The phytotoxicity in Arabidopsis of the plant-derived phenylpropanoid pathway inhibitor 3,4-(methylenedioxy)cinnamic acid (MDCA) is based on the perturbation of auxin homeostasis.
Abstract
The phenylpropanoid 3,4-(methylenedioxy)cinnamic acid (MDCA) is a plant-derived compound first extracted from roots of Asparagus officinalis and further characterized as an allelochemical. Later on, MDCA was identified as an efficient inhibitor of 4-COUMARATE-CoA LIGASE (4CL), a key enzyme of the general phenylpropanoid pathway. By blocking 4CL, MDCA affects the biosynthesis of many important metabolites, which might explain its phytotoxicity. To decipher the molecular basis of the allelochemical activity of MDCA, we evaluated the effect of this compound on Arabidopsis thaliana seedlings. Metabolic profiling revealed that MDCA is converted in planta into piperonylic acid (PA), an inhibitor of CINNAMATE-4-HYDROXYLASE (C4H), the enzyme directly upstream of 4CL. The inhibition of C4H was also reflected in the phenolic profile of MDCA-treated plants. Treatment of in vitro grown plants resulted in an inhibition of primary root growth and a proliferation of lateral and adventitious roots. These observed growth defects were not the consequence of lignin perturbation, but rather the result of disturbing auxin homeostasis. Based on DII-VENUS quantification and direct measurement of cellular auxin transport, we concluded that MDCA disturbs auxin gradients by interfering with auxin efflux. In addition, mass spectrometry was used to show that MDCA triggers auxin biosynthesis, conjugation, and catabolism. A similar shift in auxin homeostasis was found in the c4h mutant ref3-2, indicating that MDCA triggers a cross talk between the phenylpropanoid and auxin biosynthetic pathways independent from the observed auxin efflux inhibition. Altogether, our data provide, to our knowledge, a novel molecular explanation for the phytotoxic properties of MDCA.
Plants growing in a tight community are in continuous competition for space, light, water, and nutrients. Potential survival strategies include optimizing plant architecture and maximizing growth rate, allowing the plant to capture light and receive nutrients and water more efficiently, while placing neighboring plants in an unfavorable position (Einhellig, 1995; Weir et al., 2004). Besides developmental shifts, plants release an array of secondary metabolites (allelochemicals) into the rhizosphere to negatively affect the growth and reproduction of neighboring, competitor plants (Putnam, 1988; Bertin et al., 2003). Despite a lot of research effort having been devoted to allelopathic chemical warfare over the past decades, it remains a difficult study object due to the complexity of plant-plant interactions (Zeng, 2014). Nevertheless, the significance of allelochemicals in structuring plant communities and preserving biodiversity has been fully recognized by the scientific community. Moreover, allelochemicals show the potential to be used as an environmentally friendly alternative for weed control to improve agricultural productivity (Zeng, 2014).
Strictly speaking, the term “allelochemical” refers to a compound produced and released by one organism to affect the growth and development of susceptible species (Weir et al., 2004). In practice, compounds derived from plant extracts or exudates are often cataloged as allelochemicals based on their inhibitory effect on seed germination and/or growth of other plant species in an artificial setup. Despite their importance, the molecular mode of action of a given allelochemical compound has rarely been studied in detail; however, toxicity is relatively easily demonstrated, identifying its molecular target is far more challenging. An interesting example is the phenylpropanoid 3,4-(methylenedioxy)cinnamic acid (MDCA), which was isolated from lyophilized root tissues of Asparagus [Asparagus officinalis L.; Hartung et al. (1990)]. It was suggested to be an allelochemical based on its inhibitory effect on root and shoot growth of Lepidium sativum (Hartung et al., 1990). Independent studies revealed that MDCA acts as an efficient competitive inhibitor of 4-COUMARATE-CoA LIGASE (4CL), the enzyme converting hydroxycinnamates to their corresponding CoA-esters (Knobloch and Hahlbrock, 1977; Chakraborty et al., 2009). This conversion is an early step in the general phenylpropanoid pathway leading to a wide array of metabolites, including coumarins, stilbenes, salicylic acid, flavonoids, and monolignols (Vogt, 2010). Given that inhibition of 4CL in this metabolic pathway will have far-reaching effects on plant growth and development (Voelker et al., 2010), it is tempting to link the proposed phytotoxicity of MDCA to this metabolic block.
Here, we evaluate whether the phytotoxicity of MDCA is a direct consequence of the inhibition of 4CL or if MDCA targets also other biological processes in Arabidopsis (Arabidopsis thaliana). We found that MDCA causes strong developmental defects in Arabidopsis seedlings at early developmental stages. Convincing evidence was found that MDCA affects the homeostasis of the plant signaling compound auxin. Our results provide an alternative explanation for the molecular mechanism underlying the phytotoxic properties of MDCA, and suggest that these multiple modes of action make it an attractive candidate as an environmental agrochemical or synergist.
RESULTS
MDCA Affects Plant Growth and Development in a Dose-Dependent Manner
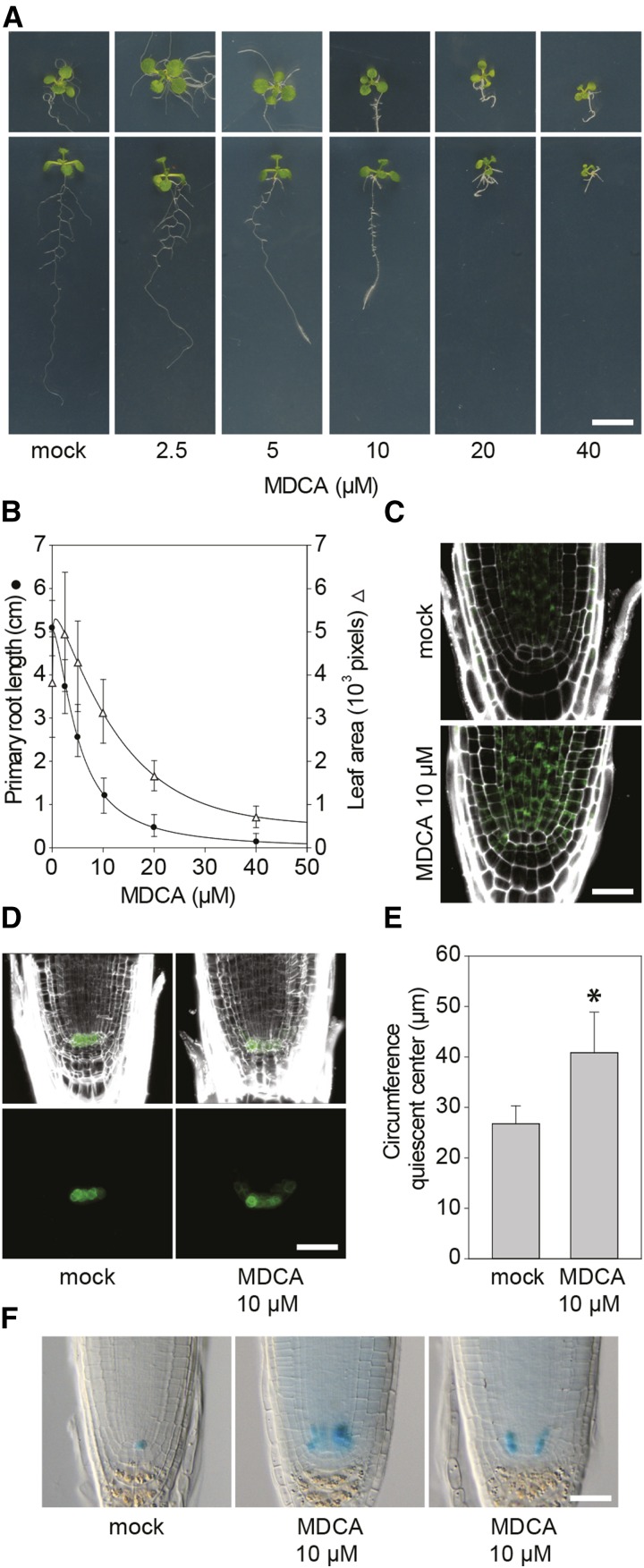
MDCA was put forward as an allelochemical based on the inhibition of L. sativum root and shoot growth when used at a concentration of 260 μM or higher (Hartung et al., 1990). In an attempt to support the idea that MDCA has allelopathic properties, we tested the effect of MDCA on germination of Arabidopsis seeds. None of the tested concentrations (0 µM to 200 µM) had an inhibitory effect on germination as scored by radical emergence (Supplemental Fig. S1A). However, a clear restrictive effect on the postembryonic development of the plants was observed at the different concentrations tested (Supplemental Fig. S1B). To obtain profound insight into the effect of MDCA on plant growth and development, the experiment was repeated using lower MDCA concentrations (0 µM to 40 µM). Twelve d after germination (DAG), both the primary root and rosette growth of the seedlings were analyzed (Fig. 1A). Compared to the control, the MDCA-treated plants had more lateral roots and the roots displayed a marked agravitropic response. In addition, a dose-dependent decrease of both the primary root length and leaf area was observed (Fig. 1B). The MDCA-concentration required to reduce the primary root length by 50% (IC50-root) was determined to be 5.07 μM, while no further development of the primary root was observed from germination onwards at 40 μM MDCA. The effect of MDCA on leaf development was less severe and higher concentrations were needed to reduce the leaf area by half (IC50-leaf of 18.30 µM). Noteworthy, the decline in leaf area could be an indirect effect caused by the severe reduction in primary root length at these relatively high MDCA concentrations.
Figure 1.
Effect of MDCA on growth and development of Arabidopsis. A, Root/rosette phenotype of seedlings (12 DAG) grown on 0.5 × MS-medium supplemented with MDCA (n > 20; scale bar: 1 cm). B, MDCA dose response curves for primary root length (circle; sigmoidal-logistic, four parameters) and leaf area (triangle; Weibull, five parameters; n > 20). Error bars represent sds. C, Confocal images showing KNOLLE promoter activity (green) in the primary root tip of pKNOLLE:KNOLLE-GFP seedlings. Seedlings were germinated 7 d on 0.5 × MS-medium before being transferred to 0.5 × MS-medium supplemented with 10 μM MDCA for 5 d (n = 5; scale bar: 15 μm). D, Confocal images showing QC broadening (green) in the primary root tip of pWOX5:GFP seedlings. Seedlings were germinated 7 d on 0.5 × MS-medium before being transferred to 0.5 × MS-medium supplemented with 10 μM MDCA for 5 d (n = 5; scale bar: 15 μm). PI was used in (C) and (D) as counterstain to visualize the cell wall. E, Quantification of the QC-region by measuring the circumference of pWOX5-driven GFP-expressing cells in the primary root tip (n = 5). Error bars represent sds. The asterisk in (E) represents significant difference in circumference of pWOX5-driven GFP expressing cells between 10 μM MDCA-treated and mock-treated plants (0.001 < P value < 0.05) as determined by Dunnett’s test. F, Light microscopic images showing ERF115 promoter activity in the primary root tip of pERF115:GUS seedlings. Seedlings were germinated 3 d at 24°C on 0.5 × MS-medium supplemented with or without (mock) 10 μM MDCA (n > 128; scale bar: 15 μm).
A reduction in primary root length could point toward a perturbed activity of the apical root meristem, and more detailed analysis of this region revealed a prominent broadening of the main root tip upon MDCA treatment. To test whether this lateral expansion is a direct consequence of an increase in meristematic cell division activity, we treated a pKNOLLE:KNOLLE-GFP translational fusion-line with MDCA (Lukowitz et al., 1996). KNOLLE is a marker for cell plate formation, and under normal conditions the corresponding gene is expressed in the actively dividing cells of the main root tip. A 5-d treatment of Arabidopsis seedlings (3 DAG) with 10 μM MDCA led to an accumulation of fluorescence in the main root tip (Fig. 1C). No fluorescence was observed in the initial QC-cells, which is in line with the known low mitotic activity of these cells (Heyman et al., 2014). As the QC regulates the meristematic region tightly by maintaining the surrounding cells as stem cells, we reasoned that MDCA would induce broadening of the meristem in the QC region. Accordingly, the QC marker line pWOX5:GFP (Haecker et al., 2004) showed a radial expansion of the GFP-signal in the QC area (a 1.48-fold increase in circumference; Fig. 1, D and E) upon MDCA treatment compared to mock-treated plants. The radial expansion of WOX5-positive cells indicates that MDCA either stimulates QC cell division activity or gives the cells surrounding the QC a QC-identity. The former contradicts the absence of strong mitotic activity of the QC cells as observed in the KNOLLE reporter line. However, a putative positive effect of MDCA on QC cell division activity could have gone unnoticed as QC cells self-renew at a low proliferation rate (Dolan et al., 1993). To reveal the effect of MDCA on QC cell proliferation, a reporter line was used in which GUS was driven by the promoter of the transcription factor ERF115, which is a rate-limiting factor for QC cell division (Heyman et al., 2013). In mock-treated Arabidopsis seedlings (3 DAG), ERF115 promoter activity was observed in one or several of the QC cells in 32.6% of the root tips (60/184; Fig. 1F). An additional 34 root tips (18.5%) had GUS activity outside the QC-region. The addition of 10 μM MDCA to the growth medium led to an increase in the number of seedlings with ectopic pERF115-driven GUS expression (52/128; 40.6%; Fig. 1F). Interestingly, pERF115:GUS-positive QC cells were only observed in two of the 128 MDCA-treated seedlings, suggesting that MDCA has a negative impact on QC cell division activity. Together, this suggests that the MDCA-mediated lateral expansion of QC-identity in the stem cell niche is due to changes in cell identity of QC-neighboring cells rather than activation of QC division.
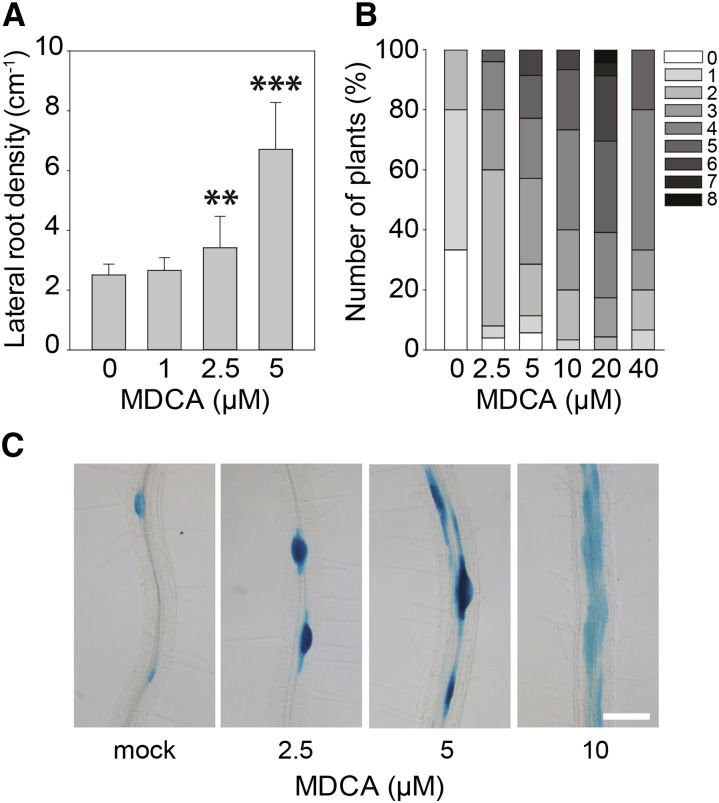
In contrast to its inhibitory effect on primary root growth and leaf development, MDCA stimulated lateral root formation and adventitious rooting in a dose-dependent manner (Fig. 2, A and B). A 1.36-fold and 2.67-fold increase in lateral root density (LRD) was observed in 12-d-old plants treated with 2.5 µM and 5 µM MDCA, respectively. To evaluate the potential effect of MDCA on the spatial activation of the cell cycle in the pericycle and thereby on the spacing between lateral roots, the cell cycle reporter line pCYCB1:GUS was used (Colón-Carmona et al., 1999). In mock-treated plants, GUS-expression was restricted to the cells actively dividing at the lateral root initiation sites. MDCA treatment (0 µM to 10 µM) resulted in a dose-dependent increase in the number of pericycle cells expressing GUS, which is in line with the observed increase in LRD along the primary root (Fig. 2C). The observed perturbation in longitudinal spacing of lateral root primordia lead in some extreme cases to the outgrowth of fasciated lateral roots.
Figure 2.
Effect of MDCA on lateral and adventitious root growth of Arabidopsis. A, Lateral root density of seedlings 12 DAG grown on 0.5 × MS-medium supplemented with MDCA (n > 15). Error bars represent sds and asterisks were used to indicate statistically significant differences compared to the corresponding mock-treated control sample as determined by Dunnett’s test P values: *P < 0.05, **P < 0.001, ***P < 0.0001. B, Number of adventitious roots of seedlings 12 DAG grown on 0.5 × MS-medium supplemented with MDCA. Plants were germinated (after a short light-pulse of 4 h) for 7 d in darkness and subsequently transferred to light for 5 d to stimulate adventitious rooting. Adventitious root numbers (percentages) are represented in grayscale (n > 30). C, Light microscopic images of lateral root primordia as shown by pCYCB1:GUS expression of seedlings 12 DAG grown on 0.5 × MS-medium supplemented with MDCA (n = 5; scale bar: 0.05 cm).
In conclusion, the data suggested that MDCA reduces primary root growth of Arabidopsis seedlings by affecting the stem cell niche and the meristem at the main root apex, while stimulating lateral root formation by increasing cell division activity along the pericycle.
Phenolic Profiling Supports Inhibition of the Core Phenylpropanoid Pathway by MDCA
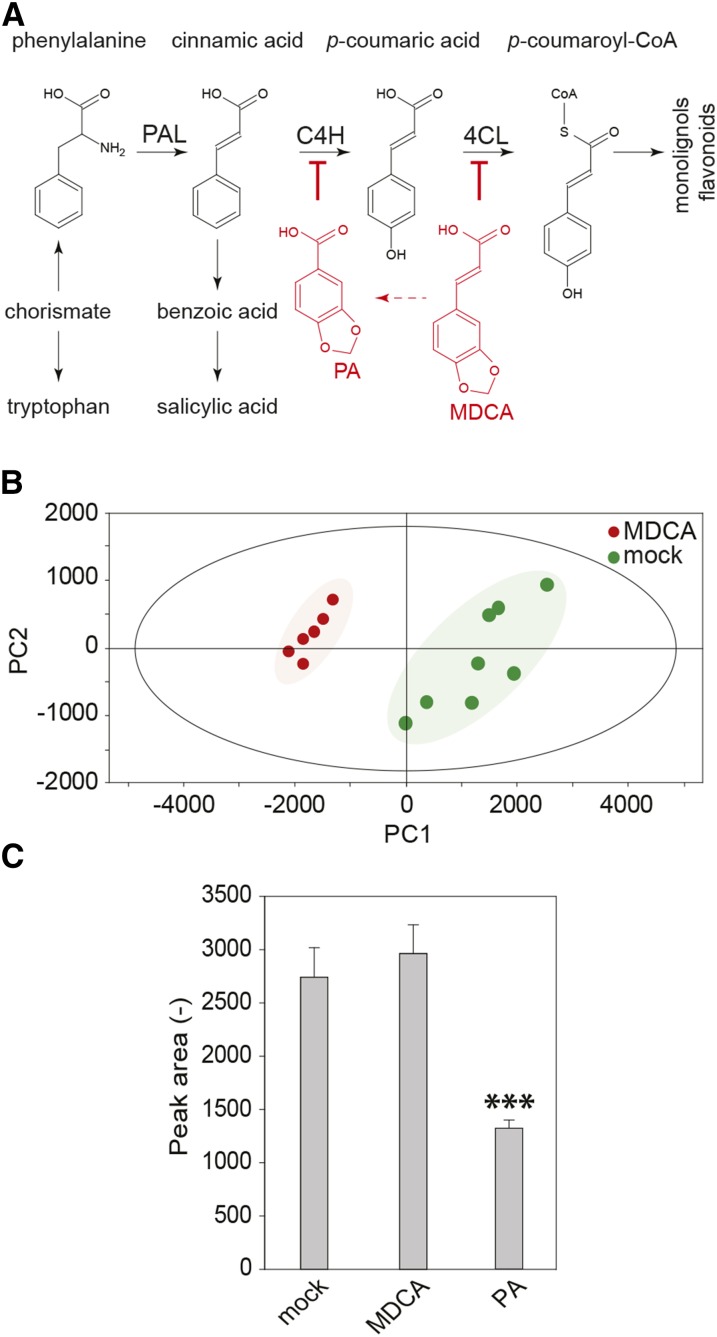
Previous in vitro assays based on heterologous systems provided evidence that MDCA acts as a competitive inhibitor of 4CL (Knobloch and Hahlbrock, 1977; Chakraborty et al., 2009; Fig. 3A). As this enzyme catalyzes a key step of the general phenylpropanoid pathway, we used phenolic profiling to evaluate how the carbon flux over this pathway is redirected in seedlings upon treatment with MDCA. To this end, the methanol-soluble metabolites of 12-d-old in vitro grown Arabidopsis seedlings treated with 10 μM MDCA were analyzed by ultra-high-pressure liquid chromatography-mass spectrometry (UHPLC-MS). This method allows the detection of several classes of metabolites (e.g. oligolignols, flavonoids, and glucosinolates) and their corresponding derivatives or pathway intermediates (Fig. 3A). A total of 1247 peaks was detected over the whole experiment. Principal component analysis revealed that MDCA-treated samples separated from control samples based on a combination of the first and second principal components, indicating metabolic shifts upon MDCA treatment (Fig. 3B). Univariate statistical analysis was applied to select peaks with significantly different intensity in the MDCA-treated samples. Among the 383 obtained peaks, 247 were increased in abundance, whereas 136 were reduced upon MDCA treatment. The remarkably high number of differentials might reflect the developmental shift caused by MDCA, adding differentials to the compound lists that are only indirectly related to the treatment.
Figure 3.
Phenolic profiling of MDCA treated Arabidopsis seedlings. A, The general phenylpropanoid pathway leading to a wide array of secondary metabolites. PA is a chemical inhibitor of C4H. MDCA is a chemical inhibitor of 4CL. The red arrow depicts the in planta processing of MDCA towards PA. B, Principal component analysis scores plot of 10 μM MDCA-treated (red) and mock-treated (green) seedlings 12 DAG. PC1 and PC2 explained 73.74% and 10.15% of the variation, respectively. Each symbol corresponds with a biological replicate representing 10 seedlings (n > 6). C, The enzymatic conversion of CA toward p-coumaric acid by C4H and potential chemical inhibition of C4H by MDCA using microsomes of yeast (Saccharomyces cerevisiae) expressing C4H of Arabidopsis. PA was used as a positive control. The product (p-coumaric acid) was detected by UHPLC-MS analysis (n = 6). Error bars represent sds. Three asterisks in (C) represent statistically significant differences compared to mock-treated microsomes (P value < 0.0001) as determined by Dunnett’s test.
Of the 10 highest accumulating compounds in MDCA-treated plants (Supplemental Fig. S2), five were conjugates of MDCA, indicating that MDCA is heavily processed by the plant. Four other compounds of the top-10 list were conjugates of cinnamic acid (CA), indicative for the accumulation of CA in MDCA-treated plants. CA is the substrate of C4H, the enzyme preceding 4CL in the phenylpropanoid pathway, and the accumulation of CA conjugates is indicative for an inhibition of C4H in MDCA-treated plants. However, MDCA was unable to inhibit Arabidopsis C4H activity in a heterologous expression system (Fig. 3C), making it unlikely that the accumulation of CA conjugates is due to inhibition of C4H by MDCA. Here, the metabolomics data provided insight into the underlying molecular mechanism. Among the compounds that had accumulated in MDCA-treated plants were several conjugates of piperonylic acid (PA), a compound structurally related to MDCA (Fig. 3 and Supplemental Fig. S3). Based on the fact that PA or PA-derived conjugates were never found in Arabidopsis, and on the previous observation that MDCA is heavily processed in the plant (Supplemental Fig. S2), we concluded that the presence of PA-derived compounds is the consequence of the in planta processing of MDCA. Interestingly, PA is known as a competitive inhibitor of C4H (Schalk et al., 1998; Fig. 3C), explaining the observed accumulation of CA conjugates. Despite the clear indications for a PA-mediated inhibition of C4H, a potential inhibition of 4CL by MDCA could not be excluded at this point. The top-10 list of compounds for which the concentration dropped upon MDCA treatment (Supplemental Fig. S2) holds several glycosylated forms of ferulic acid-containing phenolic dimers, glucosinolates, and flavonoids. A subsequent targeted search for related metabolites revealed an MDCA-dependent reduction in glycosylated oligolignols and p-coumaric acid-derived phenylpropanoids (Supplemental Fig. S4). The observed shift in the phenolic profile of MDCA-treated seedlings is in line with the inhibition of an enzyme catalyzing an early step in the core phenylpropanoid pathway.
Lignin Reduction Is Not at the Basis of the MDCA-Induced Developmental Defects
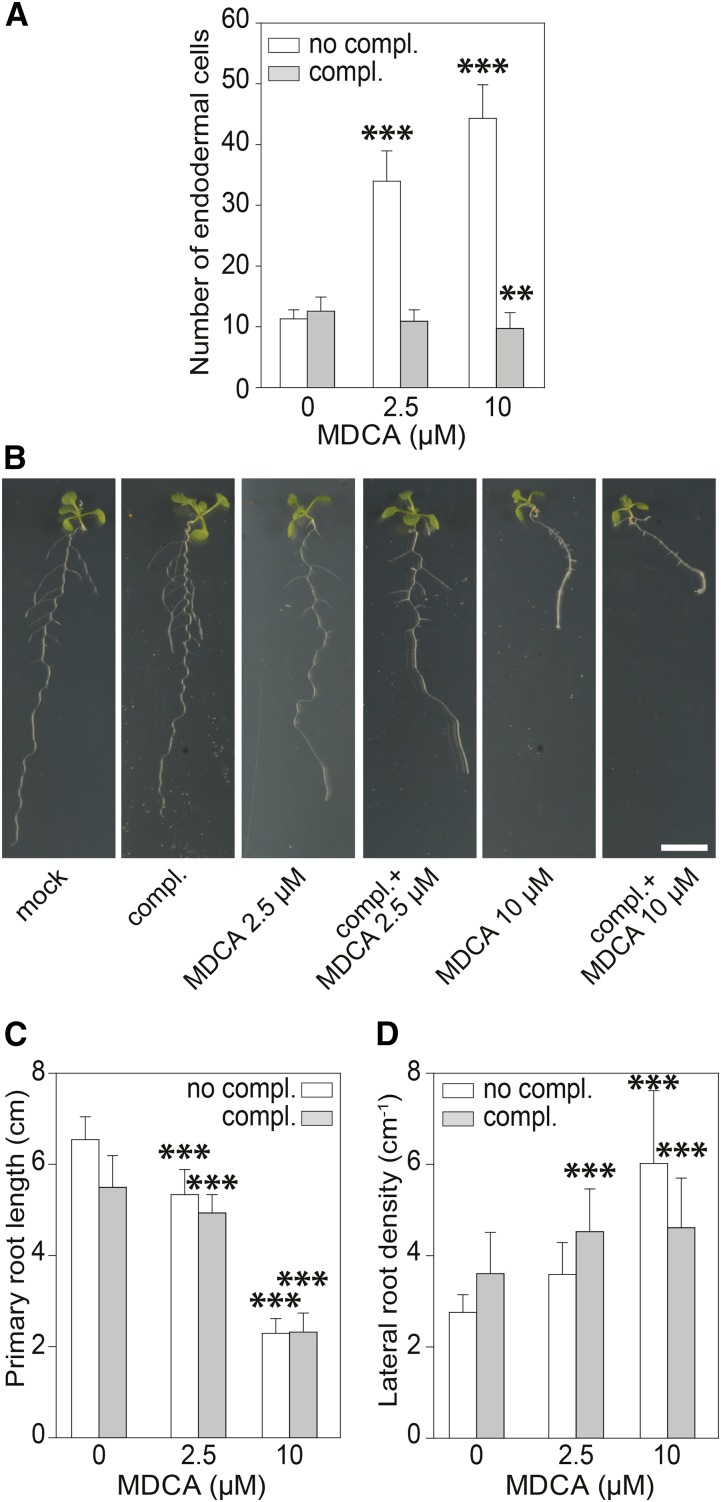
The MDCA-induced reduction of glycosylated oligolignols is considered indicative for a drop in lignin deposition (Vanholme et al., 2012). To provide supporting evidence for this assumption, we evaluated the effect of MDCA on the formation of the Casparian strip in the main root (Naseer et al., 2012). For this assay, Arabidopsis roots are incubated in propidium iodide (PI), a dye that diffuses in the apoplast of root tissue. At the endodermis, this diffusion is blocked by the lignin-rich Casparian strip, leaving the cell walls of the stele unstained. Only when the Casparian strip is impaired will PI pass the endodermis and stain the underlying cell layers. The number of endodermal cells from the elongation zone to the region where PI is excluded from the stele is used as a measure for Casparian strip integrity and, hence, lignification. In 7-d-old Arabidopsis seedlings treated with 2.5 μM and 10 μM MDCA, the cell count was significantly higher compared to that of the control, indicating a delay in Casparian strip development and an inhibition in lignin deposition (Fig. 4A; Supplemental Fig. S5). Similar results were found with inhibitors targeting other steps of the lignin biosynthetic pathway (Naseer et al., 2012; Van de Wouwer et al., 2016). The formation of the Casparian strip was rescued by supplying MDCA-treated plants with a mixture of coniferyl and sinapyl alcohol (both end-products of the monolignol biosynthetic pathway, and building blocks of lignin), confirming that MDCA affects the biosynthesis of lignin (Fig. 4A; Supplemental Fig. S5).
Figure 4.
The impact of the MDCA-induced lignin reduction on the plant phenotype. A, Effect of MDCA treatment on the Casparian strip formation (white; no complementation) in Arabidopsis seedlings 5 DAG and complementation by exogenous application of two monolignols (gray; complementation): 50 μM of each coniferyl alcohol and sinapyl alcohol, which allows for the formation of a functional Casparian strip (n = 10). See text for additional explanation on this experiment. B, Root phenotype of seedlings (12 DAG) grown on 0.5 × MS-medium supplemented with MDCA and monolignols (n > 20; scale bar: 1 cm). (C and D) Combined effect of monolignols and MDCA on the primary root length of Arabidopsis seedlings 12 DAG (n > 20) and lateral root density of seedlings 12 DAG (n > 15). Seedlings were grown on 0.5 × MS medium supplemented with 0, 2.5, or 10 μM MDCA. Monolignols (coniferyl alcohol and sinapyl alcohol; 50 μM each) were added to the tissue culture medium, where mentioned (complementation). Error bars represent sds and asterisks were used to indicate statistically significant differences compared to the corresponding mock-treated control sample, as determined by Dunnett’s test P values: *P < 0.05, **P < 0.001, ***P < 0.0001.
Having convincing evidence that lignin deposition is indeed affected in MDCA-treated seedlings, we wondered whether the reduced lignin deposition could be at the basis of the observed developmental abnormalities. The underlying idea is that lignin is considered a signature for cell differentiation (Barros et al., 2015) and deferring lignin deposition could extend or perturb the developmental program, resulting in growth defects (Bonawitz and Chapple, 2013). If this were the case, restoring lignin deposition by the addition of monolignols should also rescue the MDCA-induced developmental root phenotype. To examine whether the MDCA-related plant phenotype is a consequence of lignin depletion, we grew Arabidopsis on medium containing coniferyl and sinapyl alcohol and different concentrations of MDCA (0, 2.5, or 10 μM). Twelve DAG the seedlings were analyzed. Interestingly, phenotypes linked to MDCA, such as the reduction of the primary root length and the proliferation of lateral roots, could not be restored by adding monolignols (Fig. 4, B–D).
Based on these results, we concluded that MDCA indeed reduces lignin deposition in Arabidopsis seedlings, but this reduction is not at the basis of the observed developmental defects caused by MDCA.
Salicylic Acid Has No Major Role in the MDCA-Induced Developmental Defects
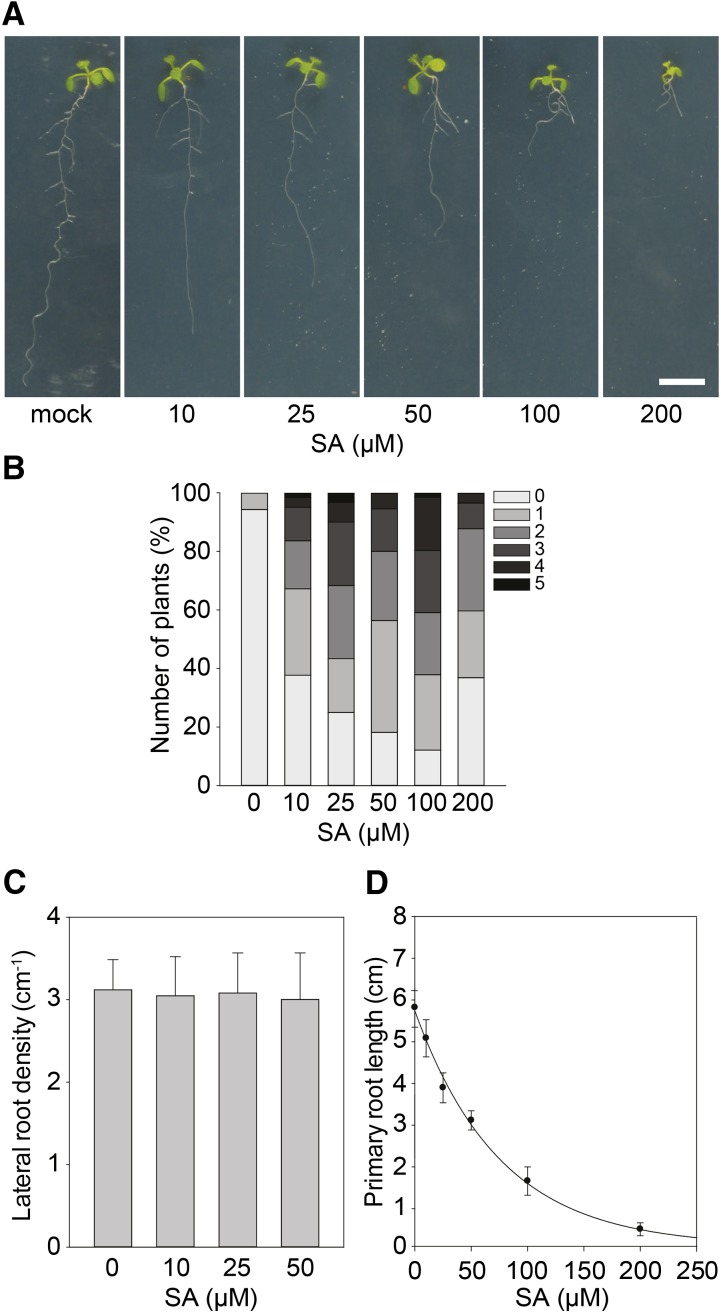
Treatment of seedlings with MDCA significantly affected the relative abundance of different phenylpropanoid intermediates and derivatives, as shown by the phenolic profiling presented above. A relative shift in the abundance of some of these metabolites might interfere with specific metabolic or signaling pathways and lead to growth inhibition (Bonawitz and Chapple, 2013). Recently, some of the observed growth defects in mutants perturbed in lignin biosynthesis have been linked with the accumulation of salicylic acid (SA), a plant hormone involved in pathogen defense that is biosynthetically related to phenylpropanoids and that can be synthesized from CA (Gallego-Giraldo et al., 2011). Therefore, the MDCA-mediated inhibition of the phenylpropanoid pathway downstream of CA could theoretically lead to the accumulation of SA (Boerjan et al., 2003). SA levels were quantified by UHPLC-MS in 12 DAG Arabidopsis seedlings grown on medium with 10 μM MDCA. Interestingly, MDCA-treated seedlings showed a mild, but significant increase in SA levels (1.35-fold increase; Supplemental Fig. S6). As SA-levels were close to the detection limit, we further evaluated whether this increase in SA-levels could underlie the MDCA-induced phenotypes by checking to what extent SA-treatment of Arabidopsis seedlings could phenocopy the effect of MDCA treatment (Fig. 5A). While stimulation of adventitious rooting was only observed at relatively high concentrations (>10 μM of SA; Fig. 5B), SA had no effect on LRD (Fig. 5C) and its IC50-root (IC50-root of 51.00 μM; Fig. 5D) was one order-of-magnitude higher than that of MDCA (IC50-root of 5.07 μM; Fig. 1B).
Figure 5.
Effect of SA on growth and development of Arabidopsis. A, Root phenotype of seedlings (12 DAG) grown on 0.5 × MS-medium supplemented with SA (n > 20; scale bar: 1 cm). B, Number of adventitious roots of seedlings 12 DAG grown on 0.5 × MS-medium supplemented with SA. Plants were germinated (after a short light-pulse of 4 h) for 7 d in darkness and subsequently transferred to light for 5 d to stimulate adventitious rooting. Adventitious root numbers (percentages) are represented in grayscale (n > 30). C, Lateral root density of seedlings (12 DAG) grown on 0.5 × MS-medium supplemented with SA (n > 15). Error bars represent sds. No significant differences were obtained between the mock-treated control sample and SA-treated samples as determined by Dunnett’s test P values > 0.05. D, SA dose response curve for primary root growth (sigmoidal-logistic, four parameters; n > 20). Error bars represent sds.
Therefore, we concluded that it is unlikely that SA (or a corresponding conjugate) is the bioactive compound causing the root developmental defects observed in MDCA-treated plants, and that SA has no major role in the MDCA-induced developmental defects.
MDCA Triggers Auxin Biosynthesis
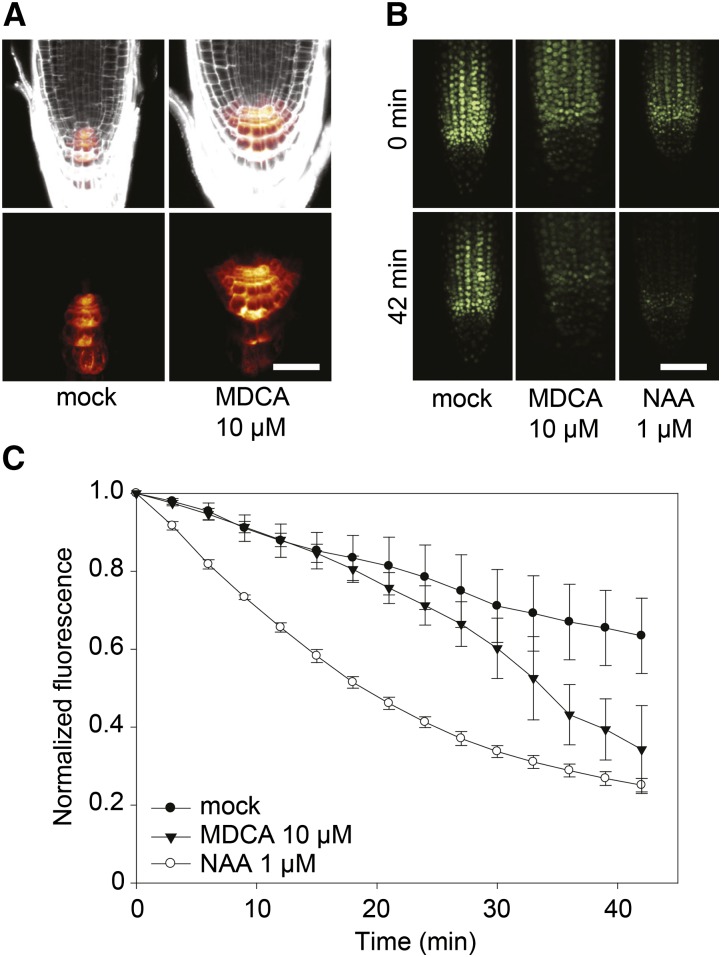
As the aberrant root phenotype caused by MDCA resembles typical auxin-induced phenotypes (e.g. reduction of the primary root length and induction of lateral and adventitious roots), the auxin-responsive reporter DR5rev:GFP was used to evaluate whether MDCA alters the auxin response (Ulmasov et al., 1997). Seven DAG Arabidopsis seedlings treated for 4 d with 10 μM MDCA displayed a significant increase in fluorescence in the main root tip compared to mock-treated plants (Fig. 6A), suggesting that free auxin levels are increased in MDCA-treated plants. To detect dynamic changes in endogenous free auxin distribution at high spatial and temporal resolution, the DII-VENUS auxin sensor line was used (Brunoud et al., 2012). A time-lapse analysis of DII-VENUS fluorescence was performed on the main root tip of Arabidopsis seedlings 7 DAG. When 1 μM naphthalene-1-acetic acid (NAA; a synthetic auxin) was added to the root, DII-VENUS fluorescence dropped to 25% of its initial intensity after 42 min, which is in line with previously published data (Brunoud et al., 2012). Replacing NAA by 10 μM MDCA resulted in a drop of DII-VENUS fluorescence to 37% of its initial intensity over the same time interval. Both auxin-sensitive sensors indicate that MDCA depends on the auxin-signaling cascade for its activity (Fig. 6, B and C).
Figure 6.
Activity of auxin reporters in MDCA-treated Arabidopsis seedlings. A, Confocal images of MDCA-induced activation of the auxin-responsive DR5 promoter in the primary root tip of pDR5rev:GFP seedlings. Seedlings were germinated 7 d on 0.5 × MS-medium before being transferred to 0.5 × MS-medium supplemented with 10 μM MDCA for 5 d (n = 5; scale bar: 35 μm). PI was used as counterstain to visualize the cell walls. Color-code depicts (red; low to white; high) pDR5rev:GFP signal intensity. B, Confocal images of DII-VENUS degradation in the primary root tip of DII-VENUS-YFP seedlings. Seedlings were germinated 7 d on 0.5 × MS-medium before being transferred to 0.5 × MS-medium with 10 μM MDCA or 1 μM NAA. Individual root tips were imaged at the onset of the experiment and after 42 min (n = 3; scale bar: 50 μm). C, Time-course of DII-VENUS fluorescence in the primary root tips of seedlings grown as described above. Fluorescence was quantified every 3 min over a 42-min interval. The fluorescence was normalized against the initial value for each treatment. Error bars represent sds.
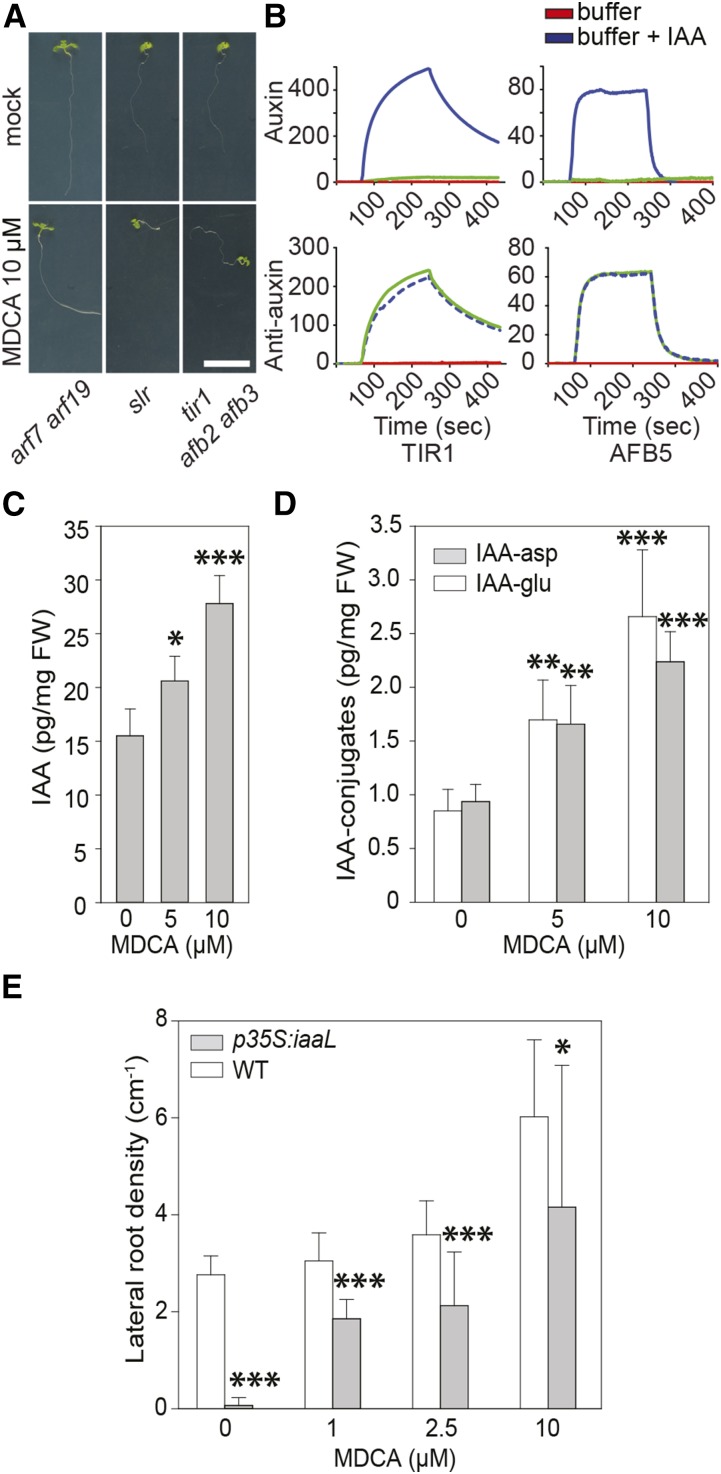
The canonical auxin-signaling cascade is activated when auxin interacts with the TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX (TIR1/AFB) receptors, whereupon AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) auxin signaling repressors are degraded and the expression of downstream target genes (e.g. the DR5rev-driven GFP) is activated (Gray et al., 1999). To verify whether MDCA activates this auxin response pathway, we studied the effect of MDCA on Arabidopsis mutants defective in auxin signaling including the solitary root-1 (slr) gain-of-function mutant, the arf7 arf19 double loss-of-function mutant, and the tir1 afb2 afb3 triple loss-of-function mutant (Fig. 7A; Fukaki et al., 2002; Dharmasiri et al., 2005; Okushima et al., 2007). MDCA failed to induce lateral root formation in all mutants tested, suggesting that lateral root formation induced by MDCA is dependent on events close to the TIR1 or TIR1-related receptor complex. This could indicate that MDCA act as an auxin analog and activates an auxin response by interacting directly with TIR1. Interestingly, MDCA has some of the structural requirements for primary auxin activity as stipulated by Went (1949), including a ring system with a double bond and a side chain with the carboxylic group separated from the ring by at least one carbon atom. Regardless of these structural and physicochemical similarities between MDCA and IAA (the endogenous ligand of TIR1), both compounds show particular differences making it unlikely for MDCA to act as an auxin (Supplemental Fig. S7, A–C). For example, the unsubstituted indole ring of IAA is hydrophobic while the unsubstituted dioxole group of MDCA is polar due to the presence of two oxygen atoms. In addition, adjacent to its carboxylic group, IAA has a freely rotatable CH2 group, while the transalkene bond of MDCA is rigid, and does not rotate to position the carboxylic group correctly from the plane of the ring to interact with the polar residues of the TIR1 receptor complex in a way similar to IAA. Molecular docking of MDCA in the auxin receptor pocket of TIR1 indeed showed a clear difference in its orientation as compared to IAA as modeled by Tan et al. (2007) (Supplemental Fig. S7, D and E).
Figure 7.
The importance of auxin in MDCA-mediated developmental defects in seedlings. A, Root phenotype of arf7 arf19, slr, and tir1 afb2 afb3 mutant seedlings (12 DAG) grown on 0.5 × MS-medium supplemented with 10 μM MDCA (n > 25; scale bar: 1 cm). B, SPR sensorgrams showing the auxin-dependent interaction between TIR1 or AFB5 with IAA7/14 DII. Each sensorgram shows the binding with IAA (blue), an auxin-free injection (red), plus the data for MDCA (green). For auxin activity assays, MDCA (50 μM) was mixed with TIR1 or AFB5 prior to injection over DII peptide. For antiauxin assays, MDCA (50 μM) was mixed with TIR1 or AFB5 plus 5 μM IAA prior to injection. Dashed lines were used to increase the visibility. (C and D) Free IAA levels and IAA-amino acid conjugate (IAA-Asp and IAA-Glu) levels of seedlings (12 DAG) grown on 0.5 × MS-medium supplemented with different concentrations of MDCA. E, Each biological replicate represents 10 seedlings that were pooled and analyzed (n = 6). Error bars represent sds and asterisks were used to indicate statistically significant differences between wild type and p35S:iaaL at a given concentration as determined by Dunnett’s test P values: *P < 0.05, **P < 0.001, ***P < 0.0001.
To find supporting evidence for the in silico predictions and further evaluate whether MDCA activates auxin signaling by directly interacting with the TIR1/AFB receptor, surface plasmon resonance (SPR; Lee et al., 2014) was used to measure the interaction kinetics of recombinant TIR1 and AFB5 with an immobilized IAA7 degron peptide. The choice of the interaction partners was based on the requirement of both proteins for the MDCA-induced lateral root proliferation, as deduced from the experiment with the auxin signaling mutants. A strong readout was obtained with the positive control IAA, indicative for a strong agonist, whereas no evidence for a specific binding of MDCA to TIR1 or AFB5 was found (Fig. 7B). It is possible that some ligands bind to TIR1/AFB5 and prevent subsequent binding of the receptor-ligand complex to the degron peptide. Since such antagonists could give auxin-hypersensitive phenotypes (Leyser et al., 1996), we tested MDCA also for antiauxin activity but no activity was found (Fig. 7B). These findings support the prediction that MDCA is not able to replace auxin in the TIR1 binding pocket and, thus, cannot be considered as an auxin analog interacting with the TIR1-related auxin receptors.
As MDCA-induced developmental effects depend on the canonical auxin-signaling cascade whereas MDCA itself is not an auxin analog, we hypothesized that MDCA affects free auxin concentrations in planta. To test this hypothesis, endogenous auxin levels were quantified in Arabidopsis seedlings germinated on 0.5 × MS media containing MDCA (5 μM or 10 μM) and harvested 12 DAG. Compared to mock-treated plants, the MDCA treatment resulted in elevated free IAA levels as well as increased levels of the IAA-conjugates IAA-Glu and IAA-Asp (Fig. 7, C and D). In addition, the concentrations of most of the measured auxin precursors were also significantly higher in the MDCA-treated plants (tryptamine, indole-3-acetamide, indole-3-acetonitrile, indole-3-acetaldoxime, and indole-3-acetaldehyde), indicating that MDCA strongly boosts the auxin biosynthetic pathway (Supplemental Fig. S8). We subsequently checked whether the aberrant lateral root phenotype induced by MDCA could be reduced by artificially lowering free IAA levels in planta by the constitutive overexpression of a bacterial IAA LYS SYNTHETASE (iaaL), coding for an enzyme that inactivates free IAA by conjugating it to Lys (Romano et al., 1991). In contrast to MDCA-treated wild-type plants, MDCA-treated p35S:iaaL plants showed less lateral roots (Fig. 7E), indicating that the increase in lateral roots induced by MDCA is mediated by free IAA.
The observed activation of auxin biosynthesis could either be a consequence of MDCA-mediated perturbation of the phenylpropanoid pathway or an off-target effect. To uncouple these processes, we measured auxin concentrations in the c4h mutant ref3-2 (Schilmiller et al., 2009). Blocking the phenylpropanoid pathway in such an early step could result in the accumulation of upstream compounds, including Trp, the precursor of IAA. This approach allowed us to study putative crosstalk between the auxin biosynthetic and phenylpropanoid pathways independent of MDCA. C4H was preferred over 4CL as target, because the phenolic profile of MDCA-treated seedlings was consistent with inhibition of C4H rather than 4CL. In addition, we showed earlier processing of MDCA toward PA in planta (Supplemental Fig. S3), suggesting the inhibition of C4H in MDCA-treated plants. Auxin profiling performed on leaves of 2-months-old ref3-2 mutants revealed shifts in free auxin as well as auxin intermediates, conjugates, and catabolites resulting in a profile similar to the one observed in MDCA-treated plants (Supplemental Fig. S9). This indicates that auxin biosynthesis is affected as a result of the perturbation of the phenylpropanoid pathway.
In conclusion, the above data suggest that MDCA affects plant growth and development by increasing endogenous auxin (IAA) concentrations in Arabidopsis seedlings.
MDCA Affects Auxin Transport
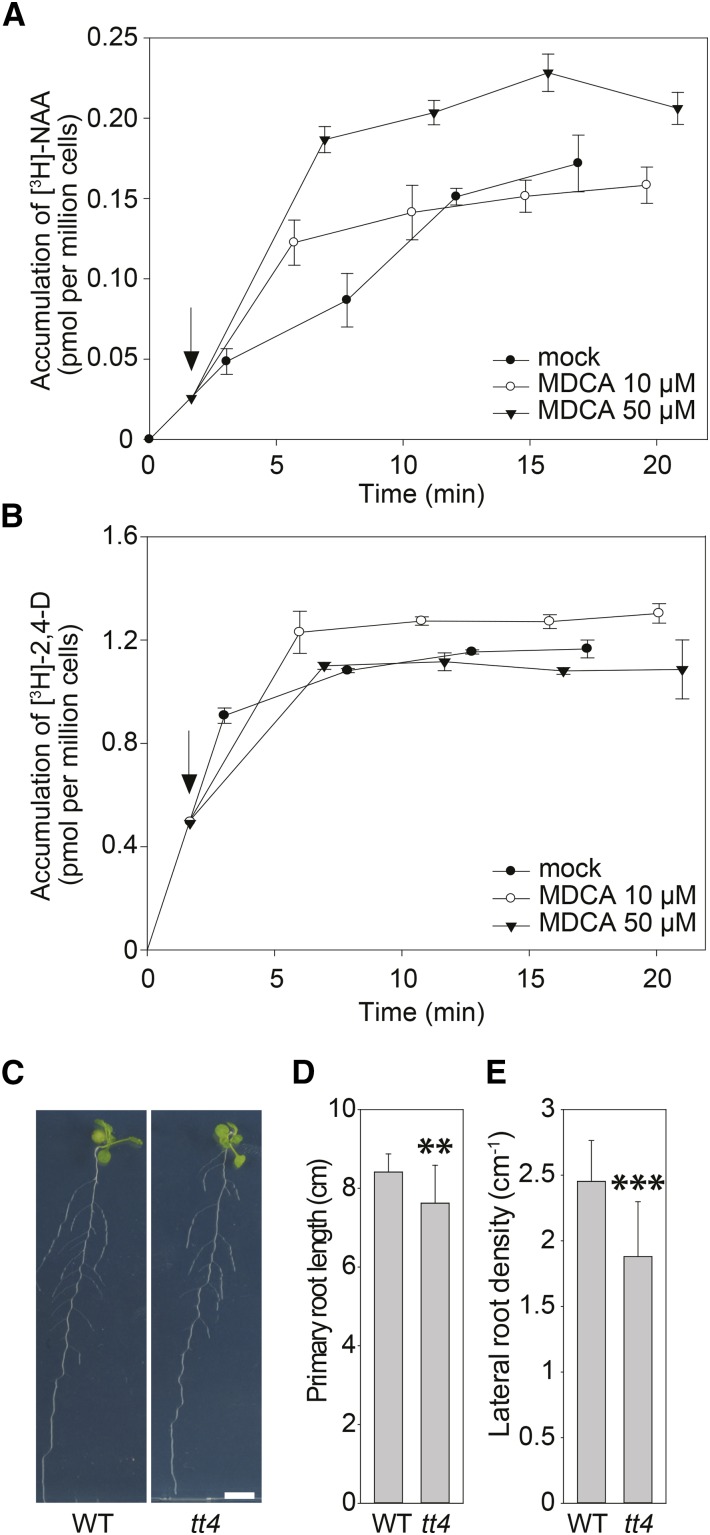
Although the increase in free IAA levels is in agreement with the observed response of the DII-VENUS reporter, it is unlikely that the rapid drop in DII-VENUS fluorescence is due to the activation of the auxin biosynthetic pathway. In addition, the phenylpropanoid pathway is not activated in the main root tip where the degradation of the DII-VENUS signal was followed over time (Bell-Lelong et al., 1997). These observations question the suggested cross talk with the phenylpropanoid pathway and indicates that other processes might be involved in the MDCA-induced alteration of free auxin levels in the main root tip, such as the redistribution of available auxin over the root by perturbation of intercellular auxin transport. In contrast to the activation of auxin biosynthesis, the inhibition of auxin transport is likely to be a much faster response. To test whether MDCA affects auxin transport, we performed [3H]-NAA accumulation assays in tobacco BY-2 cell cultures (Petrásek et al., 2003, 2006). In tobacco cells, NAA enters the cells mainly by diffusion and it is an excellent substrate for the active auxin efflux (Delbarre et al., 1996). Therefore, a change in intracellular accumulation of radioactively labeled [3H]-NAA over time provides a measure of the rate of auxin efflux from the cells. When treated with 1-naphthylphthalamic acid (NPA), to inhibit cellular auxin efflux, NAA accumulates inside the cells above the level of untreated cells (Delbarre et al., 1996). Interestingly, intracellular NAA concentrations also increased when NPA was replaced by 50 μM MDCA, indicating that MDCA affects cellular auxin accumulation by blocking auxin efflux (Fig. 8A). Similar results were obtained using Arabidopsis cell suspensions (Supplemental Fig. S10). In contrast to NAA, the transport of 2,4-dichlorophenoxy acetic acid (2,4-D) into tobacco BY-2 cells is largely dependent on auxin uptake carriers making 2,4-D an excellent marker for the auxin influx activity. When added to BY-2 cell cultures 2,4-D rapidly accumulated in the cells, and MDCA had a mild, but positive effect on this profile; however, the mild accumulation likely reflects the partial inhibition of the active cellular efflux of 2,4-D under these conditions as a similar profile was described for the auxin efflux blocker NPA as well (Hosek et al., 2012). In Arabidopsis cell cultures, 2,4-D seems to be a good substrate for both influx and efflux auxin carriers (Seifertová et al., 2014) and, accordingly, the positive effect of MDCA on the accumulation of 2,4-D is more pronounced here than in BY-2 cells (Supplemental Fig. S10). Taken together, these results indicate that MDCA affects the auxin efflux from cells, but not the influx into cells (Fig. 8, A and B).
Figure 8.
The impact of MDCA on auxin transport and root development. A and B, Effect of MDCA on the net accumulation of A, [3H]-NAA or B, [3H]-2,4-D in 2-d-old suspension-cultured tobacco BY-2 cells (20 min uptake period). Arrows point at time of application of MDCA. Error bars in (A) and (B) represent sds. C, Root phenotype of wild type and tt4-mutant seedlings (12 DAG) grown on 0.5 × MS-medium (n > 20; scale bar: 1 cm). D, Primary root length of wild-type and tt4-mutant seedlings (12 DAG) grown on 0.5 × MS-medium (n > 20). E, Lateral root density of wild-type and tt4-mutant seedlings (12 DAG) grown on 0.5 × MS-medium (n > 15). (D and E) Error bars represent sds and asterisks represent significant differences between wild-type and tt4-mutant seedlings as determined by Dunnett’s test. Dunnett’s test P values: *P < 0.05, **P < 0.001, ***P < 0.0001. WT = wild type.
Flavonoids, as one group of products of the phenylpropanoid pathway, are obvious candidates to link the perturbation of the core phenylpropanoid pathway with the observed inhibition in auxin transport. Indeed, particular flavonoids have been described to alter auxin transport (Brown et al., 2001; Buer and Muday, 2004; Peer and Murphy, 2007; Yin et al., 2014) and phenolic profiling showed that the levels of all detected flavonoids dropped upon MDCA treatment. Intriguingly, the phenotype of the transparent testa-4 (tt4) mutant, which is hampered in the first enzymatic step of flavonoid biosynthesis (CHALCONE SYNTHASE) and hence is depleted of flavonoids, resembles that of MDCA-treated plants at the seedling stage, including a reduction in primary root length and an increase in LRD (Brown et al., 2001). However, these root phenotypes were difficult to confirm in a follow-up study and turned out to depend strongly on the experimental conditions (Buer and Djordjevic, 2009). To check whether the drop in flavonoids could be the cause of the MDCA-induced phenotype, the tt4 mutant was grown under the same growth conditions as those used to study the MDCA-dependent growth defects (Fig. 8C). Under these conditions, the tt4 mutant showed a mild but significant reduction in both primary root length and LRD, as compared to wild-type Arabidopsis (Fig. 8, D and E). This phenotype differs considerably from the MDCA-induced lateral root proliferation, indicating that, although the drop in flavonoids could be responsible for an indirect additive effect in MDCA-treated plants, it is not the preliminary cause of the altered LRD phenotype in MDCA-treated seedlings.
Altogether, these data suggest that MDCA disturbs auxin homeostasis by two independent mechanisms. The perturbation of auxin biosynthesis is a consequence of the MDCA-mediated inhibition of the phenylpropanoid pathway, whereas the inhibition of auxin transport is a direct effect of MDCA.
DISCUSSION
MDCA is a naturally occurring compound, initially extracted from root tissues of Asparagus (Hartung et al., 1990) and further characterized as an allelochemical (Weir et al., 2004). Later on, MDCA was identified as an efficient chemical inhibitor of 4CL, a key enzyme of the general phenylpropanoid pathway leading toward a broad range of secondary metabolites (Knobloch and Hahlbrock, 1977; Funk and Brodelius, 1992; Hartung et al., 1990; Funk and Brodelius, 1994; Chakraborty et al., 2009). Substantial inhibition of the phenylpropanoid pathway can be lethal for the plant (Bonawitz and Chapple, 2013), potentially explaining the allelochemical properties of MDCA (Bertin et al., 2003). Besides its assumed role in suppressing growth of neighboring plants, it was also proposed to lead to autotoxicity, manifested by the poor reestablishment of Asparagus plants in soil where it had been grown before (Laufer and Garrison, 1977; Yang, 1982; Young and Chou, 1984; Hartung et al., 1990).
When tested on Arabidopsis seedlings, severe morphological alterations of the root architecture were observed. In an attempt to link the developmental defects to the described inhibitory activity on the phenylpropanoid pathway, phenolic profiling was performed on MDCA-treated seedlings. Interestingly, the observed shift in the profile suggested an inhibition of the phenylpropanoid pathway upstream of p-coumaric acid, but downstream of CA, indicating that the perturbation occurs at the level of C4H. This conclusion contradicts studies describing MDCA as a competitive inhibitor of 4CL (Knobloch and Hahlbrock, 1977; Chakraborty et al., 2009), with only mild inhibitory activity on C4H (Schalk et al., 1998). However, the suggested inhibition of C4H can be explained by the in planta processing of MDCA to PA, which is a well-known inhibitor of C4H (Schalk et al., 1998). Despite the difficulty to conclude whether MDCA or PA is at the basis of the observed perturbation of the phenylpropanoid pathway, treating seedlings with MDCA clearly inhibits in planta lignin deposition as proven by the Casparian strip assay. However, no evidence was found supporting the notion that the reduction in lignin deposition was causative to the observed aberrant root phenotype.
The remarkable accumulation of intermediates upstream of C4H induced by MDCA suggested cinnamic acid-derived SA as a potential candidate to explain the observed developmental defects. In Medicago sativa, a correlation between SA concentration and the level of lignin reduction was found (Gallego-Giraldo et al., 2011; Lee et al., 2011), whereas the growth defects of a lignin biosynthetic mutant in Arabidopsis was partially restored by reducing the elevated SA levels in planta (Gallego-Giraldo et al., 2011). We therefore tested for SA accumulation in MDCA-treated plants. Mass spectrometry revealed a mild, but significant increase upon MDCA treatment. However, replacing MDCA for SA in the culture medium did not phenocopy the MDCA-induced developmental defects in Arabidopsis seedlings. Hence, no strong evidence was found for a role for SA in the MDCA-induced growth defects.
Rather than focusing on the perturbed phenylpropanoid pathway, we reasoned that MDCA itself could have additional targets, not necessarily related to its inhibitory activity on this pathway. Supporting evidence for an alternative target was found in an old study describing the activity of MDCA in a bioassay for auxin-type growth regulators (Aberg, 1961). Based on the growth stimulation of oat and wheat roots, this compound was cataloged as an antiauxin; however, the underlying molecular mechanism was never resolved (Aberg, 1961). Interestingly, the observed reduction in primary root length and the increased lateral root proliferation observed on the MDCA-treated Arabidopsis seedlings hinted for a link to auxin homeostasis (Overvoorde et al., 2010). Also, the induction of cell division activity in the root tip and the broadening of the QC region were indicative of an auxin imbalance, because auxin tightly regulates the cellular organization of the root meristem (Petricka et al., 2012; Takatsuka and Umeda, 2014).
The link with auxin was later underpinned by showing the inability of MDCA to stimulate lateral root proliferation in the auxin signaling mutants slr, arf7 arf19, and tir1 afb2 afb3, demonstrating the necessity of functional auxin signaling for the root-related MDCA-induced phenotypes to occur. Subsequent receptor binding assays showed that MDCA itself is not an auxin analog, suggesting that it acts upstream of TIR1, most likely affecting auxin levels. This was confirmed by measuring the levels of auxin and auxin-related compounds in MDCA-treated plants. Besides an increase in free IAA, the concentrations of IAA conjugates and degradation products, as well as intermediates of the auxin biosynthetic pathway, were increased in MDCA-treated plants. We hypothesized that the MDCA-mediated increase in auxin content could be a direct consequence of blocking the phenylpropanoid pathway at the level of C4H or 4CL. The resulting accumulation of compounds upstream of these enzymatic steps could affect the shikimate pathway, which provides Phe to the phenylpropanoid pathway. Importantly, besides Phe, the shikimate pathway leads to the precursors of the other two aromatic acids: Tyr and Trp (Vogt, 2010). This makes it tempting to speculate that interfering with the phenylpropanoid pathway leads to an increase in Trp, the precursor of IAA. The interaction between phenylpropanoid and auxin biosynthesis was suggested before, when swellings at branch junctions were observed in the ref3-2 mutant (Schilmiller et al., 2009). Although no additional evidence was available, increases in auxin concentration in the cauline leaves that subtend the swollen nodes were considered the cause of the growth defects. Our auxin measurements performed on this mutant are consistent with this hypothesis.
In addition to the link with auxin biosynthesis, the initial trigger altering auxin levels inside the plant could be the inhibition of polar auxin transport. The fast responses observed in the cellular auxin transport assay (both in BY-2 and Arabidopsis cells) as well as in the DII-VENUS experiment support such a direct response, independent of auxin biosynthesis. To explain further the molecular mechanism, we investigated whether flavonoids could act as mediators of the MDCA-related auxin efflux inhibition. Not only was the abundance of these phenylpropanoid-derived compounds dramatically altered in MDCA-treated plants, some have been described as endogenous auxin transport inhibitors (Peer and Murphy, 2007; Yin et al., 2014). Although the latter has been disputed (Li et al., 2010), recent evidence suggests that e.g. rhamnosylated flavonols (namely kaempferol 3-O-rhamnoside-7-O-rhamnoside) are active compounds inhibiting polar auxin transport and/or modifying auxin homeostasis in Arabidopsis shoots (Yin et al., 2014 and Kuhn et al., 2016). Nevertheless, the instant effect of the auxin efflux inhibition in BY-2 cells upon the addition of MDCA is supportive for a fast and most likely direct effect on auxin transport. In addition, the reduction in flavonoid levels is difficult to link with the induction of root proliferation in MDCA-treated plants as the flavonoid depleted tt4-mutant showed a reduction in LRD in our experimental setup. As MDCA affects different molecular processes, we cannot exclude a potential effect of flavonoid reduction that is masked by the MDCA-mediated induction of auxin biosynthesis. Nevertheless, the flavonoid data presented in this study together with the controversy concerning the role of flavonoids in the control of polar auxin transport, suggest that lateral root formation caused by MDCA is largely independent from flavonoids.
An intriguing question is whether MDCA has a more general physiological role in planta besides its potential activity as an allelochemical. The fact that this molecule has so far only been found in A. officinalis L. is not supportive for a universal role in plant development. However, this can be merely due to the fact that it was never searched for in other plant species. Plants produce an enormous number of metabolites of which only a small fraction has been characterized in detail (Fernie et al., 2004). In addition, most of the current metabolic profiling projects deliberately avoid the many unidentified compounds and focus only on the known ones, adding no information to the list of identified or characterized plant metabolites (Fernie et al., 2004). Even if the presence of MDCA itself cannot be demonstrated in other plants, we cannot exclude that other plants make structural analogs that act in a similar way as MDCA. From this perspective, the dioxole group of MDCA is of particular interest. This functional group is found in other plant-derived bioactive molecules belonging to different chemical classes, including (iso)flavonoids (e.g. pisatin), lignans (e.g. sesamin and kobusin), and alkaloids (e.g. sanguinarine, berberin, aristolochic acid, and narciclasine; Bailey, 1970; Trifunovic et al., 2003; Ono et al., 2006; Evidente et al., 1983; Gardiner et al., 2008; Na et al., 2011; Hu et al., 2012; Nakagawa et al., 2012; Hara and Kurita, 2014). Interestingly, the bioactivity of berberin and coptisin has been linked to the dioxole group as a structural analog, but without this functional group (i.e. palmatine), it turned out to be inactive (Nakagawa et al., 2012). Unfortunately, information concerning the molecular targets of the listed compounds is limited, making it nearly impossible to deduce a general in planta target for the dioxole-containing molecules. In animals, cytochrome P450 (CYP450) enzymes were put forward as targets for dioxole-containing compounds (Lafite et al., 2007), making it tempting to extrapolate this to MDCA, especially as several CYP450 enzymes are key in both the phenylpropanoid and auxin biosynthetic pathway. However, MDCA has been shown to inhibit 4CL (Knobloch and Hahlbrock, 1977; Chakraborty et al., 2009), which is not a CYP450 enzyme. Although the phenolic profile of MDCA-treated plants is supportive for an MDCA-mediated inhibition of the CYP450 enzyme C4H in the plant, this is most likely the indirect effect of the metabolism of MDCA into PA, a known C4H-inhibitor (Schalk et al., 1998). Further exploring the similarities and differences caused by the methylenedioxy group containing bioactive molecules could help in unraveling their molecular mechanism. In this perspective, it is interesting to mention narciclasine, a dioxole-containing alkaloid isolated from Narcissus tazetta bulbs (Na et al., 2011; Hu et al., 2012). Similar to MDCA, narciclasine is considered a potential allelochemical affecting postembryonic plant development by inhibiting auxin responses and modulating polar auxin transport in the target plant (Na et al., 2011; Hu et al., 2012).
In conclusion, we found that MDCA not only perturbs the phenylpropanoid pathway but also affects auxin homeostasis by triggering auxin biosynthesis and interfering with polar auxin transport. The inhibition of the phenylpropanoid pathway combined with the modified auxin homeostasis is most likely at the basis of its property as allelochemical. The activity of MDCA represents an example of polypharmacology, a little explored mechanism in which active pharmaceuticals exert their influence by multiple, instead of single sites of action (Reddy and Zhang, 2013). The relative importance of both processes in the phytotoxicity of MDCA remains unknown, but the effectiveness of hormone perturbation agrees well with the fact that phytohormonal superauxins have been among the most successful herbicides used in agriculture for decades (Grossmann, 2010), making MDCA an attractive candidate to become an agrochemical or synergist.
MATERIALS AND METHODS
Plants, Chemicals, and Growth Conditions
All experiments whereby the effect of MDCA on the plant phenotype was studied were performed with Arabidopsis [Arabidopsis thaliana Columbia 0 (Col-0)] unless otherwise stated. The transgenic lines were in the same ecotype: p35S:iaaL, DII-VENUS, DR5rev:GFP, pCYCB1:GUS, pWOX5:GFP, pKNOLLE:KNOLLE-GFP, pERF115:GUS, tir1 afb2 afb3, slr, arf7 arf19 (Romano et al., 1991; Lukowitz et al., 1996; Colón-Carmona et al., 1999; Fukaki et al., 2002; Friml et al., 2003; Dharmasiri et al., 2005; Okushima et al., 2007; Brunoud et al., 2012; Heyman et al., 2013). Seeds were vapor-phase-sterilized and grown on 0.5 × MS-medium. The medium was supplemented with one of following compounds: PA, MDCA, NAA, and SA (Sigma Aldrich). After sowing, seeds were incubated at 4°C for at least 2 d, whereupon they were placed in the growth chamber under a 16-h-light/8-h-dark photoperiod regime (long d photoperiod) at 21°C (24°C for the pERF115:GUS line). Chemical compounds were dissolved in DMSO and added to the autoclaved medium prior to pouring the plates. For the Casparian strip method, PI (Sigma Aldrich) was used to counterstain the cell wall. The adventitious rooting assay was performed by placing plates in the dark for 7 d (after a short light pulse of 4 h). Plates were then exposed to light for 7 d under long d photoperiod. The number of adventitious roots (above the root-shoot junction) was counted on each seedling. The ref3-2 mutants used for auxin profiling were grown for 8 weeks on soil under long d photoperiod.
Plant Phenotyping
To quantify growth parameters and check for aberrant phenotypes, seeds were grown on round (square) plates for leaf (primary root) growth analysis, respectively. Twelve DAG plates were scanned using the Scanmaker 9800XL and root length was measured using the ImageJ software. For leaf area measurements, pictures were converted to black and white figures, after which the leaf area was measured using the ImageJ software. For each compound, the inhibitory concentration (IC50) was calculated, by plotting a dose-response curve in SigmaPlot. The dose-response curve giving the highest R2-value was used. The emerged lateral roots and adventitious roots were counted using a binocular microscope.
Confocal Microscopy and Casparian Strip Assays
Visualization and quantification of the appearance of Casparian strips was performed as previously described in Naseer et al. (2012). Arabidopsis Col-0 seeds were germinated on 0.5 × MS-Medium agar plates after 2 d in darkness at 4°C. Seedlings were grown vertically on 0.5 × MS-Medium agar supplied with 2.5 µM or 10 µM MDCA. When needed, 100 µM of a monolignol mixture containing coniferyl and sinapyl alcohol (1:1) was added. For visualization of the apoplastic barrier, seedlings were incubated in the dark for 2 min in a fresh solution of 10 µg/mL PI and rinsed in water twice. Confocal laser scanning microscopy was performed on a LSM5 Exciter confocal microscope (Carl Zeiss). An excitation window of 543 nm was used and detection was done using LP560. For quantification, “onset of elongation” was defined as the point where an endodermal cell in a median optical section was more than twice its width. From this point onwards, cells in the file were counted until PI could no longer diffuse through the apoplastic barrier, indicating that the Casparian strip was formed. For each treatment, 10 seedlings were used.
Microscopy
Root cell walls were counterstained with 30 µM PI for Arabidopsis plants transformed with pKNOLLE:KNOLLE-GFP, pWOX5:GFP, and DR5rev:GFP constructs. The excitation energy 488 nm was from an argon laser. The PI fluorescence emission was collected between 550 nm and 650 nm, and that of GFP/YFP between 500 nm and 550 nm. All images were captured with an inverted LSM 710 META confocal microscope equipped with 20× air objectives (Carl Zeiss). GUS assays were performed and inspected using differential interference contrast optics.
Time-Lapse DII-Venus
Seven-d-old Arabidopsis seedlings grown on vertical 0.5 × MS plates were used to analyze the effect of MDCA on the DII:VENUS reporter. At the initiation of the time-lapse, three seedlings (biological repeats) were transferred to glass-bottomed dishes and covered with media containing 1 µM NAA and 10 µM MDCA. The time series started 5 min after the seedlings had been placed in contact with the media and captured over 42 min (every 3 min) with a LSM 710 META confocal microscope (Carl Zeiss; 20× air objective). Images were analyzed by using the Fiji software by means of total signal from z-projection of defined region (always the same area).
UHPLC-MS
Arabidopsis seedlings were flash-frozen in liquid N2 and ground in 2-mL Eppendorf tubes using a Retsch mill (1 min, 20 Hz, 5-mm bead). For metabolite profiling, 10 to 20 seedlings of each treatment were pooled as one biological repeat. Samples were extracted with 1 mL methanol and the lyophilized pellets were subjected to liquid/liquid extraction (80 μL water/80 μL cyclohexane). A 15 μL aliquot of each sample was used for LC-MS analysis. The LC-MS system consists of an Acquity UPLC system (Waters Corporation) connected to a Synapt HDMS quadrupole time-of-flight mass spectrometer (Waters MS Technologies). Chromatographic separation was performed on an Acquity UPLC BEH C18 column (2.1 × 150 mm, 1.7 μm; Waters Corporation) using a water-acetonitrile gradient as described in Vanholme et al. (2012). The eluent was directed to the mass spectrometer equipped with an electrospray ionization source and lockspray interface for accurate mass measurements. The MS source parameters were as follows: capillary voltage, 2.5 kV; sampling cone, 37 V; extraction cone, 3.5 V; source temperature, 120°C; desolvation temperature, 400°C; cone gas flow, 50 L h−1; and desolvation gas flow, 550 L h−1. The collision energy for the trap and transfer cells was 6 V and 4 V, respectively. For data acquisition, the dynamic range enhancement mode was activated. Full-scan data were recorded in negative centroid V-mode; the mass range between m/z 100 and 1000, with a scan speed of 0.2 s scan−1, was recorded with Masslynx software (Ver. 4.1; Waters Corporation). Leucin-enkephalin (250 pg μL−1; solubilized in water: acetonitrile 1:1 [v/v] with 0.1% [v/v] formic acid) was used for lock mass calibration, with scanning every 10 s with a scan time of 0.5 s. Data processing was done with Progenesis QI ver. 2.1 (Nonlinear Dynamics). Peak areas were normalized with the function “normalize to external standard”, with dry weight (mg) of the pellet remaining after methanol extraction used as “external standard”. The peaks were further filtered on (1) their detection in all replicates of at least one treatment and (2) to have an average normalized peak intensity area higher than 500 in either MDCA- or mock-treated seedlings. Next, t tests were applied on the 1238 peaks that fulfilled these criteria. A total of 860 peaks had a P value < 0.01. Of these, 247 peaks were at least 10-fold increased in MDCA-treated seedlings and 136 were at least 10-fold reduced. The top-10 UP list was selected from the 247 increased peaks as those with the highest normalized peak area in MDCA-treated seedlings, whereas the top-10 DOWN were selected from the 136 decreased peaks as those with the highest normalized peak area in mock-treated seedlings. For the detection of SA all UPLC-MS parameters were the same as above, except (1) a mass range between m/z 100 and 4000 was used; (2) the dynamic range enhancement mode was not activated; and (3) the enhanced duty cycle mass was set at 137.02. In addition, peak integration was done with Targetlynx (Waters Corporation) using standard settings and the peak area of m/z 137.02 eluting at 10.4 min (corresponding to SA) was divided by the dry weight of the pellet remaining after methanol extraction in milligrams.
Auxin Metabolite Profiling
Auxin measurements were performed on Arabidopsis seedlings or leaves in case of the ref3-2 mutant. Samples were flash-frozen in liquid N2. For auxin metabolite profiling, 10 seedlings from each treatment were pooled to form one biological replicate. Extraction and purification of auxin and its metabolites were done as described previously with minor modifications (Novák et al., 2012). Frozen samples were homogenized using a MixerMill ball grinder (Retsch) and extracted in 1 mL 50 mM sodium phosphate buffer (pH 7.0) containing 1% sodium diethyldithiocarbamate, with the addition of deuterium and 13C6-labeled internal standards. The pH was adjusted to 2.7 with 1 M hydrochloric acid, and the extracts were purified on Oasis HLB columns (30 mg; Waters Corporation), conditioned with 1 mL methanol, 1 mL water, and 0.5 mL sodium phosphate buffer (pH 2.7). After sample application, the column was washed with 2 mL 5% methanol and then eluted with 2 mL 80% methanol. Eluates were evaporated to dryness and dissolved in 20 μL of mobile phase prior to mass spectrometry analysis using a 1290 Infinity LC system and a 6460 Triple Quad LC/MS system (Agilent Technologies; Novák et al., 2012).
Auxin Accumulation Assays
The assays were performed as described before in Petrásek et al. (2003). Auxin accumulation was measured in tobacco BY-2 cells (Nicotiana tabacum L. cv Bright Yellow 2; Nagata et al., 1992) or Arabidopsis T87 cells (ecotype Columbia 0; Axelos et al., 1992) 48 h (BY-2) or 96 h (T87) after subcultivation in 0.5 mL aliquots of cell suspension (target working cell density 7 × 105 cells × mL−1 (BY-2) as precisely determined by counting in the Fuchs-Rosenthal hemocytometer, and 20 mg FW × mL−1 (T87)). Cultivation medium was removed by filtration on 20 μm mesh nylon filters and cells were resuspended in uptake buffer (20 mM MES, 10 mM Suc, 0.5 mM CaSO4, pH adjusted to 5.7 with KOH) and equilibrated for 45 min on the orbital shaker at 27°C in darkness. Equilibrated cells were collected by filtration, resuspended in fresh uptake buffer, and incubated with continuous orbital shaking for another 90 min under the same conditions. Radiolabeled auxins [3H]-naphthalene-1-acetic acid ([3H]-NAA) and [3H]-2,4-dichlorophenoxy acetic acid ([3H]-2,4-D); specific (molar) radioactivity 20 Ci/mmol each; American Radiolabeled Chemicals) were added to the cell suspension to give a final concentration of 2 nM. At indicated time points, aliquots of cell suspension were sampled and accumulation of radiolabeled auxins was terminated by rapid filtration under reduced pressure on cellulose filters (22 mm in diameter). Cell cakes with filters were transferred into scintillation vials, extracted with ethanol (UV-spectroscopy grade) for 30 min and radioactivity was determined by liquid scintillation counting (Tri-Carb 2900TR Scintillation Counter; Packard Instrument). Counting efficiency was determined by automatic external standardization and counts were corrected for quenching automatically. Then counts were corrected for remaining surface radioactivity by subtracting counts of aliquots collected immediately after addition of radiolabeled auxin. Inhibitors were added as required from stock solutions to give appropriate final concentration and proper controls (solvent) were applied. All accumulation measurements were done at least in triplicate. Recorded accumulation values were recalculated to 1 million cells (BY-2) or to 10 mg FW (T87).
Auxin-Binding and Antiauxin Experiments Using Surface Plasmon Resonance
Auxin receptor proteins AtTIR1 and AtAFB5 were expressed in insect cells (T. ni High5) and purified as described previously in Calderón Villalobos et al. (2012) and Lee et al. (2014). The biotinylated degron peptide representing Aux/IAA7/14 was purchased from ThermoFisher Scientific and immobilized on streptavidin-coated SPR chips (GE Healthcare). SPR experiments were run as described previously in Calderón Villalobos et al. (2012) and Lee et al. (2014). Briefly, compounds were added to purified receptor proteins from stock solutions in DMSO to give working concentrations that were 50 μM unless stated otherwise (DMSO 0.1% final). Controls lacking auxin/compound and controls containing IAA (50 μM) were run as references at the start and end of every set of sensorgrams on every protein preparation. Compounds were run in three separate experiments, with characteristic results shown. For antiauxin runs, receptor proteins were mixed with 5 μM IAA plus compound at 50 μM. An antiauxin effect was then determined if the compound competed with IAA, reducing the amplitude of TIR1/AFB5 binding on the sensorgram.
Microsome Assay
The microsome assay was performed as described in Van de Wouwer et al. (2016).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Effect of MDCA on germination of Arabidopsis.
Supplemental Figure S2. Metabolites with an altered abundance in 10 μΜ MDCA-treated Arabidopsis seedlings.
Supplemental Figure S3. UHPLC-MS based detection of PA + hexose and PA + malate in MDCA-treated Arabidopsis seedlings.
Supplemental Figure S4. Metabolites with an altered abundance in 10 μΜ MDCA-treated Arabidopsis seedlings.
Supplemental Figure S5. The impact of the MDCA-induced lignin reduction on the plant phenotype.
Supplemental Figure S6. Detection of SA in 10 μΜ MDCA-treated Arabidopsis seedlings.
Supplemental Figure S7. Chemical and physical characteristics and docking of MDCA for TIR1.
Supplemental Figure S8. Concentration of auxin and auxin metabolites after MDCA treatment of Arabidopsis seedlings.
Supplemental Figure S9. Concentration of auxin and auxin metabolites in the c4h mutant ref3-2.
Supplemental Figure S10. The impact of MDCA on auxin transport in suspension-grown Arabidopsis T87 cells.
Supplementary Material
Acknowledgments
We thank Victor Flors (Plant Physiology, Department of CAMN, Castellón 12071, Spain) and John Dean (DePaul University, Department of Biological Sciences, Chicago, IL 60614) for providing us with an aliquot of salicylic acid glucoside. In addition, we are grateful to Ottoline Leyser (Sainsbury Laboratory, University of Cambridge, Cambridge CB2 1LR) and Jeffrey Heyman (VIB Department of Plant Systems Biology, Universiteit Ghent) for providing us with p35S:iaaL and pERF115:GUS Arabidopsis seeds, respectively. Finally, we thank Wim Grunewald and Toon Babylon for technical support and Karel Spruyt for imaging assistance.
Footnotes
Articles can be viewed without a subscription.
References
- Aberg B. (1961) Studies on plant growth regulators XVIII. Some β-substituted acrylic acids. Ann. Roy. Agric. Coll. Sweden 27: 99–123 [Google Scholar]
- Axelos M, Curie C, Mazzolini L, Bardet C, Lescure B (1992) A protocol for transient gene-expression in Arabidopsis thaliana protoplasts isolated from cell suspension cultures. Plant Physiol Biochem 30: 123–128 [Google Scholar]
- Bailey JA. (1970) Pisatin production by tissue cultures of Pisum sativum L. J Gen Microbiol 61: 409–415 [Google Scholar]
- Barros J, Serk H, Granlund I, Pesquet E (2015) The cell biology of lignification in higher plants. Ann Bot (Lond) 115: 1053–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Lelong DA, Cusumano JC, Meyer K, Chapple C (1997) Cinnamate-4-hydroxylase expression in Arabidopsis. Regulation in response to development and the environment. Plant Physiol 113: 729–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin C, Yang XH, Weston LA (2003) The role of root exudates and allelochemicals in the rhizosphere. Plant Soil 256: 67–83 [Google Scholar]
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Chapple C (2013) Can genetic engineering of lignin deposition be accomplished without an unacceptable yield penalty? Curr Opin Biotechnol 24: 336–343 [DOI] [PubMed] [Google Scholar]
- Brown DE, Rashotte AM, Murphy AS, Normanly J, Tague BW, Peer WA, Taiz L, Muday GK (2001) Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol 126: 524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, Beeckman T, Kepinski S, Traas J, Bennett MJ, Vernoux T (2012) A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482: 103–106 [DOI] [PubMed] [Google Scholar]
- Buer CS, Djordjevic MA (2009) Architectural phenotypes in the transparent testa mutants of Arabidopsis thaliana. J Exp Bot 60: 751–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Muday GK (2004) The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 16: 1191–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderón Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, Zheng N, Napier R, et al. (2012) A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 8: 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty M, Karun A, Mitra A (2009) Accumulation of phenylpropanoid derivatives in chitosan-induced cell suspension culture of Cocos nucifera. J Plant Physiol 166: 63–71 [DOI] [PubMed] [Google Scholar]
- Colón-Carmona A, You R, Haimovitch-Gal T, Doerner P (1999) Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Delbarre A, Muller P, Imhoff V, Guern J (1996) Comparison of mechanisms controlling uptake and accumulation of 2,4-dichlorophenoxy acetic acid, naphthalene-1-acetic acid, and indole-3-acetic acid in suspension-cultured tobacco cells. Planta 198: 532–541 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M (2005) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B (1993) Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84 [DOI] [PubMed] [Google Scholar]
- Einhellig FA. (1995) Mechanism of action of allelochemicals in allelopathy. Allelopathy 582: 96–116 [Google Scholar]
- Evidente A, Cicala MR, Randazzo G, Riccio R, Calabrese G, Liso R, Arrigoni O (1983) Lycorine structure activity relationships. Phytochemistry 22: 2193–2196 [Google Scholar]
- Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L (2004) Metabolite profiling: from diagnostics to systems biology. Nat Rev Mol Cell Biol 5: 763–769 [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M (2002) Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J 29: 153–168 [DOI] [PubMed] [Google Scholar]
- Funk C, Brodelius PE (1992) Phenylpropanoid metabolism in suspension cultures of Vanilla planifolia Andr.: IV. Induction of vanillic acid formation. Plant Physiol 99: 256–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk C, Brodelius PE (1994) Vanilla planifolia Andrews: in vitro biosynthesis of vanillin and other phenylpropanoid derivatives. In Medicinal and Aromatic Plants VI, Biotechnology in Agriculture and Forestry 26: 377–402 [Google Scholar]
- Gallego-Giraldo L, Escamilla-Trevino L, Jackson LA, Dixon RA (2011) Salicylic acid mediates the reduced growth of lignin down-regulated plants. Proc Natl Acad Sci USA 108: 20814–20819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner J, Andreeva Z, Barton D, Ritchie A, Overall R, Marc J (2008) The phospholipase A inhibitor, aristolochic acid, disrupts cortical microtubule arrays and root growth in Arabidopsis. Plant Biol (Stuttg) 10: 725–731 [DOI] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M (1999) Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev 13: 1678–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann K. (2010) Auxin herbicides: current status of mechanism and mode of action. Pest Manag Sci 66: 113–120 [DOI] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T (2004) Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131: 657–668 [DOI] [PubMed] [Google Scholar]
- Hara M, Kurita I (2014) The natural alkaloid sanguinarine promotes the expression of heat shock protein genes in Arabidopsis. Acta Physiol Plant 36: 3337–3343 [Google Scholar]
- Hartung AC, Nair MG, Putnam AR (1990) Isolation and characterization of phytotoxic compounds from asparagus (Asparagus officinalis L.) roots. J Chem Ecol 16: 1707–1718 [DOI] [PubMed] [Google Scholar]
- Heyman J, Cools T, Vandenbussche F, Heyndrickx KS, van Leene J, Vercauteren I, Vanderauwera S, Vandepoele K, De Jaeger G, van der Straeten D, de Veylder L (2013) ERF115 controls root quiescent center cell division and stem cell replenishment. Science 342: 860–863 [DOI] [PubMed] [Google Scholar]
- Heyman J, Kumpf RP, de Veylder L (2014) A quiescent path to plant longevity. Trends Cell Biol 24: 443–448 [DOI] [PubMed] [Google Scholar]
- Hosek P, Kubes M, Lanková M, Dobrev PI, Klíma P, Kohoutová M, Petrásek J, Hoyerová K, Jirina M, Zazímalová E (2012) Auxin transport at cellular level: new insights supported by mathematical modelling. J Exp Bot 63: 3815–3827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Yang L, Na X, You J, Hu W, Liang X, Liu J, Mao L, Wang X, Wang H, Bi Y (2012) Narciclasine inhibits the responses of Arabidopsis roots to auxin. Planta 236: 597–612 [DOI] [PubMed] [Google Scholar]
- Knobloch KH, Hahlbrock K (1977) 4-Coumarate:CoA ligase from cell suspension cultures of Petroselinum hortense Hoffm. Partial purification, substrate specificity, and further properties. Arch Biochem Biophys 184: 237–248 [DOI] [PubMed] [Google Scholar]
- Kuhn BM, Errafi S, Bucher R, Dobrev P, Geisler M, Bigler L, Zažímalová E, Ringli C (2016) 7-Rhamnosylated flavonols modulate homeostasis of the plant hormone auxin and affect plant development. J Biol Chem 291: 5385–5395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafite P, Dijols S, Zeldin DC, Dansette PM, Mansuy D (2007) Selective, competitive and mechanism-based inhibitors of human cytochrome P450 2J2. Arch Biochem Biophys 464: 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer GA, Garrison SA (1977) Effect of asparagus tissue on seed-germination and asparagus seedling growth; possible allelopathic interactions. HortScience 12: 385 [Google Scholar]
- Lee S, Sundaram S, Armitage L, Evans JP, Hawkes T, Kepinski S, Ferro N, Napier RM (2014) Defining binding efficiency and specificity of auxins for SCF(TIR1/AFB)-Aux/IAA co-receptor complex formation. ACS Chem Biol 9: 673–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Chen F, Gallego-Giraldo L, Dixon RA, Voit EO (2011) Integrative analysis of transgenic alfalfa (Medicago sativa L.) suggests new metabolic control mechanisms for monolignol biosynthesis. PLOS Comput Biol 7: e1002047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser HMO, Pickett FB, Dharmasiri S, Estelle M (1996) Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J 10: 403–413 [DOI] [PubMed] [Google Scholar]
- Li X, Bonawitz ND, Weng JK, Chapple C (2010) The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell 22: 1620–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Mayer U, Jürgens G (1996) Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 84: 61–71 [DOI] [PubMed] [Google Scholar]
- Na X, Hu Y, Yue K, Lu H, Jia P, Wang H, Wang X, Bi Y (2011) Narciclasine modulates polar auxin transport in Arabidopsis roots. J Plant Physiol 168: 1149–1156 [DOI] [PubMed] [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S (1992) Tobacco BY-2 cell line as the Hela cell in the cell biology of higher plants. Int Rev Cytol 132: 1–30 [Google Scholar]
- Nakagawa A, Takahashi H, Kojima S, Sato N, Ohga K, Cha BY, Woo JT, Nagai K, Horiguchi G, Tsukaya H, Machida Y, Machida C (2012) Berberine enhances defects in the establishment of leaf polarity in asymmetric leaves1 and asymmetric leaves2 of Arabidopsis thaliana. Plant Mol Biol 79: 569–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseer S, Lee Y, Lapierre C, Franke R, Nawrath C, Geldner N (2012) Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc Natl Acad Sci USA 109: 10101–10106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák O, Hényková E, Sairanen I, Kowalczyk M, Pospíšil T, Ljung K (2012) Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J 72: 523–536 [DOI] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono E, Nakai M, Fukui Y, Tomimori N, Fukuchi-Mizutani M, Saito M, Satake H, Tanaka T, Katsuta M, Umezawa T, Tanaka Y (2006) Formation of two methylenedioxy bridges by a Sesamum CYP81Q protein yielding a furofuran lignan, (+)-sesamin. Proc Natl Acad Sci USA 103: 10116–10121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde P, Fukaki H, Beeckman T (2010) Auxin control of root development. Cold Spring Harb Perspect Biol 2: a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA, Murphy AS (2007) Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci 12: 556–563 [DOI] [PubMed] [Google Scholar]
- Petrásek J, Cerná A, Schwarzerová K, Elckner M, Morris DA, Zazímalová E (2003) Do phytotropins inhibit auxin efflux by impairing vesicle traffic? Plant Physiol 131: 254–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrásek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertová D, Wisniewska J, Tadele Z, Kubes M, Covanová M, Dhonukshe P, Skupa P, et al. (2006) PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918 [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Winter CM, Benfey PN (2012) Control of Arabidopsis root development. Annu Rev Plant Biol 63: 563–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam AR. (1988) Allelochemicals from plants as herbicides. Weed Technol 2: 510–518 [Google Scholar]
- Reddy AS, Zhang S (2013) Polypharmacology: drug discovery for the future. Expert Rev Clin Pharmacol 6: 41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano CP, Hein MB, Klee HJ (1991) Inactivation of auxin in tobacco transformed with the indoleacetic acid-lysine synthetase gene of Pseudomonas savastanoi. Genes Dev 5: 438–446 [DOI] [PubMed] [Google Scholar]
- Schalk M, Cabello-Hurtado F, Pierrel MA, Atanossova R, Saindrenan P, Werck-Reichhart D (1998) Piperonylic acid, a selective, mechanism-based inactivator of the trans-cinnamate 4-hydroxylase: a new tool to control the flux of metabolites in the phenylpropanoid pathway. Plant Physiol 118: 209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Stout J, Weng JK, Humphreys J, Ruegger MO, Chapple C (2009) Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J 60: 771–782 [DOI] [PubMed] [Google Scholar]
- Seifertová D, Skůpa P, Rychtář J, Laňková M, Pařezová M, Dobrev PI, Hoyerová K, Petrášek J, Zažímalová E (2014) Characterization of transmembrane auxin transport in Arabidopsis suspension-cultured cells. J Plant Physiol 171: 429–437 [DOI] [PubMed] [Google Scholar]
- Takatsuka H, Umeda M (2014) Hormonal control of cell division and elongation along differentiation trajectories in roots. J Exp Bot 65: 2633–2643 [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LIA, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 448: 640–645 [DOI] [PubMed] [Google Scholar]
- Trifunovic S, Vajs V, Tesevic V, Djokovic D, Milosavljevic S (2003) Lignans from the plant species Achillea lingulata. J Serb Chem Soc 68: 277–280 [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Wouwer D, Vanholme R, Decou R, Goeminne G, Audenaert D, Nguyen L, Höfer R, Pesquet E, Vanholme B, Boerjan W (2016) Chemical genetics uncovers novel inhibitors of lignification, including p-iodobenzoic acid targeting CINNAMATE-4-HYDROXYLASE. Plant Physiol 172: 198–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Storme V, Vanholme B, Sundin L, Christensen JH, Goeminne G, Halpin C, Rohde A, Morreel K, Boerjan W (2012) A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell 24: 3506–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker SL, Lachenbruch B, Meinzer FC, Jourdes M, Ki C, Patten AM, Davin LB, Lewis NG, Tuskan GA, Gunter L, Decker SR, Selig MJ, et al. (2010) Antisense down-regulation of 4CL expression alters lignification, tree growth, and saccharification potential of field-grown poplar. Plant Physiol 154: 874–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T. (2010) Phenylpropanoid biosynthesis. Mol Plant 3: 2–20 [DOI] [PubMed] [Google Scholar]
- Weir TL, Park SW, Vivanco JM (2004) Biochemical and physiological mechanisms mediated by allelochemicals. Curr Opin Plant Biol 7: 472–479 [DOI] [PubMed] [Google Scholar]
- Went FW. (1949) Phytohormones; structure and physiological activity. Arch Biochem 20: 131–136 [PubMed] [Google Scholar]
- Yang HJ. (1982) Autotoxicity of Asparagus officinalis L. J Am Soc Hortic Sci 107: 860–862 [Google Scholar]
- Yin R, Han K, Heller W, Albert A, Dobrev PI, Zažímalová E, Schäffner AR (2014) Kaempferol 3-O-rhamnoside-7-O-rhamnoside is an endogenous flavonol inhibitor of polar auxin transport in Arabidopsis shoots. New Phytol 201: 466–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young CC, Chou TC (1984) Autotoxication in residues of Asparagus officinalis L. Plant Soil 85: 385–393 [Google Scholar]
- Zeng RS. (2014) Allelopathy—the solution is indirect. J Chem Ecol 40: 515–516 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.