AtORM1 and AtORM2 negatively regulate serine palmitoyltransferase, and altered expression differentially affects functionally distinct ceramide synthase activities to maintain sphingolipid homeostasis.
Abstract
Sphingolipid synthesis is tightly regulated in eukaryotes. This regulation in plants ensures sufficient sphingolipids to support growth while limiting the accumulation of sphingolipid metabolites that induce programmed cell death. Serine palmitoyltransferase (SPT) catalyzes the first step in sphingolipid biosynthesis and is considered the primary sphingolipid homeostatic regulatory point. In this report, Arabidopsis (Arabidopsis thaliana) putative SPT regulatory proteins, orosomucoid-like proteins AtORM1 and AtORM2, were found to interact physically with Arabidopsis SPT and to suppress SPT activity when coexpressed with Arabidopsis SPT subunits long-chain base1 (LCB1) and LCB2 and the small subunit of SPT in a yeast (Saccharomyces cerevisiae) SPT-deficient mutant. Consistent with a role in SPT suppression, AtORM1 and AtORM2 overexpression lines displayed increased resistance to the programmed cell death-inducing mycotoxin fumonisin B1, with an accompanying reduced accumulation of LCBs and C16 fatty acid-containing ceramides relative to wild-type plants. Conversely, RNA interference (RNAi) suppression lines of AtORM1 and AtORM2 displayed increased sensitivity to fumonisin B1 and an accompanying strong increase in LCBs and C16 fatty acid-containing ceramides relative to wild-type plants. Overexpression lines also were found to have reduced activity of the class I ceramide synthase that uses C16 fatty acid acyl-coenzyme A and dihydroxy LCB substrates but increased activity of class II ceramide synthases that use very-long-chain fatty acyl-coenzyme A and trihydroxy LCB substrates. RNAi suppression lines, in contrast, displayed increased class I ceramide synthase activity but reduced class II ceramide synthase activity. These findings indicate that ORM mediation of SPT activity differentially regulates functionally distinct ceramide synthase activities as part of a broader sphingolipid homeostatic regulatory network.
Sphingolipids play critical roles in plant growth and development as essential components of endomembranes, including the plasma membrane, where they constitute more than 40% of the total lipid (Sperling et al., 2005; Cacas et al., 2016). Sphingolipids also are highly enriched in detergent-insoluble membrane fractions of the plasma membrane that form microdomains for proteins with important cell surface activities, including cell wall biosynthesis and hormone transport (Cacas et al., 2012, 2016; Perraki et al., 2012; Bayer et al., 2014). In addition, sphingolipids, particularly those with very-long-chain fatty acids (VLCFAs), are integrally associated with Golgi-mediated protein trafficking that underlies processes related to the growth of plant cells (Bach et al., 2008, 2011; Markham et al., 2011; Melser et al., 2011). Furthermore, sphingolipids function through their bioactive long-chain base (LCB) and ceramide metabolites to initiate programmed cell death (PCD), important for mediating plant pathogen resistance through the hypersensitive response (Greenberg et al., 2000; Liang et al., 2003; Shi et al., 2007; Bi et al., 2014; Simanshu et al., 2014).
Sphingolipid biosynthesis is highly regulated in all eukaryotes. In plants, the maintenance of sphingolipid homeostasis is vital to ensure sufficient sphingolipids for growth (Chen et al., 2006; Kimberlin et al., 2013) while restricting the accumulation of PCD-inducing ceramides and LCBs until required for processes such as the pathogen-triggered hypersensitive response. Serine palmitoyltransferase (SPT), which catalyzes the first step in LCB synthesis, is generally believed to be the primary control point for sphingolipid homeostasis (Hanada, 2003). SPT synthesizes LCBs, unique components of sphingolipids, by catalyzing a pyridoxal phosphate-dependent condensation of Ser and palmitoyl (16:0)-CoA in plants (Markham et al., 2013). Similar to other eukaryotes, the Arabidopsis (Arabidopsis thaliana) SPT is a heterodimer consisting of LCB1 and LCB2 subunits (Chen et al., 2006; Dietrich et al., 2008; Teng et al., 2008). Research to date has shown that SPT is regulated primarily by posttranslational mechanisms involving physical interactions with noncatalytic, membrane-associated proteins that confer positive and negative regulation of SPT activity (Han et al., 2009, 2010; Breslow et al., 2010). These proteins include a 56-amino acid small subunit of SPT (ssSPT) in Arabidopsis, which was recently shown to stimulate SPT activity and to be essential for generating sufficient amounts of sphingolipids for pollen and sporophytic cell viability (Kimberlin et al., 2013).
Evidence from yeast and mammalian research points to a more critical role for proteins termed ORMs (for orosomucoid-like proteins) in sphingolipid homeostatic regulation (Breslow et al., 2010; Han et al., 2010). The Saccharomyces cerevisiae Orm1p and Orm2p negatively regulate SPT through reversible phosphorylation of these polypeptides in response to intracellular sphingolipid levels (Breslow et al., 2010; Han et al., 2010; Roelants et al., 2011; Gururaj et al., 2013; Muir et al., 2014). Phosphorylation/dephosphorylation of ORMs in S. cerevisiae presumably affects the higher order assembly of SPT to mediate flux through this enzyme for LCB synthesis (Breslow, 2013). In this sphingolipid homeostatic regulatory mechanism, the S. cerevisiae Orm1p and Orm2p are phosphorylated at their N termini by Ypk1, a TORC2-dependent protein kinase (Han et al., 2010; Roelants et al., 2011). The absence of this phosphorylation domain in mammalian and plant ORM homologs brings into question the nature of SPT reversible regulation by ORMs in other eukaryotic systems (Hjelmqvist et al., 2002).
Sphingolipid synthesis also is mediated by the N-acylation of LCBs by ceramide synthases to form ceramides, the hydrophobic backbone of the major plant glycosphingolipids, glucosylceramide (GlcCer) and glycosyl inositolphosphoceramide (GIPC). Two functionally distinct classes of ceramide synthases occur in Arabidopsis, designated class I and class II (Chen et al., 2008). Class I ceramide synthase activity resulting from the Longevity Assurance Gene One Homolog2 (LOH2)-encoded ceramide synthase acylates, almost exclusively, LCBs containing two hydroxyl groups (dihydroxy LCBs) with 16:0-CoA to form C16 ceramides, which are used primarily for GlcCer synthesis (Markham et al., 2011; Ternes et al., 2011; Luttgeharm et al., 2016). Class II ceramide synthase activities resulting from the LOH1- and LOH3-encoded ceramide synthases are most active in the acylation of LCBs containing three hydroxyl groups (trihydroxy LCBs) with VLCFA-CoAs, including primarily C24 and C26 acyl-CoAs (Markham et al., 2011; Ternes et al., 2011; Luttgeharm et al., 2016). Class II (LOH1 and LOH3) ceramide synthase activity is essential for producing VLCFA-containing glycosphingolipids to support the growth of plant cells, whereas class I (LOH2) ceramide synthase activity is nonessential under normal growth conditions (Markham et al., 2011; Luttgeharm et al., 2015b). It was speculated recently that LOH2 ceramide synthase functions, in part, as a safety valve to acylate excess LCBs for glycosylation, resulting in a less cytotoxic form (Luttgeharm et al., 2015b; Msanne et al., 2015). Recent studies have shown that the Lag1/Lac1 components of the S. cerevisiae ceramide synthase are phosphorylated by Ypk1, and this phosphorylation stimulates ceramide synthase activity in response to heat and reduced intracellular sphingolipid levels (Muir et al., 2014). This finding points to possible coordinated regulation of ORM-mediated SPT and ceramide synthase activities to regulate sphingolipid homeostasis, which is likely more complicated in plants and mammals due to the occurrence of functionally distinct ceramide synthases in these systems (Stiban et al., 2010; Markham et al., 2011; Ternes et al., 2011; Luttgeharm et al., 2016).
RNA interference (RNAi) suppression of ORM genes in rice (Oryza sativa) has been shown to affect pollen viability (Chueasiri et al., 2014), but no mechanistic characterization of ORM proteins in plants has yet to be reported. Here, we describe two Arabidopsis ORMs, AtORM1 and AtORM2, that suppress SPT activity through direct interaction with the LCB1/LCB2 heterodimer. We also show that strong up-regulation of AtORM expression impairs growth. In addition, up- or down-regulation of ORMs is shown to differentially affect the sensitivity of Arabidopsis to the PCD-inducing mycotoxin fumonisin B1 (FB1), a ceramide synthase inhibitor, and to differentially affect the activities of class I and II ceramide synthases as a possible additional mechanism for regulating sphingolipid homeostasis.
RESULTS
Two Functional Homologs of Mammalian ORMDLs Interact Physically with and Inhibit Arabidopsis SPT
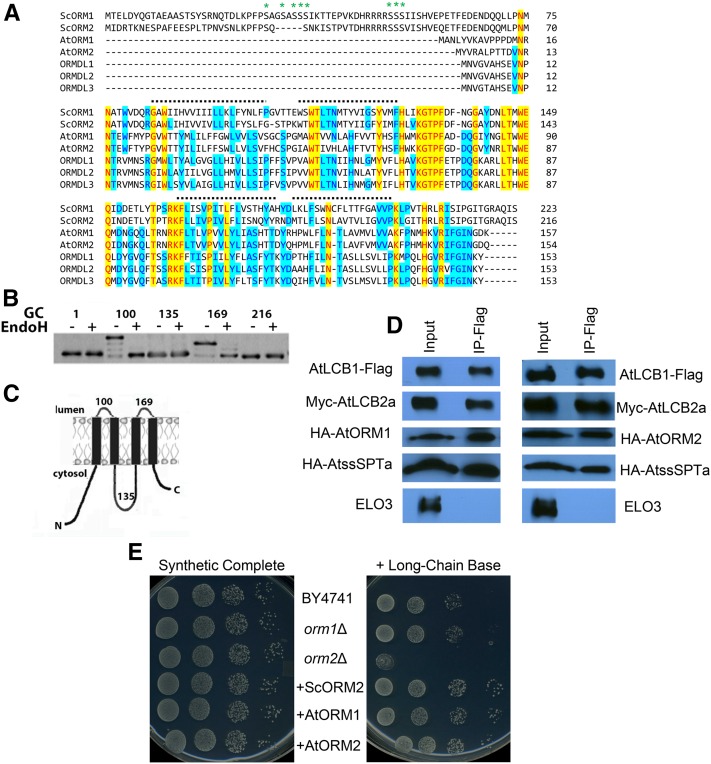
Two genes, designated AtORM1 (At1g01230) and AtORM2 (At5g42000), encoding 157- and 154-amino acid polypeptides, respectively, were identified in homology searches using human ORMDLs as a query (Hjelmqvist et al., 2002). The amino acid sequences of the Arabidopsis polypeptides share 81% identity and have predicted homologs throughout the plant kingdom. Arabidopsis ORM1 (AtORM1) and ORM2 (AtORM2) share 38% to 43% identity with human ORMDL1, ORMDL2, and ORMDL3. AtORM1 and AtORM2 also share 35% to 39% identity with S. cerevisiae ORM1 and ORM2 but, interestingly, lack the N-terminal phosphorylation domain found in the yeast ORMs (Fig. 1A). Notably, this N-terminal domain also is absent from ORMDL proteins from human and other mammals. Similar to the mammalian ORMDL proteins, AtORM1 and AtORM2 are predicted to have two, three, or four transmembrane domains based on in silico analyses using the TopPred II, SOSUI, TMPred, MEMSAT, and DAS filter programs (von Heijne, 1992; Hirokawa et al., 1998). However, glycosylation cassettes (GCs) inserted at key sites along the yeast ORM2 protein support the presence of four transmembrane domains with the N and C termini residing in the cytosol (Fig. 1B). Given the significant homology between the yeast, plant, and human ORM proteins, it is likely that all members of the ORM family of proteins have the topology depicted in Figure 1C.
Figure 1.
Arabidopsis ORMs interact physically with the Arabidopsis core SPT components and complement an S. cerevisiae ORM2 knockout mutant. A, Amino acid sequence alignment for ORM polypeptides from S. cerevisiae (ScORM1 and ScORM2), Arabidopsis (AtORM1 and AtORM2), and Homo sapiens (ORMDL1, ORMDL2, and ORMDL3). The alignments show the N-terminal extension, found only in yeast, responsible for the reversible phosphorylation (at residues marked with asterisks) that modulates SPT activity (Breslow et al., 2010). The dotted lines mark the positions of four potential transmembrane domains identified by hydropathy analyses. B, Topology mapping of ScORM2. GCs were inserted after the indicated amino acids, and the GC-tagged proteins were expressed in yeast. Increased mobility following treatment of microsomes with endoglycosidase H (EndoH) revealed that the GCs at residues 100 and 169 are glycosylated and, therefore, reside in the lumen of the endoplasmic reticulum (ER). C, Model of ORM protein topology. The figure shows the experimentally determined membrane topology of ScORM2 as shown in B. D, Coimmunoprecipitation of FLAG-tagged AtLCB1 in yeast expressing AtLCB1-FLAG, AtLCB2a-Myc, AtssSPTa-HA, and either AtORM1-HA or AtORM2-HA. Solubilized yeast microsomes were incubated with anti-FLAG beads, and protein was eluted with FLAG peptide. Solubilized microsomes (Input) and the eluent (IP-Flag) were analyzed by SDS-PAGE, and the polypeptides were detected by immunoblotting. ELO3 was used as a negative control. E, AtORM1 and AtORM2 complement the sensitivity of the S. cerevisiae orm2Δ mutant to exogenous LCBs. Shown are plates containing synthetic complete medium lacking or supplemented with the LCB phytosphingosine at a concentration of 15 µm. As shown, plates contain dilutions of the parental cell line BY4741, S. cerevisiae orm1Δ mutant, S. cerevisiae orm2Δ mutant, and S. cerevisiae orm2Δ complemented with the wild-type copy of the S. cerevisiae ORM2 (+ScORM2), AtORM1 (+AtORM1), or AtORM2 (+AtORM2).
To test whether the AtORM polypeptides interact physically with AtSPT, FLAG-tagged AtLCB1 was expressed along with Myc-AtLCB2a, hemagglutinin (HA)-tagged AtssSPTa, and HA-tagged AtORM1 or AtORM2 in a yeast mutant that lacks endogenous SPT due to knockout of the yeast LCB1 and TSC3 genes. Pull-down assays using anti-FLAG antibodies with solubilized microsomes of cells expressing these polypeptides resulted in the detection of not only AtLCB1 but also AtLCB2a, AtssSPTa, AtORM1, or AtORM2 but no detection of ELO3, an ER polypeptide not known to associate with SPT (Fig. 1D). These results are consistent with the physical interaction of AtORM1 and AtORM2 with the core SPT complex.
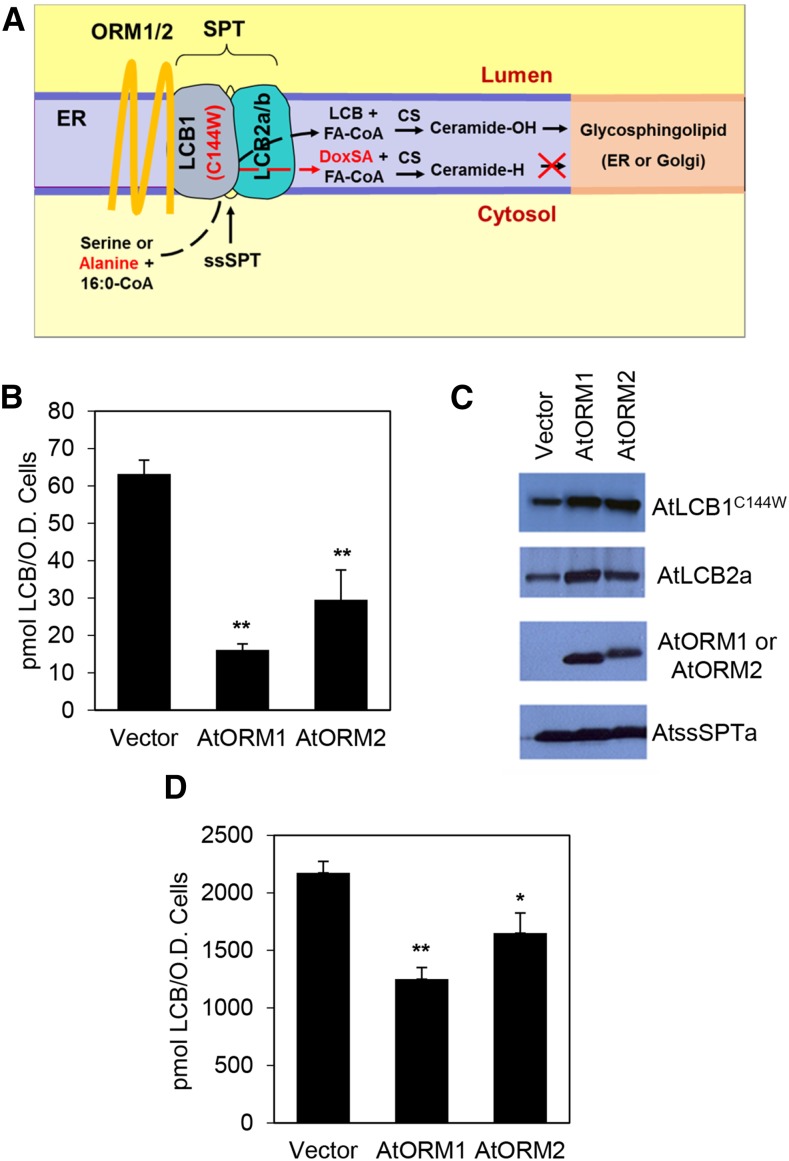
To examine their ability to function as suppressors of SPT activity, Arabidopsis ORM1 and ORM2 were expressed in the S. cerevisiae orm2Δ mutant. The orm2Δ mutant in yeast has a strong sensitivity to excess LCB, and the inclusion of 15 µm phytosphingosine in medium is toxic to this mutant (Han et al., 2010). Expression of AtORM1 or AtORM2 in the orm2Δ mutant rescued the sensitivity of the mutant to exogenous LCB, consistent with the ability of the Arabidopsis proteins to suppress SPT activity (Fig. 1E). In addition, Arabidopsis ORM1 and ORM2 were coexpressed along with AtLCB1C144W, AtLCB2a, and AtssSPTa in the yeast lcb1Δtsc3Δ mutant strain lacking endogenous SPT. AtLCB1C144W contains a single amino acid substitution analogous to the HSAN1-causing mutation in human LCB1 (Gable et al., 2010). SPT containing the AtLCB1C144W subunit condenses both Ser and Ala with palmitoyl-CoA to form normal LCB (d18:0) and deoxy-LCB (deoxy-sphinganine [DoxSA]; m18:0), respectively (Fig. 2A). Deoxy-LCBs cannot be degraded due to the missing hydroxyl group and serve as an in situ read out of SPT activity. When expressed in this background, Arabidopsis ORM1 and ORM2 markedly decreased the amount of deoxy-LCB produced, indicating that they act as SPT inhibitors (Fig. 2, B and C). The inhibitory effect of the AtORMs also was observed using native AtSPT expressed in a yeast mutant lacking endogenous SPT (Fig. 2D).
Figure 2.
AtORM1 and AtORM2 are negative regulators of SPT activity. A, Schematic representation of the core SPT complex consisting of AtLCB1, AtLCB2a/b, AtssSPT, and AtORM. The complex resides in the ER membrane and catalyzes a condensation reaction between Ser and palmitoyl-CoA to produce LCB. The C144W (HSAN1) mutation in AtLCB1 allows SPT to use Ala as well as Ser as a substrate. The deoxy-LCB produced with Ala is referred to as DoxSA and lacks the hydroxyl group that is needed for LCB degradation and for conversion to the glycosphingolipid GlcCer in the ER or GIPC in the Golgi. B, In vivo AtSPT activity was measured in a yeast mutant that lacks endogenous SPT. Cells expressing AtLCB1C144W, AtLCB2a, or AtssSPTa with or without AtORM1 or AtORM2 were used to demonstrate an inhibitory effect on SPT activity. The activity is measured by the accumulation of DoxSA, produced by the AtLCB1C144W-containing mutant SPT enzyme. The DoxSA product is not produced naturally and is not degraded. Values shown are averages of three independent assays ± sd. **, P < 0.01. C, Immunoblot of yeast microsomes expressing AtLCB1C144W-FLAG, AtLCB2a-Myc, AtORM1/2-HA, and AtssSPTa-HA. Anti-FLAG, anti-Myc, and anti-HA antibodies were used for detection. The results show that the C144W mutation in AtLCB1 does not affect the interaction of SPT components and interacting proteins. D, In vivo AtSPT activity measured in cells of yeast lacking endogenous SPT but expressing AtLCB1, AtLCB2a, AtssSPTa (Vector), and either AtORM1 or AtORM2. The activity is measured through the accumulation of total LCB produced. Values shown are averages of three independent assays ± sd. *, P < 0.05; and **, P < 0.01.
AtORM1 and AtORM2 Polypeptides Are ER Associated, and AtORM1 and AtORM2 Are Constitutively Expressed
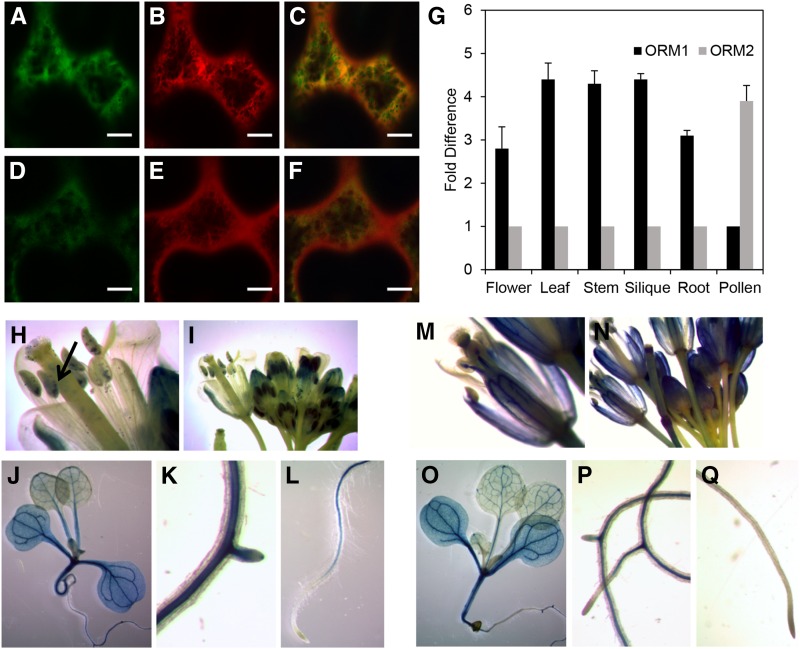
The subcellular localization of ORM1 and ORM2 polypeptides was visualized using N-terminal and C-terminal yellow fluorescent protein (YFP) tags with transient expression in Nicotiana benthamiana (Fig. 3, A–F). Tagged ORM1 and ORM2 colocalized with the ER marker mCherry-HDEL (Fig. 3, C and F), consistent with the known ER localization of AtLCB1, AtLCB2a/2b, and AtssSPTa/b (Chen et al., 2006; Dietrich et al., 2008; Teng et al., 2008; Kimberlin et al., 2013), but also were detected in other subcellular locations, including the cytosol (Fig. 3, A, C, D, and F).
Figure 3.
Subcellular localization of AtORM1 and AtORM2 polypeptides, and gene expression of AtORM1 and AtORM2 in Arabidopsis. A to F, Subcellular localization of AtORM1-YFP or AtORM2-YFP fusion coexpressed with the ER marker mCherry fusion construct. All constructs were transiently expressed in N. benthamiana through Agrobacterium tumefaciens infiltration and viewed by confocal microscopy. Green color in A and D shows AtORM1 and AtORM2 localization, respectively. The red color in B and E indicates ER marker localization. The yellow color seen in C and F indicates the colocalization of AtORM1 and AtORM2 polypeptides, respectively, with the ER marker. G, Relative expression of AtORM1 and AtORM2. Tissues were collected from wild-type Columbia-0 (Col-0), and quantitative PCR (qPCR) was used to determine AtORM1 and AtORM2 transcript levels. Protein phosphatase 2A subunit A3 (PP2AA3) was used as a reference gene. Values shown are means ± sd for three independent measurements and indicate relative fold increase of AtORM1 or AtORM2 compared with AtORM2 or AtORM1, respectively. H to Q, AtORM1 and AtORM2 promoter::GUS expression analysis. An approximately 1-kb region upstream of the AtORM1 (H–L) or AtORM2 (M–Q) start codon was fused to the GUS gene and analyzed for expression in various organs and tissues as described previously (Jefferson et al., 1987). The arrow in H indicates the expression of AtORM1 in developing embryos inside immature siliques.
To assess the in planta contributions of each gene to SPT inhibition, transcript levels of AtORM1 and AtORM2 were measured in different organs of Arabidopsis. Our analyses revealed that the AtORM1 transcript was approximately 3- to 4-fold more abundant in all tissues tested except pollen, whereas the AtORM2 transcript was approximately 4-fold more abundant (Fig. 3G). Our results are broadly consistent with those for these genes in the AtGenExpress public microarray database (Supplemental Fig. S1).
Using AtORM1 and AtORM2 promoter::GUS fusion constructs, the location of in planta expression was examined. Promoters for both genes conferred expression in vegetative tissues, with GUS staining most pronounced in vascular tissues. Differences in promoter activity for AtORM1 and AtORM2 were observed in floral tissues and roots. Based on the intensity of GUS staining, AtORM1 promoter conferred higher expression in anthers and developing embryos than the AtORM2 promoter (Fig. 3, H–J), while AtORM2 promoter conferred greater expression than the AtORM1 promoter in filaments, petals, sepals, pistils, and siliques (Fig. 3, M–O). Both AtORM1 and AtORM2 promoters yielded GUS expression in mature pollen grains and in roots. However, the AtORM1 promoter, but not the AtORM2 promoter, conferred detectable GUS expression in lateral root buds (Fig. 3, K, L, P, and Q).
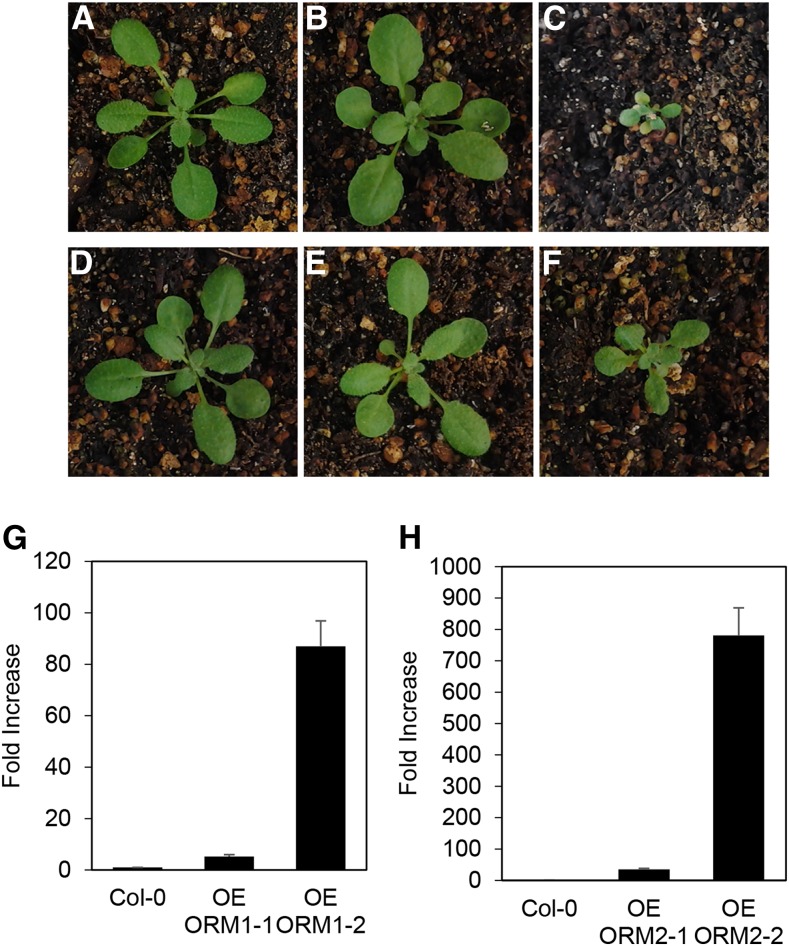
Overexpression of AtORM1 and AtORM2 Results in Dwarfed Growth
To examine the in planta functions of AtORM1 and AtORM2, three available T-DNA mutant lines for these genes were initially characterized: SALK_046054 (predicted T-DNA insertion in the first intron of AtORM1), GK-143A01 (predicted T-DNA insertion in the first exon of AtORM1), and SAIL_1286_D09 (predicted T-DNA insertion in the 5′ untranslated region of AtORM2). Homozygous lines were identified for each of these mutants, but full-length transcripts were still detected, indicating that these lines are not null mutants for the AtORM1 and AtORM2 genes (Supplemental Fig. S2). As an alternative approach for the characterization of the in planta functions of AtORM1 and AtORM2, overexpression and RNAi suppression lines for these genes were created in Arabidopsis Col-0. Overexpression lines were prepared by placing the AtORM1 or AtORM2 cDNA under the control of the cauliflower mosaic virus 35S promoter. Homozygous lines were screened by qPCR to identify those with increased expression. A portion of the lines with confirmed overexpression of AtORM1 or AtORM2 were visually indistinguishable from Col-0 plants, including those with approximately 5-fold overexpression of AtORM1 and approximately 35-fold overexpression of AtORM2 in leaves (Fig. 4, B and E). However, a second class of overexpression lines for both genes was observed with strong dwarfing. These lines included those with greater than 80-fold overexpression of AtORM1 and approximately 800-fold overexpression of AtORM2 in leaves relative to expression levels of these genes in Col-0 leaves (Fig. 4, C and F). These results suggest that a threshold of AtORM1 and AtORM2 protein levels can be reached to suppress SPT activity sufficiently to reduce growth, as observed previously for AtLCB1 RNAi suppression (Chen et al., 2006) and reduced AtssSPTa expression (Kimberlin et al., 2013). Sphingolipidomic profiling revealed little quantitative difference between amounts of sphingolipids in Col-0 and AtORM overexpression lines and virtually no difference in amounts of sphingolipids between AtORM overexpression lines with or without a growth phenotype, especially in the GIPC and GlcCer classes (Supplemental Figs. S3–S6), similar to previous findings with AtssSPT overexpression or RNAi suppression (Kimberlin et al., 2013). It was notable that concentrations of the LCB t18:0 were elevated in ceramide and GIPC molecular species in dwarfed AtORM1 and AtORM2 plants compared with Col-0 control plants (Supplemental Fig. S3). AtORM RNAi suppression lines showed significant suppression of both AtORM1 and AtORM2 transcript (Supplemental Fig. S7). Consistent with a role of AtORMs as SPT repressors, SPT activity assayed from root microsomes of RNAi suppression lines showed increased SPT activity (Supplemental Fig. S8). No reductions in growth and no significant quantitative differences were seen in the sphingolipidome, particularly in GIPC and GlcCer amounts, in AtORM RNAi lines relative to the Col-0 controls (Supplemental Fig. S4–S6).
Figure 4.
Phenotypes associated with AtORM1 and AtORM2 overexpression. A to F, High levels of AtORM1 and AtORM2 expression result in reduced plant size, early leaf senescence, and early plant death. Representative images of plants grown under the same conditions and of comparable age are shown. The wild type (Col-0) is shown in A and D. AtORM1 overexpression line 1 (OE ORM1-1) is shown in B, while AtORM2 overexpression line 1 (OE ORM2-1) is shown in E. AtORM1 overexpression line 2 (OE ORM1-2) is shown in C, while AtORM2 overexpression line 2 (OE ORM2-2) is shown in F. The phenotype correlates with ORM expression level, as lines with the strongest overexpression display a phenotype while relatively weaker overexpression lines do not show a noticeable growth phenotype. G and H, Expression levels of AtORM1 (G) and AtORM2 (H) in overexpressing lines (OE). Tissue was collected from wild-type Col-0 and lines overexpressing AtORM1 and AtORM2 grown under standard conditions. qPCR was used to determine relative AtORM1 and AtORM2 transcript levels by comparison with Col-0. PP2AA3 was used as a reference gene. Values shown are means ± sd for three independent measurements and indicate relative fold increase of AtORM1 or AtORM2 compared with wild-type levels.
Modulation of AtORM Expression Affects Sensitivity to FB1
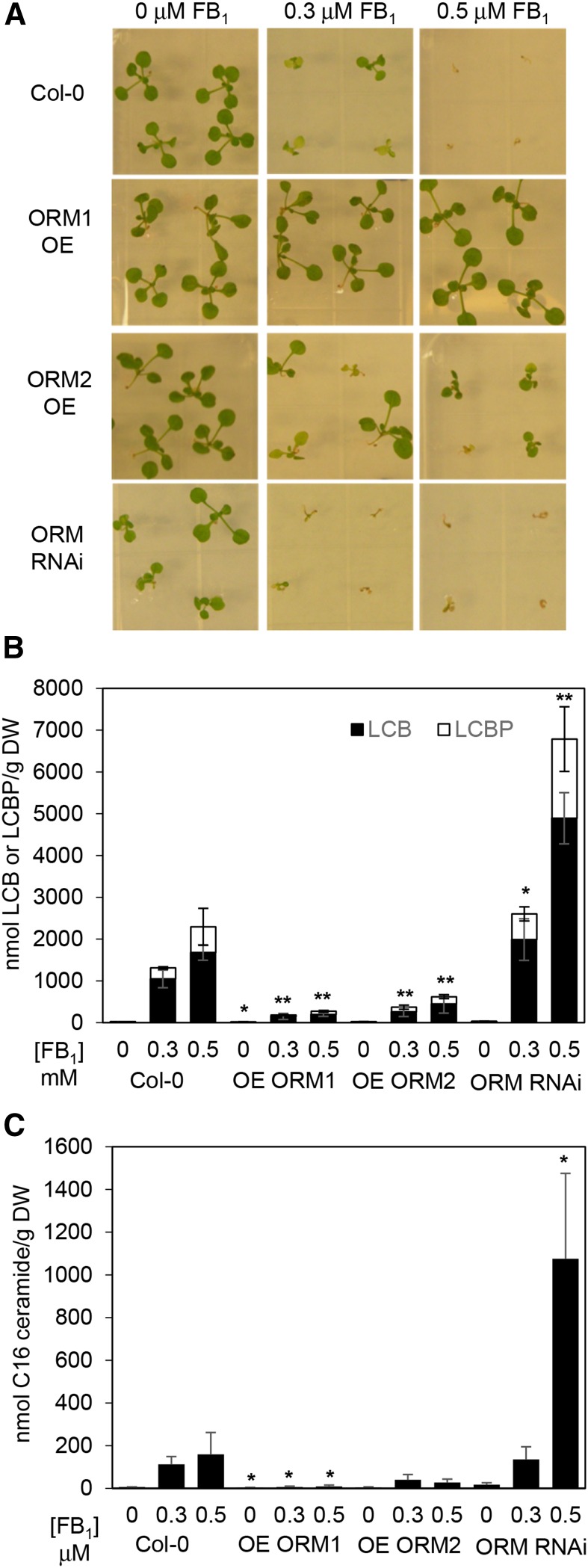
The PCD-inducing mycotoxin FB1, a ceramide synthase inhibitor, has been used routinely as a means of perturbing sphingolipid homeostasis for the study of SPT activity. From these studies, reduced SPT activity, such as that achieved by the suppression of AtssSPTa, results in enhanced resistance to FB1 due presumably to reduced accumulation of cytotoxic LCBs (Kimberlin et al., 2013). Conversely, increased SPT activity, such as that obtained by the overexpression of AtssSPTa, results in enhanced sensitivity to FB1 due presumably to the accumulation of cytotoxic LCBs (Kimberlin et al., 2013). Consistent with the suppression of SPT activity, AtORM1 and AtORM2 overexpression lines displayed increased resistance to FB1, along with decreases in the accumulation of free LCBs and LCB phosphates (Fig. 5, A and B). These lines were viable at 0.5 μm FB1, a concentration that was toxic to wild-type Arabidopsis (Fig. 5A). This phenotype was observed in a range of overexpression lines, including those with and without the dwarfing phenotype described above (Supplemental Fig. S8). Conversely, AtORM RNAi suppression lines were not viable on medium containing 0.3 μm FB1, but no loss of viability was observed in wild-type plants at this concentration (Fig. 5A). RNAi suppression lines of AtORM were observed to have a large increase in free LCBs and LCB phosphates when grown on FB1, which was particularly accentuated relative to AtORM overexpression lines and wild-type Col-0 on medium containing 0.5 µm FB1 (Fig. 5B). Mirroring alterations in LCB concentrations in transgenic and wild-type plants in response to FB1, concentrations of ceramides containing C16 fatty acids were lower in lines with overexpression of ORM and higher in RNAi suppression lines (Fig. 5C). These results indicate that altered ORM expression is an effective way of modulating plant sensitivity to FB1 and indicate that AtORMs act as inhibitors of SPT.
Figure 5.
Modulation of AtORM1 and AtORM2 expression alters sensitivity to FB1 and LCB as well as C16 ceramide accumulation. A, Altered AtORM1 and AtORM2 expression affects the sensitivity of plants to FB1, a competitive inhibitor of ceramide synthase. Seeds were sown on Linsmaier and Skoog (LS) agar plates supplemented with FB1 at 0.3 and 0.5 μm as indicated. The wild type (Col-0) is extremely sensitive to FB1 at 0.5 μm and less affected at 0.3 μm. Up-regulation of AtORM1 (ORM1) and AtORM2 (ORM2) by transgenic overexpression (OE) causes an FB1-resistant phenotype, whereas RNAi suppression of AtORM by RNAi causes an FB1-sensitive phenotype (ORM RNAi). Images were taken 14 d after seeds were sown and are representative of three independent experiments. B, Altered AtORM1 and AtORM2 affects the accumulation of cytotoxic free LCB and LCB phosphate (LCBP) levels in response to FB1 treatment. Wild-type plants show increased total LCB levels when treated with FB1. Compared with the wild type, AtORM1 and AtORM2 overexpression plants display FB1 resistance and reduced total LCB level. Alternatively, AtORM RNAi suppression plants display FB1 sensitivity and increased total LCB level. Electrospray ionization-tandem mass spectrometry analyses were performed with three independent biological replicates ± sd. Plants were grown on LS plates ± FB1 for 2 weeks before tissue collection. DW, Dry weight. *, P < 0.05; and **, P < 0.01. C, Total C16 ceramide levels are affected by the modulation of AtORM expression. AtORM1 overexpression plants show decreased accumulation of C16 ceramide when compared with wild-type plants comparatively grown on LS plates ± FB1, while ORM RNAi plants show increased accumulation of C16 ceramide. Analyses were performed as described in B with three independent biological replicates ± sd. *, P < 0.05.
Modulation of AtORM Expression Impacts Ceramide Synthase Activity
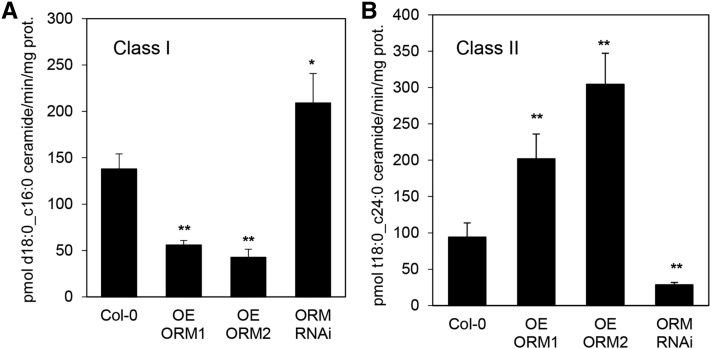
Recent studies in S. cerevisiae have suggested a role for ceramide synthase in the maintenance of sphingolipid homeostasis through a phosphorylation/dephosphorylation mechanism similar to that used for reversible modulation of ORMs (Muir et al., 2014). In contrast to S. cerevisiae, Arabidopsis has two functionally distinct ceramide synthase classes: class I and class II. This difference likely results in more complexity of potential ceramide synthase regulation of sphingolipid homeostasis in Arabidopsis. To gain insights into the coordinated regulation of ORMs and ceramide synthases, class I ceramide synthase was assayed in microsomes from roots of AtORM1 and AtORM2 overexpression lines and RNAi lines using 16:0-CoA and d18:0, the preferred substrates of this enzyme class. Class II ceramide synthase activity was assayed using 24:0-CoA and t18:0, preferred substrates of this enzyme class. Lower class I ceramide synthase activity was detected in microsomes of AtORM1 and AtORM2 overexpression lines, and increased activity was detected in microsomes from AtORM RNAi suppression lines (Fig. 6A). Conversely, class II ceramide synthase activity was decreased in microsomes from AtORM RNAi suppression lines but increased in AtORM1 and AtORM2 overexpression lines (Fig. 6B). These results suggest preferential metabolic flux through class I ceramide synthase for the synthesis of C16 fatty acid-containing ceramides with increased SPT activity resulting from disrupted ORM mediation of SPT in AtORM RNAi lines and preferential flux through class II ceramide synthases for the production of VLCFA-containing ceramides when SPT activity is limited by suppression from increased ORMs in AtORM1 and AtORM2 overexpression lines (Fig. 7). These findings are consistent with the increased concentrations of C16 fatty acid-containing ceramides detected in AtORM RNAi lines in response to FB1 treatment (Fig. 5C).
Figure 6.
Ceramide synthase activity is altered by the modulation of AtORM1 and AtORM2 expression. A, Altered AtORM1 and AtORM2 expression affects class I ceramide synthase activity. Class I ceramide synthase activity was assayed on microsomal protein prepared from hydroponically grown root tissue using 16:0-CoA and d18:0 as substrates. Increases in AtORM1 and AtORM2 overexpression (OE ORM1, OE ORM2) resulted in decreases in class I ceramide synthase activity, while RNAi suppression of AtORM (ORM RNAi) resulted in an increase in activity of this enzyme. Analyses were performed with three independent biological replicates ± sd. *, P < 0.05; and **, P < 0.01. B, Increases in AtORM1 (OE ORM1) and AtORM2 (OE ORM2) expression resulted in increases in class II ceramide synthase activity, while RNAi suppression of AtORM resulted in a decrease in activity of this enzyme class. Activity assays were conducted with microsomes from hydroponically grown root tissue using 24:0-CoA and t18:0 substrates. Analyses were performed with three independent biological replicates ± sd. **, P < 0.01.
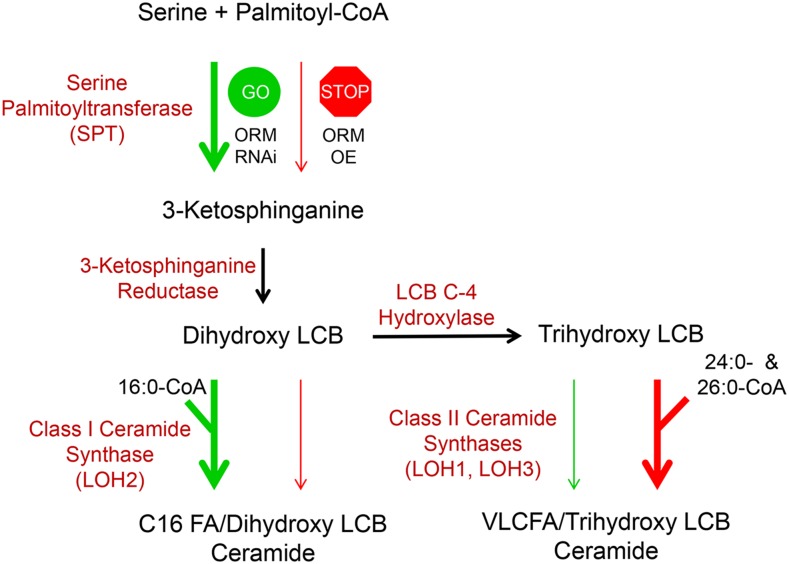
Figure 7.
Model of the coordinated regulation of sphingolipid synthesis by AtORM1 and AtORM2 and ceramide synthase activity. The model represents the core synthesis pathway of ceramides in Arabidopsis. SPT catalyzes a condensation reaction between Ser and palmitoyl-CoA, leading to the production of 3-ketosphinganine, which is reduced to dihydroxy LCB (d18:0) through 3-ketosphinganine reductase activity. Dihydroxy LCBs can be used by class I ceramide synthase (LOH2) along with 16:0-CoA to produce C16 ceramides. Alternatively, dihydroxy LCBs can be hydroxylated by LCB C-4 hydroxylase to form trihydroxy LCBs (t18:0) that are used by class II ceramide synthases (LOH1 and LOH3) along with very long-chain fatty acyl-CoAs, primarily 24:0- and 26:0-CoA, to produce ceramides containing VLCFAs. In the model shown, modulation of AtORM expression leads to alterations in ceramide synthase activity. AtORM RNAi suppression, indicated by GO, results in increased SPT activity and increased generation of LCBs, with a concomitant increase in class I activity and a decrease in class II activity. Conversely, overexpression (OE) of AtORM1 and AtORM2, indicated by STOP, results in decreased SPT activity and reduced LCB generation, with a concomitant decrease in class I activity and an increase in class II activity to ensure the production of sufficient levels of ceramides with VLCFAs to support growth. A role of LCB C-4 hydroxylase activity in mediating relative flux through class I and II ceramide synthases, particularly in response to enhanced LCB synthesis, also is possible.
DISCUSSION
The findings reported here demonstrate the occurrence of two ORM proteins in Arabidopsis that share significant identity with human and yeast ORM polypeptides. We show that these polypeptides interact physically with a complex that includes AtLCB1/AtLCB2a and AtssSPTa. Consistent with a function as suppressors of SPT activity, the expression of AtORM1 and AtORM2 rescued the sensitivity of the yeast orm2Δ mutant to exogenous LCBs. Also consistent with a function as a suppressor of SPT activity, AtORM1 and AtORM2 inhibited SPT activity when overexpressed with the core Arabidopsis SPT components in a yeast mutant deficient in the native SPT and ORMs. Furthermore, overexpression of AtORM1 and AtORM2 resulted in enhanced resistance to FB1, a phenotype also observed with reduced SPT activity associated with suppressed expression of AtLCB1 (Shi et al., 2007), AtLCB2 (Saucedo-García et al., 2011), or AtssSPT (Kimberlin et al., 2013). Moreover, RNAi-mediated down-regulation of AtORM expression resulted in increased sensitivity to FB1, a phenotype also observed with increased SPT activity associated with the up-regulation of AtssSPT expression (Kimberlin et al., 2013). Altered AtORM expression also was accompanied by either enhanced accumulation of LCBs (AtORM RNAi lines) or reduced accumulation of LCBs (AtORM overexpression lines) relative to Col-0 controls in response to FB1 treatment. These findings are consistent with altered SPT regulation in the ORM transgenic lines and with the function of AtORM1 and AtORM2 as negative regulators of SPT activity in Arabidopsis.
Also observed in parallel with these changes was enhanced accumulation of ceramides containing C16 fatty acids (AtORM RNAi lines) or reduced accumulation of ceramides containing C16 fatty acids (AtORM overexpression lines). These findings are consistent with the enhanced sequestration of LCBs as ceramides via the activity of ceramide synthase I (LOH2), which uses primarily C16 fatty acyl-CoAs as substrates, in response to deregulated SPT in AtORM RNAi lines. Also consistent with this, LOH2 activity was found to be increased significantly in microsomes from AtORM RNAi lines but decreased by more than 2-fold in AtORM overexpression lines compared with the Col-0 control. Unexpectedly, the opposite effect was observed in microsomes of these plants for class II ceramide synthase activity, which uses primarily trihydroxy LCBs and very-long-chain fatty acyl-CoAs as substrates: microsomes from AtORM RNAi lines had more than 3-fold less class II ceramide synthase activity, while microsomes for AtORM overexpression lines had more than 2-fold more activity of this enzyme class relative to the control. These findings point to coordinated control of the activities of the two functionally distinct ceramide synthase classes in response to altered ORM-mediated flux through SPT. As shown in Figure 7, we propose that the LOH2 ceramide synthase, a nonessential enzyme under normal growth conditions, provides a safety valve for excess LCB production, which is particularly accentuated in response to FB1. As we have proposed previously, increased flux through LOH2 ceramide synthase may be coupled with GlcCer synthase activity to glucosylate C16 fatty acid-containing ceramides to a less cytotoxic form. Conversely, the down-regulation of SPT activity by AtORM overexpression may result in the directing of LCB flux toward the synthesis of ceramides containing trihydroxy LCBs and VLCFAs, via class II ceramide synthases, to support growth. One notable observation of this and similar studies is that perturbations in sphingolipid homeostasis are accentuated in the presence of FB1. One possibility is that the C16 fatty acid-containing ceramides resulting from LOH2 activity in FB1-treated plants deregulate SPT or dilute possible sphingolipid signals (e.g. ceramides containing VLCFAs) that typically provide homeostatic regulation of SPT.
How the apparent coordinated mediation of SPT and ceramide synthase activities by ORM is regulated is currently under investigation. One possibility is that class I and II ceramide synthases are differentially regulated by phosphorylation. It is known, for example, that the activity of the yeast Lag1/Lac1 ceramide synthase is subject to regulation by phosphorylation at residues Ser-23 and Ser-24 in Lag1 and Lac1 by Ypk1 (Muir et al., 2014) and at residues Ser-393, Ser-395, and Ser-397 near their C termini by casein kinase2 (Fresques et al., 2015). In the latter case, phosphorylation of Lag1 and Lac1 affects the localization of these polypeptides by disrupting their ER retrieval (Fresques et al., 2015). Similar to the yeast ceramide synthase, the AtLOH1 ceramide synthase has four experimentally confirmed phosphorylated Sers (Ser-295, Ser-300, Ser-302, and Ser-304; Nühse et al., 2004), and AtLOH2 has two experimentally confirmed phosphorylated Sers (Ser-289 and Ser-291; Nühse et al., 2004; Sugiyama et al., 2008). Like the yeast Lag1 and Lac1 polypeptides, these phosphorylated Ser residues are in the vicinity of the C termini of AtLOH1 and AtLOH2, near presumptive ER retrieval sequences. Although not experimentally verified, AtLOH3 also has Ser residues analogous to those in AtLOH1 near its C terminus. The influence of this phosphorylation on ceramide synthase activity and possible changes in phosphorylation of these polypeptides in response to altered ORM regulation of SPT are currently under study. Overall, our findings indicate that the regulation of sphingolipid homeostasis extends beyond SPT and is more complex in eukaryotic organisms with multiple, functionally distinct ceramide synthases than that found in S. cerevisiae, which has only one ceramide synthase type (Mullen et al., 2012).
Although our results clearly show that AtORM1 and AtORM2 function to suppress SPT activity, still unclear in plants and mammals is how ORMs reversibly regulate SPT in response to changes in intracellular sphingolipid levels. Absent from plant and mammalian ORMs is the N-terminal Ser-rich domain found in the yeast ScORM1 and ScORM2 polypeptides. Phosphorylation of Ser residues in this domain by Ypk1 relieves SPT inhibition in response to low intracellular sphingolipid levels (Breslow et al., 2010; Han et al., 2010; Roelants et al., 2011), and dephosphorylation of these residues by Cdc55 protein phosphatase 2A results in ORM suppression of SPT in response to excess intracellular sphingolipid levels (Sun et al., 2012). The lack of the regulatory domain in plant and mammalian ORMs does not preclude that other residues are subject to reversible posttranslational modifications (e.g. phosphorylation/dephosphorylation). Other possibilities include changes in ORM turnover rates or ORM localization in response to intracellular sphingolipid levels. With regard to the latter possibility, our localization studies showed that AtORM polypeptides are present primarily, but not exclusively, in the ER. As described above, changes in ER localization have been proposed as a component of yeast ceramide synthase activity (Fresques et al., 2015). More extensive studies, beyond the scope of those in this report, are currently under way to address this central question of how SPT is regulated to maintain intracellular homeostasis in plants and animals.
MATERIALS AND METHODS
Yeast Growth and Expression Plasmids
Yeast (Saccharomyces cerevisiae) strain TDY9113 (Mata tsc3Δ:NAT lcb1Δ:KAN ura3 leu2 lys2 trp1Δ) lacking endogenous SPT was used for the expression and characterization of Arabidopsis (Arabidopsis thaliana) SPT subunits and ORM proteins. The mutant was cultured in medium containing 15 μm phytosphingosine and 0.2% (w/v) Tergitol Nonidet P-40. The AtORM1 (At1g01230) and AtORM2 (At5g42000) open reading frames were amplified by PCR and inserted into pPR3-N (Dualsystems Biotech) for expression with N-terminal HA tags. pAL2-URA was constructed for divergent constitutive expression of AtLCB1-FLAG (or AtLCB1C144W-FLAG) and Myc-tagged AtLCB2a by replacing the GAL1 and GAL10 promoters of pESC-URA (Stratagene) with the yeast LCB2 and ADH promoters, respectively. The AtssSPTa cDNA open reading frame was inserted after the 3× HA tag in pADH1 (Kohlwein et al., 2001; Kimberlin et al., 2013).
Immunoprecipitation
Immunoprecipitation was conducted as described (Kimberlin et al., 2013) with minor modifications. Microsomal membrane proteins were prepared from yeast cells expressing FLAG-tagged AtLCB1 (or AtLCB1C144W), Myc-tagged AtLCB2a, HA-tagged AtssSPTa, and HA-tagged AtORM1 or AtORM2. Microsomal membrane proteins were solubilized in 1.5% digitonin at 4°C for 2.5 h and incubated with Flag-beads (Sigma-Aldrich) overnight. The bound proteins were eluted in immunoprecipitation buffer (50 mm HEPES-KOH, pH 6.8, 150 mm potassium acetate, 2 mm magnesium acetate, 1 mm calcium chloride, and 15% glycerol) containing 0.25% digitonin and 200 μg mL−1 FLAG peptide, resolved on a 4% to 12% BisTris NuPAGE gel (Invitrogen), and detected by immunoblotting with anti-HA (Covance; 1:5,000 dilution), anti-Myc (Sigma-Aldrich; 1:3,000 dilution), and anti-FLAG (GenScript; 1:5,000 dilution) antibodies.
LCB Extraction and Analysis
LCBs were extracted, derivatized, and analyzed by reverse-phase HPLC as described previously (Gable et al., 2010). For measurement of LCBs and DoxSA formation, the areas under HPLC peaks, measured in luminescence units, were normalized to the internal C17 sphingosine standard, and the amounts of LCBs and 1-DoxSA were determined using standard curves for each of the LCBs and are reported as nmol LCBs per optical density of cells measured at 600 nm (Gable et al., 2010).
SPT Assay
Plant microsomes were prepared and SPT activity was assayed as described (Kimberlin et al., 2013), except that 50 μm palmitoyl-CoA and 20 µm bovine serum albumin were used for the Arabidopsis microsomal SPT assays. SPT activity was measured using the substrates [3H]Ser and 16:0-CoA as described (Han et al., 2009).
Yeast Complementation
Synthetic complete medium was used to grow yeast (BY4741). Synthetic complete medium was supplemented with 15 μm phytosphingosine and 0.1% Tergitol. Yeast knockout mutants, orm1Δ and orm2Δ, were obtained from the S. cerevisiae knockout library kindly provided by Jaekwon Lee. AtORM1 and AtORM2 cDNAs were cloned into the centromeric plasmid pSH15 using the XhoI and PstI restriction sites using primers P1 to P4 (Supplemental Table S1). The plasmid, pSH15, containing native S. cerevisiae ORM2 was a gift from Dr. Amy Chang. Cells were grown at 30°C and then normalized to an optical density at 600 nm = 0.1 before being serially diluted and plated.
Plant Material and Growth Conditions
All Arabidopsis Col-0 lines used in this study were stratified at 4°C for 4 d and maintained at 22°C with a 16-h-light (100 µmol m−2 s−1)/8-h-dark cycle. Plants sown on LS agar plates were surface sterilized before stratification. Plants that were grown hydroponically were maintained on modified Hoagland solution as described previously in custom-made hydroponics tanks (Conn et al., 2013).
Arabidopsis Transformation and Selection
Binary vectors were transformed into Agrobacterium tumefaciens GV3101 by electroporation. The floral dip method was used to create transgenic plants in Arabidopsis Col-0 (Clough and Bent, 1998). A green light-emitting diode flashlight and a Red 2 camera filter were used to identify transformed seeds by fluorescence of the DsRed marker protein.
RNA Isolation and qPCR
For expression analyses of AtORM1 and AtORM2, RNA extraction was done using the RNeasy Plant Kit (Qiagen) according to the manufacturer’s protocol. RNA (1 µg) was treated with DNaseI (Invitrogen) according to the manufacturer’s protocol. Treated RNA was then reverse transcribed to cDNA with the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer’s protocol. For tissue-specific expression analysis, 6- to 8-week-old Col-0 plants were used as sources of plant material. qPCR was performed on cDNA using the Bio-Rad MyiQ iCycler qPCR instrument. Values shown are averages of three independent measurements ± sd. SYBR Green was used as the fluorophore in a qPCR supermix (Qiagen). QuantiTect (Qiagen) primer sets P5 to P7 (Supplemental Table S1) were used for relative quantification. PP2AA3 (At1g13320) was used as an internal reference gene. Reverse transcription-PCR analysis of homozygous T-DNA mutant lines was performed on cDNA using primers P8 to P11 (Supplemental Table S1).
Analysis of Promoter GUS-Expressing Plants
To generate the AtORM1 promoter::GUS and AtORM2 promoter::GUS constructs, an approximately 1-kb region upstream of the start codon was PCR amplified from genomic DNA (oligonucleotides P12–P15; Supplemental Table S1) and cloned into a pBinGlyRed2 vector containing the GUS gene using the BamHI and EcoRI restriction sites. This vector was then transformed into A. tumefaciens C58, and cells harboring the binary vector were used to transform wild-type Arabidopsis as described previously (Clough and Bent, 1998). GUS staining solution was composed of 20 mm sodium phosphate (monobasic), 30 mm sodium phosphate (dibasic), 2 mm potassium ferricyanide, and 2 mm potassium ferrocyanide, along with 1 μL mL−1 Triton X-100 and 1 mg mL−1 5-bromo-4-chloro-3-indolyl-β-d-glucuronide. Tissues were preincubated in chilled 90% acetone for 10 min, then vacuum infiltrated with chilled GUS staining solution for 10 min. The tissues were incubated overnight at 37°C and then cleared with 100% ethanol followed by 70% ethanol. Images of GUS-analyzed tissue were taken with an Olympus AX70 optical microscope.
Subcellular Localization of ORM1 and ORM2
YFP fusion proteins with AtORM1 and AtORM2 were prepared by amplification of the AtORM1 and AtORM2 open reading frames using gene-specific primers (P16–P19; Supplemental Table S1). PCR products were cloned into the 35S-pFAST-eYFP vector using the SacI and KpnI restriction sites generating C-terminal YFP fusion constructs. A. tumefaciens-mediated infiltration of Nicotiana benthamiana leaves was performed with ORM1-YFP and ORM2-YFP constructs separately and in conjunction with the ER marker CD3-959 (HDEL-mCherry). Sequential imaging was performed using a Nikon A1 confocal imaging system mounted on a Nikon Eclipse 90i microscope. Excitation/emission wavelengths for YFP and mCherry were 488 nm/500 to 550 nm and 561.6 nm/570 to 620 nm, respectively, as described previously (Kimberlin et al., 2013).
Arabidopsis Mutant Genotyping
T-DNA insertion mutants were acquired from the Arabidopsis Biological Resource Center and the GABI-Kat collections. The REDextract-N-Amp Tissue PCR kit (Sigma-Aldrich) was used to extract genomic DNA from leaf tissue. Genotyping was performed by PCR using gene-specific and T-DNA-specific primer sets P20 to P28 (Supplemental Table S1).
Membrane Topology Mapping of ScORM2
The topology of ScORM2 protein was mapped based on the previously described methodology (Han et al., 2004). In brief, NheI restriction sites were created before codons for amino acids 2, 100, 135, and 169 and an XbaI site before the stop codon (217) by use of the QuikChange mutagenesis kit (Agilent Technology). A GC encoding the 53-amino acid domain of invertase (Suc2p) was inserted into the NheI or XbaI site. The microsomal proteins were isolated from the strains transformed with the GC topology reporter plasmids and treated with endoglycosidase H. The proteins were resolved using a 4% to 12% BisTris NuPAGE gel system (Invitrogen).
FB1 Screening of Arabidopsis ORM1 and ORM2 Overexpression and RNAi lines
ORM-overexpressing plants were generated by transforming Col-0 with the cauliflower mosaic virus 35S promoter:ORM cDNA constructs. AtORM1 and AtORM2 cDNAs were cloned into the binary vector pBinGlyRed3-35S using the EcoRI/XbaI restriction sites (P29–P32; Supplemental Table S1). ORM RNAi lines were generated by overexpressing a hairpin composed of an AtORM1 or AtORM2 gene fragment. The ORM gene fragments were cloned into the pINTRON vector using the XbaI, XhoI, SpeI, and HindIII restriction sites forming a hairpin using primers P33 to P36 (Supplemental Table S1). The pINTRON fragment containing the hairpin was amplified using primers P37 and P38 (Supplemental Table S1) and cloned into pBinGlyRed3-35S using the EcoRI/XbaI restriction sites. Arabidopsis (Col-0) plants were transformed with these constructs, and the resulting transformants were selected and screened by qPCR for AtORM1 and AtORM2 expression. Sensitivity screening relative to a wild-type control was done at 0, 0.3, and 0.5 µm FB1 (Sigma-Aldrich) concentrations in LS medium. Sensitivity was determined by plant growth rate and germination at the varying FB1 concentrations (Kimberlin et al., 2013). Plants were grown for 2 weeks on FB1 before analysis.
Plant Microsomal Membrane Isolation
Microsomal membrane isolation from hydroponically grown 4-week-old Arabidopsis roots was performed as described previously (Lynch and Fairfield, 1993), and protein concentration was measured using the bicinchoninic acid method (Smith et al., 1985; Lynch and Fairfield, 1993).
Generation of the AtLCB1C144W Mutant
The C144W mutation was introduced into AtLCB1 by changing codon 144 TGT to TGG by QuikChange mutagenesis (Stratagene) using oligonucleotide primers P39 and P40 (Supplemental Table S1). The mutation was confirmed by sequencing.
Ceramide Synthase Assays
Ceramide synthase assays were performed on microsomal extracts (10 μg of protein) derived from hydroponically grown root using a previously described methodology (Luttgeharm et al., 2015a, 2016). Reaction substrates for class I ceramide synthase assays were d18:0 LCB and 16:0-CoA, while substrates for class II ceramide synthase assays were t18:0 LCB and 24:0-CoA. After incubation and extraction, sphingolipids produced in the assay were analyzed by mass spectrometry as described (Luttgeharm et al., 2015a, 2016).
Sphingolipid Analysis
Sphingolipids were extracted from 2 to 15 mg of lyophilized plant material and analyzed by liquid chromatography-tandem mass spectrometry as described previously (Markham and Jaworski, 2007; Kimberlin et al., 2013).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Gene expression levels for AtORM1 (At1g01230) and AtORM2 (At5g42000) in different tissue types.
Supplemental Figure S2. Genotyping of T-DNA mutants for AtORM1 and AtORM2.
Supplemental Figure S3. Sphingolipid profile of AtORM1 and AtORM2 overexpression lines that display the dwarf phenotype.
Supplemental Figure S4. Ceramide profile for Col-0 and AtORM overexpression and RNAi lines grown on LS medium ± FB1.
Supplemental Figure S5. GlcCer profile for Col-0 and AtORM overexpression and RNAi lines grown on LS medium ± FB1.
Supplemental Figure S6. GIPC profile for Col-0 and AtORM overexpression and RNAi lines grown on LS medium ± FB1.
Supplemental Figure S7. qPCR analysis of ORM RNAi lines.
Supplemental Figure S8. SPT activity from root microsomes of wild-type and transgenic plants.
Supplemental Table S1. Oligonucleotide primers used in the reported studies.
Supplementary Material
Acknowledgments
We thank Amy Chang (University of Michigan) for providing plasmid pSH15 containing S. cerevisiae ORM2 and Jaekwon Lee (University of Nebraska-Lincoln) for providing S. cerevisiae knockout lines for ORM1 and ORM2.
Glossary
- VLCFA
very-long-chain fatty acid
- PCD
programmed cell death
- GlcCer
glucosylceramide
- GIPC
glycosyl inositolphosphoceramide
- RNAi
RNA interference
- GC
glycosylation cassette
- ER
endoplasmic reticulum
- DoxSA
deoxy-sphinganine
- Col-0
Columbia-0
- qPCR
quantitative PCR
- HA
hemagglutinin
- LS
Linsmaier and Skoog
Footnotes
This work was supported by the National Science Foundation (grant no. MCB–1158500 to J.M.S., T.M.D., and E.B.C.).
Articles can be viewed without a subscription.
References
- Bach L, Gissot L, Marion J, Tellier F, Moreau P, Satiat-Jeunemaître B, Palauqui JC, Napier JA, Faure JD (2011) Very-long-chain fatty acids are required for cell plate formation during cytokinesis in Arabidopsis thaliana. J Cell Sci 124: 3223–3234 [DOI] [PubMed] [Google Scholar]
- Bach L, Michaelson LV, Haslam R, Bellec Y, Gissot L, Marion J, Da Costa M, Boutin JP, Miquel M, Tellier F, et al. (2008) The very-long-chain hydroxy fatty acyl-CoA dehydratase PASTICCINO2 is essential and limiting for plant development. Proc Natl Acad Sci USA 105: 14727–14731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer EM, Mongrand S, Tilsner J (2014) Specialized membrane domains of plasmodesmata, plant intercellular nanopores. Front Plant Sci 5: 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi FC, Liu Z, Wu JX, Liang H, Xi XL, Fang C, Sun TJ, Yin J, Dai GY, Rong C, et al. (2014) Loss of ceramide kinase in Arabidopsis impairs defenses and promotes ceramide accumulation and mitochondrial H2O2 bursts. Plant Cell 26: 3449–3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK. (2013) Sphingolipid homeostasis in the endoplasmic reticulum and beyond. Cold Spring Harb Perspect Biol 5: a013326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK, Collins SR, Bodenmiller B, Aebersold R, Simons K, Shevchenko A, Ejsing CS, Weissman JS (2010) Orm family proteins mediate sphingolipid homeostasis. Nature 463: 1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacas JL, Buré C, Grosjean K, Gerbeau-Pissot P, Lherminier J, Rombouts Y, Maes E, Bossard C, Gronnier J, Furt F, et al. (2016) Revisiting plant plasma membrane lipids in tobacco: a focus on sphingolipids. Plant Physiol 170: 367–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacas JL, Furt F, Le Guédard M, Schmitter JM, Buré C, Gerbeau-Pissot P, Moreau P, Bessoule JJ, Simon-Plas F, Mongrand S (2012) Lipids of plant membrane rafts. Prog Lipid Res 51: 272–299 [DOI] [PubMed] [Google Scholar]
- Chen M, Han G, Dietrich CR, Dunn TM, Cahoon EB (2006) The essential nature of sphingolipids in plants as revealed by the functional identification and characterization of the Arabidopsis LCB1 subunit of serine palmitoyltransferase. Plant Cell 18: 3576–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Markham JE, Dietrich CR, Jaworski JG, Cahoon EB (2008) Sphingolipid long-chain base hydroxylation is important for growth and regulation of sphingolipid content and composition in Arabidopsis. Plant Cell 20: 1862–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chueasiri C, Chunthong K, Pitnjam K, Chakhonkaen S, Sangarwut N, Sangsawang K, Suksangpanomrung M, Michaelson LV, Napier JA, Muangprom A (2014) Rice ORMDL controls sphingolipid homeostasis affecting fertility resulting from abnormal pollen development. PLoS ONE 9: e106386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Conn SJ, Hocking B, Dayod M, Xu B, Athman A, Henderson S, Aukett L, Conn V, Shearer MK, Fuentes S, et al. (2013) Protocol: optimising hydroponic growth systems for nutritional and physiological analysis of Arabidopsis thaliana and other plants. Plant Methods 9: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich CR, Han G, Chen M, Berg RH, Dunn TM, Cahoon EB (2008) Loss-of-function mutations and inducible RNAi suppression of Arabidopsis LCB2 genes reveal the critical role of sphingolipids in gametophytic and sporophytic cell viability. Plant J 54: 284–298 [DOI] [PubMed] [Google Scholar]
- Fresques T, Niles B, Aronova S, Mogri H, Rakhshandehroo T, Powers T (2015) Regulation of ceramide synthase by casein kinase 2-dependent phosphorylation in Saccharomyces cerevisiae. J Biol Chem 290: 1395–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gable K, Gupta SD, Han G, Niranjanakumari S, Harmon JM, Dunn TM (2010) A disease-causing mutation in the active site of serine palmitoyltransferase causes catalytic promiscuity. J Biol Chem 285: 22846–22852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg JT, Silverman FP, Liang H (2000) Uncoupling salicylic acid-dependent cell death and defense-related responses from disease resistance in the Arabidopsis mutant acd5. Genetics 156: 341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gururaj C, Federman RS, Chang A (2013) Orm proteins integrate multiple signals to maintain sphingolipid homeostasis. J Biol Chem 288: 20453–20463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G, Gable K, Yan L, Natarajan M, Krishnamurthy J, Gupta SD, Borovitskaya A, Harmon JM, Dunn TM (2004) The topology of the Lcb1p subunit of yeast serine palmitoyltransferase. J Biol Chem 279: 53707–53716 [DOI] [PubMed] [Google Scholar]
- Han G, Gupta SD, Gable K, Niranjanakumari S, Moitra P, Eichler F, Brown RH Jr, Harmon JM, Dunn TM (2009) Identification of small subunits of mammalian serine palmitoyltransferase that confer distinct acyl-CoA substrate specificities. Proc Natl Acad Sci USA 106: 8186–8191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Lone MA, Schneiter R, Chang A (2010) Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc Natl Acad Sci USA 107: 5851–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K. (2003) Serine palmitoyltransferase, a key enzyme of sphingolipid metabolism. Biochim Biophys Acta 1632: 16–30 [DOI] [PubMed] [Google Scholar]
- Hirokawa T, Boon-Chieng S, Mitaku S (1998) SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14: 378–379 [DOI] [PubMed] [Google Scholar]
- Hjelmqvist L, Tuson M, Marfany G, Herrero E, Balcells S, Gonzalez-Duarte R (2002) ORMDL proteins are a conserved new family of endoplasmic reticulum membrane proteins. Genome Biol 3: RESEARCH0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberlin AN, Majumder S, Han G, Chen M, Cahoon RE, Stone JM, Dunn TM, Cahoon EB (2013) Arabidopsis 56-amino acid serine palmitoyltransferase-interacting proteins stimulate sphingolipid synthesis, are essential, and affect mycotoxin sensitivity. Plant Cell 25: 4627–4639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlwein SD, Eder S, Oh CS, Martin CE, Gable K, Bacikova D, Dunn T (2001) Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol Cell Biol 21: 109–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Yao N, Song JT, Luo S, Lu H, Greenberg JT (2003) Ceramides modulate programmed cell death in plants. Genes Dev 17: 2636–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttgeharm KD, Cahoon EB, Markham JE (2015a) A mass spectrometry-based method for the assay of ceramide synthase substrate specificity. Anal Biochem 478: 96–101 [DOI] [PubMed] [Google Scholar]
- Luttgeharm KD, Cahoon EB, Markham JE (2016) Substrate specificity, kinetic properties and inhibition by fumonisin B1 of ceramide synthase isoforms from Arabidopsis. Biochem J 473: 593–603 [DOI] [PubMed] [Google Scholar]
- Luttgeharm KD, Chen M, Mehra A, Cahoon RE, Markham JE, Cahoon EB (2015b) Overexpression of Arabidopsis ceramide synthases differentially affects growth, sphingolipid metabolism, programmed cell death, and mycotoxin resistance. Plant Physiol 169: 1108–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch DV, Fairfield SR (1993) Sphingolipid long-chain base synthesis in plants: characterization of serine palmitoyltransferase activity in squash fruit microsomes. Plant Physiol 103: 1421–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham JE, Jaworski JG (2007) Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun Mass Spectrom 21: 1304–1314 [DOI] [PubMed] [Google Scholar]
- Markham JE, Lynch DV, Napier JA, Dunn TM, Cahoon EB (2013) Plant sphingolipids: function follows form. Curr Opin Plant Biol 16: 350–357 [DOI] [PubMed] [Google Scholar]
- Markham JE, Molino D, Gissot L, Bellec Y, Hématy K, Marion J, Belcram K, Palauqui JC, Satiat-Jeunemaître B, Faure JD (2011) Sphingolipids containing very-long-chain fatty acids define a secretory pathway for specific polar plasma membrane protein targeting in Arabidopsis. Plant Cell 23: 2362–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melser S, Molino D, Batailler B, Peypelut M, Laloi M, Wattelet-Boyer V, Bellec Y, Faure JD, Moreau P (2011) Links between lipid homeostasis, organelle morphodynamics and protein trafficking in eukaryotic and plant secretory pathways. Plant Cell Rep 30: 177–193 [DOI] [PubMed] [Google Scholar]
- Msanne J, Chen M, Luttgeharm KD, Bradley AM, Mays ES, Paper JM, Boyle DL, Cahoon RE, Schrick K, Cahoon EB (2015) Glucosylceramides are critical for cell-type differentiation and organogenesis, but not for cell viability in Arabidopsis. Plant J 84: 188–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir A, Ramachandran S, Roelants FM, Timmons G, Thorner J (2014) TORC2-dependent protein kinase Ypk1 phosphorylates ceramide synthase to stimulate synthesis of complex sphingolipids. eLife 3: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TD, Hannun YA, Obeid LM (2012) Ceramide synthases at the centre of sphingolipid metabolism and biology. Biochem J 441: 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nühse TS, Stensballe A, Jensen ON, Peck SC (2004) Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 16: 2394–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perraki A, Cacas JL, Crowet JM, Lins L, Castroviejo M, German-Retana S, Mongrand S, Raffaele S (2012) Plasma membrane localization of Solanum tuberosum remorin from group 1, homolog 3 is mediated by conformational changes in a novel C-terminal anchor and required for the restriction of potato virus X movement. Plant Physiol 160: 624–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants FM, Breslow DK, Muir A, Weissman JS, Thorner J (2011) Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 108: 19222–19227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo-García M, Guevara-García A, González-Solís A, Cruz-García F, Vázquez-Santana S, Markham JE, Lozano-Rosas MG, Dietrich CR, Ramos-Vega M, Cahoon EB, et al. (2011) MPK6, sphinganine and the LCB2a gene from serine palmitoyltransferase are required in the signaling pathway that mediates cell death induced by long chain bases in Arabidopsis. New Phytol 191: 943–957 [DOI] [PubMed] [Google Scholar]
- Shi L, Bielawski J, Mu J, Dong H, Teng C, Zhang J, Yang X, Tomishige N, Hanada K, Hannun YA, et al. (2007) Involvement of sphingoid bases in mediating reactive oxygen intermediate production and programmed cell death in Arabidopsis. Cell Res 17: 1030–1040 [DOI] [PubMed] [Google Scholar]
- Simanshu DK, Zhai X, Munch D, Hofius D, Markham JE, Bielawski J, Bielawska A, Malinina L, Molotkovsky JG, Mundy JW, et al. (2014) Arabidopsis accelerated cell death 11, ACD11, is a ceramide-1-phosphate transfer protein and intermediary regulator of phytoceramide levels. Cell Rep 6: 388–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC (1985) Measurement of protein using bicinchoninic acid. Anal Biochem 150: 76–85 [DOI] [PubMed] [Google Scholar]
- Sperling P, Franke S, Lüthje S, Heinz E (2005) Are glucocerebrosides the predominant sphingolipids in plant plasma membranes? Plant Physiol Biochem 43: 1031–1038 [DOI] [PubMed] [Google Scholar]
- Stiban J, Tidhar R, Futerman AH (2010) Ceramide synthases: roles in cell physiology and signaling. Adv Exp Med Biol 688: 60–71 [DOI] [PubMed] [Google Scholar]
- Sugiyama N, Nakagami H, Mochida K, Daudi A, Tomita M, Shirasu K, Ishihama Y (2008) Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol Syst Biol 4: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Miao Y, Yamane Y, Zhang C, Shokat KM, Takematsu H, Kozutsumi Y, Drubin DG (2012) Orm protein phosphoregulation mediates transient sphingolipid biosynthesis response to heat stress via the Pkh-Ypk and Cdc55-PP2A pathways. Mol Biol Cell 23: 2388–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng C, Dong H, Shi L, Deng Y, Mu J, Zhang J, Yang X, Zuo J (2008) Serine palmitoyltransferase, a key enzyme for de novo synthesis of sphingolipids, is essential for male gametophyte development in Arabidopsis. Plant Physiol 146: 1322–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ternes P, Feussner K, Werner S, Lerche J, Iven T, Heilmann I, Riezman H, Feussner I (2011) Disruption of the ceramide synthase LOH1 causes spontaneous cell death in Arabidopsis thaliana. New Phytol 192: 841–854 [DOI] [PubMed] [Google Scholar]
- von Heijne G. (1992) Membrane protein structure prediction: hydrophobicity analysis and the positive-inside rule. J Mol Biol 225: 487–494 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.