A Proline Dehydrogenase1 promoter:luciferase screen and transcriptome of the proline synthesis mutant p5cs1-4 find coordination between proline and multiple redox-related metabolic pathways.
Abstract
Proline (Pro) accumulation is one of the most prominent changes in plant metabolism during drought and low water potential; however, the regulation and function of Pro metabolism remain unclear. We used a combination of forward genetic screening based on a Proline Dehydrogenase1 (PDH1) promoter-luciferase reporter (PDH1pro:LUC2) and RNA sequencing of the Pro synthesis mutant p5cs1-4 to identify multiple loci affecting Pro accumulation in Arabidopsis (Arabidopsis thaliana). Two mutants having high PDH1pro:LUC2 expression and increased Pro accumulation at low water potential were found to be alleles of Cytochrome P450, Family 86, Subfamily A, Polypeptide2 (CYP86A2) and Long Chain Acyl Synthetase2 (LACS2), which catalyze two successive steps in very-long-chain fatty acid (VLCFA) synthesis. Reverse genetic experiments found additional VLCFA and lipid metabolism-related mutants with increased Pro accumulation. Altered cellular redox status is a key factor in the coordination of Pro and VLCFA metabolism. The NADPH oxidase inhibitor diphenyleneiodonium (DPI) induced high levels of Pro accumulation and strongly repressed PDH1pro:LUC2 expression. cyp86a2 and lacs2 mutants were hypersensitive to diphenyleneiodonium but could be reverted to wild-type Pro and PDH1pro:LUC2 expression by reactive oxygen species scavengers. The coordination of Pro and redox metabolism also was indicated by the altered expression of chloroplast and mitochondria electron transport genes in p5cs1-4. These results show that Pro metabolism is both influenced by and influences cellular redox status via previously unknown coordination with several metabolic pathways. In particular, Pro and VLCFA synthesis share dual roles to help buffer cellular redox status while producing products useful for stress resistance, namely the compatible solute Pro and cuticle lipids.
Pro accumulation occurs in many plant species and can be induced by several types of abiotic stresses, including drought (Szabados and Savouré, 2010; Verslues and Sharma, 2010). Altered Pro metabolism also can have effects on plant development (Funck et al., 2012; Mattioli et al., 2012; Kavi Kishor et al., 2015). Of the many stimuli that can influence Pro, it is drought and low water potential (ψw) that induce the highest levels of Pro accumulation. There has been considerable effort to modify Pro metabolism to enhance drought resistance (Zhu et al., 1998; Nanjo et al., 1999; Roosens et al., 2002; Sawahel and Hassan, 2002; de Ronde et al., 2004; Su and Wu, 2004; Gleeson et al., 2005). Such efforts have been impeded by unresolved questions regarding why plants accumulate Pro and how it contributes to drought tolerance. Substantial evidence indicates a role for Pro in osmotic adjustment based on both the high levels of Pro accumulation and observations that it accumulates predominantly in the relatively small volume of the cytoplasm and organelles (Voetberg and Sharp, 1991; Bussis and Heineke, 1998). However, osmotic adjustment is not the only role of Pro accumulation. More recent data have emphasized the connection of Pro to redox status (Sharma et al., 2011; Ben Rejeb et al., 2014, 2015) and demonstrated that both Pro synthesis and catabolism are essential to promote drought tolerance (Sharma et al., 2011; Bhaskara et al., 2015). The overall picture that emerges is that both the level of Pro accumulated as well as the turnover of Pro and its connection to broader metabolic status must be considered to fully understand the role of Pro metabolism in drought resistance (Szabados and Savouré, 2010; Bhaskara et al., 2015).
Pro is synthesized by the conversion of Glu to the intermediate Δ1-pyrroline-5-carboxylate (P5C) by P5C synthetase (P5CS), and P5C is then converted to Pro by P5C reductase (P5CR; Szabados and Savouré, 2010; Verslues and Sharma, 2010). P5CS utilizes NADPH as a cofactor (Zhang et al., 1995). P5CR can utilize both NADH and NADPH; however, recent work indicates that NADPH is the predominant source of reductant used by P5CR in vivo (Giberti et al., 2014). Arabidopsis (Arabidopsis thaliana) has two P5CS genes that are not functionally equivalent (Székely et al., 2008). P5CS1 is induced by drought and related stresses at both the transcript and protein levels and is thought to be the rate-limiting step for stress-induced Pro synthesis. p5cs1 mutants have greatly reduced Pro accumulation during low ψw or salt stress but have normal growth and morphology under unstressed conditions (Székely et al., 2008; Sharma et al., 2011; Bhaskara et al., 2015). Despite many stress-related studies that have measured or manipulated P5CS1 gene expression, how P5CS1-mediated Pro synthesis is regulated and interacts with other metabolic pathways is still unclear.
Pro catabolism occurs in the mitochondria via proline dehydrogenase (PDH), which converts Pro to P5C, and P5C dehydrogenase (P5CDH), which converts P5C back to Glu, thus forming the Pro cycle (Szabados and Savouré, 2010; Verslues and Sharma, 2010). A unique feature of PDH is that it donates electrons directly to the mitochondrial electron transport chain; however, mechanistic details and the identity of the electron acceptor are not firmly established in plants (Szabados and Savouré, 2010; Verslues and Sharma, 2010). Of the two Arabidopsis genes that encode PDH, PDH1 is more highly expressed and is thought to have a predominant role in Pro catabolism (Funck et al., 2010). PDH1 expression is down-regulated by drought stress in much of the plant (Peng et al., 1996; Yoshiba et al., 1997; Sharma and Verslues, 2010). Conversely, PDH1 is induced by exogenous Pro, and this induction involves a cis-element containing the sequence ACTCAT, referred to as the proline response element (ProRE; Satoh et al., 2002). The ProRE is bound by several bZIP transcription factors that can up-regulate PDH1 in response to exogenous Pro, starvation, and perhaps other signals (Satoh et al., 2004; Weltmeier et al., 2006; Dietrich et al., 2011). The upstream factors that regulate the activity of these bZIPs are not known. The up-regulation of PDH1 in response to Pro presents an unresolved paradox. During drought stress, a high level of Pro accumulates; yet, rather than be induced by this high level of Pro, PDH1 is down-regulated in most plant tissues (Miller et al., 2005). Thus, PDH1 is regulated by incompletely understood mechanisms that likely reflect both stress and metabolic signals. All these factors make PDH1 an excellent marker to study the intersection of stress signaling and metabolic regulation.
We used a forward genetic screen based on the expression of a PDH1 promoter:Luciferase construct combined with mapping by sequencing to identify factors affecting PDH1 expression and Pro accumulation. This screen found an unexpected effect of very-long-chain fatty acid (VLCFA) synthesis on Pro accumulation. Several lines of evidence indicate that reduced VLCFA synthesis affects Pro accumulation via effects on redox status rather than signaling functions of VLCFA metabolism enzymes or lipid metabolism intermediates. In parallel, mRNA sequencing of p5cs1-4, which has reduced Pro synthesis, found substantial effects on chloroplast and mitochondria gene expression consistent with a connection of Pro to redox metabolism in both organelles. Together, these results show the extensive coordination of Pro metabolism with multiple metabolic pathways related to cellular redox status.
RESULTS
A Mutant Screen Based on the PDH1 Promoter Identifies Mutants of Cytochrome P450, Family 86, Subfamily A, Polypeptide2 and Long Chain Acyl Synthetase2 with Increased Pro Accumulation at Low ψw
A forward genetic screen was developed to identify factors affecting PDH1 expression and Pro accumulation. A 1.5-kb fragment of the PDH1 promoter and 5′ untranslated region (UTR) was used to drive the expression of the Luciferase2 reporter (PDH1pro:LUC2). Several Arabidopsis (Columbia-0 [Col-0] accession) transgenic lines containing single-locus insertions of the PDH1pro:LUC2 reporter were isolated. A single line having 5-fold down-regulation of PDH1pro:LUC2 in response to low ψw, essentially identical to the endogenous PDH1 (Fig. 1; Sharma and Verslues, 2010), was selected for further study. This unmutagenized PDH1pro:LUC2 line is referred to as wild type (W.T.) in subsequent figures. After ethyl methanesulfonate (EMS) mutagenesis, screening in the M1 and M2 generations (Supplemental Fig. S1) identified mutants with high or low PDH1pro:LUC2 expression. Candidate genes for a number of high-PDH1pro:LUC2 mutants were identified using a mapping-by-sequencing approach, and two such mutants are reported here.
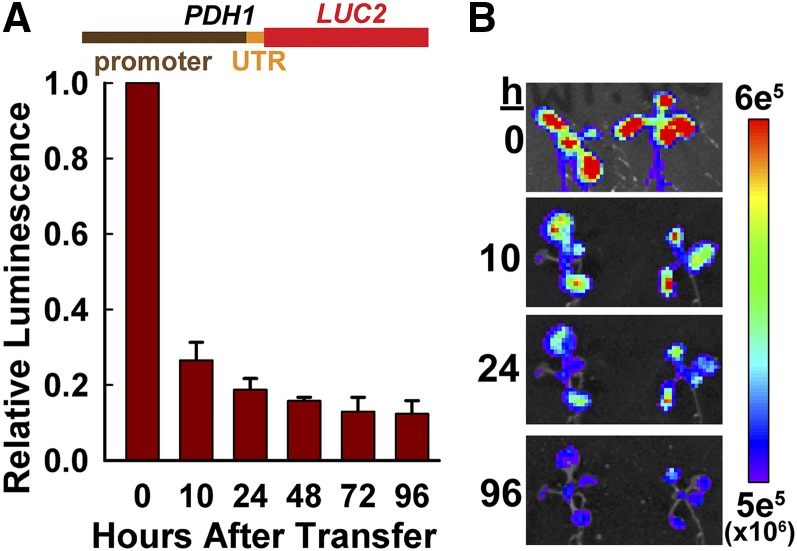
Figure 1.
A PDH1pro:LUC2 reporter responsive to low ψw. A, The PDH1 promoter and 5′ UTR were fused to the LUC2 coding region and used to generate transgenic plants. Quantitation of PDH1pro:LUC2 luminescence in shoot tissue of seedlings transferred to low ψw (−1 MPa) for the indicated lengths of time is shown. Data are means ± se (n = 6). B, Representative false-color images of luminescence intensity from the experiments reported in A. The intensity scale for the images is shown at right.
Mutants 4442-4 and 4255-1 were phenotypically similar, with each one having 3-fold increased PDH1pro:LUC2 expression in both control and stress treatments (Fig. 2, A and B). Low ψw could still repress the expression of PDH1pro:LUC2 in 4442-4 and 4255-1, but not to the level seen in the wild type. Also, both mutants had Pro accumulation nearly twice that of the wild type at −1.2 MPa (Fig. 2C) but did not differ from the wild type in the Pro content of unstressed plants. Expression of the endogenous PDH1 was increased approximately 3-fold in 4442-4 under unstressed conditions (Fig. 2D) and agreed well with the increased PDH1pro:LUC2 expression. However, at low ψw, PDH1 expression decreased to a level similar to that in the wild type, despite the higher PDH1pro:LUC2 expression (Fig. 2D). A similar difference between PDH1pro:LUC2 expression and endogenous PDH1 at low ψw also was observed in other high-Pro, high-PDH1pro:LUC2 mutants in our collection (S. Mukuri, S. Shinde, and P.E. Verslues, unpublished data).
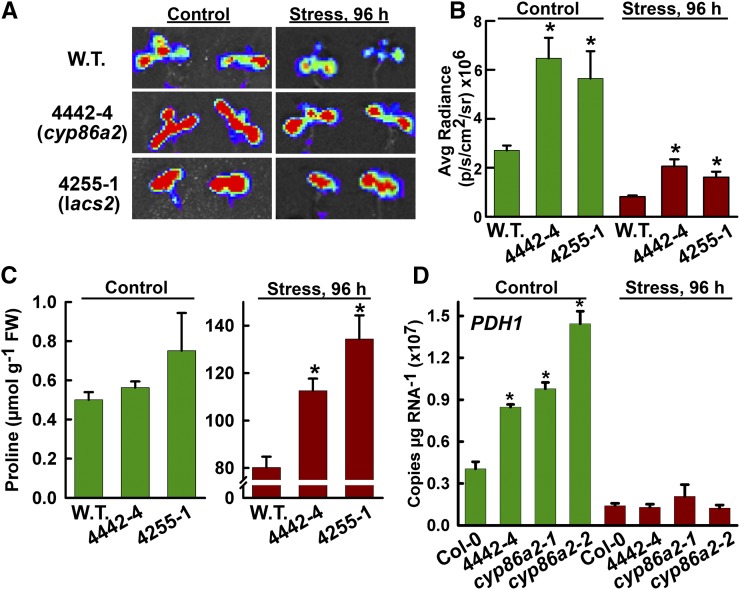
Figure 2.
Mutants 4442-4 and 4255-1 have increased PDH1pro:LUC2 activity and increased Pro accumulation at low ψw. A, Representative false-color luminescence images of the wild type (W.T.) and mutants in the unstressed control and after 96 h at −1 MPa low-ψw treatment. B, Quantification of PDH1pro:LUC2 activity in 4442-4 and 4255-1. Data are means ± se (n = 4–15). Significant differences (P ≥ 0.05) compared with the wild type in the same treatment are marked with asterisks. Luminescence intensities are given in photons (p) per second (s) per cm2 per steradian (sr). C, Pro accumulation in unstressed control (−0.25 MPa) or low-ψw stress (−1.2 MPa) for the wild type and mutants. FW, Fresh weight. Data are means ± se (n = 3–9). Significant differences (P ≥ 0.05) compared with the wild type in the same treatment are marked with asterisks. Note that the wild type is the unmutagenized PDH1pro:LUC2 line for all experiments in A to C. D, PDH1 gene expression in Col-0 wild type, 4442-4, and two T-DNA alleles of cyp86a2 in the unstressed control treatment or at 96 h after transfer to −1.2-MPa stress. Data are means ± se (n = 3). Significant differences (P ≥ 0.05) compared with the wild type in the same treatment are marked with asterisks. The expression of additional Pro metabolism genes in these mutants is shown in Supplemental Figure S4.
For 4442-4, sequencing of bulked segregants and next-generation mapping (Austin et al., 2011) identified several candidate genes (Supplemental Fig. S2A). However, only mutants of Cytochrome P450, Family 86, Subfamily A, Polypeptide 2 (cyp86a2; At4g00360) had elevated Pro levels similar to 4442-4 (Fig. 3A). Mutants of the two other candidate genes did not differ from the wild type in Pro accumulation (Supplemental Fig. S2B). For 4255-1, Long Chain Acyl Synthetase2 (LACS2-1; At1g49430) was identified as the main candidate gene (Supplemental Fig. S2C), and lacs2-1 (Schnurr et al., 2004) had increased Pro accumulation similar to 4255-1 (Fig. 3A). Genetic complementation found that F1 seedlings of 4442-4 crossed to cyp86a2-1 as well as 4255-1 crossed to lacs2-1 had the same high PDH1pro:LUC2 expression as the homozygous mutant (Fig. 3B). This indicated that 4442-4 was an allele of cyp86a2 and 4255-1 was an allele of lacs2.
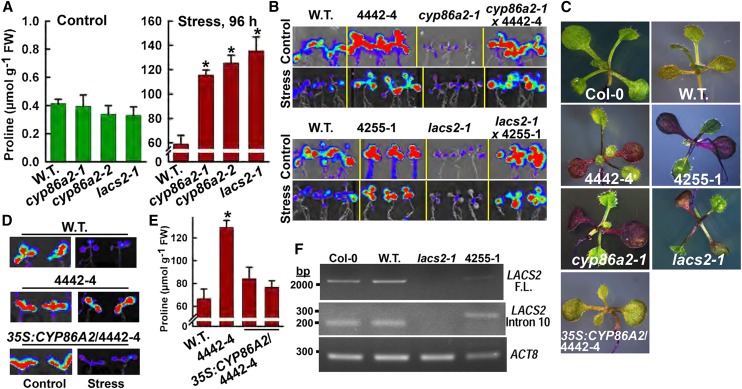
Figure 3.
4442-4 and 4255-1 are alleles of cyp86a2 and lacs2, respectively. A, Pro accumulation in two cyp86a2 T-DNA alleles and lacs2-1 under control and −1.2-MPa low ψw stress treatments. FW, Fresh weight. Data are means ± se (n = 4–6) from two experiments. Significant differences (P ≥ 0.05) compared with the wild type (W.T.) in the same treatment are marked with asterisks. B, PDH1pro:LUC2 imaging in F1 seedlings of crosses of 4442-4 and 4255-1 with cyp86a2-1 and lacs2-1, respectively, along with wild-type PDH1pro:LUC2 and mutant seedlings as controls. C, Toluidine Blue staining using seedlings of Col-0, wild-type PDH1pro:LUC2, as well as mutants and a transgenic complemented line of 4442-4. D, PDH1pro:LUC2 imaging of wild-type PDH1pro:LUC2, 4442-4, and 4442-4 complemented with 35S:CYP86A2. Stress treatment was −1 MPa for 96 h. E, Stress-induced (−1.2 MPa, 96 h) Pro accumulation in 4442-4 and two independent T3 lines of 4442-4 complemented with 35S:CYP86A2. Data are means ± se (n = 4–6) from two experiments. Significant differences (P ≥ 0.05) compared with the wild type in the same treatment are marked with asterisks. F, Reverse transcription (RT)-PCR analysis of Col-0, wild-type PDH1pro:LUC2, lacs2-1, and 4255-1 using primers to amplify the full-length LACS2 RNA or the region around intron 10. ACTIN8 (ACT8) was used as a reference gene.
Both LACS2 and CYP86A2 are endoplasmic reticulum-localized enzymes that catalyze successive steps in VLCFA synthesis: the esterification of C16 and C18 fatty acids to CoA and subsequent ω-hydroxylation of these fatty acids for the synthesis of cutin monomers (Li-Beisson et al., 2013). EMS mutants 4442-4 and 4255-1 stained with Toluidine Blue (Fig. 3C), indicating that they share the previously reported cuticle defects of cyp86a2 and lacs2 (Schnurr et al., 2004; Xiao et al., 2004). We also transgenically complemented 4442-4 with 35S:CYP86A2 (Fig. 3, C–E) and showed that introducing the cyp86a2-1 and lacs2-1 T-DNA mutants into the PDH1pro:LUC2 background produced the same high PDH1pro:LUC2 expression phenotype as the 4442-4 and 4255-1 EMS mutants (Supplemental Fig. S3). 4442-4 has a point mutation that changes Gly-37 to Asp (G-to-A transition at position 110 of the CYP86A2 coding sequence). This mutation lies in the catalytic domain (amino acids 5–511 based on the National Center for Biotechnology Information’s conserved domain database search), consistent with 4442-4 being a null allele of CYP86A2. For 4255-1, the mutation was in a cryptic splice site at the LACS2 intron 10-exon 11 junction (G-to-A transition at position 18,293,272 of chromosome 1). Primers spanning intron 10 amplified a larger fragment in 4255-1 than in the wild type (Fig. 3F), consistent with retention of the 84-bp intron 10. This alters the downstream reading frame, thus preventing the translation of functional LACS2 protein.
Further characterization of 4442-4 and 4255-1 as well as cyp86a2-1 and lacs2-1 T-DNA mutants found that the protein abundance and apparent Mr of PDH1 and P5CS1 were not altered in any of the mutants under either stress or control conditions (Fig. 4A). Thus, cyp86a2 and lacs2 did not affect the posttranslational regulation (e.g. redox-related posttranslational modifications; see below) of these two key enzymes of Pro metabolism. P5CS1 gene expression, as well as the expression of other Pro metabolism genes, also was not substantially altered in cyp86a2 mutants (Supplemental Fig. S4). Further experiments with cyp86a2 mutants found that they had increased Pro across a range of low ψw severities, with even a mild stress of −0.5 MPa leading to twice as much Pro accumulation in cyp86a2 as in the wild type (Fig. 4B). Interestingly, cyp86a2 had the same Pro accumulation as the wild type in response to salt stress or exogenous abscisic acid (Fig. 4B). Thus, the effect of disrupting VLCFA synthesis on Pro was specific to low ψw and occurred via an unknown mechanism that did not involve altered gene expression or modification of the two key Pro metabolism enzymes P5CS1 and PDH1.
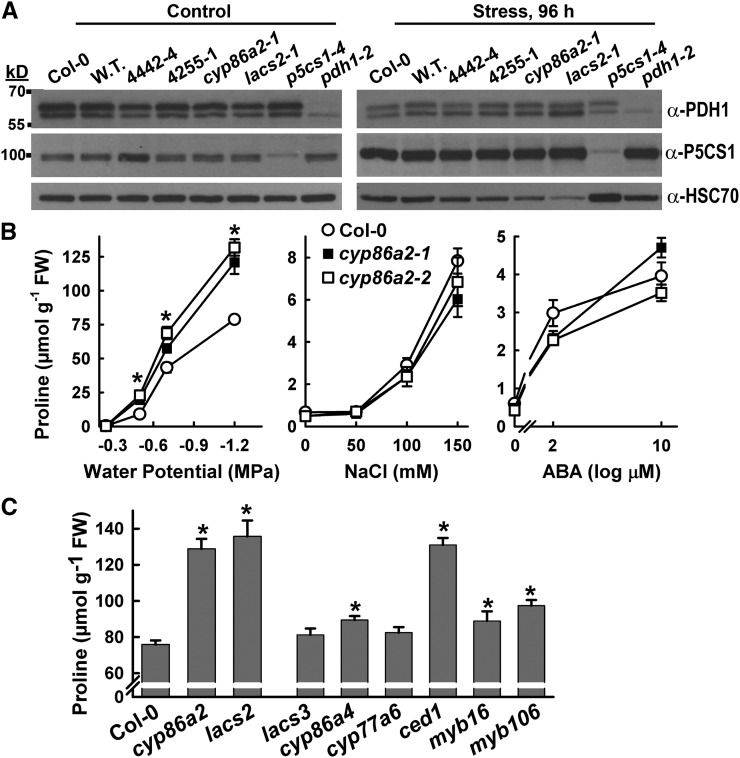
Figure 4.
Effects of cyp86a2 and lacs2 mutants on Pro metabolism and the identification of additional cuticle lipid-related mutants with increased Pro accumulation at low ψw. A, Western blots of PDH1 and P5CS1 protein levels in Col-0, wild-type PDH1pro:LUC2 (W.T.), and cyp86a2 and lacs2 mutants. p5cs1-4 and pdh1-2 were included to show the specificity of the antisera. Blots were stripped and reprobed with antisera recognizing HSC70 as a loading control. A total of 50 μg of protein was loaded in each lane. B, Effects of a range of low-ψw, salt, or abscisic acid (ABA) treatments on Pro accumulation of the Col-0 wild type and cyp86a2 mutants. FW, Fresh weight. Data are means ± se (n = 8–12) from two experiments. Significant differences (P ≥ 0.05) compared with the wild type in the same treatment are marked with asterisks. C, Stress-induced (−1.2 MPa, 96 h) Pro accumulation in cuticle metabolism mutants. Data are means ± se (n = 11–24) from two experiments. Significant differences (P ≥ 0.05) compared with the wild type are marked with asterisks. For ced1, cyp77a6, and cyp86a4, the data shown are combined from two to three T-DNA alleles that had identical phenotypes. Additional information and RT-PCR verification of T-DNA mutants can be found in Supplemental Figure S10.
Additional Mutants Affecting VLCFA Synthesis or Cuticle Deposition Also Have Increased Pro Accumulation
The fact that both CYP86A2 and LACS2 were identified in our mutant screen suggested that the high Pro accumulation and increased PDH1pro:LUC2 expression may not be due to a specific signaling or enzymatic function of the CYP86A2 and LACS2 proteins. As VLCFAs have been shown to have a signaling function in controlling development (Nobusawa et al., 2013), there were several possible ways in which VLCFA metabolism could influence Pro accumulation and PDH1pro:LUC2 expression. We hypothesized that reduced flux through VLCFA synthesis, reduced levels of a product downstream of CYP86A2, or increased levels of a lipid species upstream of LACS2 could cause the Pro-related phenotypes of cyp86a2 and lacs2 mutants. As one way to test these possibilities, we isolated T-DNA mutants of other genes involved in VLCFA metabolism or its regulation. Of these, a mutant of BODYGUARD/9-CIS EPOXYCAROTENOID DEFECTIVE1 (CED1), a cuticle lipid transporter found previously to affect the abiotic stress response (Wang et al., 2011b), had increased Pro accumulation similar to cyp86a2 and lacs2-1 (Fig. 4C). Because CED1 functions downstream of cyp86a2 and lacs2, these data indicated that it was not a reduced level of a lipid intermediate downstream of CYP86A2 that increased Pro accumulation. Rather, the blockage of VLCFA metabolism itself seemed a more likely cause. Consistent with this hypothesis, mutants of other VLCFA-related genes had moderate, but significant, increases in Pro accumulation (Fig. 4C), likely reflecting the extent that VLCFA synthesis was disrupted. CYP86A4 and CYP77A6 are most active in flowers, with a lesser effect on leaf cuticle (Li-Beisson et al., 2009), and had moderately increased or unaffected Pro accumulation (Fig. 4C). Similarly, MYB16 and MYB106 have overlapping functions, such that each single mutant causes only a partial blockage of flux through VLCFA synthesis (Oshima et al., 2013), and, thus, only a moderate effect on Pro accumulation. Other mutants or inhibitor treatments affecting lipid metabolism upstream of LACS2 also had high Pro levels (see below), further indicating that reduced metabolic flux through lipid metabolism was the most likely cause of increased Pro accumulation in cyp86a2 and lacs2-1.
Cellular Redox Status Is a Factor in Coordinating Pro and VLCFA Metabolism
Pro and VLCFA synthesis lead to different products and occur in different cellular compartments. However, a commonality is that both Pro and VLCFA synthesis consume NADPH and regenerate NADP+ (Sharma et al., 2011; Li-Beisson et al., 2013; Giberti et al., 2014; Ben Rejeb et al., 2015) and, thus, can influence cellular redox status. To determine whether altered redox status was involved in the high-Pro and high-PDH1pro:LUC2 expression of cyp86a2 and lacs2, dithiothreitol (DTT) was used to increase reductant load, while the reactive oxygen species (ROS) scavengers ascorbic acid (AsA) and N,N'-dimethylthiourea (DMTU) or the NADPH oxidase inhibitor diphenyleneiodonium (DPI) were used to decrease reductant load and interfere with ROS signaling.
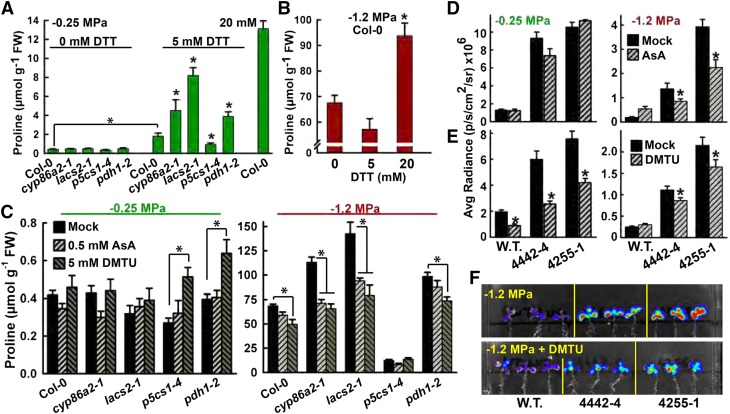
Treatment of unstressed Col-0 (wild type) with 5 or 20 mm DTT increased Pro accumulation 4- or 30-fold, respectively (Fig. 5A). This was in agreement with previous results (Kolbe et al., 2006). cyp86a2-1 and lacs2-1 had greater response of Pro accumulation to DTT (Fig. 5A). This was consistent with these mutants having altered redox status or less capacity to buffer redox status. Pro accumulation of pdh1-2 also was more responsive to DTT, while p5cs1-4 was less responsive, indicating that both Pro synthesis and Pro catabolism were involved in the response to DTT (Fig. 5A). We also observed that 20 mm, but not 5 mm, DTT increased Pro accumulation at low ψw (Fig. 5B). Note that we did not analyze Pro in mutants treated with 20 mm DTT or mutants treated with a combination of DTT and low ψw, because the mutants were more sensitive to these treatments and exhibited bleaching and seedling death. Conversely, the ROS scavengers AsA and DMTU suppressed low-ψw-induced Pro accumulation of cyp86a2-1 and lacs2-1 back to the wild-type level (Fig. 5C). AsA and DMTU also significantly inhibited the increased PDH1pro:LUC2 expression of 4442-4 and 4255-1 at low ψw (Fig. 5, D and F). DMTU also blocked the increased PDH1pro:LUC2 expression of unstressed 4442-4 and 4255-1 (Fig. 5E).
Figure 5.
Effects of DTT and the ROS scavengers AsA and DMTU on Pro accumulation and PDH1pro:LUC2 activity. A, Effects of DTT treatment on the Pro accumulation of seedlings at high ψw. FW, Fresh weight. Data are means ± se (n = 10–44) combined from three experiments. Significant differences (P ≥ 0.05) compared with the wild type in the same treatment or between 0 and 5 mm DTT treatment of the wild type are marked with asterisks. B, Effects of DTT treatment at −1.2 MPa on Pro accumulation. Data are means ± se (n = 10–44) combined from three experiments. Significant differences (P ≥ 0.05) compared with the wild type are marked with asterisks. C, Effects of AsA and DMTU on Pro levels at −0.25 and −1.2 MPa. Data are means ± se (n = 10–44) combined from three experiments. Significant differences (P ≥ 0.05) compared with the mock treatment are marked with asterisks. D, Effects of AsA on PDH1pro:LUC2 activity in the wild type, 4442-4, and 4255-1 at high ψw (−0.25 MPa) or low ψw (−1.2 MPa). Data are means ± se (n = 10–44) combined from three experiments. Significant differences (P ≥ 0.05) compared with the mock treatment are marked with asterisks. E, Effects of DMTU on PDH1pro:LUC2 activity in the wild type, 4442-4, and 4255-1. Data are means ± se (n = 10–44) combined from three experiments. Significant differences (P ≥ 0.05) compared with the mock treatment are marked with asterisks. Luminescence intensities in D and E are given in photons (p) per second (s) per cm2 per steradian (sr). F, False-color imaging of PDH1pro:LUC2 activity in representative seedlings from the experiments reported in E.
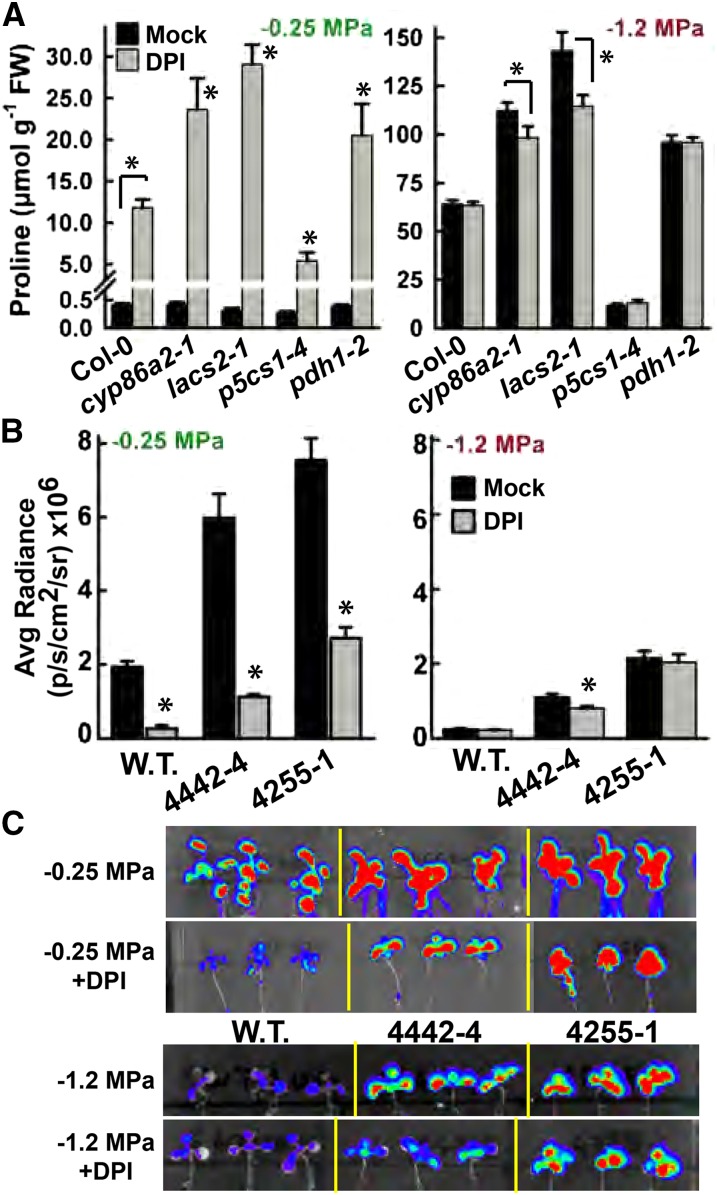
Also consistent with a role of redox or ROS in Pro regulation, blocking NADPH oxidase activity using the inhibitor DPI caused a nearly 30-fold increase in Pro content of Col-0 seedlings at high ψw (Fig. 6A). DPI treatment of unstressed wild-type seedlings (−0.25 MPa) completely suppressed PDH1pro:LUC2 expression to the level typically seen at low ψw (Fig. 6B). An even greater response to DPI was seen in cyp86a2-1, lacs2-1, and pdh1-2, as Pro content increased 50- to 60-fold over the untreated control and PDH1pro:LUC2 activity was suppressed to the wild-type level (Fig. 6, A and B; −0.25 MPa data). Conversely, the response to DPI was less in p5cs1-4, indicating that Pro synthesis via P5CS1 was required for DPI-induced Pro accumulation. At low ψw, where Pro levels are already high and PDH1pro:LUC2 expression is lower, DPI had a lesser effect (Fig. 6, A and B).
Figure 6.
Effects of DPI on Pro accumulation and PDH1pro:LUC2 activity. A, Effects of DPI (2.5 μm) on Pro accumulation of seedlings at high ψw (−0.25 MPa) or low ψw (−1.2 MPa). FW, Fresh weight. Data are means ± se (n = 14–18) combined from three experiments. Significant differences (P ≥ 0.05) compared with the wild type (Col-0) in the same treatment or between mock and DPI treatments of Col-0 are marked with asterisks. B, Effects of DPI on PDH1pro:LUC2 activity in the wild type (W.T.), 4442-4, and 4255-1 at high ψw (−0.25 MPa) or low ψw (−1.2 MPa). Data are means ± se (n = 10–44) combined from three experiments. Significant differences (P ≥ 0.05) compared with the mock treatment are marked with asterisks. Luminescence intensities are given in photons (p) per second (s) per cm2 per steradian (sr). C, False-color imaging of PDH1pro:LUC2 activity in representative seedlings from the experiments reported in B.
Despite the DPI data, measurements of NADP+-NADPH ratio found no significant differences between the wild type, cyp86a2, and lacs2-1 (Supplemental Fig. S5). There was an accumulation of NADPH in all genotypes at low ψw, consistent with previous data (Sharma et al., 2011); however, we did not find a significant difference in the NADP+-NADPH ratio. We also did not see any differences in the amount or localization of ROS staining between the wild type, cyp86a2-1, and lacs2-1 (Supplemental Fig. S6), indicating that uncontrolled ROS buildup did not occur under our stress conditions. These assays do not rule out the involvement of NADP+-NADPH ratio, increased ROS, or ROS signaling in the cyp86a2 and lacs2-1 phenotypes. However, when combined with the AsA, DMTU, and DPI data, these measurements do suggest that any changes in ROS or pyridine nucleotide redox status may be specific to certain subcellular compartments. This is perhaps consistent with changes in VLCFA synthesis in the endoplasmic reticulum being communicated to Pro metabolism in the cytoplasm and mitochondria by specific, but unknown, signaling mechanisms. In this way, redox status may be buffered before damaging bulk changes in redox metabolites or ROS occur. The redox status of the chloroplast also is very likely to be involved in such redox communication and also may influence Pro accumulation (see below).
RNA Sequencing of p5cs1-4 Shows the Influence of Pro Synthesis on the Redox Metabolism of Chloroplast and Mitochondria
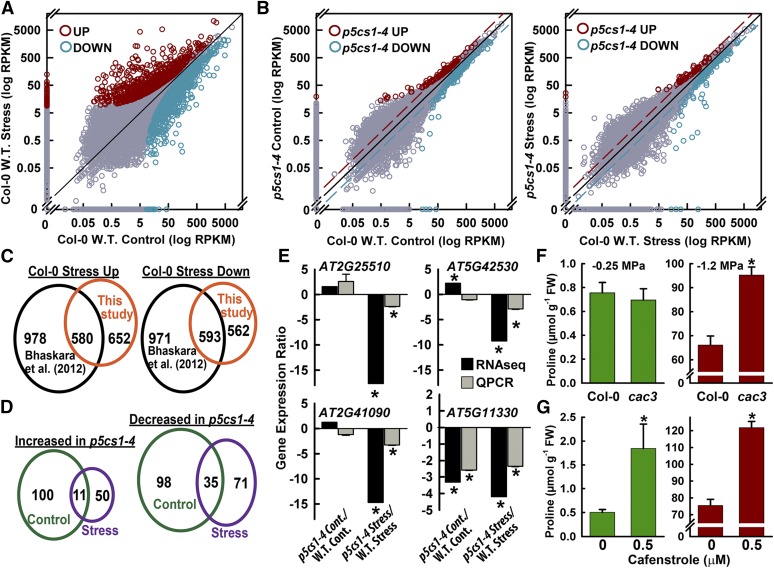
To further test the relationship between Pro- and redox-related metabolism, as well as more generally investigate the effects of reduced Pro synthesis, mRNA sequencing of p5cs1-4 was conducted. p5cs1-4 lacks P5CS1 protein expression (Fig. 4A) and has greatly reduced Pro accumulation and reduced growth at low ψw (Székely et al., 2008; Sharma et al., 2011; Kesari et al., 2012; Bhaskara et al., 2015). For the Col-0 wild type, RNA sequencing (Fig. 7A; Supplemental Table S1) found 1,232 genes with significantly increased expression at low ψw, with an enrichment of GO terms associated with abiotic stress (Supplemental Table S2). Conversely, 1,155 genes were down-regulated (Supplemental Table S3). As one way to assess the quality of this data set, we compared it with microarray data from Bhaskara et al. (2012), who used the same 96-h, −1.2-MPa stress treatment. Approximately half the genes found to be stress up- or down-regulated in Col-0 by RNA sequencing also were found to be up- or down-regulated in the microarray analysis of Bhaskara et al. (2012; Fig. 7B). This is a substantial overlap between the two data sets, when one considers that the RNA sequencing analysis detects genes not present on the ATH1 chips used by Bhaskara et al. (2012) and that, for high-expression genes, RNA sequencing can be more effective in detecting small changes in expression. Conversely, even large fold-change expression differences of genes with overall low expression (low read counts) did not pass the statistical cutoffs used in analyzing the RNA sequencing data.
Figure 7.
RNA sequencing of the Col-0 wild type (W.T.) and p5cs1-4 shows the effects of Pro on chloroplast and mitochondria metabolism and identifies additional lipid metabolism loci affecting Pro accumulation. A, Plot of reads assigned per kilobase of target per million mapped reads (RPKM) values for Col-0 in the unstressed control versus Col-0 after 96 h of low-ψw (−1.2 MPa) treatment. Significantly up- or down-regulated genes are indicated by red or blue circles, respectively, while other data points are plotted in gray. A complete listing of RPKM values can be found in Supplemental Table S1, and lists of significantly up- or down-regulated genes as well as significantly enriched Gene Ontology (GO) terms can be found in Supplemental Tables S2 and S3. B, Plots of RPKM values for p5cs1-4 versus the Col-0 wild type in the unstressed control treatment and after 96 h of low-ψw treatment. Data presentation is as described for A. A complete listing of RPKM values can be found in Supplemental Table S1, and lists of significantly up- or down-regulated genes in p5cs1-4 in both control and stress treatments along with a listing of significantly enriched GO terms can be found in Supplemental Tables S4 to S7. Dashed lines indicate two-fold difference in RPKM. C, Comparison of low-ψw up- or down-regulated genes identified by RNA sequencing with those identified by previous microarray analysis of the same stress treatment. D, Comparison of genes up- or down-regulated in p5cs1-4 in the control and low-ψw stress treatments. E, Comparison of fold change in gene expression detected by RNA sequencing versus fold change detected by quantitative PCR (qPCR) for selected genes down-regulated in p5cs1-4. qPCR data are means ± se (n = 5–6) from two experiments. Ratios significantly different (P ≥ 0.05) from 1 based on a one-sided Student’s t test are indicated with asterisks. F, Pro levels in unstressed control treatment (−0.25 MPa) or after 96 h of low-ψw treatment (−1.2 MPa) for the Col-0 wild type and the cac3 mutant. FW, Fresh weight. Data are means ± se (n = 15–18) from three experiments. A significant difference (P ≥ 0.05) compared with the wild type in the same treatment is marked with an asterisk. G, Effects of the KCS inhibitor cafenstrole on Pro accumulation at high or low ψw. Data are means ± se (n = 15–18) from three experiments. Significant differences (P ≥ 0.05) compared with the wild type in the same treatment are marked with asterisks.
For p5cs1-4, 111 genes were significantly up-regulated and 132 were down-regulated compared with wild-type Col-0 in the unstressed high-ψw treatment. At low ψw, 61 genes were significantly up-regulated and 106 genes were down-regulated in p5cs1-4 compared with wild-type Col-0 (Fig. 7, B and D; Supplemental Tables S4–S7). There were three main patterns of interest in the p5cs1-4 gene expression data. The most prominent pattern was that genes differentially expressed in p5cs1-4 were strongly enriched for functions in chloroplast and mitochondrial redox metabolism (see significantly enriched GO categories in Supplemental Tables S4–S7). This included a striking prevalence of chloroplast-encoded genes among the genes having higher expression in unstressed p5cs1-4 (14 chloroplast-encoded genes out of 111 genes significantly up-regulated; Supplemental Table S4). The portion of chloroplast genes up-regulated in unstressed p5cs1-4 was significantly higher (enriched) compared with the portion of chromosome-encoded genes (two-tailed P < 1 × 10−16 by Fisher’s exact test). The genes up-regulated in p5cs1-4 at low ψw also had a significant enrichment of chloroplast-encoded genes (seven out of 61 up-regulated genes; Supplemental Table S6; two-tailed P < 1 × 10−8 by Fisher’s exact test). In unstressed p5cs1-4, the majority of the most highly up-regulated genes (greater than 5-fold) were chloroplast-encoded genes (Supplemental Table S4). The effects of p5cs1-4 on photosynthesis gene expression were concentrated in the light reactions, while there was little effect of p5cs1-4 on Calvin cycle- or photorespiration-related genes (MapMan analysis; Supplemental Fig. S7). In addition to these genes up-regulated in p5cs1-4, GO analysis found strong and statistically significant enrichment of photosynthesis-related functions among the genes down-regulated in p5cs1-4 (Supplemental Tables S5 and S7).
Also striking was the observation that more than 30 genes encoding chloroplast or mitochondrial NAD(P)H dehydrogenases, as well as genes involved in splicing or processing mitochondrial NAD genes, had higher RPKM values in p5cs1-4, while only three such genes had lower RPKM in p5cs1-4 (Supplemental Table S10; note that this analysis included all genes with fold change greater than 2 to include genes with large fold changes that could not be called significant changes because of low read counts in the wild type). MapMan analysis also showed that p5cs1-4 had a notably different expression pattern of genes encoding mitochondrial function genes, particularly genes in electron transport complex I, compared with the wild type (Supplemental Fig. S8). Also, p5cs1-4 had altered expression of a number of other oxidation reduction-related genes of less clear function (such as At5g11330; Fig. 7E). Together, these data indicate substantially altered chloroplast and mitochondrial electron transport in p5cs1-4 at both high and low ψw.
The second pattern of interest was the overall similar numbers of genes and similar GO enrichment profiles of up- or down-regulated genes in p5cs1-4 for both control and stress treatments (Supplemental Tables S4–S7). There were differences in the individual genes significantly increased or decreased by p5cs1-4 in control and stress treatments (Fig. 7D). This may be mainly a stochastic effect, as many genes narrowly missed our statistical cutoff in either the control or stress treatment. The genes up- or down-regulated in p5cs1-4 included both genes whose expression was affected by stress in the wild type and further altered in p5cs1-4 as well as genes with altered expression in p5cs1-4 but not affected by low ψw in the wild type (Supplemental Tables S8 and S9).
The third pattern of interest was the relative absence of amino acid and nitrogen metabolism functions among genes differentially expressed in p5cs1-4. The most strongly enriched GO terms in p5cs1-4 up- or down-regulated genes did not include any terms related to amino acid or nitrogen metabolism (Supplemental Tables S4–S7; GO terms with P ≤ 0.001). Only a few GO terms related to Glu metabolism, ammonia transport, or ammonia assimilation were found, and these were overall less highly enriched (Supplemental Tables S4–S7), and the number of genes represented in these categories was dwarfed by the number of genes in other functional categories. Even when all the genes with 2-fold or greater RPKM differences between p5cs1-4 and the wild type were included, MapMan analysis showed that very few genes mapped to amino acid and nitrogen metabolism pathways compared with greater numbers of genes in chloroplast light reactions and mitochondrial electron transport (Supplemental Fig. S9). These observations are consistent with the conclusions of Less and Galili (2008), who found that transcriptional regulation of Pro metabolism differed from that of other amino acid metabolism pathways. Thus, the p5cs1-4 transcriptome analysis, as well as the cyp86a2 and lacs2 mutant experiments described above, indicated that Pro metabolism is coordinated extensively with cellular redox status. This included chloroplast and mitochondrial electron transport as well as lipid metabolism. At the same time, the data indicated much less connection of Pro to other aspects of carbon-nitrogen metabolism.
We also used the Signature tool in Genevestigator to find publicly available microarray data with similar patterns of differentially expressed genes to p5cs1-4 under either control or stress conditions. For p5cs1-4 in the unstressed control treatment, this analysis found several data sets related to light signaling (phyA and cop1 mutants) or different light treatments as well as salicylic acid (SA) treatment (Supplemental Fig. S10). For p5cs1-4 at low ψw, there was similarity to gene expression changes observed in SA synthesis and signaling mutants (e.g. sid2-1, npr1, and ssi2) as well as other pathogen signaling mutants (Supplemental Fig. S11). The similarities between p5cs1-affected and light-affected gene expression were consistent with the relationship to chloroplast and photosynthesis gene expression described above. The similarities to pathogen- and SA-related gene expression were consistent with recent data on the role of Pro metabolism in pathogen resistance (Cecchini et al., 2011; Senthil-Kumar and Mysore, 2012). However, it should be noted that these similarities in gene expression were somewhat limited, and p5cs1-4 had an overall pattern of gene expression that was distinct from any of the data sets included in the Genevestigator analysis.
p5cs1-4 mRNA Sequencing Identifies Additional Lipid Metabolism Genes That Influence Pro Accumulation
Given our findings of altered Pro accumulation in cyp86a2 and lacs2-1 mutants, we searched the p5cs1-4 RNA sequencing data for genes related to VLCFAs, cuticle or wax synthesis, or other genes related to fatty acid metabolism that had different read counts in p5cs1-4 compared with the wild type (Supplemental Table S11). Interestingly, ACCD (Atcg00500), which encodes a subunit of the acetyl-CoA carboxylase complex responsible for the first step of fatty acid synthesis in plastids (Li-Beisson et al., 2013), had 14-fold higher RPKM in stressed p5cs1-4 compared with the wild type (this difference was marginally nonsignificant in our statistical analysis). The acetyl-coenzyme A carboxylase (ACC) complex consists of several nucleus-encoded subunits in addition to the chloroplast-encoded ACCD. We isolated a T-DNA mutant of the nucleus-encoded ACC α-subunit (CAC3; At2g38040) as a way to decrease the activity of the ACC complex. The cac3 mutant had a nearly 30% increase in Pro accumulation at low ψw (Fig. 7F).
RPKM differences also were found for several genes encoding 3-ketoacyl-coenzyme A synthases (KCS). In this case, the fact that several KCS genes had putatively altered expression (Supplemental Table S11) suggested that functional redundancy may mask phenotypes of single kcs mutants. Instead, we used the KCS inhibitor cafenstrole, which has been shown to block KCS activity (Trenkamp et al., 2004; Nobusawa et al., 2013). Cafenstrol treatment increased the Pro content of unstressed seedlings nearly 3-fold and increased Pro accumulation at low ψw by more than 40% (Fig. 7G). Both KCS and the ACC complex act upstream or in a different branch of VLCFA metabolism compared with LACS2 and CYP86A2. Thus, these results added further evidence that blocking flux through lipid metabolism at any of a number of steps leads to increased Pro accumulation at low ψw. The cac3 and cafenstrole data also indicated that the buildup of lipid metabolism intermediates upstream of LACS2 was not responsible for the high-Pro phenotypes of cyp86a2-1 and lacs2-1.
DISCUSSION
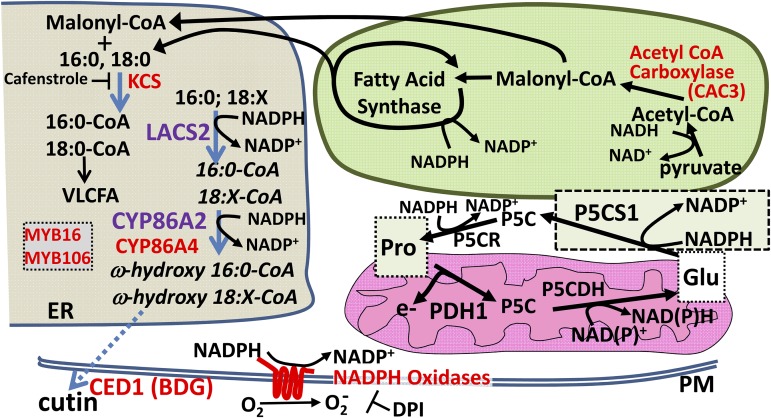
We present two major lines of evidence that show how Pro metabolism is integrated into cellular metabolism, particularly the relationship of Pro to lipid metabolism and the relationship to redox metabolism of mitochondria and chloroplast. First, cyp86a2 and lacs2-1, as well as disruptions of lipid metabolism or transport at other points (ACC complex, KCS, CED1, CYP86A4, MYB16, and MYB106), all led to increased Pro accumulation at low ψw (summarized in Fig. 8). The high-Pro and high-PDH1pro:LUC2 phenotypes of cyp86a2 and lacs2-1 could be partially or completed abolished by treatment with ROS scavengers. Similarly, cyp86a2-1 and lacs2-1 were more sensitive to the activation of Pro accumulation by reductant (DTT) or inhibited NADPH oxidase activity (DPI). Together, these data indicated that an effect of altered VLCFA synthesis on redox status, rather than signaling function of CYP86A2, LACS2, or VLCFAs themselves, was the cause of increased PDH1pro:LUC2 expression and Pro accumulation. In a complementary line of experiments, we discovered that disrupted Pro synthesis in p5cs1-4 altered the expression of chloroplast and mitochondrial genes related to redox metabolism. Two main conclusions emerge from these data. The first is that Pro metabolism is both influenced by and influences cellular redox status via previously unknown coordination with multiple metabolic pathways. A second conclusion is that Pro and lipid metabolism share dual roles under stress to help buffer cellular redox status while producing Pro and cuticle lipids useful for drought tolerance.
Figure 8.
Summary diagram showing the core pathway of Pro metabolism in relation to lipid, cuticle, and redox-related genes found to affect Pro accumulation. Multiple genes and pharmacological treatments disrupting lipid metabolism or NADPH oxidase activity were found to affect Pro accumulation and PDH1pro:LUC2 expression. This included CYP86A2 and LACS2 identified in the PDH1pro:LUC2 forward genetic screen; CYP86A4, CED1, MYB16, and MYB106 identified by reverse genetics; CAC3 and KCS genes identified by p5cs1-4 RNA sequencing; as well as strong effects of the NADPH oxidase inhibitor DPI. The reactions catalyzed by these proteins are spread across several compartments, including endoplasmic reticulum (ER), chloroplast, and plasma membrane (PM). The commonality among these pathways and Pro metabolism itself is an effect on redox status by the consumption of NADPH and the regeneration of NADP+, either directly, as for P5CS1, P5CR, LACS2, CYP86A2, and NADPH oxidases, or indirectly by modifying the flux through lipid synthesis, as for CAC3, KCS, CED1, MYB16, and MYB106. This commonality, along with data obtained with ROS scavengers and DTT treatment, indicates that redox status is a key factor linking these pathways together. Consistent with this, RNA sequencing data showed that p5cs1-4 had substantial alterations in gene expression-related chloroplast and mitochondrial redox metabolism, further indicating the relationship of Pro to redox status. The similar effects of p5cs1-4 on gene expression at both high and low ψw indicated the importance of Pro metabolism, and likely a high rate of flux through the Pro cycle of synthesis and catabolism, even in unstressed plants where Pro level is low. P5CS1 is drawn in its own box to indicate that its subcellular localization is unclear. How changes in redox state are communicated between these subcellular compartments is unknown.
We selected the PDH1 promoter for forward genetic screening because it could serve as a sensor for alterations in both stress signaling and metabolic state. Both cyp86a2 (4442-4) and lacs2 (4255-1) mutants had increased PDH1pro:LUC2 expression, but not increased Pro, in the unstressed control. In the wild-type background, PDH1pro:LUC2 expression of unstressed seedlings could be fully repressed to the stress level simply by the addition of the NADPH oxidase inhibitor DPI. The high PDH1pro:LUC2 expression of cyp86a2 and lacs2 mutants likewise could be repressed by DPI. Together, these data show that PDH1pro:LUC2 indeed acted as a sensitive indicator of altered redox and metabolic state even under conditions where Pro level itself was not changed. At low ψw, DPI, AsA, and DMTU had lesser, but still significant, effects, presumably because other stress-related signals also were acting on PDH1pro:LUC2 expression and Pro accumulation. The PDH1pro:LUC2 system can be further used as a new tool to understand unknown redox, metabolic, and stress signaling mechanisms.
It was of interest that there was not a complete correspondence between the expression of the PDH1pro:LUC2 reporter and endogenous PDH1 at low ψw. The two agreed very well in unstressed plants. At low ψw, however, the endogenous PDH1 was fully repressed despite the high Pro accumulation in cyp86a2 and lacs2, while the PDH1pro:LUC2 reporter was only partially repressed. It is possible that the PDH1pro:LUC2 construct may not incorporate all elements (such as the 3′ UTR or PDH1 coding region) responsible for the stress repression of PDH1. Alternatively, there may be position-dependent regulation of the PDH1 promoter that does not occur at the PDH1pro:LUC2 locus. While fortuitous, this partial decoupling of PDH1 and PDH1pro:LUC2 at low ψw allowed a larger range of mutants affecting Pro accumulation to be isolated. The PDH1pro:LUC2 system, and modifications of it that include more or differing sections of PDH1, can be used as a system to explore the stress repression of PDH1 more fully.
The cyp86a2 and lacs2 mutants had altered Pro accumulation despite no difference in P5CS1 or PDH1 protein level or any change in expression of the core Pro metabolism genes. How Pro and VLCFA metabolism are coordinated is part of a larger question of how redox state can be communicated between different subcellular compartments such as the endoplasmic reticulum, chloroplast, and mitochondria. Such mechanisms are little known, and Pro metabolism is a promising system in which to address such questions. However, we must first more definitively answer fundamental questions, such as where Pro is synthesized (cytoplasm or chloroplast). It is also interesting that mutants of PDH1, CYPP86A2, and LACS2 all have pathogen resistance phenotypes (Xiao et al., 2004; Bessire et al., 2007; Cecchini et al., 2011; Senthil-Kumar and Mysore, 2012). Also, there were similarities between genes differentially expressed in p5cs1-4 and genes differentially expressed in defense signaling and SA-related mutants (Supplemental Figs. S10 and S11). For pdh1 mutants, their altered pathogen resistance is due to Pro- and PDH1-dependent mitochondrial ROS production. Possibly, altered Pro levels and redox status are involved in the pathogen resistance phenotypes of cyp86a2 and lacs2 and are at least partially responsible for the gene expression changes in p5cs1-4.
RNA sequencing of p5cs1-4 further supported the coordination of Pro and lipid metabolism and showed a striking effect of p5cs1-4 on the expression of genes involved in redox metabolism of the mitochondria and chloroplast. These results are consistent with those of Lovell et al. (2015), who conducted quantitative trait locus mapping using a recombinant inbred line population constructed using Arabidopsis accessions Tsu and Kas, which have contrasting levels of Pro accumulation during drought. They found a significant cytoplasmic effect on Pro accumulation, and the most likely variation underlying this affect was in mitochondrial NADH Dehydrogenase Subunit9 (NAD9). In our data, NAD9 had 10-fold higher RPKM in p5cs1-4 compared with the wild type at high ψw and 3-fold higher at low ψw (Supplemental Table S10). In addition, more than 15 other NADs and related genes had similar patterns of higher RPKM in p5cs1-4 (Supplemental Table S10). It is possible that NAD expression is up-regulated to compensate for the reduced Pro level (and presumably reduced Pro catabolism) in p5cs1-4. These data underscore the importance of the Pro cycle to feed reductant into the mitochondria and are consistent with other evidence showing the importance of mitochondrial metabolism in drought resistance (Pastore et al., 2007; Giraud et al., 2008; Zsigmond et al., 2008; Atkin and Macherel, 2009; Rasmusson and Wallström, 2010; Skirycz et al., 2010; Schertl et al., 2014).
Under both high and low ψw, there was a striking enrichment of chloroplast-encoded genes among genes differentially expressed in p5cs1-4. This included genes involved in chloroplast protein translation as well as genes involved directly in the light reactions and NADPH metabolism. How and why disrupting Pro metabolism had such a striking effect on chloroplast gene expression is not as immediately clear as the connection of Pro to mitochondrial metabolism. However, the gene expression differences we saw in p5cs1-4 are broadly consistent with the idea that Pro metabolism may have a role in consuming NADPH and regenerating NADP+ to provide a continued supply of electron acceptors for chloroplast electron transport (Sharma et al., 2011) and with observations that Pro accumulation and Pro metabolism gene expression are modulated by light (Hayashi et al., 2000; Abrahám et al., 2003). The fact that effects of p5cs1-4 on chloroplast gene expression could be seen even in unstressed plants implies that flux through the cycle of Pro synthesis and catabolism is high enough to impact chloroplast function even when Pro levels are low. Further interpretation of these data is limited by the dearth of knowledge of metabolic flux through Pro metabolism, the unclear subcellular localization of P5CS1, and the lack of data on whether Pro metabolism mutants have reduced photosynthetic efficiency. In chloroplasts, enzyme activities are coordinated with redox status via the modification of protein disulfide bond status by ferredoxins and thioredoxins (Meyer et al., 2012). It will be of interest to further investigate how redox metabolism in the chloroplast could be coordinated with Pro synthesis that likely (although not certainly) occurs in the cytoplasm. Likewise, it has been suggested the mammalian P5CS may be regulated by thioredoxins (Liang et al., 2013). Whether similar regulation exists in plants has not been investigated, to our knowledge. Our p5cs1-4 transcriptome data make further experiments to uncover the specific mechanisms underlying the redox-Pro and Pro-photosynthesis connections even more compelling.
To our knowledge, there are little or no data directly comparable to our transcriptome analysis of p5cs1-4. Perhaps the most closely related work is that of Satoh et al. (2002), who reported genes induced by exogenous Pro. We may expect that these genes would be less expressed in stressed p5cs1-4, which has greatly decreased Pro levels compared with the wild type. Indeed, five out of the 20 Pro-inducible genes with a ProRE reported by Satoh et al. (2002) were less expressed in p5cs1-4 (Supplemental Table S8); however, three such genes were up-regulated in p5cs1-4 (Supplemental Table S9). Satoh et al. (2002) also found other genes responsive to Pro but lacking a ProRE. However, the identity of these genes was not reported, precluding further comparison with our data. Whether PDH1 and the ProRE are regulated directly by Pro level via some unknown Pro-sensing mechanism or respond indirectly to other metabolic or redox-related signaling factors, as suggested by our analysis of cyp86a2 and lacs2 mutants, will be a question of interest as we characterize additional mutants from the PDH1pro:LUC2 screen.
We observed that the NADPH oxidase inhibitor DPI caused a massive increase in the Pro content of unstressed seedlings and suppressed PDH1pro:LUC2 expression in the wild type as well as in cyp86a2 and lacs2 mutants. These data indicate how redox status, possibly including the redox status of the NADPH pool or ROS signaling, may be involved in adjusting Pro metabolism to match cellular redox and metabolic state. However, our result differs from that of Ben Rejeb et al. (2015), who saw no effect of DPI on the Pro accumulation of unstressed plants. Their experiments were conducted on medium containing Suc, which could have altered metabolite levels and redox status and thus obscured the DPI Pro response.
The hypothesis that Pro metabolism is regulated by redox or metabolic factors as well as abiotic stress has been proposed and discussed in several places (Szabados and Savouré, 2010; Verslues and Sharma, 2010; Sharma et al., 2011; Servet et al., 2012; Liang et al., 2013; Ben Rejeb et al., 2014; Giberti et al., 2014; Zhang and Becker, 2015), but experimental data to support such hypotheses are limited. The PDH1pro:LUC2 mutant screen has revealed an unexpected coordination of Pro and lipid metabolism, with altered cellular redox status being one of the factors allowing the coordinated regulation of these pathways. Likewise, the gene expression data set reported here greatly strengthens hypotheses that the Pro cycle is a significant determinant of chloroplast and mitochondria metabolism even when the Pro level is low. Our study demonstrates the need to mechanistically understand the redox-dependent regulation of Pro metabolism. For example, do Pro metabolism proteins undergo redox-sensitive posttranslational modifications that affect their activity, or does the transcriptional regulation of PDH1 transcription include redox-sensitive factors? The PDH1pro:LUC2 screen and the characterization of additional mutants promise to provide further new insights into Pro metabolism, its regulation, and its roles in stress resistance.
MATERIALS AND METHODS
Construction of PDH1pro:LUC2, Mutagenesis, and Screening
A 1,511-bp fragment encompassing the PDH1 promoter out to −1,389 bp and the 5′ UTR (122 bp) of the Arabidopsis (Arabidopsis thaliana) PDH1 transcript was amplified using primers to add PstI and SalI cloning sites (primer sequences are given in Supplemental Table S12). This promoter fragment was chosen because it was similar in length to the PDH1 promoter fragment we used previously to analyze PDH1 promoter:GUS expression (Sharma et al., 2011) and contains the ProRE described by Satoh et al. (2002) as well as several other potential cis-elements (Nakashima et al., 1998). This fragment was 112 bp shorter than the PDH1 promoter fragment we used previously for promoter:GUS analysis (Sharma et al., 2011) to avoid a SalI site that interfered with cloning. The amplified fragment was ligated into pJET1.2 blunt cloning vector (Thermo Fisher Scientific) and confirmed by sequencing. The PDH1 promoter fragment was then cloned into pCAB2-LUC2-pCAMBIA1390 (Wang et al., 2011a) containing the pGL4.10-LUC2 (Promega) Luciferase2 sequence. pCAB2-LUC2-pCAMBIA1390 was SalI/PstI digested and gel purified to remove the CAB2 promoter and then ligated with the PstI-PDH1 promoter-SalI fragment. The resulting plasmid was transformed into Agrobacterium tumefaciens strain GV3101 by electroporation and used to transform the Col-0 wild type by the floral dip method. T2 lines having a 3:1 segregation ratio consistent with a single-locus insertion were selected, and homozygous T3 plants were confirmed by antibiotic screening. These lines were further tested for similar repression of PDH1pro:LUC2 during low ψw and the induction of PDH1pro:LUC2 expression upon stress release as the endogenous PDH1. A single transgenic line was selected for mutagenesis. Subsequent genome sequencing confirmed that this line has insertion of the PDH1pro:LUC2 construct adjacent to position 11,887,500 of chromosome 1.
Approximately 60,000 T3 seeds of this line were EMS mutagenized following standard protocols (Weigel and Glazebrook, 2006). Mutagenized seeds were sown in large pots (approximately 60–70 seed per pot). M1 seeds from each pot were collected together as a pool, resulting in more than 900 pools of mutagenized seeds. From each of these pools, 100 seeds were sterilized and plated in a grid pattern on 140-mm-diameter plates on which the agar had been overlaid with nylon mesh to facilitate seedling transfer. The medium used was our laboratory standard medium of one-half-strength Murashige and Skoog salts with 2 mm MES (pH 5.7) buffer but no sugar (Sharma et al., 2011). Each plate also included three to five unmutagenized PDH1pro:LUC2 seeds for comparison. After stratification for 4 d, plates were incubated vertically in a growth chamber (23°C and light intensity of 100 μmol m−1 s−1) for 7 d. On day 7, plates were sprayed with 1 mm luciferin and imaged using a Xenogen Ivis system (Perkin-Elmer). Seedlings having high or low luciferase activity were marked. Seedlings were then transferred to low-ψw polyethylene glycol (PEG)-infused agar plates (−1 MPa) for 4 d, and luciferase activity was imaged again. PEG-agar plates were prepared as described previously (Sharma et al., 2011), with the relative volumes of agar and PEG overlay adjusted for the larger plates.
Seedlings having high or low luciferase activity either before or after the stress treatment were transferred to soil. Seeds from these plants (M2 generation) were then used for a secondary screen conducted in the same manner but with five to eight seedlings per line imaged and luciferase activity quantified (using the Xenogen Ivis software). Several hundred putative mutants were rescreened in the M2 generation, and more than 30 lines with consistently high ProDH1pro:LUC2 activity and a smaller set of mutants with low ProDH1pro:LUC2 (approximately seven, although some are still being verified) were selected for further analysis. Mutants having the greatest difference in luciferase activity compared with the wild type were selected for further analysis, including backcrosses, complementation testing, crosses to Landsberg erecta for mapping, and further phenotypic analysis.
Whole-Genome Sequencing, Single-Nucleotide Polymorphism Calling, and Next-Generation Mapping
Mutants having high PDH1pro:LUC2 were crossed to Landsberg erecta, and the resulting F2 seeds were plated as described above. Mutants in which the high-PDH1pro:LUC2 phenotype segregated in a 3:1 ratio consistent with a single recessive locus were considered suitable for sequencing analysis (in most cases, the recessive nature of the mutation also was confirmed during backcrossing to unmutagenized PDH1pro:LUC2). Genomic DNA was extracted from a pool of 80 to 100 homozygous mutant seedlings selected from the F2 population based on their high PDH1pro:LUC2 expression. DNA was extracted using the Qiagen Plant DNeasy kit, and the purity of the DNA extracts was tested by absorbance measurements and agarose gel analysis.
For whole-genome sequencing, six mutant DNA pools were prepared using Illumina TruSeq DNA library construction. Briefly, 1 μg of each DNA pool was sheared to fragments of 150 to 500 bp using a Covaris M220 focused ultrasonicator with the 200-bp program. The sheared DNA was size enriched by double-sided solid-phase reversible immobilization using an Agencourt AmPure XP kit. A TruSeq DNA LT Sample Prep Kit (Illumina) was used to perform end repair, A tailing, and adaptor ligation. The barcoded samples were then amplified by eight cycles of PCR and cleaned up by AmPure XP beads (Beckman Agencourt). The absolute concentrations and profiles of the libraries were determined by Qubit (Invitrogen) and BioAnalyzer High Sensitivity DNA chips (Agilent), respectively. The final library sizes range from 250 to 750 bp, with a major size of 350 bp. The molar concentrations of the libraries were derived by qPCR normalization using the KAPA NGS Library qPCR kit (KAPA Biosystems). Multiplexed sequencing was conducted on one lane of PE2*101 on an Illumina HiSeq2500. The Illumina CASAVA 1.8.2 pipeline was applied for barcode demultiplexing and generating fastq files. The Illuminia sequencing and initial data processing were conducted by the High Throughput Genomics Core facility of the Biodiversity Research Center, Academia Sinica.
After sequencing, reads were mapped to The Arabidopsis Information Resource 10 genome (ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR10_genome_release/) using Bowtie2 (Langmead and Salzberg, 2012) and BLAT (Kent, 2002), and single-nucleotide polymorphism calling was conducted by the RackJ software package (http://rackj.sourceforge.net/). Mapping results in BAM format were processed into pileup files using Samtools version 0.1.16 and then emap files as instructed on the Next Generation Mapping Web site (http://bar.utoronto.ca/NGM/). The processed emap files were analyzed using the Next Generation Mapping analysis tool (http://bar.utoronto.ca/NGM/) described by Austin et al. (2011).
Phenotypic Analyses and T-DNA Mutants
Seeds of T-DNA mutants were obtained from the Arabidopsis Biological Resource Center, and homozygous plants were confirmed by PCR genotyping using primers from the Signal (www.signal.salk.edu) database. RT-PCR was used to confirm the absence of transcript in each new mutant (Supplemental Fig. S12; genotyping and RT-PCR primers are given in Supplemental Table S12). Seeds for lacs2-1 and lacs3-1 were provided by the laboratory of Dr. John Browse. The Pro metabolism mutants p5cs1-4 and pdh1-2 were verified previously by our laboratory (Sharma and Verslues, 2010; Sharma et al., 2011). For transgenic complementation of 4442-4, the full-length cDNA sequence of CYP86A2 was amplified from total RNA (primers are given in Supplemental Table S12) and cloned into pDONOR 207. For plant expression, the clone was moved into pEG100 (Earley et al., 2006) and transformed into A. tumefaciens GV3101, and the 4442-4 EMS mutant was transformed by the floral dip method.
Free Pro was quantified using a ninhydrin assay (Bates et al., 1973) adapted to 96-well plate format (Verslues, 2010). For pharmacological treatments, seedlings were pretreated with AsA (Sigma), DMTU (ACROS Organics), or DPI (Sigma) for 24 h and then transferred to low ψw (−1.2 MPa, 96 h) with the same concentration of the chemical. Cafenstrole (Wako Chemical) treatment was performed in the same manner but without pretreatment before transfer to low ψw. Western blotting of P5CS1 and PDH1 was performed using antisera generated by our laboratory as described previously (Kesari et al., 2012; Bhaskara et al., 2015). For Toluidine Blue staining, whole seedlings or leaves were submerged in an aqueous solution of 0.05% (w/v) Toluidine Blue for 2 to 5 min. Excess stain was removed by rinsing with sterile distilled water before being photographed using a Lumar v12 stereoscope (Zeiss). For RT-PCR and quantitative RT-PCR analysis, total RNA was extracted from seedlings using the RNeasy plant mini kit (Qiagen). Quantitative PCR was performed using KAPA SYBR FAST Master Mix on a QuantStudio 12K Flex Real-Time PCR System (Applied Biosystems). NADP and NADPH were measured using an analysis kit (BioAssay Systems) with sample collection and extraction performed as described previously (Sharma et al., 2011). ROS staining used 2′,7′-dichlorodihydrofluorescein diacetate dye with excitation at 488 nm and emission at 500 to 550 nm, with images analyzed on a Zeiss LSM510 Meta microscopy system.
mRNA Sequencing
mRNA sequencing was carried out by the High Throughput Genomics Core facility of the Biodiversity Research Center, Academia Sinica, following the protocols provided by Illumina, including mRNA sequencing library preparation, addition of MID barcodes to the double-stranded cDNA fragments, and volume adjustment. Using the Illumina mRNA-seq kit, 10 to 15 μg of total RNA was used for the isolation of poly(A) RNA by oligo(dT) beads and subjected to cation-catalyzed fragmentation for 4 min at 94°C. The mRNA fragments were then converted into double-stranded cDNA by random priming, and the ends were repaired and A tailed. Multiplexing barcodes were then added to the DNA fragment ends with modifications from the paired-end genomic DNA library prep kit. The ligation products were size selected on agarose gels (200–400 bp), subjected to 18 cycles of PCR, and cleaned up by AmPure beads (Beckman Agencourt). The absolute concentrations of the libraries were determined fluorometrically by Qubit (Invitrogen) and BioAnalyzer High Sensitivity DNA chips (Agilent).
Mixing of the differentially barcoded samples for each lane was based on qPCR-derived relative concentration for the amplifiable fraction of each library. Briefly, the relative amplifiable concentration of each library was estimated by real-time PCR analyses (Roche Light Cycler 480) using the KAPA NGS Library qPCR kit by regression to the curve generated using the included standard of known concentration. Correlations between output clusters and qPCR concentrations were derived by plotting the relative concentrations of previously sequenced PhiX control and libraries against the output cluster numbers. The relative concentrations of the MID libraries were then estimated by regression. The amount (fmol) of each library to be mixed in a pool for each lane was determined based on the proportion each would account for in the projected output clusters (i.e. estimating the fmol required per library to generate 300,000/8 = 37,500 raw clusters per tile if the lane to be used for sequencing contained a pool of eight barcodes). The sequencing was performed on an Illumina Genome Analyzer IIx in the High Throughput Sequencing Core Facility of Academia Sinica, and 73-nucleotide reads were obtained. Base calling and demultiplexing were performed using CASAVA and bclfastq1.6.
RPKM values were calculated using the RackJ software package (http://rackj.sourceforge.net/) with reads mapped to The Arabidopsis Information Resource 10 genome using BLAT (Kent, 2002). RPKM values of control and treatment samples were compared using Z statistics as described by Lan et al. (2013). GO enrichment was computed using the TopGO elim method (Alexa et al., 2006) using GOBU with its MultiView plugin (Lin et al., 2006).
Accession Numbers
mRNA sequencing results are available under Gene Expression Omnibus accession number GSE75933.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Example of primary screen plate images using EMS-mutagenized PDH1pro:LUC2.
Supplemental Figure S2. Next-generation mapping analysis and identification of CYP86A2 and LACS2 as the causative mutated genes in the 4442-2 and 4255-1 EMS mutants.
Supplemental Figure S3. PDH1pro:LUC2 crossed to cyp86a2-1 and lacs2-1 T-DNA mutants shows the same high-PDH1pro:LUC2 phenotype as the 4244-2 and 4255-2 EMS mutants.
Supplemental Figure S4. Expression of Pro synthesis and catabolism genes in cyp86a2 mutants.
Supplemental Figure S5. NADP+ and NADPH analysis of the wild type, cyp86a2-1, and lacs2-1.
Supplemental Figure S6. ROS levels of the wild type, cyp86a2-1, and lacs2-1.
Supplemental Figure S7. MapMan analysis of photosynthesis-related gene expression in stress- and control-treated wild-type plants as well as p5cs1-4 compared with the wild type in unstressed and low-ψw (−1.2 MPa, 96 h) treatments.
Supplemental Figure S8. MapMan analysis of mitochondrial electron transport-related gene expression in stress- and control-treated wild-type plants as well as p5cs1-4 compared with the wild type in unstressed and low-ψw (−1.2 MPa, 96 h) treatments.
Supplemental Figure S9. MapMan analysis of amino acid metabolism-related gene expression in p5cs1-4 compared with the wild type in unstressed and low-ψw (−1.2 MPa, 96 h) treatments.
Supplemental Figure S10. Differentially expressed genes in p5cs1-4 control versus wild-type control compared with public microarray data using the Genevestigator Signature analysis tool.
Supplemental Figure S11. Differentially expressed genes in p5cs1-4 stress versus wild-type stress compared with public microarray data using the Genevestigator Signature analysis tool.
Supplemental Figure S12. RT-PCR check of lipid- and cuticle-related T-DNA mutants.
Supplemental Table S1. RPKM values from RNA sequencing analysis of wild-type Col-0 and p5cs1-4 under either control (−0.25 MPa) or stress (transfer to −1.2 MPa for 96 h) conditions.
Supplemental Table S2. Genes with increased expression in the Col-0 wild type at 96 h after transfer to low-ψw (−1.2 MPa) stress, and GO terms significantly enriched in the up-regulated genes.
Supplemental Table S3. Genes with decreased expression in the wild type at 96 h after transfer to low-ψw (−1.2 MPa) stress, and significantly enriched GO terms among the down-regulated genes.
Supplemental Table S4. Genes with increased expression in unstressed p5cs1-4 compared with unstressed wild-type Col-0, along with significantly enriched GO terms.
Supplemental Table S5. Genes with decreased expression in unstressed p5cs1-4 compared with unstressed wild-type Col-0, along with significantly enriched GO terms.
Supplemental Table S6. Genes with increased expression in stressed (−1.2 MPa, 96 h) p5cs1-4 compared with stressed wild-type Col-0, along with significantly enriched GO terms.
Supplemental Table S7. Genes with decreased expression in stressed (−1.2 MPa, 96 h) p5cs1-4 compared with stressed wild-type Col-0, along with significantly enriched GO terms.
Supplemental Table S8. Both stress-responsive and nonresponsive genes are among those up-regulated in p5cs1-4.
Supplemental Table S9. Both stress-responsive and nonresponsive genes are among those down-regulated in p5cs1-4.
Supplemental Table S10. Chloroplast and mitochondrial NAD(P)H dehydrogenase genes with putatively altered expression in p5cs1-4.
Supplemental Table S11. Lipid and wax metabolism genes with putatively altered expression in p5cs1-4.
Supplemental Table S12. Primer sequences used in this study.
Supplementary Material
Acknowledgments
We thank the laboratory of Shu-Hsing Wu for the pCAB2-LUC2-pCAMBIA1390 plasmid; John Browse (Washington State University) for the lacs2 and lacs3 mutants; Mei-Yeh Liu and the High Throughput Genomics Core, Biodiversity Research Center, Academia Sinica, for sequencing services; Mei-Jane Fang for assistance with luciferase imaging and microscopy; Ang-Hsi Lin for mutant screening; and Srilakshmi Mukiri and Trent Z. Chang for laboratory assistance.
Glossary
- ProRE
proline response element
- VLCFA
very-long-chain fatty acid
- UTR
untranslated region
- Col-0
Columbia-0
- EMS
ethyl methanesulfonate
- DTT
dithiothreitol
- ROS
reactive oxygen species
- AsA
ascorbic acid
- DMTU
N,N'-dimethylthiourea
- DPI
diphenyleneiodonium
- GO
Gene Ontology
- RPKM
reads assigned per kilobase of target per million mapped reads
- SA
salicylic acid
- PEG
polyethylene glycol
- qPCR
quantitative PCR
- RT
reverse transcription
Footnotes
This work was supported the Taiwan Ministry of Science and Technology (grant no. NSC 102–2628–B–001–003 to P.E.V.) and by an Academia Sinica Career Development Award to P.E.V.
Articles can be viewed without a subscription.
References
- Abrahám E, Rigó G, Székely G, Nagy R, Koncz C, Szabados L (2003) Light-dependent induction of proline biosynthesis by abscisic acid and salt stress is inhibited by brassinosteroid in Arabidopsis. Plant Mol Biol 51: 363–372 [DOI] [PubMed] [Google Scholar]
- Alexa A, Rahnenführer J, Lengauer T (2006) Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607 [DOI] [PubMed] [Google Scholar]
- Atkin OK, Macherel D (2009) The crucial role of plant mitochondria in orchestrating drought tolerance. Ann Bot (Lond) 103: 581–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin RS, Vidaurre D, Stamatiou G, Breit R, Provart NJ, Bonetta D, Zhang J, Fung P, Gong Y, Wang PW, et al. (2011) Next-generation mapping of Arabidopsis genes. Plant J 67: 715–725 [DOI] [PubMed] [Google Scholar]
- Bates LS, Waldren RP, Teare ID (1973) Rapid determination of free proline for water stress studies. Plant Soil 39: 205–207 [Google Scholar]
- Ben Rejeb K, Abdelly C, Savouré A (2014) How reactive oxygen species and proline face stress together. Plant Physiol Biochem 80: 278–284 [DOI] [PubMed] [Google Scholar]
- Ben Rejeb K, Lefebvre-De Vos D, Le Disquet I, Leprince AS, Bordenave M, Maldiney R, Jdey A, Abdelly C, Savouré A (2015) Hydrogen peroxide produced by NADPH oxidases increases proline accumulation during salt or mannitol stress in Arabidopsis thaliana. New Phytol 208: 1138–1148 [DOI] [PubMed] [Google Scholar]
- Bessire M, Chassot C, Jacquat AC, Humphry M, Borel S, Petétot JMC, Métraux JP, Nawrath C (2007) A permeable cuticle in Arabidopsis leads to a strong resistance to Botrytis cinerea. EMBO J 26: 2158–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara GB, Nguyen TT, Verslues PE (2012) Unique drought resistance functions of the Highly ABA-Induced clade A protein phosphatase 2Cs. Plant Physiol 160: 379–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara GB, Yang TH, Verslues PE (2015) Dynamic proline metabolism: importance and regulation in water limited environments. Front Plant Sci 6: 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussis D, Heineke D (1998) Acclimation of potato plants to polyethylene glycol-induced water deficit. II. Contents and subcellular distribution of organic solutes. J Exp Bot 49: 1361–1370 [Google Scholar]
- Cecchini NM, Monteoliva MI, Alvarez ME (2011) Proline dehydrogenase contributes to pathogen defense in Arabidopsis. Plant Physiol 155: 1947–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ronde JA, Cress WA, Krüger GHJ, Strasser RJ, Van Staden J (2004) Photosynthetic response of transgenic soybean plants, containing an Arabidopsis P5CR gene, during heat and drought stress. J Plant Physiol 161: 1211–1224 [DOI] [PubMed] [Google Scholar]
- Dietrich K, Weltmeier F, Ehlert A, Weiste C, Stahl M, Harter K, Dröge-Laser W (2011) Heterodimers of the Arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress. Plant Cell 23: 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Funck D, Eckard S, Müller G (2010) Non-redundant functions of two proline dehydrogenase isoforms in Arabidopsis. BMC Plant Biol 10: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funck D, Winter G, Baumgarten L, Forlani G (2012) Requirement of proline synthesis during Arabidopsis reproductive development. BMC Plant Biol 12: 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giberti S, Funck D, Forlani G (2014) Δ1-Pyrroline-5-carboxylate reductase from Arabidopsis thaliana: stimulation or inhibition by chloride ions and feedback regulation by proline depend on whether NADPH or NADH acts as co-substrate. New Phytol 202: 911–919 [DOI] [PubMed] [Google Scholar]
- Giraud E, Ho LHM, Clifton R, Carroll A, Estavillo G, Tan YF, Howell KA, Ivanova A, Pogson BJ, Millar AH, et al. (2008) The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol 147: 595–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson D, Lelu-Walter MA, Parkinson M (2005) Overproduction of proline in transgenic hybrid larch (Larix × leptoeuropaea (Dengler)) cultures renders them tolerant to cold, salt and frost. Mol Breed 15: 21–29 [Google Scholar]
- Hayashi F, Ichino T, Osanai M, Wada K (2000) Oscillation and regulation of proline content by P5CS and ProDH gene expressions in the light/dark cycles in Arabidopsis thaliana L. Plant Cell Physiol 41: 1096–1101 [DOI] [PubMed] [Google Scholar]
- Kavi Kishor PB, Hima Kumari P, Sunita MSL, Sreenivasulu N (2015) Role of proline in cell wall synthesis and plant development and its implications in plant ontogeny. Front Plant Sci 6: 544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. (2002) BLAT: the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesari R, Lasky JR, Villamor JG, Des Marais DL, Chen YJC, Liu TW, Lin W, Juenger TE, Verslues PE (2012) Intron-mediated alternative splicing of Arabidopsis P5CS1 and its association with natural variation in proline and climate adaptation. Proc Natl Acad Sci USA 109: 9197–9202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbe A, Oliver SN, Fernie AR, Stitt M, van Dongen JT, Geigenberger P (2006) Combined transcript and metabolite profiling of Arabidopsis leaves reveals fundamental effects of the thiol-disulfide status on plant metabolism. Plant Physiol 141: 412–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan P, Li W, Lin WD, Santi S, Schmidt W (2013) Mapping gene activity of Arabidopsis root hairs. Genome Biol 14: R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Less H, Galili G (2008) Principal transcriptional programs regulating plant amino acid metabolism in response to abiotic stresses. Plant Physiol 147: 316–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Zhang L, Natarajan SK, Becker DF (2013) Proline mechanisms of stress survival. Antioxid Redox Signal 19: 998–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Pollard M, Sauveplane V, Pinot F, Ohlrogge J, Beisson F (2009) Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proc Natl Acad Sci USA 106: 22008–22013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, DeBono A, Durrett TP, et al. (2013) Acyl-lipid metabolism. The Arabidopsis Book 11: e0161 doi/10.1199/tab.0161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WD, Chen YC, Ho JM, Hsiao CD (2006) GOBU: toward an integration interface for biological objects. J Inf Sci Eng 22: 19–29 [Google Scholar]
- Lovell JT, Mullen JL, Lowry DB, Awole K, Richards JH, Sen S, Verslues PE, Juenger TE, McKay JK (2015) Exploiting differential gene expression and epistasis to discover candidate genes for drought-associated QTLs in Arabidopsis thaliana. Plant Cell 27: 969–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattioli R, Biancucci M, Lonoce C, Costantino P, Trovato M (2012) Proline is required for male gametophyte development in Arabidopsis. BMC Plant Biol 12: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer Y, Belin C, Delorme-Hinoux V, Reichheld JP, Riondet C (2012) Thioredoxin and glutaredoxin systems in plants: molecular mechanisms, crosstalks, and functional significance. Antioxid Redox Signal 17: 1124–1160 [DOI] [PubMed] [Google Scholar]
- Miller G, Stein H, Honig A, Kapulnik Y, Zilberstein A (2005) Responsive modes of Medicago sativa proline dehydrogenase genes during salt stress and recovery dictate free proline accumulation. Planta 222: 70–79 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Satoh R, Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K (1998) A gene encoding proline dehydrogenase is not only induced by proline and hypoosmolarity, but is also developmentally regulated in the reproductive organs of Arabidopsis. Plant Physiol 118: 1233–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanjo T, Kobayashi M, Yoshiba Y, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (1999) Antisense suppression of proline degradation improves tolerance to freezing and salinity in Arabidopsis thaliana. FEBS Lett 461: 205–210 [DOI] [PubMed] [Google Scholar]
- Nobusawa T, Okushima Y, Nagata N, Kojima M, Sakakibara H, Umeda M (2013) Synthesis of very-long-chain fatty acids in the epidermis controls plant organ growth by restricting cell proliferation. PLoS Biol 11: e1001531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima Y, Shikata M, Koyama T, Ohtsubo N, Mitsuda N, Ohme-Takagi M (2013) MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development in Arabidopsis and Torenia fournieri. Plant Cell 25: 1609–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore D, Trono D, Laus MN, Di Fonzo N, Flagella Z (2007) Possible plant mitochondria involvement in cell adaptation to drought stress. A case study: durum wheat mitochondria. J Exp Bot 58: 195–210 [DOI] [PubMed] [Google Scholar]
- Peng Z, Lu Q, Verma DPS (1996) Reciprocal regulation of delta 1-pyrroline-5-carboxylate synthetase and proline dehydrogenase genes controls proline levels during and after osmotic stress in plants. Mol Gen Genet 253: 334–341 [DOI] [PubMed] [Google Scholar]
- Rasmusson AG, Wallström SV (2010) Involvement of mitochondria in the control of plant cell NAD(P)H reduction levels. Biochem Soc Trans 38: 661–666 [DOI] [PubMed] [Google Scholar]
- Roosens NH, Al Bitar F, Loenders K, Angenon G, Jacobs M (2002) Overexpression of ornithine-delta-aminotransferase increases proline biosynthesis and confers osmotolerance in transgenic plants. Mol Breed 9: 73–80 [Google Scholar]
- Satoh R, Fujita Y, Nakashima K, Shinozaki K, Yamaguchi-Shinozaki K (2004) A novel subgroup of bZIP proteins functions as transcriptional activators in hypoosmolarity-responsive expression of the ProDH gene in Arabidopsis. Plant Cell Physiol 45: 309–317 [DOI] [PubMed] [Google Scholar]
- Satoh R, Nakashima K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2002) ACTCAT, a novel cis-acting element for proline- and hypoosmolarity-responsive expression of the ProDH gene encoding proline dehydrogenase in Arabidopsis. Plant Physiol 130: 709–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawahel WA, Hassan AH (2002) Generation of transgenic wheat plants producing high levels of the osmoprotectant proline. Biotechnol Lett 24: 721–725 [Google Scholar]
- Schertl P, Cabassa C, Saadallah K, Bordenave M, Savouré A, Braun HP (2014) Biochemical characterization of proline dehydrogenase in Arabidopsis mitochondria. FEBS J 281: 2794–2804 [DOI] [PubMed] [Google Scholar]
- Schnurr J, Shockey J, Browse J (2004) The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell 16: 629–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil-Kumar M, Mysore KS (2012) Ornithine-delta-aminotransferase and proline dehydrogenase genes play a role in non-host disease resistance by regulating pyrroline-5-carboxylate metabolism-induced hypersensitive response. Plant Cell Environ 35: 1329–1343 [DOI] [PubMed] [Google Scholar]
- Servet C, Ghelis T, Richard L, Zilberstein A, Savoure A (2012) Proline dehydrogenase: a key enzyme in controlling cellular homeostasis. Front Biosci (Landmark Ed) 17: 607–620 [DOI] [PubMed] [Google Scholar]
- Sharma S, Verslues PE (2010) Mechanisms independent of abscisic acid (ABA) or proline feedback have a predominant role in transcriptional regulation of proline metabolism during low water potential and stress recovery. Plant Cell Environ 33: 1838–1851 [DOI] [PubMed] [Google Scholar]
- Sharma S, Villamor JG, Verslues PE (2011) Essential role of tissue-specific proline synthesis and catabolism in growth and redox balance at low water potential. Plant Physiol 157: 292–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A, De Bodt S, Obata T, De Clercq I, Claeys H, De Rycke R, Andriankaja M, Van Aken O, Van Breusegem F, Fernie AR, et al. (2010) Developmental stage specificity and the role of mitochondrial metabolism in the response of Arabidopsis leaves to prolonged mild osmotic stress. Plant Physiol 152: 226–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Wu R (2004) Stress-inducible synthesis of proline in transgenic rice confers faster growth under stress conditions than that with constitutive synthesis. Plant Sci 166: 941–948 [Google Scholar]
- Szabados L, Savouré A (2010) Proline: a multifunctional amino acid. Trends Plant Sci 15: 89–97 [DOI] [PubMed] [Google Scholar]
- Székely G, Abrahám E, Cséplo A, Rigó G, Zsigmond L, Csiszár J, Ayaydin F, Strizhov N, Jásik J, Schmelzer E, et al. (2008) Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J 53: 11–28 [DOI] [PubMed] [Google Scholar]
- Trenkamp S, Martin W, Tietjen K (2004) Specific and differential inhibition of very-long-chain fatty acid elongases from Arabidopsis thaliana by different herbicides. Proc Natl Acad Sci USA 101: 11903–11908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE. (2010) Quantification of water stress-induced osmotic adjustment and proline accumulation for Arabidopsis thaliana molecular genetic studies. In Sunkar R, ed, Plant Stress Tolerance: Methods and Protocols. Humana Press, New York, pp 301–316 [DOI] [PubMed] [Google Scholar]
- Verslues PE, Sharma S (2010) Proline metabolism and its implications for plant-environment interaction. The Arabidopsis Book 8: e0140 doi/10.1199/tab.0140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voetberg GS, Sharp RE (1991) Growth of the maize primary root at low water potentials. III. Role of increased proline deposition in osmotic adjustment. Plant Physiol 96: 1125–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wu JF, Nakamichi N, Sakakibara H, Nam HG, Wu SH (2011a) LIGHT-REGULATED WD1 and PSEUDO-RESPONSE REGULATOR9 form a positive feedback regulatory loop in the Arabidopsis circadian clock. Plant Cell 23: 486–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Xiong L, Li W, Zhu JK, Zhu J (2011b) The plant cuticle is required for osmotic stress regulation of abscisic acid biosynthesis and osmotic stress tolerance in Arabidopsis. Plant Cell 23: 1971–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis: A laboratory manual. Cold Spring Harbor Laboratory Press, NY [Google Scholar]
- Weltmeier F, Ehlert A, Mayer CS, Dietrich K, Wang X, Schütze K, Alonso R, Harter K, Vicente-Carbajosa J, Dröge-Laser W (2006) Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. EMBO J 25: 3133–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Goodwin SM, Xiao Y, Sun Z, Baker D, Tang X, Jenks MA, Zhou JM (2004) Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO J 23: 2903–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiba Y, Kiyosue T, Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K (1997) Regulation of levels of proline as an osmolyte in plants under water stress. Plant Cell Physiol 38: 1095–1102 [DOI] [PubMed] [Google Scholar]
- Zhang CS, Lu Q, Verma DPS (1995) Removal of feedback inhibition of delta 1-pyrroline-5-carboxylate synthetase, a bifunctional enzyme catalyzing the first two steps of proline biosynthesis in plants. J Biol Chem 270: 20491–20496 [DOI] [PubMed] [Google Scholar]
- Zhang L, Becker DF (2015) Connecting proline metabolism and signaling pathways in plant senescence. Front Plant Sci 6: 552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu BC, Su J, Chan MC, Verma DPS, Fan YL, Wu R (1998) Overexpression of a Δ1-pyrroline-5-carboxylate synthetase gene and analysis of tolerance to water- and salt-stress in transgenic rice. Plant Sci 139: 41–48 [Google Scholar]
- Zsigmond L, Rigó G, Szarka A, Székely G, Otvös K, Darula Z, Medzihradszky KF, Koncz C, Koncz Z, Szabados L (2008) Arabidopsis PPR40 connects abiotic stress responses to mitochondrial electron transport. Plant Physiol 146: 1721–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.