Two MYB transcription factors regulate CBF4 in opposing manners, with implications for the understanding of cold acclimation and freezing tolerance in Medicago truncatula.
Abstract
Cold acclimation is an important process by which plants respond to low temperature and enhance their winter hardiness. C-REPEAT BINDING FACTOR1 (CBF1), CBF2, and CBF3 genes were shown previously to participate in cold acclimation in Medicago truncatula. In addition, MtCBF4 is transcriptionally induced by salt, drought, and cold stresses. We show here that MtCBF4, shown previously to enhance drought and salt tolerance, also positively regulates cold acclimation and freezing tolerance. To identify molecular factors acting upstream and downstream of the MtCBF4 transcription factor (TF) in cold responses, we first identified genes that are differentially regulated upon MtCBF4 overexpression using RNAseq Digital Gene Expression Profiling. Among these, we showed that MtCBF4 directly activates the transcription of the COLD ACCLIMATION SPECIFIC15 (MtCAS15) gene. To gain insights into how MtCBF4 is transcriptionally regulated in response to cold, an R2R3-MYB TF, MtMYB3, was identified based on a yeast one-hybrid screen as binding directly to MYB cis-elements in the MtCBF4 promoter, leading to the inhibition of MtCBF4 expression. In addition, another MYB TF, MtMYB61, identified as an interactor of MtMYB3, can relieve the inhibitory effect of MtMYB3 on MtCBF4 transcription. This study, therefore, supports a model describing how MtCBF4 is regulated by antagonistic MtMYB3/MtMYB61 TFs, leading to the up-regulation of downstream targets such as MtCAS15 acting in cold acclimation in M. truncatula.
The productivity of a crop is affected by various environmental stresses, such as cold and freezing. After cereals, legumes are the most significant source of human food and forage worldwide (Graham and Vance, 2003), and knowledge of the genes and networks involved in abiotic stress responses is required to develop stress-tolerant legume crops. Many plants increase their freezing tolerance via a process known as cold acclimation, which involves undergoing a period of low but nonfreezing temperatures (Thomashow, 1999). Pennycooke et al. (2008) reported that Medicago truncatula does not cold acclimate. However, Zhang et al. (2011) demonstrated that M. truncatula is able to increase its freezing tolerance after being subjected to cold-acclimating conditions. The different experimental conditions and methods used in these two studies may partly explain the different results (Pennycooke et al., 2008; Zhang et al., 2011). By examining physiological markers of cold acclimation in M. truncatula, Thapa (2008) also independently demonstrated that this species undergoes cold acclimation. Thus, understanding the regulatory mechanisms underlying cold acclimation in the M. truncatula model legume is relevant for the breeding of freezing tolerance in legumes.
The mechanisms through which legumes resist freezing stresses have already been studied at the physiological and molecular levels. A DEHYDRATION-RESPONSIVE ELEMENT BINDING FACTOR1/C-REPEAT BINDING FACTOR (DREB1/CBF) transcription factor (TF), GmDREB3, was cloned in soybean (Glycine max), and overexpression of GmDREB3 in transgenic Arabidopsis (Arabidopsis thaliana) plants enhanced cold, drought, and high-salt stress tolerance (Chen et al., 2009). Expression of GmDREBa, GmDREBb, and GmDREBc is induced by salt, drought, and cold stresses, and these TFs specifically bind to dehydration-responsive elements (DREs; Li et al., 2005). Fourteen DREB1-type TFs were identified by Kidokoro et al. (2015), and most of them were induced by abiotic stress. RD29A:GmDREB1F;1-transformed Arabidopsis enhanced cold, drought, high-salt, and heat stress tolerance (Kidokoro et al., 2015). Two MYB TFs, GmMYB76 and GmMYB177, also were shown to enhance freezing tolerance in Arabidopsis by inducing the expression of freezing tolerance-responsive genes (Liao et al., 2008). In alfalfa (Medicago sativa), cold induces a calcium influx, which affects protein phosphorylation and the expression levels of cold acclimation-specific genes such as CAS15 and CAS18 (Mohapatra et al., 1989; Monroy et al., 1993a, 1993b; Wolfraim et al., 1993; Monroy and Dhindsa, 1995). In M. truncatula and Medicago falcata, cold induces the expression of CBF1 to CBF3 and CAS genes (Pennycooke et al., 2008; Zhang et al., 2011). The expression level of MtCBF4 is induced by drought, salinity, and cold stress, and ectopic MtCBF4 expression in Arabidopsis enhances drought and salt tolerance, as well as salt tolerance when transiently expressed in M. truncatula roots (Li et al., 2011). The molecular mechanisms linking DREB/CBF TFs to stress tolerance in legumes, including cold acclimation and freezing tolerance, nevertheless remain mostly unknown.
In Arabidopsis, however, the DREB1/CBF transcriptional regulations essential for cold acclimation have been studied in more detail. AtCBF1 to AtCBF3 TFs bind DRE cis-elements and induce cold-responsive (COR) gene expression (Gilmour et al., 1998; Shinwari et al., 1998; Maruyama et al., 2004; Vogel et al., 2005) and, consequently, improve freezing tolerance (Gilmour et al., 2004). In addition, CBF gene expression is tightly regulated by calcium. Accordingly, mutation of the Ca2+/H+ antiporter CALCIUM EXCHANGER1 enhances freezing tolerance following cold acclimation and increases CBF gene expression (Catalá et al., 2003). The calmodulin-binding transcriptional activator TF family also is involved in the regulation of CBF genes by binding to the CM2 motif in the AtCBF2 promoter and increasing its expression (Doherty et al., 2009). AtCBF3 expression is induced by a basic helix-loop-helix TF, INDUCER OF CBF EXPRESSION1 (ICE1), and is negatively regulated by MYB15 (Chinnusamy et al., 2003; Agarwal et al., 2006). Accordingly, HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 (HOS1), a negative regulator of ICE1 that mediates ICE1 ubiquitination and degradation, also is a negative regulator of CBF3 (Dong et al., 2006). In contrast, ICE1 sumoylation by the SUMO E3 ligase SIZ1 (for SAP and MIZ1) enhances CBF expression (Miura et al., 2007). Considering the hormonal regulation of Arabidopsis CBF genes, jasmonate positively regulates the expression of CBFs by repressing JAZ1/4 TFs and then increasing the transcriptional activity of ICE1 (Hu et al., 2013). In addition, OST1, a protein kinase involved in abscisic acid (ABA) signaling, phosphorylates and stabilizes ICE1 and suppresses HOS1-mediated ICE1 degradation, thus positively regulating CBFs (Ding et al., 2015). Finally, ethylene represses AtCBF1 to AtCBF3 expression via EIN3 (Shi et al., 2012).
In this study, we showed using both overexpression and mutants that the MtCBF4 TF enhances M. truncatula freezing tolerance and directly and positively regulates the expression of the cold acclimation gene MtCAS15. A specific MYB TF, MtMYB3, was identified as binding directly to the MtCBF4 promoter and repressing its expression under normal conditions. In addition, a cold-inducible MYB TF, MtMYB61, interacts with the DNA-binding domain of MtMYB3, likely releasing the inhibition of MtCBF4 expression by MtMYB3 in response to cold stress. This consequently allows the expression of MtCAS15 and other MtCBF4 downstream targets to increase, ultimately enhancing cold acclimation and freezing tolerance in M. truncatula.
RESULTS
MtCBF4 Plays a Positive Role in Cold Acclimation and Freezing Tolerance in M. truncatula
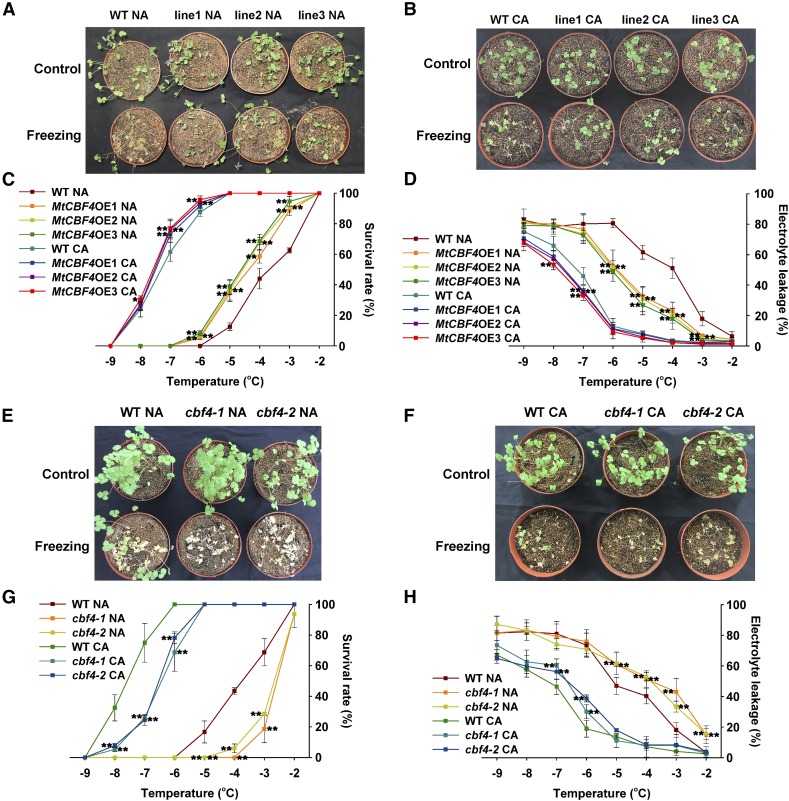
To determine the function of MtCBF4 in freezing tolerance, MtCBF4 was overexpressed in M. truncatula R108 plants. Three stable transgenic lines were selected to determine MtCBF4 expression by quantitative reverse transcription (qRT)-PCR and western blot (Supplemental Fig. S1, A and B). No major dwarf phenotype was detected in MtCBF4-overexpressing plants (Fig. 1, A and B). We examined differences in the level of freezing tolerance between the wild type and MtCBF4-overexpressing lines with and without acclimation to low temperatures. The survival rates of the three lines of nonacclimated MtCBF4-overexpressing plants were significantly higher than those of the wild type at the freezing temperatures of −3°C, −4°C, −5°C, and −6°C (Fig. 1, A and C). Electrolyte leakage, which is an indicator of the degree of cell membrane damage, was lower in MtCBF4-overexpressing lines than in wild-type plants at −4°C, −5°C, and −6°C freezing temperatures (Fig. 1D). After cold acclimation, both transgenic and wild-type plants displayed enhanced freezing tolerance. The survival rates of transgenic plants were slightly higher than that of wild-type plants, and the electrolyte leakage values in MtCBF4-overexpressing lines were lower at −6°C and −7°C (Fig. 1, B–D). These results indicate that MtCBF4 overexpression enhances freezing tolerance more strongly in nonacclimated plants than in cold-acclimated plants.
Figure 1.
MtCBF4 enhances the freezing tolerance of M. truncatula plants. A, Freezing phenotypes of nonacclimated (NA) wild-type (WT) plants and three MtCBF4-overexpressing (MtCBF4OE) lines. Five-week-old plants were grown under a long-day photoperiod at 22°C, and then at −4°C for 1 h, followed by a recovery at 22°C for 4 d (n > 30). B, Freezing phenotypes of cold-acclimated (CA) wild-type plants and three MtCBF4OE lines. Three-week-old plants were grown under a long-day photoperiod at 22°C, then for 2 weeks at 4°C and at −7°C for 1 h, followed by a recovery at 22°C for 4 d (n > 30). C, Survival rates of plants in A and B for the indicated freezing temperatures. Mean values and sd were calculated from the results of three independent experiments. Asterisks indicate significant differences between MtCBF4OE lines and the wild type (Kruskal-Wallis nonparametric test: **, P < 0.01 and *, P < 0.05). D, Electrolyte leakage of excised leaves from plants in A and B for the indicated freezing temperatures. Mean values and sd were calculated from the results of three independent experiments. Asterisks indicate significant differences between MtCBF4OE lines and the wild type (Kruskal-Wallis nonparametric test: **, P < 0.01). E, Freezing phenotypes of nonacclimated wild-type plants and cbf4-1 and cbf4-2 mutants. Plants were grown and stressed as in A (n > 30). F, Freezing phenotypes of cold-acclimated wild-type plants and cbf4-1 and cbf4-2 mutants. Plants were grown and stressed as in B (n > 30). G, Survival rate of plants in E and F for the indicated freezing temperatures. Mean values and sd were calculated from the results of three independent experiments. Asterisks indicate significant differences between cbf4-1 and cbf4-2 mutants and the wild type (Kruskal-Wallis nonparametric test: **, P < 0.01). H, Electrolyte leakage of excised leaves from plants in E and F for the indicated freezing temperatures. Mean values and sd were calculated from the results of three independent experiments. Asterisks indicate significant differences between cbf4-1 and cbf4-2 mutants and the wild type (Kruskal-Wallis nonparametric test: **, P < 0.01).
To further identify the function of CBF4 in cold acclimation, we screened a Tnt1 insertion mutant collection (Cheng et al., 2014). Two allelic mutants were identified, cbf4-1 and cbf4-2, with insertions located in the coding sequence (Supplemental Fig. S1C). These two cbf4 mutant alleles displayed freezing-sensitive phenotypes compared with the wild type. Under nonacclimated conditions, the cbf4 mutants displayed decreased survival rates and increased electrolyte leakage than those of wild-type plants at −3°C, −4°C, and −5°C (Fig. 1, E, G, and H). After cold acclimation, these cbf4 mutants also exhibited lower survival rates and higher electrolyte leakage than wild-type plants (−6°C and −7°C; Fig. 1, F–H). Together with the overexpression data, we conclude that MtCBF4 positively regulates freezing tolerance under nonacclimated and cold-acclimated conditions.
MtCBF4 Regulates Genes Linked to Stresses and Development
To identify downstream targets and pathways regulated by MtCBF4, we performed an RNAseq Digital Gene Expression Profiling analysis of the three 35S:MtCBF4 lines compared with the wild type. A total of 26,777,628 and 28,319,270 raw reads were generated from MtCBF4-overexpressing and control (wild-type) samples, respectively. After removing reads containing adapters, poly-N, and low-quality reads, 26,559,842 and 28,117,504 cleaned reads remained in MtCBF4 overexpression and control samples, respectively. Using stringent criteria (P < 0.01), we determined that 179 genes were up-regulated (fold change > 1.5) and 63 genes were down-regulated (fold change < 0.67) in the MtCBF4-overexpressing lines (Supplemental Table S1). Among these, several genes are linked to cold stress responses. For example, MtCOR413 (Medtr4g068520) is involved in maintaining membrane function and integrity under low-temperature stress to limit damaging cell membranes (Breton et al., 2003; Okawa et al., 2008). MtLTI6A (for low temperature-induced protein; Medtr7g111380) and MtLTI6B (Medtr7g111350) regulate the membrane potential (Medina et al., 2001). MtCAS15 (Medtr7g086340) is induced by low temperature in Medicago spp., and a positive correlation between freezing tolerance and its expression level was identified in Medicago spp. (Monroy et al., 1993a; Pennycooke et al., 2008; Zhang et al., 2011). In addition, MtLTI65 (AC235674_16) and MtY3G1 (Medtr2g014050; a late embryogenesis abundant protein) also were reported to participate in cold stress in previous studies (Nordin et al., 1993; Hincha and Thalhammer, 2012).
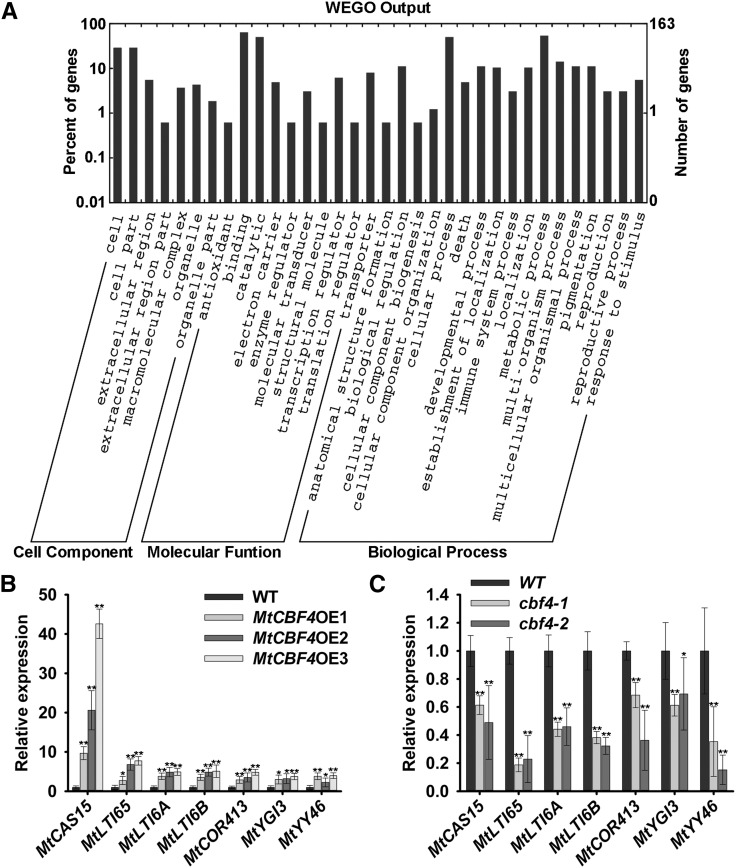
An enrichment analysis of protein functional categories differentially expressed was conducted based on Gene Ontology (GO; Fig. 2A; Supplemental Fig. S2). This revealed that, in addition to abiotic stress responses, MtCBF4 mainly affects biotic responses and developmental processes.
Figure 2.
MtCBF4 regulates various genes linked to developmental and environmental responses. A, Classification of differentially expressed genes based on GO annotation. B, Expression of cold-related genes in the wild type (WT) and the MtCBF4-overexpressing (MtCBF4OE) lines grown at 22°C. Mean values and sd were calculated from the results of three independent experiments (Kruskal-Wallis nonparametric test: **, P < 0.01 and *, P < 0.05). C, Expression of cold-related genes in cbf4-1, cbf4-2, and wild-type plants grown at 22°C. Mean values and sd were calculated from the results of three independent experiments (Kruskal-Wallis nonparametric test: **, P < 0.01 and *, P < 0.05).
The regulation of selected genes associated with freezing tolerance was analyzed by qRT-PCR in plants with altered MtCBF4 expression: MtCAS15, MtLTI65, MtLTI6A, MtLTI6B, and MtCOR413, as well as two WD40 repeat-like protein genes, MtYGI3 (CU651566_11) and MtYY46 (Medtr5g035980; Zhu et al., 2008). MtCBF4-overexpressing plants exhibited higher expression levels of these seven abiotic stress-related genes, and the most highly up-regulated gene was MtCAS15 (Fig. 2B). In contrast, these genes were significantly down-regulated in cbf4-1 and cbf4-2 compared with wild-type plants (Fig. 2C). These results suggest that CBF4 regulates several genes previously linked to cold acclimation and that MtCBF4 overexpression likely improves freezing tolerance by regulating the expression of multiple cold-response genes.
MtCBF4 Directly Binds the Promoter of MtCAS15
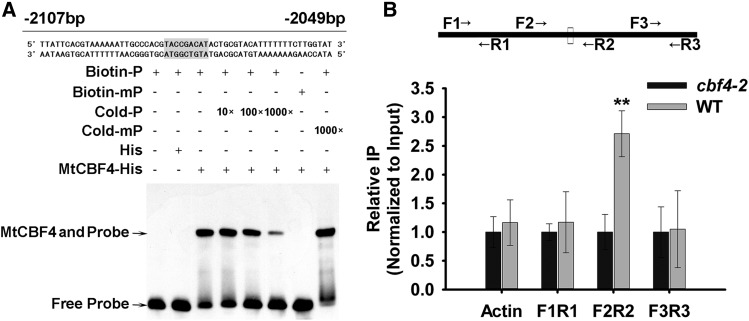
MtCAS15 is one of the most significantly up-regulated genes in MtCBF4-overexpressing transgenic plants, as indicated by the qRT-PCR analyses. Therefore, we used an electrophoretic mobility shift assay (EMSA) to determine whether MtCBF4 binds directly to the MtCAS15 promoter. The MtCBF4 protein indeed bound a wild-type probe containing the MtCAS15 promoter DRE cis-element (TACCGACAT), and this binding efficiently competed with an unlabeled probe (Fig. 3A). However, MtCBF4 did not bind a mutant probe containing a mutated DRE cis-element (Fig. 3A; Supplemental Table S2), and an unlabeled mutant probe did not compete with the binding of MtCBF4 to the wild-type probe. Altogether, these results indicate that MtCBF4 can bind to a DRE cis-element in the MtCAS15 promoter in vitro.
Figure 3.
MtCBF4 binds to the promoter of MtCAS15. A, EMSA to test MtCBF4 binding to DRE cis-elements in the promoter of MtCAS15. A purified His-C terminus of MtCBF4 fusion protein (200 ng) was incubated with 20 fm of a biotin-labeled probe or a mutant probe (see “Materials and Methods”). For the competition assay, a nonlabeled probe with different concentrations (from 10 to 1,000 times) or a nonlabeled mutant probe (1,000 times) was added. The highlighted areas represent the DRE cis-element. The numbers of base pairs upstream of ATG are indicated on the primer sequence. B, ChIP assay showing the binding of MtCBF4 to the promoter of MtCAS15. Wild-type (WT) plants and the anti-CBF4 antibody were used for the ChIP assay, and cbf4-2 mutant plants were used as a negative control. Three pairs of primers were used for PCR: the F2/R2 pair covering the MtCAS15 promoter region containing the DRE cis-element, the F1/R1 pair located upstream of the DRE cis-element, and the F3/R3 pair located downstream of the DRE cis-element. MtACTIN was used as a negative control. Mean values and sd were calculated from the results of three independent experiments (Kruskal-Wallis nonparametric test: **, P < 0.01).
To determine whether MtCBF4 binds to the DRE cis-element of the MtCAS15 promoter in vivo, a chromatin immunoprecipitation (ChIP)-quantitative PCR assay was performed using wild-type compared with cbf4-2 mutant plants. The anti-CBF4 antibody was used to immunoprecipitate. The MtCAS15 promoter fragment F2R2 (primers MtCAS15-ChIP-qPCRF2 and MtCAS15-ChIP-qPCRR2) containing the DRE cis-element was enriched in wild-type immunoprecipitate compared with the cbf4-2 mutant as control plants, whereas promoter fragments lacking the DRE cis-element, F1R1 (primers MtCAS15-ChIP-qPCRF1 and MtCAS15-ChIP-qPCRR1) and F3R3 (primers MtCAS15-ChIP-qPCRF3 and MtCAS15-ChIP-qPCRR3), which are upstream and downstream, respectively, of F2R2, were not enriched. Furthermore, an MtACTIN fragment used as an additional negative control was not enriched in the wild-type immunoprecipitate (Fig. 3B). These results indicate that MtCBF4 protein can bind to the MtCAS15 promoter in vivo, suggesting that, in M. truncatula, MtCBF4 directly and positively regulates at least MtCAS15.
A MYB Transcription Factor, MtMYB3, Directly Binds the Promoter of MtCBF4
To understand how MtCBF4 was transcriptionally induced in response to cold, we searched the MtCBF4 promoter region sequence (2 kb upstream of the start codon) using the Plant cis-Acting Regulatory DNA Elements database (Higo et al., 1999). Different putative cis-elements were identified, including nine predicted binding sites for MYB TFs. Two types of these MYB cis-elements possess two sites in the promoter, and the others possess only one. Each of the MYB cis-elements was inserted into pAbAi vector and then transformed into yeast strain Saccharomyces cerevisiae Y1HGOLD. The basal expression of the six vectors containing MYB cis-elements except the MYB-binding site (TAGTTA) was not suppressed by 1,000 ng mL−1 aureobasidin A, likely because of these MYB cis-elements being recognized by endogenous yeast transcription factors. Therefore, only the MYB-binding site (TAGTTA) was used for the yeast one-hybrid screen of the library. Fifteen candidate genes were identified in the screen, including two MYB TFs. Following confirmation analyses of the two MYB candidates in a one-to-one experimental design, only the Medtr1g086530 gene (J. Craig Venter Institute database, Mt genome version 4.0) encoding a putative R2R3-MYB TF was confirmed and referred to as MtMYB3. MtMYB3 shows nuclear localization in onion (Allium cepa) epidermal cells (Supplemental Fig. S3A) and displays transcriptional activity in yeast (Saccharomyces cerevisiae YRG-2; Supplemental Fig. S3B).
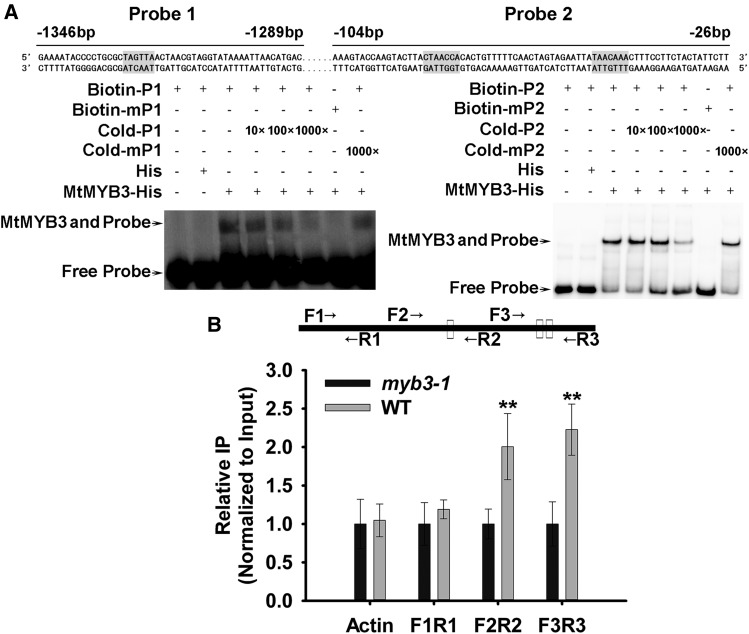
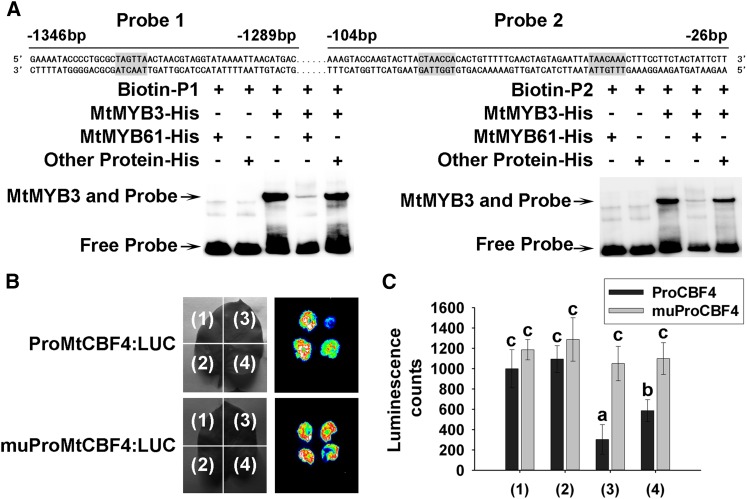
To determine whether MtMYB3 binds directly to the MtCBF4 promoter in vitro, an EMSA was performed using a His-tagged MtMYB3 protein. MtMYB3 bound two different sites on the MtCBF4 promoter, P1 (containing the predicted MYB-binding sequence TAGTTA) and P2 (containing CTAACCA and TAACAAA sequences). However, MtMYB3 did not bind to the other predicted MYB-binding sites (Supplemental Fig. S4D). When MYB cis-elements were mutated, MtMYB3 binding was abolished (Fig. 4A; Supplemental Table S2). These results indicate that the MtMYB3 TF can bind specifically to the MtCBF4 promoter in vitro.
Figure 4.
MtMYB3 binds to the promoter of MtCBF4. A, EMSA to test MtMYB3 binding to MYB cis-elements in the promoter of MtCBF4. A purified His-C terminus of MtMYB3 fusion protein (200 ng) was incubated with 20 fm of a biotin-labeled probe or a mutant probe (see “Materials and Methods”). For the competition assay, a nonlabeled probe with different concentrations (from 10 to 1,000 times) or a nonlabeled mutant probe (1,000 times) was added. The highlighted areas represent the MYB cis-elements. The numbers of base pairs upstream of ATG are indicated on the primer sequences. B, ChIP assay showing the binding of MtMYB3 with the promoter of MtCBF4. Wild-type (WT) plants as well as the anti-MYB3 antibody were used for the ChIP assay, and myb3-1 mutant seedlings were used as a negative control. Three pairs of primers were used: the F2/R2 and F3/R3 pairs covering the MtCBF4 promoter region containing the MYB cis-elements and the F1/R1 pair located upstream of the MYB cis-elements. MtACTIN was used as a negative control. Mean values and sd were calculated from the results of three independent experiments (Kruskal-Wallis nonparametric test: **, P < 0.01).
The ChIP experiment between the wild type and the myb3-1 mutant was performed to determine whether MtMYB3 also bound the MtCBF4 promoter in vivo. The anti-MYB3 antibody was used. The result demonstrates that fragments F2R2 (primers MtCBF4-ChIP-qPCRF2 and MtCBF4-ChIP-qPCRR2) and F3R3 (primers MtCBF4-ChIP-qPCRF3 and MtCBF4-ChIP-qPCRR3), which contain MYB cis-elements, were enriched in the wild-type immunoprecipitate compared with myb3-1 mutant plants, whereas F1R1 (the promoter region upstream of the predicted MYB cis-elements; primers MtCBF4-ChIP-qPCRF1 and MtCBF4-ChIP-qPCRR1) was not enriched (Fig. 4B). Again, MtACTIN amplification, used as an additional negative control, was not enriched in the wild-type immunoprecipitate. Taken together, these results indicate that MtCBF4 is a direct target of MtMYB3.
MtMYB3 Plays a Negative Role in Cold Acclimation
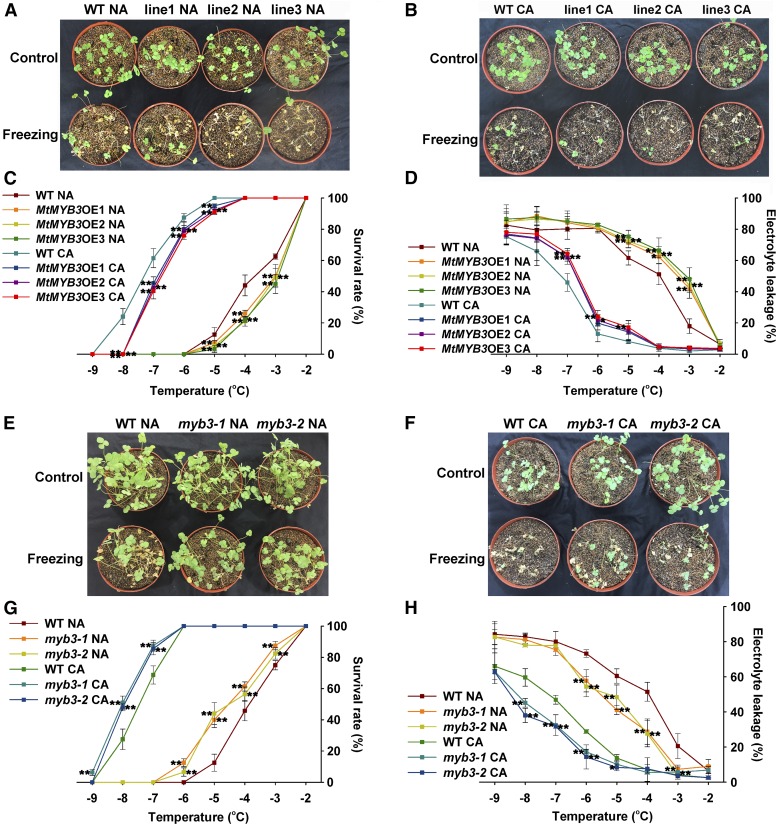
To substantiate the role of MtMYB3 in cold acclimation, we obtained 35S:MtMYB3-3xFLAG transgenic M. truncatula R108 plants and analyzed the expression levels of the transgene by qRT-PCR and western blotting (Supplemental Fig. S4, A and B); then, MtMYB3-overexpressing and wild-type plants were exposed to a freezing stress with or without acclimation to low temperatures. Under the condition of nonacclimation, the survival rates of MtMYB3-overexpressing plants were decreased significantly, and the electrolyte leakage values were increased consistently compared with those of wild-type plants at −3°C, −4°C, and −5°C (Fig. 5, A, C, and D). After cold acclimation, the survival rates of MtMYB3-overexpressing plants also were decreased substantially, and the electrolyte leakage values were higher than those of wild-type plants at −5°C, −6°C, and −7°C (Fig. 5, B–D). Therefore, we concluded that MtMYB3 overexpression confers reduced freezing tolerance in M. truncatula.
Figure 5.
MtMYB3 decreases the freezing tolerance of M. truncatula plants. A, Freezing phenotypes of nonacclimated (NA) wild-type (WT) plants and MtMYB3-overexpressing (MtMYB3OE) lines. Five-week-old plants were grown under a long-day photoperiod at 22°C, and then at −4°C for 1 h, followed by a recovery at 22°C for 4 d (n > 30). B, Freezing phenotypes of cold-acclimated (CA) wild-type plants and MtMYB3OE lines. Three-week-old plants were grown under a long-day photoperiod at 22°C, then for 2 weeks at 4°C and at −7°C for 1 h, followed by a recovery at 22°C for 4 d (n > 30). C, Survival rates of plants in A and B for the indicated freezing temperatures. Mean values and sd were calculated from the results of three independent experiments. Asterisks indicate significant differences between MtMYB3OE lines and the wild type (Kruskal-Wallis nonparametric test: **, P < 0.01). D, Electrolyte leakage of excised leaflets from plants in A and B for the indicated freezing temperatures. Mean values and sd were calculated from the results of three independent experiments. Asterisks indicate significant differences between MtMYB3OE lines and the wild type (Kruskal-Wallis nonparametric test: **, P < 0.01 and *, P < 0.05). E, Freezing phenotypes of nonacclimated wild-type plants and myb3-1 and myb3-2 mutants. Plants were grown and stressed as in A (n > 30). F, Freezing phenotypes of cold-acclimated wild-type plants and myb3-1 and myb3-2 mutants. Plants were grown and stressed as in B (n > 30). G, Survival rates of plants in E and F for the indicated freezing temperatures. Mean values and sd were calculated from the results of three independent experiments. Asterisks indicate significant differences between myb3-1 and myb3-2 mutants and the wild type (Kruskal-Wallis nonparametric test: **, P < 0.01). H, Electrolyte leakage of excised leaflets from plants in E and F for the indicated freezing temperatures. Mean values and sd were calculated from the results of three independent experiments. Asterisks indicate significant differences between myb3-1 and myb3-2 mutants and the wild type (Kruskal-Wallis nonparametric test: **, P < 0.01 and *, P < 0.05).
Two Tnt1 insertions were identified in the MtMYB3 gene, each located in the first exon (Supplemental Fig. S4C). Both myb3-1 and myb3-2 mutants showed higher survival rates and lower electrolyte leakage than those of wild-type plants at −3°C, −4°C, −5°C, and −6°C under non-cold-acclimated conditions (Fig. 5, E, G, and H). After cold acclimation, these myb3 mutants also displayed higher survival rates and lower electrolyte leakage (−7°C and −8°C; Fig. 5, F–H). Therefore, we concluded that the freezing tolerance was enhanced in myb3 mutants compared with wild-type plants. These results, together with the overexpression data, demonstrate that MtMYB3 negatively regulates cold acclimation.
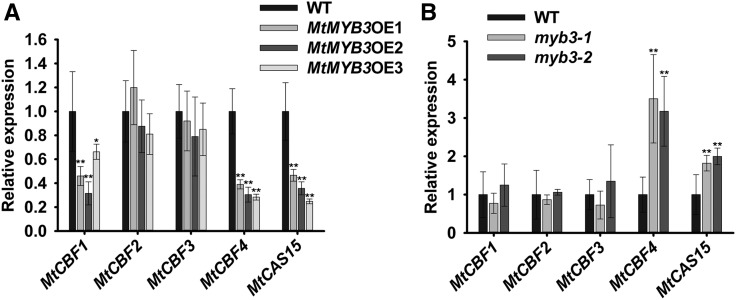
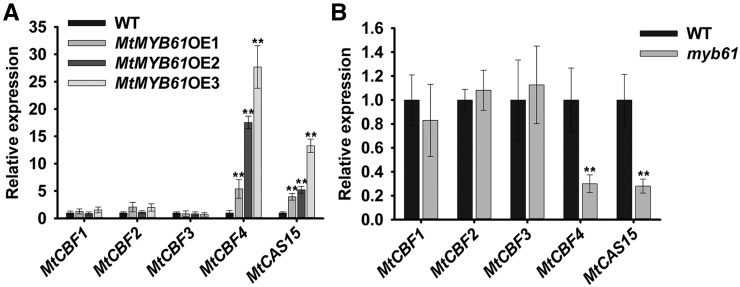
We further examined the expression of the cold stress-responsive genes MtCBF1, MtCBF2, MtCBF3, MtCBF4, and MtCAS15 in 35S:MtMYB3-3FLAG transgenic and wild-type plants via qRT-PCR. Expression levels of MtCBF1 and MtCBF4 were decreased significantly, whereas those of MtCBF2 and MtCBF3 displayed no significant change (Fig. 6A). MtCAS15, a direct target of MtCBF4, also showed decreased expression in MtMYB3-overexpressing plants. Accordingly, expression levels of MtCBF4 and MtCAS15 were enhanced in myb3-1 and myb3-2 mutants compared with wild-type plants, indicating that MtMYB3 negatively regulates the expression levels of at least MtCBF4 and MtCAS15 (Fig. 6B). MtCBF1 expression was not enhanced in the myb3 mutant, possibly because its regulation involves other TFs. Taken together, these results indicate that MtMYB3 is a negative regulator of the MtCBF4/MtCAS15 regulatory module acting in cold acclimation.
Figure 6.
MtMYB3 down-regulates the expression of MtCBF4. A, Expression of MtCBFs and MtCAS15 in wild-type (WT) plants and MtMYB3-overexpressing (MtMYB3OE) lines grown at 22°C. Mean values and sd were calculated from the results of three independent experiments (Kruskal-Wallis nonparametric test: **, P < 0.01 and *, P < 0.05). B, Expression of MtCBFs and MtCAS15 in wild-type plants and myb3-1 and myb3-2 mutants grown at 22°C. Mean values and sd were calculated from the results of three independent experiments (Kruskal-Wallis nonparametric test: **, P < 0.01).
MtMYB3 Interacts with MtMYB61 in Vitro and in Vivo
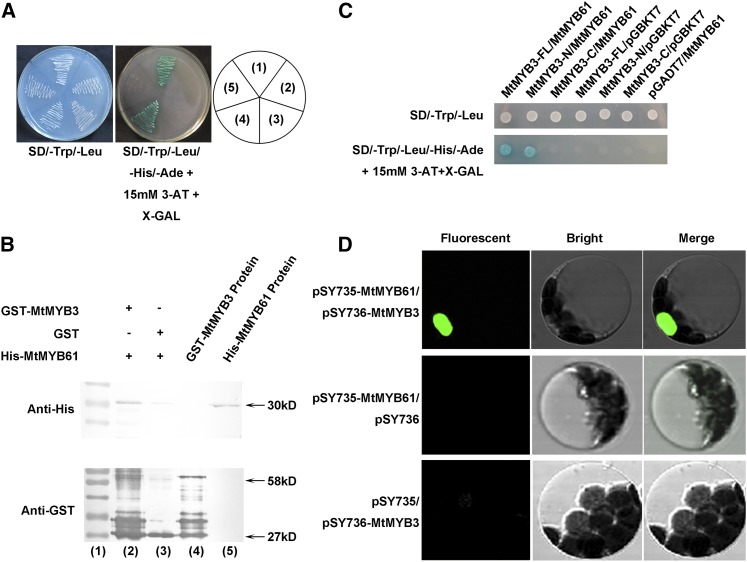
The function of MtMYB3 was identified as negatively regulating MtCBF4, which does not explain how MtCBF4 expression is induced by cold stress. To gain insights into MtMYB3-MtCBF4 regulation in relation to cold acclimation, MtMYB3 was used as a bait protein to perform a yeast two-hybrid screen (Supplemental Table S3). Interestingly, another MYB TF, MtMYB61 (Medtr1g086510; JCVI database, Mt genome version 4.0), which also belongs to the R2R3-MYB subfamily, was the most frequently retrieved candidate gene identified in the screen and, therefore, was selected for further analyses. MtMYB61 localizes in the nucleus in onion epidermal cells (Supplemental Fig. S5A) and displays transcriptional activity in yeast cells (Supplemental Fig. S5B). MtMYB3 and/or MtMYB61 were expressed simultaneously in yeast (Saccharomyces cerevisiae AH109), and HIS3, ADE2, and MEL1 reporter genes were activated only when both MYB TFs were expressed, indicating that the MtMYB3 interaction with MtMYB61 is relevant in yeast for transcriptional activation (Fig. 7A). The interaction between MtMYB3 and MtMYB61 was independently validated using a glutathione S-transferase (GST) pull-down experiment. The results showed that the MtMYB3-GST protein interacts with the MtMYB61-His protein, whereas the negative control GST protein did not show the interaction signal (Fig. 7B). We then mapped domains that were required for the MtMYB3-MtMYB61 interaction using yeast two-hybrid assays. MtMYB61 interacts with the N-terminal (amino acids 1–133) DNA-binding domain of MtMYB3 but not with the C-terminal region (amino acids 134–278; Fig. 7C).
Figure 7.
MtMYB3 interacts with MtMYB61. A, Analysis of the interaction between MtMYB3 and MtMYB61 using a yeast two-hybrid assay on a synthetic dropout (SD)/4D agar selective medium (-Trp,-Leu,-His,-Ade) with 15 mm 3-aminotriazole (3-AT). Sectors are as follows: (1) pGBKT7-MtMYB61/pGADT7-MtMYB3; (2) pGBKT7-MtMYB61/pGADT7; (3) pGBKT7/pGADT7-MtMYB3; (4) pGBKT7-53/pGADT7-RecT (+, positive control); and (5) pGBKT7/pGADT7 (−, negative control). B, Analysis of the interaction between MtMYB3 and MtMYB61 using a GST pull-down assay. A GST or a GST-MtMYB3 protein was bound to beads and then incubated with His-MtMYB61 proteins. SDS-PAGE separated the eluted proteins, then anti-His and anti-GST antibodies were used for western blot. Lanes are as follows: (1) protein size marker; (2) pull-down assay of GST-MtMYB3; (3) pull-down assay of GST; (4) GST-MtMYB3 protein (control); and (5) His-MtMYB61 proteins (control). C, A yeast two-hybrid assay was conducted to analyze the interaction between MtMYB3 full length (FL), MtMYB3-N (amino acids 1–133), MtMYB3-C (amino acids 134–278), and MtMYB61 on an SD/4D agar selective medium (-Trp,-Leu,-His,-Ade) with 15 mm 3-AT. D, Interaction between MtMYB61 and MtMYB3 in Arabidopsis protoplasts using BiFC.
A bimolecular fluorescence complementation (BiFC) assay was additionally performed to further test the interaction between MtMYB3 and MtMYB61 in vivo. MtMYB61 and MtMYB3 were cotransformed into Arabidopsis protoplasts, and yellow fluorescent protein fluorescence was detected in the nucleus using confocal laser-scanning microscopy (Fig. 7D), whereas no green fluorescence was detected in negative controls. These results indicate that MtMYB3 and MtMYB61 can interact both in vivo and in vitro.
MtMYB61 Plays a Positive Role in Cold Acclimation
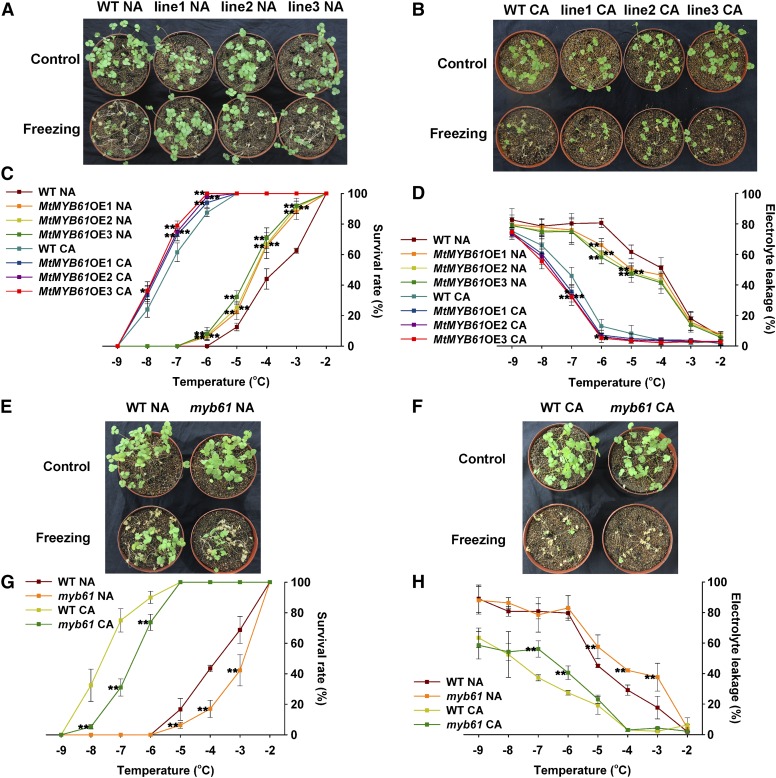
Because MtMYB3 directly regulates MtCBF4 expression, we investigated whether MtMYB61 also is involved in cold responses. As was done previously for MtCBF4 and MtMYB3, we overexpressed MtMYB61 and obtained stable transgenic M. truncatula R108 lines (Supplemental Fig. S6, A and B). In the absence of cold acclimation, MtMYB61-overexpressing plants exhibited significantly higher survival rates and lower electrolyte leakage (−5°C and −6°C) than wild-type plants (Fig. 8, A, C, and D). After cold acclimation, MtMYB61-overexpressing plants also showed increased survival rates and decreased electrolyte leakage compared with wild-type plants (Fig. 8, B–D). MtMYB61 increased the freezing tolerance of nonacclimated plants more strongly than that of cold-acclimated plants.
Figure 8.
MtMYB61 enhances the freezing tolerance of M. truncatula plants. A, Freezing phenotypes of nonacclimated (NA) wild-type (WT) plants and MtMYB61-overexpressing (MtMYB61OE) lines. Five-week-old plants were grown under a long-day photoperiod at 22°C, and then at −4°C for 1 h, followed by a recovery at 22°C for 4 d (n > 30). B, Freezing phenotypes of cold-acclimated (CA) wild-type plants and the MtMYB61OE lines. Three-week-old plants were grown under a long-day photoperiod at 22°C, then for 2 weeks at 4°C and at −7°C for 1 h, followed by a recovery at 22°C for 4 d (n > 30). C, Survival rate of plants in A and B for the indicated freezing temperatures. Mean values and sd were calculated from the results of three independent experiments. Asterisks indicate significant differences between MtMYB61OE lines and the wild type (Kruskal-Wallis nonparametric test: **, P < 0.01 and *, P < 0.05). D, Electrolyte leakage of excised leaves from plants in A and B for the indicated freezing temperatures. Mean values and sd were calculated from the results of three independent experiments. Asterisks indicate significant differences between MtMYB61OE lines and the wild type (Kruskal-Wallis nonparametric test: **, P < 0.01 and *, P < 0.05). E, Freezing phenotypes of nonacclimated wild-type plants and the myb61 mutant. Plants were grown and stressed as in A (n > 30). F, Freezing phenotypes of cold-acclimated wild-type plants and the myb61 mutant. Plants were grown and stressed as in B (n > 30). G, Survival rates of plants in E and F for the indicated freezing temperatures. Mean values and sd were calculated from the results of three independent experiments. Asterisks indicate significant differences between the myb61 mutant and the wild type (Kruskal-Wallis nonparametric test: **, P < 0.01). H, Electrolyte leakage of excised leaves from plants in E and F for the indicated freezing temperatures. Mean values and sd were calculated from the results of three independent experiments. Asterisks indicate significant differences between the myb61 mutant and the wild type (Kruskal-Wallis nonparametric test: **, P < 0.01).
A Tnt1 insertional mutant was identified, myb61, with an insertion located in the first exon (Supplemental Fig. S6C), which showed lower survival rates and higher electrolyte leakage than the wild type at −3°C, −4°C, and −5°C under nonacclimated conditions (Fig. 8, E, G, and H). After cold acclimation, myb61 also showed lower survival rates and higher electrolyte leakage (−6°C and −7°C) than wild-type plants (Fig. 8, F–H). These results, together with the overexpression data, led us to conclude that MtMYB61 positively regulates freezing tolerance.
To determine how MtMYB61 regulates cold stress responses at the molecular level with respect to the MtCBF4/MtCAS15 module, we examined the expression of MtCBFs and MtCAS15 in MtMYB61-overexpressing plants and in the myb61 mutant. MtMYB61 overexpression was correlated with increased MtCBF4 and MtCAS15 expression (Fig. 9A). Conversely, the expression levels of MtCBF4 and MtCAS15 were decreased in the myb61 mutant (Fig. 9B). In contrast, MtCBF1, MtCBF2, and MtCBF3 exhibited no major change in their transcriptional levels in MtMYB61-overexpressing or mutant plants (Fig. 9). Therefore, the positive role of MtMYB61 in freezing tolerance is correlated with the expression level of MtCBF4 and MtCAS15, suggesting an opposite function to that of the MtMYB3 TF.
Figure 9.
MtMYB61 up-regulates the expression of MtCBF4. A, Expression of MtCBFs and MtCAS15 in wild-type (WT) plants and the MtMYB61-overexpressing (MtMYB61OE) lines grown at 22°C. Mean values and sd were calculated from the results of three independent experiments (Kruskal-Wallis nonparametric test: **, P < 0.01). B, Expression of MtCBFs and MtCAS15 in wild-type plants and the myb61 mutant grown at 22°C. Mean values and sd were calculated from the results of three independent experiments (Kruskal-Wallis nonparametric test: **, P < 0.01).
MtMYB61 Inhibits MtMYB3 Binding to the MtCBF4 Promoter
Because MtMYB61 positively regulates MtCBF4/MtCAS15 expression, whereas MtMYB3 directly negatively regulates MtCBF4/MtCAS15 expression, and because MtMYB61 interacts physically with MtMYB3, we tested its potential direct interaction with the MtCBF4 promoter. EMSA revealed no detectable binding of MtMYB61 to any cis-element of the MtCBF4 promoter (Fig. 10A; Supplemental Fig. S6D), indicating that MtMYB61 may not interact directly with the MtCBF4 promoter. These results led us to hypothesize that MtMYB61 may interfere with the suppression of MtMYB3 in the expression of MtCBF4.
Figure 10.
MtMYB61 decreases the binding activity of MtMYB3 to MYB cis-elements in the promoter of MtCBF4. A, EMSA using biotin-labeled MYB cis-elements in the promoter of MtCBF4 and purified protein MtMYB3 (200 ng) incubated with (+) or without (−) MtMYB61 protein (200 ng) or with an unrelated His-C terminus of MtWRKY76 fusion protein (XM_003588780; National Center for Biotechnology Information database; 200 ng). The highlighted areas represent the MYB cis-elements. The numbers of base pairs upstream of ATG are indicated on the primer sequences. B, MtMYB61 decreased the binding activity of MtMYB3 on MYB cis-elements in the promoter of MtCBF4 in vivo. N. benthamiana leaves were transformed with the ProMtCBF4:LUC or ProMtCBF4:LUC construct bearing mutated MYB cis-elements and a vector control (1), Pro-35S:MtMYB61 (2), Pro-35S:MtMYB3 (3), or Pro-35S:MtMYB61 and Pro-35S:MtMYB3 (4). C, Quantification of LUC activity. Mean values and sd were calculated from the results of three independent experiments (Kruskal-Wallis nonparametric test: significant differences are indicated by letters, α < 0.05).
We performed EMSA using both the MtMYB61 and MtMYB3 proteins. The addition of equal amounts of MtMYB61 protein led to reduced binding of the MtCBF4 promoter by MtMYB3, a result that was not observed using a negative control protein (Fig. 10A). These results indicate that MtMYB61 inhibits the binding of MtMYB3 to MYB cis-elements in the MtCBF4 promoter in vitro.
In addition, we tested the in vivo relationships between MtMYB3 and MtMYB61 in the regulation of MtCBF4 expression using Nicotiana benthamiana transactivation assays. No difference in ProMtCBF4:LUC (for luciferase) expression was detected when a control empty vector or Pro-35S:MtMYB61 vector was used, indicating that MtMYB61 is not sufficient to modulate MtCBF4 expression. In contrast, MtMYB3 decreased MtCBF4 expression, further indicating the negative transcriptional function of this TF. The combination of the Pro-35S:MtMYB3 and Pro-35S:MtMYB61 vectors revealed that the negative effect of MtMYB3 was partially relieved in the presence of MtMYB61, leading to an intermediate activation of the reporter gene (Fig. 10, B and C). When using a ProMtCBF4:LUC construct in which the MYB cis-elements were mutated, no difference was observed in any of the conditions tested (control vector, Pro-35S:MtMYB3, Pro-35S:MtMYB61, or the combination of Pro-35S:MtMYB3 and Pro-35S:MtMYB61; Fig. 10, B and C). These results indicate that MtMYB61 is able to inhibit the negative effect of MtMYB3 on MtCBF4 expression depending on MYB cis-element.
Cold Rapidly Induces the Expression of MtMYB61 and Decreases MtMYB3 Binding to the MtCBF4 Promoter
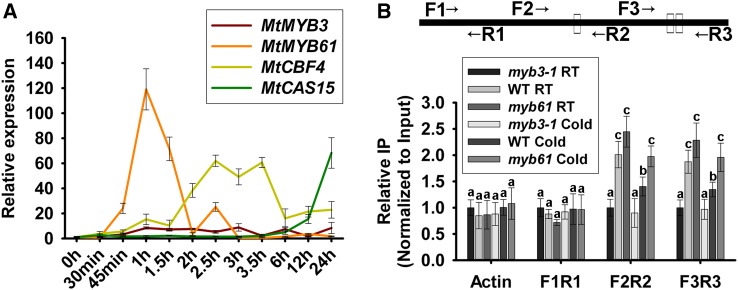
The mechanism by which MtMYB61 and MtMYB3 regulate MtCBF4 expression depending on the cold stress condition was further investigated in M. truncatula. First, qRT-PCR was performed to analyze the transcript levels of MtMYB3, MtMYB61, MtCBF4, and MtCAS15 in wild-type plants submitted to a cold stress kinetic (4°C from 0 to 24 h). MtMYB3 transcript levels showed no strong change in response to cold, whereas MtMYB61 expression was rapidly and strongly induced after cold treatment, culminating at 1 h before rapidly returning to basal level. MtCBF4 also strongly responded to cold, but more slowly, reaching its highest level between 2.5 and 3.5 h, whereas MtCAS15 transcript levels showed a strong induction at 12 and 24 h (Fig. 11A). These results indicate that MtMYB61 is induced earlier than MtCBF4 when exposed to cold stress. Thus, the kinetic is consistent with a model whereby MtMYB61 induction in response to cold may allow the induction of MtCBF4 expression, which consequently increases the transcription of MtCAS15.
Figure 11.
MtMYB61 relieves the inhibition of MtCBF4 by MtMYB3 during cold stress in M. truncatula plants. A, Transcript levels of MtMYB3, MtMYB61, MtCBF4, and MtCAS15 genes in response to a low temperature (4°C) in the wild type. Five-week-old seedlings were grown at 22°C and then at 4°C for the indicated periods of time. Mean values and sd were calculated from the results of three independent experiments. B, ChIP assay showing that the efficiency of MtMYB3 binding on the MtCBF4 promoter was affected by MtMYB61. Leaves from wild-type (WT), myb3-1, and myb61 plants, as well as an anti-MYB3 antibody, were used for a ChIP experiment. Three pairs of primers were used: F2/R2 and F3/R3 pairs covering the MtCBF4 promoter region containing MYB cis-elements and the F1/R1 pair located upstream of the MYB cis-elements. MtACTIN was used as a negative control. Mean values and sd were calculated from the results of three independent experiments (Kruskal-Wallis nonparametric test: significant differences are indicated by letters, α < 0.05). RT, Room temperature.
In addition, we determined whether the efficiency of MtMYB3 binding to the MtCBF4 promoter was affected by MtMYB61. Leaves from wild-type, myb3-1, and myb61 plants were used for a ChIP experiment. Using the same primers as before, fragments F2R2 (primers MtCBF4-ChIP-qPCRF2 and MtCBF4-ChIP-qPCRR2) and F3R3 (primers MtCBF4-ChIP-qPCRF3 and MtCBF4-ChIP-qPCRR3) contain the MYB cis-elements in the promoter of MtCBF4, and the fragment F1R1 (primers MtCBF4-ChIP-qPCRF1 and MtCBF4-ChIP-qPCRR1) is located in the promoter region upstream of the predicted MYB cis-elements. The F2R2 and F3R3 regions of the MtCBF4 promoter were significantly enriched in immunoprecipitates performed with the wild-type compared with the myb3-1 mutant (Fig. 11B). Enrichment of the F2R2 and F3R3 regions in the myb61 mutant was slightly higher than in wild-type plants. In contrast, neither the MtCBF4 promoter F1R1 region nor MtACTIN was enriched in any of the immunoprecipitates (wild type, myb61, or myb3-1). Under cold stress, the enrichment of the F2R2 and F3R3 regions was decreased significantly in the wild type but not in the myb61 mutant (Fig. 11B). These results suggest that MtMYB61 can modulate the binding of MtMYB3 to the MtCBF4 promoter in M. truncatula. Above all, these results suggest that the accumulation of MtMYB61 affects the binding of MtMYB3 to the MtCBF4 promoter, subsequently increasing the transcription of MtCBF4.
DISCUSSION
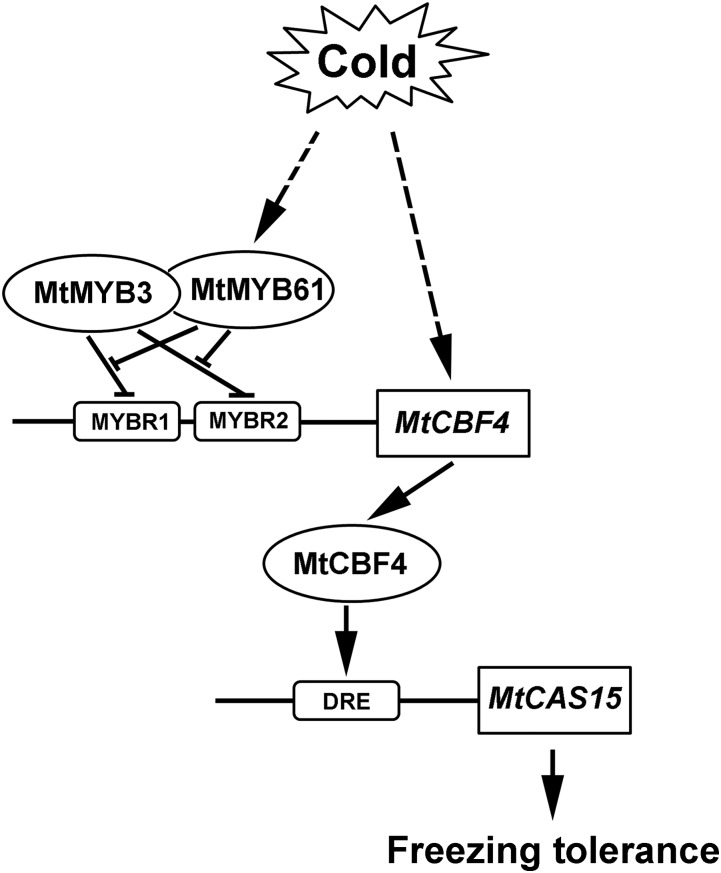
In this study, we identified an MYB/CBF transcriptional network controlling cold acclimation in a legume plant (Fig. 12). Under nonstressed conditions, the expression of MtCBF4 is repressed by the binding of MtMYB3 to its promoter. Rapidly after a cold stress, MtCBF4 expression is enhanced depending on the transcriptional induction of MtMYB61, which interacts with the DNA-binding domain of MtMYB3, relieving MtMYB3-dependent transcriptional inhibition of MtCBF4. The accumulation of MtCBF4 leads to the direct transcriptional activation of cold acclimation gene expression, such as MtCAS15, and ultimately improves plant freezing tolerance. In this model, MtCBF4/MtCAS15 transcription is precisely controlled by cold depending on the antagonism between MtMYB3 and MtMYB61. However, it remains to be determined how MtMYB61 expression is rapidly induced by cold.
Figure 12.
Model for MtMYB3/MtMY61 regulation of the MtCBF4/MtCAS15 cold acclimation signaling module in M. truncatula. Under nonstressed conditions, the expression level of MtCBF4 is repressed by MtMYB3 binding on its promoter. Cold stress rapidly induces the transcription of MtMYB61, which interacts with the DNA-binding domain of MtMYB3 and relieves the transcriptional inhibition of MtCBF4 by MtMYB3. This activates the expression of MtCBF4, which directly induces the expression of the cold acclimation-specific gene MtCAS15.
MtCBF4 was identified from microarray data previously; we named it MtCBF4 because previous studies in M. truncatula had already identified and characterized MtCBF1, MtCBF2, and MtCBF3 (Li et al., 2011). Therefore, we named MtCBF4 in accordance with the order of gene discovery in M. truncatula. The expression of MtCBF4 is induced by cold, salt, drought, and ABA, and MtCBF4 overexpression increases salt and drought tolerance in transgenic Arabidopsis plants (Li et al., 2011). It is possible, therefore, that the transcriptional network identified here could be extrapolated to other abiotic stress responses and ABA regulation. The phenotypes of MtCBF4-overexpressing and mutant plants reveal that this TF regulates cold acclimation in M. truncatula, consistent with previous studies showing that freezing tolerance in this species depends on cold-acclimating conditions (Zhang et al., 2011). Among the different genes that are activated in MtCBF4-overexpressing plants, at least MtCAS15 is directly up-regulated by MtCBF4. In Arabidopsis, COR genes are induced by cold stress via CBF TFs and are positively associated with freezing tolerance (Gilmour et al., 1998; Maruyama et al., 2004; Vogel et al., 2005). In M. truncatula, we show here that a CBF directly activates the expression level of a CAS gene and enhances freezing tolerance, suggesting that the regulation of cold acclimation genes by CBFs is evolutionarily conserved. MtCBF4 showed slightly higher sequence identity with AtCBF4 (57%) than with AtCBF1 to AtCBF3 (53%; Li et al., 2011). Phylogenetic analyses revealed that DREB1/CBF proteins from Arabidopsis, M. truncatula, and soybean fall into four groups: MtCBF2 to MtCBF4 and GmCBF1 and GmCBF2 were clustered in one group; AtCBF1 to AtCBF4 were clustered in another group, MtCBF1, GmCBF3, AtDDF1, and AtDDF2 were clustered in the third group; and GmDREB1 and GmDREB2 were clustered in the fourth group (Li et al., 2011). Although the function of MtCBF4 was similar to those of AtCBFs, which AtCBF is homologous to MtCBF4 is unclear.
Transcriptomic analyses of CBF4-overexpressing plants reveal that this TF also affects the expression of genes associated with biotic responses. Overlaps between abiotic and biotic stress responses have been reported previously. MtNAC969 plays a negative role in salt stress responses and promotes nitrogen fixation and/or delays nodule senescence (de Zélicourt et al., 2012). SIZ1 up-regulates the expression of CBF3 through sumoylation, stabilizes ICE1 (Miura et al., 2007), and affects the accumulation of salicylic acid and PAD4-mediated pathogen resistance signaling (Lee et al., 2007). DEAR1 represses the expression of DREB1 as well as PR gene expression and the accumulation of salicylic acid, thereby limiting the growth of the Pseudomonas syringae pathogen (Tsutsui et al., 2009). MtCBF4, therefore, may be another example of a TF mediating cross talk between biotic and abiotic stress signaling pathways.
This study additionally identified that MtCBF4 expression in response to cold is directly, rapidly, and tightly regulated by two opposing R2R3-MYB TFs interacting with each other: the MtMYB3 repressor and the cold-induced MYB61 inducer. Overexpression and mutant studies correspondingly indicated that both MYB TFs have opposing functions in cold acclimation and freezing tolerance. The fact that the expression of MtCBF1 to MtCBF3 genes generally is not affected when MtMYB3 or MtMYB61 is altered, in contrast to MtCBF4 and MtCAS15 expression, suggests that this regulatory network may be specific for the MtCBF4 transcriptional regulation. The promoters of MtCBF1 to MtCBF4 were analyzed, and the MYB cis-element TAGTTA bound by MYB3 was specifically found in the promoters of MtCBF1 and MtCBF4 but not in the promoters of MtCBF2 and MtCBF3. Another MYB cis-element, CTAACCA, bound by MYB3 also was specific in the MtCBF4 promoter and was not found in the other MtCBF promoters, although the MtCBF1 and MtCBF2 promoters also possessed the related A/TAACCA sequence. These differences may partially explain why MtMYB3 specifically regulates MtCBF4.
Phylogenetic analyses show that the MYB genes that participate in abiotic stress (Li et al., 2015) from Arabidopsis, Malus domestica, Oryza sativa, Triticum aestivum, N. benthamiana, Vitis vinifera, Solanum tuberosum, Chrysanthemum spp., soybean, Avicennia marina, Leymus chinensis, Pyrus communis, Craterostigma plantagineum, Saccharum officinarum, and M. truncatula were divided into four groups (Supplemental Fig. S7). The genes that clustered together have similar functions. The TAGTTA and CTAACCA sequences found in the MtCBF4 promoter as being bound by MtMYB3 also may bound by AtMYB2 (Abe et al., 2003), which, interestingly, is in the same clade as MtMYB3 (type 1). In addition, the closest soybean homolog of MtMYB3 and MtMYB61, which are themselves both closely related (type 1), is GmMYB76, which was linked previously to cold stress responses (Liao et al., 2008). GmMYB76 can enhance freezing and salt tolerance in transgenic Arabidopsis. Another similar characteristic is that GmMYB76 can form heterodimers with GmMYB177, but limited information is known about the role of this gene in soybean or the associated molecular mechanisms. These similarities strongly suggest that the regulatory network identified here is conserved in the M. truncatula model legume and soybean, highlighting the relevance of this work for breeding improved freezing tolerance in legume crops. The fact that another two abiotic stress-related MYB-encoding genes in M. truncatula, MtMYB634 and MtMYB636, are closely related to AtMYB15 (type 3) suggests that this pathway also may exist in legumes (Agarwal et al., 2006; Liao et al., 2008). Together with the distantly related cold-related GmMYB92 (type 4; Liao et al., 2008), this suggests that several MYB regulatory modules relevant for cold responses coexist in legumes, and their relevance, relative contribution, and potential cross talk in the regulation of plant freezing tolerance remain to be determined. Several features distinguish MtMYB3 and MtMYB61 compared with AtMYB15, despite these phylogenetically distantly related R2R3 MYB TFs all having the ability to bind directly to the promoter of CBF genes. First, AtMYB15 expression is induced rapidly by cold stress, similar to MtMYB61, but in contrast to MtMYB3. Second, AtMYB15 represses AtCBF1, AtCBF2, and AtCBF3 transcription (Agarwal et al., 2006), similar to MtMYB3 for MtCBF4, whereas MtMYB61 activates MtCBF4 expression. Thus, the cold regulation of MYB/CBF regulatory modules is complex and seems to rely on a species-specific combination of TFs.
The basic helix-loop-helix TF AtICE1 is a positive regulator of AtCBF3 and interacts physically with MYB15 (Chinnusamy et al., 2003; Agarwal et al., 2006). This observation suggests that other TFs than MYBs also may participate in regulating CBF gene expression in legumes. Accordingly, MtICE1 (Medtr7g083900.1; JCVI database, Mt genome version 4.0), the protein most closely related to AtICE1, was shown to interact with MtCAS31, indicating that the ICE1-CBF pathway may be conserved in Medicago spp. (Xie et al., 2012).
During the cold acclimation of M. truncatula plants, the MtMYB61 expression level increases quickly in response to low temperatures. Based on transient transformations in N. benthamiana, cold stress does not seem to have a major impact on the strength of the MtMYB3-MtMYB61 protein interaction (Supplemental Fig. S8). As the ChIP assay showed increased MtMYB3 binding to the MYB cis-elements of the MtCBF4 promoter in the myb61 mutant compared with the wild type, this suggests that the interaction of MtMYB61 with the DNA-binding domain of MtMYB3 inhibits its binding to the MtCBF4 promoter under cold stress. Thus, we propose that MtCBF4 expression is repressed by MtMYB3 under nonstressed conditions and that the cold-induced MtMYB61 interaction with MtMYB3 relieves this inhibition (Fig. 12). However, it remains open that other transcription factors are required to enhance MtCBF4 transcription in response to cold. Deciphering the complexity of such regulatory modules and their possible conservation across plant species, including agronomically relevant crops, may be a source for genetic improvements to freezing tolerance.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Two genotypes of Medicago truncatula, Jemalong A17 and R108 (Li et al., 2011), were used in this work.
The pMDC32-MtCBF4-3×FLAG, pMDC32-MtMYB3-3×FLAG, and pMDC32-MtMYB61-3×FLAG plasmid constructs were generated using corresponding full-length genes cloned from M. truncatula Jemalong A17 cDNA. All primers used for these constructs are listed in Supplemental Table S4 (pMDC32-MtCBF4-F, pMDC32-MtCBF4-R, pMDC32-MtMYB3-F, pMDC32-MtMYB3-R, pMDC32-MtMYB61-F, and pMDC32-MtMYB61-R). These vectors were introduced into the Agrobacterium tumefaciens EHA105 strain for transformation into M. truncatula R108 (Cosson et al., 2006). Transgenic plants (T3 generation) were used for phenotyping. The Tnt1 insertional mutant lines were screened from the Noble Foundation using a reverse genetic PCR approach: NF9694, cbf4-1; NF7967, cbf4-2; NF13136, myb3-1; NF19047, myb3-2; and NF14253, myb61 (Tadege et al., 2008; Cheng et al., 2014).
M. truncatula seeds were sterilized and seedling germinated as described previously (de Lorenzo et al., 2007). Germinated seedlings with approximately 2-cm-long roots were transplanted into a soil:vermiculite (1:2, v/v) mixture and grown for 5 weeks in a greenhouse at 22°C under a 16-h photoperiod. For cold acclimation experiments, 3-week-old M. truncatula plants were exposed to low temperature (4°C and 12 h of light) for 2 weeks.
For protoplasts assays, the Arabidopsis (Arabidopsis thaliana) Columbia ecotype was germinated and grown in vitro on Murashige and Skoog medium at 22°C under a 16-h-light/8-h-dark photoperiod at 100 mmol m–2 s–1 for 1 week, then transferred to a soil:vermiculite (1:1, v/v) mixture to be grown at 22°C under a 12-h photoperiod for 4 weeks in a greenhouse.
Nicotiana benthamiana was germinated and grown on plates containing Murashige and Skoog medium at 22°C under a 16-h-light/8-h-dark photoperiod at 100 mmol m–2 s–1 for 1 week, then transplanted to a soil:vermiculite (1:1, v/v) mixture and grown in a greenhouse at 22°C under a 16-h photoperiod for 5 weeks.
Freezing Tolerance and Electrolyte Leakage Assays
The freezing tolerance assay in M. truncatula was executed as described previously (Pennycooke et al., 2008) with several modifications. Briefly, 5-week-old whole plants grown in a soil:vermiculite (1:2, v/v) mixture with or without cold acclimation were subsequently placed in a freezing chamber (RuMED4001; Shi et al., 2012) that was set to maintain a temperature of 0°C for 1 h. The temperature was then decreased at a rate of 1°C h−1 in the dark. The plants were maintained for 1 h at the designated temperature from −2°C to −9°C and transferred to 4°C overnight, then transferred back to 22°C with light (16-h daylength). Survival rates and electrolyte leakage parameters were measured after 4 d as described below.
The values of electrolyte leakage were detected as described previously (Pennycooke et al., 2008) with several modifications. Briefly, five compound leaves from independent plants were put into 50-mL conical tubes. Three independent biological replicates (three different tubes) were analyzed for each overexpression or mutant line and the wild-type control. Twenty-five milliliters of deionized water was added to each tube. The tubes were placed under a vacuum for 15 min and then shaken for 1 h at 250 rpm before conductivity (C1) was measured using an electrical conductivity meter (HI8733; Hanna Instruments; www.hannainst.com). The tubes were subsequently placed in boiling water for 15 min, allowed to recover at 22°C, and shaken, and total potential conductivity (C2) was then measured. The electrical conductivity values also were determined for deionized and distilled water (C0) to determine background levels, and ion leakage was represented as follows: (C1 − C0)/(C2 −C0).
RNA Extraction, Reverse Transcription-PCR, and qRT-PCR Analysis
Total RNA was extracted from leaves collected from 5-week-old plants using the TRIzol reagent (Invitrogen) and then reverse transcribed to cDNA via Moloney murine leukemia virus reverse transcriptase (Promega), and real-time reverse transcription-PCR analyses were performed with the CFX-96 Real-Time System (Bio-Rad) using the SYBR Premix ExTaq (TaKaRa). The MtACTIN gene was used as a reference gene (Li et al., 2011), and relative expression levels were calculated as described previously in these studies. All primers related to this experiment are listed in Supplemental Table S4.
cDNA Library Preparation and Sequencing
Five-week-old MtCBF4-overexpressing transgenic plants and wild-type control plants were used for high-throughput RNAseq Digital Gene Expression Profiling. Three micrograms of RNA per sample was used to generate sequencing libraries using the NEBNext Ultra RNA Library Prep Kit (Illumina, New England Biolabs) according to the manufacturer’s recommendations. Index codes were used to attach the sequences to each sample, and then the clustering of index-coded samples proceeded on the cBot Cluster Generation System via the TruSeq SR Cluster Kit v3-cBot-HS (Illumina) following the manufacturer’s instructions. The libraries then were sequenced on the Illumina HiSeq 2000 platform, and 50-bp single-end reads were obtained. The reference genome of M. truncatula and gene annotation files (version 4.0) were downloaded from the JCVI genome Web site (http://www.jcvi.org/medicago/). Single-end cleaned reads were aligned to the reference genome through TopHat using the default parameters (Trapnell et al., 2009). Most of the cleaned reads (74.07%–75.44%) could be aligned to the reference genome, and 69.8% to 70.98% of the cleaned reads were uniquely mapped. The DESeq R package was used for the differential expression analyses of the two genotypes (three biological replicates each; Anders and Huber, 2010), and the Benjamini-Hochberg approach was used to adjust the resulting P values for false discovery rate. Genes with an adjusted P < 0.01 were assigned as expressed differentially. GO enrichment analysis of differentially expressed genes between wild-type and MtCBF4 overexpression plants was implemented using the GOseq R package (Young et al., 2010). The criterion considered for significantly enriched GO terms is P < 0.01.
Purification of Fusion Proteins in Escherichia coli and EMSA
The MtCBF4, MtMYB3, and MtMYB61 proteins were fused with a His tag and expressed in the E. coli BL21 strain using the pET-30a (+) vector (Yang et al., 2011). Expression of the three fusion proteins was induced with isopropyl β-d-thiogalactoside, and recombinant proteins were purified through nickel affinity chromatography following the manufacturer’s instructions. The LightShift Chemiluminescent EMSA kit (Thermo) were used for EMSA experiments performed according to the manufacturer’s instructions. Probes consisting of DNA fragments containing DRE or MYB cis-elements, as well as mutated probes, were generated using the primers listed in Supplemental Table S2.
ChIP
ChIP was performed as described previously (Saleh et al., 2008) with some modifications. Leaves from 5-week-old plants grown at 22°C in a soil-vermiculite mixture were cross-linked twice in 1% formaldehyde diluted by buffer I (Saleh et al., 2008) for 15 min each under vacuum, and the reaction was stopped by treatment with 0.125 m Gly under another vacuum for 5 min. After the leaves were washed three times with distilled water, chromatin was isolated and sheared as described previously (Saleh et al., 2008). The anti-CBF4 and anti-MYB3 polyclonal antibodies were used to immunoprecipitate the target proteins (MtCBF4 and MtMYB3). Quantitative PCR was performed to identify enriched DNA fragments in the immunoprecipitates compared with inputs. For cold treatment, plants were grown at 4°C for 2 h before leaves were collected. The primer pairs used in these assays are listed in Supplemental Table S4. MtCBF4 (full length) and a truncated MtMYB3 (amino acids 167–278) were expressed in the E. coli BL21 strain using the pET-30a (+) vector, purified through nickel affinity chromatography, and used to produce the anti-CBF4 and anti-MYB3 antibodies, respectively. These antibodies were produced by Beijing Protein Innovation (http://www.proteomics.org.cn). Briefly, 400 ng of antigen proteins was first injected into rabbits and then injected another three times (200 ng each time) to boost immunization. The arterial blood serum was collected, and the antibody was purified using Protein A Agarose (GE Healthcare; 17-0402-01).
Yeast One-Hybrid Assay
Total RNA used for library construction was isolated from whole M. truncatula Jemalong A17 plants. Library construction was performed by Invitrogen. The titer of the library was 3.4 × 106 colony-forming units mL−1. Triplicate repeats of the fragment containing this MYB-binding site (TAGTTA) were used to construct the pAbAi-MYB cis-element bait vector for the yeast one-hybrid screen of the library. The artificial fragment sequence is shown in Supplemental Table S2 (Y1HMYB-F and Y1HMYB-R). Yeast one-hybrid screens were performed following the manufacturer’s protocol (Clontech; www.clontech.com; protocol no. PT4087-1).
Yeast Two-Hybrid Assay
An M. truncatula Jemalong A17 cDNA library was constructed using 4-week-old plants that were subjected to cold, salt, or drought stress in our laboratory, as described previously (Li et al., 2011). pGBKT7-MtMYB3 was generated to be used as the bait vector and to screen the M. truncatula cDNA library, following the manufacturer’s protocol (Clontech). MtMYB3 also was linked to the pGADT7 prey vector, and MtMYB61 was linked to the pGBKT7 bait vector. All primers used in this experiment are listed in Supplemental Table S4 (pGBKT7-MtMYB61-F, pGBKT7-MtMYB61-R, pGADT7-MtMYB3-F, and pGADT7-MtMYB3-R). The two vectors were cotransformed into the yeast (Saccharomyces cerevisiae AH109) strain AH109 according to the manufacturer’s protocol (Clontech), and colonies were selected on synthetic dropout (SD)/-Trp-Leu medium following incubation at 30°C. The obtained colonies were assayed on SD/-Leu/-Trp/-Ade/-His + 15 mm 3-AT, and the α-Gal activity was determined.
To map the MtMYB3-MtMYB61 interaction site, the C-terminal and N-terminal regions of MtMYB3 were cloned into the prey vector. The interaction between the MtMYB3 prey vector (pGADT7-N-MtMYB3 or pGADT7-C-MtMYB3) and the bait vector pGBKT7-MtMYB61 was analyzed as described above. All primers used in this experiment are listed in Supplemental Table S4 (pGBKT7-MtMYB61N-F, pGBKT7-MtMYB61N-R, pGADT7-MtMYB3C-F, and pGADT7-MtMYB3C-R).
GST Pull-Down Assay
GST, GST-MtMYB3, and His-MtMYB61 were expressed in E. coli BL21, and the recombinant proteins were purified by GST-agarose affinity chromatography as described previously (Yang et al., 2011). A GST pull-down assay was conducted as described previously (Yang et al., 2011). All primers used in this experiment are listed in Supplemental Table S4 (pGEX-4T-1-MtMYB3GST-F, pGEX-4T-1-MtMYB3GST-R, pET30a-MtMYB61His-F, and pET30a-MtMYB61His-R).
BiFC, Subcellular Localization in Arabidopsis Protoplasts, and Transactivation Analyses in Yeast
For BiFC assays and subcellular localization analyses, Arabidopsis protoplasts were prepared as described previously (Xie et al., 2012). Transactivation analyses in yeast (Saccharomyces cerevisiae YRG-2) were performed as described previously (Li et al., 2011). All primers used in this experiment are listed in Supplemental Table S4 (pSY735-MtMYB61-F, pSY735-MtMYB61-R, pSY736-MtMYB3-F, and pSY736-MtMYB3-R).
Transient Expression Assays in N. benthamiana Leaves
The ProMtCBF4:LUC reporter construct was generated using a 2,500-bp MtCBF4 promoter sequence fused with the LUC reporter gene using the BamHI and SalI sites of the pCAMBIA1381Z vector. All primers used in this experiment are listed in Supplemental Table S4 (pCAMBIA1381-ProMtCBF4LUC-F and pCAMBIA1381-ProMtCBF4LUC-R). The pMDC32-Pro-35S:MtMYB3:3xFLAG and pMDC32-Pro-35S:MtMYB61:3xFLAG or pMDC32-Pro-35S:MtMYB3:3xFLAG and Pro-35S:MtMYB61:3xFLAG plasmids were coinfiltrated with the pCAMBIA1381Z-ProCBF4-LUC vector into N. benthamiana plants using the A. tumefaciens EHA105 strain, as described previously (Li et al., 2013).
Infiltrated plants were incubated at 22°C for 48 h before imaging with a digital CCD camera (Carestream In-vivo FX PRO; Rauch et al., 2009). A low-light cooled CCD imaging apparatus was used to capture LUC images and determine the luminescence intensity. Leaves were sprayed with 100 mm luciferin and placed in the dark for 10 min prior to luminescence detection. At least five independent LUC quantifications were assessed, with similar results.
Phylogenetic Analysis
The protein sequences of MYB TFs associated in abiotic stress (Li et al., 2015) were aligned and clustered using ClustalW 2.1 (Larkin et al., 2007). The phylogeny of these protein sequences was constructed by EvolView (Zhang et al., 2012).
Accession Numbers
The raw data from the RNAseq Digital Gene Expression Profiling analysis have been deposited in the National Center for Biotechnology Information database under accession number PRJNA284671.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. qRT-PCR and western-blot analysis of Pro-35S:MtCBF4 transgenic M. truncatula plants.
Supplemental Figure S2. Functional category enrichment analysis of differentially expressed genes based on GO terms.
Supplemental Figure S3. Subcellular localization and transcriptional activation analysis of MtMYB3.
Supplemental Figure S4. qRT-PCR and western-blot analysis of Pro-35S:MtMYB3 transgenic M. truncatula plants.
Supplemental Figure S5. Subcellular localization and transcriptional activation analysis of MtMYB61.
Supplemental Figure S6. qRT-PCR and western-blot analysis of Pro-35S:MtMYB61 transgenic M. truncatula plants.
Supplemental Figure S7. Phylogenetic analysis of selected MYB transcription factors involved in abiotic stress from Arabidopsis, M. domestica, O. sativa, T. aestivum, N. benthamiana, V. vinifera, S. tuberosum, Chrysanthemum spp., soybean, A. marina, L. chinensis, P. communis, C. plantagineum, S. officinarum, and M. truncatula.
Supplemental Figure S8. Cold does not affect the interaction between MtMYB3 and MtMYB61.
Supplemental Table S1. List of differentially expressed genes between wild-type and 35S:MtCBF4 transgenic plants
Supplemental Table S2. Oligonucleotide probes used in EMSA and the artificial fragment sequence used in the yeast one-hybrid assay.
Supplemental Table S3. List of yeast two-hybrid assay clones identified with MtMYB3 protein as bait.
Supplemental Table S4. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Jean Marie Prosperi and Magalie Delalande (BRC for M. truncatula, Unité Mixte de Recherche 1097, Institut National de la Recherche Agronomique) for providing seeds of M. truncatula Jemalong A17 and R108 as well as Dr. Shouyi Chen and Dr. Chuanyou Li (Institute of Genetics and Developmental Biology) and Dr. Yan Guo, Dr. Shuhua Yang, and Dr. Dongtao Ren (China Agricultural University) for providing vectors.
Glossary
- TF
transcription factor
- DRE
dehydration-responsive element
- ABA
abscisic acid
- qRT
quantitative reverse transcription
- GO
Gene Ontology
- EMSA
electrophoretic mobility shift assay
- ChIP
chromatin immunoprecipitation
- BiFC
bimolecular fluorescence complementation
- 3-AT
3-aminotriazole
- SD
synthetic dextrose
Footnotes
This work was supported by the National Natural Science Foundation of China (grant no. 31371689), the National Basic Research Program of China (973 Program; grant no. 2012CB215301), and Hi-Tech Research and Development (863 Program; grant no. 2011AA100209).
Articles can be viewed without a subscription.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, Zhu JK (2006) A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem 281: 37636–37645 [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton G, Danyluk J, Charron JBF, Sarhan F (2003) Expression profiling and bioinformatic analyses of a novel stress-regulated multispanning transmembrane protein family from cereals and Arabidopsis. Plant Physiol 132: 64–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalá R, Santos E, Alonso JM, Ecker JR, Martínez-Zapater JM, Salinas J (2003) Mutations in the Ca2+/H+ transporter CAX1 increase CBF/DREB1 expression and the cold-acclimation response in Arabidopsis. Plant Cell 15: 2940–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Xu Z, Xia L, Li L, Cheng X, Dong J, Wang Q, Ma Y (2009) Cold-induced modulation and functional analyses of the DRE-binding transcription factor gene, GmDREB3, in soybean (Glycine max L.). J Exp Bot 60: 121–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Wang M, Lee HK, Tadege M, Ratet P, Udvardi M, Mysore KS, Wen J (2014) An efficient reverse genetics platform in the model legume Medicago truncatula. New Phytol 201: 1065–1076 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson V, Durand P, d’Erfurth I, Kondorosi A, Ratet P (2006) Medicago truncatula transformation using leaf explants. Methods Mol Biol 343: 115–127 [DOI] [PubMed] [Google Scholar]
- de Lorenzo L, Merchan F, Blanchet S, Megías M, Frugier F, Crespi M, Sousa C (2007) Differential expression of the TFIIIA regulatory pathway in response to salt stress between Medicago truncatula genotypes. Plant Physiol 145: 1521–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zélicourt A, Diet A, Marion J, Laffont C, Ariel F, Moison M, Zahaf O, Crespi M, Gruber V, Frugier F (2012) Dual involvement of a Medicago truncatula NAC transcription factor in root abiotic stress response and symbiotic nodule senescence. Plant J 70: 220–230 [DOI] [PubMed] [Google Scholar]
- Ding Y, Li H, Zhang X, Xie Q, Gong Z, Yang S (2015) OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev Cell 32: 278–289 [DOI] [PubMed] [Google Scholar]
- Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF (2009) Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21: 972–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong CH, Agarwal M, Zhang Y, Xie Q, Zhu JK (2006) The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc Natl Acad Sci USA 103: 8281–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Fowler SG, Thomashow MF (2004) Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol Biol 54: 767–781 [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF (1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J 16: 433–442 [DOI] [PubMed] [Google Scholar]
- Graham PH, Vance CP (2003) Legumes: importance and constraints to greater use. Plant Physiol 131: 872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hincha DK, Thalhammer A (2012) LEA proteins: IDPs with versatile functions in cellular dehydration tolerance. Biochem Soc Trans 40: 1000–1003 [DOI] [PubMed] [Google Scholar]
- Hu Y, Jiang L, Wang F, Yu D (2013) Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25: 2907–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidokoro S, Watanabe K, Ohori T, Moriwaki T, Maruyama K, Mizoi J, Myint Phyu Sin Htwe N, Fujita Y, Sekita S, Shinozaki K, et al. (2015) Soybean DREB1/CBF-type transcription factors function in heat and drought as well as cold stress-responsive gene expression. Plant J 81: 505–518 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, Yoo CY, Baek D, Kim DH, Jeong JC, et al. (2007) Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J 49: 79–90 [DOI] [PubMed] [Google Scholar]
- Li CN, Ng CKY, Fan LM (2015) MYB transcription factors, active players in abiotic stress signaling. Environ Exp Bot 114: 80–91 [Google Scholar]
- Li D, Zhang Y, Hu X, Shen X, Ma L, Su Z, Wang T, Dong J (2011) Transcriptional profiling of Medicago truncatula under salt stress identified a novel CBF transcription factor MtCBF4 that plays an important role in abiotic stress responses. BMC Plant Biol 11: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhao B, Yuan D, Duan M, Qian Q, Tang L, Wang B, Liu X, Zhang J, Wang J, et al. (2013) Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression. Proc Natl Acad Sci USA 110: 3167–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XP, Tian AG, Luo GZ, Gong ZZ, Zhang JS, Chen SY (2005) Soybean DRE-binding transcription factors that are responsive to abiotic stresses. Theor Appl Genet 110: 1355–1362 [DOI] [PubMed] [Google Scholar]
- Liao Y, Zou HF, Wang HW, Zhang WK, Ma B, Zhang JS, Chen SY (2008) Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res 18: 1047–1060 [DOI] [PubMed] [Google Scholar]
- Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, Shimada Y, Yoshida S, Shinozaki K, Yamaguchi-Shinozaki K (2004) Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J 38: 982–993 [DOI] [PubMed] [Google Scholar]
- Medina J, Catalá R, Salinas J (2001) Developmental and stress regulation of RCI2A and RCI2B, two cold-inducible genes of Arabidopsis encoding highly conserved hydrophobic proteins. Plant Physiol 125: 1655–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM (2007) SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra SS, Wolfraim L, Poole RJ, Dhindsa RS (1989) Molecular cloning and relationship to freezing tolerance of cold-acclimation-specific genes of alfalfa. Plant Physiol 89: 375–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroy AF, Castonguay Y, Laberge S, Sarhan F, Vezina LP, Dhindsa RS (1993a) A new cold-induced alfalfa gene is associated with enhanced hardening at subzero temperature. Plant Physiol 102: 873–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroy AF, Dhindsa RS (1995) Low-temperature signal transduction: induction of cold acclimation-specific genes of alfalfa by calcium at 25°C. Plant Cell 7: 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroy AF, Sarhan F, Dhindsa RS (1993b) Cold-induced changes in freezing tolerance, protein phosphorylation, and gene expression (evidence for a role of calcium). Plant Physiol 102: 1227–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordin K, Vahala T, Palva ET (1993) Differential expression of two related, low-temperature-induced genes in Arabidopsis thaliana (L.) Heynh. Plant Mol Biol 21: 641–653 [DOI] [PubMed] [Google Scholar]
- Okawa K, Nakayama K, Kakizaki T, Yamashita T, Inaba T (2008) Identification and characterization of Cor413im proteins as novel components of the chloroplast inner envelope. Plant Cell Environ 31: 1470–1483 [DOI] [PubMed] [Google Scholar]
- Pennycooke JC, Cheng H, Stockinger EJ (2008) Comparative genomic sequence and expression analyses of Medicago truncatula and alfalfa subspecies falcata COLD-ACCLIMATION-SPECIFIC genes. Plant Physiol 146: 1242–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch D, Gross S, Harding J, Niewiesk S, Lairmore M, Piwnica-Worms D, Ratner L (2009) Imaging spontaneous tumorigenesis: inflammation precedes development of peripheral NK tumors. Blood 113: 1493–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Shi Y, Tian S, Hou L, Huang X, Zhang X, Guo H, Yang S (2012) Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 24: 2578–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinwari ZK, Nakashima K, Miura S, Kasuga M, Seki M, Yamaguchi-Shinozaki K, Shinozaki K (1998) An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem Biophys Res Commun 250: 161–170 [DOI] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, Cayrel A, Endre G, Zhao PX, Chabaud M, et al. (2008) Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J 54: 335–347 [DOI] [PubMed] [Google Scholar]
- Thapa B (2008) Understanding cold acclimation in Medicago truncatula. PhD thesis. Iowa State University, Ames [Google Scholar]
- Thomashow MF. (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui T, Kato W, Asada Y, Sako K, Sato T, Sonoda Y, Kidokoro S, Yamaguchi-Shinozaki K, Tamaoki M, Arakawa K, et al. (2009) DEAR1, a transcriptional repressor of DREB protein that mediates plant defense and freezing stress responses in Arabidopsis. J Plant Res 122: 633–643 [DOI] [PubMed] [Google Scholar]
- Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF (2005) Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 41: 195–211 [DOI] [PubMed] [Google Scholar]
- Wolfraim LA, Langis R, Tyson H, Dhindsa RS (1993) cDNA sequence, expression, and transcript stability of a cold acclimation-specific gene, cas18, of alfalfa (Medicago falcata) cells. Plant Physiol 101: 1275–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Zhang R, Qu Y, Miao Z, Zhang Y, Shen X, Wang T, Dong J (2012) Overexpression of MtCAS31 enhances drought tolerance in transgenic Arabidopsis by reducing stomatal density. New Phytol 195: 124–135 [DOI] [PubMed] [Google Scholar]
- Yang XY, Chen ZW, Xu T, Qu Z, Pan XD, Qin XH, Ren DT, Liu GQ (2011) Arabidopsis kinesin KP1 specifically interacts with VDAC3, a mitochondrial protein, and regulates respiration during seed germination at low temperature. Plant Cell 23: 1093–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene Ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gao S, Lercher MJ, Hu S, Chen WH (2012) EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res 40: W569–W572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LL, Zhao MG, Tian QY, Zhang WH (2011) Comparative studies on tolerance of Medicago truncatula and Medicago falcata to freezing. Planta 234: 445–457 [DOI] [PubMed] [Google Scholar]
- Zhu J, Jeong JC, Zhu Y, Sokolchik I, Miyazaki S, Zhu JK, Hasegawa PM, Bohnert HJ, Shi H, Yun DJ, et al. (2008) Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proc Natl Acad Sci USA 105: 4945–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.