Physcomitrella patens heterotrimeric G-proteins affect gametophore elongation and are essential for sporophyte formation, suggesting their unique role in life cycle completion in early land plants.
Abstract
In this study, we report the functional characterization of heterotrimeric G-proteins from a nonvascular plant, the moss Physcomitrella patens. In plants, G-proteins have been characterized from only a few angiosperms to date, where their involvement has been shown during regulation of multiple signaling and developmental pathways affecting overall plant fitness. In addition to its unparalleled evolutionary position in the plant lineages, the P. patens genome also codes for a unique assortment of G-protein components, which includes two copies of Gβ and Gγ genes, but no canonical Gα. Instead, a single gene encoding an extra-large Gα (XLG) protein exists in the P. patens genome. Here, we demonstrate that in P. patens the canonical Gα is biochemically and functionally replaced by an XLG protein, which works in the same genetic pathway as one of the Gβ proteins to control its development. Furthermore, the specific G-protein subunits in P. patens are essential for its life cycle completion. Deletion of the genomic locus of PpXLG or PpGβ2 results in smaller, slower growing gametophores. Normal reproductive structures develop on these gametophores, but they are unable to form any sporophyte, the only diploid stage in the moss life cycle. Finally, the mutant phenotypes of ΔPpXLG and ΔPpGβ2 can be complemented by the homologous genes from Arabidopsis, AtXLG2 and AtAGB1, respectively, suggesting an overall conservation of their function throughout the plant evolution.
In all known eukaryotes, cellular signaling involves heterotrimeric GTP-binding proteins (G-proteins), which consist of Gα, Gβ, and Gγ subunits (Cabrera-Vera et al., 2003). According to the established paradigm, when Gα is GDP-bound, it forms a trimeric complex with the Gβγ dimer and remains associated with a G-protein coupled receptor. Signal perception by the receptor facilitates GDP to GTP exchange on Gα. GTP-Gα dissociates from the Gβγ dimer, and both these entities can transduce the signal by interacting with different effectors. The duration of the active state is determined by the intrinsic GTPase activity of Gα, which hydrolyzes bound GTP into GDP and inorganic phosphate (Pi), followed by the reassociation of the inactive, trimeric complex (Siderovski and Willard, 2005).
In plants, G-protein signaling has been studied in only a few angiosperms to date at the functional level, although the proteins exist in the entire plant lineage (Hackenberg and Pandey, 2014; Urano and Jones, 2014; Hackenberg et al., 2016). Interestingly, while the overall biochemistry of the individual G-protein components and the interactions between them are conserved between plant and metazoan systems, deviations from the established norm are also obvious. For example, the repertoire of canonical G-proteins is significantly limited in plants; the human genome codes for 23 Gα, 5 Gβ, and 12 Gγ proteins, whereas most plant genomes, including those of basal plants, typically encode 1 canonical Gα, 1 Gβ, and three to five Gγ proteins (Urano and Jones, 2014). The only exceptions are some polyploid species, such as soybean, which have retained most of the duplicated G-protein genes (Bisht et al., 2011; Choudhury et al., 2011). Moreover, even in plants that possess only a single canonical Gα and Gβ protein, for example Arabidopsis (Arabidopsis thaliana) and rice, the phenotypes of plants lacking either one or both proteins are relatively subtle. The mutant plants exhibit multiple developmental and signaling defects but are able to complete the life cycle without any major consequences. These observations have questioned the significance of G-protein mediated signaling pathways in plants.
Interestingly, plants also possess certain unique variants of the classical G-protein components such as the type III Cys-rich Gγ proteins and extra-large GTP-binding (XLG) proteins, which add to the diversity and expanse of the G-protein signaling networks (Roy Choudhury et al., 2011; Chakravorty et al., 2015; Maruta et al., 2015). The XLG proteins are almost twice the size of typical Gα proteins, with the C-terminal region that codes for Gα-like domain and an extended N-terminal region without any distinctive features. Plant XLGs are encoded by entirely independent genes and therefore are different from the mammalian extra-long versions of Gα proteins such as XLαs and XXLαs, which are expressed due to the use of alternate exons (Abramowitz et al., 2004). Three to five copies of XLG proteins are present in the genome of most angiosperms. At the functional level, the XLG proteins have been characterized only from Arabidopsis, to date, where recent studies suggest that the proteins compete with canonical Gα for binding with the Gβγ dimers and may form functional trimeric complexes (Chakravorty et al., 2015; Maruta et al., 2015). The XLG and Gβγ mutants of Arabidopsis seem to function in the same pathways during the regulation of a subset of plant responses, for example primary root length and its regulation by abscisic acid (ABA); the root waving and skewing responses; sensitivity to Glc, salt, and tunicamycin; and sensitivity to certain bacterial and fungal pathogens (Ding et al., 2008; Pandey et al., 2008; Chakravorty et al., 2015; Maruta et al., 2015). However, many of the phenotypes of Arabidopsis Gα and Gβγ mutants are also distinct from that of the xlg triple mutants. For example, compared to the wild-type plants, the canonical G-protein mutants exhibit altered response to gibberellic acid, brassinosteroids, and auxin and show changes in leaf shape, branching, flowering time, and stomatal densities (Ullah et al., 2003; Chen et al., 2004; Pandey et al., 2006; Zhang et al., 2008; Nilson and Assmann, 2010). The xlg triple mutants behave similarly to wild-type plants in all these aspects of development and signaling. Moreover, whether the XLG proteins are authentic GTP-binding and -hydrolyzing proteins and the extent to which they directly participate in G-protein-mediated signaling pathways remains confounding (Chakravorty et al., 2015; Maruta et al., 2015). Even in plants with a limited number of G-protein subunits such as Arabidopsis, one Gα and three XLGs potentially compete for a single Gβ protein, and the analysis of null mutants is not straightforward, that is, it is not possible to delineate whether the phenotypes seen in the Gα null mutants are truly due to the lack of Gα and/or because of an altered stoichiometry or availability of Gβ for the XLG proteins.
As a bryophyte, Physcomitrella patens occupies a unique position in the evolutionary history of plants. It lacks vasculature but exhibits alteration between generations, which is dominated by a gametophytic (haploid) phase and a short sporophytic (diploid) phase (Cove et al., 2009). Many of the pathways related to hormone signaling, stress responses, and development are conserved between angiosperms and P. patens (Cove et al., 2009; Sun, 2011; Komatsu et al., 2013; Yasumura et al., 2015). It is also an intriguing example in the context of the G-protein signaling, because its fully sequenced genome does not encode a canonical Gα gene, although genes coding for the Gβ and Gγ proteins exist. A single gene for a potential XLG homolog also exists in the P. patens genome. This unique assortment of proteins predicts several alternative scenarios for G-protein signaling in P. patens. For example, the P. patens Gβγ proteins might be nonfunctional due to the loss of canonical Gα and are left in the genome as evolutionary artifacts. Alternatively, the Gβγ proteins of P. patens might maintain functionality regardless of the existence of a canonical Gα protein in pathways not regulated via classic G-protein signaling modes. Finally, a more likely scenario could be that the potential XLG protein can substitute for the Gα function in P. patens.
To explore these possibilities and understand better the conserved and unique mechanisms of G-protein signaling pathways in plants and their significance, we examined the role of G-protein subunits in P. patens. We provide unambiguous evidence for the genetic coupling of XLG and Gβ proteins in controlling P. patens development. In contrast to all other plant species analyzed to date, where G-proteins are not essential for growth and survival, the XLG or one of the Gβ proteins is required for the sporophyte formation and life cycle completion in P. patens. Furthermore, one of the Arabidopsis XLG proteins, XLG2, and the canonical Gβ protein AGB1 can functionally complement the P. patens mutant phenotypes. These data provide new insights in the evolutionary breadth and the spectrum of signaling pathways regulated by G-proteins in plants.
RESULTS
Heterotrimeric G-Proteins in P. patens
All plant species analyzed to date, with the possible exception of chlorophyte algae (Hackenberg and Pandey, 2014; Hackenberg et al., 2016), possess Gα, Gβ, and Gγ subunits of the heterotrimeric G-proteins. The proteins share an extremely high degree of sequence similarity within the plant lineages, making their identification possible by homology-based searches. Analysis of the P. patens genome using BlastP search with Arabidopsis G-protein sequences identified two Gβ and two Gγ proteins; however, no sequences matching to the canonical Gα protein were identified. Instead, the Blast searches did reveal the presence of one gene coding an XLG. We named this sequence PpXLG (Pp1s147_153V6.1). Similar searches were able to identify both canonical and extra-large Gα protein homologs in algae and additional bryophytes and lycophytes, implying that a canonical Gα coding sequence is indeed missing from the P. patens genome.
Conceptually translated sequence of PpXLG codes for an 1,168-amino acid protein, which is significantly longer than the XLG proteins identified so far from angiosperms (e.g. Arabidopsis XLGs code for 848 to 888 amino acid proteins). The C-terminal region of the protein shows homology to Gα protein and possesses a long N-terminal extension, typical of XLG proteins of other plants (Fig. 1A). Sequence comparison of PpXLG with the Arabidopsis XLG proteins (AtXLGs) shows 35% to 40% similarity between them. The C-terminal region of PpXLG shows 30% similarity with AtGPA1. However, the comparison of the putative GTP/GDP-binding motif suggests higher similarity of PpXLG with XLG proteins rather than with the Gα proteins (Supplemental Fig. S1).
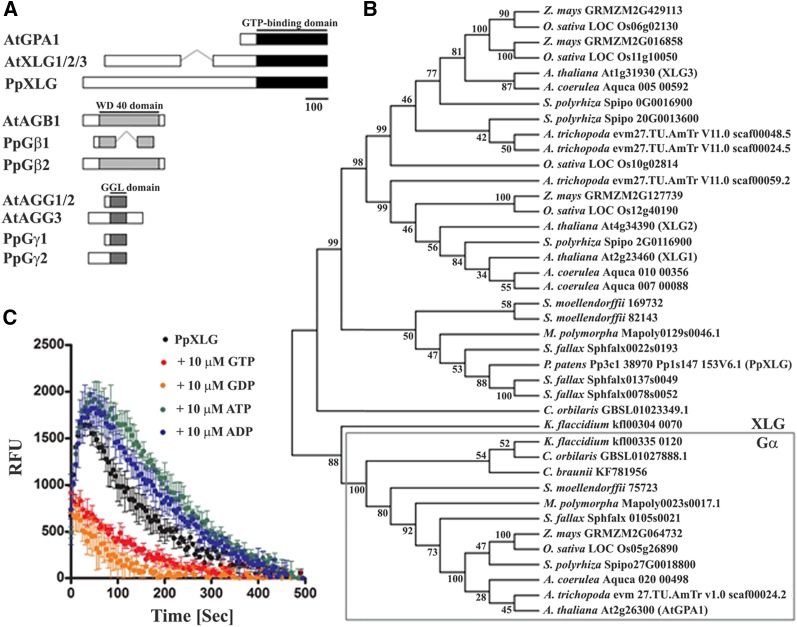
Figure 1.
Heterotrimeric G-proteins in P. patens. A, Schematic representation of heterotrimeric G-proteins and XLG proteins in Arabidopsis and the predicted domain architecture in P. patens. Conserved domains are labeled in black and gray. B, Phylogenetic relationship analysis of Gα proteins and XLG proteins of selected species representing different clades of the plant lineage. The evolutionary history was inferred by using the maximum likelihood method based on the JTT matrix-based model (Jones et al., 1992). Branches corresponding to partitions reproduced in <50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. Evolutionary analyses were conducted in MEGA6 (Tamura et al., 2013). C, GTP-binding and hydrolysis activity of the Gα-like domain of PpXLG. The Gα-like domain of PpXLG was recombinantly expressed in E. coli and used in fluorophore conjugated BODIPY GTP-FL assays (Roy Choudhury et al., 2013). Specific binding and hydrolysis was detectable without (black) and after addition of 10 µM ATP (green) or ADP (blue), whereas 10 µM GTP (red) or GDP (orange) clearly suppressed binding of BODIPY GTP-FL.
Phylogenetic relationship analysis of the representative Gα and XLG proteins of plant species from across the major lineages separates them in two clusters (Fig. 1B). Charophyte algae such as Coleochaeta orbicularis and Klebsormidium flaccidiium possess both XLG and Gα encoding genes in their genomes, suggesting the divergence of both these classes of genes early during evolution in the plant lineage. Intriguingly, the potential K. flaccidiium XLG (kfl00304_0070), despite its excessive length of 2,619 amino acids, shows close evolutionary relationship with both types of Gα proteins by being associated with the Gα cluster in phylogenetic analysis. Among XLG proteins, PpXLG, together with the putative XLG homologs identified from the bryophytes Marchantia polymorpha (Mapoly0129s0046.1) and Sphagnum fallax (Sphfalx0022s0193, Sphfalx0137s0049, Sphfalx0078s0052) and the lycophyte Selaginella moellendorffii (169732 and 82143) represents the most basal land plant group identified to date. These proteins are positioned as a phylogenetic out-group in relation to other XLG proteins from angiosperms (Fig. 1B). Notably, in contrast to P. patens, other species in this group also possess potential homologs of canonical Gα proteins.
The two Gβ homologs identified in P. patens were named PpGβ1 (Pp1s28_162V6.1) and PpGβ2 (Pp1s7_401V6.2). The proteins exhibited 60% and 62% similarity with the Arabidopsis AGB1, respectively (Supplemental Fig. S2). However, PpGβ1 lacks parts of the N- and C-terminal regions as well as a portion of the central WD 40-domain found in PpGβ2 and AtAGB1 (Fig. 1A).
The two Gγ-like sequences from the P. patens genome were named PpGγ1 (Pp1s39_119V6.1) and PpGγ2 (Pp1s22_182V6.1). The proteins showed the highest similarity with Arabidopsis AGG2 (28.4% and 26.5%, respectively), whereas the overall similarity with AtAGG1 and AtAGG3 was in the range of 20% to 23%. Such low sequence similarity within Gγ proteins is typical due to their relatively small sizes and variable sequences (Roy Choudhury et al., 2011). The proteins do possess the highly conserved central region represented by the GGL domain (Fig. 1A). Interestingly, PpGγ1 does not have a prenylation motif, which is present in PpGγ2 (Supplemental Fig. S3A). Both these types of Gγ proteins, represented by the presence or absence of a classical prenylation motif at their C terminus, are found in most other land plants as well (Roy Choudhury et al., 2011). The two PpGγ proteins lack a transmembrane helical domain and Cys-rich region found in the Type III Gγ proteins (Choudhury et al., 2011). Consequently, PpGγ proteins as well as Gγ proteins from other bryophytes were associated with the nonhelical domain possessing Gγ protein cluster containing AtAGG1 and AtAGG2, rather than with the AtAGG3 cluster (Supplemental Fig. S3B).
PpXLG Binds and Hydrolyzes GTP
Although recent homology modeling of XLG proteins suggests their high degree of similarity with the canonical Gα protein, GPA1 (Chakravorty et al., 2015), the catalytic regions of XLG proteins are somewhat divergent and some of the key amino acids proposed to be required for the classical GTP-binding and -hydrolytic activity are missing from this region (Temple and Jones, 2007). The AtXLG proteins have been shown to exhibit GTP-binding and -hydrolyzing activities, although they require the presence of calcium instead of magnesium, which is typical of canonical Gα proteins (Heo et al., 2012). We evaluated the GTP binding and hydrolysis of the C-terminal region of PpXLG protein using a real-time in vitro assay (Roy Choudhury et al., 2013). Incubation of the recombinantly expressed C-terminal region of PpXLG with BODIPY-labeled GTP resulted in the typical increase in fluorescence due to the binding of the fluorophore-conjugated GTP, followed by the slow extinction of the signal caused by the hydrolysis of the nucleotide triphosphate (Fig. 1C). The specificity of binding and hydrolysis for GTP was demonstrated by competitive substrate and product inhibition. A significant decrease in fluorescence was observed by the addition of 100-fold excess of unlabeled GTP or GDP but not by the addition of equal amount of unlabeled ATP or ADP. Incidentally, PpXLG protein was active in the presence of magnesium, as is seen for other Gα-like proteins and not calcium, as was reported for AtXLG proteins (Heo et al., 2012). Overall, these data support that PpXLG displays the biochemical characteristics of a Gα protein.
Specific Subunits of G-Proteins Affect Gametophyte Development in P. patens
Deletion mutants lacking the genomic locus of each of the G-protein subunits (ΔPpXLG, ΔPpGβ1, ΔPpGβ2, ΔPpGγ1, and ΔPpGγ2) were generated using homologous recombination (Cove et al., 2009). Each of the mutants was confirmed for the presence of the targeted deletion at the corresponding locus by PCR analysis as shown for ΔPpXLG (Supplemental Fig. S4) and ΔPpGβ2 (Supplemental Fig. S5). In addition, we also generated a double mutant lacking the genomic loci of both PpGγ1 and PpGγ2 (ΔPpGγ double). All the mutants were analyzed together with the wild-type P. patens accession Gransden (Gd) for overall growth and development.
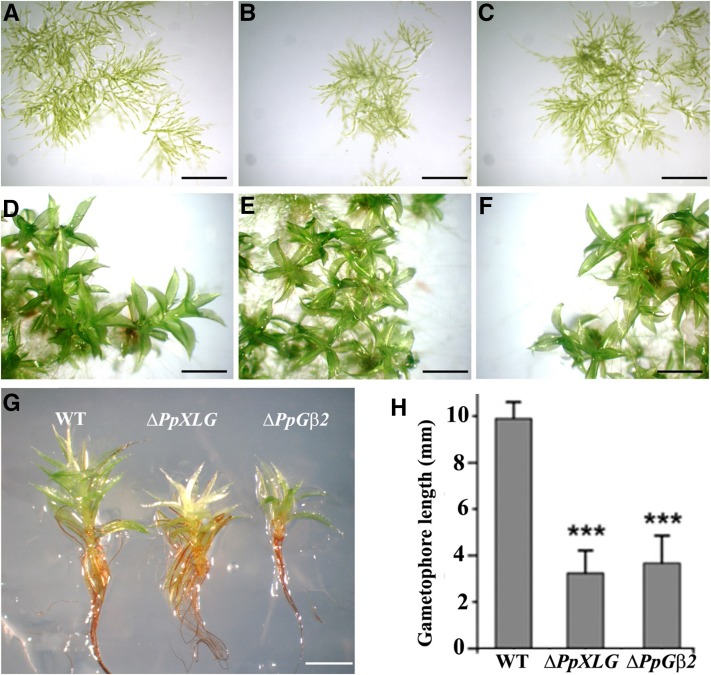
While the filamentous protonemata tissue (Fig. 2, A–C) and early gametophores (Fig. 2, D–F) developed normally for each of the genotypes, during later stages the ΔPpXLG and ΔPpGβ2 mutants showed clear phenotypic differences from the wild-type plants. The ΔPpXLG and ΔPpGβ2 gametophores did not elongate as much as the wild-type plants under normal growth conditions (Fig. 2G). These also had a reduced number of leaves possibly due to the shorter gametophore, which potentially did not allow for the development of additional leaf layers. Both these mutants’ gametophores also exhibited irregular leaf arrangement and relatively slimmer leaves compared to the wild-type plants. After 6 weeks of growth, ΔPpXLG and ΔPpGβ2 showed substantial growth arrest of gametophores resulting in a significantly reduced biomass and a pronounced dwarf phenotype (Fig. 2H). No differences in phenotypes were observed in ΔPpGβ1, ΔPpGγ1, ΔPpGγ2, and ΔPpGγ double mutants compared to the wild-type plants under identical growth conditions, implying the specific role of PpXLG and PpGβ2 in controlling gametophore development.
Figure 2.
Development of wild-type (WT), ΔPpXLG, and ΔPpGβ2 P. patens plants. Protonemata growth of 1-week-old P. patens (A) wild-type (WT; B), ΔPpXLG (C), and ΔPpGβ2 and 24-d-old gametophores of WT (D), ΔPpXLG (E), and ΔPpGβ2 (F) on BCD medium. G, WT, ΔPpXLG, and ΔPpGβ2 plants at 44 d showing impaired elongation of mutants. Scale bar = 500 µm. H, Quantification of gametophore length after 6 weeks of growth on BCD medium (n = 12). Mean value and SD are plotted. Significance was calculated using t test for unequal variances (P < 0.001).
Gametophore development involves regulation by different phytohormones (Katsumata et al., 2011; Coudert et al., 2013, 2015), and reduced elongation of gametophores is reminiscent of an altered auxin response. Moreover, G-protein mutants in vascular plants exhibit altered sensitivity to a variety of phytohormones (Ashikari et al., 1999; Ullah et al., 2003; Chen et al., 2004; Pandey et al., 2006). Therefore, we tested whether the reduced gametophore elongation is related to the changes in phytohormone accumulation or signaling in ΔPpXLG and ΔPpGβ2 mutants. The endogenous level of acidic plant hormones in gametophores was measured by mass spectrometry-based analysis. The response to different hormones was determined by cultivating the wild-type and mutant moss on media containing different plant hormones. The endogenous levels of ABA, salicylic acid, and indole-3-acetic acid (IAA) did not show significant differences between ΔPpXLG, ΔPpGβ2, and wild-type gametophores (Supplemental Fig. S6A). Similarly, neither ΔPpXLG nor ΔPpGβ2 showed any obvious differences compared to the wild-type plants when cultivated on media with exogenous ABA, ethylene precursor 1-aminocyclopropane-1-carboxylic acid, 6-Benzylaminopurine, or IAA (Supplemental Fig. S6B), even though the effect of these hormones on overall moss growth and development was obvious. These data suggest that the changes in the phytohormone levels or signaling are not significantly affected by G-protein mutations in moss and are likely not the reason for altered gametophore development.
PpXLG and PpGβ2 Are Essential for Sporophyte Formation in P. patens
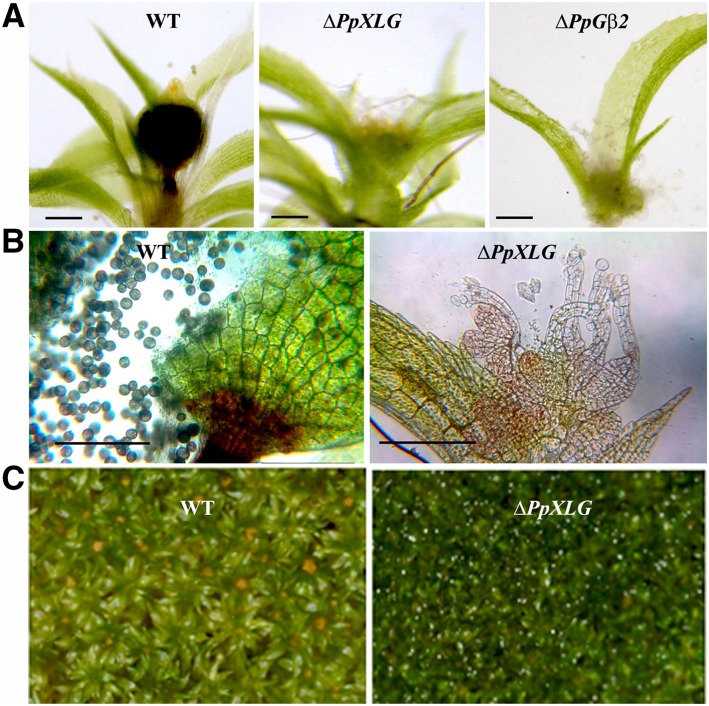
The most striking developmental defect observed in the ΔPpXLG and ΔPpGβ2 plants was their inability to form sporophytes, the only diploid stage in the moss life cycle (Fig. 3A). When grown on low nitrogen media and transferred to the sporulation-inducing conditions, the apical region of wild-type gametophores developed distinct sporophytes that possessed viable spores. In contrast, the ΔPpXLG and ΔPpGβ2 mutants exhibited an open, exposed female archegonia and male antheridia (Fig. 3B). No sporophytes were ever observed on ΔPpXLG or ΔPpGβ2 gametophores, even after extended growth period (Fig. 3C). All additional mutants displayed phenotypes similar to the wild-type plants and formed normal sporophytes in response to sporulation induction.
Figure 3.
Sporophyte formation and morphology of the reproductive organs in wild-type (WT) and mutant plants. A, Apical portion of gametophores after sporulation induction showing sporophytes on WT and the nonsporophyte phenotype of ΔPpXLG and ΔPpGβ2 mutants. Scale bar = 200 µm. B, Apical portion of gametophores of WT possessing a sporophyte and ΔPpXLG of the same age. WT sporophytes possess spores, whereas ΔPpXLG comprises open, nonfertilized archegonia. Scale bar = 200 µm. C, No sporophytes were ever formed on ΔPpXLG or ΔPpGβ2 (not shown) even after extended period of growth.
To determine if the inability to form sporophytes was related to the lack of or altered development of female (archegonia) or male (antheridia) reproductive organs in the ΔPpXLG and ΔPpGβ2 plants, we analyzed the morphology of their apical regions, together with the sporophyte-bearing wild-type plants of the same age. Both reproductive organs were unambiguously present without any obvious morphological alteration in ΔPpXLG and ΔPpGβ2 plants (Fig. 4). This suggests a defect either in the fertility of one or both reproductive organs, or during the process of fertilization itself. Regardless of the mechanism involved, these data clearly establish that PpXLG and PpGβ2 are essential for sporophyte formation and, consequently, life cycle completion in P. patens.
Figure 4.
Reproductive organ development in wild-type (WT) and mutant P. patens plants. Dissected gametophores apices after 5 weeks growth on sporulation media with reduced nitrogen content. WT and mutants possess normal antheridia (A) and archegonia (B). No difference in the morphology of either of the reproductive organs was observed. Scale bar = 100 µm.
Inability to Form Sporophyte Is Not Related to Female Sterility
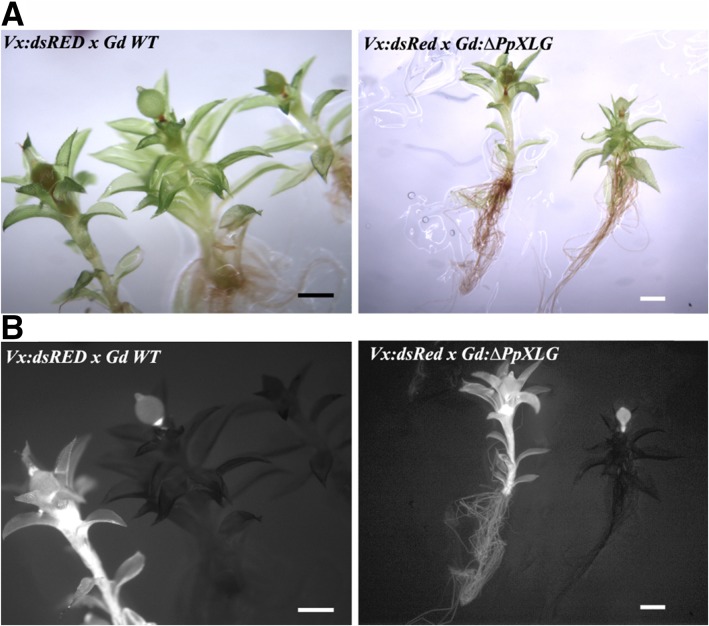
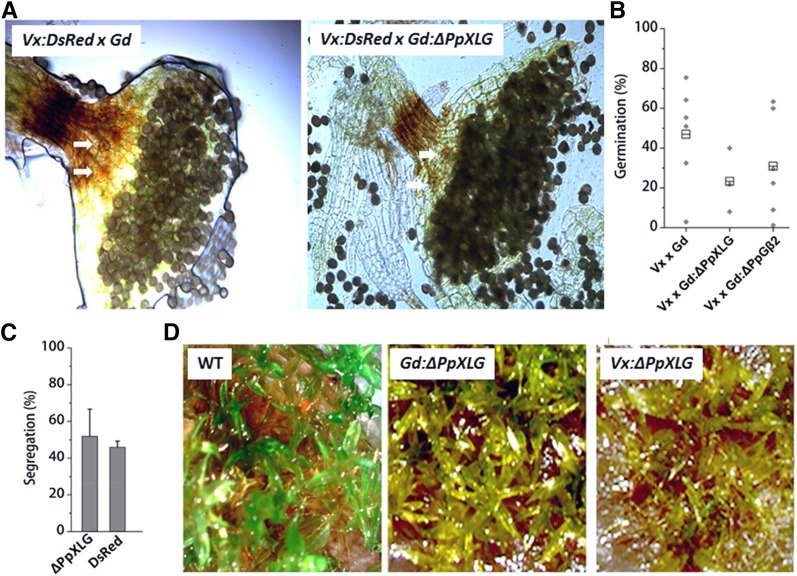
To evaluate whether the sporophyte deficient phenotype is due to the sterility of reproductive organs, we performed crossing experiments using ΔPpXLG or ΔPpGβ2 with a red fluorescent protein (dsRED) tagged line of P. patens accession Villersexel (Vx) described previously (Perroud et al., 2011). Both ΔPpXLG and ΔPpGβ2 were able to form sporophytes when grown together with Vx:dsRED in sporulation assays (Fig. 5A; Supplemental Fig. S7A), indicating that female fertility was not affected by either of the deletions. We confirmed the origin of the pollinating sperm cell, responsible for the sporulation event, by visualizing dsRED fluorescence in the corresponding individual plants. Cross-fertilization of ΔPpXLG or ΔPpGβ2 with Vx:dsRED produced two different populations of sporophyte-carrying gametophores: the nonfluorescent gametophores carrying fluorescent sporophytes represent crosses between Vx:dsRED as sperm cell donor and Gd:ΔPpXLG (Fig. 5B) or Gd:ΔPpGβ2 (Supplemental Fig. S7B) hosting the inseminated egg cell, whereas self-fertilization of Vx:dsRED resulted in plants possessing both fluorescent gametophores and sporophytes (Fig. 5B). We repeated this experiment twice independently and never observed any nonfluorescent sporophytes that would represent self-fertilization of ΔPpXLG or ΔPpGβ2. These observations confirm that sporulation occurred only when insemination with Vx:dsRED sperm cells took place. In contrast, wild-type accession Gd (Fig. 5B) as well as ΔPpGβ1, ΔPpGγ1, ΔPpGγ2, ΔPpGγ1/ΔPpGγ2 (Supplemental Fig. S7) formed nonfluorescent sporophytes on nonfluorescent gametophores, indicating self-fertilization in addition to the fluorescent sporophytes derived from crosses with Vx:dsRED. These data unequivocally support that the inability to form sporophytes is not due to a defect in the female reproductive organ in ΔPpXLG or ΔPpGβ2.
Figure 5.
Phenotypes of crosses between Gd wild-type (WT) and Vx:DsRed. A, White light and (B) dsRED fluorescence microscopy of crossed Vx:dsRED and Gd WT as well as Vx:dsRED and Gd:ΔPpXLG. Fluorescent sporophytes were formed on ΔPpXLG, confirming insemination by Vx:dsRED spores and no defect in female fertility of the mutants. Scale bar = 1 mm.
To address the possibility that the lack of formation of sporophytes in ΔPpXLG or ΔPpGβ2 is due to the male sterility of these mutants, we performed crosses with Gd individuals as sperm cell donors. However, these inseminations were never successful in any of the experiments, including those using crosses with wild-type Gd plants. This is in agreement with the previous observations (Perroud et al., 2011) and is likely due the strong potency of the Vx accession as male parent compared to the Gd accession. Therefore, male sterility as a reason for the lack of sporophytes on ΔPpXLG or ΔPpGβ2 gametophores, although highly likely, can neither be confirmed nor excluded at this point.
Spores Lacking PpXLG or PpGβ2 Are Normal
Sporophytes of recombinant Vx:dsRED × Gd:ΔPpXLG and Vx:dsRED × Gd:ΔPpGβ2 showed no obvious differences in morphology compared to the wild-type Vx:dsRED × wild-type Gd outcross. All sporophytes possessed stomata at the lower part of the capsule and, more importantly, produced viable spores (Fig. 6A). Spores of the recombinant Vx:dsRED × Gd:ΔPpXLG and Vx:dsRED × Gd:ΔPpGβ2 crosses germinated with widely distributed efficiencies averaging in the range of 20% to 30% (Fig. 6B). Spores from wild-type Vx × Gd crosses germinated with relatively higher frequencies, but also showed a wide range of germination efficiency common for outcrosses between Vx and Gd accessions, as reported previously (Perroud et al., 2011).
Figure 6.
Phenotypes of sporophytes formed on ΔPpXLG by crossing with Vx:DsRed. A, Dissected sporophytes of Vx:dsRED × GdWT and Vx:dsRED × Gd:ΔPpXLG crosses showing stomata (white arrows) and spores. B, Germination efficiency determined by 9 replicates of approximately 100 spores. The average values are plotted as gray diamonds. Total mean values are indicated as boxes. C, Segregation of genetic marker as determined via PCR or dsRED fluorescence microscopy. Mean and SD of 5 replicates each with 30 descendants are shown. D, Top view of P. patens plants wild type (WT) Gd, Gd:ΔPpXLG, and a representative recombinant cross Vx:dsRED × Gd:ΔPpXLG grown on reduced nitrogen media. WT plants form sporophytes, whereas no sporophytes were formed on either Gd:ΔPpXLG or Vx:ΔPpXLG.
To analyze a potential role of PpXLG during early stages of spore development, we tested ΔPpXLG spores’ viability. We screened recombinant descendants for carrying the ΔPpXLG locus via PCR with specific primers and identified ΔPpXLG in about 50% descendants. Similarly, dsRED segregated with a 1:1 ratio, as determined by fluorescence microscopy, indicating independent Mendelian inheritance for both these genomic loci (Fig. 6C). We then randomly selected ΔPpXLG descendants from five different spores derived from two independent sporophytes and tested them for growth and gametophyte formation. Similar to what was seen with the ΔPpXLG and ΔPpGβ2 mutants, the protonemata tissue of recombinant Vx × Gd:ΔPpXLG developed normally, indicating no effect of ΔPpXLG during germination and protonemata development in moss. However, the gametophores of recombinant Vx × Gd:ΔPpXLG were impaired in growth, showing severe dwarfism and did not form any sporophytes (Fig. 6D) in three independent experiments. These data confirm that the severe developmental defect caused by the lack of PpXLG exists not only in the Gd accession, but also in a recombinant Vx × Gd background.
Arabidopsis XLG2 and AGB1 Complement Corresponding Moss Mutants
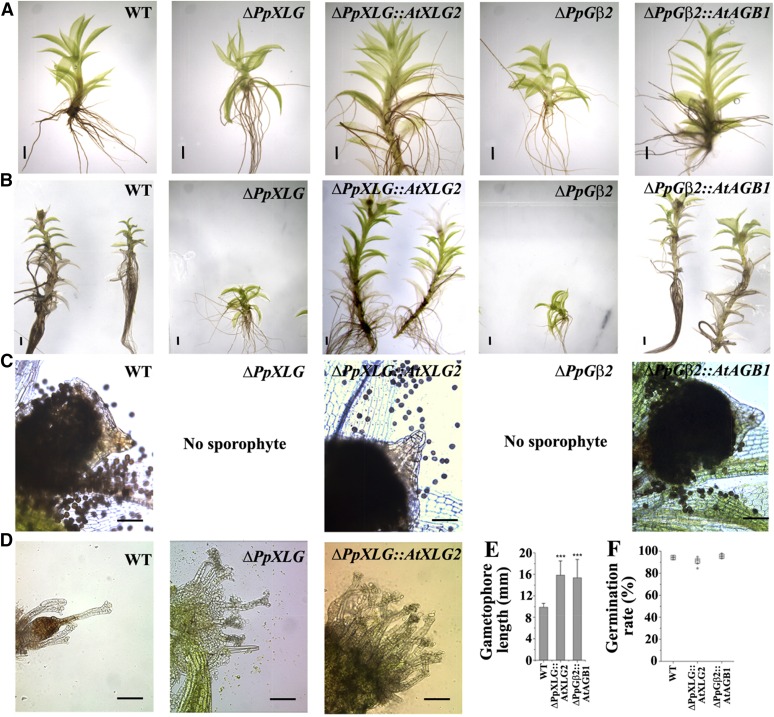
PpXLG and PpGβ were identified as homologs of Arabidopsis G-proteins, which are separated by millions of years of evolution. The two species also have a profoundly different life cycle, that is, a dominant gametophytic phase in moss versus dominant sporophytic phase in Arabidopsis. To ascertain that the phenotypes observed in the ΔPpXLG and ΔPpGβ2 mutants are truly due to the loss of these genes and not due to any nonspecific recombination events and to establish that PpXLG and PpGβ are true functional homologs of G-proteins found in higher plants, cross-species complementation analyses were performed. The coding sequences of AtXLG2, which shows highest sequence similarity with PpXLG, and AtAGB1 were stably transformed in the ΔPpXLG and ΔPpGβ2 mutants, respectively, by targeting the site-directed insertion into the deleted P. patens genomic loci. The specific incorporation of the Arabidopsis homologous genes was verified by PCR (Supplemental Fig. S8), and ΔPpXLG::AtXLG2 and ΔPpGβ2::AtAGB1 plants were analyzed for the complementation of mutant phenotypes. ΔPpXLG::AtXLG2 and ΔPpGβ2::AtAGB1 gametophores were significantly longer than those of the corresponding background deletion mutants ΔPpXLG or ΔPpGβ2 and are similar to (or even longer than) the wild-type gametophores (Figs. 7A and 6, B and E). In addition, introduction of AtXLG2 and AtAGB1 restored the ability to develop functional sporophytes in ΔPpXLG and ΔPpGβ2, respectively (Fig. 7C), although a subset of ΔPpXLG::AtXLG2 and ΔPpGβ2::AtAGB1 still lacked sporophytes and exhibited open, exposed arrangement of archegonia seen in ΔPpXLG and ΔPpGβ2. Wild-type Gd, on the other hand, only rarely possessed gametophores without a sporophyte, and individuals lacking a sporophyte comprised less diffuse archegonia, covered by the apical gametophore leaves (Fig. 7D). The sporophytes of both ΔPpXLG::AtXLG2 and ΔPpGβ2::AtAGB1 carried viable spores, which germinated with 90% to 95% efficiency comparable to those of wild type (Fig. 7F). These results confirm that the Arabidopsis XLG2 and AGB1 are true functional homologs of PpXLG and PpGβ2 despite the proteins being involved in regulating novel developmental processes in P. patens.
Figure 7.
Arabidopsis XLG2 and AGB1 complement ΔPpXLG and ΔPpGβ2 phenotypes. AtXLG2 and AtAGB1 CDS introduced in the deleted genomic locus of ΔPpXLG or ΔPpGβ2, respectively, restored the growth defect of 6-week-old (A) and 12-week-old (B), postsporulation induction gametophores. C, ΔPpXLG::AtXLG2 and ΔPpGβ2::AtAGB1 also regained the capability to form sporophytes with viable spores similar to WT. D, Dissected apices of nonsporulated gametophores possess an open archegonia structure in ΔPpXLG::AtXLG2 similar to ΔPpXLG and ΔPpGβ2 but different from the hidden archegonia structure of WT. Scale bar = 1 mm (A and B) and 200 µm (C and D). E, Gametophore length of WT and complemented plants was quantified from 6-week-old plants (n = 12) using the Image J software. Mean value and SD are plotted. Significance was calculated using t test for unequal variances (P = 0.01). F, Germination efficiency was determined by 9 replicates of approximately 100 spores from 5 sporophytes. Mean values for each sporophyte are plotted in gray diamonds. Total average values are indicated as boxes.
DISCUSSION
In this study, we present a functional description of heterotrimeric G-proteins in a nonvascular plant. G-protein signaling system, which exists in the entire plant lineage, has been functionally characterized only from a few angiosperms to date. Genetic and physiological analysis of Arabidopsis, rice, soybean, and maize mutants suggests that G-proteins regulate a broad spectrum of signaling pathways in plants (Urano and Jones, 2014). The most obvious developmental defects exist in the Gα mutants of rice and maize (Ashikari et al., 1999; Bommert et al., 2013), which show severe dwarfism and alterations in inflorescence architecture, and in soybean where G-proteins regulate nodule formation (Roy Choudhury and Pandey, 2015). In Arabidopsis, where G-proteins have been characterized in most detail, changes in the overall plant development, morphology, and response to environment are observed due to the lack of one or more G-protein subunits; however, none of these impair the ability of the plant to complete its life cycle (Urano and Jones, 2014). In contrast, the analysis of G-proteins from P. patens identifies their essential role in life cycle completion in this species.
This study was designed in the context of the unique position of moss in the evolutionary history of plants as well as the unusual assortment of the G-protein subunits that exist in P. patens. The presence of a single XLG-like protein and the absence of a canonical Gα (Fig. 1) offers a distinctive opportunity to establish the role of XLG protein as a bona fide Gα protein and its ability to function in the same genetic pathway as the Gβ protein, without an influence on the altered protein stoichiometry or competitive binding by multiple Gα proteins with a single Gβ. Our data confirm that the PpXLG exhibits biochemical properties of classic Gα proteins (Fig. 1C) and works in the same genetic pathway with the canonical PpGβ to regulate critical developmental events (Figs. 2 and 3). Although we have characterized only a single line for each of the deletions, the following observations support the idea that the phenotypes are specific to the gene(s) of interest and are not due to the nonspecific recombinations. First, the deletion of two different genes that are expected to function together results in identical phenotypes of the mutants. Moreover, the phenotypes were observed in the next generation, and a 1:1 segregation was observed in the recombinant spores (Fig. 6). Finally, our data are further strengthened by the fact that homologous genes from Arabidopsis, AtXLG2 and AtAGB1, are able to complement the phenotypes of ΔPpXLG and ΔPpGβ2 mutants, respectively (Fig. 7). Incidentally, the AtXLG2 and AtAGB1 single or double mutants of Arabidopsis do not exhibit any reproductive defects and are able to continue their life cycle normally. It remains to be ascertained whether this specialized function of PpXLG and PpGβ2 in P. patens is due to its gametophyte-dominant life cycle or if there are additional, yet unidentified proteins in advanced plant lineages that compensate for the life cycle-affecting functions of these proteins. Further analysis of the roles of G-proteins from additional basal plants will help answer some of these questions.
Intriguingly, the PpXLG and PpGβ2 proteins seem to act independently of a Gγ protein, which typically is an integral part of a heterotrimeric complex and is thought to be required for the canonical or extra-large G-protein containing trimers in Arabidopsis (Chakravorty et al., 2015; Maruta et al., 2015). One possibility is that there are still unidentified Gγ-like proteins present in the P. patens genome that participate in heterotrimer formation with PpXLG:PpGβ2 complex. Although our analysis did not reveal any additional candidates, the identification of Gγ proteins is relatively difficult due to their short sequence lengths and low similarity with each other (Roy Choudhury et al., 2011). Therefore, additional, significantly variable Gγ proteins may still remain to be identified. Alternatively, it is also possible that the PpXLG:PpGβ2 dimer can function without a Gγ protein and acts in a nonconventional manner. One of the main roles of Gγ proteins is to target the heterotrimeric complex to the plasma membrane (Chakravorty et al., 2015). However, in plants, the G-protein subunits are also localized in intracellular compartments and may signal from locations where the membrane targeting by a Gγ protein is not required (Weiss et al., 1993, 1994; Ding et al., 2008; Chakravorty et al., 2015). The lack of one or both Gγ proteins identified in the P. patens genome, PpGγ1 or PpGγ2, or the additional Gβ protein, PpGβ1, had no effect on overall growth and development of plants (Supplemental Fig. S7). The role of these three proteins is not known at this time; however, it is certainly not due to these being pseudogenes. Each of the G-protein genes in P. patens is transcriptionally active and can be amplified from the protonemata tissue (our data). This is also supported by the analysis of publically available transcriptome data. RNAseq-based expression analysis using meristem, sporophyte, and whole plant shows the expression of each of the genes, although PpGβ1 and PpGγ2 are expressed at a relatively lower level compared to other genes (Frank and Scanlon, 2015a, 2015b).
The shorter gametophore phenotype of ΔPpXLG and ΔPpGβ2 (Fig. 2) is reminiscent of the dwarfism observed for the COMPACT PLANT2 (CT2) mutant in maize and dwarf1 (d1) mutation in rice, both of which correspond to the lack of a functional Gα protein in these species (Ashikari et al., 1999; Bommert et al., 2013). However, these mutations have no effect on the life cycle completion in these plants. The dwarf phenotype of monocot Gα mutants has been attributed to the defects in the CLAVATA signaling pathway in maize (Bommert et al., 2013) and altered sensitivity to gibberellic acid and brassinosteroids in rice (Ashikari et al., 1999). Gibberellic acid and brassinosteroid signaling pathways seem not to be fully functional in P. patens (Hirano et al., 2007; Yasumura et al., 2007; Rensing et al., 2008). Additionally, none of the P. patens mutants exhibited any major changes in the endogenous hormone levels or their response to any of the exogenously applied hormones tested (Supplemental Fig. S6). Therefore, a predominant role of phytohormones in regulating G-protein-dependent gametophore elongation in P. patens, similar to internode elongation in rice, seems unlikely.
The inability to form sporophytes in P. patens could result from either the lack of or abnormal development of antheridia and/or archegonia, or from their inability to fertilize because of a defect in cell-cell communication or signal transduction pathways. Our data clearly show that both the reproductive organs are present and morphologically normal in ΔPpXLG or ΔPpGβ2 mutants (Fig. 4). Additionally, successful crossing of Vx:dsRED plants with ΔPpXLG or ΔPpGβ2 confirms that female fertility was not affected by these deletions (Fig. 5). The descending recombinant sporophytes developed normally and hosted viable spores. The analysis of the segregation of the ΔPpXLG allele indicated no elevated embryonic lethality (Fig. 6). Consequently, the gene dosage of a single PpXLG and PpGβ2 copy was sufficient to form a vital zygote and subsequently develop functional sporophytes, the only stages in the P. patens life cycle with diploid chromosomes. In other words, the lack of either PpXLG or PpGβ2 in the haploid female germline can be overcome by a copy of the gene present in the male antherozoid to form a functional diploid zygote and consequently functional sporophytes.
Because back-crossing with ΔPpXLG or ΔPpGβ2 as sperm cell donor was not successful due to the dominance of Vx:dsRED as a male parent (Perroud et al., 2011) and all attempts to inseminate Gd wild type with recombinant Vx:dsRED × ΔPpXLG at different time points were also unsuccessful, the question whether the lack of PpXLG or PpGβ2 in the male gamete may result in their inability to form sporophyte cannot be answered at this time. This could potentially be due to the nonsynchronous development of both populations because similar to Gd:ΔPpXLG, the recombinant Vx:dsRED × Gd:ΔPpXLG also showed severe growth inhibition, which likely prevented them to act as effective male parent. Therefore, even though our results point to a defect in male fertility as a cause of the lack of sporophyte formation, secondary morphological effects as a consequence of the growth retardation cannot be excluded as reason for the lack of sexual reproduction.
An alternative hypothesis could be that both male and female reproductive organs are normal and functional, but there is a defect in cell-cell communication and signal transduction during fertilization. Since almost nothing is currently known about such signaling events during P. patens fertilization or about the role of G-protein subunits in sperm motility or fertilization in any plant species, further studies focused toward characterization of these mechanisms would be helpful to identify the roles of PpXLG and PpGβ2 during this event. So far, the lack of sporophyte formation has been reported for deletion mutation of two genes involved in the general gene expression regulation processes. DICER-LIKE4 (PpDCL4; Arif et al., 2012) and DNA METHYLTRANSFERASE 1 (PpMET1; Yaari et al., 2015) are key players during transacting small RNA interference and DNA methylation, respectively. Mutations in either of these genes result in lack of sporophyte formation. However, unlike ΔPpXLG and ΔPpGβ2, ΔPpMET1 displays normal gametophore development and the reason why it cannot form sporophyte is unknown. ΔPpDCL4, on the other hand, exhibits abnormal morphology all through development and is female sterile.
Taken together, our data confirm that the XLG protein homologs, which are present in the entire plant lineage, are functionally linked to the canonical G-proteins and can act in the same genetic pathway as a Gβ protein to regulate critical growth and development processes. This has enormous significance for the diversity and expanse of G-protein signaling in plants. Our data also suggest that early in plant evolution, G-proteins were pivotal for plant reproduction and survival. Whether the proteins progressed to more modulatory roles in advanced plant lineages or new, yet-to-be identified proteins evolved to function together with the G-proteins to compensate for their obligate requirement will be uncovered in future studies. Although the nature of the signal transduced by G-proteins during the regulation of P. patens development remains unsolved, the finding that homologous Arabidopsis genes can complement corresponding mutants (Fig. 7) provides us with new insights in the evolutionary conservation of the G-protein complex in the plant lineage.
While the morphological details of the moss sporophyte, its developmental time line, and the transcriptional profile are relatively well described (Cove et al., 2009; Vidali and Bezanilla, 2012; Hiss et al., 2014; Frank and Scanlon, 2015a, 2015b; Ortiz-Ramirez et al., 2016), the mechanistic details and signal transduction pathways involved during sporophyte formation are largely unknown. Our results provide a clue to the types of proteins that might be involved in the signaling event that takes place during sporophyte formation. Future studies geared toward identifying the downstream components of G-proteins in P. patens and their action mechanisms will help uncover the details of this critical developmental and signaling event.
MATERIALS AND METHODS
Sequence Retrieval and Alignment
Putative homologs of Physcomitrella patens G-protein genes were identified by BlastP searches using the Arabidopsis (Arabidopsis thaliana) and rice G-protein genes as queries on Phytozome database (www.phytozome.jgi.gov). CDS sequences corresponding to PpGα/XLG (Pp1s147_153V6.1), PpGβ1 (Pp1s28_162V6.1), PpGβ2 (Pp1s7_401V6.2), PpGγ1 (Pp1s39_119V6.1), and PpGγ2 (Pp1s22_182V6.1) were verified by amplification with specific primer from cDNA and subsequent sequencing. All primers used in this study are listed in Supplemental Table S1. Gene sequences for additional species were acquired from the Phytozome and the Marchantia genome project (marchantia.info). Conceptually translated sequences were used for sequence alignments using Clustal X (Sievers et al., 2011) and phylogenic analysis using MEGA 6 (Tamura et al., 2013). The evolutionary history was inferred by using the Maximum Likelihood method based on the JTT matrix-based model (Jones et al., 1992). The bootstrap consensus tree inferred from 1,000 replicates is taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in <50% bootstrap replicates are collapsed. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model and then selecting the topology with superior log likelihood value. The analysis involved 40 protein sequences. There were a total of 3,296 positions in the final dataset.
Plant Material and Growth Conditions
The Gd wild-type strain of P. patens (Hedw.) Bruch & Schimp was used in this study. The plants were grown in a growth chamber at 25°C and a 16-h-light/8-h-dark regime with a photosynthesis photon flux density of approximately 60 µmol m−2 ⋅ s−1. Cultivation and vegetative propagation were carried out as described previously (Cove et al., 2009). Plants were grown on BCD medium for phenotypic analysis, on BCD medium supplemented with 5 mm (di)ammonium tartrate (Sigma, St Louis, MO) for fast propagation or protoplast isolation, and on BCD medium with reduced potassium nitrate content (400 μM) for sporulation or crossing. All media were supplemented with 0.7% micropropagation agar type II (Caisson Labs, Smithfield, UT). Crossing experiments for the analysis of female sterility were performed with red fluorescent protein (dsRED) tagged line of P. patens accession Vx (Perroud et al., 2011).
Plasmid Construction
Deletion constructs were designed to remove the full open reading frame of the PpXLG, PpGβ1, PpGβ2, PpGγ1, and PpGγ2 loci. The flanking regions of corresponding genes (according the individual gene model annotated in phytozome 8.1) encompassing 1,000 bp upstream and downstream of the coding region were amplified from the genomic DNA extracted from P. patens protonemata as previously described (Cove et al., 2009) and cloned between AvrII/XhoI and AscI/NotI restriction sites of the pBHRF plasmids. For generating ΔPpGγ1 ΔPpGγ2 double mutants, a corresponding pBNRF construct was generated for PpGγ1. Arabidopsis genes were amplified from the cDNA synthesized from the total RNA extracted from Arabidopsis leaf tissue using Trizol reagent (Life Technologies). One µg of total RNA after treatment with Turbo DNA free was used for first-strand synthesis using SuperScript III RT reverse transcriptase and oligo(dT) primer (Life Technologies). Arabidopsis complementation constructs were designed to insert AtXLG2 or AtAGB1 coding sequence in the deleted P. patens genomic locus of ΔPpXLG or ΔPpGβ2, respectively. Sequence verified AtXLG2 or AtAGB1 CDS were cloned between SbfI/SalI or NheI/XhoI of pBNRF plasmids comprising PpXLG or PpGβ2 flanking regions similar to the pBHRF constructs described above.
Generation of Loss-of-Function Mutants and Complementation
Deletion as well as complementation mutants were generated by polyethylene glycol-mediated transformation of protoplasts as described previously (Cove et al., 2009). Briefly, 7-d-old protonemata were treated with 0.5% Driselase (D8037; Sigma) in 8.5% mannitol for 45 min. Protoplasts were isolated by filtration and washed twice in 8.5% mannitol with 10 mm CaCl2 (Perroud and Quatrano, 2006). Transformation was performed using 15 μg open-end DNA fragments derived from plasmid DNA digestion. Successful transformants were selected by growth on media supplemented with 25 μg L−1 Hygromycin or G418 (Sigma). Of 25, 12, 14, 15, and 6 (PpXLG, PpGβ1, PpGβ2, PpGγ1, and PpGγ2) antibiotic-resistant transformants, 3, 4, 3, 10, and 4, respectively, were tested for the loss of the corresponding CDS sequence and the insertion at the targeted locus by PCR genotyping after genomic DNA isolation as described previously (Cove et al., 2009). For ΔPpXLG and ΔPpGβ2, one positive transformation line was selected and used for selection marker cassette removal by transient expression of a cre/lox recombinase as described previously (Demko et al., 2014). Loss of resistance was assessed by growth deficiency on 25 μg L−1 Hygromycin or G418 and verified by PCR and sequencing of the genomic locus (Supplemental Figs. S4 and S5). ΔPpXLG and ΔPpGβ2 were used for transformation with AtXLG2 and AtAGB1 CDS complementation constructs. Integration at the PpXLG or PpGβ2 locus was verified by PCR of selection marker resistant transformants, and one line was selected for further analysis (Supplemental Fig. S8).
Sporulation Assay
For sporulation assays, P. patens was grown on modified BCD media with reduced potassium nitrate content (400 μM) for 5 weeks at 25°C before transfer to 15°C. The plants were moisturized twice after 2 and 3 weeks posttransfer and incubated until sporophytes were visible. Sporulation assays were repeated three times for each mutant. Crossing experiments were performed similar to sporulation assays, except both the accessions to be used for crossing were grown together in the same container. Sporophyte fluorescence was observed using a Nikon SMZ 1500 stereomicroscope (Nikon Instruments, Melville, NY). Almost mature sporophytes (light brown) were harvested at approximately 2 to 3 weeks after last watering and sterilized before germination on BCD media as described previously (Cove et al., 2009). Presporulation was investigated before induction with water at 15°C and was observed using Nikon eclipse E800 microscope (Nikon Instruments).
Recombinant Protein Purification and In Vitro GTP-Binding and Hydrolysis Assays
The Gα-like domain of PpXLG (aa 631-1169) was cloned in pET28a vector and expressed in Escherichia coli and used for in vitro BODIPY GTP-FL assays (Choudhury et al., 2013). For competition assays, nonfluorescent GTP, GDP, ATP, or ADP (10 µM) was added to the reaction mix. Fluorescence (excitation 485 nm, emission 520 nm) was recorded using Tecan Infinite 200 PRO (Tecan Group, Maennedorf, Switzerland). BSA was used as negative control and signal was normalized to the last measurement after 600 s.
Plant Hormone Detection and Analysis
P. patens tissue (protonemata/gametophore) was harvested from 21-d-old plants and lyophilized for 16 h before grinding in the presence of liquid N2. Fifty mg ground plant material was used for acidic hormone quantification. Salicylic acid, ABA, and IAA were assayed using Liquid chromatography–mass spectrometry (LC-MS/MS) method as described previously (Hackenberg and Pandey, 2014). A mixture of deuterium labeled standards (D5SA, D6ABA, D2JA, D5IAA) at 2.5 μm each was spiked at the beginning of the extraction in each sample. Samples were homogenized twice in 900 µL MeOH/ACN (1:1, v/v) and once in 200 μL of 30% methanol followed by analysis with LC-MS/MS (Shimadzu LC system with AB Sciex 4000 QTRAP mass spectrometer and TurboIonSpray electrospray ion source using C18 column (Onyx, 4.6 mm × 100 mm, Phenomenex).. The gradient was from 60% 0.1% (v/v) acetic acid in Milli-Q water to 100% of 90% acetonitrile (v/v) with 0.1% acetic acid (v/v). Hormones were detected using MRM transitions and quantified to standard samples.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers Pp1s147_153V6.1 (PpGα/XLG), Pp1s28_162V6.1 (PpGβ1), Pp1s7_401V6.2 (PpGβ2 ), Pp1s39_119V6.1 (PpGγ1), and Pp1s22_182V6.1 (PpGγ2).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Amino acid sequence alignment of the Gα-like proteins in Arabidopsis and P. patens.
Supplemental Figure S2. Amino acid sequence alignment and phylogenetic relationship analysis of the Gβ-like proteins from selected plant species.
Supplemental Figure S3. Amino acid sequence alignment and phylogenetic relationship analysis of the Gγ-like proteins from selected plant species.
Supplemental Figure S4. Scheme and genotypic analysis of PpXLG locus in wild-type and mutant P. patens plants.
Supplemental Figure S5. Scheme and genotypic analysis of PpGβ2 locus in wild-type and mutant P. patens plants.
Supplemental Figure S6. Phytohormone level and growth of on exogenously added phytohormones of wild-type, ΔPpXLG, and PpGβ2 plants.
Supplemental Figure S7. Analysis of fluorescent gametophyte and sporophyte of wild-type, mutant, and complemented P. patens plants.
Supplemental Figure S8. Integration of Arabidopsis XLG2 and AGB1 at the ΔPpXLG and ΔPpGβ2 loci.
Supplemental Table S1. List of primers used in this study.
Glossary
- ABA
abscisic acid
- IAA
3-acetic acid
Footnotes
Articles can be viewed without a subscription.
References
- Abramowitz J, Grenet D, Birnbaumer M, Torres HN, Birnbaumer L (2004) XLalphas, the extra-long form of the alpha-subunit of the Gs G protein, is significantly longer than suspected, and so is its companion Alex. Proc Natl Acad Sci USA 101: 8366–8371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arif MA, Fattash I, Ma Z, Cho SH, Beike AK, Reski R, Axtell MJ, Frank W (2012) DICER-LIKE3 activity in Physcomitrella patens DICER-LIKE4 mutants causes severe developmental dysfunction and sterility. Mol Plant 5: 1281–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikari M, Wu J, Yano M, Sasaki T, Yoshimura A (1999) Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the alpha-subunit of GTP-binding protein. Proc Natl Acad Sci USA 96: 10284–10289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisht NC, Jez JM, Pandey S (2011) An elaborate heterotrimeric G-protein family from soybean expands the diversity of plant G-protein networks. New Phytol 190: 35–48 [DOI] [PubMed] [Google Scholar]
- Bommert P, Je BI, Goldshmidt A, Jackson D (2013) The maize Gα gene COMPACT PLANT2 functions in CLAVATA signalling to control shoot meristem size. Nature 502: 555–558 [DOI] [PubMed] [Google Scholar]
- Cabrera-Vera TM, Vanhauwe J, Thomas TO, Medkova M, Preininger A, Mazzoni MR, Hamm HE (2003) Insights into G protein structure, function, and regulation. Endocr Rev 24: 765–781 [DOI] [PubMed] [Google Scholar]
- Chakravorty D, Gookin TE, Milner MJ, Yu Y, Assmann SM (2015) Extra-Large G proteins (XLGs) expand the repertoire of subunits in Arabidopsis heterotrimeric G protein signaling. Plant Physiol 169: 512–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Pandey S, Huang J, Alonso JM, Ecker JR, Assmann SM, Jones AM (2004) GCR1 can act independently of heterotrimeric G-protein in response to brassinosteroids and gibberellins in Arabidopsis seed germination. Plant Physiol 135: 907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudert Y, Dievart A, Droc G, Gantet P (2013) ASL/LBD phylogeny suggests that genetic mechanisms of root initiation downstream of auxin are distinct in lycophytes and euphyllophytes. Mol Biol Evol 30: 569–572 [DOI] [PubMed] [Google Scholar]
- Coudert Y, Palubicki W, Ljung K, Novak O, Leyser O, Harrison CJ (2015) Three ancient hormonal cues co-ordinate shoot branching in a moss. eLife 4: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cove DJ, Perroud PF, Charron AJ, McDaniel SF, Khandelwal A, Quatrano RS (2009) The moss Physcomitrella patens: a novel model system for plant development and genomic studies. Cold Spring Harb Protoc 2009: pdb emo115 [DOI] [PubMed]
- Demko V, Perroud PF, Johansen W, Delwiche CF, Cooper ED, Remme P, Ako AE, Kugler KG, Mayer KF, Quatrano R, et al. (2014) Genetic analysis of DEFECTIVE KERNEL1 loop function in three-dimensional body patterning in Physcomitrella patens. Plant Physiol 166: 903–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Pandey S, Assmann SM (2008) Arabidopsis extra-large G proteins (XLGs) regulate root morphogenesis. Plant J 53: 248–263 [DOI] [PubMed] [Google Scholar]
- Frank MH, Scanlon MJ (2015a) Cell-specific transcriptomic analyses of three-dimensional shoot development in the moss Physcomitrella patens. Plant J 83: 743–751 [DOI] [PubMed] [Google Scholar]
- Frank MH, Scanlon MJ (2015b) Transcriptomic evidence for the evolution of shoot meristem function in sporophyte-dominant land plants through concerted selection of ancestral gametophytic and sporophytic genetic programs. Mol Biol Evol 32: 355–367 [DOI] [PubMed] [Google Scholar]
- Hackenberg D, McKain M, Lee S-G, Roy Choudhury S, McCann T, Schreier S, Harkess A, Pires JC, Wong GK-S, Jez J, et al. (2016) Gα:RGS protein pairs maintain functional compatibility and conserved interaction interfaces throughout evolution despite frequent loss of RGS proteins in plants. New Phytol (In press) [DOI] [PubMed] [Google Scholar]
- Hackenberg D, Pandey S (2014) Heterotrimeric G proteins in green algae: an early innovation in the evolution of the plant lineage. Plant Signal Behav 9: e28457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JB, Sung S, Assmann SM (2012) Ca2+-dependent GTPase, extra-large G protein 2 (XLG2), promotes activation of DNA-binding protein related to vernalization 1 (RTV1), leading to activation of floral integrator genes and early flowering in Arabidopsis. J Biol Chem 287: 8242–8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Nakajima M, Asano K, Nishiyama T, Sakakibara H, Kojima M, Katoh E, Xiang H, Tanahashi T, Hasebe M, et al. (2007) The GID1-mediated gibberellin perception mechanism is conserved in the Lycophyte Selaginella moellendorffii but not in the Bryophyte Physcomitrella patens. Plant Cell 19: 3058–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiss M, Laule O, Meskauskiene RM, Arif MA, Decker EL, Erxleben A, Frank W, Hanke ST, Lang D, Martin A, et al. (2014) Large-scale gene expression profiling data for the model moss Physcomitrella patens aid understanding of developmental progression, culture and stress conditions. Plant J 79: 530–539 [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8: 275–282 [DOI] [PubMed] [Google Scholar]
- Katsumata T, Fukazawa J, Magome H, Jikumaru Y, Kamiya Y, Natsume M, Kawaide H, Yamaguchi S (2011) Involvement of the CYP78A subfamily of cytochrome P450 monooxygenases in protonema growth and gametophore formation in the moss Physcomitrella patens. Biosci Biotechnol Biochem 75: 331–336 [DOI] [PubMed] [Google Scholar]
- Komatsu K, Suzuki N, Kuwamura M, Nishikawa Y, Nakatani M, Ohtawa H, Takezawa D, Seki M, Tanaka M, Taji T, et al. (2013) Group A PP2Cs evolved in land plants as key regulators of intrinsic desiccation tolerance. Nat Commun 4: 2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta N, Trusov Y, Brenya E, Parekh U, Botella JR (2015) Membrane-localized extra-large G proteins and Gbg of the heterotrimeric G proteins form functional complexes engaged in plant immunity in Arabidopsis. Plant Physiol 167: 1004–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilson SE, Assmann SM (2010) The alpha-subunit of the Arabidopsis heterotrimeric G protein, GPA1, is a regulator of transpiration efficiency. Plant Physiol 152: 2067–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Ramirez C, Hernandez-Coronado M, Thamm A, Catarino B, Wang M, Dolan L, Feijo JA, Becker JD (2016) A transcriptome atlas of Physcomitrella patens provides insights into the evolution and development of land plants. Mol Plant 9: 205–220 [DOI] [PubMed] [Google Scholar]
- Pandey S, Chen JG, Jones AM, Assmann SM (2006) G-protein complex mutants are hypersensitive to abscisic acid regulation of germination and postgermination development. Plant Physiol 141: 243–256 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pandey S, Monshausen GB, Ding L, Assmann SM (2008) Regulation of root-wave response by extra large and conventional G proteins in Arabidopsis thaliana. Plant J 55: 311–322 [DOI] [PubMed] [Google Scholar]
- Perroud PF, Cove DJ, Quatrano RS, McDaniel SF (2011) An experimental method to facilitate the identification of hybrid sporophytes in the moss Physcomitrella patens using fluorescent tagged lines. New Phytol 191: 301–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroud PF, Quatrano RS (2006) The role of ARPC4 in tip growth and alignment of the polar axis in filaments of Physcomitrella patens. Cell Motil Cytoskeleton 63: 162–171 [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Roy Choudhury S, Bisht NC, Thompson R, Todorov O, Pandey S (2011) Conventional and novel Gγ protein families constitute the heterotrimeric G-protein signaling network in soybean. PLoS One 6: e23361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy Choudhury S, Pandey S (2015) Phosphorylation-dependent regulation of G-protein cycle during nodule formation in soybean. Plant Cell 27: 3260–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy Choudhury S, Westfall CS, Hackenberg D, Pandey S (2013) Measurement of GTP-binding and GTPase activity of heterotrimeric Gα proteins. Methods Mol Biol 1043: 13–20 [DOI] [PubMed] [Google Scholar]
- Siderovski DP, Willard FS (2005) The GAPs, GEFs, and GDIs of heterotrimeric G-protein alpha subunits. Int J Biol Sci 1: 51–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP. (2011) The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol 21: R338–R345 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple BR, Jones AM (2007) The plant heterotrimeric G-protein complex. Annu Rev Plant Biol 58: 249–266 [DOI] [PubMed] [Google Scholar]
- Ullah H, Chen JG, Temple B, Boyes DC, Alonso JM, Davis KR, Ecker JR, Jones AM (2003) The beta-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell 15: 393–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Jones AM (2014) Heterotrimeric G protein-coupled signaling in plants. Annu Rev Plant Biol 65: 365–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L, Bezanilla M (2012) Physcomitrella patens: a model for tip cell growth and differentiation. Curr Opin Plant Biol 15: 625–631 [DOI] [PubMed] [Google Scholar]
- Weiss CA, Garnaat CW, Mukai K, Hu Y, Ma H (1994) Isolation of cDNAs encoding guanine nucleotide-binding protein beta-subunit homologues from maize (ZGB1) and Arabidopsis (AGB1). Proc Natl Acad Sci USA 91: 9554–9558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss CA, Huang H, Ma H (1993) Immunolocalization of the G protein alpha subunit encoded by the GPA1 gene in Arabidopsis. Plant Cell 5: 1513–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaari R, Noy-Malka C, Wiedemann G, Auerbach Gershovitz N, Reski R, Katz A, Ohad N (2015) DNA METHYLTRANSFERASE 1 is involved in (m)CG and (m)CCG DNA methylation and is essential for sporophyte development in Physcomitrella patens. Plant Mol Biol 88: 387–400 [DOI] [PubMed] [Google Scholar]
- Yasumura Y, Crumpton-Taylor M, Fuentes S, Harberd NP (2007) Step-by-step acquisition of the gibberellin-DELLA growth-regulatory mechanism during land-plant evolution. Curr Biol 17: 1225–1230 [DOI] [PubMed] [Google Scholar]
- Yasumura Y, Pierik R, Kelly S, Sakuta M, Voesenek LA, Harberd NP (2015) An ancestral role for CONSTITUTIVE TRIPLE RESPONSE1 proteins in both ethylene and abscisic acid signaling. Plant Physiol 169: 283–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Hu G, Cheng Y, Huang J (2008) Heterotrimeric G protein alpha and beta subunits antagonistically modulate stomatal density in Arabidopsis thaliana. Dev Biol 324: 68–75 [DOI] [PubMed] [Google Scholar]