SVR9 and its homolog SVR9L1 encode functionally redundant chloroplast translation initiation factors essential for chloroplast and leaf development in Arabidopsis.
Abstract
Chloroplast development requires the coordinated expressions of nuclear and chloroplast genomes, and both anterograde and retrograde signals exist and work together to facilitate this coordination. We have utilized the Arabidopsis yellow variegated (var2) mutant as a tool to dissect the genetic regulatory network of chloroplast development. Here, we report the isolation of a new (to our knowledge) var2 genetic suppressor locus, SUPPRESSOR OF VARIEGATION9 (SVR9). SVR9 encodes a chloroplast-localized prokaryotic type translation initiation factor 3 (IF3). svr9-1 mutant can be fully rescued by the Escherichia coli IF3 infC, suggesting that SVR9 functions as a bona fide IF3 in the chloroplast. Genetic and molecular evidence indicate that SVR9 and its close homolog SVR9-LIKE1 (SVR9L1) are functionally interchangeable and their combined activities are essential for chloroplast development and plant survival. Interestingly, we found that SVR9 and SVR9L1 are also involved in normal leaf development. Abnormalities in leaf anatomy, cotyledon venation patterns, and leaf margin development were identified in svr9-1 and mutants that are homozygous for svr9-1 and heterozygous for svr9l1-1 (svr9-1 svr9l1-1/+). Meanwhile, as indicated by the auxin response reporter DR5:GUS, auxin homeostasis was disturbed in svr9-1, svr9-1 svr9l1-1/+, and plants treated with inhibitors of chloroplast translation. Genetic analysis established that SVR9/SVR9L1-mediated leaf margin development is dependent on CUP-SHAPED COTYLEDON2 activities and is independent of their roles in chloroplast development. Together, our findings provide direct evidence that chloroplast IF3s are essential for chloroplast development and can also regulate leaf development.
One of the most remarkable achievements in nature is the evolution of the semiautonomous organelles including chloroplasts and mitochondria in eukaryotic cells via endosymbiosis (Dyall et al., 2004; Jensen and Leister, 2014). Current-day chloroplasts are estimated to contain approximately 3000 proteins, similar to their cyanobacterial ancestors (Leister, 2003). However, during the endosymbiotic process, the vast majority of genes encoding chloroplast proteins has been transferred to the nucleus and became part of the nuclear genome, and consequently higher plants have evolved regulatory pathways that exert control over these nuclear genes for chloroplast proteins (Woodson and Chory, 2008). The fine coordination between the nucleus and the chloroplast has been fulfilled via two-way communications from the nucleus to the chloroplast (anterograde) and also from the chloroplast to the nucleus (retrograde; Nott et al., 2006; Woodson and Chory, 2012; Chi et al., 2013).
Much of what we know regarding retrograde signaling stemmed from the regulation of the expressions of nuclear genes for chloroplast proteins by the functional and developmental states of the chloroplast. In wild-type Arabidopsis (Arabidopsis thaliana) seedlings, carotenoid biosynthesis inhibitor norflurazon (NF) can induce photobleaching of the chloroplast and trigger the massive down-regulation of nuclear genes for chloroplast proteins such as CHLOROPHYLL A/B BINDING PROTEIN (Nott et al., 2006; Chi et al., 2013). Taking advantage of this NF-triggered retrograde response, a series of genomes uncoupled (gun) mutants that retained CHLOROPHYLL A/B BINDING PROTEIN expressions upon NF treatment have been identified (Susek et al., 1993; Koussevitzky et al., 2007). Based on the GUN and other work, the chlorophyll biosynthetic precursor, Mg-protoporphyrin IX, has been shown as one molecule that potentially serves as the signal molecule in the NF-triggered retrograde signaling, although there are reports arguing against this notion (Mochizuki et al., 2008; Moulin et al., 2008). AP2 type transcription factor ABA INSENSITIVE4 has been identified as a nuclear transcription repressor that executes retrograde signaling downstream of chloroplast GUNs (Koussevitzky et al., 2007). Additional retrograde signals and signaling components have also been identified. A screen with low concentration of NF and light intensity has identified additional happy on norflurazon mutants (Saini et al., 2011). A chloroplast envelope-anchored plant homeodomain transcription factor PTM may be involved in signal transduction from the plastid to the nucleus and PTM was shown to be able to bind the promoter of ABA INSENSITIVE4 and activate its expression (Sun et al., 2011). The physiological states of the chloroplast can also trigger signaling to the nucleus (Wilson et al., 2009; Leister, 2012). For example, the redox state of the photosynthetic electron transfer chain has long been thought of as a generator of chloroplast-derived signals (Pesaresi et al., 2009; Kindgren et al., 2012). Reactive oxygen species such as singlet oxygen generated from chloroplasts are also able to trigger nuclear gene expression responses, via an EXECUTER1 and 2-mediated pathway (Wagner et al., 2004; Lee et al., 2007; Galvez-Valdivieso and Mullineaux, 2010; Ramel et al., 2012). Recently, a plastid isoprenoid biosynthetic intermediate, methylerythritol phosphate, was shown to be a new retrograde signal and able to trigger the expression of stress related nuclear genes such as HYDROPEROXIDE LYASE (Xiao et al., 2012). Plastid gene expression has also been proposed as a source of retrograde signals (Gray et al., 2003). Chloroplast translation inhibitors such as lincomycin could trigger the down-regulation of photosynthesis-associated nuclear genes, which may be part of the broader plastid gene expression-mediated retrograde pathway (Pesaresi et al., 2006).
Besides these retrograde signaling pathways that often use the expressions of nuclear genes for chloroplast proteins as the readout, there is also evidence suggesting other modes of retrograde regulation, for example, the influence on overall plant growth and development by the functional and developmental states of the chloroplast (Hricová et al., 2006; Fleischmann et al., 2011; Tiller and Bock; 2014). In the Arabidopsis immutans mutant and the tomato (Lycopersicon esculentum (L.) Mill.) ghost mutant, both defective in the plastid alternative oxidase and displaying distinctive green-white leaf variegation phenotypes, there are conspicuous leaf mesophyll cell developmental defects in white leaf sectors (Josse et al., 2000; Aluru et al., 2001). Some Arabidopsis albino mutants such as pale cress and cla1 were also shown to have leaves that lack the characteristic palisade tissue (Reiter et al., 1994; Mandel et al., 1996). In contrast to the rapid progressing of the overall field of plastid retrograde signaling, our understanding of the regulation of plant and leaf development by the developmental and functional states of the chloroplast has remained descriptive.
We have used the Arabidopsis leaf variegation mutant var2 as a tool to probe the genetic regulation of chloroplast development (Liu et al., 2010b, 2013; Qi et al., 2016). In this work, we report the identification of a var2 genetic suppressor gene SUPPRESSOR OF VARIEGATION9 (SVR9). Molecular cloning and functional complementation tests established that SVR9 and its homolog SVR9-LIKE1 (SVR9L1) act redundantly as chloroplast translation initiation factor 3 (IF3). Down-regulation of chloroplast IF3s’ activities not only caused chloroplast development defects, but also led to a series of leaf developmental abnormalities including more serrated leaf margin, disorganized mesophyll cells, and altered cotyledon venation patterns. We further demonstrated that auxin homeostasis is disturbed in mutants defective in chloroplast IF3s or in plants treated with chloroplast translation inhibitors. Genetic evidence suggests that the NAC family transcription factor CUP-SHAPED COTYLEDON2 (CUC2) is involved in the chloroplast IF3-mediated leaf margin development (Nikovics et al., 2006). Our results suggest that chloroplast translation initiation factors SVR9 and SVR9L1 are required not only for chloroplast development and var2-mediated leaf variegation, but also are involved in the coordination of leaf and chloroplast development.
RESULTS
The Isolation and Cloning of an Extragenic var2 Suppressor Locus SVR9
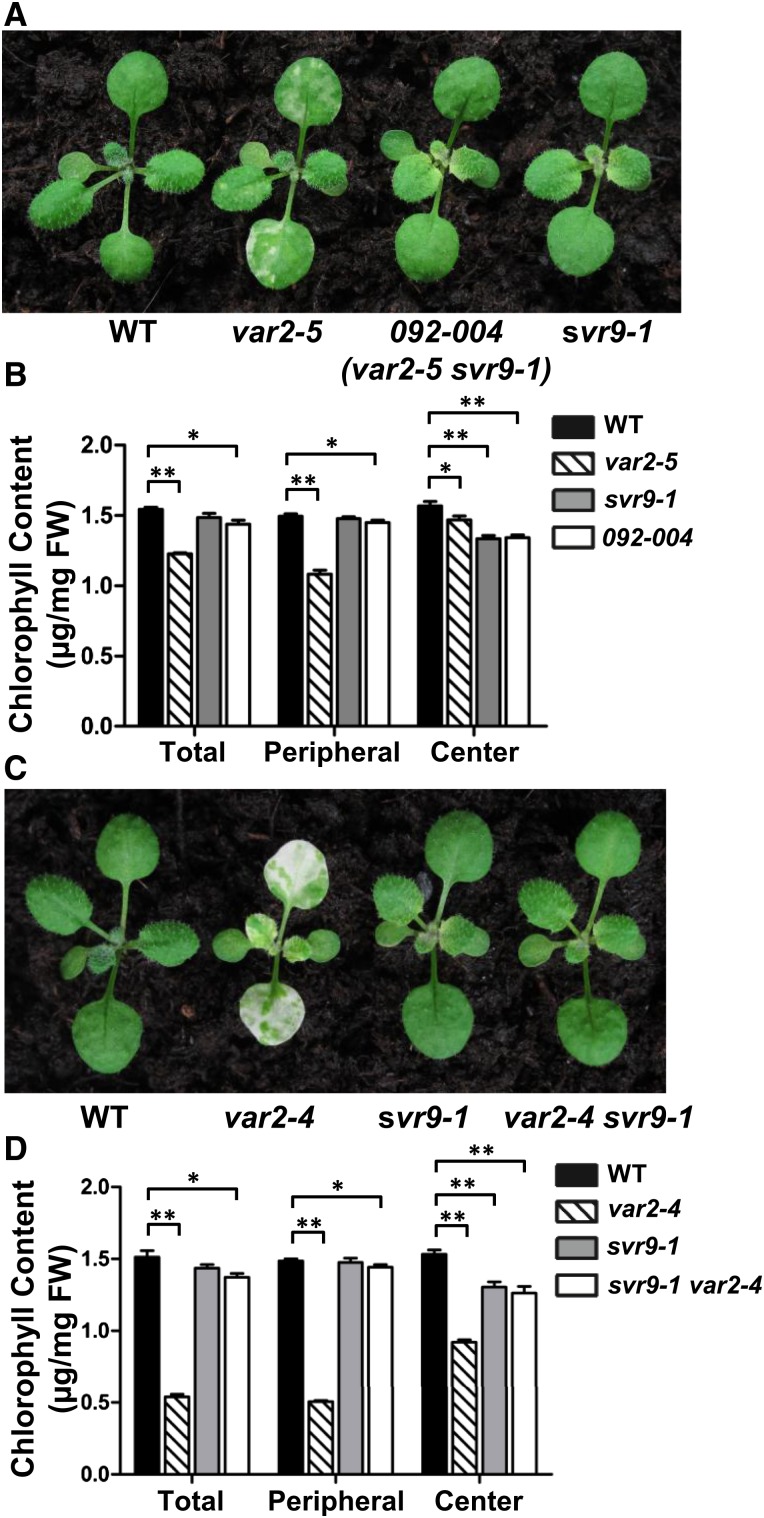
We have isolated and characterized genetic suppressors of the var2 leaf variegation mutant through an activation tagging screen in the var2-5 mutant background (Liu et al., 2010b; Putarjunan et al., 2013). Here we report the isolation of a new var2-5 suppressor mutant designated 092-004 (Fig. 1A). Following our naming system, the suppressor locus in 092-004 was named SVR9, and the genotype of 092-004 was var2-5 svr9-1 and the suppressor single mutant was svr9-1. Phenotypically, svr9-1 showed a distinctive virescent phenotype, i.e. young and emerging leaf tissues displayed pronounced chlorosis while these yellow tissues gradually turned green as mutant leaves matured (Fig. 1A). Consistent with the virescent appearance, chlorophylls accumulated at lower levels in young and emerging tissues, but increased in mature tissues in svr9-1 and 092-004 (Fig. 1B). The suppression of var2 leaf variegation by svr9-1 was not allele-specific as svr9-1 can suppress a stronger var2 allele, var2-4 (Fig. 1, C and D).
Figure 1.
Suppression of var2 leaf variegation by svr9-1. A, Representative 15-d-old wild-type, var2-5, 092-004 (var2-5 svr9-1), and svr9-1 plants. C, Representative 15-d-old wild type, var2-4, svr9-1, and var2-4 svr9-1 double mutant. B and D, Chlorophyll contents of indicated tissues in plants shown in (A) and (C), respectively. Total: Entire rosettes excluding the cotyledons; peripheral: the first two true leaves; center: rosettes excluding the cotyledons and the first two true leaves. Chlorophyll measurements were normalized on a fresh tissue weight basis. Data were presented as mean ± sd of three biological replicates. *0.01 < P < 0.05; **P < 0.01. WT, Wild type.
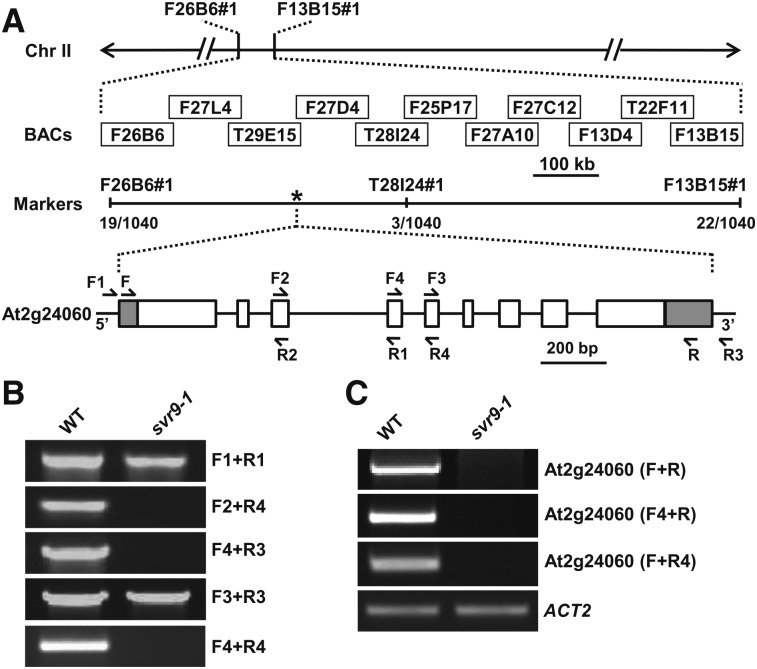
Initial cosegregation analyses judged by herbicide Basta resistance indicated that svr9-1 was likely linked with T-DNA, but we were not able to identify the T-DNA insertion site via various techniques. We thus utilized a map-based cloning procedure to identify SVR9 (Fig. 2). Bulked segregant analysis and subsequent fine mapping narrowed SVR9 to an approximately 415 kb region on chromosome 2 (Fig. 2A). Based on the svr9-1 phenotype, we reasoned that SVR9 likely codes for a chloroplast-localized protein. DNA sequences of potential nuclear genes for chloroplast proteins in the interval were examined. Surprisingly, PCRs failed to amplify genomic regions around intron 4 of At2g24060 in svr9-1, suggesting At2g24060 genomic sequences may be altered in svr9-1 (Fig. 2B). Consistent with an abnormality at genomic DNA level, At2g24060 transcripts were not detected in svr9-1 (Fig. 2C). These results indicated that At2g24060 is disrupted in svr9-1.
Figure 2.
Cloning of SVR9. A, Schematic representation of the positional cloning procedure used to identify svr9-1 mutation. Primers of the molecular markers designed in this study were listed in Supplemental Table S2. A total of 570 F2 plants (1140 chromosomes) was used in fine mapping. Numbers of recombinants were marked under each molecular marker. The asterisk indicated the position of SVR9 (At2g24060). In the gene model, boxes represent exons, and lines represent introns. 5′ and 3′ untranslated regions were shaded. Arrows represent the positions of the primers used in (B) and (C). B, PCR analyses using various primer combinations indicated that At2g24060 genomic regions were disrupted in svr9-1. C, Semiquantitative RT-PCR analyses of the accumulations of SVR9 transcripts in wild type and svr9-1 using indicated primer combinations. Expression of ACT2 was used as an internal control.
SVR9 Encodes a Chloroplast Translation IF3
To confirm that the virescent phenotype of svr9-1 and the suppression of var2-5 were indeed due to a potential disruption of At2g24060, we carried out complementation tests. Independent transgenic lines overexpressing At2g24060 under the 35S promoter were able to reverse the virescent phenotype of svr9-1 and restore the leaf variegation phenotype in 092-004, thus confirming that At2g24060 is SVR9 (Fig. 3A).
Figure 3.
Complementation of svr9-1 and 092-004. A, Leaf-color phenotypes of 2-week-old wild type, svr9-1, var2-5, 092-004, and representative lines overexpressing At2g24060 in svr9-1 or 092-004 background. B, Leaf-color phenotypes of 2-week-old wild type, svr9-1, var2-5, 092-004, and representative lines overexpressing CTPSVR9-infC in svr9-1 or 092-004 background. WT, Wild type.
SVR9/At2g24060 was annotated to encode a putative chloroplast protein of 312 amino acids. SVR9 shows high homologies with translation IF3 proteins in prokaryotes (Supplemental Fig. S1). To test whether the virescent phenotype of svr9-1 was indeed caused by the lack of a chloroplast IF3, we carried out functional complementation tests using the Escherichia coli IF3 protein infC (Sacerdot et al., 1982). First, we established that the putative chloroplast transit peptide of SVR9 (cTPSVR9, N-terminal 1-71 amino acid residues of SVR9) was sufficient to guide GFP into the chloroplast (Supplemental Fig. S2). Then a binary vector containing a chimeric gene with the cTPSVR9 fused at the N terminus of E. coli infC under the control of the 35S promoter was generated (P35S:cTPSVR9-infC) and transformed into svr9-1. Independent transgenic plants showed green leaf coloration, indicating that cTPSVR9-infC can functionally replace SVR9 (Fig. 3B). Moreover, when P35S:cTPSVR9-infC was introduced into 092-004, transgenic plants showed leaf variegation phenotype resembled that of var2-5, suggesting also a functional complementation (Fig. 3B). These results indicate that E. coli infC can substitute for SVR9 and the phenotype of svr9-1 and the suppression of var2-5 leaf variegation by svr9-1 were caused by the lack of a chloroplast translation IF3.
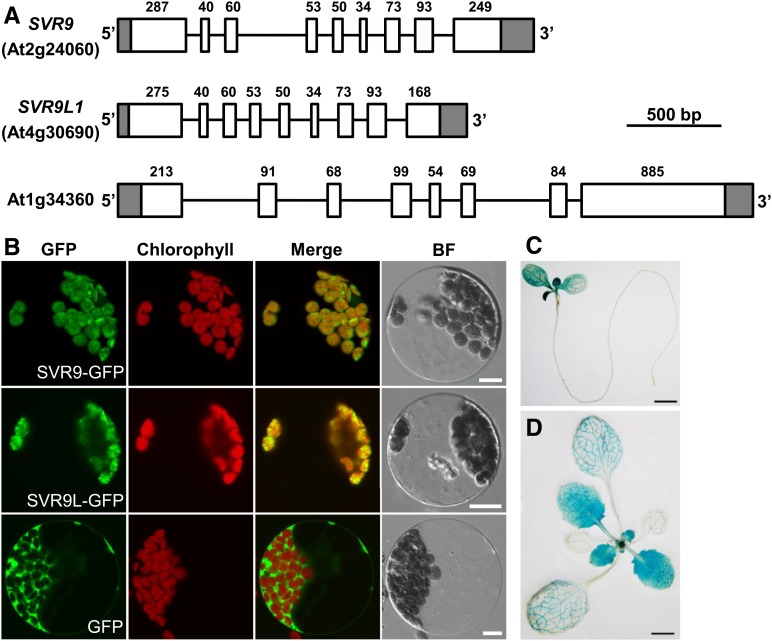
Phylogenetic analysis of SVR9-LIKE proteins in various photosynthetic organisms showed that higher plant chloroplasts often contain more than one IF3 coding gene (Supplemental Fig. S3). In Arabidopsis, two other genes, At4g30690 and At1g34360, also potentially code for prokaryotic IF3-like proteins (Fig. 4A; Nesbit et al., 2015). Of the three genes, SVR9 and At4g30690 code for putative chloroplast proteins and share the same gene structures, and approximately 78% identities in amino acid sequences (Fig. 4A; Supplemental Fig. S1 and Supplemental Fig. S3). We thus named At4g30690 as SVR9-LIKE1 (SVR9L1). In contrast, At1g34360 codes for a mitochondrial protein and its gene structure was different from that of SVR9 or At4g30690 (Fig. 4A; Nesbit et al., 2015). To confirm the chloroplast localizations of SVR9 and SVR9L1, constructs expressing C-terminal GFP fusion proteins SVR9-GFP and SVR9L1-GFP were generated and used to transform Arabidopsis wild-type leaf protoplasts. When GFP alone is expressed, green fluorescent signals were mainly detected in the cytosol (Fig. 4B). On the other hand, protoplasts expressing SVR9-GFP or SVR9L1-GFP showed signals that clearly aligned with chlorophyll autofluorescence, confirming the chloroplast localization of both proteins (Fig. 4B). Taken together, our results demonstrate that SVR9 and SVR9L1 encode the chloroplast translation IF3 proteins.
Figure 4.
SVR9 and SVR9-LIKE genes in Arabidopsis. A, Gene models of three Arabidopsis genes coding for putative prokaryotic type IF3 proteins. Gene models were drawn as in Fig. 2A. Numbers of nucleotides of each exon excluding the lengths of untranslated regions were labeled on top of each exon. B, Transient expression of SVR9-GFP, SVR9L1-GFP, or GFP alone in wild-type Arabidopsis leaf protoplasts. A single representative protoplast was shown for each transformation. Merged images from GFP and chlorophyll channels are shown in the “Merge” lane. BF, Bright field. Bars, 10 μm. C and D, Tissue expression patterns of SVR9. Illustrated are GUS staining of 6-d-old (C) and 2-week-old (D) transgenic plants expressing the transcriptional fusion of PSVR9:GUS.
Lastly, we examined the tissue expression pattern of SVR9. An approximately 1.9 kb promoter region upstream of SVR9 start codon was transcriptionally fused with the GUS gene and introduced into wild-type Arabidopsis. Histochemical GUS staining was performed in PSVR9:GUS lines at various developmental stages. In general, strong GUS activities were detected in photosynthetic tissues while GUS levels were below the detection limit in roots (Fig. 4C). In addition, SVR9 showed stronger expressions in young, newly emerged leaves as compared to the gradually reducing expressions in older leaves (Fig. 4, C and D). These observations suggest that the expression of SVR9 is regulated by leaf developmental stages and the need for SVR9 activities is probably higher in actively growing young tissues, consistent with the virescent mutant phenotypes.
Genetic Interactions among svr9-2, svr9l1-1, and var2
To further dissect the functions of SVR9 and SVR9L1, and their interactions with var2, we sought for additional mutant alleles of these two genes. For SVR9, we obtained a SAIL T-DNA insertion line SAIL_172_F02 and confirmed that the T-DNA was located in the last exon of SVR9, 175 bp upstream of the stop codon, and this allele was named svr9-2 (Supplemental Fig. S4). For SVR9L1, we obtained a SALK T-DNA insertion line SALK_140431C, which harbored a T-DNA in the sixth intron, and this mutant allele was named svr9l1-1 (Supplemental Fig. S4). In contrast to the virescent phenotype of svr9-1, svr9-2 showed an overall wild-type-like phenotype and reduced accumulation of SVR9 transcripts, suggesting that it is a weaker allele of SVR9 (Supplemental Fig. S4). Similarly, svr9l1-1 mutants were also indistinguishable from wild type (Supplemental Fig. S4). Double mutants of svr9-2 and svr9l1-1 also resembled wild type (Supplemental Fig. S4).
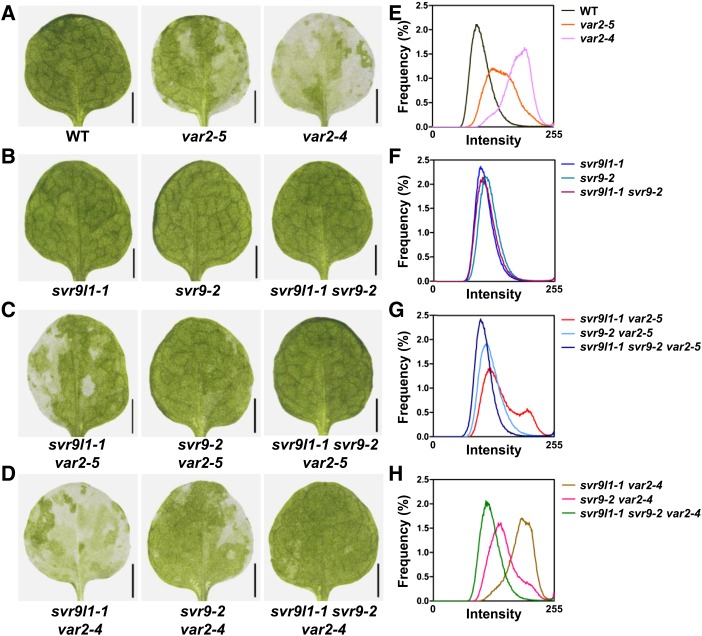
We next examined the genetic interactions between svr9-2, svr9l1-1, and two var2 mutant alleles, var2-4 and var2-5 (Supplemental Fig. S5). To evaluate the extent of leaf variegation in a quantitative manner, images of the first true leaves from each genotype were converted to grayscale and distributions of the pixel intensity values of these images were generated and compared. Green sectors of a leaf will have a lower pixel intensity value while white sectors contain pixels with higher intensity values. Uniform leaf color will give rise to a distribution with a relatively narrow peak. In contrast, variegated leaf images have broader peaks and contain more pixels of higher intensity values. Analysis of wild-type, var2-5, and var2-4 leaf images showed that they each display a distinctive pattern of pixel intensity histogram that correlates well with the degree of variegation (Fig. 5, A and E). On the other hand, svr9l1-1, svr9-2, and svr9l1-1 svr9-2 leaves are not only visually indistinguishable from wild type but also have pixel intensity histograms similar to that of wild type (Fig. 5, B and F). Double mutants of svr9l1-1 with var2-5 or var2-4 showed leaf variegation that resembled var2-5 and var2-4, respectively, suggesting that svr9l1-1 alone does not significantly alter var2 leaf variegation (Fig. 5, C and D). In contrast, double mutants of svr9-2 with var2-5 or var2-4 showed reduced leaf variegation than var2-5 and var2-4, respectively, suggesting that a weaker allele of SVR9 can partially suppress var2 leaf variegation (Fig. 5, C and D). The stronger effects of svr9-2 compared to svr9l1-1 in suppressing var2 variegation were also clearly reflected by the shifts observed in the histograms of svr9-2 var2 and svr9l1-1 var2 double mutant leaf images (Fig. 5, G and H). Interestingly, leaf variegation was almost absent in svr9l1-1 svr9-2 var2 triple mutants (Fig. 5, C, D, G, and H). The fact that var2 leaf variegation was suppressed to a stronger degree in the svr9l1-1 svr9-2 background than in the svr9-2 mutant alone suggested an additive effect of svr9l1-1 and svr9-2 on suppressing var2 leaf variegation. Together, these data suggest that despite their overall wild-type appearances, svr9l1-1, svr9-2, and svr9l1-1 svr9-2 mutants possess molecular phenotypes, and these phenotypes can modify var2 leaf variegation.
Figure 5.
Genetic interactions among var2, svr9-2, and svr9l1-1. A to D, The first true leaves of representative 2-week-old wild type, var2-5, and var2-4 (A); svr9l1-1, svr9-2, and svr9l1-1 svr9-2 double mutant (B); svr9l1-1 var2-5 double mutant, svr9-2 var2-5 double mutant, and svr9l1-1 svr9-2 var2-5 triple mutant (C); and svr9l1-1 var2-4 double mutant, svr9-2 var2-4 double mutant, and svr9l1-1 svr9-2 var2-4 triple mutant (D). E to H, Quantitative comparison of leaf variegation based on the frequency distributions of the pixel intensity values of leaf images shown in (A) to (D). WT, Wild type.
Chloroplast Translation IF3 Activities Are Essential for Plant Development
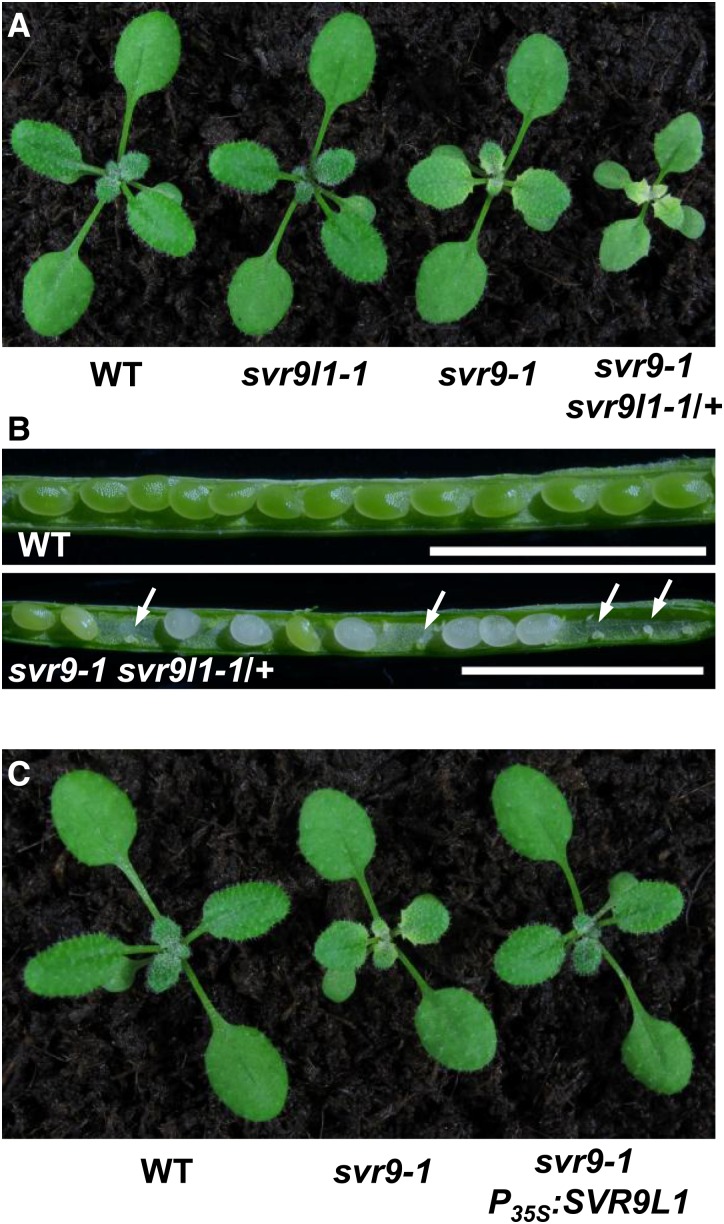
Given the high homology between SVR9 and SVR9L1, we next tested the functional relationship between these two genes. As svr9-2 svr9l1-1 double mutants resembled wild type, we sought to obtain svr9-1 svr9l1-1 double mutants. However, extensive genotyping did not identify double mutants homozygous for both svr9-1 and svr9l1-1. We did recover mutant plants that were homozygous for svr9-1 but heterozygous for svr9l1-1 (referred to as svr9-1 svr9l1-1/+ hereafter), which showed a stronger virescent phenotype and smaller stature than svr9-1 alone (Fig. 6A). Genotyping the selfed progeny of svr9-1 svr9l1-1/+ did not yield the expected double mutants either. In the siliques of svr9-1 svr9l1-1/+ plants, we observed an increased occurrence of abortive seeds (Fig. 6B). In addition, we generated transgenic plants in svr9-1 background that expressed SVR9L1 under the control of the 35S promoter. The overexpression of SVR9L1 effectively rescued the virescent phenotype of svr9-1 (Fig. 6C). These data suggest that: (1) SVR9 and SVR9L1 are functionally redundant; (2) svr9-1 svr9l1-1 is likely lethal; and (3) chloroplast IF3 activities are essential for chloroplast and plant development.
Figure 6.
Functional redundancy of SVR9 and SVR9L1. A, Overall phenotypes of 2-week-old wild type, svr9l1-1, svr9-1, and svr9-1 svr9l1-1/+. B, Seed settings of wild type and svr9-1 svr9l1-1/+. Developing siliques of wild type and svr9-1 svr9l1-1/+ at the same stage were dissected and photographed with a stereoscope. White arrows indicated the abolished embryos in svr9-1 svr9l1-1/+ silique. Bars, 2.5 mm. C, Overall phenotypes of 2-week-old wild type, svr9-1, and a representative line overexpressing SVR9L1 in the svr9-1 background. WT, Wild type.
Compromised Chloroplast Translation IF3 Activities Lead to Leaf Developmental Defects
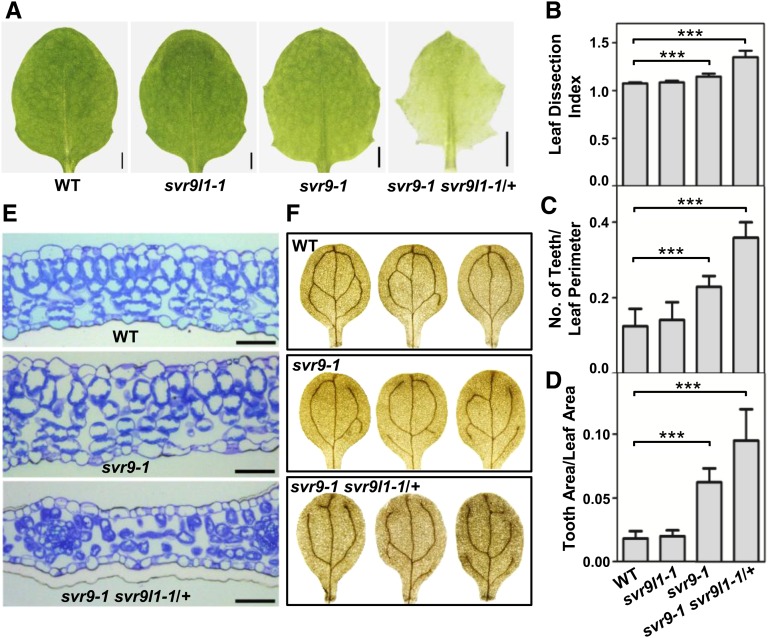
During the investigation of mutant alleles of SVR9 and SVR9L1 and their genetic interactions, we noticed a number of leaf developmental defects in svr9-1 and svr9-1 svr9l1-1/+. First, leaf margin development was perturbed in these mutants. svr9-1 displayed a more serrated leaf margin compared to that of wild type (Fig. 7A). Leaf margins of svr9l1-1 resembled wild type, consistent with its wild-type appearance (Fig. 7A). However, svr9-1 svr9l1-1/+ plants showed an even more pronounced leaf margin serration compared to either of the single mutants (Fig. 7A). The increased leaf serrations from svr9-1 to svr9-1 svr9l1-1/+ was further validated by the quantitative analysis of leaf shape based on three different parameters, the leaf dissection index (perimeter2/4π × leaf area), the number of teeth/leaf perimeter, and the tooth area/leaf area (Royer et al., 2008; Bilsborough et al., 2011; Fig. 7, B–D). These analyses suggest that decreased activities of chloroplast IF3s promote the initiation and the outgrowth of leaf lobes. However, we did not observe major leaf serration changes in svr8-2 (a null mutant lacking the chloroplast ribosomal protein L24), suggesting that not all mutants of chloroplast translation display abnormal leaf margin development (Supplemental Fig. S6; Liu et al., 2013).
Figure 7.
Leaf development phenotypes in svr9-1, svr9l1-1, and svr9-1 svr9l1-1/+ mutants. A, Photographs of the third true leaves of representative 2-week-old wild type, svr9l1-1, svr9-1, and svr9-1 svr9l1-1/+. Bars, 1 mm. B to D, Quantitative comparisons of leaf shapes in wild type, svr9l1-1, svr9-1, and svr9-1 svr9l1-1/+ based on the leaf dissection index (perimeter2/4π × leaf area) (B), the number of teeth/leaf perimeter (C), and the tooth area/leaf area (D). All measurements were performed on the third true leaves of 2-week-old plants. Ten leaves of each genotype were included in the statistical analysis. Data were presented as mean ± sd. ***P < 0.001. E, Cross sections of the basal part of the first true leaves of 10-d-old wild type, svr9-1, and svr9-1 svr9l1-1/+. Bars, 50 μm. F, Common cotyledon vein patterns observed in wild type, svr9-1, and svr9-1 svr9l1-1/+. WT, Wild type.
Second, we examined the leaf anatomy of these mutants. Cross sections of wild-type leaves reveal the organized arrangement of palisade and spongy mesophyll (Fig. 7E). In svr9-1, palisade mesophyll can still be distinguished but is less organized than that of wild type (Fig. 7E). Leaf mesophyll was more dramatically affected in svr9-1 svr9l1-1/+, in that the distinction between palisade and spongy mesophyll became obscure and intercellular air space appeared to be increased (Fig. 7E).
Third, we discovered that leaf vascular development is also altered in svr9-1 and svr9-1 svr9l1-1/+ mutants, as indicated by the cotyledon venation patterns. We used the number of closed areoles in mature cotyledons as an indicator of leaf vascular development (Sieburth, 1999). In wild type, cotyledons with two, three, and four areoles were common. In svr9-1, although the predominant portions of cotyledons show two or three areoles, cotyledons with only one areole were identified in svr9-1 (Table I; Fig. 7F). The defects were more conspicuous in svr9-1 svr9l1-1/+. The percentage of three or four areoles were drastically decreased in svr9-1 svr9l1-1/+ (Table I; Fig. 7F). Moreover, cotyledons with no closed areoles can be identified in svr9-1 svr9l1-1/+ (Table I; Fig. 7F). Abnormal leaf vascular development is not generally associated with defects of chloroplast development as we did not observe major cotyledon venation changes in an albino mutant svr4-2 we reported earlier (Supplemental Table S1; Yu et al., 2011).
Table I. Quantification of cotyledon vein patterns in wild type, svr9-1, and svr9-1 svr9l1-1/+.
Percentages of different types of areoles are indicated in the parentheses. N.A., Not applicable.
| Genotype | Totala | Cotyledon Vein Patterns | |||||
|---|---|---|---|---|---|---|---|
| Zero areole | One areole | Two areoles | Three areoles | Four areoles | Five areoles | ||
| Wild type | 328 | N.A. | N.A. | 134 (40.8%) | 135 (41.2%) | 59 (18%) | N.A. |
| svr9-1 | 429 | N.A. | 13 (3%) | 164 (38.2%) | 183 (42.7%) | 68 (15.9%) | 1 (0.2%) |
| svr9-1 svr9/1-1/+ | 317 | 3 (1%) | 67 (21.1%) | 177 (55.8%) | 69 (21.8%) | 1 (0.3%) | N.A. |
Total number of cotyledons examined.
Taken together, our data show that the down-regulated chloroplast translation IF3 activities alter leaf margin, mesophyll, and vascular development.
Chloroplast Translation Defects Alter Auxin Homeostasis
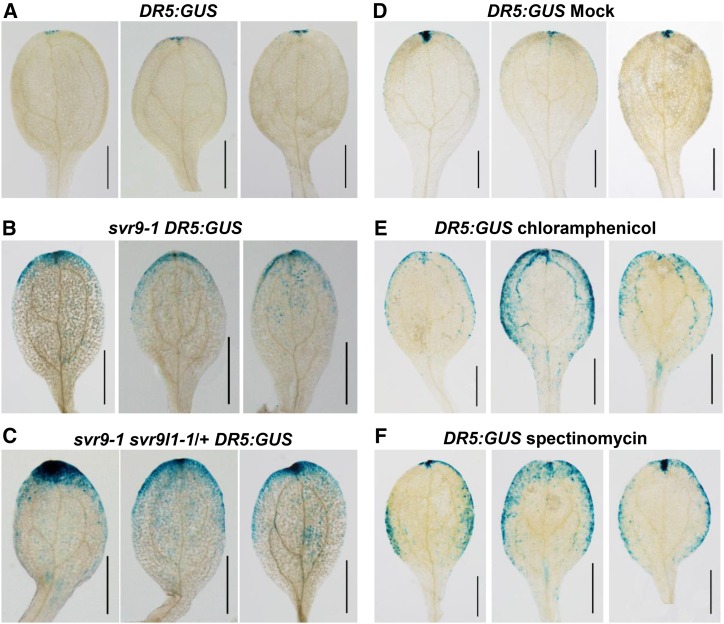
Phytohormone auxin has been implicated in the regulation of leaf margin and vascular development (Scarpella et al., 2006; Bilsborough et al., 2011). Given the margin and vascular defects of svr9-1 and svr9-1 svr9l1-1/+, we explored whether auxin homeostasis was altered in the mutants. To monitor auxin homeostasis, we crossed an auxin reporter line DR5:GUS into svr9-1 and svr9-1 svr9l1-1/+ mutant backgrounds, respectively. GUS staining of DR5:GUS reporter line seedlings revealed auxin maxima at cotyledon tips, consistent with previous reports (Ulmasov et al., 1997). In contrast, stronger and more expanded DR5:GUS signals were observed in svr9-1 background compared to that of the DR5:GUS reference plants (Fig. 8, A and B). Clear signals can be observed along the leaf margin in svr9-1 DR5:GUS plants (Fig. 8B). A stronger increase in the DR5:GUS staining was observed in the svr9-1 svr9l1-1/+ background (Fig. 8C). If the mutations of chloroplast IF3s can lead to abnormal auxin homeostasis, one would expect that the inhibition of chloroplast translation might also cause similar alterations in auxin homeostasis. To test this hypothesis, we treated DR5:GUS reporter plants with chemical inhibitors of prokaryotic translation, chloramphenicol and spectinomycin, respectively. Compared to plants subjected to the mock treatments, both chloramphenicol and spectinomycin treatments led to stronger and more diffused distributions of auxin signals (Fig. 8, D–F). These data suggest that disturbances in chloroplast translation induced by mutations or pharmacological treatments can affect auxin homeostasis.
Figure 8.
Repression of chloroplast translation alters DR5:GUS expression pattern. Images of three representative cotyledons were shown for each genotype or treatment. A to C, Comparison of DR5:GUS expression patterns in wild-type, svr9-1, and svr9-1 svr9l1-1/+ backgrounds. DR5:GUS activities were assayed in cotyledons of 6-d-old seedlings grown on solid 1/2 MS medium. D to F, Effects of chloroplast translation inhibitors on DR5:GUS expression. Six-d-old wild-type background DR5:GUS seedlings maintained in liquid 1/2 MS culture were treated with 5 mm chloramphenicol, 5 mm spectinomycin, or equal amounts of ethanol (mock treatment) for 24 h before assaying DR5:GUS activities. Bars, 1 mm.
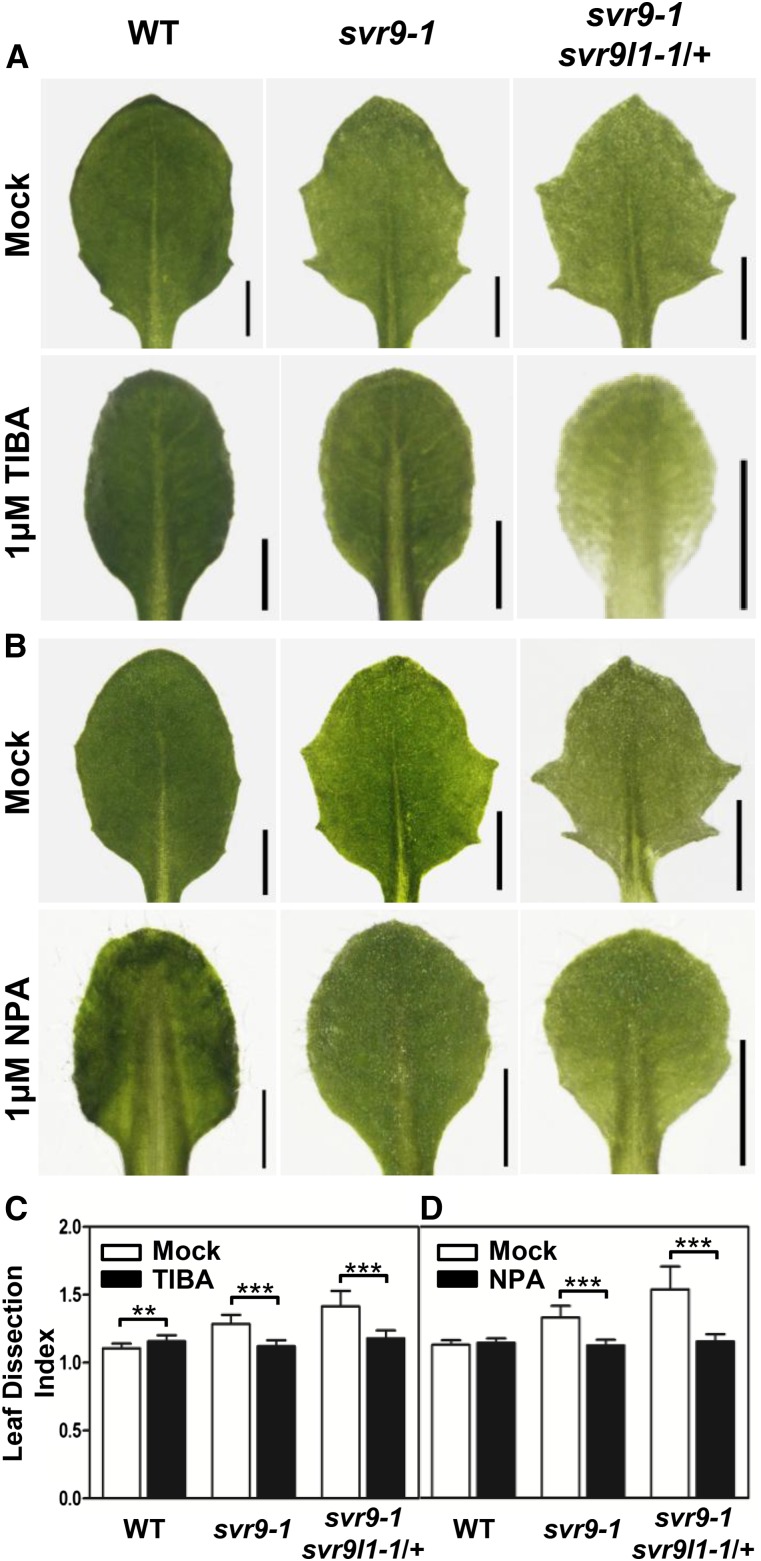
We next tested whether auxin homeostasis can impact the development of leaf margins of svr9-1 and svr9-1 svr9l1-1/+. Mutant plants were treated with auxin transport inhibitor 2,3,5-triiodobenzoic acid (TIBA) or N-1-naphthylphthalamic acid (NPA). Compared with mock treatments, svr9-1 and svr9-1 svr9l1-1/+ lost the serrated leaf margins upon TIBA or NPA treatments (Fig. 9, A and B). Consistent with the visual observations, statistical analysis showed that both TIBA and NPA treatments greatly reduced the leaf dissection index of svr9-1 and svr9-1 svr9l1-1/+ (Fig. 9, C and D). These results indicate that artificially altering auxin homeostasis by drugs could modulate svr9-1 and svr9-1 svr9l1-1/+ mutant leaf margins.
Figure 9.
Effects of auxin transport inhibitors on leaf margin development. A and B, Wild type, svr9-1, and svr9-1 svr9l1-1/+ were grown on solid 1/2 MS medium supplemented with 1 μm TIBA (A), 1 μm NPA (B), or equal amounts of DMSO (mock treatment) for 14 d. The third true leaves from representative plants of each genotype are illustrated. Bars, 1 mm. C and D, Quantification of the effects of TIBA (C) and NPA (D) on leaf shape by leaf dissection index measurements. Data were presented as mean ± sd. **0.001 < P < 0.01; ***P < 0.001. WT, Wild type.
CUC2 Is Involved in the Regulation of Leaf Margin Development Mediated by Chloroplast Translation IF3s
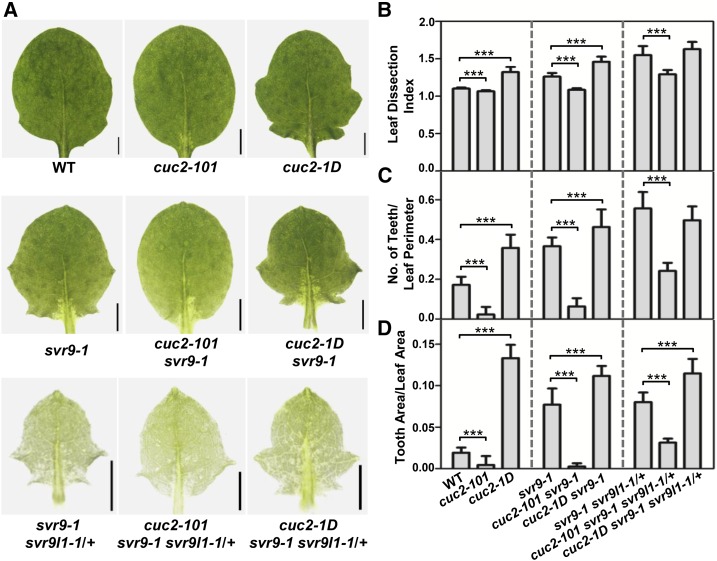
It is well established that NAC family transcription factor CUC2 is a central regulator in auxin-mediated leaf margin development (Nikovics et al., 2006; Kawamura et al., 2010; Bilsborough et al., 2011). To dissect genetically the regulation of leaf margin by chloroplast IF3s, we investigated the genetic interactions among SVR9, SVR9L1, and CUC2. We obtained a CUC2 loss-of-function mutant SALK_035713 (named cuc2-101 hereafter) and a gain-of-function mutant cuc2-1D (Larue et al., 2009). Consistent with previous reports, cuc2-101 showed a smooth leaf margin while cuc2-1D displayed a more serrated leaf margin (Fig. 10A; Nikovics et al., 2006; Larue et al., 2009). We next crossed svr9-1 and svr9-1 svr9l1-1/+ into the cuc2-101 and cuc2-1D background, respectively. Overall, the presence of cuc2-101 mutation greatly suppressed the leaf dissection index, the number of teeth/leaf perimeter, and the tooth area/leaf area in svr9-1 (Fig. 10, A–D). Similar suppression of leaf serrations by cuc2-101 was also observed in svr9-1 svr9l1-1/+, albeit to a less extent (Fig. 10, A–D). On the other hand, the addition of cuc2-1D to svr9-1 increased leaf serration based on the three parameters we monitored (Fig. 10, B–D). In svr9-1 svr9l1-1/+ background, introduction of cuc2-1D mutation has no obvious impact on the leaf dissection index and the number of teeth/leaf perimeter, but cuc2-1D did increase the tooth area/leaf area in svr9-1 svr9l1-1/+ (Fig. 10, B–D). This is consistent with previous notion that increased CUC2 expression promotes leaf marginal serrations mostly by increasing the lobe outgrowth (Kawamura et al., 2010). Our observations suggest that svr9-1 and svr9-1 svr9l1-1/+-mediated leaf margin development involves CUC2.
Figure 10.
Genetic interactions between cuc2 mutants and svr9-1, svr9l1-1 mutants. A, Photographs of the third true leaves in representative 2-week-old wild type, cuc2-101, cuc2-1D, svr9-1, cuc2-101 svr9-1, cuc2-1D svr9-1, svr9-1 svr9l1-1/+, cuc2-101 svr9-1 svr9l1-1/+, and cuc2-1D svr9-1 svr9l1-1/+. B to D, Quantitative comparisons of leaf margins of indicated genotypes. Leaf margin quantifications were based on the leaf dissection index (B), the number of teeth/leaf perimeter (C), and the tooth area/leaf area (D). All leaf shape parameters were obtained as in Fig. 7. Data were presented as mean ± sd. ***P < 0.001. WT, Wild type.
Interestingly, despite the genetic interaction between CUC2 and SVR9/SVR9L1 on the leaf shape characteristics, neither cuc2 mutants affects the virescent leaf color phenotype of svr9-1 or svr9-1 svr9l1-1/+ (Fig. 10A; Supplemental Fig. S7). Together the genetic interactions between SVR9 and CUC2 suggest that SVR9 and SVR9L1-mediated leaf margin development is dependent on CUC2 activities and is independent of their roles in chloroplast development.
DISCUSSION
Chloroplast Translation IF3 Activities Are Essential for Chloroplast and Plant Development
We are interested in using var2 as a tool to gain insight into the genetic regulation of chloroplast development (Liu et al., 2010b). In this study, we isolated a new var2 genetic suppressor locus termed SVR9. We showed that SVR9 encodes a chloroplast protein bearing high homology to prokaryotic translation IF3. In addition, svr9-1 mutant phenotypes were fully complemented by a bacterial IF3 directed by the chloroplast transit peptide of SVR9. Taken together, we showed conclusively that SVR9 codes for a bona fide chloroplast translation IF3.
In Arabidopsis, there are three genes that are annotated to code for prokaryotic type translation IF3 (Nesbit et al., 2015). Besides SVR9, we showed that a close homolog of SVR9, which we named SVR9-LIKE1 (SVR9L1), is also a chloroplast protein, while the third gene product is a mitochondrial protein (Nesbit et al., 2015). This complement of prokaryotic type IF3s is consistent with the endosymbiotic origins of the two organelles. For chloroplast-localized SVR9 and SVR9L1, multiple pieces of evidence suggest that they share redundant functions in chloroplast development. SVR9 and SVR9L1 share the same gene structures and high homology at amino acid sequence level (Fig. 4A; Supplemental Fig. S1), suggesting that they may have evolved through gene duplication. Functionally, svr9-1 can be fully rescued by the overexpression of SVR9L1 (Fig. 6C). Moreover, genetic interaction studies revealed that the combined activities of SVR9 and SVR9L1 are essential for chloroplast and plant development, as we were not able to recover double homozygous mutants for svr9-1 and svr9l1-1. Based on the severity of svr9-1 and svr9l1-1 mutant phenotypes, it is likely that SVR9 contributes more to the overall chloroplast IF3 activities while SVR9L1 plays a relatively minor role, and the genetic relationship between SVR9 and SVR9L1 indicates an unequal functional redundancy (Briggs et al., 2006).
Three prokaryotic translation initiation factors, IF1, IF2, and IF3, are employed by the 70S ribosome, and IF3 has been proposed to be responsible for sensing translation initiation regions of mRNAs and ensuring initiation fidelity (Lomakin et al., 2006; Milon et al., 2008). Our results indicate that chloroplast translation IF3 activities, contributed by both SVR9 and SVR9L1 in Arabidopsis chloroplasts, are indispensable for chloroplast and plant development. Previous studies have indicated that chloroplast translation initiation factor IF2 activities are also essential for plant development (Miura et al., 2007). These data suggest that the proper initiation of translation in the chloroplast is essential for chloroplast biogenesis.
The Suppression of var2 Leaf Variegation by SVR9 and SVR9L1 Mutations
Previously we have proposed a threshold model to explain the mechanisms of leaf variegation mediated by VAR2/AtFtsH2 (Yu et al., 2004, 2005). The core of our model is that the development of functional chloroplasts requires a threshold level of thylakoid FtsH complexes/activities. Moreover, we envisioned that this threshold would be dynamic, and changing functional or developmental status of the chloroplast may necessitate different thresholds. Based on the model, we should be able to identify functional or developmental processes that, when disrupted, may modify the threshold and the need for FtsH activities. To this end, we carried out var2 genetic suppressor screens. In these genetic suppressors of var2, we propose that the suppressor mutations lead to disturbed functional or developmental chloroplast states that lower the endogenous threshold for VAR2/AtFtsH2. Through both our work with the SVR loci and work from other groups, a definitive link between the threshold and some aspects of chloroplast translation has been established (Miura et al., 2007; Yu et al., 2008; Liu et al., 2010a, 2010c, 2013). However, a key aspect of the threshold model, i.e. a multipoint correlation of the changing functional or developmental states of the chloroplast with the corresponding changing needs for FtsH activities, remained hypothetical. In this study, through multiple mutant alleles and mutant combinations of SVR9 and SVR9L1, we were able to create a series of chloroplast developmental states and explore this important aspect of the threshold hypothesis. In wild-type background where chloroplast IF3s are at full capacity, VAR2/AtFtsH2 activities are necessary for chloroplast development and the lack of VAR2/AtFtsH2 causes leaf variegation. The svr9l1-1 mutation slightly reduces IF3 activities, but functional VAR2/AtFtsH2 is still required and leaf variegation is not suppressed. However, in svr9-2, the reduction of IF3 activities is greater than in svr9l1-1. The state generated by svr9-2 may lower the threshold need for VAR2/AtFtsH2 and leaf variegation reduces. IF3 activities are further reduced in svr9l1-1 svr9-2 double mutants, and the mutant background of svr9l1-1 sv9-2 generated a state that the need for VAR2/AtFtsH2 activities is no longer necessary for chloroplast development, thus a full suppression of leaf variegation. When IF3 activities is further reduced in svr9-1 and svr9-1 svr9l1-1/+ mutants, the lack of IF3 activities itself starts to impair chloroplast development and mutant plants display a clear visual phenotype, and this functional state bypasses the need for VAR2/AtFtsH2, as observed in our previous var2 suppressor mutants (Liu et al., 2010b; Putarjunan et al., 2013).
Several messages can be gathered from these data. First, our genetic data indicate that the full wild-type level of IF3 activities is not necessary for chloroplast development under lab conditions, as svr9l1-1, svr9-2, and svr9l1-1 svr9-2 resembled wild type. However, a key message from our data is that a superficial wild-type appearance does not necessarily mean a lack of phenotypes at molecular level. In this sense, genetic suppressor screens can be a very sensitive and powerful approach to uncover subtle molecular alterations in mutants. Second, the different mutants and mutant combinations in our study generated different chloroplast states where VAR2/AtFtsH2 functions may be partially or completely bypassed, supporting our previous hypothesis that the threshold for FtsH activities can vary.
The Interaction between Chloroplast and Leaf Development
The chloroplast is the site for photosynthesis, as well as the venue for the synthesis of numerous metabolic and regulatory molecules such as phytohormones, and the proper functioning of the chloroplast is essential for plant growth and development. It is thus not surprising that the functional or developmental states of the chloroplast can impact processes outside of the chloroplast. Previous studies have established multiple regulatory pathways that coordinate the states of the chloroplast with gene expressions in the nucleus. In addition, it is widely known that plant leaf development is also linked with the states of the chloroplast. However, we lack detailed information regarding the relationship between leaf development and the chloroplast.
In this study, we uncovered a range of leaf developmental phenotypes in SVR9 and SVR9L1 mutants that may be linked with altered auxin homeostasis and auxin-regulated pathways. First, we discovered that the cotyledon venation patterns were abnormal in svr9-1 and svr9-1 svr9l1-1/+ mutants. Overall, we observed that mutant cotyledons showed a trend of reduced complexity of vascular development as indicated by the numbers of areoles (Fig. 7F; Table I). It is generally believed that auxin is a central regulator of leaf vascular development, thus the venation pattern changes in the mutants may reflect an altered auxin homeostasis (Sieburth, 1999; Cheng et al., 2007). Second, we observed that leaf margins are more serrated in svr9-1 and svr9-1 svr9l1-1/+ mutants than those of the wild type (Fig. 7, A–D). The induction of leaf serrations are dictated by a feedback loop of auxin maxima established by PIN1 convergence points and CUC2 activities (Bilsborough et al., 2011). The expression of CUC2 promotes the formation of PIN1 convergence points and in turn the auxin maxima along the leaf margin where CUC2 expressions are then repressed by auxin (Bilsborough et al., 2011). This feedback regulation of CUC2, PIN1, and auxin level eventually leads to the alternate presence of CUC2 expressions at leaf indentations and auxin maxima at leaf protrusions (Bilsborough et al., 2011; Byrne, 2012). Third, we observed alterations in auxin homeostasis in svr9-1 and svr9-1 svr9l1-1/+ mutants as indicated by the changes in DR5:GUS expression patterns (Fig. 8, A–C). Changes in auxin homeostasis can also be observed in plants treated with chloroplast translation inhibitors (Fig. 8, D–F). Treatments with auxin transport inhibitors can alter the leaf margin phenotypes of svr9-1 and svr9-1 svr9l1-1/+ mutants (Fig. 9). These data suggest that auxin may play an important role in leaf developmental processes regulated by chloroplast IF3s. Surprisingly, we did not observe altered leaf margins in svr8-2, a null mutant of the chloroplast ribosomal protein L24 (Supplemental Fig. S6; Liu et al., 2013). It is possible that when individual components of the chloroplast translation system were mutated, chloroplast translation is affected in such a manner that may or may not trigger the abnormal development of leaf margins. This raises the interesting possibility that specific aspects of chloroplast translation are associated with leaf margin development. Last but not least, our genetic data support the notion that chloroplast IF3s SVR9 and SVR9L1 regulate leaf margin development at least partially via the CUC2-mediated pathway because a lack of CUC2 greatly hampered but did not completely abolish the manifestation of leaf margin phenotypes of svr9-1 and svr9-1 svr9l1-1/+ (Fig. 10). The genetic interaction results further suggest that there may be other factors involved in the output of the retrograde signals from the svr9-1 svr9l1-1/+ mutant chloroplasts (Fig. 10). Based on our data, it is clear that certain functional states of the chloroplast, as generated by the lack of SVR9 and SVR9L1, are able to regulate leaf development. It is possible that these states of the chloroplast are capable of altering the auxin homeostasis, which in turn regulates CUC2 activities. It is also possible that these states influence auxin and CUC2 separately. Although the precise mechanisms remain to be elucidated, it is tempting to speculate that these functional states may generate retrograde signals that coordinate the states of the chloroplast with leaf development.
CONCLUSION
Taken together, our data indicate that chloroplast translation IF3 activities are essential for plant and chloroplast development. Mutations in the chloroplast translation IF3s can suppress the leaf variegation phenotype of var2 mutants. Moreover, our data indicate that the functional or developmental states of the chloroplast can regulate leaf development, particularly the processes that are associated with auxin. Currently we do not know whether this regulation involves the known retrograde signaling pathways or represents a new route of signaling, and future research will undoubtedly expand our understanding of the intricate relationship between the chloroplast and the rest of the plant cell.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) strains used in this study are of the Columbia-0 background. svr9-1 was isolated in this study. The T-DNA insertion lines SAIL_172_F02 (svr9-2), SALK_140431C (svr9l1-1), SALK_035713 (cuc2-101), and cuc2-1D were obtained from the Arabidopsis Biological Resource Center; svr4-2 and svr8-2 have been described in Yu et al. (2011) and Liu et al. (2013). Seeds were sown on commercial soil mix (Pindstrup, Denmark) or on half-strength Murashige and Skoog (1/2 MS) medium supplemented with 1% Suc, and stratified for 2 d at 4°C before placed in growth rooms. When solid medium was needed, 0.8% agar (w/v) was added to the 1/2 MS medium. Arabidopsis plants were maintained at approximately 22°C under continuous illumination (approximately 100 µmol m−2 s−1).
Chlorophyll Content Measurements
Fresh plant tissues were harvested, weighed, and finely ground in liquid N2. Total chlorophyll was extracted with 95% ethanol. Chlorophyll contents were determined as described in Lichtenthaler (1987). Chlorophyll measurements of different samples were normalized on a fresh tissue weight basis. Each sample consisted of leaf tissues pooled from two to four individual plants. Mean chlorophyll content of leaf tissues of each genotype was calculated from three independent pooled leaf samples. Differences in chlorophyll content between wild type and mutants were evaluated with P values generated by t test.
Map-Based Cloning
The SVR9 locus was initially mapped by the bulked segregation analysis using a pool of 95 F2 mutant seedlings from a cross between svr9-1 and Landsberg erecta (Lukowitz et al., 2000). A mapping population consisting of 570 F2 mutant plants was used to further fine map the svr9-1 mutation. Primers of the molecular markers generated in this study were listed in Supplemental Table S2. Primers used to amplify various At2g24060 genomic fragments were listed in Supplemental Table S2.
Semiquantitative RT-PCR Analysis
Total cellular RNAs were purified using Trizol RNA reagent (Life Technologies) following the manufacturer’s instructions and stored at −80°C. For semiquantitative RT-PCR analysis, first strand cDNAs were synthesized from 1 μg DNase I-treated total RNAs using a PrimeScript reverse transcription kit (Takara). The gene-specific primers were listed in Supplemental Table S2.
Functional Complementation of svr9-1
All primers used in vector construction were listed in Supplemental Table S2. To complement svr9-1, full-length cDNAs of SVR9/At2g24060 and SVR9L1/At4g30690 were amplified using primers 24060F and 24060R, and 30690F and 30690R, respectively. The amplified fragments were cloned into pBluescript KS+ (pBS) and sequenced before subcloned into a binary vector pBI111L (Yu et al., 2004). The resulting constructs were named P35S:At2g24060 and P35S:At4g30690, respectively.
To express Escherichia coli IF3 infC in Arabidopsis, the coding sequence of infC was amplified from E. coli genomic DNA using primers E. coli IF3-F and E. coli IF3-R and cloned into pBS. The coding sequence of the cTP region of SVR9 (1–71 amino acids of SVR9) was amplified with cTPF and cTPR and cloned into pBS. After sequencing pBS-infC and pBS-SVR9cTP, the infC and SVR9cTP were sequentially subcloned into pBI111L to have SVR9cTP fused in frame at the N-terminal of infC. The resulting construct was named P35S:cTPSVR9-infC.
Each of the binary vectors was transformed into Agrobacterium tumefaciens by electroporation, and the floral dip method was used for Arabidopsis transformation (Clough and Bent, 1998). T1 transgenic lines were screened on solid 1/2 MS medium containing 50 mg × L−1 kanamycin.
Protoplast Transient Expression Assays
To transiently express C-terminal GFP fusion proteins SVR9cTP-GFP, SVR9-GFP, and SVR9L1-GFP in leaf protoplasts, the coding sequences of SVR9cTP, SVR9, and SVR9L1 were cloned into pTF486 to fuse in frame with GFP coding sequences (Yu et al., 2008). Fresh prepared wild-type Arabidopsis mesophyll protoplasts were transformed with each of the transient expression vectors following the procedures described in Yoo et al. (2007). After transformation, protoplasts were incubated overnight and examined with confocal microcopy using the Apo TIRF 60× Oil objective (N.A. 1.49; Nikon A1). GFP and chlorophyll autofluorescence signals were monitored through 525/50 and 700/75 emission filters, respectively.
Histochemical GUS Staining
To generate SVR9 promoter-β-glucuronidase (GUS) transcriptional fusion construct, a 1913-bp genomic DNA fragment upstream of the start codon of SVR9 gene was amplified using primers 24060PF-XbaI2 and 24060PR-BamHI2 and cloned into pCB308 (Xiang et al.,1999). The resulting construct was used to transform wild-type Arabidopsis plant. PSVR9:GUS lines were screened based on Basta resistance. GUS activities were assayed in T1 and T2 generations (Jefferson, 1987).
To test the impact of svr9-1 and svr9l1-1 mutations on DR:GUS expression patterns, DR5:GUS reporter gene was introduced to svr9-1 and svr9-1svr9l1-1/+ via genetic crossing. DR5:GUS activities in different genetic backgrounds were assayed in 6-d-old seedlings.
Quantification of Leaf Variegation
The first true leaves of two-week-old Arabidopsis seedlings were hand-cut and mounted in water. Images of each individual leaf were captured using a stereoscope (SMZ25; Nikon) equipped with a CCD camera (DS-U3; Nikon). All leaves were photographed with the same settings. To quantify the extent of variegation, leaf images were first converted to grayscale using the ImageJ software. Pixels constituted the leaf blade were selected and intensity values of these pixels were obtained using the NIS-Elements software (Nikon). Frequency was calculated as number of pixels of each intensity value (ranging from 0 to 255)/number of pixels constituting the leaf blade area. Frequency distributions of pixel intensity values of each leaf image were generated using the GraphPad Prism software. The quantification of leaf variegation was repeated three times. Similar patterns of frequency distributions of pixel intensity values were obtained for each repeat.
Leaf Margin Analysis
Water-mounted individual leaves were photographed using a stereoscope (SMZ25; Nikon) equipped with a CCD camera (DS-U3; Nikon). Three parameters commonly used in leaf margin analysis, including the leaf dissection index (perimeter2/4π × leaf area), the number of teeth/leaf perimeter, and the tooth area/leaf area, were used in this study to quantify the differences in leaf margin (Royer et al., 2008; Bilsborough et al., 2011). The leaf blade region (excluding leaf petiole) was used in quantitative analyses. Leaf area and perimeter measurements were performed using the ImageJ software. Graphs were generated by the GraphPad Prism software. Differences between means of each leaf shape parameter were evaluated by P values generated by t test.
Analysis of Leaf Cross Sections and Cotyledon Vein Patterns
Basal parts of the first true leaves of 10-d-old wild type, svr9-1, and svr9-1 svr9l1-1/+ were hand-cut and infiltrated with fixation solution (4% (v/v) glutaraldehyde in 0.1 mm sodium phosphate buffer, pH6.8). Fixed tissues were dehydrated and embedded in Technovit 7100 resin (EMS). Semithin sections (10 μm) prepared with a microtome (RM2265; Leica) were stained with 1% (v/v) toluidine blue O and observed with a Leica DM5000B microscope equipped with a DFC425C CCD camera.
To observe cotyledon veins, cotyledons of 10-d-old plants were hand-cut and decolored in 70% ethanol until the veins became clearly visible. Decolored cotyledons were then photographed with a digital camera mounted on a stereo microscope. Cotyledon vein patterns were categorized based on the number of areoles formed (Sieburth, 1999)
Chemical Treatments
All chemicals used in this study were purchased from Sigma-Aldrich unless otherwise specified. NPA and TIBA were dissolved in dimethyl sulfoxide (DMSO). Wild-type, svr9-1, and svr9-1 svr9l1-1/+ seeds were germinated and grown on solid 1/2 MS medium supplemented with 1 μM NPA or 1 μM TIBA. Plants grown on solid 1/2 MS medium with an equal concentration of DMSO served as controls. Images of the third true leaves in two-week-old plants were examined.
To test DR5:GUS expression in response to chloroplast translation inhibitors, DR5:GUS lines of different genetic backgrounds were germinated and grown on liquid 1/2 MS medium for 6 d before adding chloramphenicol or spectinomycin to a final concentration of 5 mM. Equal amounts of ethanol were added as the mock treatment. After 24 h of treatment, seedlings were harvested and assayed for GUS activities. Plant liquid cultures were shaken at 120 rpm to keep the seedlings floating.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Information Resource or GenBank/EMBL databases under the following accession numbers: SVR9, At2g24060; SVR9L1, At4g30690; VAR2/AtFtsH2, At2g30950; CUC2, At5g53950, E. coli infC, NP_416233.1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Protein sequence alignments of SVR9/At2g24060, SVR9L1/At4g30690, At1g34360, and E. coli infC.
Supplemental Figure S2. Chloroplast localization of cTPSVR9-GFP.
Supplemental Figure S3. Phylogenetic analysis of prokaryotic IF3-like proteins from E. coli and representative photosynthetic species.
Supplemental Figure S4. Identification of svr9-2 and svr9l1-1.
Supplemental Figure S5. Whole plant phenotypes of representative 2-week-old plants of the same genotypes shown in Figure 5, A–D.
Supplemental Figure S6. Comparison of leaf margin development in wild type, svr9-1, and svr8-2.
Supplemental Figure S7. Whole plant phenotypes of representative 2-week-old plants of the same genotypes shown in Figure 10A.
Supplemental Table S1. Comparison of cotyledon vein patterns of wild type and svr4-2.
Supplemental Table S2. Primers used in this study.
Supplementary Material
References
- Aluru MR, Bae H, Wu D, Rodermel SR (2001) The Arabidopsis immutans mutation affects plastid differentiation and the morphogenesis of white and green sectors in variegated plants. Plant Physiol 127: 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilsborough GD, Runions A, Barkoulas M, Jenkins HW, Hasson A, Galinha C, Laufs P, Hay A, Prusinkiewicz P, Tsiantis M (2011) Model for the regulation of Arabidopsis thaliana leaf margin development. Proc Natl Acad Sci USA 108: 3424–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs GC, Osmont KS, Shindo C, Sibout R, Hardtke CS (2006) Unequal genetic redundancies in Arabidopsis—a neglected phenomenon? Trends Plant Sci 11: 492–498 [DOI] [PubMed] [Google Scholar]
- Byrne ME. (2012) Making leaves. Curr Opin Plant Biol 15: 24–30 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2007) Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W, Sun X, Zhang L (2013) Intracellular signaling from plastid to nucleus. Annu Rev Plant Biol 64: 559–582 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dyall SD, Brown MT, Johnson PJ (2004) Ancient invasions: from endosymbionts to organelles. Science 304: 253–257 [DOI] [PubMed] [Google Scholar]
- Fleischmann TT, Scharff LB, Alkatib S, Hasdorf S, Schöttler MA, Bock R (2011) Nonessential plastid-encoded ribosomal proteins in tobacco: a developmental role for plastid translation and implications for reductive genome evolution. Plant Cell 23: 3137–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez-Valdivieso G, Mullineaux PM (2010) The role of reactive oxygen species in signalling from chloroplasts to the nucleus. Physiol Plant 138: 430–439 [DOI] [PubMed] [Google Scholar]
- Gray JC, Sullivan JA, Wang JH, Jerome CA, MacLean D (2003) Coordination of plastid and nuclear gene expression. Philos Trans R Soc Lond B Biol Sci 358: 135–144, discussion 144–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hricová A, Quesada V, Micol JL (2006) The SCABRA3 nuclear gene encodes the plastid RpoTp RNA polymerase, which is required for chloroplast biogenesis and mesophyll cell proliferation in Arabidopsis. Plant Physiol 141: 942–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA. (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Report 5: 387–405 [Google Scholar]
- Jensen PE, Leister D (2014) Chloroplast evolution, structure and functions. F1000Prime Rep 6: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josse EM, Simkin AJ, Gaffé J, Labouré AM, Kuntz M, Carol P (2000) A plastid terminal oxidase associated with carotenoid desaturation during chromoplast differentiation. Plant Physiol 123: 1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura E, Horiguchi G, Tsukaya H (2010) Mechanisms of leaf tooth formation in Arabidopsis. Plant J 62: 429–441 [DOI] [PubMed] [Google Scholar]
- Kindgren P, Kremnev D, Blanco NE, de Dios Barajas López J, Fernández AP, Tellgren-Roth C, Kleine T, Small I, Strand A (2012) The plastid redox insensitive 2 mutant of Arabidopsis is impaired in PEP activity and high light-dependent plastid redox signalling to the nucleus. Plant J 70: 279–291 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Larue CT, Wen J, Walker JC (2009) A microRNA-transcription factor module regulates lateral organ size and patterning in Arabidopsis. Plant J 58: 450–463 [DOI] [PubMed] [Google Scholar]
- Lee KP, Kim C, Landgraf F, Apel K (2007) EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 10270–10275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D. (2003) Chloroplast research in the genomic age. Trends Genet 19: 47–56 [DOI] [PubMed] [Google Scholar]
- Leister D. (2012) Retrograde signaling in plants: from simple to complex scenarios. Front Plant Sci 3: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls, carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Liu X, Rodermel SR, Yu F (2010a) A var2 leaf variegation suppressor locus, SUPPRESSOR OF VARIEGATION3, encodes a putative chloroplast translation elongation factor that is important for chloroplast development in the cold. BMC Plant Biol 10: 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yu F, Rodermel S (2010b) An Arabidopsis pentatricopeptide repeat protein, SUPPRESSOR OF VARIEGATION7, is required for FtsH-mediated chloroplast biogenesis. Plant Physiol 154: 1588–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yu F, Rodermel S (2010c) Arabidopsis chloroplast FtsH, var2 and suppressors of var2 leaf variegation: a review. J Integr Plant Biol 52: 750–761 [DOI] [PubMed] [Google Scholar]
- Liu X, Zheng M, Wang R, Wang R, An L, Rodermel SR, Yu F (2013) Genetic interactions reveal that specific defects of chloroplast translation are associated with the suppression of var2-mediated leaf variegation. J Integr Plant Biol 55: 979–993 [DOI] [PubMed] [Google Scholar]
- Lomakin IB, Shirokikh NE, Yusupov MM, Hellen CU, Pestova TV (2006) The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. EMBO J 25: 196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible WR (2000) Positional cloning in Arabidopsis: why it feels good to have a genome initiative working for you. Plant Physiol 123: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MA, Feldmann KA, Herrera-Estrella L, Rocha-Sosa M, León P (1996) CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J 9: 649–658 [DOI] [PubMed] [Google Scholar]
- Milon P, Konevega AL, Gualerzi CO, Rodnina MV (2008) Kinetic checkpoint at a late step in translation initiation. Mol Cell 30: 712–720 [DOI] [PubMed] [Google Scholar]
- Miura E, Kato Y, Matsushima R, Albrecht V, Laalami S, Sakamoto W (2007) The balance between protein synthesis and degradation in chloroplasts determines leaf variegation in Arabidopsis yellow variegated mutants. Plant Cell 19: 1313–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Tanaka R, Tanaka A, Masuda T, Nagatani A (2008) The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc Natl Acad Sci USA 105: 15184–15189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin M, McCormac AC, Terry MJ, Smith AG (2008) Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc Natl Acad Sci USA 105: 15178–15183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbit AD, Whippo C, Hangarter RP, Kehoe DM (2015) Translation initiation factor 3 families: what are their roles in regulating cyanobacterial and chloroplast gene expression? Photosynth Res 126: 147–159 [DOI] [PubMed] [Google Scholar]
- Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P (2006) The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18: 2929–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Jung HS, Koussevitzky S, Chory J (2006) Plastid-to-nucleus retrograde signaling. Annu Rev Plant Biol 57: 739–759 [DOI] [PubMed] [Google Scholar]
- Pesaresi P, Hertle A, Pribil M, Kleine T, Wagner R, Strissel H, Ihnatowicz A, Bonardi V, Scharfenberg M, Schneider A, Pfannschmidt T, Leister D (2009) Arabidopsis STN7 kinase provides a link between short- and long-term photosynthetic acclimation. Plant Cell 21: 2402–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P, Masiero S, Eubel H, Braun HP, Bhushan S, Glaser E, Salamini F, Leister D (2006) Nuclear photosynthetic gene expression is synergistically modulated by rates of protein synthesis in chloroplasts and mitochondria. Plant Cell 18: 970–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putarjunan A, Liu X, Nolan T, Yu F, Rodermel S (2013) Understanding chloroplast biogenesis using second-site suppressors of immutans and var2. Photosynth Res 116: 437–453 [DOI] [PubMed] [Google Scholar]
- Qi Y, Zhao J, An R, Zhang J, Liang S, Shao J, Liu X, An L, Yu F (2016) Mutations in circularly permuted GTPase family genes AtNOA1/RIF1/SVR10 and BPG2 suppress var2-mediated leaf variegation in Arabidopsis thaliana. Photosynth Res 127: 355–367 [DOI] [PubMed] [Google Scholar]
- Ramel F, Birtic S, Ginies C, Soubigou-Taconnat L, Triantaphylidès C, Havaux M (2012) Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci USA 109: 5535–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter RS, Coomber SA, Bourett TM, Bartley GE, Scolnik PA (1994) Control of leaf and chloroplast development by the Arabidopsis gene pale cress. Plant Cell 6: 1253–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer DL, McElwain JC, Adams JM, Wilf P (2008) Sensitivity of leaf size and shape to climate within Acer rubrum and Quercus kelloggii. New Phytol 179: 808–817 [DOI] [PubMed] [Google Scholar]
- Sacerdot C, Fayat G, Dessen P, Springer M, Plumbridge JA, Grunberg-Manago M, Blanquet S (1982) Sequence of a 1.26-kb DNA fragment containing the structural gene for E. coli initiation factor IF3: presence of an AUU initiator codon. EMBO J 1: 311–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini G, Meskauskiene R, Pijacka W, Roszak P, Sjögren LL, Clarke AK, Straus M, Apel K (2011) ‘Happy on norflurazon’ (hon) mutations implicate perturbance of plastid homeostasis with activating stress acclimatization and changing nuclear gene expression in norflurazon-treated seedlings. Plant J 65: 690–702 [DOI] [PubMed] [Google Scholar]
- Scarpella E, Marcos D, Friml J, Berleth T (2006) Control of leaf vascular patterning by polar auxin transport. Genes Dev 20: 1015–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE. (1999) Auxin is required for leaf vein pattern in Arabidopsis. Plant Physiol 121: 1179–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Feng P, Xu X, Guo H, Ma J, Chi W, Lin R, Lu C, Zhang L (2011) A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat Commun 2: 477. [DOI] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J (1993) Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799 [DOI] [PubMed] [Google Scholar]
- Tiller N, Bock R (2014) The translational apparatus of plastids and its role in plant development. Mol Plant 7: 1105–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Przybyla D, Op den Camp R, Kim C, Landgraf F, Lee KP, Würsch M, Laloi C, Nater M, Hideg E, Apel K (2004) The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306: 1183–1185 [DOI] [PubMed] [Google Scholar]
- Wilson PB, Estavillo GM, Field KJ, Pornsiriwong W, Carroll AJ, Howell KA, Woo NS, Lake JA, Smith SM, Harvey Millar A, von Caemmerer S, Pogson BJ (2009) The nucleotidase/phosphatase SAL1 is a negative regulator of drought tolerance in Arabidopsis. Plant J 58: 299–317 [DOI] [PubMed] [Google Scholar]
- Woodson JD, Chory J (2008) Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet 9: 383–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Chory J (2012) Organelle signaling: how stressed chloroplasts communicate with the nucleus. Curr Biol 22: R690–R692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver DJ (1999) A mini binary vector series for plant transformation. Plant Mol Biol 40: 711–717 [DOI] [PubMed] [Google Scholar]
- Xiao Y, Savchenko T, Baidoo EE, Chehab WE, Hayden DM, Tolstikov V, Corwin JA, Kliebenstein DJ, Keasling JD, Dehesh K (2012) Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 149: 1525–1535 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yu F, Liu X, Alsheikh M, Park S, Rodermel S (2008) Mutations in SUPPRESSOR OF VARIEGATION1, a factor required for normal chloroplast translation, suppress var2-mediated leaf variegation in Arabidopsis. Plant Cell 20: 1786–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Park SS, Liu X, Foudree A, Fu A, Powikrowska M, Khrouchtchova A, Jensen PE, Kriger JN, Gray GR, Rodermel SR (2011) SUPPRESSOR OF VARIEGATION4, a new var2 suppressor locus, encodes a pioneer protein that is required for chloroplast biogenesis. Mol Plant 4: 229–240 [DOI] [PubMed] [Google Scholar]
- Yu F, Park S, Rodermel SR (2004) The Arabidopsis FtsH metalloprotease gene family: interchangeability of subunits in chloroplast oligomeric complexes. Plant J 37: 864–876 [DOI] [PubMed] [Google Scholar]
- Yu F, Park S, Rodermel SR (2005) Functional redundancy of AtFtsH metalloproteases in thylakoid membrane complexes. Plant Physiol 138: 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.