Abstract
Sigma-1 receptor (S1R) is a unique pluripotent modulator of living systems and has been reported to be associated with a number of neurological diseases including pathological pain. Intrathecal administration of S1R antagonists attenuate the pain behavior of rodents in both inflammatory and neuropathic pain models. However, the S1R localization in the spinal cord shows a selective ventral horn motor neuron distribution, suggesting the high likelihood of S1R in the dorsal root ganglion (DRG) mediating the pain relief by intrathecally administered drugs. Since primary afferents are the major component in the pain pathway, we examined the mouse and rat DRGs for the presence of the S1R. At both mRNA and protein levels, qRT-PCR and Western confirmed that the DRG contains greater S1R expression in comparison to spinal cord, cortex, or lung but less than liver. Using a custom-made highly specific antibody, we demonstrated the presence of a strong S1R immuno-fluorescence in all rat and mouse DRG neurons co-localizing with the NSE marker, but not in neural processes or GFAP-positive glial satellite cells. In addition, S1R was absent in afferent terminals in the skin and in the dorsal horn of the spinal cord. Using immuno-electron microscopy, we showed that S1R is detected in the nuclear envelope and endoplasmic reticulum of DRG cells. In contrast to other cells, S1R is also located directly at the plasma membrane of the DRG neurons. The presence of S1R in the nuclear envelope of all DRG neurons suggests an exciting potential role of S1R as a regulator of neuronal nuclear activities and/or gene expression, which may provide insight towards new molecular targets for modulating nociception at the level of primary afferent neurons.
Graphical Abstract
Introduction
The Sigma-1 receptor (S1R), originally proposed as a subtype of the opioid receptor based on benzomorphan opiate binding (Martin et al., 1976), is now known to be a distinct non-opioid receptor protein of 223 amino acids. Its structure was recently solved to be a single transmembrane protein (Schmidt et al, 2016), in contrast to the earlier prediction of a two transmembrane domain protein. The protein primary sequence is unique in the mammalian genome, showing closest similarity to the yeast sterol isomerase (Moebius et al., 1997).
S1R is localized in significant amounts in the endoplasmic reticulum (ER) and is also found in the ER associated mitochondrial membrane (Hayashi and Su, 2007), in the plasma membrane (Kourrich et al., 2013), and more recently is reported to be present in the nuclear envelope (Tsai et al., 2015b).
S1R endogenous ligands include steroids, the trace amine dimethyltryptamine (DMT) (Fontanilla et al., 2009), the lipid sphingosine (Ramachandran et al., 2009), and myristic acid (Tsai et al., 2015a). S1R also exhibits high affinity binding to exogenous hallucinogenic ligands such as cocaine, ketamine, and haloperidol, suggesting a potential involvement of this receptor in drug addiction. The exact role of S1R in an organism has yet to be elucidated. However, accumulating evidence of pleotropic modulation of many targets in the plasma membrane, cytosol, and nuclear envelope has led to the suggestion that the S1R is a “pluripotent modulator” of the cell (Su et al., 2016).
In the central nervous system, the S1R protein is expressed in the granular layer of the olfactory bulb, central gray zone, motor nuclei of the hind brain, and in various hypothalamic nuclei (Alonso et al., 2000). At the spinal cord level, a careful comparison between wildtype and S1R-KO mice using a custom-made well-characterized antibody demonstrated a strong immunoreactive signal in the ventral motoneurons (Mavlyutov et al., 2010). Consistent with the broad expression of S1R in the central nervous system, many reports have suggested an association of this receptor with a variety of nervous system diseases including amyotrophic lateral sclerosis, Alzheimer's disease, Parkinson's disease, and depression (Su et al., 2016). Another well-described neurologic phenotype in in which S1R may have a potential role is inflammatory and neuropathic pain, an association which is well supported by genetic and pharmacological manipulations (Cendán et al., 2005; Kim et al., 2006; Entrena et al., 2009; Gris et al., 2015). The likely role of S1R in pain is also suggested by an unbiased expression screening of genes regulated by the sciatic nerve axotomy model of neuropathic pain, which demonstrated a 2 to 5-fold increase in the expression of S1R in the dorsal root ganglion (DRG) (Xiao et al., 2002). In fact, the S1R is emerging as a novel target in the therapeutic intervention for pain (Zamanillo et al., 2013; Davis, 2015; Gris et al., 2015).
Although the exact role of S1R in pathological pain remains unknown, its abundance in the DRG is interesting. The DRG is the anatomical location that houses the cell soma of the primary afferent sensory neurons and is a critical organ for nociceptive signal processing. The DRG has also been an emerging target for intervention since its location in the peripheral nervous system allows more accessibility relative to the other sites in the central nervous system (Sapunar et al., 2012). A recent report indicated abundant expression of the S1R in the DRG (Bangaru et al., 2013). However, we sought to reexamine the cellular and subcellular anatomical distribution of S1R in the DRG of both rats and mice using a highly-specific well-characterized S1R-antibody (Ramachandran et al., 2007).
Experimental Procedures
Animals
All studies were approved (Protocol M02512, and M02569) by the local institutional animal care use committee, and all animals were treated in accordance with published NIH standards. Male Sprague Dawley rats weighing 300 g were purchased from Envigo Lab (Envigo, Madison, WI, USA). Oprs1 mutant (+/−) B6;129S5-Sigmar1Gt(OST422756)Lex/Mmucd mouse litters on a C57BL/6J × 129s/SvEv mixed background were purchased from the Mutant Mouse Regional Resource Center (#011750, MMRRC, UC Davis, CA, USA). All mice and rats were maintained on a normal 12-hour light/dark cycle and handled in accordance with animal care and use guidelines of the University of Wisconsin, Madison. Animals were maintained on a 4% fat diet (Harkland Teklad, Madison, WI, USA) with food and water available ad libitum. Two of two month old male rats, two S1R wildtype and two S1R knockout mice were used for histological evaluation with confocal and immunoelectron microscopy. For confocal microscopy a minimum of four sections were stained with each antibody. For immunoelectron microscopy a minimum of dozen ultrathin sections were examined per animal.
Intrathecal Injections
Mice were deeply anesthetized by isoflurane and 10 μl of AAV2/8-eGFP (5.6×1013 vg/ml) (University of Iowa Viral Vector Core, Iowa City, IA, USA) were delivered to mice between L4 and L5 spinal segments intrathecally using a 30-gauge needle. The injection was administered by gripping gently the rodent iliac crest and inserting the needle at a 45° angle in the central point between hip bones. For analysis of GFP expression in the spinal cord and DRG, animals were euthanatized and intracardially perfused with 4% paraformaldehyde (PFA) in 0.1M phosphate buffer (PB) 4 weeks after injection.
Intraplantar and Tongue injections
For intraplantar injections, under 2% isoflurane anesthesia, a Hamilton syringe was inserted into the plantar surface of the paw and 1 μl of 1 μg/μl of Cy3 fluorescently-conjugated receptor-binding domain of tetanus toxin was injected. The Cy3-TBD was prepared in a stapling reaction essentially as described (Darios et al., 2010) by mixing SNAP25-Cy3, synaptobrevin-TBD and a syntaxin SNARE helix peptide at equimolar ratios in 100 mM NaCl, 20 mM HEPES, 0.4% n-octylglucoside, pH 7.4. SNAP25-Cy3 was prepared by conjugating Cy3-NHS ester to the free cysteines of recombinant rat SNAP25. For injection into the tongue, mice were anesthetized with pentobarbital (30 mg/kg of body weight) and 1 μl of the same reagent was injected. Animals were placed in individual cages and 12 hours later were intra-cardially perfused with 4% PFA, tissue dissected, post-fixed for 4 hours and processed for histology.
Western Blot
For liver and lung, tissues from the periphery of the organ devoid of large blood vessels or airway were harvested. Cortical tissue was from the frontal cortex. Approximately 30 mg of tissue isolated from all organs were disrupted in RIPA buffer using a pestle in an Eppendorf tube, followed by further homogenization using 20 G needle and syringe. After sonication, the tissue lysates were incubated on ice for 30 minutes to extract the protein, clarified by centrifugation, and 20 μg of total protein homogenate quantified by BCA assay were loaded per lane onto a 10% acrylamide gel. The gel was run under constant 150 volts for 1 hour and dry transferred onto a nitrocellulose membrane. Membranes were blocked with 5% milk/TBST for 1 hour, and incubated overnight with primary antibodies at 1: 1000 mouse anti-S1R (#SC-166392, Santa Cruz Biotechnology, Dallas, TX, USA), 1: 5,000 mouse anti-β-actin (#A1978, Sigma-Aldrich, St. Louis, MO, USA), 1: 10,000 mouse anti-GAPDH (#CB1001, EMD Millipore, Billerica, MA, USA) in 1% milk/TBST solution. Membranes were washed and incubated with 1: 5,000 of goat anti-mouse HRP (#31430, Thermo Fisher Scientific, Waltham, MA, USA) for 1 hour at room temperature before imaging.
qRT-PCR
Total RNA from non-fixed tissues after cold saline perfusion was extracted using QI shredder and RNeasy Kits (Qiagen, Hilden, Germany) following the manufacturer's instructions. The concentration of extracted RNA in samples was determined using UV absorption with a NanoDrop2000 spectrophotometer (Thermo Fisher Scientific). cDNA was prepared by using a two-step qRT-PCR assay, in which a first strand cDNA was synthesized from 300 ng of total RNA using iScript first strand synthesis kit (Bio-Rad, Hercules, CA, USA) containing both random hexamer and oligo d(T) primers following the manufacturer's instructions. The prepared cDNA was then diluted to ½ its original concentration, and 1 μl of cDNA was used for qRT-PCR using EvaGreen qRT Master mix kit (Biotium, Hayward, CA, USA). Validated PCR primer pairs (Qiagen) for S1R and 18s rRNA were used for the PCR reactions.
Anti-S1R antibody
An affinity-purified rabbit anti-S1R antibody provided by Dr. Arnold Ruoho (University of Wisconsin) was used. Briefly, a maltose-binding protein S1R fusion protein expressed and purified from E.coli was sent to a commercial vendor (Covance, Denver, CO, USA) for the preparation of crude polyclonal antibody. The antibody was further purified on an affinity column as described (Ramachandran et al., 2007).
Immunohistochemistry and confocal microscopy
General anesthesia was induced by delivery of 3-4% isoflurane in oxygen at 3 L/ min, followed by a pentobarbital/phenytoin injection. Once deeply anesthetized, animals were intra-cardially perfused with saline followed by 4% PFA in 0.1 M PB (pH 7.4). Bilateral DRGs from L3-L6 as well as the corresponding spinal cord segment were identified by lumbar enlargement, and by tracing the nerve roots that correspond to the segmental level of interest, and dissected in situ after exposure of cord and DRG by laminectomy. Harvested tissues were post-fixed in 4% paraformaldehyde for 7 h, and then cryo-protected by soaking in phosphate-buffered 30% sucrose solution overnight at 4 °C. Cryo-sections of 10 μm each were cut from DRGs frozen in the optimum cutting temperature (O.C.T.) embedding medium (# 4583, Sakura Finetek USA, Inc., Torrance, CA, USA). Sections on slides were permeabilized with 1% Triton X-100 in PBS for 20 min, blocked with 10% normal goat serum for 2 h at room temperature, and then incubated with primary antibodies overnight at 4 °C. The following primary antibodies at the indicated dilution were used: rabbit anti-Sigma-1 receptor, 1:100 (Ramachandran et al., 2007), chicken anti-GFP, 1:1000 (#GFP-1020, Aves, Tigard, OR, USA), mouse anti-NeuN, 1:200 (#MAB377, Sigma-Aldrich), mouse anti-GFAP, 1:50 (#Ab4648, AbCam, Cambridge, UK), mouse anti-NSE, 1:200 (#05-291, Upstate Biotechnology, Lake Placid, NY, USA), mouse anti-NF200, 1:400 (#N0142, Sigma-Aldrich).
After rinsing the sections 3×, fluorescently conjugated secondary antibodies, 1: 1,000 (Thermo Fisher) were applied at room temperature for 2 h. Sections were then rinsed 3×, counterstained with 4’,6-diamidino-2-phenylindole (DAPI) for 5 min, and then mounted in the Prolong Diamond mounting medium (Thermo Fisher) and cover-slipped. The slides were left in the dark overnight and sealed using clear nail polish (Electron Microscopy Sciences, Hatfield, PA, USA). Images were taken with a Nikon A1R laser confocal microscope (Nikon, Tokyo, Japan), supplied with a green 488 nm Argon laser and a red 561 nm DPSS laser through an Apo60X VC oil-immersion objective with NIS elements software. Z-stacks were collected at 0.5 μm each. Final images were processed using the ImageJ program.
Immuno-electron microscopy
Immuno-EM experiments were performed following our previously published methods. Both mice and rats were intra-cardially perfused with 4% PFA and 0.2% glutaraldehyde in 0.1 M PB. DRGs were dissected by laminectomy and post-fixed in the same fixative overnight. DRGs were embedded into 3% agarose and 100 μm thick sections were cut using a Leica VT 100S vibratome. Sections were quenched in 1% sodium borohydrate for 30 min, rinsed with PBS and permeabilized in 0.05% Triton X-100 for 15 min, and then blocked in normal goat serum for 1 h. Sections were incubated with primary anti-S1R antibody (1:100 in PBS) for 48 h at 4 °C. Immunostaining was further revealed with ABC peroxidase kit (Vector Laboratories, Burlingame, CA, USA) and a mixture of 0.02% diaminobenzidine and 0.01% H2O2 in 50 mM Tris, pH 7.6 for 10 min. The sections were then rinsed and post-fixed with 2% glutaraldehyde for 30 min, followed by washing 3× in 100 mM Tris-maleic acid. Electron-dense polymer of diaminobenzidine was further intensified by a mixture of 2.6% hexamethyltetramine, 0.2% silver nitrate, and 0.2% sodium tetraborate all in 100 mM Tris-maleic acid buffer pH 7.4 for 10 min at 60 °C in the dark. The sections were rinsed in nanopure H2O and 0.01 M PBS and placed in 0.05% solution of gold chloride for 5 min. To wash away unbound silver particles, samples were first treated with 3% sodium thiosulfate for 2 min, and then washed 3× in water. The samples were post-fixed with 1% osmium tetroxide/1% potassium ferrocyanide for 1 h, rinsed and stained en bloc with 1% uranyl acetate, followed by dehydration in graded series of ethanol and washed twice with propylene oxide (5 min each time). Samples were further infiltrated in Epon resin/propylene oxide (1:1 ratio), and then in pure Epon, and finally polymerized between two glass slides. Thin sections of 70 nm were cut using a Leica EM UC7 ultra-microtome, counterstained in 8% uranyl acetate, and viewed and imaged with Phillips CM120 STEM electron microscope.
Results
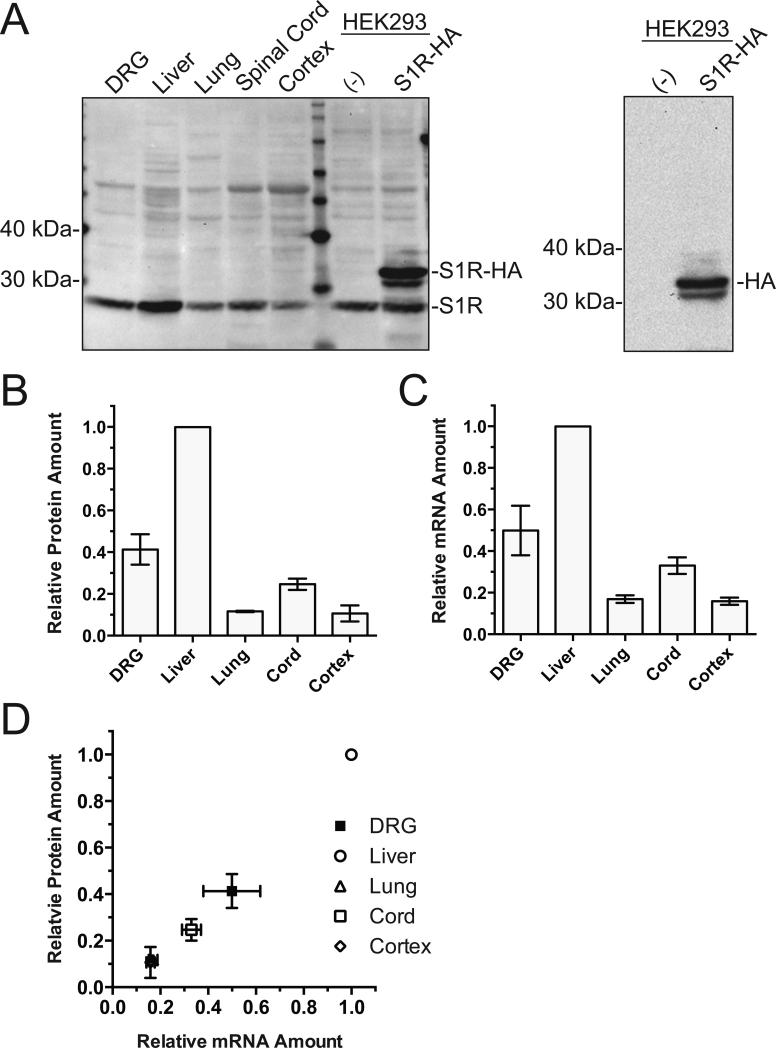
We first compared the level of S1R expression in different organs from the rat. Western blot and quantitative RT-PCR (qRT-PCR) assessment of S1R protein and mRNA levels from different organs demonstrated high expression of the protein and mRNA in the DRG (Figure 1A-C). The protein loading for the Western blot was normalized by BCA quantitation of the tissue lysate since the conventional endogenous house-hold protein normalizers such as GAPDH and β-actin are quite variable between different organs (Ferguson et al., 2005; Dittmer and Dittmer, 2006; Data not shown). The qRT-PCR was normalized by 18s ribosomal RNA (Suzuki et al., 2000; Kuchipudi et al., 2012). The S1R and 18s ribosomal RNA qRT-PCR assays were confirmed to be within the linear range of amplification. Of the tissues examined, the order of abundance of S1R protein was liver > DRG > spinal cord > lung ~ brain cortex in general agreement with a prior report (Hayashi and Su 2007). A strong correlation between mRNA and protein levels in different organs (Figure 1D) suggested that differential S1R expression in different organs is generally under transcriptional control.
Figure 1.
Examination of S1R protein and mRNA expression in different rat organs. A. Western blot of S1R protein expression by tissue types loaded with 20 μg of total protein per lane. Our custom S1R antibody is not compatible with Western blot; therefore, a commercial mouse anti-S1R antibody (Santa Cruz, #SC-166392) was used. HEK cell lysate non-transfected or transfected with rat-S1R-HA shows both endogenous and overexpressed S1R. Reblot of the same HEK cell membrane with anti-HA antibody (right panel) confirms the identity of the immunoreactive band as rat-S1R-HA. Some non-specific bands as well as a band with a molecular mass consistent with S1R dimer were detected on the Western blots with this antibody despite the reducing condition employed which should eliminate non-covalent dimer formation. B. Densitometry quantification of Western blots showing S1R protein expression in various rat tissues (n=3). The densitometric value from different organs were normalized by the value for liver. C. qRT-PCR quantification of S1R mRNA indicated as fold-difference calculated from ΔΔCT by tissue type normalized to liver (n=3). 18s ribosomal RNA served as the internal control. D. Relative tissue S1R protein expression quantified by western blot densitometry is plotted against mRNA of S1R in corresponding tissues. One-way ANOVA indicated significant difference (P<0.0001 for both) in protein and mRNA expression between tissues. The correlation coefficient r2=0.97 (P<0.0001) for protein vs. mRNA abundance indicated significant correlation rejecting the null hypothesis of no correlation. All values are mean ± S.E.M.
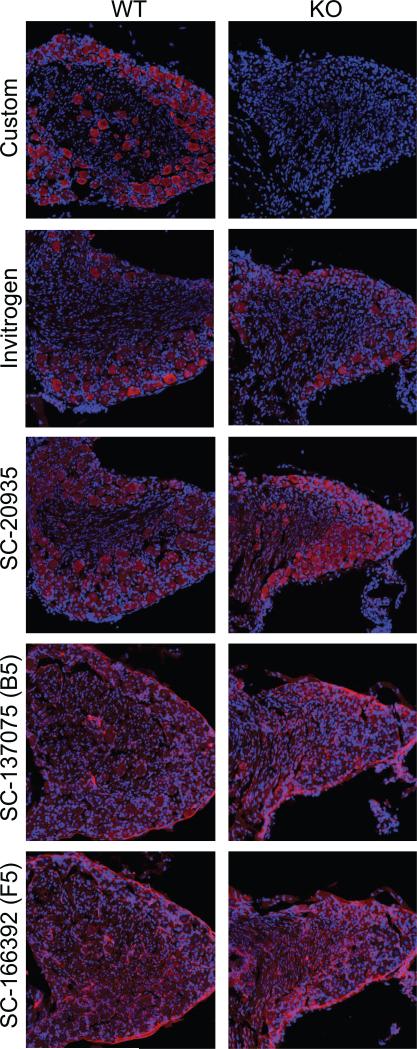
Preliminary to our study of the cellular and subcellular S1R protein expression in the DRG, we sought to validate the specificity of our anti-S1R antibody by immunostaining DRG from S1R wildtype and knockout mice. With our antibody, immunostaining of wildtype mice DRG showed a robust and specific immunoreactive (IR) signal while no such signal was observed in the DRG from S1R knockout mice. We performed the same analysis using four commercially available anti-S1R antibodies and noted that none of these antibodies showed the specificity of our custom antibody documented by the complete lack of non-specific immunoreactive signals in the S1R knockout DRG (Figure 2).
Figure 2.
Determination of the specificity of 5 different anti-S1R antibodies (antibodies from Santa Cruz Biotechnology #SC-20935, #SC-137075, #SC-166392, and from Thermo-Fisher/Invitrogen #42-3300) using S1R KO mouse DRG tissue as a negative control. S1R-IR (red) and DAPI (blue). DRG tissue sections obtained from wild type or S1R KO mice were immuno-stained as described in Methods. Scale bar = 50 μm.
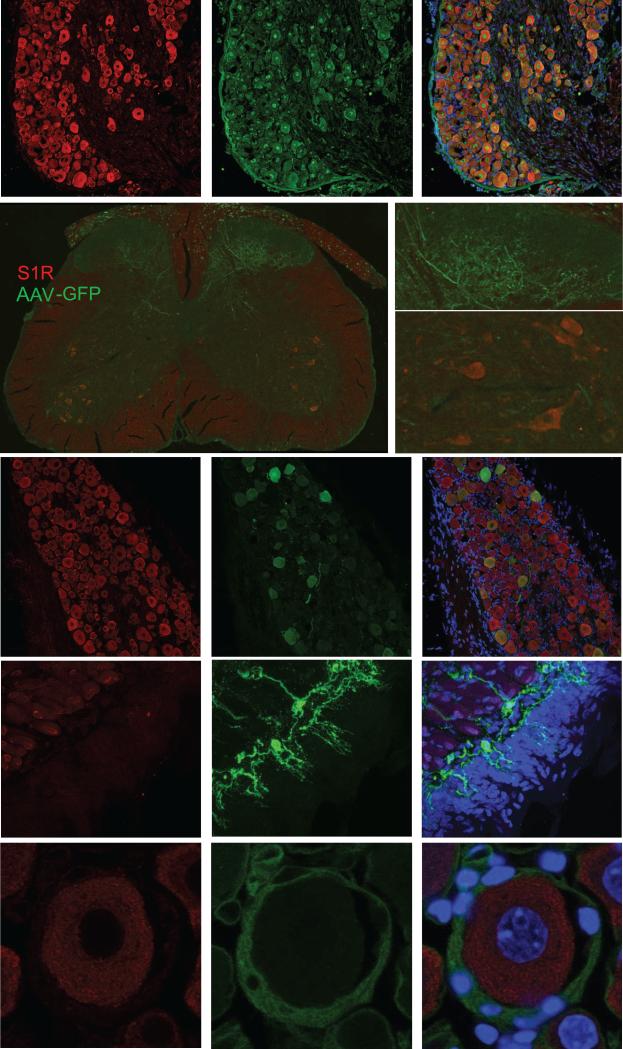
Confocal microscopy localized the intense S1R-IR to the cell bodies of neurons in the DRGs. The S1R-IR overlapped with the NeuN-IR, which is a pan-neuronal marker indicating broad expression of S1R in all neuronal DRG cells (Figure 3A). Neuron-Specific Enolase (NSE) marking all neurons co-localized with S1R-IR, and neurofilament 200 (NF200) marking large diameter DRG neurons, were largely positive for the S1R as well (Data not shown).
Figure 3.
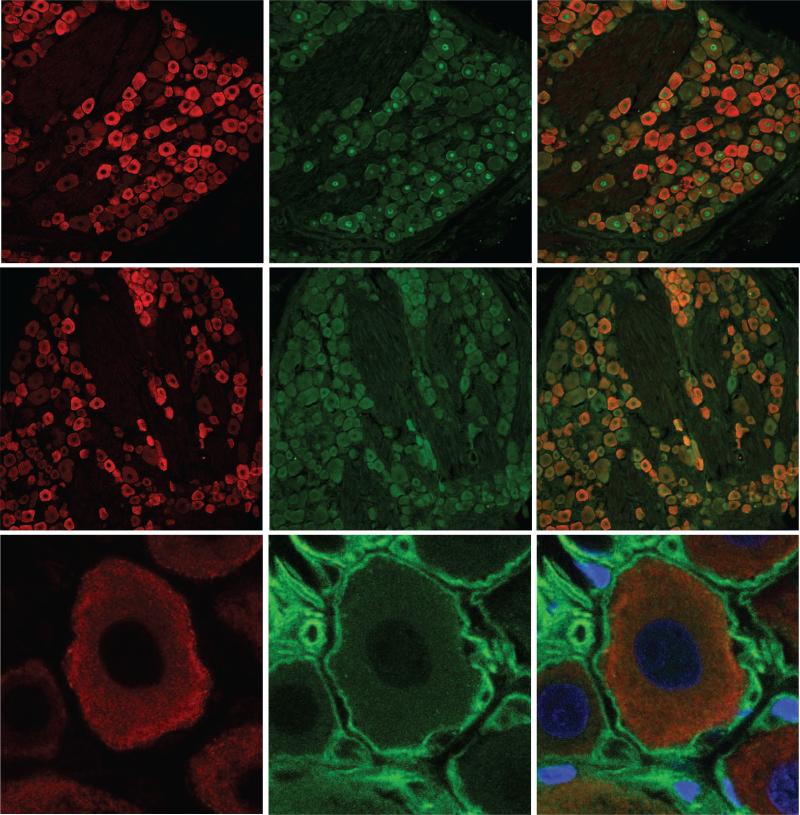
S1R is abundant in somas of mouse DRG neurons, but not detected in the corresponding neuronal processes and satellite cells. A. S1R (red) and neuronal marker NeuN (green) detection in mice DRG sections. All NeuN positive cells are also positive for S1R indicated by the overlap image. B. Tissue from a mouse 4 weeks after intrathecal injection of AAV-eGFP showed selective eGFP (green) expression in processes from the DRG neurons projecting into the dorsal horn of the spinal cord. The enlarged view of the spinal cord shows no localization of S1R (red) with the fluorescent processes in the dorsal horn. In contrast, the lower enlarged panel of this figure shows that the ventral horn motoneurons exhibit high amounts of S1R. C. Cell bodies of AAV-eGFP injected DRG neurons also co-localize with positive stain for S1R (arrows). In contrast, green axons of DRG neurons appeared to be negative for S1R (arrowheads). D. In the mouse tongue, S1R is not detected in sensory processes labeled by fluorescent Tetanus Toxin Binding domain (TBD, green). Neuronal processes indicated by the arrows appear to be negative for S1R. E. Satellite cells labeled with GFAP (green) do not express detectable amounts of S1R. Scale bar: A = 50 μm; B = 200 μm; C = 50 μm; D = 10 μm; E = 10 μm.
To determine whether the neuronal processes also expressed the S1R, we selectively labeled the DRG soma and processes by transducing the DRG neurons with adeno-associated virus (AAV) expressing the diffusible eGFP reporter (AAV-eGFP). Four weeks after intrathecal administration of AAV-eGFP robust expression of the eGFP reporter could be seen in the DRG cell bodies and their processes extending into the dorsal horn of the spinal cord as initially reported by (Storek et al., 2008). S1R-IR was not detected in eGFP-positive neuronal processes in the dorsal horn of the spinal cord (Figure 3B). However, consistent with our previous observations, S1R-IR was detected in motoneurons in the ventral horn (Figure 3B). S1R-IR was also not found in neuronal processes within the DRG, but was co-localized only with the soma of eGFP positive neurons located in the ganglion (Figure 3C). To check for the expression of S1R in sensory terminals, we injected fluorescently conjugated receptor-binding domain of the tetanus toxin in the tongue (Figure 3D) or under the skin (Data not shown) since this toxin selectively labels neuronal processes (Ferrari et al., 2013). These data indicate that S1R expression is enriched or restricted to the soma of DRG neurons, but not in their peripheral sensory ending or in the afferent axons.
Soma of DRG neurons are known to be encapsulated by specialized glial satellite cells. They secrete a variety of supportive factors for the maintenance of DRG neurons and also provide electrical insulation of DRG soma, preventing the spread of electrical impulses from one cell to another. Since specialized glial satellite cells are important cellular component of DRG, we examined whether S1R is expressed in the DRG satellite cells. Satellite cells are selectively detected by the anti-GFAP antibody, and a clear ring of cells non-overlapping with the neuronal marker can be identified. A careful examination of DRG sections co-stained with anti-S1R and anti-GFAP antibodies failed to detect S1R-IR in satellite cells in mice (Figure 3E). Similar to mice, rat tissue showed a consistent staining pattern with an abundant amount of S1R expression in NeuN and NSE-positive DRG neurons while mainly lacking signal in GFAP-positive satellite cells (Figure 4), in contrast to an earlier report where a different antibody against S1R was used (Bangaru et al., 2013).
Figure 4.
S1R is abundant in somas of all types of rat DRG neurons, but not in satellite cells. A. S1R (red), neuronal marker NeuN (green), and merged image of a rat DRG section. All NeuN positive cell nuclei co-localize with S1R. Similar to mouse DRG, all neurons in the rat DRG express S1R. However, the intensity of staining for S1R is more variable in rat DRG neurons. Arrows point to neurons with greater staining for S1R, while arrowheads point to neurons with relatively lower staining for S1R. B. S1R (red) and Neuron Specific Enolase (NSE, green). All NSE labeled neurons co-localize with S1R staining. C. Satellite cells labeled with GFAP (green) do not express detectable amounts of S1R. Scale bar: A and B = 50 μm, C = 10 μm.
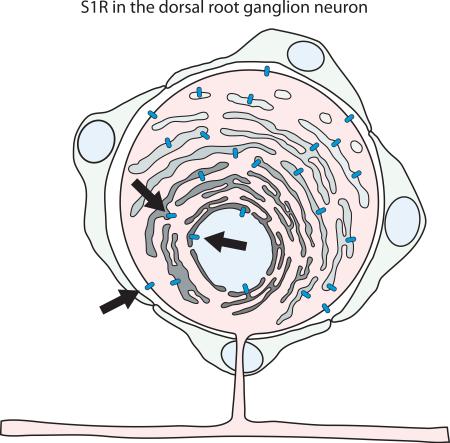
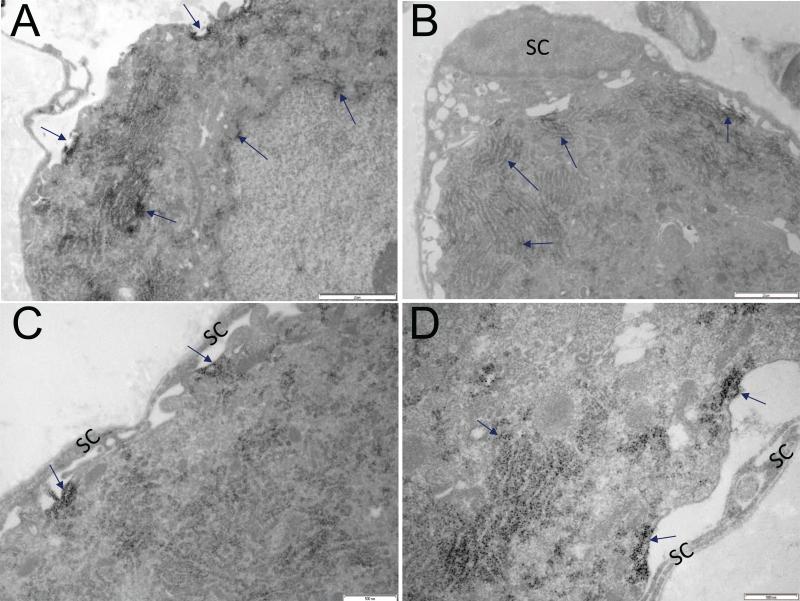
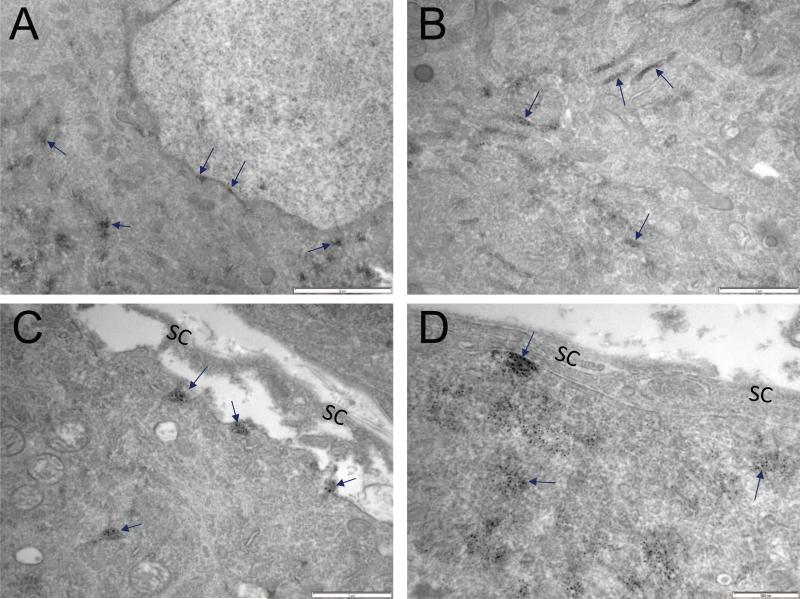
The resolution limit of confocal microscopy is insufficient to determine the precise subcellular localization of S1R, and so we extended the study using immuno-electron microscopy. Similar to results obtained with confocal microscopy, immuno-electron microscopy did not show S1R in satellite cells. In DRG neurons, S1R was detected in the nuclear envelope, endoplasmic reticulum, and the plasma membrane. This pattern of S1R subcellular localization was similar in both rat and mouse DRG (Figures 5 & 6).
Figure 5.
Subcellular distribution of S1R in mouse DRGs A. Arrows point to S1R detected in plasma membrane, endoplasmic reticulum and nuclear envelope. Dark precipitate indicates the presence of S1R. B-D. S1R is not detected in satellite cells (SC) and their processes. Arrows point to S1R localization to the plasma membrane and endoplasmic reticulum. Scale bar: A and B = 2 μm; C and D = 500 nm.
Figure 6.
Subcellular distribution of S1R in rat DRG. A. Arrows point to S1R detected in nuclear envelope and endoplasmic reticulum. Dark precipitate indicates localization of S1R. B. Arrows point to S1R in the endoplasmic reticulum. C, D. Very little if any S1RIR is detected in satellite cells of rat DRGs. Arrows indicate S1R localization to the plasma membrane and endoplasmic reticulum of DRG somata. Scale bar: A = 2 μm; B = 1 μm; C = 1 μm; D = 500 nm.
Discussion
Our survey of rat organs confirmed a significant level of S1R protein and mRNA expression in the DRG. The abundance of S1R mRNA in the DRG demonstrated in the present study by qRT-PCR agrees with the intense in situ hybridization signal seen in the neonatal mouse DRG (Allen Institute Mouse Spinal Cord Gene Expression Atlas, http://mousespinal.brain-map.org/imageseries/show.html?id=100040415). Expression of S1R appeared to be generally regulated at the transcriptional level, since the amount of protein expression did correlate with the abundance of mRNA in the different organs examined.
The 5’-UTR of both mouse and human S1R genes contains consensus binding sequences for many transcription factors including activator protein-1 and -2, nuclear factor-kB, -1/L, -GMa, and -GMb, GATA-1 and Zeste just within 1kb upstream of the transcription start site (Seth et al., 1997; Prasad et al., 1998). Upregulation of the S1R transcript via the pERK/eIF2α-ATF4 pathway during ER stress has been reported (Mitsuda et al., 2011), and the selective serotonin uptake inhibitor fluvoxamine also upregulates the S1R transcript through direct activation of ATF4 without involving pERK (Omi et al., 2014). However, the transcriptional regulation of S1R in normal non-stressed or pharmacologically stimulated tissue has not been investigated extensively. S1R protein has a long half-life of about 72 hrs (Hayashi & Su, 2007) resulting in the control of protein abundance solely through regulation of protein synthesis difficult. Despite our data supporting transcriptional regulation of protein abundance between different organs additional mechanisms of regulation such as control of subcellular protein targeting and protein half-life are likely important as well.
Characterization of commercially available anti-S1R antibodies using DRG sections from wildtype and knockout mice showed that none of the four commercially available anti-S1R antibodies gave specific immunohistochemical staining under the condition tested. It is worrisome that the commercially available antibodies tested gave strong but non-specific IR signal, a result that casts doubt on the reliability of many immunohistochemical results in the literature performed using these antibodies. We characterized the cellular and subcellular expression of S1R in the DRG using a custom anti-S1R antibody with greater specificity. Examination of S1R expression with confocal light microscopy demonstrated robust IR in all DRG neurons, showing good overlap between S1R-IR and NeuN-IR or NSE-IR. The S1R-IR was restricted to the neuronal soma with no signal detected in the sensory endings or the afferent axons of the DRG neurons. Likewise, we did not see significant S1R-IR in satellite cells. Immuno-electron microscopy demonstrated subcellular localization of S1R-IR most abundantly in the endoplasmic reticulum, but with an unambiguous presence in the nuclear envelope and in the plasma membrane of the DRG soma. The overall pattern of S1R-IR was the same in mouse and rat.
This pattern of subcellular S1R expression is consistent with other cell types examined at the EM level (i.e. motoneurons and retinal ganglion cells) (Mavlyutov et al., 2015 a and b) except that S1R-IR is observed in the plasma membrane in the DRG soma. In the other cell types, the S1R-IR is found near the plasma membrane (i.e. subsurface cisternae), but not in the plasma membrane itself. DRG neurons, motor neurons, and retinal ganglion cells share morphological similarities in terms of the presence of a large soma with long neurites, but the DRG neuron is unique in that they are encapsulated by glial satellite cells. The glial satellite cells are thought to support the DRG neurons by functionally providing insulation against cell-to-cell spread of electrical activity and nutrition from growth factors and cytokines (Hanani, 2005; Costa and Neto, 2015). Whether the satellite cells additionally enable translocation of the S1R to the plasma membrane is unknown.
It is remarkable that DRGs express abundant S1R and that S1R knockout mice exhibit relief from pathological pain. While it is speculative at this point to imply that the S1R in the DRG is involved in pain signaling, primary sensory afferent neurons are the gateway for nociception and a possible anatomical location mediating the pain phenotype. What is the role of S1R expressed in the DRG in pathological pain? S1R expression in the DRG is restricted mostly to the neuronal soma, with insignificant amounts detected in the sensory ending or the afferent axons. Therefore, we can exclude the possibility that S1R can directly regulate excitability of the sensory axons through direct modulation of ion channels already in the plasma membrane. However, the S1R is considered to be an ER-resident chaperone protein (Hayashi and Su, 2007; Su et al., 2010) and like other chaperone proteins, it is likely to regulate protein folding, proteolysis, and subcellular protein trafficking (Hartl et al., 2011). Such chaperone function, as well as the regulation of inter-organelle calcium signaling and oxidative stress (Wilson and González-Billault, 2015), could indirectly alter the excitability of the sensory axon by the S1R even when its presence is restricted to the neuron soma.
It was recently shown that S1R is activated by cocaine and translocates from the ER into the nuclear envelope where it forms a complex with the inner nuclear membrane protein emerin (Tsai et al., 2015b). This complex can further recruit histone deacetylase 2 and transcription factor 3, processes that may regulate gene expression. The role of S1R as a transcriptional regulator is a developing concept. A large difference in the expression of many genes (Tsai et al., 2012) was demonstrated in cultured primary hippocampal neurons after knockdown of the S1R by siRNA. It is plausible that the S1R localized to the nuclear envelope could regulate the expression of other genes, which can directly or indirectly affect the electrical activity of DRG neurons and thus pain transduction. A systematic search for the differences in gene expression in the DRGs from S1R-wildtype and knockout mice might elucidate how S1R restricted to the neuronal soma could alter the pain phenotype.
Highlights.
- Sigma-1 receptor is abundant in DRG neuronal soma, but not in satellite cells.
- Sigma-1 receptor is absent in processes of DRG neurons.
- In DRG neurons, S1R targets endoplasmic reticulum, nuclear envelope, and plasma membrane.
Acknowledgements
Supported by NIH grant (R01 GM107054) to Dr. Jay Yang. We thank Dr. Yoshi Matsuoka for intrathecal injection of viral constructs into mice and Dr. Arnold Ruoho (U Wisconsin) for providing the custom antibody.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors have read and have abided by the statement of ethical standards for manuscripts submitted to Neuroscience. All authors listed above have approved the final article.
References
- Alonso G, Phan V, Guillemain I, Saunier M, Legrand A, Anoal M, Maurice T. Immunocytochemical localization of the sigma 1 receptor in the adult rat central nervous system. Neuroscience. 2000;97:155–170. doi: 10.1016/s0306-4522(00)00014-2. [DOI] [PubMed] [Google Scholar]
- Bangaru ML, Weihrauch D, Tang QB, Zoga V, Hogan Q, Wu H. Sigma-1 receptor expression in sensory neurons and the effect of painful peripheral nerve injury. Mol Pain. 2013;9:47. doi: 10.1186/1744-8069-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cendán CM, Pujalte JM, Portillo-Salido E, Montoliu L, Baeyens JM. Formalin-induced pain is reduced in sigma 1 receptor knockout mice. Eur J Pharmacol. 2005;511:73–74. doi: 10.1016/j.ejphar.2005.01.036. [DOI] [PubMed] [Google Scholar]
- Costa FAL, Neto FLM. Satellite glial cells in sensory ganglia: its role in pain. Brazilian J Anesthesiol English Ed. 2015;65:73–81. doi: 10.1016/j.bjan.2013.07.013. [DOI] [PubMed] [Google Scholar]
- Darios F, Niranjan D, Ferrari E, Zhang F, Soloviev M, Rummel A, Bigalke H, Suckling J, Ushkaryov Y, Naumenko N, Shakirzyanova A, Giniatullin R, Maywood E, Hastings M, Binz T, Davletov B. SNARE tagging allows stepwise assembly of a multimodular medicinal toxin. Proc Natl Acad Sci. 2010;107:18197–18201. doi: 10.1073/pnas.1007125107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MP. Sigma-1 receptors and animal studies centered on pain and analgesia. Expert Opin Drug Discov. 2015;0441:1–16. doi: 10.1517/17460441.2015.1051961. [DOI] [PubMed] [Google Scholar]
- Dittmer A, Dittmer J. β-Actin is not a reliable loading control in Western blot analysis. Electrophoresis. 2006;27:2844–2845. doi: 10.1002/elps.200500785. [DOI] [PubMed] [Google Scholar]
- Entrena JM, Cobos EJ, Nieto FR, Cendán CM, Gris G, Del Pozo E, Zamanillo D, Baeyens JM. Sigma-1 receptors are essential for capsaicin-induced mechanical hypersensitivity: Studies with selective sigma-1 ligands and sigma-1 knockout mice. Pain. 2009;143:252–261. doi: 10.1016/j.pain.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Ferguson RE, Carroll HP, Harris A, Maher ER, Selby PJ, Banks RE, Banks R. Housekeeping proteins: A preliminary study illustrating some limitations as useful references in protein expression studies. Proteomics. 2005;5:566–571. doi: 10.1002/pmic.200400941. [DOI] [PubMed] [Google Scholar]
- Ferrari E, Gu C, Niranjan D, Restani L, Rasetti-Escargueil C, Obara I, Geranton SM, Arsenault J, Goetze TA, Harper CB, Nguyen TH. Synthetic self-assembling clostridial chimera for modulation of sensory functions. Bioconjug Chem. 2013;24:1750–1759. doi: 10.1021/bc4003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanilla D, Johannessen M, Hajipour AR, Cozzi NV, Jackson MB, Ruoho AE. The hallucinogen N,N-dimethyltryptamine (DMT) is an endogenous sigma-1 receptor regulator. Science. 2009;323:934–937. doi: 10.1126/science.1166127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gris G, Cobos EJ, Zamanillo D, Portillo-Salido E. Sigma-1 receptor and inflammatory pain. Inflamm Res. 2015:377–381. doi: 10.1007/s00011-015-0819-8. [DOI] [PubMed] [Google Scholar]
- Hanani M. Satellite glial cells in sensory ganglia: From form to function. Brain Res Rev. 2005;48:457–476. doi: 10.1016/j.brainresrev.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Su TP. Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca2+ signaling and cell survival. Cell. 2007;131:596–610. doi: 10.1016/j.cell.2007.08.036. [DOI] [PubMed] [Google Scholar]
- Kim HW, Kwon YB, Roh DH, Yoon SY, Han HJ, Kim KW, Beitz AJ, Lee JH. Intrathecal treatment with sigma1 receptor antagonists reduces formalin-induced phosphorylation of NMDA receptor subunit 1 and the second phase of formalin test in mice. Br J Pharmacol. 2006;148:490–498. doi: 10.1038/sj.bjp.0706764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourrich S, Hayashi T, Chuang JY, Tsai SY, Su TP, Bonci A. Dynamic interaction between sigma-1 receptor and Kv1.2 shapes neuronal and behavioral responses to cocaine. Cell. 2013;152:236–247. doi: 10.1016/j.cell.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchipudi SV, Tellabati M, Nelli RK, White GA, Perez BB, Sebastian S, Slomka MJ, Brookes SM, Brown IH, Dunham SP, Chang KC. 18S rRNA is a reliable normalisation gene for real time PCR based on influenza virus infected cells. Virol J. 2012;9:230. doi: 10.1186/1743-422X-9-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin WR, Eades CG, Thompson JA, Huppler RE, Gilbert PE. The effects of morphine and nalorphine-like drugs in the nondependent and morphine-dependent chronic spinal dog. J Pharmacol Exp Ther. 1976;197:517–532. [PubMed] [Google Scholar]
- Mavlyutov TA, Guo LW, Epstein ML, Ruoho AE. Role of the Sigma-1 receptor in amyotrophic lateral sclerosis (ALS). J Pharmacol Sci. 2015a;127:10–16. doi: 10.1016/j.jphs.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavlyutov TA, Epstein M, Guo LW. Subcellular localization of the sigma-1 receptor in retinal neurons — an electron microscopy study. Sci Rep. 2015b;5:1–11. doi: 10.1038/srep10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavlyutov TA, Epstein ML, Andersen KA, Ziskind-Conhaim L, Ruoho AE. The sigma-1 receptor is enriched in postsynaptic sites of C-terminals in mouse motoneurons. An anatomical and behavioral study. Neuroscience. 2010;167:247–255. doi: 10.1016/j.neuroscience.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda T, Omi T, Tanimukai H, Sakagami Y, Tagami S, Okochi M, Kudo T, Takeda M. Sigma-1Rs are upregulated via PERK/eIF2ahpha/ATF4 pathway and execute protective function in ER stress. Biochem Biophys Res Commun. 2011;415:519–525. doi: 10.1016/j.bbrc.2011.10.113. [DOI] [PubMed] [Google Scholar]
- Moebius FF, Reiter RJ, Hanner M, Glossmann H. High affinity of sigma 1-binding sites for sterol isomerization inhibitors: evidence for a pharmacological relationship with the yeast sterol C8-C7 isomerase. Br J Pharmacol. 1997;121:1–6. doi: 10.1038/sj.bjp.0701079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omi T, Tanimukai H, Kanayama D, Sakagami Y, Tagami S, Okochi M, Morihara T, Sato M, Yanagida K, Kitasyoji A, Hara H, Imaizumi K, Maurice T, Chevallier N, Marchal S, Takeda M, Kudo T. Fluvoxamine alleviates ER stress via induction of Sigma-1 receptor. Cell Death Dis. 2014;5:e1332. doi: 10.1038/cddis.2014.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad PD, Li HW, Fei YJ, Ganapathy ME, Fujita T, Plumley LH, Yang-Feng TL, Leibach FH, Ganapathy V. Exon-intron structure, analysis of promoter region, and chromosomal localization of the human type 1 sigma receptor gene. J Neurochem. 1998;70:443–451. doi: 10.1046/j.1471-4159.1998.70020443.x. [DOI] [PubMed] [Google Scholar]
- Ramachandran S, Chu UB, Mavlyutov TA, Pal A, Pyne S, Ruoho AE. The sigma1 receptor interacts with N-alkyl amines and endogenous sphingolipids. Eur J Pharmacol. 2009;609:19–26. doi: 10.1016/j.ejphar.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Lu H, Prabhu U, Ruoho AE. Purification and characterization of the guinea pig sigma-1 receptor functionally expressed in Escherichia coli. Protein Expr Purif. 2007;51:283–292. doi: 10.1016/j.pep.2006.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapunar D, Kostic S, Banozic A, Puljak L. Dorsal root ganglion - A potential new therapeutic target for neuropathic pain. J Pain Res. 2012;5:31–38. doi: 10.2147/JPR.S26603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HR, Zheng S, Gurpinar E, Koehl A, Manglik A, Kruse AC. Crystal structure of the human S1 receptor. Nature. 2016;532:527–530. doi: 10.1038/nature17391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth P, Leibach FH, Ganapathy V. Cloning and structural analysis of the cDNA and the gene encoding the murine type 1 sigma receptor. Biochem Biophys Res Commun. 1997;241:535–540. doi: 10.1006/bbrc.1997.7840. [DOI] [PubMed] [Google Scholar]
- Storek B, Reinhardt M, Wang C, Janssen WGM, Harder NM, Banck MS, Morrison JH, Beutler AS. Sensory neuron targeting by self-complementary AAV8 via lumbar puncture for chronic pain. Proc Natl Acad Sci. 2008;105:1055–1060. doi: 10.1073/pnas.0708003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su TP, Hayashi T, Maurice T, Buch S, Ruoho AE. The sigma-1 receptor chaperone as an inter-organelle signaling modulator. Trends Pharmacol Sci. 2010;31:557–566. doi: 10.1016/j.tips.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su TP, Su TC, Nakamura Y, Tsai SY. The Sigma-1 Receptor as a Pluripotent Modulator in Living Systems. Trends Pharmacol Sci. 2016;37:262–278. doi: 10.1016/j.tips.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Higgins PJ, Crawford DR. Control selection for RNA quantitation. Biotechniques. 2000;29:332–337. doi: 10.2144/00292rv02. [DOI] [PubMed] [Google Scholar]
- Tsai SY, Pokrass MJ, Klauer NR, Nohara H, Su TP. Sigma-1 receptor regulates Tau phosphorylation and axon extension by shaping p35 turnover via myristic acid. Proc Natl Acad Sci. 2015a;112:6742–6747. doi: 10.1073/pnas.1422001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SY, Rothman RK, Su TP. Insights into the sigma-1 receptor chaperone's cellular functions: A microarray report. Synapse. 2012;66:42–51. doi: 10.1002/syn.20984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SY, Chuang JY, Tsai MS, Wang XF, Xi ZX, Hung JJ, Chang WC, Bonci A, Su TP. Sigma-1 receptor mediates cocaine-induced transcriptional regulation by recruiting chromatin-remodeling factors at the nuclear envelope. Proc Natl Acad Sci. 2015b;112:E6562–6570. doi: 10.1073/pnas.1518894112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C, González-Billault C. Regulation of cytoskeletal dynamics by redox signaling and oxidative stress: implications for neuronal development and trafficking. Front Cell Neurosci. 2015;9:381. doi: 10.3389/fncel.2015.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao HS, Huang QH, Zhang FX, Bao L, Lu YJ, Guo C, Yang L, Huang WJ, Fu G, Xu SH, Cheng XP, Yan Q, Zhu ZD, Zhang X, Chen Z, Han ZG, Zhang X. Identification of gene expression profile of dorsal root ganglion in the rat peripheral axotomy model of neuropathic pain. Proc Natl Acad Sci. 2002;99:8360–8365. doi: 10.1073/pnas.122231899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanillo D, Romero L, Merlos M, Vela JM. Sigma 1 receptor: A new therapeutic target for pain. Eur J Pharmacol. 2013;716:78–93. doi: 10.1016/j.ejphar.2013.01.068. [DOI] [PubMed] [Google Scholar]