Abstract
The existence of dispersal syndromes contrasting disperser from resident phenotypes within populations has been intensively documented across taxa. However, how such suites of phenotypic traits emerge and are maintained is largely unknown, although deciphering the processes shaping the evolution of dispersal phenotypes is a key in ecology and evolution. In this study, we created artificial populations of a butterfly, in which we controlled for individual phenotypes and measured experimentally the roles of selection and genetic constraints on the correlations between dispersal-related traits: flight performance and wing morphology. We demonstrate that (i) trait covariations are not due to genetic correlations, (ii) the effects of selection are sex-specific, and (iii) both divergent and stabilizing selection maintain specific flight performance phenotypes and wing morphologies. Interestingly, some trait combinations are also favoured, depending on sex and fitness components. Moreover, we provide evidence for the role of (dis)assortative mating in the evolution of these dispersal-related traits. Our results suggest that dispersal syndromes may have high evolutionary potential, but also that they may be easily disrupted under particular environmental conditions.
Keywords: phenotypic variability, stabilizing versus disruptive selection, correlational selection, fitness landscape, genetic correlation, non-random mating
1. Introduction
Individuals are characterized by mosaics of traits that compose their phenotypes. It has long been demonstrated in nature that these traits are often correlated at the individual level (e.g. [1–3]). This implies that specialized phenotypes composed by suites of correlated morphological, physiological, behavioural, and/or life-history traits assembled in syndromes are frequent [4]. Famous examples are plant defence syndromes [5], pollination syndromes [6], migratory syndromes [7], behavioural syndromes [4], and dispersal syndromes [8].
The observation that phenotypic syndromes are frequent across taxa and biological functions raises the question of their origin. Covariations between traits have been proposed to result either from environmental or genetic constraints (non-selected pleiotropic effects of genes), i.e. the constraint hypothesis, or from selection when a particular combination of traits work well together, i.e. the adaptive hypothesis [9–11]. Unravelling the genetic bases of trait covariation has important implications for its evolution. Indeed, genetically linked traits are obligatory co-evolving traits, while non-genetically linked traits can evolve independently. Therefore, deciphering the mechanisms responsible for the existence of trait correlations and the processes maintaining phenotypic architectures is key to studying the evolvability of many biological functions. Although not recent, the question of the origin of phenotypic syndromes still lacks empirical responses. This is mainly because it is often difficult to determine in the same biological system the genetic and environmental part of variation in traits involved in syndromes together with the investigation of the fitness consequences of syndromes [12].
Dispersal, defined as movement potentially leading to gene flow [13], is a key process in ecology and evolution, from population and community regulation, to adaptation and speciation [14]. Dispersal is crucial for multiple facets of an individual's life because it limits competition with parents, kin, or conspecifics and allows escape from unsuitable environmental conditions (e.g. [15,16]). This means that it is key in organisms' response to global change [17]. According to these important ecological and evolutionary roles, theory predicts that dispersal can be either selected or counter-selected (due to its inherent costs [18]) in particular environmental and/or social conditions [19]. Thus, contrasted dispersal-related phenotypes are susceptible to coexist within a species, and contrasted selective pressures can generate individual heterogeneity in dispersal within populations.
The existence of dispersal syndromes, in which resident and disperser individuals are characterized by suites of correlated dispersal-related traits, have been intensively described across many taxa, from unicellular organisms to animals and plants (e.g. [20–22]). Indeed, distinct specialized phenotypes can coexist and persist in the long term not only at the interspecific level (e.g. [23]), but also at the intra-specific level (e.g. [24,25]). Among the most emblematic examples are heterotypic species for which individuals with distinct locomotory apparatus coexist within populations (e.g. plant fruits with or without a flight apparatus (e.g. [26]), or insects with or without functional hind wings [27]).
The long-standing debate on the origin of covariations between phenotypic traits [9–11] has been recently synthetized in the context of dispersal syndromes [8]. It has been pointed out the necessity of distinguishing between the roles played by proximal causes (i.e. genetic correlations, environmentally induced trait covariations, and effects of dispersal on the expression of other traits) and ultimate causes (i.e. divergent selection on dispersal phenotypes, dispersal plasticity, and eco-evolutionary feedbacks) potentially responsible for the emergence of dispersal syndromes. However, this task is not easy because it requires in the same biological system the determination of the genetic and environmental part of variation in dispersal traits involved in syndromes, and the investigation of the fitness consequences of dispersal syndromes [12]. This explains why much more emphasis has been devoted to the study of the mechanisms that maintain the integrity of dispersal polymorphisms rather than on those that generate variability. Empirical and theoretical works indeed showed that dispersal symmetry [28] or asymmetry [29], dominance [30], assortative mating [31], and balancing selection [32] can all maintain dispersal polymorphisms. One of the most famous examples refers to balancing selection acting in the butterfly Melitea cinxia on the locus encoding the phosphoglucose isomerase (PGI), a metabolic enzyme involved in glycolysis [33]. Heterozygotes at this locus tend to have higher fitness than homozygotes [34], and it has been shown that this gene is involved in dispersal strategies [35]. That being said, it remains generally unresolved if and how selection acts to maintain not only values of a specific dispersal-related trait, but also the combinations of traits that determine dispersers and residents within populations (see however [36,37]). This is mostly because the proximal and ultimate causes of the emergence and the maintenance of dispersal syndromes are poorly investigated at the intra-specific level, although meta-analyses and phylogenetic studies have recently shed light on this topic at the inter-specific level [23,38–40]. Three main questions are at the research front in the field of dispersal and echo back to the more general debate on the evolution of correlated traits raised above: (i) do correlations between traits involved in dispersal syndromes evolve as consequences of a common genetic basis? (ii) Does correlational selection operate to maintain covariations between dispersal-related traits? (iii) By which mechanisms does selection operate to maintain dispersal syndromes over the long term?
In this study, we tackled these questions using an empirical approach by testing for the existence of selective and/or genetic effects responsible for the emergence and maintenance of a butterfly dispersal syndrome. We further tested for the mechanistic role of assortative mating in the maintenance of the variability of these traits. Non-random mating can impact on phenotypes' distributions because of their influence on phenotypes' transmission. Indeed, disassortative and assortative mating are mechanisms that may lead to or evolve in response to stabilizing and disruptive selection, respectively (see review in [41]). To do so, we used the Large White butterfly Pieris brassicae as the model species. We focused our work on flight performance and wing morphology because (i) significant correlation was consistently measured between these two traits in two distinct experiments involving unrelated P. brassicae populations ([42,43], see also in another butterfly species in [44]), (ii) they are both commonly involved in butterflies' dispersal syndromes [45,46].
Pieris brassicae is homotypic for its locomotory apparatus, meaning that it is composed of a mixture of individuals along a mobility and wing morphology gradient [47]. We have recently shown that P. brassicae's flight performance covaries with wing morphology [42,43], exploratory behaviour [42], orientation at emergence [48], dynamics of copulations (unpublished), and dispersal [43]. Especially, we demonstrated that disperser individuals in experimental metapopulations had longer wings and higher flight performances than resident individuals. Therefore, P. brassicae presents a dispersal syndrome involving morphological, physiological, behavioural, and life-history traits. It has also been shown that flight performance increases with latitude using individuals sampled along a south–north gradient in France. This means that mobility is likely to be selected at the species level [47]. Here, we first determined the distributions of flight performance and wing morphology in a natural population of P. brassicae to describe the variation of these traits. Second, we tested on a breeding pool of individuals whether specific flight performance values and wing morphologies impacted several fitness components under semi-natural conditions. We characterized what kind of selection was at work on these traits: direct or correlational; disruptive, stabilizing, or directional. Third, based on full pedigrees, we built the matrix of additive genetic variances and covariances (G-matrix) between these traits to test for the existence of genetic constraints at the basis of the observed phenotypic covariations. Finally, we tested for a potential role of (dis)assortative mating in the evolution of the dispersal syndrome relating the patterns of dispersal-related trait associations between sexual partners with fitness components. We also tested for the effect of other phenotypic traits commonly involved in butterflies' partner choice like wing melanization or body morphology [49,50].
2. Material and methods
(a). Biological material and breeding conditions
To determine the natural distribution of flight performance and wing morphology in P. brassicae, we collected eggs from a cabbage field in Ariège (southwestern France) in summer 2010 and bred a cohort of 87 individuals (from three different clutches) in common garden conditions. Eggs and larvae were kept in a climate chamber under controlled photoperiod (14 light (L) : 10 dark (D) cycle) and temperature (23 ± 1°C during light and 18 ± 1°C during dark) conditions. The larvae were fed ad libitum with fresh cabbage and kept in 40 × 20 × 10 cm boxes. Flight performance was measured on adults following the protocol detailed below and wing length was measured with a calliper.
To measure the effects of selection on dispersal phenotypes, we used a second set of individuals issued from our laboratory breeding. This breeding was established in August 2011 using eggs originating from three locations in Ariège (southwestern France, mean distance between locations: 10 km) and one place in Vaucluse (Southeastern France). At least six egg clutches were used from each locality. In P. brassicae, fertilized females usually lay few egg clutches composed of up to hundreds of eggs for the first clutch, and of fewer eggs (often only a few dozen) for the subsequent clutches. We only collected large clutches, to minimize the possibility of collecting several egg clutches from a single female. Therefore, our breeding was likely established from at least 24 founding females. After two generations (from August to October 2011) during which adults were placed within a semi-natural common garden and larvae bred under fully controlled conditions (the same as above), we artificially induced a diapausing period to pupae (from December 2011 to spring 2012, 20 ± 1°C during light periods and 12 ± 1°C during dark periods). In spring 2012, we stopped diapause and gradually led adults to emergence throughout the experiment. These adults were maintained and bred under common laboratory conditions until their release in the mating cages (see below). We obtained a total of 210 butterflies on which we performed the following sequence of tests. During the first 24 h after emergence, each butterfly was placed in a 1 × 1 × 1 m breeding cage with a water source and nectariferous flowers. Males and females were separated to prevent sexual interactions. On the day after emergence, butterflies were individually tested for flight performance. On the same day, they were weighed and scanned to obtain morphological measurements (see below). Based on the distribution of flight performance of these 210 butterflies, we selected 80 adults (40 males and 40 females; thereafter called ‘parental generation’) so as to maximize the variance in flight performance and minimize the relatedness between individuals (individuals were issued from 10 clutches from 10 distinct females). This parental generation was then split into four replicates of 20 individuals (10 males and 10 females in each replicate) ensuring the continuum of flight performances needed in each artificial population. This density allows high survival rates in the semi-natural cages used to follow mating behaviours [51] and represents the maximal density allowing efficient observation of matings and oviposition behaviour by experimenters (see below). Larvae resulting from these artificial populations were reared under the exact same conditions as their parents and the resulting offspring were reared under common laboratory conditions (n = 214). Tests on these adults were identical to those performed on the parental generation.
(b). Measures of phenotypic traits
It has previously been demonstrated that both flight performance and wing length are components of P. brassicae's dispersal syndrome [43,51]. We have also demonstrated that wing surface is related to flight performance [42]. These three variables were thus used in this study.
We measured individual flight performance through a validated behavioural test monitoring flight performance in stressful conditions [42,43,47,52], which consisted of introducing each butterfly into a shaken plastic chamber and measuring their time in flight (see the electronic supplementary material, for further information).
We then studied morphological traits using digital images. Butterflies were anaesthetized with nitric oxide in a 10 × 10 × 10 cm box (Inject + Matic Sleeper TAS®). Each individual was weighed using a scale with a precision of 0.1 mg (Precisa 80A-200 M) and subsequently fixed between two transparent plastic sheets and placed within a slightly opened scanner (Epson Perfection 2480 Photo, mode PROFESSIONAL). The resulting images were then analysed using ImageJ [53] to measure wing length and wing surface. One of our aims was to test for the role of assortative mating in the evolution of the dispersal syndrome of P. brassicae. Several traits linked to body and wing characteristics can influence butterfly mating ([49,50]; see mating experiments below). Therefore, we also measured total body length and percentage of melanized area on the dorsal side of the left forewing [54] on the basis of images. For each individual, we quantified melanized areas as those that were below a threshold of grey intensity (i.e. 120 on a scale of 0 = black to 255 = white). After the scanning procedure, butterflies were individually marked on both hindwings with a colour pen allowing rapid identification during mating experiments. We compared the melanized areas from the dorsal and ventral sides of both the left and right forewings, as well as the lengths of the two forewings, in a subsample of 20 butterflies to control melanization and wing length measurements for each individual. Each pair of measurements had a correlation coefficient greater than or equal to 0.87. In addition, we performed 20 repetitions of melanization and morphology measures for a single individual to ensure that our established values were reliable (standard deviation was 2.7 for the overall sample and 0.018 for the repeated measurements of melanization; respectively, 1.57 and 0.0002 for body length; 1.5 and 0.0004 for wing length).
Hence, six phenotypic variables were measured, hereafter called flight performance, wing length, wing surface, body length, body mass, and wing melanization.
(c). Mating experiments and fitness measurements
In April and May 2012, 10 males and 10 females were released into each of four replicated 10 × 10 × 2 m outdoor cages, which are part of the Metatron experimental platform [51]. Butterflies were released at the same hour for the four replicates, and the replicates were all run on sunny days. To stimulate reproduction and provide opportunities for egg laying, a fresh cabbage was placed at the centre of each cage. For each replicate, we recreated artificial populations for which the variance in flight performance was maximized, by releasing individuals selected on the basis of their scores on the flight performance test. Individuals spent on average 2 days in the laboratory (range = 1–6 days, but only four individuals were kept more than 4 days). There was no difference in the distribution of variables within each replicate based on sex, with the exception of melanization and wing length in males (ANOVA, both p-values = 0.011). Butterflies were monitored daily from 9.00 to 18.00. Every 30 min, we identified each butterfly in the cages and checked for mating and egg laying. Mating and egg-laying durations were indeed never shorter than 30 min (authors' personal observation). Using this procedure, egg clutches were unambiguously attributed to their corresponding parents and were reared under the conditions described in the Biological material and breeding conditions section.
Four different components of fitness were estimated during this experiment:
(1) lifespan was measured as the time individuals lived after their release in the experimental mating cages;
(2) mating success scored if an individual mated or not;
(3) egg production scored if a mated individual produced eggs or not; and
(4) offspring production scored if a mated individual produced imagoes or not.
(d). Analyses of selection on syndrome traits
We first estimated the correlations among pairs of traits for each individual and for each sex using Spearman's correlations to assess whether flight performance and wing morphology were effectively correlated in our sample. Then, we placed our study in the general framework of the measurement of selection on correlated characters developed by Lande & Arnold [10]. Briefly, multiple regressions with fitness components as the response variable and correlated phenotypic traits as explanatory variables are used to detect the presence of selection and estimate its intensity [10]. Significant linear terms indicate the presence of linear selection (i.e. directional selection), significant quadratic terms indicate the presence of nonlinear selection (i.e. stabilizing or disruptive selection), and significant cross product terms indicate the presence of selection on combinations of traits (i.e. correlational selection). A limitation of this general framework is that the strength and significance of nonlinear selection may be underestimated, especially in the presence of correlative selection [55,56]. To overcome these limitations, we used Partial Least-Squares regressions (PLS) to analyse the data. PLS regression is a robust modelling method for data analysis, especially when the effect of a great number of correlated explanatory variables is investigated from a restricted number of observations [57]. This means that PLS regression is specifically pertinent in the context of selection analyses for traits assembled in syndromes because a complete model including numerous correlated traits and their interactions can be fitted without the need to summarize variables with Principal Component Analyses (in order to limit multi-collinearity which may drastically bias the coefficient estimations, [58]). Moreover, it allows reliable estimations of the significance and intensity of selective effects using very small sample sizes, as found in this study. The basic principles of PLS regressions and the detailed procedure used in this study can be found in the electronic supplementary material.
(e). Heritability and genetic correlations
To assess genetic covariances and correlations between the six phenotypic traits, we estimated the G-matrix (matrix of genetic variances and covariances) using multivariate animal models [59]. Animal models have proved very useful to estimate heritabilities and genetic correlations compared with parent–offspring regressions when sample sizes are limited as in our study [60]. Additive genetic effect was added as a random effect, and sex was included as a fixed effect as sexual differences in morphology are known in P. brassicae [42,47]. Phenotypic traits were standardized to a mean of 0 and a variance of 1 to avoid that our conclusions be based on traits with larger means [61,62]. We thus used the following model:
with wing length, wing surface, flight performance, melanization, body mass, and body length being the standardized ‘phenotypic traits’ to explain and ‘animal’ the individual effect linked to the pedigree to assess additive genetic variance. The procedure used to run Animal Models is detailed in the electronic supplementary material.
(f). Detection of assortative mating
To assess the presence of non-random mating, we built six models corresponding to the six phenotypic traits in which males' trait values were expressed as a function of females' trait values using Generalized Linear Mixed Models (GLMM) with the lmerTest R package [63]. p-values were calculated by means of the Satterthwaite method with the replicate (cage) as a random variable. To rule out the possibility that the observed association of phenotypes between sexual partners could have been generated by purely stochastic processes, mate resemblance was compared to a theoretical distribution of trait association between randomly chosen partners. We generated 1 000 sets of 33 pairs randomly chosen among the males and females that reproduced within each replicate of the experiment. For each replicate, we performed Spearman's correlations between trait values of paired males and females and compared the observed ρ to the distribution of the simulated ρ. Assortative mating was detected if 95% of the simulated values were inferior to the observed value. Similarly, if 95% of the simulated values were superior to the observed ρ, disassortative mating was detected. All traits were standardized prior to analyses by subtracting the mean and dividing this value by the standard deviation of their respective distributions. For (dis)assorted traits, we tested for the role of selection in the maintenance of these non-random associations between sexual partners. To do so, we built normalized similarity indexes consisting of the differences between the trait values of males and females. We checked for multi-collinearity between indexes and then used Ridge regressions, because they allow coping with strong collinearity between explanatory variables [64,65], provided that models are simple. Two models were built with egg production and offspring production as response variables and similarity indexes as explanatory variables.
All statistical analyses were performed using the software R v. 3.1 [66].
3. Results
(a). Trait distributions and correlations between traits
We assessed the frequency distribution of flight performance and wing length in a cohort of 87 individuals from a natural population. The flight performance distribution was not normal (W = 0.84, p < 0.001, Shapiro test) but rather (albeit not perfectly) bimodal (electronic supplementary material, figure S1a), which means that most individuals could be allocated to one of two opposite classes (low or high flight performance). By contrast, wing length followed a normal distribution (W = 0.99, p = 0.46, Shapiro test; electronic supplementary material, figure S1b). The shapes of distributions were similar for both sexes.
To create our artificial populations, we selected 80 individuals among a pool of 210 butterflies from our breeding, for which we ensured that the distribution of flight performance and wing length were similar to those of the natural population, i.e. bimodal and normal (see the electronic supplementary material, figure S2). Correlations between the six measured phenotypic traits are shown in the electronic supplementary material, table S1 for each sex. Melanization was the only trait uncorrelated to any other trait. Among significant correlations, some were found in both sexes and others were sex-specific. Wing length and wing surface were positively correlated in both sexes, as were body length and body mass. Three pairs of traits (exclusively morphological traits: wing surface and body length, wing surface and body mass, wing length and body length) showed significant positive correlations in females. In males, two pairs of traits linked to dispersal showed significant negative correlations. Flight performance was indeed correlated to wing length and wing surface. A similar sex-specific correlation between flight performance and wing length was also observed in the natural population (Spearman's ρ = 0.126, p = 0.39 for females and Spearman's ρ = 0.477, p = 0.001 for males).
(b). Selection analyses on dispersal traits
To assess individual fitness, we recorded lifespan, mating success, and fecundity by collecting eggs and raising offspring until adult emergence. All PLS were significant and explained between 41.1 and 100% of variance in fitness components, with male dispersal-related traits explaining a higher part of variation in the four fitness components than female dispersal-related traits (electronic supplementary material, table S2). However, among the four fitness components, only three were significantly affected by dispersal-related traits: lifespan, mating success, and offspring production (table 1). These analyses revealed that both linear and nonlinear selections are acting on dispersal-related traits. Indeed, intermediate male flight performance favoured lifespan, while extreme flight performance favoured male mating success (figure 1) and offspring production. Extreme flight performance favoured female lifespan, while only low flight performance favoured female offspring production. Wing length had a significant quadratic effect on male mating success, with intermediate males having a higher probability of mating compared with males with extreme wing lengths. Male offspring production increased for two trait combinations: high flight performance/small wings and low flight performance/long wings. Finally, the age at release had a strong significant effect on lifespan, with the oldest butterflies being those that survived the longest. Wing surface had no significant effect on fitness.
Table 1.
Sex-specific magnitude of the significant effect of dispersal traits on fitness. This table details the regression coefficients associated for each significant variable with their 95% confidence intervals and p-values. These regression coefficients were doubled for quadratic variables [67] and adjusted following [68] to obtain estimators of selection coefficients [10]. For male offspring production, it was impossible to estimate selection coefficients likely due to a limited sample size and/or a very high predictive power of our model, which led PLS regression to compute large coefficient values.
| fitness component | significant variables | regression coefficient | p-value | selection coefficient | selection type |
|---|---|---|---|---|---|
| male survival | flight performance2 age at release | −0.052 [−0.111;0.007] 0.207 [0.105;0.264] |
0.047 0.0002 |
−0.104 0.207 |
stabilizing selection directional selection |
| female survival | flight performance2 age at release | 0.099 [−0.003;0.017] 0.233 [0.122;0.198] |
0.03 0.011 |
0.198 0.233 |
disruptive selection directional selection |
| male mating success | flight performance2 with length2 | 3.22 [0.841;214.99] −2.008 [−155.371; −0.892] |
0.01 0.002 |
0.341 −0.213 |
disruptive selection stabilizing selection |
| male offspring production | flight performance × wing length flight performance2 |
−149.02 [−216.185;0.04] 57.56 [−1.154;88.009] |
0.031 0.05 |
n.a. n.a. |
correlational selection disruptive selection |
| female offspring production | flight performance | −4.73 [−112.597;2.052] | 0.05 | −0.189 | directional selection |
Figure 1.
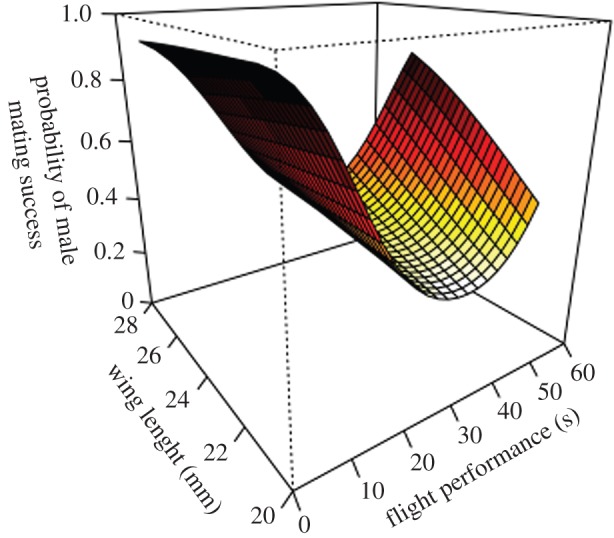
Fitness landscape of male mating success. The relationship was built on the basis of PLS results for the mean wing surface and age at release within the male population, and for weighted means of each replicate/family combinations. In this figure, we only represent the complex fitness landscape presenting two significant variables with estimable selection coefficient. (Online version in colour.)
(c). Heritability and genetic correlations
The flight performance and morphology of adult offspring were compared with parental values to assess their heritability using animal models with sex as a covariate. Only wing surface was not heritable, whereas the five other phenotypic traits were significantly heritable: heritability rates were high for wing melanization and wing length, intermediate for flight performance and body mass, and low for body length (electronic supplementary material, table S3a). Sex was retained in the selection of model terms for three traits: females were more melanized than males and also displayed higher body mass and body length compared with males. With regards to genetic covariances and correlations, only the positive genetic correlation between wing length and wing surface was significant (see the electronic supplementary material, table S3b).
(d). Mating preference
Thirty-three of the 40 released females mated (each mating only once), while 26 of the 40 males mated (seven of them mated twice). Correlations between traits of mating partners are shown in the electronic supplementary material, figure S3. Males and females of the same pair had significantly divergent body lengths (ρ = −0.464, p = 0.004), wing lengths (ρ = −0.368, p = 0.021), and wing surfaces (ρ = −0.319, p = 0.031), reflecting disassortative mating for these traits. By contrast, males and females of the same pair had significantly similar flight performances (ρ = 0.575, p < 0.001), reflecting assortative mating for flight performance. No correlation between mating partners' phenotypes was observed for body mass and wing melanization. Ridge regressions showed that only disassortative mating on the basis of wing length significantly affected a fitness component (offspring production was favoured when sexual partners had different wing lengths, t-value = 3.396; p = 0.0007).
4. Discussion
Wing length and flight performance are key phenotypic traits in butterflies because they are involved in many biological functions: e.g. in mate location, male–male competition, food search, thermoregulation ability, or escape behaviour. Recent meta-analyses showed that they are also key traits in butterflies' dispersal ability [45,46]. This was experimentally validated in P. brassicae that exhibits a dispersal syndrome in which dispersers have longer wings and a higher flight performance than residents [43]. Such correlations between phenotypic traits can be the consequence of constraints, either genetic or environmental (i.e. without any selection effects on phenotypes), or can result from divergent and/or correlational selection (i.e. phenotypic syndromes are adaptive) [9–11]. Here, we measured four fitness components of butterflies with known phenotypes issued from our breeding and placed under semi-natural conditions to decipher the mechanisms by which a dispersal syndrome can emerge and evolve. We used a complete methodology including animal models and PLS regressions. Notably, PLS appeared to be a powerful tool to study selection on correlated traits analogous to the Lande and Arnold method [10] but with the advantages of robustness for small sample sizes and to provide an unbiased coefficient estimation despite the presence of multi-collinearity among explanatory variables (which is highly valuable in the context of phenotypic syndromes). We showed that complex and sex-specific selective effects, and not heritable genetic constraints, can shape the evolution of P. brassicae's trait covariations. We hereafter mostly discuss these results in the context of evolution of dispersal syndromes, but also point out their importance for other aspects of butterflies' ecology when relevant.
(a). Association between phenotypic traits
Our results revealed complex associations between some of these traits. The positive correlations between body length and mass, and between wing length and surface observed both in males and females are well known as universal scaling relationships (e.g. [69]). The other, sex-dependent associations are more intriguing. First, we showed that body length was positively related to wing length and wing surface in females only, while body length and wing morphology were not genetically correlated. In insects, female fecundity scales positively with body size (e.g. [70] for insects, [71] for butterflies). We suggest that egg load is such a strong constraint on flight that it generates an obligatory positive scaling relationship between female body size and wing size.
We also report that two pairs of traits involved in P. brassicae's dispersal syndrome were correlated in males: flight performance was negatively related to wing length and wing surface. We determined that wing length and wing surface were highly genetically correlated, i.e. they share a common genetic basis likely responsible for the covariation we observed at the phenotypic level. However, only wing length was significantly heritable. Therefore, wing length, and not wing surface, is probably one of the main drivers of wing morphology evolution in our model species. As well, we did not detect any impact of wing surface on fitness components. Therefore, we will hereafter only discuss the role of wing length and flight performance on dispersal syndrome evolution. Wing length and flight performance are two traits under selection in flying insects (e.g. [72,73]), which is expected given their role in many insect biological functions. We confirm that selection influences the evolution of these two traits in P. brassicae. Interestingly, we found that the effects of selection were sex-specific and differed between fitness components.
(b). Sex-specific selection
Sex-specific selection, where fitness landscapes are different between males and females, and sexually antagonistic selection, where the signs of the covariance between a trait and fitness differ between males and females, have been extensively documented (see reviews in [74]) and can occur in the context of dispersal. Indeed, differences in dispersal rates between males and females, i.e. sex-biased dispersal, have been described in many taxa including butterflies (e.g. [75,76]). In P. brassicae, females tend to disperse more than males [43]. However, we did not evidence selective effects that favoured dispersers' trait values specifically in females as compared to males: high (and low) flight performance favoured female lifespan, but also male mating success and offspring production. Furthermore, females with longer wings had no benefit compared to other females, while long-winged males with low flight performance were favoured. Therefore, the dispersal bias toward females in P. brassicae does not seem to be linked to female-specific phenotypic attributes, which is in accordance with the absence of a significant interactive effect between sex and phenotypic traits on dispersal decisions in this species [43]. We suggest that phenotypic adaptations to dispersal are the same in the two sexes, although dispersal rates are different, and that other sex-specific ecological processes linked to males' and females' lifestyles also influence the evolution of wing length and flight performance (for example, intra- and inter-sex interactions, or resource use strategy). This hypothesis is in accordance with the observations that correlations between flight performance and wing length are highly consistent in males ([42,43], this study), while the same correlation is less frequently observed in females [43]. The existence of sex-specific syndromes has rarely been investigated except in the context of plant morphology ([6], see however [77]); here, we provide an example of a sex-specific syndrome in the context of animal dispersal.
(c). Flight performance, wing length, and fitness components
We measured the effect of flight performance and wing length on four fitness components. Among them, egg production was unaffected by phenotypic traits. The study of the three other fitness components (female lifespan, male mating success, and male offspring production) revealed that disruptive selection generally drives the evolution of flight performance. This means that both low and high performers (i.e. potentially residents and dispersers) have a selective advantage compared with intermediate phenotypes, which may be responsible for the long-term maintenance of dispersal polymorphisms in this species. Strong disruptive selection can lead to strong divergence between populations, and even speciation [78]. This process could thus result in the isolation of high and low performers in P. brassicae. However, a stabilizing effect on flight performance was also detected on male lifespan, which should limit the distribution of the flight performance trait towards extreme values. At broader spatial and temporal scales, it is also likely that fluctuating environmental conditions favouring either low or high performers (balancing selection) prevent strong divergence between extreme flight performance phenotypes within P. brassicae. Besides, probability of offspring production was lower for females with high flight performance, which would suggest the existence of an oogenesis-flight syndrome in P. brassicae, i.e. a negative correlation between reproduction (here offspring production) and flight performance [79]. All in all, our results suggest that the effects of selection on this dispersal-related trait could promote the existence of dispersers and residents without provoking a strong divergence between them.
We have evidenced a positive effect of the age of release on lifespan in both sexes, meaning that individuals that spent a longer time under laboratory conditions survived longer within semi-natural cages than those that spent less time. Butterflies living their first days of life under optimal laboratory conditions may endure reduced physiological costs than individuals living their first days under more costly semi-natural conditions (e.g. variation in weather conditions, predation risk), explaining this result.
We also detected significant correlational selection favouring short-winged males with high flight performance and long-winged males with low flight performance. This result shows that particular combinations of these two traits can be the target of selection, and thus influence the emergence and maintenance of dispersal syndromes. Interestingly, wing length and flight performance were genetically uncorrelated, i.e. the emergence of P. brassicae's dispersal syndrome is probably not due to genetic constraints. A corollary is that this syndrome is susceptible to be disrupted rapidly if selective pressures on these traits vary intensively. This might explain why a positive correlation between wing length and flight performance was reported in our previous studies [42,43] and in our natural sample, while we found a negative correlation in our experimental sample. We also suspect that such differences across individual pools with distinct histories reflect a high evolutionary potential of P. brassicae's syndrome. This hypothesis would deserve further experimental investigation, which could be more generally performed in the general context of phenotypic syndromes.
By contrast, we found a disruptive effect of correlational selection on wing length (short-winged males with high flight performance and long-winged males with low flight performance were favoured) that is opposed to the stabilizing effect we evidenced measuring male mating success. The evolution of trait correlations can thus be complex and driven by several, and sometimes discordant, selective effects.
(d). (Dis)assortative mating on dispersal-related traits
In this study, we have also tested for the mechanistic role of (dis)assortative mating in the maintenance of dispersal polymorphisms. We found significant assortative mating on the basis of flight performance, and significant disassortative mating on the basis of wing length, wing surface, and body length. In butterflies, several studies have showed the key role of wing colour patterns in the choice of sexual partners (e.g. [50,80]). Our study highlights that wing morphology and flight performance may also be cues for non-random associations between sexual partners. To what extent these traits may serve as signals for mate choice or drive non-random mating patterns without choice (for example, if flight performance influences the probability to encounter partners) is still an open question that surely deserves further investigation. Assortative mating on the basis of flight performance has the potential to accelerate the spread of resident and disperser phenotypes in populations of P. brassicae through the fitness benefits we have demonstrated above. Interestingly, we also found that a direct fitness reward to disassortment was found for wing length, because the dissimilarity in wing lengths between sexual partners was associated with a higher number of offspring, which should favour intermediate wing lengths. Clearly, the existence of adaptive disperser and resident phenotypes in populations tends to increase the frequency of short- and long-winged individuals. But other selective pressures and/or morphological constraints also skew the distribution of wing length towards normality in P. brassicae, as we highlighted with the detection of disassortative mating on wing length, which may explain the high frequency of individuals with intermediate wing length observed in our natural population.
5. Conclusion
Determining the potential causes of the emergence and long-term maintenance of phenotypic syndromes is a fundamental, but challenging question in evolutionary ecology. In a butterfly, we have determined that the correlations between phenotypic traits involved in a dispersal syndrome can arise independently of genetic constraints, and that they may be adaptive. Furthermore, nonlinear and linear selection can act independently and concomitantly to shape the evolution of traits involved in syndromes, by favouring specific trait values or specific combinations of traits. Determining the processes that shape the evolution of dispersal syndromes can be challenging because we showed that the effect of selection can be sex-specific and divergent between fitness components. Interestingly, such observations suggest a high evolutionary potential of dispersal syndromes, meaning that correlations between phenotypic traits can emerge and may be rapidly disrupted in response to specific environmental conditions. Such observations may have important implications for species' abilities to respond to changing environments, and should therefore be the focus of future experiments. Finally, we strongly suggest that future work on fitness landscapes should integrate several fitness proxies and appropriate methodology to study correlated traits, in order to correctly understand and predict the evolution of phenotypic syndromes.
Supplementary Material
Acknowledgements
The authors warmly thank Julien Cote and Staffan Jacob for helpful discussions on this manuscript.
Data accessibility
The data supporting this article are available in the Dryad Digital Repository, with identifier doi:10.5061/dryad.s73t1 [81].
Authors' contributions
D.L., N.L., and M.B. conceived the study. D.L., N.L., and O.C. performed the experiments. All authors participated to the statistical analyses at the exception of PLS regressions performed by R.B. and animal models performed by S.D. D.L., N.L., and M.B. wrote the first draft of the manuscript, all authors contributed substantially to revisions.
Competing interests
We have no competing interests.
Funding
This study was funded by ANR through the DIAME (open call, 2007), MOBIGEN (6th extinction call, 2009), INDHET (open call, 2012) and GEMS (young researchers, 2013) projects. This work is part of the Labex TULIP (ANR-10-LABX-41). D.L. thanks FNRS-F.S.R. and University of Louvain for salary funding.
References
- 1.Searle SR. 1961. Phenotypic, genetic and environmental correlations. Biometrics 17, 474–480. ( 10.2307/2527838) [DOI] [Google Scholar]
- 2.Roff DA. 1992. The evolution of life histories: theory and analysis. New York, NY: Chapman and Hall. [Google Scholar]
- 3.Conner JK, Cooper IA, La Rosa RJ, Pérez SG, Royer AM. 2014. Patterns of phenotypic correlations among morphological traits across plants and animals. Phil. Trans. R. Soc. B 369, 20130246 ( 10.1098/rstb.2013.0246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sih A, Bell A, Johnson JC. 2004. Behavioral syndromes: an ecological and evolutionary overview. Trends Ecol. Evol. 19, 372–378. ( 10.1016/j.tree.2004.04.009) [DOI] [PubMed] [Google Scholar]
- 5.Agrawal AA, Fishbein M. 2006. Plant defense syndromes. Ecology 87, S132–S149. ( 10.1890/0012-9658(2006)87%5B132:PDS%5D2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 6.Fenster CB, Armbruster WS, Wilson P, Dudash MR, Thomson JD. 2004. Pollination syndromes and floral specialization. Annu. Rev. Ecol. Evol. Syst. 35, 375–403. ( 10.1146/annurev.ecolsys.34.011802.132347) [DOI] [Google Scholar]
- 7.Dingle H. 2006. Animal migration: is there a common migratory syndrome? J. Ornithol. 147, 212–220. ( 10.1007/s10336-005-0052-2) [DOI] [Google Scholar]
- 8.Ronce O, Clobert J. 2012. Dispersal syndromes. In Dispersal ecology and evolution (eds Clobert J, Baguette M, Benton TG, Bullock JM), pp. 119–138. Oxford, UK: Oxford University Press. [Google Scholar]
- 9.Lande R. 1979. Quantitative genetic analysis of multivariate evolution, applied to brain: body size allometry. Evolution 33, 402–416. ( 10.2307/2407630) [DOI] [PubMed] [Google Scholar]
- 10.Lande R, Arnold SJ. 1983. The measurement of selection on correlated characters. Evolution 37, 1210–1226. ( 10.2307/2408842) [DOI] [PubMed] [Google Scholar]
- 11.Bell AM. 2005. Behavioural differences between individuals and two populations of stickleback (Gasterosteus aculeatus). J. Evol. Biol. 18, 464–473. ( 10.1111/j.1420-9101.2004.00817.x) [DOI] [PubMed] [Google Scholar]
- 12.Cote J, Dreiss A, Clobert J. 2008. Social personality trait and fitness. Proc. R. Soc. B 275, 2851–2858. ( 10.1098/rspb.2008.0783) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ronce O. 2007. How does it feel to be like a rolling stone? Ten questions about dispersal evolution. Annu. Rev. Ecol. Evol. Syst. 38, 231–253. ( 10.1146/annurev.ecolsys.38.091206.095611) [DOI] [Google Scholar]
- 14.Kokko H, López-Sepulcre A. 2006. From individual dispersal to species ranges: perspectives for a changing world. Science 313, 789–791. ( 10.1126/science.1128566) [DOI] [PubMed] [Google Scholar]
- 15.Cousens R, Dytham C, Law R. 2008. Dispersal in plants. A population perspective. Oxford, UK: Oxford University Press. [Google Scholar]
- 16.Clobert J, Baguette M, Benton TG, Bullock JM. 2012. Dispersal ecology and evolution. Oxford, UK: Oxford University Press. [Google Scholar]
- 17.Travis JMJ, et al. 2013. Dispersal and species’ responses to climate change. Oikos 122, 1532–1540. ( 10.1111/j.1600-0706.2013.00399.x) [DOI] [Google Scholar]
- 18.Bonte D, et al. 2012. Costs of dispersal. Biol. Rev. 87, 290–312. ( 10.1111/j.1469-185X.2011.00201.x) [DOI] [PubMed] [Google Scholar]
- 19.Duputié A, Massol F. 2013. An empiricist's guide to theoretical predictions on the evolution of dispersal. Interface Focus 3, 20130028 ( 10.1098/rsfs.2013.0028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moles AT, Westoby M. 2006. Seed size and plant strategy across the whole life cycle. Oikos 113, 91–105. ( 10.1111/j.0030-1299.2006.14194.x) [DOI] [Google Scholar]
- 21.Fjerdingstad EJ, Schtickzelle N, Manhes P, Gutierrez A, Clobert J. 2007. Evolution of dispersal and life history strategies – Tetrahymena ciliates. BMC Evol. Biol. 7, 133 ( 10.1186/1471-2148-7-133) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bestion E, Clobert J, Cote J. 2015. Dispersal response to climate change: scaling down to intraspecific variation. Ecol. Lett. 18, 1226–1233. ( 10.1111/ele.12502) [DOI] [Google Scholar]
- 23.Stevens VM, et al. 2014. A comparative analysis of dispersal syndromes in terrestrial and semi-terrestrial animals. Ecol. Lett. 17, 1039–1052. ( 10.1111/ele.12303) [DOI] [PubMed] [Google Scholar]
- 24.Clobert J, Le Galliard J-F, Cote J, Meylan S, Massot M. 2009. Informed dispersal, heterogeneity in animal dispersal syndromes and the dynamics of spatially structured populations. Ecol. Lett. 12, 197–209. ( 10.1111/j.1461-0248.2008.01267.x) [DOI] [PubMed] [Google Scholar]
- 25.Cote J, Clobert J, Brodin T, Fogarty S, Sih A. 2010. Personality-dependent dispersal: characterization, ontogeny and consequences for spatially structured populations. Phil. Trans. R. Soc. B 365, 4065–4076. ( 10.1098/rstb.2010.0176) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheptou P-O, Carrue O, Rouifed S, Cantarel A. 2008. Rapid evolution of seed dispersal in an urban environment in the weed Crepis sancta. Proc. Natl Acad. Sci. USA 105, 3796–3799. ( 10.1073/pnas.0708446105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zera AJ, Denno RF. 1997. Physiology and ecology of dispersal polymorphism in insects. Annu. Rev. Entomol. 42, 207–230. ( 10.1146/annurev.ento.42.1.207) [DOI] [PubMed] [Google Scholar]
- 28.Tallmon DA, Luikart G, Waples RS. 2004. The alluring simplicity and complex reality of genetic rescue. Trends Ecol. Evol. 19, 489–496. ( 10.1016/j.tree.2004.07.003) [DOI] [PubMed] [Google Scholar]
- 29.Zakas C, Hall DW. 2012. Asymmetric dispersal can maintain larval polymorphism: a model motivated by Streblospio benedicti. Int. Comp. Biol. 52, 197–212. ( 10.1093/icb/ics055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fronhofer EA, Kubisch A, Hovestadt T, Poethke HJ. 2011. Assortative mating counteracts the evolution of dispersal polymorphisms. Evolution 65, 2461–2469. ( 10.1111/j.1558-5646.2011.01312.x) [DOI] [PubMed] [Google Scholar]
- 31.Langellotto GA, Denno RF, Ott JR. 2000. A trade-off between flight capability and reproduction in males of a wing-dimorphic insect. Ecology 81, 865–875. ( 10.1890/0012-9658(2000)081%5B0865:ATOBFC%5D2.0.CO;2) [DOI] [Google Scholar]
- 32.Zera AJ, Brisson JA. 2012. Quantitative, physiological, and molecular genetics of dispersal/migration. In Dispersal ecology and evolution (eds Clobert J, Baguette M, Benton TG, Bullock JM), pp. 63–75. Oxford, UK: Oxford University Press. [Google Scholar]
- 33.Wheat CW. 2012. Dispersal genetics: emerging insights from fruitflies, butterflies and beyond. In Dispersal ecology and evolution (eds Clobert J, Baguette M, Benton TG, Bullock JM), pp. 95–107. Oxford, UK: Oxford University Press. [Google Scholar]
- 34.Orsini L, Wheat CW, Haag CR, Kvist J, Frilander MJ, Hanski I. 2009. Fitness differences associated with Pgi SNP genotypes in the Glanville fritillary butterfly (Melitaea cinxia). J. Evol. Biol. 22, 367–375. ( 10.1111/j.1420-9101.2008.01653.x) [DOI] [PubMed] [Google Scholar]
- 35.Hanski I. 2011. Eco-evolutionary spatial dynamics in the Glanville fritillary butterfly. Proc. Natl Acad. Sci. USA 108, 14 397–14 404. ( 10.1073/pnas.1110020108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Talavera M, Arista M, Ortiz PL. 2012. Evolution of dispersal traits in a biogeographical context: a study using the heterocarpic Rumex bucephalophorus as a model. J. Ecol. 100, 1194–1203. ( 10.1111/j.1365-2745.2012.01999.x) [DOI] [Google Scholar]
- 37.Korsten P, van Overveld T, Adriaensen F, Matthysen E. 2013. Genetic integration of local dispersal and exploratory behaviour in a wild bird. Nat. Comm. 4, 2362 ( 10.1038/ncomms3362) [DOI] [PubMed] [Google Scholar]
- 38.Leslie AB, Beaulieu JM, Crane PR, Donoghue MJ. 2013. Explaining the distribution of breeding and dispersal syndromes in conifers. Proc. R. Soc. B 280, 20131812 ( 10.1098/rspb.2013.1812) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buoro M, Carlson SM. 2014. Life-history syndromes: integrating dispersal through space and time. Ecol. Lett. 17, 756–767. ( 10.1111/ele.12275) [DOI] [PubMed] [Google Scholar]
- 40.Willis CG, Hall JC, Rubio de Casas R, Wang TY, Donohue K. 2014. Diversification and the evolution of dispersal ability in the tribe Brassiceae (Brassicaceae). Ann. Bot. 114, 1675–1686. ( 10.1093/aob/mcu196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang Y, Bolnick D, Kirkpatrick M. 2013. Assortative mating in animals. Am. Nat. 181, E125–E138. ( 10.1086/670160) [DOI] [PubMed] [Google Scholar]
- 42.Ducatez S, Legrand D, Chaput-Bardy A, Stevens VM, Fréville H, Baguette M. 2012. Inter-individual variation in movement: is there a mobility syndrome in the large white butterfly Pieris brassicae? Ecol. Entom. 37, 377–385. ( 10.1111/j.1365-2311.2012.01375.x) [DOI] [Google Scholar]
- 43.Legrand D, Trochet A, Moulhérat S, Calvez O, Stevens VM, Ducatez S, Clobert J, Baguette M. 2015. Ranking the ecological causes of dispersal in a butterfly. Ecography 38, 822–831. ( 10.1111/ecog.01283) [DOI] [Google Scholar]
- 44.Berwaerts K, Van Dyck H, Aerts P. 2002. Does flight morphology relate to flight performance? An experimental test with the butterfly Pararge aegeria. Funct. Ecol. 16, 484–491. ( 10.1046/j.1365-2435.2002.00650.x) [DOI] [Google Scholar]
- 45.Sekar S. 2012. A meta-analysis of the traits affecting dispersal ability in butterflies: can wingspan be used as a proxy? J. Anim. Ecol. 81, 174–184. ( 10.1111/j.1365-2656.2011.01909.x) [DOI] [PubMed] [Google Scholar]
- 46.Stevens VM, Trochet A, Van Dyck H, Clobert J, Baguette M. 2012. How is dispersal integrated in life histories: a quantitative analysis using butterflies. Ecol. Lett. 15, 74–86. ( 10.1111/j.1461-0248.2011.01709.x) [DOI] [PubMed] [Google Scholar]
- 47.Ducatez S, Baguette M, Trochet A, Chaput-Bardy A, Legrand D, Stevens VM, Fréville H. 2012. Flight endurance and heating rate vary with both latitude and habitat connectivity in a butterfly species. Oikos 122, 601–611. ( 10.1111/j.1600-0706.2012.20947.x) [DOI] [Google Scholar]
- 48.Larranaga N, Baguette M, Calvez O, Trochet A, Ducatez S, Legrand D. 2013. Intra- and inter-individual variation in flight direction in a migratory butterfly co-vary with individual mobility. J. Exp. Biol. 216, 3156–3163. ( 10.1242/jeb.082883) [DOI] [PubMed] [Google Scholar]
- 49.Rutowski RL. 2010. Evidence for mate choice in a sulphur butterfly (Colias eurytheme). Ethology 70, p103–114. (doi:10.1111/j.1439-0310.1985.tb00504.x) [Google Scholar]
- 50.Ellers J, Boggs CL. 2003. The evolution of wing color: male mate choice opposes adaptive wing color divergence in Colias butterflies. Evolution 57, 1100–1106. ( 10.1111/j.0014-3820.2003.tb00319.x) [DOI] [PubMed] [Google Scholar]
- 51.Legrand D, et al. 2012. The Metatron: an experimental system to study dispersal and metaecosystems for terrestrial organisms. Nat. Methods 9, 828–833. ( 10.1038/nmeth.2104) [DOI] [PubMed] [Google Scholar]
- 52.Ducatez S, Baguette M, Stevens VM, Legrand D, Fréville H. 2012. Complex interactions between paternal and maternal effects: parental experience and age at reproduction affect fecundity and offspring performance in a butterfly. Evolution 66, 3558–3569. ( 10.1111/j.1558-5646.2012.01704.x) [DOI] [PubMed] [Google Scholar]
- 53.Abramoff MD, Magalhães PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics Int. 11, 36–42. [Google Scholar]
- 54.Chaput-Bardy A, Ducatez S, Legrand D, Baguette M. 2014. Fitness costs of thermal reaction norms for wing melanisation in the large white butterfly (Pieris brassicae). PLoS ONE 9, e90026 ( 10.1371/journal.pone.0090026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Phillips PC, Arnold SJ. 1989. Visualizing multivariate selection. Evolution 43, 1209–1222. ( 10.2307/2409357) [DOI] [PubMed] [Google Scholar]
- 56.Blows MW, Brooks R. 2003. Measuring non-linear selection. Am. Nat. 162, 815–820. ( 10.1086/378905) [DOI] [PubMed] [Google Scholar]
- 57.Wold S, Sjöström M, Eriksson L. 2001. PLS-regression: a basic tool of chemometrics. Chem. Intell. Lab. Syst. 58, 109–130. ( 10.1016/S0169-7439(01)00155-1) [DOI] [Google Scholar]
- 58.Graham MH. 2003. Confronting multicollinearity in ecological multiple regression. Ecology 84, 2809–2815. ( 10.1890/02-3114) [DOI] [Google Scholar]
- 59.Wilson AJ, Réale D, Clements MN, Morrissey MM, Postma E, Walling CA, Kruuk LE, Nussey DH. 2010. An ecologist's guide to the animal model. J. Anim. Ecol. 79, 13–26. ( 10.1111/j.1365-2656.2009.01639.x) [DOI] [PubMed] [Google Scholar]
- 60.de Villemereuil P, Gimenez O, Doligez B. 2013. Comparing parent–offspring regression with frequentist and Bayesian animal models to estimate heritability in wild populations: a simulation study for Gaussian and binary traits. Meth. Ecol. Evol. 4, 260–275. ( 10.1111/2041-210X.12011) [DOI] [Google Scholar]
- 61.Hadfield JD, Nutall A, Osorio D, Owens IPF. 2007. Testing the phenotypic gambit: phenotypic, genetic and environmental correlations of colour. J. Evol. Biol. 20, 549–557. ( 10.1111/j.1420-9101.2006.01262.x) [DOI] [PubMed] [Google Scholar]
- 62.Agrawal AF, Stinchcombe JR. 2009. How much do genetic covariances alter the rate of adaptation? Proc. R. Soc. B 276, 1183–1191. ( 10.1098/rspb.2008.1671) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuznetsova A, Brockhoff PB, Christensen RHB. 2014. lmerTest. R package, version 2.0-6. See http://cran.r-project.org/web/packages/lmerTest/.
- 64.Hoerl AE, Kennard RW. 1970. Ridge regression: biased estimation for nonorthogonal problems. Technometrics 12, 55–67. ( 10.1080/00401706.1970.10488634) [DOI] [Google Scholar]
- 65.McDonald GC. 2009. Ridge regression. WIREs: Comp. Stat. 1, 93–100. ( 10.1002/wics.14) [DOI] [Google Scholar]
- 66.R Core Team. 2013. R: a language and environment for statistical computing. Vienna, Austria: (http://www.R-project.org) [Google Scholar]
- 67.Janzen FJ, Stern HS. 1998. Logistic regression for empirical studies of multivariate selection. Evolution 52, 1564–1571. ( 10.2307/2411330) [DOI] [PubMed] [Google Scholar]
- 68.Stinchcombe JR, Agrawal AF, Hohenlohe PA, Arnold SJ, Blows MW. 2008. Estimating nonlinear selection using quadratic selection coefficients: double or nothing? Evolution 62, 2435–2440. ( 10.1111/j.1558-5646.2008.00449.x) [DOI] [PubMed] [Google Scholar]
- 69.Schmidt-Nielsen K. 1984. Why is animal size so important? Cambridge, UK: Cambridge University Press. [Google Scholar]
- 70.Honěk A. 1993. Intraspecific variation in body size and fecundity in insects: a general relationship. Oikos 66, 483–492. ( 10.2307/3544943) [DOI] [Google Scholar]
- 71.Berger D, Walters R, Gotthard K. 2008. What limits insect fecundity? Body size- and temperature-dependent egg maturation and oviposition in a butterfly. Funct. Ecol. 22, 523–529. ( 10.1111/j.1365-2435.2008.01392.x) [DOI] [Google Scholar]
- 72.Woiwod P, Reynolds DR, Thomas CD. 2001. Insect movement: mechanisms and consequences. New York, NY: CABI Publishing. [Google Scholar]
- 73.Gyulavári HA, Therry L, Dévai G, Stoks R. 2014. Sexual selection on flight endurance, flight-related morphology and physiology in a scrambling damselfly. Evol. Ecol. 28, 639–654. ( 10.1007/s10682-014-9703-1) [DOI] [Google Scholar]
- 74.Bonduriansky R, Chenoweth SF. 2009. Intralocus sexual conflict. Trends Ecol. Evol. 24, 280–288. ( 10.1016/j.tree.2008.12.005) [DOI] [PubMed] [Google Scholar]
- 75.Baguette M, Vansteenwegen C, Convi I, Nève G. 1998. Sex-biased density-dependent migration in a metapopulation of the butterfly Proclossiana eunomia. Acta Oecol. 19, 17–24. ( 10.1016/S1146-609X(98)80004-0) [DOI] [Google Scholar]
- 76.Lawson Handley LJ, Perrin N. 2007. Advances in our understanding of mammalian sex-biased dispersal. Mol. Ecol. 16, 1559–1578. ( 10.1111/j.1365-294X.2006.03152.x) [DOI] [PubMed] [Google Scholar]
- 77.Fresneau N, Kluen E, Brommer JE. 2014. A sex-specific behavioral syndrome in a wild passerine. Behav. Ecol. 25, 359–367. ( 10.1093/beheco/aru008) [DOI] [Google Scholar]
- 78.Rueffler C, Van Dooren TJM, Leimar O, Abrams PA. 2006. Disruptive selection and then what? Trends Ecol. Evol. 21, 238–245. ( 10.1016/j.tree.2006.03.003) [DOI] [PubMed] [Google Scholar]
- 79.Johnson CG. 1969. Migration and dispersal of insects by flight. London, UK: Methuen. [Google Scholar]
- 80.Kemp DJ. 2008. Female mating biases for bright ultraviolet iridescence in the butterfly Eurema hecabe (Pieridae). Behav. Ecol. 19, 1–8. ( 10.1093/beheco/arm094) [DOI] [Google Scholar]
- 81.Legrand D, Larranaga N, Bertrand R, Ducatez S, Calvez O, Stevens VM, Baguette M. 2016. Data from: Evolution of a butterfly dispersal syndrome. Dryad Digital Repository. ( 10.5061/dryad.s73t1) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting this article are available in the Dryad Digital Repository, with identifier doi:10.5061/dryad.s73t1 [81].


