repeated numerous times during the process of transcriptional activation by the SMADs, with the likelihood that the histone acetylation is also reversed by HDACs. In another study, a different model of TIF1γ action was proposed, which does not involve its E3 ubiquitin ligase activity at all (Xi et al. 2011). In that study in mouse ESCs, TIF1γ was shown to interact with phosphorylated SMAD2/3 in the absence of SMAD4 and to be recruited to upstream regions of the mesendoderm genes, Gsc and Mixl1, by virtue of an interaction between the PHD finger-bromodomain with H3 tails that are unmethylated at Lys4, acetylated at Lys18, and trimethylated at Lys9. This was shown to cause displacement of the H3K9me3-interacting protein HP1 and subsequent remodeling of the chromatin template to allow activated SMAD2/3–SMAD4 complexes to bind to their enhancer sites at these target genes (Xi et al. 2011). This model leaves open several questions, most specifically how the TIF1γ–SMAD2/SMAD3 complex is initially recruited in a sequence-specific manner, and then how this leads to chromatin remodeling to allow the canonical SMAD2/3–SMAD4 complexes to bind.
In ESCs, genes that are up-regulated in response to NODAL/activin signaling are frequently trimethylated in the basal state at H3K27, a modification that is catalyzed by the polycomb complex PRC2 (Muller and Verrijzer 2009). This mark is often found in conjunction with H3K4me3, and, as a result, these regions of chromatin are referred to as bivalent domains and thought to reflect an inactive yet poised/primed state (Voigt et al. 2013). In ChIP-seq studies in hESCs and mESCs, NODAL/activin induction of specific genes, such as Nodal itself and the mesendodermal genes Eomes and Gsc, has been shown to be associated with loss of H3K27me3 (Dahle et al. 2010; Kim et al. 2011). Demethylation of H3K27me3 in response to activin/NODAL has been thought to be mediated by the enzyme JMJD3 (jumonji domain containing 3), as SMAD2/3 can bind JMJD3 in immunoprecipitations, and JMJD3 is recruited on activin signaling to the SMAD2/3-binding sites in the regulatory regions of the Gsc and Eomes genes (Kim et al. 2011). However, the timing of JMJD3 recruitment relative to the removal of the H3K27me3 mark has not been investigated, nor is it yet known how general this mechanism is. SMAD signaling also controls methylation of H3K4. In hESCs, NODAL/activin signaling through SMAD2/3 cooperates with the pluripotency factor NANOG to recruit DPY30, which is a subunit of the histone methyltransferase complex COMPASS to promote H3K4 trimethylation at promoters of pluripotency and mesendoderm genes (Bertero et al. 2015).
Finally, a completely novel mechanism of transcriptional activation was uncovered where TGF-β was found to activate CDKN2B by inducing active demethylation of the promoter DNA (Thillainadesan et al. 2012). CDKN2B is repressed and methylated in the absence of signal by a complex comprising the corepressor complex CoREST (Laugesen and Helin 2014), together with the oncoprotein ZNF217 and the DNA methylase DNMT3A. TGF-β triggers active DNA demethylation in a process that depends on loss of ZNF217/CoREST/DNMT3A and recruitment of SMAD2/3, the HAT CBP, and the DNA glycosylase TDG with the deaminase, AID. DNA demethylation is thought to occur via the base excision repair machinery. This is the first example of such a mechanism, and it will be intriguing to discover whether other genes are regulated in a similar manner.
Corepressors
Transcriptional repression can be achieved by reversing histone modifications elicited by coactivators, for example, by recruitment of HDACs to remove activating histone acetylations, as well as by inducing repressive histone marks. Recruitment of HDACs can occur through interactions with the SMADs themselves, as in the example given above, in which phosphorylated SMAD3 recruits HDAC4/5 to mediate transcriptional repression of BGLAP (Kang et al. 2005). In other cases, the SMADs interact with corepressors to bring about transcriptional repression. One of the earliest examples of such a corepressor was TGIF (TG-interacting factor), which was shown to be recruited by SMAD2 to TGF-β/NODAL/activin-responsive elements and repressed transcription (Wotton et al. 1999). It mediated transcriptional repression by recruiting HDACs and inhibiting the ability of activated SMAD2 to bind p300. The idea that TGIF had a role in limiting NODAL responses was strengthened when it was shown that mice lacking Tgif1 and Tgif2 fail in gastrulation, a defect that can be partially rescued by reducing levels of NODAL (Powers et al. 2010). Moreover, conditional deletion of Tgif1/2 results in defects in left–right asymmetry, which were also alleviated by reducing the dose of NODAL (Powers et al. 2010). Other examples of corepressors that function with the SMADs are Evi-1, which cooperates with carboxy-terminal-binding protein (CtBP) to repress TGF-β, BMP, and activin-induced transcription (Izutsu et al. 2001; Alliston et al. 2005), and ZNF451, which blocks the ability of SMAD3/4 to recruit p300 in response to TGF-β (Feng et al. 2014). The corepressor responsible for transcriptional repression may also be part of a larger repressive chromatin-remodeling complex. One recent example of this concerns the TSO complex described above, which inhibits mesendodermal gene expression and buffers pluripotency gene expression in human ESCs (Beyer et al. 2013). In this case, a proteomic approach was used to identify the NuRD complex as being responsible for this transcriptional repression.
PERSPECTIVES
In summary, the main message that emerges from this review is how versatile the SMADs are in their nuclear functions. The TGF-β family ligands have very pleiotropic effects and this is likely because of the fact that the SMADs can cooperate with an impressive array of transcription factors, coactivators, and corepressors (Feng and Derynck 2005; Ross and Hill 2008). It is interesting to speculate that key to this versatility may be the short DNA-binding site, because it allows the SMADs to derive the specificity for its functions primarily from the interacting factors, rather than from nucleotide sequence per se.
We are now in a position to appreciate the range of SMAD partners that cooperate in their function as transcriptional regulators, and also some of the underlying mechanisms used by the SMADs. Expression profiling and ChIP-seq experiments have identified SMAD target genes and SMAD-binding sites on a genome-wide scale in different cellular contexts, and some chromatin and transcriptional signatures associated with differentiation processes driven by the SMAD pathways (e.g., see Kim et al. 2011; Mullen et al. 2011; Beyer et al. 2013). However, this work has for the most part focused on autocrine signaling or inductions with ligands that were performed on a time scale of days, rather than minutes or hours, which is the sort of temporal resolution required. What we currently lack is knowledge of the exact sequence of events that occur from the activated SMADs arriving in the nucleus to the induction or repression of transcription of target genes. We need to understand the chromatin landscape that allows the SMADs to bind to specific sites in the first place and how this is set up in a cell-type-specific manner. It will then be crucial to know what enzymes the SMADs bring in to remodel that landscape, and in what order, to execute appropriate transcriptional responses of the correct amplitude and duration.
ACKNOWLEDGMENTS
I thank members of the Hill laboratory for useful comments on the manuscript. I thank Raphael Chaleil for preparing panels C through E in Figure 1. Work in the Hill laboratory is funded by the Francis Crick Institute (Grant No. FCI01), which receives its core funding from Cancer Research UK, UK Medical Research Council, and the Wellcome Trust. Hill laboratory work is also funded by the European Commission Network of Excellence EpiGeneSys (HEALTH-F4-2010-257082).
Footnotes
Editors: Rik Derynck and Kohei Miyazono
Additional Perspectives on The Biology of the TGF-β Family available at www.cshperspectives.org
REFERENCES
- Affolter M, Pyrowolakis G, Weiss A, Basler K. 2008. Signal-induced repression: The exception or the rule in developmental signaling? Dev Cell 15: 11–22. [DOI] [PubMed] [Google Scholar]
- Agricola E, Randall RA, Gaarenstroom T, Dupont S, Hill CS. 2011. Recruitment of TIF1γ to chromatin via its PHD finger-bromodomain activates its ubiquitin ligase and transcriptional repressor activities. Mol Cell 43: 85–96. [DOI] [PubMed] [Google Scholar]
- Alliston T, Choy L, Ducy P, Karsenty G, Derynck R. 2001. TGF-β-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. EMBO J 20: 2254–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alliston T, Ko TC, Cao Y, Liang YY, Feng XH, Chang C, Derynck R. 2005. Repression of bone morphogenetic protein and activin-inducible transcription by Evi-1. J Biol Chem 280: 24227–24237. [DOI] [PubMed] [Google Scholar]
- Arora K, Dai H, Kazuko SG, Jamal J, O’Connor MB, Letsou A, Warrior R. 1995. The Drosophila schnurri gene acts in the Dpp/TGF β signaling pathway and encodes a transcription factor homologous to the human MBP family. Cell 81: 781–790. [DOI] [PubMed] [Google Scholar]
- Attisano L, Wrana JL. 2013. Signal integration in TGF-β, WNT, and Hippo pathways. F1000Prime Rep 5: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baburajendran N, Palasingam P, Narasimhan K, Sun W, Prabhakar S, Jauch R, Kolatkar PR. 2010. Structure of Smad1 MH1/DNA complex reveals distinctive rearrangements of BMP and TGF-β effectors. Nucleic Acids Res 38: 3477–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baburajendran N, Jauch R, Tan CY, Narasimhan K, Kolatkar PR. 2011. Structural basis for the cooperative DNA recognition by Smad4 MH1 dimers. Nucleic Acids Res 39: 8213–8222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel C, Paro R. 2011. Silencing chromatin: Comparing modes and mechanisms. Nat Rev Genet 12: 123–135. [DOI] [PubMed] [Google Scholar]
- Bertero A, Madrigal P, Galli A, Hubner NC, Moreno I, Burks D, Brown S, Pedersen RA, Gaffney D, Mendjan S, et al. 2015. Activin/Nodal signaling and NANOG orchestrate human embryonic stem cell fate decisions by controlling the H3K4me3 chromatin mark. Genes Dev 29: 702–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer TA, Weiss A, Khomchuk Y, Huang K, Ogunjimi AA, Varelas X, Wrana JL. 2013. Switch enhancers interpret TGF-β and Hippo signaling to control cell fate in human embryonic stem cells. Cell Rep 5: 1611–1624. [DOI] [PubMed] [Google Scholar]
- Blitz IL, Cho KW. 2009. Finding partners: How BMPs select their targets. Dev Dyn 238: 1321–1331. [DOI] [PubMed] [Google Scholar]
- Calo E, Wysocka J. 2013. Modification of enhancer chromatin: What, how, and why? Mol Cell 49: 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacko BM, Qin B, Correia JJ, Lam SS, de Caestecker MP, Lin K. 2001. The L3 loop and C-terminal phosphorylation jointly define Smad protein trimerization. Nat Struct Biol 8: 248–253. [DOI] [PubMed] [Google Scholar]
- Chen X, Rubock MJ, Whitman M. 1996. A transcriptional partner for MAD proteins in TGF-β signalling. Nature 383: 691–696. [DOI] [PubMed] [Google Scholar]
- Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. 1997. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature 389: 85–89. [DOI] [PubMed] [Google Scholar]
- Chen CR, Kang Y, Siegel PM, Massagué J. 2002. E2F4/5 and p107 as Smad cofactors linking the TGFβ receptor to c-myc repression. Cell 110: 19–32. [DOI] [PubMed] [Google Scholar]
- Cordenonsi M, Montagner M, Adorno M, Zacchigna L, Martello G, Mamidi A, Soligo S, Dupont S, Piccolo S. 2007. Integration of TGF-β and Ras/MAPK signaling through p53 phosphorylation. Science 315: 840–843. [DOI] [PubMed] [Google Scholar]
- Dahle O, Kumar A, Kuehn MR. 2010. Nodal signaling recruits the histone demethylase Jmjd3 to counteract polycomb-mediated repression at target genes. Sci Signal 3: ra48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H, Hogan C, Gopalakrishnan B, Torres-Vazquez J, Nguyen M, Park S, Raftery LA, Warrior R, Arora K. 2000. The zinc finger protein schnurri acts as a Smad partner in mediating the transcriptional response to decapentaplegic. Dev Biol 227: 373–387. [DOI] [PubMed] [Google Scholar]
- de Caestecker MP, Yahata T, Wang D, Parks WT, Huang S, Hill CS, Shioda T, Roberts AB, Lechleider RJ. 2000. The Smad4 activation domain (SAD) is a proline-rich, p300-dependent transcriptional activation domain. J Biol Chem 275: 2115–2122. [DOI] [PubMed] [Google Scholar]
- Deheuninck J, Luo K. 2009. Ski and SnoN, potent negative regulators of TGF-β signaling. Cell Res 19: 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. 1998. Direct binding of Smad3 and Smad4 to critical TGFβ-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J 17: 3091–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Zhang Y, Feng XH. 1998. Smads: Transcriptional activators of TGF-β responses. Cell 95: 737–740. [DOI] [PubMed] [Google Scholar]
- Ding N, Yu RT, Subramaniam N, Sherman MH, Wilson C, Rao R, Leblanc M, Coulter S, He M, Scott C, et al. 2013. A vitamin D receptor/SMAD genomic circuit gates hepatic fibrotic response. Cell 153: 601–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissi MH, Li X, Sheu TJ, Zuscik MJ, Schwarz EM, Puzas JE, Rosier RN, O’Keefe RJ. 2003. Runx2/Cbfa1 stimulation by retinoic acid is potentiated by BMP2 signaling through interaction with Smad1 on the collagen X promoter in chondrocytes. J Cell Biochem 90: 1287–1298. [DOI] [PubMed] [Google Scholar]
- Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, Martello G, Stinchfield MJ, Soligo S, Morsut L, et al. 2009. FAM/USP9x, a deubiquitinating enzyme essential for TGFβ signaling, controls Smad4 monoubiquitination. Cell 136: 123–135. [DOI] [PubMed] [Google Scholar]
- Elston R, Inman GJ. 2012. Crosstalk between p53 and TGF-β signalling. J Signal Transduct 2012: 294097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng XH, Derynck R. 2005. Specificity and versatility in TGF-β signaling through Smads. Annu Rev Cell Dev Biol 21: 659–693. [DOI] [PubMed] [Google Scholar]
- Feng XH, Zhang Y, Wu RY, Derynck R. 1998. The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for smad3 in TGF-β-induced transcriptional activation. Genes Dev 12: 2153–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Wu H, Xu Y, Zhang Z, Liu T, Lin X, Feng XH. 2014. Zinc finger protein 451 is a novel Smad corepressor in transforming growth factor-β signaling. J Biol Chem 289: 2072–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuda NJ, Ardehali MB, Lis JT. 2009. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 461: 186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Steffen J, Laughon A. 2005. Dpp-responsive silencers are bound by a trimeric Mad-Medea complex. J Biol Chem 280: 36158–36164. [DOI] [PubMed] [Google Scholar]
- Germain S, Howell M, Esslemont GM, Hill CS. 2000. Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev 14: 435–451. [PMC free article] [PubMed] [Google Scholar]
- Gomis RR, Alarcon C, He W, Wang Q, Seoane J, Lash A, Massagué J. 2006. A FoxO–Smad synexpression group in human keratinocytes. Proc Natl Acad Sci 103: 12747–12752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieder NC, Nellen D, Burke R, Basler K, Affolter M. 1995. schnurri is required for Drosophila Dpp signaling and encodes a zinc finger protein similar to the mammalian transcription factor PRDII-BF1. Cell 81: 791–800. [DOI] [PubMed] [Google Scholar]
- Hata A, Lo RS, Wotton D, Lagna G, Massagué J. 1997. Mutations increasing autoinhibition inactivate tumour suppressors Smad2 and Smad4. Nature 388: 82–87. [DOI] [PubMed] [Google Scholar]
- Inman GJ, Hill CS. 2002. Stoichiometry of active smad-transcription factor complexes on DNA. J Biol Chem 277: 51008–51016. [DOI] [PubMed] [Google Scholar]
- Itoh S, Ericsson J, Nishikawa J, Heldin CH, ten Dijke P. 2000. The transcriptional co-activator P/CAF potentiates TGF-β/Smad signaling. Nucleic Acids Res 28: 4291–4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izutsu K, Kurokawa M, Imai Y, Maki K, Mitani K, Hirai H. 2001. The corepressor CtBP interacts with Evi-1 to repress transforming growth factor β signaling. Blood 97: 2815–2822. [DOI] [PubMed] [Google Scholar]
- Janknecht R, Wells NJ, Hunter T. 1998. TGF-β-stimulated cooperation of smad proteins with the coactivators CBP/p300. Genes Dev 12: 2114–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahata K, Hayashi M, Asaka M, Hellman U, Kitagawa H, Yanagisawa J, Kato S, Imamura T, Miyazono K. 2004. Regulation of transforming growth factor-β and bone morphogenetic protein signalling by transcriptional coactivator GCN5. Genes Cells 9: 143–151. [DOI] [PubMed] [Google Scholar]
- Kang Y, Chen CR, Massagué J. 2003. A self-enabling TGFβ response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol Cell 11: 915–926. [DOI] [PubMed] [Google Scholar]
- Kang JS, Alliston T, Delston R, Derynck R. 2005. Repression of Runx2 function by TGF-β through recruitment of class II histone deacetylases by Smad3. EMBO J 24: 2543–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Habas R, Katsuyama Y, Naar AM, He X. 2002. A component of the ARC/Mediator complex required for TGFβ/Nodal signalling. Nature 418: 641–646. [DOI] [PubMed] [Google Scholar]
- Kim SW, Yoon SJ, Chuong E, Oyolu C, Wills AE, Gupta R, Baker J. 2011. Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. Dev Biol 357: 492–504. [DOI] [PubMed] [Google Scholar]
- Koinuma D, Tsutsumi S, Kamimura N, Imamura T, Aburatani H, Miyazono K. 2009a. Promoter-wide analysis of Smad4 binding sites in human epithelial cells. Cancer Sci 100: 2133–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koinuma D, Tsutsumi S, Kamimura N, Taniguchi H, Miyazawa K, Sunamura M, Imamura T, Miyazono K, Aburatani H. 2009b. Chromatin immunoprecipitation on microarray analysis of Smad2/3 binding sites reveals roles of ETS1 and TFAP2A in transforming growth factor β signaling. Mol Cell Biol 29: 172–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korchynskyi O, ten Dijke P. 2002. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J Biol Chem 277: 4883–4891. [DOI] [PubMed] [Google Scholar]
- Kunwar PS, Zimmerman S, Bennett JT, Chen Y, Whitman M, Schier AF. 2003. Mixer/Bon and FoxH1/Sur have overlapping and divergent roles in Nodal signaling and mesendoderm induction. Development 130: 5589–5599. [DOI] [PubMed] [Google Scholar]
- Laugesen A, Helin K. 2014. Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell 14: 735–751. [DOI] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Takacs CM, Bazzini AA, Divito KR, Fleming ES, Giraldez AJ. 2013. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503: 360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichsenring M, Maes J, Mossner R, Driever W, Onichtchouk D. 2013. Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science 341: 1005–1009. [DOI] [PubMed] [Google Scholar]
- Lenhart KF, Holtzman NG, Williams JR, Burdine RD. 2013. Integration of Nodal and BMP signals in the heart requires FoxH1 to create left-right differences in cell migration rates that direct cardiac asymmetry. PLoS Genet 9: e1003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Scolan E, Zhu Q, Wang L, Bandyopadhyay A, Javelaud D, Mauviel A, Sun L, Luo K. 2008. Transforming growth factor-β suppresses the ability of Ski to inhibit tumor metastasis by inducing its degradation. Cancer Res 68: 3277–3285. [DOI] [PubMed] [Google Scholar]
- Levine M. 2011. Paused RNA polymerase II as a developmental checkpoint. Cell 145: 502–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy L, Howell M, Das D, Harkin S, Episkopou V, Hill CS. 2007. Arkadia activates Smad3/Smad4-dependent transcription by triggering signal-induced SnoN degradation. Mol Cell Biol 27: 6068–6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Black BL, Derynck R. 2001. TGF-β inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3. Genes Dev 15: 2950–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Lin X, Cai Z, Zhang Z, Han C, Jia S, Meng A, Wang Q. 2011. Global identification of SMAD2 target genes reveals a role for multiple co-regulatory factors in zebrafish early gastrulas. J Biol Chem 286: 28520–28532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lönn P, van der Heide LP, Dahl M, Hellman U, Heldin CH, Moustakas A. 2010. PARP-1 attenuates Smad-mediated transcription. Mol Cell 40: 521–532. [DOI] [PubMed] [Google Scholar]
- Luo K. 2004. Ski and SnoN: Negative regulators of TGF-β signaling. Curr Opin Genet Dev 14: 65–70. [DOI] [PubMed] [Google Scholar]
- Macias MJ, Martin-Malpartida P, Massagué J. 2015. Structural determinants of Smad function in TGF-β signaling. Trends Biochem Sci 40: 296–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J. 2012. TGFβ signalling in context. Nat Rev Mol Cell Biol 13: 616–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa K, Shinozaki M, Hara T, Furuya T, Miyazono K. 2002. Two major Smad pathways in TGF-β superfamily signalling. Genes Cells 7: 1191–1204. [DOI] [PubMed] [Google Scholar]
- Mizutani A, Koinuma D, Tsutsumi S, Kamimura N, Morikawa M, Suzuki HI, Imamura T, Miyazono K, Aburatani H. 2011. Cell-type-specific target selection by combinatorial binding of Smad2/3 proteins and hepatocyte nuclear factor 4 in HepG2 cells. J Biol Chem 286: 29848–29860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa M, Koinuma D, Tsutsumi S, Vasilaki E, Kanki Y, Heldin CH, Aburatani H, Miyazono K. 2011. ChIP-seq reveals cell-type-specific binding patterns of BMP-specific Smads and a novel binding motif. Nucleic Acids Res 39: 8712–8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa M, Koinuma D, Miyazono K, Heldin CH. 2013. Genome-wide mechanisms of Smad binding. Oncogene 32: 1609–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. 2011. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell 147: 565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Verrijzer P. 2009. Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr Opin Genet Dev 19: 150–158. [DOI] [PubMed] [Google Scholar]
- Nagano Y, Mavrakis KJ, Lee KL, Fujii T, Koinuma D, Sase H, Yuki K, Isogaya K, Saitoh M, Imamura T, et al. 2007. Arkadia induces degradation of SnoN and c-Ski to enhance transforming growth factor-β signaling. J Biol Chem 282: 20492–20501. [DOI] [PubMed] [Google Scholar]
- Nakahiro T, Kurooka H, Mori K, Sano K, Yokota Y. 2010. Identification of BMP-responsive elements in the mouse Id2 gene. Biochem Biophys Res Commun 399: 416–421. [DOI] [PubMed] [Google Scholar]
- Nicol R, Stavnezer E. 1998. Transcriptional repression by v-Ski and c-Ski mediated by a specific DNA binding site. J Biol Chem 273: 3588–3597. [DOI] [PubMed] [Google Scholar]
- Ober EA, Field HA, Stainier DY. 2003. From endoderm formation to liver and pancreas development in zebrafish. Mech Dev 120: 5–18. [DOI] [PubMed] [Google Scholar]
- Overstreet JM, Samarakoon R, Meldrum KK, Higgins PJ. 2014. Redox control of p53 in the transcriptional regulation of TGF-β1 target genes through SMAD cooperativity. Cell Signal 26: 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouponnot C, Jayaraman L, Massagué J. 1998. Physical and functional interaction of SMADs and p300/CBP. J Biol Chem 273: 22865–22868. [DOI] [PubMed] [Google Scholar]
- Powers SE, Taniguchi K, Yen W, Melhuish TA, Shen J, Walsh CA, Sutherland AE, Wotton D. 2010. Tgif1 and Tgif2 regulate Nodal signaling and are required for gastrulation. Development 137: 249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrowolakis G, Hartmann B, Muller B, Basler K, Affolter M. 2004. A simple molecular complex mediates widespread BMP-induced repression during Drosophila development. Dev Cell 7: 229–240. [DOI] [PubMed] [Google Scholar]
- Qin H, Chan MWY, Liyanarachchi S, Balch C, Potter D, Souriraj IJ, Cheng ASL, Agosto-Perez FJ, Nikonova EV, Yan PS, et al. 2009. An integrative ChIP-chip and gene expression profiling to model SMAD regulatory modules. BMC Syst Biol 3: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall RA, Germain S, Inman GJ, Bates PA, Hill CS. 2002. Different Smad2 partners bind a common hydrophobic pocket in Smad2 via a defined proline-rich motif. EMBO J 21: 145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S, Hill CS. 2008. How the Smads regulate transcription. Int J Biochem Cell Biol 40: 383–408. [DOI] [PubMed] [Google Scholar]
- Ross S, Cheung E, Petrakis TG, Howell M, Kraus WL, Hill CS. 2006. Smads orchestrate specific histone modifications and chromatin remodeling to activate transcription. EMBO J 25: 4490–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmierer B, Tournier AL, Bates PA, Hill CS. 2008. Mathematical modeling identifies Smad nucleocytoplasmic shuttling as a dynamic signal-interpreting system. Proc Natl Acad Sci 105: 6608–6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J, Le HV, Shen L, Anderson SA, Massagué J. 2004. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117: 211–223. [DOI] [PubMed] [Google Scholar]
- Shi Y, Wang YF, Jayaraman L, Yang H, Massagué J, Pavletich NP. 1998. Crystal structure of a Smad MH1 domain bound to DNA: Insights on DNA binding in TGF-β signaling. Cell 94: 585–594. [DOI] [PubMed] [Google Scholar]
- Silvestri C, Narimatsu M, von Both I, Liu Y, Tan NB, Izzi L, McCaffery P, Wrana JL, Attisano L. 2008. Genome-wide identification of Smad/Foxh1 targets reveals a role for Foxh1 in retinoic acid regulation and forebrain development. Dev Cell 14: 411–423. [DOI] [PubMed] [Google Scholar]
- Simonsson M, Kanduri M, Gronroos E, Heldin CH, Ericsson J. 2006. The DNA binding activities of Smad2 and Smad3 are regulated by coactivator-mediated acetylation. J Biol Chem 281: 39870–39880. [DOI] [PubMed] [Google Scholar]
- Slagle CE, Aoki T, Burdine RD. 2011. Nodal-dependent mesendoderm specification requires the combinatorial activities of FoxH1 and Eomesodermin. PLoS Genet 7: e1002072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehling-Hampton K, Laughon AS, Hoffmann FM. 1995. A Drosophila protein related to the human zinc finger transcription factor PRDII/MBPI/HIV-EP1 is required for dpp signaling. Development 121: 3393–3403. [DOI] [PubMed] [Google Scholar]
- Stroschein SL, Wang W, Zhou S, Zhou Q, Luo K. 1999. Negative feedback regulation of TGF-β signaling by the SnoN oncoprotein. Science 286: 771–774. [DOI] [PubMed] [Google Scholar]
- Sun Y, Liu X, Ng-Eaton E, Lodish HF, Weinberg RA. 1999. SnoN and Ski protooncoproteins are rapidly degraded in response to transforming growth factor β signaling. Proc Natl Acad Sci 96: 12442–12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thillainadesan G, Chitilian JM, Isovic M, Ablack JN, Mymryk JS, Tini M, Torchia J. 2012. TGF-β-dependent active demethylation and expression of the p15ink4b tumor suppressor are impaired by the ZNF217/CoREST complex. Mol Cell 46: 636–649. [DOI] [PubMed] [Google Scholar]
- Thuault S, Tan EJ, Peinado H, Cano A, Heldin CH, Moustakas A. 2008. HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J Biol Chem 283: 33437–33446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu DC, DiBiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, et al. 2011. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell 147: 577–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuneyoshi N, Tan EK, Sadasivam A, Poobalan Y, Sumi T, Nakatsuji N, Suemori H, Dunn NR. 2012. The SMAD2/3 corepressor SNON maintains pluripotency through selective repression of mesendodermal genes in human ES cells. Genes Dev 26: 2471–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varelas X, Sakuma R, Samavarchi-Tehrani P, Peerani R, Rao BM, Dembowy J, Yaffe MB, Zandstra PW, Wrana JL. 2008. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat Cell Biol 10: 837–848. [DOI] [PubMed] [Google Scholar]
- Vincent T, Neve EP, Johnson JR, Kukalev A, Rojo F, Albanell J, Pietras K, Virtanen I, Philipson L, Leopold PL, et al. 2009. A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-β mediated epithelial–mesenchymal transition. Nat Cell Biol 11: 943–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P, Tee WW, Reinberg D. 2013. A double take on bivalent promoters. Genes Dev 27: 1318–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield LM, Hill CS. 2013. Beyond TGFβ: Roles of other TGFβ superfamily members in cancer. Nat Rev Cancer 13: 328–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Wei X, Zhu T, Zhang M, Shen R, Xing L, O’Keefe RJ, Chen D. 2007. BMP-2 activates Smad6 gene transcription through bone-specific transcription factor Runx2. J Biol Chem 282: 10742–10748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A, Charbonnier E, Ellertsdottir E, Tsirigos A, Wolf C, Schuh R, Pyrowolakis G, Affolter M. 2010. A conserved activation element in BMP signaling during Drosophila development. Nat Struct Mol Biol 17: 69–76. [DOI] [PubMed] [Google Scholar]
- Wong C, Rougier-Chapman EM, Frederick JP, Datto MB, Liberati NT, Li JM, Wang XF. 1999. Smad3-Smad4 and AP-1 complexes synergize in transcriptional activation of the c-Jun promoter by transforming growth factor β. Mol Cell Biol 19: 1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotton D, Lo RS, Lee S, Massagué J. 1999. A Smad transcriptional corepressor. Cell 97: 29–39. [DOI] [PubMed] [Google Scholar]
- Wu JW, Fairman R, Penry J, Shi Y. 2001. Formation of a stable heterodimer between Smad2 and Smad4. J Biol Chem 276: 20688–20694. [DOI] [PubMed] [Google Scholar]
- Wu JW, Krawitz AR, Chai J, Li W, Zhang F, Luo K, Shi Y. 2002. Structural mechanism of Smad4 recognition by the nuclear oncoprotein Ski: Insights on Ski-mediated repression of TGF-β signaling. Cell 111: 357–367. [DOI] [PubMed] [Google Scholar]
- Xi Q, He W, Zhang XH, Le HV, Massagué J. 2008. Genome-wide impact of the BRG1 SWI/SNF chromatin remodeler on the transforming growth factor β transcriptional program. J Biol Chem 283: 1146–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Q, Wang Z, Zaromytidou AI, Zhang XH, Chow-Tsang LF, Liu JX, Kim H, Barlas A, Manova-Todorova K, Kaartinen V, et al. 2011. A poised chromatin platform for TGF-β access to master regulators. Cell 147: 1511–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi K, Goto D, Hamamoto T, Takenoshita S, Kato M, Miyazono K. 1999. Alternatively spliced variant of Smad2 lacking exon 3. Comparison with wild-type Smad2 and Smad3. J Biol Chem 274: 703–709. [DOI] [PubMed] [Google Scholar]
- Yao LC, Blitz IL, Peiffer DA, Phin S, Wang Y, Ogata S, Cho KW, Arora K, Warrior R. 2006. Schnurri transcription factors from Drosophila and vertebrates can mediate Bmp signaling through a phylogenetically conserved mechanism. Development 133: 4025–4034. [DOI] [PubMed] [Google Scholar]
- Yoon SJ, Wills AE, Chuong E, Gupta R, Baker JC. 2011. HEB and E2A function as SMAD/FOXH1 cofactors. Genes Dev 25: 1654–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. 2011. Pioneer transcription factors: Establishing competence for gene expression. Genes Dev 25: 2227–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, Kern SE. 1998. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell 1: 611–617. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Feng XH, Derynck R. 1998. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-β-induced transcription. Nature 394: 909–913. [DOI] [PubMed] [Google Scholar]
- Zhang YW, Yasui N, Ito K, Huang G, Fujii M, Hanai J, Nogami H, Ochi T, Miyazono K, Ito Y. 2000. A RUNX2/PEBP2α A/CBFA1 mutation displaying impaired transactivation and Smad interaction in cleidocranial dysplasia. Proc Natl Acad Sci 97: 10549–10554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE. 2011. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet 12: 7–18. [DOI] [PubMed] [Google Scholar]