Although nitrate stimulates NRT1-1 transcription in all root tissues, it represses protein accumulation in lateral primordial, explaining that NRT1-1 controls lateral root development only in low nitrate medium.
Abstract
Plants are able to modulate root growth and development to optimize their nitrogen nutrition. In Arabidopsis (Arabidopsis thaliana), the adaptive root response to nitrate (NO3−) depends on the NRT1.1/NPF6.3 transporter/sensor. NRT1.1 represses emergence of lateral root primordia (LRPs) at low concentration or absence of NO3− through its auxin transport activity that lowers auxin accumulation in LR. However, these functional data strongly contrast with the known transcriptional regulation of NRT1.1, which is markedly repressed in LRPs in the absence of NO3−. To explain this discrepancy, we investigated in detail the spatiotemporal expression pattern of the NRT1.1 protein during LRP development and combined local transcript analysis with the use of transgenic lines expressing tagged NRT1.1 proteins. Our results show that although NO3− stimulates NRT1.1 transcription and probably mRNA stability both in primary root tissues and in LRPs, it acts differentially on protein accumulation, depending on the tissues considered with stimulation in cortex and epidermis of the primary root and a strong repression in LRPs and to a lower extent at the primary root tip. This demonstrates that NRT1.1 is strongly regulated at the posttranscriptional level by tissue-specific mechanisms. These mechanisms are crucial for controlling the large palette of adaptive responses to NO3− mediated by NRT1.1 as they ensure that the protein is present in the proper tissue under the specific conditions where it plays a signaling role in this particular tissue.
Nitrogen (N) is an essential element throughout plant growth and development as it is a constituent of many biomolecules (Hawkesford et al., 2012). Plants can use both inorganic and organic sources of N from the soil, but nitrate (NO3−) is the main form taken up and assimilated by most crop plants (Nacry et al., 2013). Soil NO3− concentration is often the major nutrient factor limiting the growth and yield of crop plants, as it fluctuates dramatically in both time and space. From few µm, it can reach the range of 1 to 10 mm following fertilizer application or a burst of nitrification (Crawford and Glass, 1998; Miller et al., 2007). To cope with this, plants are able to quickly modulate their NO3− acquisition efficiency according to changes both in their own internal N status and in the external NO3− availability (Forde 2002). In particular, low N status of the whole plant leads to a fast upregulation of root NO3− uptake systems (Lejay et al., 1999; Nacry et al., 2013) and a stimulation of root growth for improved foraging of the soil (Forde 2002; Hermans et al., 2006). In addition, plants also sense NO3− per se, and the presence of this nutrient in the external medium activates specific signaling pathways affecting the expression of hundreds of genes (Bouguyon et al., 2012). In particular, NO3− induces the expression of many root NO3− transporter genes and accelerates the emergence and growth of lateral roots (LRs) specifically in the root portions in contact with a local high NO3− concentration. This is of major significance, as it contributes to preferential colonization and exploitation of the NO3−-rich areas of the soil (Drew, 1975; Burns, 1991; Gansel et al., 2001; Forde, 2002; Zhang et al., 2007; Gojon et al., 2009; Ruffel et al., 2011; Nacry et al., 2013; Mounier et al., 2014; O’Brien et al., 2016).
Compelling evidence indicates that the membrane NO3− transporter NRT1.1 (CHL1/NPF6.3), first identified as an influx carrier participating in the root uptake of NO3− (Tsay et al., 1993; Liu and Tsay, 2003), plays a crucial role in the sensing and signaling mechanisms triggering many adaptive responses to changes in external NO3− availability (Gojon et al., 2009, 2011; Nacry et al., 2013; Forde, 2014; Bouguyon et al., 2015). In most cases, the signaling action of NRT1.1 appears to be independent from its NO3− transport activity, leading to the proposal that it acts as a NO3− transceptor, with a dual transporter/sensor function (Remans et al., 2006; Walch-Liu and Forde, 2008; Ho et al., 2009; Wang et al., 2009; Krouk et al., 2010; Gojon et al., 2011; Bouguyon et al., 2015). The role of NRT1.1 as a NO3− sensor is complex because it is able to activate different physiological or developmental responses to NO3− through several independent sensing/signaling mechanisms that can be uncoupled by point mutations in the protein (Bouguyon et al., 2015). In particular, NRT1.1 is required for correct regulation of many NO3−-responsive genes, including those involved in NO3− transport and assimilation that are induced by NO3− itself (Ho et al., 2009; Wang et al., 2009). The exact mechanism of NRT1.1-dependent gene induction by NO3− is still unclear, but it has been recently proposed to involve phospholipase C and calcium as a second messenger (Riveras et al., 2015). In addition, NRT1.1 is a master player in the NO3− regulation of root system architecture as it governs LR growth in response to NO3− (Remans et al., 2006; Krouk et al., 2010; Mounier et al., 2014; Bouguyon et al., 2015).
LRs form postembryonically from pericycle cells and develop through a series of well-defined stages, from the initiation of a LR primordium (LRP; stage I) to its emergence out of the parent root (stage VIII; Casimiro et al., 2003; Malamy, 2005; Malamy and Benfey, 1997; Péret et al., 2009). In this process, auxin has been shown to play a central role, controlling each step of LRP development (Dubrovsky et al., 2008; Benková et al., 2003; Bhalerao et al., 2002; Fukaki et al., 2007; Fukaki and Tasaka, 2009; Ljung et al., 2001). Within the LRP, an auxin gradient is progressively established with a maximum at the apex that is required for proper growth of the LRP (Benková et al., 2003). A “fountain model” was proposed for auxin flow in LRP, postulating that auxin coming from the primary root enters LRP through the inner cell layers (acropetal flow), reaches the apex where it accumulates, and is redirected to the main root via the outer cell layers (basipetal flow; Benková et al., 2003).
Within this general scheme, we recently proposed a model for the mechanism by which NRT1.1 regulates LR growth in response to NO3− (Krouk et al., 2010; Bouguyon et al., 2015). Briefly, using heterologous expression systems, we found that NRT1.1 not only transports NO3− but also auxin. The auxin influx activity of NRT1.1 is, however, inhibited by NO3− in a concentration-dependent manner. On the other hand, we observed that NRT1.1 acts as a repressor of LRP emergence at low NO3− availability, and that this repressive effect is associated with a strong NRT1.1-dependent inhibition of the DR5 auxin reporter activity in the LRPs. Taking into account the perfect match between the tissue localization of the NRT1.1 protein in the LRPs with that of the basipetal transport route for auxin, we postulated that the role of NRT1.1 is to facilitate auxin basipetal transport out of LRPs in the absence of NO3− or at low NO3− concentration, thus lowering auxin levels in these organs and consequently slowing down their outgrowth. At high external NO3− concentration (>∼0.5 mm), the NRT1.1-dependent auxin transport activity is inhibited, reducing the basipetal export auxin out of the LRPs. Auxin then accumulates in LRPs, stimulating their development. This model was further strengthened by the fact that point mutations in the NRT1.1 protein (T101A or P492L) that markedly reduce or suppress NRT1.1-dependent auxin transport lead to the same alterations of the LR growth as those recorded in NRT1.1 null (chl1) mutants (Bouguyon et al., 2015).
However, this model suffers from a major paradox that requires clarification to understand how NRT1.1 precisely controls LR growth in response to NO3−. Indeed, there is a strong discrepancy between regulation and function of NRT1.1 in LRPs, as NRT1.1 gene is strongly inducible by NO3− both at whole root level (Tsay et al., 1993; Wang et al., 1998; Okamoto et al., 2003) and in LRPs (Guo et al., 2001, 2002; Remans et al., 2006; Mounier et al., 2014), whereas it has the most striking effect on LR growth in the absence of NO3− (Krouk et al., 2010; Bouguyon et al., 2015).
In this work, we thus aimed at investigating the spatiotemporal pattern of NRT1.1 protein expression in LRPs to determine whether this pattern can reconcile regulation and function of this protein during LR development. As shown by the results detailed below, our work establishes that NRT1.1 is subject to a complex posttranscriptional regulation at both mRNA and protein levels, which restores the consistency between expression and function of the protein. In particular, we found that despite the fact that NRT1.1 is a NO3−-inducible gene, the expression of the NRT1.1 protein is markedly downregulated by NO3− in LRPs, but not in other root tissues such as epidermis and cortex. This is fully consistent with the hypothesis that NRT1.1 plays a role in controlling growth of LRP only in the absence or at low concentration of NO3−, whereas in other root tissues, it acts as a sensor in response to NO3− provision.
RESULTS
NO3− Prevents Expression of the NRT1.1 Protein in LRPs
In our previous report (Krouk et al., 2010), we only studied the tissue localization of NRT1.1 in emerged LRPs or young LRs. Therefore, to dissect the role of NRT1.1 during LRP development, we investigated its expression pattern at the protein level in LRPs at various developmental stages until emergence in seedlings grown in presence or absence of NO3− or with Gln as an alternative N source. In addition to the pNRT1.1::NRT1.1-GFP lines described previously (Krouk et al., 2010), we generated additional transgenic lines in chl1-5 background expressing a pNRT1.1::NRT1.1-GFPLoop where the GFP has been introduced in the cytosolic loop (E269) instead of the C-terminal part of NRT1.1, in order to make sure that the insertion site of the GFP does not affect its expression pattern. The pNRT1.1::NRT1.1-GFPLoop transgene fully complements both root development phenotype (Supplemental Fig. S1) and chlorate resistance (data not shown) displayed by the chl1-5 mutant, showing that it encodes a functional protein.
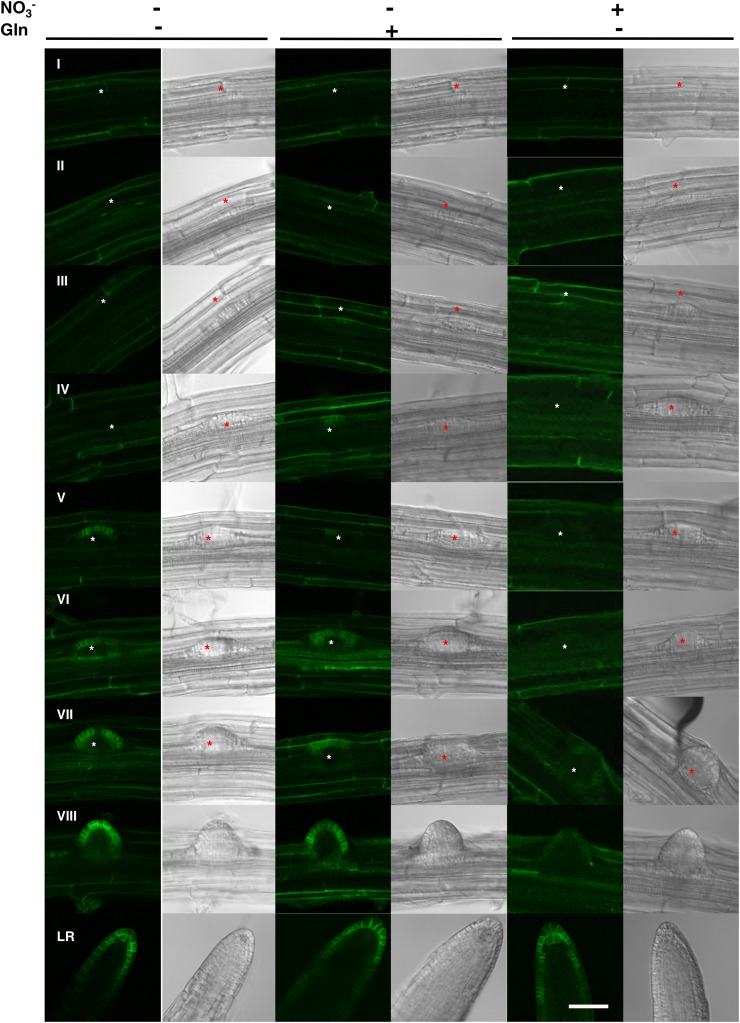
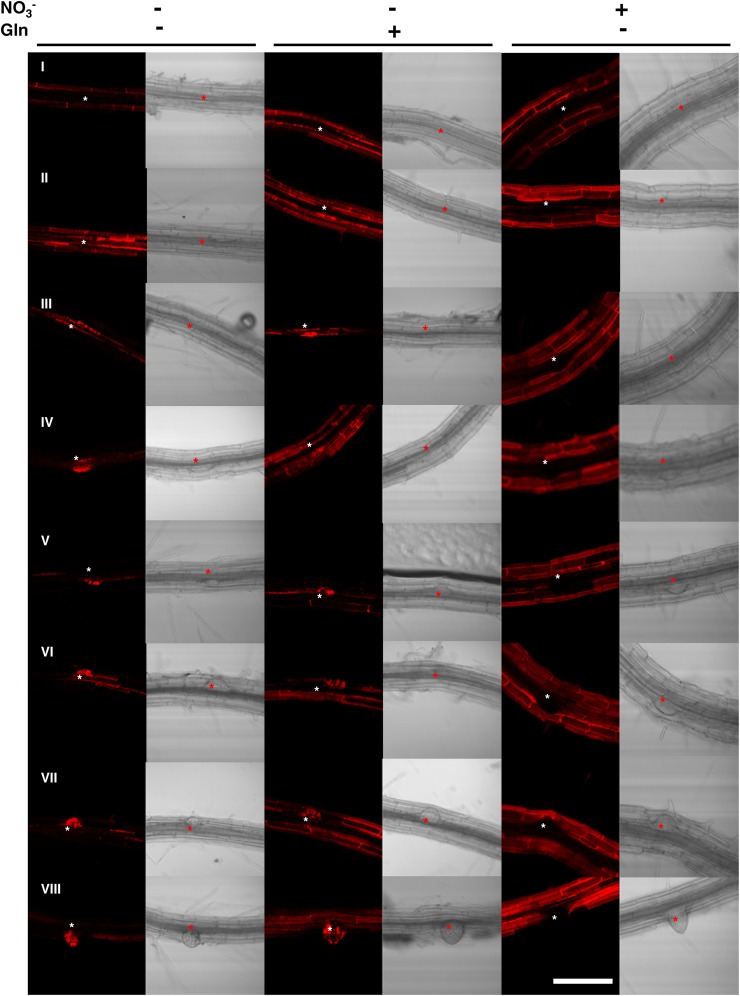
When seedlings of one pNRT1.1::NRT1.1-GFPLoop line (pNRT1.1::NRT1.1-GFPLoop16-3), were grown on NO3−-supplemented medium (Fig. 1, right), NRT1.1-GFP signal was surprisingly only observed in LRPs just before emergence (stage VII) and in newly emerged LRs. When visible, the signal appeared in the outermost cell layer of the LRP tip and remains restricted in this cell layer after emergence. This localization pattern is consistent with the observations of Krouk et al., (2010). Unexpectedly, when seedlings were cultivated on N-free medium, a strong NRT1.1-GFP signal was observed from a much earlier LRP developmental stage as compared to what was recorded on NO3− (Fig. 1, left). Indeed, GFP signal appeared from stage IV onward (Fig. 1, left, IV to LR). The same expression pattern was recorded in Gln-fed plants, showing that upregulation of NRT1.1-GFP expression at early LRP development stages is not due to N deficiency but specifically to the lack of NO3− (Fig. 1, middle). Even at the emergence stage, the GFP signal detected in N-starved or Gln-fed plants appeared to be much stronger than in NO3−-fed plants. Only in elongating LRs, the NRT1.1-GFP signal appeared to become less dependent of the N treatment. Interestingly, in the absence of NO3−, even if NRT1.1-GFP signal was observed at earlier stages during LRP development, it remained always localized in the outermost cell layer of the LRP as it is the case in presence of NO3−. Altogether, these data are consistent with the hypothesis that NO3− per se acts as a repressor of NRT1.1 protein accumulation in LRPs, postponing its expression almost until emergence.
Figure 1.
NRT1.1-GFP protein accumulation in LRPs is repressed by NO3−. NRT1.1-GFP localization in LRPs of pNRT1.1::NRT1.1-GFPLoop16-3 plants grown for 8 d on basal medium without N (left) or supplemented with 0.5 mm Gln (middle) or 1 mm NO3− (right). Each line corresponds to a LRP developmental stage from initiation (I) to fully emerged LR as described by Malamy and Benfey (1997). The asterisk visualizes the location of the LRP. The pictures shown are representative of >20 primordia from >10 plants of three independent experiments. White bar represents 50 μm.
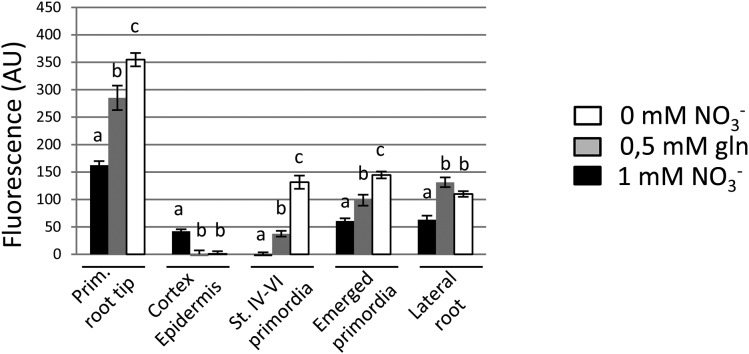
To make sure that these data are actually representative of the regulation of NRT1.1 and to determine if the repressive effect of NO3− on NRT1.1 expression is specific of LRPs or is a more general pattern in the root, we quantified NRT1.1-GFP signals at different locations in the root (i.e. primary root tip, cortex, and epidermis of the primary root, LRPs at two different developmental stages, and elongating LRs) in five independent lines expressing either NRT1.1Loop or NRT1.1Cter translational fusions (Fig. 2; Supplemental Figs. S2, S3, and S4). These data confirmed that NO3− provision represses NRT1.1-GFP expression in the LRPs, as compared to either Gln supply or N deprivation. Furthermore, this was found to be also true at the primary root tip. However, this is not a general pattern, first because N treatments did not significantly affect NRT1.1-GFP expression in elongating LRs and second because NO3− strongly stimulated NRT1.1-GFP expression in epidermis and cortex of the primary root. Altogether, these observations show a marked differential regulation of NRT1.1 protein expression by NO3−, depending on the root tissues.
Figure 2.
NRT1.1-GFP accumulation is differentially regulated by N in root tissues. Fluorescence quantification in different root tissues of 8-d-old plants form pNRT1.1::NRT1.1-GFPLoop16-3 transgenic line expressing NRT1.1-GFP protein fusion in the absence or presence of NO3− (1 mm) or 0.5 mm Gln. Fluorescence was quantified respectively in primary root tip, cortex, and epidermis of primary root (above the first emerged primordium), unemerged primordia stage IV to VI, emerged primordia, and growing LRs (±0.5 mm). Values are the mean of eight to 12 plants from two independent experiments and are normalized to Col (error bars are se). For each tissue, data were analyzed through one-way ANOVA, followed by a Tukey’s test as a post hoc analysis, and are statistically significant at P < 0.05.
NRT1.1 mRNA Accumulation Is Posttranscriptionally Controlled by NO3−
The repressive effect of NO3− on NRT1.1-GFP protein accumulation during almost the whole pre-emergence LRP development is a strongly intriguing observation because it is at odds with the numerous reports showing that the NRT1.1 gene expression is induced by NO3− (Tsay et al., 1993; Wang et al., 1998; Okamoto et al., 2003), even in LRPs from the earliest stages of development where pNRT1.1 was shown to be activated by NO3− (Guo et al., 2001, 2002; Remans et al., 2006; Krouk et al., 2010; Mounier et al., 2014). This suggests the occurrence of posttranscriptional regulatory mechanisms uncoupling protein expression from promoter activity.
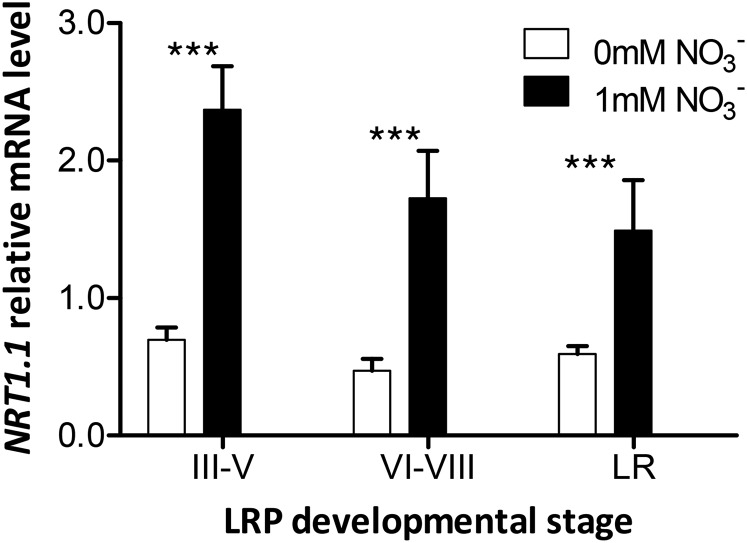
To delineate at which level the potential regulation occurs, we isolated LRP-enriched samples from roots of plants grown with or without NO3− using the gravistimulation method (Lucas et al., 2008) and assessed NRT1.1 mRNA accumulation by quantitative real-time PCR (qRT-PCR). Gravistimulated root fragments were surgically harvested after three time points to obtain (1) early LRPs (32 h or 40 h for seedlings grown on NO3−-supplemented or NO3−-depleted medium, respectively), (2) late LRPs (40 h or 48 h), and (3) fully emerged LRPs (48 h or 64 h). In our hands, gravistimulation succeeded in inducing LRP initiation at the bending site in more than 95% of seedlings whatever the medium (data not shown), with over 90% of LRPs between stages III and V, VI and VIII, and fully emerged for the previously indicated time points. Relative NRT1.1 mRNA accumulation was much higher (between three and five times) in LRP-enriched samples from seedlings grown on NO3− than on NO3−-free medium (Fig. 3). In addition, in presence of NO3−, NRT1.1 mRNA accumulation was higher in early developmental stages than after emergence.
Figure 3.
NRT1.1 mRNA accumulation is stimulated by NO3− in LRP. LRP-enriched root samples were harvested from 7-d-old seedlings grown on basal medium containing no N or 1 mm NO3− before gravistimulation by rotating the plates at 90°. The part of the primary root containing the LRP (curved 4-mm segment) was surgically separated 28 h, 32 h, and 40 h after gravistimulation on NO3−-containing medium and 32 h, 40 h, and 48 h after gravistimulation on N-free medium (corresponding respectively to stages III–V, VI–VIII, and emerged LR) and stored in liquid nitrogen. The NRT1.1 mRNA level was measured by qRT-PCR and normalized using CLATHRIN as an internal standard. Differences with NO3− treatments are statistically significant at ***P < 0.001.
These data showing opposite responses of NRT1.1 mRNA and NRT1.1 protein to NO3− in LRPs suggest that this ion has a specific negative effect on NRT1.1 protein synthesis and/or stability. However, even if the gravistimulation method is highly efficient for harvesting LRP-enriched samples, it is not possible to rule out the hypothesis that part of the NRT1.1 mRNA quantified in the above experiments was extracted from other tissues than LRPs. Therefore, to unambiguously suppress the transcriptional level of regulation by NO3−, we used transgenic lines expressing an mCherry-tagged version of NRT1.1 under control of the estrogen receptor-based transactivator XVE inducible expression system (Bouguyon et al., 2015).
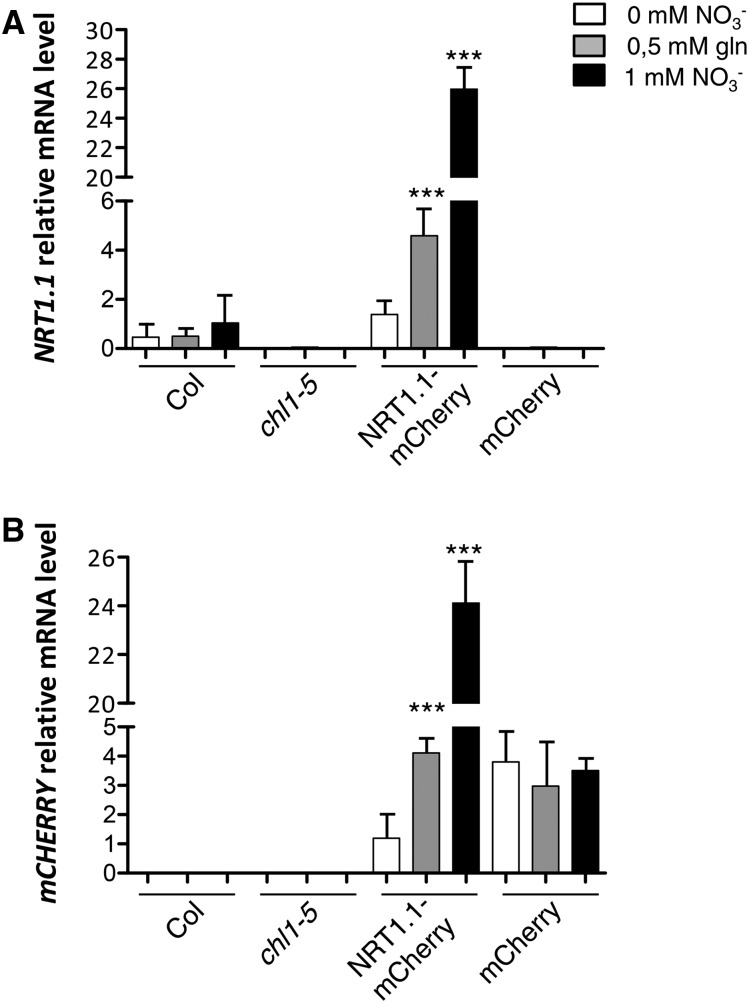
In our growth culture conditions, as previously reported by Zuo et al., (2000), estradiol concentrations up to 5 µm did not affect RSA (Supplemental Fig. S5). As expected, upon induction, NRT1.1-mCherry mRNA level was dramatically increased up to 25 times in transgenic lines as compared to NRT1.1 expression in ecotype Columbia (Col; Fig. 4), whereas no detectable expression was recorded in either uninduced seedlings (data not shown), or in chl1-5 KO mutant or in control transgenic lines expressing the mCherry alone (Fig. 4A). Surprisingly, NRT1.1-mCherry mRNA accumulation was dramatically decreased when NO3− was absent from the culture medium and only very partially restored in presence of Gln. This response cannot be explained by an alteration of the promoter activity due to the N source because no difference in mCherry transcript accumulation was recorded in transgenic control line (Fig. 4B). Taken together, these results demonstrate that NRT1.1 transcript accumulation is strongly posttranscriptionally stimulated by NO3− and to a much lower extent by reduced N metabolites.
Figure 4.
NRT1.1::mCherry mRNA accumulation is regulated by NO3− at the posttranscriptional level. NRT1.1, NRT1.1::mCherry, and mCherry mRNA levels measured in roots of seedlings grown on basal medium without N (white boxes) or supplemented with 0.5 mm Gln (gray boxes) or 1 mm NO3− (black boxes) for 10 d and transferred on the same media supplemented with 5 µm of β-estradiol for 48 h. The transcript accumulation levels were measured using primers specific of NRT1.1 (A) or mCherry (B) CDS by qRT-PCR and normalized using CLATHRIN as an internal standard. Col, chl1-5, NRT1.1::mCherry, and mCherry, respectively, correspond to Col wild-type genotype, chl1-5 null NRT1.1 mutant, transgenic line expressing the NRT1.1-mCherry construct under the control of the estradiol-inducible promoter in chl1-5 background (pNRT1.1::NRT1.1-mcherry chl1-5 7-4), and transgenic line expressing the mCherry construct under the control of the estradiol inducible promoter in chl1-5 background. Differences with N treatments are statistically significant at ***P < 0.001.
NRT1.1 Protein Expression in LRPs Is Controlled by a NO3−-Dependent Signal
In control transgenic plants grown on NO3−, mCherry protein was homogeneously distributed in all root tissues including LRPs (Supplemental Fig. S6). As expected, very similar expression patterns were observed in seedlings grown in absence of N or on Gln (data not shown). In contrast, NRT1.1-mCherry expression pattern strongly differed between tissues and depending on the N source (Fig. 5; Supplemental Fig. S7). In NO3−-fed plants, NRT1.1-mCherry expression was restricted to outermost cell layers, epidermis, cortex, and endodermis with the highest expression level in cortex (Fig. 5, right). Strikingly, NRT1.1-mCherry was never observed in LRPs. On N-free medium, expression is restricted to internal cell layers and LRPs into which NRT1.1-mCherry signal was particularly strong as early as stages III and IV (Fig. 5, right). In Gln-supplemented medium, the observed NRT1.1-mCherry signal was intermediate both in terms of expression pattern and signal intensity, as compared to seedlings grown on N-free and NO3−-supplemented conditions (Fig. 5, middle). Similar expression patterns were observed in four independent lines expressing NRT1.1-mCherry (Supplemental Fig. S7). As on NO3−-free medium, when seedlings were grown on Gln, NRT1.1-mCherry was detected in young LRPs as early as stage III and onward. The overall signal intensity in LRPs on NO3−-containing medium appears to be reduced compared to seedlings grown on N-free medium, and patchy expression is detected as early as stage V. Collectively, the data obtained with the various NRT1.1- mCherry lines (Fig. 5; Supplemental Fig. S7) are fully consistent with those obtained with NRT1.1-GFP (Figs. 1 and 2), and show that NO3− acts as a repressor of NRT1.1 protein accumulation in LRPs, independently of any transcriptional regulation. Furthermore, they also confirm that this repressive role of NO3− on NRT1.1 protein expression is tissue specific, because NRT1.1-mCherry signals were at the opposite strongly induced by NO3− in epidermis and cortex, as it was the case for NRT1.1-GFP.
Figure 5.
NRT1.1-mCherry protein accumulation in LRPs is repressed by NO3−. NRT1.1-mCherry localization in LRP of the transgenic line expressing NRT1.1-mCherry under the control of a β-estradiol inducible promoter (pNRT1.1::NRT1.1-mcherry chl1-5 7-4) grown for 10 d on basal medium without N (left) or supplemented with 0.5 mm Gln (middle) or 1 mm NO3− (right), and transferred on the same media supplemented with 5 µm of β-estradiol for 48 h. Each line corresponds to a LRP developmental stage from initiation (I) to emergence (VIII) as described by Malamy and Benfey (1997). The asterisk visualizes the location of the LRP. The pictures shown are representative of >20 primordia from >10 plants of two independent experiments. White bar represents 50 μm.
Repression of NRT1.1 Protein Accumulation in LRPs by NO3− Promotes Local Auxin Accumulation and Favors LRP Outgrowth
In our previous reports (Krouk et al., 2010; Bouguyon et al., 2015), we proposed that NRT1.1 controls LRP emergence by modulating local auxin gradients in these organs, resulting in an enhanced auxin accumulation, and thus a stimulated emergence, in presence of NO3−. However, we also reported that neither NO3− nor NRT1.1 affects LRP initiation or auxin accumulation in very young LRPs. Therefore, the finding that NO3− regulates NRT1.1 protein expression in LRPs prompted us to investigate in more detail the temporal pattern of auxin accumulation/signaling in LRPs in response to the N source, using the auxin biosensor DR5::VENUS (Brunoud et al., 2012) in both wild type and chl1-5 mutant backgrounds. This recently developed marker was used because its nuclear localization offers a cellular resolution and more accurate comparison between tissues or plant culture conditions.
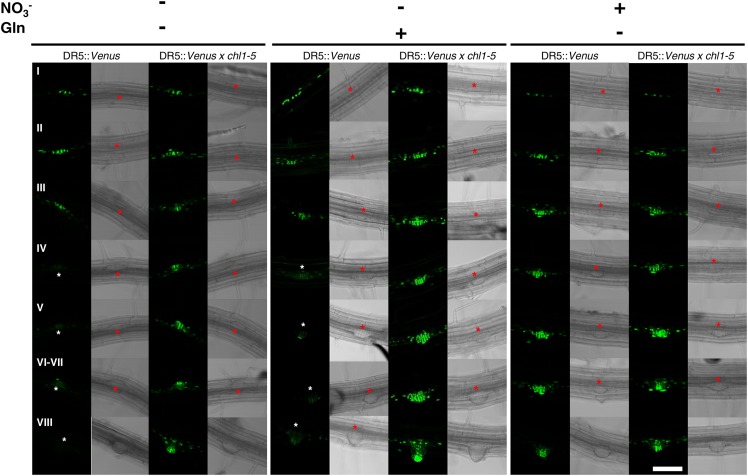
In seedlings grown on NO3−, DR5 activity was observed as early as the first division of pericycle cells and remained very high during all LRP developmental stages, regardless of the wild-type or chl1-5 genetic background (Fig. 6, right). In seedlings grown on N-free medium, if no differences were observed between wild-type and chl1-5 for DR5::VENUS expression at early LRP developmental stages (stages I–III; Figure 6, left), marked differences appeared at later stages. Indeed, DR5 activity became barely detectable in all LRPs from stage IV and onward for wild-type seedlings, whereas it remained high in chl1-5 mutant background (Fig. 6, left). A similar pattern of DR5::VENUS expression was observed in seedlings grown in presence of Gln—as compared to N-deprived (Fig. 6, central), although the expression level is generally slightly higher than in N-deprived plants.
Figure 6.
NRT1.1 represses DR5::VENUS expression in LRPs in NO3−-free medium. VENUS localization in LRP of wild-type and chl1-5 seedlings grown for 10 d on basal medium without N (left) or supplemented with 0.5 mm Gln (middle) or 1 mm NO3− (right). Each line corresponds to a LRP developmental stage from initiation (I) to emergence (VIII) as described by Malamy and Benfey (1997). The asterisk visualizes the location of the LRP. The pictures shown are representative of >20 primordia from >10 plants of two independent experiments. White bar represents 50 μm.
Taken together, the above data indicate that in LRPs, NRT1.1 and VENUS protein expression display striking mutually exclusive expression patterns. Indeed, VENUS expression is high when NRT1.1 protein is not expressed, which occurs in three situations: (1) in the presence of NO3− at all stages of development in both wild-type and chl1-5 backgrounds, (2) in the absence of NO3− at early stages of development in both wild-type and chl1-5 backgrounds, and (3) in the absence of NO3− at late stages of development in chl1-5 background. Conversely, VENUS signal vanishes when NRT1.1 starts to be expressed, i.e. in the absence of NO3− at late stages of development in wild-type background. As this mutual exclusion of NRT1.1 and VENUS in LRPs cannot be explained by a repressive effect of auxin on NRT1.1 expression (Supplemental Fig. S8), it fully supports our previous proposal that NRT1.1 prevents auxin accumulation in LRPs in response to the absence, or the low availability, of NO3− (Krouk et al., 2010).
DISCUSSION
Since the first reports suggesting that NRT1.1 may have another function in addition to NO3− transport (Guo et al., 2001), which could be NO3− signaling (Muños et al., 2004), many studies have strengthened the conclusion that this protein is a major component of the NO3− sensing system in Arabidopsis (Arabidopsis thaliana; Krouk et al., 2006; Remans et al., 2006; Walch-Liu and Forde 2008; Ho et al., 2009; Wang et al., 2009; Krouk et al., 2010; Hachiya et al., 2011; Mounier et al., 2014; Bouguyon et al., 2015; Riveras et al., 2015). These studies have also highlighted the complexity of the sensing function of NRT1.1, which triggers a wide range of different physiological and developmental responses, under very different conditions of N nutrition. This is strikingly illustrated by the observations that NRT1.1 has a predominant action in the absence of NO3− to regulate LRP emergence, whereas at the opposite, it induces the expression of NO3− transport and assimilation genes in response to NO3−. We hypothesized that this functional complexity relies on several independent NO3− sensing/signaling mechanisms at the level of NRT1.1 itself (Bouguyon et al., 2015). Accordingly, the mechanism recently proposed to account for the NRT1.1-dependent NO3− induction of gene expression, i.e. phospholipase C and calcium-dependent transduction pathway (Riveras et al., 2015), is markedly different from that explaining the action of NRT1.1 on NO3− regulation of LRP emergence, i.e. NO3−-regulated auxin transport activity (Krouk et al., 2010).
Our work now provides evidence that the sensing/signaling function of NRT1.1 also involves an additional level of complexity, through powerful tissue-specific posttranscriptional regulatory mechanisms, which ultimately determine where and when the NRT1.1 protein is present at the plasma membrane. Our data on the posttranscriptional regulation of NRT1.1 also reconciles many separate and sometimes contradictory observations into a comprehensive model of how NRT1.1 acts to modulate LRP development.
As mentioned earlier, the role of NRT1.1 in regulating LRP development only in the absence of NO3−, or at low NO3− availability, seemed in marked contradiction with the fact that NRT1.1 gene expression is strongly induced by NO3− (Tsay et al., 1993; Wang et al., 1998; Okamoto et al., 2003). However, the estradiol-inducible system that has been used to get rid of the NO3− transcriptional regulation unraveled a much more complex N-dependent regulation of NRT1.1 transcript accumulation. Whereas, as reported by Zuo et al., (2000), a high and constitutive mCherry mRNA level is observed in induced transgenic lines expressing the unfused mCherry protein, regardless of the growth medium, the accumulation of NRT1.1-mCherry mRNA is markedly increased by NO3−. This suggests that NO3− may stabilize the native NRT1.1 mRNA, in addition to promoting its synthesis at the transcriptional level. The underlying mechanism remains elusive. Indeed, as the NRT1.1-mCherry transgene contains NRT1.1 coding DNA sequence (CDS) CDS only, the regulatory sequences involved in posttranscriptional control of NRT1.1 mRNA accumulation do not rely on either untranslated regions or introns. Moreover, based on an in silico screen, no miRNA target sequence was identified in NRT1.1 CDS, suggesting that regulation of NRT1.1 mRNA accumulation does not involve specific targeting by known noncoding RNAs.
The regulation of NRT1.1 expression appears even more complex when considering the protein level, especially in LRPs. In these organs, NRT1.1 promoter activity is very intense (Guo et al., 2001, 2002; Remans et al., 2006; Mounier et al., 2014), and NRT1.1 mRNA accumulation is not only very high at all stages of development but is also stimulated by NO3− supply (Fig. 3). Most importantly, our work reveals that a posttranscriptional regulation occurs at the protein level to ultimately yield to the opposite response, i.e. repression of NRT1.1 by NO3−. As a matter of fact, we were not able to detect any significant NRT1.1-GFP or NRT1.1-mCherry protein expression in LRPs prior emergence in NO3−-fed plants, whereas on NO3−-free medium (N-free or Gln supplemented medium), the protein starts to accumulate much earlier (from stages III and IV onward). Interestingly, such repression of NRT1.1 protein expression by NO3− is not seen in all root tissues. It was found to some extent at the primary root tip (Fig. 2; Supplemental Figs. S3, S4, and S7), but neither in elongating laterals roots, nor in epidermis and cortex where NRT1.1-GFP or NTR1.1- mCherry signal is, on the contrary, highest in presence of NO3−.
It is not possible from our data to determine whether the posttranscriptional inhibitory effect of NO3− on NRT1.1 protein expression in LRPs is associated with inhibited synthesis and/or increased degradation. Translational regulation of gene expression has been reported in root tissues in response to stresses. For instance, translational state of mRNAs was found to be dramatically disturbed by a variety of suboptimal environmental conditions including water stress (Kawaguchi et al., 2004), Suc starvation (Nicolaï et al., 2006), hypoxia (Mustroph et al., 2009), saline stress (Matsuura et al., 2010), cadmium (Sormani et al., 2011), light (Juntawong and Bailey-Serres, 2012), or heat stress (Yángüez et al., 2013). However, in response to stress, most genes are underrepresented in translatome, with the noticeable exception of those involved in stress response and sensing/signaling (Mustroph et al., 2009; Juntawong and Bailey-Serres, 2012). Thus, the fact that NRT1.1 protein is more expressed in response to N starvation in LRPs would be consistent with its translational upregulation as a sensing/signaling protein. Alternatively, an interesting parallel can be made with the results recently reported by Jabnoune et al., (2013) on the regulation of PHO1;2 in rice by a cis-natural antisense transcript. To date, no NRT1.1 cis-natural antisense transcript has been identified. However, Luo et al., (2013) based on RNA seq analysis, identified several tags that may indicate that a similar mechanism is also involved in NRT1.1 translational regulation.
Remarkably, the spatiotemporal pattern of NRT1.1 protein abundance in LRPs is fully consistent with those of the NO3− regulation of local DR5 activity and of LRP development. This allows to resolve another previous paradox, because the changes in auxin accumulation in LRPs in response to NRT1.1 mutation were reported to occur at a relatively late stage of LRP development (Krouk et al., 2010) and were thus far from being consistent with the temporal pattern of the NRT1.1 gene expression that is activated as soon as the LRP are initiated (Guo et al., 2001). First, the lack of NRT1.1-GFP or NRT1.1-mCherry signal in LRPs prior to stages III and IV, regardless of the N source, is consistent with the observation that, in our conditions, NO3− affects neither DR5 activity in LRPs nor growth of these LRPs at early stages of their development (Krouk et al., 2010). Second, the earlier and stronger NRT1.1 protein expression in LRPs after stage IV of plants grown without NO3− (as compared with NO3−-fed plants) is in agreement with a predominant role of NRT1.1 in regulating LRP outgrowth in the absence or at low availability of NO3− (Krouk et al., 2010; Bouguyon et al., 2015). Moreover, this work further supports our previous proposal that NRT1.1 exerts a repressive effect on LRP development by preventing auxin accumulation in these organs. First, we showed that NRT1.1-GFP or NRT1.1-mCherry expression patterns and DR5::VENUS expression pattern in LRPs are mutually exclusive (compare Figs. 1, 5, and 6), and second that DR5::VENUS expression is always high in chl1-5 mutant background (Fig. 6). The finding that NO3− represses both NRT1.1-GFP and NRT1.1-mCherry protein accumulation in LRPs indicates that NO3− promotes auxin accumulation in LRPs and thus their development, through a dual action: first by inhibiting the auxin transport capacity of NRT1.1 as shown previously (Krouk et al., 2010), and second by preventing NRT1.1 protein expression in LRPs. These data allow to refine our previous model (Krouk et al., 2010) and highlight stages III and IV as a key checkpoint at which NRT1.1 protein begins to regulate LRP outgrowth in response to NO3− availability. At low NO3−, NRT1.1 protein will start to be expressed and to prevent auxin accumulation in LRPs, thus hampering both the normal progression of the LRP developmental program. At high NO3−, NRT1.1 expression is largely postponed until emergence and its auxin transport activity is repressed. As a consequence, auxin accumulates in LRPs, therefore stimulating LRP outgrowth and the modifications of the overlying tissues required for this outgrowth.
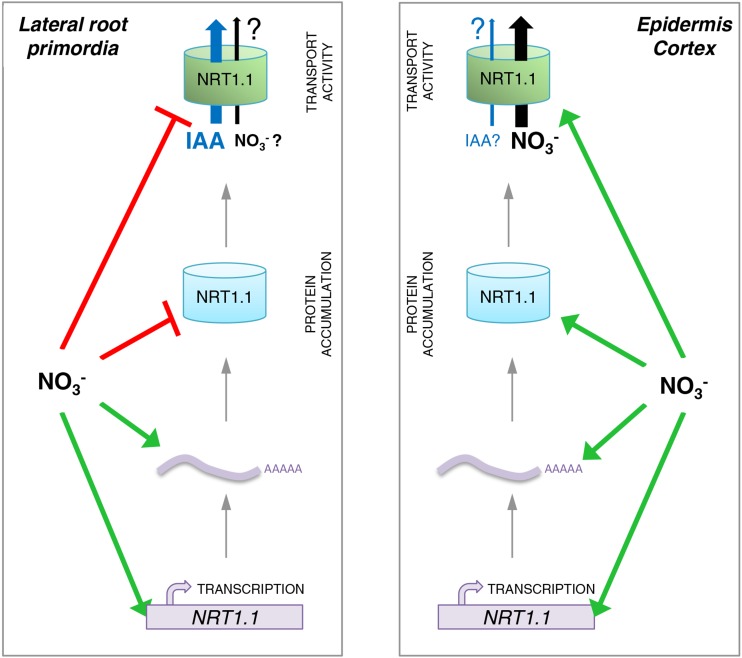
Overall, our work suggests that although NRT1.1 mRNA accumulation is always under transcriptional and posttranscriptional stimulation by NO3−, the posttranscriptional regulation at the protein level is crucial for coordinating the presence of NRT1.1 with its specific signaling function, depending on the tissular context (Fig. 7). In LRPs, this regulation favors NRT1.1 expression under low NO3− or NO3−-free conditions, which is consistent with a predominant role of NRT1.1 as an auxin carrier controlling LRP outgrowth under the same conditions (Krouk et al., 2010; Mounier et al., 2014; Bouguyon et al., 2015). In contrast, NRT1.1 protein expression is promoted by NO3− in epidermis and cortex, which fits with the role of this protein as an influx NO3− transporter, and a sensor activating NO3− signaling pathways unrelated to LR development such as those leading to the induction of NO3− transport and assimilation genes (Ho et al., 2009; Wang et al., 2009; Riveras et al., 2015). The functional complexity of NRT1.1 has been recently highlighted (Bouguyon et al., 2015). Our data show that this complexity not only relies on a high functional versatility of the protein, but also on complex tissue-specific regulatory mechanisms ensuring that this transceptor is appropriately expressed to trigger a response relevant to the tissue and the experimental conditions.
Figure 7.
Schematic model for NO3−-dependent transcriptional and posttranscriptional regulations of NRT1.1 in LRPs or other root tissues. The model depicts four different levels of regulation of NRT1.1 expression by NO3−, depending on the root tissues: promoter activity, mRNA accumulation, protein accumulation, and transport activity. This model postulates that NO3− stimulates NRT1.1 promoter activity (Guo et al., 2001, 2002; Remans et al., 2006; Mounier et al., 2014) and increases mRNA stability (this work) inducing an overaccumulation of mRNA both in LRPs (left) and in epidermis and cortex of the primary root (right) but differentially affects protein accumulation and activity between these tissues. On the one hand, NO3− represses NRT1.1 protein accumulation (this work) and NRT1.1 auxin transport activity (Krouk et al., 2010) in LRPs, whereas it stimulates NRT1.1 protein accumulation in epidermis and cortex (this work). These differential regulations are consistent with different functions of NRT1.1 between LRPs and other root tissues. In LRPs, NRT1.1 mostly acts in response to lack of NO3−, as an auxin transporter preventing LR emergence (Krouk et al., 2010). Accordingly, NO3− stimulates LR emergence by repressing NRT1.1 protein accumulation and auxin transport activity, which leads to auxin accumulation in the primordia that in turn accelerates their development. In epidermis or cortex of the primary root (right), NRT1.1 mostly acts in response to NO3− supply, as an influx transporter involved in NO3− uptake (Tsay et al., 1993) and as a NO3− sensor, in particular triggering expression of other genes of the NO3− assimilation pathway (Ho et al., 2009; Wang et al., 2009). Accordingly, NO3− not only stimulates NRT1.1 mRNA accumulation at the transcriptional and posttranscriptional levels, but also favors protein accumulation and NO3− uptake from the external medium.
MATERIALS AND METHODS
Plant Material
The Arabidopsis (Arabidopsis thaliana) accession used in this study was Col-0. The pDR5::VENUS (Brunoud et al., 2012) tagged line was used and crossed with the chl1-5 mutant (Tsay et al., 1993). Homozygous plants for both chl1-5 mutation and tagged line were screened on F2 and F3 offsprings by PCR for chl1-5 deletion as described by Mounier et al., (2014) and fluorescence.
The NRT1.1 tagged lines were pNRT1.1::NRT1.1-GFP (Krouk et al., 2010) in chl1-5 background and NRT1.1-GFPLoop, which was obtained as described below. The pNRT1.1::NRT1.1 (6.184-kb genomic fragment, including the 1.533-kb 5′ untranslated region and promoting sequence upstream of the ATG the genomic sequence of NRT1-1 and 427 bp downstream of the stop codon) was amplified by PCR (P1: tttgttctcgctcttccaca and P2: tcgagagacaattgagccagt) and cloned in pENTR/D/TOPO entry vector (Invitrogen) and then introduced in the pGWB1 binary vector (no promoter, no tag) obtained from Tsuyoshi Nakagawa (Research Institute of Molecular Genetics, Shimane University, Matsue, Japan) by LR recombination (Invitrogen). The GFP coding sequence (738 bp) has been amplified using XhoI restriction site containing primers (P3: acgctcgagatggtgagcaagggcgagg and P4: acgctcgagcttgtacagctcgtccatgc) and cloned by restriction in the XhoI restriction site of pNRT1.1::NRT1.1(E269) cloned in the pGWB1 vector. All steps were performed according to the manufacturer's instructions. Prior to transformation of Agrobacterium, the expression construct was sequenced. Binary vector containing the GFP fusion construct was introduced into Agrobacterium tumefaciens strain GV3101. Arabidopsis chl1-5 mutant plants were transformed by dipping the flowers in the presence of Silwet L77 (Clough and Bent, 1998). Transgenic seedlings were selected on a medium containing 30 mg L−1 of hygromycin. For further analyses, T1 segregation ratios were analyzed to select transformants with one T-DNA insertion and to isolate T3-homozygous plants. Functionality of the construct was tested by restoring chlorate sensitivity (data not shown) and wild-type LR growth of transgenic seedlings.
Transgenic Arabidopsis line expressing NRT1.1-mCherry under control of a β-estradiol inducible promoter in chl1-5 mutant and Col background were obtained as described by Bouguyon et al. (2015; Supplementary Material and Methods). The pER8 binary vector (Zuo et al., 2000) was used as backbone. Functionality of the construct was tested by restoring chlorate sensitivity (data not shown), wild-type LR growth of transgenic seedlings and expressing mCherry in chl1-5 background.
Plant Culture and Root Growth Analysis
Seedlings were grown and root system architecture was analyzed as described by Krouk et al., (2010). Basal culture medium without N was supplemented with the appropriate concentration of KNO3 or Gln for each experiment as described in figure legends. Sterile β-estradiol water diluted solutions was mixed to tempered (60°C) culture medium to reach 5 µm final concentration.
For sampling of LRP-enriched root portions, LR induction was performed on 7-d-old seedlings by rotating the plates at 90°. The part of the primary root containing the LRP (curved 4-mm segment) was surgically separated and stored in liquid nitrogen 28, 32, and 40 h after bending on NO3−-containing medium and 32, 40, and 48 h after bending on NO3−-free medium (corresponding respectively to stages III–V, VI–VIII, and emerged). β-Estradiol (Sigma) stock solution was dissolved in dimethyl sulfoxide.
RNA Analysis
Root samples were frozen in liquid nitrogen and disrupted for 1 min at 30 oscillations s−1 in a Retch mixer mill MM301 homogenizer. Total RNA was extracted using TRIzol reagent (Invitrogen), DNase treated (Qiagen), and purified using an RNeasy MinElute Cleanup kit (Qiagen), and reverse transcription was achieved with 4 μg of RNAs with ThermoScript RT-PCR System for First-Strand cDNA Synthesis (Invitrogen) using an anchored oligo(dT)20 primer. Accumulation of transcript was measured by qRT-PCR (LightCycler 480; Roche Diagnostics) using the Light Cycler 480 SYBR Green 1 Master kit (Roche Diagnostics). All steps were performed according manufacturer’s recommendations. The gene expression was normalized using CLATHRIN as an internal standard. The specific primers used are summarized in Table I.
Table I. Primers used for qRT-PCR.
| Description | Arabidopsis Genome Initiative | Primer | Sequence (5′-3′) |
|---|---|---|---|
| CLATHRIN | AT4G24550 | Forward | AGCATACACTGCGTGCAAAG |
| Reverse | TCGCCTGTGTCACATATCTC | ||
| NRT1.1 | AT1G12110 | Forward | GCACATTGGCATTAGGCTTT |
| Reverse | CTCAATCCCCACCTCAGCTA | ||
| mCherry | Forward | CCCCGTAATGCAGAAGAAGA | |
| Reverse | TCCAACTTGATGTTGACGTTGT |
Confocal Microscopy
For confocal microscopy observations, seedlings were mounted on slides in osmosed water. The microscope was a LSM 510 Axiovert 200M with a Plan-Apochromat 20×/0.75 objective (Zeiss) under control of the LSM 510 software supplied by the constructor. GFP/VENUS was excited at 488 nm by an argon source and re-emitted light was filtered by a passing band of 505 to 530 nm. mCherry was excited with the 543-nm line of a helium-neon laser and detected via a 585-nm long-pass filter (red). Laser intensity was adjusted for each line, and pinhole was set at 1 unit, resolution at 1024 × 1024. GFP quantifications were done with an Axiovert 200M microscope (Zeiss) and coolsnapHQ camera (Photometrics Scientific) and images analyzed with the MetaFluor V7.7.8.0 software (Molecular Devices). GFP was excited with a HC-495 filter and detected via a 525/45 bandpass filter (green). mCherry quantifications were done with an Olympus BH2 microscope (Olympus) and Hamamatsu ORCA-ER camera (Hamamatsu, Japan) and images analyzed with ImageJ software (Schindelin et al.; 2012). mCherry was excited with a 510 to 550 filter and detected via a 590 long-pass filter. The data represent the mean pixel values in a ROI, to which were subtracted the background values measured in the part of the root not expressing reporter and the measures in the same tissues of GFP-negative plants or estradiol-uninduced lines.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. NRT1.1-GFPLoop fusion protein fully complements the chl1-5 root development phenotype.
Supplemental Figure S2. NRT1.1-GFP accumulation in LRPs is repressed by NO3−.
Supplemental Figure S3. NRT1.1-GFP accumulation is stimulated by NO3− in primary root epidermis and cortex but repressed in primary root tip.
Supplemental Figure S4. NRT1.1-GFP accumulation is differentially regulated by N in root tissues.
Supplemental Figure S5. Estradiol does not modify root system architecture.
Supplemental Figure S6. mCherry protein accumulation in roots of the chl1-5 mutant.
Supplemental Figure S7. NRT1.1-mCherry accumulation is differentially regulated by N in root tissues.
Supplemental Figure S8. NRT1.1-GFP and NRT1.1-mCherry accumulation is enhanced by auxin in LRPs.
Supplementary Material
Acknowledgments
We thank Carine Alcon for the help with analysis of confocal images, Xavier Dumont for assistance with Arabidopsis transformations, staff members of the Institut de Biologie Intégrative des Plantes for technical assistance with biological material culture, and students and trainees for assistance with laboratory work. Confocal observations were made at the Montpellier RIO Imaging facility.
Glossary
- LRP
lateral root primordium
- LR
lateral root
Footnotes
This work was supported by the Agropolis Foundation (RHIZOPOLIS project to A.G. and P.N., and RTRA 2009-2011 project to F.P.-W.), the Knowledge Biobase Economy European project (KBBE-005-002 Root enhancement for crop improvement to M.P. and P.N.), and the European EURoot project (FP7-KBBE-2011-5 to J.R., A.G., and P.N.).
References
- Bhalerao RP, Eklöf J, Ljung K, Marchant A, Bennett M, Sandberg G (2002) Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J 29: 325–332 [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Bouguyon E, Brun F, Meynard D, Kubeš M, Pervent M, Leran S, Lacombe B, Krouk G, Guiderdoni E, Zažímalová E, et al. (2015) Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat Plants 1: 15015. [DOI] [PubMed] [Google Scholar]
- Bouguyon E, Gojon A, Nacry P (2012) Nitrate sensing and signaling in plants. Semin Cell Dev Biol 23: 648–654 [DOI] [PubMed] [Google Scholar]
- Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, Beeckman T, Kepinski S, Traas J, Bennett MJ, et al. (2012) A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482: 103–106 [DOI] [PubMed] [Google Scholar]
- Burns IG. (1991) Short and long term effects of a change in the spatial distribution of nitrate in the root zone on N uptake, growth and root development of young lettuce plants. Plant Cell Environ 14: 21–23 [Google Scholar]
- Casimiro I, Beeckman T, Graham N, Bhalerao R, Zhang H, Casero P, Sandberg G, Bennett MJ (2003) Dissecting Arabidopsis lateral root development. Trends Plant Sci 8: 165–171 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crawford NM, Glass A (1998) Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci 10: 389–395 [Google Scholar]
- Drew MC. (1975) Comparison of effects of a localised supply of phosphate, nitrate, ammonium and potassium on growth of seminal root system and shoot in barley. New Phytol 75: 479–490 [Google Scholar]
- Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benková E (2008) Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc Natl Acad Sci USA 105: 8790–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG. (2002) Local and long-range signaling pathways regulating plant responses to nitrate. Annu Rev Plant Biol 53: 203–224 [DOI] [PubMed] [Google Scholar]
- Forde BG. (2014) Nitrogen signalling pathways shaping root system architecture: an update. Curr Opin Plant Biol 21: 30–36 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Okushima Y, Tasaka M (2007) Auxin-mediated lateral root formation in higher plants. Int Rev Cytol 256: 111–137 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tasaka M (2009) Hormone interactions during lateral root formation. Plant Mol Biol 69: 437–449 [DOI] [PubMed] [Google Scholar]
- Gansel X, Muños S, Tillard P, Gojon A (2001) Differential regulation of the NO3- and NH4+ transporter genes AtNrt2.1 and AtAmt1.1 in Arabidopsis: relation with long-distance and local controls by N status of the plant. Plant J 26: 143–155 [DOI] [PubMed] [Google Scholar]
- Gojon A, Krouk G, Perrine-Walker F, Laugier E (2011) Nitrate transceptor(s) in plants. J Exp Bot 62: 2299–2308 [DOI] [PubMed] [Google Scholar]
- Gojon A, Nacry P, Davidian JC (2009) Root uptake regulation: a central process for NPS homeostasis in plants. Curr Opin Plant Biol 12: 328–338 [DOI] [PubMed] [Google Scholar]
- Guo FQ, Wang R, Chen M, Crawford NM (2001) The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is activated and functions in nascent organ development during vegetative and reproductive growth. Plant Cell 13: 1761–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F-Q, Wang R, Crawford NM (2002) The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is regulated by auxin in both shoots and roots. J Exp Bot 53: 835–844 [DOI] [PubMed] [Google Scholar]
- Hachiya T, Mizokami Y, Miyata K, Tholen D, Watanabe CK, Noguchi K (2011) Evidence for a nitrate-independent function of the nitrate sensor NRT1.1 in Arabidopsis thaliana. J Plant Res 124: 425–430 [DOI] [PubMed] [Google Scholar]
- Hawkesford M, Horst W, Kichey T, Lambers H, Schjoerring J, Skrumsager Møller I, White P (2012) Functions of macronutrients. In Marschner P, ed, Marschner’s Mineral Nutrition of Higher Plants, Ed 3 Elsevier, Amsterdam, pp 135–189 [Google Scholar]
- Hermans C, Hammond JP, White PJ, Verbruggen N (2006) How do plants respond to nutrient shortage by biomass allocation? Trends Plant Sci 11: 610–617 [DOI] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF (2009) CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Jabnoune M, Secco D, Lecampion C, Robaglia C, Shu Q, Poirier Y (2013) A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. Plant Cell 25: 4166–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P, Bailey-Serres J (2012) Dynamic light regulation of translation status in Arabidopsis thaliana. Front Plant Sci 3: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R, Girke T, Bray EA, Bailey-Serres J (2004) Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J 38: 823–839 [DOI] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K, et al. (2010) Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell 18: 927–937 [DOI] [PubMed] [Google Scholar]
- Krouk G, Tillard P, Gojon A (2006) Regulation of the high-affinity NO3− uptake system by NRT1.1-mediated NO3− demand signaling in Arabidopsis. Plant Physiol 142: 1075–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejay L, Tillard P, Lepetit M, Olive Fd, Filleur S, Daniel-Vedele F, Gojon A (1999) Molecular and functional regulation of two NO3- uptake systems by N- and C-status of Arabidopsis plants. Plant J 18: 509–519 [DOI] [PubMed] [Google Scholar]
- Liu KH, Tsay YF (2003) Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J 22: 1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K, Bhalerao RP, Sandberg G (2001) Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J 28: 465–474 [DOI] [PubMed] [Google Scholar]
- Lucas M, Godin C, Jay-Allemand C, Laplaze L (2008) Auxin fluxes in the root apex co-regulate gravitropism and lateral root initiation. J Exp Bot 59: 55–66 [DOI] [PubMed] [Google Scholar]
- Luo C, Sidote DJ, Zhang Y, Kerstetter RA, Michael TP, Lam E (2013) Integrative analysis of chromatin states in Arabidopsis identified potential regulatory mechanisms for natural antisense transcript production. Plant J 73: 77–90 [DOI] [PubMed] [Google Scholar]
- Malamy JE. (2005) Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ 28: 67–77 [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Matsuura H, Ishibashi Y, Shinmyo A, Kanaya S, Kato K (2010) Genome-wide analyses of early translational responses to elevated temperature and high salinity in Arabidopsis thaliana. Plant Cell Physiol 51: 448–462 [DOI] [PubMed] [Google Scholar]
- Miller AJ, Fan X, Orsel M, Smith SJ, Wells DM (2007) Nitrate transport and signalling. J Exp Bot 58: 2297–2306 [DOI] [PubMed] [Google Scholar]
- Mounier E, Pervent M, Ljung K, Gojon A, Nacry P (2014) Auxin-mediated nitrate signalling by NRT1.1 participates in the adaptive response of Arabidopsis root architecture to the spatial heterogeneity of nitrate availability. Plant Cell Environ 37: 162–174 [DOI] [PubMed] [Google Scholar]
- Muños S, Cazettes C, Fizames C, Gaymard F, Tillard P, Lepetit M, Lejay L, Gojon A (2004) Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1. Plant Cell 16: 2433–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJ, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacry P, Bouguyon E, Gojon A (2013) Nitrogen acquisition by roots: physiological and developmental mechanisms ensuring plant adaptation to a fluctuating resource. Plant Soil 370: 1–29 [Google Scholar]
- Nicolaï M, Roncato MA, Canoy AS, Rouquié D, Sarda X, Freyssinet G, Robaglia C (2006) Large-scale analysis of mRNA translation states during sucrose starvation in Arabidopsis cells identifies cell proliferation and chromatin structure as targets of translational control. Plant Physiol 141: 663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien JA, Vega A, Bouguyon E, Krouk G, Gojon A, Coruzzi G, Gutiérrez RA (2016) Nitrate transport, sensing, and responses in plants. Mol Plant 9: 837–856 [DOI] [PubMed] [Google Scholar]
- Okamoto M, Vidmar JJ, Glass AD (2003) Regulation of NRT1 and NRT2 gene families of Arabidopsis thaliana: responses to nitrate provision. Plant Cell Physiol 44: 304–317 [DOI] [PubMed] [Google Scholar]
- Péret B, De Rybel B, Casimiro I, Benková E, Swarup R, Laplaze L, Beeckman T, Bennett MJ (2009) Arabidopsis lateral root development: an emerging story. Trends Plant Sci 14: 399–408 [DOI] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Filleur S, Diatloff E, Mounier E, Tillard P, Forde BG, Gojon A (2006) The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc Natl Acad Sci USA 103: 19206–19211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveras E, Alvarez JM, Vidal EA, Oses C, Vega A, Gutiérrez RA (2015) The calcium ion is a second messenger in the nitrate signaling pathway of Arabidopsis. Plant Physiol 169: 1397–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel S, Krouk G, Ristova D, Shasha D, Birnbaum KD, Coruzzi GM (2011) Nitrogen economics of root foraging: transitive closure of the nitrate-cytokinin relay and distinct systemic signaling for N supply vs. demand. Proc Natl Acad Sci USA 108: 18524–18529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sormani R, Delannoy E, Lageix S, Bitton F, Lanet E, Saez-Vasquez J, Deragon JM, Renou JP, Robaglia C (2011) Sublethal cadmium intoxication in Arabidopsis thaliana impacts translation at multiple levels. Plant Cell Physiol 52: 436–447 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay YF, Schroeder JI, Feldmann KA, Crawford NM (1993) The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72: 705–713 [DOI] [PubMed] [Google Scholar]
- Walch-Liu P, Forde BG (2008) Nitrate signalling mediated by the NRT1.1 nitrate transporter antagonises L-glutamate-induced changes in root architecture. Plant J 54: 820–828 [DOI] [PubMed] [Google Scholar]
- Wang R, Liu D, Crawford NM (1998) The Arabidopsis CHL1 protein plays a major role in high-affinity nitrate uptake. Proc Natl Acad Sci USA 95: 15134–15139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Xing X, Wang Y, Tran A, Crawford NM (2009) A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiol 151: 472–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yángüez E, Castro-Sanz AB, Fernández-Bautista N, Oliveros JC, Castellano MM (2013) Analysis of genome-wide changes in the translatome of Arabidopsis seedlings subjected to heat stress. PLoS One 8: e71425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Rong H, Pilbeam D (2007) Signalling mechanisms underlying the morphological responses of the root system to nitrogen in Arabidopsis thaliana. J Exp Bot 58: 2329–2338 [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH (2000) Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.