Highlights
-
•
We reviewed single nucleotide polymorphisms for oral pre-cancer susceptibility.
-
•
All of them were pathway based candidate gene association studies.
-
•
The current level of evidence is very limited.
-
•
Integrated characterization of germline/somatic alterations in oral cancer & pre-cancer is needed.
Keywords: Oral potentially malignant disorders, Susceptibility, SNP, Polymorphisms
Summary
Background
Oral cancers are preceded by oral potentially malignant disorders (OPMD). Understanding genetic susceptibility for OPMD risk could provide an opportunity for risk assessment of oral cancer through early disease course. We conducted a review of single nucleotide polymorphism (SNP) studies for OPMD risk.
Methods
We identified all relevant studies examining associations of SNPs with OPMD (leukoplakia, erythroplakia and oral sub-mucous fibrosis) conducted world-wide between January, 2000 and February, 2016 using a combined keyword search on PubMed. Of these, 47 studies that presented results as odds ratios and 95% CI were considered for full review.
Results
The majority of eligible studies that explored candidate gene associations for OPMD were small (N < 200 cases), limiting their scope to provide strong inference for any SNP identified to date in any population. Commonly studied SNPs were genes of carcinogen metabolism (n = 18 studies), DNA repair (n = 11 studies), cell cycle control (n = 8 studies), extra-cellular matrix alteration (n = 8 studies) and immune-inflammatory (n = 6 studies) pathways. Based on significant associations as reported by two or more studies, suggestive markers included SNPs in GSTM1 (null), CCND1 (G870A), MMP3 (-1171; promotor region), TNFα (-308; rs800629), XPD (codon 751) and Gemin3 (rs197412) as well as in p53 (codon 72) in Indian populations. However, an equal or greater number of studies reported null or mixed associations for SNPs in GSTM1 (null), p53 (codon 72), XPD (codon 751), XRCC (rs25487 C/T), GSTT1 (null) and CYP1A1m1 (MspI site).
Conclusion
Candidate gene association studies have not yielded consistent data on risk loci for OPMD. High-throughput genotyping approaches for OPMD, with concurrent efforts for oral cancer, could prove useful in identifying robust risk-loci to help understand early disease course susceptibility for oral cancer.
Introduction
Oral cancer is a growing public health problem in the high incidence zones of Asia (i.e. Sri Lanka, India, Pakistan and China-Taiwan), as well as in certain parts of Western and Eastern Europe, Latin America, the Caribbean and the Pacific Islands [1]. Recent trends also suggest increasing incidence in the US and in other parts of Europe including the United Kingdom [2], [3]. A poor 5-year survival (3.1–3.3%) attributed to advanced stages of diagnosis has been shown to improve with early detection (54.3–60.2%) [4]. Oral cancer is believed to be preceded by oral potentially malignant disorders (OPMD), a well-established pre-cancer stage, that can be visually detected in the oral cavity [5], [6].
Oral potentially malignant disorders are early clinical features that are thought to undergo histopathological and molecular changes en-route to invasive oral cancer [5], [7]. OPMD (leukoplakia, erthyroplakia and oral sub-mucous fibrosis) are primarily caused by life-style risk exposures (tobacco smoking, smokeless tobacco use, betel quid chewing and alcohol), [5], [8], [9] but it is possible that inter-individual and inter-population differences in risk [5] could be partially explained by different distributions of genetic variants (including single nucleotide polymorphisms, SNPs) [10], [11] that may cause variation in the ability to metabolize carcinogens and/or effective repair of the damage caused by them [12]. Identifying genetic factors that render individuals susceptible to OPMD risk could have practical significance in terms of identifying potential biomarkers for long term risk assessment for the development of oral cancer [13].
Systematic analyses of candidate gene association studies have suggested that SNPs in genes involved in carcinogen metabolism, DNA repair, cell cycle control, extracellular matrix alteration and folate metabolism could be associated with increased susceptibility for oral cancer [11], with varying susceptibility for different ethnic groups [11]. However, false-positive report probability (FPRP) analysis of these candidate gene association studies based on study power and prior probability found no true oral cancer susceptibility variants [11].
Here, we conduct a review of candidate gene SNP association studies for OPMD (leukoplakia, erythroplakia and oral sub-mucous fibrosis, which have generally been related to lifestyle risk exposures such as tobacco, betel quid and alcohol), to summarize existing evidence on genetic variants (SNPs) for OPMD risk and to ascertain knowledge gaps to inform future research on potential biomarkers for risk assessment of oral cancer development through early disease course susceptibility in high-risk populations (based on use of tobacco, betel quid and alcohol).
Methods
We conducted a literature search in PubMed to identify genetic association studies of single nucleotide polymorphisms (SNPs) for OPMD conducted world-wide and published in the English language using the following key words in titles and abstracts: “polymorphism” OR “mutation” OR “SNP” OR “gene mutation” OR “gene polymorphism” OR “gene alteration” AND “ pre-cancer” OR “premalignant” OR “potentially malignant” OR “leukoplakia” OR “erythroplakia” OR “OSMF” OR “sub-mucous fibrosis” OR “dysplasia” AND “oral” OR “head” OR “neck” AND Humans. The search was conducted for publications between 2000 and 2016 with the last retrieval done on 29th of February 2016. A preliminary review of abstracts was conducted to determine study relevance. A set of eligibility criteria was applied at this stage: (1) Genetic association studies for single nucleotide polymorphisms in OPMD (2) article in English (3) include human subjects (not in vitro or in animals). Studies that met these eligibility criteria were obtained for further review of the full-text article. Final inclusion was made for case control studies with two specific criteria:
-
(i)
Biopsy-confirmed cases and unrelated healthy controls
-
(ii)
Studies which reported odds ratios with associated 95% confidence intervals.
In addition to the electronic search of keywords, we also searched the reference list of all identified relevant studies. If two or more studies examined overlapping study populations, all studies were retained if they reported on different SNPs. If no additional SNPs were evaluated, studies of smaller sample size were excluded.
Data extraction
The following information was extracted from each study when possible and applicable, using a standard data collection form with the following elements: first author, year of publication, population ethnicity, sample size, age of study subjects, clinical and pathological description of OPMD examined (such as leukoplakia, oral sub-mucous fibrosis, erythroplakia and histopathological features such as hyperplasia and dysplasia), genes studied and function of the gene. Information was collected on SNPs and odds ratios with 95% confidence intervals for observed associations (Table 1; online material only).
Results
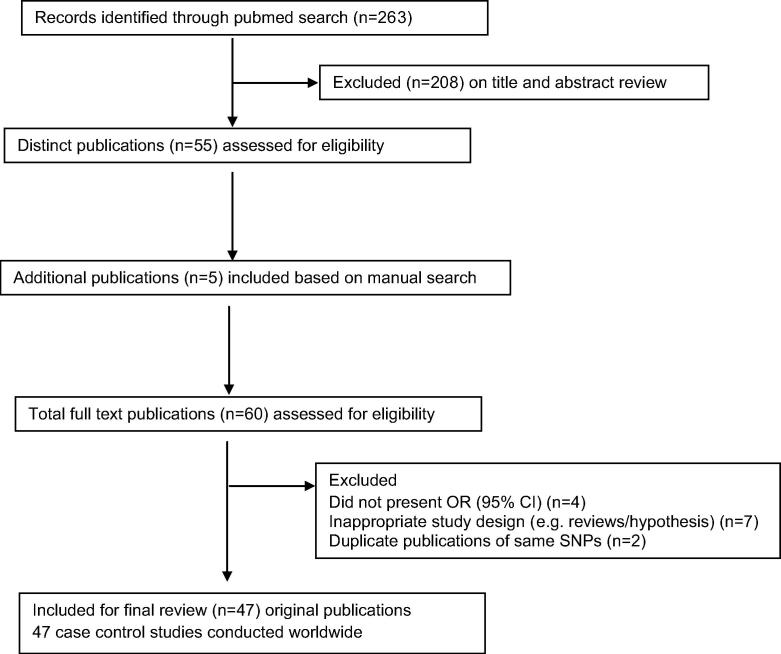
A total of 263 articles were retrieved using the combined key term search on Pubmed. A review of abstracts yielded 51 original articles and 4 review articles that met the eligibility criteria for further review of the full-text articles. Five more original research articles were identified to meet the inclusion criteria from a manual search of the reference list of the 55 included original articles and review studies. A total of 47 eligible original research studies conducted world-wide were included for final review (Fig. 1).
Fig. 1.
Summary of evidence search and selection for Single nucleotide polymorphisms and OPMDs (up to 29th February 2016).
All eligible studies had biopsy-confirmed cases and healthy unrelated controls. Heterogeneity existed among the studies in terms of sample size and reporting of results, with less emphasis on standardized loci information and replication. A majority of the reviewed studies had small sample sizes and thus were underpowered for reliably detecting risk alleles with a low to moderate prevalence (20% or less) and effect size (RR < 1.5) [10], [14].
Table 1 (online material) summarizes key characteristics of the reviewed studies. The majority of included studies (n = 39 out of 47 studies) were small (Ncases < 200). Over three-fourths (82.9%) of the studies were conducted on Asian populations (53.2% Indians and 29.7% Taiwanese) and the rest (17.1%) on Caucasians, Hispanics, African-Americans and Brazilians. The most commonly studied SNPs were in genes of carcinogen metabolism (n = 18 studies), DNA repair (n = 11 studies), cell cycle control (n = 8 studies), extra-cellular matrix alteration (n = 8 studies) and immune-inflammatory (n = 6) pathways.
Suggestive markers of increased susceptibility for OPMD risk based on significant associations as reported by at least 2 or more studies worldwide included GSTM1 null genotype, CCND1 (G870A) with risk allele A, MMP3 (-1171; promotor region) with risk allele 5A, TNFα (308; rs800629) with risk allele A and XPD (codon 751) with risk allele C as well as p53 (codon72) with risk allele C in Indian populations. However, an equal or more number of studies reported null associations for GSTM1 (null), p53 (codon72) and XPD (codon 751). The C allele of rs197412 in Gemin3, a micro RNA processing gene, was associated with reduced risk of OPMD based on significant associations as reported by at least 2 or more studies. Markers that showed mixed associations included XRCC1 (rs25487 C/T; codon399) with allele T, GSTT1 (null genotype) and CYP1A1 m1 (MspI site).
There were few studies conducted on similar loci from across the world, limiting our ability to compare these findings across populations. However, increased susceptibility for OPMD risk with SNPs in GSTM1 (null), CCND1 (G870A), XPD (codon 751) and MMP3 (-1171; promotor region) was seen equally across the majority of populations (Asians, Caucasians, Brazilians and others). Risk associated with a SNP in p53 (codon 72) was reported in Indian populations only. The C allele of Gemin3 (rs197412 C/T) was found to be associated with reduced risk for OPMD in Indian and Caucasian populations. Frequencies of allele or genotype for increased susceptibility, reduced risk or mixed associations for OPMD in different control populations are compared in Table 2.
Table 2.
A comparison of allele or genotype frequencies for increased susceptibility, reduced risk or mixed associations for OPMD in different control populations based on reviewed studies.
| Susceptible gene/SNP loci | Risk allele/genotype | Risk allele/genotype frequency in controls (%) |
||
|---|---|---|---|---|
| Asians | Caucasians and othersa | Brazilians | ||
| GSTM1 | Null genotype | 18–60 (Indians) | NA | 33.8 |
| 49.4 (Taiwanese) | ||||
| p53codon 72 | Heterozygous (Arg/Pro) | 46–54 (Indians); 54.3 (Taiwanese) | 38.7 | NA |
| Homozygous (Arg/Arg) | 22–24 (Indians); 20 (Taiwanese) | 11.7 | ||
| CCND1 G870A | Heterozygous (G/A) | 49 (Indians) | 47.8 | NA |
| Homozygous (A/A) | 23 (Indians) | 18.7 | ||
| MMP3 | 5A allele | 7 (Indians and Taiwanese) | NA | NA |
| TNFα308b |
Heterozygous AG | 17.6–29.7 (Taiwanese) | 12.3–43.3 | NA |
| Homozygous AA | 0.7–7.8 (Taiwanese) | 0–1 | ||
| Gene/SNP loci with reduced risk | Allele with reduced risk | Allele/genotype frequency in controls (%) | ||
| Gemin(rs197412)b | C | 38 (Indians); 10–59.4 (Asians) | 45.8–69 | NA |
| Gene/SNP loci with mixed associations | Allele/genotype with mixed associations | Allele/genotype frequency in controls (%) | ||
| XRCC1 (rs25487) | T | 20.7–56.5 (Indians) | NA | NA |
| CYP1A1 m1 (MspI site) | Heterozygous (±) | 35 to 47.5 (Indians); 54.1 (Taiwanese) | NA | NA |
| Homozygous (−/−) | 4–27.5 (Indians); 9.6 (Taiwanese) | |||
| GSTT1 | Null genotype | 6.2–75 (Indians); 61.2 (Taiwanese) | NA | 22.5 |
African-Americans, Hispanics and Africans.
Also extracted from National Center for Biotechnology Information. Available at http://www.ncbi.nlm.nih.gov/SNP/.
Discussion
We conducted a review of SNP association studies regarding risk of OPMD (leukoplakia, erythroplakia and oral sub-mucous fibrosis) to understand the state of evidence with respect to genetic determinants of risk. With no previous GWAS data available in OPMD in Indian or other populations, the selection of studied SNPs was largely driven through pathway exploration. These included genes involved in pathways of carcinogen metabolism, DNA repair, cell cycle control, extra-cellular matrix alteration and immune-inflammation.
In our review of 47 eligible studies, six SNPs in GSTM1 (null) [15], [16], [17], [18], CCND1 (G870A) [19], [20], MMP3 (-1171; promotor region) [21], [22], TNFα (-308; rs800629) [23], [24], XPD (codon 751) [25], [26] and Gemin3 (rs197412) [27], [28] were identified as suggestive markers for OPMD susceptibility in populations worldwide and an additional SNP in p53 (codon72) for Indian populations [29], [30]. However, we found an equal or more number of studies reporting null associations for SNPs in GSTM1 (null), XPD (codon 751) and p53 (codon72) [19], [31], [32], [33], [34], [35], [36], [37], [38], [39] and mixed associations for SNPs in XRCC1 (rs25487 C/T) [19], [32], [40], [41], GSTT1 (null) [16], [17], [31], [32], [33] and CYP1A1m1 (MspI site) [16], [31], [42], [43] in different populations, leaving no strong loci for follow up.
Most reviewed studies examined risk of leukoplakia, erythroplakia and oral sub-mucous fibrosis, except for two studies which also included a sub-set of lichen planus samples [23], [44]. Small sample sizes with respect to different sub-types of OPMD, lack of validation efforts and limited comparisons among different OPMD (e.g., studies have been conducted only in OSMF samples for MMP3 (-1171; promotor region)) restrict our scope to draw conclusions about difference in susceptibility for different OPMD with respect to the presence of a particular SNP.
Increased susceptibility for OPMD risk with SNPs in GSTM1 (null), CCND1 (G870A), XPD (codon 751) and MMP3 (-1171; promotor region) was common to majority of populations (Asians, Caucasians, Brazilians and others). However, the risk associated with SNP in p53 (codon 72) was restricted to Indian populations. It is possible that the high prevalence of SNP in p53 (codon 72) in Indian population (Table 2) may be partly responsible for higher incidence of OPMD in Indian population [6]. This draws some support from the fact that p53 is the most commonly inactivated tumour suppressor gene in the development of oral cancer [7]. However, it is also possible that this is a chance association. Gemin3 (rs197412 C/T) was found to be associated with reduced risk for OPMD in Indian and Caucasian populations. However, validation studies in similar or other populations are scarce which restrict our scope for valid comparisons and these results have to be interpreted with great caution.
All eligible studies included in the review were of case-control design limiting further comparisons based on study design. Regarding genotyping methods, 40.4% of the studies (19/47) used Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP). The rest used PCR-DNA direct sequencing (n = 7), Taqman assays (n = 7), multiplex PCR/PCR (n = 5) or Illumina Goldengate assay (n = 1) or SNPlex assay (n = 1) and a few studies (n = 7) used more than one method for different SNPs such as different PCR methods including RFLP, single strand conformation polymorphism, polyacrylamide gel analysis and multiplex or Taqman assays. Studies that utilized more than one method on a subset or on all samples confirmed the validity of the different methods of genotyping such as direct sequencing, PCR-RFLP and Taqman assays [27], [38], [45], [46].
The reviewed studies on OPMD were subject to several limitations. Most studies lacked sufficient sample size, and hence power to detect low-to moderate risk associations (particularly with respect to sub-types of OPMD); reporting of results including risk allele/genotype frequencies was not standardized; and all reported studies lacked validation efforts. Further, very small sample sizes could also have over-estimated the magnitude of true associations in addition to their inability to detect true associations and report false associations [10], [47]. Finally, with few exceptions, the candidate gene approach has generally not reliably identified the correct target loci [48]. Thus, it is not possible to indicate strong inference for any SNP identified to date in any population.
The current level of evidence from candidate gene studies for genetic susceptibility to OPMD susceptibility is limited. Although there are no published GWAS data for OPMD [11], [49], GWA studies of cancers of the upper aero-digestive tract (UADT; Oral, pharynx, laryngeal, oesophageal cancers) have identified variants at 12q24 (rs4767364) in the ALDH2 gene, 4q21 (rs1494961) in the HELQ gene, rs1042758 (ADH1C), rs1229984 (ADH1B), and rs1573496 (ADH7) as being significantly associated with risk of all UADT cancers including oral cavity cancers [50], [51].
High-throughput genotyping strategies with sufficient numbers of each sub-type of OPMD might be a better strategy to identify robust risk loci for potential early identification of susceptible individuals. The genome-wide association study (GWAS) approach has successfully identified hundreds of risk loci in germline DNA for various cancers. There are no GWAS data published to date for OPMD, and no published large-scale GWAS data for oral cancer [11], [49], [52], although efforts are under way for oral cancer. The integrated characterization of germline and somatic alterations for OPMD and oral cancer, with well-annotated information on sub-types and a sophisticated analysis for combined and unique risk loci could help to identify susceptibility markers during early disease course and predict disease progression [7], [53]. Such an effort is likely to be relevant to public health prevention and promotion in high incidence zones of oral cancer such as in South Asia.
Conflict of interest
None declared.
Ethical approval
Not required as we utilized already published reports.
Funding Acknowledgements
This work was supported by a Wellcome Trust Capacity Strengthening Strategic Award Extension phase to the Public Health Foundation of India and a consortium of UK universities (WT084754/Z/08/A).
Footnotes
Table 1 data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.oraloncology.2016.08.005.
Contributor Information
Krithiga Shridhar, Email: g.krithiga@phfi.org.
Aastha Aggarwal, Email: aastha.aggrawal@phfi.org.
Gagandeep Kaur Walia, Email: gagandeep.k.walia@phfi.org.
Smriti Gulati, Email: smriti5gulati@gmail.com.
A.V. Geetha, Email: geetha.nambiar@phfi.org.
D. Prabhakaran, Email: dprabakaran@ccdcindia.org.
Preet K. Dhillon, Email: preet.dhillon@phfi.org.
Preetha Rajaraman, Email: rajarama@mail.nih.gov.
Appendix A. Supplementary material
Summary characteristics of reviewed case-control studies on single nucleotide polymorphisms for OPMDs risk.
References
- 1.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, et al. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet]. Lyon, France: International Agency for Research on Cancer; 2013. Available from: http://globocan.iarc.fr [accessed 29th February 2016].
- 2.Bessell A., Glenny A.M., Furness S., Clarkson J.E., Oliver R., Conway D.I. Interventions for the treatment of oral and oropharyngeal cancers: surgical treatment. Cochrane Database Syst Rev. 2011;9 doi: 10.1002/14651858.CD006205.pub3. [DOI] [PubMed] [Google Scholar]
- 3.Chaturvedi A.K., Anderson W.F., Lortet-Tieulent J., Curado M.P., Ferlay J., Franceschi S. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J Clin Oncol. 2013;31(36):4550–4559. doi: 10.1200/JCO.2013.50.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sankaranarayanan R., Swaminathan R., Brenner H., Chen K., Chia K.S., Chen J.G. Cancer survival in Africa, Asia, and Central America: a population-based study. Lancet Oncol. 2010;11(2):165–173. doi: 10.1016/S1470-2045(09)70335-3. [DOI] [PubMed] [Google Scholar]
- 5.Napier S.S., Speight P.M. Natural history of potentially malignant oral lesions and conditions: an overview of the literature. J Oral Pathol Med. 2008;37(1):1–10. doi: 10.1111/j.1600-0714.2007.00579.x. [DOI] [PubMed] [Google Scholar]
- 6.Rajaraman P., Anderson B.O., Basu P., Belinson J.L., Cruz A.D., Dhillon P.K. Recommendations for screening and early detection of common cancers in India. Lancet Oncol. 2015;16(7):e352–e361. doi: 10.1016/S1470-2045(15)00078-9. [DOI] [PubMed] [Google Scholar]
- 7.Lingen M.W., Pinto A., Mendes R.A., Franchini R., Czerninski R., Tilakaratne W.M. Genetics/epigenetics of oral premalignancy: current status and future research. Oral Dis. 2011;17(Suppl 1):7–22. doi: 10.1111/j.1601-0825.2011.01789.x. [DOI] [PubMed] [Google Scholar]
- 8.Radoi L., Luce D. A review of risk factors for oral cavity cancer: the importance of a standardized case definition. Community Dent Oral Epidemiol. 2013;41(2):97–109. doi: 10.1111/j.1600-0528.2012.00710.x. e78-91. [DOI] [PubMed] [Google Scholar]
- 9.Guha N., Warnakulasuriya S., Vlaanderen J., Straif K. Betel quid chewing and the risk of oral and oropharyngeal cancers: a meta-analysis with implications for cancer control. Int J Cancer. 2013 doi: 10.1002/ijc.28643. [DOI] [PubMed] [Google Scholar]
- 10.Hiyama T., Yoshihara M., Tanaka S., Chayama K. Genetic polymorphisms and head and neck cancer risk (Review) Int J Oncol. 2008;32(5):945–973. [PubMed] [Google Scholar]
- 11.Liao G., Wang Y., Zhou Y.Q., Li T.W., Zeng D.Q., Zeng X. Host genetic susceptibility to oral cancer: evidence from meta-analyses and pooled analyses. Oral Dis. 2014;20(7):644–649. doi: 10.1111/odi.12184. [DOI] [PubMed] [Google Scholar]
- 12.Scully C., Field J.K., Tanzawa H. Genetic aberrations in oral or head and neck squamous cell carcinoma (SCCHN): 1. Carcinogen metabolism, DNA repair and cell cycle control. Oral Oncol. 2000;36(3):256–263. doi: 10.1016/s1368-8375(00)00007-5. [DOI] [PubMed] [Google Scholar]
- 13.Li N., Hu Q., Jiang C., Hu Y., Yuan Y., Jian X. Novel genetic biomarkers for susceptibility to oral submucous fibrosis: cytochrome P450 3A. Med Hypotheses. 2011;77(5):834–836. doi: 10.1016/j.mehy.2011.07.049. [DOI] [PubMed] [Google Scholar]
- 14.Hall I.P., Blakey J.D. Genetic association studies in Thorax. Thorax. 2005;60(5):357–359. doi: 10.1136/thx.2005.040790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y.F., Sung F.C., Tsai M.H., Hua C.H., Liu C.S., Huang Y.T. Interactions between cigarette smoking and polymorphisms of xenobiotic-metabolizing genes: the risk of oral leukoplakia. Dis Markers. 2013;34(4):247–255. doi: 10.3233/DMA-130967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh T., Gupta S., Bajpai P., Agarwal D., Agarwal M., Gupta O.P. Association of CYP1A1, GSTM1, and GSTT1 gene polymorphism with risk of oral submucous fibrosis in a section of North Indian population. Mol Biol Rep. 2012;39(10):9383–9389. doi: 10.1007/s11033-012-1802-x. [DOI] [PubMed] [Google Scholar]
- 17.Agrawal D., Gupta S., Agarwal D., Gupta O.P., Agarwal M. Role of GSTM1 and GSTT1 polymorphism: susceptibility to oral submucous fibrosis in the North Indian population. Oncology. 2010;79(3–4):181–186. doi: 10.1159/000318533. [DOI] [PubMed] [Google Scholar]
- 18.Duarte E.C., Ribeiro D.C., Gomez M.V., Ramos-Jorge M.L., Gomez R.S. Genetic polymorphisms of carcinogen metabolizing enzymes are associated with oral leukoplakia development and p53 overexpression. Anticancer Res. 2008;28(2A):1101–1106. [PubMed] [Google Scholar]
- 19.Yadav B.K., Kaur J., Srivastava A., Ralhan R. Effect of polymorphisms in XRCC1, CCND1 and GSTM1 and tobacco exposure as risk modifier for oral leukoplakia. Int J Biol Markers. 2009;24(2):90–98. doi: 10.1177/172460080902400205. [DOI] [PubMed] [Google Scholar]
- 20.Huang M., Spitz M.R., Gu J., Lee J.J., Lin J., Lippman S.M. Cyclin D1 gene polymorphism as a risk factor for oral premalignant lesions. Carcinogenesis. 2006;27(10):2034–2037. doi: 10.1093/carcin/bgl048. [DOI] [PubMed] [Google Scholar]
- 21.Chaudhary A.K., Singh M., Bharti A.C., Shukla S., Singh A.K., Mehrotra R. Synergistic effect of stromelysin-1 (matrix metalloproteinase-3) promoter (-1171 5A->6A) polymorphism in oral submucous fibrosis and head and neck lesions. BMC Cancer. 2010;10:369. doi: 10.1186/1471-2407-10-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tu H.F., Liu C.J., Chang C.S., Lui M.T., Kao S.Y., Chang C.P. The functional (-1171 5A–>6A) polymorphisms of matrix metalloproteinase 3 gene as a risk factor for oral submucous fibrosis among male areca users. J Oral Pathol Med. 2006;35(2):99–103. doi: 10.1111/j.1600-0714.2006.00370.x. [DOI] [PubMed] [Google Scholar]
- 23.Hsu H.J., Yang Y.H., Shieh T.Y., Chen C.H., Kao Y.H., Yang C.F. Role of cytokine gene (interferon-gamma, transforming growth factor-beta1, tumor necrosis factor-alpha, interleukin-6, and interleukin-10) polymorphisms in the risk of oral precancerous lesions in Taiwanese. Kaohsiung J Med Sci. 2014;30(11):551–558. doi: 10.1016/j.kjms.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiu C.J., Chiang C.P., Chang M.L., Chen H.M., Hahn L.J., Hsieh L.L. Association between genetic polymorphism of tumor necrosis factor-alpha and risk of oral submucous fibrosis, a pre-cancerous condition of oral cancer. J Dent Res. 2001;80(12):2055–2059. doi: 10.1177/00220345010800120601. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Spitz M.R., Lee J.J., Huang M., Lippman S.M., Wu X. Nucleotide excision repair pathway genes and oral premalignant lesions. Clin Cancer Res. 2007;13(12):3753–3758. doi: 10.1158/1078-0432.CCR-06-1911. [DOI] [PubMed] [Google Scholar]
- 26.Ramachandran S., Ramadas K., Hariharan R., Rejnish Kumar R., Radhakrishna Pillai M. Single nucleotide polymorphisms of DNA repair genes XRCC1 and XPD and its molecular mapping in Indian oral cancer. Oral Oncol. 2006;42(4):350–362. doi: 10.1016/j.oraloncology.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Roy R., De Sarkar N., Ghose S., Paul R.R., Ray A., Mukhopadhyay I. Association between risk of oral precancer and genetic variations in microRNA and related processing genes. J Biomed Sci. 2014;21:48. doi: 10.1186/1423-0127-21-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clague J., Lippman S.M., Yang H., Hildebrandt M.A., Ye Y., Lee J.J. Genetic variation in MicroRNA genes and risk of oral premalignant lesions. Mol Carcinog. 2010;49(2):183–189. doi: 10.1002/mc.20588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sikka S., Sikka P. Association of human papilloma virus 16 infection and p53 polymorphism among tobacco using oral leukoplakia patients: a clinicopathologic and genotypic study. Int J Prev Med. 2014;5(4):430–438. [PMC free article] [PubMed] [Google Scholar]
- 30.Mitra S., Sikdar N., Misra C., Gupta S., Paul R.R., Roy B. Risk assessment of p53 genotypes and haplotypes in tobacco-associated leukoplakia and oral cancer patients from eastern India. Int J Cancer. 2005;117(5):786–793. doi: 10.1002/ijc.21263. [DOI] [PubMed] [Google Scholar]
- 31.Anantharaman D., Chaubal P.M., Kannan S., Bhisey R.A., Mahimkar M.B. Susceptibility to oral cancer by genetic polymorphisms at CYP1A1, GSTM1 and GSTT1 loci among Indians: tobacco exposure as a risk modulator. Carcinogenesis. 2007;28(7):1455–1462. doi: 10.1093/carcin/bgm038. [DOI] [PubMed] [Google Scholar]
- 32.Anantharaman D., Samant T.A., Sen S., Mahimkar M.B. Polymorphisms in tobacco metabolism and DNA repair genes modulate oral precancer and cancer risk. Oral Oncol. 2011;47(9):866–872. doi: 10.1016/j.oraloncology.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 33.Bathi R.J., Rao R., Mutalik S. GST null genotype and antioxidants: risk indicators for oral pre-cancer and cancer. Indian J Dent Res. 2009;20(3):298–303. doi: 10.4103/0970-9290.57365. [DOI] [PubMed] [Google Scholar]
- 34.Shukla D., Dinesh Kale A., Hallikerimath S., Vivekanandhan S., Venkatakanthaiah Y. Genetic polymorphism of drug metabolizing enzymes (GSTM1 and CYP1A1) as risk factors for oral premalignant lesions and oral cancer. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2012;156(3):253–259. doi: 10.5507/bp.2012.013. [DOI] [PubMed] [Google Scholar]
- 35.Sikdar N., Paul R.R., Roy B. Glutathione S-transferase M3 (A/A) genotype as a risk factor for oral cancer and leukoplakia among Indian tobacco smokers. Int J Cancer. 2004;109(1):95–101. doi: 10.1002/ijc.11610. [DOI] [PubMed] [Google Scholar]
- 36.Majumder M., Sikdar N., Ghosh S., Roy B. Polymorphisms at XPD and XRCC1 DNA repair loci and increased risk of oral leukoplakia and cancer among NAT2 slow acetylators. Int J Cancer. 2007;120(10):2148–2156. doi: 10.1002/ijc.22547. [DOI] [PubMed] [Google Scholar]
- 37.Majumder M., Sikdar N., Paul R.R., Roy B. Increased risk of oral leukoplakia and cancer among mixed tobacco users carrying XRCC1 variant haplotypes and cancer among smokers carrying two risk genotypes: one on each of two loci, GSTM3 and XRCC1 (Codon 280) Cancer Epidemiol Biomarkers Prev. 2005;14(9):2106–2112. doi: 10.1158/1055-9965.EPI-05-0108. [DOI] [PubMed] [Google Scholar]
- 38.Lin Y.C., Huang H.I., Wang L.H., Tsai C.C., Lung O., Dai C.Y. Polymorphisms of COX-2 -765G>C and p53 codon 72 and risks of oral squamous cell carcinoma in a Taiwan population. Oral Oncol. 2008;44(8):798–804. doi: 10.1016/j.oraloncology.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 39.Ye Y., Lippman S.M., Lee J.J., Chen M., Frazier M.L., Spitz M.R. Genetic variations in cell-cycle pathway and the risk of oral premalignant lesions. Cancer. 2008;113(9):2488–2495. doi: 10.1002/cncr.23854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukherjee S., Bhowmik A.D., Roychoudhury P., Mukhopadhyay K., Ray J.G., Chaudhuri K. Association of XRCC1, XRCC3, and NAT2 polymorphisms with the risk of oral submucous fibrosis among eastern Indian population. J Oral Pathol Med. 2012;41(4):292–302. doi: 10.1111/j.1600-0714.2011.01097.x. [DOI] [PubMed] [Google Scholar]
- 41.Majumder M., Indra D., Roy P.D., Datta S., Ray J.G., Panda C.K. Variant haplotypes at XRCC1 and risk of oral leukoplakia in HPV non-infected samples. J Oral Pathol Med. 2009;38(2):174–180. doi: 10.1111/j.1600-0714.2008.00690.x. [DOI] [PubMed] [Google Scholar]
- 42.Chaudhuri S.R., Mukherjee S., Paul R.R., Haldar A., Chaudhuri K. CYP1AI and CYP2E1 gene polymorphisms may increase susceptibility to oral submucous fibrosis among betel quid chewers of eastern India. Gene. 2013;513(2):268–271. doi: 10.1016/j.gene.2012.10.043. [DOI] [PubMed] [Google Scholar]
- 43.Kao S.Y., Wu C.H., Lin S.C., Yap S.K., Chang C.S., Wong Y.K. Genetic polymorphism of cytochrome P4501A1 and susceptibility to oral squamous cell carcinoma and oral precancer lesions associated with smoking/betel use. J Oral Pathol Med. 2002;31(9):505–511. doi: 10.1034/j.1600-0714.2002.00158.x. [DOI] [PubMed] [Google Scholar]
- 44.Wu S.J., Chen Y.J., Shieh T.Y., Chen C.M., Wang Y.Y., Lee K.T. Association study between novel CYP26 polymorphisms and the risk of betel quid-related malignant oral disorders. ScientificWorldJournal. 2015:1–9. doi: 10.1155/2015/160185. 160185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shieh T.M., Tu H.F., Ku T.H., Chang S.S., Chang K.W., Liu C.J. Association between lysyl oxidase polymorphisms and oral submucous fibrosis in older male areca chewers. J Oral Pathol Med. 2009;38(1):109–113. doi: 10.1111/j.1600-0714.2008.00695.x. [DOI] [PubMed] [Google Scholar]
- 46.Wang L.H., Ting S.C., Chen C.H., Tsai C.C., Lung O., Liu T.C. Polymorphisms in the apoptosis-associated genes FAS and FASL and risk of oral cancer and malignant potential of oral premalignant lesions in a Taiwanese population. J Oral Pathol Med. 2010;39(2):155–161. doi: 10.1111/j.1600-0714.2009.00873.x. [DOI] [PubMed] [Google Scholar]
- 47.Button K.S., Ioannidis J.P., Mokrysz C., Nosek B.A., Flint J., Robinson E.S. Power failure: why small sample size undermines the reliability of neuroscience. Nat Rev Neurosci. 2013;14(5):365–376. doi: 10.1038/nrn3475. [DOI] [PubMed] [Google Scholar]
- 48.Siontis K.C., Patsopoulos N.A., Ioannidis J.P. Replication of past candidate loci for common diseases and phenotypes in 100 genome-wide association studies. Eur J Hum Genet. 2010;18(7):832–837. doi: 10.1038/ejhg.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johansson M., Roberts A., Chen D., Li Y., Delahaye-Sourdeix M., Aswani N. Using prior information from the medical literature in GWAS of oral cancer identifies novel susceptibility variant on chromosome 4--the AdAPT method. PLoS ONE. 2012;7(5):1–14. doi: 10.1371/journal.pone.0036888. e36888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.P, Clavel-Chapelon F., Palli D., Tumino R., Krogh V., Panico S. A genome-wide association study of upper aerodigestive tract cancers conducted within the INHANCE consortium. PLoS Genet. 2011;7(3):1–14. doi: 10.1371/journal.pgen.1001333. e1001333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Babron M.C., Kazma R., Gaborieau V., McKay J., Brennan P., Sarasin A. Genetic variants in DNA repair pathways and risk of upper aerodigestive tract cancers: combined analysis of data from two genome-wide association studies in European populations. Carcinogenesis. 2014;35(7):1523–1527. doi: 10.1093/carcin/bgu075. [DOI] [PubMed] [Google Scholar]
- 52.Hindorff LA, MacArthur J (European Bioinformatics Institute), Morales J (European Bioinformatics Institute), Junkins HA, Hall PN, Klemm AK, TA. M, A Catalog of Published Genome-Wide Association Studies. Available at: <www.genome.gov/gwastudies>, [accessed 8th June 2015].
- 53.Das Roy P., Sengupta D., Dasgupta A.K., Kundu S., Chaudhuri U., Thakur I. Single nucleotide polymorphism network: a combinatorial paradigm for risk prediction. PLoS ONE. 2013;8(9):e74067. doi: 10.1371/journal.pone.0074067. [DOI] [PMC free article] [PubMed] [Google Scholar]
References [54–68] are cited in Table 1
- 54.Erdei E., Luo L., Sheng H., Maestas E., White K.A., Mackey A. Cytokines and tumor metastasis gene variants in oral cancer and precancer in Puerto Rico. PLoS ONE. 2013;8(11):1–13. doi: 10.1371/journal.pone.0079187. e79187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mondal P., Datta S., Maiti G.P., Baral A., Jha G.N., Panda C.K. Comprehensive SNP scan of DNA repair and DNA damage response genes reveal multiple susceptibility loci conferring risk to tobacco associated leukoplakia and oral cancer. PLoS ONE. 2013;8(2):e56952. doi: 10.1371/journal.pone.0056952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y.T., Lin L.W., Chen C.Y., Wang C.P., Liu H.P., Houng J.Y. Polymorphism of angiotensin I-converting enzyme gene is related to oral cancer and lymph node metastasis in male betel quid chewers. Oral Oncol. 2012;48(12):1257–1262. doi: 10.1016/j.oraloncology.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 57.Majumder M., Ghosh S., Roy B. Association between polymorphisms at N-acetyltransferase 1 (NAT1) & risk of oral leukoplakia & cancer. Indian J Med Res. 2012;136(4):605–613. [PMC free article] [PubMed] [Google Scholar]
- 58.Chaudhary A.K., Pandya S., Mehrotra R., Singh M. Role of functional polymorphism of matrix metalloproteinase-2 (-1306 C/T and -168 G/T) and MMP-9 (-1562 C/T) promoter in oral submucous fibrosis and head and neck squamous cell carcinoma in an Indian population. Biomarkers. 2011;16(7):577–586. doi: 10.3109/1354750X.2011.609602. [DOI] [PubMed] [Google Scholar]
- 59.Huang S.H., Chang P.Y., Liu C.J., Lin M.W., Hsia K.T. O6-methylguanine-DNA methyltransferase gene coding region polymorphisms and oral cancer risk. J Oral Pathol Med. 2010;39(8):645–650. doi: 10.1111/j.1600-0714.2009.00880.x. [DOI] [PubMed] [Google Scholar]
- 60.Chaudhary A.K., Pandya S., Mehrotra R., Bharti A.C., Jain S., Singh M. Functional polymorphism of the MMP-1 promoter (-1607 1G/2G) in potentially malignant and malignant head and neck lesions in an Indian population. Biomarkers. 2010;15(8):684–692. doi: 10.3109/1354750X.2010.511267. [DOI] [PubMed] [Google Scholar]
- 61.Pu X., Lippman S.M., Yang H., Lee J.J., Wu X. Cyclooxygenase-2 gene polymorphisms reduce the risk of oral premalignant lesions. Cancer. 2009;115(7):1498–1506. doi: 10.1002/cncr.24157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Misra C., Majumder M., Bajaj S., Ghosh S., Roy B., Roychoudhury S. Polymorphisms at p53, p73, and MDM2 loci modulate the risk of tobacco associated leukoplakia and oral cancer. Mol Carcinog. 2009;48(9):790–800. doi: 10.1002/mc.20523. [DOI] [PubMed] [Google Scholar]
- 63.Yang H., Lippman S.M., Huang M., Jack Lee J., Wang W., Spitz M.R. Genetic polymorphisms in double-strand break DNA repair genes associated with risk of oral premalignant lesions. Eur J Cancer. 2008;44(11):1603–1611. doi: 10.1016/j.ejca.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin S.C., Lo S.S., Liu C.J., Chung M.Y., Huang J.W., Chang K.W. Functional genotype in matrix metalloproteinases-2 promoter is a risk factor for oral carcinogenesis. J Oral Pathol Med. 2004;33(7):405–409. doi: 10.1111/j.1600-0714.2004.00231.x. [DOI] [PubMed] [Google Scholar]
- 65.Lin S.C., Chung M.Y., Huang J.W., Shieh T.M., Liu C.J., Chang K.W. Correlation between functional genotypes in the matrix metalloproteinases-1 promoter and risk of oral squamous cell carcinomas. J Oral Pathol Med. 2004;33(6):323–326. doi: 10.1111/j.1600-0714.2004.00214.x. [DOI] [PubMed] [Google Scholar]
- 66.Sikdar N., Mahmud S.A., Paul R.R., Roy B. Polymorphism in CYP1A1 and CYP2E1 genes and susceptibility to leukoplakia in Indian tobacco users. Cancer Lett. 2003;195(1):33–42. doi: 10.1016/s0304-3835(03)00156-3. [DOI] [PubMed] [Google Scholar]
- 67.Chiu C.J., Chang M.L., Chiang C.P., Hahn L.J., Hsieh L.L., Chen C.J. Interaction of collagen-related genes and susceptibility to betel quid-induced oral submucous fibrosis. Cancer Epidemiol Biomarkers Prev. 2002;11(7):646–653. [PubMed] [Google Scholar]
- 68.Ralhan R., Agarwal S., Mathur M., Wasylyk B., Srivastava A. Association between polymorphism in p21(Waf1/Cip1) cyclin-dependent kinase inhibitor gene and human oral cancer. Clin Cancer Res. 2000;6(6):2440–2447. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary characteristics of reviewed case-control studies on single nucleotide polymorphisms for OPMDs risk.