Abstract
The notion of state for a system is prevalent in the quantitative sciences and refers to the minimal system summary sufficient to describe the time evolution of the system in a self-consistent manner. This is a prerequisite for a principled understanding of the inner workings of a system. Owing to the complexity of intracellular processes, experimental techniques that can retrieve a sufficient summary are beyond our reach. For the case of stochastic biomolecular reaction networks, we show how to convert the partial state information accessible by experimental techniques into a full system state using mathematical analysis together with a computational model. This is intimately related to the notion of conditional Markov processes and we introduce the posterior master equation and derive novel approximations to the corresponding infinite-dimensional posterior moment dynamics. We exemplify this state reconstruction approach using both in silico data and single-cell data from two gene expression systems in Saccharomyces cerevisiae, where we reconstruct the dynamic promoter and mRNA states from noisy protein abundance measurements.
Keywords: optimal filtering, continuous time Markov chains, chemical master equation, moment dynamics, gene expression
1. Introduction
Today's experimental techniques of molecular biology give us some insight into biological cells but provide a far from complete picture of the inner workings of such cells or even of any of their subcomponents. With the advances in quantitative single-cell technologies, the generation of calibrated models of particular cellular processes such as the expression of a gene becomes feasible [1–7]. A calibrated model can then be simulated forward to explore the a prior behaviours that one can expect to observe experimentally. However, such forward simulations are not useful if one asks which of those behaviours are compatible with the actual measurement of a particular experiment. For instance, a stochastic gene expression model can give rise to various mRNA and protein trajectories but a model alone cannot be used to determine those mRNA dynamics that are compatible with particular protein measurement trajectories. In general, the problem is to reconstruct the dynamics of experimentally inaccessible states of a process that best match the trajectories of the observable states of the process in a particular experimental run. In other words, the observations allow us to filter the a priori behaviours into compatible posterior behaviours. Mathematically, we condition the inaccessible states onto those observations. Such conditioning or filtering has a long tradition in mathematics and engineering [8,9]. The key is to obtain governing master equations, such as the Kushner and the Zakai equations [10,11], that describe the time evolution of the conditional probability distribution. The best known of such governing equations is the Kalman filter, which yields a finite parametrization of the posterior distribution by considering the case that states evolve according to a linear stochastic differential equation and measurements are Gaussian distributed [12]. In most other cases (e.g. nonlinear stochastic differential equation), no finite parametrization of the posterior distribution can be found and a plethora of approximations has been proposed in recent decades [13–15]. In comparison with the other approximation methods, such as the popular extended Kalman filter, particle filters [16,17] do not rely on any local linearization techniques while, as a Monte Carlo method, they are computationally expensive and do not scale well to high state dimensions.
In this work, we complement the chemical master equation used to describe the a priori dynamics of a stochastic reaction network with its posterior counterpart. It refactors the seminal result of Wonham obtained for the case of continuous-time Markov processes [18]. The posterior master equation and its exact posterior moment equation exhibit the same scalability problems as their traditional a priori counterparts and we present scalable approximations of the posterior process. More specifically, the main contribution of this work is a novel approximation approach obtained by specific adaptations of moment closure techniques to the posterior setting. In contrast with traditional optimal filtering, where observations of the accessible states are assumed to be available in continuous time [19], we mainly focus on the practical scenario where observations are available only at discrete time points.
2. Conditional Markov processes
We consider a well-mixed reaction system of d species and r different reaction channels. The state X(t) comprising the integer abundance of the species at time t shows Markovian dynamics where state transitions take place according to the change vectors  of each reaction and where the reaction's propensity is given by the function
of each reaction and where the reaction's propensity is given by the function  for
for  . With that, a reaction system with X(0) copies of each species at time zero will have
. With that, a reaction system with X(0) copies of each species at time zero will have
 |
copies at time t, where  are independent Poisson processes of unit rate [20]. Starting with some initial distribution
are independent Poisson processes of unit rate [20]. Starting with some initial distribution  , the distribution
, the distribution  at time t evolves according to the chemical master equation [21]. Following our nomenclature, we refer to this equation as the unconditional or prior master equation as it determines the probability over species abundances if no further information or measurements on the system are provided. The traditional way in mathematics to formalize measurements [19] is to assume some l-dimensional covariate process Y(t) of X(t), for instance of the form
at time t evolves according to the chemical master equation [21]. Following our nomenclature, we refer to this equation as the unconditional or prior master equation as it determines the probability over species abundances if no further information or measurements on the system are provided. The traditional way in mathematics to formalize measurements [19] is to assume some l-dimensional covariate process Y(t) of X(t), for instance of the form
| 2.1 |
where B is a full-rank matrix and W(t) is the standard l-dimensional Brownian motion independent of X(t). For example, for l = 1 and  one can think of a reaction system where the i-th species is fluorescently labelled and Y(t) corresponds to the integrated fluorescence intensity measured at the microscope. Observation model (2.1) could be appropriate in the context of fluorescence correlation spectroscopy [22] where a photon count trace at the photo-multipliers under high arrival rates admits a diffusion approximation of the form (2.1). Having such observations available one can ask for the probability over species abundance in the presence of that information. That is, the conditional probability
one can think of a reaction system where the i-th species is fluorescently labelled and Y(t) corresponds to the integrated fluorescence intensity measured at the microscope. Observation model (2.1) could be appropriate in the context of fluorescence correlation spectroscopy [22] where a photon count trace at the photo-multipliers under high arrival rates admits a diffusion approximation of the form (2.1). Having such observations available one can ask for the probability over species abundance in the presence of that information. That is, the conditional probability  , where τ denotes the time up to which measurements are available. Accordingly, one distinguishes between filtering and smoothing for τ = t and τ > 1, respectively. As conditioning usually reduces variance, the measurements from a single cell result in less uncertainty in the dynamic states of the reaction system than with the traditional (prior) chemical master equation. Interestingly, the process X(t) conditioned on such covariate information is still Markovian [8]. Refactoring the seminal work of Wonham for optimal filtering of continuous-time Markov chains [18], the resulting conditional chemical master equation for τ = 1 reads
, where τ denotes the time up to which measurements are available. Accordingly, one distinguishes between filtering and smoothing for τ = t and τ > 1, respectively. As conditioning usually reduces variance, the measurements from a single cell result in less uncertainty in the dynamic states of the reaction system than with the traditional (prior) chemical master equation. Interestingly, the process X(t) conditioned on such covariate information is still Markovian [8]. Refactoring the seminal work of Wonham for optimal filtering of continuous-time Markov chains [18], the resulting conditional chemical master equation for τ = 1 reads
 |
2.2 |
with  ,
,  and
and  , where expectation is taken with respect to
, where expectation is taken with respect to  . Owing to its dependency on that expectation, (2.2) is cumbersome to solve and one often resorts to an equation for the unnormalized version of
. Owing to its dependency on that expectation, (2.2) is cumbersome to solve and one often resorts to an equation for the unnormalized version of  , performing normalization after numerical integration (electronic supplementary material, S.3). Equation (2.2) is a special case of the general class of optimal filtering equations [19].
, performing normalization after numerical integration (electronic supplementary material, S.3). Equation (2.2) is a special case of the general class of optimal filtering equations [19].
In most live-cell imaging applications observation at continuous time are unrealistic due to experimental constraints such as phototoxicity and bleaching. In practice one is faced with observations at discrete times  with
with  usually admitting a conditional distribution of the form
usually admitting a conditional distribution of the form  . Given the states at observation times, the observations are assumed to be independent. The conditional probability
. Given the states at observation times, the observations are assumed to be independent. The conditional probability  with
with  satisfies the unconditional master equation
satisfies the unconditional master equation
 |
2.3 |
with  together with the reset conditions
together with the reset conditions
| 2.4 |
at the observation times  , with the normalizing constant
, with the normalizing constant  . For the smoothing case, the conditional probability
. For the smoothing case, the conditional probability  for any
for any  denoted by
denoted by  admits the factorization through the Markov property of the form
admits the factorization through the Markov property of the form
| 2.5 |
where  with
with  and normalizer
and normalizer 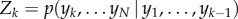 . The probability of future observations given the current system state
. The probability of future observations given the current system state  satisfies the backward master equation
satisfies the backward master equation
 |
2.6 |
with reset conditions
| 2.7 |
and terminal conditions  for all
for all  . By solving the backward and the forward conditional master equation, one can determine the smoothing probability
. By solving the backward and the forward conditional master equation, one can determine the smoothing probability  through (2.5). Examining this factorization and the fact that
through (2.5). Examining this factorization and the fact that  , we conclude that our knowledge of the underlying system state X(t) is less uncertain when all measurements up to time T are incorporated compared with the filtering case where measurements up to time t are taken into account. Differentiating (2.5) yields an evolution equation for
, we conclude that our knowledge of the underlying system state X(t) is less uncertain when all measurements up to time T are incorporated compared with the filtering case where measurements up to time t are taken into account. Differentiating (2.5) yields an evolution equation for  directly
directly
 |
2.8 |
which we term the posterior or smoothing master equation with  . It comprises novel time-varying posterior or smoothing propensity functions of the form
. It comprises novel time-varying posterior or smoothing propensity functions of the form
| 2.9 |
Hence, the prior propensities  are modulated by a time-varying fraction that steers the process towards future measurements. The expression for the posterior propensities provides the means to draw sample paths of the posterior smoothing process through a stochastic simulation scheme adapted to time-varying propensities [23,24].
are modulated by a time-varying fraction that steers the process towards future measurements. The expression for the posterior propensities provides the means to draw sample paths of the posterior smoothing process through a stochastic simulation scheme adapted to time-varying propensities [23,24].
As an illustrative example, we consider the smoothing problem for a birth–death process  with respective rates c1, c2 and with a single noise-free observation
with respective rates c1, c2 and with a single noise-free observation  at t1 = T and the deterministic initial condition X(0) = 0. This set-up corresponds to the classical bridging problem or to the problem of endpoint conditioned sampling of Markov chains [25] and can be solved explicitly for this case. We aim to compute the smoothing distribution
at t1 = T and the deterministic initial condition X(0) = 0. This set-up corresponds to the classical bridging problem or to the problem of endpoint conditioned sampling of Markov chains [25] and can be solved explicitly for this case. We aim to compute the smoothing distribution  . The prior distribution
. The prior distribution  coincides with the filtering distribution within
coincides with the filtering distribution within  and admits a representation in terms of a Poisson distribution with time-varying rate [26,27]
and admits a representation in terms of a Poisson distribution with time-varying rate [26,27]
with  and
and  . Accordingly, the solution of the backward equation (2.6) can be expressed as
. Accordingly, the solution of the backward equation (2.6) can be expressed as
With that, one computes the probability distribution  of the endpoint conditioned process through (2.5). The same result is obtained by integrating the smoothing master equation (2.8), where the posterior propensities for this example are
of the endpoint conditioned process through (2.5). The same result is obtained by integrating the smoothing master equation (2.8), where the posterior propensities for this example are
| 2.10 |
The function  is a monotone decreasing function in t and reaches zero at t = T. Hence, in order to reach the state X(T) = 0 with probability one, the posterior birth rate converges to zero while the death rate becomes unbounded as
is a monotone decreasing function in t and reaches zero at t = T. Hence, in order to reach the state X(T) = 0 with probability one, the posterior birth rate converges to zero while the death rate becomes unbounded as  . Some sample paths of the posterior birth–death process with time-varying rates (2.10) are shown in figure 1.
. Some sample paths of the posterior birth–death process with time-varying rates (2.10) are shown in figure 1.
Figure 1.

Sample paths of the posterior birth–death process (T = 50 s, c1 = 5 molecules s−1, c2 = 0.1 s−1) with time-varying propensities (2.10).
3. Posterior moment equations
The presented posterior master equations inherit the same scability problems as the original chemical master equation due to the combinatorial increase in the cardinality of  with the number of species d. Traditional approaches that can approximately capture the stochastic dynamics of the prior process are the van Kampen expansion [21] and moment closure techniques [28–30]. Similar techniques can be applied to the posterior master equations (2.3), (2.8) and also to the backward equation (2.6). For the latter, a linear noise approximation was performed in [31]. We follow the moment closure approach and subsequently derive novel approximate posterior moment dynamics. Throughout we will consider propensity functions of the form
with the number of species d. Traditional approaches that can approximately capture the stochastic dynamics of the prior process are the van Kampen expansion [21] and moment closure techniques [28–30]. Similar techniques can be applied to the posterior master equations (2.3), (2.8) and also to the backward equation (2.6). For the latter, a linear noise approximation was performed in [31]. We follow the moment closure approach and subsequently derive novel approximate posterior moment dynamics. Throughout we will consider propensity functions of the form  with
with  the stochastic rate constant of the reaction j and
the stochastic rate constant of the reaction j and  any polynomial function of bounded degree. Using a multi-index
any polynomial function of bounded degree. Using a multi-index  with
with  for
for 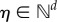 [30], we can compactly write
[30], we can compactly write  with the shorthand
with the shorthand  . Similar to the moment expansion for the traditional master equation [30], one can develop a moment expansion of the filtering master equation (2.3). Denoting its moment of order η by
. Similar to the moment expansion for the traditional master equation [30], one can develop a moment expansion of the filtering master equation (2.3). Denoting its moment of order η by  the generally infinite set of moment equations reads
the generally infinite set of moment equations reads
 |
3.1 |
with the reset conditions at the observation time  following the Kallianpur–Striebel formula [19]
following the Kallianpur–Striebel formula [19]
| 3.2 |
In terms of computational complexity, solving the moment equations together with (3.2) does not show any advantage compared with directly solving (2.3) because (3.2) still involves the filtering distribution  . The main contribution of this work is to propose an approximate approach to the reset condition (3.2) for the posterior moments.
. The main contribution of this work is to propose an approximate approach to the reset condition (3.2) for the posterior moments.
4. Approximate posterior moments dynamics
In the following, we derive novel approximate posterior moment dynamics exploiting a special form of the tower property for conditional expectations
for integrable functions s and u. Additionally, we make use of traditional moment closure techniques. We begin with a univariate system (i.e. d = 1) and assume that observations are subject to additive noise  , where wi are i.i.d. random variables with bounded moments up to order four. For
, where wi are i.i.d. random variables with bounded moments up to order four. For  , we obtain the following approximation to the reset conditions (3.2) for the posterior filtering moments (electronic supplementary material, S.6):
, we obtain the following approximation to the reset conditions (3.2) for the posterior filtering moments (electronic supplementary material, S.6):
 |
4.1 |
where the coefficients  are determined by the linear equations obtained from the tower property (4.1) (electronic supplementary material, S.6)
are determined by the linear equations obtained from the tower property (4.1) (electronic supplementary material, S.6)
 |
4.2 |
with  , which, however, involve moment
, which, however, involve moment  with order q up to 2η. To cope with this problem, one can employ moment closure techniques and approximate the higher order moments by functions of lower order moments up to order η; see, for example, [32–34]. With that, the linear set of equations can be solved for
with order q up to 2η. To cope with this problem, one can employ moment closure techniques and approximate the higher order moments by functions of lower order moments up to order η; see, for example, [32–34]. With that, the linear set of equations can be solved for  and (4.1) can serve as an approximation to (3.2). We provide explicit expressions for (4.1) in the case of normal, lognormal and modified normal moment closure techniques in the electronic supplementary material, S.6.1–6.3. It is worth noting that the update formula of the celebrated Kalman filter turns out to be a special case of our approximation approach for the case of a normal closure combined with an additive Gaussian measurement model (electronic supplementary material, S.6.1).
and (4.1) can serve as an approximation to (3.2). We provide explicit expressions for (4.1) in the case of normal, lognormal and modified normal moment closure techniques in the electronic supplementary material, S.6.1–6.3. It is worth noting that the update formula of the celebrated Kalman filter turns out to be a special case of our approximation approach for the case of a normal closure combined with an additive Gaussian measurement model (electronic supplementary material, S.6.1).
We also provide an alternative approximation to posterior variance for that case. It is natural to assume that the reset step, which incorporates the information of a measurement, leads to a variance  smaller than
smaller than  . That is, at measurement point ti,
. That is, at measurement point ti,
| 4.3 |
and hence  , where coefficient m can be obtained by using (4.1) (see electronic supplementary material, S.6.4). For a multi-variate system, we define the first and the second filtering moments
, where coefficient m can be obtained by using (4.1) (see electronic supplementary material, S.6.4). For a multi-variate system, we define the first and the second filtering moments  and
and  , respectively, together with the filtering covariance
, respectively, together with the filtering covariance  . The corresponding moment dynamics for these quantities are detailed in the electronic supplementary material, S.7. The proposed approach can be applied to other observation models such as a lognormal noise model, which will subsequently be used in a gene expression model.
. The corresponding moment dynamics for these quantities are detailed in the electronic supplementary material, S.7. The proposed approach can be applied to other observation models such as a lognormal noise model, which will subsequently be used in a gene expression model.
5. An Rauch–Tung–Striebel approximation to the smoothing moments
Based on the proposed moment approximation to the filtering distribution, one can derive approximations to the moments of the smoothing distribution leveraging existing results for the Rauch–Tung–Striebel (RTS) or the modified Bryson–Frazier (MBF) smoother [35]. Here, we consider the RTS smoother because the RTS smoother has some desired properties such as stability and several other smoothers are based upon it [35,36]. Similarly, a reaction system with d ≥ 1 denotes the first and the second smoothing moments by  and the smoothing covariance by
and the smoothing covariance by 
Let us consider the first-order reaction networks. The prior mean  of such a system is given by (equation (S48) with
of such a system is given by (equation (S48) with  in the electronic supplementary material, S.5.1)
in the electronic supplementary material, S.5.1)
| 5.1 |
For the gene expression model introduced below, one gets
 |
5.2 |
Let  be the state transition matrix corresponding to
be the state transition matrix corresponding to  , that is,
, that is,  and
and  , which yields
, which yields  for all
for all  . According to (2.5), the posterior smoothing and filtering moments have to coincide at the endpoint tN. Consequently, we initialize our approximation to
. According to (2.5), the posterior smoothing and filtering moments have to coincide at the endpoint tN. Consequently, we initialize our approximation to  by the approximate filtering moments at tN obtained through (4.1) and (4.2). The resulting RTS smoother (see electronic supplementary material, S.8) is then given by the backward equation
by the approximate filtering moments at tN obtained through (4.1) and (4.2). The resulting RTS smoother (see electronic supplementary material, S.8) is then given by the backward equation
 |
with smoother gain  and
and  .
.
6. Application to gene expression models
Consider the standard two-state gene expression model consisting of six reactions that involve four different species: G1 and G0 for the active and inactive promoter, respectively, M for mRNA and P for the expressed protein
Through the conservation of active and inactive promoters, the state of the system can be represented by  , where X1, X2 and X3 are the amounts of G1, M and P, respectively. A key problem in gene expression is to reconstruct the inaccessible states such as the mRNA abundance from noisy measurements of the protein abundance dynamics (e.g. through fluorescent labelling). Such state reconstruction is of particular interest for transient induction of genes, where the time-varying inducer can be modelled by a time-varying promoter activation rate c1 = c1(t). Throughout this section, we assume lognormal measurement noise on the protein dynamics. That is,
, where X1, X2 and X3 are the amounts of G1, M and P, respectively. A key problem in gene expression is to reconstruct the inaccessible states such as the mRNA abundance from noisy measurements of the protein abundance dynamics (e.g. through fluorescent labelling). Such state reconstruction is of particular interest for transient induction of genes, where the time-varying inducer can be modelled by a time-varying promoter activation rate c1 = c1(t). Throughout this section, we assume lognormal measurement noise on the protein dynamics. That is,
with  ,
,  and the corresponding observation matrix
and the corresponding observation matrix  . In the following, we show that for synthetic data and for real single-cell data from Saccharomyces cerevisiae the proposed method allows one to reconstruct robustly the mRNA abundance and true protein abundance from such noisy measurement trajectories. Before the next observation instant ti, the propagation of the moments is given by the prior moment equations (see, for example, [37] and also equation (S48) with
. In the following, we show that for synthetic data and for real single-cell data from Saccharomyces cerevisiae the proposed method allows one to reconstruct robustly the mRNA abundance and true protein abundance from such noisy measurement trajectories. Before the next observation instant ti, the propagation of the moments is given by the prior moment equations (see, for example, [37] and also equation (S48) with  in the electronic supplementary material, S.5.1)
in the electronic supplementary material, S.5.1)
 |
where  and b1 are now time dependent due to the time-varying promoter activation rate c1. At measurement times ti the following reset conditions are applied:
and b1 are now time dependent due to the time-varying promoter activation rate c1. At measurement times ti the following reset conditions are applied:
 |
where  and
and  are given by
are given by
 |
6.1. In silico experiments
We first apply our proposed approximate approach to the gene expression model using simulated non-stationary data and, for simplicity, assume a constant reaction rate c1. Based on the obtained filtering moments, we compute the RTS approximate for the smoothing moments. For reference, the filtering and the smoothing moments are also computed exactly by integration of the corresponding conditional master equation. The comparison between the approximate moments and the exact ones is given in figure 2, where the prior moments are also given for comparison. Note that the measurement noise standard deviation σ = 0.2 is larger than those in the experimental data used in this study, which were identified as σ = 0.15 in the Msn2 case and σ = 0.125 in the GEV case below, respectively. The approximation to the filtering moments and to the smoothing moments is in good agreement with the exact results. Moreover, one can observe that the actual mRNA and protein dynamics corresponding to the actual measured data points can accurately be tracked by the respective posterior means. From the above discussion, it is evident that the traditional prior dynamics cannot provide this single-trajectory resolution. In the electronic supplementary material, S.7.3, we apply the proposed moment approximation to another in silico problem that exhibits bimodal distributions. It indicates that the posterior distribution of multi-modal systems can often be unimodal due to the conditioning and can hence be approximated well by low-order moment equations.
Figure 2.
Comparison between the proposed approximate moment dynamics versus the exact results for the gene expression model (c1 = 0.02 s−1, c2 = 0.03 s−1, c3 = 1 s−1, c4 = 0.05 s−1, c5 = 0.005 s−1, c6 = 0.02 s−1, ti − ti−1 = 50 s, σ = 0.2) and comparison between posterior moments and prior moments: (a) filtering moments and (b) smoothing moments with simulated data.
6.2. Gene expression systems in yeast
We apply our proposed state reconstruction approach to two different inducible gene expression systems in Saccharomyces cerevisiae. Both systems can be described by the above gene expression model with a time-varying promoter activation rate caused by the nuclear translocation of an inducer.
In the first system, a microfluidic device is used to control the nuclear–cytoplasmic translocation dynamics of the transcription factor Msn2 by modulating the levels of the small molecule 1-NM-PP1 to control the expression of a fluorescent reporter protein (see [6] for a detailed description; subsequently referred to as the Msn2 system). The second system is an artificial gene expression system centred around the chimeric transcription factor GAL4DBD.ER.VP16 (GEV). The GEV translocation is again modulated using a microfluidic device controlling the supply of the hormone β-oestradiol. The nuclear transcription factor GEV activates the transcription of genes under a GAL1 promoter, where we placed a fluorescent reporter protein as a readout (see [7] for a detailed description; subsequently referred to as the GEV system). In both model systems, fluorescent time-lapse microscopy is used to monitor the nuclear–cytoplasmic translocation of the respective transcription factor fused to a fluorescent protein, as well as the expression of a fluorescent protein induced by the respective transcription factor in individual cells.
The two case studies are successively complicated. In the Msn2 case, we associate for every single-cell trace a separate parameter set. This is feasible due to a sufficient number of observations for each trace. The problem thus corresponds to the in silico study from above and no extrinsic noise model is assumed that couples parameter sets from different traces. In most scenarios, however, one needs to pool together heterogeneous traces in order to achieve sufficient estimation accuracy. To illustrate this complication we show, for the GEV case, how an extrinsic noise model can be incorporated into the proposed filtering or smoothing method. The extension is in line with the observation that the GEV expression variability shows a large extrinsic component [7].
6.3. Approximate state reconstruction for Msn2 system
To demonstrate the effectiveness of the proposed method, we reconstruct the mRNA dynamics based on the noisy fluorescent readout of the protein level. For the first case, we used single-cell traces from [6] to estimate the parameters of the kinetic expression model and the measurement noise model using the algorithm given in [7]. The temporal profile of the promoter activation rate is estimated from the nuclear concentration of the transcription factor (see also [6,7] for details). We remark that this induction happens quite rapidly (figure 3). Generally, it is challenging to perform state estimation for such fast varying systems.
Figure 3.
Comparison between the proposed approximate filtering moments and the exact moments based on two exemplary trajectories from the experimental dataset of [6]: (a,b) protein and mRNA dynamics corresponding to trajectory A; (c,d) protein and mRNA dynamics corresponding to trajectory B.
Figure 3 shows the reconstruction results for two exemplary single-cell traces of the dataset in [6]. Applying finite-state-projection techniques [38,39], we were also able to compute the exact moments from solving posterior master equations (2.3) and (2.8) as a reference. However, for larger systems, solving the corresponding master equation quickly becomes computationally infeasible—motivating our approximate approach. Figure 3 indicates that the proposed approximation to the filtering moments works well. As it only involves solving a set of ordinary differential equations with reset conditions the approach is scalable to large reaction systems.
Similarly, we applied the proposed smoothing algorithm to the single-cell trajectories. In particular, we used the presented RTS approximation to the smoothing moments. However, it is observed that the traditional RTS approximation does not always work for the considered experimental data. Figure 4 shows that the approximation works well for trajectory B while it fails for trajectory A where one can observe a collapse of the smoothing covariance for the approximate method even though the involved approximate filtering moments are accurate (cf. figure 3). This indicates that novel approximate methods for the smoothing moments are needed to overcome the limitations of traditional RTS schemes.
Figure 4.
Comparison between the RTS approximations and the exact smoothing moments based on two exemplary trajectories from the experimental dataset of [6]: (a,b) protein and mRNA dynamics corresponding to trajectory A; (c,d) protein and mRNA dynamics corresponding to trajectory B.
6.4. State reconstruction in the presence of extrinsic noise
Apart from the inherent randomness of biomolecular reactions, gene expression was shown to exhibit a substantial degree of extrinsic noise [5,40–42], stemming from various factors in a cell's microenvironment. The considered GEV system showed significant extrinsic noise [7] and we therefore minimally extend the standard gene expression system by a random protein translation rate c4 assumed to be a gamma distribution characterized by shape and rate parameters a and b. In contrast with the Msn2 case study, where we assumed the model parameters to be given, we aim to estimate states and parameters—for the latter in particular the population heterogeneity captured by (a, b). Treating a parameter as just another state that also follows a certain prior distribution, we aim to quantify the gain in certainty about states as observations are acquired. More specifically, before receiving any data we assume a prior heterogeneity characterized by values  and
and  . Hence, the prior moments of an average cell in the population compute to
. Hence, the prior moments of an average cell in the population compute to  , where the outer expectation is over
, where the outer expectation is over  . In order to compute these prior moments, we employ the marginal moments of [3], which treats the unknown reaction rate c4 as a dummy species by rewriting the translation reaction as
. In order to compute these prior moments, we employ the marginal moments of [3], which treats the unknown reaction rate c4 as a dummy species by rewriting the translation reaction as  with unit rate. The resulting prior moments for the model of the GEV system are given in figure 5.
with unit rate. The resulting prior moments for the model of the GEV system are given in figure 5.
Figure 5.
Comparison between the prior marginal moments, RTS approximations to the posterior marginal moments and the exact posterior moments based on one exemplary trajectory from the experimental dataset of [7]: (a,b) means of protein and mRNA; (c,d) standard deviations of protein and mRNA.
After receiving the data, the prior heterogeneity can be turned into a posterior by conditioning on all L recorded time traces. To obtain such a posterior over a and b, we employ Markov chain Monte Carlo techniques to sample
 |
with  the measured trace corresponding to cell m, measured at N time points. The mean value of this distribution serves as a Bayesian point estimate
the measured trace corresponding to cell m, measured at N time points. The mean value of this distribution serves as a Bayesian point estimate  that we can then use to determine the posterior moments. When receiving a new single-cell trajectory
that we can then use to determine the posterior moments. When receiving a new single-cell trajectory  one can now ask for its most likely mRNA and protein dynamics given this posterior heterogeneity. That is, we aim to compute
one can now ask for its most likely mRNA and protein dynamics given this posterior heterogeneity. That is, we aim to compute  , where the outer expectation is over
, where the outer expectation is over  . To approximate these posterior moments, we combine our proposed smoothing approach with [3] to obtain a posterior marginal moment equation. Thereby, we follow again the RTS ansatz and use the exact moments obtained from integrating the smoothing master equation (2.8) as a reference. Conceptually, the resulting marginal moments are equivalent to averaging the traditional smoothing moments for random c4 drawn from the gamma distribution. The results for one exemplary single-cell trajectory are given in figure 5. It is observed that the RTS approximation to the smoothing moments is accurate for the considered data of the GEV system. Also evident from the comparison in figure 5 is the significant reduction of variance of the posterior moments with respect to the prior moment dynamics.
. To approximate these posterior moments, we combine our proposed smoothing approach with [3] to obtain a posterior marginal moment equation. Thereby, we follow again the RTS ansatz and use the exact moments obtained from integrating the smoothing master equation (2.8) as a reference. Conceptually, the resulting marginal moments are equivalent to averaging the traditional smoothing moments for random c4 drawn from the gamma distribution. The results for one exemplary single-cell trajectory are given in figure 5. It is observed that the RTS approximation to the smoothing moments is accurate for the considered data of the GEV system. Also evident from the comparison in figure 5 is the significant reduction of variance of the posterior moments with respect to the prior moment dynamics.
7. Conclusion
Our capacity to decipher the inner workings of a cellular process strongly depends on the dimensionality of the available molecular readout. For time-resolved single-cell analysis the number of simultaneous readouts remains limited and biologists are trained to qualitatively infer the behaviour of unobservable states of the process. However, with the rise of the computational models that can quantitatively capture the behaviour processes, one can now improve on this qualitative inference. We remark, again, that estimating the most likely latent state of the process for a given observation is different from just computing the solution to a calibrated model. The theory of optimal filtering offers the general solution to the problem on how to combine data with a dynamic model to predict such states. However, it is known that solving the exact filtering or smoothing problem is computationally costly and we show that for a biochemical network it is at least as costly as integrating the chemical master equation.
To this end, we develop an approximate but scalable approach to filtering by exploring the fundamental relationship [43] and combining it with traditional moment closure techniques. We verify the effectiveness of the proposed method through single-cell experimental data and through in silico experiments. Based on the approximation to the filtering moments obtained by the method, one can further compute the RTS approximation to the smoothing moment. Although the RTS approximation often works well (see figures 4c,d and 5), it also show significant deviations (e.g. figure 4), even when the proposed approximation to the filtering moments performs well (figure 3). This lack of robustness indicates that a novel approximation to the smoothing moments needs to be developed for the case of stochastic reaction networks with lognormal measurement noise.
We observe that the imposed conditioning often yields posterior processes with confined, unimodal distributions even if the corresponding prior process exhibits multi-modal distributions. Hence, approximations through moment closure or state-space truncation of posterior processes for complex systems such as multi-stable or oscillatory networks [44] appears feasible and is a promising research question. Moreover, the proposed method can be extended to a hybrid framework (e.g. [45–47]) where a diffusion approximation can be performed for some states and reactions. This is especially interesting for multi-scale cellular processes, for instance in signal transduction coupled to gene expression where different abundance scales of molecules are involved. The proposed state reconstruction approach can profit from such a hybridization and would lead to even more scalable algorithms. Because it is well known that the moment closure techniques can also fail [33,48], theoretical analysis, such as the computation of error bounds, for the special case of posterior moments is a promising future research topic. The developed moment approximations of the filtering distribution can also be used for the stochastic decoupling of networks as proposed in [49].
8. Methods and experimental protocols
8.1. Mathematical methods and algorithms
Details on mathematical derivations of the posterior master equations, the posterior moment equations and details on the discussed case studies together with more corresponding simulation results are given in the electronic supplementary material. The Matlab codes used to generate all results in this paper are available at http://www.bcs.tu-darmstadt.de/media/bcs/Reconstructing_dynamic_molecular_states.zip.
8.2. Calibrating YFP fluorescence to absolute numbers of molecules
The previous work [6] quantified induction of Msn2 target genes as the mean fluorescence intensity per pixel in arbitrary units (arb. units) through a fast-maturing YFP reporter protein, mCitrineV163A. However, in order to apply the state reconstruction approach, it is necessary to calibrate these measurements to obtain absolute numbers of YFP molecules per cell. A calibration relationship was developed by measuring the mCitrineV163A fluorescence of five yeast proteins of known abundance [50] using the same exposure conditions as in the original study [6]. The five yeast genes were: YGP117C (1280 molecules per cell), TMA108 (5110 molecules per cell), HOG1 (6780 molecules per cell), TDA1 (10 200 molecules per cell) and CAR1 (42 800 molecules per cell). Each gene was C-terminally tagged with mCitrineV163A (in a pKT-vector; available from AddGene as no. 64685) followed by a HIS-marker by transforming a polymerase chain reaction-generated mCitrineV163A-HIS construct into the original haploid S288C Saccharomyces cerevisiae strain used by Ghaemmaghami et al. [51] (EY0986, MAT a, ATCC201388, his31, leu20, met150, ura30, S288C) and selecting on SD-HIS plates. To minimize experimental variability, we picked and measured four independent clones for each of the five genes. We used the untagged wild-type strain EY0986 to determine the autofluorescence background. To measure fluorescence intensity from each clone, we closely followed the protocol described by Ghaemmaghami et al. Briefly, we picked a single colony to inoculate a flask containing yeast extract peptone dextrose medium. Cells were grown overnight until an OD600 of approximately 0.7. Cells were fixed by incubating 0.9 ml of culture with 0.1 ml 10% buffered formalin solution (Sigma-Aldrich HT5011) for 5 min with occasional mixing. Cells were spun down and washed with 0.1 M KH2PO4 pH 8.5 and then 1.2 M sorbitol in KH2PO4 pH 8.5. Cells were re-suspended in 20 µl 1.2 M sorbitol in KH2PO4 pH 8.5. Then 2 µl of this solution was loaded onto a microscope slide, a coverslip added and sealed with nail polish. Cells were then immediately imaged using the exact same exposure conditions as described in [6]. Images were then analysed and fluorescence quantified as previously described [6,51]. After quantifying the fluorescence intensity per cell for each of the five genes, we then fitted a simple line to the data and found that each YFP molecule contributed about 100.8 arb. units fluorescence per cell under our excitation settings [51]. We therefore divided the total fluorescence per cell from the previous dataset [6] to obtain the total number of YFP molecules per cell.
8.3. Fluorescence microscopy and image analysis in pGAL1 Y-Venus expression
The experiments were performed on the same epifluorescence microscope (Eclipse Ti, Nikon Instruments), 60× (NA1.4) oil objective and specific (CFP/YFP/mCherry) excitation and emission filters located in an incubation chamber set to maintain 30°C. Imaging conditions and parameters were kept constant for all experiments. Single colonies of the respective yeast strain were picked, inoculated in synthetic (SD) medium and grown overnight at 30°C. The saturated cultures were then diluted and grown in log phase for at least two doubling times (more than 4 h). Before they were loaded into the imaging chambers, the cell suspensions were diluted again (OD600 = 0.01) and briefly sonicated. Single-cell traces were recorded by fluorescence microscopy with a 30 min induction pulse of 25, 50 and 100 nM β-oestradiol. The pulses were done by switching between two hydrostatic pressure (1 p.s.i.) driven flows (SD-full and SD-full + β-oestradiol) using a three-way solenoid valve (The Lee Company) connected to the cell chamber (µ-Slide VI; Ibidi). All microscopy images were analysed with the YeastQuant platform. The GEV relocation and Venus expression time-lapse movies were segmented on the basis of the nuclear CFP image from the HTA2-CFP marker. The expression of the Y-Venus protein was quantified as the total intensity in the cell. The expression levels of the YFP-tagged proteins were measured with illumination conditions similar to those used for the Y-Venus imaging. See [7] for a detailed description.
Supplementary Material
Supplementary Material
Acknowledgements
The authors would like to thank the referees for their comments and Leo Bronstein for helpful discussions on the topic.
Authors' contributions
H.K. conceived the study, designed the study, coordinated the study and helped draft the manuscript; L.H. conceived the novel approximate approach, carried out the mathematical derivation and computation and helped draft the manuscript; L.P. carried out the computation; C.Z. carried out the computation; M.U. carried out the molecular laboratory work, participated in data analysis and helped draft the manuscript; A.S.H. carried out the molecular laboratory work, participated in data analysis and helped draft the manuscript. All authors gave final approval for publication.
Competing interests
The authors declare no competing interests.
Funding
This work was supported by SystemsX.ch Transfer Project TF-196. H.K. acknowledges the support of the LOEWE Research Priority Program CompuGene.
References
- 1.Shen H, et al. 2006. Automated tracking of gene expression in individual cells and cell compartments. J. R. Soc. Interface 3, 787–794. ( 10.1098/rsif.2006.0137) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suter DM, Molina N, Gatfield D, Schneider K, Schibler U, Naef F. 2011. Mammalian genes are transcribed with widely different bursting kinetics. Science 332, 472–474. ( 10.1126/science.1198817) [DOI] [PubMed] [Google Scholar]
- 3.Zechner C, Ruess J, Krenn P, Pelet S, Peter M, Lygeros J, Koeppl H. 2012. Moment-based inference predicts bimodality in transient gene expression. Proc. Natl Acad. Sci. USA 109, 8340–8345. ( 10.1073/pnas.1200161109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hao N, O'Shea EK. 2012. Signal-dependent dynamics of transcription factor translocation controls gene expression. Nat. Struct. Mol. Biol. 19, 31–39. ( 10.1038/nsmb.2192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowsher CG, Swain PS. 2012. Identifying sources of variation and the flow of information in biochemical networks. Proc. Natl Acad. Sci. USA 109, E1320–E1328. ( 10.1073/pnas.1119407109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen AS, O'Shea EK. 2013. Promotor decoding of transcription factor dynamics involves a trade-off between noise and control of gene expression. Mol. Syst. Biol. 9, 704 ( 10.1038/msb.2013.56) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zechner C, Unger M, Pelet S, Peter M, Koeppl H. 2014. Scalable inference of heterogeneous reaction kinetics for pooled single-cell recordings. Nat. Methods 11, 197–202. ( 10.1038/nmeth.2794) [DOI] [PubMed] [Google Scholar]
- 8.Stratonovich RL. 1968. Conditional Markov processes and their application to the theory of optimal control. New York, NY: American Elsevier Pub. Co. [Google Scholar]
- 9.Crisan D, Rozovskii B. 2011. The Oxford handbook of nonlinear filtering. New York, NY: Oxford University Press. [Google Scholar]
- 10.Kushner HJ. 1964. On the differential equations satisfied by conditional probability densities of Markov processes with applications. J. SIAM Control Ser. A 2, 106–119. [Google Scholar]
- 11.Zakai M. 1969. On the optimal filtering of diffusion processes. Zeitschrift für Wahrscheinlichkeitstheorie und Verwandte Gebiete 11, 230–243. ( 10.1007/BF00536382) [DOI] [Google Scholar]
- 12.Kalman RE. 1960. A new approach to linear filtering and prediction problems. J. Basic Eng. 82, 35–45. ( 10.1115/1.3662552) [DOI] [Google Scholar]
- 13.Särkkä S. 2013. Bayesian filtering and smoothing. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 14.Julier SJ, Uhlmann JK. 2004. Unscented filtering and nonlinear estimation. Proc. IEEE 92, 401–422. ( 10.1109/JPROC.2003.823141) [DOI] [Google Scholar]
- 15.Hoteit I, Luo X, Pham D-T. 2012. Particle Kalman filtering: a nonlinear Bayesian framework for ensemble Kalman filters. Mon. Weather Rev. 140, 528–542. ( 10.1175/2011MWR3640.1) [DOI] [Google Scholar]
- 16.Gordon N, Salmond D, Smith AFM. 1993. Novel approach to non-linear and non-Gaussian Bayesian state estimation. Proc. Inst. Elect. Eng. F 140, 107–113. [Google Scholar]
- 17.Del Moral P. 2013. Mean field simulation for Monte Carlo integration. Boca Raton, FL: CRC Press. [Google Scholar]
- 18.Wonham WM. 1964. Some applications of stochastic differential equations to optimal nonlinear filtering. SIAM J. Control 2, 347–369. [Google Scholar]
- 19.Bain A, Crisan D. 2009. Fundamentals of stochastic filtering. New York, NY: Springer. [Google Scholar]
- 20.Ethier SN, Kurtz TG. 1986. Markov processes: characterization and convergence. Hoboken, NJ: John Wiley & Sons. [Google Scholar]
- 21.Van Kampen NG. 2007. Stochastic processes in physics and chemistry, 3rd edn Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- 22.Qian H. 1990. On the statistics of fluorescence correlation spectroscopy. Biophys. Chem. 38, 49–57. ( 10.1016/0301-4622(90)80039-A) [DOI] [PubMed] [Google Scholar]
- 23.Anderson DF. 2007. A modified next reaction method for simulating chemical systems with time dependent propensities and delays. J. Chem. Phys. 127, 214107 ( 10.1063/1.2799998) [DOI] [PubMed] [Google Scholar]
- 24.Lewis PAW, Shedler GS. 1979. Simulation of nonhomogeneous Poisson processes by thinning. Naval Res. Logistics Q. 26, 403–413. ( 10.1002/nav.3800260304) [DOI] [Google Scholar]
- 25.Hobolth A, Stone EA. 2009. Simulation from endpoint-conditioned, continuous-time Markov chains on a finite state space, with applications to molecular evolution. Ann. Appl. Stat. 3, 1204–1231. ( 10.1214/09-AOAS247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gardiner CW. 2009. Stochastic methods—a handbook for the natural and social sciences, 4th edn Berlin, Germany: Springer. [Google Scholar]
- 27.Jahnke T, Huisinga W. 2007. Solving the chemical master equation for monomolecular reaction systems analytically. J. Math. Biol. 54, 1–26. ( 10.1007/s00285-006-0034-x) [DOI] [PubMed] [Google Scholar]
- 28.Krishnarajah I, Cook A, Marion G, Gibson G. 2009. Novel moment closure approximations in stochastic epidemics. Bull. Math. Biol. 67, 855–873. ( 10.1016/j.bulm.2004.11.002) [DOI] [PubMed] [Google Scholar]
- 29.Engblom S. 2006. Computing the moments of high dimensional solutions of the master equation. Appl. Math. Compt. 180, 498–515. ( 10.1016/j.amc.2005.12.032) [DOI] [Google Scholar]
- 30.Gillespie CS. 2009. Moment-closure approximations for mass-action models. IET Syst. Biol. 3, 52–58. ( 10.1049/iet-syb:20070031) [DOI] [PubMed] [Google Scholar]
- 31.Ruttor A, Opper M. 2009. Efficient statistical inference for stochastic reaction processes. Phys. Rev. Lett. 103, 230601 ( 10.1103/PhysRevLett.103.230601) [DOI] [PubMed] [Google Scholar]
- 32.Singh A, Hespanha JP. 2007. A derivative matching approach to moment-closure approximations. Bull. Math. Biol. 69, 1909–1925. ( 10.1007/s11538-007-9198-9) [DOI] [PubMed] [Google Scholar]
- 33.Hausken K, Moxnes JF. 2011. Systematization of a set of closure techniques. Theor. Popul. Biol. 80, 175–184. ( 10.1016/j.tpb.2011.07.001) [DOI] [PubMed] [Google Scholar]
- 34.Lakatos E, Ale A, Kirk PDW, Stumpf MPH. 2015. Multivariate moment closure techniques for stochastic kinetic models. J. Chem. Phys. 143, 094107 ( 10.1063/1.4929837) [DOI] [PubMed] [Google Scholar]
- 35.Crassidis JL, Junkins JL. 2012. Optimal estimation of dynamic systems, 2nd edn Boca Raton, FL: Chapman & Hall/CRC. [Google Scholar]
- 36.Bierman GJ. 1973. Fixed interval smoothing with discrete measurements. Int. J. Control 18, 65–75. ( 10.1080/00207177308932487) [DOI] [Google Scholar]
- 37.Lestas I, Paulsson J, Ross N, Vinnicombe G. 2008. Noise in gene regulatory networks. IEEE Trans. Autom. Contr. 53, 189–200. ( 10.1109/TAC.2007.911347) [DOI] [Google Scholar]
- 38.Munsky B, Khammash M. 2006. The finite state projection algorithm for the solution of the chemical master equation. J. Chem. Phys. 124, 044104 ( 10.1063/1.2145882) [DOI] [PubMed] [Google Scholar]
- 39.Wolf V, Goel R, Mateescu M, Henzinger TA. 2010. Solving the chemical master equation using sliding windows. BMC Syst. Biol. 4, 42 ( 10.1186/1752-0509-4-42) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swain PS, Elowitz MB, Siggia ED. 2002. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc. Natl Acad. Sci. USA 99, 12 795–12 800. ( 10.1073/pnas.162041399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hilfinger A, Paulsson J. 2011. Separating intrinsic from extrinsic fluctuations in dynamic biological systems. Proc. Natl Acad. Sci. USA 109, 12 167–12 172. ( 10.1073/pnas.1018832108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruess J, Milias-Argeitis A, Lygeros J. 2013. Designing experiments to understand the variability in biochemical reaction networks. J. R. Soc. Interface 10, 20130588 ( 10.1098/rsif.2013.0588) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gillespie DT. 1977. Exact stochastic simulation of coupled chemical reactions. J. Chem. Phys. 81, 2340–2361. ( 10.1021/j100540a008) [DOI] [PubMed] [Google Scholar]
- 44.Koeppl H, Hafner M, Ganguly A, Mehrotra A. 2011. Deterministic characterization of phase noise in biomolecular oscillators. Phys. Biol. 8, 055008 ( 10.1088/1478-3975/8/5/055008) [DOI] [PubMed] [Google Scholar]
- 45.Ganguly A, Altintan D, Koeppl H. 2015. Jump-diffusion approximation of stochastic reaction dynamics: error bounds and algorithms. SIAM J. Multiscale Model. Simul. 13, 1390–1419. ( 10.1137/140983471) [DOI] [Google Scholar]
- 46.Hasenauer J, Wolf V, Kazeroonian A, Theis FJ. 2014. Method of conditional moments (MCM) for the chemical master equation: a unified framework for the method of moments and hybrid stochastic-deterministic models. J. Math. Biol. 69, 687–735. ( 10.1007/s00285-013-0711-5) [DOI] [PubMed] [Google Scholar]
- 47.Menz S, Latorre JC, Schütte C, Huisinga W. 2012. Hybrid stochastic deterministic solution of the chemical master equation. SIAM J. Multiscale Model. Simul. 10, 1232–1262. ( 10.1137/110825716) [DOI] [Google Scholar]
- 48.Schnoerr D, Sanguinetti G, Grima R. 2014. Validity conditions for moment closure approximations in stochastic chemical kinetics. J. Chem. Phys. 141, 084103 ( 10.1063/1.4892838) [DOI] [PubMed] [Google Scholar]
- 49.Zechner C, Koeppl H. 2014. Uncoupled analysis of stochastic reaction networks in fluctuating environments. PLoS Comput. Biol. 10, e1003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. 2003. Global analysis of protein expression in yeast. Nature 425, 737–741. ( 10.1038/nature02046) [DOI] [PubMed] [Google Scholar]
- 51.Hansen AS, Hao N, O'Shea EK. 2015. High-throughput microfluidics to control and measure signaling dynamics in single yeast cells. Nat. Protoc. 10, 1181–1197. ( 10.1038/nprot.2015.079) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






