AMOS1 modulates P homeostasis in response to P deficiency through ABA-antagonized ethylene signaling.
Abstract
Plastid intramembrane proteases in Arabidopsis (Arabidopsis thaliana) are involved in jasmonic acid biosynthesis, chloroplast development, and flower morphology. Here, we show that Ammonium-Overly-Sensitive1 (AMOS1), a member of the family of plastid intramembrane proteases, plays an important role in the maintenance of phosphate (P) homeostasis under P stress. Loss of function of AMOS1 revealed a striking resistance to P starvation. amos1 plants displayed retarded root growth and reduced P accumulation in the root compared to wild type (Col-0) under P-replete control conditions, but remained largely unaffected by P starvation, displaying comparable P accumulation and root and shoot growth under P-deficient conditions. Further analysis revealed that, under P-deficient conditions, the cell wall, especially the pectin fraction of amos1, released more P than that of wild type, accompanied by a reduction of the abscisic acid (ABA) level and an increase in ethylene production. By using an ABA-insensitive mutant, abi4, and applying ABA and ACC exogenously, we found that ABA inhibits cell wall P remobilization while ethylene facilitates P remobilization from the cell wall by increasing the pectin concentration, suggesting ABA can counteract the effect of ethylene. Furthermore, the elevated ABA level and the lower ethylene production also correlated well with the mimicked P deficiency in amos1. Thus, our study uncovers the role of AMOS1 in the maintenance of P homeostasis through ABA-antagonized ethylene signaling.
Phosphorus (P) is indispensible for all living organisms. P functions in a variety of central cellular processes, including the biosynthesis of organic compounds, bio-energy generation, and cell signaling (Poirier and Bucher, 2002). P, however essential, is highly dilute and immobile in soils due to its unique chemical properties. To survive P limitation, plants employ sophisticated mechanisms to facilitate P acquisition and mobilization, which include remodeling of root architecture, enhancing organic acid excretion, and increasing the expression of transporters and acid phosphatases (Fang et al., 2009). A key role of ethylene in the morphological and physiological responses to P deficiency has also been established (Lopez-Bucio et al., 2003; Zhang et al., 2003). Although knowledge of P acquisition and the P-signaling system has increased dramatically in recent years (Baek et al., 2013; Chandrika et al., 2013; Lv et al., 2014; Wang et al., 2014a, 2014b; Ayadi et al., 2015; Chen et al., 2015; Lambers et al., 2015), the regulatory networks underlying plant adaptation to P deficiency still remain to be clarified.
The site-2 protease (S2P) is a membrane-targeting protease (Rawson et al., 1997). In mammals, S2Ps function in signaling pathways involved in fundamental cellular processes, such as lipid metabolism and the endoplasmic-reticulum (ER) stress response through proteolytic activation of downstream transcription factors such as SREBPs and AFT6, releasing them from the membrane (Rawson, 2013). These processes are essential under stress. Since their discovery in mammals, S2P-like proteins have also been found in bacteria, fungi, and plants (Rudner et al., 1999; Bolter et al., 2006; Bien et al., 2009). The Arabidopsis (Arabidopsis thaliana) genome harbors four S2P homologs: AtS2P1/AraSP (At2g32480), AtS2P2 (At1g05140), AtAMOS1/AtEGY1 (At5g35220), and AtEGY2 (At5g05740; Adam, 2015). Although their substrates have yet to be determined, the S2P homologs in Arabidopsis have been implicated in jasmonic acid biosynthesis, chloroplast development, and flower morphology (Adam, 2015). However, whether S2Ps may be involved in P nutrition has not been examined. Among the four S2P homologs, AtAMOS1/AtEGY1 is of particular interest. In a previous study, AtAMOS1 was demonstrated to recruit abscisic acid signaling to regulate global gene expression under ammonium stress (Li et al., 2012), suggesting a central role for the S2P homolog AtAMOS1 in macronutrient physiology. Here, we explore the role of amos1, an ammonium-hypersensitive mutant, in response to P deficiency. Additionally, we examine the involvement of the phytohormones abscisic acid (ABA) and ethylene in remobilizing P from the cell wall during the P-deficiency response. Our findings unravel the critical role of the plastid metalloprotease AMOS1 in the maintenance of P homeostasis under P stress, through recruitment of ABA-antagonized ethylene signaling.
RESULTS
Loss of Function of AMOS1 Confers Resistance to P Deficiency
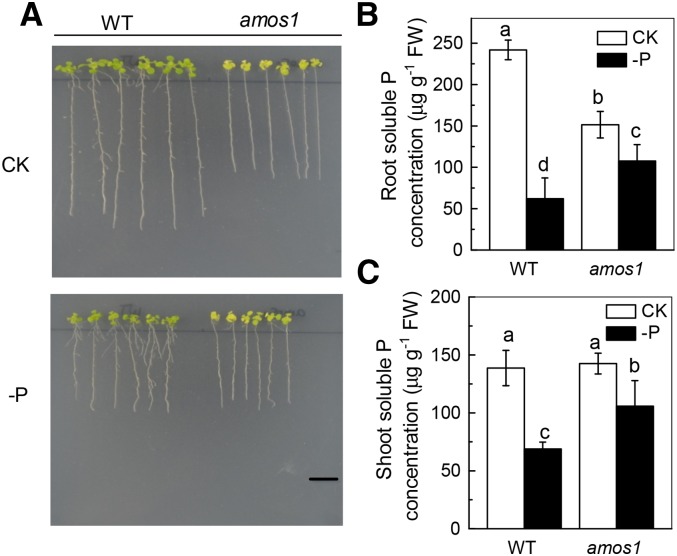
To investigate the response of the Arabidopsis ammonium-hypersensitive mutant amos1 to P deficiency, wild type and amos1 plants were grown in either P-replete or deficient media for 7 d. As shown in Figure 1A and Supplemental Figure S1, P deficiency led to reduced root and shoot growth of wild type while there was essentially no influence on the growth of amos1, indicating that amos1 was more resistant to P deficiency than wild type. However, as amos1 showed reduced root growth under P-replete condition compared to wild type (Fig. 1A), another question was whether the relatively superior response of amos1 under P-deficient conditions was due to the mutant’s already poorer growth (and presumably P acquisition) when grown under P-replete conditions. For this reason, the response of amos1 to nitrogen (N) and potassium (K) deficiency was also tested, and, rather than showing resistance, amos1 still grew poorly both under N-deficient and K-deficient conditions, as seen in root length and biomass (Supplemental Fig. S2), underscoring that amos1 is uniquely resistant to P deficiency. Determination of soluble P concentration in the root showed that amos1 accumulated less P in the root than wild type when grown under control conditions; however, upon P starvation, soluble P in both root and shoot in amos1 was higher than that in wild type (Fig. 1, B and C), indicating that amos1 can reutilize P more efficiently.
Figure 1.
Impact of phosphate withdrawal on wild type and amos1. The seeds of wild type and amos1 were surface-sterilized and germinated on complete nutrient medium, then seedlings of identical root length (1 cm) were transferred to either complete nutrient medium (CK) or medium without P (−P) for 7 d (A). For determination of soluble P, 6-week-old wild-type and amos1 seedlings were subjected to P-sufficient or -deficient solution for 7 d, prior to determination of the soluble P concentration in the root (B) and shoot (C). Scale bar = 1 cm. Values are means ± sd (sd), n = 4. Significant differences (P < 0.05) are denoted by different letters on the bars.
Cell Wall P in Roots Contributes to P Recycling in amos1
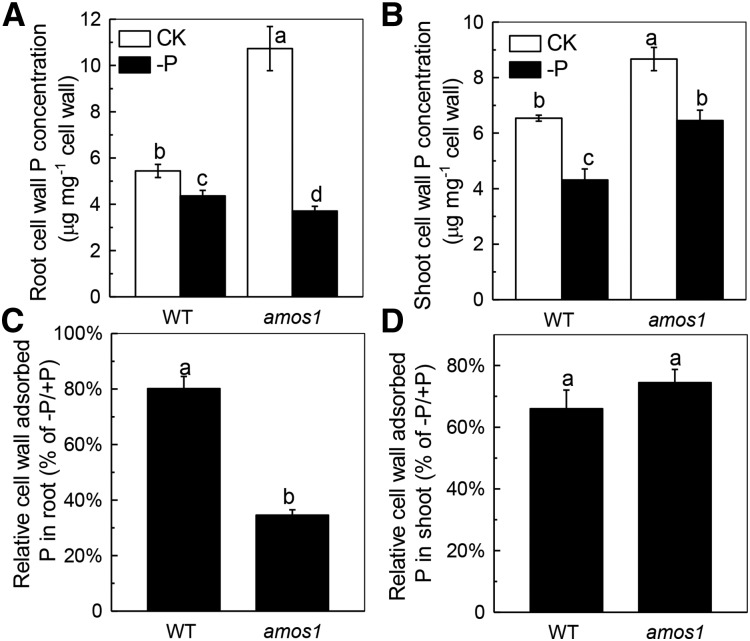
We next set out to answer the question where the P originated in the amos1 root or shoot when grown under P-deficient conditions. It was previously demonstrated that nearly 50% of total P is retained in the cell wall and that the cell wall can contribute greatly to cellular P reutilization in rice (Oryza sativa; Zhu et al., 2015). To verify whether accessing of the P reservoir in the cell wall was associated with the resistance of amos1 to P deficiency, the size of the P pool adsorbed in the cell wall of wild type and amos1 was determined under both +P and –P conditions. Interestingly, although P concentration was much higher in amos1 shoots than roots under P deficiency, there was almost no alteration in shoot P reutilization as indicated by shoot cell wall P (Fig. 2, B and D). Thus, the elevated P in amos1 shoots under P deficiency was most likely due to P translocation from the roots, and amos1 roots may act as the P source. As expected, a greater quantity of P was adsorbed in the cell wall fraction of amos1 roots under P-replete conditions, while a significantly smaller P pool was found in the root cell walls of amos1 under P-deficient conditions compared to wild type (Fig. 2A). Thus, the ratio of P retained (–P/+P) in the amos1 root cell wall was much lower than in the wild type (Fig. 2C), indicating that more P was released from the root cell wall of amos1, allowing for sustained growth of amos1 under P deficiency.
Figure 2.
P retention in the cell wall. Seedlings were grown hydroponically under P-replete or P-deficient conditions for 7 d, and cell wall-adsorbed P was measured in the root (A) and shoot (B). The relative cell wall-adsorbed P in the root (C) and shoot (D) was calculated from the P concentration in the root or shoot cell wall under P deficiency divided by the P concentration in the root or shoot cell wall under P-replete conditions, respectively. Values are means ± sd, n = 4. Significant differences (P < 0.05) are denoted by different letters on the bars.
Cell Wall Pectin Contributes to Efficient P Recycling in amos1
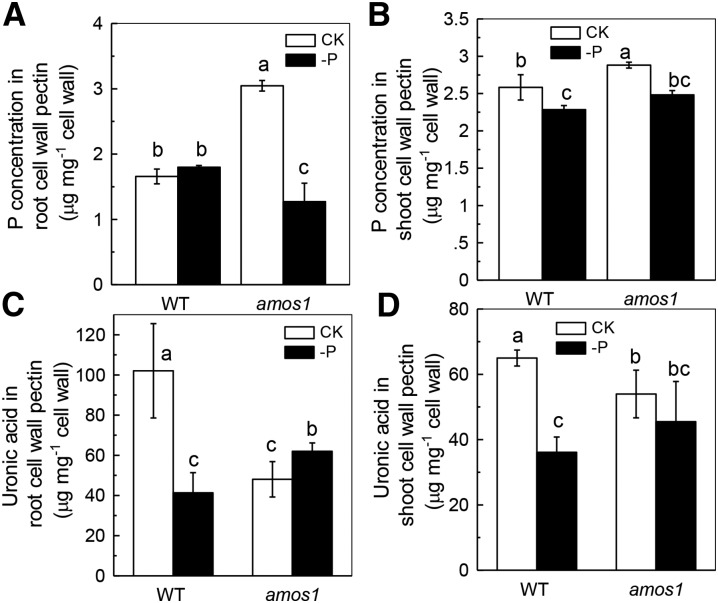
Previous studies showed that P deficiency decreases pectin concentration in the root cell wall of Arabidopsis (Zhu et al., 2012) and that pectin contributes greatly to cell wall P reutilization in rice (Zhu et al., 2015). Since the cell wall matrix acts as an important repository for P and a potential source for P reutilization, we tested whether pectin plays a role in the P-deficiency response in Arabidopsis. As expected, more P was adsorbed in the pectin fraction of the amos1 root cell wall under P-replete conditions while less P was adsorbed under P-deficient conditions compared to wild type, indicating greater P-release potential in the pectin fraction of amos1 root cell walls (Fig. 3A). In agreement with this, an increase in the pectin concentration of the amos1 root cell wall was found under P-deficient versus P-replete conditions, while the pectin concentration was decreased in the wild-type root cell wall under P-deficient conditions (Fig. 3C). However, the almost complete absence of any change in the shoot cell wall pectin concentration and its P retention in both the wild type and amos1 (−P versus +P), again, excludes the involvement of the shoot in the greater degree of P reutilization observed in amos1 (Fig. 3, B and D), Thus, we focused on roots instead of shoots in following examinations.
Figure 3.
The concentration of P in cell wall pectin and pectin. Seedlings (after germination) were subjected to P-sufficient and -deficient nutrient solutions for 7 d, and cell wall polysaccharides from roots and shoots were fractionated into pectin, then cell wall P concentration was deposited in pectin from the root (A) or shoot (B), and uronic acid in the root (C) or shoot (D) was determined accordingly. Values are means ± sd, n = 4. Significant differences (P < 0.05) are denoted by different letters on the bars.
Expression of Genes Involved in P Uptake, Translocation, and Allocation in P-Deficient Plants
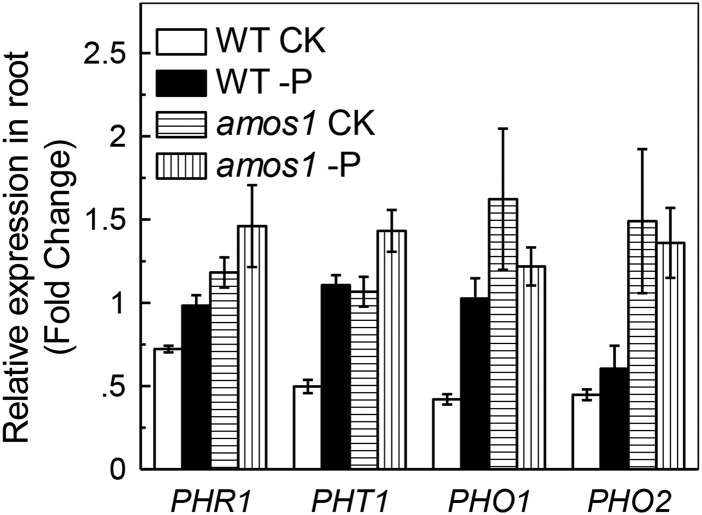
To determine the transcriptional response to P starvation, quantitative real-time PCR (RT-PCR) was subsequently performed following P starvation for 7 d. As revealed by RT-PCR, the wild-type root showed increased transcription of PHR1, PHT1, PHO1, and PHO2 after P starvation (Fig. 4), although the expression of all these genes was consistently higher in amos1. To our surprise, amos1 roots already displayed elevated transcription of PHR1, PHT1, PHO1, and PHO2, even in the absence of P starvation (Fig. 4), suggesting that loss of function of AMOS1 itself mimicked the stress imposed by P deficiency, consistent with the retarded root growth under P-replete control conditions that was indistinguishable from that under P-deficient conditions (Fig. 1A).
Figure 4.
Relative transcription of genes involved in P acquisition, and allocation in roots of amos1 and wild type. Seedlings were grown in P-replete or -deficient solution for 7 d.
Involvement of ABA in P Recycling from the Cell Wall in amos1
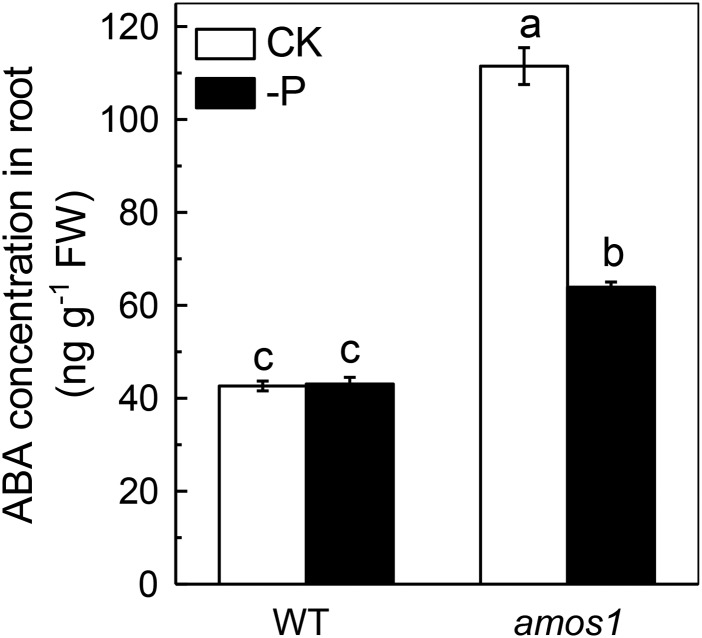
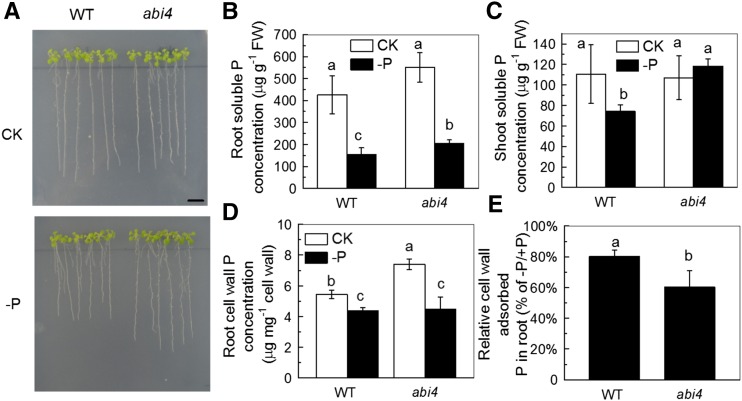
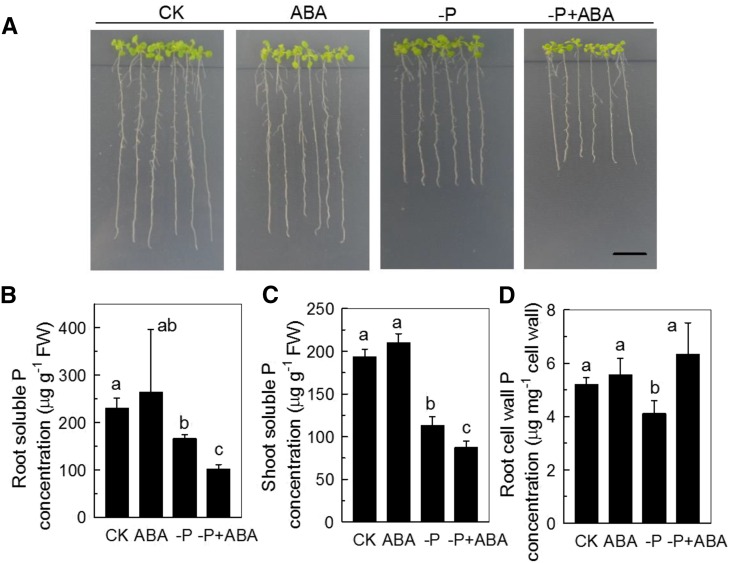
ABA was shown to function downstream of AMOS1 in its response to ammonium hypersensitivity (Li et al., 2012). We now ask whether ABA was also involved in the response to P deficiency in amos1. As shown in Figure 5, P deficiency significantly reduced the root ABA concentration of amos1, while there was almost no influence on wild type, indicating that ABA may be involved in the resistance to P deficiency of amos1. Examination of a loss-of-function mutant in a key transcription factor for ABA signaling, ABSCISIC ACID INSENSITIVE4 (abi4), and exogenous application of ABA confirmed this hypothesis; abi4 phenocopied amos1, showed improved growth (Fig. 6A), significantly greater root length, higher root and shoot biomass (Supplemental Fig. S3), significantly higher soluble P pools in roots and shoots (Fig. 6, B and C), and less root cell wall P retention under P-deficient conditions (Fig. 6, D and E), while exogenous ABA application retarded root and shoot growth (Fig. 7A; Supplemental Fig. S4), decreased soluble P in roots and shoots (Fig. 7, B and C), and increased cell wall-adsorbed P under the −P treatment (Fig. 7D).
Figure 5.
ABA accumulation in roots of seedlings of wild-type and amos1 under P-sufficient or -deficient conditions. Values are means ± sd, n = 4. Significant differences (P < 0.05) are denoted by different letters on the bars.
Figure 6.
Effect of phosphate depletion on soluble P and P retention in the cell wall in wild-type and abi4 seedlings. Seeds of wild type and abi4 were surface-sterilized and germinated on solid complete medium, then seedlings of identical root length (1 cm) were transferred to either solid complete medium (CK) or medium without P (−P) for 7 d (A). For determination of soluble P, 6-week-old wild type and abi4 seedlings were subjected to P-sufficient or -deficient solution for 7 d, prior to determination of the soluble P concentration in the root (B) and shoot (C). The relative cell wall-adsorbed P in the root (D) and shoot (E) were calculated as described in Figure 3. Scale bar = 1 cm. Values are means ± sd, n = 4. Significant differences (P < 0 0.05) are denoted by different letters on the bars.
Figure 7.
The effect of ABA on soluble P and on cell wall-retained P. A, Seedlings of WT were subjected to P-replete or -deficient solid medium, with or without 0.05 μm ABA for 7 d. For determination of soluble P concentration and cell wall P, seedlings were subjected to P-replete or -deficient solution, with or without 0.05 μm ABA for 7 d. B and C denote soluble P concentration in root and shoot respectively. D indicates cell wall-retained P in the root. Scale bar = 1 cm. Values are means ± sd, n = 4. Significant differences (P < 0.05) are denoted by different letters on the bars.
ABA Antagonizes Ethylene Signaling in P Recycling from the Cell Wall in amos1
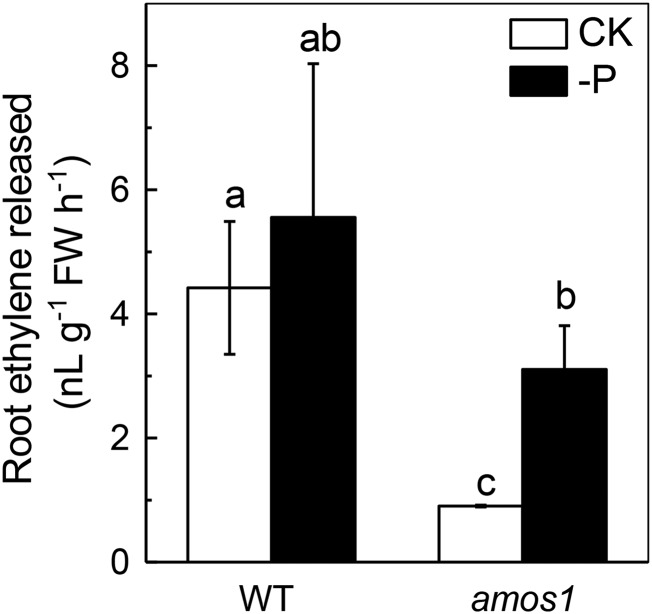
Lastly, we asked the question how ABA interferes with P remobilization from the cell wall in Arabidopsis. A recent study showed that the transcription of ACS4 and ACS8 genes, involved in ethylene biosynthesis, is inhibited by ABI4 (Dong et al., 2016), and more recently, Zhu et al. (2016) found that ethylene acts upstream of cell wall pectin to increase P remobilization from the cell wall in rice; therefore, we examined ethylene production in wild type and amos1 root. As expected, significantly enhanced ethylene production was observed in amos1 roots under P-deficient versus P-replete conditions compared to wild type (Fig. 8), indicating that ABA antagonizes ethylene signaling in the P-remobilization mechanism from the cell wall, which was furthermore confirmed by the exogenous application of the ethylene precursor ACC. As shown in Supplemental Figure S5, after ACC was applied exogenously under the P-deficient and P-replete conditions, production of ethylene was enhanced, accompanied by an increase in the cell wall pectin concentration; thus, more P was released from the cell wall pectin fraction, and, as a result a higher root-soluble P concentration was available, which was in agreement with Zhu et al. (2016). All the above results indicate that ABA counteracts the effect of ethylene in remobilizing the root cell wall P, and this conclusion was further supported by the fact that the high ABA level and the low ethylene production correlated well with the lower cell wall P reutilization in the amos1 mutant under P-replete conditions.
Figure 8.
Ethylene production in roots of amos1 and wild-type seedlings. Seedlings of amos1 and wild type were grown hydroponically under P-replete or -deficient conditions for 7 d, followed by determination of ethylene using GC-MS. Values are means ± sd, n = 4. Significant differences (P < 0.05) are denoted by different letters on the bars.
DISCUSSION
To overcome low P availability, plants have adopted a series of biochemical, physiological, and molecular strategies, including enhanced acid phosphatase (APase) activity and elevated transcription of genes essential for P acquisition (Goldstein et al., 1988; Devaiah et al., 2007; Zhou et al., 2008; Niu et al., 2013; Wang et al., 2014c). In addition, many nonspecific, shock-induced, gene-expression changes are also classically observed under P deficiency (Hammond et al., 2003). In addition to improved exploitation of P resources in the soil, plans can also remobilize previously acquired and internally sequestered P during periods of P deficiency (Raghothama and Karthikeyan, 2005). This can involve the plant-internal engagement of acid phosphatases, ribonucleases, and phosphohydrolases (Yun and Kaeppler, 2001; Gong et al., 2011). A recently uncovered mechanism engaged under P starvation in rice is associated with remobilization of P from storage pools in the root cell wall, and more specifically, the cell wall pectin fraction (Zhu et al., 2015). In the current study in Arabidopsis, resistance of P starvation also shows clear association with the cell wall, and in particular the pectin fraction, in the loss-of-function mutant amos1. To elucidate the possible mechanisms underlying cell wall-based P recycling in amos1, we examined the involvement of the phytohormones ABA and ethylene and show them to be fundamental to plant resistance to P starvation and the accessing of the P pool in the root system. Our work, thus, provides an important mechanistic extension to the earlier study in rice, in establishing not only the importance of cell wall P remobilization, but its regulation through phytohormone signaling.
The mature plant cell wall consists largely of cellulose and matrix polysaccharides, including pectin and hemicelluloses (Cosgrove, 2005). Cell wall polysaccharides of groundnut roots have been shown to exhibit P-solubilizing activity after growth on low P substrates (Ae et al., 1990). Polygalacturonic acids easily form complexes with metals, such as Fe3+ or Al3+, via phosphate ligand exchange, facilitating the desorption of P from soil minerals (Nagarajah et al., 1970). The carboxyl-acid groups, in particular of the d-GalUA component of cell wall pectin, have a strong affinity for Fe3+, and such metal binding has been linked to P release (Gessa et al., 1997). More recently, P deficiency was shown to lead to a decrease in the pectin and hemicellulose 1 fractions of cell walls in Arabidopsis in the context of cadmium exposure, and pectin was shown to enhance P remobilization in rice under P deficiency (Zhu et al., 2012, 2015, 2016). These studies indicate that cell wall polysaccharides become involved in the recycling of P deposited in the plant cell wall when external P becomes scarce. Our study establishes the role of the P repository in cell wall pectin in amos1 under P deficiency, with greatly enhanced P release from the pectin fraction in the cell walls of amos1 under P deficiency compared to P-replete conditions (Fig. 3A), accompanied by significant increases in the pectin concentration of the amos1 root cell wall (Fig. 3B). As a result, less P was retained in the amos1 cell wall under P deficiency when compared with wild type (Fig. 2), with elevated soluble P concentration in both roots and shoots of amos1, rendering it more resistant to P deficiency than wild type (Fig. 1; Supplemental Fig. S1). It is interesting that the expression of PHR1, PHT1, PHO1, and PHO2 was consistently higher in amos1 root compared with wild type in the absence of P starvation (Fig. 4), further indicating that the loss of function of AMOS1 itself mimicks the stress imposed by P deficiency; as a result, amos1 displayed retarded growth under P-replete (control) conditions (Fig. 1; Supplemental Fig. S1). However, amos1 also showed reduced root and shoot growth under −K and −N conditions, which is consistent with the retarded growth under P-replete conditions (Fig. 1; Supplemental Figs. S1 and S2), indicating that amos1 is uniquely resistant to P deficiency.
The finding that amos1 accumulates more ABA under P-replete than under P-deficient conditions (Fig. 5) led us to hypothesize that ABA may be involved in the response to P deficiency in amos1. Our results here demonstrate that ABA is a negative regulator in the response to P deficiency, as exogenous application of ABA resulted in higher sensitivity while the loss-of-function mutant in a key transcription factor for ABA signaling, abi4, exhibited higher resistance to P deficiency (Figs. 6 and 7). This is in accordance with earlier reports that ABA is engaged negatively in the response to P deficiency as evidenced in the repressed transcription of several At4 family members, a group of highly conserved P-deficiency-induced riboregulators in Arabidopsis (Shin et al., 2006). In addition, Arabidopsis mutants exhibiting lowered sensitivity to ABA or impaired ABA biosynthesis, abi2-1 and aba1, respectively, displayed reduced P-starvation response (PSR) gene expression when grown under low P supply (Trull et al., 1997; Ciereszko and Kleczkowski, 2002). However, defects in ABA signaling in these mutants did not affect other aspects typically related to P deficiency, such as enhanced phosphatase activity or altered root-to-shoot biomass ratio (Trull et al., 1997). Together, these data indicate that ABA is involved as a negative regulator in discrete, but not all, branches of the signaling responses engaged under P deficiency. The role of ABA in P deficiency is therefore complex, and the underlying physiological and molecular mechanisms remain elusive. Here, we demonstrate that ABA can exacerbate sensitivity to P deficiency in Arabidopsis by reducing P remobilization from the cell wall, as seen in amos1, abi4, and wild type supplied with exogenous ABA. This negative role of ABA in P remobilization from plant-internal P repositories, thus, appears to constitute one of the branch points at which ABA affects plant response to P deficiency.
Several studies have also established the role of the gaseous hormone ethylene in the response to P starvation (Nagarajan and Smith, 2012). Several genomic studies have demonstrated that P deficiency alters expression of genes involved in both ethylene biosynthesis and signaling in the root (Thibaud et al., 2010; Chacon-Lopez et al., 2011). By means of modification of the cell wall, ethylene was shown to mediate resistance against fungal colonization (Lloyd et al., 2011), and, in a previous study, ethylene was shown to be a likely signal in P remobilization from the cell wall (Zhu et al., 2016). Thus, the fact that amos1 accumulated more ABA (Fig. 5) and less ethylene compared to wild type under normal conditions (Fig. 8) suggests ABA may act against ethylene in the context of P deficiency. More recently, ABA was also found to inhibit ethylene biosynthesis via ABI4 (Dong et al., 2016). In agreement with these data, less ethylene was detected in the ABA-accumulating mutant amos1 in our current study (Fig. 8), and this was furthermore consistent with enhanced P retention in the root cell wall, indicating that ABA may play a negative role in P remobilization from the cell wall.
The fact that the ammonium-responsive AMOS1 is also involved in the maintenance of cellular P homeostasis raised the possibility that the N-signaling pathway engages in crosstalk with P signaling. Indeed, several lines of evidence suggest the existence of such crosstalk: First, an E3 ubiquitin ligase, nitrogen limitation adaptation (NLA), was reported to serve as a regulator of both N and P homeostasis in Arabidopsis (Kant et al., 2011). Second, transcriptomic analysis has shown that several P-starvation-responsive genes identified previously (Müller et al., 2007), MYB75, NF-YA10, and Phosphoenolpyruvate Carboxylase Kinase (PPCK), are down-regulated in amos1 by excessive ammonium treatment (Li et al., 2012). Third, Li et al. (2012) demonstrated that ABA signaling is recruited by AMOS1 under ammonium stress, while, in the current study, ABA signaling was found to be employed by AMOS1 under P deficiency. However, it has yet to be determined whether ethylene is involved under ammonium stress and whether N-acquisition-related genes alter their expression in response to P deficiency.
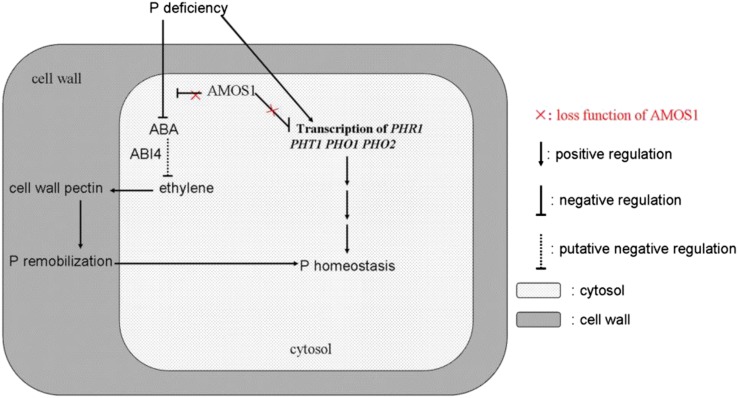
In conclusion, we demonstrate here, for the first time, that the plastid intramembrane protease AMOS1 is involved in the regulation of P remobilization from the root cell wall under P deficiency via recruitment of ABA-antagonized ethylene signaling in Arabidopsis (Fig. 9). Our study, thus, supports the importance of P remobilization from the plant cell wall under P deficiency, reveals a novel regulatory player in this process, and furnishes new evidence on the distinct roles of ABA and ethylene signaling during the important and widespread macronutritional stress of P deficiency.
Figure 9.
Schematic representation of P remobilization in amos1. Solid arrows and solid lines with perpendicular lines indicate positive and negative regulation patterns, respectively. Dashed lines indicate connections as verified by another study (Dong et al., 2016). P deficiency suppresses ABA production and increases transcription of genes responsible for P acquisition and transportation, which are negatively regulated by AMOS1. Ethylene biosynthesis was relieved of transcriptional repression mediated by ABI4, a downstream element of ABA signaling, due to a reduction in ABA levels. Enhanced ethylene production promotes cell wall pectin formation, which enables the release of P from the root cell wall, thus maintaining P homeostasis in amos1 under P deficiency.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) wild type (Columbia ecotype, Col-0) and the mutants amos1 and abi4 in the Col-0 background were used in the current study. The nutrient solution was composed of macronutrients as follows, in mM: KNO3, 6.0; Ca(NO3)2, 4.0; MgSO4, 1; NH4H2PO4, 0.1; and micronutrients as follows, in μM: Fe(III)-EDTA, 50; H3BO3, 12.5; MnSO4, 1; CuSO4, 0.5; ZnSO4, 1; H2MoO4, 0.1; NiSO4 0.1, according to Murashige and Skoog (1962). Seeds were stored in an eppendorf tube in the dark at 4°C for 2 to 3 d, followed by surface-sterilization and sowing on solid complete medium (nutrient solution supplemented with 8% agar). For the P-deficiency (−P) assay on solid culture, NH4H2PO4 was replaced by the same concentration of NH4NO3. For the potassium-deficiency (−K) assay on solid culture, KNO3 was omitted. For the nitrogen-deficiency assay (−N) on solid culture, 4 mm CaCl2 and 3 mm K2HPO4 were used. The same solid medium containing all nutrient elements as described above served as the control in −P, −K, and −N assays, respectively. Seedlings were maintained in a growth chamber at 23°C ± 1°C under a light intensity of 140 µmol m−2 s−1 and a photoperiod of 16 h of light and 8 h of dark.
For hydroponic culture, seedlings were first planted on the above solid Murashige and Skoog medium for 2 weeks, and the young plantlets were placed on vermiculite for an additional 3 weeks in controlled-environment chambers. Seedlings of similar rosette diameter were then transferred to the nutrient solutions containing the above-described Murashige and Skoog salts for another week. Then, plants were subjected to the following treatments: +P (complete nutrient solution), −P (P withdrawn from the control solution), and +P and −P with or without ABA and ACC (0.05 μm ABA or 1 μm ACC were added where indicated).
Determination of Soluble P Concentration
Fresh weights of root and leaves of Arabidopsis were determined, followed by tissue homogenization with mortar and pestle in liquid nitrogen. Inorganic phosphate was extracted with 4 mL of 5% (v/v) sulphuric acid (5 M) solution. After centrifugation at 12,000 g, 400 μL of supernatant was transferred and mixed with 200 μL of 15% (w/v) freshly prepared ascorbic acid (pH 5.0) dissolved in ammonium molybdate. The mixture was incubated at 37°C for 30 min, and the A650 was recorded. P concentration was calculated by normalization to fresh weight.
Cell Wall Extraction and Fractionation, and Pectin Measurement
Extraction of crude cell wall materials from root and shoot, and subsequent fractionation of cell wall components, were carried out as previously described (Zhu et al., 2015).
Uronic acid concentration in pectin was determined using GalUA (Sigma) as a standard (Blumenkratz and Asboe-Hansen, 1973). Briefly, 200 μL pectin extracts were treated with 1 mL 98% H2SO4 (containing 0.0125 m Na2B4O7·10H2O) and incubated at 100°C for 5 min. After chilling, 20 μL m-hydroxy-diphenyl (0.15%) was applied to the solution and kept for 20 min at room temperature, followed by measuring the A520 with spectrophotometry.
P Retention in the Cell Wall
A total of 5 mg cell wall material was placed into a 2-mL eppendorf tube to which 2 n HCl had been added. Thatze solution was shaken on a rotary shaker for 3 d. Subsequently, supernatant was collected by centrifugation at 23,000 g and determined by inductively coupled plasma atomic emission spectroscopy (ICP-AES; Fisons ARL Accuris, Ecublens, Switzerland). The P-adsorption ability of cell wall material was assessed according to the P concentration in the –P plants compared to that in the +P plants.
Quantitative Real-Time PCR Analysis
Total RNA was isolated from root using TRIzol reagent (Invitrogen). 1 µg of total RNA served as a template, and cDNA was reversely transcribed using the PrimeScript RT reagent kit (Takara). For the quantitative analysis of gene expression, 1 µL of 10-fold-diluted cDNA was used as the template, performed with SYBR Premix ExTaq (TaKaRa) using the following primer pairs: for TUBULIN, forward: 5′-AAGTTCTGGGAAGTGGTT-3′ and reverse: 5′-CTCCCAATGAGTGACAAA-3′; AtPHR1, forward: 5′-ACAGCAATAACGGAACGGGCAAG-3′ and reverse: 5′-GCTCTTTCACTACCGCCAAGACTG-3′; AtPHT1, forward: 5′-GCCAAGGTAGACGCAGGATA-3′ and reverse: 5′-AACCTCAG CCTCACCAGAGA-3′; AtPHO1, forward: 5′-TACGCGAGAGAAA ACAACGA-3′ and reverse: 5′-TTCCGGAGAACCAAATTGTC-3′; AtPHO2, forward: 5′-TTTTACACAAGCCACCAAAGC-3′ and reverse: 5′-TCACGAGCATGTCCAACAA-3′. Each cDNA sample was run in triplicate. Relative quantification values for each target gene were calculated by the 2-ΔΔCT method (Livak and Schmittgen, 2001) using TUBULIN as a reference gene.
Measurement of Ethylene and ABA Production
Determination of ethylene production was conducted as previously described (Li et al., 2013). Endogenous ABA accumulation was quantified by the ELISA as described in Yang et al. (2001).
Data Analysis
All histograms in “Results” are shown as the mean ± sd for one set of representative data from four independent replicates. Data were subjected to one-way ANOVA, and the means were compared by Student’s t test. Bars with different letters mark statistical significance at the P < 0.05 level.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Effect of phosphate bioavailability on whole-plant growth in Arabidopsis.
Supplemental Figure S2. Effect of potassium and nitrogen bioavailability on whole-plant growth in Arabidopsis.
Supplemental Figure S3. Effect of phosphate bioavailability on whole-plant growth of wild-type and abi4.
Supplemental Figure S4. The influence of ABA on whole-plant growth.
Supplemental Figure S5. The influence of ACC on whole-plant growth.
Supplementary Material
Acknowledgments
We thank the Chinese Academy of Sciences, the National Natural Science Foundation of China, China Postdoctoral Science Foundation, and the Natural Sciences and Engineering Research Council of Canada for funding our research. We are also grateful to two anonymous reviewers for valuable comments to improve the quality of our work.
References
- Adam Z. (2015) Plastid intramembrane proteolysis. Biochim Biophys Acta 1847: 910–914 [DOI] [PubMed] [Google Scholar]
- Ae N, Arihara J, Okada K, Yoshihara T, Johansen C (1990) Phosphorus uptake by pigeon pea and its role in cropping systems of the Indian subcontinent. Science 248: 477–480 [DOI] [PubMed] [Google Scholar]
- Ayadi A, David P, Arrighi JF, Chiarenza S, Thibaud MC, Nussaume L, Marin E (2015) Reducing the genetic redundancy of Arabidopsis PHOSPHATE TRANSPORTER1 transporters to study phosphate uptake and signaling. Plant Physiol 167: 1511–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Park HC, Kim MC, Yun DJ (2013) The role of Arabidopsis MYB2 in miR399f-mediated phosphate-starvation response. Plant Signal Behav 8: e23488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien CM, Chang YC, Nes WD, Kwon-Chung KJ, Espenshade PJ (2009) Cryptococcus neoformans Site-2 protease is required for virulence and survival in the presence of azole drugs. Mol Microbiol 74: 672–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenkrantz N, Asboe-Hansen G (1973) New method for quantitative determination of uronic acids. Anal Biochem 54: 484–489 [DOI] [PubMed] [Google Scholar]
- Bölter B, Nada A, Fulgosi H, Soll J (2006) A chloroplastic inner envelope membrane protease is essential for plant development. FEBS Lett 580: 789–794 [DOI] [PubMed] [Google Scholar]
- Chacón-López A, Ibarra-Laclette E, Sánchez-Calderón L, Gutiérrez-Alanis D, Herrera-Estrella L (2011) Global expression pattern comparison between low phosphorus insensitive 4 and WT Arabidopsis reveals an important role of reactive oxygen species and jasmonic acid in the root tip response to phosphate starvation. Plant Signal Behav 6: 382–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrika NNP, Sundaravelpandian K, Yu SM, Schmidt W (2013) ALFIN-LIKE 6 is involved in root hair elongation during phosphate deficiency in Arabidopsis. New Phytol 198: 709–720 [DOI] [PubMed] [Google Scholar]
- Chen J, Wang Y, Wang F, Yang J, Gao M, Li C, Liu Y, Liu Y, Yamaji N, Ma JF, Paz-Ares J, Nussaume L, et al. (2015) The rice CK2 kinase regulates trafficking of phosphate transporters in response to phosphate levels. Plant Cell 27: 711–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciereszko I, Kleczkowski LA (2002) Effects of phosphate deficiency and sugars on expression of rab18 in Arabidopsis: hexokinase-dependent and okadaic acid-sensitive transduction of the sugar signal. Biochim Biophys Acta 1579: 43–49 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Devaiah BN, Karthikeyan AS, Raghothama KG (2007) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol 143: 1789–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Yu Y, Li S, Wang J, Tang S, Huang R (2016) Abscisic acid antagonizes ethylene production through the ABI4-mediated transcriptional repression of ACS4 and ACS8 in Arabidopsis. Mol Plant 9: 126–135 [DOI] [PubMed] [Google Scholar]
- Fang ZY, Shao C, Meng YJ, Wu P, Chen M (2009) Phosphate signaling in Arabidopsis and Oryza sativa. Plant Sci 176: 170–180 [Google Scholar]
- Gessa C, Deiana S, Premoli A, Ciurli A (1997) Redox activity of caffeic acid towards iron(III) complexed in a polygalacturonate network. Plant Soil 190: 289–299 [Google Scholar]
- Goldstein AH, Baertlein DA, McDaniel RG (1988) Phosphate starvation inducible metabolism in Lycopersicon esculentum I. Excretion of acid phosphatase by tomato plants and suspension-cultured cells. Plant Physiol 87: 711–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong YM, Guo ZH, He LH, Li JS (2011) Identification of maize genotypes with high tolerance or sensitivity to phosphorus deficiency. J Plant Nutr 34: 1290–1302 [Google Scholar]
- Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swarup R, Woolaway KE, White PJ (2003) Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol 132: 578–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Peng M, Rothstein SJ (2011) Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in arabidopsis. PLoS Genet 7: e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambers H, Finnegan PM, Jost R, Plaxton WC, Shane MW, Stitt M (2015) Phosphorus nutrition in Proteaceae and beyond. Nat Plants 1: 15109. [DOI] [PubMed] [Google Scholar]
- Li B, Li Q, Xiong L, Kronzucker HJ, Krämer U, Shi W (2012) Arabidopsis plastid AMOS1/EGY1 integrates abscisic acid signaling to regulate global gene expression response to ammonium stress. Plant Physiol 160: 2040–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Li B, Dong G, Feng X, Kronzucker HJ, Shi W (2013) Ammonium-induced shoot ethylene production is associated with the inhibition of lateral root formation in Arabidopsis. J Exp Bot 64: 1413–1425 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lloyd AJ, William Allwood J, Winder CL, Dunn WB, Heald JK, Cristescu SM, Sivakumaran A, Harren FJM, Mulema J, Denby K, Goodacre R, Smith AR, et al. (2011) Metabolomic approaches reveal that cell wall modifications play a major role in ethylene-mediated resistance against Botrytis cinerea. Plant J 67: 852–868 [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L (2003) The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 6: 280–287 [DOI] [PubMed] [Google Scholar]
- Lv Q, Zhong Y, Wang Y, Wang Z, Zhang L, Shi J, Wu Z, Liu Y, Mao C, Yi K, Wu P (2014) SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell 26: 1586–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R, Morant M, Jarmer H, Nilsson L, Nielsen TH (2007) Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol 143: 156–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nagarajah S, Posner AM, Quirk JP (1970) Competitive adsorption of phosphate with polygalacturonate and other organic anions on kaolinite and oxide surfaces. Nature 228: 83–85 [DOI] [PubMed] [Google Scholar]
- Nagarajan VK, Smith AP (2012) Ethylene’s role in phosphate starvation signaling: more than just a root growth regulator. Plant Cell Physiol 53: 277–286 [DOI] [PubMed] [Google Scholar]
- Niu Y, Chai R, Dong H, Wang H, Tang C, Zhang Y (2013) Effect of elevated CO₂ on phosphorus nutrition of phosphate-deficient Arabidopsis thaliana (L.) Heynh under different nitrogen forms. J Exp Bot 64: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y, Bucher M (2002) Phosphate transport and homeostasis in Arabidopsis. In Somerville CR, Meyerowitz EM eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG, Karthikeyan AS (2005) Phosphate acquisition. Plant Soil 274: 37–49 [Google Scholar]
- Rawson RB. (2013) The site-2 protease. Biochim Biophys Acta 1828: 2801–2807 [DOI] [PubMed] [Google Scholar]
- Rawson RB, Zelenski NG, Nijhawan D, Ye J, Sakai J, Hasan MT, Chang TY, Brown MS, Goldstein JL (1997) Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol Cell 1: 47–57 [DOI] [PubMed] [Google Scholar]
- Rudner DZ, Fawcett P, Losick R (1999) A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc Natl Acad Sci USA 96: 14765–14770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Shin HS, Chen R, Harrison MJ (2006) Loss of At4 function impacts phosphate distribution between the roots and the shoots during phosphate starvation. Plant J 45: 712–726 [DOI] [PubMed] [Google Scholar]
- Thibaud MC, Arrighi JF, Bayle V, Chiarenza S, Creff A, Bustos R, Paz-Ares J, Poirier Y, Nussaume L (2010) Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J 64: 775–789 [DOI] [PubMed] [Google Scholar]
- Trull MC, Guiltinan MJ, Lynch JP, Deikman J (1997) The responses of wild-type and ABA mutant Arabidopsis thaliana plants to phosphorus starvation. Plant Cell Environ 20: 85–92 [Google Scholar]
- Wang L, Lu S, Zhang Y, Li Z, Du X, Liu D (2014c) Comparative genetic analysis of Arabidopsis purple acid phosphatases AtPAP10, AtPAP12, and AtPAP26 provides new insights into their roles in plant adaptation to phosphate deprivation. J Integr Plant Biol 56: 299–314 [DOI] [PubMed] [Google Scholar]
- Wang S, Zhang S, Sun C, Xu Y, Chen Y, Yu C, Qian Q, Jiang DA, Qi Y (2014a) Auxin response factor (OsARF12), a novel regulator for phosphate homeostasis in rice (Oryza sativa). New Phytol 201: 91–103 [DOI] [PubMed] [Google Scholar]
- Wang Z, Ruan W, Shi J, Zhang L, Xiang D, Yang C, Li C, Wu Z, Liu Y, Yu Y, Shou H, Mo X, et al. (2014b) Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc Natl Acad Sci USA 111: 14953–14958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang J, Wang Z, Zhu Q, Wang W (2001) Hormonal changes in the grains of rice subjected to water stress during grain filling. Plant Physiol 127: 315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun SJ, Kaeppler SM (2001) Induction of maize acid phosphatase activities under phosphorus starvation. Plant Soil 237: 109–115 [Google Scholar]
- Zhang YJ, Lynch JP, Brown KM (2003) Ethylene and phosphorus availability have interacting yet distinct effects on root hair development. J Exp Bot 54: 2351–2361 [DOI] [PubMed] [Google Scholar]
- Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P (2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146: 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XF, Lei GJ, Jiang T, Liu Y, Li GX, Zheng SJ (2012) Cell wall polysaccharides are involved in P-deficiency-induced Cd exclusion in Arabidopsis thaliana. Planta 236: 989–997 [DOI] [PubMed] [Google Scholar]
- Zhu XF, Wang ZW, Wan JX, Sun Y, Wu YR, Li GX, Shen RF, Zheng SJ (2015) Pectin enhances rice (Oryza sativa) root phosphorus remobilization. J Exp Bot 66: 1017–1024 [DOI] [PubMed] [Google Scholar]
- Zhu XF, Zhu CQ, Zhao XS, Zheng SJ, Shen RF (2016) Ethylene is involved in root phosphorus remobilization in rice (Oryza sativa) by regulating cell-wall pectin and enhancing phosphate translocation to shoots. Ann Bot (Lond) mcw04410.1093/aob/mcw044 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.