Abstract
Calculating internal dose from therapeutic radionuclides currently relies on estimates made from multiple radiation exposure measurements, converted to absorbed dose in specific organs using the Medical Internal Radiation Dose (MIRD) schema. As an alternative biodosimetric approach, we utilized gene expression analysis of whole blood from patients receiving targeted radiotherapy. Collected blood from patients with relapsed or refractory neuroblastoma who received 131I-labeled metaiodobenzylguanidine (131I-mIBG) at the University of California San Francisco (UCSF) was used to compare calculated internal dose with the modulation of chosen gene expression. A total of 40 patients, median age 9 years, had blood drawn at baseline, 72 and 96 h after 131I-mIBG infusion. Whole-body absorbed dose was calculated for each patient based on the cumulated activity determined from injected mIBG activity and patient-specific time-activity curves combined with 131I whole-body S factors. We then assessed transcripts that were the most significant for describing the mixed therapeutic treatments over time using real-time polymerase chain reaction (RT-PCR). Modulation was evaluated statistically using multiple regression analysis for data at 0, 72 and 96 h. A total of 10 genes were analyzed across 40 patients: CDKN1A; FDXR; GADD45A; BCLXL; STAT5B; BAX; BCL2; DDB2; XPC; and MDM2. Six genes were significantly modulated upon exposure to 131I-mIBG at 72 h, as well as at 96 h. Four genes varied significantly with absorbed dose when controlling for time. A gene expression biodosimetry model was developed to predict absorbed dose based on modulation of gene transcripts within whole blood. Three transcripts explained over 98% of the variance in the modulation of gene expression over the 96 h (CDKN1A, BAX and DDB2). To our knowledge, this is a novel study, which uses whole blood collected from patients treated with a radiopharmaceutical, to characterize biomarkers that may be useful for biodosimetry. Our data indicate that transcripts, which have been previously identified as biomarkers of external exposures in ex vivo whole blood and in vivo radiotherapy patients, are also good early indicators of internal exposure. However, for internal sources of radiation, the biokinetics and physical decay of the radionuclide strongly influence the gene expression.
INTRODUCTION
Dose estimation after the administration of therapeutic radionuclides presents unique challenges. The practice of calculating internal radiation dose is done by either bioassay (such as urine or fecal samples) or by estimation based on activity administered, using biokinetic models as provided by the Committee on Medical Internal Radiation Dose (MIRD). While dosimeters can be used to measure dose externally, the development of a dosimeter to assess internal dose is needed. We hypothesized that the use of transcriptional assays to monitor changes at the molecular level and measurement of dose-response characteristics after irradiation might be a feasible approach towards characterizing internal biodosimetry (1).
Over the last two decades genome-scale tools have helped to identify large-scale changes in gene expression profiles within minutes to hours after irradiation, and have shown that a broad variety of biological pathways are modulated (2–5). These studies have proven useful for predicting panels of transcripts that may be beneficial for biodosimetry (3, 4, 6–15). However, most of these studies have been primarily focused on external irradiation. Recently, several groups have looked at internal radiation exposures with an emphasis on the mouse model (16–19). Internal radiation studies in humans have been focused on biomarkers of multiple organ damage, but have not been specifically directed toward looking for DNA damage-related responses in peripheral blood (20–23), where biomarkers of dose response may circulate (10, 15, 24–31). Characterization of known radiation-responsive transcript profiles for therapeutic radionuclides may also be useful in both the clinic and triage setting as biodosimeters.
One radiopharmaceutical of interest is iodine-131 conjugated with metaiodobenzylguanidine (131I-mIBG), which is used in patients with advanced neuroblastoma that have poor outcomes. MIBG is a norepinephrine analogue that is quickly taken up by neuronal-derived cancer cells expressing the norepinephrine transporter (32, 33). A total of 90% of all neuroblastoma tumors are mIBG-avid (32, 34), such that targeted radiotherapy with 131I-mIBG can be used in the majority of these patients. For patients with relapsed or refractory high-risk neuroblastoma, targeted radiotherapy is often the therapy of choice. Recent efforts have focused on combining 131I-mIBG with potential radiosensitizers, such as vorinostat (35, 36) or irinotecan (37). Currently, there are also cooperative group studies underway evaluating whether incorporating 131I-mIBG into upfront treatment is beneficial in cancer treatment.
When 131I-mIBG is used as part of targeted radiotherapy, the mIBG is rapidly taken up by neuroblastoma tumors with a peak at approximately 6 h (32). This rapid clearing from the blood causes approximately 10% to remain in the blood stream within a few hours after treatment (32). It has also been shown that approximately 15% immediately exits the body through the bladder (38–40). Therefore, during the course of treatment, the patient is being exposed to an exponentially decreasing radioactive source that is both eliminated from the body and radioactively decaying. While the majority of the biodosimetry studies have been done using external radiation (2–14, 29, 41), patients receiving radiopharmaceuticals may be suitable models for internal biodosimetry studies.
The current study provides the opportunity to assess biological response to ionizing radiation from an exponentially decaying source from within the human body. The findings from this work may prove useful during triage after a “dirty bomb” explosion in a densely populated area, or where the release of a radioactive isotope would present more of an internal concern. In this study, we combined physical and biological dosimetry for the first time in patients receiving a targeted radionuclide therapy. We assessed expression of a panel of radiation response genes in the peripheral blood. Our goal was to describe the change in expression of these genes after 131I-mIBG therapy and to determine the extent to which expression of these genes correlated with estimated whole-body radiation dose.
MATERIALS AND METHODS
Patient Recruitment and 131I-mIBG Therapy
Patients were recruited for this pilot evaluation of novel radiation biomarkers from among individuals with relapsed or refractory neuroblastoma treated with 131I-mIBG at the University of California San Francisco School of Medicine (UCSF; Table 1). Patients with relapsed and refractory neuroblastoma receiving mIBG treatment at UCSF were eligible to participate. All patients had MIBG-avid neuroblastoma by 123I-mIBG diagnostic scans and were required to be older than 24 months of age. Based on availability of specific clinical trials, patients received either 131I-mIBG as a single agent or 131I-mIBG with either vorinostat or with vincristine and irinotecan. Patients were treated with 131I-mIBG infusion over 90–120 min at UCSF, with the usual thyroid blockade and bladder protection using a bladder catheter (37). All patients consented to gene expression studies. This study was approved by the UCSF Committee on Human Research and the Institutional Review Boards at University of California Davis and Lawrence Livermore National Laboratory.
TABLE 1.
Patient Characteristics
| Number of patients | |
|---|---|
| Total number of patients | 40 |
| Sex | |
| Female | 10 |
| Male | 30 |
| Concomitant radiation sensitizer | |
| Irinotecan + vincristine: | 19 |
| Vorinostat | 7 |
| None | 14 |
| Disease status prior to treatment | |
| Relapsed | 18 |
| Refractory | 22 |
| Age at treatment | |
| Median | 7 years |
| Range | 3–30 years |
| Patient weight | |
| Mean | 31.2 kg |
| SD | 21.2 |
| 131I-mIBG administered activity | |
| First treatment | |
| Mean | 17.232 mCi/kg |
| SD | 1.5 |
| Second treatment | |
| Mean | 17.8 mCi/kg |
| SD | 1.2 |
Blood Sample Processing
A total of 2.5 ml per time point of peripheral blood was drawn using PAXgene™ RNA blood tubes (QIAGEN®, Valencia, CA) at baseline, 72 and 96 h after 131I-mIBG treatment. Blood tubes were kept at −80°C for 30 days after treatment to ensure all 131I had decayed below background levels prior to isolation and analysis. The RNA was then extracted from whole blood using the PAX Gene Kit following the manufacturer's instructions. RNA was stored at −80°C until ready for use. RNA was quantified using a NanoDrop ND 1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and Qubit® Fluorometer (Invitrogen™, Carlsbad, CA). RNA quality was checked using an Experion™ analyzer (Bio-Rad, Hercules, CA).
Gene Selection for Biodosimetry
Table 2 list genes that were selected based on prior performance in previously conducted and published gene expression experiments that emphasized robust dose-responsive transcripts (26, 28, 29, 42, 43) for validation in our model system. GAPDH was chosen as the endogenous reference gene. Genes were representative of tumor suppressor protein p53 (TP53 or p53) activation of the DNA damage response pathway as well as associated signaling pathways. The apoptosis pathways included FDXR, BBC3, BCL2, BCLXL, BAX and BIM, while the cell cycle arrest domain included CDKN1A and GADD45A. The DNA repair domain was represented by XPC and DDB2.
TABLE 2.
Selected Transcripts of Interest
| Gene | Name | Primer no. |
|---|---|---|
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase | H202758991_g1 |
| CDKN1A | Cyclin-dependent kinase inhibitor 1A (p21) | Hs0035578_m1 |
| FDXR | Ferrodixin reductase | Hs00244586_m1 |
| GADD45A | Growth arrest and DNA-damage-inducible alpha | Hs00169255_m1 |
| DDB2 | Damage-specific DNA binding protein 2 (XPE) | Hs03044953_m1 |
| XPC | Xeroderma pigmentosum, complementation group C | Hs00190295_m1 |
| MDM2 | E3 ubiquitin protein ligase | Hs00234753_m1 |
| BCL2 | B-cell CLL/lymphoma 2 | Hs99999018_m1 |
| BCLXL | BCL2-like 1 | Hs00236329_m1 |
| BAX | BCL2-associated X protein | Hs99990001_m1 |
| STAT5B | Signal transducer and activator of transcription 5B | Hs00273500_m1 |
Generation of cDNA and Real-Time PCR
For RT-PCR analysis, 200 ng of RNA was converted to single-strand cDNA using the High Capacity cDNA archive kit (Applied Biosystems®, Foster City, CA). RNA was combined with buffer, enzyme mix and RNase-free water. The reactions for cDNA synthesis were conducted using a thermocycler. Incubation was done for 1 h at 37°C, then 5 min at 95°C. cDNA was held at 4°C until it was removed and stored in a freezer between −15°C and −25°C. The cDNA was then preamplified with TaqMan® PreAmp Master Mix (Applied Biosystems). A pooled mix of TaqMan assay primers was combined with the preamp master mix. Because patients had low white blood cell levels after their treatment, 14 cycles of preamplification were done in a thermocycler to ensure that enough material was available to quantify the entire transcript panel of interest. The cycles lasted 10 min at 95°C, followed by 14 cycles of 15 s at 95°C and 4 min at 60°C.
Real-time PCR (RT-PCR) was used to quantify the expression of genes for each patient. Each reaction was performed in a volume of 20 μl and repeated in triplicate using SharkaTAQ™ Stable QPCR Master Mix (Frontier Genomics, Auke Bay, AK) and Applied Biosystems TaqMan primers (Table 2) using a 7900HT Fast Real-Time PCR System. The following PCR program was used: 10 min at 95°C, followed by 40 cycles of alternating 15 s at 95°C and 1 min at 60°C. Results from RT-PCR were in the form of cycle threshold (Ct) values, which is the cycle when amplification curves show logarithmic growth characteristics.
Physical Dosimetry Calculations
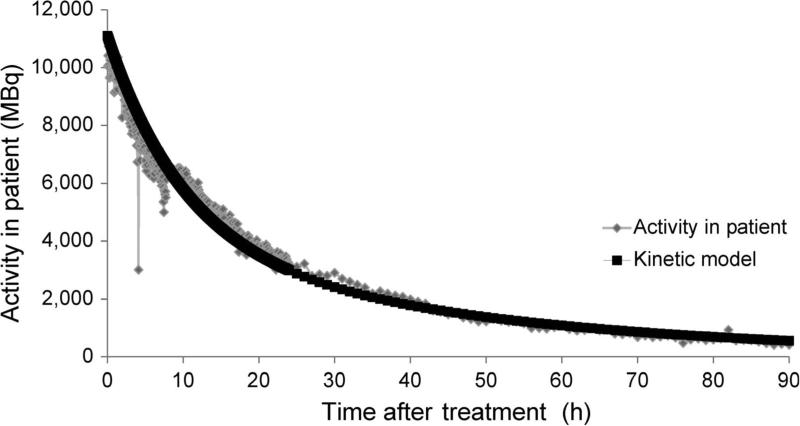
Mean absorbed dose (referred to as absorbed dose) accumulated up to each blood-collection point for each patient was calculated using standard MIRD schema similarly as previously described by Buckley et al. (44). To determine the absorbed dose for each patient during treatment, the total retention of injected activity of mIBG was first determined using a Fluke 451P Pressurized Ion Chamber radiation detector that was fixed to the ceiling above the patient for the duration of the patient's treatment. The exposure rate emitted from the patient was measured every 3 min. The measured decay curve was normalized to injected activity to provide time-activity estimates over time for each patient (example shown in Fig. 2).
FIG. 2.
An example of exponential decay of 131I within a single patient. 131I-mIBG, as well as other radionuclides, are eliminated by two major processes within the human body: physical decay and biological decay (elimination). The amount of activity in the patient is derived from comparing the amount of activity injected to the peak dose rate given off by the patient. Due to patient movement, distance from the radiation detector changes, resulting in dose-rate fluctuations. The data is drawn on a time plot and used to estimate total dose. The black line represents the calculated amount of activity remaining in the patient at a given time based on the kinetic model. Diamond-shaped symbols represent the calculated activity in the patient based on dose rate, which is measured by the meter mounted on the ceiling above the patient.
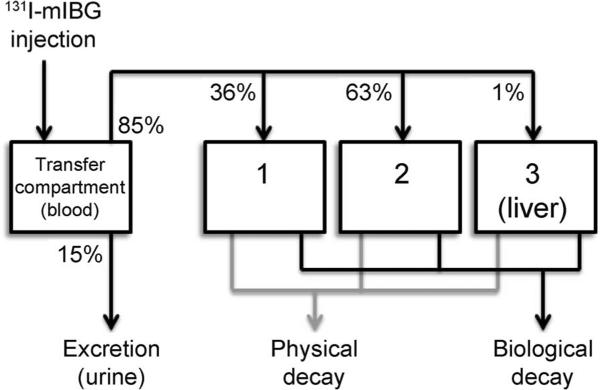
A three-compartment model (Fig. 1) was applied to the measured time-activity data for three patients using their emitted exposure rate data from the radiation detector above the bed. The effective decay constants, C1, C2 and C3, representing the combined effect of physical and biological decay, were obtained for three patients where the decay constants were fitted mathematically based on the physical dosimetry data analysis from the patients, then averaged among the three patients. These effective decay constants were then used to calculate Ã, or the time-integrated activity, at a given time after 131I-mIBG treatment, using the following equation:
| (1) |
Equation (1) is the time-activity curve that describes the biokinetic activity of 131I-mIBG in a patient. Each respective λ is the effective decay constant for each compartment, t is the time after treatment, Ao is the initial activity injected and à is the resulting time-integrated activity at a given t. Prior empirical testing at UCSF showed that 15% of the initial injected activity is lost in the first 3 h after treatment is started, and can be assumed to be lost immediately to the urinary system and through the catheter.
FIG. 1.
A three-compartment 131I-mIBG biokinetic model. This biokinetic model represents Eq. (1). The model is used for calculating dose in a patient undergoing 131I-mIBG treatment. After treatment, 15% of the original injection goes straight to the bladder and is excreted. Of the remaining 85%, which is distributed among mIBG-avid tissues in the body, 1% remains in the liver until it physically decays completely, while the other 99% is shared among other tissues and processed through the two other compartments.
The mean absorbed dose, DWB←WB (in Gy) was calculated as the product of the cumulated activity and the MIRD whole-body to whole-body S value.
| (2) |
As shown by Amundson et al. (43), SWB←WB values were corrected for the weight of each child, using Eq. (3). Since these are calculated using an adult as a reference individual, S values were recalculated using the following equation to take into account the differences in size for each child in the study:
| (3) |
where mpatient is the mass of the patient in kilograms. This equation was generated by interpolating the SWB←WB values from the MIRD phantoms for a newborn; for 1-, 5-, 10- and 15-year-old children; and for an adult, each of which has a specific mass (44, 45).
Absorbed doses calculated as above are referred to as kinetic model-based doses (KM dose) in this study.
Statistical Methods
Fold change was calculated with the 2−ΔΔCt method as described by Livak and Schmittgen (46), using Ct values as determined from the RT-PCR analysis using absolute quantification. A Ct value is inversely proportional to the amount of material in a sample; the higher the Ct the less amount of material. Multiple linear regression was used to assess the effect of time since treatment, absorbed dose and concomitant radiation sensitizer exposure on gene expression. Welch t tests, which assumed unknown variance, were performed to compare differences between serial treatments for patients who received more than one treatment. To determine which variables contributed most to the variance explanation in dose-dependent fold change, backward stepwise regression analysis was performed using P < 0.05 for variable entry. A significance level of α = 0.05 was used for all tests. A P value of 0.05 was used for determining significance. All analyses were conducted using the R environment (47).
RESULTS
Patient Characteristics
Characteristics of the 40 patients who participated in this pilot study are shown in Table 1. There were 30 males and 10 females. A total of 14 patients received mIBG treatment only, while 19 patients received vincristine/irinotecan and 7 received vorinostat in combination with mIBG. Nine patients received a second mIBG treatment approximately 6 weeks after the initial treatment. Our analysis focused only on the effects in patients that were associated with radiation dose and time for mIBG treatment within the first 96 h after exposure to demonstrate the utility of our approach. In addition, 6 of the patients lacked complete dosimetry information after the initial exposure and these were used for testing our exposure model.
Modulation of Gene Expression after 131I-mIBG Treatment
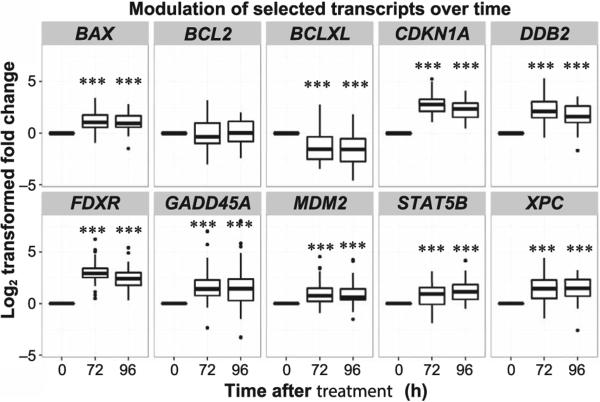
All 10 transcripts of interest gave measurable baseline signals in the RT-PCR assays. The expression levels of 9 transcripts were significantly altered compared to controls after mIBG treatment (Supplementary Table S1; http://dx.doi.org/10.1667/RR14263.1.S1). Figure 3 shows the change in translation over the course of 72 and 96 h from baseline. The transcripts CDKN1A, FDXR, GADD45A, STAT5B, BAX, XPC, MDM2 and DDB2 were significantly upregulated at 72 h as well as at 96 h while BCLXL was significantly downregulated at 72 h. MDM2 was the only transcript that was upregulated at 72 h and returned to baseline within the first 96 h. Interestingly, the BCLXL transcript was more significantly downregulated with a lower fold change at the 96 h time point compared to 72 h postirradiation. The BCL2 transcript did not show modulated expression in our study.
FIG. 3.
Box-whisker plots show the modulation of selected transcripts over time. This collection of plots shows the changes in gene expression at each time interval after 131I-mIBG treatment. The x-axis shows the time points sampled (0–96 h), while the y-axis shows the transformed log2 fold change of expression levels. The transcripts BAX, CDKN1A, DDB2, FDXR, GADD45A, MDM2, STAT5B and XPC show time-dependent upregulation while BCLXL shows time-dependent downregulation (paired Student's t test vs. 0 h after treatment, ***P < 0.001). BCL2 did not have a time-dependent response to radiation.
Significance of Differential Transcript Response Varied with Dose and Time
Multiple regression analyses were performed to determine the significance of the effects of dose and elapsed time after treatment on each transcript. All fold changes used were log2 transformed to normalize the expression around 0 for statistical testing. Concomitant drug treatment (vorinostat or vincristine/irinotecan) was used as a control variable, but was not included in the final analysis, to keep the focus on the effects of dose from the mIBG treatment alone. Table 3 shows the results of where the values are the β for each parameter while values in parentheses are standard error. The β values represent slopes of the regression line where a positive value represents increasing expression while a negative value represents decreasing expression. Due to the varying modulation of transcripts between 72 and 96 h, these results demonstrated a time dependency that needs to be taken into account for generating dose-related models.
TABLE 3.
Multiple Regression Analysis Results for Gene Expression Based on Dose and Time
| Intercept | Total absorbed dose | 72 h | 96 h | |
|---|---|---|---|---|
| CDKN1A | 0.29 (0.15) | −6E – 4 (3E – 3) | 2.96b (0.94) | 2.47a (1.03) |
| FDXR | −0.22 (0.19) | −4E – 3 (4E – 3) | 4.15c (1.16) | 3.74b (1.27) |
| GADD45A | −0.31 (0.32) | −3E – 3 (8E – 3) | 2.35 (1.94) | 2.43 (2.14) |
| BCLXL | 0.80c (0.23) | 9E – 3 (5E – 3) | −3.46a (1.46) | −3.92a (1.61) |
| STAT5B | −0.23 (0.17) | −3E – 4 (3E – 3) | 0.86 (1.05) | 1.31 (1.16) |
| BAX | −0.14 (0.15) | −8E – 3a (4E – 4) | 3.19b (0.96) | 3.28b (1.06) |
| BCL2 | −0.41 (0.21) | −2E – 3 (5E – 5) | 0.36 (1.32) | 0.58 (1.46) |
| XPC | −0.50a (0.19) | −9E – 3 (5E – 3) | 3.68b (1.21) | 3.89b (1.33) |
| DDB2 | −0.36 (0.24) | −6E – 3 (6E – 3) | 3.82a (1.48) | 3.26a (1.63) |
| MDM2 | 0.01 (0.23) | −2E – 3 (6E – 3) | 1.72 (1.52) | 1.76 (1.66) |
P < 0.05
P < 0.01
P < 0.001.
A Best-Fit Gene Expression Model Defines the Three Transcripts that Best Predict Internal Dose
Based on the multiple regression analysis results, genes that presented significant differences in expression and time as a continuous variable, rather than as a factor, were analyzed in a step-wise regression analysis to determine which of these genes best described the linear change in dose over time. The lowest Akaike Information Criterion (AIC) determined the equation that best fit the data. The result of the regression showed that the response of three transcripts over the time measured described 98.7% of the variance in dose across all patients. CDKN1A, BAX, DDB2 and time after 131I-mIBG treatment contributed to describing the variance in the dose that a patient receives. This model could then be used to predict dose as a calibration equation. Values listed in Table 4 were used with Eq. (2) to determine the gene expression model-based dose.
| (4) |
The β is the slope of the regression line of each gene, which describes the linear change in dose and time in relationship to the differential gene expression.
TABLE 4.
Best-Fit Model for GE Dose Prediction
| β value | Standard error | P value | |
|---|---|---|---|
| CDKN1A | 11.16 | 3.38 | 0.0015 |
| BAX | −19.44 | 5.48 | 0.0007 |
| DDB2 | 13.09 | 4.09 | 0.002 |
| Hour | 2.75 | 0.10 | < 2E–16 |
The β values in Eq. (2) were multiplied by the associated genes’ log2 transformed fold changes (or elapsed hours since 131I-mIBG treatment) and then summed together to give the gene expression model-based dose result. These values are referred to as the gene expression model dose (GE dose). To compare how useful a gene expression model was, a model using time as the only independent variable was used to estimate doses as well [see Eq. (3)]. Using this time-only model, at 72 h the predicted dose was 2.21 Sv, and at 96 h the predicted dose was 2.94 Sv.
| (5) |
Gene Expression Model Predicts Absorbed Dose for 131I-mIBG Patients with Incomplete Dose Information
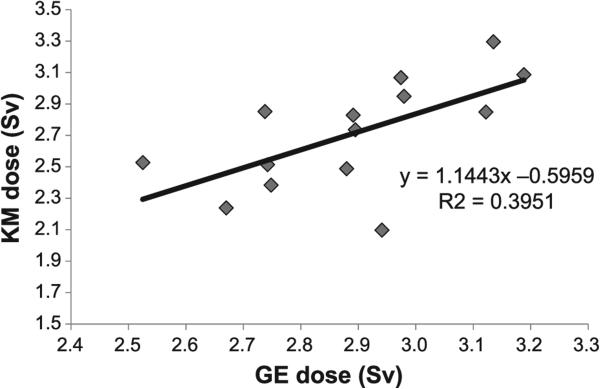
The best-fit model was used to estimate GE dose on six patients based only on gene expression fold change and time after 131I-mIBG treatment. These six patients were selected for model fitting because they lacked complete dose information other than the total for mCi/kg of 131I-mIBG injected at the start of treatment. Results are shown in Table 5. On average, the gene expression model estimated the GE dose within an absolute 8.5% of the KM dose while the model using only the hour as a predictor estimated the dose within an absolute 10.5%. Thirteen out of fourteen of the GE dose estimates were within the 95% prediction interval, while the time-only model had 11 out of 14 of the calculated doses within the 95% prediction interval (results not shown). The two models for absorbed dose (KM and GE) within a patient were compared with one another. The KM dose and GE dose were significantly correlated, suggesting that each model has similar prediction qualities for measuring the absorbed dose from a patient (Fig. 4).
TABLE 5.
Comparison of KM Dose vs. GE Dose Using Best-Fit Model Equation
| Patient | Hour | KM dose (Sv) | GE dose (Sv) | Prediction interval (95%) (Sv) | Absolute percentage difference between KM and GE doses |
|---|---|---|---|---|---|
| 1 | 72 | 2.64 | 2.49 | 2.00–2.97 | 5.71% |
| 1 | 96 | 2.91 | 3.05 | 2.57–3.53 | 4.74% |
| 1a | 72 | 2.63 | 2.32 | 1.84–2.80 | 11.86% |
| 1a | 96 | 2.91 | 2.99 | 2.51–3.47 | 2.81% |
| 2 | 72 | 2.56 | 2.11 | 1.63–2.60 | 17.66% |
| 2 | 96 | 2.83 | 2.84 | 2.33–3.39 | 0.44% |
| 3 | 72 | 2.78 | 2.86 | 2.33–3.39 | 2.86% |
| 3 | 96 | 3.07 | 3.43 | 2.92–3.94 | 11.84% |
| 4 | 72 | 2.43 | 2.47 | 1.98–2.95 | 1.68% |
| 4 | 96 | 2.68 | 2.89 | 2.41–3.38 | 8.04% |
| 5 | 72 | 2.82 | 1.94 | 1.45–2.44 | 31.19% |
| 5 | 96 | 3.12 | 2.93 | 2.45–3.41 | 6.14% |
| 6 | 72 | 2.77 | 2.54 | 2.05–3.04 | 8.11% |
| 6 | 96 | 3.05 | 2.87 | 2.38–3.35 | 6.12% |
Second treatment for patient.
FIG. 4.
Kinetic mode-based dose (KM dose) vs. gene expression-based dose (GE dose). This figure shows the relationship between GE dose from the best-fit model (Eq. 2) and the KM dose using the retention function (Eq. 1). KM dose and GE dose are collinear (R2 = 0.395, P = 0.016) and represent similar prediction qualities for measuring absorbed dose from a patient.
Multiple mIBG Treatments did not Affect Transcript Responses
Residual treatment may be a complication due to altered transcript levels and needs to be accounted for in the study. Nine patients received two rounds of 131I-mIBG therapy within this study with no shorter than one month elapsing between the treatments. Welch's two-sample t tests were performed to judge whether there were changes in gene expression based on prior exposure from mIBG therapy. There were no significant differences between the two treatments started at different times based on altered gene expression. Thus individuals with prior treatment could be considered as single-dosed patients.
DISCUSSION
Our results showed that previously identified DNA damage response and apoptosis transcripts for external radiation exposure will work as early indicators for internal exposure to 131I using peripheral white blood cells. Our study also demonstrates that biodosimetry of internal radiation dose in human chemo-radiotherapeutic patients is a potential option during, as well as after treatment with internal radiotherapeutics such as mIBG, when it is possible to obtain a blood sample prior to treatment to be used as a pretreatment control. In this study we found that white blood cell gene expression after internal radiation exposure was correlated with the kinetics of dose decay rather than total absorbed dose. Interestingly, our study suggests that prior treatments were not a confounding factor as shown by analyzing nine patients with prior mIBG treatment over the time period studied. This may indicate that baseline levels for the transcripts we studied are reset within the time frame between treatments, four weeks, for our patient population. Importantly, we confirmed that a predictive model is possible based on significant differential transcript modulation associated with time after administration of a radiopharmaceutical.
These findings illustrate the potential use of DNA damage-related transcript analysis in humans, in the absence of physical dosimetry, for events involving large uncontrolled releases of radionuclides in which human and environmental uptake is probable, e.g., dirty bombs or nuclear reactor accidents. In such scenarios, since both external and internal radiation exposure would be of some consequence, developing biodosimetry for internal radiation dose could have utility for establishing triage in affected areas. Additional experiments will be necessary to elucidate the low-dose effects and differentiate them from the higher doses measured in this current pilot study. Our data demonstrate that a small panel of responsive transcripts may be useful in such circumstances at early times to rapidly sort out the 131I-exposed from unexposed individuals at doses in the range of approximately 2–3 Gy.
Using RNA in peripheral white blood cells as a biodosimeter for internal exposures is plausible for several reasons: 1. Obtaining a blood sample for testing is a relatively noninvasive procedure; 2. Blood circulates throughout the body and can be used to estimate whole-body doses; and 3. The white blood cells are well characterized in terms of DNA damage and sensitivities to radiation exposures. However, because beta-emitters (such as 131I) deposit most of their emitted energy locally, e.g., within the blood, specific organs or at the tumor site, the majority of the dose is not necessarily deposited homogenously throughout the whole body. Therefore, using whole-body dose estimates as the measurement of comparison to gene expression might not be the best choice. The dose estimate can be difficult and requires consideration of biological and physical decay, as was shown in this study. There are also other issues such as self-shielding (i.e., aggregate particles absorbing emitted energy), blood flow velocity and the continually changing blood vessel geometry, which may affect the modeling (48). However, when samples are taken at regular intervals over a longer period of time (approximately 168 h) and the activity concentration of 131I-mIBG is analyzed, estimation of total activity in the blood, and therefore dose, could be calculated. A study conducted by A. A. Yalow derived the following empirical equation:
| (6) |
Where Dblood is the absorbed dose to the blood, t1 is the effective half-life of mIBG retention in the blood (h), and B1 and B2 are the concentration of mIBG in the first and last measurements, respectively (49). Flower et al. (49) calculated an absorbed dose to the blood of 0.04–0.17 mGy/MBq, whereas in this study, we calculated the absorbed dose to the blood to be between 0.01 and 0.10 mGy/MBq with a range of doses to the blood of 0.95–1.47 Gy. However, since the blood was never tested at 168 h, nor was any blood tested for activity content, the dose estimates were not used for any calculations. If patients were better monitored and sampled, it might be possible to calculate dose and develop an accurate and specific biodosimetric model, as conducted in our experiments.
We found that the genes, previously identified as potential external biodosimeters, were responsive to internal radiation exposures in humans at similar levels of expression. These results were similar to findings by Paul et al., using a mouse model and gene expression arrays (19). Significant modulation was identified and detected up to 96 h after treatment using CDKN1A, FDXR, BCLXL, BAX, XPC and DDB2 transcripts. This similarity suggests that these genes may be robust biomarkers on different types of radiation exposure routes (external and internal). Classifying the gene expression within cells after irradiation is of the upmost importance when choosing a model that best reflects the amount of energy deposited in a given biological system. In the future, it may be ideal to use array or sequencing technologies to better understand what is happening mechanistically in the peripheral blood of these cancer patients.
One notable difference between our study and previous studies (10, 50) was that the amount of modulation (fold change) in our study was attenuated for comparable total doses. Paul and Amundson reported that FDXR expression increased by about 12-fold 6 h after 2 Gy irradiation with little change after 24 h (10). This is in contrast to our study, which showed that the difference between 72 h (8.48-fold change) and 96 h (6.05-fold change) was approximately 2.4-fold. Manning et al. reported that FDXR increased significantly after 24 h to an approximate fold change of over 37 (50). This is much higher than our average fold change of 8.48 at 72 h. However, due to constraints on patient sampling, it is unknown if FDXR would be higher after 24 h in our study. Nevertheless, assuming FDXR modulation increases with increased dose, the increased dose rate at 24 h would lead one to predict a higher fold change at 24 h. This prediction necessitates further exploration into both the time dependence of gene expression for our selected panel of transcripts and how they change for internal exposures.
Nearly all genes tested were upregulated in this study and showed peak expression at 72 h with a descent in expression at 96 h. This behavior was also documented in a recent study by Tucker et al. (15) where the changes in CDKN1A ΔCt for 2.5 Gy dose at 1 day vs. 2 days were carefully examined. Based on our study and Tucker's study, there are two behaviors of gene expression that require consideration in biodosimetry models.
The first behavior of concern is the dose-dependent response where greater dose causes more expression of that gene. As has been shown in studies, CDKN1A in particular appears to increase in expression linearly with increasing dose until it reaches a threshold around 6 Gy and then plateaus (15, 42, 51). This affects dose prediction, since doses greater than 6 Gy cannot be predicted with certainty (51). Our study, however, contradicts this. Our doses continued to increase over time, albeit due to an exponentially decaying dose rate. Nonetheless, as our dose increased, gene expression became less significant or plateaued.
The second behavior of concern is the time-dependent response: as the time lapse increases after radiation exposure, the expression of the gene should dampen. Making this more complicated are the effects of an exponentially decaying source of radiation that is persistent rather than acute. Our study took this into account by including time elapsed as a part of the equation for predicting whole-body dose. The true cause of the time dependency in our study is most likely directly related to the exponentially decaying but ever-present radiation field acting on the white blood cells. But the entire mechanism requires further research to develop a model for comparing internal dose to external dose using similar decay parameters.
Despite increasing knowledge of genomics, there is still a need for further information on how individualized panels of biomarkers perform in unique populations and under different exposure conditions. For example, how does a dose-responsive biomarker respond to internal dose in young individuals? To address this question, our study selected DNA damage-related responsive transcripts identified in peripheral blood, where biomarkers of dose response are known to circulate. For example, CDKN1A is very responsive to radiation and studies have shown that its increase in behavior is close to linear with increasing dose. However, these prior studies primarily address acute, external doses, which should be useful for determining doses from whole-body radiation therapy and, possibly, criticality accidents. For targeted radiotherapy or nuclear accidents, these studies are less useful. Instead, a study like this one is needed that takes into account the exponential decay of activity after an uptake of a radionuclide. In this study, CDKN1A had a peak response at 72 h with a subsequent decay at 96 h (as did FDXR and most other genes in the study), which indicates additional data are needed to identify the longevity of the gene expression.
Fold change was used for comparison of expression in this study, but the ΔCT value might be a simpler estimate if one can precisely control for the input quality and amount of RNA. The disadvantage of using fold change is that to calculate a difference one must have a base value. In the case of a nuclear incident, the base value would be unknown and so the fold change model would not work. A ΔCT value compared across up and down genes may be a better indicator of exposure. More extensive testing and predictive models would need to be developed to take advantage of the ΔCT value. With further study, a base CT value for specific genes could be decided as a threshold, from which a quick analysis could determine if an individual had been exposed to higher levels of radiation. Nonetheless, we were able to develop a model that predicted internal dose with good precision using only three transcripts.
In summary, this study shows that even at 96 h after radiopharmaceutical treatment, the modulation in gene expression is still significant enough to discriminate between exposed and nonexposed samples using a selected gene transcript panel. We also showed how time dependency of gene expression becomes very important for internal dose estimation. Taking into account the modulation of multiple genes enables us to generate a prediction equation that could reasonably predict a patient's or exposed individual's dose in comparison to calculated methods. In addition to serving as biomarkers for internal radiation dose, these biomarkers have been incorporated into an ongoing randomized phase II clinical trial to compare different mIBG treatment regimens and to identify the paths for benefiting patient outcome in the future.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Angela Evans for technical help with sample processing. This project was supported by the National Institutes of Health/National Cancer Institute (grant no. R01CA172067). Funding was also provided by the U.S. Department of Energy under contract no. DE-AC52-07NA27344, with funding from the U.S. DOE Low Dose Radiation Research Program, grant no. KP110202. In addition, support was provided by NIH T32 grant no. 5T32CA128583-05 awarded to UCSF Benioff Children's Hospital, and by the Campini Foundation, Alex's Lemonade Stand Foundation (MIBG Infrastructure Grant) and NIH/NCRR UCSF-CTSI grant no. UL 1 TR000004 and Columbia University NIH/NIAID pilot grant U19 AI067773.
Footnotes
SUPPLEMENTARY INFORMATION
Table S1. Models for assessing cancer risks.
REFERENCES
- 1.Swartz HM, Williams BB, Flood AB. Overview of the principles and practice of biodosimetry. Radiat Environ Biophys. 2014;53:221–32. doi: 10.1007/s00411-014-0522-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Snyder AR, Morgan WF. Radiation-induced chromosomal instability and gene expression profiling: searching for clues to initiation and perpetuation. Mutat Res. 2004;568:89–96. doi: 10.1016/j.mrfmmm.2004.06.048. [DOI] [PubMed] [Google Scholar]
- 3.Park W-Y, Hwang C-I, Im C-N, Kang M-J, Woo J-H, Kim J-H, et al. Identification of radiation-specific responses from gene expression profile. Oncogene. 2002;21:8521–8. doi: 10.1038/sj.onc.1205977. [DOI] [PubMed] [Google Scholar]
- 4.Yin E, Nelson DO, Coleman MA, Peterson LE, Wyrobek AJ. Gene expression changes in mouse brain after exposure to low-dose ionizing radiation. Int J Radiat Biol. 2003;79:759–75. doi: 10.1080/09553000310001610961. [DOI] [PubMed] [Google Scholar]
- 5.Amundson S, Bittner M, Chen Y. Fluorescent cDNA microarray hybridization reveals complexity and heterogeneity of cellular genotoxic stress responses. Oncogene. 1999;18:3666–72. doi: 10.1038/sj.onc.1202676. [DOI] [PubMed] [Google Scholar]
- 6.Amundson SA, Do KT, Vinikoor L, Koch-Paiz CA, Bittner ML, Trent JM, et al. Stress-specific signatures: expression profiling of p53 wild-type and -null human cells. Oncogene. 2005;24:4572–9. doi: 10.1038/sj.onc.1208653. [DOI] [PubMed] [Google Scholar]
- 7.Gruel G, Lucchesi C, Pawlik A, Frouin V. Novel microarray-based method for estimating exposure to ionizing radiation. Radiat Res. 2006;166:746–56. doi: 10.1667/RR0260.1. [DOI] [PubMed] [Google Scholar]
- 8.Kimura S, Ishidou E, Kurita S, Suzuki Y, Shibato J, Rakwal R, et al. DNA microarray analyses reveal a post-irradiation differential time-dependent gene expression profile in yeast cells exposed to X-rays and gamma-rays. Biochem Biophys Res Commun. 2006;346:51–60. doi: 10.1016/j.bbrc.2006.05.126. [DOI] [PubMed] [Google Scholar]
- 9.Zhou T, Chou J, Simpson D. Profiles of global gene expression in ionizing-radiation-damaged human diploid fibroblasts reveal synchronization behind the G1 checkpoint in a G0-like state. Environ Health Perspect. 2006;114:553–9. doi: 10.1289/ehp.8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paul S, Amundson S. Development of gene expression signatures for practical radiation biodosimetry. Int J Radiat Oncol. 2008;71:1236–44. doi: 10.1016/j.ijrobp.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jen K-Y, Cheung VG. Transcriptional response of lymphoblastoid cells to ionizing radiation. Genome Res. 2003;13:2092–100. doi: 10.1101/gr.1240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otomo T, Hishii M, Arai H, Sato K, Sasai K. Microarray analysis of temporal gene responses to ionizing radiation in two glioblastoma cell lines: up-regulation of DNA repair genes. J Radiat Res. 2004;45:53–60. doi: 10.1269/jrr.45.53. [DOI] [PubMed] [Google Scholar]
- 13.Rieger K, Hong W. Toxicity from radiation therapy associated with abnormal transcriptional responses to DNA damage. Proc Natl Acad Sci U S A. 2004;101:6635–40. doi: 10.1073/pnas.0307761101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Short SC, Buffa FM, Bourne S, Koritzinsky M, Wouters BG, Bentzen SM. Dose- and time-dependent changes in gene expression in human glioma cells after low radiation doses. Radiat Res. 2007;168:199–208. doi: 10.1667/RR0940.1. [DOI] [PubMed] [Google Scholar]
- 15.Tucker JD, Joiner MC, Thomas RA, Grever WE, Bakhmutsky MV, Chinkhota CN, et al. Accurate gene expression-based biodosimetry using a minimal set of human gene transcripts. Int J Radiat Oncol. 2014;88:933–9. doi: 10.1016/j.ijrobp.2013.11.248. [DOI] [PubMed] [Google Scholar]
- 16.Langen B, Rudqvist N, Parris TZ, Schüler E, Helou K, Forssell-Aronsson E. Comparative analysis of transcriptional gene regulation indicates similar physiologic response in mouse tissues at low absorbed doses from intravenously administered 211At. J Nucl Med. 2013;54:990–8. doi: 10.2967/jnumed.112.114462. [DOI] [PubMed] [Google Scholar]
- 17.Schüler E, Parris TZ, Rudqvist N, Helou K, Forssell-Aronsson E. Effects of internal low-dose irradiation from 131I on gene expression in normal tissues in Balb/c mice. EJNMMI Res. 2011;1:29. doi: 10.1186/2191-219X-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schüler E, Rudqvist N, Parris TZ, Langen B, Spetz J, Helou K, et al. Time- and dose rate-related effects of internal (177)Lu exposure on gene expression in mouse kidney tissue. Nucl Med Biol. 2014;41:825–32. doi: 10.1016/j.nucmedbio.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 19.Paul S, Ghandhi SA, Weber W, Doyle-Eisele M, Melo D, Guilmette R, et al. Gene expression response of mice after a single dose of (137)cs as an internal emitter. Radiat Res. 2014;182:380–9. doi: 10.1667/RR13466.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abend M, Pfeiffer RM, Ruf C, Hatch M, Bogdanova TI, Tronko MD, et al. Iodine-131 dose dependent gene expression in thyroid cancers and corresponding normal tissues following the Chernobyl accident. PLoS One. 2012;7:e39103. doi: 10.1371/journal.pone.0039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ory C, Ugolin N, Schlumberger M, Hofman P, Chevillard S. Discriminating gene expression signature of radiation-induced thyroid tumors after either external exposure or internal contamination. Genes (Basel) 2011;3:19–34. doi: 10.3390/genes3010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maenhaut C, Detours V, Dom G, Handkiewicz-Junak D, Oczko-Wojciechowska M, Jarzab B. Gene expression profiles for radiation-induced thyroid cancer. Clin Oncol (R Coll Radiol) 2011;23:282–8. doi: 10.1016/j.clon.2011.01.509. [DOI] [PubMed] [Google Scholar]
- 23.Bauchinger M, Schmid E, Braselmann H. Cytogenetic evaluation of occupational exposure to external g-rays and internal 241Am contamination. Mutat Res. 1997;395:173–8. doi: 10.1016/s1383-5718(97)00166-6. [DOI] [PubMed] [Google Scholar]
- 24.Ropolo M, Balia C, Roggieri P, Lodi V, Nucci MC, Violante FS, et al. The micronucleus assay as a biological dosimeter in hospital workers exposed to low doses of ionizing radiation. Mutat Res. 2012;747:7–13. doi: 10.1016/j.mrgentox.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 25.Knops K, Boldt S, Wolkenhauer O, Kriehuber R. Gene expression in low-and high-dose-irradiated human peripheral blood lymphocytes: Possible applications for biodosimetry. Radiat Res. 2012;178:304–12. doi: 10.1667/rr2913.1. [DOI] [PubMed] [Google Scholar]
- 26.Budworth H, Snijders AM, Marchetti F, Mannion B, Bhatnagar S, Kwoh E, et al. DNA repair and cell cycle biomarkers of radiation exposure and inflammation stress in human blood. PLoS One. 2012;7:e48619. doi: 10.1371/journal.pone.0048619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao BS, Natarajan AT. Retrospective biological dosimetry of absorbed radiation. Radiat Prot Dosimetry. 2001;95:17–23. doi: 10.1093/oxfordjournals.rpd.a006516. [DOI] [PubMed] [Google Scholar]
- 28.Paul S, Smilenov LB, Amundson SA. Widespread decreased expression of immune function genes in human peripheral blood following radiation exposure. Radiat Res. 2013;180:575–83. doi: 10.1667/RR13343.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amundson SA, Grace M, McLeland C. Human in vivo radiation-induced biomarkers Gene expression changes in radiotherapy patients. Cancer Res. 2004;64:6368–71. doi: 10.1158/0008-5472.CAN-04-1883. [DOI] [PubMed] [Google Scholar]
- 30.Ivey RG, Moore HD, Voytovich UJ, Thienes CP, Lorentzen TD, Pogosova-Agadjanyan EL, et al. Blood-based detection of radiation exposure in humans based on novel phospho-Smc1 ELISA. Radiat Res. 2011;175:266–81. doi: 10.1667/RR2402.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ory C, Ugolin N, Hofman P, Schlumberger M, Likhtarev IA, Chevillard S. Comparison of transcriptomic signature of post-Chernobyl and postradiotherapy thyroid tumors. Thyroid. 2013;23:1390–400. doi: 10.1089/thy.2012.0318. [DOI] [PubMed] [Google Scholar]
- 32.Vöö S, Bucerius J, Mottaghy FM. I-131-MIBG therapies. Methods. 2011;55:238–45. doi: 10.1016/j.ymeth.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 33.DuBois SG, Matthay KK. I-131 Metaiodobenzylguanidine therapy in children with advanced neuroblastoma. Q J Nucl Med Mol Imaging. 2013;57:53–65. [PubMed] [Google Scholar]
- 34.DuBois SG, Matthay KK. Radiolabeled metaiodobenzylguanidine for the treatment of neuroblastoma. Nucl Med Biol. 2008;35:S35–48. doi: 10.1016/j.nucmedbio.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.More S, Itsara M, Yang X, Geier E. Vorinostat increases expression of functional norepinephrine transporter in neuroblastoma in vitro and in vivo model systems. Clin Cancer Res. 2011;17:2339–49. doi: 10.1158/1078-0432.CCR-10-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mueller S, Yang X, Sottero T, Gragg A. Cooperation of the HDAC inhibitor vorinostat and radiation in metastatic neuroblastoma: efficacy and underlying mechanisms. Cancer Lett. 2011;306:223–9. doi: 10.1016/j.canlet.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DuBois SG, Chesler L, Groshen S, Hawkins R, Goodarzian F, Shimada H, et al. Phase I study of vincristine, irinotecan, and 131I-metaiodobenzylguanidine for patients with relapsed or refractory neuroblastoma: a new approaches to neuroblastoma therapy trial. Clin Cancer Res. 2012;18:2679–86. doi: 10.1158/1078-0432.CCR-11-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matthay KK, Panina C, Huberty J, Price D, Glidden D V, Tang HR, et al. Correlation of tumor and whole-body dosimetry with tumor response and toxicity in refractory neuroblastoma treated with (131)I-MIBG. J Nucl Med. 2001;42:1713–21. [PubMed] [Google Scholar]
- 39.Matthay KK, DeSantes K, Hasegawa B, Huberty J, Hattner RS, Ablin A, et al. Phase I dose escalation of 131I-metaiodobenzylguanidine with autologous bone marrow support in refractory neuroblastoma. J Clin Oncol. 1998;16:229–36. doi: 10.1200/JCO.1998.16.1.229. [DOI] [PubMed] [Google Scholar]
- 40.Koral KF, Huberty JP, Frame B, Matthay KK, Maris JM, Regan D, et al. Hepatic absorbed radiation dosimetry during I-131 metaiodobenzylguanidine (MIBG) therapy for refractory neuroblastoma. Eur J Nucl Med Mol Imaging. 2008;35:2105–12. doi: 10.1007/s00259-008-0873-3. [DOI] [PubMed] [Google Scholar]
- 41.Paul S, Smilenov LB, Elliston CD, Amundson SA. Radiation dose-rate effects on gene expression in a mouse biodosimetry model. Radiat Res. 2015;184:24–32. doi: 10.1667/RR14044.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wyrobek A, Manohar C. Low dose radiation response curves, networks and pathways in human lymphoblastoid cells exposed from 1 to 10 cGy of acute gamma radiation. Mutat Res. 2011;722:119–30. doi: 10.1016/j.mrgentox.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 43.Amundson SA, Myers TG, Fornace AJ. Roles for p53 in growth arrest and apoptosis: putting on the brakes after genotoxic stress. Oncogene. 1998;17:3287–99. doi: 10.1038/sj.onc.1202576. [DOI] [PubMed] [Google Scholar]
- 44.Buckley SE, Chittenden SJ, Saran FH, Meller ST, Flux GD. Whole-body dosimetry for individualized treatment planning of 131I-MIBG radionuclide therapy for neuroblastoma. J Nucl Med. 2009;50:1518–24. doi: 10.2967/jnumed.109.064469. [DOI] [PubMed] [Google Scholar]
- 45.Cristy M, Eckerman KF. ORNL Health and Safety Research Division Report No. ORNL/TM-8381 V1-V7. Oak Ridge National Laboratory; Oak Ridge, TN: 1987. Specific absorbed fractions of energy at various ages from internal photon sources. ( http://bit.ly/2aNbfJw) [Google Scholar]
- 46.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 47.R Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. [Google Scholar]
- 48.Hänscheid H, Fernández M, Eberlein U, Lassmann M. Self-irradiation of the blood from selected nuclides in nuclear medicine. Phys Med Biol. 2014;59:1515–31. doi: 10.1088/0031-9155/59/6/1515. [DOI] [PubMed] [Google Scholar]
- 49.Flower MA, Fielding SL. Radiation dosimetry for therapy of neuroblastoma radiation dosimetry for neuroblastoma I-mIBG therapy of neuroblastoma. Phys Med Biol. 1996;41:1933–40. doi: 10.1088/0031-9155/41/10/006. [DOI] [PubMed] [Google Scholar]
- 50.Manning G, Kabacik S, Finnon P, Bouffler S, Badie C. High and low dose responses of transcriptional biomarkers in ex vivo X-irradiated human blood. Int J Radiat Biol. 2013;89:512–22. doi: 10.3109/09553002.2013.769694. [DOI] [PubMed] [Google Scholar]
- 51.Tucker JD, Divine GW, Grever WE, Thomas RA, Joiner MC, Smolinski JM, et al. Gene expression-based dosimetry by dose and time in mice following acute radiation exposure. PLoS One. 2013;8:e83390. doi: 10.1371/journal.pone.0083390. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.