Summary
Cholesterol is an essential component of the cellular membranes and, by extension, of the HIV envelope membrane, which is derived from the host cell plasma membrane. Depletion of the cellular cholesterol has a inhibitory effect on HIV assembly, reduces infectivity of the produced virions, and makes the cell less susceptible to HIV infection. It is not surprising that the virus has evolved to gain access to cellular proteins regulating cholesterol metabolism. One of the key mechanisms used by HIV to maintain high levels of cholesterol in infected cells is Nef-mediated inhibition of cholesterol efflux and the cholesterol transporter responsible for this process, ABCA1. In this article, we describe methods to investigate these effects of HIV-1 infection.
Keywords: HIV-1, Nef, cholesterol efflux, ABCA1, confocal microscopy, image analysis
1. Introduction
Replication of enveloped viruses assembling at the plasma membrane of infected cells, such as HIV, critically depends on cholesterol, and depletion of cholesterol in HIV-infected cell affects viral production and infectivity (1). HIV assembles at the plasma membrane domains enriched in cholesterol and sphingolipids called lipid rafts (2). Lipid rafts are also used by HIV as entry gates into the target cells (3). In general, high levels of lipid rafts on the plasma membrane are beneficial for HIV replication. It is not surprising that HIV evolved to regulate the abundance of lipid rafts. The mechanism used by the virus for this purpose is inhibition of the cellular ATP binding cassette transporter A1 (ABCA1), which controls cholesterol efflux from cells to apoA-I acceptor in the blood. Activity of ABCA1 is inversely correlated to the abundance of lipid rafts on the plasma membrane: stimulation of ABCA1 expression reduces the number of lipid rafts, whereas inhibition of ABCA1 activity increases lipid raft abundance (4, 5). To inhibit ABCA1 activity, HIV relies on its protein Nef, which blocks ABCA1 exit from the endoplasmic reticulum (ER) and stimulates its degradation (6, 7). To accomplish this effect, Nef interacts with ER chaperone calnexin, which regulates folding and maturation of glycosylated proteins, and disrupts the interaction between calnexin and ABCA1, thus abrogating the processing of ABCA1 in ER (8). Importantly, this activity of Nef affects not only infected cells, but also bystander cells, which take up Nef released from infected cells into the bloodstream (9, 10). The side effect of this viral activity is increased risk of atherosclerosis and cardiovascular disease in HIV-infected patients (11). The protocols described in this chapter provide effective and reliable methods to analyze and visualize ABCA1 and measure the rate of cholesterol efflux from HIV-infected cells.
2. Materials
Prepare all solutions using deionized water and analytical grade reagents. Prepare and store all reagents at room temperature (unless indicated otherwise).
2.1. Cholesterol efflux
Human monocytic cell line THP-1.
24-well plates.
RPMI-1640. Store at 4°C.
Fetal Bovine Serum (FBS). Store at 4°C.
Pen/Strep. Store at 4°C.
L-glutamine. Store at 4°C.
Phorbol 12-myristate-13-acetate (PMA). Prepare 10 mg/ml stock solution in DMSO, aliquot and store at −20°C. Prepare working solution by diluting the stock solution in PBS to 100 µg/ml; it can be stored at −20°C for 1–2 months.
[3H]cholesterol. Store at 4°C.
Purified ApoA-I, store at −20°C (see Note 1).
Ultrapure water.
Ethanol.
LXR agonist TO-901317. Prepare 100 mg/ml (200 mM) stock solution in EtOH, store at −70°C; working solution is prepared by diluting stock solution in PBS to 100 µM.
PBS.
Lysis buffer (1% Triton X-100 in RPMI-1640).
Scintillation fluid.
2.2. Confocal microscopy and image analysis
HeLa-ABCA1-GFP cell line (a kind gift of Dr. A. Remaley (12)).
Nef expression vector pT7consnefhis6 (NIH AIDS Research and Reference Reagent Program).
Dulbecco's Modified Eagle's Medium (DMEM). Store at 4°C.
Selective DMEM medium: DMEM supplemented with 10% FBS, 100-fold diluted pen/strep, 100-fold diluted L-Glu, 0.15 mg/ml G418, and 0.2 mg/ml Hygromycin B. Store at 4°C.
Fetal Bovine Serum (FBS). Store at 4°C.
Pen/Strep. Store at −20°C.
L-glutamine (L-Glu). Store at −20°C.
G418 Sulfate (G418) (50 mg/ml solution). Store at 4°C.
Hygromycin B (50 mg/ml solution). Store at 4°C.
PBS.
Polyclonal rabbit anti-Nef antisera (NIH AIDS Research and Reference Reagent Program). Store at 4°C.
Mouse monoclonal anti-Calnexin - ER membrane marker antibody. Store at 4°C.
Alexa Fluor® 647 Goat Anti-Rabbit IgG (H+L) Antibody. Store at 4°C.
DyLight 550 Goat Anti-Mouse IgG Antibody. Store at 4°C.
DAPI dilactate. Store at 4°C.
Goat IgG. Store at 4°C.
Albumin, from bovine serum (BSA). Store at 4°C.
Formaldehyde Solution.
Triton X-100.
Ethanol.
Ultrapure water.
6-well plates.
Coverslips (cover glasses; thickness no. 1½).
Microscope Slides (3”×1”×1 mm).
Mounting medium Fluoromount-G.
Blocking solution: 20 µl Goat IgG per ml of 1% BSA in PBS (filtered through 0.2 µm filter). Prepare fresh before use.
3. Methods
3.1. Cholesterol efflux analysis
This protocol describes measurement of cholesterol efflux from human monocytic cell line, THP-1, differentiated into macrophage-like cells by PMA treatment. The format we usually use is the 24-well plate format, but it can be formatted to 12 or 48-well plates and the assay can be adapted for different cell types. The assay measures efflux of radioactively labelled cholesterol pre-loaded into the target cells to the apoA1 acceptor. This process is controlled by ABCA1, which is induced in cells by stimulation with LXR agonist TO-901317 (see Note 2). The procedure allows one to measure either the capacity of cells to release cholesterol to extracellular acceptors or the capacity of the acceptor to accept cholesterol released from cells. For the former, the acceptor should be added in saturating concentration, for the latter, the concentration of the acceptor should be approximately half of the saturating concentration. The assay consists of the following steps: i) loading cells with labelled cholesterol; ii) equilibrating labelled cholesterol among all intracellular cholesterol pools; and iii) quantitating the transfer of labelled cholesterol from cells to the acceptor.
Resuspend cells from stock culture and count them. Plate cells into 24-well plates (see Note 3) at the final density of 0.3×106 cells per well in 1 ml of complete RPMI-1640 supplemented with 10% FBS, 1% Pen/Strep, and 2 mM L-Glutamine. The density can be adjusted to accommodate requirements of treatment prior to the efflux assay (e.g. transfections, treatment with an inhibitor or activator, infection with a virus, etc.).
Dispense the required amount of [3H]cholesterol into a 1.5 ml microfuge tube (0.5 μCi or 19 kBq per well is required for a typical assay) (see Note 4). Dilute [3H]cholesterol solution in complete media to the final concentration 5 μCi/ml and add labelled cholesterol into each well (100 μl/well, final volume per well is 1 ml). Add PMA to a final concentration of 100 ng/ml, and incubate cells for 72 hours in cell culture incubator (37°C, 5% CO2).
Remove media containing [3H]cholesterol. Wash cells gently with PBS. Repeat washing three times.
Prepare serum-free medium with 1 μM TO-901317, add 1 ml to each well. Incubate for 18 hours in the cell culture incubator.
Wash cells gently with PBS.
Add 500 μl of apoA-I solution in serum-free medium (final apoA-I concentration − 30 µg/ml for saturating efflux, or 15 µg/ml for non-saturating) (see Note 5).
One set of wells is used to determine background efflux; add to these wells serum-free medium without apoA-I.
Incubate cells for 2 hours in cell culture incubator (37°C, 5% CO2). Duration of the efflux incubation may vary from 30 min to 8 h if required, but for shorter incubations the amount of radioactivity and/or number of cells may need to be increased.
After incubation collect media into 1.5 ml microfuge tubes. Spin at 10,000×g for 5–10 min at room temperature to remove cellular debris.
Transfer 100 μl of media into 7 ml scintillation vial. Add 5 ml of Insta-gel Plus and vortex mixture. Store remaining samples at 4°C.
Add 1 ml of lysis buffer to each well, shake the plate on a shaker, and collect the lysate. Check cells under microscope to ensure that all cells have been removed. If some cells remain, add 100 μl of lysis buffer and scrape the wells.
Transfer 100 μl of cell lysates into 7 ml scintillation vials. Add 5 ml of Insta-gel Plus and vortex the mixture.
Count radioactivity in the media collected and the cells on a scintillation counter for dpm of [3H] cholesterol.
The rate of cholesterol efflux is usually expressed as a proportion of cholesterol moved from cells to the acceptor. The following formula is used:
The specific efflux is calculated as a difference between the efflux in the presence and absence of an acceptor (blank): Specific cholesterol efflux (%) = Cholesterol efflux to the acceptor (%) − Cholesterol efflux without acceptor (%)
3.2. Confocal microscopy and image analysis
This protocol describes the technique for visualization of ABCA1 by fluorescent microscopy in HeLa cells stably expressing ABCA1-GFP (12) and transfected with HIV-1 Nef, but can be used for any HIV-infected or transfected cells (see Note 6). The analysis described involves staining for Nef and calnexin (an ER chaperone), and counterstaining of the nuclei. The procedure helps to identify cellular components interacting with ABCA1 and allows to follow and to analyze changes in ABCA1 distribution upon such interactions. The presented example elucidates changes in the interaction of ABCA1 with one of its cellular counterparts – calnexin – when HIV Nef is expressed. Since calnexin also serves as a marker for Endoplasmatic Reticulum (ER), the altered ABCA1 distribution and its retention in ER can be demonstrated.
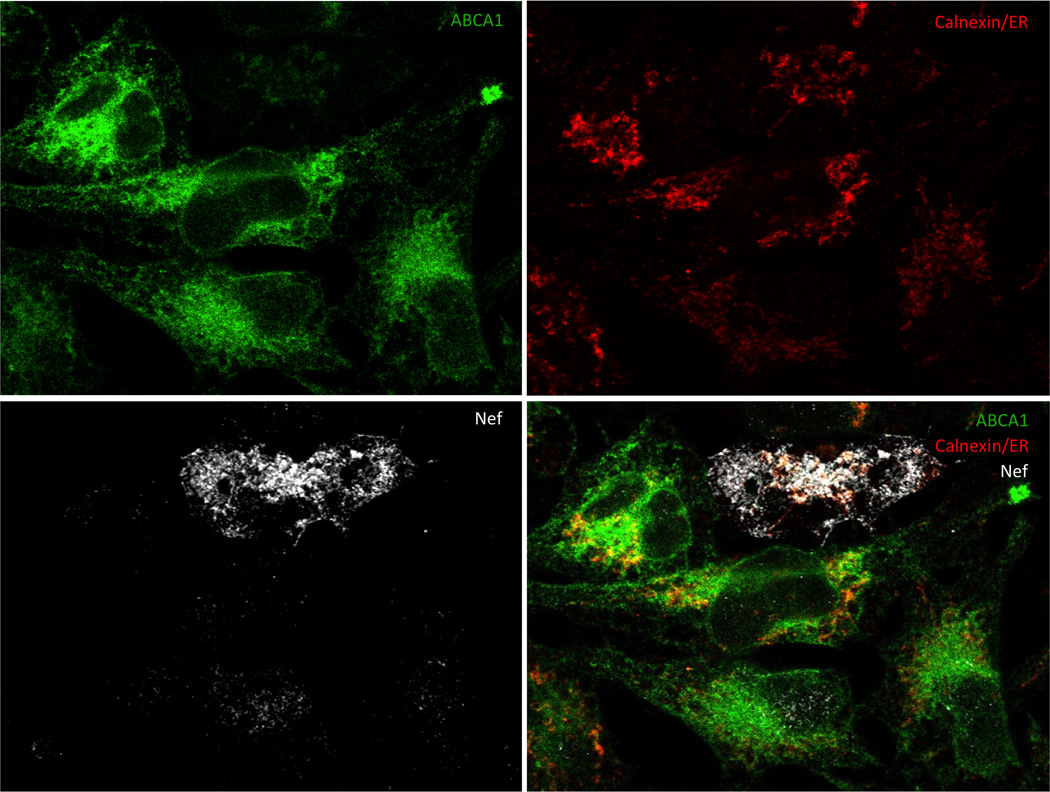
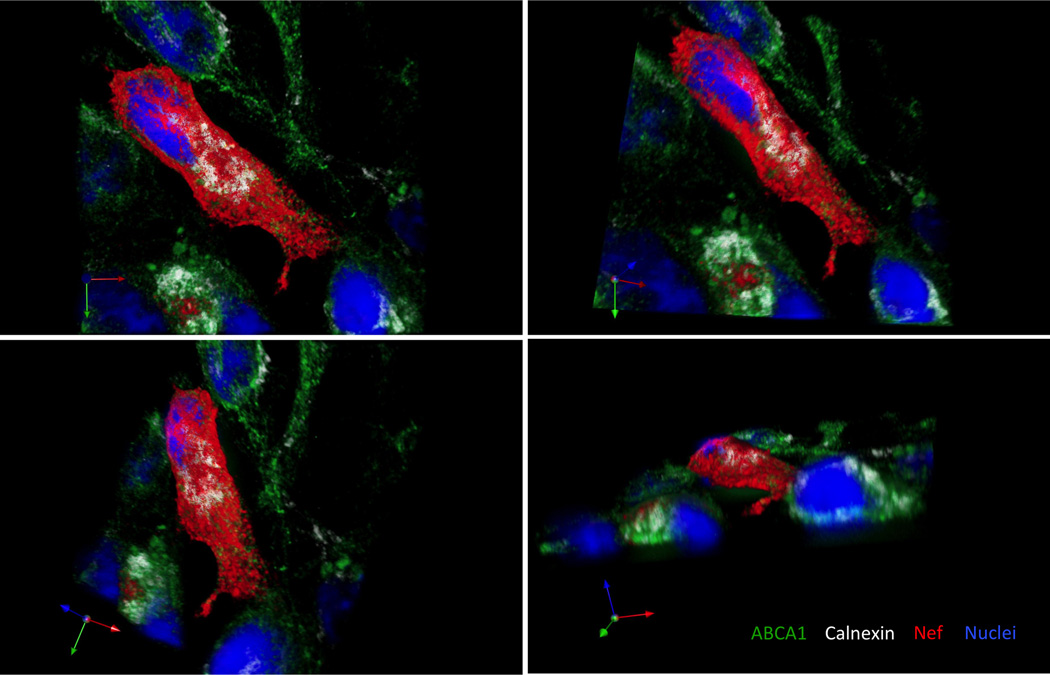
Confocal microscope can generate either 2D or 3D images. Both serve as valuable tools for analysis of events on cellular and subcellular level. Creation of a tile-scan image using 2D imaging allows to analyze large area in a single optical plane and to choose a representative section for presentation and further analysis. An example of such a section is in Figure 1 where a cell expressing HIV Nef (white) is surrounded by Nef negative cells. ABCA1 (green) is markedly suppressed in the Nef positive cell and remaining ABCA1 is almost exclusively localized in the areas positive for calnexin (red). In the surrounding cells not expressing Nef, only a smaller fraction of ABCA1 is co-localizing (yellow to orange) with calnexin, while a substantial proportion of ABCA1 is localized outside of ER, mainly on the cellular membrane, where it can perform its functions associated with cholesterol efflux. A better understanding of subcellular localization of molecules of interest is provided by a 3D image (Fig. 2). Using appropriate software, a 3D image can be rotated into positions that best reveal relationships between molecules of interest.
Clean and sterilize the coverslips by dipping them into 70% ethanol.
Air dry the coverslips (remove the majority of ethanol by vacuum).
Place the sterilized coverslips into 6 well plates.
Plate HeLa-ABCA1-GFP cells to approximately 40% confluency in selective DMEM medium.
Incubate the seeded plates in CO2 incubator at 37°C overnight. Optimal cell culture confluency for transfection is 60%–80%.
Prepare Transfection Mix 1 (100 µl per well) by adding Nef-expressing plasmid DNA (2 µg per well) to DMEM media (no FBS, antibiotics or supplements added) in an Eppendorf tube. Pipette twice up and down.
Prepare Transfection Mix 2 (100 µl per well) by adding 6 µl of Metafectene into 94 µl of DMEM media (no FBS, antibiotics or supplements added) in an Eppendorf tube. Pipette twice up and down.
Add Transfection Mix 1 forcefully into Transfection Mix 2 (do not apply any mixing procedure after Mix 1 and 2 are combined since it may destroy the DNA/lipid complex).
Incubate 20 minutes at room temperature.
During the incubation time, replace culture medium in 6-well plates with cells with 3 ml of fresh selective DMEM medium/well.
Transfer 200 µl of DNA/lipid complex drop-wise to each well using a 200 µl tip with its sharp end cut off. Target the drops over the area of the coverslip.
Incubate the plates with transfected cells in CO2 incubator at 37°C overnight.
The next day, replace culture medium with 3 ml of fresh selective DMEM medium/well and continue incubation in CO2 incubator at 37°C.
48 hours (see Note 7) post transfection, aspirate the culture medium and wash the cultures twice with warm PBS (37°C).
Fix cells with 3.5% formaldehyde solution (formaldehyde solution prepared in PBS from the 37% concentrate) for 10 minutes at room temperature.
Wash the fixed cells three times with PBS. The plates can be stored at 4°C overnight.
Permeabilize fixed cells with 0.1% Triton X-100 (in PBS) for 5 min using 2 ml Triton solution/well.
Wash the permeabilized cell three times with PBS.
Block with 2 ml of Blocking solution/well for 30 min at room temperature (cover plates with aluminum foil). The plates can be left with blocking solution at +4°C overnight.
Drain the coverslips (on the paper towel and using vacuum) and transfer them on the parafilm in a large Petri dish (use syringe needle and flat end forceps to manipulate the coverslips).
Incubate with 200 µl (for 18 mm x18 mm coverslip) of primary antibody (mixture of anti-Nef rabbit serum and anti-Calnexin-ER membrane marker mouse monoclonal antibody, both diluted 1:750 in PBS, and ChromPure Goat IgG diluted 1:30 in PBS) at room temperature for 60 minutes (use wet paper towel to keep humid conditions in the Petri dish; perform incubation in the dark).
Wash three times with PBS in a 6-well plate.
Drain the coverslips and transfer them back on the parafilm in a large Petri dish.
Incubate with 200 µl (for 18 mm x18 mm coverslip) of secondary antibody (goat anti rabbit Alexa-Fluor 647 antibody and goat anti mouse DyLight 550 antibody, both diluted 1:500 in PBS, and ChromPure Goat IgG diluted 1:30 in PBS) at room temperature for 30 minutes (use wet paper towel to keep humid conditions in the Petri dish; perform incubation in the dark).
Wash three times with PBS in a 6-well plate.
Stain with DAPI (diluted 1:10,000 from the stock solution; 2 ml/well) for 5 min.
Wash three times with PBS in a 6-well plate. At this step, it is possible to leave coverslips in the plates with PBS at +4°C overnight.
Drain the coverslips (on the paper towel and using vacuum).
Mount the coverslips on microscope slides with mounting medium.
Leave the slides to settle in the dark at room temperature overnight.
Use the slides for imaging or store them in the dark at +4°C.
High-content and 3-dimentional image data acquisition is performed on a Zeiss Cell Observer Spinning Disk Confocal instrument, equipped with two Photometrics Evolve Delta high-speed (512×512) EM CCD cameras on Axio Observer Z1 microscope equipped with scanning stage with piezo z-motor, which ensures generation of highly reliable 3D image sets. Stitching and multi-position modules are part of the Zen Blue software.
3D image sets are acquired with Plan Apochromat 150×/1.35 objective lens, allowing generation of 512×512 images at pixel resolution of 0.089 µm. 16-bit image format is used for intensity registration, as pixel integration within 25% or above of the available dynamic range is considered satisfactory. This approach allows for detection efficiency of GFP/ABCA1, not possible even on advanced point-scan confocal microscopes.
For 3D analyses, the z-step is set at 0.250 µm to optimize for the z-point spread and optical section thickness. Multiple positions are selected and memorized. The instrument then executes capturing of image sets channel by channel for each individual position in z-dimension (Fig. 2).
For 2D analyses, single optical plane is captured but large squared area is selected, to produce tile-scan image set with overlap between adjacent image frames at 15–25%. This overlap is used for subsequent stitching using precise correlation in the overlapping areas.
Four channels are generated using the following parameters: i) channel 1, representing Alexa Fluor 568, encoding for calnexin is generated by excitation with 561 nm diode laser and the emission filtered by 629/62 nm band filter; ii) channel 2, representing GFP/ABCA1 is created by excitation with 488 nm diode laser while the emission is filtered by 535/30 nm band emission filter; iii) channel 3, representing nuclear stain DAPI is recorded using excitation of 405 nm diode laser and the emission is filtered with 450/50 nm emission filter; iv) channel 4, representing the Alexa Fluor 647 encoding Nef is generated by 633 nm diode laser excitation and the emission filtered with 690/50 nm emission filter. The entire z-stack is captured channel by channel (Fig. 2).
Image analysis is performed using Volocity software from 2D or 3D image sets.
For visualization of the co-localization, voxel overlap is first determined by establishing legitimate intensity threshold for each channel. Using interactive stepwise threshold and false color feedback, elimination of the dark currents serves as a threshold point. Intersecting the thresholds of the two channels determines the voxel overlap. For voxel co-localization the so called positive co-localization is used, which is the overlap of the voxels that bear an intensity value larger than the mean value (over threshold) of the given channel. The co-localized voxels are visualized as a separate channel. Visualization is achieved by 3D rendering using transparency mode. The co-localization is compared by plotting the co-localization co-efficiency.
For two 2D analyses the cells having high and no Nef expression are selected by free hand tool from a large tiled and stitched image. The large size of these images allows extraction of several hundred cells for each group. The average intensity for Nef, ABCA1 and calnexin can be plotted and compared.
Figure 1. A 2D image of HeLa-ABCA1-GFP cells transfected with HIV-1 Nef.
Calnexin and HIV-1 Nef were visualized using anti-Calnexin-ER membrane marker mouse monoclonal antibody followed by goat anti-mouse DyLight 550 antibody (red), and by anti-Nef rabbit serum followed by goat anti rabbit Alexa-Fluor 647 antibody (white), respectively. Cellular nuclei were stained by DAPI dilactate (blue). ABCA1 is co-expressed with GFP (green). Co-localization of ABCA1 and calnexin in Nef-negative cells is visualized as yellow colored areas in the overlay. ABCA1 abundance is greatly reduced in the Nef-positive cell, reflecting the inhibitory effect of Nef on this cholesterol transporter.
Figure 2. A 3D image of HeLa-ABCA1-GFP cells transfected with HIV-1 Nef.
Calnexin and HIV-1 Nef were visualized using anti-Calnexin-ER membrane marker mouse monoclonal antibody followed by goat anti mouse DyLight 550 antibody (white), and by anti-Nef rabbit serum followed by goat anti rabbit Alexa-Fluor 647 antibody (red), respectively. Cellular nuclei were stained by DAPI dilactate (blue). ABCA1 is co-expressed with GFP (green). Analysis of the 3D image was performed in Volocity allowing for precise spatial determination of localization of molecules of interest within a cell.
Supplementary Material
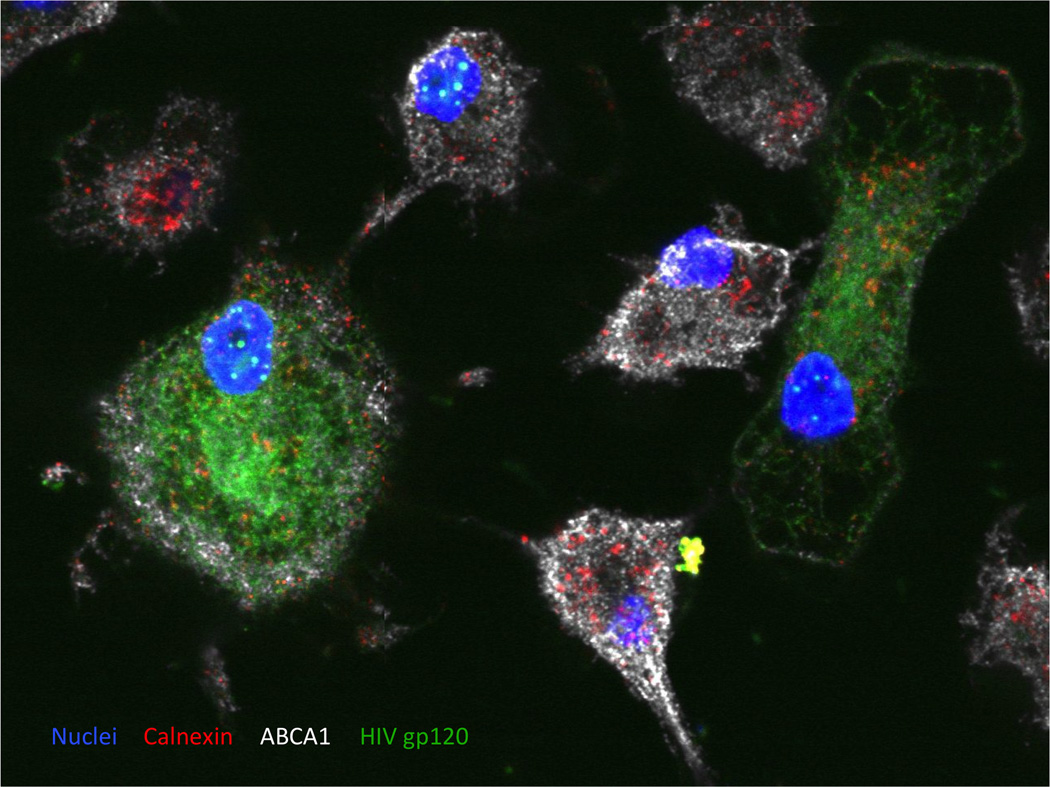
Figure 3. A 2D image of monocyte-derived macrophages infected with HIV-1.
Peripheral blood monocytes were differentiated into the macrophages and infected with VSV-G pseudotyped HIV-1 NL4-3. HIV-1 envelope protein was stained using anti-HIV gp120 sheep serum followed by Alexa Fluor® 488 Donkey Anti-Sheep IgG (H+L) Antibody (green). Cellular proteins Calnexin and ABCA1 were visualized using mouse monoclonal anti-Calnexin-ER membrane marker antibody followed by goat anti-mouse DyLight 550 antibody (red) and by anti-ABCA1 rabbit polyclonal antibody followed by goat anti rabbit Alexa-Fluor 647 antibody (white), respectively. Cellular nuclei were stained by DAPI dilactate (blue). Abundance of ABCA1 is reduced in HIV-infected macrophages.
Acknowledgments
This study was supported by NIH grants R01HL093818, R01HL101274, R21AI108533, P30AI087714, P30HD040677, S10OD010710, 1S10RR025565.
Footnotes
As acceptors for cholesterol efflux, one can use isolated HDL, plasma and serum (human or other species). However, apoA-I is a specific acceptor for ABCA1-mediated efflux, whereas HDL, plasma and serum accept cholesterol delivered by a variety of transporters.
When the experiment is performed to evaluate ABCA1 activity, TO-901317 stimulation can be omitted. However, cholesterol efflux may be too low in such cases, and one may need to titrate the dose of TO-901317. No TO-901317 stimulation is needed when working with cells transfected with ABCA1-expressing vector.
The number of wells should be sufficient for quadruplicate determinations for each experimental condition.
Commercially available [3H]cholesterol comes as a solution in either toluene or ethanol. If [3H]cholesterol is a toluene solution, dry it completely with N2 in a toluene-resistant tube and resuspend in absolute ethanol at a final concentration of 1 μCi/μl. Vortex and mix well. If [3H]cholesterol is an ethanol solution, it can be used immediately.
Other acceptors that can be used are high density lipoprotein (HDL) (final concentration 50 µg/ml saturating, or 25–30 µg/ml non-saturating), plasma or serum (final concentration 5% saturating, 1–2% non-saturating).
Other cell types, infected with HIV or transfected with vectors expressing HIV proteins, can be analyzed by this method. An example is presented in Fig. 3, which shows monocyte-derived macrophages infected with HIV-1. HIV-1 Env was stained using anti-HIV gp120 sheep serum as a primary and Alexa Fluor® 488 conjugated donkey anti-sheep IgG as a secondary antibody, ABCA1 was stained using anti-ABCA1 rabbit polyclonal antibody as a primary and Alexa Fluor® 647 conjugated goat anti-rabbit IgG as a secondary antibody and the endoplasmic reticulum (ER) chaperone calnexin was stained using mouse monoclonal anti-calnexin - ER membrane marker antibody as primary and goat anti mouse DyLight 550 conjugated antibody as secondary). Calnexin regulates folding and maturation of glycosylated proteins, including ABCA1 and gp160, in the ER. Nuclei were counterstained with DAPI.
Transfected cells can be incubated for 72 hrs in some cases to obtain better expression.
References
- 1.Waheed AA, Freed EO. Lipids and membrane microdomains in HIV-1 replication. Virus Res. 2009;143:162–176. doi: 10.1016/j.virusres.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ono A, Freed EO. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc Natl Acad Sci U S A. 2001;98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manes S, del Real G, Lacalle RA, et al. Membrane raft microdomains mediate lateral assemblies required for HIV-1 infection. EMBO Rep. 2000;1:190–196. doi: 10.1093/embo-reports/kvd025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koseki M, Hirano K-i, Masuda D, et al. Increased lipid rafts and accelerated lipopolysaccharide-induced tumor necrosis factor-{alpha} secretion in Abca1-deficient macrophages. J. Lipid Res. 2007;48:299–306. doi: 10.1194/jlr.M600428-JLR200. [DOI] [PubMed] [Google Scholar]
- 5.Landry YD, Denis M, Nandi S, et al. ATP-binding Cassette Transporter A1 Expression Disrupts Raft Membrane Microdomains through Its ATPase-related Functions. J. Biol. Chem. 2006;281:36091–36101. doi: 10.1074/jbc.M602247200. [DOI] [PubMed] [Google Scholar]
- 6.Cui HL, Grant A, Mukhamedova N, et al. HIV-1 Nef mobilizes lipid rafts in macrophages through a pathway that competes with ABCA1-dependent cholesterol efflux. J Lipid Res. 2012;53:696–708. doi: 10.1194/jlr.M023119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brichacek B, Darwish C, Popratiloff A, et al. HIV-1 Infection of Macrophages Induces Retention of Cholesterol Transporter ABCA1 in the Endoplasmic Reticulum. AIDS Res Hum Retroviruses. 2014 doi: 10.1089/aid.2014.0156. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jennelle L, Hunegnaw R, Dubrovsky L, et al. HIV-1 Protein Nef Inhibits Activity of ATP Binding Cassette Transporter A1 by Targeting Endoplasmic Reticulum Chaperone Calnexin. J Biol Chem. 2014 doi: 10.1074/jbc.M114.583591. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asztalos BF, Mujawar Z, Morrow MP, et al. Circulating Nef induces dyslipidemia in simian immunodeficiency virus-infected macaques by suppressing cholesterol efflux. J Infect. Dis. 2010;202:614–623. doi: 10.1086/654817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui HL, Ditiatkovski M, Kesani R, et al. HIV protein Nef causes dyslipidemia and formation of foam cells in mouse models of atherosclerosis. FASEB J. 2014;28:2828–2839. doi: 10.1096/fj.13-246876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowe SM, Westhorpe CL, Mukhamedova N, et al. The macrophage: the intersection between HIV infection and atherosclerosis. J. Leukoc. Biol. 2010;87:589–598. doi: 10.1189/jlb.0809580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neufeld EB, Remaley AT, Demosky SJ, et al. Cellular Localization and Trafficking of the Human ABCA1 Transporter. J.Biol.Chem. 2001;276:27584–27590. doi: 10.1074/jbc.M103264200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.