Abstract
The genetic integrity of living organisms is constantly threatened by environmental and endogenous sources of DNA damaging agents that can induce a plethora of chemically modified DNA lesions. Unrepaired DNA lesions may elicit cytotoxic and mutagenic effects and contribute to the development of human diseases including cancer and neurodegeneration. Understanding the deleterious outcomes of DNA damage necessitates the investigation about the effects of DNA adducts on the efficiency and fidelity of DNA replication and transcription.
Conventional methods for measuring lesion-induced replicative or transcriptional alterations often require time-consuming colony screening and DNA sequencing procedures. Recently, a series of mass spectrometry (MS)-based strategies have been developed in our laboratory as an efficient platform for qualitative and quantitative analyses of the changes in genetic information induced by DNA adducts during DNA replication and transcription. During the past few years, our laboratory has used these MS-based methods for assessing the replicative or transcriptional blocking and miscoding properties of more than 30 distinct DNA adducts. When combined with genetic manipulation, these methods have also been successfully employed for revealing the roles of various DNA repair proteins or translesion synthesis DNA polymerases in modulating the adverse effects of DNA lesions on transcription or replication in mammalian and bacterial cells.
In this Account, we focus on the development of MS-based approaches for determining the effects of DNA adducts on DNA replication and transcription in vitro as well as in bacterial and mammalian cells. We also highlight their applications to lesion bypass, mutagenesis and repair studies of three representative types of DNA lesions, i.e., methylglyoxal-induced N2-(1-carboxyethyl)-2'-deoxyguanosine (N2-CEdG), oxidatively induced 8,5'-cyclopurine-2'-deoxynucleosides, and regioisomeric alkylated thymidine lesions. Specially, we discuss the similar and distinct effects of the minor-groove DNA lesions including N2-CEdG and O2-alkylated thymidine lesions as well as the major-groove O4-alkylated thymidine lesions on DNA replication and transcription machinery. The MS-based strategies described herein should be generally applicable for quantitative measurement of the biological consequences and repair of other DNA lesions in vitro and in cells.
Graphical abstract

Introduction
The genomes of living organisms are continuously challenged by external or internal sources that can induce a battery of DNA damage products. If left unrepaired, these DNA lesions may elicit many deleterious consequences including genome instability and mutations, which may eventually confer risks for cancer, neurodegeneration and other human diseases.1,2 Understanding the biological significance of DNA damage necessitates the investigation about how DNA lesions compromise the flow of genetic information during DNA replication and transcription.
Many in vitro and in vivo studies have been carried out for determining lesion-induced perturbations on replication or transcription by utilizing DNA templates containing a site-specifically incorporated and structurally defined DNA adduct. Traditionally, the assessments of the biological consequences and repair of DNA lesions often involve extensive colony selection and Sanger sequencing procedures.1,3-5 Essigmann and coworkers introduced an elegant lesion bypass and mutagenesis assay, where no phenotypic selection is required and the nature of the misincorporation events during replication of single-stranded lesion-containing M13 plasmids in Escherichia coli cells can be quantitatively determined by restriction endonuclease digestion and postlabeling analysis.3,6 In addition, liquid chromatography-tandem mass spectrometry (LC-MS/MS) was employed as an efficient approach for sequencing the primer extension products from the replication of various DNA lesions in vitro.7,8 Inspired by these methods, our laboratory developed an MS-based experimental system for determining the blocking and mutagenic potentials of DNA lesions during replication in E. coli cells.9 We also extended the application of MS-based method for assessing how structurally defined DNA lesions situated in double-stranded plasmids perturb DNA replication and transcription, and how they are repaired in mammalian cells.10,11 To date, our laboratory has successfully utilized these MS-based assays for assessing the effects of more than 30 structurally defined DNA adducts on the perturbation of the efficiency and fidelity of DNA replication and/or transcription in vitro and in vivo.8-29 Furthermore, we have employed these methods to define the respective roles of various translesion synthesis (TLS) DNA polymerases and DNA repair proteins in the replicative and transcriptional alterations induced by DNA lesions. 8-27,29
In this Account, we place emphasis on the development of MS-based strategies for assessing quantitatively the biological consequences of chemically modified nucleosides during replication and transcription. We also present examples of their applications to lesion mutagenesis, bypass and repair studies in vitro as well as in bacterial and mammalian cells, and highlight some of our major discoveries made in this line of research during the past few years.
MS-based in vitro replication assay
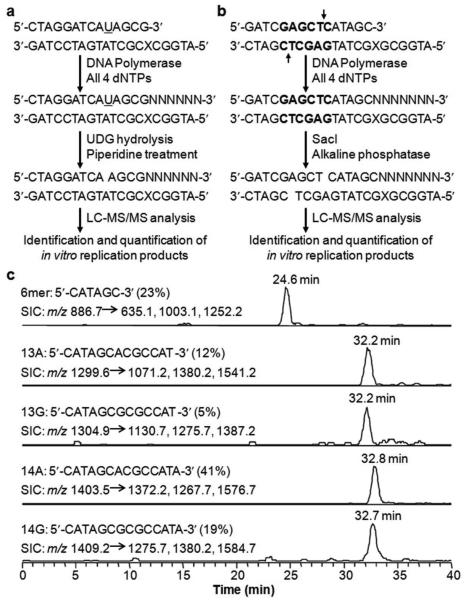
Guengerich and coworkers first reported the application of LC-MS/MS for analyzing in vitro primer extension products of 1,N2-ethenoguanine-bearing substrates.7 Central to this method is the use of a uracil-containing primer, where the removal of uracil in the extended replication products with uracil DNA glycosylase and cleavage of the resultant abasic site with hot piperidine allow for the generation of short oligodeoxyribonucleotides (ODNs) that are amenable to LC-MS/MS analysis (Figure 1a).7,8 Alternatively, specific restriction recognition sequence(s) can be introduced into the primer-template complex, and the in vitro replication products can be digested with restriction enzyme(s) to produce short ODN fragments for LC-MS/MS analysis (Figure 1b).25,26
Figure 1.
Experimental procedures for two MS-based in vitro replication assays (a, b), and representative selected-ion chromatograms revealing the distributions of SacI-treated primer extension products from human Pol η-mediated replication of O2-EtdT-bearing substrate in vitro (c) The uracil site is underlined, and the SacI site is highlighted in bold. ‘X’ indicates a lesion or unmodified base. Adapted from ref. 25. Copyright 2013 American Chemical Society.
The LC-MS/MS quantification of replication products was initially based on the relative abundances of ions of the corresponding ODNs observed in the mass spectra under the assumption that different ODNs have the same ionization efficiency.7 However, the hydrophobicity and free energy of desolvation can vary with the lengths and nucleobase compositions of the ODNs.8 To correct for the varied ionization efficiencies of different ODNs, an improved LC-MS/MS method was developed for more accurate identification and quantification of replication products, with the help of calibration curves constructed from synthetic ODNs representing the replication products of interest.8 This improved quantification method has been successfully used for in vitro replication studies of several DNA lesions.8,25,26 Representative selected-ion chromatograms revealing the distributions of replication products from human DNA polymerase η (Pol η)-mediated primer extension reaction for O2-ethylthymidine (O2-EtdT)-bearing substrate is shown in Figure 1c.25 The LC-MS/MS quantification results revealed that O2-EtdT is not a strong block to human Pol η in vitro, as manifested by the low amount (23%) of unextended primer (6-mer) in the total replication products. In addition, O2-EtdT can direct appreciable frequencies of dGMP misincorporation during Pol η-mediated replication in vitro.25
MS-based assay for replication studies in E. coli cells
The MS-based replication assay has also been employed for investigating the genotoxicity and mutagenicity of DNA lesions in E. coli cells, where LC-MS/MS is used for identifying, and sometimes quantifying, the progeny products emanating from the replication of lesion-bearing DNA templates.9,12-14,19,20,24 The experimental system begins with the ligation of lesion-bearing or lesion-free control ODNs into the EcoRI-linearized M13 genome with the help of two scaffolds flanking the lesion site (Figure 2a). The scaffolds and unligated linear plasmid DNA are degraded by the 3′✧5′ exonuclease activity of T4 DNA polymerase. The lesion-bearing or lesion-free control genomes are then premixed with a non-lesion competitor genome and co-transfected into E. coli cells that are proficient or deficient in specific repair proteins or TLS polymerases. The region of interest in the progeny genome is PCR amplified with a pair of primers spanning the site where the lesion was initially incorporated. The resultant PCR products are subsequently digested with appropriate restriction enzymes to generate short ODN fragments for LC-MS/MS analysis (Figure 2b).9,12,19
Figure 2.
A schematic diagram illustrating the procedures for the construction of lesion-bearing M13 genome (a) and an outline of MS-based replication assay in E. coli cells (b). The BbsI and MluCI sites are highlighted in bold. ‘X’ indicates a lesion or unmodified base.
Compared with the control genome, the competitor genome has three more nucleotides between the two restriction sites used for MS-based replication assay in E. coli cells (Figure 2b). This design facilitates the output from one biological experiment to be analyzed for determining quantitatively the degrees to which a structurally defined DNA lesion inhibits DNA replication and induces mutations during replication. The replicative bypass efficiency, which characterizes the extent to which a DNA lesion impedes DNA replication, is determined by normalizing the ratio of total lesion signal to the competitor signal against that obtained for the lesion-free control. The effect of a DNA lesion on replication fidelity is quantified by the signal intensity of mutant restriction fragment over the total signal intensity of all restriction fragments (mutant and unmutated) arising from the replication of lesion-bearing genome.9,12,19
The LC-MS/MS method has been successfully applied for the identification and quantification of restriction fragments arising from the replication of several DNA adducts, including a guanine-cytosine (G[8-5]C) intrastrand cross-link lesion in E. coli cells.9,12 More recently, polyacrylamide gel electrophoresis (PAGE) has also been incorporated into the workflow of MS-based in vivo replication assay.12-14,19,20,24 As an example, it was shown that the restriction fragments with a single nucleotide difference arising from replication of O2-EtdT-containing plasmid in E. coli cells can be readily resolved from each other with 30% native PAGE analysis.19 In this context, PAGE analysis can be directly used for quantifying the restriction fragments of interest, while LC-MS and MS/MS are employed for the identifications of the restriction fragments. As displayed in Figure 3, the identities of A→C, A→G and A→T mutations opposite the O2-EtdT site were supported by LC-MS and MS/MS results.19
Figure 3.
LC-MS and MS/MS for monitoring the identities of restriction fragments arising from the replication of O2-EtdT-bearing plasmid in E. coli cells. (a) High resolution “ultra zoom-scan” ESI-MS revealed the presence of the [M-3H]3− ions of 10-mer restriction fragment d(AATTATAGCN), with ‘N’ being A (wild-type) or C/G/T (A→C/G/T mutation). (b) A representative product-ion spectrum (MS/MS) of the [M-3H]3− ion (m/z 1002.2) of the mutant sequence d(AATTATAGCC). Shown above the spectrum is a scheme summarizing the observed [an − Base] and wn fragment ions, and nomenclature follows that described previously.44 Adapted with permission from ref. 19. Copyright 2014 Oxford University Press.
MS-based assays for replication studies in mammalian cells
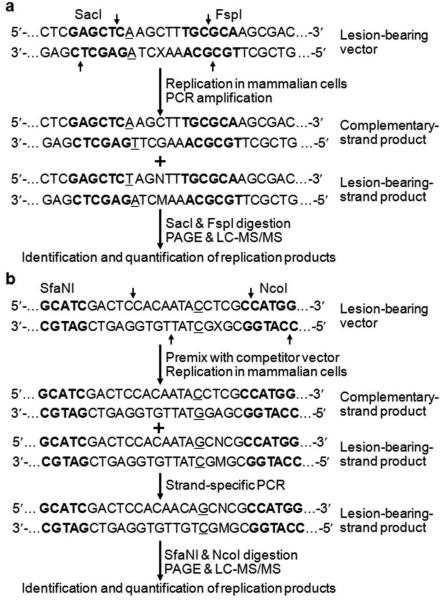
The MS-based assay has also been extended to assess the effects of DNA lesions on DNA replication in mammalian cells.10,15,27,28 As illustrated in Figure 4a, a lesion-bearing or lesion-free double-stranded plasmid is allowed to replicate in mammalian cells. The progenies of the plasmid are isolated from the host cells, and the residual unreplicated plasmid DNA is removed by DpnI digestion. The progeny genomes are then PCR amplified with a pair of primers spanning the initial lesion site and several DpnI recognition sites, and the resulting PCR products are restriction digested for PAGE and LC-MS/MS analyses to identify and quantify the ODN fragments of interest, as described above.15
Figure 4.
Experimental procedures for two MS-based replication assays in mammalian cells (a, b). The A/A and C/C mismatch sites are underlined, and the SacI, FspI, SfaNI and NcoI sites are highlighted in bold.
This method has been successfully employed for assessing the cytotoxic and mutagenic properties of several DNA adducts, including the methylglyoxal-induced N2-(1-carboxyethyl)-2'-deoxyguanosine (N2-CEdG) in mammalian cells.10,15,27,28 One challenging step in this experimental system is the construction of double-stranded vectors containing site-specifically inserted DNA lesions. However, the use of gapped vector-based strategy enables the facile preparation of site-specifically modified plasmids.10,15 For example, Yuan et al.15 used this strategy to routinely generate the N2-CEdG-bearing vectors at an overall yield of 30%, with the help of the nicking enzyme Nt.BstNBI that has two tandem recognition sites in the parent pTGFP-Hha10 vector. A complementary experimental strategy, based on the extension of a synthetic lesion-bearing primer on a circular single-stranded phage DNA, can be used for the generation of lesion-containing double-stranded vectors if the parent vectors do not carry appropriate tandem nicking enzyme recognition sites.30 Since the lesion-bearing strand is sometimes replicated less efficiently than its complementary undamaged strand in mammalian cells, the introduction of a mismatch near the lesion site in the site-specifically modified vectors facilitates the independent assessment of the products arising from the replication of the lesion-bearing and lesion-free complementary strands.10,15 In this respect, the complementary strand signal (i.e. a restriction fragment of PCR products arising from the replication of the complementary undamaged strand of lesion-bearing vector) can serve as an internal standard for determining the effects of the DNA lesion on replication efficiency in mammalian cells.15
A modified version of replication assay, i.e. strand-specific PCR-based competitive replication and adduct bypass (SSPCR-CRAB) method, has also been developed for lesion bypass and mutagenesis studies in mammalian cells (Figure 4b).17 There are three salient points for this modified assay: (i) lesion-bearing or lesion-free control plasmids are co-transfected with a non-lesion competitor vector at a given molar ratio into mammalian cells; (ii) the progeny genomes arising from the replication of the lesion-carrying strand, but not its complementary undamaged strand, is selectively amplified by PCR for subsequent analysis; (iii) the competitor signal is used as an internal standard for gauging the ability of DNA lesion to inhibit DNA replication in mammalian cells. This assay is particularly useful for those DNA lesions that strongly block DNA replication, under which circumstances the replication product(s) emanating from the lesion-situated strand is much less abundant than that from the opposite undamaged strand, rendering it difficult to determine reliably the bypass efficiencies and mutation frequencies. With the use of the SSPCR-CRAB assay and by adjusting the molar ratio of lesion-bearing over competitor templates, the restriction fragments formed from the replication of the template housing a strongly blocking lesion (e.g. 8,5'-cyclopurine-2'-deoxynucleoside) can be readily identified and quantified.17
MS-based assays for transcription studies in vitro and in vivo
The majority of cells in the body are quiescent or slowly replicating, but they need to maintain active transcription for normal physiological processes; thus, it is of great concern to determine how DNA adducts alter the flow of genetic information at the level of transcription.1 Building upon the aforementioned replication assays, our laboratory recently developed a competitive transcription and adduct bypass (CTAB) method for quantitative measurement of transcriptional inhibition and mutagenesis induced by DNA adducts in vitro and in mammalian cells (Figure 5).11,31 Briefly, lesion-containing or lesion-free control templates are co-transcribed with a lesion-free competitor template in vitro or in mammalian cells. The resulting runoff RNA products are reverse transcribed, PCR amplified and restriction digested to produce small ODN fragments for PAGE and LC-MS/MS analyses.11,31
Figure 5.
Outline of the CTAB assay. ‘X’ indicates a lesion or unmodified base , which is located on the transcribed strand of the TurboGFP gene downstream of the CMV and T7 promoters. The arrowheads indicate the +1 transcription start sites of CMV and T7 promoters. Adapted with permission from ref. 23. Copyright 2015 Oxford University Press.
A critical step in the CTAB method is the site-specific incorporation of DNA lesions into double-stranded vectors, which can be achieved using the aforementioned experimental strategies.15,31 As a general rule, all transcription templates used for the CTAB assay should be devoid of mammalian replication origins, which ensures that the observed changes in transcription are exclusively induced from encountering of DNA lesions with the machinery of transcription, but not replication.31 Up to now, we have used the CTAB assay for assessing the effects of 14 distinct DNA adducts on transcription mediated by single-subunit T7 RNA polymerase (RNAP) and multi-subunit mammalian RNAP II.11,16,21-23 The CTAB assay should also be amenable to investigating how DNA lesions perturb transcription mediated by RNAPs from other organisms, by placing DNA lesions downstream of specific promoter sequences of these RNAPs. Moreover, our method has been successfully used to examine the roles of specific repair proteins on the transcriptional alterations induced by DNA lesions in cells by manipulating their gene expression.11,16,21-23
Applications of MS-based replication and transcription assays
A number of discoveries have been made using the above-mentioned MS-based strategies for assessing the effects of DNA lesions on replication or transcription in vitro and in cells.8-28 In this section, we highlight their applications to replicative or transcriptional bypass, mutagenesis and repair studies of three representative types of DNA lesions (Figure 6). Specially, we discuss the similar and distinct effects of these DNA lesions on replication and transcription, and we compare and contrast the recognition of minor-groove DNA lesions formed on the N2 position of guanine and O2 position of thymine by DNA replication and transcription machinery.
Figure 6.
Chemical structures of representative DNA lesions discussed in this Account.
Alkylated thymidine lesions
Alkylation at the N3, O2, and O4 positions of thymidine in DNA is known to occur in mammalian cells and tissues and has been linked to cigarette smoking.32,33 We recently examined the cytotoxic and mutagenic properties of N3-, O2- or O4-EtdT during replication in E. coli cells, as well as their effects on DNA transcription mediated by T7 RNAP or human RNAP II in vitro and in human cells.19,22 The results showed that N3-EtdT and O2-EtdT, but not O4-EtdT, are strong blocks to DNA replication in E. coli cells (Figure 7a). Similarly, N3-EtdT and O2-EtdT substantially inhibit DNA transcription in vitro and in certain human cells. Additionally, O4-EtdT induces a high frequency of A→G mutation opposite the lesion site, whereas N3-EtdT and O2-EtdT evoke promiscuous nucleotide misincorporations during DNA replication or transcription (Figures 3 and 7b). These observations may be attributed to the unique chemical properties of the three regioisomeric ethylated thymidines. In this vein, alkylation of thymine at the O4 position may facilitate its preferential pairing with guanine, whereas the addition of an alkyl group to the N3 or O2 position of thymine may render the nucleobase unfavorable in pairing with any of the canonical nucleobases.19,22 It was also demonstrated that Pol V plays a major role in the error-prone bypass of all three regioisomeric EtdT lesions, though Pol IV is also involved in the misincorporation of dCMP opposite N3-EtdT and O2-EtdT.19 Furthermore, when located on the template DNA strand, N3-EtdT, but not O2-EtdT or O4-EtdT, was found to be an efficient substrate for transcription-coupled nucleotide excision repair (TC-NER) in human cells.22
Figure 7.
Replicative bypass efficiencies (a) and mutagenic potentials (b) of N3-, O2- and O4-EtdT in E. coli cells. Adapted with permission from ref. 19. Copyright 2014 Oxford University Press.
We have also systematically examined how the addition of various alkyl groups (from methyl to butyl, Figure 8a) to the O2 or O4 position of thymidine affects the efficiency and fidelity of DNA replication in E. coli cells.20,29 All O2-alkyldT lesions exhibit promiscuous miscoding properties, whereas the O4-alkyldT lesions only direct the misincorporation of guanine nucleotide opposite the lesion site, which may be attributed to their distinct chemical properties as discussed above. In addition, all O2-alkyldT lesions strongly inhibit DNA replication, with the replication bypass efficiency decreasing with the size of the alkyl group; however, all O4-alkyldT lesions, except for O4-sBudT, are not strong impediments to DNA replication in E. coli cells (Figure 8b, c). Moreover, the small O2-MedT is replicated less efficiently than the bulky O4-sBudT, suggesting that the minor-groove thymidine lesions pose a greater challenge for the E. coli DNA replication machinery than their major-groove counterparts.19,28
Figure 8.
Chemical structures of the examined alkyl groups (a), and replicative bypass efficiencies of O2- and O4-alkyldT lesions (b, c) in E. coli cells. Adapted with permission from refs. 20 and 29. Copyright 2014 and 2015 Oxford University Press.
Methylglyoxal-induced N2-CEdG
Methylglyoxal is produced endogenously as a byproduct of the ubiquitous glycolysis pathway and induces the formation of R- and S-diastereomers of the minor-groove N2-CEdG as the major stable DNA adducts.12,34 In contrast to the strong blocking effects and promiscuous miscoding properties of the minor-groove O2-alkyldT lesions, Yuan et al.12 found that the bypass efficiencies for the minor-groove R- and S-N2-CEdG were ~39% and 75%, respectively, and the two lesions could only induce very low (~0.5%) frequencies of G→T transition mutation in wild-type E. coli cells. While Pol V is the major polymerase for bypassing the O2-alkyldT lesions, Pol IV is the only SOS-induced polymerase involved in bypassing the two diastereomers of N2-CEdG in E. coli cells 12. Similarly, both R- and S-N2-CEdG are not strong blocks to mammalian DNA replication machinery (Figure 9a). In addition, depletion of Pol κ (a Pol IV homolog) or Pol ι can lead to elevated frequencies of G→T and G→A mutations (Figure 9c), suggesting that both Pol κ and Pol ι are required for the accurate bypass of N2-CEdG lesions in mammalian cells.15 The lack of strong perturbation on the efficiency or fidelity of DNA replication for the N2-CEdG lesions might be attributed to the fact that alkylation of the N2 position of guanine does not alter markedly the Watson-Crick base pairing property of guanine. In addition, structural and biochemical studies support that DinB family DNA polymerases (Pol IV in E. coli and Pol κ in mammals) are capable of accommodating readily the minor-groove N2-dG modifications into their active site.12,35 While the active site of DinB family polymerases might be able to accommodate the O2-alkyldT lesions, the inability of the modified thymine to form favorable base pairing with any of four canonical nucleobases may prevent productive nucleotide incorporation opposite the lesions. This may provide a structural rationale for the distinct recognition of the minor-groove lesions formed on O2 of thymine and N2 of guanine by the DNA replication machinery.
Figure 9.
Bypass efficiencies (a, b) and mutagenic potentials (c) of N2-CEdG lesions during replication and transcription in mammalian cells. The 293T cell line is a derivative of human embryonic kidney 293 cell line and contains the SV40 T-antigen. GM04429 and GM00637 are NER-deficient (lacking XPA) and repair-proficient human cell lines, respectively. N2-CEdG lesions are not mutagenic during replication in wild-type mammalian cells, but they can induce G→T and G→A mutations in mammalian cells that are deficient in Pol κ or Pol ι. Adapted with permission from refs. 11 and 15. Copyright 2012 Nature Publishing Group (Figure 9b). Figure 9a was adapted from the data originally published in J. Biol. Chem. Yuan, B. et al. The roles of DNA polymerases κ and ι in the error-free bypass of N2-carboxyalkyl-2′-deoxyguanosine lesions in mammalian cells. J. Biol. Chem. 2011; Vol 286: 17503-17511. © the American Society for Biochemistry and Molecular Biology.
In contrast to our results for the O2-EtdT, neither diastereomer of N2-CEdG lesions is capable of inducing detectable mutant transcripts in vitro or in mammalian cells (Figure 9c). On the other hand, both R- and S-N2-CEdG exhibit strong inhibitory effects on transcription and may trigger TC-NER in mammalian cells (Figure 9b), which parallels the findings made for the O2-EtdT lesion.11,15 These studies together suggest that RNA polymerases, unlike the DinB family translesion synthesis DNA polymerases, may not effectively tolerate minor-groove modifications.
Oxidatively induced 8,5'-cyclopurine-2'-deoxynucleosides
Reactive oxygen species can react with DNA to produce a wide spectrum of DNA damage products including 8,5'-cyclo-2'-deoxyadenosine (cdA) and 8,5'-cyclo-2'-deoxyguanosine (cdG).36 It was found that the (5'S) diastereomers of cdA and cdG are able to strongly block DNA replication and induce substantial frequencies of base substitutions at the lesion site in E. coli and mammalian cells (Figure 10a, c).17,37 Additionally, E. coli Pol V as well as mammalian Pol η, Pol ι and Pol ζ are involved in the efficient bypass of S-cdA and S-cdG in vivo.17,37 Analogous to their blocking effects on replication, S-cdA and S-cdG are found to be strong impediments to transcription mediated by T7 RNAP or human RNAP II in vitro and in human cells (Figure 10b).11 Different from replicative mutagenesis, transcriptional bypass of S-cdA and S-cdG does not lead to detectable misincorporation events directly opposite the lesion site. In contrast, S-cdA and S-cdG can induce a type of mutant transcript that contains a single-nucleotide deletion immediately downstream of the lesion site during T7 RNA polymerase-mediated transcription in vitro. In NER-deficient human cells, transcriptional bypass of S-cdA and S-cdG results in the misincorporation of an adenine opposite the next nucleotide downstream of the lesion site (Figure 10c).11 These findings again underscore the differential recognition of these lesions by the DNA replication and transcription machinery.
Figure 10.
Bypass efficiencies (a, b) and mutagenic potentials (c) of S-cdA and S-cdG during replication and transcription in mammalian cells. Adapted with permission from refs. 11 and 17. Copyright 2012 Nature Publishing Group (Figure 10b). Figure 10a was adapted from the data originally published in J. Biol. Chem. You, C. et al. Translesion synthesis of 8,5'-cyclopurine-2'-deoxynucleosides by DNA polymerases η, ι, and ζ. J. Biol. Chem. 2013; Vol 288: 28548-28556. © the American Society for Biochemistry and Molecular Biology.
Conclusions
In this Account, we have reviewed the development and application of MS-based strategies for assessing the biological consequences of DNA lesions during replication and transcription. These methods are quite effective and obviate the exhaustive colony selection and sequencing steps that most conventional methods entail. Because of the use of the entire population of progeny products for the analyses, these methods provide statistically reliable and quantitative data for gauging the bypass and miscoding properties of DNA lesions. In addition, the incorporation of LC-MS/MS into the workflow enables accurate identification of mutant products arising from replicative or transcriptional bypass of DNA lesions. In this respect, the LC-MS/MS approach can be used to interrogate nucleotide misincorporation or indel events at or near the original lesion site rather than examining only the mutation at the lesion site in the restriction endonuclease and postlabeling (REAP) assay 3,6,9,11,12,15,20. The MS-based methods are also quite sensitive and can be reliably used for determining a mutation frequency that is ~0.5%, with the standard deviation being ~0.1% that is comparable to the reproducibility of other methods such as the REAP assay 6,9,12,38. These MS-based methods have been successfully used in lesion bypass, mutagenesis, and repair studies of single and tandem nucleobase lesions, intrastrand cross-links, as well as epimeric 2-deoxyribose lesions.8-29 They should also be generally applicable for the quantitative measurement of the biological consequences and repair of other types of DNA lesions, such as interstrand cross-links and DNA-protein cross-links. However, it should be noted that these MS-based assays do not generally provide information regarding the kinetics for nucleotide insertion. In addition, these methods require the presence of appropriate restriction sites flanking the lesion, and thus they are not suitable for the study of DNA lesions located in some specific DNA sequence contexts that cannot be restriction digested to generate short ODNs for LC-MS/MS analysis.
Next-generation sequencing-based technologies have been employed for the high-throughput interrogation of the genotoxicity and mutagenicity of DNA lesions during replication or transcription;37-39 nevertheless, these assays are sometimes limited by the lack of advanced computational infrastructure and specialized expertise to handle massive amount of sequencing data.40 Additionally, pyrosequencing was recently applied for a rapid evaluation of the ability of a DNA lesion to induce moderate to high levels of base substitutions during replication; however, this method could not be reliably used for assessing frameshift mutations.41
Most existing lesion bypass and mutagenesis assays, including the MS-based methods described herein, require the use of single- or double-stranded vectors containing site-specific DNA adducts as episomes in the host cells.1,3,4,9,10,31 These vector-based model systems have provided a plethora of useful information for gauging the blocking and mutagenic potentials of DNA lesions in cells. However, lesion-induced changes in genetic information revealed by these vector-based studies may differ, to some extent, from their effects on replication or transcription when they are located on chromosomes in cells. Livneh and coworkers42 recently reported the use of a phase integrase to stably integrate a lesion-containing vector into chromosomal DNA for studying the mechanism of DNA damage tolerance in mammalian cells. It can be envisaged that, in combination with the integrase-based method, the MS-based strategies are amenable for examining how structurally defined DNA lesions compromise replication and transcription, as well as how these lesions are repaired in cells at the chromosomal level in the future.
Further development and applications of lesion bypass and mutagenesis assays will improve our understanding of the mechanisms of carcinogenesis and facilitate the development of new drugs in cancer chemotherapy. In this vein, while DNA damage-induced replicative mutagenesis has been widely considered as a major culprit in cancer development, transcriptional mutagenesis may also play an important role in tumor development, especially in quiescent or slowly replicating cells.1,2 It has been proposed that lesion-induced transcription infidelity in non-dividing cells may result in the generation of a substantial pool of mutant proteins allowing a cell-growth switch, which may lead to error-prone replication of the original damaged DNA and induce a heritable change through a process known as retromutagenesis.1 In addition, it was recently suggested that RNA transcripts could be used as templates for DNA double-strand break (DSB) repair in highly transcribed genomic loci in non-dividing cells.43 It is therefore possible that lesion-induced mutant transcripts may act as templates for DNA DSB repair and thus may ultimately cause a permanent alteration in the DNA sequence under certain conditions.
Acknowledgments
This work summarized in this Account has been supported in part by the National Institutes of Health (DK082779, ES019873, ES025121 and CA101864).
Biographies
Changjun You received his Ph.D. in Biochemistry and Molecular Biology from Huazhong Agricultural University under the supervision of Professors Qifa Zhang and Changyin Wu. He is currently conducting postdoctoral research with Professor Yinsheng Wang at the University of California-Riverside.
Yinsheng Wang obtained his Ph.D. in Chemistry from Washington University in St. Louis in 2001. He is currently a professor of Chemistry and the Director for the Environmental Toxicology Graduate Program at UC Riverside. Yinsheng’s research concentrates on the use of mass spectrometry, along with molecular biology and/or synthetic organic chemistry, for understanding the occurrence and biological consequences of DNA adducts and post-translational modifications of proteins. Yinsheng was the recipient for the inaugural Chemical Research in Toxicology Young Investigator Award in 2012 and the 2013 Biemann Medal.
Footnotes
Footnotes
The authors declare no competing financial interest.
References
- (1).Bregeon D, Doetsch PW. Transcriptional mutagenesis: causes and involvement in tumour development. Nat. Rev. Cancer. 2011;11:218–227. doi: 10.1038/nrc3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat. Rev. Cancer. 2011;11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Delaney JC, Essigmann JM. Biological properties of single chemical-DNA adducts: a twenty year perspective. Chem. Res. Toxicol. 2008;21:232–252. doi: 10.1021/tx700292a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ziv O, Diamant N, Shachar S, Hendel A, Livneh Z. Quantitative measurement of translesion DNA synthesis in mammalian cells. Methods Mol. Biol. 2012;920:529–542. doi: 10.1007/978-1-61779-998-3_35. [DOI] [PubMed] [Google Scholar]
- (5).Livingston AL, O'Shea VL, Kim T, Kool ET, David SS. Unnatural substrates reveal the importance of 8-oxoguanine for in vivo mismatch repair by MutY. Nat. Chem. Biol. 2008;4:51–58. doi: 10.1038/nchembio.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Delaney JC, Smeester L, Wong C, Frick LE, Taghizadeh K, Wishnok JS, Drennan CL, Samson LD, Essigmann JM. AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo. Nat. Struct Mol Biol. 2005;12:855–860. doi: 10.1038/nsmb996. [DOI] [PubMed] [Google Scholar]
- (7).Zang H, Goodenough AK, Choi JY, Irimia A, Loukachevitch LV, Kozekov ID, Angel KC, Rizzo CJ, Egli M, Guengerich FP. DNA adduct bypass polymerization by Sulfolobus solfataricus DNA polymerase Dpo4: analysis and crystal structures of multiple base pair substitution and frameshift products with the adduct 1,N2-ethenoguanine. J. Biol. Chem. 2005;280:29750–29764. doi: 10.1074/jbc.M504756200. [DOI] [PubMed] [Google Scholar]
- (8).Gu C, Wang Y. In vitro replication and thermodynamic studies of methylation and oxidation modifications of 6-thioguanine. Nucleic Acids Res. 2007;35:3693–3704. doi: 10.1093/nar/gkm247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Hong H, Cao H, Wang Y. Formation and genotoxicity of a guanine-cytosine intrastrand cross-link lesion in vivo. Nucleic Acids Res. 2007;35:7118–7127. doi: 10.1093/nar/gkm851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Yuan B, O'Connor TR, Wang Y. 6-Thioguanine and S6-methylthioguanine are mutagenic in human cells. ACS Chem. Biol. 2010;5:1021–1027. doi: 10.1021/cb100214b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).You C, Dai X, Yuan B, Wang J, Brooks PJ, Niedernhofer LJ, Wang Y. A quantitative assay for assessing the effects of DNA lesions on transcription. Nat. Chem. Biol. 2012;8:817–822. doi: 10.1038/nchembio.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Yuan B, Cao H, Jiang Y, Hong H, Wang Y. Efficient and accurate bypass of N2-(1-carboxyethyl)-2'-deoxyguanosine by DinB DNA polymerase in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 2008;105:8679–8684. doi: 10.1073/pnas.0711546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Yuan B, Wang Y. Mutagenic and cytotoxic properties of 6-thioguanine, S6-methylthioguanine, and guanine-S6-sulfonic acid. J. Biol. Chem. 2008;283:23665–23670. doi: 10.1074/jbc.M804047200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Yuan B, Jiang Y, Wang Y. Efficient formation of the tandem thymine glycol/8-oxo-7,8-dihydroguanine lesion in isolated DNA and the mutagenic and cytotoxic properties of the tandem lesions in Escherichia coli cells. Chem. Res. Toxicol. 2010;23:11–19. doi: 10.1021/tx9004264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Yuan B, You C, Andersen N, Jiang Y, Moriya M, O'Connor TR, Wang Y. The roles of DNA polymerases κ and ι in the error-free bypass of N2-carboxyalkyl-2'-deoxyguanosine lesions in mammalian cells. J. Biol. Chem. 2011;286:17503–17511. doi: 10.1074/jbc.M111.232835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).You C, Dai X, Yuan B, Wang Y. Effects of 6-thioguanine and S6-methylthioguanine on transcription in vitro and in human cells. J. Biol. Chem. 2012;287:40915–40923. doi: 10.1074/jbc.M112.418681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).You C, Swanson AL, Dai X, Yuan B, Wang J, Wang Y. Translesion synthesis of 8,5'-cyclopurine-2'-deoxynucleosides by DNA polymerases η, ι, and ζ. J. Biol. Chem. 2013;288:28548–28556. doi: 10.1074/jbc.M113.480459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Liang Q, Dexheimer TS, Zhang P, Rosenthal AS, Villamil MA, You C, Zhang Q, Chen J, Ott CA, Sun H, Luci DK, Yuan B, Simeonov A, Jadhav A, Xiao H, Wang Y, Maloney DJ, Zhuang Z. A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses. Nat. Chem. Biol. 2014;10:298–304. doi: 10.1038/nchembio.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Zhai Q, Wang P, Wang Y. Cytotoxic and mutagenic properties of regioisomeric O2-, N3- and O4-ethylthymidines in bacterial cells. Carcinogenesis. 2014;35:2002–2006. doi: 10.1093/carcin/bgu085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Zhai Q, Wang P, Cai Q, Wang Y. Syntheses and characterizations of the in vivo replicative bypass and mutagenic properties of the minor-groove O2-alkylthymidine lesions. Nucleic Acids Res. 2014;42:10529–10537. doi: 10.1093/nar/gku748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).You C, Ji D, Dai X, Wang Y. Effects of Tet-mediated oxidation products of 5-methylcytosine on DNA transcription in vitro and in mammalian cells. Sci. Rep. 2014;4:7052. doi: 10.1038/srep07052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).You C, Wang P, Dai X, Wang Y. Transcriptional bypass of regioisomeric ethylated thymidine lesions by T7 RNA polymerase and human RNA polymerase II. Nucleic Acids Res. 2014;42:13706–13713. doi: 10.1093/nar/gku1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).You C, Wang J, Dai X, Wang Y. Transcriptional inhibition and mutagenesis induced by N-nitroso compound-derived carboxymethylated thymidine adducts in DNA. Nucleic Acids Res. 2015;43:1012–1018. doi: 10.1093/nar/gku1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Amato NJ, Zhai Q, Navarro DC, Niedernhofer LJ, Wang Y. In vivo detection and replication studies of alpha-anomeric lesions of 2'-deoxyribonucleosides. Nucleic Acids Res. 2015;43:8314–8324. doi: 10.1093/nar/gkv725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Andersen N, Wang P, Wang Y. Replication across regioisomeric ethylated thymidine lesions by purified DNA polymerases. Chem. Res. Toxicol. 2013;26:1730–1738. doi: 10.1021/tx4002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Andersen N, Wang J, Wang P, Jiang Y, Wang Y. In-vitro replication studies on O2-methylthymidine and O4-methylthymidine. Chem. Res. Toxicol. 2012;25:2523–2531. doi: 10.1021/tx300325q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Ji D, You C, Wang P, Wang Y. Effects of tet-induced oxidation products of 5-methylcytosine on DNA replication in mammalian cells. Chem. Res. Toxicol. 2014;27:1304–1309. doi: 10.1021/tx500169u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Ji D, Wang Y. Facile enzymatic synthesis of base J-containing oligodeoxyribonucleotides and an analysis of the impact of base J on DNA replication in cells. PLoS One. 2014;9:e103335. doi: 10.1371/journal.pone.0103335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Wang P, Amato NJ, Zhai Q, Wang Y. Cytotoxic and mutagenic properties of O4-alkylthymidine lesions in Escherichia coli cells. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv941. DOI: 10.1093/nar/gkv941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Bregeon D, Doetsch PW. Reliable method for generating double-stranded DNA vectors containing site-specific base modifications. Biotechniques. 2004;37:760–762. doi: 10.2144/04375ST01. 764, 766. [DOI] [PubMed] [Google Scholar]
- (31).You C, Wang Y. Quantitative measurement of transcriptional inhibition and mutagenesis induced by site-specifically incorporated DNA lesions in vitro and in vivo. Nat. Protoc. 2015;10:1389–1406. doi: 10.1038/nprot.2015.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Anna L, Kovacs K, Gyorffy E, Schoket B, Nair J. Smoking-related O4-ethylthymidine formation in human lung tissue and comparisons with bulky DNA adducts. Mutagenesis. 2011;26:523–527. doi: 10.1093/mutage/ger011. [DOI] [PubMed] [Google Scholar]
- (33).Chen HJ, Lee CR. Detection and simultaneous quantification of three smoking-related ethylthymidine adducts in human salivary DNA by liquid chromatography tandem mass spectrometry. Toxicol. Lett. 2014;224:101–107. doi: 10.1016/j.toxlet.2013.10.002. [DOI] [PubMed] [Google Scholar]
- (34).Synold T, Xi B, Wuenschell GE, Tamae D, Figarola JL, Rahbar S, Termini J. Advanced glycation end products of DNA: quantification of N2-(1-carboxyethyl)-2'-deoxyguanosine in biological samples by liquid chromatography electrospray ionization tandem mass spectrometry. Chem. Res. Toxicol. 2008;21:2148–2155. doi: 10.1021/tx800224y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol. Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- (36).Chatgilialoglu C, Ferreri C, Terzidis MA. Purine 5',8-cyclonucleoside lesions: chemistry and biology. Chem. Soc. Rev. 2011;40:1368–1382. doi: 10.1039/c0cs00061b. [DOI] [PubMed] [Google Scholar]
- (37).Yuan B, Wang J, Cao H, Sun R, Wang Y. High-throughput analysis of the mutagenic and cytotoxic properties of DNA lesions by next-generation sequencing. Nucleic Acids Res. 2011;39:5945–5954. doi: 10.1093/nar/gkr159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Chang SC, Fedeles BI, Wu J, Delaney JC, Li D, Zhao L, Christov PP, Yau E, Singh V, Jost M, Drennan CL, Marnett LJ, Rizzo CJ, Levine SS, Guengerich FP, Essigmann JM. Next-generation sequencing reveals the biological significance of the N2,3-ethenoguanine lesion in vivo. Nucleic Acids Res. 2015;43:5489–5500. doi: 10.1093/nar/gkv243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Nagel ZD, Margulies CM, Chaim IA, McRee SK, Mazzucato P, Ahmad A, Abo RP, Butty VL, Forget AL, Samson LD. Multiplexed DNA repair assays for multiple lesions and multiple doses via transcription inhibition and transcriptional mutagenesis. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E1823–1832. doi: 10.1073/pnas.1401182111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Rizzo JM, Buck MJ. Key principles and clinical applications of "next-generation" DNA sequencing. Cancer Prev. Res. 2012;5:887–900. doi: 10.1158/1940-6207.CAPR-11-0432. [DOI] [PubMed] [Google Scholar]
- (41).Minko IG, Earley LF, Larlee KE, Lin YC, Lloyd RS. Pyrosequencing: applicability for studying DNA damage-induced mutagenesis. Environ. Mol. Mutagen. 2014;55:601–608. doi: 10.1002/em.21882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Izhar L, Ziv O, Cohen IS, Geacintov NE, Livneh Z. Genomic assay reveals tolerance of DNA damage by both translesion DNA synthesis and homology-dependent repair in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E1462–1469. doi: 10.1073/pnas.1216894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Keskin H, Shen Y, Huang F, Patel M, Yang T, Ashley K, Mazin AV, Storici F. Transcript-RNA-templated DNA recombination and repair. Nature. 2014;515:436–439. doi: 10.1038/nature13682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).McLuckey SA, Van Berkel GJ, Glish GL. Tandem mass spectrometry of small, multiply charged oligonucleotides. J. Am. Soc. Mass Spectrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]