Arabidopsis accessions show different patterns of integrating root responses to phosphate starvation and salt stress.
Abstract
Nutrient availability and salinity of the soil affect the growth and development of plant roots. Here, we describe how inorganic phosphate (Pi) availability affects the root system architecture (RSA) of Arabidopsis (Arabidopsis thaliana) and how Pi levels modulate responses of the root to salt stress. Pi starvation reduced main root length and increased the number of lateral roots of Arabidopsis Columbia-0 seedlings. In combination with salt, low Pi dampened the inhibiting effect of mild salt stress (75 mm) on all measured RSA components. At higher salt concentrations, the Pi deprivation response prevailed over the salt stress only for lateral root elongation. The Pi deprivation response of lateral roots appeared to be oppositely affected by abscisic acid signaling compared with the salt stress response. Natural variation in the response to the combination treatment of salt and Pi starvation within 330 Arabidopsis accessions could be grouped into four response patterns. When exposed to double stress, in general, lateral roots prioritized responses to salt, while the effect on main root traits was additive. Interestingly, these patterns were not identical for all accessions studied, and multiple strategies to integrate the signals from Pi deprivation and salinity were identified. By genome-wide association mapping, 12 genomic loci were identified as putative factors integrating responses to salt stress and Pi starvation. From our experiments, we conclude that Pi starvation interferes with salt responses mainly at the level of lateral roots and that large natural variation exists in the available genetic repertoire of accessions to handle the combination of stresses.
To optimize their performance in constantly changing conditions, plants need to adjust their developmental program to their environment. A plant’s final phenotype is highly dependent on external signals, and the level of plasticity can facilitate responses to stresses (Pierik and Testerink, 2014). Recently, the importance of root adaptations has received increasing attention. Root morphology can be affected by nutrient availability (Giehl et al., 2014), osmotic stress (Malamy, 2005), salinity (Galvan-Ampudia and Testerink, 2011), and light (Kellermeier et al., 2014). Root development is controlled by auxin and cytokinin signaling (Petricka et al., 2012) and is modulated by external stimuli through other hormones and alterations in auxin or cytokinin sensitivity (Jung and McCouch, 2013). Modulation of root system architecture (RSA) by environmental cues is a result of independent changes of individual RSA traits that may exhibit different sensitivities to the same factor (Gruber et al., 2013; Julkowska and Testerink, 2015). For example, under salt stress, the growth of the main root (MR) of Arabidopsis (Arabidopsis thaliana) Columbia-0 (Col-0) is affected more severely than lateral root (LR) formation and elongation (Julkowska et al., 2014). The responses of both MR and LRs are guided by abscisic acid (ABA) but through different mechanisms (Duan et al., 2013). Upon salt exposure, increased ABA synthesis causes the stabilization of DELLA proteins (Achard et al., 2006). In this way, ABA, through the suppression of GA3 signaling, inhibits cell divisions in the root tip. However, at the later phases of salt stress, low ABA concentrations promote MR growth recovery (Geng et al., 2013), while in LRs, ABA maintains the quiescent state at early stages of their development (Duan et al., 2013). The elongation of LRs in salt conditions also is dependent on the activity of ABA-independent SnRK2.10 protein kinase (McLoughlin et al., 2012).
In natural habitats, plants are usually exposed to multiple stress factors, and one stress can modulate the effect of the other. Nutrient levels have been proposed to influence salt-induced changes in RSA (Duan et al., 2013; Pierik and Testerink, 2014). Contradicting reports on the effect of salinity on lateral root density (LRD) might be explained by the different nutrient concentrations used in basal media (Duan et al., 2013; Julkowska et al., 2014). However, these modulations have not been narrowed down to the level of a particular nutrient, and there is no consensus on the direction of this interaction. One of the most scarce nutrients for plant demands is inorganic phosphate (Pi; Lynch, 2011), which has a significant effect on root morphology (Gruber et al., 2013). Low Pi availability reduces MR growth, but at the same time, LR formation and elongation as well as root hair formation increase (Williamson et al., 2001; López-Bucio et al., 2002; Al-Ghazi et al., 2003; Müller and Schmidt, 2004). In saline soils, phosphate ions tend to form insoluble precipitates that are not available to plants, exposing them to the combination of salt stress and Pi starvation (Russell and Wild, 1988; Naidu and Rengasamy, 1993). Phosphate homeostasis can be modulated by salt stress; for example, in some species, including cotton (Gossypium hirsutum), melon (Cucumis melo), and lupin (Lupinus albus), salt stress reduced phosphate uptake, while an opposite trend was observed for maize (Zea mays; Navarro et al., 2001). Increased phosphate levels had a negative effect on salt tolerance of soybean (Glycine max; Phang et al., 2009).
Arabidopsis accessions of various origins are a useful resource for genetic studies on responses to stress. Exploring natural variation has contributed to the identification of quantitative trait loci controlling RSA development under control and abiotic stress conditions (Mouchel et al., 2004; Rosas et al., 2013; Meijón et al., 2014; Slovak et al., 2014). Root growth under potassium, iron, or phosphate starvation has been linked to allelic polymorphisms (Reymond et al., 2006; Pineau et al., 2012; Kellermeier et al., 2013), whereas salt-induced changes in RSA have partially been explained by differences in the sensitivity to ABA within Arabidopsis accessions (Julkowska et al., 2014). However, natural variation in root responses to combinations of stresses has not been studied before.
In this study, we present a detailed description of how salt stress and phosphate starvation signals affect root architecture. By comparing the growth dynamics of MR and LRs in the Arabidopsis accession Col-0, we show that responses to salt are modulated by Pi starvation (double stress). A comprehensive analysis of a panel of 330 Arabidopsis accessions revealed that not all the components of RSA are affected in the same way by double stress. We classified RSA traits regarding their responses to a combination of salinity and Pi starvation into four major patterns. The combination of salt and phosphate starvation reduced LR development in a similar way to salt stress alone; while the growth of MR was reduced more severely compared with single stress conditions. MR apical zone size was affected in the same manner for salt, Pi deprivation, and their combination, while the basal zone size response showed an intermediate level. We show that the contrasting types of regulation of LR elongation by salt and Pi starvation could be explained by a differential effect of ABA. Finally, with genome-wide association mapping (GWAS), we identified 12 candidate loci putatively involved in responses to the combination of salt and Pi starvation. Our results demonstrate how salt-induced changes in RSA can be dependent on Pi availability and identify putative genetic components in the integration of salt stress and Pi starvation signals.
RESULTS
Phosphate Starvation Modulates MR Growth, LR Emergence, and Elongation in a Different Manner
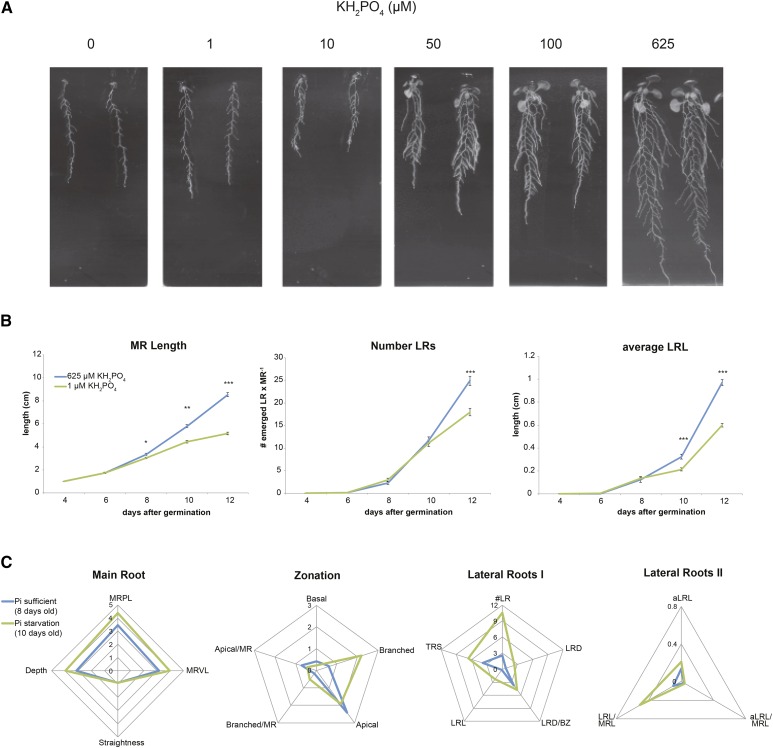
Pi availability is known to influence root growth and development. Due to limited ways of quantifying the RSA of Arabidopsis in soil, most of the reports use agar medium-based plate assays. However, various experimental setups have resulted in different phenotypes (Ristova and Busch, 2014). In order to define the most suitable conditions to capture RSA changes caused by Pi starvation, a dose-response experiment was conducted. Four-day-old seedlings of Arabidopsis accession Col-0 were transferred from control medium (Pi sufficient; 625 μm KH2PO4) to Pi-free medium supplemented with different KH2PO4 concentrations (0, 1, 10, 50, 100, and 625 μm; Fig. 1A). For KH2PO4 concentrations lower than 625 μm (control), starvation symptoms were observed, including the inhibition of root and shoot growth. Surprisingly, 10 μm KH2PO4 restricted MR growth more than 1 μm or total starvation (0 μm). Remarkably, for the other Pi-insufficient concentrations used, the level of MRL inhibition was similar, suggesting its high sensitivity to Pi availability. On the other hand, modulations of LR emergence and growth were observed only at KH2PO4 concentrations below 10 μm.
Figure 1.
Quantification of Arabidopsis root growth and architecture dynamics reveals phosphate-dependent changes in LR traits. Four-day-old seedlings of Arabidopsis accession Col-0 were transferred to 0.5× Murashige and Skoog medium containing 0, 1, 10, 50, 100, or 625 μm KH2PO4. A, Root phenotypes of 14-d-old seedlings. B, Growth dynamics of main root length (MRL), lateral root number (#LR), and average lateral root length (aLRL) from 2 to 8 d after transfer to Pi-sufficient medium (625μm KH2PO4; blue lines) or Pi starvation medium (1 μL of KH2PO4; green lines). Error bars represent se. Values presented are averages for 16 replicates. Statistical comparison was done by Student’s t test for each time point separately, and significant differences between conditions are denoted with asterisks (***, P < 0.001; **, P < 0.01; and *, P < 0.05). C, Quantification of RSA traits on Pi-sufficient medium (625 μm KH2PO4; blue lines) or Pi starvation medium (1 μL of KH2PO4; green lines). Trait descriptions can be found in Table I. RSA traits were quantified from 8- and 10-d-old seedlings for control and Pi starvation, respectively. Values presented are averages for 16 replicates.
To study the dynamics of the Pi starvation effect on root growth in our plate assay, we selected medium containing 0, 1, and 10 μm KH2PO4 as the Pi starvation conditions and followed MRL, number of LRs, and average LR length from the day of the transfer to 12 d after germination (Fig. 1B; Supplemental Fig. S1A). For all Pi-deficient conditions applied, MR growth was found to slow down earlier than LR growth (Fig. 1B; Supplemental Fig. S1A). The almost complete cessation of MR growth 10 d after germination observed only in Pi-deficient conditions suggests Pi to be involved in MR growth maintenance.
In order to get a more detailed picture of the Pi starvation impact on RSA, we quantified 17 RSA traits (Table I) at a single time point. Four-day-old seedlings were transferred to either control (Pi sufficient; containing 625 μm KH2PO4) or Pi starvation (1 μm KH2PO4) medium. When comparing control and Pi-starved seedlings from the same day, it has to be noted that early restriction of the MR growth induced by Pi starvation may lead to underestimation of its effect on root zonation and LR traits. Therefore, RSA traits were quantified at 8 or 10 d after germination for control or Pi starvation, respectively, with EZ-Rhizo software (Armengaud et al., 2009). With this approach, we were able to minimize the differences in MRL between Pi-sufficient and Pi-deprived seedlings (Fig. 1C). The number, density, and total length of LRs as well as total root size were higher on Pi starvation than on the control conditions (Table I; Fig. 1C). Root zonation changed from apical oriented toward a more branched phenotype. (Table I; Fig. 1C). This experimental setup allowed us to compensate for the inhibition of MR growth and capture the changes in RSA induced by Pi starvation.
Table I. Overview of RSA traits measured for control and Pi starvation conditions.
Average values and se of the traits are shown for Arabidopsis accession Col-0. Values presented are averages for 16 replicates. Statistical analysis was performed with Student’s t test. Significant differences are denoted with asterisks (***, P < 0.001; **, P < 0.01; and *, P < 0.05).
| Category | RSA Trait | Description | Unit | Control (8 d) |
Pi Starved (10 d) |
||
|---|---|---|---|---|---|---|---|
| Average | se | Average | se | ||||
| MR | MRPL | Main root path length | cm | 3.455 | 0.102 | 4.355*** | 0.122 |
| MRVL | Main root vector length | cm | 3.146 | 0.088 | 3.947*** | 0.106 | |
| Straightness | MRPL/MRVL | Ratio | 0.911 | 0.003 | 0.907 | 0.003 | |
| Depth | Depth | cm | 3.134 | 0.089 | 3.937*** | 0.104 | |
| Zonation | Basal | Basal zone length | cm | 0.421 | 0.040 | 0.231** | 0.034 |
| Branched | Branched zone length | cm | 0.612 | 0.111 | 2.191*** | 0.092 | |
| Apical | Apical zone length | cm | 2.422 | 0.115 | 1.933** | 0.082 | |
| Branched/MR | Branched zone length per main root path length | Ratio | 0.173 | 0.028 | 0.503*** | 0.015 | |
| Apical/MR | Apical zone length per main root path length | Ratio | 0.703 | 0.029 | 0.443*** | 0.013 | |
| LR | #LR | Number of lateral roots | No. | 2.643 | 0.372 | 10.529*** | 0.763 |
| LRD | Lateral root density per main root cm | No. cm−1 | 0.748 | 0.086 | 2.401*** | 0.146 | |
| LRD/BZ | Lateral root density per branched zone cm | No. cm−1 | 3.730 | 0.517 | 4.760 | 0.223 | |
| aLRL | Average lateral root length | cm | 0.135 | 0.030 | 0.207* | 0.015 | |
| LRL | Lateral root length | cm | 0.343 | 0.070 | 2.256*** | 0.266 | |
| LRL/MRL | Lateral root length per main root length | Ratio | 0.094 | 0.017 | 0.504*** | 0.047 | |
| aLRL/MRL | Average lateral root length per main root length | Ratio | 0.038 | 0.008 | 0.047 | 0.003 | |
| TRS | Total root size | cm | 3.798 | 0.165 | 6.611*** | 0.376 | |
The Availability of Phosphate Modulates RSA Responses to Salt
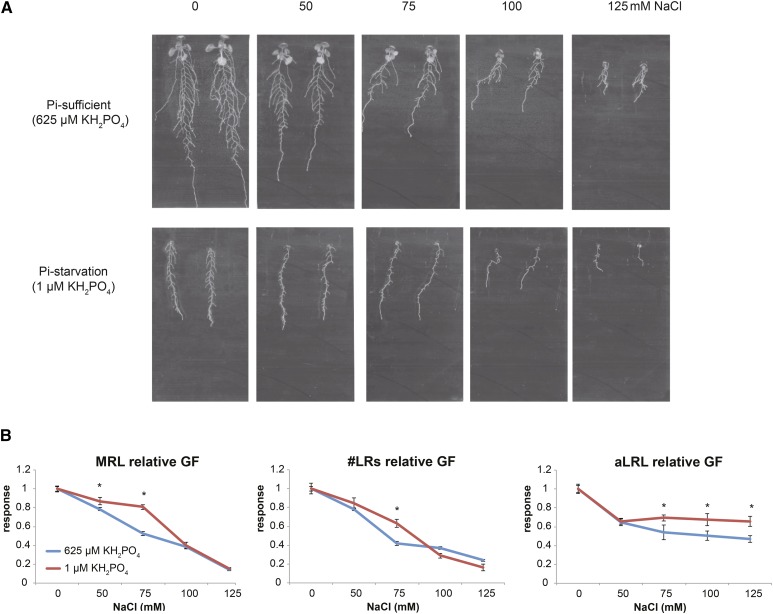
The importance of nutrient levels for RSA responses to salinity was proposed recently (Duan et al., 2013; Pierik and Testerink, 2014). In order to verify how Pi availability affects root development on salt, five NaCl concentrations (0, 50, 75, 100, and 125 mm) were combined with Pi-sufficient (625 μm KH2PO4) or Pi starvation (0, 1, and 10 μm KH2PO4) medium (Fig. 2A). The same trend was observed for all three Pi starvation conditions (Supplemental Fig. S1), and 1 μm KH2PO4 was selected as a representative concentration for further experiments. To compare the impact of Pi levels on the salt sensitivity of MR growth and LR emergence and elongation, we applied the ROOT-FIT model (Julkowska et al., 2014). This model uses a set of quadratic functions to compare salt sensitivity between MRL, #LR, and aLRL. The increases in #LR and aLRL at all conditions applied could be described with quadratic functions (Supplemental Table S1). However, Pi starvation led to linear MR growth (Supplemental Table S1; Supplemental Fig. S2). To be able to compare the effects of different stresses, we used a modified ROOT-FIT model to describe each RSA trait separately with the appropriate function that can best fit RSA under all conditions tested: control, Pi starvation, salt, and their combination. Thus, MRL sensitivity was fitted with a linear function, and #LR and aLRL were fitted with quadratic functions (Supplemental Table S1).
Figure 2.
Pi starvation modulates the effect of salinity on MR and LR growth parameters. Four-day-old seedlings of Arabidopsis accession Col-0 were transferred to Pi-sufficient medium (625 μm KH2PO4) or Pi starvation medium (1 μm KH2PO4) supplemented with 0, 50, 75, 100, or 125 mm NaCl. A, Root phenotypes of 14-d-old seedlings. B, Relative MRL, #LR, and aLRL growth factors (GF). The growth factors for individual values of MRL, #LR, and aLRL were calculated according to a quadratic model (ROOT-FIT) for #LR and aLRL and with a linear function for MRL. The relative growth factors were calculated separately for each Pi concentration as a ratio of the growth factor on each salt concentration divided by the value on medium without salt. Graphs present average values for 16 replicates. Error bars represent se. Statistical comparison was done by two-way ANOVA followed by lsd posthoc test, and significant differences are denoted with asterisks (P < 0.05).
Calculated relative growth factors (normalized to 0 mm NaCl of the same Pi level) allowed us to compare the effect of salinity on growth rates of MR and LRs and its dependency on Pi availability (Fig. 2B; Supplemental Table S1). The inhibitory effect of salt was alleviated when Pi was limited to 50 and 75 mm NaCl for MRL and 75 mm for #LR. At higher salt concentrations (100 and 125 mm), Pi starvation did not influence the salt-induced inhibition of MR growth and LR emergence, suggesting that only below a certain threshold can Pi starvation diminish the inhibitory effect of salinity. Interestingly, for aLRL, the relative growth factors at 75, 100, and 125 mm NaCl were increased by Pi starvation. This suggests that LR elongation is less sensitive to the combination of Pi starvation and high salt concentrations than LR emergence.
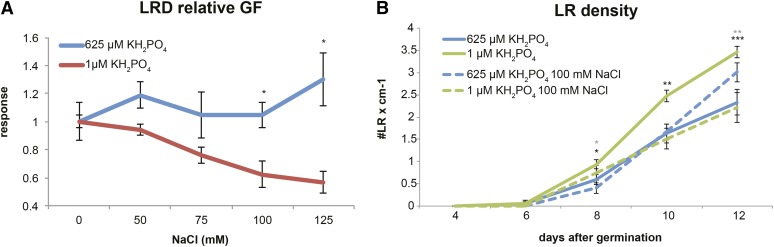
Although salt stress did not affect LRD in Pi-sufficient medium, the application of 100 and 125 mm NaCl under Pi-deficient conditions did decrease LRD (Fig. 3A). On both Pi regimes, the dynamics of LRD fitted the quadratic model (Supplemental Table S1). The calculated rate of changes in LRD was clearly not affected by salt in Pi-sufficient medium, while a gradual decrease was observed under low Pi availability (Fig. 3). Pi starvation alone, on the other hand, increased LRD (Fig. 3B). Together, these data show that the effect of salt on LRD is highly dependent on Pi availability.
Figure 3.
Salt decreases LRD in a dose-dependent manner under Pi-deficient conditions but not on Pi-sufficient medium. A, The effect of Pi levels on LRD was estimated by calculating relative growth factors (GF) for five tested NaCl concentrations on two Pi regimes. The relative growth factors were calculated according to a quadratic model (ROOT-FIT) separately for each Pi concentration as a ratio of growth factors on each salt concentration divided by the value on medium without salt. Graphs present average values for 16 replicates. Statistical comparison was done by two-way ANOVA followed by lsd posthoc test, and significant differences are denoted with asterisks (P < 0.05). B, Increase in LRD from 2 to 8 d after transfer to Pi-sufficient medium (625 μm KH2PO4; blue line) or Pi starvation medium (1 μL of KH2PO4; green line) supplemented with 0 mm (solid lines) or 100 mm (dashed lines) NaCl. Values presented are averages for 16 replicates. Statistical comparison was done by Student’s t test for each time point separately, and significant differences between conditions are denoted with asterisks (***, P < 0.001; **, P < 0.01; and *, P < 0.05). Error bars represent se.
Natural Variation in RSA Integration of Salt and Phosphate Starvation Responses
Natural variation in root morphology adjustments was reported recently for salt (Julkowska et al., 2014) as well as for Pi starvation (Chevalier et al., 2003; Reymond et al., 2006), yet never for their combination. To describe natural variation in responses to the joint effect of salt stress and Pi starvation, changes in RSA of 330 Arabidopsis accessions from the HapMap population (Atwell et al., 2010) were followed. Four-day-old seedlings were transferred to control (625 μm KH2PO4), salt (75 mm NaCl), Pi starvation (1 μm KH2PO4), or double stress (1 μm KH2PO4 and 75 mm NaCl) medium. Root growth was monitored from 6 to 14 d after germination (Supplemental Fig. S3). Seventeen RSA traits (Table I) were quantified with EZ-Rhizo software (Armengaud et al., 2009) for all accessions at 8 and 10 d after transfer for control and all stress conditions, respectively. In order to get an overview of the effect of the double stress on the whole HapMap population, for each individual RSA trait, we calculated the average from 330 accessions studied. The impact of each stress (salt, Pi starvation, and double stress) was analyzed by checking the statistical significance of difference for each individual RSA trait on stress conditions from the control (Supplemental Table S3). All studied RSA traits were affected by at least one stress condition. #LR decreased in the presence of salt for Col-0 but was not influenced at the level of the whole population (Fig. 2B; Supplemental Table S3). LRD, not affected by salinity for Col-0, decreased in the case of the HapMap population (Fig. 2B; Supplemental Table S3). This implies that the picture obtained by studying Col-0 is not representative for a wider selection of accessions. For the HapMap population, double stress had no effect on MRPL, apical zone size, and LRD, even though both salt and Pi starvation alone affected these traits. Lack of the influence of double stress on #LR and total root size was consistent with the same trend for salt stress. Interestingly, we did not find any RSA traits that were affected only by double stress (Supplemental Table S3).
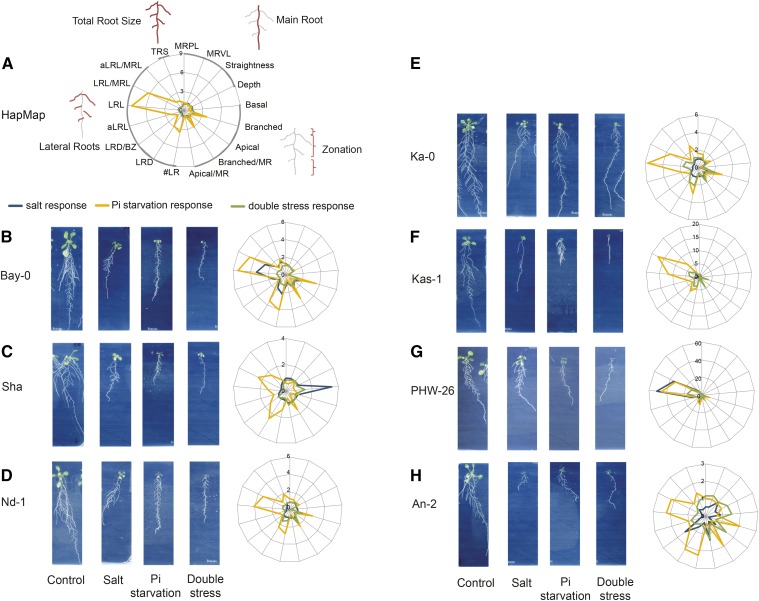
To be able to compare the relative effects of all treatments for each accession separately as well as for the HapMap population, values of the RSA traits were normalized to the corresponding values under control conditions. The differences in RSA responses between each tested condition for the entire population and for selected accessions are presented by spider webs (Fig. 4). These illustrate that overall responses of the HapMap population show a high impact of low Pi availability on LR parameters, reflected by an 8 times higher response of #LR to Pi starvation than to salt or double stress (Fig. 4A; Supplemental Table S3). All of the accessions studied showed an increase in #LR on Pi starvation, but only half of them were able to maintain this response in double stress conditions.
Figure 4.
Natural variation in RSA responses to phosphate starvation, salt stresses, and their combination. A, Radial graph presenting average responses to salt, Pi starvation, and double stress of the whole population studied. B to H, Phenotypes of 14-d-old seedlings under control and stress conditions. Four-day-old seedlings were transferred to control (625 μm KH2PO4), salt (625 μm KH2PO4 and 75 mm NaCl), phosphate starvation (1 μm KH2PO4), or double stress (1 μm KH2PO4 and 75 mm NaCl) medium. Radial (spider web) graphs present responses to salt, Pi starvation, and double stress of the HapMap population (A) or accessions Bay-0 (B), Sha (C), Nd-1 (D), Ka-0 (E), Kas-1 (F), PHW-26 (G), and An-2 (H). Responses were calculated by dividing the value of an RSA trait by the average of the trait on control condition. The legend for all the graphs is presented in A.
We selected three accessions that were described previously to have differential responses to the single stresses Pi starvation (Bayreuth [Bay-0] and Shahdara [Sha]) or salt stress (Niederzenz [Nd-1]; Reymond et al., 2006; Julkowska et al., 2014). Sha and Bay-0 show different responses of the MR to Pi starvation (Reymond et al., 2006). Despite using different media and germinating them on control conditions before applying stress, we confirmed that the reduction of MR was more severe for Sha than for Bay-0 on Pi starvation as well as on a double stress (Fig. 4, B and C). Interestingly, the MR of Sha was inhibited the most by Pi starvation, while on double stress, this effect was masked and the response was similar to that for salt. MR growth of Bay-0 responded to salt and Pi starvation in a similar way, but the response to their combined effect was lower than in the case of single stresses. This suggests that Bay-0 and Sha possess different adaptive strategies not only to Pi starvation but also for salt on two different Pi concentrations. Nd-1, identified previously to decrease MRL, #LR, and aLRL in the presence of salt, showed a positive response for these traits on Pi starvation, while the double stress response value was in between the single stress ones (Fig. 4D).
We also selected two accessions with responses to Pi starvation that deviated from the rest of the population. Despite the huge inhibitory effect that Pi starvation has on the MR of most accessions, Karnten (Ka-0; Fig. 4E) was able to maintain MR growth on Pi starvation as well as on double stress, while a decrease in MR was observed in the case of salt stress. Another type of extreme phenotype was represented by Kashmir (Kas-1), showing the typical Pi starvation brushy root with highly inhibited MR and numerous LRs, not maintained on double stress (Fig. 4F).
Additionally, we selected two accessions with contrasting responses to double stress. The spider web representation of the responses of PHW-26 illustrates that all LR traits increased, but the degree of the response varied (Fig. 4G). On the contrary, the response of Antwerpen An-2 to double stress did not resemble changes made by individual stresses and showed a unique pattern of response (Fig. 4H).
Our results suggest that, next to the natural variation in responses to the single action of salinity and Pi starvation, how RSA responds to their combined effect differs between accessions. The different shapes of the spider webs illustrate the myriad various stress integration responses for all RSA components. The RSA modulation induced by double stress could not have been predicted by studying these two stresses separately.
RSA Responses to the Combination of Salt and Phosphate Starvation Can Be Classified into Four Patterns
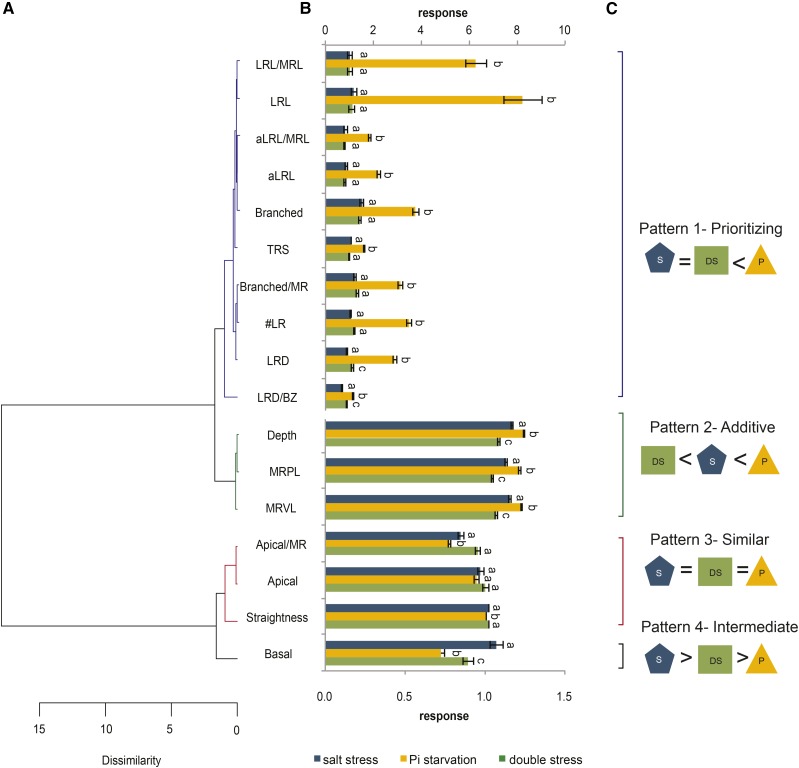
To analyze the RSA changes to the combined action of salt and Pi starvation, we classified responses to single and double stresses at the level of the whole population studied. Based on hierarchical clustering of the average HapMap population responses, we classified separate RSA traits based on the integration of salt stress and Pi starvation into four patterns. The effect of salt stress and double stress was similar for all of the LR-related traits, while Pi starvation alone resulted in a positive response for these traits (Fig. 5, B and C; Supplemental Table S4). It seems that the inhibitory effect of salt on LRs is dominant and Pi starvation cannot overcome it. In that way, roots tend to prioritize their response to salt (pattern 1). An additive effect was found for traits describing MR features, for which the responses to double stress were lower than those to salt or Pi starvation alone (pattern 2; Fig. 5, B and C; Supplemental Table S4). Apical zone size and straightness responded in a similar way to both single and double stresses (pattern 3; Fig. 5, B and C; Supplemental Table S4). Finally, double stress had an intermediate effect on the basal zone relative to the responses to salt and Pi starvation (pattern 4; Fig. 5, B and C; Supplemental Table S4). Interestingly, we did not find any traits for which the double stress response would resemble the response to Pi starvation alone. This approach enabled us to discover that individual RSA components integrate salt stress and Pi starvation in a different way, with a dominant effect of salinity on LRs for most of the accessions tested.
Figure 5.
RSA responses to the combined effect of salt and phosphate starvation can be classified into four major patterns. Four-day-old seedlings were transferred to control (625 μm KH2PO4), salt (625 μm KH2PO4 and 75 mm NaCl), Pi starvation (1 μm KH2PO4), or double stress (1 μm KH2PO4 and 75 mm NaCl) medium. For each treatment, values of the RSA traits at day 10 were normalized for the corresponding values under control conditions at day 8. For each trait, averages for whole HapMap population were calculated for every stress condition tested. A, Dendrogram of responses to salt, Pi starvation, and double stress obtained by hierarchical clustering by the Ward linkage method. B, Responses of 17 RSA traits to salt stress, Pi starvation, and double stress. Values represent averages for the whole population studied. Error bars represent se. Statistical analysis was done for each trait separately with one-way ANOVA followed by the Bonferroni multiple comparison test (P < 0.05). Different letters denote significant differences between conditions within the same trait. Traits from the top cluster are plotted on the top axis, and the others are plotted on the bottom axis. C, Four patterns of responses to double stress. Pattern 1 (prioritizing) clusters traits that show a response to double stress (DS) resembling the response to salt (S) but different from the response to Pi starvation (P). Pattern 2 (additive) represents traits with different responses for all treatments. Pattern 3 (similar) represents traits that showed the same responses to salt, Pi starvation, and double stresses. Pattern 4 (intermediate) traits show a response to double stress between those of the single stresses.
To determine whether the four identified patterns would be representative for all accessions, we next clustered all the individual accessions based on their response to salt, Pi starvation, and double stress for each individual RSA trait. Indeed, the majority of the accessions integrated salt and Pi starvation signals according to the pattern found for the whole HapMap population (Supplemental Fig. S4; Supplemental Table S5). Interestingly, we also identified rare accessions with different integration patterns: NFA (NFA-10), PAR-5, Rschew (Rsch-4), and Tabor (Ta-0) UKSE06-272 prioritized the response to Pi starvation over salt for lateral root length (LRL; Supplemental Table S5).
Root Responses to Double Stress Partly Rely on RSA Phenotypes on Salt and Pi Starvation
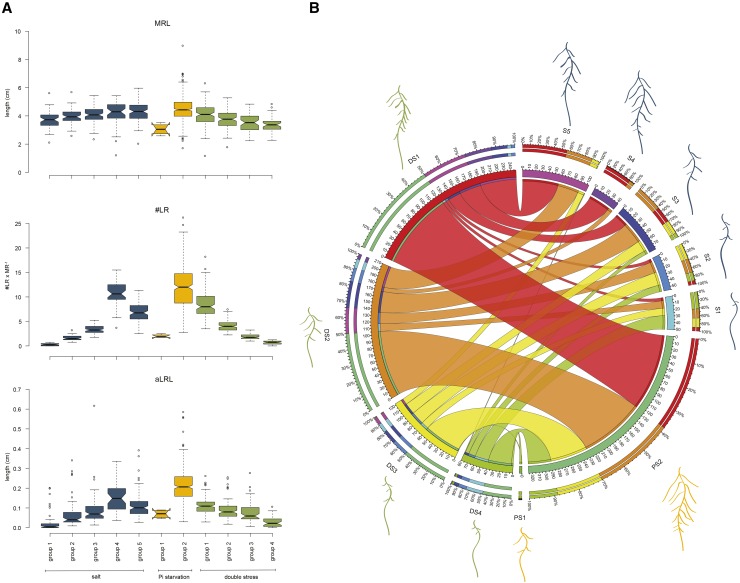
Next, we asked to what extent RSA on double stress is dependent on single stress phenotypes. Accession-specific correlations between double stress and single stresses were studied for each RSA trait (Supplemental Table S6). All phenotypes, except apical zone size as a portion of MRL, showed a significant correlation between double stress and both single stresses. The strongest correlations were observed for total root size, MRL, #LR, and aLRL, and their values were similar for salt stress and Pi starvation. This indicates that responses to double stress are related to phenotypes on single stresses. Therefore, we clustered 330 accessions based on MRL, #LR, and aLRL separately for salt, Pi starvation, and double stress. Five groups of accessions were identified for salt conditions (Fig. 6A), with the majority of accessions showing high MRL and intermediate #LR and aLRL (group 5), and the smallest number of accessions clustered in group 4, representing the highest MRL, #LR, and aLRL. For Pi starvation, two groups were identified. Only four accessions, Ostra Mocklo Ömö 2-1, Rsch-4, Caen (Cen-0), and T1040, showed a distinct phenotype of low MRL, #LR, and aLRL (group 1); all others responded to Pi starvation with higher #LR and aLRL (group 2). Four groups were found for double stress, with most of the accessions in group 1 showing the highest total root size for this condition.
Figure 6.
Natural variation reveals the relationship between RSA on double stress and single stresses. A total of 330 accessions were clustered with the Ward linkage method based on their MRL, #LR, and aLRL on salt, Pi starvation, and double stress. A, MRL, #LR, and aLRL of the identified groups. The box plots present median trait values observed for all accessions within a group. The whiskers extend to data points that are less than 1.5× from the interquartile range (IQR) away from the first and third quartiles. Notches represent 1.58 × IQR/square root (n) and give 95% confidence that two medians differ. B, Relationships between individual groups found for double stress (DS1–DS4), salt (S1–S5), or Pi starvation (PS1 and PS2). The outer circle shows the percentage of accessions that were found in clusters from other conditions, represented by colors corresponding to the inner circle. The size of each circle corresponds to the number of accessions found in each group. Ribbon size encodes the number of accessions from single stress clusters that were found in each double stress group. Graphical visualization of average RSA is presented for each group shown in A.
We examined the relationship between the groups on double stress compared with salt or Pi starvation (Fig. 6B; Supplemental Table S7). Accessions with the most developed RSA on salt (group 4) were mostly found with similar RSA on double stress (group 1). In accordance, the highest proportion of members of salt groups 1 and 2 with the lowest MRL, #LR, and aLRL belonged to double stress groups 4 and 3, respectively. Accessions with poorly developed RSA on Pi starvation (group 2) were likely to have even fewer and shorter LRs on double stress (groups 3 and 4). Together, these findings suggest that, in general, root phenotypes on salt and Pi starvation determine the phenotypes on double stress. However, for almost 30% of accessions developing high #LR and aLRL on Pi starvation (group 2), this ability was masked in double stress conditions (groups 3 and 4). Moreover 10 accessions (Belmonte-4-94, Coimbra Co-2, LI-OF-095, MIB-22, MIB-84, N (N4), PAR-5, TDr-1, Tottarp-2, and UKNW06-460) showed high overall root size on both salt (group 5) and Pi starvation (group 2) but were not able to maintain it when two stresses were combined (group 3; Supplemental Table S7).
Accessions described previously as salt tolerant (Tsu [Tsu-0], Burren [Bur-0], and Wildbad [Wl-0]) were found in the same salt and double stress RSA response groups as those shown previously to be salt sensitive (Col-0 and Cape Verde Islands [Cvi-0]; Rus et al., 2006; Katori et al., 2010). Moreover, we tested the correlation of our data from MRL, #LR, and aLRL with earlier reports on rosette size and water content in salt conditions (Supplemental Table S8; Julkowska et al., 2016). The only significant, but still very low (r2 = 0.23), correlation was found between #LR on double stress and rosette size in salt stress. Nevertheless, it has to be noted that the setup of these experiments differed greatly from our setup.
Identification of the Genetic Components of Responses to Double Stress by GWAS
Phenotypic data of 17 RSA trait responses to salt, Pi starvation, and double stress obtained for 330 accessions were used for association mapping with a panel of 250,000 single-nucleotide polymorphism (SNP) markers (Atwell et al., 2010). GWAS was performed with the scan_GLS algorithm (Kruijer et al., 2015), which includes correction for population structure (Cao et al., 2011). Together, 22 SNPs were found to be associated with double stress responses, the majority of these associated with the trait LRL. Linkage disequilibrium (LD) was calculated for each SNP, and gene selection was extended with markers in LD with mapped SNPs (Kooke et al., 2016). Eleven loci containing 19 genes were mapped specifically with responses to double stress, and one locus containing 17 genes in LD with the identified SNP was found to be associated with responses to salt, Pi starvation, and double stress (Table II). Among the selected candidates, four protein kinases were found: At5g11400 and At5g11410 in the locus associated with responses to salt, Pi starvation, and double stress, and At2g44830 and SERK2 as putative factors guiding responses specific for double stress. SERK2 is Leu-rich repeat receptor-like kinase that has been shown to be involved in brassinosteroid signaling (Gou et al., 2012). The identified loci also included regulators of ABA signaling (SOAR1; Mei et al., 2014) and auxin biosynthesis (YUC4; Cheng et al., 2006) as well as genes engaged with sugar signaling (TPPC; Schluepmann et al., 2004), cell division (CYCA2;2; Vandepoele et al., 2002), microtubule functioning (At2g188876), mRNA metabolism (At5g28220 and At5g11412), and endosomal sorting (ISTL1; Buono et al., 2016).
Table II. Candidate loci associated with responses to the combination of salt and Pi starvation identified with GWAS.
The candidate genes were selected based on LOD score [−log10(α × P value)] and different thresholds of minor allele frequency (MAF) of the SNP associated with the RSA phenotype. For loci containing SNPs in LD with an associated marker, the SNP initially associated with the RSA trait is indicated with an asterisk.
| Locus | Chromosome | Marker | Position | LOD | Phenotype | MAF | Gene | Gene Description | Remarks |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | m13374 | 7,841,617 | 6.85 | aLRL | 0.01 | AT1G22210 | TPPC, TREHALOSE-6-PHOSPHATE PHOSPHATASE C | Specific for responses to double stress |
| LRL/MRL | |||||||||
| 2 | m22386 | 12,461,353 | 5.86 | LRL | 0.1 | AT1G34210 | SERK2, SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE2 | ||
| m22387 | 12,461,560 | 6.50 | 0.1 | ||||||
| m22389 | 12,461,977 | 6.07 | 0.1 | ||||||
| m22392 | 12,462,722 | 6.07 | 0.1 | ||||||
| m22399 | 12,464,518 | 6.02 | 0.1 | ||||||
| m22400 | 12,464,689 | 6.02 | 0.1 | AT1G34220 | IST1-LIKE1, ISTL1 | ||||
| m22403 | 12,465,407 | 6.02 | 0.1 | ||||||
| m22415 | 12,470,273 | 6.46 | 0.1 | AT1G34240 | hAT-like transposase family | ||||
| m22422 | 12,472,127 | 6.27 | 0.1 | AT1G34245 | EPF2, EPIDERMAL PATTERNING FACTOR2 | ||||
| 3 | 2 | m64345 | 8,047,170 | 6.12 | MRPL | 0.1 | AT2G18540 | mIC-like cupins superfamily protein; functions in nutrient reservoir activity | |
| 5.96 | MRVL | 0.1 | |||||||
| 4 | m64548 | 8,152,277 | 6.14 | LRL | 0.01 | ||||
| m64560 | 8,155,450 | 6.01 | 0.01 | AT2G18820 | Non-LTR retrotransposon family (LINE) | ||||
| m64583 | 8,166,012 | 6.85 | 0.01 | ||||||
| 5 | m64591 | 8,169,986 | 5.89 | LRL | 0.05 | AT2G18876 | Encodes a microtubule-associated protein | ||
| 6 | m64600 | 8,177,544 | 5.71 | LRL | 0.01 | AT2G18880 | VEL2, VERNALIZATION5/VIN3-LIKE2 | ||
| 7 | m68418 | 10,952,801 | 7.16 | MRVL | 0.1 | AT2G25710 | HCS1, HOLOCARBOXYLASE SYNTHASE1 | ||
| 6.97 | MRPL | 0.1 | |||||||
| 8 | m78909 | 18,487,046 | 5.64 | aLRL | 0.1 | AT2G44830 | Protein kinase superfamily protein | ||
| 9 | 3 | m82851 | 1,350,879 | 6.12 | LRL | 0.01 | AT3G04903 | Encodes a defensin-like (DEFL) family protein | |
| 10 | 4 | ml38169 | 7,153,336 | 5.86 | LRL | 0.1 | AT4G11900 | S-Locus lectin protein kinase family protein | |
| 11 | 5 | At5G11280 | Unknown | Common for responses to double stress, salt, and Pi starvation | |||||
| At5G11290 | Encodes a plant protein of unknown function | ||||||||
| At5G11300 | CYCA2;2, MITOTIC-LIKE CYCLIN3B from Arabidopsis | ||||||||
| At5GU310 | SOAR1, SUPPRESSOR OF THE ABAR OVEREXPRESSOR1; negative regulator of ABA signaling | ||||||||
| At5G11320 | AtYUC4, YUC4, YUCCA4 | ||||||||
| At5G 11330 | FAD/NAD(P)-binding oxidoreductase family protein | ||||||||
| At5G11340 | Acyl-CoA N-acyltransferases (NAT) superfamily protein | ||||||||
| At5G11350 | DNase l-like superfamily protein | ||||||||
| At5G11360 | lnterleukin-1 receptor-associated kinase4 protein | ||||||||
| At5G11370 | FBD/Leu-rich repeat domain-containing protein | ||||||||
| At5G11380 | 1-DEOXY-d-XYLULOSE 5-PHOSPHATE (DXP) SYNTHASE3 | ||||||||
| At5G11390 | WIT1, WPP DOMAIN-INTERACTING PROTEIN1 | ||||||||
| At5G 11400 | Protein kinase superfamily protein | ||||||||
| At5G11410 | Protein kinase superfamily protein | ||||||||
| At5G11412 | RNA-binding (RRM/RBD/RNP motifs) family protein | ||||||||
| At5G11420 | Unknown | ||||||||
| ml67447 | 3,649,667 | 6.97 | MRPL | 0.01 | At5G11430* | SPOC domain/transcription elongation factor S-ll protein | |||
| 6.42 | MRVL | ||||||||
| AT5G28210 | mRNA-capping enzyme family protein | Specific for responses to double stress | |||||||
| ml81370 | 10,180,694 | 5.99 | aLRL/MRL | 0.01 | AT5G28220* | Protein prenylyltransferase superfamily protein | |||
| 12 | AT5G28232 | Mutator-like transposase family | |||||||
| AT5G28235 | Ulpl protease family protein | ||||||||
| AT5G28240 | Similar to Ulpl protease family protein | ||||||||
| AT5G28263 | Mutator-like transposase family |
ABA Sensitivity Underlies the Different Responses of LRs to Salt and Phosphate Starvation
One of the important components of salt signaling in roots is ABA (De Smet et al., 2003; Duan et al., 2013). Despite the huge overlap between transcripts regulated by Pi starvation and ABA (Woo et al., 2012), the exact influence of ABA on responses to Pi limitation has never been confirmed (for review, see Chiou and Lin, 2011), and this question was never addressed to roots specifically. In order to verify whether the opposite effect of salt and Pi starvation (Supplemental Table S3) on LR growth can be due to different ABA sensitivity and to elucidate whether ABA can take part in integrating the combined effects of salinity and Pi limitations, we tested the RSA of the abi1-1 mutant on salt, Pi starvation, and double stress. The abi1-1 mutation leads to constitutive activation of the protein phosphatase ABA-INSENSITIVE1 (ABI1), causing complete abolishment of ABA responses (Bertauche et al., 1996; Leung et al., 1997).
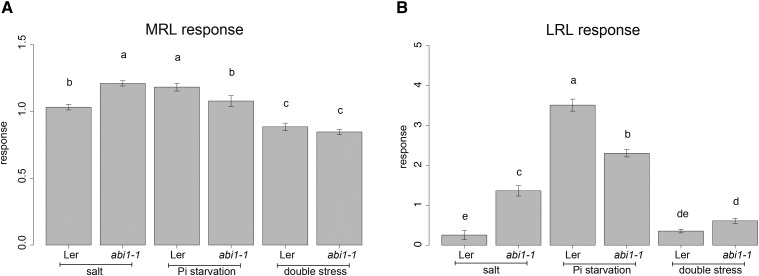
In control conditions, abi1-1 developed shorter MRL in comparison with the background line Landsberg erecta (Ler; Supplemental Fig. S5A). Consistent with previous studies, abi1-1 seedlings failed to inhibit LR growth in response to salt stress (Fig. 7B; Duan et al., 2013). On the other hand, abi1-1 developed shorter LR under Pi-deficient conditions (Fig. 7), suggesting that ABA has a positive role in maintaining LR elongation in limited Pi conditions. Double stress had an additive effect on MRL for both Ler and abi1-1, since their MR was shorter on double stress than on individual stresses (Supplemental Fig. S3A). Ler seemed to prioritize its LRL responses to salt (Fig. 7B; Supplemental Fig. S5B), since the increase in LRL made by Pi starvation was completely masked in the presence of salt. In the case of abi1-1, the LRL response to double stress also was suppressed (Supplemental Fig. S5, A and B). Taking into account the differences in MRL observed already in control conditions, the responses of MRL to double stress were similar for Ler and abi1-1 (Supplemental Fig. S5A). The latter showed a slight increase in LRL responses compared with Ler, but this difference was not significant (Fig. 7B). This suggests that the contrasting effect of salt and Pi starvation on LRs might be explained by differential ABA influence, but the integration of responses to the combination of these stresses does not rely on ABA signaling or requires multiple factors.
Figure 7.
ABA inhibits LR growth in response to salt and promotes LR growth on Pi starvation. Four-day-old seedlings of Ler and abi1-1 were transferred to control (625 μm KH2PO4), salt (625 μm KH2PO4 and 75 mm NaCl), phosphate starvation (1 μm KH2PO4), or double stress (1 μm KH2PO4 and 75 mm NaCl) medium. MRL (A) and LRL (B) were quantified from 8- and 10-d-old seedlings for control and stress, respectively. Responses were calculated by dividing the value of an RSA trait by the average of the trait on control conditions. Values presented are averages for 18 replicates. Error bars represent se. Statistical comparison was done by two-way ANOVA followed by lsd posthoc test (P < 0.05). Different letters indicate significant differences.
DISCUSSION
Root growth and development is strongly dependent on a plant’s environment. Surviving in heterogenous soil conditions relies on root plasticity that enables a plant to forage for scarce nutrients or water but also to avoid unfavorable factors such as salinity (Galvan-Ampudia et al., 2013; Giehl and von Wirén, 2014). Root morphology changes caused by environmental factors were identified previously by studying the effect of individual stresses. In their natural habitats, plants are exposed to multiple stresses, and responses to them in general differ from the responses to single stresses or a sum of the individual stresses. Transcriptomics and proteomics studies were the first to reveal the complexity of the response to multiple stresses (Rizhsky et al., 2002; Koussevitzky et al., 2008; Rasmussen et al., 2013; Rivero et al., 2014; Sewelam et al., 2014). Besides the transcriptional responses to multiple stresses, their physiological consequences have been studied only recently. RSA modulation by multiple nutrient deficiencies was found to be controlled by signaling modules controlling nutrient interactions (Kellermeier et al., 2014). The existence of marker RSA traits for individual nutrients and their combinations was proposed (Kellermeier et al., 2014).
Here, we first describe the RSA modulations in response to Pi starvation alone and then reveal how Pi levels can modulate responses to salt. Phosphate availability is known to influence root growth and development. The tendency to accumulate Pi in upper parts of the soil results in the establishment of shallow root systems, with strongly inhibited MR and numerous, long LRs (Péret et al., 2011). In Col-0 Arabidopsis seedlings, phosphate starvation reduces MR growth and increases the elongation of LRs, while there is no agreement on its effect on LRD (Williamson et al., 2001; Linkohr et al., 2002; López-Bucio et al., 2002; Al-Ghazi et al., 2003). In the experimental setup we chose here, all LR traits (#LR, LRL, and LRD) increased under phosphate-limited conditions (Fig. 1C; Table I). We observed that, 4 d after germination, the growth of MR on low-Pi medium slows down, suggesting that the Pi reservoir in 4-d-old seedlings is consumed by that time (Fig. 1B). Similar results were observed previously (Svistoonoff et al., 2007), where MR growth was arrested 2 d after transfer when 3-d-old seedlings were transferred to Pi-deficient medium. Low Pi availability decreases cell elongation and proliferation in the root tip, where Pi levels are sensed (Sánchez-Calderón et al., 2005; Svistoonoff et al., 2007). The similar magnitude of MR inhibition found in our assays for severe and mild Pi starvation, together with the almost complete restriction of MR growth 8 d after transfer (Fig. 1B), support earlier studies suggesting that Pi is one of the most important external factors regulating MR growth (Gruber et al., 2013; Kellermeier et al., 2014).
Modulation of LR growth is one of the most important aspects of nutrient-foraging strategies (Giehl and von Wirén, 2014). The increase in LR development under Pi-deficient conditions was explained by the up-regulation of the auxin receptor TRANSPORT INHIBITOR RESPONSE1 (TIR1), leading to higher sensitivity to this hormone (Pérez-Torres et al., 2008). LR responses to Pi starvation have been shown to be independent of MR growth arrest (López-Bucio et al., 2002; Pérez-Torres et al., 2008). By following LR growth in a time course, we observed a negative impact of Pi starvation on the number and length of LRs from 6 and 8 d after transfer, respectively (Fig. 1B). This suggests that there is a certain hierarchy of responses to Pi deprivation, with MR affected first, followed by changes in LRL, and finally decrease in #LR. It has to be noted that precise quantification of RSA is feasible only for young seedlings; therefore, we cannot exclude entirely that our observations are affected by developmental stage. Applying an experimental setup where Pi-starved plants were quantified 2 d later than control seedlings allowed us to capture the increase not only in #LR and aLRL but also in LRD or apical zone size at a similar MRL (Fig. 1C), consistent with most previous studies (Figs. 1C and 3B; Williamson et al., 2001; Linkohr et al., 2002; López-Bucio et al., 2002). This supports the reliability of LRD as a marker trait for Pi starvation. Moreover, the high impact of Pi deprivation on LR traits was observed not only for Col-0 but also at the level of the whole population of Arabidopsis accessions we studied (Fig. 4A). Pi starvation increased #LR and LRD of all the accessions studied. We found that increasing concentrations of NaCl had a negative effect on LRD, but only when levels of Pi were limited (Fig. 3), explaining seemingly contradictory findings (Zolla et al., 2010; Julkowska et al., 2014). Pi starvation also could partially alleviate the negative effect of salt on MRL, #LR, and aLRL (Fig. 2). Interestingly, LR emergence and elongation showed different sensitivity to the integration of their response to the combination of Pi limitation and salt stress.
By studying responses to salt, Pi starvation, and their combination at the level of whole RSA, we identified substantial natural variations in RSA modulation induced by different stresses (Fig. 4). A strong effect of Pi starvation on MR growth together with a decrease in #LR was reported previously for 50% of the 73 accessions studied by Chevalier et al. (2003). Our experimental setup was optimized to capture the remodeling of RSA rather than changes made by growth inhibition. Despite quantifying MR growth 2 d later than for control conditions, still 12% of the Arabidopsis HapMap population showed reduction of the MRL, but none of the accessions studied showed decreases in both MRL and #LR. The typical Pi starvation brushy phenotype with highly restricted MR growth and numerous LRs was confirmed in crops to be the best ideotype for Pi foraging in soil (Giehl and von Wirén, 2014). This kind of morphological adaptation was observed for Kas-1 (Fig. 4F). Interestingly, this accession also was identified as having an RSA strategy correlating with lower sodium uptake (Julkowska et al., 2014), which may indicate its adaptive potential to multiple stresses. The different degrees of MR restriction by Pi starvation of Bay-0 and Sha were explained previously by the variation in the LPR1 gene (Reymond et al., 2006). Moreover, on double stress, Sha prioritized salt responses, while Bay-0 showed an additive effect of salt and Pi deprivation (Fig. 4, B and C). Using Bay-0 × Sha recombinant inbred line populations may provide quantitative trait loci controlling specific combinations of salinity and Pi starvation.
In general, RSA phenotypes on combined stress were partly dependent on their RSA on salt and Pi starvation (Fig. 6B). The ability to maintain higher total root size on single stresses in most cases resulted in more complex RSA on double stress, while accessions with shorter MR and LRs on salt conditions also were not able to develop high total root size on double stress conditions. Interestingly, few accessions with complex RSA on both single stresses showed a severe decrease in MRL, #LR, and LRLs on double stress. The contribution of RSA to salt tolerance is still not fully understood. An RSA response type with short LRs was linked to lower sodium uptake, based on a study of 10 accessions (Julkowska et al., 2014), but no data are available for larger populations. We found no correlations of our RSA data with published results on the effects of salt on rosette size, survival, or Na+ levels in the leaf (Supplemental Table S8; Rus et al., 2006; Katori et al., 2010; Julkowska et al., 2016). However, differences in experimental setups and NaCl concentrations used could be the main reason for the lack of correlations. Further research collecting data from RSA and shoot performance from a single experimental setup would be crucial to address this issue.
Next, we asked whether a plant would prioritize responses to one of the stresses. Analysis done at the level of the HapMap population of 330 natural accessions revealed that LR traits are generally influenced by the combined effect of Pi starvation and salinity, similar to their response to salt stress alone (Fig. 5), implying that salt stress dominates the LR response. We also found that the length of the apical root zone was modulated by double stress, similar to the effect of Pi deprivation and salt (Fig. 5). Apical zone size has been found to be under the strong control of Pi levels, and combining Pi starvation with other nutrient deficiencies did not have any additional effect (Kellermeier et al., 2014). Our results suggest that it is also not affected by salinity (Fig. 5). In the HapMap population studied here, all MR traits showed an additive response to the combination of salt and Pi starvation (Fig. 5). Although the RSA phenotypes on double stress correlated with single stress phenotypes (Fig. 6), the majority of changes induced by double stress would not have been captured either by studying responses to the single action of these stresses or by studying only the Col-0 accession. It has to be noted that these patterns were observed for the 330 accessions that we analyzed and may differ for other populations and may be specific for our experimental conditions imposing severe Pi starvation and mild salt stress.
The identified patterns were representative for the majority of the accessions, but still we were able to identify rare accessions with responses that were not found at the level of the whole HapMap population (Supplemental Table S5; Supplemental Fig. S4). Further studies on accessions identified to prioritize responses to Pi starvation in terms of LR elongation could help us understand how the inhibitory effect of salt could be overcome. Studying LR traits exhibiting a salt-prioritizing pattern can contribute to our understanding of how responses to Pi starvation can be masked by salt stress, while MR traits, showing an additive effect, can help to explain how signals forming these two stresses can be amplified and reveal possible cross talk between the two signaling pathways. Through GWAS, we mapped 12 loci putatively involved in responses to the combination of salt and Pi starvation. Importantly, genes mapped with traits from the prioritizing pattern (LRL, aLRL/MRL, and aLRL) did not overlap with those associated with an additive pattern (MRPL and MRVL), supporting separate mechanisms of stress integration for MR and LRs (Table II). Interestingly, we found one locus associated with both single and double stress responses of MR traits, suggesting the involvement of the same factors in MR growth restriction induced by salt and Pi starvation alone, while for the double stress, their function may be accelerated, which could explain the additive effect found for this trait (Fig. 5). Growth dynamics data suggest the existence of different mechanisms of MR growth retardation for salt stress and Pi starvation (Figs. 1 and 2; Supplemental Fig. S1), but we cannot exclude the possibility of the involvement of the same genetic factors. Complex regulation of responses to multiple stresses also has been shown for RSA modulations by other nutrient combinations as well as for multiple abiotic and biotic stresses at the transcriptional level (Rasmussen et al., 2013; Kellermeier et al., 2014). The validation and further characterization of identified candidates will contribute to understanding mechanisms of multiple stress integration for MR and LRs.
Different levels of sensitivity to ABA of MR and LRs have been shown (Duan et al., 2013). Here, we showed that contrasting responses of LRs to salinity and Pi starvation could be the result of a differential role of ABA (Fig. 7B). In the presence of salt, ABA promotes a quiescent phase at the very early stages of LR formation, while at the latter stages, the same hormone maintains the recovery of LR growth (Duan et al., 2013; Geng et al., 2013). We observed that on Pi starvation medium, ABA had a positive effect on the growth of LRs (Fig. 7B). However, it has to be noted that although the abi1-1 response to Pi starvation was lower than that of the Ler wild type, it did show a significant response compared with the control, suggesting possible cross talk with other pathways. Possibly, ABA signaling functions as a negative feedback loop in auxin-dependent LR growth on Pi starvation (Pérez-Torres et al., 2008). LRL of abi1-1 responded to double stress in an additive manner, while Ler prioritized responding to salt (Supplemental Fig. S5B). Taken together, our data suggest that the integration of signals from Pi starvation and salt may partially rely on ABA. So far, responses to Pi starvation were considered mostly as independent from ABA (Chiou and Lin, 2011). However, many Pi deprivation-responsive genes also are under the control of ABA (Woo et al., 2012), and ABA transport through xylem in bean (Phaseolus vulgaris), as well as stomata sensitivity in cotton, were increased by Pi starvation (Radin, 1984; Jaschke et al., 1997). Our results show that LR responses to low Pi also may be regulated by ABA.
CONCLUSION
The work presented here illustrates how root responses to salinity are dependent on Pi availability. Using a modified ROOT-FIT model, we show that the effect of mild salinity stress on Col-0 MR growth, LR emergence, and LR elongation can be rescued partially by Pi starvation. A detailed analysis of RSA for the whole HapMap population revealed complex RSA modulation by the combination of salt and Pi starvation, where LRs prioritize responses to salt and MR growth is affected more severely by double stress than by single stresses. We identified candidate genomic loci putatively involved in responses to the combination of salt and Pi starvation. Our study suggests that MR growth inhibition can be guided by different mechanisms for salt and Pi starvation and that LRs may exhibit different ABA sensitivities on salt stress and Pi starvation. Further research using the natural variation in RSA plasticity presented here and validation of candidates mapped by GWAS could lead to a better understanding of the control of root adaptation to stresses.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of accessions from the Arabidopsis (Arabidopsis thaliana) HapMap population were available from previous research (Julkowska et al., 2016). In order to ensure the same seed quality, all lines were propagated together under long-day conditions (21°C, 70% humidity, and 16/8-h light/dark cycle) with 8 weeks of vernalization at 4°C from week 3 after sowing. Accessions that did not germinate or flower or that showed poor growth in the following experiments were excluded from further analysis. The final data set consisted of 330 accessions (Supplemental Table S2).
Seeds were surface sterilized with 20 mL of thin bleach and 600 μL of 37.5% HCl for 3 h followed by 1.5 h in laminar flow to evaporate chlorine gas. Seeds were stratified in 0.2% Bactoagar at 4°C in the dark for 72 h. Seeds were germinated on one-half-strength Murashige and Skoog medium (Caisson Labs) supplied with 0.5% Suc, 0.1% MES monohydrate, and 1% Bactoagar (Difco), pH 5.8 (adjusted with KOH). Seeds were germinated on vertically positioned plates (70° angle) under long-day conditions (21°C, 70% humidity, and 16/8-h light/dark cycle). Four-day-old seedlings were transferred to square petri dishes containing 40 mL of medium. Each plate contained four seedlings of two different genotypes. The number of replicates was as indicated for each experiment. For phosphate starvation medium, Murashige and Skoog basal salt without phosphate (Caisson Labs) supplemented with 0, 1, 10, 50, or 100 μm KH2PO4 was used. Phosphate-rich medium was obtained with 0.5× Murashige and Skoog medium (Caisson Labs) containing 625 μm KH2PO4. Salt stress medium contained 50, 75, 100, or 125 mm NaCl. All plates were dried for 1.5 h. Plates were placed in the growth chamber in random manner.
Stress-Induced Modulations of Root System Architecture
Plates were scanned every second day up to day 10 after transfer with an Epson Perfection V700 scanner at 200 dots per inch (dpi) resolution. Phenotypes were analyzed from images from different days as indicated per experiment. The entire HapMap population was screened in seven separate experiments, with Col-0, Bay-0, Mz-0, Sha, and Ws used as internal controls. RSA was quantified with EZ-Rhizo software (Armengaud et al., 2009). All data sets were cleared from outliers or accessions that showed high variation between replicates, and the final data set consisted of 330 accessions listed in Supplemental Table S2. Statistical analyses were done with Graph Pad Prism version 5.0a or R Studio software using the tests indicated separately per experiment. Accession-specific correlations between the average value of each RSA trait on double stress and salt or Pi starvation as well as between MRL, #LR, and aLRL and previously published data were calculated with Pearson correlation. Hierarchical clustering of 330 accessions based on their MRL, #LR, and aLRL on salt, Pi starvation, and double stress was performed with the Ward linkage method in R software (ward.d2). The same dissimilarity cutoff value was used for each condition. Relationships between clusters found on salt or Pi starvation with those from double stress were visualized with the Circos online tool (http://circos.ca/; Krzywinski et al., 2009).
Descriptive Model of Stress-Induced Modulations of RSA
Four-day-old seedlings of Col-0 were transferred on medium supplemented with different concentrations of NaCl (0, 50, 75, 100, and 125 mm) and 1 or 625 μm KH2PO4. Plates were scanned every second day up to day 10 after transfer with an Epson Perfection V700 scanner at 200 dpi resolution. RSA was quantified with EZ-Rhizo software (Armengaud et al., 2009). All data sets were cleared from outliers. Statistical analyses were done with GraphPad Prism version 5.0a and Excel.
Changes in MRL, #LR, aLRL, and LRD were calculated with quadratic functions from the ROOT-FIT model as described (Julkowska et al., 2014). Additionally, MRL was described with a linear function: MRL = MRLSTART + GROWTHMR × t, where MRLSTART is the MRL 4 d after germination, t is the time in days after transfer, and GROWTHMR is growth rate (cm d−2). The growth factors for the linear model were calculated per seedling using the linest function in Excel and then averaged. Relative growth factors were calculated by dividing individual growth factors for each NaCl concentration separately by average growth rate for 0 mm NaCl at the same Pi regime and then averaged. The fit of the obtained growth functions was tested on an average of raw data measured by calculating the coefficient of determination (Supplemental Table S1).
Patterns of RSA Responses to Single Stresses and Pi Starvation
Four-day-old seedlings were transferred to control (625 μm KH2PO4 and 0 mm NaCl), salt (625 μm KH2PO4 and 75 mm NaCl), Pi starvation (1 μm KH2PO4 and 0 mm NaCl), and double stress (625 μm KH2PO4 and 75 mm NaCl) media. Plates were scanned every second day up to day 10 after transfer with an Epson Perfection V700 scanner at 200 dpi resolution. RSA was quantified with EZ-Rhizo software (Armengaud et al., 2009) from images obtained 4 d after transfer for control medium and 6 d after transfer in the case of stress media. All data sets were cleared from outliers. Statistical analyses were done with GraphPad Prism version 5.0a. Responses to salt, Pi starvation, and double stress were calculated for every RSA trait measured per each accession as a ratio of the raw value from a particular stress and the average of the corresponding value on the control condition and then averaged for the whole population. The obtained values were used for hierarchical clustering analyses performed in R software with the Ward linkage method (ward.d2). Four major patterns of integrating salt and Pi starvation signals were described as follows: prioritizing when S = DS < P; additive when DS < S < P; similar when S = DS = P; and intermediate when S > DS > PS, where DS is the combined effect of salt and Pi starvation (double stress), S represents salt, and P stands for Pi starvation. Hierarchical clustering of all the accessions based on their response to salt, Pi starvation, and double stress for each individual RSA trait was performed with the Ward linkage method.
GWAS
Averages of each RSA trait response were associated with a publicly available panel of 250,000 SNPs (Atwell et al., 2010). GWAS was performed with the scan_GLS algorithm using the EMMA-X model (Kruijer et al., 2015). Multiple GWAS were used with α of 0.01 or 0.05 with all SNPs available as well as with the subset of SNPs with minimal MAF of 0.01, 0.05, and 0.1. Two corrections for multiple testing were applied: BT = 1, where threshold is determined by –log10(α × P value); and BT = 4, where the number of markers was replaced by the number of effective tests approach, as described (Gao, 2011). The selection of candidate loci was performed based on LOD score, MAF, and trait heritability. The threshold for association LOD score [−10log(P value)] was determined with the Gao correction (Gao, 2011). Traits with heritability less than 0.2 were excluded from further analysis, and only SNPs with MAF greater that 0.01 were taken into account. For each selected marker, LD was checked with the LD-SNP tool with an LD cutoff of 0.8 (Kooke et al., 2016). Identified associated SNPs were assigned to the closest gene based on The Arabidopsis Information Resource 10, and the selection of candidate genes was extended to the LD region. Selected candidate genes are presented in Table II.
Arabidopsis Biological Resource Center numbers of all analyzed accessions can be found in Supplemental Table S2. Loci identifiers are listed in Table II and refer to The Arabidopsis Information Resource 10 database.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Growth dynamics of MRL, #LR, and aLRL from 2 to 8 d after transfer to Pi-sufficient medium or multiple Pi starvation media supplemented with 0 or 75 mm NaCl.
Supplemental Figure S2. MRL fits a quadratic model on Pi-sufficient medium, while on Pi starvation, the linear model presents the best fit.
Supplemental Figure S3. RSA of Col-0 on control, salt, Pi starvation, and combined stress.
Supplemental Figure S4. Natural variation within patterns of salt stress and Pi starvation integration.
Supplemental Figure S5. Insensitivity to ABA results in altered MR and LR growth on salt, Pi starvation, and double stress.
Supplemental Table S1. Values of RSA growth factors and r2 values used in the ROOT-FIT model.
Supplemental Table S2. Arabidopsis accessions screened for RSA responses to salt, Pi starvation, and their combined effect.
Supplemental Table S3. Overview of RSA traits measured.
Supplemental Table S4. Four major patterns of responses to double stress.
Supplemental Table S5. Accession-specific patterns of salt and Pi starvation integration.
Supplemental Table S6. Accession-specific correlations between the average value of each RSA trait on double stress and salt or Pi starvation.
Supplemental Table S7. Accessions found in each group derived from hierarchical clustering of 330 accessions based on their MRL, #LR, and aLRL on salt, Pi starvation, and double stress.
Supplemental Table S8. Accession-specific correlations between MRL, #LR, and aLRL on salt conditions described here and projected rosette area, fresh weight, dry weight, and water content reported by Julkowska et al. (2016).
Acknowledgments
We thank Jose Dinneny for seeds of Ler and abi1-1, Jiorgos Kourelis and Yu-Jia Chu for technical assistance, and Huub Hoefsloot for advice on data analysis.
Glossary
- RSA
root system architecture
- Col-0
Columbia-0
- MR
main root
- LR
lateral root
- ABA
abscisic acid
- Pi
inorganic phosphate
- MRL
main root length
- #LR
lateral root number
- aLRL
average lateral root length
- LRD
lateral root density
- GWAS
genome-wide association mapping
- SNP
single-nucleotide polymorphism
- LRL
lateral root length
- LD
Linkage disequilibrium
- dpi
dots per inch
- MAF
minor allele frequency
- LOD
log of the odds
- MRPL
main root path length
- MRVL
main root vector length
Footnotes
This work was supported by the Netherlands Organization for Scientific Research (grant no. ALW 846.11.002) and STW Perspectief 10987.
Articles can be viewed without a subscription.
References
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Al-Ghazi Y, Muller B, Pinloche S, Tranbarger TJ, Nacry P, Rossignol M, Tardieu F, Doumas P (2003) Temporal responses of Arabidopsis root architecture to phosphate starvation: evidence for the involvement of auxin signalling. Plant Cell Environ 26: 1053–1066 [Google Scholar]
- Armengaud P, Zambaux K, Hills A, Sulpice R, Pattison RJ, Blatt MR, Amtmann A (2009) EZ-Rhizo: integrated software for the fast and accurate measurement of root system architecture. Plant J 57: 945–956 [DOI] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. (2010) Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertauche N, Leung J, Giraudat J (1996) Protein phosphatase activity of abscisic acid insensitive 1 (ABI1) protein from Arabidopsis thaliana. Eur J Biochem 241: 193–200 [DOI] [PubMed] [Google Scholar]
- Buono RA, Paez-Valencia J, Miller ND, Goodman K, Spitzer C, Spalding EP, Otegui MS (2016) Role of SKD1 regulators LIP5 and IST1-LIKE 1 in endosomal sorting and plant development. Plant Physiol 171: 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Günther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, et al. (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43: 956–963 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20: 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier F, Pata M, Nacry P, Doumas P, Rossignol M (2003) Effects of phosphate availability on the root system architecture: large-scale analysis of the natural variation between Arabidopsis accessions. Plant Cell Environ 26: 1839–1850 [Google Scholar]
- Chiou TJ, Lin SI (2011) Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol 62: 185–206 [DOI] [PubMed] [Google Scholar]
- De Smet I, Signora L, Beeckman T, Inzé D, Foyer CH, Zhang H (2003) An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J 33: 543–555 [DOI] [PubMed] [Google Scholar]
- Duan L, Dietrich D, Ng CH, Chan PM, Bhalerao R, Bennett MJ, Dinneny JR (2013) Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 25: 324–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvan-Ampudia CS, Julkowska MM, Darwish E, Gandullo J, Korver RA, Brunoud G, Haring MA, Munnik T, Vernoux T, Testerink C (2013) Halotropism is a response of plant roots to avoid a saline environment. Curr Biol 23: 2044–2050 [DOI] [PubMed] [Google Scholar]
- Galvan-Ampudia CS, Testerink C (2011) Salt stress signals shape the plant root. Curr Opin Plant Biol 14: 296–302 [DOI] [PubMed] [Google Scholar]
- Gao X. (2011) Multiple testing corrections for imputed SNPs. Genet Epidemiol 35: 154–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y, Wu R, Wee CW, Xie F, Wei X, Chan PM, Tham C, Duan L, Dinneny JR (2013) A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell 25: 2132–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giehl RF, Gruber BD, von Wirén N (2014) It’s time to make changes: modulation of root system architecture by nutrient signals. J Exp Bot 65: 769–778 [DOI] [PubMed] [Google Scholar]
- Giehl RF, von Wirén N (2014) Root nutrient foraging. Plant Physiol 166: 509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou X, Yin H, He K, Du J, Yi J, Xu S, Lin H, Clouse SD, Li J (2012) Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet 8: e1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber BD, Giehl RF, Friedel S, von Wirén N (2013) Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol 163: 161–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaschke WD, Peuke AD, Pate JS, Hartung W (1997) Transport, synthesis and catabolism of abscisic acid (ABA) in intact plants of castor bean (Ricinus communis L.) under phosphate deficiency and moderate salinity. J Exp Bot 48: 1737–1747 [Google Scholar]
- Julkowska MM, Hoefsloot HC, Mol S, Feron R, de Boer GJ, Haring MA, Testerink C (2014) Capturing Arabidopsis root architecture dynamics with ROOT-FIT reveals diversity in responses to salinity. Plant Physiol 166: 1387–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julkowska MM, Klei K, Fokkens L, Haring MA, Schranz ME, Testerink C (2016) Natural variation in rosette size under salt stress conditions corresponds to developmental differences between Arabidopsis accessions and allelic variation in the LRR-KISS gene. J Exp Bot 67: 2127–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julkowska MM, Testerink C (2015) Tuning plant signaling and growth to survive salt. Trends Plant Sci 20: 586–594 [DOI] [PubMed] [Google Scholar]
- Jung JK, McCouch S (2013) Getting to the roots of it: genetic and hormonal control of root architecture. Front Plant Sci 4: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katori T, Ikeda A, Iuchi S, Kobayashi M, Shinozaki K, Maehashi K, Sakata Y, Tanaka S, Taji T (2010) Dissecting the genetic control of natural variation in salt tolerance of Arabidopsis thaliana accessions. J Exp Bot 61: 1125–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermeier F, Armengaud P, Seditas TJ, Danku J, Salt DE, Amtmann A (2014) Analysis of the root system architecture of Arabidopsis provides a quantitative readout of crosstalk between nutritional signals. Plant Cell 26: 1480–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermeier F, Chardon F, Amtmann A (2013) Natural variation of Arabidopsis root architecture reveals complementing adaptive strategies to potassium starvation. Plant Physiol 161: 1421–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooke R, Kruijer W, Bours R, Becker F, Kuhn A, van de Geest H, Buntjer J, Doeswijk T, Guerra J, Bouwmeester H, et al. (2016) Genome-wide association mapping and genomic prediction elucidate the genetic architecture of morphological traits in Arabidopsis thaliana. Plant Physiol 170: 2187–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S, Suzuki N, Huntington S, Armijo L, Sha W, Cortes D, Shulaev V, Mittler R (2008) Ascorbate peroxidase 1 plays a key role in the response of Arabidopsis thaliana to stress combination. J Biol Chem 283: 34197–34203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruijer W, Boer MP, Malosetti M, Flood PJ, Engel B, Kooke R, Keurentjes JJ, van Eeuwijk FA (2015) Marker-based estimation of heritability in immortal populations. Genetics 199: 379–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J (1997) The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkohr BI, Williamson LC, Fitter AH, Leyser HM (2002) Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J 29: 751–760 [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L (2002) Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol 129: 244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP. (2011) Root phenes for enhanced soil exploration and phosphorus acquisition: tools for future crops. Plant Physiol 156: 1041–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE. (2005) Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ 28: 67–77 [DOI] [PubMed] [Google Scholar]
- McLoughlin F, Galvan-Ampudia CS, Julkowska MM, Caarls L, van der Does D, Laurière C, Munnik T, Haring MA, Testerink C (2012) The Snf1-related protein kinases SnRK2.4 and SnRK2.10 are involved in maintenance of root system architecture during salt stress. Plant J 72: 436–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei C, Jiang SC, Lu YF, Wu FQ, Yu YT, Liang S, Feng XJ, Portoles Comeras S, Lu K, Wu Z, et al. (2014) Arabidopsis pentatricopeptide repeat protein SOAR1 plays a critical role in abscisic acid signalling. J Exp Bot 65: 5317–5330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijón M, Satbhai SB, Tsuchimatsu T, Busch W (2014) Genome-wide association study using cellular traits identifies a new regulator of root development in Arabidopsis. Nat Genet 46: 77–81 [DOI] [PubMed] [Google Scholar]
- Mouchel CF, Briggs GC, Hardtke CS (2004) Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes Dev 18: 700–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Schmidt W (2004) Environmentally induced plasticity of root hair development in Arabidopsis. Plant Physiol 134: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidu R, Rengasamy P (1993) Ion interactions and constraints to plant nutrition in Australian sodic soils. Aust J Soil Res 31: 801–819 [Google Scholar]
- Navarro JM, Botella MA, Cerdá A, Martinez V (2001) Phosphorus uptake and translocation in salt-stressed melon plants. J Plant Physiol 158: 375–381 [Google Scholar]
- Péret B, Clément M, Nussaume L, Desnos T (2011) Root developmental adaptation to phosphate starvation: better safe than sorry. Trends Plant Sci 16: 442–450 [DOI] [PubMed] [Google Scholar]
- Pérez-Torres CA, López-Bucio J, Cruz-Ramírez A, Ibarra-Laclette E, Dharmasiri S, Estelle M, Herrera-Estrella L (2008) Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell 20: 3258–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka JJ, Winter CM, Benfey PN (2012) Control of Arabidopsis root development. Annu Rev Plant Biol 63: 563–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phang TH, Shao G, Liao H, Yan X, Lam HM (2009) High external phosphate (Pi) increases sodium ion uptake and reduces salt tolerance of ‘Pi-tolerant’ soybean. Physiol Plant 135: 412–425 [DOI] [PubMed] [Google Scholar]
- Pierik R, Testerink C (2014) The art of being flexible: how to escape from shade, salt, and drought. Plant Physiol 166: 5–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineau C, Loubet S, Lefoulon C, Chalies C, Fizames C, Lacombe B, Ferrand M, Loudet O, Berthomieu P, Richard O (2012) Natural variation at the FRD3 MATE transporter locus reveals cross-talk between Fe homeostasis and Zn tolerance in Arabidopsis thaliana. PLoS Genet 8: e1003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radin JW. (1984) Stomatal responses to water stress and to abscisic acid in phosphorus-deficient cotton plants. Plant Physiol 76: 392–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S, Barah P, Suarez-Rodriguez MC, Bressendorff S, Friis P, Costantino P, Bones AM, Nielsen HB, Mundy J (2013) Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiol 161: 1783–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond M, Svistoonoff S, Loudet O, Nussaume L, Desnos T (2006) Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant Cell Environ 29: 115–125 [DOI] [PubMed] [Google Scholar]
- Ristova D, Busch W (2014) Natural variation of root traits: from development to nutrient uptake. Plant Physiol 166: 518–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero RM, Mestre TC, Mittler R, Rubio F, Garcia-Sanchez F, Martinez V (2014) The combined effect of salinity and heat reveals a specific physiological, biochemical and molecular response in tomato plants. Plant Cell Environ 37: 1059–1073 [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Mittler R (2002) The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol 130: 1143–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas U, Cibrian-Jaramillo A, Ristova D, Banta JA, Gifford ML, Fan AH, Zhou RW, Kim GJ, Krouk G, Birnbaum KD, et al. (2013) Integration of responses within and across Arabidopsis natural accessions uncovers loci controlling root systems architecture. Proc Natl Acad Sci USA 110: 15133–15138; erratum [DOI] [PMC free article] [PubMed] [Google Scholar]; Rosas U, Cibrian-Jaramillo A, Ristova D, Banta JA, Gifford ML, Fan AH, Zhou RW, Kim GJ, Krouk G, Birnbaum KD, et al. (2015) Proc Natl Acad Sci USA 112: E2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus A, Baxter I, Muthukumar B, Gustin J, Lahner B, Yakubova E, Salt DE (2006) Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet 2: e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell EJ, Wild A (1988) Russell’s Soil Conditions and Plant Growth. Longman Scientific & Technical, Burnt Mill, Harlow, Essex, UK [Google Scholar]
- Sánchez-Calderón L, López-Bucio J, Chacón-López A, Cruz-Ramírez A, Nieto-Jacobo F, Dubrovsky JG, Herrera-Estrella L (2005) Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana. Plant Cell Physiol 46: 174–184 [DOI] [PubMed] [Google Scholar]
- Schluepmann H, van Dijken A, Aghdasi M, Wobbes B, Paul M, Smeekens S (2004) Trehalose mediated growth inhibition of Arabidopsis seedlings is due to trehalose-6-phosphate accumulation. Plant Physiol 135: 879–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewelam N, Oshima Y, Mitsuda N, Ohme-Takagi M (2014) A step towards understanding plant responses to multiple environmental stresses: a genome-wide study. Plant Cell Environ 37: 2024–2035 [DOI] [PubMed] [Google Scholar]
- Slovak R, Göschl C, Su X, Shimotani K, Shiina T, Busch W (2014) A scalable open-source pipeline for large-scale root phenotyping of Arabidopsis. Plant Cell 26: 2390–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T (2007) Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet 39: 792–796 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouzé P, Rombauts S, Inzé D (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14: 903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson LC, Ribrioux SP, Fitter AH, Leyser HM (2001) Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol 126: 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo J, MacPherson CR, Liu J, Wang H, Kiba T, Hannah MA, Wang XJ, Bajic VB, Chua NH (2012) The response and recovery of the Arabidopsis thaliana transcriptome to phosphate starvation. BMC Plant Biol 12: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla G, Heimer YM, Barak S (2010) Mild salinity stimulates a stress-induced morphogenic response in Arabidopsis thaliana roots. J Exp Bot 61: 211–224 [DOI] [PMC free article] [PubMed] [Google Scholar]