Small HSPs are important for heat tolerance, interacting with and protecting an overlapping set of heat-sensitive proteins in protein translation together with HSP101.
Abstract
The ubiquitous small heat shock proteins (sHSPs) are well documented to act in vitro as molecular chaperones to prevent the irreversible aggregation of heat-sensitive proteins. However, the in vivo activities of sHSPs remain unclear. To investigate the two most abundant classes of plant cytosolic sHSPs (class I [CI] and class II [CII]), RNA interference (RNAi) and overexpression lines were created in Arabidopsis (Arabidopsis thaliana) and shown to have reduced and enhanced tolerance, respectively, to extreme heat stress. Affinity purification of CI and CII sHSPs from heat-stressed seedlings recovered eukaryotic translation elongation factor (eEF) 1B (α-, β-, and γ-subunits) and eukaryotic translation initiation factor 4A (three isoforms), although the association with CI sHSPs was stronger and additional proteins involved in translation were recovered with CI sHSPs. eEF1B subunits became partially insoluble during heat stress and, in the CI and CII RNAi lines, showed reduced recovery to the soluble cell fraction after heat stress, which was also dependent on HSP101. Furthermore, after heat stress, CI sHSPs showed increased retention in the insoluble fraction in the CII RNAi line and vice versa. Immunolocalization revealed that both CI and CII sHSPs were present in cytosolic foci, some of which colocalized with HSP101 and with eEF1Bγ and eEF1Bβ. Thus, CI and CII sHSPs have both unique and overlapping functions and act either directly or indirectly to protect specific translation factors in cytosolic stress granules.
As sessile organisms, plants require robust mechanisms to sense and respond to adverse growth conditions during their life cycle. A well-defined response of plants, as well as other organisms, to high temperatures involves the enhanced production of molecular chaperones, including the heat shock proteins HSP100 and HSP70, which are ATPases, and the small heat shock proteins (sHSPs; Kotak et al., 2007). These HSPs and other molecular chaperones are believed to prevent or reverse the inactivation and aggregation of heat-sensitive proteins, thereby contributing to restoring cellular protein homeostasis disrupted by environmentally challenging conditions (Basha et al., 2012; Doyle et al., 2013). The sHSPs are proposed to be a first line of defense, interacting with denaturing proteins to prevent their further aggregation and presenting them to the ATP-dependent HSP100 and HSP70 chaperones that can disaggregate and refold nonnative proteins (Haslbeck and Vierling, 2015). However, despite considerable evidence to support this model for sHSP function, a full understanding of the importance of sHSPs to plant heat tolerance and what critical cellular components they may protect is lacking.
Although sHSPs are found in all kingdoms of life, they are uniquely diverse in land plants, with 11 or more gene families found in monocots and eudicots (Waters, 2013). sHSP families are distinguished by conserved sequence features and intracellular localization; they are found in virtually all membrane-bound plant cell compartments, the chloroplast, mitochondrion, endoplasmic reticulum, peroxisome, and nucleus, along with several families of sHSPs in the cytosol (for review, see Santhanagopalan et al., 2015). All sHSPs have monomer Mr values between 12 to 42 kD and constitute a signature α-crystallin domain flanked by a variable-length, divergent N-terminal arm and a short C-terminal tail (de Jong et al., 1998). The majority of sHSPs assemble into oligomers of 12 to over 24 subunits, specific to the protein (Delbecq and Klevit, 2013). In higher plants, the most extensive structural and biochemical characterization is available for two classes of cytosolic sHSPs, class I (CI) and class II (CII), which become highly abundant during heat stress, accumulating together to over 1% of total cell protein within a few hours (Derocher et al., 1991; van Montfort et al., 2001; Basha et al., 2010). The CI and CII proteins likely evolved through gene duplication over 400 million years ago, as both classes are found already in mosses (Waters and Vierling, 1999). Although both CI and CII sHSPs form dodecameric oligomers, the two classes do not coassemble to form heterooligomers, but they will heterooligomerize with members of the same class from the same or different species (Basha et al., 2010). Thus, CI and CII cytosolic sHSPs have evolved to be structurally distinct, but how they may have diverged functionally is not known.
Studies of the CI and CII plant sHSPs have made major contributions to the current model for sHSP chaperone activity. Recombinant, dodecameric CI and CII sHSPs form high-Mr complexes with heat-sensitive denaturing substrates (Lee et al., 1997; Basha et al., 2004b, 2010). Model heat-sensitive substrates such as firefly luciferase can be readily renatured and reactivated from sHSP complexes by the HSP70 chaperones and cochaperones present in wheat germ or reticulocyte lysate extracts or even by the prokaryotic HSP70 homolog DnaK and cochaperones (Lee et al., 1997; Lee and Vierling, 2000). Addition of the disaggregase ClpB, the prokaryotic homolog of the HSP100 chaperones, accelerates the release and reactivation of substrate from sHSP-substrate complexes (Mogk et al., 2003).
A number of in vivo experiments are consistent with these in vitro biochemical studies. CI and CII proteins from extracts of heat stress plant tissues migrate on nondenaturing gels in the size range of dodecamers (Helm et al., 1997; Kirschner et al., 2000; Smykal et al., 2000). After severe stress, CI and CII sHSPs are observed in cytosolic aggregates (Kirschner et al., 2000) that have been termed heat stress granules (Nover and Scharf, 1997; Weber et al., 2008), and a portion of the sHSPs transit to an insoluble cell fraction presumably in association with heat-damaged proteins (Lee et al., 2005). Notably, recovery of sHSPs from the insoluble fraction is impaired in Arabidopsis (Arabidopsis thaliana) mutants of the disaggregase HSP101 (Lee et al., 2005), which is essential for heat acclimation (Hong and Vierling, 2000; Queitsch et al., 2000). Analogous studies with yeast (Cashikar et al., 2005; Haslbeck et al., 2005) and Escherichia coli (Mogk et al., 2003) sHSPs and ATP-dependent chaperones parallel these findings in plants, supporting the chaperone model for sHSP function.
CI sHSPs typically comprise the largest family of sHSP genes in higher plants (Waters, 2013), and there are six CI genes in Arabidopsis. The CII gene family is typically smaller, with only two CII genes in Arabidopsis (Scharf et al., 2001; Siddique et al., 2008). The absence of T-DNA insertion lines for all of the Arabidopsis CI genes (probably due in part to the small size of these intronless genes) and the tandem chromosomal arrangement of the Arabidopsis CII genes (Waters et al., 2008) have so far limited genetic studies of mutants that might aid in defining the functions of these cytosolic sHSPs. Individual mutants of three CI sHSPs were reported to have wild-type growth at different temperatures and after heat shock at 40°C, although dark-grown seedlings showed reduced hypocotyl elongation compared with the wild type after acclimation at 38°C followed by heat stress at 45°C (Dafny-Yelin et al., 2008). The same study reported that double mutants of the CI proteins could not be obtained. However, only single alleles were tested, and no complementation experiments were performed. Further evidence for the stress-protective role of CI and CII sHSPs in plants has primarily involved constitutive expression or overexpression of a single sHSP in different plant species, including Arabidopsis, rice (Oryza sativa), maize (Zea mays), and carrot (Daucus carota), as well as others (Malik et al., 1999; Ahn and Zimmerman, 2006; Jiang et al., 2009; Sun et al., 2012; Zhou et al., 2012; Mu et al., 2013; Wang et al., 2015). A very restricted set of stress-resistant phenotypes has been reported for sHSP-overexpressing transgenic materials, and these studies have not provided mechanistic insight into sHSP function or distinguished differences between CI and CII sHSPs.
To gain insight into the in vivo function of CI and CII sHSPs and potentially differentiate their specific roles during heat stress, we developed transgenic Arabidopsis RNA interference (RNAi) lines to knock down the expression of the CI and CII sHSP families, along with lines that overexpress a CI or CII sHSP, and evaluated their phenotypes at different growth stages under different heat stress regimes. Arabidopsis lines expressing affinity-tagged CI or CII proteins also were created and used to recover proteins associated with sHSPs during heat stress, and the behavior of specific sHSP-interacting proteins was evaluated in the RNAi lines and an HSP101 null mutant. In total, the results indicate that CI and CII sHSPs have both unique and overlapping functions and act in conjunction with HSP101 to either directly or indirectly protect specific translation factors in cytosolic stress granules.
RESULTS
CI and CII sHSPs Are Important for Heat Stress Tolerance
To examine the roles of CI and CII sHSPs in vivo, RNAi lines were constructed against either the CI (six members) or CII (two members) sHSP family. In addition, CI HSP17.4 and CII HSP17.6 were transformed into wild-type plants under the control of the constitutive cauliflower mosaic virus (CaMV) 35S promoter, creating CI and CII sHSP overexpression (OE) lines, respectively. The empty RNAi and overexpression vectors also were transformed as controls. At 22°C, no CI or CII sHSPs were detected in any of the lines except in the corresponding OE lines (Fig. 1A). Both CI and CII sHSPs accumulate at 38°C in wild-type plants, the empty vector controls, and the null mutant of the chaperone HSP101 (hsp101), originally annotated as hot1-3 (Hong and Vierling, 2001). In RNAi plants, the accumulation of CI sHSPs was estimated to be repressed at least 60% in different independently transformed plants under the heat stress conditions tested. RNAi-mediated repression of CII sHSPs was more effective, showing up to an estimated 95% decrease in total CII sHSP protein levels (Fig. 1A; Supplemental Fig. S1A). Knocking down CI sHSPs did not influence total CII sHSP abundance and vice versa (Supplemental Fig. S1A). In addition, none of the sHSP RNAi or OE lines showed significant differences in growth rate, seed yield, or flowering time and otherwise appeared morphologically like the wild type when grown under optimal conditions (Supplemental Fig. S2, A–D).
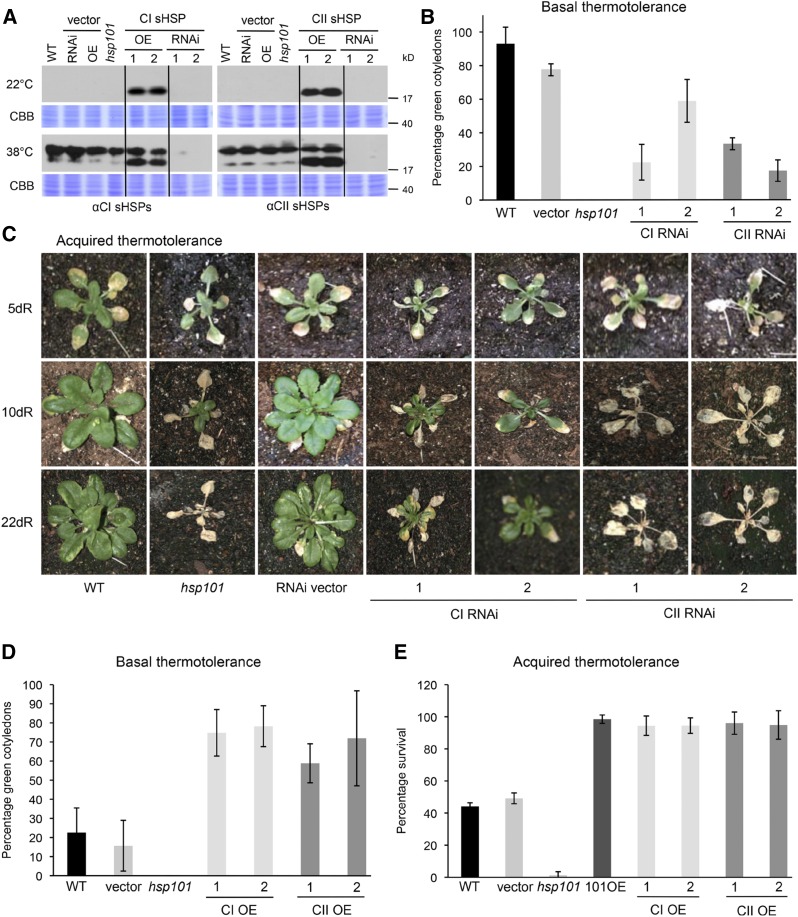
Figure 1.
CI and CII sHSPs are important for basal and acquired thermotolerance. A, Two independent CI and CII RNAi lines and two independent OE lines display corresponding sHSP protein levels. Total protein extracts were obtained from 2.5-d-old wild-type (WT), vector, hsp101, CI or CII OE, and CI or CII RNAi seedlings maintained at 22°C or treated at 38°C for 90 min and allowed to recover at 22°C for 2 h. Samples were separated using SDS-PAGE and analyzed by immunoblotting using the polyclonal antibodies indicated. Staining with Coomassie Brilliant Blue (CBB) was used as a loading control. B, CI and CII sHSPs are important for basal thermotolerance in Arabidopsis seedlings. Dark-grown seedlings (2.5-d) were maintained at 22°C or heat stressed at 43°C for 90 min in the dark, followed by recovery for 24 h at 22°C in illuminated conditions, at which time the percentage of green cotyledons was scored. Results are from a single experiment (n = 18 per line) and are consistent with observations from three experiments. Error bars represent sd. C, CI and CII sHSPs are important for acquired thermotolerance. Soil-grown, 10-d-old wild type, hsp101, vector, and CI and CII RNAi plants were pretreated at 38°C for 90 min, recovered for 2 h, and exposed to 45°C for 10 h. The development of the plants was monitored 5, 10, and 22 d after the heat stress. D, Constitutive expression of CI and CII sHSPs increases basal thermotolerance. Dark-grown seedlings (2.5-d) were maintained at 22°C or heat stressed at 43°C for 105 min in the dark, followed by recovery for 24 h at 22°C in illuminated conditions, at which time the percentage of green cotyledons was scored. Error bars represent sd (n = 24 per line). E, Overexpressing CI and CII sHSPs enhance acquired thermotolerance. The wild type, vector, hsp101, two independent OE lines of CI or CII sHSPs, and an HSP101 OE line were grown on plates for 10 d. Plants were acclimated at 38°C, recovered for 2 h, and exposed to 45°C for 90 min. The percentage of surviving seedlings was determined on day 7. Error bars represent sd (n = 60 per line).
The effect of reduced CI or CII sHSP levels on basal thermotolerance was tested by heat stressing dark-grown seedlings at 43°C for 90 min without prior acclimation and analyzing the percentage of seedlings subsequently capable of developing green cotyledons after transfer to the light compared with unstressed seedlings (Fig. 1B). All unstressed seedlings showed 100% greening after 24 h of illumination. Heat-stressed wild-type seedlings showed 90% greening 24 h after the transfer to light, while both CI and CII sHSP RNAi seedlings showed a reduced percentage of greening. Thus, both CI and CII sHSPs are important for basal thermotolerance in this assay.
Acquired thermotolerance was tested in soil-grown plants (Fig. 1C). Ten-day-old CI and CII sHSP RNAi plants, along with wild-type plants, the vector control, and the hsp101 mutant, were acclimated at 38°C, returned to 22°C for 2 h, and then heat stressed at 45°C for 10 h in the growth chamber under illuminated conditions. Plants were photographed 5, 10, and 22 d after the stress was applied. Two independently transformed CI RNAi lines exhibited reduced growth, and several leaves showed partial or complete necrosis. CII sHSP RNAi lines showed a more severe heat sensitivity and were essentially not able to recover growth, similar to hsp101 plants.
The ability of the constitutive expression of CI and CII sHSPs to enhance basal and acquired thermotolerance also was tested (Fig. 1, D and E). Dark-grown seedlings of CI and CII OE lines were heat stressed at 43°C for 105 min without prior acclimation, and OE lines of both sHSPs showed increases in the percentage of seedlings that developed green cotyledons after heat stress (Fig. 1D). Thus, constitutively expressing sHSPs effectively increased basal thermotolerance, consistent with the loss of tolerance seen in this assay with the RNAi plants. Although sHSPs accumulate to high levels at 38°C, which is the temperature we used for acclimation in tests of acquired thermotolerance, we nonetheless tested the additive effect of the constitutively expressing sHSPs on seedling survival rates after acclimation and severe heat stress. For comparison, a previously generated transgenic line that overexpresses Arabidopsis HSP101 also was included (Queitsch et al., 2000). Wild-type, vector, hsp101, and CI, CII, and HSP101 OE plants were grown on plates, heat acclimated (38°C for 1.5 h, plus 2 h at 22°C), and then stressed at 45°C for 1.5 h (Fig. 1E). The percentage of seedlings that survived the heat stress was determined after 7 d. Wild-type and vector control seedlings showed 45% to 50% survival after heat stress, while none of the hsp101 seedlings survived. In contrast, CI and CII sHSP OE lines, as well as the HSP101 OE line, showed a survival rate of close to 100% (Fig. 1E). Not only were the survival rates higher after heat stress, but also all OE lines grew significantly better (Supplemental Fig. S3A), showing that constitutive expression or overexpression of sHSPs increases heat stress tolerance to a similar extent to HSP101 overexpression. In total, these data support a clear and distinct role for CI and CII sHSPs in heat tolerance.
Capturing Proteins Associated with sHSPs during Heat Stress
To better understand the mechanism by which CI and CII sHSPs provide enhanced heat tolerance, sHSP-interacting proteins were identified using coaffinity purification with both CI and CII sHSPs. Transgenic Arabidopsis lines were created to express a CI or CII sHSP with a C-terminal StrepII affinity tag (Strep), being driven by the sHSP native, heat-inducible promoter. In addition, because CI and CII sHSPs are not expressed at detectable levels under nonstressed conditions, transgenic lines were created that expressed CI and CII Strep-tagged proteins under the control of the constitutive CaMV 35S promoter, which facilitates the identification of proteins that interact with either sHSP class specifically during heat stress.
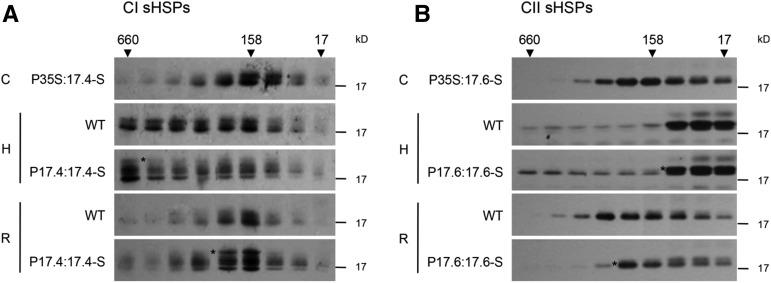
It has been shown previously that the addition of a Strep tag to the C terminus of an sHSP from Synechocystis sp. PCC 6803 fully complements an sHSP deletion strain of this cyanobacterium (Basha et al., 2004a). Additionally, the oligomeric state and in vitro chaperone activity of purified CI sHSPs are unchanged by the addition of a C-terminal Strep tag (Friedrich et al., 2004). Therefore, C-terminally fused Strep-tagged CI and CII sHSPs were constructed and transformed to Arabidopsis. Constitutive expression of both the CI and CII sHSP-Strep recombinant protein increased the basal thermotolerance, similar to their nontagged equivalents (Supplemental Fig. S3B). To test further if the introduced sHSP-Strep behaved similarly to untagged, native sHSPs in vivo, the oligomeric state of the wild-type and Strep-tagged proteins was monitored by size-exclusion chromatography of native leaf protein extracts followed by immunoblot analysis (Fig. 2). In the absence of stress, the constitutively expressed sHSPs and the CI and CII sHSP-Strep proteins elute as oligomers centered around 200 kD, as do their untagged counterparts, consistent with the known dodecameric structure of CI and CII sHSPs (Basha et al., 2010; Fig. 2). In extracts from heat-stressed plants, the CI sHSPs elute in higher Mr fractions, as seen in vitro when CI sHSPs are bound to heat-denatured model substrates (Basha et al., 2010, 2012); presumably, in vivo, the CI sHSPs are associated with substrates or partner proteins. The CII sHSPs behave somewhat differently in vivo; only a portion elutes in higher Mr fractions, while a majority elutes in fractions smaller than the dodecameric size of the native protein. On recovery from heat stress, both the CI and CII wild-type and Strep-tagged sHSPs again elute at a position corresponding to their native molecular mass around 200 kD. In total, in all cases, the Strep-tagged sHSPs behaved like their wild-type counterparts. In addition to this biochemical analysis, all plants expressing the Strep-tagged proteins showed normal growth phenotypes. Based on these data and previous work cited above, we concluded that the sHSP-Strep proteins behave like the endogenous sHSPs and could be used for affinity experiments.
Figure 2.
The C-terminal sHSP-Strep proteins behave similarly to wild-type sHSPs in vivo. Fourteen-day-old seedlings of the wild type (WT) or transgenic lines carrying either the CI or CII sHSP-Strep (S) driven by the corresponding native promoter were either control (C) or heat (H) treated (38°C for 2 h, allowed to recover at 22°C for 2 h, and then given another heat treatment for 30 min at 45°C) or heat treated and then allowed to recover for 12 h at 22°C (R). Total soluble proteins were extracted in native buffer and fractionated by size-exclusion chromatography. Fractions were collected, and an equal proportion of each fraction was analyzed by immunoblotting. Elution standards from the column are indicated in kD. Asterisks indicate the locations of the sHSP-Strep bands.
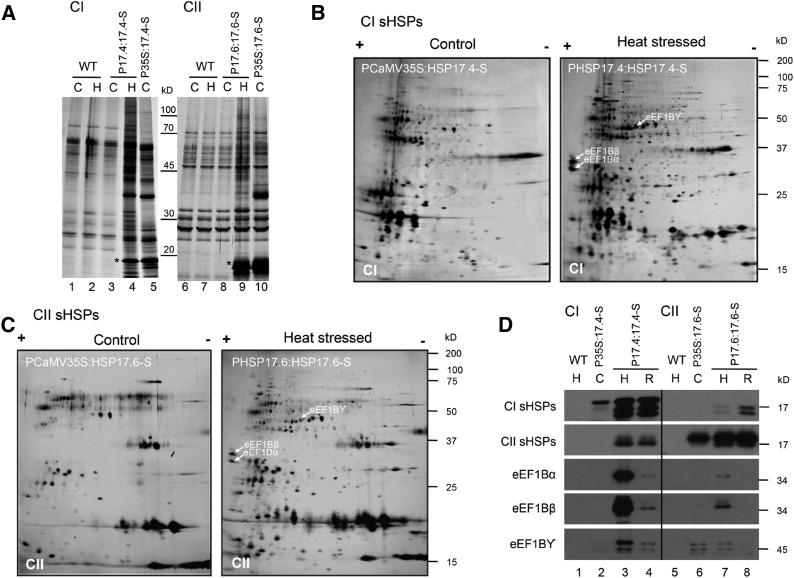
Coaffinity purifications from native protein extracts were conducted on wild-type plants and plants expressing the sHSP-Strep either constitutively or under the control of the native, heat-inducible promoter (Fig. 3A). No differences in recovered proteins were observed in experiments with wild-type plants comparing control and heat-stressed conditions (lanes 1, 2, 6, and 7), showing that heat stressing plants did not influence the background of proteins recovered with the Strep-affinity resin. The same protein banding pattern was observed in unheated plants carrying the heat-inducible sHSP-Strep constructs (lanes 3 and 8). In contrast, after heat stress, a number of proteins were recovered in the CI and CII sHSP affinity isolates (lanes 4 and 9), with significantly more proteins bound to CI sHSPs. Constitutively expressed sHSP-Strep also bound to a number of proteins under control conditions (lanes 5 and 10), but clearly to fewer proteins, and with only partial apparent overlap with proteins bound during heat stress.
Figure 3.
CI and CII sHSPs bind to putative substrates specifically during heat stress. CI and CII sHSP-Strep constructs, either under the control of PCaMV35S (P35S:17.4-S and P35S:17.6-S, respectively) or their heat-induced, native promoter (P17.4:17.4-S and P17.6:17.6-S, respectively), were transformed into Arabidopsis. A, Many proteins copurified with the sHSPs in a heat-dependent fashion. Total soluble protein extracts were obtained from 14-d-old seedlings that were kept at 22°C (control [C]) or heat treated for 2 h at 38°C, recovered for 2 h, and then exposed to 45°C for 30 min (H). Extracts were subjected to Strep-Tactin affinity isolation, eluted with SDS sample buffer, separated by SDS-PAGE, and stained with silver. The asterisks indicate the sHSP-Strep protein that was used for the affinity purification. B, 2D-PAGE of CI sHSP-interacting proteins corresponding to samples in lanes 5 (control) and 4 (heat stressed) in A, visualized by silver staining. Proteins enriched in the heat-stressed sample were excised, and their identities were determined using mass spectrometry (Supplemental Fig. S4; Supplemental Tables S1 and S2). C, CII sHSPs interact with a partially overlapping set of proteins compared with CI sHSPs. 2D SDS-PAGE shows CII sHSP-interacting proteins corresponding to samples in lanes 10 (control) and 9 (heat stressed) in A, visualized by silver staining. Proteins enriched in the heat-stressed sample were excised, and their identities were determined using mass spectrometry (Supplemental Fig. S4; Supplemental Tables S1 and S2). D, A subset of identified, sHSP-interacting proteins primarily bind CI sHSPs specifically during heat stress. Polyclonal antibodies were raised against each of the subunits of eEF1B (α, β, and γ) that were identified as interacting with the sHSPs. Immunoblot analysis was conducted on the eluates of affinity-isolated proteins from wild-type (WT), PCaMV35S:HSP17.4 or PCaMV35S:17.6-Strep, and PHSP17.4:HSP17.4 or PHSP17.6:17.6-Strep plants that were maintained at 22°C (C), acclimated and heat stressed as above (H), or allowed to recover for 3 h after heat stress (R). Immunoblots were processed with the antibodies indicated.
Identifying sHSP-Associated Proteins
To identify proteins specifically associated with the sHSPs during heat stress, the affinity isolates from samples in Figure 3A, lanes 4 and 5 for CI and lanes 9 and 10 for CII, sHSPs were separated using 2D-PAGE (Fig. 3, B and C). Protein spots that were more abundant in heat-stressed plant samples and observed in two or more independent experiments were excised and analyzed by mass spectrometry. A total of 36 CI-associated and 16 CII-associated proteins yielded peptide identifications as listed in Supplemental Tables S1 and S2 and indicated in Supplemental Figure S4, A and B. A major fraction of the sHSP-associated proteins is involved in translation: 12 for CI sHSPs and seven for CII sHSPs, all of which overlap with the CI-associated proteins, including translation elongation factor eEF1B (subunits α, β, and γ), translation initiation factor eIF4A (three isoforms), and translationally controlled tumor protein. In all experiments, more proteins were recovered associated with CI sHSPs than CII sHSPs, consistent with the observation that less CII protein was seen in higher Mr complexes (Fig. 2, A versus B). Other proteins associated with CI sHSPs included HSP70, other CI sHSPs, peptidyl prolyl isomerases, glutathione S-transferases, CDC48, jacalin-related lectins, and some metabolic enzymes (e.g. Fru bisphosphate aldolase and ascorbate peroxidase). Of these, only the jacalin-related lectin (At3g16460) and ascorbate peroxidase also were positively identified among the CII-associated proteins. None of these proteins are strongly heat regulated, indicating that their association with sHSPs during heat stress and not under control conditions is not due simply to increased levels of these proteins. Thus, it appears from both the size-exclusion analysis and affinity purification experiments that CI sHSPs are more tightly associated with translation factors and other proteins during heat stress.
To verify some of the observed associations and to further study the behavior of sHSP-associated proteins during heat stress, we chose to focus on translation factor eEF1B subunits. Each of the two homologs of the three eEF1B subunits (α1 and α2, β1 and β2, and γ1 and γ2) were expressed as recombinant proteins and used to raise polyclonal antibodies. Antibodies raised against the eEF1B α-, β-, or γ-subunits recognized both isoforms.
Immunoblot analysis using eEF1B α-, β-, and γ-antibodies was conducted on affinity isolates from wild-type and CI or CII sHSP-Strep plants (Fig. 3D). As expected, no sHSPs or eEF1B subunits were detected in the wild-type samples (lanes 1 and 5). Isolates from control plants constitutively expressing either CI or CII sHSPs contain only the corresponding sHSP-Strep protein (lanes 2 and 6). In contrast, after heat stress, the CI sHSP-Strep isolate contains all three eEF1B subunits, along with the endogenous CI sHSPs, which could be associated either through heterooligomerization or indirectly through binding to interacting proteins (lane 3). Coisolation of a small amount of CII sHSPs also is observed, although CI and CII proteins are not known to heterooligomerize (Basha et al., 2010). Notably, interaction with the eEF1B subunits, but not the sHSPs, is decreased significantly after recovery from heat stress (Fig. 3D, lane 4), consistent with the shift of the sHSPs out of a high-Mr form back to the native approximately 200-kD oligomeric state (Fig. 2A). All three eEF1B subunits also were detectable in Strep-tagged CII sHSP isolates (lane 7) and decreased during recovery (lane 8), but the interaction was not nearly as strong as that observed for CI sHSPs, as expected. Similar to the CI, the CII sHSP-Strep also pulled down the endogenous CII sHSPs expressed during heat stress (lanes 7 and 8). CII sHSP interaction with the eEF1B subunits was reduced after heat stress recovery (lane 8). As noted above, increased association of these proteins with the sHSPs during heat stress is not due to an increase in the level of eEF1B subunits; none of these proteins accumulates to higher levels during heat stress (Supplemental Fig. S1, A and B). These data confirm the identification of the eEF1B subunits as proteins that interact directly or indirectly with the sHSPs during heat stress.
CI and CII sHSPs Are Important for the Resolubilization of eEF1B Subunits during Recovery from Heat Stress
To obtain additional evidence that sHSP interaction with eEF1B subunits is significant in vivo, the solubility of eEF1B subunits during heat stress and recovery was examined in wild-type and sHSP CI and CII RNAi plants. Because sHSPs are proposed to prevent the heat-induced, irreversible aggregation of proteins, we hypothesized that sHSP substrates should become at least partially insoluble during heat stress and that more substrate would be found in the insoluble fraction after heat stress or during recovery in the RNAi lines compared with the wild type.
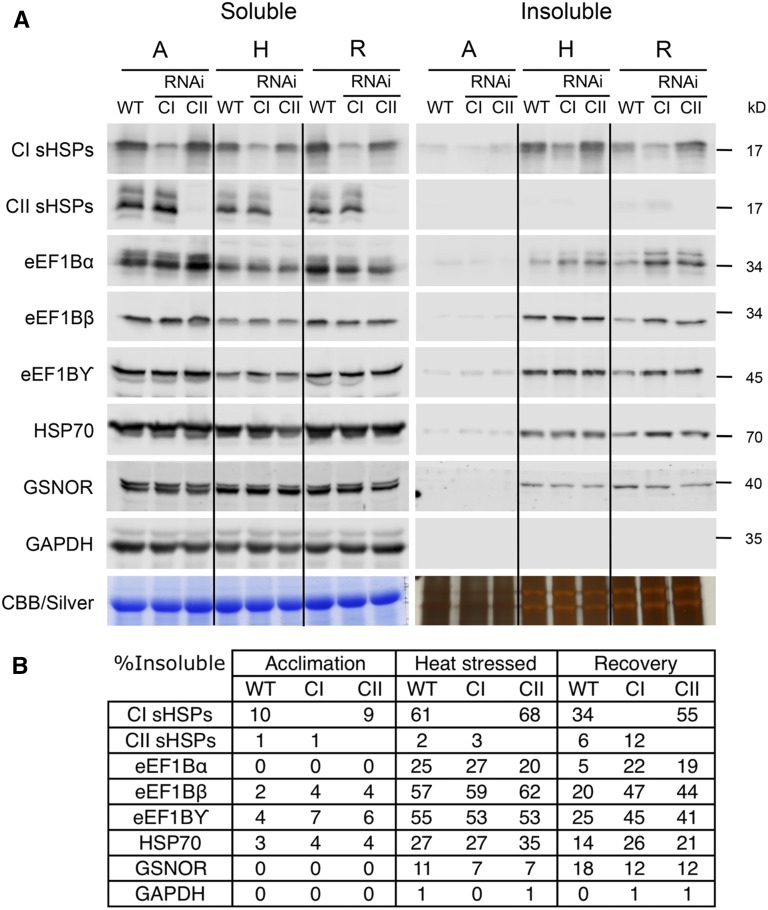
To test this hypothesis, the insoluble cell fraction of wild-type and CI and CII sHSP RNAi seedlings was tested for the accumulation of sHSPs, eEF1B subunits, HSP70, and two control proteins, S-nitrosoglutathione reductase (At5g43940; GSNOR) and cytosolic glyceraldehyde 3-phosphate dehydrogenase (At3g04120; GAPDH; Fig. 4). Soluble, insoluble, and total protein fractions were analyzed by immunoblotting, and the percentage of insoluble protein was estimated directly after acclimation, following a subsequent severe heat stress, or after 5 h of recovery from the severe stress treatment (Fig. 4; Supplemental Fig. S1A). None of the proteins tested was significantly insoluble after acclimation. Directly after heat stress, approximately 60% and somewhat less than 70% of the total CI sHSPs accumulated in the insoluble fraction in wild-type and CII sHSP RNAi plants, respectively. As expected, the increase in the insoluble fraction coincides with a reduction in the soluble fraction. After recovery, approximately 55% of the CI sHSPs remained insoluble in the CII RNAi line, compared with only an estimated 34% in the wild type. CII sHSPs remained largely soluble during heat stress, although more CII sHSPs were insoluble in the CI sHSP RNAi line (approximately 12%) after recovery in comparison with the wild type (approximately 6%). Thus, both sHSP classes hyperaccumulate in the insoluble fraction when the other class is knocked down, although CII sHSPs do not become insoluble to a similar extent as CI sHSPs. The insolubility of sHSPs under these conditions in vivo is consistent with interaction with other thermally labile proteins, as sHSPs alone do not become insoluble in vitro after similar temperature treatments (Lee et al., 1997; Basha et al., 2010).
Figure 4.
Reduction of CI, and to a lesser extent CII, sHSPs leads to increased retention of specific proteins in the insoluble cell fraction after heat stress. The solubility of several sHSP-interacting proteins was determined during acclimation (A), heat stress (H), and recovery (R). A, The soluble and insoluble protein fractions were isolated from 14-d-old wild-type (WT) plants and CI and CII sHSP RNAi lines. Samples were loaded using an equivalent percentage of the fractions and separated using SDS-PAGE. Immunoblot analysis was conducted using the antibodies indicated. Loading controls were stained using Coomassie Brilliant Blue (Soluble) or silver (Insoluble). B, The percentage of insoluble protein was determined for each protein by quantifying the soluble and insoluble protein fractions. The data were repeated in three biological replicates. Immunoblots for the total protein fraction are shown in Supplemental Figure S1A, and the values for an independent biological replicate are shown in Supplemental Table S4.
Considering potential sHSP-interacting proteins, a proportion of all eEF1B subunits became insoluble during heat stress, suggesting that these proteins are thermally labile or associated with other thermally labile components. The percentage of insoluble eEF1B subunits after this treatment varied between 20% and 60% for the different subunits, and the amount of insoluble protein was slightly higher in both the CI and CII sHSP RNAi plants compared with the wild type (Fig. 4). Following recovery, a much larger percentage of the eEF1B subunits remained insoluble in both the CI and CII sHSP RNAi lines compared with the wild type, with both RNAi lines showing a similar level of insoluble protein. Depending on the fraction that remains soluble during heat stress, it can be hard to discern subtle differences in the amount of protein returning to the soluble fraction during recovery, but there was a higher amount of most eEF1B subunits in the soluble fraction after heat stress recovery. We also examined the solubility of cytosolic HSP70, because HSP70 is required to refold proteins bound to sHSPs (Lee and Vierling, 2000) and was associated with the CI sHSPs. A greater proportion of cytosolic HSP70 remained insoluble in both CI and CII sHSP RNAi lines compared with the wild type after heat stress recovery, which was most clear in the CI sHSP RNAi line. The solubility of other proteins also was tested to determine if the sHSP dependence of resolubilization specifically occurs with eEF1B subunits or if it is due to general disaggregation activity in the cell. One protein that becomes insoluble during heat stress but was not identified to interact with either class of sHSPs is GSNOR. No difference in solubility during heat stress and recovery in either sHSP RNAi line compared with the wild type was observed. This indicates that knocking down either sHSP class does not simply change the cellular environment but specifically impacts a subset of proteins. Furthermore, although GAPDH was detected in the CII sHSP affinity purification, its solubility was not affected by heat stress, indicating that proteins do not randomly become trapped in protein aggregates.
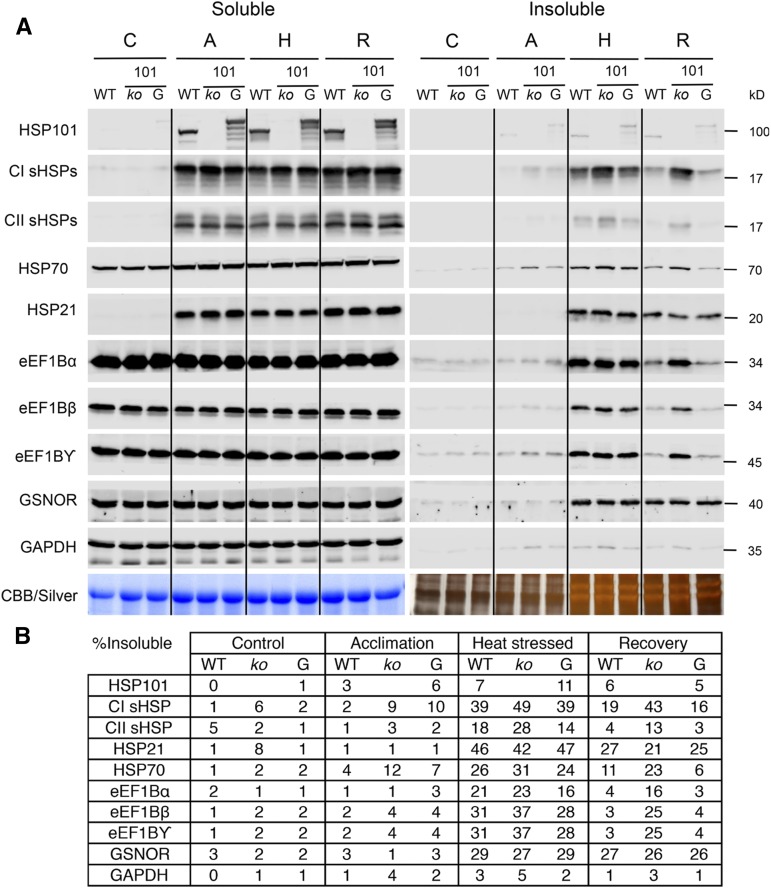
The Solubility of sHSP-Associated Proteins Requires HSP101
We have shown previously that sHSPs remain insoluble following heat stress in HSP101 mutants (Lee et al., 2005), and in vitro, the E. coli HSP101 homolog, ClpB, works together with the prokaryotic HSP70 system to disaggregate and refold sHSP-bound substrates (Mogk et al., 2003). To test further the possibility that the sHSP-interacting proteins could be sHSP substrates, we examined whether their solubility would be affected in the absence of the HSP101 disaggregase. The solubility of the sHSPs, eEF1B subunits, and a number of control proteins (chloroplast-targeted HSP21, GSNOR, and GAPDH) was tested in the wild type, the hsp101 null mutant, and the hsp101 mutant complemented with HSP101 translationally fused to GFP (Fig. 5). The HSP101-GFP fusion protein fully complements the hsp101 phenotype (Supplemental Fig. S5A) and migrates at the expected size by SDS-PAGE (Supplemental Fig. S5B). Seedlings were unheated, acclimated, heat stressed at 45°C for 1 h (a severe but nonlethal stress for the hsp101 mutant; Supplemental Fig. S1C), or allowed to recover at room temperature for 5 h after heat stress, as for Figure 4 (Fig. 5). Levels of HSP101 and all sHSPs were very low in control conditions and were induced but remained soluble during acclimation. Although HSP70 is expressed in control conditions, HSP70 levels increased approximately 45% after acclimation. No significant effect of heat treatments on the total amount of protein accumulated was observed for eEF1B subunits, GSNOR, or GAPDH (Supplemental Fig. S1, A and B). HSP101 remained soluble during all treatments, indicating that HSP101 is not thermally labile and does not stably interact with sHSPs or sHSP-interacting proteins. A proportion of HSP70 became insoluble during heat stress, similar to the observation in Figure 4 and consistent with detection in CI affinity isolates, and remained insoluble in the hsp101 mutant after heat stress recovery. Both during heat stress and after recovery, more CI and CII sHSPs accumulated in the insoluble fraction in the hsp101 mutant, as expected from previous experiments (Lee et al., 2005). In addition, a much larger amount of eEF1B subunits remained in the insoluble protein fraction during recovery in the hsp101 mutant, consistent with the conclusion that these proteins are heat labile and require sHSP cooperation with HSP101 for solubility.
Figure 5.
CI sHSP-interacting proteins remain insoluble during recovery from heat stress in an hsp101 null mutant. The wild type (WT), the hsp101 null mutant (ko), and the hsp101 mutant complemented with HSP101-GFP (G) were heat stressed, and the total, soluble, and insoluble protein fractions were isolated. A, Samples were loaded using an equivalent percentage of the soluble and insoluble fractions and separated using SDS-PAGE. Immunoblot analysis was conducted using the antibodies indicated. Loading controls were stained using Coomassie Brilliant Blue (Soluble) or silver (Insoluble). B, The percentage of insoluble protein was determined for each protein by quantifying the soluble and insoluble protein fractions. The data were repeated in four biological replicates. Immunoblots for the total protein fraction are shown in Supplemental Figure S1A, and the values for an independent biological replicate are shown in Supplemental Table S4.
To determine if the retention of proteins in the insoluble fraction in the hsp101 plants is due to a general inability of the mutant to cope with heat stress, rather than a specific effect on these sHSP-interacting proteins, we also examined the solubility of GSNOR, which becomes insoluble during heat stress (Fig. 4), and of HSP21, which is located exclusively in plastids (Chen and Vierling, 1991). A little under half of the HSP21 became insoluble during heat stress, which was reduced to approximately 25% after 5 h of recovery from heat stress. Notably, the absence of HSP101 had no effect on the abundance of GSNOR or HSP21 in the insoluble fraction during heat stress or recovery. Altogether, the data support the conclusion that HSP101 is specifically important for the disaggregation of cytosolic sHSPs and their interacting proteins.
CI and CII sHSPs Are Recruited to Distinct Cytosolic Foci during Acclimation
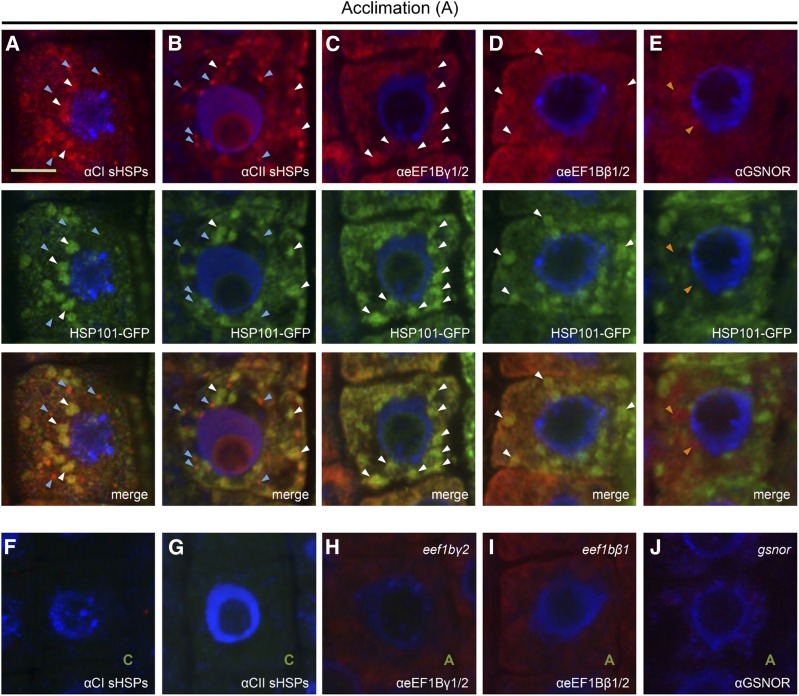
To address further the extent to which HSP101 and CI and CII sHSPs interact with eEF1B subunits in vivo, the localization of these proteins was examined. Immunolocalization experiments were conducted using CI and CII sHSP and eEF1Bβ and eEF1Bγ polyclonal antibodies to determine where in the cell and to what extent colocalization occurs and to determine if the CI and CII sHSPs behave similarly during heat stress. We focused on the behavior of proteins during the heat acclimation treatment, as we were not able to obtain consistent immunolocalization data with severely heat-stressed tissues (Fig. 6). To observe HSP101, we took advantage of the HSP101-GFP line used in Figure 5. Imaging was performed in roots to more readily study intracellular protein localization; roots of sHSP RNAi lines exhibit a reduced growth phenotype after heat stress (Supplemental Fig. S2E). Both CI sHSPs and HSP101-GFP were detected in punctate structures after acclimation (Fig. 6A, white arrowheads), and all HSP101-GFP structures colocalized with CI sHSP structures. However, in addition, CI sHSPs also accumulated in smaller punctate structures that did not colocalize with HSP101-GFP (blue arrowheads). HSP101-GFP also colocalized with CII sHSPs (Fig. 6B, white arrowheads), but there were more, smaller punctate structures containing CII sHSPs that did not colocalize with HSP101-GFP (blue arrowheads). This indicates that CI and CII show related localization patterns, but there is more overlap between CI sHSPs and HSP101-GFP-containing structures. Unheated control plants, which do not express detectable sHSPs or HSP101, were used to verify the antibody specificity (Fig. 6, F and G) and did not show any signal in the red or green channel with the microscope settings used. HSP101-GFP also formed analogous punctate structures during heat acclimation in live tissue, which was not observed for free yellow fluorescent protein (YFP; Supplemental Fig. S5C), supporting the physiological significance of the structures observed by immunolocalization.
Figure 6.
CI and CII sHSPs and eEF1B β- and γ-subunits colocalize with HSP101-GFP. Arabidopsis roots expressing HSP101-GFP were heat acclimated (90 min, 38°C plus 2 h, 21°C), fixed, and processed for immunolocalization using the indicated polyclonal antibodies. HSP101-GFP was localized based on GFP fluorescence, and all other proteins were detected using secondary antibodies coupled to Alexa 594. A, CI sHSPs localize to small bright cytosolic foci (blue arrowheads) and to larger aggregates (white arrowheads) that colocalize with HSP101-GFP. B, CII sHSP localization was similar to CI sHSPs, but there are a larger number of small bright cytosolic foci with CII sHSPs (blue arrowheads). C and D, eEF1Bγ and eEF1Bβ show partial colocalization with HSP101-GFP-containing aggregates. E, GSNOR is more uniformly distributed in the cell and did not colocalize with HSP101-GFP-containing aggregates (orange arrowheads). F and G, Plants kept in control conditions were treated similarly to those in A and B and did not show any detectable signal in the red and green channels, verifying the specificity of the signal. H and I, The eEF1Bγ2 and eEF1Bβ1 T-DNA insertion lines displayed a reduced signal with the corresponding antibodies. J, The gsnor mutant did not show any detectable signal when using the GSNOR antibody, verifying the specificity of the signal.
We then examined the localization of the sHSP-interacting proteins eEF1Bβ and eEF1Bγ compared with the localization of GSNOR, which shows no sHSP interaction. Both eEF1Bβ and eEF1Bγ colocalize in punctate structures with HSP101-GFP (Fig. 6, C and D, white arrowheads), and no additional punctate structures were observed that did not colocalize with HSP101-GFP, indicating that CI and CII punctate structures that are not colocalized with HSP101 (Fig. 6, A and B, blue arrowheads) are not associated with eEF1B subunits. The specificity of the eEF1B antibodies was verified using T-DNA insertion lines for both homologs of eEF1Bβ and eEF1Bγ (Supplemental Fig. S6A). The eef1bγ1 and eef1bβ2 plants showed a clear reduction in antibody reactivity on immunoblots (Supplemental Fig. S6B), and both the β and γ mutants showed reductions in fluorescence signal with the corresponding antibodies (Fig. 6, H and I), indicating that the immune reactivity seen in wild-type plants represents the eEF1B subunits. As an additional control, immunolocalization of GSNOR also was performed; GSNOR becomes insoluble during heat stress, but the solubility is not dependent on either sHSPs or HSP101. GSNOR did appear in some condensed areas (orange arrowheads) but did not colocalize with HSP101-GFP (Fig. 6E). A gsnor protein null mutant (Lee et al., 2008) was used to verify the specificity of the GSNOR immune signal (Fig. 6J).
DISCUSSION
Although sHSPs are ubiquitous stress proteins that are well documented to prevent the irreversible aggregation of model, heat-sensitive proteins in vitro (Basha et al., 2012; Haslbeck and Vierling, 2015), the in vivo activities of sHSPs are far from clear. This is especially true in land plants, which express multiple classes of sHSPs that are targeted to different cellular compartments and that often constitute several isoforms. Using RNAi, it was possible here to examine the effects of dramatically reducing the heat-induced accumulation of either CI or CII cytosolic sHSPs in stable transgenic lines of Arabidopsis. Reduction of CI sHSPs did not impact the level of CII sHSPs and vice versa, and the HSP70 and HSP101 chaperones also accumulated to wild-type levels in the RNAi lines. However, the severe reduction of either the CI or CII sHSPs was sufficient to compromise the ability of dark-grown seedlings to green after direct heat stress and for soil-grown plants to recover from extended heat treatment after acclimation. The phenotype of the CII RNAi plants was generally more severe, which we suggest is due to the more effective reduction of CII proteins, rather than to any difference in the relative requirement of the different sHSPs for survival. Thus, these two classes of sHSPs clearly perform nonredundant functions, as predicted from their evolutionary history. A previous report indicating that complete loss of only a single CI sHSP reduces the heat tolerance of hypocotyl elongation suggests that there could even be specific requirements for individual CI paralogs (Dafny-Yelin et al., 2008). However, this requires further investigation.
Despite the apparent requirement of both CI and CII HSPs for optimal heat tolerance, as demonstrated by the RNAi experiments, constitutive expression of either class alone enhanced the greening of dark-grown seedlings after direct heat stress (basal thermotolerance), consistent with previous reports using different assays to test the heat stress tolerance of plants constitutively expressing CI or CII sHSPs (Sun et al., 2012; Zhou et al., 2012; Wang et al., 2015). We also observed that, even though endogenous sHSPs are highly expressed in heat-acclimated plants, the individual sHSP OE lines had a still higher survival rate than the wild type after heat stress following acclimation. These results, along with the RNAi data, indicate that both sHSP classes have a dose-dependent effect on heat stress tolerance and protective functions independent of each other.
We note that relatively severe heat treatments were required to observe significant, immediate differences in the growth phenotypes of plants with reduced or increased sHSP expression. It remains untested whether treatments at more moderate high temperatures, which also induce sHSPs (e.g. 30°C or higher; Derocher et al., 1991), would have more subtle effects on the growth and reproduction of plants with these altered levels of sHSPs. However, regulated combinatorial enhancement of sHSP and HSP101 expression in specific target tissues could be a valuable strategy for enhancing plant heat tolerance, given the increase in incidents of severe high temperature (Lesk et al., 2016).
Both CI and CII sHSPs exhibit what is now considered to be classical chaperone activity when recombinant sHSPs are assayed in vitro; they capture denaturing, heat-sensitive proteins in large sHSP-substrate complexes that remain soluble, preventing the formation of large insoluble substrate aggregates, which presumably would be detrimental if formed in cells (Basha et al., 2012). However, to date, in vitro assays have failed to distinguish any class-specific differences in substrate protection, although recombinant CI and CII sHSPs exhibit distinct biochemical properties, will not form heterooligomers (Basha et al., 2010), and, as shown here, it is clear they have nonredundant roles in vivo. Tripp et al. (2009) reported that the expression of CI sHSPs, but not CII sHSPs, could enhance the protection of introduced firefly luciferase during heat stress of tomato (Solanum lycopersicum) protoplasts, suggesting that CII sHSPs might have an entirely different function from chaperone activity. It should be noted, however, that CII sHSPs from different species interact with and protect luciferase in vitro (Basha et al., 2010). Altogether, it is clear that in vitro or in vivo assays with heterologous, heat-sensitive proteins do not capture essential aspects of sHSP function.
To probe the in vivo function of CI and CII sHSPs, we identified proteins associated with either class specifically during heat stress. Size fractionation of plant cell extracts demonstrated that CI and CII sHSPs elute at an increased size after severe heat stress, consistent with heat-dependent interaction and the protection of other cellular components. Interestingly, the CII proteins also appeared in fractions smaller than their expected dodecameric size after heat stress, a behavior not observed for CI sHSPs in the same extracts, and the CII affinity purifications recovered fewer proteins. While the significance of this differential behavior is unclear, it further distinguishes proteins of the two sHSP classes. A striking result from affinity purification of both sHSPs from these extracts was the recovery of proteins involved directly in translation, including initiation factor eIF4A, an RNA helicase (three isoforms), and the three subunits (α, β, and γ) of elongation factor eEF1B, which is the GDP-exchange factor for eEF1A (Janssen and Möller, 1988). We validated the significance of sHSP interaction with eEF1B subunits in vivo in four distinct ways. First, using subunit-specific antibodies to probe affinity isolates, the interactions were confirmed to occur after heat stress and to decline during recovery, with greater interaction with CI than with CII sHSPs. Second, compared with the wild type, more of the eEF1B subunits remained in the insoluble fraction during heat stress recovery in the RNAi lines, consistent with a requirement of the sHSPs for the optimal solubility of these proteins. Third, the eEF1B subunits also were retained in the insoluble fraction during heat stress recovery in an HSP101 mutant, supporting the idea that these proteins are in an aggregated state that must be resolved by the HSP101 disaggregase, which is known to cooperate with sHSPs (Cashikar et al., 2005; Haslbeck et al., 2005). Finally, immunocytochemistry colocalized the eEF1B β- and γ-subunits with both the CI and CII sHSPs and with HSP101 in cytosolic foci after heat stress. Thus, there is a direct or indirect functional interaction of CI and CII sHSPs with eEF1B.
Interestingly, it was reported recently that eEF1Bγ becomes insoluble during heat stress in Saccharomyces cerevisiae and mammalian cells (Grousl et al., 2013; Cherkasov et al., 2015; Wallace et al., 2015), and we found previously that EF-Ts, the prokaryotic nucleotide-exchange factor for elongation factor EF-Tu, was recovered in affinity isolates with HSP16.6 from the cyanobacterium Synechocystis sp. 6803 (Basha et al., 2004a). Thus, the sensitivity of this translation component to stress (or involvement in translational regulation during stress), along with its association with sHSPs, may extend from prokaryotes to eukaryotes. In addition, in yeast and mammalian cells, eEF1Bγ colocalized with the stress granule marker PolyA-Binding Protein1 (Cherkasov et al., 2015; Wallace et al., 2015). Stress granules are a type of cytoplasmic mRNP that forms when translation initiation is impaired by a wide variety of different stress conditions, but their exact composition can vary depending on the stress (Buchan et al., 2011). They harbor nontranslating mRNAs and are distinct from P-bodies, other cytosolic mRNPs where mRNA degradation occurs (for review, see Franks and Lykke-Andersen, 2008). sHSPs in plants have long been known to be associated with heat stress-induced cytoplasmic structures termed heat stress granules by Nover et al. (1989). Plant heat stress granules have been proposed to be distinct from both stress granules and P-bodies (Weber et al., 2008). However, it is significant that, in addition to eEF1B and eIF4A, other proteins recovered in association with CI sHSPs were identified recently as components of yeast and/or human stress granules (Rinnerthaler et al., 2013; Cherkasov et al., 2015; Wallace et al., 2015; Walters et al., 2015; Jain et al., 2016), including eIF5A, a translationally controlled tumor protein (also recovered with CII sHSPs) that is putatively an important component of the target of rapamycin signaling pathway (Berkowitz et al., 2008), aminoacyl tRNA synthetase complex-interacting multifunctional protein1, and Obg-like ATPase1 (all proteins involved in translation), the chaperones HSP70, peptidyl prolyl isomerases, and CDC48, as well as profilin2 and S-adenosyl-Met synthase (Supplemental Table S3). We question the nature of the distinction between heat stress granules and stress granules and conclude that it is very likely that at least some of the sHSP-containing structures we observed include mRNA. Indeed, work in yeast and Drosophila spp. has concluded that chaperones are associated with stress granules and are necessary for the resumption of translation after severe heat stress (Cherkasov et al., 2013). While our experiments point to a clear interaction between sHSPs, specific translation factors, and HSP101, we also observed distinct cytosolic foci of CI and CII sHSPs that did not colocalize with either the identified translation factors or HSP101. The relationship of these structures to the stress granules remains to be determined.
CONCLUSION
CI and CII sHSPs are both important for tolerance to severe heat stress and interact with a partially overlapping set of heat-aggregating proteins during heat stress, many of which are known components of mRNA-containing stress granules in yeast and humans. The association of sHSPs with many stress granule proteins, their apparent requirement for the complete solubility of these proteins, along with colocalization in cytosolic foci with HSP101 suggest that these chaperones are involved in processes required for the resumption of normal translation activity following heat stress. While it is tempting to suggest that the proteins found associated with sHSPs represent specific heat-sensitive sHSP substrates, further work is required to demonstrate direct interactions and to connect the in vitro chaperone model of sHSP activity with these in vivo data.
MATERIALS AND METHODS
Plant Growth Conditions and Heat Treatments
Seeds for Arabidopsis (Arabidopsis thaliana) plants grown on soil were stratified before planting in 0.1% agar at 4°C for 2 to 3 d, or alternatively, stratified at 4°C for 3 d after planting on damp soil. Plants were grown in growth chambers on a 16/8-h day/night cycle at 21°C/18°C with 80 μmol m−2 s−1 photons on average with regular watering.
Seeds that were sown on plates were surface sterilized in a 1.6-L desiccator using 20 mL of household bleach and 600 μL of 40% HCl for 3 h. Alternatively, seeds were surface sterilized in 70% ethanol for 1 min followed by 50% bleach for 10 min with occasional inversion. Seeds were washed six times with sterile water to remove residual bleach. Seeds were sown on 5-mm-thick PNS plates [5 mm KNO3, 2 mm MgSO4, 2 mm Ca(NO3), 50 μm FeEDTA, 2.5 mm KPO4 (pH 5.5), 70 μL of H3BO3, 14 μm MnCl2, 0.5 μm CuSO4, 1 μm ZnSO4, 0.2 μm Na2MoO4, 10 μm NaCl, and 0.01 μm CoCl2] supplemented with 0.8% agar and 0.5% Suc.
All heat stress treatments were conducted in a calibrated hot air incubator in the dark unless indicated otherwise. Seedlings or adult plants were shielded from incubator air currents to reduce any variability of temperature within the incubator. The duration and severity of the heat stress are specified for each experiment. The internal temperature of plates that were transferred from to 21°C to 45°C typically ramped up to 40°C after 10 min, reached an internal temperature of 44°C after 17 min, and stabilized at 45°C after 22 min.
SDS-PAGE and Immunoblot Analysis
Protein samples for immunoblot analysis of denatured proteins were prepared by grinding tissue directly in 1× SDS sample buffer (2% [w/v] SDS, 12% [v/v] glycerol, 5% [v/v] β-mercaptoethanol, 62.5 mm Tris, pH 6.8, and 0.0025% [w/v] Bromophenol Blue) in a 5:1 (v/w) buffer:tissue ratio, and protein concentrations were determined using a Coomassie Brilliant Blue dye-binding assay (Ghosh et al., 1988). Proteins were separated by SDS-PAGE on 7.5% to 15% (w/v) polyacrylamide gels. For immunoblot analysis, proteins were transferred to nitrocellulose (Bio-Rad) and processed for either fluorescent or chemiluminescent detection. Primary antibodies were all raised in rabbits and are available at Agrisera unless stated otherwise. Antibody dilutions and Agrisera order numbers were as follows: CI sHSPs (1:3,000; AS07 254); CII sHSPs (1:3,000; AS07 255), eEF1Bα (1:3,000; AS10 679), eEF1Bβ (1:3,000; AS10 677), eEF1Bγ (1:3,000; AS10 676), HSP70 (1:5,000; AS08 371), GSNOR (1:1,000; AS09 647), GAPC (Dr. Ming-Che Shih, University of Iowa; 1:5,000), N-ter HSP101 (1:5,000; AS07 253), C-ter HSP101 (1:1,000; AS08 287), and HSP21 (1:1,000; AS08 285). Blots prepared for fluorescent detection were incubated with IRDye 700CW donkey α-rabbit antibody (1:20,000) and detected using a Li-Cor Odyssey CLx. For chemiluminescent detection, blots were incubated with ECL donkey α-rabbit IgG (1:10,000; GE Healthcare), visualized using Pierce ECL immunoblot substrate, and detected using a G:Box (Syngene).
Root Growth and Hypocotyl Elongation Assay
Seedlings were grown on PNS plates in illuminated or dark conditions for the root growth or hypocotyl elongation assay, respectively. For the root growth assay, seeds were stratified for 1 to 3 d on the plates and placed at an angle approximately 15° from vertical in the growth chamber on a 16/8-h day/night cycle at 21°C/18°C with 80 μmol m−2 s−1 photons on average. After 4 d of growth, heat treatments were applied as described, and the location of the root tip was marked. Plates were returned to the growth chamber, and further root growth was measured after 9 d. For the hypocotyl elongation assays, plates were wrapped in aluminum foil and seeds were stratified at 4°C for 3 d before being placed vertically in a dark cabinet at room temperature. After 2.5 d of growth, the location of the hypocotyl tip was marked by briefly removing the foil, and then heat treatments were applied as described. After an additional 2.5 d of growth, plates were photographed, and hypocotyl elongation after the treatment was measured using ImageJ (http://imagej.nih.gov/ij/).
Construction of Plant Transformation Vectors
CI and CII sHSP-Strep and OE Lines
Plant transformation vectors were created to express the Arabidopsis CI HSP17.4 (At3g46230) with a C-terminal affinity tag driven by either its native promoter or a CaMV 35S promoter, along with a vector to constitutively express the native, untagged protein. A clone containing the promoter (720 bp), the 5′ untranslated region (UTR), the genomic coding sequence, and the 3′ UTR (249 bp) of CI HSP17.4 was kindly provided by Dr. Y. Komeda (Takahashi and Komeda, 1989). A StrepII tag (Trp-Ser-His-Pro-Gln-Phe-Glu-Lys) was added C terminally of the coding region using around-the-world PCR with primers 1 and 2 (Supplemental Table S4), resulting in PHSP17.4:HSP17.4-Strep:3′UTR, which was digested using XbaI and SalI and ligated into the pBin19 binary vector (kanamycin [Kan] resistant) also digested with XbaI and SalI. PCaMV35S:HSP17.4-Strep was created by amplifying the coding region and the Strep tag from the above construct, using primers 3 and 4, and ligating into the PRT plasmid (Amp resistant; Töpfer et al., 1987) containing a CaMV 35S promoter with EcoRI and XbaI. The PCaMV:HSP17.4-Strep:3′UTR fragment was moved into theBin19 binary vector (Kan resistant) using KpaI and SacI sites. To express HSP17.4 constitutively without a Strep tag, PCaMV:HSP17.4:3′UTR was created by inserting the coding region of the HSP17.4 cDNA into the PRT vector using EcoRI and XbaI sites and further processing as explained for the PCaMV:HSP17.4-Strep:3′UTR cloning. The same set of plasmids also was created for CII HSP17.6II (AT5G12020). The promoter (1,382 bp), 5′ UTR, genomic sequence, and 3′ UTR (464 bp) of HSP17.6II were amplified from genomic DNA using primers 5 and 6 and cloned into the pBSKS vector (Amp resistant; Stratagene) in the SmaI position. The Strep tag was added C terminally using around-the-world PCR using primers 7 and 9 and was inserted into HindIII and BamHI sites in Bin19 (Kan resistant). The construct containing PCaMV35S:HSP17.6 was kindly provided by Marc Kirschner (Forreiter et al., 1997). The insert including the terminator was digested using HindIII and ligated in the pBin19 vector (Kan resistance). The Strep tag was then added using primers 8 and 9, and PCaMV:HSP17.6-Strep:3′UTR was digested using HindIII and ligated into pBin19 (Kan resistant). All constructs were verified by DNA sequencing.
CI and CII sHSP Double-Stranded RNA Lines
To obtain small interfering RNA lines, vectors to express double-stranded RNA were designed for both the CI and CII sHSP gene families. Conserved gene regions were selected for CI HSP17.4 (bp 95–589) and for CII HSP17.6II (bp 63–553) and amplified using primers 10 and 11 or 12 and 13, respectively. The inserts were digested with AscI and SwaI for insertion in the 5′→3′ orientation and with BamHI and SpeI for insertion in the 3′→5′ orientation and then ligated into the pFGC1008 vector (ABRC stock no. CD3-446; Kan resistant).
HSP101-GFP
Plasmids containing either the promoter region of HSP101 (At1g74310; 734 bp) or the HSP101 coding sequence were obtained from Ung Lee (University of Arizona; Hong and Vierling, 2000). GFP was amplified from pMDC83 (Curtis and Grossniklaus, 2003) using primers 14 and 15 to add flanking XhoI and XbaI restriction sites. The fragment was digested with XhoI and XbaI and inserted C terminally of the HSP101 promoter. The XhoI fragment flanking the HSP101 coding sequence was excised with XhoI and ligated between the HSP101 promoter and GFP, creating PHSP101:HSP101(CDS)-GFP. This construct was digested using KpnI and XbaI and ligated into pBIN19 (Kan resistant), in front of the CaMV 35S poly(A) transcription terminator.
Generation of Transgenic Plants
Vectors were transformed via the Agrobacterium tumefaciens strain GV3103 into wild-type Columbia-0 for all sHSP constructs or into the HSP101 protein null mutant hot1-3 (Hong and Vierling, 2001) in the case of HSP101-GFP by floral dip transformation (Clough and Bent, 1998). Plants transformed using pBIN19 were selected using 50 μg mL−1 Kan, and plants transformed using pFGC1008 were selected using 30 μg mL−1 hygromycin. Expression levels of the sHSPs or HSP101-GFP were confirmed by immunoblot analysis using the corresponding antibodies. PCaMV35S:YFP was obtained from the University of Amsterdam (van Leeuwen et al., 2007).
Cloning and Expression of eEF1Bα1, eEF1Bβ1, and eEF1Bγ1
The cDNA clones of eEF1Bα1 (At5g12110), eEF1Bβ1 (At1g30230), and eEF1Bγ1 (At1g57720) were obtained from the Arabidopsis Biological Resource Center. The coding regions were amplified while adding 5′ and 3′ flanking restriction sites: NdeI and BamHI for eEF1Bα1 (primers 16 and 17) and NdeI and XhoI for eEF1Bβ1 and eEF1Bγ1 (primers 18/19 and 20/21, respectively). The PCR products were digested, ligated into the expression vector pJC20, and transformed to DH5α Escherichia coli cells. After sequence confirmation, clones were transformed into Rosetta II E. coli cells for protein production. Cells containing the plasmids of interest were grown in 2xYT at 24°C to an OD600 of 1.6 (approximately 36 h), at which point 1 mm isopropylthio-β-galactoside was added to induce protein expression for 12 h. Cells were spun down at 3,020g for 15 min and resuspended in TE buffer (25 mm Tris-HCl, 1 mm, EDTA, 5 mm ε-caproic acid, and 1 mm benzamidine). Cells were lysed using eight 30-s cycles of sonication followed by 30-s rest periods on ice. Cell lysates were spun for 15 min 10,000g, and eEF1Bα1, eEF1Bβ1, and eEF1Bγ1 remained soluble. Lysates were subjected to ammonium sulfate precipitation. The eEF1Bα1, eEF1Bβ1, and eEF1Bγ1 proteins were enriched in the 0% to 50% (w/v) ammonium sulfate fraction. Fractions containing the proteins were dialyzed in 50 mm Tris-HCl and 1 mm EDTA, pH 7.5, applied onto an 8-cm-long × 1.5-cm-radius DEAE-Sepharose column at 4°C, and eluted using 0 to 1 m NaCl. The protein-enriched fractions were pooled and dialyzed as described before, concentrated, applied to an S-200 HR Sephacryl size-exclusion column, and eluted with 25 mm NaPO4 buffer and 100 mm NaCl, pH 7.5. Antibodies against the purified proteins were raised in rabbits by Agrisera.
Strep-Tactin Affinity Chromatography and 2D Electrophoresis
Fifteen-day-old, light-grown seedlings of the indicated genotypes were grown on plates and heat stressed for 30 min at 38°C, recovered for 2 h at 22°C, and then exposed to 45°C for 30 min. Seedlings were ground in IP buffer (25 mm HEPES, 200 mm NaCl, 0.5% Triton X-100, 1 mm benzamidine, 5 mm ε-aminocaproic acid, and 1 mm EDTA, pH 7.5) in a 5:1 ratio (buffer volume:tissue weight). Samples were centrifuged at 16,100g for 15 min at 4°C, and total soluble protein was quantified using the Coomassie Brilliant Blue dye-binding assay (Ghosh et al., 1988). A total of 3 mg of soluble protein was incubated for 2 h at 4°C with 30 μL of Strep-Tactin Sepharose (IBA; catalog no. 2-1201-010) that had been preequilibrated in IP buffer. Resin was washed with IP buffer as described (Basha et al., 2004a). Elution of proteins from the Strep-Tactin resin for SDS-PAGE analysis was performed by boiling the resin with 60 μL of 1× SDS sample buffer. For silver staining, 30 μL per lane was separated, while 3 μL was separated for immunoblot analysis. For 2D-PAGE analysis, the samples were eluted with isoelectric focusing rehydration buffer, according to the manufacturer’s instructions, in 7 m urea, 2 m thiourea, 2% CHAPS, 2% IPG focusing buffer pH 3–10 NL (Amersham Biotech), and 3 mg mL−1−1 DTT. The isoelectric focusing dimension separation was performed using 3–10 NL strips (18 cm; Amersham Biotech) as described (Basha et al., 2004a). The running conditions were 8 h at 150 V, 5 h at 500 V, 5 h at 1,000 V, 5 h at 2,500 V, and then 20 h at 3,500 V. The SDS-PAGE dimension used 11% to 17% (w/v) polyacrylamide gradient gels. Gels were silver stained as described (Rabilloud et al., 1988).
HPLC-Tandem Mass Spectrometry of sHSP-Interacting Proteins
Proteins of interest were excised from 2D gels and digested with trypsin, and peptides were prepared for mass spectrometry as described (Basha et al., 2004a). Peptide extracts were introduced onto a 100-μm i.d. × 5-cm C18 column using an autosampler and separated with a 25-min gradient of 2% to 100% acetonitrile in 0.5% formic acid. The column eluate was directed into a Thermo Finnigan LCQ Deca ion trap mass spectrometer. The mass range scanned was 400 to 1,500 atomic mass units (amu), and data-dependent scanning was used to select the top-three most abundant ions in a parent scan for tandem mass spectrometry. Tandem mass spectrometry scans were searched using SEQUEST against a database of Arabidopsis sequences from TAIR (June 2005 version). The gene names were updated using the nomenclature annotated by TAIR (March 2016 version). The search allowed for static modification of Cys (57 amu; iodoacetamidation), and differential modification of Met (16 amu; oxidation) was considered. X correlation cutoffs of 2.0 for 2+ ions, 3.0 for 3+ ions, and delta Xcorr > 0.05 were applied, and data were sorted using DTASelect (Tabb et al., 2002).
Immunolocalization
Arabidopsis seedlings were grown vertically on PNS plates in the light for 6 d. Seedlings were exposed to 38°C for 90 min, allowed to recover for 2 h at room temperature, and subsequently fixed using 4% paraformaldehyde in PBS, pH 7.2, for 1 h with gentle shaking. Seedlings were transferred to Superfrost Plus adhesive slides (Fisherbrand), rinsed with water, and dried for 1 h at 38°C. A hydrophobic barrier was created encompassing the roots using a Super Pap pen (Daido Sangyo). Seedlings were rehydrated in PBS, and cell walls were digested using a 2% Driselase cocktail (Sigma-Aldrich) for 45 min at 37°C. Seedlings were washed using PBS and permeabilized with 10% dimethyl sulfoxide and 3% Nonidet P-40 (Sigma-Aldrich) for 1 h at room temperature. The seedlings were washed and blocked using 2% BSA in PBS for 2 h at 37°C. All primary antibodies were diluted 1:400 (or 1:200 for anti-GSNOR) in 2% BSA, and 300 μL was put on the slides and incubated overnight at 4°C. The next day, samples with primary antibody were incubated for an additional 1 h at 37°C prior to being washed and incubated in donkey anti-rabbit antibody conjugated to Alexa 594 (Life Science Technologies) at a 1:600 dilution in 2% BSA and incubated for 3 h at 37°C. 4′,6-Diamino-phenylindole was used as a nuclear counterstain at a 1:1,000 dilution in PBS, and following washing, seedlings were covered with Citifluor. Images were obtained with a Fluoview 1000MPE, IX81 motorized inverted research microscope (Olympus) equipped with a Hamamatsu C8484-05G camera. All immunolocalization images were acquired with a Plapon 60× oil, numerical aperture 1.42 lens. The excitation/emission wavelengths were as follows: 4′,6-diamino-phenylindole, 405/461 nm; GFP, 473/520 nm; and Alexa 594, 559/618 nm.
To visualize HSP101-GFP, living tissue was imaged by exposing roots to 10 μg mL−1 propidium iodide in water before mounting on slides, and images were acquired using a UPLSAPO 60× water, numerical aperture 1.20 lens. The excitation/emission wavelengths used were as follows: GFP, 473/510 nm; and propidium iodide, 559/619 nm. Line Kalman was used as an averaging factor, and the images were processed using FV10-ASW and ImageJ.
Preparation of Soluble and Insoluble Protein Fractions
Two-week-old seedlings grown on PNS plates were either untreated (control) or heat stressed as described. For each sample, 0.7 g of plant tissue was harvested, and a crude protein extract was prepared using 1 mL of protein isolation buffer (25 mm HEPES, pH 7.5, 200 mm NaCl, 0.5 mm EDTA, 5 mm ε-amino-N-caproic acid, and 1 mm benzamidine). Crude protein samples were never frozen and were kept on ice whenever possible throughout the procedure. Protein concentrations were determined using a Coomassie Brilliant Blue dye-binding assay (Ghosh et al., 1988). After grinding the sample using a mortar and pestle, samples were further homogenized with a Cole-Parmer PTFE glass tissue grinder for 1 min on ice. The crude protein extract was transferred to a microcentrifuge tube, and 500 μL was used for separation into soluble and insoluble fractions, while 300 μL was added to 100 μL of 4× SDS sample buffer (8% [w/v] SDS, 46% [v/v] glycerol, 20% [v/v] β-mercaptoethanol, 250 mm Tris, pH 6.8, and 0.01% [w/v] Bromophenol Blue) to monitor total protein content. For fractionation, samples were spun in a tabletop centrifuge at 16,100g for 15 min. The supernatant was removed, and 300 μL was added to 100 μL of 4× SDS sample buffer for immunoblot analysis. To facilitate washing of the insoluble fraction, 0.1 g of quartz salt (Sigma-Aldrich) was added to the pellet fraction, and samples were washed six times with 1 mL of protein isolation buffer. For each wash, the pellet was resuspended by pipetting and vortexing, and the samples were centrifuged subsequently at 16,100g for 15 min. After the washes, the insoluble fraction was washed once more in 25 mm Tris-HCl, pH 7.5. The pellet fraction was resuspended in 2× sample buffer, creating a total volume of 500 μL. Samples were spun for 30 s at 1,500g, and the soluble fraction was transferred to a new microcentrifuge tube.
Isolating Homozygous T-DNA Insertion Lines
Homozygous lines carrying a T-DNA insertion in the coding region of eEF1Bβ1 (Salk_046102; primers 22 and 23), eEF1Bβ2 (Salk_107994; primers 24 and 25), eEF1Bγ1 (Salk_047484; primers 26 and 27), and eEF1Bγ2 (Salk_033274; primers 28 and 29) were isolated. Primer 30 was used as the border primer, and immunoblot analysis was conducted to determine the effect of the T-DNA insertion on the expression of the corresponding protein (Supplemental Fig. S6).
Size-Exclusion Chromatography
Seedlings were grown on plates for 15 d in the light and then ground in a Cole-Parmer PTFE glass tissue grinder using a 2.5:1 (w/v) tissue:buffer ratio containing 25 mm NaPO4, 200 mm NaCl, 1 mm benzamidine, 5 mm aminocaproic acid, and 1 mm EDTA, pH 7.5. Samples were centrifuged at 16,100g for 15 min at 4°C. A total of 100 μL of the supernatant (protein concentration approximately 3 mg mL−1) was loaded onto a Bio-Rad 250 size-exclusion column and eluted with 25 mm NaPO4 and 200 mm NaCl, pH 7.5. Elution fractions (11 fractions of 0.5 mL) were collected, phenol precipitated as described (Faurobert et al., 2007), and solubilized in 100 μL of 1× SDS sample buffer. Samples (20 µL) were analyzed using 11% to 17% gradient SDS-PAGE and immunoblot analysis.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: HSP17.4 (At3g46230), HSP17.6II (At5g12020), HSP101 (At1g74310), eEF1Bα1 (At5g12110), eEF1Bβ1 (At1g30230), and eEF1Bγ1 (At1g57720).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Abundance of sHSP-interacting proteins remains unaltered during heat stress, and hsp101 mutant seedlings are able to survive the heat stress treatment.
Supplemental Figure S2. CI and CII sHSP RNAi and OE lines develop normally under control conditions and show reduced root growth after heat stress.
Supplemental Figure S3. Overexpression of sHSPs increases heat stress tolerance.
Supplemental Figure S4. sHSP-interacting proteins for which peptide data were obtained by mass spectrometry.
Supplemental Figure S5. PHSP101:HSP101-GFP is heat induced, complements the hsp101 null phenotype, and accumulates in cytosolic foci during heat stress.
Supplemental Figure S6. Location of T-DNA insertions in eEF1Bβ1 and eEF1Bβ2, eEF1Bγ1 and eEF1Bγ2, and the specificity of the EF1B β- and γ-antibodies.
Supplemental Figure S7. CI sHSPs accumulate in punctate structures in the CII sHSP RNAi during heat stress acclimation.
Supplemental Table S1. CI sHSP-interacting proteins.
Supplemental Table S2. CII sHSP-interacting proteins.
Supplemental Table S3. CI/CII sHSP-interacting proteins for which the orthologs are present in stress granules in S. cerevisiae and human cells.
Supplemental Table S4. Percentage of insoluble proteins from an independent experiment.
Supplemental Table S5. Primers.
Supplementary Material
Acknowledgments
We thank Dr. Y. Komeda from the University of Tokyo and Marc Kirschner from Goethe University for supplying the vectors harboring CI and CII sHSP coding regions, respectively; Dr. Joop Vermeer at the University of Amsterdam for the P35S:YFP line; Dr. M.C. Shih at the University of Iowa for the GAPDH antibody; and Matthew Grimes for his honor’s thesis studies of eEF1Bγ.
Glossary
- CI
class I
- CII
class II
- RNAi
RNA interference
- OE
overexpression
- CaMV
cauliflower mosaic virus
- UTR
untranslated region
- Kan
kanamycin
- amu
atomic mass units
Footnotes
This work was supported by the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (grant no. 96–351003232), the National Science Foundation (grant no. DBI 0820047), the Department of Energy (grant no. DE–SC0006646), the National Institutes of Health (grant no. RO1 GM42761), and the Massachusetts Life Sciences Center (New Faculty Research Award to E.V.).
Articles can be viewed without a subscription.
References
- Ahn YJ, Zimmerman JL (2006) Introduction of the carrot HSP17.7 into potato (Solanum tuberosum L.) enhances cellular membrane stability and tuberization in vitro. Plant Cell Environ 29: 95–104 [DOI] [PubMed] [Google Scholar]
- Basha E, Jones C, Wysocki V, Vierling E (2010) Mechanistic differences between two conserved classes of small heat shock proteins found in the plant cytosol. J Biol Chem 285: 11489–11497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basha E, Lee GJ, Breci LA, Hausrath AC, Buan NR, Giese KC, Vierling E (2004a) The identity of proteins associated with a small heat shock protein during heat stress in vivo indicates that these chaperones protect a wide range of cellular functions. J Biol Chem 279: 7566–7575 [DOI] [PubMed] [Google Scholar]
- Basha E, Lee GJ, Demeler B, Vierling E (2004b) Chaperone activity of cytosolic small heat shock proteins from wheat. Eur J Biochem 271: 1426–1436 [DOI] [PubMed] [Google Scholar]
- Basha E, O’Neill H, Vierling E (2012) Small heat shock proteins and α-crystallins: dynamic proteins with flexible functions. Trends Biochem Sci 37: 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz O, Jost R, Pollmann S, Masle J (2008) Characterization of TCTP, the translationally controlled tumor protein, from Arabidopsis thaliana. Plant Cell 20: 3430–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Yoon JH, Parker R (2011) Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J Cell Sci 124: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashikar AG, Duennwald M, Lindquist SL (2005) A chaperone pathway in protein disaggregation: Hsp26 alters the nature of protein aggregates to facilitate reactivation by Hsp104. J Biol Chem 280: 23869–23875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Vierling E (1991) Analysis of conserved domains identifies a unique structural feature of a chloroplast heat shock protein. Mol Gen Genet 228: 328. [PubMed] [Google Scholar]
- Cherkasov V, Grousl T, Theer P, Vainshtein Y, Glässer C, Mongis C, Kramer G, Stoecklin G, Knop M, Mogk A, et al. (2015) Systemic control of protein synthesis through sequestration of translation and ribosome biogenesis factors during severe heat stress. FEBS Lett 589: 3654–3664 [DOI] [PubMed] [Google Scholar]
- Cherkasov V, Hofmann S, Druffel-Augustin S, Mogk A, Tyedmers J, Stoecklin G, Bukau B (2013) Coordination of translational control and protein homeostasis during severe heat stress. Curr Biol 23: 2452–2462 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafny-Yelin M, Tzfira T, Vainstein A, Adam Z (2008) Non-redundant functions of sHSP-CIs in acquired thermotolerance and their role in early seed development in Arabidopsis. Plant Mol Biol 67: 363–373 [DOI] [PubMed] [Google Scholar]
- de Jong WW, Caspers GJ, Leunissen JA (1998) Genealogy of the alpha-crystallin–small heat-shock protein superfamily. Int J Biol Macromol 22: 151–162 [DOI] [PubMed] [Google Scholar]
- Delbecq SP, Klevit RE (2013) One size does not fit all: the oligomeric states of αB crystallin. FEBS Lett 587: 1073–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derocher AE, Helm KW, Lauzon LM, Vierling E (1991) Expression of a conserved family of cytoplasmic low molecular weight heat shock proteins during heat stress and recovery. Plant Physiol 96: 1038–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SM, Genest O, Wickner S (2013) Protein rescue from aggregates by powerful molecular chaperone machines. Nat Rev Mol Cell Biol 14: 617–629 [DOI] [PubMed] [Google Scholar]
- Faurobert M, Pelpoir E, Chaïb J (2007) Phenol extraction of proteins for proteomic studies of recalcitrant plant tissues. Methods Mol Biol 355: 9–14 [DOI] [PubMed] [Google Scholar]
- Forreiter C, Kirschner M, Nover L (1997) Stable transformation of an Arabidopsis cell suspension culture with firefly luciferase providing a cellular system for analysis of chaperone activity in vivo. Plant Cell 9: 2171–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J (2008) The control of mRNA decapping and P-body formation. Mol Cell 32: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich KL, Giese KC, Buan NR, Vierling E (2004) Interactions between small heat shock protein subunits and substrate in small heat shock protein-substrate complexes. J Biol Chem 279: 1080–1089 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Gepstein S, Heikkila JJ, Dumbroff EB (1988) Use of a scanning densitometer or an ELISA plate reader for measurement of nanogram amounts of protein in crude extracts from biological tissues. Anal Biochem 169: 227–233 [DOI] [PubMed] [Google Scholar]
- Grousl T, Ivanov P, Malcova I, Pompach P, Frydlova I, Slaba R, Senohrabkova L, Novakova L, Hasek J (2013) Heat shock-induced accumulation of translation elongation and termination factors precedes assembly of stress granules in S. cerevisiae. PLoS ONE 8: e57083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M, Miess A, Stromer T, Walter S, Buchner J (2005) Disassembling protein aggregates in the yeast cytosol: the cooperation of Hsp26 with Ssa1 and Hsp104. J Biol Chem 280: 23861–23868 [DOI] [PubMed] [Google Scholar]
- Haslbeck M, Vierling E (2015) A first line of stress defense: small heat shock proteins and their function in protein homeostasis. J Mol Biol 427: 1537–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm KW, Lee GJ, Vierling E (1997) Expression and native structure of cytosolic class II small heat-shock proteins. Plant Physiol 114: 1477–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SW, Vierling E (2000) Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc Natl Acad Sci USA 97: 4392–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SW, Vierling E (2001) Hsp101 is necessary for heat tolerance but dispensable for development and germination in the absence of stress. Plant J 27: 25–35 [DOI] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R (2016) ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164: 487–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen GM, Möller W (1988) Kinetic studies on the role of elongation factors 1 beta and 1 gamma in protein synthesis. J Biol Chem 263: 1773–1778 [PubMed] [Google Scholar]
- Jiang C, Xu J, Zhang H, Zhang X, Shi J, Li M, Ming F (2009) A cytosolic class I small heat shock protein, RcHSP17.8, of Rosa chinensis confers resistance to a variety of stresses to Escherichia coli, yeast and Arabidopsis thaliana. Plant Cell Environ 32: 1046–1059 [DOI] [PubMed] [Google Scholar]
- Kirschner M, Winkelhaus S, Thierfelder JM, Nover L (2000) Transient expression and heat-stress-induced co-aggregation of endogenous and heterologous small heat-stress proteins in tobacco protoplasts. Plant J 24: 397–411 [DOI] [PubMed] [Google Scholar]
- Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf KD (2007) Complexity of the heat stress response in plants. Curr Opin Plant Biol 10: 310–316 [DOI] [PubMed] [Google Scholar]
- Lee GJ, Roseman AM, Saibil HR, Vierling E (1997) A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J 16: 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GJ, Vierling E (2000) A small heat shock protein cooperates with heat shock protein 70 systems to reactivate a heat-denatured protein. Plant Physiol 122: 189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee U, Wie C, Escobar M, Williams B, Hong SW, Vierling E (2005) Genetic analysis reveals domain interactions of Arabidopsis Hsp100/ClpB and cooperation with the small heat shock protein chaperone system. Plant Cell 17: 559–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee U, Wie C, Fernandez BO, Feelisch M, Vierling E (2008) Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. Plant Cell 20: 786–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesk C, Rowhani P, Ramankutty N (2016) Influence of extreme weather disasters on global crop production. Nature 529: 84–87 [DOI] [PubMed] [Google Scholar]
- Malik MK, Slovin JP, Hwang CH, Zimmerman JL (1999) Modified expression of a carrot small heat shock protein gene, hsp17. 7, results in increased or decreased thermotolerance. Plant J 20: 89–99 [DOI] [PubMed] [Google Scholar]
- Mogk A, Schlieker C, Friedrich KL, Schönfeld HJ, Vierling E, Bukau B (2003) Refolding of substrates bound to small Hsps relies on a disaggregation reaction mediated most efficiently by ClpB/DnaK. J Biol Chem 278: 31033–31042 [DOI] [PubMed] [Google Scholar]
- Mu C, Zhang S, Yu G, Chen N, Li X, Liu H (2013) Overexpression of small heat shock protein LimHSP16.45 in Arabidopsis enhances tolerance to abiotic stresses. PLoS ONE 8: e82264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf KD (1997) Heat stress proteins and transcription factors. Cell Mol Life Sci 53: 80–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Neumann D (1989) Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol Cell Biol 9: 1298–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Hong SW, Vierling E, Lindquist S (2000) Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12: 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabilloud T, Carpentier G, Tarroux P (1988) Improvement and simplification of low-background silver staining of proteins by using sodium dithionite. Electrophoresis 9: 288–291 [DOI] [PubMed] [Google Scholar]
- Rinnerthaler M, Lejskova R, Grousl T, Stradalova V, Heeren G, Richter K, Breitenbach-Koller L, Malinsky J, Hasek J, Breitenbach M (2013) Mmi1, the yeast homologue of mammalian TCTP, associates with stress granules in heat-shocked cells and modulates proteasome activity. PLoS ONE 8: e77791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhanagopalan I, Basha E, Ballard KN, Bopp NE, Vierling E (2015) Model chaperones: small heat shock proteins from plants. In Tanguay RM, Hightower LE, eds, The Big Book on Small Heat Shock Proteins. Springer International Publishing, Cham, Switzerland, pp 119–153 [Google Scholar]
- Scharf KD, Siddique M, Vierling E (2001) The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing alpha-crystallin domains (Acd proteins). Cell Stress Chaperones 6: 225–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique M, Gernhard S, von Koskull-Döring P, Vierling E, Scharf KD (2008) The plant sHSP superfamily: five new members in Arabidopsis thaliana with unexpected properties. Cell Stress Chaperones 13: 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smykal P, Masin J, Hrdy I, Konopasek I, Zarsky V (2000) Chaperone activity of tobacco HSP18, a small heat-shock protein, is inhibited by ATP. Plant J 23: 703–713 [DOI] [PubMed] [Google Scholar]
- Sun L, Liu Y, Kong X, Zhang D, Pan J, Zhou Y, Wang L, Li D, Yang X (2012) ZmHSP16.9, a cytosolic class I small heat shock protein in maize (Zea mays), confers heat tolerance in transgenic tobacco. Plant Cell Rep 31: 1473–1484 [DOI] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR III (2002) DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res 1: 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Komeda Y (1989) Characterization of two genes encoding small heat-shock proteins in Arabidopsis thaliana. Mol Gen Genet 219: 365–372 [DOI] [PubMed] [Google Scholar]
- Töpfer R, Matzeit V, Gronenborn B, Schell J, Steinbiss HH (1987) A set of plant expression vectors for transcriptional and translational fusions. Nucleic Acids Res 15: 5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp J, Mishra SK, Scharf KD (2009) Functional dissection of the cytosolic chaperone network in tomato mesophyll protoplasts. Plant Cell Environ 32: 123–133 [DOI] [PubMed] [Google Scholar]
- van Leeuwen W, Vermeer JE, Gadella TW Jr, Munnik T (2007) Visualization of phosphatidylinositol 4,5-bisphosphate in the plasma membrane of suspension-cultured tobacco BY-2 cells and whole Arabidopsis seedlings. Plant J 52: 1014–1026 [DOI] [PubMed] [Google Scholar]
- van Montfort RL, Basha E, Friedrich KL, Slingsby C, Vierling E (2001) Crystal structure and assembly of a eukaryotic small heat shock protein. Nat Struct Biol 8: 1025–1030 [DOI] [PubMed] [Google Scholar]
- Wallace EW, Kear-Scott JL, Pilipenko EV, Schwartz MH, Laskowski PR, Rojek AE, Katanski CD, Riback JA, Dion MF, Franks AM, et al. (2015) Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 162: 1286–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RW, Muhlrad D, Garcia J, Parker R (2015) Differential effects of Ydj1 and Sis1 on Hsp70-mediated clearance of stress granules in Saccharomyces cerevisiae. RNA 21: 1660–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AQ, Yu XH, Mao Y, Liu Y, Liu GQ, Liu YS, Niu XL (2015) Overexpression of a small heat-shock-protein gene enhances tolerance to abiotic stresses in rice. Plant Breed 134: 384–393 [Google Scholar]
- Waters ER. (2013) The evolution, function, structure, and expression of the plant sHSPs. J Exp Bot 64: 391–403 [DOI] [PubMed] [Google Scholar]
- Waters ER, Aevermann BD, Sanders-Reed Z (2008) Comparative analysis of the small heat shock proteins in three angiosperm genomes identifies new subfamilies and reveals diverse evolutionary patterns. Cell Stress Chaperones 13: 127–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters ER, Vierling E (1999) The diversification of plant cytosolic small heat shock proteins preceded the divergence of mosses. Mol Biol Evol 16: 127–139 [DOI] [PubMed] [Google Scholar]
- Weber C, Nover L, Fauth M (2008) Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J 56: 517–530 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Chen H, Chu P, Li Y, Tan B, Ding Y, Tsang EW, Jiang L, Wu K, Huang S (2012) NnHSP17.5, a cytosolic class II small heat shock protein gene from Nelumbo nucifera, contributes to seed germination vigor and seedling thermotolerance in transgenic Arabidopsis. Plant Cell Rep 31: 379–389 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.