Abstract
Field-based, high-throughput phenotyping enables the detailed characterization of plant populations under relevant conditions, providing valuable biological insight into the life history of plants.
Establishing the connection between genotype and phenotype is currently one of the most significant challenges facing modern plant biology. Although spectacular advances in next-generation DNA sequencing have allowed genomic data to become commonplace throughout biology, progress has been much slower in translating this discrete DNA base pair information into an accurate description of phenotypic variation. The limiting factor for quantifying phenotypes, particularly in the context of agricultural and native plant populations, has been the lack of phenotypic information of a scale, density, and accuracy comparable to DNA sequencing data. The extensive collection of phenotypic data for many physiological and developmental traits from many individuals remains onerous (Furbank and Tester, 2011). As a result, for large populations, there is often a focus on traits that are easy or inexpensive to measure, while more costly or difficult-to-score phenotypes are studied in only a few individuals. This is especially true for traits that are complex in nature, meaning that they have polygenic inheritance and varying responses to the environment. Complex traits are of primary interest not only because they represent the majority of important agronomic crop traits but also because they govern key biological processes that influence overall plant productivity and adaptability in plant populations (Lynch and Walsh, 1998). Success in deciphering these processes could translate into increased genetic gains in plant breeding, elucidation of the mechanisms impacting important ecophysiological traits, and improved crop management decisions to further maximize yield and quality. In light of these potential benefits, the aim of this Update article is to present the basic principles of phenomics, summarize the current state of field-based phenotyping, and highlight key challenges and limitations. In addition, the areas of data collection and management, environmental characterization, and crop growth models (CGMs) are presented as topics where further consideration is needed to capitalize on advancements in phenotyping technologies.
Phenomics, or high-throughput phenotyping, which emerged in recent years in response to limited phenotyping capacity, is the use of sensor and imaging technologies that permits the rapid, low-cost measurement of many phenotypes across time and space with less labor; it can include laboratory, greenhouse, and field-based applications. In model plant species with small physical stature, such as Arabidopsis (Arabidopsis thaliana), large populations can be evaluated under controlled environmental conditions. However, the use of controlled environmental systems is not scalable for many areas of interest. Native species often need to be evaluated in their natural environment and over a broad geographic and climatic distribution, and agricultural crop trials must simultaneously evaluate thousands of potential cultivars. Furthermore, these controlled systems are unable to replicate the environmental variables of a field environment that influence complex traits such as grain yield or drought tolerance. Therefore, field-based, high-throughput phenotyping (FB-HTP) capacity is desperately needed to understand phenotypic variation relevant to a broad range of research areas such as food and nutritional security, anthropogenic effects on the environment, and ecological community interactions.
ADVANCES
Plants are intrinsically related to their environment, and observed phenotypes are a direct product of this interaction. Therefore, the ability to study and quantify phenotypes under real-world conditions is essential to the basic understanding, as well as improvement, of ecophysiological traits.
Recent technological developments have enabled progress in plant phenotyping, but areas such as root phenotyping are still lacking the needed instrumentation in order to capitalize on these developments.
Extraction of high-dimensional phenotype data from images is becoming more commonplace with advancements in image-processing software. This is leading to the discovery of novel phenotypes not identified previously but that are more related to underlying physiological processes.
Developments in envirotyping and crop growth modeling can provide a useful framework for understanding plant development.
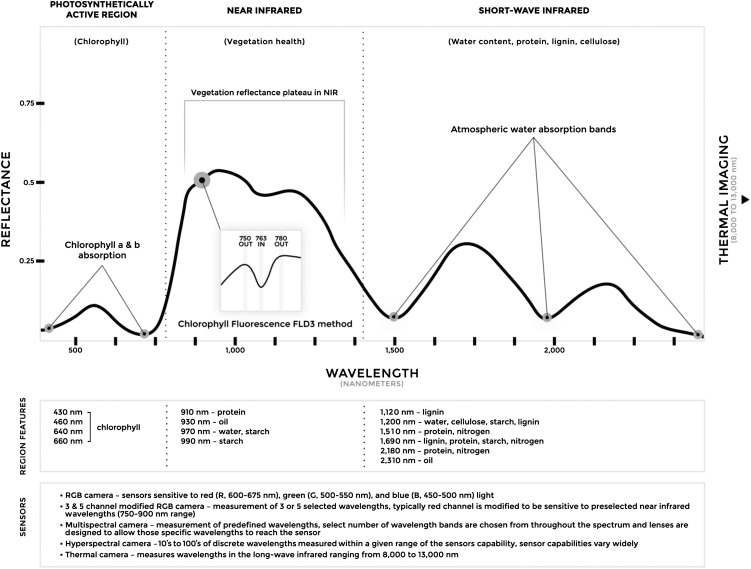
The physical basis for most nondestructive, proximal sensing systems is the quantification of absorption, transmission, or reflectance characteristics of the electromagnetic radiation (EM) spectrum’s interaction with the plant canopy surface (Mulla, 2013; Araus and Cairns, 2014). The EM spectrum, specifically the wavelengths between 400 and 2,500 nm, can be broken down into three major parts that offer information about plant status, structural properties, and biochemical composition (Fig. 1). These three subregions are composed of (1) the photosynthetically active region (400–700 nm PAR), in which photosynthetic pigments, namely chlorophylls a and b, strongly absorb light; (2) the near-infrared region (700–1,400 nm), in which healthy plant tissue is highly reflective; and (3) the shortwave infrared region (1,400–2,500 nm), in which water and biomolecules contribute to reflectance characteristics (Jones and Vaughan, 2010; Homolová et al., 2013). In addition to these regions, thermal infrared, typically 8 to 13 μm when used for remote sensing, can provide information about canopy temperature (Jones, 2004). The variation present in these spectral traits give rise to ecological, species-, and genotype-specific phenotypes.
Figure 1.
Typical spectral reflectance curve for healthy vegetation. The unique spectral signature of vegetation in the wavelength range of 350 to 2,500 nm allows it to be differentiated from other types of land features. The shape of the reflectance spectrum is influenced by the chlorophyll content, health, water content, and biochemical composition of the vegetation (Curran, 1989; Jones and Vaughan, 2010), which then can be used to help identify the type of vegetation and diagnose its status. Thermal sensing of vegetation is valuable for the detection of drought stress and closure of stomata. There are five common types of sensors that are used to measure spectral variation, with differences among them in the specific targeted wavelengths. The inset image depicts the spectra underlying solar-induced chlorophyll fluorescence based on application of the Fraunhofer line discrimination principle using three spectral bands (FLD3). Measurements of chlorophyll fluorescence can be used to detect the early stages of biotic or abiotic stress before the appearance of visible symptoms. NIR, Near-infrared region.
Robust sensors mounted on a field-deployable vehicle are imperative for FB-HTP. Although the aim of this review is not to summarize specific sensor technologies (for summary, see Jones and Vaughan, 2010; Sankaran et al., 2015), a brief list is provided for orientation. The most common types of canopy sensors include digital imaging via red-green-blue cameras; multispectral, including color-infrared modified digital cameras; hyperspectral; thermal; fluorescence; and three-dimensional (3D; time-of-flight and stereo cameras as well as light detection and ranging). The choice of vehicle for positioning sensors directly impacts the scale of research that can be conducted as well as the achievable sensor resolution and associated costs (for review of vehicles, considerations, and limitations, see White et al., 2012; Sankaran et al., 2015). For phenotyping large ecological systems, satellites and manned aircraft remain the only practical option (Kerr and Ostrovsky, 2003; Asner et al., 2012), but small unmanned aircraft systems (UASs) are now a viable alternative for smaller, geographically distinct areas (Kim et al., 2013). Finally, for small-scale experimental field studies, several vehicle options include high-clearance tractors, cable gantries, cranes, linear move irrigation systems, and small UASs in addition to stationary observation platforms (Haberland, 2010; Andrade-Sanchez et al., 2013; Busemeyer et al., 2013a; Chapman et al., 2014; Liebisch et al., 2015).
PLANT CANOPY PHENOTYPING
Canopy reflectance can provide insight into overall plant health as well as specific physiological processes. To date, most FB-HTP and remote sensing applications have focused on using vegetative indices to infer overall plant status (for review, see Bannari et al., 1995; Govender et al., 2009), with normalized difference vegetation index (Tucker, 1979) being the most well known. Although these indices generally can be informative, they use less than 1% of available spectra and lack the ability to give detailed information on physiological processes. In contrast, canopy spectroscopy, using either multispectral or hyperspectral sensors, can assess canopy chemistry and composition by capturing more of the spectra. Several research groups have demonstrated that hyperspectral data (400–2,500 nm) can be utilized to nondestructively infer leaf chemical properties in various species (Ustin et al., 2009; Asner et al., 2011, 2015), with specific applications including estimation of canopy nitrogen (Martin et al., 2008) and lignin content (Wessman et al., 1988). Spectral signatures arising from these various leaf biochemical compounds can give further insight into community-level phenotypes, including species diversity, ecosystem composition, and threats to native plants by the spread of invasive species (Turner et al., 2003; Bradley, 2014).
However, spectral signatures of canopies are jointly influenced by environmental factors and the biochemical composition of the plant (Gates et al., 1965; Curran, 1989), making it difficult to understand how different wavelengths across the measured spectrum translate into biological meaning and function. In particular, the complex interaction of light with the canopy surface and the associated effects of light scatter pose a significant challenge. Attempts have been made to handle this heterogeneity with the seminal Scattering by Arbitrarily Inclined Leaves and Leaf Optical Properties Spectra models, but these canopy reflectance models oversimplify the structural characteristics of leaves into a single parameter (Verhoef, 1984; Jacquemoud and Baret, 1990). An additional layer of complexity is the corresponding geometry and position of the sensor itself, which can introduce systematic error into the analysis of the spectral data and, therefore, must be properly accounted for in the model (Lobell et al., 2002). Despite these limitations, the use of canopy reflectance data at large scales provided by satellite and manned aerial systems has proven valuable. However, far higher spatial resolution is needed for the study of phenotypes at the experimental plot level.
With recent technological developments, the ability to use smaller, more portable sensors in combination with vehicles, whether ground or aerially based, permits the collection of canopy data with higher spatial resolution. In addition, most developed FB-HTP platforms incorporate sets of multiple sensors, creating complementary streams of data that, when combined, provide more information than what individual sensors alone can achieve (termed sensor fusion). In one of the first demonstrations of FB-HTP, Montes et al. (2011) developed a platform carrying light curtains (measure of canopy height) and spectral reflectance sensors (canopy reflectance) to predict aboveground biomass accumulation in maize (Zea mays). Their results showed that the combination of data from both sensors was more predictive than data from either sensor alone. Such combined sensor data were used successfully by Busemeyer et al. (2013b) to phenotype × Triticosecale Wittmack L. for harvestable biomass at multiple developmental stages. More recently, Deery et al. (2014) developed an advanced platform combining numerous high-precision sensors that are capable of capturing details of plant physical structure such as canopy leaf angle and can produce 3D surface reconstructed images of plants.
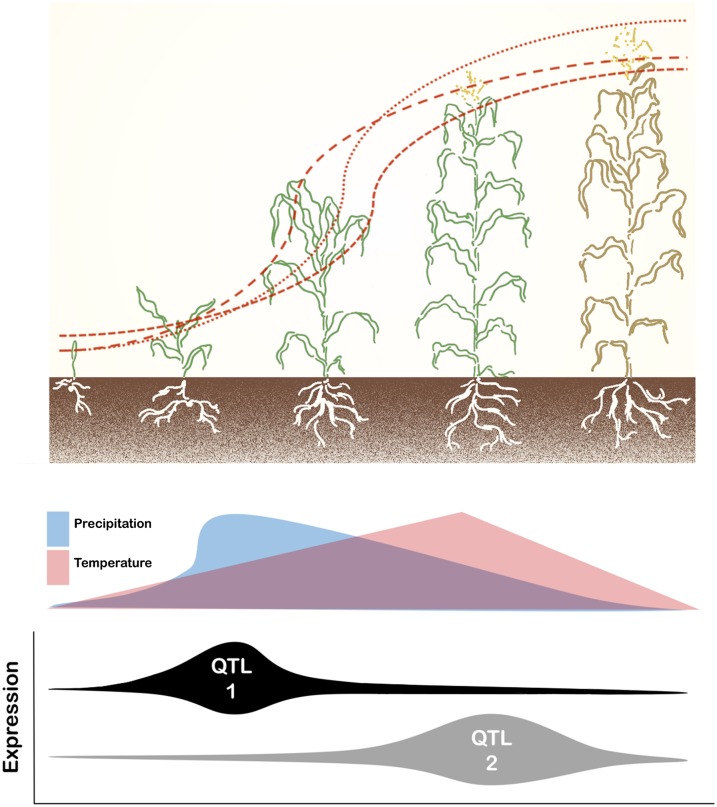
FB-HTP systems have been used to study the temporal nature of trait development and its interaction with environmental effects. Historically, canopy temperature has been used to assess the water status of plants, as plants with available soil moisture exhibit decreased leaf surface temperatures relative to atmospheric conditions (Ehrler, 1973; Jackson et al., 1981). Recently, Pauli et al. (2016) demonstrated the use of FB-HTP to study the physiological processes underlying heat and drought responses in an upland cotton (Gossypium hirsutum) population grown under contrasting irrigation regimes. They were able to assess the dynamic nature of multiple canopy traits in response to rapidly changing field conditions within one day and over the growing season. The longitudinal collection of phenotypic data enabled the detection of quantitative trait loci (QTL) with temporal expression patterns coinciding with plant growth stages (Fig. 2). Further supporting the ability of FB-HTP to monitor plant development under managed stress, Sharma and Ritchie (2015) used a similar system to identify differential growth characteristics among cotton cultivars evaluated under varying irrigation levels. The novel ability of FB-HTP to capture the temporal changes of a phenotype provides the critical pivot needed to focus on developmental quantitative genetics in order to grasp how trait manifestation is a function of many QTL whose effects fluctuate with time (Wu et al., 1999). Because the expression dynamics of individual QTL vary and are modulated further by phenological stage as well as their interaction with the environment, it is not possible to capture this dynamism with single time point analyses. Therefore, recognizing the individual behavior of QTL may permit genotype optimization with respect to target environments or in identifying how ecosystem disturbances impact the genetics of natural populations.
Figure 2.
Relationship of plant growth stages, environmental patterns, and QTL expression. FB-HTP facilitates the study of plants at distinct developmental stages or continuously throughout the season, thus allowing the logistic growth pattern of plants to be modeled. As a result, the genetic mapping of QTL can be performed with phenotype data obtained by successive measurement of a trait, such as height, throughout plant development, thereby enabling the expression dynamics of identified QTL to be monitored in the context of changing environmental conditions.
Although its application in FB-HTP is still in its infancy, fluorescence imaging, which quantifies the light reemitted from chlorophyll a molecules via chlorophyll fluorescence, can provide information about the photosynthetic apparatus under various abiotic and biotic stresses (Maxwell and Johnson, 2000). Furthermore, changes in fluorescence readings often precede the manifestation of any visible symptoms (Lichtenthaler and Miehé, 1997), making it useful for the early detection of abiotic and biotic stress (Massacci et al., 2008; Jedmowski and Brüggemann, 2015; Ni et al., 2015). The use of solar-induced chlorophyll fluorescence, based on the Fraunhofer line discrimination principle (Plascyk, 1975; Meroni et al., 2009), was recently implemented successfully using UAS-based hyperspectral imagery (Zarco-Tejada et al., 2013) for the early detection of Verticillium wilt in a production olive (Olea europaea) orchard (Calderón et al., 2013). As an extension of this technology to ecological studies, the relationship of chlorophyll fluorescence to gross primary production has enabled the assessment of temporal changes in ecosystem health and productivity in response to climatic change (Damm et al., 2015; Yang et al., 2015). Although fluorescence imaging is promising, it still suffers from the same limitations as other spectral imaging techniques, including inconsistent or uneven illumination, wind disturbances under field conditions, and being unable to discriminate the underlying cause of the stress.
PLANT DISEASE AND PEST PHENOTYPING
The plant canopy’s role as a vital aboveground interface with the environment makes identifying and quantifying disturbances due to biotic factors a critical aspect of FB-HTP. The outcomes of early and accurate detection of plant diseases and pests in the field have a global economic impact. The presence of many biotic diseases, including those caused by viruses, bacteria, fungi, oomycetes, and insect pests, can be difficult to detect until disease progression is advanced, pathogen load is high, and symptoms are abundant. At this late stage, disease is extremely difficult to control (Fry, 1982). Methods for the direct and indirect assessment of pathogen presence in the field were reviewed recently (Mahlein et al., 2012; Fang and Ramasamy, 2015; Mutka and Bart, 2015), and these methods are widely employed (Ward et al., 2004; Lievens and Thomma, 2005; Mirmajlessi et al., 2015). The early detection of plant diseases prior to their spread remains the larger challenge. Various techniques such as fluorescence, visible and infrared spectroscopy, and hyperspectral imaging have been tested for their ability to detect biotic stress with differing levels of success (Sankaran et al., 2010; Mutka and Bart, 2015). Although the assessment of pathogen presence remains the detection method with the highest sensitivity and specificity, a robust method for the indirect detection of biotic stress in general would provide a complementary early warning system.
The task of biotic stress identification is conceptually simple: identify the plant phenotypic signatures that are specifically displayed during response to biotic stress. However, symptoms of abiotic and biotic stresses phenotypically overlap, thus complicating these analyses. Altered water content, thermal characteristics, and changes in red-green-blue profiles are among the major phenotypic alterations elicited by both biotic and abiotic stress. For example, vascular bacterial and fungal pathogens can obstruct water transport within the plant, leading to symptoms phenotypically similar to those of drought stress (Yadeta and Thomma, 2013). Efforts to identify phenotypic indicators of biotic stress have approached this challenge by using FB-HTP in conjunction with traditional field surveys to locate diseased areas. In addition to on-ground phenomic methods, it may be possible to use data collected from satellites to assess plant health on a larger, ecological scale (Zhang et al., 2014). These experiments are challenging because of the complexity and unpredictability of field systems, but the technology holds great promise for the identification and management of both current and emerging plant diseases.
Since biotic and abiotic stress induce distinct early events that lead to phenotypic changes, research aimed at developing methods of distinguishing the differences between biotic and abiotic stress responses has high potential for success. Consequently, many researchers are conducting experiments under controlled conditions with a known inoculum and an ability to apply additional abiotic and/or multiplexed biotic stresses in predictable ways. As an example, fluorescence spectroscopy was used to distinguish citrus (Citrus spp.) plants experiencing mechanical injury, drought, variegated chlorosis, or bacterial infection (Lins et al., 2006; Marcassa et al., 2006; Belasque et al., 2008; Bock et al., 2008). Granum et al. (2015) investigated the ability to detect early stages of infection of avocado (Persea americana) by the fungus Rosellinia necatrix using fluorescence and thermal imaging. Additional research within controlled environments to understand these complex interactions and their phenotypic outputs, with particular focus on in planta changes that occur during early stages of infection, can be expected to eventually translate to in-field early detection systems for plant diseases.
PLANT ROOT PHENOTYPING
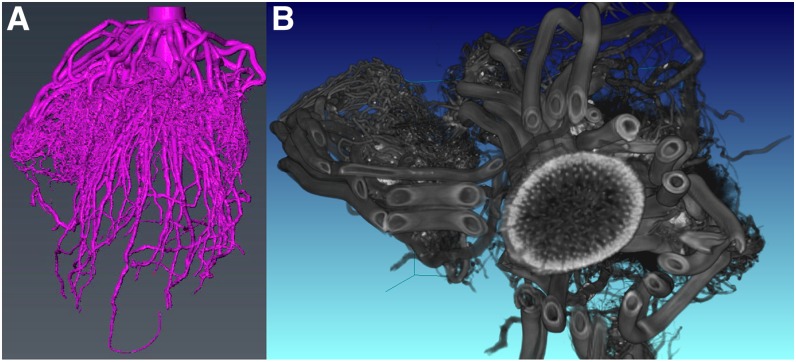
High-quality and high-throughput root phenotyping in the field has long been a valuable but elusive target for plant scientists. In many ways, the trench and monolith excavation methods developed in the early 1900s (Weaver, 1926) are still state of the art, although they are now assisted by modern tools, especially digital images (Fig. 3). High-throughput soil coring (Wasson et al., 2014), root crown excavation (Das et al., 2015), and minirhizotron analysis (Maeght et al., 2013) are common ways in which roots are studied in the field, but each provides limited information relative to the actual root structure. Thus, there remains a lack of a coherent view of root phenotypes and their genetic and environmental conditioning (Topp et al., 2016). An ideal technology would allow the explicit in situ visualization of root architecture as it develops through time and likely would involve sensors that were either aboveground or located belowground with minimum invasiveness. The fundamental problem is that roots are largely composed of water and organic matter, which are usually abundant in the soil. Furthermore, most soils have structural inhomogeneities, and other properties, such as water content and temperature, change over time. These factors create significant difficulties for identifying consistent patterns in digital data that can separate root and soil.
Figure 3.
Adding another dimension to the analysis of field-excavated roots. 3D reconstructions are shown for a maize root crown that was excavated, washed, and imaged by x-ray computed tomography. The scan took less than 5 min. A, A segmented volume from which root architectural features can be explicitly measured in 3D space, such as total root length, angles, tortuosity, branching topology, etc. B, A two-dimensional slice of the raw x-ray computed tomography volume showing individual branches from a whorl of nodal roots and information internal to the stem.
While none of these challenges for root phenotyping will likely be solved in the near term, there have been promising advances in sensor development, image analysis, and signal processing. In particular, the application of geophysical survey methods that use magnetic and electrical field information for the nondestructive analysis of soil features has some promise for root phenotyping. Ground-penetrating radar (GPR) uses radio waves (typically microwaves) to discriminate objects based on differences in their electrical permittivity (for review, see Guo et al., 2013). An antenna is used to transmit and receive EM pulses into the ground, which interact with subsurface objects in ways characteristic of their electrical properties. By moving the array across the ground in proscribed transects, a reflectance profile of the subsurface, called a radargram, can be made (Zenone et al., 2008). Roots can generate a characteristic hyperbolic pattern, which can be used to generate two-dimensional images or even 3D models of root systems (Zhu et al., 2014).
There are several limitations to GPR becoming an effective tool for field root phenotyping, especially for fine roots (Hirano et al., 2009), given the fundamental tradeoff between penetration ability and object resolution. Longer wavelengths can penetrate farther but can resolve less detail, and vice versa. Furthermore, electrically conductive materials that have high reflectance to radar, such as wet-clay soils, greatly reduce root discrimination. Soil temperature and the amount of nonliving organic material also are known to influence GPR signal; therefore, extensive calibration must be undertaken, especially for nonuniform soils. Even in ideal conditions (relatively dry, sandy soils), careful comparisons with excavated root systems have not shown strong correlations, especially for roots growing downward (Zenone et al., 2008; Guo et al., 2015a). To date, GPR has not been shown to be capable of detecting fine (approximately less than 2 mm) roots, which may constitute the majority of root length for many plants (Pierret et al., 2005; Brown et al., 2009). GPR may currently be useful to provide a rough estimate of biomass and root distribution in some scenarios, but care should be taken because many important contexts in which we wish to understand root biology (e.g. water content, root angle variation, and interactions with soil structure) also affect our observation ability.
Electrical resistivity tomography (ERT) uses differential electrical properties of materials to map aspects of the soil. ERT has been used to monitor soil water content in a maize field (Beff et al., 2013) as well as tree root biomass distribution (Amato et al., 2008), but it has limited resolution that requires substantial embedded infrastructure and cannot be easily scaled to entire fields. Nonetheless, ERT has shown some potential to improve signal processing for GPR by mapping background soil properties at sites of interest (Zenone et al., 2008). Electrical capacitance also has been exploited to estimate root biomass in plants (for review, see Dietrich et al., 2013) and even for QTL identification in maize (Messmer et al., 2011); however, these relationships are only sometimes consistent (Dietrich et al., 2013). The concept of mapping soil qualities, rather than root structure per se (Werban et al., 2013; Hartemink and Minasny, 2014), provides a potentially powerful ally to direct root detection methods, although many fundamental connections between the two have yet to be established. As machine learning and other computationally intensive statistical methods improve, so will our ability to accurately classify and quantify biological structures and processes from highly multivariate and noisy data (Singh et al., 2012; Aanensen et al., 2014; Smith et al., 2015).
CHALLENGES OF FB-HTP
Based on the limited number of success stories presented herein, it becomes apparent that FB-HTP (and phenomics in general) remains in its infancy, with numerous challenges facing it. Perhaps the most restrictive factor to widespread implementation of FB-HTP is the lack of any commercially available platforms that offer a turn-key solution to address a plethora of biological questions. As the examples referenced within highlight, most FB-HTP platforms are constructed by cross-disciplinary research groups consisting of agricultural engineers, plant breeders, and computer scientists, which demonstrates the amount of expertise required to construct even simple platforms that are one-off solutions. These required resources, including large budgets and specialized personnel, generally are not available to most researchers, thereby putting the needed tools of FB-HTP out of reach for many. Additionally, these purpose-developed platforms are specific to the research question being addressed; therefore, they are not commonly transferable to other crops, field designs, or research settings. This poses a significant limitation, as it prevents the extensive testing of these types of tools that is desired and needed by the larger community.
Looking forward, it is challenging to predict which type of platform will best be able to serve the needs of the research community. Any platform that is to be successful must be easy to use and deploy in the field, inexpensive, utilize robust sensors, have a sizable user community, and have acceptable technical performance under field conditions. The best way to meet these criteria is to develop tools for FB-HTP that are open source so that the largest number of researchers are able to contribute to the design, development, and testing of these needed resources. Currently, UASs embody some of these features because of the extensive research and design invested in them, due to their pervasive application outside of basic research, but they still have a major limitation in that they are only able to phenotype plant canopies. Because of this limitation and others, there is still a strong need for ground-based vehicles that can capitalize on their proximity to plants to increase data resolution as well as capture finer scale phenotypes and permit the transition beyond canopy-level phenotypes. Although the future hardware demands of FB-HTP remain unclear, the required tools and resources will need to become universal, and the most realistic way to achieve that is through low-cost, open-source technology.
Technical limitations aside, the question remains of what value is the vast amount of data that are being generated and how biological meaning will be extracted from these data to answer important questions. Such considerations become especially relevant with regard to the large amount of image data being generated (and video in the near future). Although imaging technologies are relatively new to plant biology, they have been transformative in other fields, including medical and geochemical, and have led to substantial new understandings and capabilities. This is largely because imaging and sensor data allow the capture of multivariate information that may not be perceived by humans but is nonetheless essential for understanding plant biology. This can be exemplified by Chen et al. (2014), who used high-throughput image analysis to gain insight into drought stress physiology. Using an open-source image-analysis pipeline, the authors were able to extract nearly 400 phenotypic traits from images, but more importantly, they were able to derive new traits of physiological importance that captured growth dynamics as well as stress-responsive traits, something not possible with low-throughput phenotyping. Examples of this type highlight the potential for high-dimensional data to reveal new phenotypes that serve to further the basic understanding of plant biology. However, as data of this nature are utilized, a healthy balance must be struck between validating phenotypic data from FB-HTP with state-of-the-art conventional measurements and exploring how novel, data-derived traits can be of value for uncovering biological phenomena.
There are still many issues that stand as hurdles to FB-HTP becoming an accepted technology in the plant science field. Primary among these challenges are the development of resources to carry out this work and how to leverage this information for insight into plant biology. With these considerations in mind, three areas important to the success of phenomics, and FB-HTP in particular, are data management and documentation, characterization of the environment in which plant phenotypes arise, and, most importantly, application of these data for understanding the mechanisms governing plant growth and development.
PLANT PHENOTYPE DATA CAPTURE, MANAGEMENT, AND UTILIZATION
A promising development in FB-HTP is the ability to accurately capture and record phenotype data from field-grown plants using smartphones and tablets. The scope of phenotyping with mobile apps can be broadly classified into on-site experiments conducted by scientists and survey-level or crowd-sourced information to gather information on larger ecological or agriculture landscapes. The data types, computational requirements, and image-processing challenges for these two purposes are largely similar, with minor distinctions in user interface, data structure, and data organization. Plant breeder-led trials generally consist of thousands of distinct entries from a common location, while farmer surveys or surveys of natural populations target hundreds of locations with limited data collection at each site. In order to facilitate data collection and storage, a number of software apps have been developed ranging from research-specific tools to applications for farm- and landscape-level surveys. Field Book is an example of an application developed to specifically meet the requirements of plant breeding programs where detailed data collection is necessary (Rife and Poland, 2014). In contrast, iNaturalist (http://www.inaturalist.org) is an application developed to capitalize on crowdsourcing and is used to catalog biodiversity, identify endangered species, and monitor invasive species. For FB-HTP, improved data management and electronic data capture constitute a critical foundation, and numerous apps have been developed to meet this need (IBreedIT [https://play.google.com/store/apps/details?id=com.ibreedit.mobile.live], Daventi [https://itunes.apple.com/us/app/fieldapp/id457953534?mt=8&ign-mpt=uo%3D4], Phenome Networks [http://phenome-networks.com/phenome-one/application], PhenoType [https://play.google.com/store/apps/details?id=com.phenotype.phenotype], and Diversity Arrays Technology [https://play.google.com/store/apps/details?id=com.diversityarrays.kdsmart]). There also is a growing number of Web apps to curate and link data gathered in the field and, therefore, increase the utility and value of collected data (Aanensen et al., 2014).
In order to be useful, these apps rely on data collection using standard methods, protocols, and formatting. This is due to the fact that, regardless of how they are collected, phenotype data are inherently complex, given that they can range from populations to individual plants to specific tissues and can be recorded on comparative, discrete, or continuous scales. Because the data are essentially infinite in both diversity and scale, data documentation, integration, representation, and accessibility are critical aspects that present significant challenges (for review, see Deans et al., 2015). In order to be broadly useful, large-scale, high-dimensional data sets must be represented in such a way that data can be easily aggregated and extracted to address biologically questions. In addition, data collection methods must be reported at a level to enable experimental reproducibility. Central to these issues is the proper reporting of experimental metadata, the information that describes experimental design, sampling methods, environmental conditions, instrumentation, and analysis (Krajewski et al., 2015). To determine which types of metadata should be reported for a given experiment to make it both discoverable and reusable, Minimal Information About a Plant Phenotyping Experiment standards (http://cropnet.pl/phenotypes; Krajewski et al., 2015) should be followed, and methods used to measure phenotypes should be detailed at a level that enables reproducibility (Sandve et al., 2013). For example, the maize Genomes to Fields initiative has collected phenotype data and has released detailed methods that not only outline how to collect maize phenotypic data under field conditions but also the requisite metadata to enable discovery, aggregation, analysis, and reuse (available at http://www.genomes2fields.org/; see About and then Project Overview and Scope). This information includes detailed descriptions of what traits were measured (plant height, ear height, stand count, etc.), units of measurement, and protocols for how to measure traits, including when the data should be collected, so that all of the project’s phenotypic data can be aggregated regardless of where and by whom they were collected.
To avoid confusion due to simple naming issues and maximize the potential to integrate data across experiments, environments, or even species, the use of well-established nomenclatures, controlled vocabularies, and ontologies for both metadata documentation and phenotypic descriptions is needed. Further impetus to report data using these community standards lies in the fact that many online data repositories rely on them for data representation. However, while mechanisms to report experimental design metadata based on shared vocabularies are comparatively mature, methods to accurately describe phenotypic descriptions are just now emerging. Oellrich et al. (2015) report a proof of concept wherein plant phenotypes for six species were curated as entity quality (EQ) statements to enable analysis by semantic reasoning. For example, dwarf plants are generally short (due to reduced internode length) with leaves that are both reduced in length and increased in width. Using EQ statements, the first of these three aspects of the dwarf phenotype would be reported as “internode length reduced.” Their analyses of the resulting data demonstrated that, once plant phenotype data were represented as ontology-based EQ statements, semantic reasoning could be used to discover shared biology and increase analytical power.
In summary, proper metadata documentation accompanying FB-HTP data is critical for data discovery, analysis, and reuse. To enable cooperative plant phenotyping activities, the International Plant Phenotyping Network (IPPN; http://www.plant-phenotyping.org/) was recently formed. The IPPN provides leadership as the foremost body coordinating multinational efforts for plant phenotyping across many species and represents numerous stakeholders from all sectors of the research community. One goal of this organization is to develop and disseminate standards that aid in the distribution and sharing of phenotypic data. The IPPN represents a useful starting place to get a foothold on the status of existing plant phenotyping efforts, standards, and data sets.
ENVIROTYPING
Although it is well known that plants grow in heterogenous environments, analytical methodologies tend to oversimplify the contribution of the environment to the phenotype. This weakens our understanding of how ecosystems or even individual plants respond to changes in the environment. Furthermore, the long-term effects of environmental conditions, such as temperature and water availability, on phenological development are significant; environmental conditions greatly influence the initiation and duration of vegetative, reproductive, and maturation stages of plants (Bahuguna and Jagadish, 2015; Mickelbart et al., 2015). Due to the interrelatedness of physiological traits, plant developmental phases, and growth conditions, there is a need to gain a better understanding of the complexities of genotype-by-environment interactions. An additional component contributing to these complex interactions are the management practices (genotype-by-environment-by-management interaction) employed in both agricultural and natural system settings that can have significant, large effects on how plants interact with their environment. To this end, envirotyping, which is the detailed characterization of the environment in which plants are growing (Cooper et al., 2014), needs to be employed.
The characterization of atmospheric conditions at local levels has become relatively commonplace with the advent of portable weather stations and interpolation methods using data from regional weather stations. However, the ability to quantify the soil environment, including parameters like pH, organic matter, and moisture content, at high spatial resolution remains challenging given the heterogeneity of soils. Grid soil sampling and electromagnetic soil mapping, which measure soil conductivity to estimate properties like salinity, water content, and organic matter, have proven useful in characterizing spatial soil variability (Mallarino and Wittry, 2004; Brevik et al., 2006; Sun et al., 2013; Guo et al., 2015b). These methods are not without their limitations though; both grid soil sampling and electromagnetic mapping are typically performed once per season, making it impossible to capture the dynamic nature of the soil-water profile. In addition, these methods are too invasive for implementation in most natural systems.
To adequately characterize environmental conditions throughout the course of a season, a network of affordable, open-source sensors is the best option for field-level environmental data collection. Bitella et al. (2014) designed a novel, low-cost hardware platform that incorporates sensors for recording air temperature and relative humidity as well as soil temperature and moisture content at multiple depths. Given the low cost of these sensors, they could be spaced at close, regular intervals throughout the field, providing a high-resolution sampling grid. To further enhance the capability of stationary sensors, the incorporation of wireless networks connected to a central computer (node) could be implemented to synchronize data collection in real time (Hirafuji et al., 2004; Fukatsu and Hirafuji, 2005). By incorporating photovoltaic powered sensors and networks, envirotyping also could be carried out in remote locations for longer periods of time with minimal disruption to the natural habitat.
CROP AND PLANT MODELING
Since their inception in the 1960s, CGMs have developed into useful tools for assessing the adaptation of cropping systems, informing management choices, and guiding policy decisions. Typically, models are run using either historical or predicted weather data (based on unrelated climate models) and are then used to determine optimal solutions of input parameters such as planting date or fertilizer application. Within the last 20 years, models have been used increasingly in crop adaptation and breeding, but this requires better modeling of physiological processes (for summary, see Holzworth et al., 2014). In the context of breeding, models have been used for two purposes, first to extend envirotyping beyond the analysis of environment variables per se (i.e. using the model as an integrator that characterizes the growth environment), and second to extend the phenotyping per se at the level of the plot or plant, sometimes described as model-assisted phenotyping (Luquet et al., 2006; Rebolledo et al., 2015). A newly sought outcome from applications of CGMs is to combine their outputs of development and growth variables with knowledge from genomics and understanding of genetic architecture in order to predict the trait combinations, and eventually the allele combinations, that provide improved solutions in breeding (Technow et al., 2015).
The generation of environment proxies, such as drought indices, that then can be utilized in statistical analyses of phenotypes collected in one or more trials on a set of genotypes (Chapman et al., 2000; Chenu et al., 2011; Cooper et al., 2014). For this purpose, the models need to be capable of simulating the performance of one or more check genotype(s), with the inputs typically being daily weather data and an accurate trial-level soil characterization. The demands on model precision are relatively lower for envirotyping than for other uses: reasonable prediction of phenology and biomass production/water use demand can be sufficient to estimate drought patterns (Chapman, 2008) or accumulated stresses due to heat (Löffler et al., 2005; Zheng et al., 2015). Similar types of applications of models are made in ecological research to estimate the incidence and intensity of favorable or stressful growing conditions, again with a similar need for weather and soil data (Haxeltine and Prentice, 1996). While soil and management are perhaps the major spatial variables to consider in agricultural situations, spatial modeling of the environment in ecosystems is typically more challenging due to changes in topography, species distribution, and multispecies canopy, all of which affect the resulting microclimate and the specific influence on plant adaptation.
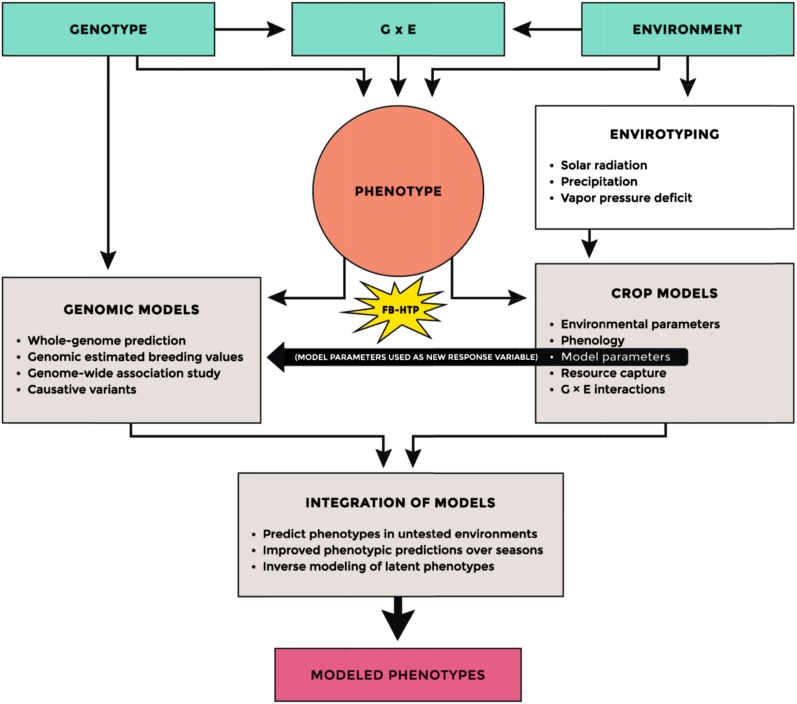
Model-assisted phenotyping can be taken to refer to multiple roles of models in relation to plant phenotyping. These roles include (1) use of models to decide which traits to measure; (2) calculation of extended phenotypes using inputs of plant phenotyping; (3) estimation of plant growth parameters from integration of multiple data sets from plant phenotyping; and (4) feedback from plant phenotyping to improve the physiological basis of models (Fig. 4). Until now, the most common use of models to assist phenotyping has likely been in association with the first role. Many examples have been associated with optimizing the phenology, especially the timing of reproductive stages, with respect to the historic or predicted future occurrence of different types of stresses (Zheng et al., 2015). Most existing models are easily suited to this purpose as long as their estimation of phenology and the damage functions for the stresses are reasonably precise.
Figure 4.
Schematic illustrating the integration of several data types for modeling phenotypes. The flow diagram shows the process in which genotype, environment (including management practices), and their interaction produce the phenotype and how FB-HTP can be integrated with envirotyping, genomic models, and crop models to predict phenotypes. Envirotyping and FB-HTP, in conjunction with results from previous experimentation, provide the multivariate data needed to develop genotype-specific crop models that capture the nonlinear dynamics of plant development under variable environmental conditions. Combining genomic models, which tend to capture additive genetic effects, with crop models may provide the ability to better incorporate nonadditive genetic effects and genotype-by-environment (G × E) interactions. The integration of these two approaches has the potential to significantly enhance the prediction accuracy of modeled phenotypes.
The most desired use of CGMs has been to propose ideotypes of trait combinations that would result in optimal adaptation for given combinations of genotype-by-environment-by-management interactions in current or future climate scenarios (Hammer et al., 2014; Asseng et al., 2015). While the models used may have been well tested for a range of environments and/or genotypes, these models are far more convincing and useful in plant phenotyping and breeding if the modeling has been augmented by physiological and genotypic experimentation. For example, Singh et al. (2012) demonstrated that the incorporation of QTL effects partially responsible for sorghum (Sorghum bicolor) root architecture into a CGM showed that narrow root angle would generally improve yield via increased access to water. The final and most convincing part of this work was that one of these major QTL was mapped across breeding populations and found to be highly associated with higher yields in trials over multiple genetic backgrounds, years, and locations. This comprehensive approach combines experimental data, physiological understanding of a trait from multiple genotypes, model simulation, and breeding trial validation to determine what traits to measure (Hammer et al., 2016). The current cutting edge of phenomics/CGM application in breeding is best described by the CGM-whole-genome prediction approach of Cooper et al. (2016), who show how a relatively simple CGM can be inserted between genomic and phenomic data to impose biological pathway constraints to the statistical prediction models.
CONCLUSION
Progress has been made in FB-HTP, and more generally phenomics, although significant barriers remain preventing its widespread implementation and utilization (see “Outstanding Questions”). Although early results seem to suggest that FB-HTP will be a valuable technology for the plant sciences, questions remain concerning whether the investment in FB-HTP is practical and justified on a larger scale. Contemplating the future, it is clear that open-source, low-cost hardware solutions need to be developed so that more researchers have access to the tools and can test and evaluate them within the context of their respective research programs; only this will determine if expenditure on these resources was warranted. Parallel to hardware improvement, increased research into data management and documentation needs to be implemented so that accurate comparisons can be carried out between different FB-HTP systems, further assisting in the maturation of this nascent technology. As with all new technologies, the initial development phase is often fraught with setbacks, challenges, and vigorous debate, but these are all healthy signs of a technology worth pursuing.
Advancements in all areas of plant science are beginning to bring field-based research into a new era, an era where genomics, envirotyping, and phenomics coalesce to unravel the complexities of plant biology. Continued progress in artificial intelligence coupled with autonomous vehicles will permit the generation of data sets of unprecedented dimensionality as robots begin to collect data nearly around the clock. These advancements in combination with aerial phenotyping, a rapidly maturing technology, and exhaustive envirotyping will create a fully integrated field site whereby trait development and expression can be evaluated in the context of dynamic environmental conditions. By achieving this goal, the transition to studying developmental quantitative genetics becomes possible, thereby allowing the interpretation of genetic effects in relation to the environment and life cycle in a way not previously possible. Building on this framework, CGMs could incorporate these components into their machinery, helping to further the understanding of plant development and its relationship with the environment, which will permit a deeper knowledge of the complexities of trait development in response to changing climatic conditions. Essential to this will be the continued work to integrate CGMs with whole-genome prediction methods to enable the robust prediction of phenotypes in a number of environments, including those in which genotypes have not been tested previously. This should not only assist in the breeding of superior, stress-resilient cultivars but also enable the identification of those natural populations that could be at risk from climatic variation and their interaction with other anthropomorphic disturbances.
OUTSTANDING QUESTIONS
There is a need to evaluate whether the data generated by high-throughput phenotyping is capturing true biological signal and if that signal is worth the investment of resources. Will it provide better information or deeper knowledge?
Will latent phenotypes be more useful than those defined by humans?
Developmental quantitative genetics needs to become the focus of future field-based research so that gene/QTL expression can be understood within the context of both environmental conditions and phenological stages.
The incorporation of CGMs and envirotyping with high-throughput phenotyping will be crucial to understanding the dynamic interaction between plants and their environment. This will help elucidate the physiological mechanisms responsible for observed phenotypes as well as improve the prediction of phenotypes.
Acknowledgments
We thank Daniel Ilut, Elodie Gazave, Christine Diepenbrock, and Maryn Carlson for expert comments on the article and Trevor Rife for contributions on mobile field apps. We apologize for not citing original work of many colleagues because of space constraints.
Glossary
- FB-HTP
field-based, high-throughput phenotyping
- EM
electromagnetic radiation
- 3D
three-dimensional
- UAS
unmanned aircraft system
- QTL
quantitative trait locus
- GPR
ground-penetrating radar
- ERT
electrical resistivity tomography
- EQ
entity quality
- IPPN
International Plant Phenotyping Network
- CGM
crop growth model
Footnotes
This work was supported by Cotton Incorporated Fellowship (to D.P.) and Core Project Funds (to M.A.G.), Cornell University start-up funds (to M.A.G.), the National Science Foundation (grant no. IOS–1238187 to J.P. and M.A.G.), and the Iowa State University Plant Sciences Institute Faculty Scholars Program (to C.J.L-D.).
Articles can be viewed without a subscription.
References
- Aanensen DM, Huntley DM, Menegazzo M, Powell CI, Spratt BG (2014) EpiCollect+: linking smartphones to web applications for complex data collection projects. F1000 Res 3: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato M, Basso B, Celano G, Bitella G, Morelli G, Rossi R (2008) In situ detection of tree root distribution and biomass by multi-electrode resistivity imaging. Tree Physiol 28: 1441–1448 [DOI] [PubMed] [Google Scholar]
- Andrade-Sanchez P, Gore MA, Heun JT, Thorp KR, Carmo-Silva AE, French AN, Salvucci ME, White JW (2013) Development and evaluation of a field-based high-throughput phenotyping platform. Funct Plant Biol 41: 68–79 [DOI] [PubMed] [Google Scholar]
- Araus JL, Cairns JE (2014) Field high-throughput phenotyping: the new crop breeding frontier. Trends Plant Sci 19: 52–61 [DOI] [PubMed] [Google Scholar]
- Asner GP, Knapp DE, Boardman J, Green RO, Kennedy-Bowdoin T, Eastwood M, Martin RE, Anderson C, Field CB (2012) Carnegie Airborne Observatory-2: increasing science data dimensionality via high-fidelity multi-sensor fusion. Remote Sens Environ 124: 454–465 [Google Scholar]
- Asner GP, Martin RE, Anderson CB, Knapp DE (2015) Quantifying forest canopy traits: imaging spectroscopy versus field survey. Remote Sens Environ 158: 15–27 [Google Scholar]
- Asner GP, Martin RE, Knapp DE, Tupayachi R, Anderson C, Carranza L, Martinez P, Houcheime M, Sinca F, Weiss P (2011) Spectroscopy of canopy chemicals in humid tropical forests. Remote Sens Environ 115: 3587–3598 [Google Scholar]
- Asseng S, Ewert F, Martre P, Rotter RP, Lobell DB, Cammarano D, Kimball BA, Ottman MJ, Wall GW, White JW, et al. (2015) Rising temperatures reduce global wheat production. Nat Clim Chang 5: 143–147 [Google Scholar]
- Bahuguna RN, Jagadish KSV (2015) Temperature regulation of plant phenological development. Environ Exp Bot 111: 83–90 [Google Scholar]
- Bannari A, Morin D, Bonn F, Huete AR (1995) A review of vegetation indices. Remote Sens Rev 13: 95–120 [Google Scholar]
- Beff L, Günther T, Vandoorne B, Couvreur V, Javaux M (2013) Three-dimensional monitoring of soil water content in a maize field using electrical resistivity tomography. Hydrol Earth Syst Sci 17: 595–609 [Google Scholar]
- Belasque J Jr, Gasparoto MC, Marcassa LG (2008) Detection of mechanical and disease stresses in citrus plants by fluorescence spectroscopy. Appl Opt 47: 1922–1926 [DOI] [PubMed] [Google Scholar]
- Bitella G, Rossi R, Bochicchio R, Perniola M, Amato M (2014) A novel low-cost open-hardware platform for monitoring soil water content and multiple soil-air-vegetation parameters. Sensors (Basel) 14: 19639–19659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C, Parker P, Cook A, Gottwald T (2008) Visual rating and the use of image analysis for assessing different symptoms of citrus canker on grapefruit leaves. Plant Dis 92: 530–541 [DOI] [PubMed] [Google Scholar]
- Bradley B. (2014) Remote detection of invasive plants: a review of spectral, textural and phenological approaches. Biol Invasions 16: 1411–1425 [Google Scholar]
- Brevik EC, Fenton TE, Lazari A (2006) Soil electrical conductivity as a function of soil water content and implications for soil mapping. Precis Agric 7: 393–404 [Google Scholar]
- Brown AL, Day FP, Stover DB (2009) Fine root biomass estimates from minirhizotron imagery in a shrub ecosystem exposed to elevated CO2. Plant Soil 317: 145–153 [Google Scholar]
- Busemeyer L, Mentrup D, Möller K, Wunder E, Alheit K, Hahn V, Maurer HP, Reif JC, Würschum T, Müller J, et al. (2013a) BreedVision: a multi-sensor platform for non-destructive field-based phenotyping in plant breeding. Sensors (Basel) 13: 2830–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busemeyer L, Ruckelshausen A, Möller K, Melchinger AE, Alheit KV, Maurer HP, Hahn V, Weissmann EA, Reif JC, Würschum T (2013b) Precision phenotyping of biomass accumulation in triticale reveals temporal genetic patterns of regulation. Sci Rep 3: 2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderón R, Navas-Cortés JA, Lucena C, Zarco-Tejada PJ (2013) High-resolution airborne hyperspectral and thermal imagery for early detection of Verticillium wilt of olive using fluorescence, temperature and narrow-band spectral indices. Remote Sens Environ 139: 231–245 [Google Scholar]
- Chapman SC. (2008) Use of crop models to understand genotype by environment interactions for drought in real-world and simulated plant breeding trials. Euphytica 161: 195–208 [Google Scholar]
- Chapman SC, Cooper M, Hammer GL, Butler DG (2000) Genotype by environment interactions affecting grain sorghum. II. Frequencies of different seasonal patterns of drought stress are related to location effects on hybrid yields. Aust J Agric Res 51: 209–222 [Google Scholar]
- Chapman SC, Merz T, Chan A, Jackway P, Hrabar S, Dreccer MF, Holland E, Zheng B, Ling TJ, Jimenez-Berni J (2014) Pheno-copter: a low-altitude, autonomous remote-sensing robotic helicopter for high-throughput field-based phenotyping. Agronomy 4: 279–301 [Google Scholar]
- Chen D, Neumann K, Friedel S, Kilian B, Chen M, Altmann T, Klukas C (2014) Dissecting the phenotypic components of crop plant growth and drought responses based on high-throughput image analysis. Plant Cell 26: 4636–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenu K, Cooper M, Hammer GL, Mathews KL, Dreccer MF, Chapman SC (2011) Environment characterization as an aid to wheat improvement: interpreting genotype-environment interactions by modelling water-deficit patterns in north-eastern Australia. J Exp Bot 62: 1743–1755 [DOI] [PubMed] [Google Scholar]
- Cooper M, Messina CD, Podlich D, Totir LR, Baumgarten A, Hausmann NJ, Wright D, Graham G (2014) Predicting the future of plant breeding: complementing empirical evaluation with genetic prediction. Crop Pasture Sci 65: 311–336 [Google Scholar]
- Cooper M, Technow F, Messina C, Gho C, Totir LR (2016) Use of crop growth models with whole-genome prediction: application to a maize multienvironment trial. Crop Sci 56: 1–16 [Google Scholar]
- Curran PJ. (1989) Remote sensing of foliar chemistry. Remote Sens Environ 30: 271–278 [Google Scholar]
- Damm A, Guanter L, Paul-Limoges E, Van Der Tol C, Hueni A, Buchmann N, Eugster W, Ammann C, Schaepman ME (2015) Far-red sun-induced chlorophyll fluorescence shows ecosystem-specific relationships to gross primary production: an assessment based on observational and modeling approaches. Remote Sens Environ 166: 91–105 [Google Scholar]
- Das A, Schneider H, Burridge J, Ascanio AKM, Wojciechowski T, Topp CN, Lynch JP, Weitz JS, Bucksch A (2015) Digital imaging of root traits (DIRT): a high-throughput computing and collaboration platform for field-based root phenomics. Plant Methods 11: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans AR, Lewis SE, Huala E, Anzaldo SS, Ashburner M, Balhoff JP, Blackburn DC, Blake JA, Burleigh JG, Chanet B, et al. (2015) Finding our way through phenotypes. PLoS Biol 13: e1002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deery D, Jimenez-Berni J, Jones H, Sirault X, Furbank R (2014) Proximal remote sensing buggies and potential applications for field-based phenotyping. Agronomy 4: 349–379 [Google Scholar]
- Dietrich RC, Bengough AG, Jones HG, White PJ (2013) Can root electrical capacitance be used to predict root mass in soil? Ann Bot (Lond) 112: 457–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrler WL. (1973) Cotton leaf temperatures as related to soil water depletion and meteorological factors. Agron J 65: 404–409 [Google Scholar]
- Fang Y, Ramasamy RP (2015) Current and prospective methods for plant disease detection. Biosensors (Basel) 5: 537–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry W. (1982) Disease management in practice. In Principles of Plant Disease Management. Academic Press, New York, pp 303–329 [Google Scholar]
- Fukatsu T, Hirafuji M (2005) Field monitoring using sensor-nodes with a web server. Journal of Robotics and Mechatronics 17: 164–172 [Google Scholar]
- Furbank RT, Tester M (2011) Phenomics: technologies to relieve the phenotyping bottleneck. Trends Plant Sci 16: 635–644 [DOI] [PubMed] [Google Scholar]
- Gates DM, Keegan HJ, Schleter JC, Weidner VR (1965) Spectral properties of plants. Appl Opt 4: 11–20 [Google Scholar]
- Govender M, Dye P, Weiersbye I, Witkowski E, Ahmed F (2009) Review of commonly used remote sensing and ground-based technologies to measure plant water stress. Water SA 35: 741–752 [Google Scholar]
- Granum E, Pérez-Bueno ML, Calderón CE, Ramos C, De Vicente A, Cazorla FM, Barón M (2015) Metabolic responses of avocado plants to stress induced by Rosellinia necatrix analysed by fluorescence and thermal imaging. Eur J Plant Pathol 142: 625–632 [Google Scholar]
- Guo L, Chen J, Cui X, Fan B, Lin H (2013) Application of ground penetrating radar for coarse root detection and quantification: a review. Plant Soil 362: 1–23 [Google Scholar]
- Guo L, Wu Y, Chen J, Hirano Y, Tanikawa T, Li W, Cui X (2015a) Calibrating the impact of root orientation on root quantification using ground-penetrating radar. Plant Soil 395: 289–305 [Google Scholar]
- Guo Y, Huang J, Shi Z, Li H (2015b) Mapping spatial variability of soil salinity in a coastal paddy field based on electromagnetic sensors. PLoS ONE 10: e0127996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland JA. (2010) AgIIS, Agricultural Irrigation Imaging System, design and application. PhD thesis. University of Arizona, Tucson, AZ [Google Scholar]
- Hammer G, Messina C, Van Oosterom E, Chapman S, Singh V, Borrell A, Jordan D, Cooper M (2016) Molecular breeding for complex adaptive traits: how integrating crop ecophysiology and modelling can enhance efficiency. In Yin X, Struik PC, eds, Crop Systems Biology. Springer, pp 147–162 [Google Scholar]
- Hammer GL, McLean G, Chapman S, Zheng B, Doherty A, Harrison MT, Van Oosterom E, Jordan D (2014) Crop design for specific adaptation in variable dryland production environments. Crop Pasture Sci 65: 614–626 [Google Scholar]
- Hartemink AE, Minasny B (2014) Towards digital soil morphometrics. Geoderma 230: 305–317 [Google Scholar]
- Haxeltine A, Prentice IC (1996) BIOME3: An equilibrium terrestrial biosphere model based on ecophysiological constraints, resource availability, and competition among plant functional types. Global Biogeochem Cycles 10: 693–709 [Google Scholar]
- Hirafuji M, Fukatsu T, Haoming H (2004) Full-wireless field monitoring server for advanced sensor-network. In Proceedings of the AFITA/WCCA Joint Congress on IT in Agriculture. pp 692–697 [Google Scholar]
- Hirano Y, Dannoura M, Aono K, Igarashi T, Ishii M, Yamase K, Makita N, Kanazawa Y (2009) Limiting factors in the detection of tree roots using ground-penetrating radar. Plant Soil 319: 15–24 [Google Scholar]
- Holzworth DP, Huth NI, Devoil PG, Zurcher EJ, Herrmann NI, McLean G, Chenu K, Van Oosterom EJ, Snow V, Murphy C, et al. (2014) APSIM: evolution towards a new generation of agricultural systems simulation. Environ Model Softw 62: 327–350 [Google Scholar]
- Homolová L, Malenovský Z, Clevers JGPW, García-Santos G, Schaepman ME (2013) Review of optical-based remote sensing for plant trait mapping. Ecol Complex 15: 1–16 [Google Scholar]
- Jackson RD, Idso SB, Reginato RJ, Pinter PJJ (1981) Canopy temperature as a crop water stress indicator. Water Resour Res 17: 1133–1138 [Google Scholar]
- Jacquemoud S, Baret F (1990) PROSPECT: a model of leaf optical properties spectra. Remote Sens Environ 34: 75–91 [Google Scholar]
- Jedmowski C, Brüggemann W (2015) Imaging of fast chlorophyll fluorescence induction curve (OJIP) parameters, applied in a screening study with wild barley (Hordeum spontaneum) genotypes under heat stress. J Photochem Photobiol B 151: 153–160 [DOI] [PubMed] [Google Scholar]
- Jones HG. (2004) Application of thermal imaging and infrared sensing in plant physiology and ecophysiology. Adv Bot Res 41: 107–163 [Google Scholar]
- Jones HG, Vaughan RA (2010) Remote Sensing of Vegetation. Oxford University Press, New York [Google Scholar]
- Kerr JT, Ostrovsky M (2003) From space to species: ecological applications for remote sensing. Trends Ecol Evol 18: 299–305 [Google Scholar]
- Kim J, Lee S, Ahn H, Seo D, Park S, Choi C (2013) Feasibility of employing a smartphone as the payload in a photogrammetric UAV system. ISPRS J Photogramm Remote Sens 79: 1–18 [Google Scholar]
- Krajewski P, Chen D, Ćwiek H, van Dijk ADJ, Fiorani F, Kersey P, Klukas C, Lange M, Markiewicz A, Nap JP, et al. (2015) Towards recommendations for metadata and data handling in plant phenotyping. J Exp Bot 66: 5417–5427 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK, Miehé JA (1997) Fluorescence imaging as a diagnostic tool for plant stress. Trends Plant Sci 2: 316–320 [Google Scholar]
- Liebisch F, Kirchgessner N, Schneider D, Walter A, Hund A (2015) Remote, aerial phenotyping of maize traits with a mobile multi-sensor approach. Plant Methods 11: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievens B, Thomma BP (2005) Recent developments in pathogen detection arrays: implications for fungal plant pathogens and use in practice. Phytopathology 95: 1374–1380 [DOI] [PubMed] [Google Scholar]
- Lins E, Belasque J, Gasparoto MC, Bagnato VS, Marcassa LG (2006) Fluorescence spectroscopy for detection of citrus canker in orange plantation. In Frontiers in Optics. Optical Society of America, JWD55 [Google Scholar]
- Lobell DB, Asner GP, Law BE, Treuhaft RN (2002) View angle effects on canopy reflectance and spectral mixture analysis of coniferous forests using AVIRIS. Int J Remote Sens 23: 2247–2262 [Google Scholar]
- Löffler CM, Wei J, Fast T, Gogerty J, Langton S, Bergman M, Merrill B, Cooper M (2005) Classification of maize environments using crop simulation and geographic information systems. Crop Sci 45: 1708–1716 [Google Scholar]
- Luquet D, Dingkuhn M, Kim H, Tambour L, Clement-Vidal A (2006) EcoMeristem, a model of morphogenesis and competition among sinks in rice. 1. Concept, validation and sensitivity analysis. Funct Plant Biol 33: 309–323 [DOI] [PubMed] [Google Scholar]
- Lynch M, Walsh B (1998) Genetics and Analysis of Quantitative Traits. Sinauer, Sunderland, MA [Google Scholar]
- Maeght JL, Rewald B, Pierret A (2013) How to study deep roots: and why it matters. Front Plant Sci 4: 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlein AK, Oerke EC, Steiner U, Dehne HW (2012) Recent advances in sensing plant diseases for precision crop protection. Eur J Plant Pathol 133: 197–209 [Google Scholar]
- Mallarino AP, Wittry DJ (2004) Efficacy of grid and zone soil sampling approaches for site-specific assessment of phosphorus, potassium, pH, and organic matter. Precis Agric 5: 131–144 [Google Scholar]
- Marcassa L, Gasparoto M, Belasque J Jr, Lins E, Nunes FD, Bagnato V (2006) Fluorescence spectroscopy applied to orange trees. Laser Phys 16: 884–888 [Google Scholar]
- Martin ME, Plourde LC, Ollinger SV, Smith ML, McNeil BE (2008) A generalizable method for remote sensing of canopy nitrogen across a wide range of forest ecosystems. Remote Sens Environ 112: 3511–3519 [Google Scholar]
- Massacci A, Nabiev SM, Pietrosanti L, Nematov SK, Chernikova TN, Thor K, Leipner J (2008) Response of the photosynthetic apparatus of cotton (Gossypium hirsutum) to the onset of drought stress under field conditions studied by gas-exchange analysis and chlorophyll fluorescence imaging. Plant Physiol Biochem 46: 189–195 [DOI] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN (2000) Chlorophyll fluorescence: a practical guide. J Exp Bot 51: 659–668 [DOI] [PubMed] [Google Scholar]
- Meroni M, Rossini M, Guanter L, Alonso L, Rascher U, Colombo R, Moreno J (2009) Remote sensing of solar-induced chlorophyll fluorescence: review of methods and applications. Remote Sens Environ 113: 2037–2051 [Google Scholar]
- Messmer R, Fracheboud Y, Bänziger M, Stamp P, Ribaut JM (2011) Drought stress and tropical maize: QTLs for leaf greenness, plant senescence, and root capacitance. Field Crops Res 124: 93–103 [Google Scholar]
- Mickelbart MV, Hasegawa PM, Bailey-Serres J (2015) Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat Rev Genet 16: 237–251 [DOI] [PubMed] [Google Scholar]
- Mirmajlessi SM, Destefanis M, Gottsberger RA, Mänd M, Loit E (2015) PCR-based specific techniques used for detecting the most important pathogens on strawberry: a systematic review. Syst Rev 4: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes JM, Technow F, Dhillon BS, Mauch F, Melchinger AE (2011) High-throughput non-destructive biomass determination during early plant development in maize under field conditions. Field Crops Res 121: 268–273 [Google Scholar]
- Mulla DJ. (2013) Twenty five years of remote sensing in precision agriculture: key advances and remaining knowledge gaps. Biosystems Eng 114: 358–371 [Google Scholar]
- Mutka AM, Bart RS (2015) Image-based phenotyping of plant disease symptoms. Front Plant Sci 5: 734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Liu Z, Huo H, Li ZL, Nerry F, Wang Q, Li X (2015) Early water stress detection using leaf-level measurements of chlorophyll fluorescence and temperature data. Remote Sens 7: 3232–3249 [Google Scholar]
- Oellrich A, Walls RL, Cannon EK, Cannon SB, Cooper L, Gardiner J, Gkoutos GV, Harper L, He M, Hoehndorf R, et al. (2015) An ontology approach to comparative phenomics in plants. Plant Methods 11: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli D, Andrade-Sanchez P, Carmo-Silva AE, Gazave E, French AN, Heun J, Hunsaker DJ, Lipka AE, Setter TL, Strand RJ, et al. (2016) Field-based high-throughput plant phenotyping reveals the temporal patterns of quantitative trait loci associated with stress-responsive traits in cotton. G3 (Bethesda) 6: 865–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierret A, Moran CJ, Doussan C (2005) Conventional detection methodology is limiting our ability to understand the roles and functions of fine roots. New Phytol 166: 967–980 [DOI] [PubMed] [Google Scholar]
- Plascyk JA. (1975) The MK II Fraunhofer Line Discriminator (FLD-II) for airborne and orbital remote sensing of solar-stimulated luminescence. Opt Eng 14: 339-0 [Google Scholar]
- Rebolledo MC, Dingkuhn M, Courtois B, Gibon Y, Clément-Vidal A, Cruz DF, Duitama J, Lorieux M, Luquet D (2015) Phenotypic and genetic dissection of component traits for early vigour in rice using plant growth modelling, sugar content analyses and association mapping. J Exp Bot 66: 5555–5566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rife TW, Poland JA (2014) Field Book: an open-source application for field data collection on Android. Crop Sci 54: 1624–1627 [Google Scholar]
- Sandve GK, Nekrutenko A, Taylor J, Hovig E (2013) Ten simple rules for reproducible computational research. PLOS Comput Biol 9: e1003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran S, Khot LR, Espinoza CZ, Jarolmasjed S, Sathuvalli VR, Vandemark GJ, Miklas PN, Carter AH, Pumphrey MO, Knowles NR, et al. (2015) Low-altitude, high-resolution aerial imaging systems for row and field crop phenotyping: a review. Eur J Agron 70: 112–123 [Google Scholar]
- Sankaran S, Mishra A, Ehsani R, Davis C (2010) A review of advanced techniques for detecting plant diseases. Comput Electron Agric 72: 1–13 [Google Scholar]
- Sharma B, Ritchie GL (2015) High-throughput phenotyping of cotton in multiple irrigation environments. Crop Sci 55: 958–969 [Google Scholar]
- Singh V, Van Oosterom EJ, Jordan DR, Hammer GL (2012) Genetic control of nodal root angle in sorghum and its implications on water extraction. Eur J Agron 42: 3–10 [Google Scholar]
- Smith N, Rivera L, Burford N, Bowman T, El-Shenawee M, Desouza G (2015) Towards root phenotyping in situ using THz imaging. In 40th International Conference on Infrared, Millimeter, and Terahertz Waves. IEEE, pp 1–2 [Google Scholar]
- Sun Y, Cheng Q, Lin J, Schellberg J, Schulze Lammers P (2013) Investigating soil physical properties and yield response in a grassland field using a dual‐sensor penetrometer and EM38. J Plant Nutr Soil Sci 176: 209–216 [Google Scholar]
- Technow F, Messina CD, Totir LR, Cooper M (2015) Integrating crop growth models with whole genome prediction through approximate Bayesian computation. PLoS ONE 10: e0130855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp CN, Bray AL, Ellis NA, Liu Z (2016) How can we harness quantitative genetic variation in crop root systems for agricultural improvement? J Integr Plant Biol 58: 213–225 [DOI] [PubMed] [Google Scholar]
- Tucker CJ. (1979) Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens Environ 8: 127–150 [Google Scholar]
- Turner W, Spector S, Gardiner N, Fladeland M, Sterling E, Steininger M (2003) Remote sensing for biodiversity science and conservation. Trends Ecol Evol 18: 306–314 [Google Scholar]
- Ustin SL, Gitelson AA, Jacquemoud S, Schaepman M, Asner GP, Gamon JA, Zarco-Tejada P (2009) Retrieval of foliar information about plant pigment systems from high resolution spectroscopy. Remote Sens Environ (Suppl 1) 113: S67–S77 [Google Scholar]
- Verhoef W. (1984) Light scattering by leaf layers with application to canopy reflectance modeling: the SAIL model. Remote Sens Environ 16: 125–141 [Google Scholar]
- Ward E, Foster SJ, Fraaije BA, McCartney HA (2004) Plant pathogen diagnostics: immunological and nucleic acid‐based approaches. Ann Appl Biol 145: 1–16 [Google Scholar]
- Wasson AP, Rebetzke GJ, Kirkegaard JA, Christopher J, Richards RA, Watt M (2014) Soil coring at multiple field environments can directly quantify variation in deep root traits to select wheat genotypes for breeding. J Exp Bot 65: 6231–6249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver J. (1926) Root Development of Field Crops. McGraw-Hill, New York [Google Scholar]
- Werban U, Bartholomeus H, Dietrich P, Grandjean G, Zacharias S (2013) Digital soil mapping: approaches to integrate sensing techniques to the prediction of key soil properties. Vadose Zone J 12: doi/10.2136/vzj2013.10.0178 [Google Scholar]
- Wessman CA, Aber JD, Peterson DL, Melillo JM (1988) Remote sensing of canopy chemistry and nitrogen cycling in temperate forest ecosystems. Nature 335: 154–156 [Google Scholar]
- White JW, Andrade-Sanchez P, Gore MA, Bronson KF, Coffelt TA, Conley MM, Feldmann KA, French AN, Heun JT, Hunsaker DJ, et al. (2012) Field-based phenomics for plant genetics research. Field Crops Res 133: 101–112 [Google Scholar]
- Wu WR, Li WM, Tang DZ, Lu HR, Worland AJ (1999) Time-related mapping of quantitative trait loci underlying tiller number in rice. Genetics 151: 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadeta KA, Thomma BP (2013) The xylem as battleground for plant hosts and vascular wilt pathogens. Front Plant Sci 4: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Tang J, Mustard JF, Lee JE, Rossini M, Joiner J, Munger JW, Kornfeld A, Richardson AD (2015) Solar-induced chlorophyll fluorescence that correlates with canopy photosynthesis on diurnal and seasonal scales in a temperate deciduous forest. Geophys Res Lett 42: 2977–2987 [Google Scholar]
- Zarco-Tejada PJ, Catalina A, González MR, Martín P (2013) Relationships between net photosynthesis and steady-state chlorophyll fluorescence retrieved from airborne hyperspectral imagery. Remote Sens Environ 136: 247–258 [Google Scholar]
- Zenone T, Morelli G, Teobaldelli M, Fischanger F, Matteucci M, Sordini M, Armani A, Ferrè C, Chiti T, Seufert G (2008) Preliminary use of ground-penetrating radar and electrical resistivity tomography to study tree roots in pine forests and poplar plantations. Funct Plant Biol 35: 1047–1058 [DOI] [PubMed] [Google Scholar]
- Zhang J, Pu R, Yuan L, Wang J, Huang W, Yang G (2014) Monitoring powdery mildew of winter wheat by using moderate resolution multi-temporal satellite imagery. PLoS ONE 9: e93107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Chapman SC, Christopher JT, Frederiks TM, Chenu K (2015) Frost trends and their estimated impact on yield in the Australian wheatbelt. J Exp Bot 66: 3611–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Huang C, Su Y, Sato M (2014) 3D ground penetrating radar to detect tree roots and estimate root biomass in the field. Remote Sens 6: 5754–5773 [Google Scholar]