Abstract
Thiazolinyl imine reductases catalyze the NADPH-dependent reduction of a thiazoline to a thiazolidine, a required step in the formation of the siderophores yersiniabactin (Yersinia spp.) and pyochelin (Pseudomonas aeruginosa). These stand-alone nonribosomal peptide tailoring domains are structural homologues of sugar oxidoreductases. Two closed structures of the thiazolinyl imine reductase from Yersinia enterocolitica (Irp3) are presented here: an NADP+-bound structure to 1.45 Å resolution and a holo structure to 1.28 Å resolution with NADP+ and a substrate analogue bound. Michaelis—Menten kinetics were measured using the same substrate analogue and the homologue from P. aeruginosa, PchG. The data presented here support the hypothesis that tyrosine 128 is the likely general acid residue for catalysis and also highlight the phosphopantetheine tunnel for tethering of the substrate to the nonribosomal peptide synthetase module during assembly line biosynthesis of the siderophore.
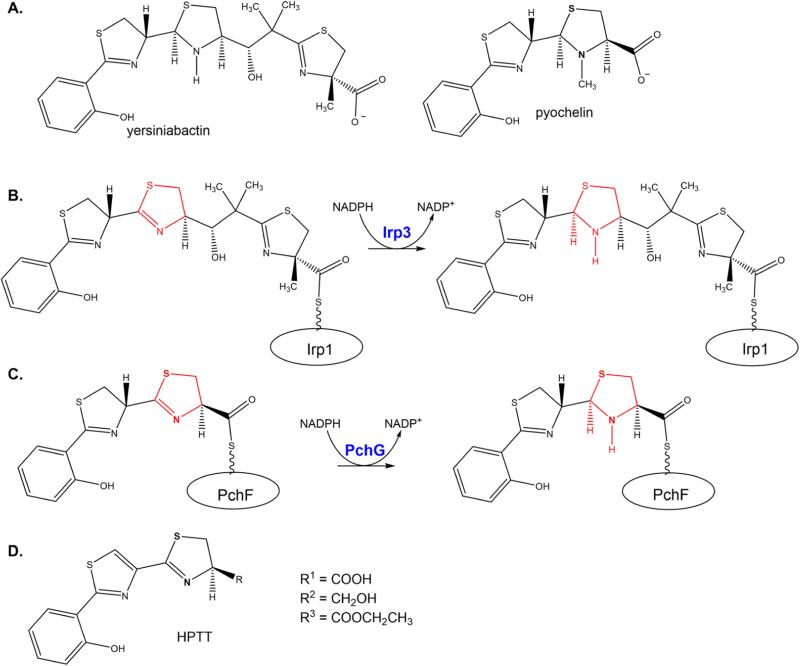
Yersiniabactin and pyochelin are highly similar salicylate-capped siderophores produced by Yersinia spp. and Pseudomonas aeruginosa, respectively, under iron-limiting conditions (Scheme 1A).1,2 Indeed, the first three cycles of the two siderophores, a hydroxyphenyl, a thiazoline, and a thiazolidine, differ by only the methyl group on the thiazolidine ring of pyochelin. Yersiniabactin further contains a methylated malonyl group and a final methylated thiazoline. Pyochelin and the first three rings of yersiniabactin are generated from salicylate and two cysteines by nonribosomal peptide synthetases (NRPS).3,4 The cysteines are modified by tailoring domains (epimerase, reductase, and methyltransferase) to generate the observed structures. Yersiniabactin is further developed by a polyketide synthase (PKS) module for addition and tailoring of the malonyl moiety, and a final NRPS module for generation of the last methylated thiazolidine from cysteine.
Scheme 1.
Yersiniabactin and Pyochelin and the Required Reductase Activities for Their Biosynthesisa
a(A) Mature yersiniabactin and pyochelin siderophores. The stereochemistry for yersiniabactin and pyochelin was taken from refs 3 and 40—44. (B) Irp3 catalyzes the reduction of the C-terminal thiazoline ring to thiazolidine while the substrate is tethered to the nonribosomal peptide synthetase Irp1 through a post-translational modification that is the basis of the thiotemplate mechanism. (C) Pyochelin, the siderophore produced by P. aeruginosa, is produced in the pathway with the stand-alone reductase PchG. PchG catalyzes the reduction of the same thiazoline ring as Irp3 while the substrate is covalently bound to the nonribosomal peptide synthetase PchF. (D) Substrate analogue HPTT.
The focus of this work is the stand-alone NADPH-dependent imine reductase tailoring enzymes common to both assembly lines. The reductase from Yersinia enterocolitica is called Irp3 (YbtU for Yersinia pestis), whereas the P. aeruginosa enzyme is PchG (Scheme 1B,C). Irp3 and PchG catalyze the reduction of the third ring, a thiazoline to a thiazolidine. In pyochelin production, the substrate for the reaction is the hydroxyphenyl-thiazoline-thiazoline covalently attached to an NRPS module through a phosphopantetheine tether. The NRPS protein, PchF, subsequently methylates the nitrogen of the newly formed thiazolidine before release of the completed siderophore.4,5 In yersiniabactin production, the substrate is tethered either to NRPS Irp2 [called high-molecular weight protein 2 (HMWP2) in the Y. pestis literature] or to NRPS/PKS hybrid Irp1 (HMWP1). Previous data are inconclusive with respect to the point at which in the assembly line Irp3 affects the reduction.6 At least two possibilities exist. First, the reduction could occur midway through the assembly line, when the three-ring precursor is attached to NRPS Irp2. This would look comparable to pyochelin biosynthesis (Scheme 1C), with the identical substrate tethered to Irp2. Conversely, the reduction could occur late in the biosynthesis, when the full chain has been synthesized, requiring only reduction and release from PKS/NRPS Irp1, as depicted in Scheme 1B. Both hypotheses have attributes. In the first case, both Irp3 and PchG, which share 26% sequence identity, would catalyze identical reactions on identical ring systems. In the latter case, the reductases would catalyze the same reaction on highly related substrates as a late step in biosynthesis just prior to methylation and release from the NRPS(/PKS) assembly line.
Previously, apo and NADP+-bound structures of Irp3 have been determined in open conformations.7 Irp3 was shown to be structurally homologous to sugar oxidoreductase and biliverdin reductase, but with significant loop insertions in the C-terminal domain. These loop insertions were hypothesized to be important in protein—protein contacts required for interaction with the assembly line proteins to which the substrate was attached. Here, we present two closed structures of Irp3 in a new crystal system: one with NADP+ bound and a second holo structure with both NADP+ and a substrate analogue bound (Scheme 1D). We also present steady state kinetic data for PchG using the same substrate analogue.
MATERIALS AND METHODS
Irp3 Protein Overexpression and Purification
The Irp3 protein with an N-terminal histidine tag was overexpressed and purified as previously described.7 The final buffer consisted of 50 mM potassium phosphate (pH 8) and 100 mM sodium citrate. The protein was concentrated to 19 mg/mL as determined by the Bradford assay and stored at −80 °C.
PchG Overexpression Plasmid
The pchG gene was amplified from P. aeruginosa PAO1 genomic DNA by polymerase chain reaction by use of Herculase polymerase (Agilent) supplemented with 0—10% dimethyl sulfoxide (DMSO). The forward primer (5′-AAT TAT ATA CAT ATG AGC GAC GTC CGT TCC GTG-3′) includes an NdeI site (underlined), whereas the reverse primer (5′-TAA ATA CTC GAG TCA CGA GGC TTG CTC CAG CAC CTG-3′) contains an XhoI site (underlined). The amplified 1075 bp fragment was digested with NdeI and XhoI, and the 1049 bp digested fragment was ligated into the pET28b plasmid (Novagen) digested with the same enzymes. The resulting plasmid encodes the pchG gene with an N-terminal histidine tag.
PchG Protein Overexpression and Purification
Escherichia coli BL21 (DE3) cells containing the PchG expression plasmid were grown in LB broth containing 50 μg/mL kanamycin at 37 °C while being shaken (225 rpm). When the OD600 was ~0.9, protein expression was induced with the addition of isopropyl β-d-thiogalactopyranoside to a final concentration of 200 μM and the temperature was reduced to 25 °C. The cells were harvested by centrifugation (6000g for 10 min at 4 °C) after ~20 h. The cell pellet was resuspended in 25 mM Tris-HCl (pH 8), 500 mM NaCl, and 5 mM imidazole (buffer A). Cells were disrupted by use of a French pressure cell (35000 psi), and cellular debris was removed by centrifugation (12000g for 60 min at 4 °C). The supernatant was applied to a chelating Sepharose fast-flow column (Amersham Biosciences) charged with nickel chloride and pre-equilibrated in buffer A. PchG protein eluted at 250 mM imidazole in a linear gradient from 5 to 500 mM in buffer A. The pooled fractions were dialyzed against 50 mM potassium phosphate (pH 8), 100 mM sodium citrate, and 10% glycerol. The concentration was determined to be 8 mg/mL by the Bradford assay and stored at −80 °C.
Preparation of Substrate Analogues
General Experimental Procedures
All chemical reagents were purchased from commercial suppliers and used without further purification. Flash column chromatography was performed on silica gel (4—63 mm) from Sorbent Technologies. Separation was also performed with a Teledyne Isco CombiFlash Rf. Microwaved reactions took place in a Biotage Microwave reactor. 1H and 13C nuclear magnetic resonance (NMR) spectra were recorded using a 500 MHz Bruker AVIII spectrometer equipped with a cryogenically cooled carbon observe probe using tetramethylsilane as an internal standard. Chemical shifts (δ) are reported in parts per million, and coupling constants (J) are reported in hertz. The high-resolution mass spectrum (HRMS) was recorded on an LCT Premier (Micromass Ltd., Manchester, U.K.) time-of-flight mass spectrometer with an electrospray ion source in either positive or negative mode. Melting points were measured with a Thomas capillary melting point apparatus and are uncorrected. Chiral analysis was conducted on an Agilent 1200 RRLC instrument with a photodiode array on a Chiralpak ID column.
2-(2-Hydroxyphenyl)thiazole-4-carbonitrile
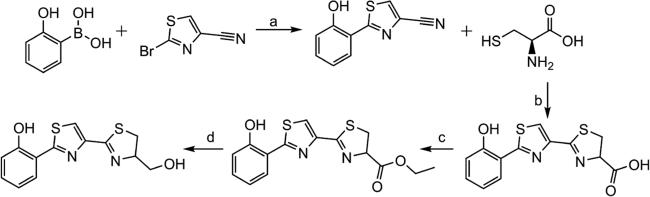
A microwave vial was charged with 2-bromo-4-cyanothiazole (1.06 mmol), 2-(hydroxyphenyl)boronic acid (1.38 mmol), and Pd(PPh3)4 (0.0529 mmol) (Scheme 2). The vial was sealed and flushed with argon for 5 min. Dimethoxyethane (8 mL) and a 2 M potassium carbonate solution (1.06 mL, 2.12 mmol) were added through the septum, and the reaction mixture was stirred at room temperature for an additional 5 min and then heated to 150 °C for 45 min using a microwave reactor. The reaction mixture was cooled to room temperature and diluted with an EtOAc/H2O mixture (2:1, 30 mL). The aqueous layer was extracted with EtOAc (2 × 30 mL). The organic layers were combined, washed with brine, and dried over Na2SO4. The solvent was removed in vacuo, and the resulting residue was purified by flash column chromatography (1:5 EtOAc/pentane) to yield a pale yellow solid (104 mg, 49%): mp 169—172 °C; 1H NMR (500 MHz, DMSO-d6) δ 11.42 (s, 1H), 8.84 (s, 1H), 8.17 (dd, J = 7.9, 1.7 Hz, 1H), 7.39 (ddd, J = 8.6, 7.2, 1.7 Hz, 1H), 7.07 (dd, J = 8.3, 1.2 Hz, 1H), 7.00 (td, J = 7.8, 1.1 Hz, 1H); 13C NMR (126 MHz, DMSO-d6) δ 164.13, 155.12, 133.20, 132.06, 127.36, 124.01, 119.60, 118.24, 116.30, 114.91.
Scheme 2.
Synthesis of Substrate Analoguesa
aReagents and conditions: (a) Pd(PPh3)4, K2CO3/DME, H2O, 150 °C; (b) MeOH, K2HPO4(aq) (pH 6.5), 60 °C; (c) SOCl2/EtOH, 40 °C; (d) Ca(BH4)2/EtOH, −20 °C.
2′-(2-Hydroxyphenyl)-4,5-dihydro[2,4′-bisthiazole]-4-carboxylic Acid (HPTT-COOH)
l-Cysteine (0.546 mmol) and 2-(2-hydroxyphenyl)thiazole-4-carbonitrile (0.273 mmol) were combined in a round-bottom flask. Methanol (5 mL) and 0.1 M potassium phosphate buffer (pH 6.5, 5 mL) were added, and the reaction mixture was heated to 60 °C and stirred for 14 h. The reaction mixture was cooled to room temperature, diluted with H2O (20 mL), and acidified with citric acid to pH ~2. The mixture was filtered through a pad of Celite and extracted with CH2Cl2 (3 × 20 mL). The organic layers were combined and dried over Na2SO4. The solvent was removed in vacuo to afford a light beige solid (58 mg, 70%). The purified product was in agreement with literature values for melting point, 1H NMR, and 13C NMR.8 Enantiopurity was determined by chiral high-performance liquid chromatography (HPLC) using a mobile phase of (A) water containing 0.05% formic acid and (B) acetonitrile, with a gradient from 2 to 80% B over 12 min, and isocratic at 80% B from 12 to 15 min (98:2 R:S).
Ethyl 2′-(2-Hydroxyphenyl)-4,5-dihydro[2,4′-bisthiazole]-4-carboxylate (HPTT-OEt)
Thionyl chloride (0.414 mmol) was added to ethanol (8.56 mmol) while it was being vigorously stirred in a −10 °C ice/water bath. The mixture was warmed to room temperature. HPTT-COOH (0.153 mmol) was added, and the reaction mixture was heated to 40 °C for 4 h. The mixture was cooled to room temperature, and the solvent was removed in vacuo. The residue was reconstituted in saturated Na2CO3 (10 mL) and extracted with CH2Cl2 (4 × 10 mL). The organic layers were combined, and the solvent was removed in vacuo to afford a yellow oil (24.8 mg, 49%): 1H NMR (500 MHz, DMSO-d6) δ 11.24 (s, 1H), 8.34 (s, 1H), 8.14 (dd, J = 8.0, 1.7 Hz, 1H), 7.35 (ddd, J = 8.5, 7.2, 1.8 Hz, 1H), 7.05 (dd, J = 8.2, 1.1 Hz, 1H), 6.99 (ddd, J = 8.2, 7.2, 1.2 Hz, 1H), 5.38 (dd, J = 9.7, 8.4 Hz, 1H), 4.20 (qd, J = 7.1, 2.4 Hz, 2H), 3.64 (ABX, JAX = 8.4 Hz, JBX = 9.8 Hz, JAB = 11.4 Hz, 2H), 1.25 (t, J = 7.1 Hz, 3H); 13C NMR (126 MHz, DMSO-d6) δ 170.96, 164.92, 163.34, 155.50, 147.07, 131.95, 127.81, 122.74, 120.08, 119.25, 116.92, 78.60, 61.61, 34.71, 14.52; HRMS [M + H]+ 335.0449 (calcd), 335.0516 (found).
2-[4-(Hydroxymethyl)-4,5-dihydro[2,4′-bisthiazol]-2′-yl]-phenol (HPTT-OH)
NaBH4 (0.236 mmol) was added to a mixture of HPTT-OEt (0.236 mmol), CaCl2·2H2O (0.109 mmol), and EtOH (1.5 mL) at −20 °C. The reaction mixture was stirred at that temperature overnight. The solution was warmed to room temperature, and saturated NH4Cl (1 mL) was added. The reaction mixture was then concentrated under reduced pressure leaving a residue. Deionized H2O (8 mL) was added, and the mixture was extracted with CH2Cl2 (3 × 15 mL). The combined organic layer was dried over Na2SO4, and the solvent was removed in vacuo to afford a crude oil. The oil was purified using the CombiFlash (4:1 to 5:1 EtOAc/CH2Cl2 gradient) to afford an off-white solid (22 mg, 31%): mp 159—160 °C; 1H NMR (500 MHz, DMSO-d6) δ 11.23 (s, 1H), 8.23 (s, 1H), 8.13 (dd, J = 8.0, 1.7 Hz, 1H), 7.34 (ddd, J = 8.6, 7.2, 1.7 Hz, 1H), 7.04 (dd, J = 8.3, 1.1 Hz, 1H), 7.02—6.96 (m, 1H), 4.99 (t, J = 5.7 Hz, 1H), 4.72 (dtd, J = 8.8, 7.2, 4.4 Hz, 1H), 3.69 (ddd, J = 10.9, 5.5, 4.4 Hz, 1H), 3.55 (ddd, J = 10.9, 6.9, 5.9 Hz, 1H), 3.45 (dd, J = 11.0, 9.0 Hz, 1H), 3.31 (dd, J = 10.9, 7.4 Hz, 1H); 13C NMR (126 MHz, DMSO-d6) δ 163.25, 162.04, 155.48, 147.74, 131.88, 127.82, 121.85, 120.08, 119.26, 116.93, 79.99, 62.77, 34.41; HRMS [M + H]+ 293.0340 (calcd), 293.0419 (found).
Crystallization of NADP+-Bound Irp3
Irp3 cocrystallization was achieved by the hanging drop method at 24 °C. Drops of 1.5 μL of purified Irp3 protein containing 6 mM NADP+ and 3 mM HPTT-COOH were mixed with equal volumes of a reservoir solution composed of 0.1 M HEPES (pH 7.5) and 20% PEG 8000. Large rhombus-shaped crystals (300 μm × 300 μm × 300 μm) formed after 3 days. The crystals were serially washed in a reservoir solution supplemented with 20% (v/v) ethylene glycol as a cryoprotectant and flash-cooled at −160 °C.
Crystallization of NADP+- and HPTT-COOH-Bound Irp3
Drops containing 1.5 μL of purified Irp3 protein containing 6 mM NADP+ and 6 mM HPTT-COOH were mixed with equal volumes of a reservoir solution of 0.2 M sodium formate (pH 7.3) and 20% PEG 3350. The crystals were serially washed in a reservoir solution supplemented with 6 mM HPTT-COOH, 6 mM NADP+, and 20% (v/v) ethylene glycol as a cryoprotectant and flash-cooled for data collection.
Data Collection and Structure Solution for NADP+-Bound Irp3
NADP+-bound Irp3 diffraction data (0.15° oscillation images for a total of 360°) were collected at the Stanford Synchrotron Radiation Lightsource (SSRL, Stanford, CA) beamline 7-1 at a wavelength of 0.9795 Å at 100 K. The exposure time per frame was 0.2 s with 96% attenuation and a crystal to detector distance of 262.3 mm. The data were indexed and scaled with XDS9 to 1.45 Å. The crystals were assigned to space group P21 with the following unit cell dimensions: a = 41.44 Å, b = 148.51 Å, c = 65.01 Å, and β = 95.62°. Data collection statistics are listed in Table 1.
Table 1.
Data Collection and Refinement Statistics for Irp3 Holo Structures
| NADP+-bound | NADP+- and HPTT-bound | |
|---|---|---|
| Data Collectiona | ||
| wavelength (Å) | 0.9795 | 1.1887 |
| space group | P21 | P21 |
| cell dimensions [a, b, c (Å); β (deg)] | 41.44, 148.51, 65.01; 95.62 | 41.39, 148.55, 65.40; 95.30 |
| resolution (Å) | 39.74–1.45 (1.53–1.45) | 39.42–1.28 (1.34–1.28) |
| R sym b | 0.088 (0.850) | 0.056 (0.488) |
| R meas | 0.095 (0.922) | 0.066 (0.588) |
| R pim | 0.036 (0.352) | 0.035 (0.322) |
| total no. of observations | 934414 (131446) | 696955 (78679) |
| total no. of unique observations | 136095 (19678) | 196862 (25144) |
| mean I/sd(I) | 12.0 (2.1) | 12.5 (2.5) |
| completeness (%) | 98.4 (97.5) | 96.7 (84.6) |
| redundancy | 6.9 (6.7) | 3.5 (3.1) |
| Wilson B factor (Å2) | 15.28 | 14.26 |
| Refinement | ||
| resolution (Å) | 39.74–1.45 (1.47–1.45) | 36.04–1.28 (1.29–1.28) |
| R cryst c | 0.159 (0.260) | 0.162 (0.286) |
| R free | 0.179 (0.280) | 0.174 (0.296) |
| total no. of unique observations | 136023 (3993) | 196796 (3750) |
| no. of non-hydrogen atoms | ||
| protein | 5630 | 5715 |
| NADP+ | 96 | 144 |
| HPTT-COOH | 40 | |
| solvent | 8 | 24 |
| water | 457 | 474 |
| root-mean-square deviation for bonds (Å) | 0.008 | 0.011 |
| root-mean-square deviation for angles (deg) | 1.147 | 1.16 |
| overall mean B factor (Å2) | 20.54 | 20.6 |
| protein | 19.98 | 19.98 |
| NADP+ | 18.35 | 21.13 |
| HPTT-COOH | 23.42 | |
| Ramachandran plot analysisd (%) | ||
| favored region | 97.08 | 97.01 |
| allowed region | 2.92 | 2.99 |
| outlier region | 0.00 | 0.00 |
Data indexed and scaled with XDS.
, where Ih is the intensity of reflection h and 〈I〉 is the mean intensity of all symmetry-related reflections.
, where Fo and Fc are observed and calculated structure factor amplitudes, respectively. Five percent of the reflections was reserved for Rfree.
Calculated with MolProbity.
Molecular replacement calculations were performed using PHASER.10 Molecule A of the NADP+-bound Irp3 structure [Protein Data Bank (PDB) entry 4GMG] with the NADP+ and waters removed was used as a search model, yielding a clear solution with a log likelihood gain of 15068. The map generated with this solution showed density in the active site corresponding to the NADP+ and a complete M—N loop compared with the previous NADP+-bound Irp3 model. Model building and refinement were performed using Coot11 and Refmac512 from the CCP4 software suite with anisotropic B factors. NADP+ was modeled in manually in Coot. Waters were added manually using the water addition function in Coot, and the positions were verified following a refinement cycle in Refmac. The last several rounds of refinement and model verification were performed using Phenix.13
Data Collection and Structure Solution for NADP+-and HPTT-COOH-Bound Irp3
Irp3 oscillation images (180° with an oscillation angle of 0.35°) were collected at SSRL beamline 7-1 at a wavelength of 1.1888 Å at 100 K. The exposure time per frame was 18.3 s with a crystal to detector distance of 114.7 mm. The data were indexed and scaled to 1.28 Å with XDS. The crystals were assigned to space group P21 with the following unit cell dimensions: a = 41.39 Å, b = 148.55 Å, c = 65.40 Å, and β = 95.30°.
Molecular replacement calculations were performed using PHASER. Molecule A of the NADP+-bound Irp3 structure determined in the previous section with the NADP+ and waters removed was used as a search model, yielding a solution with a log likelihood gain of 6113. The map generated showed extra density in the active site corresponding to NADP+ and HPTT-COOH. Iterative rounds of model building and refinement were performed with Coot and Refmac5 using anisotropic scaling. NADP+ was modeled in manually in Coot. The model and restraints for HPTT-COOH were built in JLigand14 and manually placed in the active site. Waters were added using the add waters function in Coot, and the positions were verified following each refinement cycle. The last refinement cycles and model verification were completed in Phenix.
Irp3 Crystallographic Models
The final NADP+-bound Irp3 model contains two monomers per asymmetric unit. While the crystals were grown in the presence of HPTT-COOH, none is evident in the structure. The structure contains one NADP+ per monomer, two ethylene glycol molecules from the crystallization condition, and 457 water molecules. Molecule A contains residues 5—358 (of 365), and molecule B contains residues 4—358. Ramachandran analysis as calculated by Molprobity15 showed good geometry with 97.08% of the residues in the favored region and no residues in the outlier region.
The final NADP+- and HPTT-COOH-bound Irp3 model consists of two monomers in the asymmetric unit with one NADP+ and one HPTT-COOH per monomer. The occupancy of HPTT-COOH in molecule A is 85%, and the occupancy of NADP+ is 100%. In molecule B, the occupancy of HPTT-COOH is 70% and two conformations of NADP+ were modeled at 70 and 30% occupancy (Supplemental Figure 1). Additionally, the structure contains two ethylene glycol and two formate molecules from the crystallization solution and 474 water molecules. Molecule A contains residues 5—357, and molecule B contains residues 5—358. Ramachandran analysis calculated by Molprobity showed good geometry with 97.01% of the residues in the favored region and no outliers.
The coordinates and structure factors have been deposited in the Protein Data Bank as entries 5KVQ (NADP+-bound) and 5KVS (holo).
Imine Reductase Activity Assay with HPTT-COOH
The standard assay buffer contained 50 mM potassium phosphate (pH 8) and 100 mM sodium citrate. The oxidation of NADPH to NADP+ was assessed in an assay containing 10 μM PchG, 200 μM NADPH, and 0—1 mM HPTT-COOH, HPTT-OEt, or HPTT-OH. Time point samples were removed over a 2 h period, and the protein was removed by filtration using a membrane with a 10000 molecular weight cutoff. The NADPH concentration in the filtrate was determined by the absorbance at 340 nm following HPLC separation with a BioSuite C18 PAB 3.5 μm column [4.6 mm × 250 mm (Waters)] as described previously.16 The mobile phase was an isocratic gradient of 10 mM sodium phosphate (pH 7.5) for 8 min followed by a wash step of 100% acetonitrile for 8 min with a flow rate of 1 mL/min. The same assay was performed with Irp3: no activity was detected using 10 μM enzyme measuring for 20 h with HPTT-COOH, HPTT-OEt, or HPTT-OH.
RESULTS AND DISCUSSION
Chiral Amines
A chiral amine is one in which the carbon of the carbon—nitrogen bond is stereogenic.17 An estimated 40% of all pharmaceutical drugs contain a chiral amine in their structures.18,19 There has been a tremendous push for the development of enantiomerically pure chiral amines after the U.S. Food and Drug Administration and the European Committee for Proprietary Medicinal Products required the characterization of each enantiomer of the pharmaceutical product. Different enantiomers of pharmaceutically active compounds have been shown to produce widely different biological responses.20 One of the most recognized examples is thalidomide in which the R(+) tertiary amine enantiomer has sedative properties whereas the S(−) enantiomer is a teratogen resulting in birth defects.21 Importantly, thalidomide readily enantiomerizes in vivo to a racemic mixture. The production of large quantities of pure chiral amines has been a challenge for the production of pharmaceuticals and agrochemicals.
To overcome the limitations of synthesizing enatiomerically pure amines, biocatalytic transformations are becoming popular. Biocatalysis provides cost-effective, sustainable reactions at ambient temperatures.22 The biocatalytic reduction of imines and other chiral amines is still challenging because many natural imine reductases are very specific for their substrate, such as dihyrofolate reductase,23 or the enzyme responsible for the imine reduction has not been identified. In the past several years, great strides have been made in using imine reductases in biocatalysis with the identification of a novel enzyme class of stereospecific imine reductases from Streptomyces spp.24,25 Since then, imine reductases from several bacteria have been characterized and their structures determined with broad substrate specificity and high stereoselectivity but low to moderate catalytic effciency.26−32
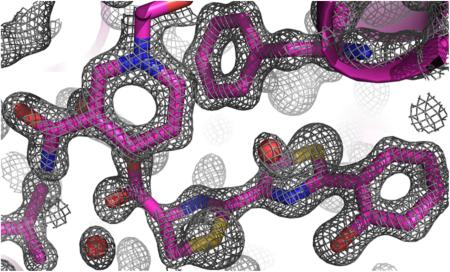
The biosynthesis of the siderophores pyochelin and yersiniabactin requires the enantiospecific reduction of a thiazoline to a thiazolidine (Scheme 1B,C). These reactions are conducted by the sequentially homologous enzymes PchG (P. aeruginosa) and YbtU (Y. pestis)/ Irp3 (Y. enterocolitica).5,6 The previously determined open apo and NADP+-bound structures of Irp3 led to the hypothesis that reduction of the C—N double bond requires transfer of a hydride from NADPH and donation of a proton from the general acid. The general acid residue was suggested to be histidine 101 but alternatively could be tyrosine 128.7 Here, two new structures of Irp3 are determined. The first is an NADP+-bound structure in a new crystal system and the second a holo structure with NADP+ and a substrate analogue bound (Figure 2). The primary structural difference between this NADP+-bound structure and the holo structure and the previously determined apo and NADP+-bound structures is the ordering of the M—N loop, which closes over the active site of the opposing monomer in the dimer (Figure 1C). Another difference is the high quality of the electron density maps: the structures here are determined to higher resolution (1.28—1.45 Å), whereas previous structures were determined to 1.85—2.31 Å resolution. This high resolution will be useful in identifying the sulfur atoms of the substrate analogue, as described below.
Figure 2.
Stereoview of the substrate analogue binding to Irp3. Simulated annealing (2Fo — Fc) omit map contoured at 1.5σ (gray cages) carved at 1.1 Å showing the density for NADP+ (green sticks) and HPTT-COOH (pale cyan sticks).
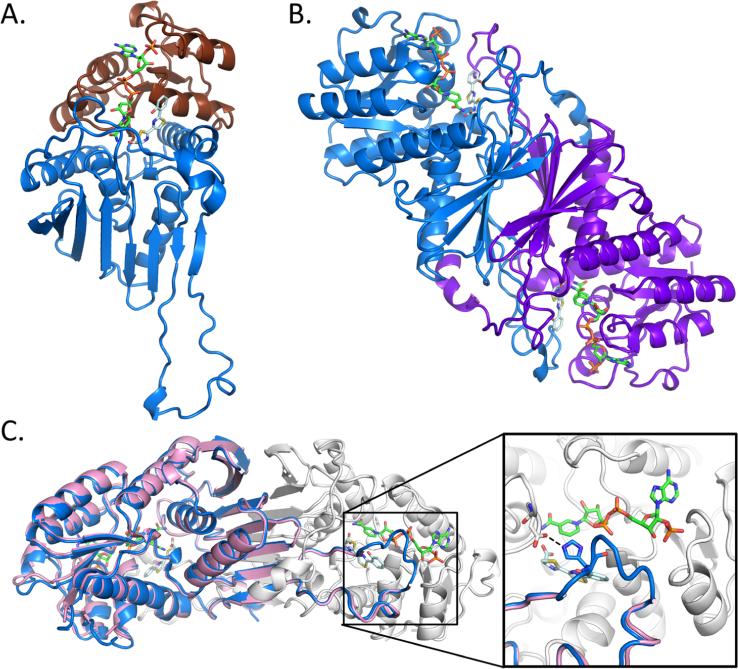
Figure 1.
Overall structure of Irp3. (A) The monomer structure of Irp3 forms two domains, the N-terminal NADP(H) binding domain (chocolate cartoon) and the C-terminal dimerization domain (blue cartoon). (B) Dimeric structure of Irp3 with monomer A colored blue and monomer B colored purple. (C) The new crystal form of Irp3 (blue and white cartoons) allows for M—N loop closure over the active site of the opposing monomer. The M—N loop was disordered in the original crystal form (pink cartoon). The inset shows a close-up of the active site and the binding site of NADP+ (green sticks) and HPTT-COOH (pale cyan sticks). The hydrogen bond between the M—N loop (histidine 257) of one monomer and the active site cavity (aspartic acid 217) of the other monomer is highlighted. Protein structure figures were generated using PyMOL.45
Structure of NADP+-Bound Irp3
Two structures of Irp3 have been reported previously:7 an apo structure (PDB entry 4GMF) and an NADP+-bound structure (PDB entry 4GMG). The crystals for these structures belonged to space group P212121 with two dimers per asymmetric unit. Here, a new crystal form for Irp3 was optimized in a different space group, P21, for an NADP+-bound structure, and for a holo structure containing both NADP+ and a substrate analogue called HPTT-COOH (Scheme 1D). Whereas the new crystal form has lower symmetry, the unit cell volume is 3.5-fold smaller and therefore contains only one dimer per asymmetric unit. Indeed, the molecules pack into the crystal making different contacts despite the mother liquors for crystal formation being remarkably similar. The new crystal form conditions lack a salt containing a divalent anion and have different percentages of PEG 3350 or PEG 8000 relative to the previously determined crystal form.
As would be expected, the NADP+-bound structure in the new space group is very similar to the previously determined NADP+-bound structure with root-mean-squared deviation values of 0.54—0.75 Å for 336—347 Cα residues (depending on the monomers being compared). In other words, the structure determined in the new crystal system gave the anticipated two-domain structure, with an N-terminal Rossmann fold for cofactor binding and a C-terminal α/β domain for dimerization (Figure 1A,B). The original Irp3 structures (PDB entries 4GMF for the apo form and 4GMG for the NADP+-bound form)7 contained a large loop connecting strands M and N (residues 250—277), of which 6—15 residues were disordered, depending on the monomer. In the new crystal form, the M—N loop is fully ordered in both monomers and each M—N loop forms a lid over the active site of the opposing monomer (Figure 1C). The M—N loop makes a hydrogen bond (His257) to the opposing monomer (Asp217) but makes no direct bonds to the NADP+ molecule.
Holo-Irp3 Structure
Patel and Walsh previously reported the release of hydroxyphenyl-thiazole-thiazoline-COOH [HPTT-COOH (Scheme 1D)] from the nonribosomal peptide synthetase PchF in the absence of reductase PchG.4 The molecule is an oxidation product of the reaction intermediate hydroxyphenyl-bisthiazolinyl-COOH that is generated during aborted synthesis. We hypothesized that HPTT-COOH would be a stable substrate analogue for PchG and Irp3, so the compound was generated by the synthesis described in Materials and Methods and depicted in Scheme 2. A holo-Irp3 structure was determined with both HPTT-COOH and NADP+ bound. The overall architectures of the NADP+-bound (determined above) and holo structures are very similar, with a root-mean-square deviation of 0.15—0.49 Å for 353 Cα residues. The cofactor and substrate analogue show exceptional electron density in the simulated annealing omit map for the structure with holes evident in the rings and more density for the sulfurs compared to the carbon and nitrogen atoms (Figure 2). The NADP+ was fully occupied in the active site; however, the HPTT-COOH occupancy was refined to 70—85% depending on the monomer. Alcohol (HPTT-OH) and ethyl ester (HPTT-OEt) analogues were also generated as described in Materials and Methods (Scheme 2). Both were used in crystallization experiments; however, structures determined from cocrystallization with NADP+ or soaking of NADP+-containing crystals did not have substrate analogue density in the active site (data not shown).
Substrate Binding
When the original structures of Irp3 were determined, they were compared to several sugar oxidoreductases of known structure and function with structural homology (root-mean-square deviations of 2—3 Å for ~250 Cα atoms).7 Substrate binding for the sugar oxidoreductases33−36 has been attributed to a loop comparable to the G-6 loop (which connects strand G to helix 6); however, this structural feature is not maintained in Irp3. Instead, the loop connecting strands J and K (residues 210—225) occupies the same three-dimensional space. The hypothesis that the J—K loop provides substrate specificity is not borne out by the structures determined here. While residues 219—225 do form a small portion of the substrate binding cavity, no obvious interactions (hydrogen bonding or ring stacking) promote specificity. Indeed, the binding cavity appears to be a predominantly hydrophobic pocket of suffcient size and shape to bind substrate. The HPTT-COOH substrate analogue forms hydrogen bonds to the nicotinamide amine and the hydroxyl of tyrosine 128 only through its carboxylate. Because the substrate in vivo would be covalently attached to the phosphopantetheine post-translational modification of the carrier protein in an NRPS module, we propose that true substrate specificity arises from the protein—protein interactions of the tailoring enzyme with the NRPS.
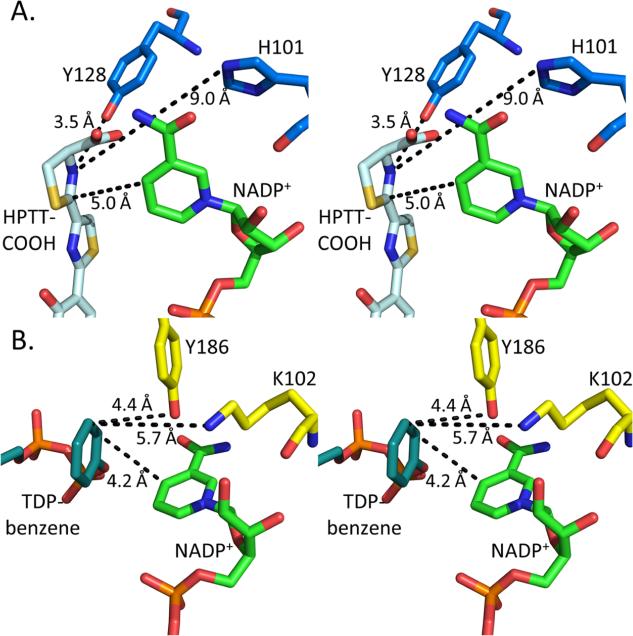
The proposed mechanism for the structurally homologous sugar oxidoreductases involves donation of a hydride from the nicotinamide ring and donation of a proton to the substrate from a general acid residue.35—38 In the sugar oxidoreductases, the active site residue involved in proton donation has been hypothesized to be either the middle residue of the EKP35,36 consensus sequence or the final residue of the GGX3DX3(Y/H) consensus sequence.33,34,37,38 In the thiazolinyl imine reductases, the EKP consensus sequence is 100EHP102, which may still able to act as a proton donor. However, the GGX3DX3(Y/H) consensus sequence is not present.7 The loop that should contain this second consensus sequence (residues 157—162) is only five residues in length and cannot perform the same role. Instead, Irp3 has a tyrosine residue (128) at the same location in three dimensions, but derived from the linker between the N- and C-terminal domains. This residue is conserved in PchG (tyrosine 124). The structure of KijD10,35 a ketoreductase from Actinomadura kijaniata in the biosynthetic pathway for l-digitoxose, contains a substrate analogue, TDP-benzene, which aligns in three dimensions with HPTT-COOH in Irp3. In KijD10, the proposed lysine general acid (K102) derived from the 101EKP103 consensus sequence is 5.7 Å from the double bond being reduced, within range of proton donation (Figure 3). Tyrosine 186 from the 177GGX3DX3(Y/H)186 consensus sequence is also within a reasonable distance for proton donation at 4.4 Å. However, in Irp3, histidine 101 in the EHP consensus sequence is 9.0 Å from the HPTT-COOH nitrogen. Tyrosine 128, on the other hand, is 3.5 Å from the nitrogen and a prime candidate for proton donation. Therefore, we propose that tyrosine 128 is the general acid residue. This mechanism is analogous to that described for dihydrofolate reductase: the hydride likely adds to the electropositive carbon, and the proton from the active site general acid adds to the nitrogen to generate the thiazolidine.4
Figure 3.
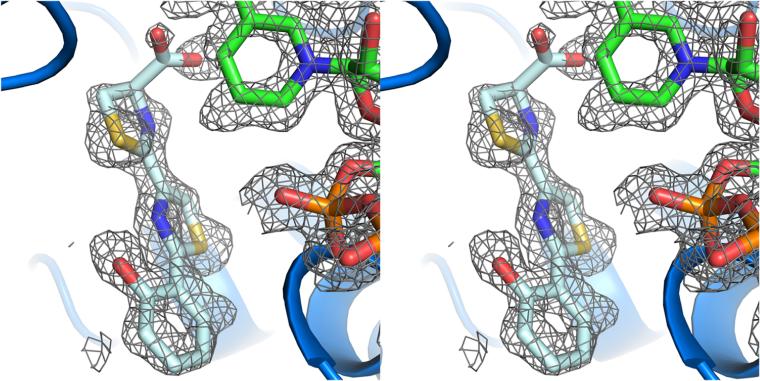
Catalytic residues of Irp3 and KijD10. Stereoimages of the active sites of (A) Irp3 and (B) KijD10 highlighting the distance from the proposed catalytic residues to the substrate analogue (HPTT-COOH, pale cyan in panel A; TDP-benzene, deap teal in panel B) and NADP+ (green). In KijD10, lysine 102 is proposed to be the general acid based on mutagenesis studies.35 There is some debate among the other sugar reductases as to the general acid, but note that lysine 102 and tyrosine 186 are similar distances from the substrate analogue. In Irp3, tyrosine 128 is at a suitable distance for being the general acid whereas histidine 101, which is structurally and sequentially equivalent to lysine 102 of KijD10, is too distant to be the likely candidate.
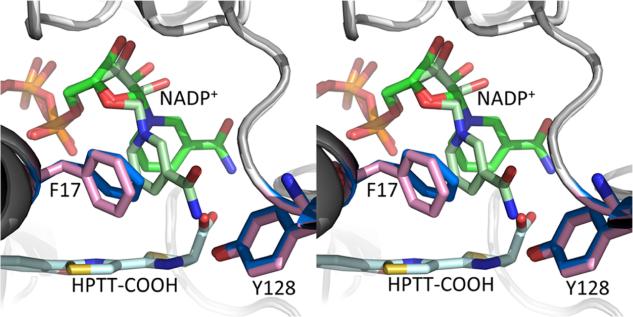
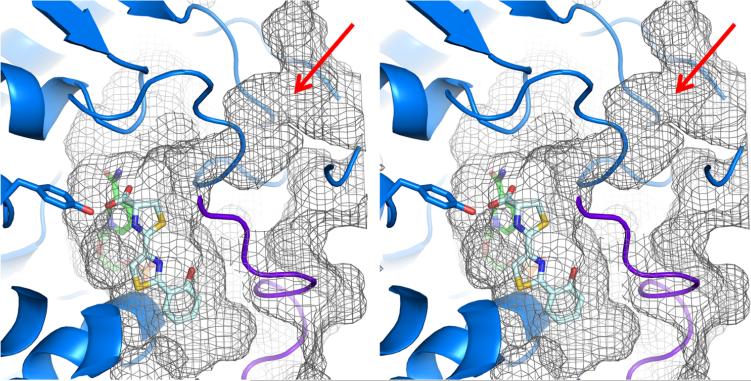
The nicotinamide ring in the NADP+ structure is ring stacking with phenylalanine 17, found in the glycine rich sequence reminiscent of NAD(P) binding motifs in the first turn of a Rossmann fold. In the holo structure, the nicotinamide ring swings away to align with the second thiazoline ring (Figure 4). The substrate binding cavity is closed on one side by NADP+ and has suffcient room for either potential Irp3 substrate (three rings or full-length) (Figure 5). Interestingly, the cavity extends farther down than the terminal hydroxyphenyl ring, which may indicate that the interactions between the carboxylate of the substrate analogue with the nicotinamide and the potential general acid (tyrosine 128) have drawn the substrate analogue away from its true binding mode. The observed conformational flexibility of the nicotinamide ring of the NADP+ alleviates concern that a binding mode lower in the pocket would be catalytically irrelevant. However, a shift of the ring system further in the binding cavity would increase the distance between either of the potential general acid residues and the site of reduction on the substrate. A tunnel is evident leading away from the carboxylate, past the ordered M′–N′ loop (M—N loop from the opposing monomer), to the surface of the protein (Figure 5, red arrow). The tunnel is of an appropriate length for the phosphopantetheine tether.
Figure 4.
Shift in the nicotinamide ring upon substrate binding. In the NADP+-bound Irp3 structure (light gray cartoon), the nicotinamide (light green sticks) is ring stacked with phenylalanine 17 (pink sticks). However, in the holo structure (dark gray cartoon), the nicotinamide of the NADP+ (dark green sticks) rotates away from phenylalanine (blue sticks) to align over the third ring in the substrate analogue (pale cyan sticks).
Figure 5.
Stereoview of active site tunnel. A tunnel from the surface of Irp3 (mesh) leads to the active site cavity (note red arrow). NADP+ is shown as green sticks, HPTT-COOH as pale cyan sticks, and tyrosine 128 as blue sticks. The M′–N′ loop (from the opposing monomer) is colored purple. The substrate is attached to an NRPS carrier domain through a phosphopantetheinyl tether that would be >20 Å long, allowing access to the active site via the tunnel. The surface topology was calculated using CASTp.46
The binding of the substrate analogue in the active site is not consistent with a siderophore ready to bind iron. Two previous structures of pyochelin have been determined with the phenolate and carboxylate oxygens and the two nitrogens bound to iron placing the sulfur atoms on the same side of the molecule. The small molecule crystal structure of ferri-pyochelin showed two pyochelin molecules bound to two iron atoms with the remaining chelation sites filled by an acetate and water molecule.30 The structure of the outer membrane receptor, FptA, with ferri-pyochelin bound showed the sulfur placement determined by sulfur SAD.29 Additionally, the small molecule structure of ferri-yersiniabactin was determined with the phenolate and carboxylate oxygens as well as the three nitrogens from the thiazoline/thiazolidine rings bound to the iron.3 In the holo-Irp3 structure, the thiazoline rings in HPTT-COOH are rotated such that the sulfur atoms are on opposite sides of the molecule as shown by the greater electron density for the sulfur atoms than for nitrogen and carbon (Figure 2).
There is one more piece of evidence that suggests that the binding mode seen in the holo crystal structure is not catalytically competent. The stereochemistry of the pyochelin and yersiniabactin products has been determined from the crystal structures mentioned above. If the hydride and proton were to add to the thiazole ring in the binding mode shown in the holo structure, the wrong stereochemistry would result. There are two hypotheses we propose that would allow the appropriate chiral amine to be generated. First, the terminal thiazoline ring may be rotated 180°, which would also orient the rings appropriately for metal binding. Second, the entire molecule may be rotated. Both cases would better orient the carboxylate toward the tunnel of the phosphopantetheine tether. However, both proposals would move the nitrogen of the chiral amine to a position more distant from either of the potential general acid residues.
Enzymatic Activity
Enzymatic NADPH oxidation to NADP+ is most commonly assayed by monitoring a decrease in absorbance at 340 nm. This assay was not possible with the HPTT-COOH substrate analogue because of the significant absorbance of HPTT-COOH at that wavelength. Therefore, an HPLC assay was developed to separate NADPH and NADP+ from HPTT-COOH and the reduced product. The decrease in absorbance at 340 nm of the NADPH peak was monitored over time. Care was taken to differentiate enzyme activity from uncatalyzed NADPH oxidation, which was slower than what could be detected over the 2 h time course of the Michaelis—Menten experiments (though readily detectable at times of >20 h). PchG, the Irp3 homologue from P. aeruginosa, showed measurable activity with a Km of 80 ± 20 μM, a kcat of 0.020 ± 0.001 min−1, and a kcat/Km of 4 ± 1 M−1 s−1. Irp3 did not demonstrate measurable activity with HPTT-COOH as the substrate. Remember that the physiological substrate for both PchG and Irp3 is attached to the phosphopantetheine of the carrier domain and therefore will not have a carboxylate in the active site. We hypothesized that substituting the carboxylate with an alcohol or an ethyl ester would overcome this problem and could make a more robust substrate. HPTT-OH and HPTT-OEt were made but showed no activity with either enzyme. However, this may have been compounded by their sparing solubility in the enzyme buffer system.
The kinetics of the yersiniabactin and pyochelin assembly lines have been determined. Yersiniabactin can be generated in 22 chemical steps by four enzymes in vitro at a rate of 1.4 min−1.6 Similarly, pyochelin is generated by four enzymes in 13 chemical steps at a rate of 2 min−1.4 The binding mode of HPTT-COOH is not suitable for production of the physiologically relevant chiral chemistry, and the compound also does not provide a kinetically valid model. Even if the PchG reductase were the rate-limiting step of the assembly line, the activity measured here is 100-fold slower than the production of pyochelin by the complete assembly line. The carboxylic acid, which forms hydrogen bonds to both the nicotinamide ring and the general acid, likely prevents effcient catalysis by preventing the appropriate binding mode. Development of a substrate analogue that replaces the carboxylic acid with a moiety that is more similar to the thioester group of the phosphopantetheine tether and yet soluble is a future goal of this work.
Comparison to Other Characterized Imine Reductases
The search for imine reductases for biocatalytic use has led to the identification of several bacterial enzymes with stereoselective activity that have been called IREDs (imine reductase). While the enzymes show strong enantioselectivity, their kinetic parameters are similar to those defined here for PchG using the ineffcient HPTT-COOH substrate analogue. Several structures of imine reductases have been determined with and without NADP+.19,32 The IREDs are intertwined dimers that shared no structural similarity with Irp3. Only one structure has been determined with a substrate bound; however, the electron density for the substrate is inconclusive, and the enantioselectivity (R vs S) is dependent on the storage of the protein.39 Irp3 and PchG have the advantage of having a known physiological substrate; however, the complex nature of the substrate has, to date, limited our ability to study the kinetic properties of these enzymes.
CONCLUSIONS
A closed NADP+-bound structure and a closed holo structure for a stand-alone reductase tailoring domain of yersiniabactin biosynthesis, Irp3, were determined to high resolution. These structures suggest that tyrosine 128 is the general acid residue for catalysis, keeping in mind the caveats related to the likely noncatalytic binding mode of the substrate analogue. These data show for the first time the tunnel for tethering of the substrate to the nonribosomal peptide synthetase carrier domain. Substrate specificity for the imine reductases of siderophore biosynthesis may be the result of protein—protein interactions between the reductase and NRPS. Slow turnover was observed with a hydroxyphenyl-thiazole-thiazoline-carboxylate substrate by the reductase from P. aeruginosa, PchG, a positive step forward for the study of these enzymes; however, this turnover did relevant rates.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to A. S. Chilton, J. T. Douglas, B. Neuenswander, and A. M. Sherwood for technical assistance. We thank the staff of the SSRL for their support and assistance.
Funding
This publication was made possible by funds from National Institutes of Health (NIH) Grants R01 AI77725 and K02 AI093675 from the National Institute for Allergy and Infectious Disease (A.L.L.), National Science Foundation Grant (NSF) CHE-1403293 (A.L.L.), and Graduate Training Program in Dynamic Aspects of Chemical Biology NIH Grant T32 GM008545 (T.A.R. and A.P.R.). Support for NMR instrumentation was provided by NIH Shared Instrumentation Grant S01 RR024664 and NSF Major Research Instrumentation Grant 0320648. Diffraction data were collected at the Stanford Synchrotron Radiation Laboratory (SSRL), a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.
ABBREVIATIONS
- HPTT
hydroxyphenyl-thiazole-thiazoline
- Irp1
polyketide synthase-nonribosomal peptide synthetase from Y. enterocolitica for the production of yersiniabactin
- Irp2
nonribosomal peptide synthetase from Y. enterocolitica for the production of yersiniabactin
- HMWP1
polyketide synthase-nonribosomal peptide synthetase from Y. petis for the production of yersiniabactin
- HMWP2
nonribosomal peptide synthetase from Y. pestis for the production of yersiniabactin
- IRED
imine reductase
- Irp3
thiazolinyl imine reductase from Y. enterocolitica
- KijD10
ketoreductase from A. kijaniata
- PchF
nonribosomal peptide synthetase from P. aeruginosa for the production of pyochelin
- PchG
thiazolinyl imine reductase from P. aeruginosa
- NADP+
nicotinamide adenine dinucleotide phosphate
- YbtU
thiazolinyl imine reductase from Y. pestis
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.6b00735.
Simulated annealing (2Fo — Fc) omit map contoured at 1.5σ carved at 1.1 Å showing the density for NADP+ and HPTT-COOH (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Bearden SW, Fetherston JD, Perry RD. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect. Immun. 1997;65:1659–1668. doi: 10.1128/iai.65.5.1659-1668.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox CD. Iron uptake with ferripyochelin and ferric citrate by Pseudomonas aeruginosa. J. Bacteriol. 1980;142:581–587. doi: 10.1128/jb.142.2.581-587.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller MC, Parkin S, Fetherston JD, Perry RD, Demoll E. Crystal structure of ferric-yersiniabactin, a virulence factor of Yersinia pestis. J. Inorg. Biochem. 2006;100:1495–1500. doi: 10.1016/j.jinorgbio.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Patel HM, Walsh CT. In vitro reconstitution of the Pseudomonas aeruginosa nonribosomal peptide synthesis of pyochelin: characterization of backbone tailoring thiazoline reductase and N-methyltransferase activities. Biochemistry. 2001;40:9023–9031. doi: 10.1021/bi010519n. [DOI] [PubMed] [Google Scholar]
- 5.Reimmann C, Patel HM, Serino L, Barone M, Walsh CT, Haas D. Essential PchG-dependent reduction in pyochelin biosynthesis of Pseudomonas aeruginosa. J. Bacteriol. 2001;183:813–820. doi: 10.1128/JB.183.3.813-820.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller DA, Luo L, Hillson N, Keating TA, Walsh CT. Yersiniabactin synthetase: a four-protein assembly line producing the nonribosomal peptide/polyketide hybrid siderophore of Yersinia pestis. Chem. Biol. 2002;9:333–344. doi: 10.1016/s1074-5521(02)00115-1. [DOI] [PubMed] [Google Scholar]
- 7.Meneely KM, Lamb AL. Two structures of a thiazolinyl imine reductase from Yersinia enterocolitica provide insight into catalysis and binding to the nonribosomal peptide synthetase module of HMWP1. Biochemistry. 2012;51:9002–9013. doi: 10.1021/bi3011016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mislin GL, Burger A, Abdallah MA. Synthesis of new thiazole analogues of pyochelin, a siderophore of Pseudomonas aeruginosa and Burkholderia cepacia. A new conversion of thiazolines into thiazoles. Tetrahedron. 2004;60:12139–12145. [Google Scholar]
- 9.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 11.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 12.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 13.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lebedev AA, Young P, Isupov MN, Moroz OV, Vagin AA, Murshudov GN. JLigand: a graphical tool for the CCP4 template-restraint library. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2012;68:431–440. doi: 10.1107/S090744491200251X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beaupre BA, Hoag MR, Carmichael BR, Moran GR. Kinetics and equilibria of the reductive and oxidative half-reactions of human renalase with alpha-NADPH. Biochemistry. 2013;52:8929–8937. doi: 10.1021/bi401185m. [DOI] [PubMed] [Google Scholar]
- 17.Nugent TC. Chiral amine synthesis: methods, developments and applications. Wiley-VCH Verlag GmbH & Co.; Weinheim, Germany: 2010. [Google Scholar]
- 18.Ghislieri D, Turner NJ. Biocatalytic Approaches to the Synthesis of Enantiomerically Pure Chiral Amines. Top. Catal. 2014;57:284–300. [Google Scholar]
- 19.Schrittwieser JH, Velikogne S, Kroutil W. Biocatalytic Imine Reduction and Reductive Amination of Ketones. Adv. Synth. Catal. 2015;357:1655–1685. [Google Scholar]
- 20.Breuer M, Ditrich K, Habicher T, Hauer B, Kesseler M, Sturmer R, Zelinski T. Industrial methods for the production of optically active intermediates. Angew. Chem. Int. Ed. 2004;43:788–824. doi: 10.1002/anie.200300599. [DOI] [PubMed] [Google Scholar]
- 21.Tian C, Xiu P, Meng Y, Zhao W, Wang Z, Zhou R. Enantiomerization mechanism of thalidomide and the role of water and hydroxide ions. Chem. - Eur. J. 2012;18:14305–14313. doi: 10.1002/chem.201202651. [DOI] [PubMed] [Google Scholar]
- 22.Huisman GW, Collier SJ. On the development of new biocatalytic processes for practical pharmaceutical synthesis. Curr. Opin. Chem. Biol. 2013;17:284–292. doi: 10.1016/j.cbpa.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 23.Schnell JR, Dyson HJ, Wright PE. Structure, dynamics, and catalytic function of dihydrofolate reductase. Annu. Rev. Biophys. Biomol. Struct. 2004;33:119–140. doi: 10.1146/annurev.biophys.33.110502.133613. [DOI] [PubMed] [Google Scholar]
- 24.Mitsukura K, Suzuki M, Shinoda S, Kuramoto T, Yoshida T, Nagasawa T. Purification and characterization of a novel (R)-imine reductase from Streptomyces sp. GF3587. Biosci., Biotechnol., Biochem. 2011;75:1778–1782. doi: 10.1271/bbb.110303. [DOI] [PubMed] [Google Scholar]
- 25.Mitsukura K, Kuramoto T, Yoshida T, Kimoto N, Yamamoto H, Nagasawa T. A NADPH-dependent (S)-imine reductase (SIR) from Streptomyces sp. GF3546 for asymmetric synthesis of optically active amines: purification, characterization, gene cloning, and expression. Appl. Microbiol. Biotechnol. 2013;97:8079–8086. doi: 10.1007/s00253-012-4629-4. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez-Mata M, Frank A, Wells E, Leipold F, Turner NJ, Hart S, Turkenburg JP, Grogan G. Structure and activity of NADPH-dependent reductase Q1EQE0 from Streptomyces kanamyceticus, which catalyses the R-selective reduction of an imine substrate. ChemBioChem. 2013;14:1372–1379. doi: 10.1002/cbic.201300321. [DOI] [PubMed] [Google Scholar]
- 27.Gand M, Muller H, Wardenga R, Hohne M. Characterization of three novel enzymes with imine reductase activity. J. Mol. Catal. B: Enzym. 2014;110:126–132. [Google Scholar]
- 28.Huber T, Schneider L, Prag A, Gerhardt S, Einsle O, Muller M. Direct Reductive Amination of Ketones: Structure and Activity of S-Selective Imine Reductases from Streptomyces. ChemCatChem. 2014;6:2248–2252. [Google Scholar]
- 29.Man H, Wells E, Hussain S, Leipold F, Hart S, Turkenburg JP, Turner NJ, Grogan G. Structure, Activity and Stereoselectivity of NADPH-Dependent Oxidoreductases Catalysing the S-Selective Reduction of the Imine Substrate 2-Methylpyrroline. ChemBioChem. 2015;16:1052–1059. doi: 10.1002/cbic.201402625. [DOI] [PubMed] [Google Scholar]
- 30.Leipold F, Hussain S, Ghislieri D, Turner NJ. Asymmetric Reduction of Cyclic Imines Catalyzed by a Whole-Cell Biocatalyst Containing an (S)-Imine Reductase. ChemCatChem. 2013;5:3505–3508. [Google Scholar]
- 31.Hussain S, Leipold F, Man H, Wells E, France SP, Mulholland KR, Grogan G, Turner NJ. An (R)-Imine Reductase Biocatalyst for the Asymmetric Reduction of Cyclic Imines. ChemCatChem. 2015;7:579–583. doi: 10.1002/cctc.201402797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grogan G, Turner NJ. InspIRED by Nature: NADPH-Dependent Imine Reductases (IREDs) as Catalysts for the Preparation of Chiral Amines. Chem. - Eur. J. 2016;22:1900–1907. doi: 10.1002/chem.201503954. [DOI] [PubMed] [Google Scholar]
- 33.Carbone V, Endo S, Sumii R, Chung RP, Matsunaga T, Hara A, El-Kabbani O. Structures of dimeric dihydrodiol dehydrogenase apoenzyme and inhibitor complex: probing the subunit interface with site-directed mutagenesis. Proteins: Struct., Funct., Genet. 2008;70:176–187. doi: 10.1002/prot.21566. [DOI] [PubMed] [Google Scholar]
- 34.Carbone V, Hara A, El-Kabbani O. Structural and functional features of dimeric dihydrodiol dehydrogenase. Cell. Mol. Life Sci. 2008;65:1464–1474. doi: 10.1007/s00018-008-7508-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kubiak RL, Holden HM. Combined structural and functional investigation of a C-3″-ketoreductase involved in the biosynthesis of dTDP-L-digitoxose. Biochemistry. 2011;50:5905–5917. doi: 10.1021/bi200514b. [DOI] [PubMed] [Google Scholar]
- 36.Thoden JB, Holden HM. Biochemical and structural characterization of WlbA from Bordetella pertussis and Chromobacterium violaceum: enzymes required for the biosynthesis of 2,3-diacetamido-2,3-dideoxy-D-mannuronic acid. Biochemistry. 2011;50:1483–1491. doi: 10.1021/bi101871f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dambe TR, Kuhn AM, Brossette T, Giffhorn F, Scheidig AJ. Crystal structure of NADP(H)-dependent 1,5-anhydro-D-fructose reductase from Sinorhizobium morelense at 2.2 A resolution: construction of a NADH-accepting mutant and its application in rare sugar synthesis. Biochemistry. 2006;45:10030–10042. doi: 10.1021/bi052589q. [DOI] [PubMed] [Google Scholar]
- 38.Kingston RL, Scopes RK, Baker EN. The structure of glucose-fructose oxidoreductase from Zymomonas mobilis: an osmoprotective periplasmic enzyme containing nondissociable NADP. Structure. 1996;4:1413–1428. doi: 10.1016/s0969-2126(96)00149-9. [DOI] [PubMed] [Google Scholar]
- 39.Aleku GA, Man H, France SP, Leipold F, Hussain S, Toca-Gonzalez L, Marchington R, Hart S, Turkenburg JP, Grogan G, Turner NJ. Stereoselectivity and structural characterization of an imine reductase (IRED) from Amycolatopsis orientalis. ACS Catal. 2016;6:3880–3889. [Google Scholar]
- 40.Johnstone TC, Nolan EM. Beyond iron: non-classical biological functions of bacterial siderophores. Dalton Trans. 2015;44:6320–6339. doi: 10.1039/c4dt03559c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlegel K, Lex J, Taraz K, Budzikiewicz H. The X-ray structure of the pyochelin Fe3+ complex. Z. Naturforsch., C: J. Biosci. 2006;61:263–266. doi: 10.1515/znc-2006-3-418. [DOI] [PubMed] [Google Scholar]
- 42.Schlegel K, Taraz K, Budzikiewicz H. The stereoisomers of pyochelin, a siderophore of Pseudomonas aeruginosa. BioMetals. 2004;17:409–414. doi: 10.1023/b:biom.0000029437.42633.73. [DOI] [PubMed] [Google Scholar]
- 43.Youard ZA, Mislin GL, Majcherczyk PA, Schalk IJ, Reimmann C. Pseudomonas fluorescens CHA0 produces enantio-pyochelin, the optical antipode of the Pseudomonas aeruginosa siderophore pyochelin. J. Biol. Chem. 2007;282:35546–35553. doi: 10.1074/jbc.M707039200. [DOI] [PubMed] [Google Scholar]
- 44.Youard ZA, Wenner N, Reimmann C. Iron acquisition with the natural siderophore enantiomers pyochelin and enantio-pyochelin in Pseudomonas species. BioMetals. 2011;24:513–522. doi: 10.1007/s10534-010-9399-9. [DOI] [PubMed] [Google Scholar]
- 45.DeLano W. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA.: 2002. [Google Scholar]
- 46.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.