A vacuolar anion channel in vascular cells of Arabidopsis contributes affects long-distance Na+ and Cl− transport rapidly after onset of salinity stress.
Abstract
The ability to control the cytoplasmic environment is a prerequisite for plants to cope with changing environmental conditions. During salt stress, for instance, Na+ and Cl− are sequestered into the vacuole to help maintain cytosolic ion homeostasis and avoid cellular damage. It has been observed that vacuolar ion uptake is tied to fluxes across the plasma membrane. The coordination of both transport processes and relative contribution to plant adaptation, however, is still poorly understood. To investigate the link between vacuolar anion uptake and whole-plant ion distribution during salinity, we used mutants of the only vacuolar Cl− channel described to date: the Arabidopsis (Arabidopsis thaliana) ALMT9. After 24-h NaCl treatment, almt9 knock-out mutants had reduced shoot accumulation of both Cl− and Na+. In contrast, almt9 plants complemented with a mutant variant of ALMT9 that exhibits enhanced channel activity showed higher Cl− and Na+ accumulation. The altered shoot ion contents were not based on differences in transpiration, pointing to a vacuolar function in regulating xylem loading during salinity. In line with this finding, GUS staining demonstrated that ALMT9 is highly expressed in the vasculature of shoots and roots. RNA-seq analysis of almt9 mutants under salinity revealed specific expression profiles of transporters involved in long-distance ion translocation. Taken together, our study uncovers that the capacity of vacuolar Cl− loading in vascular cells plays a crucial role in controlling whole-plant ion movement rapidly after onset of salinity.
Solute fluxes across the vacuolar membrane are at the center of plant performance and survival in fluctuating environmental conditions. The large central vacuole serves as a storage reservoir that accumulates and releases ions as well as metabolites according to demands. The physical and functional plasticity of the vacuole enables plants to use energy and nutrients efficiently and maintain optimal physiological conditions in the cytosol. Vacuolar storage capacity regulates intracellular ion homeostasis but also influences whole-plant ion accumulation and distribution. For example, nhx1 nhx2 mutant plants lack two major Na+ and K+/H+ antiporters at the tonoplast and show lower tissue K+ accumulation (Barragán et al., 2012). Similarly, Arabidopsis (Arabidopsis thaliana) mutants deficient in the vacuolar NO3−/H+ exchanger CLCa (De Angeli et al., 2006) have diminished nitrate (NO3−) contents in shoots and roots (Geelen et al., 2000; Monachello et al., 2009). Manipulating ion fluxes through the application of high ionic concentrations (for instance NaCl stress) can reveal the functional role of the vacuole in the coordination of ion movement at the whole-plant level.
Salinity has a negative impact on plant development, and this is based on an osmotic and an ion toxicity effect (for review, see Teakle and Tyerman, 2010; Munns and Tester, 2008). Although salt stress responses and adaptation mechanisms were investigated with a focus on Na+ toxicity and accumulation (Craig Plett and Møller, 2010), Cl− ions similarly interfere with metabolic processes, in particular in photosynthetic tissue (Tavakkoli et al., 2010, 2011; Geilfus et al., 2015; Genc et al., 2016). To mitigate the damaging effects of salinity, the movement of toxic ions across cellular membranes is tightly regulated. At the whole-plant level, plants restrict shoot ion accumulation by controlling net ion uptake and xylem loading (Britto et al., 2004; Møller and Tester, 2007; Brumós et al., 2010; Craig Plett and Møller, 2010; Teakle and Tyerman, 2010). Recirculation of ions to the roots via the phloem does not significantly contribute to the reduction of Na+ and Cl− levels in leaf tissue (Munns, 2002; Davenport et al., 2007). The ability of shoot ion exclusion is limited. Therefore, plants allocate toxic ions to specific cells or sites within the shoots to adapt to salinity. Indeed, Na+ and Cl− accumulate preferentially in old leaves (Sibole et al., 2003; Craig Plett and Møller, 2010), leaf margins (Shapira et al., 2009), and epidermal cells (Huang and Van Steveninck, 1989; Karley et al., 2000a, 2000b; James et al., 2006) to protect photosynthetically active cells. Consequently, the controlled loading of Na+ and Cl− into xylem vessels of the vasculature system in roots and shoots majorly determines long-distance ion movement such as root-to-shoot translocation and distribution within the shoots during salinity. Despite the knowledge of these physiological adaptation strategies to salt stress, the core molecular machinery underlying the regulation of ion uptake, xylem loading, and partitioning is only slowly being identified.
At the cellular level, plants are capable of sequestering toxic ions into the vacuoles to minimize harm once ions have accumulated to high concentrations during salt stress (James et al., 2006; Munns and Tester, 2008). Several transport proteins localized at the tonoplast have been suggested to facilitate intracellular ion partitioning and thereby regulate cellular ion homeostasis (Martinoia et al., 2007; Martinoia et al., 2012). However, only few transporters involved in vacuolar ion uptake during salinity have been studied. The two vacuolar cation/H+ antiporters NHX1 and NHX2 contribute to salinity tolerance in several plant species (Munns and Tester, 2008, and references therein). Subsequently, a role of these NHXs in vacuolar Na+ sequestration has been challenged by findings suggesting their cellular function in K+ compartmentation (Leidi et al., 2010; Jiang et al., 2010). With respect to intracellular Cl− uptake, two members of the channel and transporter protein family CLC (Chloride Channel), CLCc and CLCg, have been shown to be involved in salinity tolerance (Jossier et al., 2010; Nguyen et al., 2016). Besides the importance of fluxes across the tonoplast, transport proteins localized to other endomembranes such as the trans-Golgi network have been implicated in salinity adaptation mechanisms (Krebs et al., 2010; Bassil et al., 2011; Henderson et al., 2015). Yet, the limited knowledge about salt stress-related endomembrane transporters and their functional roles, especially with regards to Cl− sequestration, restricts our understanding of the contribution of intracellular ion homeostasis to long-distance ion fluxes and salinity adaptation mechanisms.
The ALMT (Aluminum-activated Malate Transporter) protein family is unique to plants and encodes channels able to mediate anion fluxes across cellular membranes (Barbier-Brygoo et al., 2011). In clade II of the Arabidopsis ALMT family, two ion channels were shown to be localized at the tonoplast and mediate anion fluxes directed to the vacuolar lumen (Kovermann et al., 2007; Meyer et al., 2011; De Angeli et al., 2013). One of them, ALMT6, transports malate (MA2−) into guard cell vacuoles in a Ca2+- and pH-dependent manner (Meyer et al., 2011). ALMT9, a channel shown to be expressed in mesophyll and guard cells, is permeable for both MA2−and Cl−, whereby its physiological function is linked to the Cl− conductivity (Kovermann et al., 2007; De Angeli et al., 2013). ALMT9-mediated Cl− currents across the tonoplast are activated by physiological concentrations of cytosolic MA2−, and mesophyll vacuoles of almt9 knock-out mutants lacking the vacuolar channel exhibit lower overall Cl− currents (De Angeli et al., 2013). Moreover, almt9 plants have reduced Cl− uptake into vacuoles of guard cells resulting in impaired light-dependent stomatal opening and reduced wilting during drought stress.
In the current study, we aimed at uncovering the consequences of impaired intracellular Cl− fluxes across the tonoplast on whole-plant ion transport during salinity. To address this issue, we used knock-out mutants of the only genuine vacuolar Cl− channel described so far, ALMT9. We found that almt9 plants show a reduced shoot accumulation of Cl− as well as Na+ after 24-h salinity, whereas mutants with enhanced ALMT9-mediated Cl− currents possess increased ion accumulation. Using transcriptome approaches, we demonstrate that genes encoding plasma membrane-localized transport proteins that contribute to long-distance ion translocation exhibit an altered transcriptional response in almt9 during salt stress. In line with this, we show that ALMT9 is highly expressed in the vasculature of shoots and roots. Collectively, the data suggest that vacuolar ion uptake is not only crucial to confer cellular tolerance during long-term salinity but also to modulate shoot ion accumulation and whole-plant ion distribution during early phases of salinity.
RESULTS
ALMT9 Is Expressed in the Vasculature and Is Up-Regulated upon NaCl Application
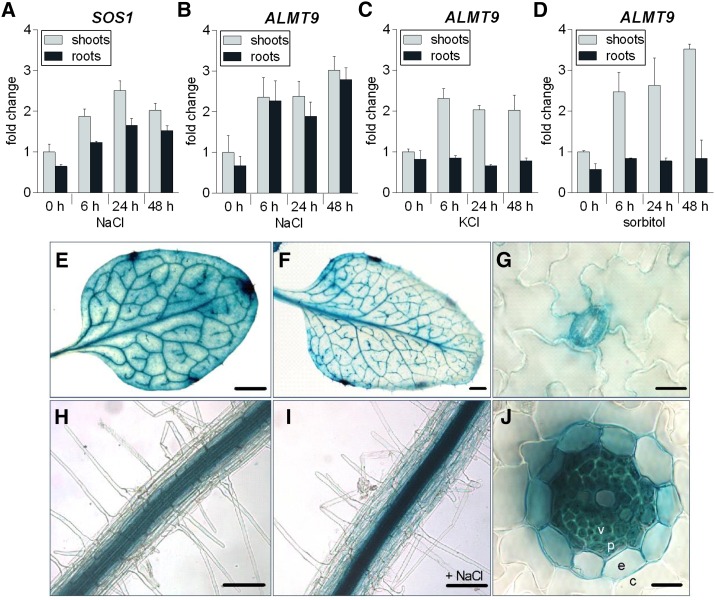
Gene regulation at the transcriptional level is commonly observed upon salt (NaCl) stress in plants (Tester and Davenport, 2003). To investigate whether ALMT9 is involved in intracellular Cl− transport during salinity, we examined its expression levels by quantitative real-time PCR (qRT-PCR) in shoots and roots of Arabidopsis wild-type plants grown in a hydroponic system in response to 100 mm NaCl for up to 48 h. The up-regulation of expression of the salt stress-inducible gene SOS1 (Shi et al., 2000, 2002) verified that plants experience salinity stress in our experimental conditions in shoots and roots at the molecular level (Fig. 1A). We found that ALMT9 was transcriptionally up-regulated in response to NaCl application in both tissues after 6 h (Fig. 1B). Over 48 h of NaCl treatment, shoot expression of ALMT9 showed a 3-fold increase when compared to the expression level prior to stress; in roots, the transcription increased 4.2 times. To discriminate whether the stimulation of ALMT9 transcription is specific to NaCl or depends on a general osmotic effect, we applied 100 mm KCl and 200 mm sorbitol to the plants. Both treatments stimulated the expression of ALMT9 in shoots (2-fold and 3.5-fold increase after 48 h, respectively), whereas no transcriptional response was detectable in root tissue (Fig. 1, C and D). These findings show that the transcriptional up-regulation of ALMT9 in roots is specific to the ionic stress of salinity.
Figure 1.
Transcriptional regulation and expression pattern analysis of ALMT9. A to D, qRT-PCR analysis of SOS1 (A) and ALMT9 (B–D) expression in hydroponically grown wild-type shoots and roots after the application of 100 mm NaCl (A and B), 100 mm KCl (C), and 200 mm sorbitol (D) for 0, 6, 24, and 48 h. The data were normalized to expression levels in shoots prior to treatment (0 h). ACT2 served as a reference gene. Data are means ± sd of n = 3 biological replicates. E to J, ALMT9 expression pattern revealed by histochemical localization of GUS activity directed by the ALMT9 promoter. E, Mesophyll cell and vasculature expression in the third rosette leaf. F, Vasculature expression in the sixth rosette leaf. G, Expression in guard cells. H, Expression in root stelar cells. I, Expression in response to 100 mm NaCl for 24 h was enhanced but remained restricted to the root stele. J, In cross-sections of roots, no expression was detected in cortex cells (c), but in the endodermis (e), the pericycle (p), and the vasculature (v). Scale bars represent 0.2 mm in E and F, 10 µm in G and J, and 100 µm in H and I.
Subsequently, we used transgenic plants carrying the GUS reporter gene under the control of the ALMT9 promoter region (ALMT9pro:GUS) to investigate the expression pattern and tissue specificity of ALMT9 during transcriptional up-regulation in response to salinity. As previously shown, ALMT9 was expressed in leaf mesophyll and guard cells (Fig. 1, E and G; Kovermann et al., 2007; De Angeli et al., 2013). The mesophyll expression was predominantly detected in mature leaves. In contrast, GUS staining was found in the vasculature throughout all developmental stages of the leaf (Fig. 1, E and F). ALMT9 promoter-driven GUS expression could also be detected in the stele of roots (Fig. 1H). A transverse section allowed to locate ALMT9 expression in vascular and pericycle cells as well as weakly in the endodermis (Fig. 1J). However, ALMT9 was not expressed in root cortex or epidermis cells. As observed in the qRT-PCR (Fig. 1B), NaCl treatment enhanced the intensity of the GUS staining (Fig. 1I), corresponding to a transcriptional up-regulation of ALMT9. Nevertheless, the tissue-specific expression pattern did not change upon salt exposure.
The almt9 Mutants Have Reduced Shoot Na+ and Cl− Accumulation during Early Salinity
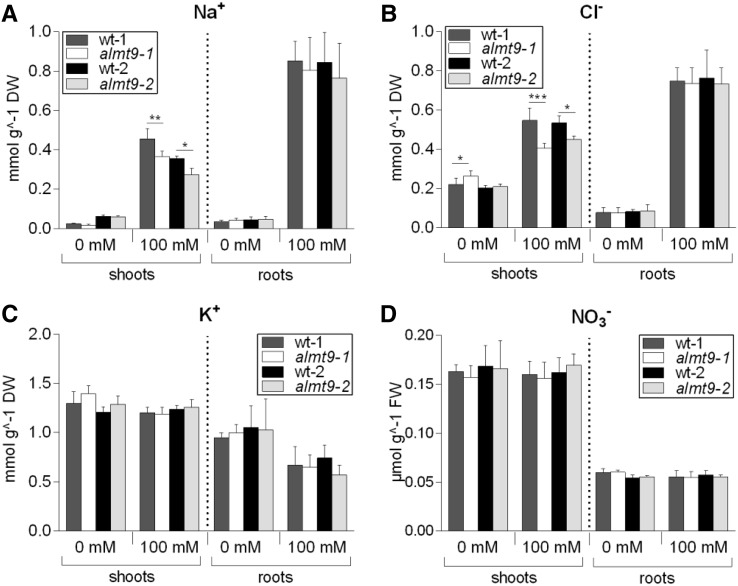
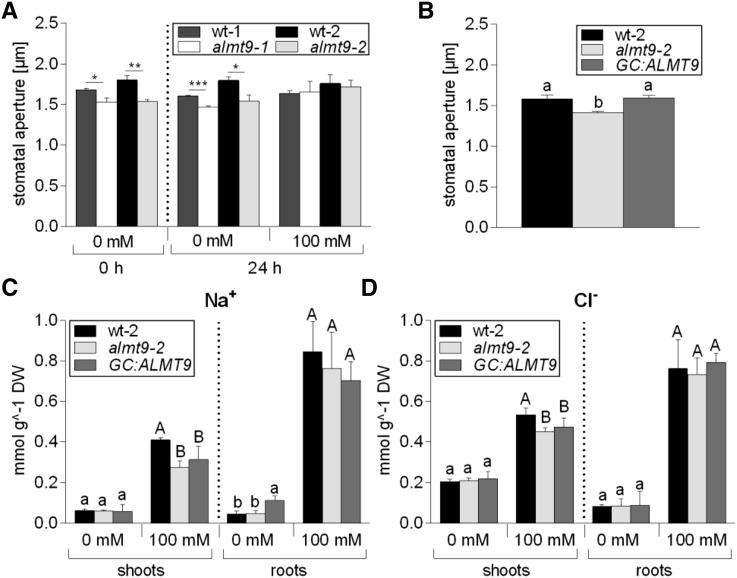
To assess whether the transcriptional up-regulation of ALMT9 in response to NaCl is associated with a physiological role of ALMT9 during salinity, we conducted ion content measurements in shoots and roots of two independent almt9 knock-out mutant lines and the corresponding wild types (De Angeli et al., 2013). A hydroponic system was used, and roots were exposed to control conditions or salt stress (100 mm NaCl), respectively, for 24 h. Under control conditions, hardly any difference in Na+ and Cl− contents were detected between almt9 mutants and wild types (Fig. 2, A and B; Supplemental Table S1). However, upon 24-h salt stress, Cl− accumulation was significantly lower in shoots of almt9 plants compared to the corresponding wild types (26 ± 4% in almt9-1 and 16 ± 3% in almt9-2). We tested whether this difference was reflected in Cl− xylem sap content, and although not significant at a P < 0.05-level, we found a substantial reduction of 32% in almt9 mutants compared to wild type (Supplemental Fig. S1). Interestingly, Na+ contents in shoots of almt9 mutants were similarly reduced upon salinity treatment (20 ± 6% in almt9-1 and 23 ± 10% in almt9-2), but root ion content was not significantly altered (Fig. 2, A and B; Supplemental Table S1). Of note, differences in shoot ion accumulation were observed specifically under NaCl stress, and no reduction in Na+ and Cl− levels were detected under 100-mm KCl or 100-mm NaNO3 treatment for 24 h (Supplemental Fig. S2). When plants were treated with 100 mm NaCl for 1 week, reduced shoot ion accumulation was no longer significant between wild-type and almt9 mutants (Supplemental Fig. S3). Taken together, these results show that almt9 knock-out mutants exhibit, besides the reduction in intracellular Cl− fluxes (De Angeli et al., 2013), an altered capacity to accumulate Na+ and Cl− in shoots during early salinity.
Figure 2.
Ion content analysis in shoots and roots of wild-type and almt9 mutants upon 24-h salt stress. The two knock-out alleles almt9-1 and almt9-2 and the corresponding wild types (wt-1 and wt-2) were grown in hydroponics, and Na+ (A), Cl− (B), K+ (C), and NO3− (D) contents were determined prior to (0 mM) and after NaCl treatment (100 mM). Data are means ± sd of n ≥ 5 biological replicates derived from two independent experiments. One-way ANOVA of each tissue and treatment and a pairwise comparison was used for statistical analysis. Asterisks indicate significant differences from the corresponding wt (*P < 0.05, **P < 0.01, ***P < 0.001). DW, Dry weight; FW, fresh weight.
In the following, we elucidated whether reduced Na+ and Cl− levels in almt9 mutants during salinity were accompanied by alterations in the accumulation of other ion species. K+ (Fig. 2C; Supplemental Table S1), Mg2+ (Supplemental Fig. S4A), NO3− (Fig. 2D; Supplemental Table S1), and MA2- (Supplemental Fig. S4B) contents showed no significant differences between both genotypes under control and salinity conditions. Also, the osmolality of the shoot press sap was indistinguishable between almt9 and wild types (Supplemental Fig. S4C). This suggests that despite reduced Na+ and Cl− contents in the shoots of almt9 mutants, these plants accumulate other solutes that maintain a shoot sap osmolality comparable to that of the wild type.
ALMT9 has high sequence similarities with several clade II members of the ALMT family (Kovermann et al., 2007). To exclude functional redundancy, we generated a transgenic hairpin RNA-expressing line in the genetic background of almt9-1 that simultaneously targets other ALMTs (clade II) for transcriptional down-regulation (Supplemental Fig. S5A). Shoots of this line did not show further reduction in Na+ or Cl− accumulation than almt9-1 (Supplemental Fig. S5, B and C), indicating that other closely related ALMT members do not contribute notably to the modulation of Na+ and Cl− shoot accumulation upon 24-h salt stress.
ALMT9-Mediated Vacuolar Cl− Currents Contribute to the Regulation of Whole-Plant Ion Accumulation
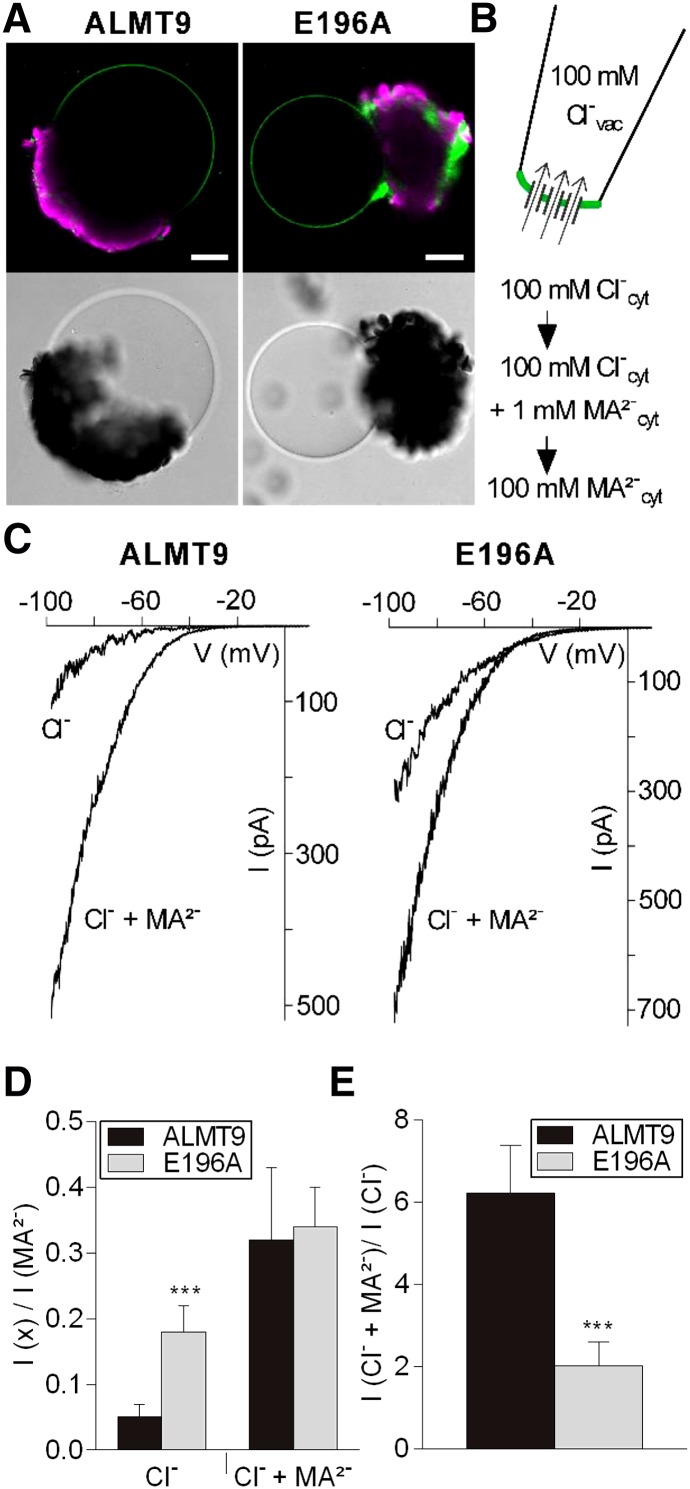
Our data show that almt9 mutants with reduced vacuolar Cl− currents (De Angeli et al., 2013) accumulate less Na+ and Cl− during salinity (Fig. 2). Hence, we wondered whether enhancing vacuolar Cl− currents would reverse or complement this effect on whole-plant ion accumulation. When modifying ion fluxes during salinity by transgenic approaches, it is fundamental to maintain cell type specificity of the transport processes (Møller et al., 2009). Therefore, instead of using ectopic overexpression of ALMT9, we aimed at identifying a mutated channel variant that exhibits increased Cl− current activity. For this purpose, we took advantage of a collection of point-mutated ALMT9 channels that was previously generated (Zhang et al., 2013). We found that the amino acid exchange E196A induced the desired alterations in ALMT9 channel properties, but did not change the channel localization at the tonoplast (Fig. 3A). By patch-clamping, it had been shown that the mutation E196A does not affect the MA2− conductivity of ALMT9 (Zhang et al., 2013). Here, we additionally examined the Cl− conductivity (Fig. 3B) and found that ALMT9E196A displayed Cl− currents and a marked inward but no outward rectification similar to ALMT9 (Fig. 3C). However, sequential cytosolic-side buffer exchanges (Fig. 3B) revealed that the ratio of Cl− to MA2− currents (ICl−/ IMA2−) differed between both channel variants (Fig. 3D). The Cl− current amplitude was 5 ± 2% of the MA2− current amplitude in ALMT9 and 18 ± 4% in ALMT9E196A (Fig. 3D). This shows that ALMT9E196A is able to mediate Cl− currents that are approximately three times higher than ALMT9-mediated Cl− currents. In addition, by determining ICl− + MA2−/ ICl−, we found that ALMT9 and ALMT9E196A were differently activated by cytosolic MA2− (Fig. 3C). ALMT9 showed a 6- ± 1-fold increase of Cl− conductivity in the presence of 1 mm MA2− at −100 mV, whereas ALMT9E196A showed a 2- ± 0.6-fold increase (Fig. 3E). However, the ratio ICl− + MA2−/ IMA2− that was 0.32 ± 0.1 and 0.34 ± 0.06 in ALMT9 and ALMT9E196A, respectively, was not significantly altered between both channel variants (Fig. 3D). These data demonstrate that the mutation E196A impacts the basal activity of ALMT9 by increasing Cl− currents across the tonoplast constitutively.
Figure 3.
Electrophysiological properties of the mutant channel ALMT9E196A. A, Fluorescence and transmission images of vacuoles released from lysed tobacco protoplasts that transiently overexpress ALMT9-GFP (left) and ALMT9E196A-GFP (right). Auto-fluorescence of chloroplasts is shown in magenta. Scale bars = 20 µm. B, Patch-clamp experimental procedure. Vacuoles were patched in excised cytosolic-side-out configuration under symmetric ionic conditions (100 mm Cl−vac/ 100 mm Cl−cyt). The cytosolic buffer was sequentially exchanged (100 mm Cl−; 100 mm Cl− + 1 mm MA2−; 100 mm MA2−) on the same membrane patch. C, Representative currents of ALMT9 (left) and ALMT9E196A (right) in presence of 100 mm Cl− (Cl−) and 100 mm Cl− + 1 mm MA2− (Cl− + MA2−) in the cytosolic buffers. Currents were evoked by a 2.5-s voltage ramp ranging from +40 mV to −100 mV. D, Relative Cl− and Cl− + MA2− currents mediated by ALMT9 and ALMT9E196A. Currents were normalized to the current amplitude measured at −100 mV in presence of 100 mm MA2− in the cytosolic solution. E, Level of MA2−-activation (ICl− + MA2−/ ICl−) of ALMT9- and ALMT9E196A-mediated Cl− currents at −100 mV. Data are means ± sd. Asterisks indicate statistically significant differences between ALMT9 (n = 5) and ALMT9E196A (n = 6) currents (*P < 0.05, **P < 0.01, ***P < 0.001; two-tailed Student’s t test).
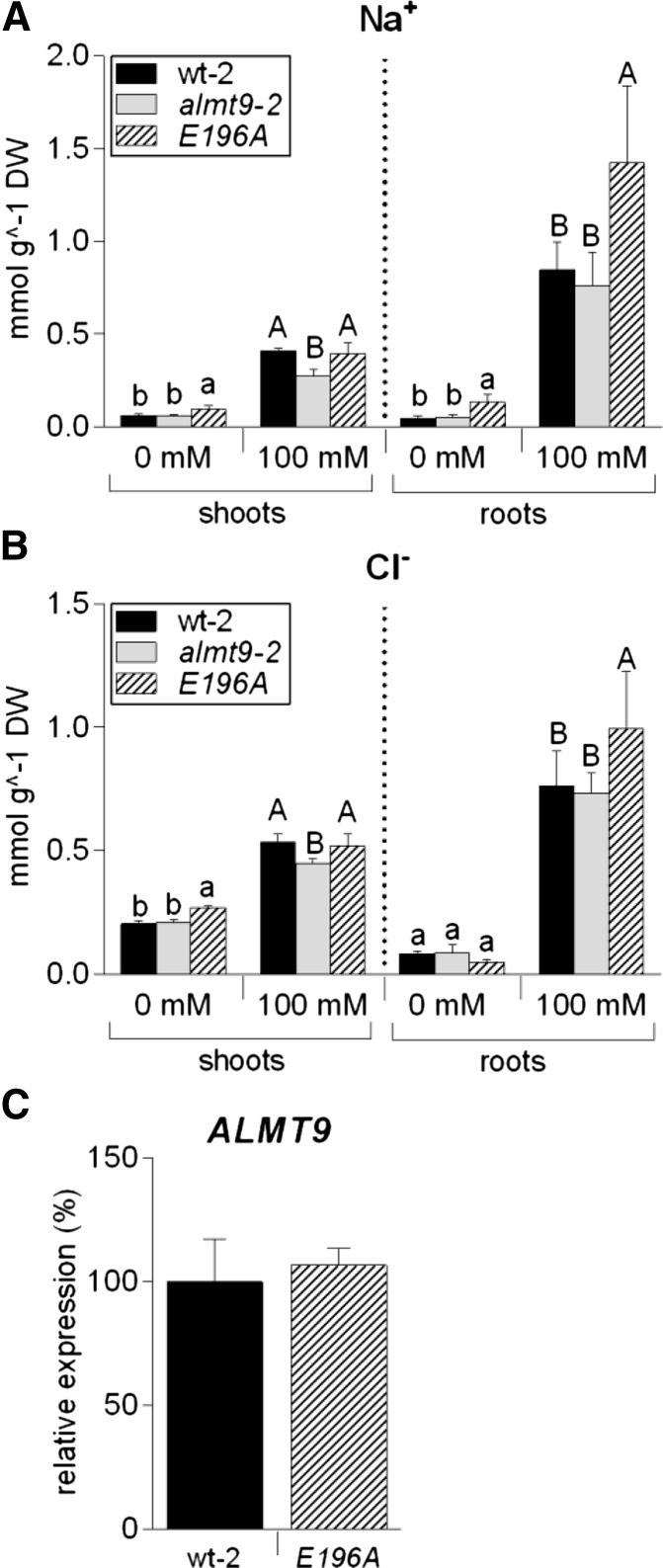
The electrophysiological measurements identified ALMT9E196A as a suitable tool to modify Cl− fluxes across the tonoplast. To test the physiological consequences of higher vacuolar Cl− currents, we expressed the mutant channel under the spatial and temporal control of the ALMT9 promoter (ALMT9pro:ALMT9E196A) in almt9-2; hereafter, this point-mutated complementation line is referred to as E196A. Determining the ion content in shoots revealed that E196A had slightly higher Na+ and Cl− concentrations than wild-type-2 and almt9-2 under control conditions (Fig. 4, A and B). In addition, the reduced shoot ion accumulation phenotype of almt9-2 was recovered in E196A under salt treatment. In roots, we found that 24-h salinity treatment induced a significantly higher ion accumulation in E196A (1.3 times more Cl− and 1.7 times more Na+) compared to wild-type-2 (Fig. 4, A and B). To exclude the possibility that differences in ion accumulation occur due to elevated ALMT9 expression, we analyzed the transcript amounts by qRT-PCR. ALMT9 expression in E196A and wild-type-2 did not differ significantly excluding a transcriptional effect on the measured ion concentrations (Fig. 4C). In summary, these results demonstrate that the magnitude of ALMT9-mediated vacuolar Cl− uptake alters ion accumulation at the whole-plant level.
Figure 4.
Na+ and Cl− content analysis in the transgenic line E196A. In plants of E196, the point-mutated channel variant ALMT9E196A is expressed under the control of the native ALMT9 promoter in the genetic background of almt9-2. Na+ (A) and Cl− (B) measurements were performed in shoot and root tissue of hydroponically grown wt-2, almt9-2, and E196A plants upon treatment with control (0 mM) or NaCl (100 mM) solutions for 24 h. The results are shown as mean ± sd of n ≥ 5 biological replicates derived from two independent experiments. For statistical analysis, one-way ANOVA of each tissue and treatment and a Tukey-Kramer multiple comparison posttest was used. Different lowercase letters indicate significant differences in ion content (P < 0.05) under control conditions, capital letters under salinity. DW, Dry weight. C, Expression analysis of ALMT9 in wt-2 and E196A using qRT-PCR. ACT2 served as a reference gene. The expression levels were normalized to wt-2. Data are means ± sd from n = 2 biological replicates.
Lower Shoot Ion Accumulation in almt9 Is Not Caused by Impaired Stomatal Opening
Transpiration rates highly impact root-to-shoot translocation of ions. In a previous study, almt9 has been shown to display impaired light-dependent stomatal opening due to reduced Cl− fluxes into the vacuoles of guard cells (De Angeli et al., 2013). Therefore, we investigated whether decreased leaf transpiration in almt9 might be responsible for the observed reduction in shoot ion translocation under salt stress. Previously, stomatal apertures of almt9 had been analyzed on peeled epidermal strips (De Angeli et al., 2013). In the current study, we performed in situ stomatal assays using hydroponically grown plants (see “Materials and Methods”) that provide a snapshot of the native stomatal aperture over a time course of NaCl treatment. Indeed, using this method in blind assays, the previously reported reduced stomatal aperture of almt9 mutant plants (De Angeli et al., 2013) was reproduced (Fig. 5A). Salinity treatment induced stomatal closure in wild-type-1 and almt9-1 after 3 h (Supplemental Fig. S6A). Upon 24-h salt stress, we observed a recovery of the stomatal aperture, which is in accordance with the observation that Na+ can be used as osmotically active solute for stomatal opening (Zhao et al., 2011; Yu and Assmann, 2015). Interestingly, the reduced stomatal opening in almt9 plants was restored upon salinity (Fig. 5A). Similarly, when we conducted the same assay using 100 mm KCl, we found no significant difference between the stomatal aperture of wild-type-1 and almt9-1 (Supplemental Fig. S6B), suggesting that the complementation was based on the increased presence of osmotica. This indicated that reduced almt9 stomatal apertures and transpiration were not responsible for decreased shoot ion accumulation under salt stress.
Figure 5.
Investigation of stomatal movement and ion content in a guard cell-specific complementation line of almt9-2. A, In situ assay of native stomatal apertures (see “Materials and Methods”) using hydroponically grown plants of the almt9-1 and almt9-2 mutant alleles and the corresponding wild types (wt-1 and wt-2). Roots were exposed to control (0 mM) or NaCl (100 mM) solutions, and the stomatal aperture was measured before (0 h) and after (24 h) treatment. Error bars correspond to SEM, which was calculated from averages of at least four biological replicates. Asterisks indicate statistically significant differences in stomatal aperture from the corresponding wt (*P < 0.05, **P < 0.01, ***P < 0.01; two-tailed Student’s t test). B, In situ assay of native stomatal apertures of wt-2, almt9-2, and the complemented line GC:ALMT9 that guard cell-specifically expresses ALMT9 in control conditions. Error bars correspond to SEM, which was calculated from n ≥ 4 biological replicates. C and D, Contents of Na+ (C) and Cl− (D) were measured in shoots and roots of hydroponically grown wt-2, almt9-2, and GC:ALMT9 plants after 24-h treatment with control (0 mM) or NaCl (100 mM) solutions. The combined results from two independent experiments are shown as mean ± sd (n ≥ 5). For statistical analysis in B to D, one-way ANOVA and a Tukey-Kramer multiple comparison were used. Different letters indicate significant differences in ion content (P < 0.05) within each tissue (lowercase in control conditions, capital letters under NaCl stress). DW, Dry weight.
To gain definite evidence that the stomatal conductance of almt9 does not contribute to lower ion accumulation, we generated an ALMT9 complementation line under the control of the guard cell-specific MYB60 (Cominelli et al., 2005) promoter (MYB60pro:ALMT9) in the genetic background of almt9-2, referred to as GC:ALMT9 (Supplemental Fig. S7). The reduced light-dependent stomatal opening of almt9-2 was rescued in GC:ALMT9 (Fig. 5B). However, GC:ALMT9 did not complement the ion accumulation phenotype of almt9 mutants (Fig. 5, C and D). This result demonstrates that impaired stomatal opening and transpiration of almt9 does not account for differences in shoot ion accumulation during early salinity.
Disturbed Intracellular Cl− Fluxes Induce a Specific Expression Pattern under Salinity
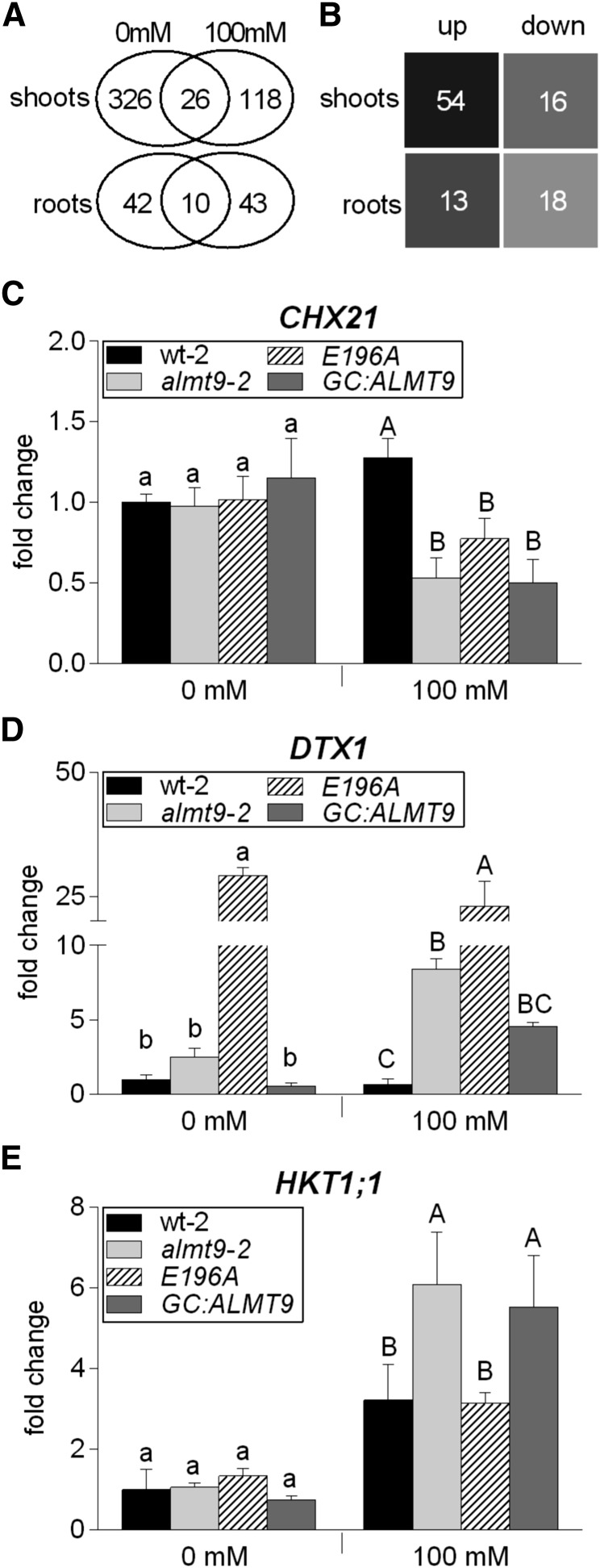
To unravel the physiological basis of the detected differences in ion accumulation and elucidate the role of vacuolar ion uptake during salinity, we conducted a transcript profile analysis by RNA-seq using wild-type-1 and almt9-1 shoots and roots under nonsaline and saline conditions (0 mm or 100 mm NaCl for 24 h). Data of three biological replicates were analyzed as described in “Materials and Methods.” To identify genes with significant differences in expression between wild-type-1 and almt9-1, we used a fold change cutoff level of 2 (log2 ratio ≥ ± 1, P < 0.01). A total of 352 genes showed a 2-fold or greater difference in expression in shoots and 52 genes in roots under control conditions, and 144 in shoots and 53 in roots under salinity (Fig. 6A; Supplemental Data Sets S1–S4).
Figure 6.
Transcriptome analysis in wild-type and almt9 shoots and roots upon salinity. A and B, RNA-seq analysis of hydroponically grown wt-1 and almt9-1 plants upon exposure to control (0 mM) or NaCl (100 mM) conditions for 24 h. A, Venn diagram showing the number of differentially expressed genes between wt-1 and almt9-1 within each tissue and treatment. The overlap between the ovals represents genes that have significant changes in gene expression under both treatments. B, Number of genes that are significantly differentially expressed between both genotypes exclusively under salinity. For selection requirements, see “Materials and Methods.” up, up-regulated genes in almt9-1; down, down-regulated genes in almt9-1. C to E, The expression levels of candidate genes were determined in wt-2, almt9-2, and the complemented lines E196A and GC:ALMT9 by qRT-PCR. The same experimental set-up as for the RNA-seq analysis was used. Transcript abundance of CHX21 (C) and DTX1 (D) was determined in shoots, transcript abundance of HKT1;1 (E) in roots. The data were normalized to the expression level of the respective gene in wt-2 under control conditions. ACT2 served as a reference gene. Each data point was derived from n ≥ 3 biological replicates and is shown as mean ± sd. In C to E, significances at P < 0.05 were analyzed by one-way ANOVA and Tukey-Kramer multiple comparison posttest for each treatment and are indicated by lettering (lowercase for control conditions, capital letters for salinity).
Strikingly, under nonsaline conditions, numerous genes that were suggested to be stress-inducible and/or involved in ABA-mediated signaling, such as the PP2C-type protein phosphatases AIP1 (Lim et al., 2012) and ABI2 (Merlot et al., 2001; Rubio et al., 2009), were differentially expressed in almt9-1 (Supplemental Data Sets S1 and S2). Besides, several genes encoding ATP-Binding Cassette (ABC)-transport proteins and genes that belong to the NRT (Nitrate Transporter) families (NRT1.8 [Li et al., 2010] in shoots and NRT2.4 [Kiba et al., 2012] in roots) exhibited changes in expression levels (Supplemental Data Sets S1 and S2). The transcriptional modification of stress- and transport-related genes indicates that the reduced vacuolar Cl− fluxes in almt9 mutants impact plants at the molecular and physiological level.
We identified only 26 genes in shoots and 10 genes in roots with a significant difference in the expression level under both control and salinity conditions (Fig. 6A). Hence, salt stress evokes distinct changes between the transcriptomes of wild-type-1 and almt9-1 mutants.
Subsequently, we closely analyzed the subset of genes with a significant up- or down-regulation in almt9-1 exclusively under salt stress but not under control conditions (Fig. 6B; Supplemental Tables S2 and S3; for selection criteria, see “Materials and Methods”). We identified 54 genes that were up-regulated in almt9-1 shoots and 16 genes that were down-regulated; 13 genes that were up-regulated in almt9-1 roots and 18 genes that were down-regulated. Several of these genes have functions associated with salt stress (Supplemental Fig. S8; Supplemental Tables S2 and S3), such as stress signal transduction, for example, ERF/AP2-type transcription factors, DDF1 and DDF2 (Magome et al., 2004, 2008); redox homeostasis, for example, ascorbate peroxidase, APX2; and hormone homeostasis, e.g. UGT74E2 (Tognetti et al., 2010). The data show that the lack of the vacuolar Cl− channel ALMT9 has a global impact on the genome-wide transcriptional response under salinity.
Among the genes showing differential transcriptional regulation exclusively upon salinity, we examined candidates that code for transporter proteins with a putative role in salinity-related processes in more detail (Fig. 6, C–E). The RNA-seq results were verified by qRT-PCR using the second knock-out allele almt9-2. We confirmed that the expression of the CHX21 gene that encodes for a putative plasma membrane Na+ transporter (Hall et al., 2006) was indistinguishable between both genotypes under control condition in shoots (Fig. 6C; Supplemental Table S2). However, upon salinity the transcript levels increased slightly in the wild type but decreased in almt9. Interestingly, we identified three Multidrug and Toxic Compound Extrusion (MATE)-related transporter genes by RNA-seq of which the expression was highly up-regulated in almt9-1 shoots upon salinity (Supplemental Table S2). We confirmed this expression pattern by qRT-PCR using one of them, DTX1 (Fig. 6D), a plasma membrane transporter that was suggested to export toxic compounds (Li et al., 2002). In the RNA-seq data of roots, the expression of HKT1;1, a well-known gene coding for a Na+-selective transport protein at the plasma membrane of root stelar cells (Davenport et al., 2007), showed high variability between the three biological replicates and was therefore not among the significantly differentially regulated genes. Since HKT1;1 has a crucial role in diminishing Na+ translocation to shoots, we further investigated its transcriptional response to salinity in both genotypes by qRT-PCR. The analysis of four biological independent experiments proved that HKT1;1 expression increased in almt9-2 roots in response to salinity approximately twice as much as in wild-type-2 roots (Fig. 6E). SOS1, another gene known to encode for a Na+ transporter involved in salinity responses (Shi et al., 2000, 2002), did not show significantly altered expression levels between the different genotypes (Supplemental Fig. S9).
Our qRT-PCR analysis of CHX21, DTX1, and HKT1;1 included the complementation lines E196A and GC:ALMT9 to untangle the molecular basis of the transcriptional disregulation of the candidate transporter genes. For all three investigated genes, GC:ALMT9 showed similar expression patterns as almt9-2 (Fig. 6, C–E), consistent with the finding that differences in ion concentration do not arise from differences in stomatal movement between wild type and almt9 during salinity (Fig. 5). The E196A plants, which have enhanced vacuolar Cl− conductivity in the vasculature and leaf mesophyll, showed a more complex transcriptional response. Indeed, E196A roots displayed the same expression profile of HKT1;1 as wild-type-2 (Fig. 6E), but had a similar expression pattern as almt9-2 regarding the shoot CHX21 expression (Fig. 6C) and showed an even more pronounced transcriptional up-regulation of DTX1 in shoots than the almt9-2 mutant (Fig. 6D).
The RNA-seq data identified further transporter genes that show changes in gene expression in almt9-1 specifically upon salinity (Supplemental Tables S2 and S3), namely ACA12 (Limonta et al., 2014), ABCB4 (Terasaka et al., 2005), SUC5 (Baud et al., 2005; Pommerrenig et al., 2013), and OCT1 (Lelandais-Brière et al., 2007; Strohm et al., 2015). Notably, no member of the ALMT, CLC, or NRT family has been identified as differentially regulated in almt9-1 in response to salinity (Supplemental Tables S2 and S3).
DISCUSSION
During salinity the sequestration of Na+ and Cl− ions into the vacuole is crucial to maintain optimal metabolic conditions in the cytosol and mitigate cellular damage. Our data provide new insights into the role of intracellular ion partitioning during salt stress. Crucially, we show that Cl− fluxes across the vacuolar membrane have consequences on fluxes across the plasma membrane and thereby influence global plant ion movement. Firstly, we present evidence that disturbed vacuolar Cl− uptake has a dramatic effect on shoot ion accumulation during early salt stress. Secondly, the data show a strong correlation between anion and cation fluxes during salinity as not only Cl− but also Na+ accumulation was affected in almt9 and E196A mutants specifically under NaCl treatment. And thirdly, our results imply that a disturbed intracellular ion homeostasis acts as a feedback-signal that regulates the expression of genes encoding for plasma membrane-localized transport proteins.
Ions are delivered to the shoot via the transpiration stream. Therefore, differences in shoot ion contents might arise from diminished transpiration rates, or from the modification of transport processes in the root stele that catalyze long-distance ion translocation to shoots. Here, we provide comprehensive evidence that stomatal movement and transpiration does not account for the differences in shoot ion accumulation between almt9 and wild-type plants during salinity. In contrast, we show that HKT1;1 shows elevated expression levels in roots of almt9 mutants in response to salinity. HKT1;1 is a transport protein at the plasma membrane of the root stele involved in xylem retrieval of Na+ from the transpiration stream and reduction of Na+ root-to-shoot transfer (Davenport et al., 2007). Knock-out mutants of HKT1;1 show elevated xylem (Sunarpi et al., 2005) and shoot (Mäser et al., 2002) Na+ levels when exposed to salt stress. In contrast, cell type-specific overexpression of HKT1;1 in stelar root cells reduces shoot Na+ contents (Møller et al., 2009). Thus, the transcriptional up-regulation of HKT1;1 in almt9 mutants might contribute to lower shoot Na+ contents. In accordance, E196A plants exhibit similar HKT1;1 expression levels in roots as the wild-type and a similar shoot ion accumulation during salinity.
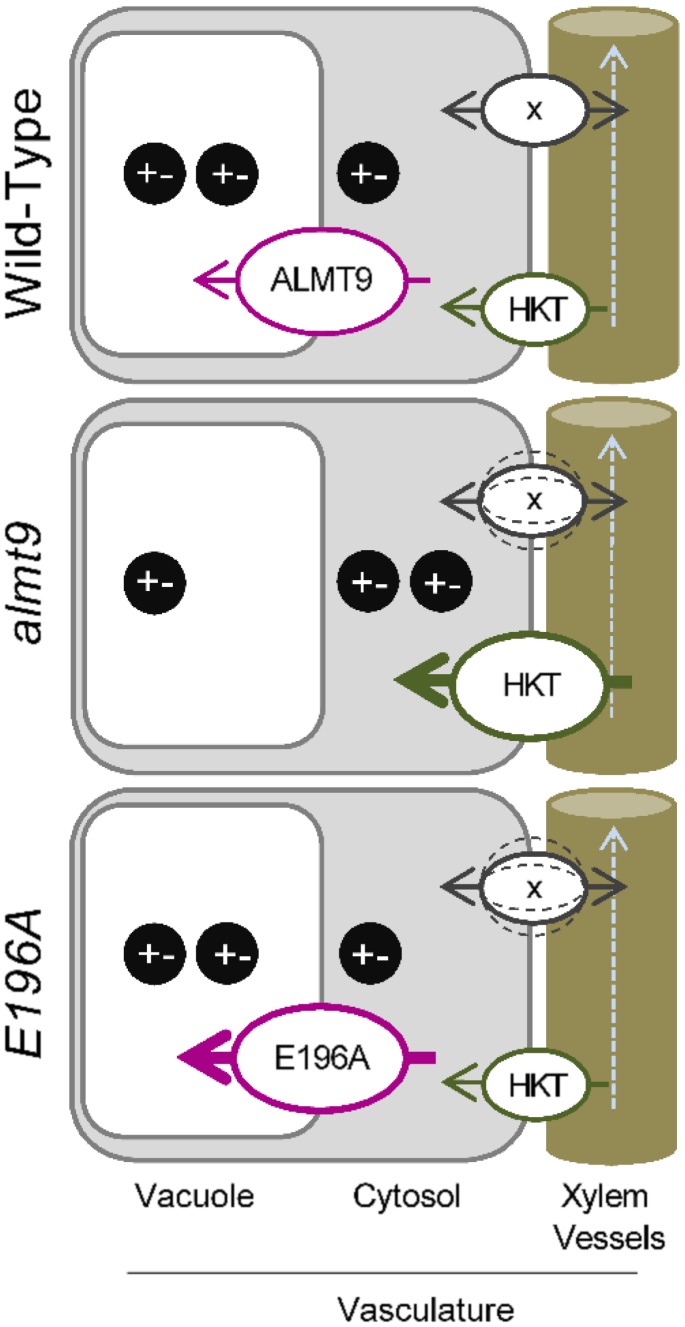
The coinciding expression pattern of ALMT9 and HKT1;1 (Møller et al., 2009) in the root stele suggests that the lack of the Cl− channel in these cells modifies the expression of transporters at the plasma membrane. The overall tissue ion content in roots was not altered in almt9 and wild-type plants. Therefore, we propose a model in which the absence of ALMT9 reduces the loading capacity of Cl− and presumably also Na+ ions into the vacuole of stelar cells (Fig. 7). A linked transport of both Cl− and Na+ is supported by the finding that NaNO3 treatment does not reduce shoot ion accumulation in the almt9 mutants which lack the Cl− channel. The reduced vacuolar ion uptake might result in a disturbed intracellular ion homeostasis which might in turn mimic an elevated salt stress that promotes the transcriptional up-regulation of HKT1;1. Consistently, E196A plants with restored or increased vacuolar Cl− uptake have a transcriptional response of HKT1;1 similar to wild-type plants (Fig. 7). The importance of intracellular ion homeostasis in regulating cellular events in response to salinity in roots was also demonstrated in the sos1-1 mutant, which is deficient in cellular Na+ extrusion and which showed a magnification of salt stress responses (Oh et al., 2010). A role of ALMT9 in regulating the vacuolar ion uptake in the root stele during salinity is in agreement with the fact that ALMT9 expression in roots is specifically up-regulated in response to NaCl, but not to other ionic or osmotic stresses. Hence, our findings show that the control of vacuolar ion contents in pericycle and xylem parenchyma cells contributes to whole-plant ion accumulation during salinity as previously suggested (Storey et al., 2003, Läuchli et al., 2008). In addition, we propose that the capacity of vacuolar ion uptake in these cells is a factor that influences root-to-shoot ion transport processes via the modification of the expression of plasma membrane-bound ion transporters in response to salt stress. How a potentially disturbed intracellular ion homeostasis leads to changes in gene expression is still an open question. It has to be noted that the vacuolar storage capacity also influences heavy metal long-distance transport. However, the reduced loading of heavy metals in the vacuoles of root stelar cells causes enhanced shoot ion accumulation (Arrivault et al., 2006, Peng and Gong, 2014). Furthermore, astonishingly, we found that NaCl treatment results in reduced shoot ion accumulation in almt9 plants whereas KCl treatment does not, indicating that long-distance Cl− transport in almt9 knock-out mutants is altered specifically in presence of NaCl. Therefore, it seems that an ion- and/or stress-dependent signaling pathway operates downstream the vacuolar ion loading capacity.
Figure 7.
Proposed model for the transcriptional regulation of transporter genes involved in whole-plant ion distribution by vacuolar ion uptake during early salinity. The expression of plasma membrane-localized transport proteins involved in long-distance ion transport is tuned by the efficiency of vacuolar Cl− fluxes in the vasculature at the onset of salinity stress. The reduced Cl− storage in the vacuoles of almt9 mutants might perturb the intracellular homeostasis of Na+ and Cl− (black circles represent positively and negatively charged ions). This might signal an enhanced NaCl stress and initiate elevated expression levels of transporters such as the Na+ transporter HKT1;1 (HKT). In accordance, wild-type and E196A plants have more efficient vacuolar Cl− uptake, and exhibit lower salt-induced HKT1;1 transcript increase. Similarly, transcript levels of other genes encoding for transporters (x) involved in the regulation of long-distance ion translocation in shoots and roots might be up- or down-regulated (dashed lines) in response to the capacity of storing Cl− in the vacuoles of the vascular system.
The disturbed intracellular ion homeostasis in the vascular system of almt9 might also affect the regulation of transport proteins in shoots (Fig. 7). Interestingly, CHX21 (Hall et al., 2006) and DTX1 (Li et al., 2002), plasma membrane transporters that were identified in the shoot RNA-seq analysis, were also associated with the modulation of xylem loading and long-distance ion transport. CHX21 was previously shown to be expressed in endodermal root cells and suggested to control root-to-shoot translocation of Na+ during salinity (Hall et al., 2006). Although we did not determine the cell type-specific distribution of Na+ ions, the potential role in the regulation of Na+ xylem content (Hall et al., 2006) together with the differential expression in almt9 mutant shoots leads us to propose that CHX genes are likely to contribute to intercellular Na+ partitioning within shoots during salinity.
In our study, the MATE-related transporter gene DTX1 as well as two further DTX members of the same subfamily (Li et al., 2002) were highly up-regulated in shoots of almt9. DTX1 has been described as a detoxification efflux carrier, and a role in exporting toxic compounds into the xylem for long-distance transport and distribution within the plant has been hypothesized (Li et al., 2002). Since also E196A shoots showed a dramatic increase in DTX1 levels even under control conditions, the transcriptional up-regulation is apparently stimulated by the disregulated vacuolar Cl− fluxes in both mutants. This observation points toward a role of DTX1 in the export of compounds that accumulate in response to impaired intracellular ion homeostasis or to an osmotic imbalance that is probable in mutants with reduced or enhanced vacuolar Cl− uptake. An implication of ALMT9 in maintaining the osmotic status in shoots under salinity is suggested by the transcriptional up-regulation in response to KCl and sorbitol specifically in this tissue. Interestingly, DTX1 and the two other identified DTX members are coexpressed (Obayashi et al., 2007; www.atted.jp) with UGT74E2 (Tognetti et al., 2010), the expression of which was also found to be highly up-regulated in almt9 shoots in the RNA-seq analysis. The hydrogen peroxide-responsive UGT74E2 encodes a UDP-glycosyltransferase that contributes to salt stress adaptation mechanisms (Tognetti et al., 2010). It will be of interest to determine the physiological substrate of these DTX transporters and investigate their functional role during salinity, which might be linked to UGT74E2 activity.
The altered expression of Na+ transporters such as HKT1;1 and CHX21 in the Cl− channel mutants almt9 and E196A substantiates the strong coupling of the transport of both ion species during salinity as also observed in the ion content measurements. Strikingly, among the transporter genes that were differentially regulated in almt9 under salinity, we did not identify genes that encode for putative anion transporters. However, transcription levels of transport-related genes under nonsaline conditions might contribute to alterations in tissue ion content during salinity as suggested for different grapevine (Vitis vinifera) species (Henderson et al., 2014). For instance, the basal expression of two genes that encode for plasma membrane-localized NRT transporters showed different levels in almt9. Some members of the NRT1/PTR (Nitrate Transporter 1/ Peptide Transporter; Léran et al., 2014) family have been found to mediate transmembrane NO3− and Cl− fluxes and were suggested to be implemented in salinity responses (Li et al., 2010, 2016; Chen et al., 2012, Taochy et al., 2015). Hence, the expression landscape of transporter genes under control conditions might alter ion fluxes constitutively and contribute potentially to differences in tissue ion accumulation during salinity.
In addition, the activity of several anion channels and transporters has been shown to be stimulated at other levels than the transcriptional, for instance by phosphorylation (Frachisse et al., 2000; Liu and Tsay, 2003; Lee et al., 2009; Wege et al., 2014), or by signals such as the pH (Frachisse et al., 2000, Colcombet et al., 2005; Meyer et al., 2011) and Ca2+ concentrations (Gilliham and Tester, 2005; Meyer et al., 2011). If the activity of transporters involved in Cl− fluxes during salinity is modified by these regulatory mechanisms, RNA-seq experiments will not identify the corresponding genes. The same applies for other Na+ transporter proteins such as SOS1, which might contribute to differences in Na+ accumulation in almt9 mutants although not being differentially regulated at the transcriptional level.
The lower ion contents in almt9 were detected during early salinity, but not upon extended exposure to salt stress. Transporter proteins that belong to the ALMT or other families might contribute to efficient vacuolar Cl− uptake in almt9 during prolonged salinity. For example, members of the CLC family are able to transport Cl− across the tonoplast (Jossier et al., 2010; Wege et al., 2010; De Angeli et al., 2013; Nguyen et al., 2015). The relative contribution of CLC-mediated Cl− currents might thereby increase with accumulating Cl− contents over time, since the slightly negative membrane potential at the tonoplast supports the passive uptake of Cl− by channels only up to a concentration gradient of 3-fold (Teakle and Tyerman, 2010). By contrast, secondary active transporters such as CLCa can account for up to 50-fold concentration gradients across the tonoplast (De Angeli et al., 2006). This indicates that plants have a set of tonoplast-localized transport proteins that are implicated in intracellular ion uptake at different time-points during salinity.
To conclude, while the role of plasma membrane-localized transport proteins in salt stress adaptation mechanisms has been studied intensively, the importance of vacuolar ion uptake in particular during early salinity remained elusive. In our study, we clearly show that the significance of ion movement across the tonoplast during salt stress exceeds a role of vacuolar ion sequestration during long-term salinity to guarantee cellular survival. The differences in shoot ion content in almt9 and E196A mutants and the transcriptional alteration of several plasma membrane-localized transporters with an established or putative role in long-distance ion translocation suggest an interplay between disturbed intracellular ion homeostasis and tissue ion fluxes. Our findings provide strong evidence that the capacity of vacuolar ion uptake in vascular tissue is a pivotal factor that contributes to the modulation of long-distance ion transport at the onset of salinity.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Salinity Treatment
The two independent T-DNA insertion lines of ALMT9 (almt9-1: WiscDsLox499H09; almt9-2: SALK_055490) and the corresponding wild types were described in a previous study (De Angeli et al., 2013). Arabidopsis (Arabidopsis thaliana) wild-type Col-0 and almt9 plants were grown in a growth chamber (21°C, 12 h light /12 h dark, 40% relative humidity) in hydroponic culture. 1/2 Murashige and Skoog (MS), pH 5.6, was used as nutrient solution. It has to be noted that 1/2 MS contains 9.4 mm KNO3, 0.6 mm K2HPO4, and 20.6 mm NH4NO3. Unless otherwise specified, treatment was applied to 5-week-old plants by adding control (0 mm supplemental NaCl) or salt stress (100 mm supplemental NaCl) solutions. Treatment was started 6 h after dawn and material was harvested after 24 h.
Construct Design and Generation of Transgenic Plants
To generate ALMT9pro:GUS, a 548-bp promoter sequence upstream of the start codon of ALMT9 was cloned into pMDC163 (Curtis and Grossniklaus, 2003). The gene silencing suppressor mutant rdr-6-11 (Peragine et al., 2004; Meyer et al., 2011) and Col-0 plants were transformed with the construct, and positive transformation events were selected by hygromycin.
The intron-containing hairpin RNA was generated using pKANNIBAL (Wesley et al., 2001). The construct was based on the ALMT5 nucleotide sequence in order to target ALMT family clade II members (Kovermann et al., 2007) for multiple down-regulation. NotI fragments were subcloned into pART27 (Gleave, 1992) for stable transformation into the almt9-1 knock-out background (transgenic line hpRNA). Positive transformation events were selected by kanamycin. The expression of ALMT family clade II members (ALMT3, ALMT4, ALMT5, ALMT6) was evaluated by qRT-PCR using 7-d-old T3 seedlings grown on plates containing 1/2 MS, pH5.6, and 1% phytoagar. The line hpRNA was selected based on the highest degree of transcriptional down-regulation of ALMT clade II members.
To generate the point-mutated complementation line ALMT9pro:ALMT9E196A (line E196A), ALMT9 and its upstream promoter region (548 bp) were amplified from wild-type genomic DNA and cloned into pPLV22 (De Rybel et al., 2011). The site-directed mutagenesis was performed as described previously (Zhang et al., 2013). The construct was transformed into almt9-2 and positive transformation events were selected by BASTA. Seven-day-old seedlings of the T3 progeny of line E196A and wild-type-2 were grown on plates containing 1/2 MS, pH5.6, and 1% phytoagar to determine ALMT9 expression by qRT-PCR.
To generate a guard cell-specific complementation line of almt9-2 (line GC:ALMT9), ALMT9 cDNA was amplified and cloned into pMDC32 (Curtis and Grossniklaus, 2003) under the control of the guard cell-specific MYB60 promoter (Cominelli et al., 2005) as described previously (Nagy et al., 2009). The construct MYB60pro:ALMT9 was transformed into almt9-2. T3 progeny of hygromycin-resistant transformants of GC:ALMT9 was used to evaluate ALMT9 expression by PCR. Mesophyll cell protoplasts of 5-week-old soil-grown wild-type-2, almt9-2, and GC:ALMT9 plants were isolated by enzymatic digestion as described in the “Patch-Clamp Measurements” section. Total RNA of mesophyll cell protoplast, and shoots and roots of plants grown on plates (1/2 MS, pH5.6, and 1% phytoagar), was extracted and cDNA was synthesized as described in the “qRT-PCR” section. The PCR conditions were set as follows: 98°C for 2 min in the first cycle; subsequently, multiple amplification cycles consisting of 30 s at 98°C, 30 s at 58°C, and 2.5 min at 72°C; the final extension was 5 min at 72°C. To amplify ALMT9, 32 amplification cycles were used for KAT1 35 cycles and for ACT2 27 cycles. KAT1 served as a control of a guard cell-specific gene (Nakamura et al., 1995) to exclude contamination of mesophyll cell protoplasts with guard cells.
Primers used for amplification and evaluation of the transgenic lines are listed in Supplemental Table S4. Arabidopsis was stable transformed via Agrobacterium tumefaciens using the floral dipping method (Clough and Bent, 1998).
qRT-PCR
Total RNA was extracted from approximately 30 mg shoot and root tissue (pool of material from two to three plants) by using the SV Total RNA Isolation System (Promega) following the manufacturer’s instructions. Then 1 µg total RNA was reverse transcribed using oligo(dT) priming and M-MLV reverse transcriptase (Promega) according to the manufacturer’s instructions. Transcript levels were determined by qRT-PCR using the 7500 Fast Real-Time PCR System (Applied Biosystems) and a SYBR Green PCR Master Mix (Applied Biosystems). Transcript levels were calculated with the standard curve method as described by Pfaffl (2001) and were normalized against the expression of the actin gene ACT2. All reactions were performed in technical triplicates that were averaged to generate one biological replicate. Primers used for the transcript analysis are listed in Supplemental Table S4.
Tissue-Specific Expression of ALMT9
T3 ALMT9pro:GUS reporter plants were grown on plates (1/2 MS, pH5.6, and 1% phytoagar) for 2 to 3 weeks. For the evaluation of the expression pattern under salinity, plants were transferred to plates containing 100 mm supplemental NaCl for 24 h. Shoots and roots were assayed for GUS activity in a staining solution containing 1 mm X-Gluc (5-bromo-4-chloro-3-indolyl-β-d-glucuronide) in 50 mm sodium phosphate buffer, pH 7.15, 0.5 mm potassium ferricyanide, 0.5 mm potassium ferrocyanide, and 0.05% Triton X-100. After vacuum infiltration, the tissue was incubated for approximately 4 h (roots) or overnight (shoots) at 37°C in the staining solution. The reaction was stopped with several washes in 50 mm sodium phosphate buffer, pH 7.15, and tissues were cleared with 70% ethanol overnight. Images were captured with a digital reflex camera from Canon and with a Leica DMR widefield fluorescence microscope (Leica Microsystems). To obtain transverse sections, roots were embedded in a 4% agarose solution and 150 µm sections were generated by vibratome. Three independent transgenic lines in the rdr6-11 background and three lines in Col-0 were analyzed for expression of the GUS reporter that each showed similar expression patterns.
Ion and Osmolality Content Quantification
Mg2+, K+, Na+, and Cl− ions were extracted and quantified as described in Munns et al., 2010. In brief, shoots and roots of three to four plants were pooled for each biological replicate. Dry weight from shoot and root tissue was determined, and ions were extracted using 0.5 m nitric acid overnight at 70°C. Cation content was determined using atomic absorption spectrometry, and Cl− was determined using the colorimetric ferricyanide method (Munns et al., 2010). All reactions were performed in technical triplicates that were averaged to generate one biological replicate. The measurements of wt-2, the knock-out mutant almt9-2, and its complementation lines GC:ALMT9 and E196A were performed in parallel. Hence, the results of the Na+ and Cl− contents measured in wt-2 and almt9-2 are presented in Figures 2, 4, and 5.
For nitrate quantification, a colorimetric assay with salicylic acid was used according to Cataldo and Maroon, 1975. In brief, nitrate was extracted from shoots and roots (pool of two plants per biological replicate) in 5 mL hot deionized water (90°C to 95°C) and incubated in an 80°C waterbath for 30 min. Cooled supernatants and standard solutions were incubated at room temperature with 5% salicylic acid (w/v) that was dissolved in 96% sulfuric acid. After 20 min, 2 m NaOH was added to raise the pH and absorbance was measured at 480 nm. All reactions were performed in technical duplicates that were averaged to generate a biological replicate.
Malate was extracted from shoot tissue as described previously (Hurth et al., 2005). Malate contents were measured with the l-Malic Acid Enzymatic BioAnalysis Kit (Roche, R-Biopharm) according to the manufacturer’s instructions. All reactions were performed in technical duplicates that were averaged to generate a biological replicate.
Osmolality was determined using the leaf sap extracts of shoots. The material underwent a cycle of freezing and thawing and was subsequently mechanically pressed. After centrifugation, the supernatant was collected and used for osmolality measurements with a micro-osmometer (Advanced Instruments, Inc., model 3320).
Xylem Sap Collection
Five-week-old hydroponically grown plants were subjected to 0 mm and 100 mm NaCl, respectively. Treatment was started at the onset of the light period. After 24 h of treatment, the stem was cut with a sharp razor blade, and a borosilicate thin wall capillary (30-0062, Harvard Apparatus) was put over each stem. Drops of xylem sap that accumulated in the capillary were collected after 4 to 6 h using a micropipette. Xylem sap of at least ten plants was pooled per genotype and treatment to generate one biological replicate. By this, material for four biological replicates per genotype and treatment was obtained. Cl− content measurement was performed as described in “Ion and Osmolality Content Quantification.” Xylem sap of the plants treated with 100 mm NaCl was diluted 1:10 using 0.5 m nitric acid.
Confocal Microscopy
The constructs ALMT9-GFP and ALMT9E196A-GFP, which have been described previously (Zhang et al., 2013), were transformed into Agrobacterium tumefaciens (GV3101) by electroporation. The Agrobacterium-mediated infiltration of 4-wk-old tobacco leaves was performed as described previously with slight modifications (Yang et al., 2001). After transient transformation, tobacco plants were grown in the greenhouse (16 h light/ 8 h dark, 25°C/ 23°C, 100–200 μmol photons m−2 s−1, 60% relative humidity) for another 2 to 3 d and then used to extract protoplasts for confocal microscopy and patch-clamp measurements.
Tobacco mesophyll protoplasts transiently overexpressing ALMT9-GFP and ALMT9E196A-GFP were extracted by enzymatic digestion as described in “Patch-Clamp Measurements.” Vacuoles were isolated by a slight osmotic shock. Images were obtained at the Imagerie-Gif platform (http://www.i2bc.paris-saclay.fr/spip.php?article278) on a Leica SP8 inverted confocal microscope with laser excitation at 488 nm and collection of emitted light at 495 to 550 nm for GFP and 600 to 650 nm for chlorophyll.
Patch-Clamp Measurements
Mesophyll protoplasts from ALMT9-GFP and ALMT9E196A-GFP overexpressing tobacco leaves were isolated by enzymatic digestion. The enzyme solution contained 0.3% (w/v) cellulase R-10, 0.03% (w/v) pectolyase Y-23, 1 mm CaCl2, 500 mm sorbitol, and 10 mm MES, pH 5.3, 550 mosmol. Protoplasts were washed twice and resuspended in the same solution without enzymes. Vacuoles were released from mesophyll protoplasts by the addition of 5 mm EDTA and a slight osmotic shock (500 mosmol, see cytosolic solution below). Transformed vacuoles exhibiting an ALMT9-GFP or ALMT9E196A-GFP signal were selected using an epifluorescence microscope. Membrane currents from tonoplast patches were recorded in the excised cytosolic-side-out configuration with the patch-clamp technique as described elsewhere (De Angeli et al., 2013). In brief, currents were recorded with an EPC8 patch-clamp amplifier and LIH8+8 AD/DA converter (HEKA Electronics, Lambrecht/Pfalz, Germany) using the Patchmaster software (HEKA Electronics). The data were analyzed with the FitMaster software (HEKA Electronics). For macroscopic current recordings, the pipette resistance was 4 to 5 MΩ. Only patches presenting a seal resistance >2 GΩ were used to perform experiments. Current recordings were filtered at 300 Hz. Currents were evoked with a voltage ramp ranging from +40 to −100 mV in 2.5 s with a holding potential of −40 mV.
The cytosolic solution contained (1) 100 mm Cl− adjusted to pH 7.5 with Bis-Tris-Propane (BTP); (2) 100 mm Cl−, 1 mM malic acid adjusted to pH 7.5 with BTP; (3) 100 mm malic acid, adjusted to pH 7.5 with BTP. The osmolality was adjusted to 500 mosmol using sorbitol. The pipette solution contained 100 mm HCl and was adjusted with BTP to pH 6. The osmolality was adjusted to 550 mosmol using sorbitol. All chemicals were purchased from Sigma-Aldrich. Liquid junction potentials were measured according to Neher, 1992 and corrected when higher than ± 2 mV. Sequential perfusion of cytosolic-side buffer was performed using a custom-built gravity-driven perfusion system.
Stomata Aperture Measurement
Before performing in situ assays of native stomatal apertures, hydroponically grown plants were transferred to an LED growth chamber and grown for 3 d at a white light intensity of 250 µmol⋅m−2⋅s−1. Counting from the first true leaf pair, the seventh leaf of each rosette was used to analyze stomata. The leaf was excised and its abaxial side pressed onto a piece of scotch tape. The mesophyll tissue was scrapped off from the adaxial side using a scalpel, and the remaining exposed abaxial epidermis was immediately rinsed three times with 10 mm MES-KOH (pH 6.1). The tape containing the epidermal strip was transferred onto a microscope slide, and randomly chosen areas covering both sides of the midvein were imaged per leaf using an inverted NIKON ECLIPSE TS 100 microscope fitted with a NIKON Plan Fluor 40x/ 0.75 objective and a NIKON Digital Sight DS-Fi1 camera. Leaves of at least four individual plants per genotype were examined in this way and genotypes were alternated during measurements. The time from leaf excision to completion of imaging was kept at maximally 5 min to ensure that the measured stomatal apertures reflected the native state and were unaffected by the process. Finally, recorded images were blind-analyzed using ImageJ (Abramoff et al., 2004), and the apertures of at least 60 stomata were measured across a minimum of six images taken per leaf. Mean apertures of a single leaf were averaged, and the mean of one biological replicate was derived from the averages of at least four leaves per genotype.
RNA Sequencing and Data Processing
Total RNA was extracted as described in “qRT-PCR.” For each tissue (shoots and roots), genotype (almt9-1 and wild-type-1), and treatment (0 mm or 100 mm supplemental NaCl for 24 h), three independent biological replicates were produced. RNA library preparation and sequencing was conducted by the Functional Genomics Center Zurich using Illumina HiSEquation 2000.
RNA-seq read alignment and expression estimation was performed with RSEM (Li and Dewey, 2011). As a reference, we used the TAIR10 genome assembly and the corresponding gene annotations provided by TAIR. Differential expression was computed with the Bioconductor package edgeR (Robinson et al., 2010) using the generalized linear model fit. A gene was considered as differentially expressed between wild-type-1 and almt9-1 by applying a threshold of 0.01 for the P value and ± 1 for the log2 ratio, corresponding to a 2-fold or greater difference in expression. In addition, we filtered out genes that had very low counts. Specifically, we did not consider a gene as expressed if it did not exceed in at least one condition a read count of 50 in the samples. Positive log2 ratios correspond to transcriptional up-regulation in almt9-1, negative log2 ratios to transcriptional down-regulation.
Among the differentially expressed genes, we further selected salinity response-specific candidates that are up-regulated in almt9-1 by requiring that (1) the gene is up- regulated under salinity (log2 ratio ≥ ± 1, P < 0.01), and (2) the gene is not up-regulated under control conditions or the log2 ratio is at least 1 lower than the log2 ratio under salinity conditions. The corresponding filtering has been applied for the down-regulated genes.
Accession Numbers
The sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: At3g18440 (ALMT9), At2g17470 (ALMT6), At1g68600 (ALMT5), At1g25480 (ALMT4), At1g18420 (ALMT3), At2g01980 (SOS1), At2g31910 (CHX21), At2g04040 (DTX1), At4g10310 (HKT1;1), At1g08810 (MYB60), At5g46240 (KAT1), and At3g18780.2 (ACT2).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Cl− content in xylem sap of wild-type-2 (wt-2) and almt9-2 mutant plants upon 24-h salt treatment.
Supplemental Figure S2. Na+ and Cl− contents in wild-type and almt9 mutant plants upon treatment with different salt compositions.
Supplemental Figure S3. Na+ and Cl− contents in wild-type and almt9 mutant plants upon prolonged exposure to salinity.
Supplemental Figure S4. Mg2+, MA2−, and osmotic contents in wild-type and almt9 mutants during 24-h salt stress.
Supplemental Figure S5. Evaluation of the transgenic hairpin-RNA-expressing line that targets clade II ALMTs for multiple transcriptional down-regulation (hpRNA).
Supplemental Figure S6. Stomatal movement in wild-type-1 and almt9-1 plants during NaCl and KCl stress.
Supplemental Figure S7. Evaluation of the guard cell-specific complementation line (GC:ALMT9) of almt9-2 by PCR.
Supplemental Figure S8. Biological functions of genes that are differentially expressed in almt9-1 exclusively under salinity.
Supplemental Figure S9. Expression analysis of SOS1 upon 24-h salt treatment.
Supplemental Table S1. Ion contents of wild-type and almt9 mutant plants upon 24-h salinity.
Supplemental Table S2. List of differentially expressed genes between wild-type-1 and almt9-1 in shoots exclusively under salinity.
Supplemental Table S3. List of differentially expressed genes between wild-type-1 and almt9-1 in roots exclusively under salinity.
Supplemental Table S4. Primers used in that study.
Supplemental Data Set S1. List of differentially expressed genes between wild-type-1 and almt9-1 in shoots under control conditions.
Supplemental Data Set S2. List of differentially expressed genes between wild-type-1 and almt9-1 in roots under control conditions.
Supplemental Data Set S3. List of differentially expressed genes between wild-type-1 and almt9-1 in shoots under salinity conditions.
Supplemental Data Set S4. List of differentially expressed genes between wild-type-1 and almt9-1 in roots under salinity conditions.
Supplementary Material
Acknowledgments
The authors kindly thank H. Brandl and C. Fabbri for supervising the atomic absorption spectrometry measurements. Furthermore, we are grateful to S. Aluri and H. Rehrauer from the Functional Genomics Centre Zurich who conducted the RNA sequencing and data analysis, respectively. We thank Rita Francisco for performing preliminary experiments. In addition, we thank Dr. Alisdair Fernie, Dr. Mutsumi Watanabe, Dr. Alexandra Florian, and Ms. Regina Wendenburg who assisted in preliminary experiments.
Glossary
- MS
Murashige and Skoog
- qRT-PCR
quantitative real-time PCR
Footnotes
U.B. and A.D.A. were supported by the Swiss National Foundation (31003A_141090/1) and U.B. additionally by a Forschungskredit of the University of Zurich. A.D.A. benefits from the support of the LabEx Saclay Plant Sciences-SPS (ANR-10-LABX-0040-SPS). C.E. was supported by the European Union’s Seventh Framework Programme for research, technological development, and demonstration under grant agreement number GA-2010-267243 – PLANT FELLOWS and a Forschungskredit of the University of Zurich.
References
- Abramoff MD, Magalhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophotonics International 11: 36–42 [Google Scholar]
- Arrivault S, Senger T, Krämer U (2006) The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J 46: 861–879 [DOI] [PubMed] [Google Scholar]
- Barbier-Brygoo H, De Angeli A, Filleur S, Frachisse JM, Gambale F, Thomine S, Wege S (2011) Anion channels/transporters in plants: from molecular bases to regulatory networks. Annu Rev Plant Biol 62: 25–51 [DOI] [PubMed] [Google Scholar]
- Barragán V, Leidi EO, Andrés Z, Rubio L, De Luca A, Fernández JA, Cubero B, Pardo JM (2012) Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24: 1127–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassil E, Ohto MA, Esumi T, Tajima H, Zhu Z, Cagnac O, Belmonte M, Peleg Z, Yamaguchi T, Blumwald E (2011) The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant Cell 23: 224–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Wuillème S, Lemoine R, Kronenberger J, Caboche M, Lepiniec L, Rochat C (2005) The AtSUC5 sucrose transporter specifically expressed in the endosperm is involved in early seed development in Arabidopsis. Plant J 43: 824–836 [DOI] [PubMed] [Google Scholar]
- Britto DT, Ruth TJ, Lapi S, Kronzucker HJ (2004) Cellular and whole-plant chloride dynamics in barley: insights into chloride-nitrogen interactions and salinity responses. Planta 218: 615–622 [DOI] [PubMed] [Google Scholar]
- Brumós J, Talón M, Bouhlal R, Colmenero-Flores JM (2010) Cl- homeostasis in includer and excluder citrus rootstocks: transport mechanisms and identification of candidate genes. Plant Cell Environ 33: 2012–2027 [DOI] [PubMed] [Google Scholar]
- Cataldo D, Maroon L, Schrader LE, Youngs VL (1975) Rapid colorimetric determination of nitrate in plant tissues by nitration of salicylic acid. Commun Soil Sci Plant Anal 6: 71–80 [Google Scholar]
- Chen CZ, Lv XF, Li JY, Yi HY, Gong JM (2012) Arabidopsis NRT1.5 is another essential component in the regulation of nitrate reallocation and stress tolerance. Plant Physiol 159: 1582–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colcombet J, Lelièvre F, Thomine S, Barbier-Brygoo H, Frachisse JM (2005) Distinct pH regulation of slow and rapid anion channels at the plasma membrane of Arabidopsis thaliana hypocotyl cells. J Exp Bot 56: 1897–1903 [DOI] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C (2005) A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol 15: 1196–1200 [DOI] [PubMed] [Google Scholar]
- Craig Plett D, Møller IS (2010) Na(+) transport in glycophytic plants: what we know and would like to know. Plant Cell Environ 33: 612–626 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport RJ, Muñoz-Mayor A, Jha D, Essah PA, Rus A, Tester M (2007) The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ 30: 497–507 [DOI] [PubMed] [Google Scholar]
- De Angeli A, Monachello D, Ephritikhine G, Frachisse JM, Thomine S, Gambale F, Barbier-Brygoo H (2006) The nitrate/proton antiporter AtCLCa mediates nitrate accumulation in plant vacuoles. Nature 442: 939–942 [DOI] [PubMed] [Google Scholar]
- De Angeli A, Zhang J, Meyer S, Martinoia E (2013) AtALMT9 is a malate-activated vacuolar chloride channel required for stomatal opening in Arabidopsis. Nat Commun 4: 1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B, van den Berg W, Lokerse A, Liao CY, van Mourik H, Möller B, Peris CL, Weijers D (2011) A versatile set of ligation-independent cloning vectors for functional studies in plants. Plant Physiol 156: 1292–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frachisse JM, Colcombet J, Guern J, Barbier-Brygoo H (2000) Characterization of a nitrate-permeable channel able to mediate sustained anion efflux in hypocotyl cells from Arabidopsis thaliana. Plant J 21: 361–371 [DOI] [PubMed] [Google Scholar]
- Geelen D, Lurin C, Bouchez D, Frachisse JM, Lelièvre F, Courtial B, Barbier-Brygoo H, Maurel C (2000) Disruption of putative anion channel gene AtCLC-a in Arabidopsis suggests a role in the regulation of nitrate content. Plant J 21: 259–267 [DOI] [PubMed] [Google Scholar]
- Geilfus CM, Mithöfer A, Ludwig-Müller J, Zörb C, Muehling KH (2015) Chloride-inducible transient apoplastic alkalinizations induce stomata closure by controlling abscisic acid distribution between leaf apoplast and guard cells in salt-stressed Vicia faba. New Phytol 208: 803–816 [DOI] [PubMed] [Google Scholar]
- Genc Y, Oldach K, Taylor J, Lyons GH (2016) Uncoupling of sodium and chloride to assist breeding for salinity tolerance in crops. New Phytol 210: 145–156 [DOI] [PubMed] [Google Scholar]
- Gilliham M, Tester M (2005) The regulation of anion loading to the maize root xylem. Plant Physiol 137: 819–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP. (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Hall D, Evans AR, Newbury HJ, Pritchard J (2006) Functional analysis of CHX21: a putative sodium transporter in Arabidopsis. J Exp Bot 57: 1201–1210 [DOI] [PubMed] [Google Scholar]
- Henderson SW, Baumann U, Blackmore DH, Walker AR, Walker RR, Gilliham M (2014) Shoot chloride exclusion and salt tolerance in grapevine is associated with differential ion transporter expression in roots. BMC Plant Biol 14: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson SW, Wege S, Qiu J, Blackmore DH, Walker AR, Tyerman SD, Walker RR, Gilliham M (2015) Grapevine and Arabidopsis cation-chloride cotransporters localize to the Golgi and trans-Golgi network and indirectly influence long-distance ion transport and plant salt tolerance. Plant Physiol 169: 2215–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CX, Van Steveninck RF (1989) Maintenance of low cl concentrations in mesophyll cells of leaf blades of barley seedlings exposed to salt stress. Plant Physiol 90: 1440–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurth MA, Suh SJ, Kretzschmar T, Geis T, Bregante M, Gambale F, Martinoia E, Neuhaus HE (2005) Impaired pH homeostasis in Arabidopsis lacking the vacuolar dicarboxylate transporter and analysis of carboxylic acid transport across the tonoplast. Plant Physiol 137: 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James RA, Munns R, von Caemmerer S, Trejo C, Miller C, Condon TA (2006) Photosynthetic capacity is related to the cellular and subcellular partitioning of Na+, K+ and Cl- in salt-affected barley and durum wheat. Plant Cell Environ 29: 2185–2197 [DOI] [PubMed] [Google Scholar]
- Jiang X, Leidi EO, Pardo JM (2010) How do vacuolar NHX exchangers function in plant salt tolerance? Plant Signal Behav 5: 792–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossier M, Kroniewicz L, Dalmas F, Le Thiec D, Ephritikhine G, Thomine S, Barbier-Brygoo H, Vavasseur A, Filleur S, Leonhardt N (2010) The Arabidopsis vacuolar anion transporter, AtCLCc, is involved in the regulation of stomatal movements and contributes to salt tolerance. Plant J 64: 563–576 [DOI] [PubMed] [Google Scholar]
- Karley AJ, Leigh RA, Sanders D (2000a) Differential ion accumulation and ion fluxes in the mesophyll and epidermis of barley. Plant Physiol 122: 835–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karley AJ, Leigh RA, Sanders D (2000b) Where do all the ions go? The cellular basis of differential ion accumulation in leaf cells. Trends Plant Sci 5: 465–470 [DOI] [PubMed] [Google Scholar]
- Kiba T, Feria-Bourrellier AB, Lafouge F, Lezhneva L, Boutet-Mercey S, Orsel M, Bréhaut V, Miller A, Daniel-Vedele F, Sakakibara H, Krapp A (2012) The Arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-starved plants. Plant Cell 24: 245–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovermann P, Meyer S, Hörtensteiner S, Picco C, Scholz-Starke J, Ravera S, Lee Y, Martinoia E (2007) The Arabidopsis vacuolar malate channel is a member of the ALMT family. Plant J 52: 1169–1180 [DOI] [PubMed] [Google Scholar]
- Krebs M, Beyhl D, Görlich E, Al-Rasheid KA, Marten I, Stierhof YD, Hedrich R, Schumacher K (2010) Arabidopsis V-ATPase activity at the tonoplast is required for efficient nutrient storage but not for sodium accumulation. Proc Natl Acad Sci USA 107: 3251–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Läuchli A, James RA, Huang CX, McCully M, Munns R (2008) Cell-specific localization of Na+ in roots of durum wheat and possible control points for salt exclusion. Plant Cell Environ 31: 1565–1574 [DOI] [PubMed] [Google Scholar]
- Lee SC, Lan W, Buchanan BB, Luan S (2009) A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci USA 106: 21419–21424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidi EO, Barragán V, Rubio L, El-Hamdaoui A, Ruiz MT, Cubero B, Fernández JA, Bressan RA, Hasegawa PM, Quintero FJ, et al. (2010) The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J 61: 495–506 [DOI] [PubMed] [Google Scholar]
- Lelandais-Brière C, Jovanovic M, Torres GA, Perrin Y, Lemoine R, Corre-Menguy F, Hartmann C (2007) Disruption of AtOCT1, an organic cation transporter gene, affects root development and carnitine-related responses in Arabidopsis. Plant J 51: 154–164 [DOI] [PubMed] [Google Scholar]
- Léran S, Varala K, Boyer JC, Chiurazzi M, Crawford N, Daniel-Vedele F, David L, Dickstein R, Fernandez E, Forde B, et al. (2014) A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci 19: 5–9 [DOI] [PubMed] [Google Scholar]
- Li B, Byrt CS, Qiu J, Baumann U, Hrmova M, Evrard A, Johnson AA, Birnbaum KD, Mayo GM, Jha D, et al. (2016) Identification of a stelar-localised transport protein that facilitates root-to-shoot transfer of chloride in Arabidopsis. Plant Physiol 170: 1014–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JY, Fu YL, Pike SM, Bao J, Tian W, Zhang Y, Chen CZ, Zhang Y, Li HM, Huang J, et al. (2010) The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22: 1633–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, He Z, Pandey GK, Tsuchiya T, Luan S (2002) Functional cloning and characterization of a plant efflux carrier for multidrug and heavy metal detoxification. J Biol Chem 277: 5360–5368 [DOI] [PubMed] [Google Scholar]
- Lim CW, Kim JH, Baek W, Kim BS, Lee SC (2012) Functional roles of the protein phosphatase 2C, AtAIP1, in abscisic acid signaling and sugar tolerance in Arabidopsis. Plant Sci 187: 83–88 [DOI] [PubMed] [Google Scholar]
- Limonta M, Romanowsky S, Olivari C, Bonza MC, Luoni L, Rosenberg A, Harper JF, De Michelis MI (2014) ACA12 is a deregulated isoform of plasma membrane Ca2+-ATPase of Arabidopsis thaliana. Plant Mol Biol 84: 387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Tsay YF (2003) Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J 22: 1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magome H, Yamaguchi S, Hanada A, Kamiya Y, Oda K (2004) dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J 37: 720–729 [DOI] [PubMed] [Google Scholar]
- Magome H, Yamaguchi S, Hanada A, Kamiya Y, Oda K (2008) The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J 56: 613–626 [DOI] [PubMed] [Google Scholar]
- Martinoia E, Maeshima M, Neuhaus HE (2007) Vacuolar transporters and their essential role in plant metabolism. J Exp Bot 58: 83–102 [DOI] [PubMed] [Google Scholar]
- Martinoia E, Meyer S, De Angeli A, Nagy R (2012) Vacuolar transporters in their physiological context. Annu Rev Plant Biol 63: 183–213 [DOI] [PubMed] [Google Scholar]
- Mäser P, Eckelman B, Vaidyanathan R, Horie T, Fairbairn DJ, Kubo M, Yamagami M, Yamaguchi K, Nishimura M, Uozumi N, et al. (2002) Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett 531: 157–161 [DOI] [PubMed] [Google Scholar]
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J 25: 295–303 [DOI] [PubMed] [Google Scholar]
- Meyer S, Scholz-Starke J, De Angeli A, Kovermann P, Burla B, Gambale F, Martinoia E (2011) Malate transport by the vacuolar AtALMT6 channel in guard cells is subject to multiple regulation. Plant J 67: 247–257 [DOI] [PubMed] [Google Scholar]
- Møller IS, Gilliham M, Jha D, Mayo GM, Roy SJ, Coates JC, Haseloff J, Tester M (2009) Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 21: 2163–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller IS, Tester M (2007) Salinity tolerance of Arabidopsis: a good model for cereals? Trends Plant Sci 12: 534–540 [DOI] [PubMed] [Google Scholar]
- Monachello D, Allot M, Oliva S, Krapp A, Daniel-Vedele F, Barbier-Brygoo H, Ephritikhine G (2009) Two anion transporters AtClCa and AtClCe fulfil interconnecting but not redundant roles in nitrate assimilation pathways. New Phytol 183: 88–94 [DOI] [PubMed] [Google Scholar]
- Munns R. (2002) Comparative physiology of salt and water stress. Plant Cell Environ 25: 239–250 [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Munns R, Wallace PA, Teakle NL, Colmer TD (2010) Measuring soluble ion concentrations (Na(+), K(+), Cl(-)) in salt-treated plants. Methods Mol Biol 639: 371–382 [DOI] [PubMed] [Google Scholar]
- Nagy R, Grob H, Weder B, Green P, Klein M, Frelet-Barrand A, Schjoerring JK, Brearley C, Martinoia E (2009) The Arabidopsis ATP-binding cassette protein AtMRP5/AtABCC5 is a high affinity inositol hexakisphosphate transporter involved in guard cell signaling and phytate storage. J Biol Chem 284: 33614–33622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura RL, McKendree WL Jr, Hirsch RE, Sedbrook JC, Gaber RF, Sussman MR (1995) Expression of an Arabidopsis potassium channel gene in guard cells. Plant Physiol 109: 371–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher E. (1992) Correction for liquid junction potentials in patch clamp experiments. Methods Enzymol 207: 123–131 [DOI] [PubMed] [Google Scholar]
- Nguyen CT, Agorio A, Jossier M, Depré S, Thomine S, Filleur S (2016) Characterization of the chloride channel-like, AtCLCg, involved in chloride tolerance in Arabidopsis thaliana. Plant Cell Physiol 57: 764–775 [DOI] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, Shibata D, Saito K, Ohta H (2007) ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res 35: D863–D869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DH, Lee SY, Bressan RA, Yun DJ, Bohnert HJ (2010) Intracellular consequences of SOS1 deficiency during salt stress. J Exp Bot 61: 1205–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JS, Gong JM (2014) Vacuolar sequestration capacity and long-distance metal transport in plants. Front Plant Sci 5: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommerrenig B, Popko J, Heilmann M, Schulmeister S, Dietel K, Schmitt B, Stadler R, Feussner I, Sauer N (2013) SUCROSE TRANSPORTER 5 supplies Arabidopsis embryos with biotin and affects triacylglycerol accumulation. Plant J 73: 392–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio S, Rodrigues A, Saez A, Dizon MB, Galle A, Kim TH, Santiago J, Flexas J, Schroeder JI, Rodriguez PL (2009) Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol 150: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira O, Khadka S, Israeli Y, Shani U, Schwartz A (2009) Functional anatomy controls ion distribution in banana leaves: significance of Na+ seclusion at the leaf margins. Plant Cell Environ 32: 476–485 [DOI] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97: 6896–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Quintero FJ, Pardo JM, Zhu JK (2002) The putative plasma membrane Na(+)/H(+) antiporter SOS1 controls long-distance Na(+) transport in plants. Plant Cell 14: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibole JV, Cabot C, Poschenrieder C, Barceló J (2003) Efficient leaf ion partitioning, an overriding condition for abscisic acid-controlled stomatal and leaf growth responses to NaCl salinization in two legumes. J Exp Bot 54: 2111–2119 [DOI] [PubMed] [Google Scholar]
- Storey R, Schachtman DP, Thomas MR (2003) Root structure and cellular chloride, sodium and potassium distribution in salinized grapevines. Plant Cell Environ 26: 789–800 [DOI] [PubMed] [Google Scholar]
- Strohm AK, Vaughn LM, Masson PH (2015) Natural variation in the expression of ORGANIC CATION TRANSPORTER 1 affects root length responses to cadaverine in Arabidopsis. J Exp Bot 66: 853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunarpi HT, Horie T, Motoda J, Kubo M, Yang H, Yoda K, Horie R, Chan WY, Leung HY, Hattori K, et al. (2005) Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J 44: 928–938 [DOI] [PubMed] [Google Scholar]
- Taochy C, Gaillard I, Ipotesi E, Oomen R, Leonhardt N, Zimmermann S, Peltier JB, Szponarski W, Simonneau T, Sentenac H, et al. (2015) The Arabidopsis root stele transporter NPF2.3 contributes to nitrate translocation to shoots under salt stress. Plant J 83: 466–479 [DOI] [PubMed] [Google Scholar]
- Tavakkoli E, Fatehi F, Coventry S, Rengasamy P, McDonald GK (2011) Additive effects of Na+ and Cl- ions on barley growth under salinity stress. J Exp Bot 62: 2189–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavakkoli E, Rengasamy P, McDonald GK (2010) High concentrations of Na+ and Cl- ions in soil solution have simultaneous detrimental effects on growth of faba bean under salinity stress. J Exp Bot 61: 4449–4459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teakle NL, Tyerman SD (2010) Mechanisms of Cl(-) transport contributing to salt tolerance. Plant Cell Environ 33: 566–589 [DOI] [PubMed] [Google Scholar]
- Terasaka K, Blakeslee JJ, Titapiwatanakun B, Peer WA, Bandyopadhyay A, Makam SN, Lee OR, Richards EL, Murphy AS, Sato F, et al. (2005) PGP4, an ATP binding cassette P-glycoprotein, catalyzes auxin transport in Arabidopsis thaliana roots. Plant Cell 17: 2922–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot (Lond) 91: 503–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognetti VB, Van Aken O, Morreel K, Vandenbroucke K, van de Cotte B, De Clercq I, Chiwocha S, Fenske R, Prinsen E, Boerjan W, et al. (2010) Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 22: 2660–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wege S, De Angeli A, Droillard MJ, Kroniewicz L, Merlot S, Cornu D, Gambale F, Martinoia E, Barbier-Brygoo H, Thomine S, et al. (2014) Phosphorylation of the vacuolar anion exchanger AtCLCa is required for the stomatal response to abscisic acid. Sci Signal 7: ra65. [DOI] [PubMed] [Google Scholar]
- Wege S, Jossier M, Filleur S, Thomine S, Barbier-Brygoo H, Gambale F, De Angeli A (2010) The proline 160 in the selectivity filter of the Arabidopsis NO(3)(-)/H(+) exchanger AtCLCa is essential for nitrate accumulation in planta. Plant J 63: 861–869 [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Yang KY, Liu Y, Zhang S (2001) Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc Natl Acad Sci USA 98: 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Assmann SM (2016) The effect of NaCl on stomatal opening in Arabidopsis wild type and agb1 heterotrimeric G-protein mutant plants. Plant Signal Behav 11: e1085275, doi: 10.1080/15592324.2015.1085275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Baetz U, Krügel U, Martinoia E, De Angeli A (2013) Identification of a probable pore-forming domain in the multimeric vacuolar anion channel AtALMT9. Plant Physiol 163: 830–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Wang YJ, Wang YL, Wang XL, Zhang X (2011) Extracellular Ca2+ alleviates NaCl-induced stomatal opening through a pathway involving H2O2-blocked Na+ influx in Vicia guard cells. J Plant Physiol 168: 903–910 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.