HSFA6b acts as a positive regulator downstream of ABA signaling, mediates salinity and drought stress responses, and is required for establishing thermotolerance.
Abstract
Heat stress response (HSR) is a conserved mechanism developed to increase the expression of heat shock proteins (HSPs) via a heat shock factor (HSF)-dependent mechanism. Signaling by the stress phytohormone abscisic acid (ABA) is involved in acquired thermotolerance as well. Analysis of Arabidopsis (Arabidopsis thaliana) microarray databases revealed that the expression of HSFA6b, a class A HSF, extensively increased with salinity, osmotic, and cold stresses, but not heat. Here, we show that HSFA6b plays a pivotal role in the response to ABA and in thermotolerance. Salt-inducible HSFA6b expression was down-regulated in ABA-insensitive and -deficient mutants; however, exogenous ABA application restored expression in ABA-deficient, but not -insensitive plants. Thus, ABA signaling is required for proper HSFA6b expression. A transcriptional activation assay of protoplasts revealed that ABA treatment and coexpression of an ABA signaling master effector, ABA-RESPONSIVE ELEMENT-BINDING PROTEIN1, could activate the HSFA6b promoter. In addition, HSFA6b directly bound to the promoter of DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN2A and enhanced its expression. Analysis of ABA responses in seed germination, cotyledon greening, and root growth as well as salt and drought tolerance in HSFA6b-null, overexpression, and dominant negative mutants revealed that HSFA6b is a positive regulator participating in ABA-mediated salt and drought resistance. Thermoprotection tests showed that HSFA6b was required for thermotolerance acquisition. Our study reveals a network in which HSFA6b operates as a downstream regulator of the ABA-mediated stress response and is required for heat stress resistance. This new ABA-signaling pathway is integrated into the complex HSR network in planta.
When plants are exposed to high temperatures, heat stress (HS) causes cellular damage, leading to severe growth retardation and possible death (Larkindale and Vierling, 2008). Organisms can survive under HS through basal thermotolerance (BT). However, organisms with acquired thermotolerance (AT) can endure lethal HS through metabolic and cellular adjustments induced during an acclimation period at a moderately high but survivable temperature before HS (Vierling, 1991; Larkindale et al., 2005). The HS response (HSR), activated by HS, is the general mechanism preventing stress-caused damage and helping organisms overcome the lethal stress (Krishna, 2004). During HSR, plants accumulate HS proteins (HSPs), which primarily function as molecular chaperones to prevent protein aggregation and facilitate appropriate refolding of the heat-damaged proteins (Parsell and Lindquist, 1993). The expression of HSPs during the HSR is primarily regulated by HS transcription factors (HSFs) that bind to the HS elements (HSEs; nGAAnnTTCn) located in the promoter of HSFs and HSPs (Rabindran et al., 1993; Schöffl et al., 1998; Nover et al., 2001). When stress is relieved, the HSR is attenuated by excess HSP70 and other proteins that repress the transcriptional activity of HSFs by binding to them and converting HSFs back to the original inactive form. AtHSBP, an HSF-binding protein in Arabidopsis (Arabidopsis thaliana), can reduce the DNA binding capacity of HSFs and function as a negative regulator of the HSR (Hsu et al., 2010).
That multiple HSFs exist among different eukaryotic organisms suggests the importance of the backup and diversification of HSFs. Vertebrates contain four HSFs, and Drosophila and Caenorhabditis elegans contain one each (Nover et al., 2001). Plants have predicted HSF families that are more diverse compared to animals: 24 HSFs and three HSF-like genes were identified in tomato, 25 in rice (Oryza sativa), 30 in maize (Zea mays), and 52 in soybean (Glycine max; Kotak et al., 2004; Scharf et al., 2012). In Arabidopsis, the HSF family is small but more defined, with 21 members: 15 are in class A, five in class B, and one in class C. The class A members, HSFAs, contain the conserved oligomerization, DNA-binding, and aromatic/hydrophobic/acidic-type activation domains, as well as nuclear localization and export signal motifs; and they function as transcription activators. The class B and C HSFs lack a defined activation domain; class B HSFs may serve as coregulators or repressors of the HSFAs, while class C HSF functions remain unclear (Boscheinen et al., 1997; Czarnecka-Verner et al., 2000, 2004; Kotak et al., 2007a; von Koskull-Döring et al., 2007; Ikeda et al., 2011). The large number of HSFs in higher plants suggests that compared with animals, plants have a more complex and highly regulated system to respond to HS for survival in a broader temperature range (Nover et al., 2001; Kotak et al., 2007a).
In Arabidopsis, HSFA1a and HSFA1b double-knockout (KO) mutants are significantly impaired in early transient mRNA accumulation of HSPs (Lohmann et al., 2004). The HSFA1a/b/d/e quadruple-KO mutant showed greatly decreased BT and AT and developmental defects; it was less tolerant to NaCl, mannitol, and H2O2 stress (Liu et al., 2011; Yoshida et al., 2011). Thus, we can conclude that class-A1 HSFs may function as the master regulators of HSR. HS-inducible HSFA2 is the dominant HSF in thermotolerant cells. Analysis of the HSFA2-KO mutant revealed that HSFA2 controls the expression of HSPs under prolonged HS and recovery (Busch et al., 2005; Schramm et al., 2006; Charng et al., 2007). In addition, HSFA2-KO plants were sensitive to oxidative stress, high light, and anoxia as compared with overexpression (OE) lines, which were more resistant to salt, osmotic, oxidative, and anoxia stresses (Ogawa et al., 2007; Banti et al., 2010). HSFA3 is induced by HS and drought, and its expression is dependent on the transcription factor DEHYDRATION-RESPONSIVE ELEMENT (DRE; A/GCCGAC core motif)-BINDING PROTEIN2A (DREB2A). DREB2A-KO and -OE lines showed reduced and increased thermotolerance, respectively, as the result of altered HSFA3 and HSP gene expression (Sakuma et al., 2006; Schramm et al., 2006, 2008; Lim et al., 2007; Yoshida et al., 2008; Qin et al., 2011). HS-induced DREB2A expression depends on HSFA1s, but HSFA1s could not mediate the drought responsiveness of DREB2A (Yoshida et al., 2011). Overexpression of HSFA2 and HSFA3 did not affect DREB2A expression; therefore, these genes do not function in regulating DREB2A expression (Nishizawa et al., 2006; Yoshida et al., 2008). HSFA1d, HSFA2, and HSFA3 are key factors in regulating oxidative-response gene ASCORBATE PEROXIDASE2 (APX2) expression under diverse stress conditions (Panchuk et al., 2002; Jung et al., 2013). APX2 is highly responsive to HS and plays an important role in the acquisition of thermotolerance (Shi et al., 2001; Panchuk et al., 2002; Larkindale and Huang, 2004; Schramm et al., 2006; Suzuki et al., 2013); a mutant with constitutively higher expression of APX2 showed enhanced tolerance to drought and high abscisic acid (ABA) levels (Rossel et al., 2006).
Other HSFAs, HSFA4a and HSFA8, may act as reactive oxygen species (ROS) sensors (Davletova et al., 2005), whereas HSFA5 acts as a specific repressor of HSFA4 isoforms to form a HSFA4-HSFA5 complex that negatively regulates this pathway. The pathway might be connected with controlled cell death triggered by pathogen infection (Baniwal et al., 2007). HSFA7a and HSFA7b are HSR factors (Liu et al., 2011). Moreover, HSFA9 acts as a master regulator of the expression of HSPs during seed development and displays a synergism between HSFA9 and ABA-responsive transcription factor ABA INSENSITIVE3 (ABI3; Kotak et al., 2007b). HSFA4c was found to be involved in root circumnutation, gravitropic response, and hormonal control of differentiation (Fortunati et al., 2008). Recently, HSFB2a was found to be involved in gametophyte development and HSFB2b was found to be important for accurate circadian rhythms following elevated temperature and salt treatment (Kolmos et al., 2014; Wunderlich et al., 2014). Consequently, a cross talk exists between HS and other abiotic stress-signaling cascades mediated by HSFs (Kotak et al., 2007a). HSFA6a and HSFA6b transcript levels are particularly expressed in response to salt, osmotic, and cold stress (von Koskull-Döring et al., 2007; Hwang et al., 2014). Overexpression of HSFA6b, but not HSFA6a, could regulate the transcription of DREB2A, which suggests nonredundant functions (Yoshida et al., 2011); however, the molecular mechanism of HSFA6b still needs to be elucidated upon.
With regards to osmotic stress signaling due to salinity, drought, and cold stress, endogenous ABA levels increase and ABA responsive transcription acts through an ABA-RESPONSIVE ELEMENT (ABRE; PyACGTGG/TC)-BINDING PROTEINS/FACTORS (AREBs/ABFs) via multiple ABREs or combinations of ABREs with coupling elements (Busk and Pagès, 1998; Fujita et al., 2011; Nakashima and Yamaguchi-Shinozaki, 2013). ABA-deficient and -insensitive mutants are sensitive to HS, whereas AREB/ABF-OE plants show enhanced thermotolerance (Larkindale and Knight, 2002; Kim et al., 2004; Larkindale et al., 2005; Suzuki et al., 2016). Thus, ABA plays a role in the thermotolerance response. Analysis of an Arabidopsis microarray database (AtGenExpress consortium; https://www.arabidopsis.org/portals/expression/microarray/ATGenExpress.jsp) showed HSFA6b had the highest expression induced by salinity, osmotic, and cold stress among all HSFs, but HSFA6a had little response to the stresses; notably, both genes were not significantly up-regulated by HS. Transcriptome analysis of the Arabidopsis AREB1/AREB2/ABF3 triple-KO mutant showed strongly reduced expression of HSFA6a and HSFA6b under salt stress, dehydration stress, and ABA treatment (Yoshida et al., 2010).
The HSR depends on a complex regulatory network involving HSF and HSP families, and HSFs and HSPs respond to various abiotic stresses besides HS (Swindell et al., 2007). However, the role of the 21 Arabidopsis HSFs under HS and non-HS conditions is not well understood, and the types of stress that interact most strongly with HSF and HSR pathways remain unclear. In this study, we investigated the function of HSFA6b genetically with T-DNA insertional KO and 35S-driven OE mutants. We used a dominant negative HSFA6b fused to an ETHYLENE-RESPONSIVE ELEMENT-BINDING FACTOR-associated amphiphilic repression-motif repression domain SRDX (Hiratsu et al., 2003) to diminish the activities of endogenous and functionally redundant factors. The results, including our microarray data, revealed that HSFA6b acts as a positive regulator downstream of ABA signaling, mediates salinity and drought-stress responses, and is required for establishing thermotolerance. Thus, HSFA6b functions in ABA signaling cascades in the complex regulatory networks of the HSR.
RESULTS
Arabidopsis Class A HS Transcription Factor HSFA6b Is Induced by Salt, Osmotic, and ABA, But Not HS
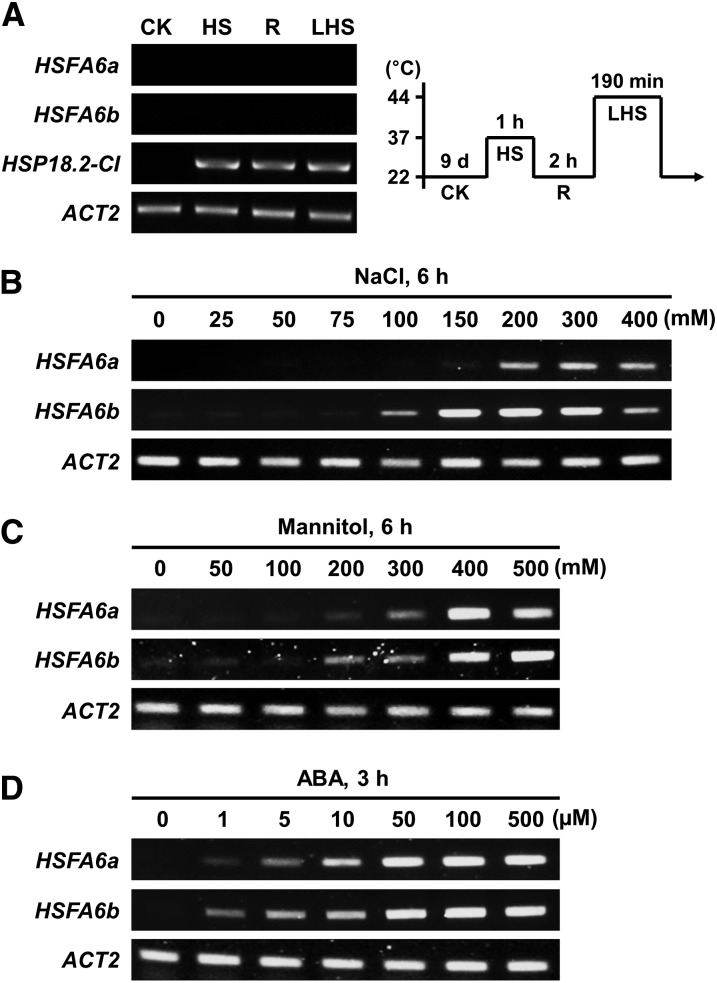
Analysis of the AtGenExpress database revealed the expression of 21 Arabidopsis HSFs responding to heat, salt, osmotic, drought, and cold stress (Supplemental Fig. S1, A and B): among the 15 class A HSF genes, HSFA6b expression was extensively induced by salt (NaCl), osmotic (mannitol), and cold, but not HS. We analyzed the gene expression of HSFA6b and its potential paralog HSFA6a in response to HS, NaCl, and mannitol as well as the osmotic stress signal mediator ABA using reverse transcription PCR (RT-PCR; Fig. 1) or real-time quantitative PCR (qRT-PCR; Supplemental Fig. S1C). HSFA6b expression was markedly induced by NaCl, mannitol, and ABA in a dose-dependent manner, but not with HS; this was supported by the microarray findings. The expression of HSFA6b occurs earlier and is stronger than that of HSFA6a for all treatments.
Figure 1.
The expression profiles of HSFA6a and HSFA6b in response to HS, salt, osmotic, and ABA stress. A to D, Nine-day-old seedlings were treated with HS or incubated with water containing NaCl, mannitol, or ABA at different concentrations and times. The transcription levels of HSFA6a and HSFA6b were analyzed by RT-PCR. The pictogram shows the HS regime (right of A): 37°C sublethal HS for 1 h (HS) → 22°C recovery for 2 h (R) → 44°C lethal HS for 190 min (LHS). 22°C treatment was a control (CK). HSP18.1-CI, used as a reference, is an HS-responsive marker gene. ACTIN2 (ACT2) was used as a loading control.
Nuclear and Cytosol Localization of HSFA6b
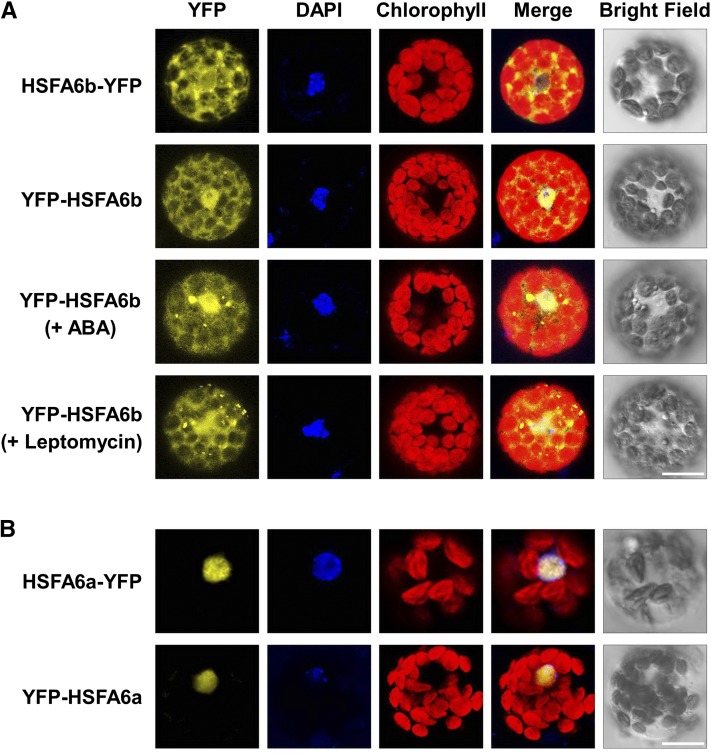
The protein structure of HSFA6b shares conserved HSF signatures with HSFA1a and HSFA2 (Nover et al., 2001), as it harbors the predicted nuclear localization and export signal peptides (Supplemental Fig. S2). HSFA6b was fused to either the N- or C-terminal yellow fluorescent protein (YFP) reporter and transiently expressed in Arabidopsis protoplasts for subcellular localization analysis (Fig. 2A). It was localized to the nucleus and cytosol, with partial but not complete translocation to the nucleus. Both 2-h treatments of 10 μm ABA and 20 nm Leptomycin B only slightly enhanced the nuclear enrichment. Meanwhile, HSFA6a, with a predicted nuclear importin but no exportin, was specifically localized to the nucleus (Fig. 2B).
Figure 2.
Transient expression profiles of HSFA6a and HSFA6b in mesophyll protoplasts. A, Confocal microscopy of Arabidopsis Col protoplasts transfected with HSFA6b fused to the N- or C-terminal YFP reporter gene, for HSFA6b-YFP or YFP-HSFA6b. YFP-HSFA6b transfected cells were also treated with 10 μm ABA or 20 nm Leptomycin B for 2 h. B, YFP-tagged HSFA6a transfection served as comparison. Blue shows nuclei stained with 4′,6-diamino-phenylindole (DAPI), and red shows chlorophyll autofluorescence. Bars = 20 μm.
HSFs form homo- or heterotrimers, resulting in altered nuclear localization, and then bind to their own promoters and/or to promoters of other HSF genes, enhancing or suppressing their promoter transcription (Miller and Mittler, 2006). We used a coimmunoprecipitation (Co-IP) and bimolecular fluorescence complementation (BiFC) assay to investigate the interaction of HSFA6b with itself and HSFA6a, as well as HS-related HSFs, such as HSFA (1a, 1b, 2, 3, 7a, 7b) and HSFB (1, 2a, 2b) in Arabidopsis protoplasts (Supplemental Fig. S3, A and B). HSFA6b neither interacts with itself, even with the 10 μm ABA treatment, nor with HSFA6a. It interacts with the well-known HSFA1a, HSFA1b, and HSFA2 in the nucleus.
Expression of HSFA6b Was Impaired in ABA-Deficient and -Insensitive Mutants under Salt Treatment
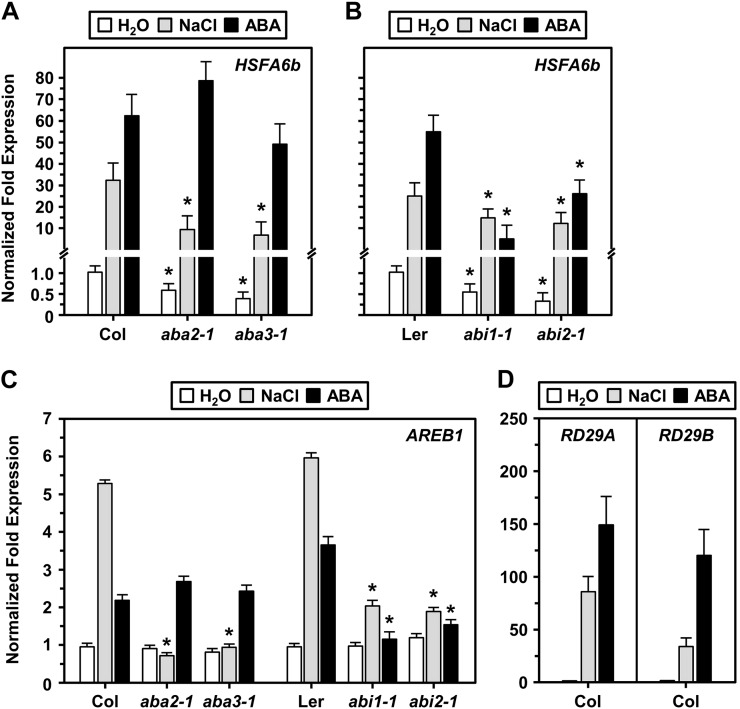
HSFA6b was the most NaCl-responsive gene among all the HSFs (Supplemental Fig. S1), which implies a potential function in salt stress signaling. We investigated the expression levels of HSFA6b and AREB1 in ABA-deficient (aba2-1 and aba3-1) and -insensitive (abi1-1 and abi2-1) mutants under a 200 mm NaCl or 20 µm ABA treatment (Fig. 3). HSFA6b expression was greatly impaired in ABA-deficient mutants under NaCl treatment; however, ABA treatment notably restored expression (Fig. 3A). HSFA6b expression was likewise equally impaired in ABA-insensitive mutants under both NaCl and ABA treatment (Fig. 3B). The AREB1 expression profile was similar to that of HSFA6b (Fig. 3C), and NaCl and drought-responsive marker genes RESPONSIVE TO DEHYDRATION29A (RD29A) and RD29B were used as references (Fig. 3D). Therefore, we suggest that NaCl-induced HSFA6b gene expression could be downstream of ABA perception and regulated by the ABA signal.
Figure 3.
The expression levels of HSFA6b and AREB1 in ABA-deficient and -insensitive mutant lines. Nine-day-old seedlings of ABA biosynthesis mutants aba2-1 and aba3-1, and ABA-insensitive mutants abi1-1 and abi2-1, incubated in H2O with 200 mm NaCl for 6 h or 20 μm ABA for 3 h. A to C, The transcription levels of HSFA6b and AREB1 were analyzed by qRT-PCR. D, NaCl and ABA-responsive marker genes RD29A and RD29B were used as references. The fold change expression was normalized relative to the level of Col or Ler H2O treatment. Data are means ± sd of three biological replicates. *Significant at P < 0.05 compared with the Col or Ler. PP2A was an internal control.
ABA Treatment and AREB1/ABF2 Coexpression Enhanced HSFA6b Promoter Activity
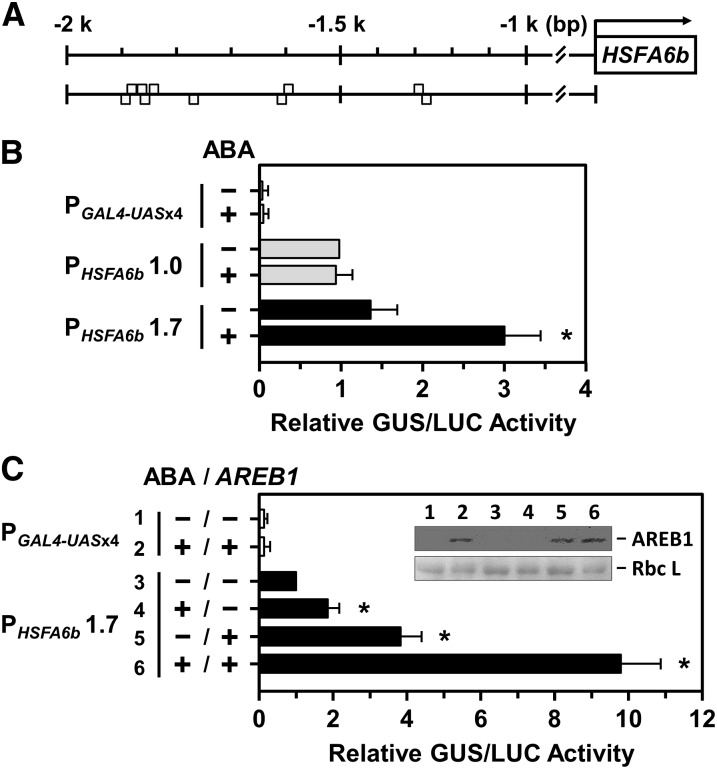
To confirm that HSFA6b expression was related to ABA signaling, we analyzed an upstream 2-kb potential promoter region of HSFA6b using Plant Promoter Analysis Navigator (PlantPAN 2.0; http://plantpan2.itps.ncku.edu.tw; Chow et al., 2016) and found that the promoter contained ABREs (Fig. 4A). After fusing the 1.7-kb promoter region to the reporter gene GUS, the protoplast transactivation assay confirmed that a 10 μm ABA treatment significantly enhanced the transcriptional activity of HSFA6b (Fig. 4B). However, a 1.0-kb upstream region of HSFA6b without ABREs, used as a reference, did not respond to ABA treatment. Coexpression with effector AREB1 (i.e. ABF2), an ABRE-dependent master transcription regulator, enhanced the transcriptional activity of HSFA6b, and in the presence of both ABA and AREB1 greatly activated HSFA6b promoter activity (Fig. 4C). Thus, the transcription of HSFA6b could be regulated in an ABA-dependent AREB/ABF-ABRE manner.
Figure 4.
The transcriptional activation of the HSFA6b promoter in response to ABA and AREB1 treatments were analyzed in mesophyll protoplasts. A, Gray boxes are predicted ABRE motifs (T/CACGTGGC) upstream of the HSFA6b promoter region at −1257 (reverse strand [−]); −1299 (forward strand [+]); −1602 (−); −1605 (+); −1776 (−); −1846 (−); −1849 (+); −1864 (+); −1869 (−); and −1872 bp (+). B and C, The transcriptional activation assay of the HSFA6b promoter. PGAL4-UASx4::GUS was a control, and 1- or 1.7-kb lengths of the HSFA6b promoter region, PHSFA6b1.0 or 1.7, respectively, were fused with GUS gene and used in the corresponding transient reporter assays. Transfected protoplasts were then treated with (+) or without (−) 10 μm ABA for 12 h and with or without effector AREB1-3XFLAG coexpression, as indicated. The AREB1-3XFLAG fusion protein, under the control of the CaMV 35S promoter, was detected by immunoblotting (α-FLAG) and the Rubisco large subunit (Rbc L) shown for equal loading (inset). The amount of relative GUS activity was normalized by luciferase (LUC) luminescence. The fold expression was normalized relative to the amount of PHSFA6b 1.0 without ABA (B), or PHSFA6b 1.7 without ABA and ARER1 (C). Data are means ± sd of three biological replicates. *Significant at P < 0.05.
HSFA6b Mutants Were Screened and Confirmed
To elucidate the function of HSFA6b required for the ABA response, HSFA6b-null mutant lines a6b-1 (Columbia [Col] ecotype background) and a6b-2 (Landsberg erecta [Ler] ecotype background) were screened and confirmed by RT-PCR (Supplemental Fig. S4A). As a reference, the HSFA6a homozygous T-DNA insertion line a6a (Col ecotype background) was screened, and RT-PCR revealed that it is a null mutant (Supplemental Fig. S4B).
For HSFA6b-OE lines, a hemagglutinin (HA) tag was fused to the N terminus of HSFA6b, with expression driven by the CaMV 35S promoter (Supplemental Fig. S4C). For HSFA6b dominant-negative effect (HSFA6b-RD) lines, an ETHYLENE-RESPONSIVE ELEMENT-BINDING FACTOR-associated amphiphilic repression-motif repression domain SRDX (Hiratsu et al., 2003) was fused to the C terminus of HA-HSFA6b, with expression driven by the 35S promoter (Supplemental Fig. S4D). RT-PCR and immunoblotting results confirmed the overexpression of transgenes in the transgenic plants HSFA6b-OE-5-6 (OE5-6) and HSFA6b-RD-3-6 (RD3-6; Supplemental Figs. S4, C and D, and S5A). These transgenic lines were generated in the ecotype Col background.
Transgenic lines with the different expression levels of transgenes HSFA6b-OE and HSFA6b-RD showed differing phenotype response to 0.75 µm ABA treatment (Supplemental Fig. S5, A and B). During early seedling growth, the overexpression of HSFA6b-OE (gain-of-function transgene) showed a higher response to ABA and overexpression of HSFA6b-RD (dominant negative effect transgene) showed reduced sensitivity to ABA. We used the representative lines OE5-6, OE13-6, and OE12-6, as well as RD3-5, RD15-3, and RD23-1 for the following experiments.
HSFA6b Is Required for Proper Responses to Salt, Osmotic, and ABA Stress
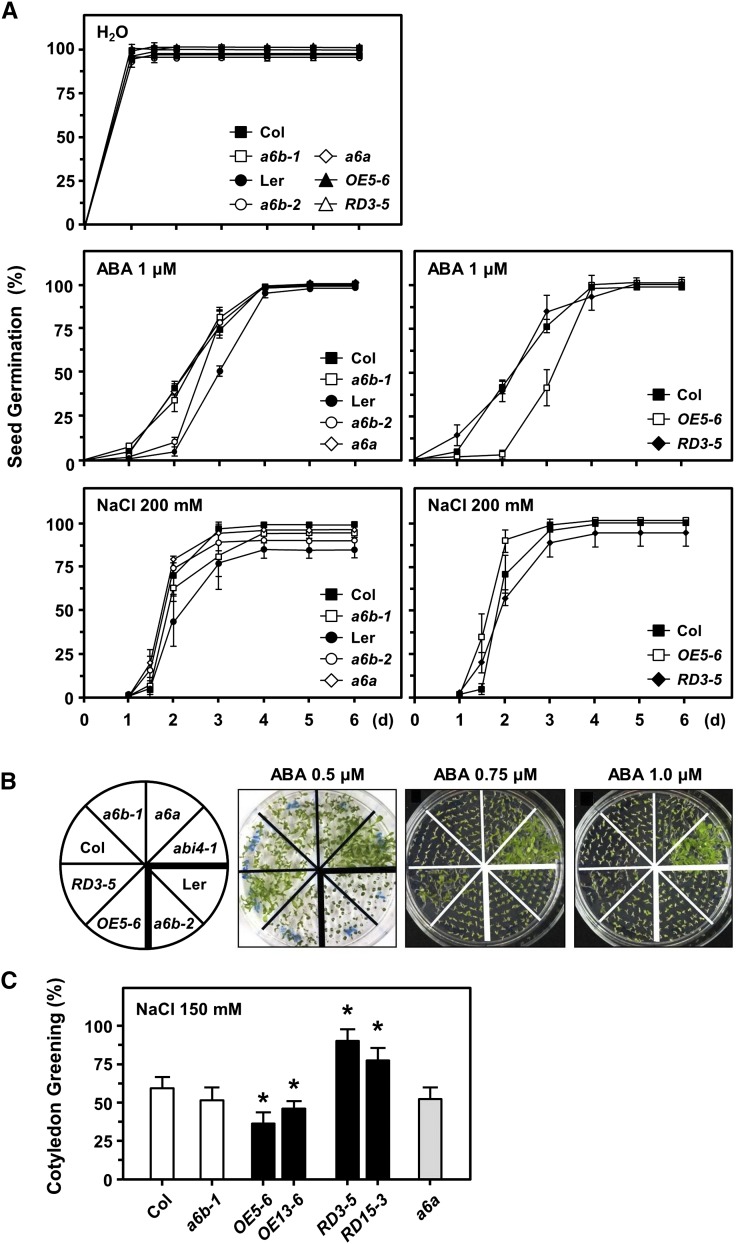
The HSFA6b-mutant lines a6b-1, a6b-2, OE5-6, and RD3-5 as well as the HSFA6a mutant a6a (used as a reference) were treated with NaCl, mannitol, and ABA in order to mimic a downstream mediator of salt, osmotic, and drought stress. ABA-inhibited seed germination, cotyledon greening, and root growth were then analyzed (Kreps et al., 2002; Fujii et al., 2007).
Seed germination did not differ between the a6b-1, a6b-2, and a6a mutants and the wild-type Col and Ler for 1 µm ABA and 200 mm NaCl treatments (Fig. 5A). Seed germination rate was markedly reduced in OE5-6 with ABA treatment at days 2 and 3 but was not affected by NaCl treatment; under both treatments, seed germination did not differ between RD3-5 and the Col.
Figure 5.
Seed germination and postgermination growth in response to salt and ABA treatments in HSFA6 mutants. A, Seed germination (%) with either 1 μm ABA or 200 mm NaCl treatment. Water treatment was used as a control. B, Seedling growth with 0.5 to 1 μm ABA on plates was photographed at day 12 after planting. abi4-1, an ABA-insensitive mutant (Col ecotype), was used as reference. C, Cotyledon greening (%) with 150 mm NaCl on plates was measured at day 12 after planting. Data are means ± sd of three biological replicates. *Significant at P < 0.05 compared with the Col.
Seeds were planted in growth medium agar plates containing 0.5, 0.75, or 1 µm ABA; postgermination seedling growth for OE5-6 was more sensitive (ABA, 0.5 µm), while growth of RD3-5 was more resistant (ABA, 0.75 µm) when compared to the Col (Fig. 5B; Supplemental Fig. S5B). As expected, ABA treatment had no effect on early seedling growth in an ABA-insensitive mutant, abi4-1 (Col ecotype background; an ABA-responsive marker in seeds) that was used as a reference. Mutants of a6b-1 and a6b-2 showed a similar phenotype as the Col and Ler, respectively.
With 150 mm NaCl treatment, the cotyledon greening ratio was lower and higher for OE (5-6 and 13-6) and RD (3-5 and 15-3) lines, respectively, when compared with the Col, while no significant difference was observed for a6b-1 (Fig. 5C), which agreed with the ABA treatment findings (Fig. 5B). The cotyledon greening ratio for a6a and the Col was similar.
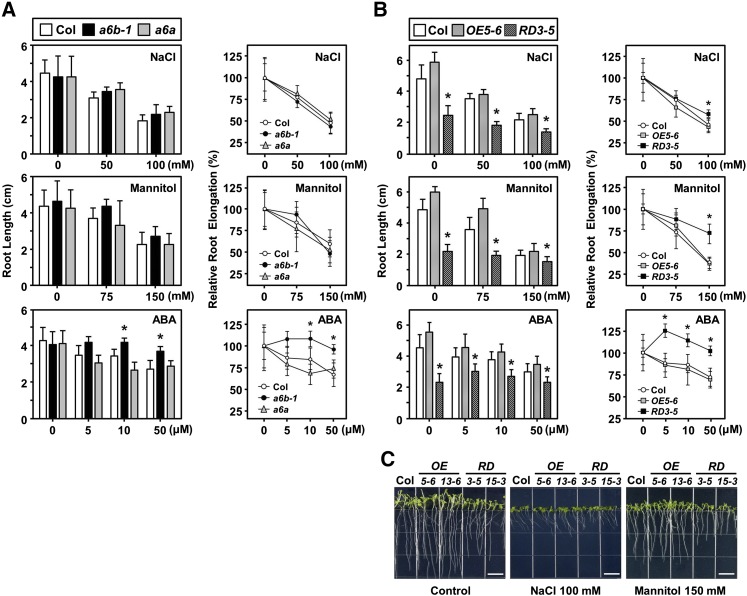
The relative root elongation rate did not differ between a6b-1 and the Col under NaCl and mannitol treatments, but a6b-1 was more resistant to ABA treatment (Fig. 6A). However, root elongation was significantly affected in RD lines (Fig. 6, B and C): With NaCl, mannitol, and ABA treatments, RD lines were more resistant growth inhibition. In addition, the ABA-mediated root growth phenotypes of a6a and the Col were similar. The two OE5-6 and 13-6 lines and two RD3-5 and 15-3 lines exhibited consistent phenotypic responses to the abiotic stresses.
Figure 6.
Root growth in response to salt, osmotic, and ABA treatment in HSFA6s mutants. A and B, Three-day-old seedlings grown on half-strength Murashige and Skoog medium were transferred to plates containing NaCl, mannitol, or ABA, as indicated. Root length (cm) and relative root elongation (%) were measured at day 9. C, Seeds were planted in half-strength Murashige and Skoog medium (control), and medium containing 100 mm NaCl or 150 mm mannitol, and then photographed at day 9. Data are means ± sd of three biological replicates. *Significant at P < 0.05 compared with the Col.
Thus, misregulation and dysfunction of HSFA6b interfered in the proper response to salt, osmotic, and ABA treatments, which implies that HSFA6b contributes to ABA-mediated stress responses.
Misregulated ABA Biosynthesis and Responsive Gene Expression, in Response to Salt and ABA, in HSFA6b Mutants
To confirm that HSFA6b is involved in ABA signaling, we investigated the effect of NaCl and ABA treatment in HSFA6b mutants by looking at the regulation of ABA biosynthesis marker genes NINE-CIS-EPOXYCAROTENOID DIOXYGENASE3 (NCED3) and ARABIDOPSIS ALDEHYDE OXIDASE3 (AAO3; Barrero et al., 2006), as well as RD29A and RD29B, which are involved in the ABA-independent and -dependent pathways, respectively (Shinozaki and Yamaguchi-Shinozaki, 2000; Huang et al., 2012b; Supplemental Fig. S6).
With NaCl treatment, the expression of NCED3 was higher than that of AAO3 in Col plants (Supplemental Fig. S6A), which concurs with previous studies (Barrero et al., 2006). NCED3 expression was suppressed in a6b-1 and RD3-5 following NaCl treatment, as compared with the Col, but the expression of AAO3 was not greatly affected in HSFA6b mutants. The expression of RD29A was not markedly affected in HSFA6b mutants following NaCl and ABA treatments, as compared with the Col (Supplemental Fig. S6B). Notably, the expression of RD29B was increased in OE5-6 and decreased in RD3-5. In addition, DREB2A expression was significantly decreased in RD3-5, but the expression profile of AREB1 was not affected in mutants. However, the expression of RD22 (Supplemental Fig. S6B), a drought-responsive marker gene mediated by the transcription activators MYB/MYC via MYB/MYC-responsive elements (MYBRE/MYCRE) in an ABA-dependent manner (Abe et al., 2003), was unaffected in HSFA6b mutants. Therefore, HSFA6b might not contribute to ABA-independent DREB1/CBF-DRE and ABA-dependent MYB/MYC-MYBRE/MYCRE regulation.
HSFA6b Participates in Salt and ABA-Mediated HSP Gene Expression
The induction and accumulation of HSPs are essential for establishing thermotolerance, and HSP gene transcription can also be induced by other abiotic stresses such as salt and drought (Supplemental Fig. S7). Intriguingly, the overexpression of DREB2A and DREB2C greatly induced HSP18.1-CI, HSP26.5-MII, and HSP70 expression, which enhanced heat and drought tolerance (Sakuma et al., 2006; Lim et al., 2007).
With 1-h 37°C HS treatment (Supplemental Fig. S8A), the expression of HSPs was up-regulated, with no difference in HSFA6b mutants when compared with the Col; in addition, the attenuation of HSP expression remained unaffected during recovery from HS (Supplemental Fig. S9A). The expression of HSFs was similar to that of HSPs in response to HS (Supplemental Fig. S8A).
Under 150 mm NaCl and 20 μm ABA treatments (Supplemental Fig. S8, B and C), the expression of HSPs was markedly suppressed in RD3-5 but effectively up-regulated in OE5-6, while the expressions of HSP18.1-CI and HSP26.5-MII were both greatly enhanced. However, the treatments had no major effect on HSF expression in OE5-6, whereas those of HSFA6a and class B HSF genes HSFB1, HSFB2a, HSFB2b, and HSFB4 were down-regulated in RD3-5.
Meanwhile, the expression of oxidative stress-responsive genes COPPER-ZINC SUPEROXIDE DISMUTASE (CuZnSOD; CSD1, CSD2, CSD3), ASCORBATE PEROXIDASE (APX1, APX2), and CATALASE (CAT1), key components of the ROS network (Davletova et al., 2005), were largely unaffected in HSFA6b mutants in response to HS, NaCl, and ABA treatments (Supplemental Fig. S9B). Notably, the expression of APX2 was significantly up-regulated and suppressed in OE5-6 and RD3-5, respectively, in response to HS (Supplemental Fig. S9B), NaCl, and ABA (Supplemental Fig. S10A). As well, superoxide dismutase (SOD) activity and H2O2 accumulation were not affected in HSFA6b mutants under the treatments (Supplemental Fig. S9, C and D).
HSFA6b Activated Transcriptional Activity of HSP18.1-CI, DREB2A, and APX2 Promoters
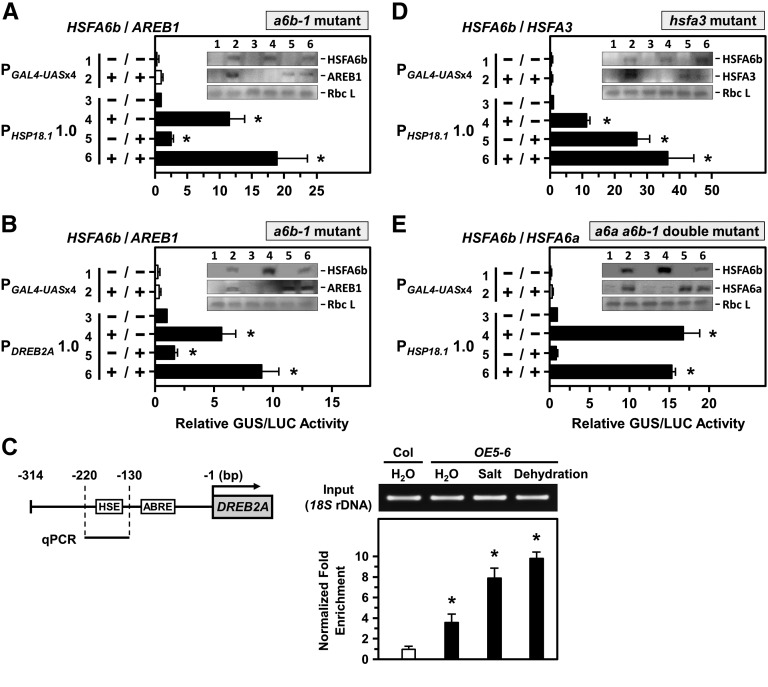
In a6b-1 mutant protoplasts, HSFA6b overexpression significantly activated the transcription of HSP18.1-CI and DREB2A promoters (Fig. 7, A and B), and coexpression of AREB1 had an additive effect on two promoters’ expressions. The interaction of HSFA6b with the HSE sequence in the DREB2A promoter (Kim et al., 2011) was confirmed by chromatin immunoprecipitation (ChIP) assay with anti-HA antibodies in OE5-6 plants (Fig. 7C). NaCl and dehydration treatments significantly enhanced the interaction. Thus, HSFA6b bound to the HSE sequence of the DREB2A promoter effectively enhanced its expression in response to the stresses.
Figure 7.
HSFA6b and AREB1 activated the transcriptional activity of the HSP18.1-CI and DREB2A promoters. A and B, a6b-1 mutant protoplasts were used for a transcriptional activation assay, as indicated in Figure 4. The 1-kb length of the HSP18.1-CI or DREB2A promoter region, respectively, was fused with GUS reporter gene. Protoplasts were transfected with or without effectors HSFA6b-3XFLAG and AREB1-3XFLAG. The effectors were analyzed by immunoblotting (insets). The fold expression was normalized relative to the level of PHSP18.1 and PDREB2A without effectors. C, HSFA6b binding to the HSE sequence of the DREB2A promoter was analyzed by ChIP assay. Schematic map of the 314-bp upstream of the DREB2A promoter region (left); ABRE (GACACGTA; −86 to −93 bp) and HSE (AGAAGATTCG; −151 to −160 bp) are in gray boxes. Seedlings were treated with 6 h 150 mm NaCl or 2.5 h dehydration. The fold enrichment of the HSE-containing region (qPCR) after ChIP was analyzed by qRT-PCR and normalized to the Col H2O treatment (right). 18S rDNA was an input control. D and E, Transcriptional activation assay in hsfa3 and a6a a6b-1 double-mutant protoplasts with or without effectors HSFA6b-3XFLAG, HSFA3-3XFLAG, and HSFA6a-3XFLAG are as described in A and B. The fold expression was normalized relative to that of the PHSP18.1 without effectors. Data are means ± sd of three biological replicates. *Significant at P < 0.05.
The DREB2A activates HSFA3 expression, and HSFA3 in turn regulates the expression of HS- and oxidative-response genes, including APX2 (Sakuma et al., 2006; Yokotani et al., 2008; Jung et al., 2013). In HSFA3-defective protoplasts (Supplemental Fig. S11A), HSFA6b overexpression significantly activated the transcription of HSP18.1-CI and APX2 promoters (Fig. 7D; Supplemental Fig. S10B), and HSFA3 coexpression had an additive effect on their expression. In HSFA6a and HSFA6b double-KO protoplasts (Supplemental Fig. S11B), we confirmed that HSFA6b functions independently of HSFA6a on HSP18.1-CI and APX2 promoter activation (Fig. 7E; Supplemental Fig. S10C).
HSFA6b Positively Regulated Thermotolerance
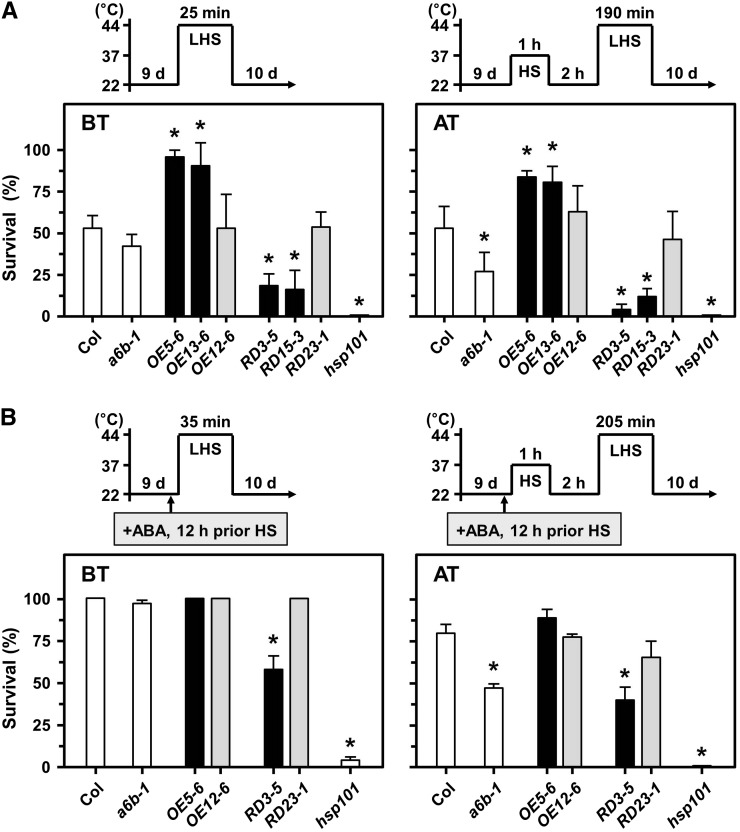
BT and AT were analyzed with a well-characterized HS-sensitive reference, the HSP101 mutant line (hsp101; At1g74310, SALK_066374; Hong and Vierling, 2000). OE5-6 and OE13-6 plants displayed significantly increased BT and AT as compared with Col plants, while a6b-1, RD3-5, and RD15-3 plants showed significantly reduced BT and AT (Fig. 8A). The transgenic lines of OE12-6 and RD23-1 (transgene expression similar to that of the Col; Supplemental Fig. S5A), used as references, showed similar thermotolerant phenotypes as the Col. RD3-5 also showed an HS-sensitive phenotype to long-term AT (Supplemental Fig. S12), with 37°C HS for 1 h, then 22°C 2-d long-term recovery followed by 44°C 155-min HS (Charng et al., 2006).
Figure 8.
Thermotolerance test in HSFA6b mutants. BT and acquired AT analysis in 9-day-old seedlings. A heat-sensitive mutant hsp101 was used as reference. Pictograms show the HS regime. A and B, Plates were incubated without or with 2.5 mL 10 μm ABA solution, respectively, 12 h before HS treatment. Survival was measured at day 10 after HS treatment. Data are means ± sd of three biological replicates. *Significant at P < 0.05 compared with the Col.
We used a 12-h ABA pretreatment before testing ABA-mediated BT and AT, and the 44°C HS period was extended to 35 and 205 min, respectively (Fig. 8B). Col plants showed ABA-enhanced survival of BT and AT as expected; however, RD3-5 plants still displayed significantly reduced BT and AT in comparison. Thus, HSFA6b was required for ABA-mediated HSR. BT, but not AT, was mildly affected in a6a as compared with Col plants (Supplemental Fig. S13).
HSFA6b Overexpression Conferred Drought and Salt Tolerance
We grew 200 seedlings in plates (9-cm diameter with 20 mL medium) for 24 d, then the growth medium was kept dehydrated (Fig. 9A, left), or seedlings were grown in high soil salinity stress by watering 2-week-old soil-grown plants with 300 mm NaCl solution over the course of 5 weeks (Fig. 9, B and C). OE5-6 plants stayed green, but RD3-5 plants showed a phenotype of anthocyanin accumulation as compared with Col plants. OE5-6 plants showed higher chlorophyll/anthocyanin ratio while the RD3-5 plants displayed a lower ratio when compared with the Col; the chlorophyll a/b ratio was not affected (Fig. 9A, right). Although no phenotype appeared at 2 weeks (Fig. 9B), the salt-responsive phenotype was observed at 5 weeks (Fig. 9C): OE5-6 showed the salt-resistant phenotype (the survival rate was 38.8% ± 6.6%), but Col, RD3-5, and a6b-1 plants did not survive (0% survival).
Figure 9.
Drought and salt tolerance test in HSFA6b mutants. A, Twenty-four-day-old plate-grown seedlings were photographed (left), then the chlorophyll a/b and chlorophyll/anthocyanin ratios were measured (right). Data are means ± sd of three biological replicates. *Significant at P < 0.05 compared with Col. B and C, Two-week-old seedlings were watered with 300 mm NaCl, then photographed at 2 and 5 weeks after treatment, respectively. The survival rates were analyzed after treatment with water containing 300 mm NaCl for 5 weeks. Arrows show surviving seedlings. One preventative data set is shown. Data are means ± sd of three biological replicates.
Transcription Profiling of HSFA6b-OE5-6 and HSFA6b-RD3-5 Transgenic Plants
We performed a genome-wide expression analysis with a microarray assay to dissect how HSFA6b regulates thermotolerance and confers drought and salt tolerance. We examined differentially expressed genes (DEGs) with levels changed >6-fold at P < 0.10 in transgenic plants OE5-6 and RD3-5 after 150 mm NaCl or 37°C HS treatment, as compared with the Col control treatment (Supplemental Table S2). The Venn diagram showed 246 NaCl- and 197 HS-induced DEGs (Supplemental Fig. S14A).
These DEGs were used for gene ontology (GO) enrichment analysis. The OE5-6 DEGs were associated with GO terms of biological process and showed a more complex network than those of RD3-5 under NaCl stress did (Supplemental Fig. S15; Supplemental Table S3A). The overrepresented GO terms were related to response to abiotic stimulus of “heat, water, and salt stress” and linked with the response to “ABA stimulus.” The enriched GO item “response to jasmonic acid (JA) stimulus” was also significantly overrepresented for OE5-6 under NaCl treatment. The results agreed with our findings that HSFA6b is downstream of ABA signaling and regulates genes enriched in abiotic stress-response networks including heat, drought, and salt stresses; in addition, HSFA6b might be involved in JA signaling. However, under HS, the networks of enriched GO terms were similar between OE5-6 and RD3-5 (Supplemental Fig. S16; Supplemental Table S3B). GO terms of “protein folding, oxidative phosphorylation, and respiratory electron transport chain” were overrepresented.
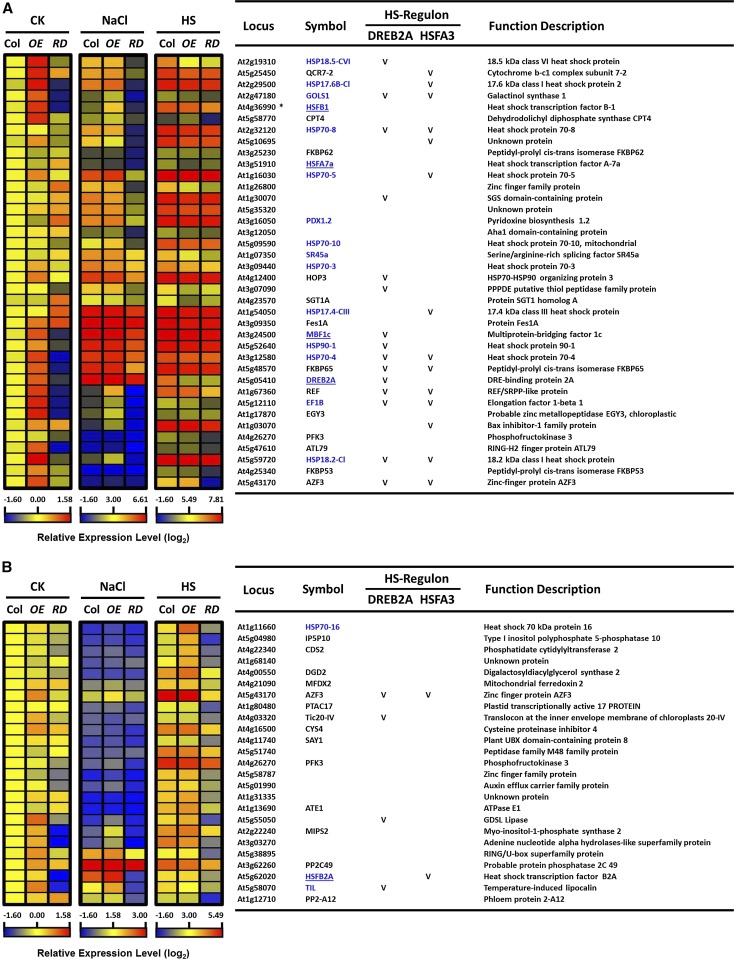
We highlighted the top 39 DEGs with >30-fold change in expression at P < 0.05 in OE5-6 and RD3-5 under NaCl or HS treatment as compared with the Col control treatment (Fig. 10A; Supplemental Table S4). We also highlighted the top 25 DEGs of HSFA6b HSR regulon in RD3-5 with an expression change of <1.5-fold compared with the Col HS (Fig. 10B). The HSFA6b regulon controlled the expression of important transcriptional regulators such as DREB2A, MUTIPROTEIN BRIDGING FACTOR1c (MBF1c; regulates HS-responsive regulon; Suzuki et al., 2011) and three HSFs (A7a, B1, B2a), as well as HSPs, zinc finger proteins, and enzymatic genes. The HSFA6b HS regulon and NaCl regulon (Supplemental Fig. S14B) showed overlap coregulation in part with DREB2A and HSFA3 HS regulons (Sakuma et al., 2006; Yoshida et al., 2008).
Figure 10.
Heat maps of the transcriptome analysis in HSFA6b mutants under salt and HS treatments. A, The top 39 DEGs with expression change >30-fold at P < 0.05 in HSFA6b mutants under salt and HS treatments as compared with wild-type control treatment. Profiles of 39 genes in the wild type (Col), OE5-6 (OE), and RD3-5 (RD) mutants after treatment with 150 mm NaCl for 6 h or 37°C HS for 1 h; 22°C treatment was a control (CK). B, The top 25 DEGs of HSFA6b HSR regulon in RD mutant had an expression change of <1.5-fold when compared with Col. The gene locus, symbol, and potential function were downloaded from The Arabidopsis Information Resource (TAIR). Normalized and averaged signals (log2) were analyzed as “heat maps” (left), and the corresponding genes are listed (right). As well, the HSFA6b HS regulon overlap with the DREB2A and HSFA3 HS regulon is indicated by a “V,” as shown in Supplemental Figure S14B. *At4g36990 and At4g36988 shared the same probeset number, 246214_at, on ATH1 GeneChip array. HS-related genes and transcription activators were highlighted in blue and underlined, respectively.
These data support our findings that HSFA6b regulates DREB2A to turn on particular downstream HS-responsive genes via the ABA signal and also might connect with the MBF1c HS-response regulon (Suzuki et al., 2011).
Thus, HSFA6b, with ABA-dependent and HS-independent expression, has a central role in regulating ABA-mediated HSR and oxidative-stress response gene expression and increasing the flexibility to modulate the compatibility of HSR regulatory networks.
DISCUSSION
Some multiple perceptions and signaling pathways in response to stress are specific, while others merge at various steps; these together confer cross protection against different environmental stresses. Abiotic stresses, especially salinity, drought, and cold, all lead to a similar physiological effect, so that signaling is transmitted through the ABA-mediated pathway that involves the expression of stress-specific and/or stress-related genes to relieve the stress (Shinozaki and Yamaguchi-Shinozaki, 2000; Huang et al., 2012b). Studies have revealed that ABA plays an important role in the acclimation of plants to a combination of salt and HS (Suzuki et al., 2016). The aba1 and abi1 mutants were more susceptible to the salt and HS combination than the wild type. In addition, the interactions of ABA with ROS signaling plays a key intermediate in the regulation of systemic acquired acclimation of plants to HS (Suzuki et al., 2013; Mittler and Blumwald, 2015). We confirmed that the transcription factor HSFA6b, whose expression depends on the ABA signal, is required for ABA-mediated HS resistance.
HSFA6b, a Nuclear Factor, Responds to ABA Signaling via the AREB/ABF-ABRE Regulon
Unlike the HSR marker genes HSP18.1-CI and HSFA2, which contain the perfect/active HSE motif (nTTCnnGAAnnTTCn/nGAAnnTTCn), HSFA6a and HSFA6b potential promoter regions only contain one aaAAtcTTCt and aGAAggaTCt sequence (Nover et al., 2001), respectively. This supports the expression data showing that neither HSFA6a nor HSFA6b respond to HS (Fig. 1; Supplemental Fig. S1). HSFA6b is a nuclear-localized factor (Fig. 2), and during ABA or Leptomycin B treatment, the nuclear localization of the YFP-tagged HSFA6b showed partial but not completed translocation to the nucleus, which implies that a limited amount of nuclear-localized HSFA6b is required for its function.
As promoters of ABA-responsive genes, ABREs serve as binding motifs for AREBs/ABFs. Reduced expression of HSFA6b in the areb1/areb2/abf3 triple mutant suggested that HSFA6b might function in the ABA-dependent pathway under salt and dehydration (Yoshida et al., 2010). We demonstrated that regulation of HSFA6b transcription could depend on ABA (Fig. 3), and the promoter translational activity assay revealed significantly induced transcription of HSFA6b by ABA and the coexpressed AREB1 (Fig. 4). Thus, HSFA6b expression is mediated through an AREB/ABF-ABRE pathway, a novel signal pathway regulating HSF gene expression independent of HS.
Phylogenetic analysis showed that HSFA6a is the closest member to HSFA6b (Scharf et al., 2012), and both were induced under abiotic stress and ABA treatments (Fig. 1). The AREB/ABF factors positively regulated HSFA6a transcription, which is required for salt and drought stresses (Hwang et al., 2014). However, transactivation assays have shown that HSFA1s function as the main positive regulators in HS-responsive gene expression via the HSE in the DREB2A promoter; also, coexpression of HSFA6b, but not HSFA6a, resulted in higher reporter gene activity than the control (Yoshida et al., 2011). We confirmed that HSFA6b expression occurs earlier and stronger than that of HSFA6a under abiotic stress and ABA treatments (Fig. 1; Supplemental Fig. S1), as well as that HSFA6b mutants affect HSFA6a expression (Supplemental Figs. S8 and S11). In addition, HSFA6a and HSFA6b expression showed a differential expression pattern in the hsfa1a/b/d/e quadruple mutant under HS stress (Supplemental Fig. S17; Liu and Charng, 2013): the results imply that HSFA1 (a, b, d) are negative regulators on HSFA6b expression but not of HSFA6a. We showed a marked difference in a6b-1 root growth in response to ABA and in AT testing as compared with a6a (Fig. 6A; Supplemental Fig. S13B), and HSFA6b functioned independently of HSFA6a on HSP18.1-CI and APX2 promoter activation (Fig. 7E; Supplemental Fig. S10C). Therefore, HSFA6b has an independent specific function in HS and oxidative stresses from HSFA6a.
HSFA6b, a Positive Regulator, Is Involved in ABA-Mediated Drought, Salt, and Thermotolerance
HSFA6b expression was highly induced by NaCl, mannitol, and ABA treatments (Fig. 1). The a6b-1 was insensitive to ABA in terms of primary root growth as compared with the wild type but displayed no significant differences under salt or drought/osmotic stress (Fig. 6A). These results suggest that contribution of HSFA6b may be small in response to these stresses or that potential functional redundancy of HSFA6 family proteins or other HSFs may compensate for the role of HSFA6b, such as HSFA1s or HSFA2 (Ogawa et al., 2007; Liu et al., 2011; Liu and Charng, 2013). Mutants of RD3-5 displayed decreased ABA inhibition sensitivity in seed germination, cotyledon greening, and root growth phenotypes, whereas OE5-6 showed enhanced ABA inhibition sensitivity (Figs. 5 and 6). These results suggest that HSFA6b has an impact on the ABA signaling pathway, and HSFA6b plays a positive regulator in ABA-mediated stress responses. HSFA6b is crucial; however, we could not determine whether these phenotypes were caused by the ectopic expression of HSFA6b or its dominant negative form.
Plants obtain thermotolerance, regulated by an HSF network, by inducing and accumulating HSPs. Overexpression of Arabidopsis HSFA2 and rice HSFA2e conferred increased thermotolerance and salt stress tolerance (Ogawa et al., 2007; Yokotani et al., 2008). ABA plays a role in thermotolerance response (Larkindale and Knight, 2002), and ABA-deficient and -insensitive mutants are sensitive to HS (Larkindale et al., 2005; Suzuki et al., 2016), with thermotolerance enhanced in AREB/ABF-OE plants (Kim et al., 2004). Thus, HSR confers tolerance to HS and other abiotic stresses such as salt, drought, and cold stress due to cross talk among stress-signaling pathways. We also confirmed that the BT, AT, and long-term AT were severely affected in HSFA6b mutants: the thermotolerance was significantly increased in OE5-6 and 13-6; consistently, RD3-5 and 13-6 showed greatly impaired establishment of thermotolerance, even with ABA pretreatment (Fig. 8; Supplemental Fig. S12). Again, HSFA6b, a pivotal positive regulator, is required for ABA-mediated HSR.
With loss of function of the major HS-responsive genes HSFA1s and HSFA2, the expression of HSP genes was reduced and thermotolerance lost. Induction and attenuation of HSR genes were not markedly affected in HSFA6b mutants in response to HS (Supplemental Figs. S8A and S9A). In addition, expression of oxidative stress-responsive marker genes and activity of ROS detoxification enzymes (SODs) were properly responded in HSFA6b mutants in response to HS (Supplemental Fig. S9, B and C). However, the NaCl and ABA-induced HSR gene expressions were enhanced in OE5-6 but largely impaired in RD3-5. Intriguingly, OE5-6 showed selective up-regulation of HSP18.1-CI and HSP26.5-MII expression, in addition to APX2 (Supplemental Figs. S8, B and C, and S9). The results confirmed that ABA-mediated HSFA6b expression is required for proper stress-related gene expression under both salt and ABA treatments.
Photosynthetic pigment content and ratios (i.e. chlorophyll/anthocyanin ratio) are indicators of stress detection and tolerance (Chalker-Scott, 1999); the chlorophyll/anthocyanin ratio is decreased in stressed plants. OE5-6 and RD3-5 mutants retained a higher and lower ratio of chlorophyll/anthocyanin, respectively, and HSFA6b overexpression conferred salt tolerance (Fig. 9). Thus, HSFA6b is required for drought and salt stress response.
Cross Talk of HSFA6 and DREB2A in HSR Genes Expression and Regulatory Networks
The physiological drought/osmotic stress responsive regulons largely depend on two major classes of cis-acting elements, ABRE and DRE. ABRE can perceive ABA-dependent signals, whereas DRE is involved in an ABA-independent pathway. DREB2A, a downstream regulator of both osmotic and HS stress, positively controls osmotic and HS-inducible gene expression (Yoshida et al., 2011). Nearly 1000 genes were down-regulated after 1-h HS treatment in the Arabidopsis hsfa1a/b/d/e quadruple mutant, with 658 genes HS-up-regulated in the wild type and 81 genes HS-up-regulated in transgenic plants overexpressing a constitutive active form of DREB2A (DREB2A-CA). In the top 100 down-regulated genes in the hsfa1a/b/d/e mutant, HSE motifs were highly enriched as compared with the whole genome, and DRE motifs were also enriched in these promoters (Yoshida et al., 2011). Of these HS-up-regulated genes in the wild type, 90 contained DRE instead of the HSE motif, the former of which is suggested to be regulated by DREB2A, downstream of the HSFA1s. For example, DREB2A harbors one HSE motif and one ABRE coupling element module but lacks the DRE motif, and its expression depends on HSFA1s under HS, but not salt or drought stress. Notably, the ABA-mediated HSFA6b positively regulates the expression of DREB2A (Yoshida et al., 2011). HSFA3 harbors one DRE but lacks HSE, and its expression depends on DREB2A; thus, overexpression of DREB2A-CA up-regulates HSFA3 but has no effect on HSFA6b expression (Yoshida et al., 2011). HSFA3 in turn up-regulates the expression of HSP18.1-CI (one DRE and two HSEs), HSP26.5-MII (one DRE and no HSE), and HSP70 (one DRE and one HSE), which have been shown to enhance thermotolerance and drought tolerance significantly (Sakuma et al., 2006).
With ABA treatment, the transcription factor AREBs/ABFs that activate DREB2A transcription depend on ABRE, resulting in only a modest accumulation of DREB2A transcript, as compared with osmotic stress. However, DREB2A expression in response to osmotic stress was largely impaired in ABA-insensitive and -deficient mutants, which highlights that in addition to ABA independence, the ABA-dependent cascade plays a positive role in the osmotic stress-responsive expression of DREB2A (Kim et al., 2011). Notably, a short-term ABA accumulation is following a 10-min HS application as reported by Suzuki et al. (2013), and then the AREBs/ABFs, DREB2A, HSP18.1-CI, and APX2 gene expression was up-regulated in 15 min, 30 to 60 min, and 1 h after the HS, respectively (Supplemental Fig. S1B). Our ChIP and transactivation assay showed that HSFA6b directly bound to the DREB2A promoter, then, when combined with AREB1 expression, additively enhanced DREB2A transcriptional activity in response to NaCl and dehydration (Fig. 7, B and C). HSFA6b overexpression significantly activated the transcription of HSP18.1-CI and APX2 promoters, and coexpression of AREB1 or HSFA3 had an additive effect on their expression (Fig. 7, A and D; Supplemental Fig. S10B). This result appears to support an HSFA6b and AREB/ABF cross talk with DREB2A transcription connected with ABA-induced thermotolerance. The existence of such multiple and complex regulons regulating AREB/ABF and DREB2A expression may allow plants to precisely and rapidly respond to abiotic stress.
The HSR genes are differentially regulated by the HSFA1s and HSFA2: HSFA1s are involved in thermotolerance and ectopic expression of HSFA2 in hsfa1a/b/d/e quadruple mutant complemented tolerance to different HS regimes, and to hydrogen peroxide, but not to salt and osmotic stresses (Liu and Charng, 2013). Our Co-IP and BiFC assays confirmed the interaction of HSFA6b with HSFA1a, HSFA1b, and HSFA2 in the nucleus (Supplemental Fig. S3). Thus, the HSFA6b might cooperate/compete with others HSF such as HSFA1s and/or HSFA2 in hetero-oligomeric complexes for its functions. The HSFA6b might have effect on DREB2A, HSFA3, and downstream gene expression (Fig. 10) plays a role in ABA-mediated thermotolerance.
The drought inducible genes were up-regulated by salt stress, but a few were cold inducible. The activators DREB1/CBF and DREB2 function separately in the cold, drought, and salt signaling pathways. Transcription of the cold-induced marker gene RD29A (one ABRE and three DREs) was through the DREB1/CBF-DRE/CRT regulon in response to low temperature and was induced under salt, drought, and ABA via ABRE and DRE (as a coupling element of ABRE) motifs cooperating mainly with AREB/ABF and DREB2A (Narusaka et al., 2003). Here, we showed that in HSFA6b mutants, RD29A expression was not affected by either NaCl or ABA treatments (Supplemental Fig. S6), so HSFA6b might not contribute to the DREB1/CBF-DRE regulon. Induced expression of RD29B (four ABREs and one DRE), which mainly depends on an ABA pathway, was largely affected in the HSFA6b mutant (Supplemental Fig. S6B). However, the expression of the ABA-dependent/drought-responsive marker gene RD22, depending on transcription factors MYB2 and MYC2 (Abe et al., 2003), was largely unaffected in HSFA6b mutants (Supplemental Fig. S6B). MYB2 and MYC2 expression was also enhanced by JA, which further indicates that abiotic and biotic stress transduction pathways are interconnected. In summary, cross talk exists in different regulons, such as ABA-dependent or -independent regulons, and in different stress responsive gene expressions (Qin et al., 2011; Huang et al., 2012b).
The top 39 DEGs of 197 DEGs (Supplemental Fig. S14) in HSFA6b overexpression (OE5-6) and dominant negative (RD3-5) mutant plants in response to HS showed increased and reduced expression of transcription activators MBF1c, HSFA7a, HSFB1, and DREB2A under salt stress, respectively, as well as specific protein and enzyme gene expression, such as previously studied in GOLS1, PDX1.2, SR45a, and EF1B (Fig. 10A). The HS-response regulon has been showed to be involved with the MBF1c HS-response regulon, in addition to the well-known network of HSFs. In addition, the top 25 DEGs of 197 DEGs of HSFA6b HSR regulon in RD3-5 showed an expression change of <1.5-fold when compared with Col HS treatment (Fig. 10B), including HSP70, HSFB2A, temperature-induced lipocalins, and enzymatic genes. Temperature-induced lipocalins are involved in the protection from abiotic stresses such as, heat, oxidative, and salt stresses (Chi et al., 2009; Levesque-Tremblay et al., 2009; Abo-Ogiala et al., 2014). HSFB2a has been reported to be required for gametophyte development (Wunderlich et al., 2014), and we proposed that it might also be required for thermotolerance acquisition. Thus, our data supports that salt and ABA-responsive HSFA6b (Figs. 1, 3, 4, and 7, B and C) triggers a downstream HS-responsive gene expression that leads to heat thermotolerance.
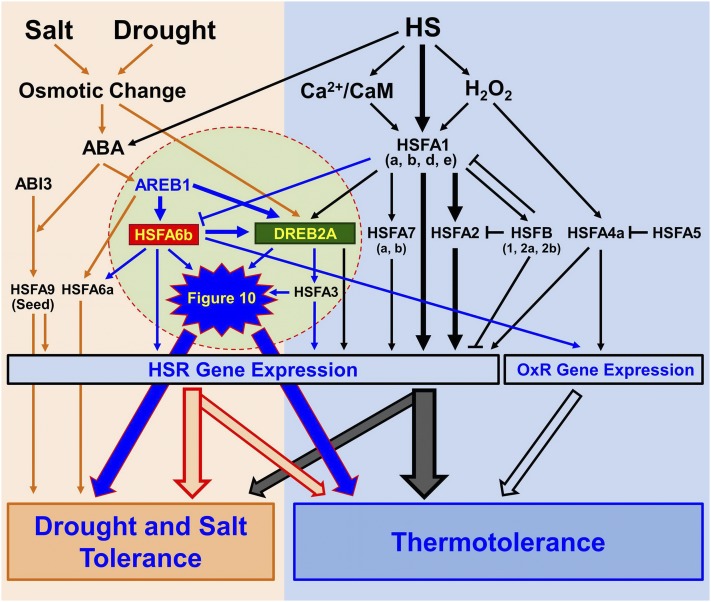
In conclusion, our data indicates that HSFA6b functions are tightly involved in ABA-mediated regulons. HSFA6b and AREB1 activate DREB2A expression in concert to mediate a cross talk between ABA-dependent and HSR gene expression. A simplified working model is shown Figure 11, which proposes a new pathway merging with the complex ABA- and HS-response networks in planta.
Figure 11.
Model of Arabidopsis HSFA6b as a hub connecting the ABA signaling pathway and ABA-mediated thermoprotection. CaM, Calmodulin; OxR, oxidative stress response. Under HS, activation of the HSFA1 (a, b, d, e) induces the expression of HSR genes including HSFA2 (the dominant factor), HSFA7 (a, b), as well as DREB2A and its downstream regulator HSFA3. HSFB (1, 2a, 2b) have roles in a negative feedback loop of HSR. HSFA4a is an H2O2 sensor, activating HSR and OxR gene expression. HSFA5 is the selective repressor of HSFA4a. HSFA1 (a, b, d) are negative regulators of HSFA6b. AREB1, HSFA6a, HSFA6b, and seed-specific HSFA9 are downstream regulators of the ABA-dependent pathway. HSFA6a is an activator of drought and salt response. HSFA6b acts synergistically with AREB1 for DREB2A expression, and then HSFA3 in turn is up-regulated, thus playing a role in HSR and OxR and mediating a cross talk between ABA-dependent and HSR regulons. The black and orange arrows show pathways that have been studied. The induction pathways highlighted in the circle and blue arrows emphasize the new ABA signaling pathway merging into the complex HSR network in Arabidopsis.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) HSFA6a-, HSFA6b-, and HSFA3-KO lines, a6a (SALK_045608; Col ecotype), a6b-1 (GK-513A02; Col ecotype), a6b-2 (GT_3_7561; Ler ecotype), and hsfa3 (SALK_01117; Col ecotype) were obtained from the Arabidopsis Biological Research Center or the Nottingham Arabidopsis Stock Center. Transgenic plants were created in the Col ecotype background by the floral-dip method (Clough and Bent, 1998) and were selected by spraying with 0.4% BASTA herbicide. Seedlings were grown at 22°C to 24°C with 16-h light at 60 to 100 μmol m−2 s−1.
RNA Preparation, cDNA Synthesis, and qRT-PCR
Total RNA was prepared with use of TRIZOL reagent (Invitrogen) and TURBO DNA-free kit (Applied Biosystems). cDNA synthesis involved cDNA reverse transcription kits (Applied Biosystems). PCR primers were designed by use of Primer3 (http://primer3.ut.ee). qRT-PCR reactions were analyzed by using the MyiQ thermocycler (Bio-Rad) with the PCR mix of iQ SYBR Green Supermix (Bio-Rad). Data analysis involved iQ5 optical system software (Bio-Rad), and the internal control for normalization was PP2AA3 (PP2A; At1g13320; Czechowski et al., 2005).
Generation of Transgenic HSFA6b Mutant Plants
For HSFA6b overexpression, the RT-PCR-amplified HSFA6b was cloned into pPE1000 (Hancock et al., 1997) using SalI and PstI to confer a HA tag at the N terminus, then subcloned into pCambia3300 (CAMBIA) with SacI for CaMV 35S promoter-driven expression. To generate the HSFA6b dominant negative mutant, HA-HSFA6b was amplified and cloned into p35S::SRDXG (Hiratsu et al., 2003) with SmaI to confer a dominant repression domain SRDX at the C terminus driven by the 35S promoter, then subcloned into binary vector pBCKH by Gateway LR reaction (Invitrogen). All gene fragments were sequenced before making the constructs.
Protoplast Preparation and Transfection
Protoplast preparation and transfection were as described previously (Yoo et al., 2007). HSFA6a and HSFA6b were cloned into p35S::EYFP (Kuo et al., 2013) to insert YFP into the N or C terminus. To generate the constructs for BiFC analysis, the RT-PCR-amplified HSFs were cloned into a pCR8/GW/TOPO vector and then recombined into pEarleyGate201-YN or pEarleyGate202-YC vectors (Lu et al., 2010). An amount of 2 × 104 protoplasts was transfected with 10 to 20 μg DNA, then incubated at 22°C for 16 to 24 h. The reconstituted YFP signals were observed by confocal microscopy (TCS SP5; Leica) as described previously (Hsu et al., 2010).
Protoplast Transactivation Assay
The potential promoters for HSFA6b (1.0 or 1.7 kb) as well as DREB2A, HSP18.1-CI, or APX2 (1.0 or 0.865 kb) were amplified and cloned into the PGAL4-UASx4::GUS vector. A CaMV 35S promoter was cloned into the PGAL4-UASx4::GUS vector, which resulted in P35S::GUS construct, to be used as a positive control. The transactivation assay involved cotransfection with a mixture of 15 μg effector (P35S::AREB1-3XFLAG, P35S::HSFA3-3XFLAG, P35S::HSFA6a-3XFLAG, or P35S::HSFA6b-3XFLAG) and reporter plasmids, using 5 μg MTC-301 luciferase plasmid as an internal control to normalize the transfection efficiencies (Ehlert et al., 2006; Hsu et al., 2010). The protoplasts were incubated for 12 to 16 h, then treated without or with 10 μm ABA for 12 h. Luciferase activity was analyzed by use of Luciferase Assay buffer according to the technical manual (Promega), and GUS activity assay was performed as described (Yoo et al., 2007). GUS activity in all samples was normalized against the luciferase internal control.
Seed Germination, Cotyledon Greening, and Root Growth Assay
Seed germination and cotyledon greening were analyzed on plates with half-strength Murashige and Skoog medium containing NaCl, mannitol, or ABA (Kreps et al., 2002; Pandey et al., 2005; Fujii et al., 2007). Germination was scored for 1 to 7 d, when the radicle had emerged from the testa, and cotyledon greening percentage was quantified at day 10. Three-day-old seedlings were transferred to NaCl-, mannitol-, or ABA-containing plates, and primary root growth was measured at days 7 to 9 by use of ImageJ (http://imagej.nih.gov/ij).
Thermotolerance, SOD Activity, H2O2, and Pigment Content Determination
Thermotolerance testing was performed as described (Charng et al., 2006; Hsu et al., 2010). Seedlings were grown at 22°C to 24°C with 16-h light for 6 to 9 d before heat treatment. Plates were sealed with plastic electric tape and submerged in a water bath, then heated at 44°C for 25 to 35 min for BT testing. For AT testing, plates were preheated at 37°C for 1 h, then recovered at 22°C for 2 h before 44°C HS for 190 to 205 min. Healthy-growing seedlings were counted 10 d after the end of HS treatment. In-gel SOD activity assay was as described (Chu et al., 2005; Huang et al., 2012a). The detection of H2O2 involved 3,3′-diaminobenzidine staining as described (Thordal-Christensen et al., 1997). Chlorophyll and anthocyanin measurement was as described (Porra et al., 1989; Neff and Chory, 1998).
Co-IP Assay
An amount of 4 × 105 protoplasts was transfected with 20 μg each of HSFA6b-3XFLAG and tester (HSFs)-YFPN plasmids, then incubated at 22°C for 16 h and harvested with GM buffer (150 mm Tris-HCl, pH 7.4). Half of total protein extracts were collected as input, and the other half were used to perform a Co-IP assay according to standard procedure (Catch and Release v2.0 Reversible Immunoprecipitation System; Millipore; 17-500). HSFA6b-3XFLAG protein was coimmunoprecipitated from total protein extracts with anti-GFP antibody (ab290; Abcom) and detected by SDS-PAGE followed by immunoblotting using α-FLAG antibody (sc-807; Santa Cruz Biotechnology).
ChIP Assay
Mature leaves (1.3 g) were collected from 4-week-old seedlings harboring HA-tagged HSFA6b (OE5-6) and incubated with 150 mm NaCl for 6 h or air-dried for 2.5 h (dehydration). ChIP involved use of the EpiQuik plant ChIP kit (P-2014-24; Epigentek). An HA-specific antibody (H3663; Sigma-Aldrich) was used to precipitate the complexes of HSFA6b-HA with DNA from chromatin. Immunoprecipitated DNA underwent PCR and qRT-PCR, and 18S rDNA was an input control (Kim et al., 2011).
Microarray Assay
Seven-day-old seedlings were treated with 150 mm NaCl for 6 h or 37°C HS for 1 h, with 22°C treatment as a control, then collected for total RNA purification by use of the RNeasy kit (Qiagen). RNA was used for cDNA synthesis, labeling, and hybridization of Affymetrix ATH1 arrays according to the Affymetrix Gene Chip Expression Analysis manual (http://www.affymetrix.com). The results were representative of two independent biological replicates, with Microarray Suite 5.0 (Affymetrix) and GeneSpring 7.3 (Silicon Genetics) used for data analysis. The ATH1 GeneChip array probesets with expression intensity <100 were filtered out in all samples. DEGs, or genes with expression change >6-fold at P < 0.10 compared with the Col control treatment, were collected. GO enrichment analysis involved with the Cytoscape 3.0.2 (http://www.cytoscape.org) plugin BiNGO categories using the GO_Full categories (Maere et al., 2005) identified by a hypergeometric test with P ≤ 5.00E-04 after Benjamini and Hochberg false discovery rate correction. The expression data from these experiments are available at Gene Expression Omnibus (GEO; accession no. GSE63372).
Statistical Analysis
Data are expressed as the mean ± sd from at least three independent biological experiments. Statistical analysis involved Student’s t test (two-tailed, unpaired). P < 0.05 was considered statistically significant.
Primers and Accession Numbers
Primers used and accession numbers are in Supplemental Table S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The expression profiles of 21 Arabidopsis HSF genes under abiotic stresses and during development.
Supplemental Figure S2. Modular structures of Arabidopsis HSFs HSFA6a and HSFA6b.
Supplemental Figure S3. Interaction of HSFA6b and different HSFs.
Supplemental Figure S4. Characterization of HSFA6b and HSFA6b T-DNA insertion, overexpression, and dominant-negative mutant lines.
Supplemental Figure S5. The expression levels of HSFA6b and ABA sensitivity in HSFA6b mutant lines.
Supplemental Figure S6. The expression levels of ABA biosynthesis and responsive genes in response to salt and ABA treatments in HSFA6b mutants.
Supplemental Figure S7. The expression profiles of Arabidopsis 18 small heat shock protein (sHSP) genes under various abiotic stresses and during development.
Supplemental Figure S8. The expression of HS-related genes in response to HS, salt, and ABA treatments in HSFA6b mutants.
Supplemental Figure S9. The expression levels of HS- and oxidative-related genes, SOD activity, and H2O2 detection in response to HS, NaCl, and ABA treatments in HSFA6b mutants.
Supplemental Figure S10. The expression level of APX2 in response to HS, NaCl, and ABA treatment in HSFA6b mutants, as well as HSFA6b-mediated HSFA3 activation of the APX2 promoter.
Supplemental Figure S11. Characterization of hsfa3 and hsfa6a hsfa6b double-mutant lines, as well as the HSFA6a and HSP18.1-CI transcription levels, in response to NaCl treatment in HSFA6b mutants.
Supplemental Figure S12. Thermotolerance test in HSFA6b mutants.
Supplemental Figure S13. Thermotolerance test in HSFA6a and HSFA6b mutants.
Supplemental Figure S14. Venn diagram of gene transcripts in response to salt or HS treatment in HSFA6b mutants.
Supplemental Figure S15. GO enrichment analysis of gene transcripts in response to salt treatment in HSFA6b mutants.
Supplemental Figure S16. GO enrichment analysis of gene transcripts in response to HS treatment in HSFA6b mutants.
Supplemental Figure S17. The expression levels of HSFA6a and HSFA6b in response to HS treatment in hsfa1a/b/d/e quadruple-KO (QK) and hsfa2 mutant lines.
Supplemental Table S1. Primers for genotyping, cloning, RT-PCR, and qRT-PCR, and accession numbers.
Supplemental Table S2. The DEGs with levels changed >6-fold at P < 0.10 in HSFA6b mutants OE5-6 and RD3-5 after 150 mm NaCl for 6 h or 37°C HS for 1 h as compared with the Col control (CK) treatment.
Supplemental Table S3. Enriched GO terms for DEGs with levels changed >6-fold at P < 0.10 in HSFA6b mutants OE5-6 and RD3-5 after 150 mm NaCl for 6 h or 37°C HS for 1 h treatment as compared with the Col control (CK) treatment.
Supplemental Table S4. The top 39 DEGs with levels changed >30-fold at P < 0.05 in HSFA6b mutants OE5-6 and RD3-5 after 150 mm NaCl for 6 h or 37°C HS for 1 h treatment as compared with the Col control (CK) treatment.
Supplementary Material
Acknowledgments
We thank Dr. Masaru Ohme-Takagi (Gene Function Research Laboratory, National Institute of Advanced Industrial Science and Technology, Japan) for providing p35S::SRDXG, Lynne Stracovsky for English editing, and the NTU Confocal Microscope Laboratory for fluorescence imaging.
Glossary
- HSR
heat stress response
- ABA
abscisic acid
- HS
heat stress
- BT
basal thermotolerance
- AT
acquired thermotolerance
- HSE
heat stress element
Footnotes
This work was supported by grants from National Taiwan University (103R892003, 104R892003, and 105R89203) and the Ministry of Science and Technology, Taiwan (102-2311-B-002-031, 103-2311-B-002-008, and 104-2311-B-002-007) to T.-L.J., and postdoctoral fellowships 102-2811-B-002-132, 103-2811-B-002-047, and 104-2811-B-002-075 to Y.-C.H.
Articles can be viewed without a subscription.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abo-Ogiala A, Carsjens C, Diekmann H, Fayyaz P, Herrfurth C, Feussner I, Polle A (2014) Temperature-induced lipocalin (TIL) is translocated under salt stress and protects chloroplasts from ion toxicity. J Plant Physiol 171: 250–259 [DOI] [PubMed] [Google Scholar]
- Baniwal SK, Chan KY, Scharf KD, Nover L (2007) Role of heat stress transcription factor HsfA5 as specific repressor of HsfA4. J Biol Chem 282: 3605–3613 [DOI] [PubMed] [Google Scholar]
- Banti V, Mafessoni F, Loreti E, Alpi A, Perata P (2010) The heat-inducible transcription factor HsfA2 enhances anoxia tolerance in Arabidopsis. Plant Physiol 152: 1471–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero JM, Rodríguez PL, Quesada V, Piqueras P, Ponce MR, Micol JL (2006) Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ 29: 2000–2008 [DOI] [PubMed] [Google Scholar]
- Boscheinen O, Lyck R, Queitsch C, Treuter E, Zimarino V, Scharf KD (1997) Heat stress transcription factors from tomato can functionally replace HSF1 in the yeast Saccharomyces cerevisiae. Mol Gen Genet 255: 322–331 [DOI] [PubMed] [Google Scholar]
- Busch W, Wunderlich M, Schöffl F (2005) Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. Plant J 41: 1–14 [DOI] [PubMed] [Google Scholar]
- Busk PK, Pagès M (1998) Regulation of abscisic acid-induced transcription. Plant Mol Biol 37: 425–435 [DOI] [PubMed] [Google Scholar]
- Chalker-Scott L. (1999) Environmental significance of anthocyanins in plant stress responses. Photochem Photobiol 70: 1–9 [Google Scholar]
- Charng YY, Liu HC, Liu NY, Chi WT, Wang CN, Chang SH, Wang TT (2007) A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol 143: 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charng YY, Liu HC, Liu NY, Hsu FC, Ko SS (2006) Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol 140: 1297–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi WT, Fung RW, Liu HC, Hsu CC, Charng YY (2009) Temperature-induced lipocalin is required for basal and acquired thermotolerance in Arabidopsis. Plant Cell Environ 32: 917–927 [DOI] [PubMed] [Google Scholar]
- Chow CN, Zheng HQ, Wu NY, Chien CH, Huang HD, Lee TY, Chiang-Hsieh YF, Hou PF, Yang TY, Chang WC (2016) PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res (D1) 44: D1154–D1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CC, Lee WC, Guo WY, Pan SM, Chen LJ, Li HM, Jinn TL (2005) A copper chaperone for superoxide dismutase that confers three types of copper/zinc superoxide dismutase activity in Arabidopsis. Plant Physiol 139: 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Czarnecka-Verner E, Pan S, Salem T, Gurley WB (2004) Plant class B HSFs inhibit transcription and exhibit affinity for TFIIB and TBP. Plant Mol Biol 56: 57–75 [DOI] [PubMed] [Google Scholar]
- Czarnecka-Verner E, Yuan CX, Scharf KD, Englich G, Gurley WB (2000) Plants contain a novel multi-member class of heat shock factors without transcriptional activator potential. Plant Mol Biol 43: 459–471 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17: 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlert A, Weltmeier F, Wang X, Mayer CS, Smeekens S, Vicente-Carbajosa J, Dröge-Laser W (2006) Two-hybrid protein-protein interaction analysis in Arabidopsis protoplasts: establishment of a heterodimerization map of group C and group S bZIP transcription factors. Plant J 46: 890–900 [DOI] [PubMed] [Google Scholar]
- Fortunati A, Piconese S, Tassone P, Ferrari S, Migliaccio F (2008) A new mutant of Arabidopsis disturbed in its roots, right-handed slanting, and gravitropism defines a gene that encodes a heat-shock factor. J Exp Bot 59: 1363–1374 [DOI] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu JK (2007) Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19: 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K (2011) ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res 124: 509–525 [DOI] [PubMed] [Google Scholar]
- Hancock KR, Phillips LD, White DW, Ealing PM (1997) pPE1000: a versatile vector for the expression of epitope-tagged foreign proteins in transgenic plants. Biotechniques 22: 861–862, 865 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Hong SW, Vierling E (2000) Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc Natl Acad Sci USA 97: 4392–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SF, Lai HC, Jinn TL (2010) Cytosol-localized heat shock factor-binding protein, AtHSBP, functions as a negative regulator of heat shock response by translocation to the nucleus and is required for seed development in Arabidopsis. Plant Physiol 153: 773–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CH, Kuo WY, Weiss C, Jinn TL (2012a) Copper chaperone-dependent and -independent activation of three copper-zinc superoxide dismutase homologs localized in different cellular compartments in Arabidopsis. Plant Physiol 158: 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GT, Ma SL, Bai LP, Zhang L, Ma H, Jia P, Liu J, Zhong M, Guo ZF (2012b) Signal transduction during cold, salt, and drought stresses in plants. Mol Biol Rep 39: 969–987 [DOI] [PubMed] [Google Scholar]
- Hwang SM, Kim DW, Woo MS, Jeong HS, Son YS, Akhter S, Choi GJ, Bahk JD (2014) Functional characterization of Arabidopsis HsfA6a as a heat-shock transcription factor under high salinity and dehydration conditions. Plant Cell Environ 37: 1202–1222 [DOI] [PubMed] [Google Scholar]
- Ikeda M, Mitsuda N, Ohme-Takagi M (2011) Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol 157: 1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HS, Crisp PA, Estavillo GM, Cole B, Hong F, Mockler TC, Pogson BJ, Chory J (2013) Subset of heat-shock transcription factors required for the early response of Arabidopsis to excess light. Proc Natl Acad Sci USA 110: 14474–14479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Mizoi J, Yoshida T, Fujita Y, Nakajima J, Ohori T, Todaka D, Nakashima K, Hirayama T, Shinozaki K, et al. (2011) An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol 52: 2136–2146 [DOI] [PubMed] [Google Scholar]
- Kim S, Kang JY, Cho DI, Park JH, Kim SY (2004) ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J 40: 75–87 [DOI] [PubMed] [Google Scholar]
- Kolmos E, Chow BY, Pruneda-Paz JL, Kay SA (2014) HsfB2b-mediated repression of PRR7 directs abiotic stress responses of the circadian clock. Proc Natl Acad Sci USA 111: 16172–16177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf KD (2007a) Complexity of the heat stress response in plants. Curr Opin Plant Biol 10: 310–316 [DOI] [PubMed] [Google Scholar]
- Kotak S, Port M, Ganguli A, Bicker F, von Koskull-Döring P (2004) Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J 39: 98–112 [DOI] [PubMed] [Google Scholar]
- Kotak S, Vierling E, Bäumlein H, von Koskull-Döring P (2007b) A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell 19: 182–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna P. (2004) Plant responses to heat stress. In Hirt H, Shinozaki K, eds, Plant Responses to Abiotic Stress. Springer, Berlin [Google Scholar]
- Kuo WY, Huang CH, Liu AC, Cheng CP, Li SH, Chang WC, Weiss C, Azem A, Jinn TL (2013) CHAPERONIN 20 mediates iron superoxide dismutase (FeSOD) activity independent of its co-chaperonin role in Arabidopsis chloroplasts. New Phytol 197: 99–110 [DOI] [PubMed] [Google Scholar]
- Larkindale J, Hall JD, Knight MR, Vierling E (2005) Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol 138: 882–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkindale J, Huang B (2004) Thermotolerance and antioxidant systems in Agrostis stolonifera: involvement of salicylic acid, abscisic acid, calcium, hydrogen peroxide, and ethylene. J Plant Physiol 161: 405–413 [DOI] [PubMed] [Google Scholar]
- Larkindale J, Knight MR (2002) Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiol 128: 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkindale J, Vierling E (2008) Core genome responses involved in acclimation to high temperature. Plant Physiol 146: 748–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque-Tremblay G, Havaux M, Ouellet F (2009) The chloroplastic lipocalin AtCHL prevents lipid peroxidation and protects Arabidopsis against oxidative stress. Plant J 60: 691–702 [DOI] [PubMed] [Google Scholar]
- Lim CJ, Hwang JE, Chen H, Hong JK, Yang KA, Choi MS, Lee KO, Chung WS, Lee SY, Lim CO (2007) Over-expression of the Arabidopsis DRE/CRT-binding transcription factor DREB2C enhances thermotolerance. Biochem Biophys Res Commun 362: 431–436 [DOI] [PubMed] [Google Scholar]
- Liu HC, Charng YY (2013) Common and distinct functions of Arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development. Plant Physiol 163: 276–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HC, Liao HT, Charng YY (2011) The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ 34: 738–751 [DOI] [PubMed] [Google Scholar]
- Lohmann C, Eggers-Schumacher G, Wunderlich M, Schöffl F (2004) Two different heat shock transcription factors regulate immediate early expression of stress genes in Arabidopsis. Mol Genet Genomics 271: 11–21 [DOI] [PubMed] [Google Scholar]
- Lu Q, Tang X, Tian G, Wang F, Liu K, Nguyen V, Kohalmi SE, Keller WA, Tsang EW, Harada JJ, et al. (2010) Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J 61: 259–270 [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Miller G, Mittler R (2006) Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann Bot (Lond) 98: 279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Blumwald E (2015) The roles of ROS and ABA in systemic acquired acclimation. Plant Cell 27: 64–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Yamaguchi-Shinozaki K (2013) ABA signaling in stress-response and seed development. Plant Cell Rep 32: 959–970 [DOI] [PubMed] [Google Scholar]
- Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J 34: 137–148 [DOI] [PubMed] [Google Scholar]
- Neff MM, Chory J (1998) Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol 118: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Yoshida E, Maruta T, Yoshimura K, Shigeoka S (2006) Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J 48: 535–547 [DOI] [PubMed] [Google Scholar]
- Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf KD (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa D, Yamaguchi K, Nishiuchi T (2007) High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J Exp Bot 58: 3373–3383 [DOI] [PubMed] [Google Scholar]
- Panchuk II, Volkov RA, Schöffl F (2002) Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol 129: 838–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey GK, Grant JJ, Cheong YH, Kim BG, Li L, Luan S (2005) ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol 139: 1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell DA, Lindquist S (1993) The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet 27: 437–496 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Qin F, Shinozaki K, Yamaguchi-Shinozaki K (2011) Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol 52: 1569–1582 [DOI] [PubMed] [Google Scholar]
- Rabindran SK, Haroun RI, Clos J, Wisniewski J, Wu C (1993) Regulation of heat shock factor trimer formation: role of a conserved leucine zipper. Science 259: 230–234 [DOI] [PubMed] [Google Scholar]
- Rossel JB, Walter PB, Hendrickson L, Chow WS, Poole A, Mullineaux PM, Pogson BJ (2006) A mutation affecting ASCORBATE PEROXIDASE 2 gene expression reveals a link between responses to high light and drought tolerance. Plant Cell Environ 29: 269–281 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K (2006) Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci USA 103: 18822–18827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöffl F, Prändl R, Reindl A (1998) Regulation of the heat-shock response. Plant Physiol 117: 1135–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf K-D, Berberich T, Ebersberger I, Nover L (2012) The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim Biophys Acta 1819: 104–119 [DOI] [PubMed] [Google Scholar]
- Schramm F, Ganguli A, Kiehlmann E, Englich G, Walch D, von Koskull-Döring P (2006) The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol Biol 60: 759–772 [DOI] [PubMed] [Google Scholar]
- Schramm F, Larkindale J, Kiehlmann E, Ganguli A, Englich G, Vierling E, von Koskull-Döring P (2008) A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J 53: 264–274 [DOI] [PubMed] [Google Scholar]
- Shi WM, Muramoto Y, Ueda A, Takabe T (2001) Cloning of peroxisomal ascorbate peroxidase gene from barley and enhanced thermotolerance by overexpressing in Arabidopsis thaliana. Gene 273: 23–27 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3: 217–223 [PubMed] [Google Scholar]
- Suzuki N, Bassil E, Hamilton JS, Inupakutika MA, Zandalinas SI, Tripathy D, Luo Y, Dion E, Fukui G, Kumazaki A, et al. (2016) ABA is required for plant acclimation to a combination of salt and heat stress. PLoS One 11: e0147625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Miller G, Salazar C, Mondal HA, Shulaev E, Cortes DF, Shuman JL, Luo X, Shah J, Schlauch K, et al. (2013) Temporal-spatial interaction between reactive oxygen species and abscisic acid regulates rapid systemic acclimation in plants. Plant Cell 25: 3553–3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Sejima H, Tam R, Schlauch K, Mittler R (2011) Identification of the MBF1 heat-response regulon of Arabidopsis thaliana. Plant J 66: 844–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Huebner M, Weber AP (2007) Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 8: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11: 1187–1194 [Google Scholar]
- Vierling E. (1991) The roles of heat shock proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 42: 579–620 [Google Scholar]
- von Koskull-Döring P, Scharf KD, Nover L (2007) The diversity of plant heat stress transcription factors. Trends Plant Sci 12: 452–457 [DOI] [PubMed] [Google Scholar]
- Wunderlich M, Gross-Hardt R, Schöffl F (2014) Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA. Plant Mol Biol 85: 541–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokotani N, Ichikawa T, Kondou Y, Matsui M, Hirochika H, Iwabuchi M, Oda K (2008) Expression of rice heat stress transcription factor OsHsfA2e enhances tolerance to environmental stresses in transgenic Arabidopsis. Planta 227: 957–967 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61: 672–685 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Ohama N, Nakajima J, Kidokoro S, Mizoi J, Nakashima K, Maruyama K, Kim J-M, Seki M, Todaka D, et al. (2011) Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol Genet Genomics 286: 321–332 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Sakuma Y, Todaka D, Maruyama K, Qin F, Mizoi J, Kidokoro S, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K (2008) Functional analysis of an Arabidopsis heat-shock transcription factor HsfA3 in the transcriptional cascade downstream of the DREB2A stress-regulatory system. Biochem Biophys Res Commun 368: 515–521 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.