Ethylene and shade share a conserved set of molecular regulators to control the plasticity of hypocotyl elongation in Arabidopsis.
Abstract
Plants have evolved shoot elongation mechanisms to escape from diverse environmental stresses such as flooding and vegetative shade. The apparent similarity in growth responses suggests a possible convergence of the signaling pathways. Shoot elongation is mediated by passive ethylene accumulating to high concentrations in flooded plant organs and by changes in light quality and quantity under vegetation shade. Here, we study hypocotyl elongation as a proxy for shoot elongation and delineate Arabidopsis (Arabidopsis thaliana) hypocotyl length kinetics in response to ethylene and shade. Based on these kinetics, we further investigated ethylene- and shade-induced genome-wide gene expression changes in hypocotyls and cotyledons separately. Both treatments induced a more extensive transcriptome reconfiguration in the hypocotyls compared with the cotyledons. Bioinformatics analyses suggested contrasting regulation of growth promotion- and photosynthesis-related genes. These analyses also suggested an induction of auxin, brassinosteroid, and gibberellin signatures and the involvement of several candidate regulators in the elongating hypocotyls. Pharmacological and mutant analyses confirmed the functional involvement of several of these candidate genes and physiological control points in regulating stress-escape responses to different environmental stimuli. We discuss how these signaling networks might be integrated and conclude that plants, when facing different stresses, utilize a conserved set of transcriptionally regulated genes to modulate and fine-tune growth.
All organisms, including plants, assess and respond to both biotic and abiotic factors in their environments (Franklin et al., 2011; Osakabe et al., 2014; Pierik and de Wit, 2014; Pierik and Testerink, 2014; Voesenek and Bailey-Serres, 2015; Quint et al., 2016). However, unlike animals, plants cannot move away from extremes in their surrounding environment but rather rely on various plastic morphological and metabolic responses. Such response traits include changes in plant architecture to escape the stress and optimize resource capture (Pierik and Testerink, 2014; Mickelbart et al., 2015). With energy reserves being invested in escape traits, plants often have lower plant biomass and crop yield (Casal, 2013). Molecular investigation of the different signaling pathways controlling these traits along with the characterization of underlying molecular components would not only enhance fundamental knowledge of stress-induced plasticity but also benefit crop improvement.
Plants are highly sensitive to changes in their light environment. Young plants growing in a canopy experience changes in light quality and quantity due to neighboring plants and compete to harvest optimum light (Casal, 2013; Pierik and de Wit, 2014). When a plant cannot outgrow its neighbors, it experiences complete vegetation shade (hereafter termed shade), which, in addition to low red:far-red (R:FR) light, is marked by a significant decline in blue light and overall light quantity. These changes initiate so-called shade-avoidance syndrome responses consisting of petiole, hypocotyl, and stem elongation, reduction of cotyledon and leaf expansion, upward movement of leaves (hyponasty), decreased branching, and increased apical dominance (Vandenbussche et al., 2005; Franklin, 2008; Casal, 2012; Pierik and de Wit, 2014). Shade-induced elongation comprises a complex network of photoreceptor-regulated transcriptional and protein-level regulation involving basic helix-loop-helix (bHLH) and homeodomain-leucine zipper (HD-ZIP) transcription factors and auxin, GA, and brassinosteroid (BR) hormone genes (Casal, 2012, 2013). Flooding often leads to partial or complete submergence of plants. Water severely restricts gas diffusion, and the consequent limited exchange of oxygen and CO2 restricts respiration and photosynthesis. Another consequence is the rapid accumulation of the volatile hormone ethylene. Ethylene is considered an important regulator of adaptive responses to flooding, including accelerated shoot elongation responses that bring leaf tips from the water layer into the air (Sasidharan and Voesenek, 2015; Voesenek and Bailey-Serres, 2015). In deepwater rice (Oryza sativa), this flooding-induced elongation response involves ethylene-mediated induction of members of the group VII ethylene response factor (ERF) family, a decline in active abscisic acid (ABA), and a consequent increase in GA responsiveness and the promotion of GA biosynthesis (Hattori et al., 2009). In submerged Rumex palustris petioles, ethylene also rapidly stimulates cell wall acidification and the transcriptional induction of cell wall modification proteins to facilitate rapid elongation (Voesenek and Bailey-Serres, 2015). Shade cues are reported to enhance ethylene production, resulting in shade-avoidance phenotypes (Pierik et al., 2004). However, these responses are mediated by ethylene concentrations of a much lower magnitude than that occurring in flooded plant organs (1 µL L−1; Sasidharan and Voesenek, 2015).
So far, it is largely unknown to what extent these growth responses to such highly diverse environmental stimuli share physiological and molecular components through time. A preliminary study in R. palustris showed that GA is a common regulator of responses to both submergence and shade (Pierik et al., 2005). Although submergence is a compound stress, rapid ethylene accumulation is considered an early and reliable flooding signal triggering plant adaptive responses. High ethylene concentrations like those that occur within submerged plant organs promote rapid shoot elongation (Voesenek and Bailey-Serres, 2015). This submergence response, which has been characterized extensively in rice and Rumex spp. (Hattori et al., 2009; van Veen et al., 2013), can be almost completely mimicked by the application of saturating (1 µL L−1) ethylene concentrations (Sasidharan and Voesenek, 2015). Saturating ethylene concentrations, therefore, were used here as a submergence mimic. Shade was given as true shade, which combines the three known key signals that trigger elongation (red and blue light depletion with relative far-red light enrichment).
A hypocotyl elongation assay in Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 (Col-0) was used as a proxy for shoot elongation under ethylene and shade in order to study to what extent ethylene and shade responses share molecular signaling components. Although ethylene suppresses Arabidopsis hypocotyl elongation in dark, high ethylene concentrations in light (as occur during submergence) stimulate hypocotyl elongation in Arabidopsis (Smalle et al., 1997; Zhong et al., 2012). Also upon simulated shade, Arabidopsis demonstrates pronounced hypocotyl elongation (Morelli and Ruberti, 2000). To capture early physiological responses and gene expression changes in response to ethylene and shade, Arabidopsis seedling hypocotyl elongation and cotyledon expansion were examined over time. The two treatments elicited characteristic hypocotyl growth kinetics. To uncover the transcriptomic changes regulating the elongation response to these signals, an organ-specific genome-wide investigation was carried out on hypocotyls and cotyledons separately at three time points corresponding to distinct hypocotyl elongation phases. Clustering analyses in combination with biological enrichment tests allowed the identification of gene clusters with expression patterns matching the hypocotyl growth trends across the three time points in both treatments. The correlation of genome-wide hypocotyl- and cotyledon-specific transcriptomic changes to publicly available microarray data on hormone treatments identified enriched hormonal signatures of auxin, BR, and GA in hypocotyl tissues and several potential growth regulatory candidate genes. Using hormone mutants and chemical inhibitors, we confirmed the combined involvement of these hormones and candidate regulators in the hypocotyl elongation response to ethylene and shade. We suggest that growth responses to diverse environmental stimuli like ethylene and shade converge on a common regulatory module consisting of both positive and negative regulatory proteins that interact with a hormonal triad to achieve a controlled fine-tuned growth response.
RESULTS
Delineation of Hypocotyl Elongation Kinetics under Ethylene and Shade in Arabidopsis Seedlings
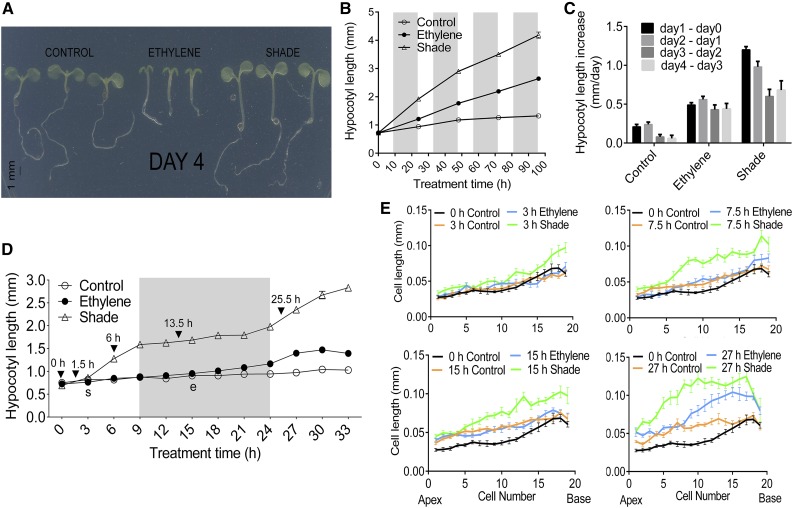
Exogenous application of ethylene (1 µL L−1) in light-grown seedlings resulted in thick yet elongated hypocotyls and smaller cotyledons as compared with untreated controls. Shade, achieved by the use of a green filter, stimulated strong hypocotyl elongation in seedlings but resulted in mildly smaller cotyledons compared with controls (Fig. 1A). Hypocotyl length increments in the two treatments relative to the control were around 2-fold under ethylene and greater than 3-fold under shade (Fig. 1, A and B). For both treatments, growth stimulation was strongest in the first 2 d of treatment (Fig. 1C) and declined on subsequent days. When combined, shade and ethylene exposure resulted in hypocotyl lengths that were intermediate to the individual treatments (Supplemental Fig. S1).
Figure 1.
Physiological responses, hypocotyl lengths, and epidermal cell length kinetics under ethylene and shade in Arabidopsis (Col-0) seedlings. One-day-old seedlings were exposed to control conditions (PAR = 140 μmol m−2 s−1, blue light = 29 μmol m−2 s−1, and R:FR light = 2.1 μmol m−2 s−1), elevated ethylene (1 µL L−1; PAR = 140 μmol m−2 s−1, blue light = 29 μmol m−2 s−1, and R:FR light = 2.1 μmol m−2 s−1), or shade (PAR = 40 μmol m−2 s−1, blue light = 3 μmol m−2 s−1, and R:FR light = 0.45 μmol m−2 s−1). A, Representative seedlings displaying typical phenotypes after 96 h of exposure to control, ethylene, or shade conditions. B, Mean hypocotyl lengths at 0, 24, 48, 72, and 96 h under control (white circles), ethylene (black circles), and shade (triangles). C, Rate of increase in hypocotyl length. Differences between mean hypocotyl lengths of subsequent time points averaged over a 1-d time interval for control, ethylene, and shade are shown. D, Detailed hypocotyl length kinetics. One-day-old seedlings were exposed to control (white circles), ethylene (black circles), and shade (triangles) and measured at 3-h time intervals. Data are means ± se (n = 60) for A to D. Shaded areas denote the 15-h dark period in the 15-h-dark/9-h-light photoperiodic growth condition. e and s denote the first points of statistically significant differences in hypocotyl length or cotyledon area relative to the control for ethylene and shade, respectively. E, Epidermal cell length kinetics. One-day-old seedlings were exposed to control, ethylene, or shade conditions. Mean cell lengths ± se (n ≥ 10) are shown for epidermal cells of the Arabidopsis hypocotyl at 0-h control (black lines) and at 3-, 7.5-, 15-, and 27-h control (orange lines), ethylene (blue lines), and shade (green lines). Apex denotes the hypocotyl-cotyledon junction, and Base denotes the hypocotyl-root junction.
To get a more detailed time line of the early elongation kinetics, we performed a follow-up experiment with 3-h measurement intervals for the first 33 h (Fig. 1D). Ethylene-mediated stimulation of hypocotyl elongation started only in the middle of the dark period after 15 h. However, under shade, longer hypocotyls were recorded already after 3 h, and this rapid stimulation continued until the start of the dark period. Interestingly, accelerated elongation was observed again at around 24 h after the start of the treatments (when the lights were switched on). Based on this time line, we determined epidermal cell lengths at time points 0, 3, 7.5, 15, and 27 h (Gendreau et al., 1997; Fig. 1E), the latter four of which are either prior to the start of accelerated growth or during it in response to ethylene and shade. The rapid stimulation of elongation under shade starts at the base of the hypocotyl (3 h) and then progresses all along the hypocotyl, with maximum elongation occurring in the middle segment, while under ethylene, accelerated elongation is observed at the middle bottom of the hypocotyl (27 h).
Organ-Specific Transcriptomics in Hypocotyl and Cotyledon under Ethylene and Shade
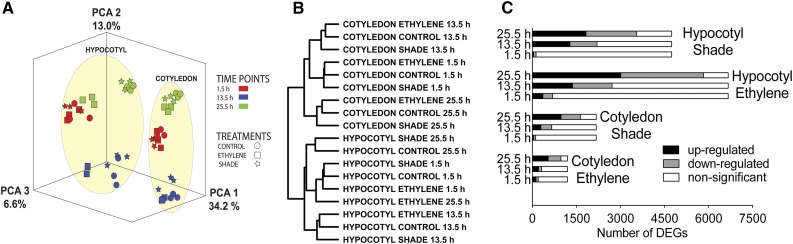
The transcriptome response to ethylene and shade in hypocotyl and cotyledon tissues was characterized using Affymetrix Arabidopsis Gene 1.1 ST arrays at three time points of hypocotyl length kinetics (1.5, 13.5, and 25.5 h; Fig. 1D). Principal component analysis (Abdi and Williams, 2010) of all replicate samples for hypocotyl and cotyledon exposed to control, ethylene, or shade conditions showed that replicate samples generally clustered together (Fig. 2A). The first principal component (34.2%) separates tissue-specific samples, whereas the second principal component (13%) showed separate clustering of the 13.5-h samples, which falls during the dark period.
Figure 2.
Overall description of microarray data. A, Principal component analysis (PCA) and hierarchical clustering were used to describe the structure in the microarray data. Expression intensities for all genes on the array for all 54 hypocotyl and cotyledon samples (three time points, three treatments, and three replicates) were projected onto the first three principal components. B, Hierarchical clustering was used to group 18 main samples (according to the mean expression intensity of three replicates for each main sample) into a dendrogram. C, Distribution of DEGs in hypocotyl and cotyledon samples at three time points in response to ethylene and shade. DEGs obtained for each time point were plotted separately as up-regulated, down-regulated (adjusted P ≤ 0.01 and log2FC greater than or less than 0), or nonsignificant (adjusted P > 0.01). Bar length denotes total DEGs obtained after combining DEGs from all three time points.
Hierarchical clustering (Eisen et al., 1998) of mean absolute expression intensities for the different main samples (combination of three replicates) revealed similar trends (Fig. 2B). Figure 2C shows the distribution of up- and down-regulated differentially expressed genes (DEGs; genes with adjusted P ≤ 0.01) in hypocotyl and cotyledon for ethylene and shade at the three harvest time points 1.5, 13.5, and 25.5 h, respectively. In both conditions and tissues, the number of both up- and down-regulated DEGs increased with time. Ethylene regulated substantially more DEGs in the hypocotyl compared with shade at all harvest time points (Fig. 2C). Data analysis identified 6,668 and 4,741 genes (hereafter termed total DEGs) that were differentially expressed in hypocotyl at one or more of the three tissue harvest time points by ethylene and shade, respectively. Interestingly, in the cotyledon at 1.5 h, the number of significant DEGs under ethylene was higher than under shade, but at the subsequent two time points, shade regulated more genes. In the cotyledon, 1,197 and 2,173 DEGs were identified that were differentially expressed at one or more of the three tissue harvest time points by ethylene and shade, respectively.
Interestingly, at 1.5 h, there was more transcriptional regulation in ethylene-exposed than in shade-exposed hypocotyls, even though subsequent hypocotyl elongation was much more rapid in shade (Figs. 1D and 2C). For ethylene-specific down-regulated DEGs at 1.5 h, the top-most enriched Gene Ontology (GO) term was cell wall organization (containing 30 genes), which suggested a repression of growth-promoting genes and a possible lack of ethylene-mediated elongation at 1.5 h (Supplemental Fig. S2). We also found six genes (AT1G65310, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE17; AT5G23870, PECTIN ACETYL ESTERASE; AT3G06770, GLYCOSIDE HYDROLASE; AT5G46240, POTASSIUM CHANNEL IN ARABIDOPSIS THALIANA1; AT1G29460, SMALL AUXIN UP-REGULATED RNA65 [SAUR65], and AT3G02170, LONGIFOLIA2) in the shade-specific up-regulated 32 genes at 1.5 h, with implied cell expansion roles and that possibly could be associated with the rapid elongation response (Philippar et al., 2004; Lee et al., 2006; Sasidharan et al., 2010; Chae et al., 2012; Nozue et al., 2015).
Different Gene Expression Clusters Contributing to Hypocotyl Growth in Ethylene and Shade
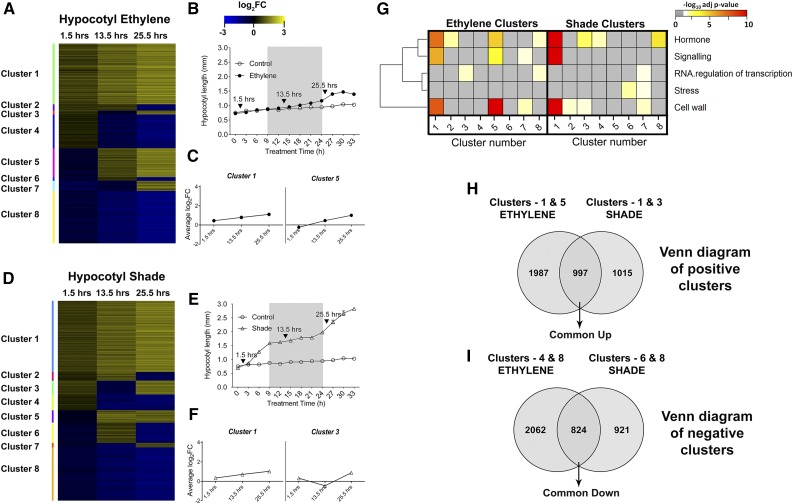
In order to find specific genes regulating the elongation phenotype under both treatments, we used temporal clustering of DEGs based on expression values. Due to distinct hypocotyl length kinetics in response to ethylene and shade (Fig. 1D), we searched for a set of temporally coexpressed genes that could potentially contribute to this treatment-specific kinetics. Time point-based clustering was performed for the 6,668 ethylene and 4,741 shade total DEGs based on the positive or negative magnitude of log2 fold change (log2FC) for DEGs at the three time points (Fig. 3, A and D). The gene expression patterns in clusters 1 and 5 across the three time points matched the ethylene hypocotyl growth kinetics closely (Fig. 3, B and C). Similarly, gene expression kinetics in clusters 1 and 3 matched the hypocotyl length kinetics in shade (Fig. 3, E and F). These growth pattern matching clusters were termed positive. All the clusters with mirror images of gene expression profiles to those of the positive clusters (clusters 8 and 4 in ethylene and clusters 8 and 6 in shade) were termed negative clusters.
Figure 3.
A to F, Temporal gene expression clusters for ethylene (A) and shade (D), hypocotyl growth curves for ethylene (B) and shade (E), and clusters with gene expression matching hypocotyl growth kinetics in ethylene (C) and shade (F). The heat map for temporal clusters (A and D) was based on the log2FC at the three microarray time points (gray areas represent dark, arrows indicate heat map time points, and treatments include control [white circles], ethylene [black circles], and shade [triangles]). Yellow denotes up-regulation, and blue denotes down-regulation. Two gene expression clusters (C and F) with mean log2FC temporal patterns resembling the hypocotyl length kinetics (B and E) were named positive clusters. G, Heat map for the hypergeometric enrichment of selected MapMan bins for temporal clusters under ethylene and shade. The horizontal axis denotes the cluster number. More intense colors indicate higher statistical significance; gray indicates nonsignificant score or absence of genes in the bin. H, Venn diagram intersection for positive clusters (clusters with gene expression patterns matching the hypocotyl length kinetics) from ethylene and shade to obtain Common Up genes. I, Venn diagram intersection for negative clusters (mirror image to positive clusters) from ethylene and shade to obtain Common Down genes.
Next, a hypergeometric overrepresentation test for selected MapMan bins (stress, hormone, signaling, RNA regulation of transcription, and cell wall) was carried out for the temporal gene clusters (Fig. 3G). Interestingly, cell wall, hormone, and signaling were highly coenriched in positive clusters (clusters 1 and 5 for ethylene and cluster 1 for shade), which hints at the coregulation of genes mapped to these terms during the transcriptomic response to ethylene and shade in the hypocotyl.
To identify growth promotion-related DEGs, we identified DEGs common to both treatments in the clusters designated previously as positive and negative clusters (Fig. 3, H and I). A total of 997 DEGs were obtained from a Venn diagram between the treatment-specific positive clusters up-regulated in at least two time points, hereafter called Common Up. Similarly, 824 DEGs shared between ethylene and shade negative clusters were down-regulated in at least two time points and hereafter are called Common Down.
Enriched functional categories in the different gene sets from Venn diagrams of positive and negative clusters were identified using the GeneCodis tool (Tabas-Madrid et al., 2012; Fig. 4). In the Common Up set, we found a variety of growth-associated GO categories, including cell wall modification, hormone (auxin and BR) signaling and metabolism, transport processes, tropisms, response to abiotic stimuli, and signal transduction. The ethylene-specific set for positive clusters was enriched for ethylene-associated terms as expected but also for various sugar metabolic, endoplasmic reticulum-related, and protein posttranslational modification-related processes. Some of the enriched GO terms in the ethylene-specific set for positive clusters also were found in the Common Up set but were caused by different genes in the same GO category, including those associated with growth, hormones, and transport processes. The shade-specific set for positive clusters showed only a few clear GO enrichments, such as trehalose metabolism, secondary cell wall biogenesis, and amino acid metabolism, but also shared some with the Common Up set, such as shade avoidance and protein phosphorylation, and with both Common Up and ethylene-specific sets for positive clusters, such as response to auxin and unidimensional growth.
Figure 4.
GO enrichment analysis using GeneCodis for positive and negative clusters. Adjusted P values for statistical significance of GO enrichment were converted into negative logarithm scores (base10; values greater than 1.3 are considered significant). Heat map colors denote this score. More intense colors indicate higher statistical significance; white indicates nonsignificant score or absence of genes in the GO terms. ER, Endoplasmic reticulum.
In the Common Down set, GO terms associated with photosynthesis, primary and secondary metabolism, response to biotic and abiotic stress, as well as photomorphogenesis were enriched. The latter is striking, given that ethylene does not alter the light environment. The ethylene-specific set for negative clusters included strong enrichment of circadian rhythm and a variety of photosynthesis- and chloroplast-associated GO terms, partially shared with the Common Down set. In the shade-specific set for negative clusters, flavonoid/anthocyanin biosynthesis and response to UV-B light and heat were enriched. The shade-specific set for negative clusters shared terms from defense-associated GO categories and cadmium, karrikin response with the Common Down set. Terms common to all three sets of negative clusters were related to photomorphogenesis and metabolic process.
Functional Characterization: Shared Components in Ethylene- and Shade-Mediated Regulation of Hypocotyl Length
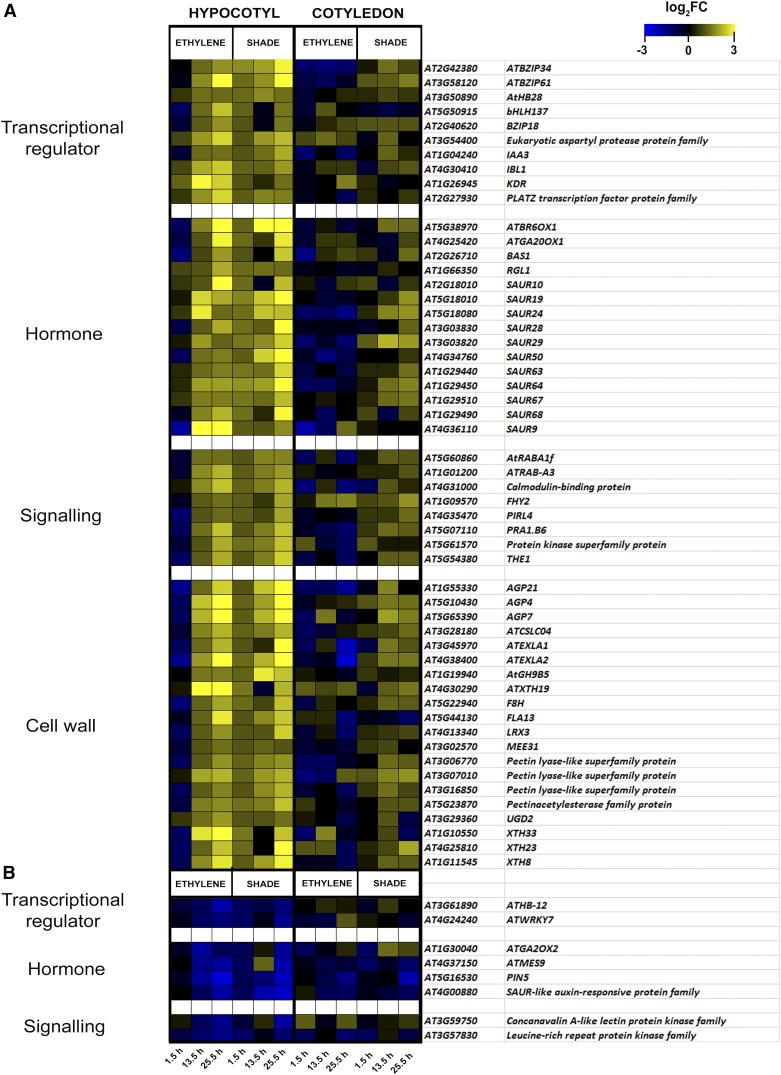
We classified Common Up and Common Down set genes into transcriptional regulators, hormone metabolism genes, signaling genes, and cell wall genes and also applied a log2FC filter (see “Log2FC Filter and Gene Classification” in “Materials and Methods”) to obtain a final list of 53 and eight genes in the two sets, respectively (Fig. 5, A and B). We selected a subset of candidate regulators for functional testing and made sure to include two transcription factors, since these may be relatively upstream in the convergence of signaling pathways. Obvious targets for these regulators would be different plant hormones, and before zooming in on these, we ran a hormonometer analysis with our transcriptome data to further divide our candidate regulators into subsets.
Figure 5.
Heat maps of Common Up (A) and Common Down (B) sets of genes classified into the categories transcriptional regulator, hormone metabolism, signaling, and cell wall genes after the application of a log2FC filter. Log2FC values at the three time points 1.5, 13.5, and 25.5 h for the ethylene and shade microarray data sets are identified in the color scheme of the heat map. Genes were categorized according to MapMan bin gene classification (shown at left).
Transcription Factor Candidates
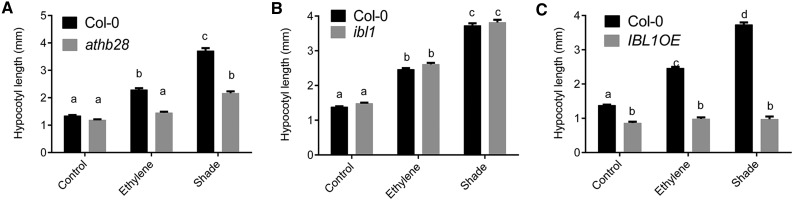
ARABIDOPSIS THALIANA HOMEOBOX PROTEIN28 (ATHB28) is a zinc finger homeodomain (ZF-HD) transcription factor that showed up to 2-fold induction in the hypocotyls in ethylene and shade. A homozygous null mutant (Supplemental Fig. S3, A–D), athb28, showed a significant reduction in hypocotyl length under both ethylene and shade (Fig. 6A), consistent with its induction upon both treatments.
Figure 6.
Hypocotyl length measurements for athb28 (A), ibl1 (B), and IBL1OE (C) following 96 h of control, ethylene, and shade treatments. Data represent means ± se (n = 30 seedlings). Different letters above the bars indicate significant differences (two-way ANOVA followed by Tukey’s HSD posthoc pairwise comparison).
Both shade and ethylene also significantly induced transcript levels of the bHLH transcription factor INCREASED LEAF INCLINATION1 BINDING bHLH1-LIKE1 (IBL1) Arabidopsis hypocotyls. However, ibl1 (SALK transfer DNA [T-DNA] insertion line) showed wild-type responses to the treatments (Fig. 6B). The bHLH transcription factors IBL1 and its homolog IBH1 have been implicated in repressing BR-mediated cellular elongation. In an ibl1 mutant, IBH1 is still present and may negatively regulate cell elongation independently of IBL1. The 35S overexpression line, IBL1OE (Zhiponova et al., 2014), had shorter hypocotyls than the wild type under control conditions. IBL1OE also lacked ethylene- and shade-induced hypocotyl elongation, implying an inhibitory role for IBL1 (Fig. 6C).
Hormone Candidates: Auxin, BR, and GA
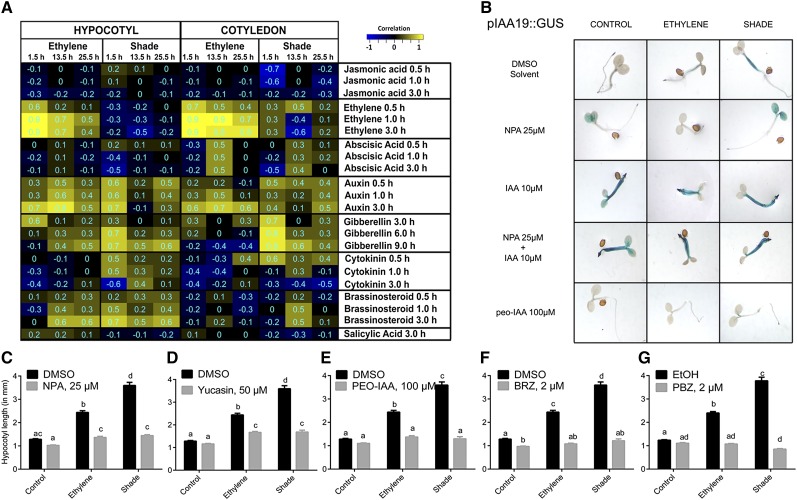
To further investigate the significant hormone-related changes among the growth-related DEGs, we analyzed our data using a hormonometer (Volodarsky et al., 2009). For both treatments, the hormonal signatures across the three time points for BR and GA most closely matched the hypocotyl elongation kinetics (Figs. 1D and 7A). The analysis also showed significant correlations with auxin responses for all data sets.
Figure 7.
A, Identification of enriched hormonal signatures in the ethylene- and shade-induced Arabidopsis transcriptome. Ethylene and shade-induced hypocotyl and cotyledon transcriptomes were analyzed for hormonal signatures using the hormonometer tool (Volodarsky et al., 2009) to establish correlations with expression data in an established hormonal transcriptome database. Positive correlations were colored yellow, and negative correlations were colored blue. Significant correlations were identified with absolute correlation values of 0.3 and higher. Numbers in the cells represent exact correlation values. Rows denote hormone treatments that are indicated by the name of the hormone and the duration of hormone treatment. Columns denote ethylene and shade transcriptomes in the hypocotyl and cotyledon at the three time points of tissue harvest. The magnitude of correlation in gene expression is indicated by the color scale at top right. B, Effects of the auxin transport inhibitors NPA (25 µm), IAA (10 µm), and NPA (25 µm) + IAA (10 µm) and the auxin perception inhibitor PEO-IAA (100 µm) on GUS staining of pIAA19:GUS lines. For NPA and PEO-IAA effects in the GUS assay, seedlings were exposed to 2 d of treatment conditions. C to G, Arabidopsis (Col-0) seedlings were treated with chemical inhibitors for auxin transport (NPA; C), auxin biosynthesis (yucasin; D), auxin perception (PEO-IAA; E), BR biosynthesis (brassinazole [BRZ]; F), and GA biosynthesis (paclobutrazol [PBZ]; G) at the indicated concentrations, and hypocotyl length was measured following 96 h of ethylene and shade. Means ± se were calculated for 30 seedlings. Different letters above the bars indicate significant differences from a two-way ANOVA followed by Tukey’s HSD posthoc pairwise comparison. DMSO, Dimethyl sulfoxide; EtOH, ethanol.
In the Common Up set, 49 genes were present that were all also auxin regulated, whereas there were 14 that were also BR regulated. In the Common Down set, 16 genes were present that can be regulated by auxin, 16 that also can be ABA regulated, and 13 that are jasmonate regulated. Interestingly, there were no genes for BR in the Common Down set. In addition, genes involved in auxin-conjugation genes (GRETCHEN HAGEN3 FAMILY PROTEIN3.17 and AT5G13370), GA-catabolizing genes (GIBBERELLIN 2-OXIDASE2 [GA2OX2], GA2OX4, and GA2OX7), and jasmonate-augmenting genes (LIPOXYGENASE1 [LOX1], LOX2, and LOX3) were down-regulated.
In order to test the possible roles of auxin, GA, and BR in mediating shade- and ethylene-induced hypocotyl elongation, we first tested the effects of pharmacological inhibitors of these hormones on shade- and ethylene-induced hypocotyl elongation. To visualize the auxin effect, we treated the pIAA19:GUS auxin response marker line with the auxin transport inhibitor 1-N-naphthylphthalamic acid (NPA). As shown in Figure 7B, application of NPA (25 µm) inhibited hypocotyl elongation and also strongly reduced staining in the hypocotyl region. This inhibition was rescued by indole-3-acetic acid (IAA; 10 µm). The auxin perception inhibitor α-(phenylethyl-2-one)-indole-3-acetic acid (PEO-IAA; 100 µm) strongly reduced staining in the whole pIAA19:GUS seedling and inhibited elongation in response to both treatments. The significant inhibition of ethylene- and shade-induced hypocotyl elongation by the different auxin inhibitors is quantitated in Figure 7, C to E. The addition of NPA, yucasin (auxin biosynthesis inhibitor), and PEO-IAA (auxin antagonist) inhibited hypocotyl elongation under both treatments, confirming that all three aspects of auxin are required for ethylene- and shade-induced hypocotyl elongation. The BR biosynthesis inhibitor brassinazole and the GA biosynthesis inhibitor paclobutrazol fully inhibited these elongation responses as well (Fig. 7, F and G).
To further validate the involvement of auxin, BR, and GA in ethylene- and shade-induced hypocotyl elongation, we tested hypocotyl elongation responses in a variety of hormone mutants, including mutants for candidate genes from Figure 5.
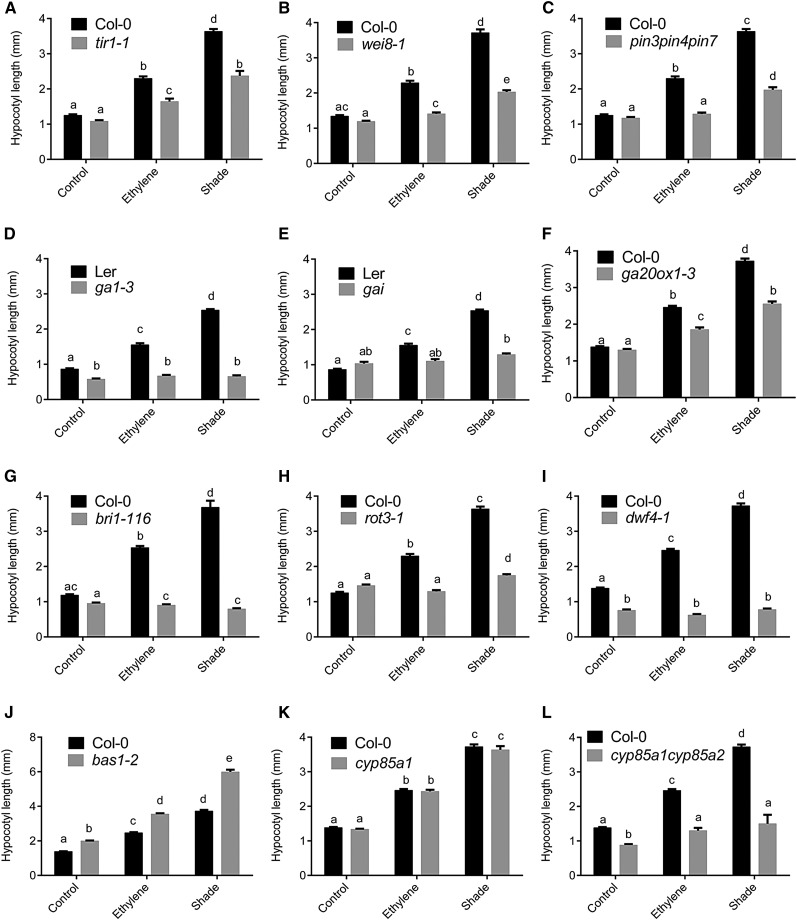
Both the auxin receptor (tir1-1) and biosynthesis (wei8-1) mutants showed significantly impaired hypocotyl elongation responses compared with the wild-type ethylene and shade responses (Fig. 8, A and B). A similar effect was seen in the auxin transport pin-formed mutant pin3pin4pin7, which had severely reduced hypocotyl elongation in both treatments (Fig. 8C).
Figure 8.
Hypocotyl elongation responses in response to ethylene and shade in auxin (A–C), GA (D–F), and BR (G–L) mutants. Means ± se were calculated for 30 seedlings. Different letters above the bars indicate significant differences from a two-way ANOVA followed by Tukey’s HSD posthoc pairwise comparison.
The GA biosynthesis (ga1-3) and GA-insensitive (gai) mutants both showed complete lack of hypocotyl elongation in both treatments (Fig. 8, D and E). We also tested the GA biosynthesis mutant ga20ox1-3, since it was identified as a Common Up gene, induced in response to both treatments (Fig. 5A). The ga20ox1-3 mutant showed a significantly reduced elongation phenotype in both treatments compared with the wild-type response (Fig. 8F).
The BR receptor (bri1-116) and biosynthesis (dwf4-1) mutants both showed severe hypocotyl elongation phenotypes and did not respond to either treatment (Fig. 8, G and I). In another biosynthesis mutant, rot3-1, while the ethylene elongation response was absent, there was a severely reduced shade response (Fig. 8H). Two BR metabolism-related genes were identified in the Common Up set (Fig. 5A): BR6OX1 and BAS1. The bas1-2 mutant showed constitutive elongation in all treatments (Fig. 8J), confirming a negative role of BR catabolism through BAS1 in hypocotyl elongation control. Although the BR biosynthetic mutant br6ox1 (cyp85a1-2) did not show any phenotypic alteration (Fig. 8K), a double mutant of BR6OX1 and BR6OX2 (cyp85a1cyp85a2) showed a complete lack of elongation in response to both ethylene and shade (Fig. 8L).
DISCUSSION
Accelerated shoot elongation is a common mode of stress escape that allows plants to grow away from stressful conditions (Pierik and Testerink, 2014). Stress escape, however, does come at an energetic cost and is only beneficial if improved conditions are achieved. Here, our goal was to establish to what extent shade and ethylene elicit similar responses through shared or distinct molecular pathways. In our study, we found distinct elongation kinetics in ethylene and shade for Arabidopsis hypocotyls, differing in both temporal regulation and the degree of response. Shade treatment evoked a rapid, strong, and persisting hypocotyl elongation, whereas ethylene initially inhibited elongation and only in the first night period started to promote hypocotyl length (Fig. 1). In both treatments, hypocotyl growth involved enhanced epidermal cell elongation. Previous studies have shown that low R:FR light induces ethylene biosynthesis in Arabidopsis (Pierik et al., 2009), and it could be argued that the shade response might act through ethylene. However, ethylene marker genes were not induced in shade, and the ethylene-insensitive ein3eil1 mutant retained a full response to shade (Supplemental Fig. S4), ruling out a role for ethylene in the shade response. Interestingly, combining shade and ethylene treatments did not lead to an additive response and instead dampened shade-induced hypocotyl elongation (Supplemental Fig. S1). The growth inhibitory effect of ethylene under shaded conditions could function similar to its effects in limiting hypocotyl elongation in the dark (i.e. via the induction of negative growth regulators such as ERF1; Zhong et al., 2012). Transcriptome characterization of the elongating hypocotyl upon exposure to single shade and ethylene stresses indicated considerable overlap between the two treatments. Thus, a large portion of DEGs under both treatments may contribute to similar processes, implying that they target shared genetic components but have treatment-specific upstream regulatory factors.
Hypocotyl Growth Promotion and Photosynthesis Repression Occur Concurrently under Ethylene and Shade
By identifying gene clusters with expression patterns closely matching the distinct ethylene and shade growth kinetics, we identified positive and negative clusters for the respective treatments. These clusters also were among the bigger clusters that contributed to most of the transcriptomic changes, suggesting that a large part of the transcriptomic response is associated with the hypocotyl growth and concurrent biological processes. Functional enrichment analysis for the Common Up set (shared between positive clusters of ethylene and shade; Fig. 4) suggested the involvement of growth-promoting genes. Cell wall genes are all involved in mediating cellular expansion in growing hypocotyls. However, they need to be controlled by either the environmental signal directly or by upstream factors in the signal transduction pathway. The Common Down set (shared between negative clusters of ethylene and shade; Fig. 4) was highly enriched in photosynthesis-related terms and proteins. The effects of ethylene on photosynthesis can be positive or negative depending on the context (Tholen et al., 2007; Iqbal et al., 2012). Low R:FR light-treated stems of tomato (Solanum lycopersicum) showed reduced expression of photosynthetic genes (Cagnola et al., 2012). This reduction was mainly due to a decrease in the expression of Calvin cycle genes, which we also observed for our Common Down set (Supplemental Table S1). In addition, under ethylene specifically, PSII and PSI genes were mostly repressed. Thus, the acceleration of hypocotyl elongation is accompanied by the repression of genes associated with nonelongation processes like metabolism and photosynthesis. This also was shown to be true for low-R:FR light-treated elongating stems of tomato (Cagnola et al., 2012). Light capture and carbon fixation are minimized and energy is apparently invested in stimulating growth (Lilley et al., 2012; Henriques et al., 2014; Sulpice et al., 2014). It would be interesting to investigate how photomorphogenic responses are associated with and influence photosynthesis and growth promotion.
The Convergence of Signaling Pathways in Response to Ethylene and Shade in the Control of Hypocotyl Elongation
In shade-avoidance responses, photoreceptors like phyB and cry1/2 would regulate the elongation phenotype via the control of phytochrome-interacting factor (PIF) levels. However, since these are photoreceptors, it seems unlikely that these proteins themselves would integrate information from the ethylene pathway as well (Li et al., 2012; Park et al., 2012). It was shown by van Veen et al. (2013) that, in the submerged petioles of R. palustris (which displays petiole elongation under complete submergence), early molecular components of light signaling (KIDARI, COP1, PIFs, and HD-ZIP IIs) are induced by ethylene independently of any change in light quality. Overexpression of PIF5, on the other hand, leads to increased ethylene production in etiolated Arabidopsis seedlings, causing inhibition of hypocotyl length (Khanna et al., 2007). Interestingly, the downstream ethylene signal transduction protein EIN3 was shown to interact physically with PIF3 (Zhong et al., 2012). Therefore, we suggest that ethylene and shade might both induce this shared gene pool by, for example, targeting (different) members of the PIF family of transcription factors. Since different PIFs likely regulate the expression of at least partly shared target genes (Leivar and Monte, 2014), this would explain our observed partial overlap in the transcriptional response to shade and ethylene. PIFs also are known to directly bind and regulate the expression of other transcription factors like homeodomain transcription factors (Kunihiro et al., 2011; Capella et al., 2015) in the control of shade-avoidance responses. Indeed, our The Arabidopsis Information Resource motif analysis hinted at the presence of significantly enriched binding signatures of PIF/MYC proteins (CACATG) as well as homeodomain proteins (TAATTA) in the upstream promoter sequences of the Common Up set genes (Kazan and Manners, 2013; Pfeiffer et al., 2014; Supplemental Fig. S5).
Several potential transcriptional regulators were identified in the narrowed down Common Up gene set. A growth-promoting role of KIDARI in regulating elongation in response to shade and ethylene was suggested previously (Hyun and Lee, 2006; van Veen et al., 2013). Up-regulation of another bHLH-encoding gene and a negative regulator of elongation, IBL1, was observed for both treatments (Fig. 6C). While PIF4 induces IBH1 and IBL1, IBH1 represses PIF4 targets (Zhiponova et al., 2014). IBH1 and its homolog IBL1 collectively regulate the expression of a large number of BR-, GA-, and PIF4-regulated genes, and this might their mode of action in shade- and ethylene-induced hypocotyl elongation. In addition to these bHLH proteins, we show that the ZF-HD transcription factor ATHB28 also is involved in regulating hypocotyl elongation under ethylene and shade (Fig. 6A). Hong et al. (2011) showed that another ZF-HD protein, MIF1, interacts strongly with four other ZF-HD proteins, including ATHB33 and ATHB28. This leads to nonfunctional MIF1-ATHB heterodimers and the inhibition of ATHB33-regulated expression and growth promotion. Transcriptomics data for 35S:MIF1 (displaying a short-hypocotyl phenotype) show down-regulation of auxin-, BR-, and GA-responsive genes and up-regulation of ABA genes (Hu and Ma, 2006). We can speculate that MIF1, on the one hand, and ATHB33 and ATHB28, on the other hand, might target the same set of hormone genes, but in an opposite manner, to control growth. What remains to be studied is how ethylene and shade regulate ZF-HD transcription factors, and this will be an important topic for future studies.
Well-established targets for the above-mentioned PIFs and homeodomain transcription factors are various aspects of auxin signaling and homeostasis, such as YUCCA biosynthetic enzymes and AUX/IAA proteins for signaling (Kunihiro et al., 2011; Li et al., 2012; Sun et al., 2012; De Smet et al., 2013). Our list of candidate genes (Fig. 5) contained the auxin-responsive transcriptional regulator IAA3 and many of the auxin-responsive SAUR genes, which have been shown to positively modulate hypocotyl elongation (Kim et al., 1998; Chae et al., 2012; Spartz et al., 2012; Sun et al., 2012) and may act individually or in concert to regulate the phenotype. With reference to elongation responses under shade in Arabidopsis seedlings, auxin seems to play a major role. An increase in free auxin levels and its transport toward epidermal cells in the hypocotyl is necessary for low-R:FR light-mediated hypocotyl elongation (Tao et al., 2008; Keuskamp et al., 2010; Zheng et al., 2016). The importance of YUCCAs and TAA1 in low-R:FR light responses has been demonstrated previously (Li et al., 2012). It is generally assumed that auxin synthesized in the cotyledons is required to regulate hypocotyl elongation in response to low-R:FR light conditions (Procko et al., 2014). Indeed, cotyledons are key regulators of hypocotyl elongation in a phytochrome-dependent way (Estelle, 1998; Tanaka et al., 2002; Endo et al., 2005; Warnasooriya and Montgomery, 2009). In our data, hormonometer analysis identified a strong induction of auxin-associated genes in the cotyledons in both treatments (Fig. 7). We speculate that the physiological regulation of hypocotyl elongation in our study depends on cotyledons via auxin dynamics. However, we also show that auxin is certainly not the only shared physiological regulator between the ethylene and shade responses.
GA20OX1 and BR6OX1 expression was up-regulated in patterns that closely matched the hypocotyl elongation profiles (Fig. 5A), and hormonometer analysis also revealed enrichments of GA and BR hormonal signatures in ethylene- and shade-exposed hypocotyls (Fig. 7). The positive role of GA in flooding-associated shoot elongation (Voesenek and Bailey-Serres, 2015) and shade avoidance (Djakovic-Petrovic et al., 2007) is well established. It is well known that GA20OX1 specifically affects plant height without having any other major phenotypic effects (Rieu et al., 2008; Barboza et al., 2013), and it has been shown to be involved in shade avoidance (Hisamatsu et al., 2005; Nozue et al., 2015). In our data, ga20ox1 knockout showed reduced elongation to shade as well as ethylene, extending its function from controlling shade avoidance to ethylene-mediated elongation responses (Fig. 8F). GA biosynthesis via GA20OX1 can be induced by BRs, suggesting a possible cross talk between the two growth-promoting hormones (Unterholzner et al., 2015). Although future studies are needed to establish if this cross talk occurs under the conditions tested here, we do confirm that BR is an important hormone involved in both responses, since several BR mutants showed disturbed elongation responses to ethylene and shade (Fig. 8, G–L). Interestingly, auxin and BRs also have partially overlapping roles in hypocotyl elongation control (Nemhauser et al., 2004; Chapman et al., 2012), further extending the cross talk toward a tripartite network. Among the BR mutants tested is the cyp85a1cyp85a2 mutant, encoding a double mutant for BR6OX2 and BR6OX1 that showed a complete lack of elongation to both treatments (Fig. 8L) similar to a brassinazole treatment (Fig. 7F). BR6OX1 was one of the direct candidate genes identified from the transcriptomics analysis (Fig. 5A). A tripartite bHLH transcription factor module consisting of IBH1, PRE, and HBI1 has been implicated previously in regulating cell elongation in response to hormonal and environmental signals (Bai et al., 2012). Several BR biosynthesis and signaling genes are direct targets of HBI1, including BR6OX1 (Fan et al., 2014), indicating the possible involvement of this bHLH regulatory module in promoting BR responses during shade and ethylene exposure.
Why is there such an elaborate network of regulators and even hormones involved in controlling unidirectional cell expansion in hypocotyl growth responses? To achieve a controlled growth, feedback loops are likely required, and cross talk between different routes is probably a necessity to deal with multiple environmental inputs simultaneously. We found BAS1 transcriptional up-regulation in hypocotyls in response to ethylene and shade. BAS1 may act to balance the hypocotyl growth promotion mediated by BRs (castasterone and brassinolide), as it inactivates both castasterone and brassinolide (Neff et al., 1999; Turk et al., 2005). In shade-avoidance research, likewise, HFR1 is induced by PIFs to suppress the growth promotion induced by the same PIF proteins, and putative control of DELLAs would modulate GA responses.
CONCLUSION
Hypocotyl elongation in response to ethylene and shade treatments is likely regulated at the upstream level by (1) a bHLH module, consisting of positive growth regulators, PIFs (PIF3 in ethylene and PIF4 and PIF5 in shade), and inhibitory factors like IBL1, and (2) a homeodomain module, where ZF-HD transcription factors like ATHB28 either may act in parallel to the bHLH transcription factors or are regulated by PIFs (similar to the induction of HD-ZIP transcription factors) to transcriptionally target genes related to the growth-promoting hormone module (auxin, BR, and GA), as hinted by promoter motif analysis. We hypothesize that, in Arabidopsis seedlings, shade and ethylene stimulate auxin synthesis in the cotyledons, which is then transported to the hypocotyl to epidermal cell layers, where it interacts with both GA and BR to coordinately induce hypocotyl elongation. This increased auxin response, indicated by elevated SAUR levels, and likely increased levels of GA and BR, as indicated by increased GA20OX1 and BR6OX expression in the hypocotyl, likely act to induce unidirectional epidermal cell wall elongation via the up-regulation of genes encoding cell wall-modifying proteins, which promote cellular expansion leading to hypocotyl elongation.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Around 30 Arabidopsis (Arabidopsis thaliana) Col-0 and mutant seeds were sown per agar plate containing 1.1 g L−1 Murashige and Skoog medium and 8 g L−1 plant agar (0.8% [w/v]; both Duchefa Biochemie). Mutants or overexpression lines used in this work were as follows: pin3pin4pin7 (Blilou et al., 2005), wei8-1 (Stepanova et al., 2008), tir1-1 (Ruegger et al., 1998), dwf4-1 (Azpiroz et al., 1998), rot3-1 (Kim et al., 1998), ga1-3 (Wilson et al., 1992), gai (Talon et al., 1990), ein3eil1 (Binder et al., 2004), bri1-116 (van Esse et al., 2012), ibl1 and IBL1OE (Zhiponova et al., 2014), cyp85a1/cyp85a2 and cyp85a1-2 (Nomura et al., 2005), bas1-2 (Turk et al., 2005), and ga20ox1-3 (Hisamatsu et al., 2005).
ibl1 (N657437), cyp85a1 (N681535), wei8-1 (N16407), and ga20ox1-3 (N669422) were obtained from the Nottingham Arabidopsis Stock Centre. bri1-116 was kindly provided by the Sacco de Vries laboratory at Wageningen University, while cyp85a1/cyp85a2 and dwf4-1 were kindly provided by the Sunghwa Choe laboratory at Seoul National University. IBL1OE overexpression lines were kindly provided by the Jenny Russinova laboratory at VIB. The bas1-2 mutant was kindly provided by the Michael Neff laboratory at Washington State University. Some GA mutants used were in the Landsberg erecta (Ler) background. All other mutants were in the Col-0 background. After stratification at 4°C for 3 d in the dark, seeds were transferred for 2 h to control light conditions (see below) and then kept in the dark (at 20°C) again for another 15 h. Subsequently, seedlings were allowed a period of 24 h of growth under control light conditions in short-day photoperiod conditions (15 h of dark/9 h of light) before being transferred to 22.4-L glass desiccators with air-tight lids for specific treatments. Col-0 genotypes were grown at 21°C ± 1°C. Ler genotypes were grown at 19°C ± 1°C (Supplemental Fig. S6).
For athb28 (GK-326G12), lines were obtained from the Nottingham Arabidopsis Stock Centre. Genotyping was performed using the following primers: for athb28, athb28_fwd (5′-CTAAGTACCGGGAATGTCAGAAG-3′), athb28_rev (5′-TAACCAACTGAGCTATTCCAGCTA-3′), and LB (Left border) primer o8474 (5′-ATAATAACGCTGCGGACATCTACATTTT-3′). To verify transcript levels, ATHB28_fwd (5′-GGAGAAGATGAAGGAATTTGCA-3′) and ATHB28_rev (5′-TGTTTCTCTTCATTGCTTGCT-3′) were used.
Treatments
Control and ethylene desiccators were kept in control light conditions (photosynthetically active radiation [PAR] = 140 μmol m−2 s−1, blue light [400–500 nm] = 29 μmol m−2 s−1, and R:FR light = 2.1 μmol m−2 s−1). Ethylene treatments were started by injecting ethylene into the desiccators (with 1 µL L−1 final concentration in the desiccator), and levels were verified with a gas chromatograph (GC955; Synspec). Shade treatment was started by putting desiccators under a single layer of Lee Fern Green Filter (Lee Hampshire; PAR = 40 μmol m−2 s−1, blue light [400–500 nm] = 3 μmol m−2 s−1, and R:FR light = 0.45 μmol m−2 s−1). For growth curve experiments, two plates with 15 seedlings per treatment per genotype distributed over two desiccators were used. For mutant analyses, one plate with 15 seedlings per treatment per genotype was used.
Imaging and Hypocotyl Length Measurements
For hypocotyl elongation assays, experiments were replicated twice. Seedling plates were collected from the desiccators. Seedlings were flattened on the agar plates to reveal the full extent of their hypocotyls, and images of the seedlings were obtained by scanning the plates using the EPSON Perfection version 370 photo scanner (Epson Europe). Hypocotyl lengths were measured from these images using ImageJ (http://rsbweb.nih.gov/ij/), and values (per data point) were obtained for n ≥ 30 to 60 seedlings. Final values for the data were obtained by taking means ± se for values from two independent experiments.
Epidermal Cell Length Measurements
Seedlings were mounted on microscope slides and covered with a coverslip. Hypocotyl epidermal cells were imaged using Olympus AX70 (20× objective, Nikon DXM1200 camera), after which cell lengths were measured using ImageJ software (http://rsbweb.nih.gov/ij/).
Microarray Tissue Harvest, RNA Isolation, and Array Hybridization
Seedlings were dissected using the BD PrecisionGlide Hypodermic 27 Gauge 1¼-Inch Gray Needle with o.d. = 0.41 mm (Becton Dickinson) to separately harvest the hypocotyl and cotyledon plus shoot apical meristem (hereafter termed cotyledon). The roots were discarded. Samples were harvested at 1.5, 13.5, and 25.5 h after the start of the treatments. For the 13.5-h time point, which occurs during the dark period, dissection was carried out under low-intensity green safelight (approximately 5 μmol m−2 s−1). To minimize the effects of green light on gene expression, seedlings were kept in the dark until dissected, and dissection was carried out in several rounds with each round involving a maximum of two to three seedlings. In total, three replicate experiments were carried out. In each replicate experiment, tissues were harvested from two independent technical replicates (each with 25 seedlings from the two technical replicate plates mentioned above). Harvested material was immediately frozen in liquid nitrogen and stored at −80°C until further use.
Frozen tissue was ground using a tissue lyser, and total RNA was isolated using the RNeasy Mini Kit (Qiagen). The Qiagen RNase-Free DNase set was used to eliminate genomic DNA contamination by performing on-column DNase digestion. Extracted RNA was verified (for quality check) and quantified using the NanoDrop ND-1000 spectrophotometer (Isogen Life Science).
RNA samples were sent to AROS Applied Biotechnology. RNA was repurified on low-elution Qiagen RNeasy columns, requantified with the NanoDrop 8000 UV-Vis Spectrophotometer, and checked for quality with the Agilent 2100 Bioanalyzer (Agilent Technologies). RNA samples with RNA integrity number values greater than 7.5 were considered for further use. Fifty nanograms of RNA from each of the two independent technical replicates was mixed in a 1:1 ratio, and the pooled sample was considered as one biological replicate for hybridization experiments. Thus, three biological replicates were obtained for the three replicate experiments. One hundred nanograms of RNA sample was processed for complementary DNA (cDNA) synthesis, and fragmentation and labeling were carried out for the RNA samples. The samples were hybridized to the Affymetrix Arabidopsis Gene 1.1 ST array plate and washed on the Affymetrix GeneAtlas system followed by scanning of arrays at AROS Applied Biotechnology (http://arosab.com/services/microarrays/gene-expression/).
Microarray Data Analysis
Scanned arrays in the form of CEL files (provided by AROS Applied Biotechnology) were checked for quality control using Affymetrix Expression Console Software and an in-house script in R and Bioconductor (http://www.r-project.org./ and http://www.bioconductor.org/; Bioconductor oligo and pd.aragene.1.1.st). Bioconductor was used for robust multiarray average normalization of raw data at the gene level to obtain summarized signal intensity values for all genes present on the array (log2 format). Principal component analysis was carried out using Affymetrix Expression Console Software (http://www.affymetrix.com/), and the dendrogram of all microarray samples according to the mean signal intensity values was generated using R (plot package). Bioconductor (Limma package) was used to carry out differential expression analysis.
Temporal Clustering and Bioinformatics Analysis
We clustered the list of total DEGs (defined as the number of DEGs that were regulated in at least one of the three time points) under ethylene and shade based on positive or negative regulation at each of the three time points. With three time points and two directions of expression (positive or negative), 23 = 8 possible trends can occur and, accordingly, as many clusters were obtained. DEGs were clustered temporally based on log2FC.
MapMan bin overrepresentation using a hypergeometric test was done using R (stats package), and adjusted P values for the statistical significance of enrichment were converted into negative logarithm scores and plotted as a heat map. A score above 1.3 was considered significant.
The GeneCodis (http://genecodis.cnb.csic.es/compareanalysis) Web tool was used for GO analysis of different sets obtained from Venn diagrams of positive and negative clusters. A score above 1.3 for the negative log of adjusted P values was considered significant.
Log2FC Filter and Gene Classification
In order to narrow down the genes for functional characterization (from the list of classified genes), we utilized a log2FC filter. To narrow down the Common Up set, a filter of log2FC < 0.5 at 1.5 h and log2FC ≥ 0.5 at both 13.5 and 25.5 h for ethylene and log2FC ≥ 0.5 at both 1.5 and 25.5 h for shade was applied. To narrow down the Common Down set, we applied a filter of log2FC > −0.5 at 1.5 h and log2FC ≤ −0.5 at both 13.5 and 25.5 h under ethylene and log2FC ≤ −0.5 at both 1.5 and 25.5 h under shade. The resulting groups of genes were then classified based on MapMan classification for the terms: RNA regulation of transcription, cell wall, signaling, and hormone metabolism. Genes from Plant TFDB (http://planttfdb.cbi.pku.edu.cn/index.php?sp=Ath) and Potsdam TFDB (http://plntfdb.bio.uni-potsdam.de/v3.0/index.php?sp_id=ATH) also were included as a source for gene classification to select for additional transcriptional regulators.
Hormone Correlational Analysis
Hormonometer software (http://genome.weizmann.ac.il/hormonometer/) was used to evaluate transcriptional similarities between the transcriptome data obtained here and the published, indexed list of those elicited by exogenous application of plant hormones. Arabidopsis gene locus identifiers were converted to Affymetrix GeneChip identifiers using the at-to-AGI converter tool (The Bio-Analytic Resource for Plant Biology; http://bar.utoronto.ca/). We used the new Affymetrix aragen1.first arrays (28,000 genes) for transcriptomics, but the hormonometer data are based on 3′ ATH1 arrays (22,000 genes). Accordingly, many locus identifiers could not be included (those that were newly incorporated in the aragene1.first arrays) in this analysis. In a few other cases where multiple ATH1 GeneChip identifiers for one locus identifier were obtained, all identifiers were retained.
Pharmacological Treatments
Auxin transport was inhibited by the use of 25 µm NPA (Duchefa Biochemie; Petrásek et al., 2003). Auxin perception was blocked by the use of 100 µm PEO-IAA (Hayashi et al., 2008). Auxin biosynthesis was blocked by the use of 50 µm yucasin (Nishimura et al., 2014). Brassinazole (2 µm; TCI Europe) was used to inhibit BR biosynthesis (Asami et al., 2000). Paclobutrazol (2 µm; Duchefa Biochemie) was used to inhibit GA biosynthesis (Rademacher, 2000). All chemicals were dissolved in appropriate solvent (dimethyl sulfoxide or ethanol) with the final solvent concentration in medium less than 0.1% to prevent toxicity due to solvents. All chemicals were applied by pipetting 150 µL of chemical solution or mock solvent as a thin film over the Murashige and Skoog agar medium on the petri plates and then allowing the solution to diffuse through the medium before starting the treatments.
GUS Staining and Imaging
For GUS assays, seedlings were transferred immediately from treatments to a GUS staining solution (1 mm 5-bromo-4-chloro-3-indolyl-d-glucoronide; Duchefa Biochem) in 100 mm sodium phosphate buffer (pH 7) along with 0.1% Triton X-100, 0.5 mm each of potassium ferrocyanide and potassium ferricyanide, and 10 mm EDTA (Merck) and kept at 37°C overnight. Seedlings were bleached in 70% ethanol for 1 d before capturing images.
Statistical Analysis and Graphing
One-way ANOVAs followed by Tukey’s honestly significant difference (HSD) posthoc tests were performed on the measurements obtained in hypocotyl length/cotyledon area kinetics to assess statistically significant differences between mean hypocotyl length/cotyledon area under ethylene or shade relative and the control at the same time point.
Two-way ANOVA followed by Tukey’s HSD posthoc test were used for pairwise multiple comparison. For hypocotyl elongation assays, statistical significance is indicated by the use of different letters. All statistical analyses were done in the R software environment. Graphs were plotted using Prism 6 software (GraphPad Software).
CEL files utilized in the organ-specific transcriptomics for hypocotyl and cotyledon tissues are posted with the Gene Expression Omnibus accession series GSE83212.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Hypocotyl elongation responses in wild-type Col-0 under control, ethylene, shade, and combination (ethylene + shade) treatments.
Supplemental Figure S2. Venn diagram of the gene intersection between up- and down-regulated DEGs of ethylene and shade separately at 1.5 h.
Supplemental Figure S3. Genotyping and transcript level verification for athb28.
Supplemental Figure S4. Hypocotyl elongation responses in the ethylene signaling mutant ein3eil1 under ethylene and shade.
Supplemental Figure S5. The Arabidopsis Information Resource motif analysis for Common Up and Common Down gene sets.
Supplemental Figure S6. Hypocotyl length of Ler under control and ethylene conditions when grown at 22°C day/20°C night and 20°C day/18°C night temperature regimes.
Supplemental Table S1. Photosynthesis gene proportions in gene sets from Venn diagrams of negative clusters.
Supplementary Material
Acknowledgments
We thank all group members of Plant Ecophysiology, Utrecht University, for help with the harvest for the transcriptomics experiment.
Glossary
- R:FR
red:far-red
- ABA
abscisic acid
- Col-0
Columbia-0
- DEGs
differentially expressed genes
- GO
Gene Ontology
- log2FC
log2 fold change
- BR
brassinosteroid
- NPA
1-N-naphthylphthalamic acid
- IAA
indole-3-acetic acid
- PEO-IAA
α-(phenylethyl-2-one)-indole-3-acetic acid
- Ler
Landsberg erecta
- PAR
photosynthetically active radiation
- HSD
honestly significant difference
Footnotes
This work was supported by the Netherlands Organization for Scientific Research (grant nos. ALW Ecogenomics 84410004 to L.A.C.J.V., ALW VENI 86312013 and ALW 82201007 to R.S., and ALW VIDI 86412003 to R.P.) and by a Utrecht University Scholarship to D.D.
Articles can be viewed without a subscription.
References
- Abdi H, Williams LJ (2010) Principal component analysis. Wiley Interdiscip Rev Comput Stat 2: 433–459 [Google Scholar]
- Asami T, Min YK, Nagata N, Yamagishi K, Takatsuto S, Fujioka S, Murofushi N, Yamaguchi I, Yoshida S (2000) Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol 123: 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiroz R, Wu Y, LoCascio JC, Feldmann KA (1998) An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell 10: 219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai MY, Fan M, Oh E, Wang ZY (2012) A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 24: 4917–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboza L, Effgen S, Alonso-Blanco C, Kooke R, Keurentjes JJB, Koornneef M, Alcázar R (2013) Arabidopsis semidwarfs evolved from independent mutations in GA20ox1, ortholog to green revolution dwarf alleles in rice and barley. Proc Natl Acad Sci USA 110: 15818–15823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder BM, Mortimore LA, Stepanova AN, Ecker JR, Bleecker AB (2004) Short-term growth responses to ethylene in Arabidopsis seedlings are EIN3/EIL1 independent. Plant Physiol 136: 2921–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Cagnola JI, Ploschuk E, Benech-Arnold T, Finlayson SA, Casal JJ (2012) Stem transcriptome reveals mechanisms to reduce the energetic cost of shade-avoidance responses in tomato. Plant Physiol 160: 1110–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella M, Ribone PA, Arce AL, Chan RL (2015) Arabidopsis thaliana HomeoBox 1 (AtHB1), a Homedomain-Leucine Zipper I (HD-Zip I) transcription factor, is regulated by PHYTOCHROME-INTERACTING FACTOR 1 to promote hypocotyl elongation. New Phytol 207: 669–682 [DOI] [PubMed] [Google Scholar]
- Casal JJ. (2012) Shade avoidance. The Arabidopsis Book 10: e0157 http://dx.doi.org/10.1199/tab.0157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ. (2013) Photoreceptor signaling networks in plant responses to shade. Annu Rev Plant Biol 64: 403–427 [DOI] [PubMed] [Google Scholar]
- Chae K, Isaacs CG, Reeves PH, Maloney GS, Muday GK, Nagpal P, Reed JW (2012) Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J 71: 684–697 [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Greenham K, Castillejo C, Sartor R, Bialy A, Sun TP, Estelle M (2012) Hypocotyl transcriptome reveals auxin regulation of growth-promoting genes through GA-dependent and -independent pathways. PLoS ONE 7: e36210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Lau S, Ehrismann JS, Axiotis I, Kolb M, Kientz M, Weijers D, Jürgens G (2013) Transcriptional repression of BODENLOS by HD-ZIP transcription factor HB5 in Arabidopsis thaliana. J Exp Bot 64: 3009–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djakovic-Petrovic T, de Wit M, Voesenek LACJ, Pierik R (2007) DELLA protein function in growth responses to canopy signals. Plant J 51: 117–126 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Nakamura S, Araki T, Mochizuki N, Nagatani A (2005) Phytochrome B in the mesophyll delays flowering by suppressing FLOWERING LOCUS T expression in Arabidopsis vascular bundles. Plant Cell 17: 1941–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estelle M. (1998) Polar auxin transport: new support for an old model. Plant Cell 10: 1775–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Bai MY, Kim JG, Wang T, Oh E, Chen L, Park CH, Son SH, Kim SK, Mudgett MB, et al. (2014) The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis. Plant Cell 26: 828–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA. (2008) Shade avoidance. New Phytol 179: 930–944 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. (2011) Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA 108: 20231–20235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Höfte H (1997) Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol 114: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori Y, Nagai K, Furukawa S, Song XJ, Kawano R, Sakakibara H, Wu J, Matsumoto T, Yoshimura A, Kitano H, et al. (2009) The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460: 1026–1030 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Tan X, Zheng N, Hatate T, Kimura Y, Kepinski S, Nozaki H (2008) Small-molecule agonists and antagonists of F-box protein-substrate interactions in auxin perception and signaling. Proc Natl Acad Sci USA 105: 5632–5637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R, Bögre L, Horváth B, Magyar Z (2014) Balancing act: matching growth with environment by the TOR signalling pathway. J Exp Bot 65: 2691–2701 [DOI] [PubMed] [Google Scholar]
- Hisamatsu T, King RW, Helliwell CA, Koshioka M (2005) The involvement of gibberellin 20-oxidase genes in phytochrome-regulated petiole elongation of Arabidopsis. Plant Physiol 138: 1106–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SY, Kim OK, Kim SG, Yang MS, Park CM (2011) Nuclear import and DNA binding of the ZHD5 transcription factor is modulated by a competitive peptide inhibitor in Arabidopsis. J Biol Chem 286: 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Ma H (2006) Characterization of a novel putative zinc finger gene MIF1: involvement in multiple hormonal regulation of Arabidopsis development. Plant J 45: 399–422 [DOI] [PubMed] [Google Scholar]
- Hyun Y, Lee I (2006) KIDARI, encoding a non-DNA binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol Biol 61: 283–296 [DOI] [PubMed] [Google Scholar]
- Iqbal N, Khan NA, Nazar R, da Silva JAT (2012) Ethylene-stimulated photosynthesis results from increased nitrogen and sulfur assimilation in mustard types that differ in photosynthetic capacity. Environ Exp Bot 78: 84–90 [Google Scholar]
- Kazan K, Manners JM (2013) MYC2: the master in action. Mol Plant 6: 686–703 [DOI] [PubMed] [Google Scholar]
- Keuskamp DH, Pollmann S, Voesenek LACJ, Peeters AJM, Pierik R (2010) Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc Natl Acad Sci USA 107: 22740–22744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Shen Y, Marion CM, Tsuchisaka A, Theologis A, Schäfer E, Quail PH (2007) The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell 19: 3915–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BC, Soh MS, Hong SH, Furuya M, Nam HG (1998) Photomorphogenic development of the Arabidopsis shy2-1D mutation and its interaction with phytochromes in darkness. Plant J 15: 61–68 [DOI] [PubMed] [Google Scholar]
- Kunihiro A, Yamashino T, Nakamichi N, Niwa Y, Nakanishi H, Mizuno T (2011) Phytochrome-interacting factor 4 and 5 (PIF4 and PIF5) activate the homeobox ATHB2 and auxin-inducible IAA29 genes in the coincidence mechanism underlying photoperiodic control of plant growth of Arabidopsis thaliana. Plant Cell Physiol 52: 1315–1329 [DOI] [PubMed] [Google Scholar]
- Lee YK, Kim GT, Kim IJ, Park J, Kwak SS, Choi G, Chung WI (2006) LONGIFOLIA1 and LONGIFOLIA2, two homologous genes, regulate longitudinal cell elongation in Arabidopsis. Development 133: 4305–4314 [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E (2014) PIFs: systems integrators in plant development. Plant Cell 26: 56–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, Cole BJ, Ivans LJ, Pedmale UV, Jung HS, et al. (2012) Linking photoreceptor excitation to changes in plant architecture. Genes Dev 26: 785–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley JLS, Gee CW, Sairanen I, Ljung K, Nemhauser JL (2012) An endogenous carbon-sensing pathway triggers increased auxin flux and hypocotyl elongation. Plant Physiol 160: 2261–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickelbart MV, Hasegawa PM, Bailey-Serres J (2015) Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat Rev Genet 16: 237–251 [DOI] [PubMed] [Google Scholar]
- Morelli G, Ruberti I (2000) Shade avoidance responses: driving auxin along lateral routes. Plant Physiol 122: 621–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Nguyen SM, Malancharuvil EJ, Fujioka S, Noguchi T, Seto H, Tsubuki M, Honda T, Takatsuto S, Yoshida S, et al. (1999) BAS1: a gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc Natl Acad Sci USA 96: 15316–15323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J (2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2: E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Hayashi K, Suzuki H, Gyohda A, Takaoka C, Sakaguchi Y, Matsumoto S, Kasahara H, Sakai T, Kato J, et al. (2014) Yucasin is a potent inhibitor of YUCCA, a key enzyme in auxin biosynthesis. Plant J 77: 352–366 [DOI] [PubMed] [Google Scholar]
- Nomura T, Kushiro T, Yokota T, Kamiya Y, Bishop GJ, Yamaguchi S (2005) The last reaction producing brassinolide is catalyzed by cytochrome P-450s, CYP85A3 in tomato and CYP85A2 in Arabidopsis. J Biol Chem 280: 17873–17879 [DOI] [PubMed] [Google Scholar]
- Nozue K, Tat AV, Kumar Devisetty U, Robinson M, Mumbach MR, Ichihashi Y, Lekkala S, Maloof JN (2015) Shade avoidance components and pathways in adult plants revealed by phenotypic profiling. PLoS Genet 11: e1004953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y, Osakabe K, Shinozaki K, Tran LSP (2014) Response of plants to water stress. Front Plant Sci 5: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Park J, Kim J, Nagatani A, Lagarias JC, Choi G (2012) Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J 72: 537–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrásek J, Cerná A, Schwarzerová K, Elckner M, Morris DA, Zazímalová E (2003) Do phytotropins inhibit auxin efflux by impairing vesicle traffic? Plant Physiol 131: 254–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A, Shi H, Tepperman JM, Zhang Y, Quail PH (2014) Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol Plant 7: 1598–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippar K, Ivashikina N, Ache P, Christian M, Lüthen H, Palme K, Hedrich R (2004) Auxin activates KAT1 and KAT2, two K+-channel genes expressed in seedlings of Arabidopsis thaliana. Plant J 37: 815–827 [DOI] [PubMed] [Google Scholar]
- Pierik R, Cuppens ML, Voesenek LACJ, Visser EJ (2004) Interactions between ethylene and gibberellins in phytochrome-mediated shade avoidance responses in tobacco. Plant Physiol 136: 2928–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R, de Wit M (2014) Shade avoidance: phytochrome signalling and other aboveground neighbour detection cues. J Exp Bot 65: 2815–2824 [DOI] [PubMed] [Google Scholar]
- Pierik R, Djakovic-Petrovic T, Keuskamp DH, de Wit M, Voesenek LACJ (2009) Auxin and ethylene regulate elongation responses to neighbor proximity signals independent of gibberellin and DELLA proteins in Arabidopsis. Plant Physiol 149: 1701–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R, Millenaar FF, Peeters AJM, Voesenek LACJ (2005) New perspectives in flooding research: the use of shade avoidance and Arabidopsis thaliana. Ann Bot (Lond) 96: 533–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R, Testerink C (2014) The art of being flexible: how to escape from shade, salt, and drought. Plant Physiol 166: 5–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko C, Crenshaw CM, Ljung K, Noel JP, Chory J (2014) Cotyledon-generated auxin is required for shade-induced hypocotyl growth in Brassica rapa. Plant Physiol 165: 1285–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Delker C, Franklin KA, Wigge PA, Halliday KJ, van Zanten M (2016) Molecular and genetic control of plant thermomorphogenesis. Nat Plants 2: 15190. [DOI] [PubMed] [Google Scholar]
- Rademacher W. (2000) Growth retardants: effects on gibberellin biosynthesis and other metabolic pathways. Annu Rev Plant Physiol Plant Mol Biol 51: 501–531 [DOI] [PubMed] [Google Scholar]
- Rieu I, Ruiz-Rivero O, Fernandez-Garcia N, Griffiths J, Powers SJ, Gong F, Linhartova T, Eriksson S, Nilsson O, Thomas SG, et al. (2008) The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J 53: 488–504 [DOI] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M (1998) The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev 12: 198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan R, Chinnappa CC, Staal M, Elzenga JTM, Yokoyama R, Nishitani K, Voesenek LACJ, Pierik R (2010) Light quality-mediated petiole elongation in Arabidopsis during shade avoidance involves cell wall modification by xyloglucan endotransglucosylase/hydrolases. Plant Physiol 154: 978–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan R, Voesenek LACJ (2015) Ethylene-mediated acclimations to flooding stress. Plant Physiol 169: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Haegman M, Kurepa J, Van Montagu M, Straeten DV (1997) Ethylene can stimulate Arabidopsis hypocotyl elongation in the light. Proc Natl Acad Sci USA 94: 2756–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inzé D, Peer WA, Murphy AS, Overvoorde PJ, Gray WM (2012) The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J 70: 978–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Doležal K, Schlereth A, Jürgens G, Alonso JM (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191 [DOI] [PubMed] [Google Scholar]
- Sulpice R, Flis A, Ivakov AA, Apelt F, Krohn N, Encke B, Abel C, Feil R, Lunn JE, Stitt M (2014) Arabidopsis coordinates the diurnal regulation of carbon allocation and growth across a wide range of photoperiods. Mol Plant 7: 137–155 [DOI] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Chu J, Li C (2012) PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet 8: e1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A (2012) GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res 40: W478–W483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talon M, Koornneef M, Zeevaart JAD (1990) Accumulation of C19-gibberellins in the gibberellin-insensitive dwarf mutant gai of Arabidopsis thaliana (L.) Heynh. Planta 182: 501–505 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Nakamura S, Mochizuki N, Nagatani A (2002) Phytochrome in cotyledons regulates the expression of genes in the hypocotyl through auxin-dependent and -independent pathways. Plant Cell Physiol 43: 1171–1181 [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholen D, Pons TL, Voesenek LACJ, Poorter H (2007) Ethylene insensitivity results in down-regulation of Rubisco expression and photosynthetic capacity in tobacco. Plant Physiol 144: 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk EM, Fujioka S, Seto H, Shimada Y, Takatsuto S, Yoshida S, Wang H, Torres QI, Ward JM, Murthy G, et al. (2005) BAS1 and SOB7 act redundantly to modulate Arabidopsis photomorphogenesis via unique brassinosteroid inactivation mechanisms. Plant J 42: 23–34 [DOI] [PubMed] [Google Scholar]
- Unterholzner SJ, Rozhon W, Papacek M, Ciomas J, Lange T, Kugler KG, Mayer KF, Sieberer T, Poppenberger B (2015) Brassinosteroids are master regulators of gibberellin biosynthesis in Arabidopsis. Plant Cell 27: 2261–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Pierik R, Millenaar FF, Voesenek LACJ, Van Der Straeten D (2005) Reaching out of the shade. Curr Opin Plant Biol 8: 462–468 [DOI] [PubMed] [Google Scholar]
- van Esse GW, van Mourik S, Stigter H, ten Hove CA, Molenaar J, de Vries SC (2012) A mathematical model for BRASSINOSTEROID INSENSITIVE1-mediated signaling in root growth and hypocotyl elongation. Plant Physiol 160: 523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veen H, Mustroph A, Barding GA, Vergeer-van Eijk M, Welschen-Evertman RAM, Pedersen O, Visser EJW, Larive CK, Pierik R, Bailey-Serres J, et al. (2013) Two Rumex species from contrasting hydrological niches regulate flooding tolerance through distinct mechanisms. Plant Cell 25: 4691–4707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voesenek LACJ, Bailey-Serres J (2015) Flood adaptive traits and processes: an overview. New Phytol 206: 57–73 [DOI] [PubMed] [Google Scholar]
- Volodarsky D, Leviatan N, Otcheretianski A, Fluhr R (2009) HORMONOMETER: a tool for discerning transcript signatures of hormone action in the Arabidopsis transcriptome. Plant Physiol 150: 1796–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnasooriya SN, Montgomery BL (2009) Detection of spatial-specific phytochrome responses using targeted expression of biliverdin reductase in Arabidopsis. Plant Physiol 149: 424–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RN, Heckman JW, Somerville CR (1992) Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol 100: 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Guo Y, Novák O, Chen W, Ljung K, Noel JP, Chory J (2016) Local auxin metabolism regulates environment-induced hypocotyl elongation. Nat Plants 2: 16025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhiponova MK, Morohashi K, Vanhoutte I, Machemer-Noonan K, Revalska M, Van Montagu M, Grotewold E, Russinova E (2014) Helix-loop-helix/basic helix-loop-helix transcription factor network represses cell elongation in Arabidopsis through an apparent incoherent feed-forward loop. Proc Natl Acad Sci USA 111: 2824–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Shi H, Xue C, Wang L, Xi Y, Li J, Quail PH, Deng XW, Guo H (2012) A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr Biol 22: 1530–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.