Physiological and gene expression analyses across field and greenhouse experiments highlight diverse gene expression patterns that produce physiologically similar responses to soil water deficits.
Abstract
Identifying the physiological and genetic basis of stress tolerance in plants has proven to be critical to understanding adaptation in both agricultural and natural systems. However, many discoveries were initially made in the controlled conditions of greenhouses or laboratories, not in the field. To test the comparability of drought responses across field and greenhouse environments, we undertook three independent experiments using the switchgrass reference genotype Alamo AP13. We analyzed physiological and gene expression variation across four locations, two sampling times, and three years. Relatively similar physiological responses and expression coefficients of variation across experiments masked highly dissimilar gene expression responses to drought. Critically, a drought experiment utilizing small pots in the greenhouse elicited nearly identical physiological changes as an experiment conducted in the field, but an order of magnitude more differentially expressed genes. However, we were able to define a suite of several hundred genes that were differentially expressed across all experiments. This list was strongly enriched in photosynthesis, water status, and reactive oxygen species responsive genes. The strong across-experiment correlations between physiological plasticity—but not differential gene expression—highlight the complex and diverse genetic mechanisms that can produce phenotypically similar responses to various soil water deficits.
Crop productivity and wild plant distributions are governed by the availability of soil moisture (Axelrod, 1972; Boyer, 1982; Ciais et al., 2005). The impact of drought and soil water deficit in agriculture is estimated to be the largest abiotic determinant of yield (Boyer, 1982; Araus et al., 2002), while drought is also considered a primary cause of speciation and adaptation in nature (Stebbins, 1952). Dehydration avoidance and other drought adaptive strategies permit plants to survive or maintain growth during periodic droughts (Blum, 1996; Chaves et al., 2003; Chaves and Oliveira, 2004). Specifically, phenotypic plasticity of stomatal conductance, water foraging, and growth traits (among many others) may effectively maintain homeostasis of leaf water potential despite soil water deficits.
Leaf water potential is a bellwether of the physiological impact of water deficit (Jones, 2007). Under drought, decreasing water availability results in reduced leaf water potentials and a sequence of physiological responses including reduced photosynthesis, growth rate, and ultimately, fitness (Taiz and Zeiger, 2014). Plants therefore seek to maintain homeostasis of leaf water potential, with the highest (least negative) values supporting the most efficient functioning of photosynthesis and other metabolic processes in most species (Lawlor and Fock, 1978; Turner and Begg, 1981; Kramer and Boyer, 1995; Cornic and Massacci, 1996; Jones, 2007). Plants that exhibit dehydration avoidance strategies compensate for soil water deficit through phenotypic plasticity of gene expression (Verslues et al., 2006; DesMarais and Juenger, 2010; DesMarais et al., 2013; Lovell et al., 2015) and downstream physiological phenotypes (Levitt, 1980), among others.
To understand plant stress responses, it is critical to determine the physiological and genetic underpinnings of drought adaptation in both field and laboratory conditions (Travers et al., 2007; Gaudin et al., 2013). A common finding among such studies is that physiological and gene expression responses to drought vary considerably depending on the severity and temporal dynamics of drying soil (Chaves et al., 2003; Barker et al., 2005; Malmberg et al., 2005; Mittler, 2006; Mishra et al., 2012). Natural soil moisture variation, which has shaped adaptive responses to drought in wild populations, is not necessarily recapitulated by controlled (often, “shock”) laboratory experiments. For example, single abiotic stresses rarely occur in isolation in the field (Mittler, 2006). Instead, wild and crop plants respond to the combination of diverse stressors such as drought, heat, and salinity, simultaneously and at both molecular (e.g. Rizhsky et al., 2002; Rizhsky et al., 2004; Suzuki et al., 2005) and physiological (e.g. Heyne and Brunson, 1940; Craufurd and Peacock, 1993; Machado and Paulsen, 2001) levels. Therefore, inquiries into evolved plant stress responses are perhaps best served by experimental conditions that emulate selective agents in the field. Given that the extent and severity of stress causes qualitatively different physiological responses, it is not surprising that several studies have found relatively weak genetic correlations between laboratory phenotypes and those collected in the field (e.g. Weinig et al., 2002; Malmberg et al., 2005; Anderson et al., 2011; Mishra et al., 2012).
Soil properties and biota can also affect plant growth and physiology (Meisner et al., 2013; Schweitzer et al., 2014), which may be exacerbated by contrasts between growth in potting mix or in native soil (Rowe et al., 2007; Heinze et al., 2016). The observation that field-grown plants have different root systems and greater total water storage than those in greenhouse pots is of particular importance to water relations (Poorter et al., 2012a). Short-term drought stress in the field may be buffered by access to larger volumes of soil and more complex root-soil-water dynamics, conditions poorly represented in most controlled settings.
The field of experimental design has been fundamentally shaped by a central problem of biology: that it is notoriously difficult to control environmental factors in the field (Jones, 2013). A classic solution is to increase biological replication, but this is generally not feasible with costly and time-sensitive physiological and genetic assays (Poorter et al., 2012b; Marchand et al., 2013). Despite these difficulties, understanding the effects of drought in field conditions is necessary because it is in these settings that yield is impacted and selection is acting to shape adaptive responses to stress. Here, we determine how the interplay between drought severity, planting condition (e.g. field, potted, greenhouse) and sampling timing impacts physiological and genomic responses to drought in the C4 perennial grass, Panicum virgatum (switchgrass). To accomplish this, we used observations collected from clonally replicated individuals of the “AP13” switchgrass genotype (derived from the Alamo cultivar), which is the genome reference for this important biofuel crop and dominant member of mesic tall grass prairie ecosystems. The Alamo cultivar is a southern lowland accession that has high vigor and performance across a variety of climatic conditions. Replicates were grown in three separate soil moisture manipulation experiments with distinct rooting environments: in medium sized pots in a greenhouse, in large containers in a field setting, and in native soil under rainout shelters. In all three of these experiments, we collected leaf-level physiological and whole-genome gene expression data from droughted and control plants.
Combined, the three experiments represent contrasts in drought experimental manipulations (i.e. the extent, timing, and duration of drought), plant characteristics (i.e. age, maturity, and size), and broadly fit with the concepts of best practice for physiological analysis of drought responses (Poorter et al., 2012b). Contrasting these experimental design considerations allows us to address how edaphic and climactic conditions impact links between gene expression and physiological phenotypic plasticity. Specifically, we assessed three fundamental questions pertaining to physiological genomics in the field: (1) How consistent is phenotypic plasticity to drought across experiments? (2) Which soil moisture deficit responses vary across sites, years, and timing of sampling? (3) How does plasticity of physiological and gene expression phenotypes covary within and across experiments? To assess these questions, we tested how leaf physiology and whole-genome gene expression responded to the effects of drought treatments, leaf water potential, and sampling time (midday and predawn). These analyses permitted inference of the number, relative effect size, and identity of differentially expressed (plastic) genes. Overall, our results suggested that differences in leaf water potential and diurnal patterns were the major drivers of gene expression variation. Furthermore, we observed consistent physiological plasticity across greenhouse dry-down and field precipitation manipulation experiments, but extreme variability in the number of differentially expressed genes.
RESULTS
Physiological and Gene Expression Variation Across Experiments
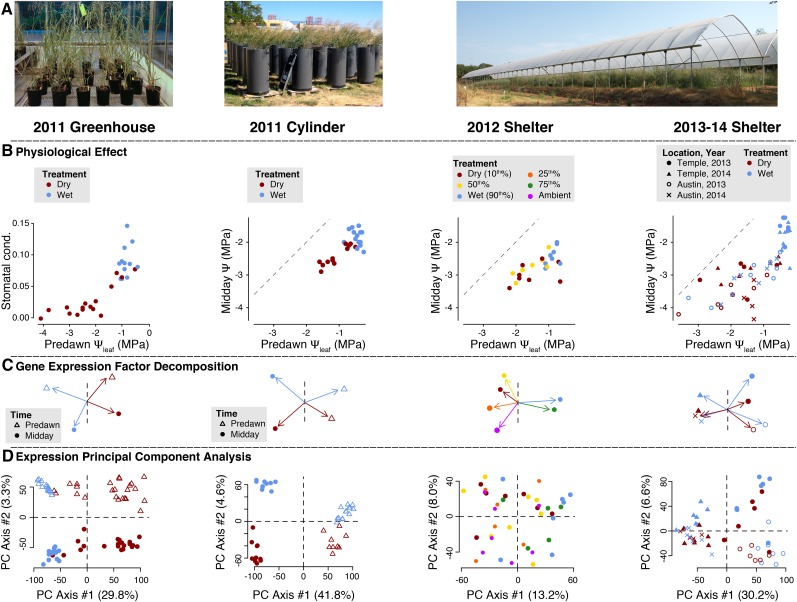
The AP13 P. virgatum accession was clonally replicated and grown in three experiments from 2010–2014: (1) a greenhouse dry-down in 3.74 L pots (“greenhouse,” expression data from this experiment was published previously, Meyer et al. [2014]), (2) 1400 L PVC cylinders in field conditions (“cylinder”), and (3) spaced plants grown directly in the field under 18 m × 73 m rainout shelters (“shelter”; Figure 1A; Aspinwall et al. [2013]). The shelter experiment was further subdivided into (3a) a 2012 experiment, where six distinct watering treatments were employed at a single field site in Temple, Texas, and (3b) experiments in 2013-2014, where two drought treatments were contrasted at sites in Austin and Temple, Texas (Fig. 1A).
Figure 1.
Physiological and gene expression responses to drought across three experiments. A, Replicates of the AP13 switchgrass genotype were grown in three separate experiments. B, Predawn leaf water potential (Ψleaf, MPa) was assessed for each plant. Midday Ψleaf measurements were paired with tissue collection for RNA for all experiments except the greenhouse, where stomatal conductance (gs) was assayed at midday instead of Ψleaf. These midday and predawn measures are plotted with independent scales for the greenhouse and remaining experiments. C, Expression matrices for genes with a significant effect of any experimental factors (time of sample collection, location and year) were used to conduct principal component analysis (PCA) decompositions. The length and direction of the vectors indicates the strength of each experimental level. A vector perpendicular to the 1st PCA axis is plotted as a dashed line. D, Finally, the principal component score for the transposed expression count matrix is plotted and grouped by the experimental factors. The percent variance explained by the first two PCA axes accompanies the axis labels. Note that in the 2012 shelter experiment expression was assayed across six treatments, but physiological phenotypes were only measured in the wet, mean, and dry treatments. Levels of replication for each experiment can be found in Table I.
Predawn leaf water potential (Ψleaf) varied considerably between watering treatments in each experiment (Table I), reflecting the physiological impacts of drought. However, plant physiology also varied with the time of sampling (cylinder and shelter experiment), location, and year (shelter experiment, Table II). Within the 2013-2014 shelter experiment, most plants grown in the Austin site exhibited more negative midday Ψleaf values than those in Temple (Fig. 1B). Consistent with reductions in midday Ψleaf, we also observed plasticity of both photosynthetic rate (A) and stomatal conductance (gs); both physiological parameters significantly declined in the drought treatments of the 2012 shelter, cylinder, and greenhouse experiments (Table II).
Table I. Summary of experiments and the effects of drought treatments.
Sample sizes (n) and mean leaf water potentials are displayed for each treatment and experiment. Accompanying each experimental factor is the number of differentially expressed genes (n DE) due to the wet-dry treatment contrast therein. *Treatment replication in 2012: dry = 7; 25th = 6; mean = 8; ambient = 7; 75th = 5; wet = 7.
| Experiment | Location | Leaf Taken At | n Wet | n Dry | Ψωεt | Ψdry | Ψ Diff. | n DE |
|---|---|---|---|---|---|---|---|---|
| Greenhouse | Greenhouse | Predawn | 14 | 24 | −0.89 | −2.42 | 1.53 | 6623 |
| Midday |
13 |
21 |
NA |
NA |
NA |
5918 |
||
| Cylinder | 1.4 m3 Cylinders | Predawn | 14 | 9 | −0.45 | −1.09 | 0.64 | 3285 |
| Midday |
10 |
10 |
−1.88 |
−2.43 |
0.55 |
5745 |
||
| 2012 Shelter |
Field |
Midday |
* |
* |
−1.32 |
−2.36 |
1.04 |
887 |
| 2013-2014 | Field | Austin, 2013 | 7 | 6 | −3.03 | −3.63 | 0.6 | 0 |
| Austin, 2014 | 7 | 4 | −3.1 | −3.79 | 0.69 | 4 | ||
| Temple, 2013 | 7 | 6 | −1.57 | −2.91 | 1.34 | 319 | ||
| Temple, 2014 | 7 | 7 | −2.04 | −2.77 | 0.73 | 154 |
Table II. ANOVA statistics from models fitting experimental treatments to physiological response variables.
Time effects were not estimable for conductance (gs) and photosynthetic rate (A), which were only measured at midday in the cylinder and greenhouse experiments. Timing of Ψleaf measurements are defined by subscripts. TypeIII F-statistics and P-values are presented along with absolute effect size, which is the proportion of differences between group means and the overall mean. Only a subset of all absolute effect sizes are presented and are indicated by the following: *Predawn, **Midday; 'Temple 2013, ''Austin 2013, ^Wet- Dry, ^^Mean- Dry
| Experiment | TypeIII Term | Phenotype | df | Abs. Effect Size | F | P |
|---|---|---|---|---|---|---|
| Cylinder | Treatment | gs | 1 | 0.99 | 78.85 | <0.001 |
| Cylinder | Treatment | A | 1 | 0.92 | 60.70 | <0.001 |
| Cylinder | Treatment | Ψ | 1 | *0.91, **0.93 | 79.72 | <0.001 |
| Cylinder | Time | Ψ | 1 | 273.9 | <0.001 | |
| Cylinder | Treatment:Time | Ψ | 1 | 0.268 | 0.6080 | |
| Greenhouse | Treatment | gs | 1 | 0.81 | 43.24 | <0.001 |
| Greenhouse | Treatment | A | 1 | 0.88 | 80.20 | <0.001 |
| Greenhouse | Treatment | Ψpredawn | 1 | 0.72 | 24.65 | <0.001 |
| Shelter 2012 | Treatment | gs | 2 | ^1.09, ^^0.17 | 19.66 | <0.001 |
| Shelter 2012 | Treatment | A | 2 | ^2.10, ^^0.52 | 19.43 | <0.001 |
| Shelter 2012 | Treatment | Ψmidday | 2 | ^0.20, ^^0.08 | 36.98 | <0.001 |
| Shelter 2012 | Treatment | Ψpredawn | 2 | ^0.48, ^^0.01 | 10.88 | 0.0010 |
| Shelter 2013-2014 | Treatment | Ψmidday | 1 | '0.48, ''0.22 | 49.89 | <0.001 |
| Shelter 2013-2014 | Location | Ψmidday | 1 | 17.85 | 0.0063 | |
| Shelter 2013-2014 | Year | Ψmidday | 1 | 2.545 | 0.1185 | |
| Shelter 2013-2014 | Treatment:Location | Ψmidday | 1 | 4.710 | 0.0357 | |
| Shelter 2013-2014 | Treatment | Ψpredawn | 1 | '1.21, ''0.56 | 20.48 | <0.001 |
| Shelter 2013-2014 | Location | Ψpredawn | 1 | 1.682 | 0.2425 | |
| Shelter 2013-2014 | Year | Ψpredawn | 1 | 1.680 | 0.2025 | |
| Shelter 2013-2014 | Treatment:Location | Ψpredawn | 1 | 7.149 | 0.0107 |
Both the physical effects of water deficit and genetic control of gene expression may drive physiological plasticity. To assess the extent of genetic responses to drought, we quantified gene expression in mature leaves in each experiment using the previously described high-throughput “TAG-seq” protocol (Meyer et al., 2011). We evaluated how distinct each treatment was with respect to the expression data using principal component analyses (PCA; Fig. 1C) and visualized PCAs of the transposed expression matrix to depict the position of each library within genetic space (Fig. 1D). Among experiments with multiple experimental factors (excluding the 2012 shelter), the first PCA axis delineated treatment differences only in the greenhouse experiment. Diurnal patterns were the strongest drivers of gene expression variation in the cylinder experiment, but were secondary to the drought treatment in the greenhouse (Fig. 1C). In the 2013-2014 shelters, differential expression across years—and to a lesser degree, sites—dominated (Fig. 1, C and D). Like in the cylinders, drought effects seemed to contribute a small proportion of expression variation in the 2013-2014 shelters. In the 2012 shelter experiment, PCA analyses clearly clustered the two wetter treatments (“wet” and “75%”) away from the drier treatments (Fig. 1C); however, gene expression variation within treatments was considerable (Fig. 1D). These results demonstrated that differential expression to drought stress was strongest in the greenhouse and least observable in the field. Consistent with the multivariate analysis of gene expression variation (Fig. 1, C and D), we observed many more genes with significant drought-induced differential expression in the greenhouse than the field (Table I).
Phenotypic and Gene Expression Assays Are As Precise in the Field As in the Greenhouse
We observed ∼5500 more differentially expressed genes (between wet and dry treatments) in the greenhouse and cylinders than we did across the shelter experiments (Table I). Here, we qualified differential expression as any case where the FDR-corrected P-value of the linear model exceeded alpha = 0.05. It is important to note that sample sizes were not identical across experiments (Table I), which may alter the power to detect differential expression. To determine if our results were biased by sample size inconsistencies, we subsampled individuals in each experiment to generate rarefaction curves. These analyses demonstrated that the observed differences among experiments were not artifacts of statistical power (Supplemental Fig. S1). This massive difference in signal was not mirrored in the physiology data, where predawn Ψleaf values varied strongly between treatments and were of similar magnitude in the greenhouse and the field (Table II). Furthermore, when both experiments were fit in a single model, there was only a marginally significant treatment-by-experiment interaction (F = 2.48, df = 4, P = 0.06). This indicates that drought treatments elicited Ψleaf responses in the same direction and with similar effect sizes across experiments.
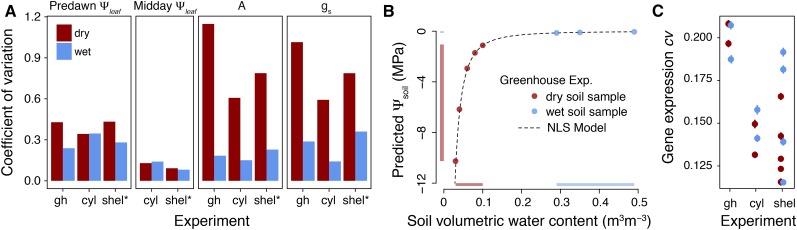
When comparing treatments, Ψleaf was more variable among droughted plants than those in wet conditions (Fig. 1B). The coefficient of variation (cv) within experiments and treatments confirms this observation (Fig. 2A); cv of dry treatment predawn Ψleaf was on average 58% greater than that of the wet treatment. Increased variability of Ψleaf in drought plants may be due to the physical properties of drying soils. Plants perceive soil moisture as total soil water potential (Ψsoil); however, as soils dry, Ψsoil exponentially declines. We modeled the progression of soil moisture decline from observed values of soil volumetric water content and Ψsoil in the greenhouse (Fig. 2B). The range of Ψsoil predictions in the dry treatment (10.3–1.1 MPa) was much greater than in the wet treatment (0.14–0.05 MPa) despite a narrower range of volumetric water content measurements (see marginal line segments in Fig. 2B). As such, implementing a consistent drought treatment in terms of Ψsoil was difficult, even in the greenhouse experiment. Our physiology data clearly mirrors the soil water potential measures: there is much more variability in the drought treatment than in well-watered conditions across all experiments (Fig. 2A).
Figure 2.
Physiological, soil and normalized expression variability across experiments and treatments. A, Coefficients of variation (cv = sd / mean) were calculated for each physiological and gene expression phenotype. Raw cv for each physiological phenotype is plotted. B, To understand the relationships between Ψsoil and soil volumetric water content, we conducted soil moisture release curves for the greenhouse potting soil where a soil sample was progressively dried and volumetric water content and Ψsoil were repeatedly measured; the range of observations for each treatment are presented by the marginal line segments. C, Mean (± se) cv across all expression traits is plotted. The experiments are abbreviated as shelter (shel), greenhouse (gh), and cylinder (cyl).
We examined the variability of our measurements of physiology and expression and (surprisingly) found that those taken in the field were less variable than those in the greenhouse. In 3/4 of the physiological phenotypes, cv in the drought treatments was greatest in the greenhouse and lowest in the cylinders. The cv of the fourth physiological phenotype, predawn Ψleaf, was nearly identical in the greenhouse (0.33), cylinder (0.34), and shelter (0.36) experiments (Fig. 2A). The cv among normalized expression phenotypes largely recapitulated the physiology data (Fig. 2C): the greenhouse experiment produced the most variable data, while the large cylinders displayed the least. The field planted individuals had similarly variable expression phenotypes as those in the cylinders (Fig. 2C).
The Effects of Drought Treatments Are Modulated by Time of Sampling
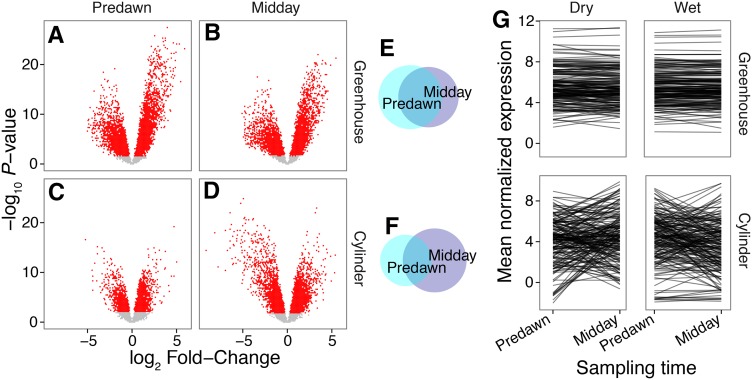
Time of sampling (predawn and midday) in the greenhouse and cylinder experiments had a substantial effect on the expression patterns of many genes (Fig. 1, C and D); however, these effects varied across experiments. For example, in the cylinder experiment, 47% (11880) of all genes differentially responded to sampling time, compared to only 30% (5278 genes) in the greenhouse (Fig. 3, A–D). These results revealed reduced diurnal expression regulation in the greenhouse, compared to plants grown in the field in cylinders.
Figure 3.
Differential gene expression due to soil water deficit is affected by the time of sampling. A–D, Differential expression between treatments was characterized via “volcano” plots, where the log2 fold change of treatment contrasts is plotted on the horizontal axis and the P-value of the associated test is on the vertical. Points were colored by whether the FDR-corrected P-value exceeded alpha = 0.05 threshold. E and F, The total number of significant genes for each of the four contrasts were plotted in Euler diagrams, where disc size is proportional to the number of genes that were significant for each treatment contrast in the greenhouse (E) and cylinder experiment (F). The corresponding number of differentially expressed genes can be found in Table I. G, To visualize the treatment*time interactions that make up these differential responses, we plotted mean normalized expression values for each of the top 100 treatment*time genes from the cylinder and greenhouse.
In addition to additive time-of-sampling effects, we detected complex interactions between drought treatments and sampling time. Drought treatment effects were directly modulated by time of sampling (treatment-by-time interactions in the linear model) in 8.4% (2,125) and 4.6% (805) of genes in the cylinder and greenhouse experiments. While expression patterns were generally conserved between predawn and midday sampling in the greenhouse (Fig. 3, A and B), the effect of the drought treatment strengthened from predawn to midday in the cylinder experiment (Fig. 3, C and D). Indeed, 67% of drought-responsive genes at midday were significantly differentially expressed in the same direction in the predawn sampling in the greenhouse (Fig. 3E), but only 30% of genes followed this pattern in the cylinders (Fig. 3F). Interestingly, the effect sizes of drought treatments were similar (mean = 2.4% smaller) at midday versus predawn in the greenhouse but 35% greater in the cylinders (Fig. 3, A–F). Such diurnal-by-treatment interactions were much stronger in the cylinders than the greenhouse. Specifically, there tended to be much stronger differential expression across sampling times in wet treatments than dry (Fig. 3, G and H; Supplemental Fig. S2). This effect was strongest in the greenhouse, where the drought treatment was extreme (Fig. 1B). Indeed, in the greenhouse, > 1.7× more genes were diurnal-regulated in wet than in dry conditions (4,544 vs. 2,548, respectively, Supplemental Fig. S2). It is possible that the extreme nature of the drought treatment in the greenhouse caused the cessation of diurnal gene regulation.
Finally, we also detected a small influence of the precise time at which plants were sampled (i.e. the order that a leaf was harvested for RNA extraction) on gene expression in the field conditions (Supplemental Fig. S3). The order of sampling was a significant predictor of Ψleaf variation in 2013-2014 (r2 = 0.12, P = 0.002), but not 2012 (r2=0.01; P > 0.1). Likewise, 21 genes differentially correlated with sampling order in 2012, but 125 did so in 2013-2014.
Paired Gene Expression and Physiology Permits Inference of Drought Effects in Variable Environments
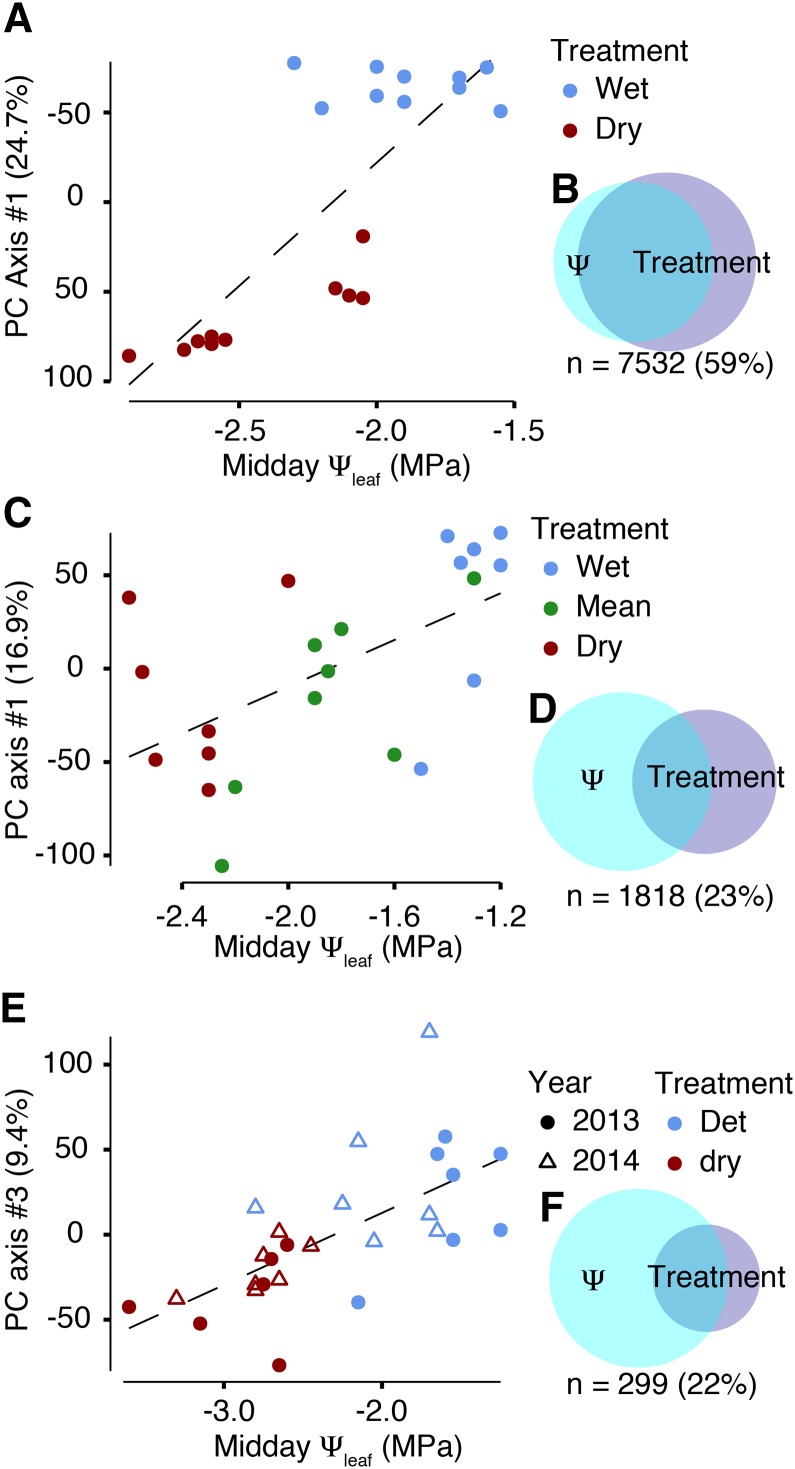
So far we have presented statistical tests between discrete watering treatments; however, due to environmental heterogeneity within soil moisture treatments (Fig. 1B), it may be more powerful and biologically relevant to look at associations between a metric of stress (e.g. Ψleaf) and physiological or gene expression phenotypic responses. Therefore, we augmented our previous comparisons with regressions of gene expression against Ψleaf measurements for all three of our experiments. In the cylinder experiment, the effects of treatment were largely recapitulated by regressing leaf water potential on expression data. For example, within the midday sampling, 83% of genes significantly associated with water potential were also detected by a contrast between treatments (Fig. 4, A and B; Table III). This consistency reflected the strong experimental effects observed during the midday harvest (Figs. 1B and 4A).
Figure 4.
Gene expression variation associated with leaf water potential. The cylinders, and the shelter experiments represent the experiments that have paired midday Ψleaf and expression assays. Principal components (PC) from the complete gene expression matrix were calculated. Of the top three PC axes, the one which is most strongly explained by midday Ψleaf is plotted. A paired Euler diagram displaying the total amount of genes differentially expressed due to treatment accompany the PCA- plots for the cylinder (A and B), 2012 experiment (C and D) and 2013-14 shelter (E and F) experiments. The total number of genes presented can be found in Table III.
Table III. The number of significant genes in each experiment.
Treatment (“Trt.”) and Ψleaf at midday sampling were characterized for 2012, 2013-14 and cylinder experiments. The number of significant genes were determined (α = 0.05) for two models: (1) ∼ treatment, (2) ∼ Ψleaf. For consistency, these models were only fit at midday for the cylinder and greenhouse data and within each site for the 2013-14 shelters.
| Experiment | Trt. | Ψleaf |
|---|---|---|
| 2012 | 887 | 965 |
| 2013-14 | 727 | 1758 |
| Temple | 665 | 309 |
| Austin | 3 | 160 |
| Cylinder at midday | 6365 | 5323 |
| Greenhouse at midday | 5584 | — |
In contrast, the regression approach and factorial treatment contrasts produced different outcomes in the shelters: fitting midday Ψleaf across treatments in 2012 increased the number of significantly differentially expressed genes by 70% (1,510 total genes compared to 887). Additionally, 965 genes responded across the gradient of midday Ψleaf, 623 (85.7%) of which were significant only when Ψleaf was the predictor (Table III). In these field experiments where a more continuous range of precipitation treatments were applied, significant gene expression across drought stress intensities was not fully captured by treatment of precipitation levels as factorial variable.
The increased power of midday Ψleaf relative to discrete treatment variables was even more pronounced in the 2013-2014 experiments, where 1,758 genes were significantly differentially expressed across Ψleaf but only 727 genes were differentially expressed across treatments (a 142% increase, Fig. 4, E and F). It is possible that this across-site effect was due to differential site characteristics, including soil quality and nutrient availability, and not drought per se. To further examine differences between the Temple and Austin shelter experiments, we split the 2013-2014 dataset by site and reanalyzed differential expression. Within sites, we observed differential expression across drought treatments among 665 genes in Temple, but only 3 in Austin. However, water potential explained differential expression of 309 (1.1×) and 160 (53×) additional genes in Temple and Austin respectively (Table III). These results indicate that utilizing measurements of physiological variation can account for expression variation that is not predicted by treatment factors alone.
Leaf-Level Physiological Responses, But Not Differential Gene Expression, Are Highly Correlated Across Experiments
The majority of genomic studies of drought have been conducted in highly controlled laboratory or greenhouse settings, which are intended to elucidate the patterns and processes of drought responses in the field. Here, we find broadly different characteristics of drought response in the shelters from those in either cylinder or greenhouse conditions. For example, across all differentially expressed genes the absolute effect size (mean absolute log2 fold change) was 75% –148% greater between treatments in the greenhouse than in the 2012 and 2013-2014 shelter experiments and the cylinder experiment; furthermore, more genes significantly responded to the drought treatments of the greenhouse (6597 genes) and cylinders (4,489) than either the 2012 (887) or 2013-2014 shelters (752). These results indicated that, despite similar phenotypic responses, field-grown plants displayed much weaker gene expression plasticity than potted greenhouse plants.
Such weaker responses could be at the gene level, in which case the direction and effect size of differential expression in the greenhouse should be highly predictive of that in the field. Alternatively, entirely different genetic responses may be present in the field. In many cases, we found strong correlations between expression in the field and the greenhouse, but r2 between log2 fold changes in the greenhouse and field never exceeded 0.35 (Supplemental Fig. S4), indicating that much of the drought responsive expression in the field was not predicted by that in the greenhouse.
While greenhouse responses to drought did not entirely predict those in the field, drought-responsive genes in the field were very clearly differentially expressed in the greenhouse. Of the 716 genes that were differentially expressed in the field and quantifiable in the greenhouse, 549 (76.7%) were also differentially expressed in the greenhouse, representing a highly significant enrichment of overlapping genes (odds ratio = 3.1, P < 0.0001, Supplemental Table S1). Furthermore, despite relatively weak predictive power across experiments, the genes found in each experiment were much more likely to be found in other experiments than would be expected by chance. In the extreme case, the overlap between significant genes in the 2012 and 2013-2014 shelter experiments is 7.1× greater than the null expectation (Supplemental Table S1). Combined, these results provide a potentially surprising result: the relatively uncontrolled field environment offered the strictest test of differential expression.
Genes and Gene Functions Related to Drought
While > 104 genes displayed phenotypic plasticity to drought, only 546 were differentially expressed in three or more of our experiments (84 were found in all experiments). Interestingly, with few exceptions, these “core” genes were consistently up- or down-regulated across all experiments (Supplemental Fig. S5). Among this set, 460 genes were homologs of annotated Arabidopsis (Arabidopsis thaliana) genes. However, of the 84 genes that were drought-responsive in all experiments, nine had no annotation, six were annotated only by protein domain/motif or general process, and two were annotated as “protein of unknown function” (PUF). These data suggest that the majority of drought-responsive genes belong to a well-known group of genes with specific assigned functions in plant biology.
As expected, we detected a significant enrichment of genes related to responses to stress and water (P < 0.0001) among the core set of genes (Supplemental Table S2). These genes included homologs of dehydrins, LEA-type proteins, aquaporins, ascorbate peroxidase, and other genes related to reactive oxygen species (ROS) detoxification, and abscisic acid (ABA)-responsive phosphatases and transcription factors. LEA proteins, dehydrin, and aquaporins are induced by drought and involved in desiccation tolerance (Supplemental Table S2 and references therein). Among the ABA-responsive gene families were PP2C genes (ABI1-2) four members of the AP2 family of transcription factors, including DREB1-2, and an ABA-responsive element-binding element (AREB). In addition, we detected genes coding GST, l-ascorbate peroxidase, ascorbate oxidoreductase, and other genes involved in reactive oxygen radical detoxification. ROS are known to accumulate during many biotic and abiotic stresses, and defense against ROS appears to be a common mechanism during drought (Supplemental Table S2 and references therein). Interestingly, two homologs of NCED9, a key enzyme in ABA biosynthesis, were also present in this core set of drought responsive genes (Supplemental Table S2 and references therein).
Other GO drought-related annotation categories were enriched in the core set of genes, including photosynthesis (which was the most highly enriched GO term, P = 9.1 × 10−9) and several annotations related to oxidation-reduction status. We detected 7 genes encoding light-harvesting complex II and many others related to C4 photosynthesis, including two Ala aminotransferases, a phosphoenolpyruvate carboxykinase, and two malate dehydrogenases. Additionally, P5CS, which encodes a key enzyme in the biosynthesis of the osmoregulator proline, was also present in the identified list of “core” drought-responsive switchgrass genes.
In the cylinder and greenhouse experiments we confirmed that genes affected by time of sampling (predawn vs. midday) were enriched for GO terms related to circadian, or light-responsive, annotations. Photosynthesis and “regulation of circadian rhythm” annotations were some of the most overrepresented categories (Supplemental Table S3).
DISCUSSION
To test the consistency of physiological and gene expression plasticity to soil moisture variation, we exposed clones of a single switchgrass genotype to drought treatments in the greenhouse and field. Pairing physiological measurements with detailed analyses of the genes that respond to drought revealed similar physiological responses but qualitatively different patterns of molecular plasticity in the field than the controlled edaphic environments of the greenhouse and cylinders. For example, plants grown in small pots in the greenhouse displayed similar leaf water potential plasticity to plants grown in the field but ∼10× more differentially expressed genes. Combined, these data indicated that many fewer differentially expressed genes were responsible for similar physiological plasticity in the field than the greenhouse.
Comparison of Soil Water Deficit Manipulations across Experiments
Soil moisture manipulation experiments are generally practiced as “dry-downs” where watering is limited or ceased and potted plants experience attenuated soil moisture. Such experiments are the basis of much of the molecular understanding of drought physiology. Alternatively, ecologists and agronomists often test drought physiological responses through precipitation exclusion (or irrigation supplementation) treatments that persist through much of the growing season, or even across years. Both approaches may be ecologically realistic. For example, many annual species grow and reproduce with the water remaining from a single rainfall event, mimicking the progressive dry-down approach. However, crop breeders or climate change biologists may seek to understand how fitness and yield can be maintained during relatively dry growing seasons across drought years. This type of “press” drought treatment imposed in the field is known to elicit different physiological, gene expression (Barker et al., 2005), and ultimately community-scale responses relative to “pulse” droughts such as those imposed by our cylinder and greenhouse experiments (Hoover et al., 2015; Hoover and Rogers, 2016).
The analyses presented here contrast not only the dry-down and field-scale approaches, but also different climatic (air temperature, vapor pressure deficit, photosynthetic photon flux density [PPFD], etc.) and edaphic (soil water retention, particle size, biotic interactions, etc.) characteristics. For example, the highly controlled aerial environment of the greenhouse elicited weaker diurnal air temperature and vapor pressure deficit progressions compared to plants grown in larger cylinders in the field (Fig. 3; Meyer et al. 2014). Consequentially, diurnal patterns of gene expression dominated drought-responsive expression profiles in the cylinders but were significantly weaker in the greenhouse, especially with regard to drought-responsive genes. Similar patterns, which have been observed among physiological characters, may be attributable to light-responsive stress pathways including phytochemical quenching cycles (Chaves et al., 2003; Mishra et al., 2012). Such temporal variation is intentionally dampened in controlled environments, such as the greenhouse or growth chamber. However, many drought responsive genes (e.g. PSI and PSII and other light-responsive pathways) are expected to modulate expression between predawn and midday conditions. As such, the cylinder experiment offers a compromise. While permitting tight control of the soil environment by using a common soil medium and uniform drying, the cylinders exposed plants to natural varying climatic features like temperature, vapor pressure deficit (VPD), and changing photoperiod cycles. Our results demonstrated that the realistic aerial environment of the cylinders increased statistical power to define the diurnal-by-drought interactions that are critical to soil moisture deficit responses (Fig. 4).
In the shelters, irrigation manipulations were implemented at the plot level over the course of multiple years. Such long timescales provided time for plants to acclimate not only to the drought treatments, but also to the edaphic characteristics of each site. Indeed, among-site differences were a major driving variable in the 2013-2014 shelter experiment. Most plants in Austin exhibited lower (more negative) midday Ψleaf than any plants in Temple (Fig. 1B). Despite similar irrigation levels, both wet and dry treatments in Austin (2014) clustered with the Temple (2014) dry treatment (Fig. 1C). It is likely that plants grown in the shallow, rocky-clay soil of the Austin site (∼0.2 m depth) experienced substantial water deficits even in the wettest irrigation treatments, while deeper soils at the Temple site (∼5 m depth) would have provided much greater buffering capacity against drought. As such, it is not necessarily surprising that we did not observe many significantly drought-induced gene expression responses at the Austin site.
Synthesis of Drought Responses across Experiments: What Factors Led to Physiological, but Not Expression, Plasticity in the Field?
Field experimentation is typically thought to require greater replication because increased environmental heterogeneity of field conditions (e.g. variation in soils, microclimate, timing of sampling, etc.) may produce more variable measurements. Our analyses generally rejected this hypothesis and demonstrated that similar ranges of environmental, physiological, and molecular heterogeneity existed within treatments across greenhouse and field sites. Instead, other factors besides residual variation must be driving the difference in gene expression—but not physiological plasticity among experiments—including (1) different mechanisms of drought responses, where short term treatments elicit different gene expression responses than long term droughts; (2) plant morphological characteristics, such as the ability to buffer soil water variation through tissue capacitance (water storage); (3) edaphic and climatic variation, such as stronger soil water potential gradients and temporally variable vapor pressure deficit in the field; or (4) physiological acclimatization.
It is possible that plants in the field have acclimated, following an initial drought responsive phase in which gene expression was similar to that of the plants studied in the greenhouse (Chaves et al., 2003). For example, the development of a larger root system may permit greater soil water foraging (e.g. Comas et al., 2013). Such acclimation responses are driven by the expression of many genes (e.g. Werner et al., 2010) and may be initiated at the time of exposure to drought. Over time, expression of such genes would no longer be required, as the necessary structures would already be in place (Maseda and Fernández, 2006). However, while acclimation may play a large role in differential physiology across experiments, it does not fully explain the massive disconnect between physiological and gene expression plasticity we observed. In concert with acclimation, different genetic pathways may modulate plant responses to press and pulse droughts. One interesting possibility is that the ecologically unrealistic shock imposed by the high rate of soil moisture reductions in the dry-down experiments may elicit programmed cell death and other shock responsive processes that are not drought-related per se. Such effects may be less important in larger field-grown plants if they are better able to buffer some of these extreme stress effects because of greater water storage and access to soil water (Maseda and Fernández, 2006). Genes that respond across different types of drought experiments seem most likely to offer the clearest picture of molecular responses to drought, as they may be less likely to represent treatment specific or shock-induced responses.
Defining Drought-Responsive Genes Across Experiments
We expected to find similar gene expression responses across experiments because several evolutionarily conserved (Rabbani et al., 2003) pathways are responsible for drought acclimatization, including cell signaling, transport and communication; plant hormone metabolism; photosynthesis; and carbohydrate biosynthesis (Schafleitner et al., 2007). However, the types of genes that responded to drought treatments varied considerably across experiments (Supplemental Table S3). This observation is not necessarily surprising, given the physiological differences between greenhouse- and field-grown plants (Chaves et al., 2003; Mittler, 2006; Mishra et al., 2012). For example, photosynthesis and energy production genes dominated the greenhouse experiment, whereas metabolism biosynthesis and stress response genes were less abundant. Similar observations have previously been reported in sunflower (Rengel et al., 2012) among other species. In contrast, longer term drought treatments disproportionally induced genes involved in other biological processes, such as membrane biogenesis, redox mechanisms, cellular biosynthesis, and metabolism (Table IV; Supplemental Tables S2, and S3; Des Marais et al. 2012). In addition, while many transcription factors or genes related to DNA metabolism were previously found in the greenhouse dataset alone (Meyer et al., 2014), very few of them were identified as significant across all four drought experiments (Table IV; Supplemental Table S2). It is possible that changes in transcription factor expression levels resulted in substantial but short-term effects in plants that experienced sudden drought in a greenhouse setting. By contrast, these effects may have been dampened in well-established and acclimatized plants in the field.
Table IV. List of “core” switchgrass genes enriched in ≥ 3/4 experiments and with known homologs involved in drought response.
The number of genes found in the core list is presented. For specific genes, references and additional information, see Supplemental Table S2.
| Functional Category | Predicted Gene Function | No. of Genes |
|---|---|---|
| Water stress response |
Dehydrins | 4 |
| LEA proteins | 3 | |
| Aquaporin |
1 |
|
| Cell rescue, abiotic stress response, and senescence |
DnaJ-like molecular chaperones | 4 |
| Other chaperones | 6 | |
| Senescence | 2 | |
| Response to biotic and abiotic stress | 3 | |
| chitinase | 1 | |
| multidrug resistance |
1 |
|
| ROS detoxification |
Glutathione S-transferase | 2 |
| l-ascorbate peroxidase | 1 | |
| ascorbate oxidoreductase |
1 |
|
| ABA response pathway |
Protein phosphatase 2C | 13 |
| AREB factor | 1 | |
| NCED9 | 2 | |
| ABA/WDS induced protein |
5 |
|
| Transcription factors |
Homeobox family | 4 |
| AP2 domain | 4 | |
| Heat shock responsive TF | 1 | |
| MYB family | 4 | |
| Zink finger family | 10 | |
| CCAAT-binding factor | 1 | |
| MADS box |
2 |
|
| Cell signaling |
Protein kinases | 23 |
| Osmotic stress potassium transporter | 1 | |
| GTP-binding | 2 | |
| Ca2+-binding transmembrane protein |
1 |
|
| C4 Photosynthesis |
Auxin response | 2 |
| Light harvesting | 7 | |
| Ala aminotransferase | 2 | |
| Phosphoenolpyruvate carboxykinase | 1 | |
| Malate dehydrogenase |
3 |
|
| Metabolism | Pro biosynthesis | 1 |
| Suc synthase | 2 | |
| β-amylase | 3 |
While transcriptional drought responses of AP13 plants varied substantially among different experiments, we detected a set of “core” responsive genes that were enriched in all four (84 genes) or 3/4 (n = 546) experiments (Supplemental Table S2). This list of genes provided a set of candidate pathways necessary to confer water deficit responses in switchgrass. Homologs of many of these genes have been documented as drought related in other species. Included in this list were dehydrin (Lopez et al., 2003) and DnaJ chaperones, which are induced under drought in many plants and may contribute to better performance under water stress (Seki et al., 2002; Nguyen et al., 2004). We found homologs of two NCED9 loci, which are key enzymes in the ABA biosynthesis pathway (Lefebvre et al., 2006) and a broad range of ABA responsive enzymes and transcription factors. These included homologs of AREB, two PP2C genes (ABI1-2), and several AP2-binding transcription factors (DREB1-2). Interestingly, AREB transcription factors have been implicated in drought-responsive transregulatory divergence in P. hallii, a close relative of switchgrass (Lovell et al., 2016). Furthermore, ABI1-2, are known to be transcriptionally up-regulated in response to ABA and control responses to drought, heat shock, and oxidative stress (Vranová et al., 2000; Merlot et al., 2001; Schweighofer et al., 2004; Schafleitner et al., 2007). DREB (drought responsive element binding proteins) and other members of the AP-2 binding gene family represent some of the best documented regulators of ABA-dependent and -independent drought responsive transcription regulatory elements (Liu et al., 1998).
It is well-documented that reduced transpiration, which accompanies drought acclimatization, may result in increased leaf temperature, light damage, and a need for transcriptional responses to both heat (Bogeat-Triboulot et al., 2007; Swarbreck et al., 2011) and ROS stress (Smirnoff, 1993; Schafleitner et al., 2007). Interestingly, we observed many genes annotated to these abiotic stress responses, including heat shock-responsive transcription factors, which corroborates the previously proposed link between thermal defense and drought response in plants (Feder and Hofmann, 1999; Meyer et al., 2014). In addition, we detected genes coding for GST, l-ascorbate peroxidase, ascorbate oxidoreductase and others involved in active oxygen radical detoxification.
Finally, we found genes from many of the a priori drought-responsive candidate groups; however, there were a few notable sets of drought-acclimatization genes that did not appear in our lists. Transcription factor families identified in the “core” set of drought responsive genes include members of the MYB-like, zinc finger, CCAAT-binding factors, Nuclear Factor Y (NF-Y), and MADS box. However, despite the apparent abundance of various types of transcription factors, the total number of identified genes and gene families with DNA binding activity was much smaller than detected previously in other studies (Schafleitner et al., 2007; Meyer et al., 2014). Specifically, we did not observe members of WRKY, NAM, TAF, NAC, and CPP1 (among others) families of transcription factors (Meyer et al., 2014; Rizhsky et al., 2002; Seki et al., 2002). It is possible that many of these transcription factors are currently mis- or unannotated due to the very preliminary nature of the Panicum virgatum genome assembly (84% of the core genes were annotated). WRKY and many other transcription factors are critical in early signaling but may not be differentially responsive over long term acclimation responses in the field.
Discussion of Best Practices in Field-Scale Physiological Genomics
We found that despite strong physiological and soil-moisture differences, plants in the field adjusted many fewer genes than potted plants in the greenhouse or plants grown in cylinders. It is clear that press droughts cause qualitatively different patterns of expression than dry-downs. These genetic differences between sustained and shock drought stress responses offer a challenge, but also a unique opportunity to study physiological diversity in the field. We encountered several significant barriers that are important to experiments addressing these differences. Particularly important are (1) the effect size of soil water deficit treatment differences, (2) fine-scale temporal expression differences, and (3) among-site variation.
In the 2012 shelter experiment we applied six different water treatments, but clustering significant genes by expression profile similarity clearly differentiated plants under wet (and to a lesser extent 75th percentile) water treatment from all other treatment levels. At the whole transcriptome level we observed very weak differences in gene expression among mean, dry, ambient, and 25th percentile water treatment conditions. Thus, under our 2012 experimental conditions in the field the observed differences in gene expression could have been captured by applying just two experimental water treatments: wet and dry. This result indicates that fewer, more distinct treatments with stronger within-treatment replication will result in more statistical power when using the experimental treatment as a factor level predictor. However, the stress gradient present across the six treatment levels proved useful as it provided a broader distribution of water potentials and improved power to detect gene expression plasticity to Ψleaf.
Time of sampling was an important factor across all experiments, ranging from within days to across years. While this is clear from contrasts between predawn and midday sampling, where > 10k genes were differentially expressed, we also found subtle differences between expression patterns at the beginning and end of sampling in any given experiment. For example, the 2012 shelter data presented here comes from a larger experiment with > 400 individuals in total. Overall sample collection for all plants took 2 h, from 11 a.m. to 1 p.m. Many genes showed linear changes in expression over the sampling period of 2 h (Supplemental Fig. S3). These data clearly demonstrated that the order and time of sample collection (and likely other microvariation factors) could affect gene expression in the field. However, only 21 genes showed significant effects of time of sampling in our experiments, which indicates that carefully planned and carried out experimental design (a narrow and consistent enough sampling window) can produce stable estimates of treatment effects that are not confounded with time-of-sampling microvariation artifacts. We corrected for the time of sampling by using the spatial and temporal block in which each individual was sampled as a random effect; where spatial and temporal blocking factors do not covary, correcting for sampling time alone can improve statistical power to define differentially expressed genes (Lovell et al., 2016).
Across experiments, we paired leaf water potential with gene expression assays. The use of physiology as a covariate for assessment of differential gene expression permitted inference of effects across harvests, years, sites, and even experiments. Combined, these results support the expectation that leaf water potential serves as a powerful proxy for the degree of drought stress experienced by individual plants. Since most plants strive to avoid the effects of drought by maintaining leaf water potential homeostasis, this variable may be a strong predictor of the perceived stress of the local environment. Whereas leaf water potential certainly confers greater power to detect differential expression and assessment of across-site drought response, using this variable as a predictor does not permit causal inference (Jones, 2007). Genes that are correlated with leaf water status may either respond to such decreases in water potential or may have led to the reduction of water potential in the first place (e.g. Fu et al., 2000). For example, ABA-sensitive genes in guard cells both cause variation in leaf water potential through stomatal regulation and respond directly to water potential (e.g. Tardieu and Davies, 1992; Speirs et al., 2013). Therefore, inference regarding leaf water potential as a predictor should be interpreted carefully, possibly corroborated with comparisons among treatments. The use of other metrics of plant water status, like absolute or relative water content, may also be useful in assessing the molecular impact of water-deficit treatments (Maseda and Fernández, 2006; Jones, 2007).
Despite the long-term nature of our field-scale droughts, our measures of drought response were snapshots; taken at a single time point when we perceived drought to have reached a critical point in the greenhouse and cylinders, or when field conditions were optimal for sampling in the shelters. Additionally, there may be subtle circadian-by-treatment interactions, which were not be captured by discrete predawn and midday sampling. By combining leaf physiology and gene expression measurements at these sampling points, we endeavored to gain a synthetic picture of how drought was affecting individuals at a whole-plant scale. Repeated sampling across natural progressions of soil wetting/drying might add further insights.
Conclusions
A major goal of modern plant biology is to better understand abiotic stress responses to improve crop plants—especially in the face of climate change (Ahuja et al., 2010; Tuteja and Gill, 2013). To do so requires that our fundamental understanding of physiology and molecular processes be translated from controlled greenhouse and laboratory experiments into the field. Methods to emulate field-like conditions in the laboratory or greenhouse settings have been developed as an alternative to traditional soil water dry-downs (Harb et al., 2010); however, even factorial combinations of stressors (Suzuki et al., 2014) may fail to capture the complex interplay of environmental variables experienced in the field. Furthermore, while it is crucial to relate findings of field studies with those performed under controlled conditions, only a few studies have been published that compare physiological traits and gene expression data in drought treatments in both field and greenhouse conditions (but see Rengel et al., 2012; Marchand et al., 2013).
Our study demonstrates how genes and phenotypic traits differentially respond to soil water deficit across greenhouse and field trials that impose different severity and duration of drought treatments. We find that the bulk of differentially regulated genes in the field are also found in the greenhouse. This indicates that the molecular and functional understanding of field grown plants is mirrored in the laboratory. However, a mechanistic understanding of how plants achieve similar physiological responses to drought across laboratory, greenhouse, and field experiments—while regulating expression of different and generally fewer genes in field environments -remains to be developed. A collection of studies, both linking controlled experiments to the field, and exploiting natural precipitation and drying (e.g. Kudoh, 2015) will likely provide critical steps toward achieving this goal. Finally, biological replication is critical to detecting physiological and gene expression variation in the field. New high-throughput tools for measuring both relevant physiological and genome-wide expression phenotypes, such as TAG-Seq and tractor-mounted imaging, may provide an excellent avenue with which to achieve the replication necessary to compare field and laboratory physiological genomic studies.
MATERIALS AND METHODS
Overview of Experiments
Experimental design and conditions for the shelter and greenhouse experiments have been published previously (Aspinwall et al., 2013; Meyer et al., 2014). Relevant details are briefly reiterated below and expanded in the online Supplemental Data (Supplemental Table S4). The levels of replication for each experiment are presented in Table I.
2010 Greenhouse Experiment
Plants were grown in 3.78 L pots at the University of TX Brackenridge Field Laboratory (Austin, Texas) in the greenhouse with mean daytime air temperature of 30°C and relative humidity of 65% (Meyer et al., 2014). Abundant watering was applied for the first 45 d of growth followed by complete withdrawal of water for the subsequent 14 d (experimental dry-down treatment group) or continued abundant watering (control group).
2011 Cylinder Experiment
Plants were grown outside at the University of Texas at Austin J.J. Pickle Research Facility and experienced natural lighting, photoperiod, humidity, and temperature changes. At sampling, the maximum air temperature was 37°C and relative humidity was 13%. Plants were grown in 1.22 m tall cylinders constructed from 0.61 m diameter gray schedule 40 polyvinyl chloride pipe with a wall thickness of 13 mm. Cylinders were arranged in 5 × 6 grid, spaced 1.2 m center-to-center and filled with Ranch Rose Mix (Geo Growers, Austin, TX). For the drought treatment, water was withheld from plants for 18 d while the control treatment continued to receive irrigation. The dry-down began on August 21. Plants were phenotyped and tissue was sampled on September 8–9, 2011.
2012-2014 Shelter Experiment
This experiment was designed to test the effects of multiple climatically realistic levels of precipitation and soil moisture on the drought responses of plants (Aspinwall et al., 2013). The treatments represented five sets of historical rainfall patterns (Aspinwall et al., 2016): the 10 driest years (“dry”), 25th, 50th, and 75th percentiles and the 10 wettest years (wet) at each site (Table II). An ambient precipitation treatment applied amounts falling at the site immediately after they occurred. The pattern of watering events in these treatments was produced using the stochastic weather generator, LARS-WG 5.5 (Semenov, 2007), calibrated with an 87 year precipitation record (Aspinwall et al., 2016). Such climatically relevant drought treatments provide a proxy for the stresses experienced over the recent history and short-term future climactic scenarios at these sites (Mearns et al., 2013; Knapp et al., 2015). Due to the drought that occurred during the 2012 growing season, the ambient treatment clustered closely with the driest treatments. While gene expression was collected for all treatments, physiology was only paired with the dry, mean, and wet treatments. In 2013 and 2014, expression data were only assayed for the wet and dry treatments.
Physiological Measurements
In each experiment, predawn Ψleaf was measured with a Scholander-type pressure chamber (PMS 1000, PMS Instruments Company, Oregon) at approximately 5:00 h local time on the uppermost fully expanded leaf of a tiller representative of the canopy. For collection, the leaf was excised from the tiller with a sharp pair of scissors slightly above the ligule and sealed in a Ziploc bag to prevent transpirational water loss until measurement (< 5 min). Midday Ψleaf was determined following the same protocol between 13:00 and 15:00 h local time.
Leaf net photosynthesis (A, µmol m-2s−1) and stomatal conductance to water vapor (gs, mmol m-2s−1) were measured using portable photosynthesis systems (LI-6400XT, LI-COR, Inc., Nebraska) on two adjacent uppermost fully expanded leaves from two separate tillers representative of the canopy. In each experiment, measurements of A and gs occurred between 11:00 and 14:30 h local time. In the greenhouse and rainout shelter experiments, leaves measured for photosynthesis were subsequently measured for midday Ψleaf. In the cylinder, experiment water status was determined for similar but independently sampled leaves.
Leaf Tissue Collection and RNA Sequencing
Tissue for RNA was collected from two leaves similar to those subjected to physiological measures as follows: (1) two tillers that were representative of the canopy were chosen, (2) the uppermost fully expanded leaves from each tiller were excised at the ligule, (3) 2 cm of the proximal portion of the excised leaf were separated from the midrib, and (4) both leaf samples were combined in a single 2 mL Eppendorf tube loaded with three stainless steel beads, immediately frozen in liquid nitrogen and transported on dry ice to the laboratory. Tissue was homogenized with a Geno/Grinder 2000. RNA was extracted with the standard Trizol protocol and treated with DNase I to remove contaminating genomic DNA. RNA-Seq library samples were prepared using a modified version of the TAG-seq protocol (Meyer et al. 2011; Supplementary methods, 1.3). In short, purified 3′ RNA was amplified and tagged prior to sequencing on the Illumina HiSeq platform. Prepared libraries were submitted to the Genomic Sequencing and Analysis Facility (University of Texas, Austin, Texas) aiming to obtain 5 million reads per sample. RNA sequencing for the greenhouse experiment was accomplished on the SOLiD platform and described in detail in (Meyer et al., 2014).
Differences in library construction and sequencing chemistry between SOLiD and Illumina systems have been implicated in producing variation in transcriptomic profiles (Tariq et al., 2011) with library construction expected to feature heavily in observed qualitative and quantitative differences (Linsen et al., 2009). Nonetheless, correlations across such samples remain reasonable (i.e. global transcriptomic profiles remain largely intact; Tyakht et al., 2014), and within-protocol (between sample) fold change estimates are thought to be robust to platform bases (i.e. differential expression analysis within a uniform protocol remains a viable assay (Toedling et al., 2012). Platform biases may influence the power of differential expression tests at the gene level (and subsequently bias our comparisons of significant genes across experimental designs) through their impact on gene level sequencing depth. However, it is known that power in gene expression count contexts is primarily driven by sample size and secondarily by sequencing depth (Ching et al., 2014). We compared differences in the power to detect differential expression among sequencing technologies and experiments (which have slightly different levels of replication). To do so, we evaluated “significance curves,” where the total number of individuals in each experiment was rarefied, allowing direct comparison of the power of an experiment when replication is identical (Supplemental Fig. S1).
Bioinformatic Analysis of RNA-Seq Data
Shelter, cylinder (Illumina) and greenhouse (SOLiD) data were processed into fastq format and poly-A tail and known TAG-SEquation 5′ adapter sequence was removed using cutadapt (Martin, 2011). The trimmed sequences were subsequently aligned to the P. virgatum V2.0 reference (http://phytozome.jgi.doe.gov; Goodstein et al. 2012). Base space reads were aligned with BWA-mem (Li and Durbin, 2009), and SOLiD were aligned with the Bowtie color space aligner (Langmead et al., 2009). Hits to genes based on the P. virgatum V2.1 annotation were assessed using the “union” mode of htseq-count (Anders et al., 2015). Multiple alignments were utilized in the sam files and nonuniquely mapping reads were excluded (see Supplemental Methods). Library preparation and sequencing effort resulted in generally similar levels of saturation of the transcriptome (Supplemental Fig. S6; Supplemental Methods).
Statistical Analysis
The LIMMA R package (Ritchie et al., 2015) was used to conduct all statistical analyses pertaining to gene expression assays and GO annotations. Various model specifications can be found in the online supplementary material, and the functions used to streamline our analyses have been written into an R package (github.com/jtlovell/limmaDE2). For each model, normalization factors were calculated to scale libraries by total counts after first excluding any genes with mean expression < 5 raw counts. These factors were used as a covariate in the “voom” normalization procedure. In addition to the normalized counts, where possible, we also used either a spatial variable or repeated measures as a blocking variable in the linear model. Gene-wise statistics were calculated via generalized least squares linear models and subsequent empirical Bayes procedures to infer variance structure across genes. P-values were FDR corrected via the Benjamini-Hochberg method via the R function, “p.adjust” (Benjamini and Hochberg, 1995). Both GO and gene overlap enrichments were inferred via Fisher’s tests.
Statistical analyses of physiological data were treated similarly to expression counts. We tested the effects of drought treatments while controlling for spatial and temporal sampling variation in mixed effects linear models implemented in the R lme4 package (see Supplemental Methods; Bates et al. 2014). Type III SS tests were calculated with the lmerTest package (Kuznetsova et al., 2013).
Multivariate tests of gene expression were conducted via principal component analyses (PCA) of normalized expression matrices in R. To determine the relative importance of each experimental factor, we culled the expression matrix to genes that were significantly differentially expressed in any factor. We subsequently applied an ANOVA decomposition of variance via PCA from the LIMMA fitted linear model (Fresno et al., 2014) on the culled expression matrix.
Accession Numbers
RNA-seq data analyzed here have been deposited in the short read archive under BioProject ID: PRJNA322529. Accession numbers and metadata are presented in Supplemental Table S5.
Supplemental Data
The following supplemental materials are available online.
Supplemental Methods. Additional physiological methods, Additional RNA extraction methods, Additional bioinformatics methods, and Additional statistical methods
Supplemental Appendix. Model specifications
Supplemental Table S1. Significance and odds ratios of significantly differentially expressed gene overlaps.
Supplemental Table S2. Gene lists and annotations of genes differentially expressed in 3 or 4 of the experiments with homologs related to drought.
Supplemental Table S3. GO enrichment across experiments.
Supplemental Table S4. Gene lists and annotations of genes differentially expressed in 3 or 4 of the experiments with homologs related to drought.
Supplemental Table S5. Accession numbers for TAG-seq reads.
Supplemental Figure S1. The number of significant genes detected per fixed replication level.
Supplemental Figure S2. The differential impact of diurnal patterns on gene expression in the wet and dry treatments of the shelter experiment.
Supplemental Figure S3. The physiological and gene expression effects of the order of sample collection.
Supplemental Figure S4. Pairwise expression correlations.
Supplemental Figure S5. Conserved expression across all experiments.
Supplemental Figure S6. Rarefaction analysis of library sequencing depth.
Supplementary Material
Acknowledgments
A. Khasanova, N. Johnson, A. Asmus, Y. Sorkin, and B. Whitaker assisted in propagating plants, planting, and maintaining the experiment. T. Logan helped prepare many of the TAG-seq libraries. Many members of the Juenger and Hawkes lab assisted in harvesting leaf tissue and measuring leaf water potentials. We thank M. Simmons, M. Bertelsen, and the Ladybird Johnson Wildflower Center for facilitating our field experiment. Computational analyses were completed on the Stampede system with allocations from the Texas Advanced Computing Center. Pre-publication switchgrass genome data (generated by J. Schmutz, J. Jenkins A. Sreedasyam, and S. Shu) were provided by the Department of Energy Joint Genome Institute. Earlier versions of this manuscript were greatly improved following comments from A. MacQueen, D. Hoover, and B. Campitelli.
Footnotes
Articles can be viewed without a subscription.
References
- Ahuja I, de Vos RCH, Bones AM, Hall RD (2010) Plant molecular stress responses face climate change. Trends Plant Sci 15: 664–674 [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JT, Lee C-R, Mitchell-Olds T (2011) Life-history QTLS and natural selection on flowering time in Boechera stricta, a perennial relative of Arabidopsis. Evolution 65: 771–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araus JL, Slafer GA, Reynolds MP, Royo C (2002) Plant breeding and drought in C3 cereals: what should we breed for? Ann Bot (Lond) 89: 925–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspinwall MJ, Fay PA, Hawkes CV, Lowry DB, Khasanova A, Bonnette J, Whitaker BK, Johnson N, Juenger TE (2016) Intraspecific variation in precipitation responses of a widespread C4 grass depend on site water limitation. J Plant Ecol 9: rtw040 [Google Scholar]
- Aspinwall MJ, Lowry DB, Taylor SH, Juenger TE, Hawkes CV, Johnson M-VV, Kiniry JR, Fay PA (2013) Genotypic variation in traits linked to climate and aboveground productivity in a widespread C₄ grass: evidence for a functional trait syndrome. New Phytol 199: 966–980 [DOI] [PubMed] [Google Scholar]
- Axelrod DI. (1972) Edaphic aridity as a factor in angiosperm evolution. Am Nat 10.2307/2459779 [Google Scholar]
- Barker T, Campos H, Cooper M, Dolan D, Edmeades G, Habben J, Schussler J, Wright D, Zinselmeier C (2005) Improving drought tolerance in maize. Plant Breed Rev 25: 173–253 [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S (2014) Fitting Linear Mixed-Effects Models using lme4. arXiv preprint arXiv.
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 10.2307/2346101 [Google Scholar]
- Blum A. (1996) Crop responses to drought and the interpretation of adaptation. Plant Growth Regul 20: 135–148 [Google Scholar]
- Bogeat-Triboulot M-B, Brosché M, Renaut J, Jouve L, Le Thiec D, Fayyaz P, Vinocur B, Witters E, Laukens K, Teichmann T, Altman A, Hausman JF, et al. (2007) Gradual soil water depletion results in reversible changes of gene expression, protein profiles, ecophysiology, and growth performance in Populus euphratica, a poplar growing in arid regions. Plant Physiol 143: 876–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer JS. (1982) Plant productivity and environment. Science 218: 443–448 [DOI] [PubMed] [Google Scholar]
- Chaves MM, Oliveira MM (2004) Mechanisms underlying plant resilience to water deficits: prospects for water-saving agriculture. J Exp Bot 55: 2365–2384 [DOI] [PubMed] [Google Scholar]
- Chaves MM, Maroco JP, Pereira JS (2003) Understanding plant responses to drought — from genes to the whole plant. Funct Plant Biol 30: 239–264 [DOI] [PubMed] [Google Scholar]
- Ching T, Huang S, Garmire LX (2014) Power analysis and sample size estimation for RNA-Seq differential expression. RNA 20: 1684–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciais P, Reichstein M, Viovy N, Granier A, Ogée J, Allard V, Aubinet M, Buchmann N, Bernhofer C, Carrara A, Chevallier F, De Noblet N, et al. (2005) Europe-wide reduction in primary productivity caused by the heat and drought in 2003. Nature 437: 529–533 [DOI] [PubMed] [Google Scholar]
- Comas LH, Becker SR, Cruz VMV, Byrne PF, Dierig DA (2013) Root traits contributing to plant productivity under drought. Front Plant Sci 4: 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornic G, Massacci A (1996) Leaf Photosynthesis Under Drought Stress. In Baker NR, ed, Photosynthesis and the Environment. Springer Netherlands, Dordrecht, pp 347–366 [Google Scholar]
- Craufurd PQ, Peacock JM (1993) Effect of Heat and Drought Stress on Sorghum (Sorghum bicolor). II. Grain Yield. Exp Agric 29: 77–86 [Google Scholar]
- Des Marais DL, Hernandez KM, Juenger TE (2013) Genotype-by-Environment Interaction and Plasticity: Exploring Genomic Responses of Plants to the Abiotic Environment. Annu Rev Ecol Evol Syst 44: 5–29 [Google Scholar]
- Des Marais DL, Juenger TE (2010) Pleiotropy, plasticity, and the evolution of plant abiotic stress tolerance. Ann N Y Acad Sci 1206: 56–79 [DOI] [PubMed] [Google Scholar]
- Des Marais DL, McKay JK, Richards JH, Sen S, Wayne T, Juenger TE (2012) Physiological genomics of response to soil drying in diverse Arabidopsis accessions. Plant Cell 24: 893–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder ME, Hofmann GE (1999) Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol 61: 243–282 [DOI] [PubMed] [Google Scholar]
- Fresno C, Balzarini MG, Fernández EA (2014) lmdme: Linear Models on Designed Multivariate Experiments in R. J Stat Softw 56: http://EconPapers.repec.org/RePEc:jss:jstsof:v:056:i07 [Google Scholar]
- Fu P, Wilen RW, Wu G-H, Robertson AJ, Gusta LV (2000) Dehydrin Gene Expression and Leaf Water Potential Differs between Spring and Winter Cereals During Cold Acclimation. J Plant Physiol 156: 394–400 [Google Scholar]
- Gaudin ACM, Henry A, Sparks AH, Slamet-Loedin IH (2013) Taking transgenic rice drought screening to the field. J Exp Bot 64: 109–117 [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harb A, Krishnan A, Ambavaram MMR, Pereira A (2010) Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol 154: 1254–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinze J, Sitte M, Schindhelm A, Wright J, Joshi J (2016) Plant-soil feedbacks: a comparative study on the relative importance of soil feedbacks in the greenhouse versus the field. Oecologia 181: 559–569 [DOI] [PubMed] [Google Scholar]
- Heyne EG, Brunson AM (1940) Genetic studies of heat and drought tolerance in maize. J Am Soc Agron 32: 803–814 [Google Scholar]
- Hoover DL, Duniway MC, Belnap J (2015) Pulse-drought atop press-drought: unexpected plant responses and implications for dryland ecosystems. Oecologia 179: 1211–1221 [DOI] [PubMed] [Google Scholar]
- Hoover DL, Rogers BM (2016) Not all droughts are created equal: the impacts of interannual drought pattern and magnitude on grassland carbon cycling. Glob Chang Biol n/a–n/a [DOI] [PubMed] [Google Scholar]
- Jones HG. (2007) Monitoring plant and soil water status: established and novel methods revisited and their relevance to studies of drought tolerance. J Exp Bot 58: 119–130 [DOI] [PubMed] [Google Scholar]
- Jones HG. (2014) Plants and Microclimate. Cambridge University Press; Cambridge, United Kingdom [Google Scholar]
- Knapp AK, Hoover DL, Wilcox KR, Avolio ML, Koerner SE, La Pierre KJ, Loik ME, Luo Y, Sala OE, Smith MD (2015) Characterizing differences in precipitation regimes of extreme wet and dry years: implications for climate change experiments. Glob Change Biol 21: 2624–2633 [DOI] [PubMed] [Google Scholar]
- Kramer PJ, Boyer JS (1995) Water Relations of Plants and Soils. Academic Press, New York, USA [Google Scholar]
- Kudoh H. (2015) Molecular phenology in plants: in natura systems biology for the comprehensive understanding of seasonal responses under natural environments. New Phytol 210: 399–412 [DOI] [PubMed] [Google Scholar]
- Kuznetsova A, Brockhoff PB, Christensen R (2013) lmerTest: Tests for random and fixed effects for linear mixed effect models (lmer objects of lme4 package). doi: 10.1111/j.2041-210x.2012.00261.x/full
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor DW, Fock DH (1978) Photosynthesis, Respiration, and Carbon Assimilation in Water-Stressed Maize at Two Oxygen Concentrations. J Exp Bot 29: 579–593 [Google Scholar]
- Lefebvre V, North H, Frey A, Sotta B, Seo M, Okamoto M, Nambara E, Marion-Poll A (2006) Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J 45: 309–319 [DOI] [PubMed] [Google Scholar]
- Levitt J. (1980) Responses of Plants to Environmental Stresses: Chilling, freezing, and high temperature stresses. Academic Press, New York, USA [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsen SEV, de Wit E, Janssens G, Heater S, Chapman L, Parkin RK, Fritz B, Wyman SK, de Bruijn E, Voest EE, Kuersten S, Tewari M, et al. (2009) Limitations and possibilities of small RNA digital gene expression profiling. Nat Methods 6: 474–476 [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez CG, Banowetz GM, Peterson CJ, Kronstad WE (2003) Dehydrin Expression and Drought Tolerance in Seven Wheat Cultivars. Crop Sci 43: 577–582 [Google Scholar]
- Lovell JT, Mullen JL, Lowry DB, Awole K, Richards JH, Sen S, Verslues PE, Juenger TE, McKay JK (2015) Exploiting differential gene expression and epistasis to discover candidate genes for drought-associated QTLs in Arabidopsis thaliana. Plant Cell 27: 969–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell JT, Schwartz S, Lowry DB, Shakirov EV, Bonnette JE, Weng X, Wang M, Johnson J, Sreedasyam A, Plott C, Jenkins J, Schmutz J, et al. (2016) Drought responsive gene expression regulatory divergence between upland and lowland ecotypes of a perennial C4 grass. Genome Res 26: 510–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado S, Paulsen GM (2001) Combined effects of drought and high temperature on water relations of wheat and sorghum. Plant Soil 233: 179–187 [Google Scholar]
- Malmberg RL, Held S, Waits A, Mauricio R (2005) Epistasis for fitness-related quantitative traits in Arabidopsis thaliana grown in the field and in the greenhouse. Genetics 171: 2013–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand G, Mayjonade B, Varès D, Blanchet N, Boniface MC, Maury P, Andrianasolo FN, Burger P, Debaeke P, Casadebaig P, et al. (2013) A biomarker based on gene expression indicates plant water status in controlled and natural environments. Plant, Cell &. Environment 36: 2175–2189 [DOI] [PubMed] [Google Scholar]
- Martin M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12 [Google Scholar]
- Maseda PH, Fernández RJ (2006) Stay wet or else: three ways in which plants can adjust hydraulically to their environment. J Exp Bot 57: 3963–3977 [DOI] [PubMed] [Google Scholar]
- Mearns LO, Sain S, Leung LR, Bukovsky MS, McGinnis S, Biner S, Caya D, Arritt RW, Gutowski W, Takle E, et al. (2013) Climate change projections of the North American Regional Climate Change Assessment Program (NARCCAP). Clim Change 120: 965–975 [Google Scholar]
- Meisner A, De Deyn GB, de Boer W, van der Putten WH (2013) Soil biotic legacy effects of extreme weather events influence plant invasiveness. Proc Natl Acad Sci USA 110: 9835–9838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J 25: 295–303 [DOI] [PubMed] [Google Scholar]
- Meyer E, Aglyamova GV, Matz MV (2011) Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-Seq procedure. Mol Ecol 20: 3599–3616 [DOI] [PubMed] [Google Scholar]
- Meyer E, Aspinwall MJ, Lowry DB, Palacio-Mejía JD, Logan TL, Fay PA, Juenger TE (2014) Integrating transcriptional, metabolomic, and physiological responses to drought stress and recovery in switchgrass (Panicum virgatum L.). BMC Genomics 15: 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra Y, Jänkänpää HJ, Kiss AZ, Funk C, Schröder WP, Jansson S (2012) Arabidopsis plants grown in the field and climate chambers significantly differ in leaf morphology and photosystem components. BMC Plant Biol 12: doi.10.1186/1471-2229-12-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. (2006) Abiotic stress, the field environment and stress combination. Trends Plant Sci 11: 15–19 [DOI] [PubMed] [Google Scholar]
- Nguyen TTT, Klueva N, Chamareck V, Aarti A, Magpantay G, Millena ACM, Pathan MS, Nguyen HT (2004) Saturation mapping of QTL regions and identification of putative candidate genes for drought tolerance in rice. Mol Genet Genomics 272: 35–46 [DOI] [PubMed] [Google Scholar]
- Poorter H, Bühler J, van Dusschoten D, Climent J, Postma JA (2012a) Pot size matters: a meta-analysis of the effects of rooting volume on plant growth. Funct Plant Biol 39: 839–850 [DOI] [PubMed] [Google Scholar]
- Poorter H, Fiorani F, Stitt M, Schurr U, Finck A, Gibon Y, Usadel B, Munns R, Atkin OK, Tardieu F, et al. (2012b) The art of growing plants for experimental purposes: a practical guide for the plant biologist. Funct Plant Biol 39: 821–838 [DOI] [PubMed] [Google Scholar]
- Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol 133: 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengel D, Arribat S, Maury P, Martin-Magniette M-L, Hourlier T, Laporte M, Varès D, Carrère S, Grieu P, Balzergue S, Gouzy J, Vincourt P, et al. (2012) A gene-phenotype network based on genetic variability for drought responses reveals key physiological processes in controlled and natural environments. PLoS One 7: e45249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Mittler R (2002) The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol 130: 1143–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R (2004) When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 134: 1683–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HI, Brown CS, Claassen VP (2007) Comparisons of Mycorrhizal Responsiveness with Field Soil and Commercial Inoculum for Six Native Montane Species and Bromus tectorum. Restor Ecol 15: 44–52 [Google Scholar]
- Schafleitner R, Gutierrez Rosales RO, Gaudin A, Alvarado Aliaga CA, Martinez GN, Tincopa Marca LR, Bolivar LA, Delgado FM, Simon R, Bonierbale M (2007) Capturing candidate drought tolerance traits in two native Andean potato clones by transcription profiling of field grown plants under water stress. Plant Physiol Biochem 45: 673–690 [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I (2004) Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci 9: 236–243 [DOI] [PubMed] [Google Scholar]
- Schweitzer JA, Juric I, Voorde TFJ, Clay K, Putten WH, Bailey JK (2014) Are there evolutionary consequences of plant–soil feedbacks along soil gradients? Funct Ecol 28: 55–64 [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, Satou M, Akiyama K, et al. (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31: 279–292 [DOI] [PubMed] [Google Scholar]
- Semenov MA. (2007) Simulation of extreme weather events by a stochastic weather generator. Clim Res 25: http://onlinelibrary.wiley.com/doi/10.1002/joc.1120/pdf [Google Scholar]
- Smirnoff N. (1993) The role of active oxygen in the response of plants to water deficit and desiccation. New Phytol 125: 27–58 [DOI] [PubMed] [Google Scholar]
- Speirs J, Binney A, Collins M, Edwards E, Loveys B (2013) Expression of ABA synthesis and metabolism genes under different irrigation strategies and atmospheric VPDs is associated with stomatal conductance in grapevine (Vitis vinifera L. cv Cabernet Sauvignon). J Exp Bot 64: 1907–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL., Jr (1952) Aridity as a Stimulus to Plant Evolution. Am Nat 86: 33–44 [Google Scholar]
- Suzuki N, Rivero RM, Shulaev V, Blumwald E, Mittler R (2014) Abiotic and biotic stress combinations. New Phytol 203: 32–43 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Rizhsky L, Liang H, Shuman J, Shulaev V, Mittler R (2005) Enhanced tolerance to environmental stress in transgenic plants expressing the transcriptional coactivator multiprotein bridging factor 1c. Plant Physiol 139: 1313–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck SM, Lindquist EA, Ackerly DD, Andersen GL (2011) Analysis of leaf and root transcriptomes of soil-grown Avena barbata plants. Plant Cell Physiol 52: 317–332 [DOI] [PubMed] [Google Scholar]
- Taiz L, Zeiger E (2010) Plant Physiology and Development, 5th edition. Sinauer Associates, Sunderland, MA, USA [Google Scholar]
- Tardieu F, Davies WJ (1992) Stomatal response to abscisic Acid is a function of current plant water status. Plant Physiol 98: 540–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq MA, Kim HJ, Jejelowo O, Pourmand N (2011) Whole-transcriptome RNAseq analysis from minute amount of total RNA. Nucleic Acids Res 39: e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toedling J, Servant N, Ciaudo C, Farinelli L, Voinnet O, Heard E, Barillot E (2012) Deep-sequencing protocols influence the results obtained in small-RNA sequencing. PLoS One 7: e32724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers SE, Smith MD, Bai J, Hulbert SH, Leach JE, Schnable PS, Knapp AK, Milliken GA, Fay PA, Saleh A, et al. (2007) Ecological genomics: making the leap from model systems in the lab to native populations in the field. Front Ecol Environ 5: 19–24 [Google Scholar]
- Turner NC, Begg JE (1981) Plant-water relations and adaptation to stress. Plant Soil 58: 97–131 [Google Scholar]
- Tuteja N, Gill SS (2013) Climate Change and Plant Abiotic Stress Tolerance. John Wiley & Sons, New York, USA [Google Scholar]
- Tyakht AV, Ilina EN, Alexeev DG, Ischenko DS (2014) RNA-Seq gene expression profiling of HepG2 cells: the influence of experimental factors and comparison with liver tissue. BMC Genomics 15: doi.10.1186/1471-2164-15-1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45: 523–539 [DOI] [PubMed] [Google Scholar]
- Vranová E, Langebartels C, Van Montagu M, Inzé D, Van Camp W (2000) Oxidative stress, heat shock and drought differentially affect expression of a tobacco protein phosphatase 2C. J Exp Bot 51: 1763–1764 [DOI] [PubMed] [Google Scholar]
- Weinig C, Ungerer MC, Dorn LA, Kane NC, Toyonaga Y, Halldorsdottir SS, Mackay TFC, Purugganan MD, Schmitt J (2002) Novel loci control variation in reproductive timing in Arabidopsis thaliana in natural environments. Genetics 162: 1875–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Nehnevajova E, Köllmer I, Novák O, Strnad M, Krämer U, Schmülling T (2010) Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell 22: 3905–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.