Mitochondrial d-lactate dehydrogenase links the last step of methylglyoxal detoxification to the mitochondrial electron transport chain through cytochrome c.
Abstract
Glycolysis generates methylglyoxal (MGO) as an unavoidable, cytotoxic by-product in plant cells. MGO scavenging is performed by the glyoxalase system, which produces d-lactate as an end product. d-Lactate dehydrogenase (d-LDH) is encoded by a single gene in Arabidopsis (Arabidopsis thaliana; At5g06580). It catalyzes in vitro the oxidation of d-lactate to pyruvate using flavin adenine dinucleotide as a cofactor; knowledge of its function in the context of the plant cell remains sketchy. Blue native-polyacrylamide gel electrophoresis of mitochondrial extracts combined with in gel activity assays using different substrates and tandem mass spectrometry allowed us to definitely show that d-LDH acts specifically on d-lactate, is active as a dimer, and does not associate with respiratory supercomplexes of the inner mitochondrial membrane. The combined use of cytochrome c (CYTc) loss-of-function mutants and respiratory complex III inhibitors showed that CYTc acts as the in vivo electron acceptor of d-LDH. CYTc loss-of-function mutants, as well as the d-LDH mutants, were more sensitive to d-lactate and MGO, indicating that they function in the same pathway. In addition, overexpression of d-LDH and CYTc increased tolerance to d-lactate and MGO. Together with fine-localization of d-LDH, the functional interaction with CYTc in vivo strongly suggests that d-lactate oxidation takes place in the mitochondrial intermembrane space, delivering electrons to the respiratory chain through CYTc. These results provide a comprehensive picture of the organization and function of d-LDH in the plant cell and exemplify how the plant mitochondrial respiratory chain can act as a multifunctional electron sink for reductant from cytosolic pathways.
In plants, d-lactate is produced as part of the detoxification of methylglyoxal (MGO), a highly reactive, cytotoxic compound formed as a side product of primary metabolism (Phillips and Thornalley, 1993; Maurino and Engqvist, 2015). MGO is formed in a paracatalytic reaction by the action of triose phosphate isomerase as well as nonenzymatically through phosphate elimination from the glycolytic intermediates glyceraldehyde 3-phosphate and dihydroxyacetone phosphate (Kalapos, 1999; Chen and Thelen, 2010). Production of MGO is increased during abiotic stresses such as salt, drought, cold, and heavy metal exposure (Yadav et al., 2005) and may contribute to the formation of reactive oxygen species (Saito et al., 2011; Hoque et al., 2012). It is assumed that Arabidopsis (Arabidopsis thaliana) possesses several enzymatic pathways to detoxify this compound (Maurino and Engqvist, 2015); at least two of them convert MGO to d-lactate. In the first pathway, MGO is metabolized to d-lactate through the glyoxalase pathway, which comprises the enzymes glyoxalase I (EC 4.4.1.5) and glyoxalase II (EC 3.1.2.6; Thornalley, 2003). In the second pathway, MGO also can be metabolized to d-lactate in a single step through the glutathione-independent cytosolic DJ-1d protein (EC 4.2.1.130; Kwon et al., 2013). This protein, a member of the PfpI/Hsp31/DJ-1 superfamily, can function as a hydrolyase that requires no cofactors (Misra et al., 1995; Subedi et al., 2011).
In plants, d-lactate is metabolized by d-lactate dehydrogenase (d-LDH; At5g06580; Engqvist et al., 2009; Wienstroer et al., 2012). In addition to d-lactate, d-LDH also can use other compounds as substrates in vitro, such as l-lactate, glycolate, and d-glycerate. However, the catalytic rates with l-lactate (kcat = 7 min−1), d-glycerate (kcat = 6 min−1), and glycolate (kcat = 0.1 min−1) are extremely low, making them unlikely in vivo substrates (Engqvist et al., 2009). In contrast, the enzyme oxidizes d-lactate (kcat = 65 min−1) at a much higher rate and with a high affinity (Km = 164 µm), resulting in a very high catalytic efficiency of d-LDH for d-lactate (Engqvist et al., 2009). d-LDH is inactive in the presence of NAD+, NADP+, or oxygen as electron acceptor. The isolated protein only showed activity in the presence of cytochrome c (CYTc) or an artificial, small molecular electron acceptor such as dichlorophenolindophenol (Engqvist et al., 2009). Although d-LDH belongs to the d-lactate CYTc oxidoreductase (EC1.1.2.4) group (Engqvist et al., 2009), which catalyzes the reaction d-lactate + 2 CYTc (oxidized) = pyruvate + 2 CYTc (reduced), experimental evidence showing that CYTc is the in vivo electron acceptor of the reaction is still lacking. This is a major shortcoming, because CYTc is known to act as a highly promiscuous electron acceptor when used in vitro. Extrapolating the in vivo specificity of d-LDH from in vitro observations, therefore, is particularly problematic.
Definitive evidence on the suborganellar localization and cellular function of plant d-LDH also is missing. CYTc is a soluble redox-active heme protein located in the mitochondrial intermembrane space (IMS) as a soluble heme protein, where it transfers electrons from complex III to complex IV as part of the cyanide-sensitive respiratory pathway (Welchen and Gonzalez, 2016). If Arabidopsis d-LDH uses CYTc as an electron acceptor, colocation in the IMS is a prerequisite. Arabidopsis d-LDH has been localized to mitochondria (Engqvist et al., 2009), which was confirmed recently by the presence of a unique orthologous protein in the mitochondrial proteome of potato (Solanum tuberosum) tuber (Salvato et al., 2014). The subcompartment of its activity has not been assessed. Where exactly d-LDH operates is critical, however, for an understanding of the pathway, interaction partners, and potential membrane transport of intermediates. Physiological data have suggested d-LDH activity in the mitochondrial matrix donating electrons to complex III in Jerusalem artichoke (Helianthus tuberosus; Atlante et al., 2005).
Here, we used a combination of biochemical, genetic, and physiological approaches, which show that Arabidopsis d-LDH localizes to the mitochondrial IMS and that d-lactate and CYTc are its specific in vivo electron donor and acceptor, respectively. Transgenic plants overexpressing both d-LDH and CYTc have enhanced capacity to detoxify d-lactate and MGO, indicating that both proteins can be limiting factors in the detoxification process.
RESULTS
Subcellular and Suborganellar Localization of d-LDH
In silico analysis of Arabidopsis d-LDH predicts an N-terminally located targeting signal, composed of a TOM20 recognition motif and an amphiphilic α-helix, for mitochondrial localization (Fukasawa et al., 2015) followed by a transmembrane α-helix span from amino acids 48 to 68 (Hofmann and Stoffel, 1993). These structural features have been described as a basis for a mitochondrial stop-transfer import mechanism (Hewitt et al., 2014). Furthermore, Arabidopsis d-LDH has been detected in mitochondria-enriched extracts by immunoblotting, and enzymatic assays using CYTc as substrate indicated mitochondrial localization indirectly (Engqvist et al., 2009). Validation by an orthogonal method, as well as direct empirical information on where in the mitochondria Arabidopsis d-LDH may operate in vivo, have been lacking. To address this, we generated C-terminal GFP fusions of the full-length coding sequence of Arabidopsis d-LDH. Transient expression of the C-terminal fusion construct d-LDH:GFP under the control of the cauliflower mosaic virus 35S (CaMV35S) promoter in wild tobacco (Nicotiana benthamiana) leaf epidermis and confocal imaging showed GFP fluorescence that colocalized with the mitochondrial marker MitoTracker Orange as well as weaker cytosolic GFP signal (Supplemental Fig. S1).
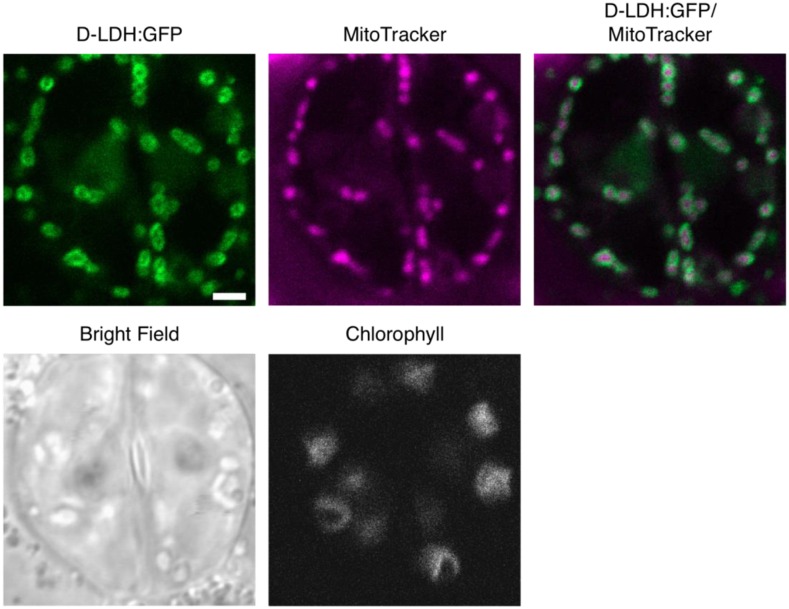
For further validation of the results obtained by transient overexpression, we next generated stable Arabidopsis transformants. Three independent lines were assessed, and the GFP signal consistently labeled small, round-to-elongated, highly dynamic subcellular structures and colocalized with MitoTracker Orange (Fig. 1). No colocalization with the chloroplastic autofluorescence was observed, and the cytosolic signal was close to background, suggesting the absence of dual organelle targeting and minimal overexpression-related artifacts (Fig. 1). These results confirm that d-LDH localizes to mitochondria.
Figure 1.
d-LDH:GFP in Arabidopsis seedlings. Arabidopsis seedlings stably expressing d-LDH:GFP and stained with MitoTracker Orange were imaged by confocal laser scanning microscopy. Representative images are shown for two guard cells of an Arabidopsis leaf. GFP, Green; MitoTracker, magenta. Bar = 2 μm.
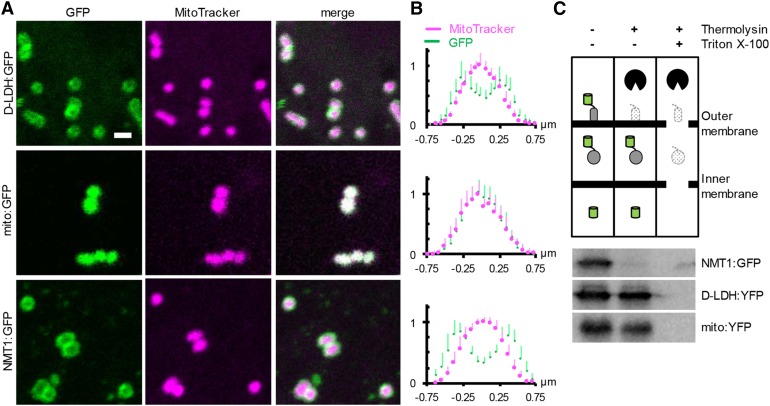
Despite clear localization of the fluorescent signals of GFP and MitoTracker to the same organelle, they showed obvious differences in shape (Figs. 1 and 2A; Supplemental Fig. S1). GFP signal appeared to resemble broad halo-like structures. To investigate if this may provide further insights into the submitochondrial localization of d-LDH:GFP, we used a recently developed quantitative approach measuring the averaged fluorescence distributions across individual mitochondria (Wagner et al., 2015). The distribution of the MitoTracker signal was comparable to that in the controls, indicating that d-LDH:GFP expression did not compromise gross mitochondrial structure or shape (Fig. 2B). Analysis of the GFP signals of 15 or more mitochondria each resulted in a fluorescence signature for d-LDH:GFP between the soluble matrix and outer membrane controls.
Figure 2.
Quantitative analysis of submitochondrial localization of d-LDH. A, Confocal laser scanning microscopy images from Arabidopsis seedlings stably expressing d-LDH:GFP (top row), Nicotiana plumbaginifolia β-ATPase mitochondrial presequence fused to GFP (mito:GFP; middle row), and NETWORK1/ELM1 fused to GFP (NMT1:GFP; bottom row) and stained with MitoTracker Orange. mito:GFP and NMT1:GFP were used as markers for the mitochondrial matrix and outer mitochondrial membrane (OMM), respectively. GFP, Green; MitoTracker, magenta. Bar = 1 µm. B, Normalized pixel intensity distributions in the GFP and MitoTracker channels plotted centrally across, and averaged over, individual mitochondria. n ≥ 15; error bars indicate sd. C, Protease protection analysis by GFP immunodetection after thermolysin treatment of intact, purified mitochondria expressing GFP or YFP fusions of the proteins used in A and B in the absence and presence of 0.1% (v/v) Triton X-100 to disrupt mitochondrial membranes. The entire blotting membrane is presented in Supplemental Figure S2. The experiment was repeated three times with consistent results.
To independently assess the possibility that a part of d-LDH may be exposed to the mitochondrial surface (i.e. the cytosolic side of the OMM), we additionally performed a set of protease protection experiments. Freshly isolated, highly intact mitochondria (greater than 90%, as determined by CYTc latency assays) were subjected to a thermolysin-proteolysis regime. The subsequent presence and size of a d-LDH:yellow fluorescent protein (YFP) fusion protein was analyzed by western blotting using a GFP antibody (Fig. 2C; Supplemental Fig. S2). The d-LDH:YFP band was lost when the protease was present and mitochondrial membranes were disrupted by Triton, but it was protected when treated with protease only. The same was observed for a mito:YFP as a soluble matrix control. In contrast, NMT1:GFP as a control for exposure to the cytosolic side of the OMM was already degraded in the sole presence of the protease.
Quantitative imaging indicates d-LDH localization between the OMM and matrix, and protease protection shows localization within the volume delimited by the OMM, consistent with d-LDH localizing to the IMS and/or the inner mitochondrial membrane (IMM). Considering the presence of a canonical mitochondrial targeting peptide and a short N-terminal hydrophobic sequence in addition, anchoring in the inner membrane with exposure to the IMS appears particularly likely.
Analysis of d-LDH by Blue Native-PAGE
A number of proteins active in the IMS, including soluble ones such as CYTc, form complexes with proteins of the respiratory chain (Braun and Schmitz, 1992; Klodmann et al., 2011; Rode et al., 2011; Vögtle et al., 2012). As our data indicated that d-LDH is an IMS protein, we aimed to analyze if it forms a part of OXPHOS supercomplexes. We first confirmed that d-LDH activity is present in wild-type isolated mitochondria and is absent in independent loss-of-function mutants of d-LDH (dldh1-1 and dldh1-2; Engqvist et al., 2009; Supplemental Fig. S3). We then extracted protein complexes and supercomplexes from mitochondria isolated from leaves and cell cultures of Arabidopsis wild-type and dldh1 plants and resolved the proteins by blue native (BN)-PAGE. The presence of d-LDH was analyzed by in gel activity assay and the identification of protein spots by mass spectrometry.
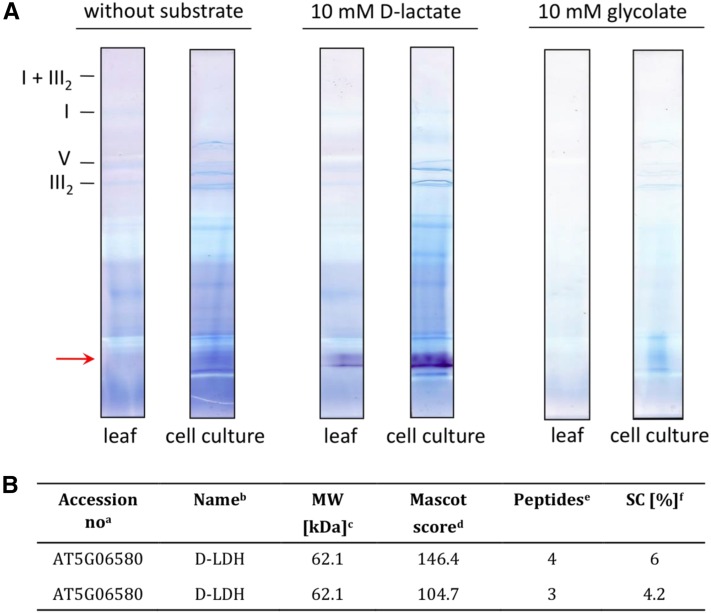
In the mitochondrial fraction obtained from leaves, as well as from cell culture, a single, well-defined enzymatic activity signal was visible using d-lactate as a substrate (Fig. 3A). The signal obtained from the mitochondrial fraction of leaves was much weaker than the signal coming from that of suspension cultured cells, suggesting that d-LDH is highly expressed in these cultured cells. No protein band showing activity with glycolate as a substrate was observed in any fraction. This indicates that Arabidopsis d-LDH does not use glycolate as a substrate and that mitochondria would not possess an oxidoreductase acting on this 2-hydroxy acid.
Figure 3.
A, In gel activity assays of mitochondrial proteins using d-lactate or glycolate as substrate. Mitochondria isolated from Arabidopsis leaves (leaf) as well as from cell suspension culture (cell culture) were solubilized with 5% (w/v) digitonin and subsequently separated on a BN gel. In gel activity assays were performed using 10 mm d-lactate or 10 mm glycolate as substrate. The position of the activity signal is marked on the left with the red arrow. Positions of membrane-bound protein complexes are given on the left (I, complex I; III2, dimeric complex III; V, ATP synthase complex; I + III2, supercomplex formed by complex I and dimeric complex III). B, d-LDH peptides identified on the BN gel by mass spectrometry. aAccession numbers of identified proteins as given by The Arabidopsis Information Resource (http://www.arabidopsis.org/). bName of the identified protein. cCalculated molecular mass of the identified proteins as deduced from the corresponding gene. dProbability score for the protein identifications based on mass spectrometry analysis and MASCOT search. eNumber of unique peptides. fSequence coverage of the protein by identified peptides.
To confirm the identity of the protein showing activity with d-lactate, two spots from the BN gel region where the activity was obtained were picked and analyzed by mass spectrometry. d-LDH (Q94AX4; At5g06580) was identified in both spots, with four and three unique peptides, respectively (Fig. 3B). On the BN gel, d-LDH runs at approximately 110 kD (Fig. 3A). This is in line with our previous results showing that the enzyme is active as a dimer (around 135 kD), while the monomer has a mass of 62 kD (Engqvist et al., 2009). Based on these results and in accordance with GelMap data (Senkler and Braun, 2012; Supplemental Fig. S4), d-LDH does not form part of OXPHOS supercomplexes, suggesting no stable interaction with the electron transport machinery.
In addition, we analyzed a possible connection of d-LDH with mitochondrial respiration by measuring the respiratory rate in dldh mutants and wild-type plants (Engqvist et al., 2009). We observed that the respiration rates of the dldh mutants were not different from those measured in wild-type plants even in the presence of KCN, which blocks flux through the CYTc-dependent pathway, only allowing oxygen consumption via AOX (Supplemental Fig. S5).
Involvement of d-LDH in MGO Detoxification
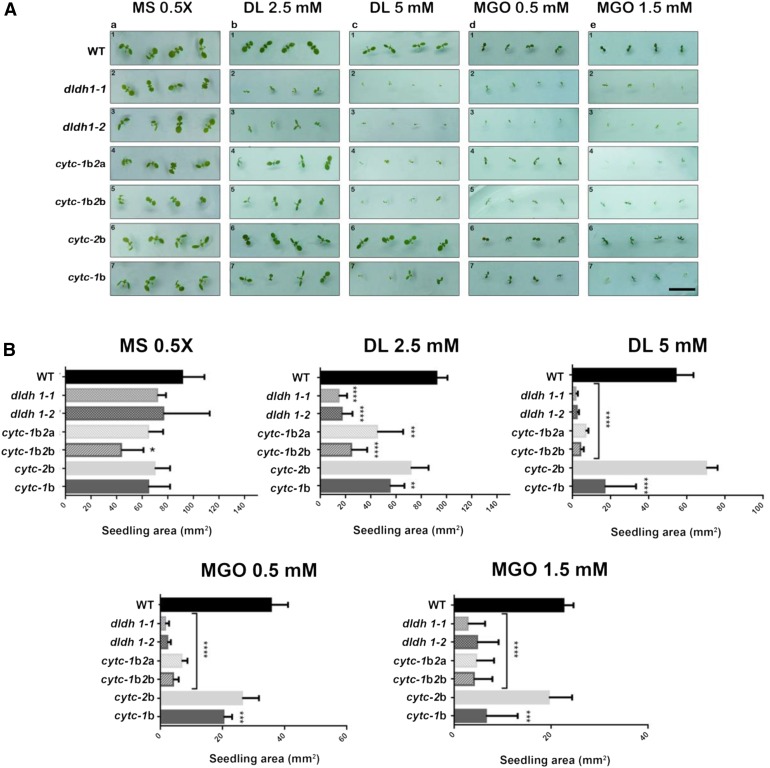
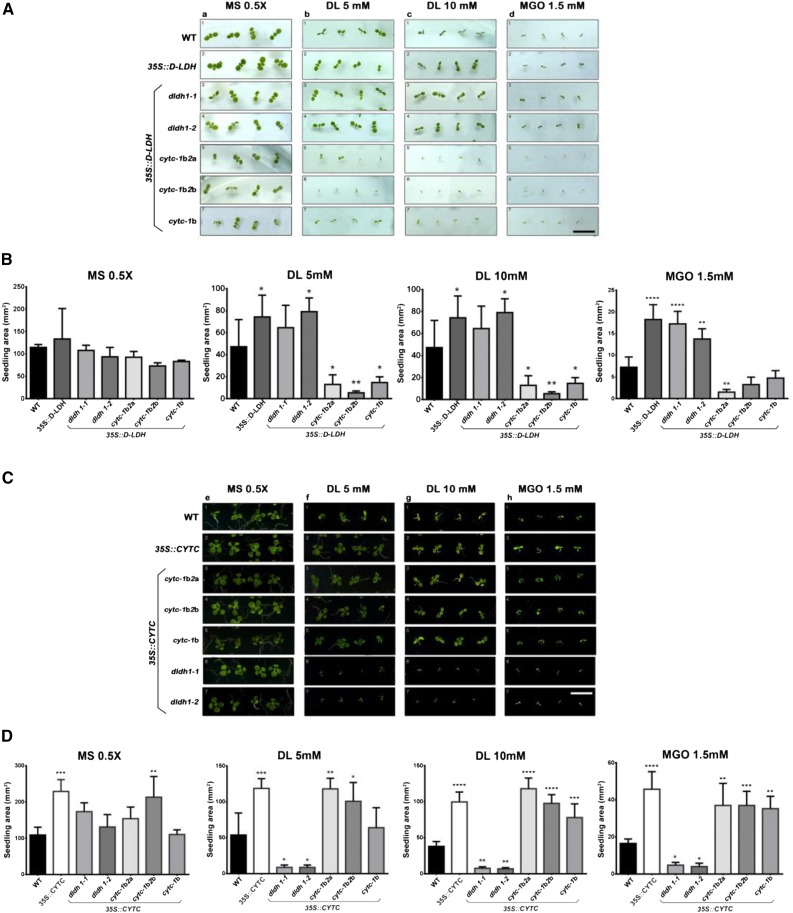
Under standard cultivation, either on soil or on plates, d-LDH loss-of-function plants (dldh1-1 and dldh1-2; Engqvist et al., 2009) showed no evident differences in development or morphology compared with the wild type (Fig. 4, A, rows 1–3, and B; Supplemental Fig. S6). Inclusion of either d-lactate (2.5–5 mm; Fig. 4, A, columns b and c, and B) or MGO (0.5–1 mm; Fig. 4, A, columns d and e, and B) to the solid growth medium impaired the growth of wild-type plants (Fig. 4A, row 1). Although the toxicity exerted by d-lactate and MGO was dose dependent, dldh mutants were more strongly affected than wild-type plants by both compounds (Fig. 4, A, rows 1–3, and B).
Figure 4.
d-LDH and CYTc deficiency increase the sensitivity of plants to d-lactate and MGO. Wild-type (WT) plants and different mutants in d-LDH, CYTc-1, and CYTc-2 were grown on plates containing 0.5× Murashige and Skoog medium alone (MS) or supplemented with the indicated concentrations of d-lactate (DL) or MGO. dldh1-1 and dldh1-2 are two different insertional mutants in the DLDH gene; cytc-1b is an insertional knockdown mutant in CYTC-1; cytc-2a and cytc-2b are knockout mutants in CYTC-2; cytc-1b2a and cytc-1b2b are the respective double mutants (Supplemental Fig. S9; for details about the mutants, see Engqvist et al. [2009] and Welchen et al. [2012]). A, Images of representative plants at day 7 after germination. Bar = 10 mm. B, Area of seedlings of the different genotypes at day 7 after germination grown under different conditions. The results are expressed as means ± sd of 100 seedlings from three different experiments. Data were analyzed by one-way ANOVA and Dunnett’s test. Asterisks indicate significant differences from wild-type plants (*, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001).
To confirm that the lack of a functional d-LDH was responsible for the higher toxicity caused by d-lactate and MGO, we expressed d-LDH under the control of the CaMV35S promoter in wild-type and dldh plants and analyzed the effect of those compounds. Increased expression of d-LDH ameliorated the growth of wild-type plants in the presence of d-lactate and MGO (Fig. 5, A, rows 1 and 2, and B) and reversed the increased sensitivity of dldh loss-of-function lines to these compounds (Fig. 5, A, rows 1, 3, and 4, and B).
Figure 5.
Effects of d-LDH and CYTc overexpression on the sensitivity to d-lactate and MGO. A and B, Wild-type (WT) plants and plants expressing d-LDH driven by the CaMV35S promoter (35S::d-LDH) in different genetic backgrounds were grown on plates containing 0.5× Murashige and Skoog medium alone (MS) or supplemented with d-lactate (DL) or MGO at the indicated concentrations. Images were taken 7 d after germination (A), and the area of seedlings was quantified (B). C and D, Plants expressing the CYTC-1 gene driven by the CaMV35S promoter (35S::CYTC) in different genetic backgrounds were grown in different media. Images were taken 14 d after germination (C), and the area of seedlings was quantified (D). Results are expressed as means ± sd of 100 seedlings from three different experiments. Data were analyzed by one-way ANOVA and Dunnett’s test. Asterisks indicate significant differences from wild-type plants (*, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001). For a schematic representation of the transgenic lines used in this experiment, see Supplemental Figure S9. Bars = 10 mm.
These results provide genetic evidence that d-LDH is required to metabolize d-lactate in vivo. Furthermore, the fact that MGO affects dldh plants more than wild-type plants indicates that d-LDH is involved in MGO detoxification.
Analysis of the Role of CYTc as an Electron Acceptor of d-LDH in Vivo
The kinetic analysis performed previously with the recombinant d-LDH indicated that this enzyme uses CYTc as an electron acceptor in vitro (Engqvist et al., 2009). In yeast and mammals, d-LDH has been described as a membrane-bound matrix-oriented enzyme that transfers electrons to complex III through ubiquinone (de Bari et al., 2002; Pallotta et al., 2004). To analyze the possibility that complex III acts as an acceptor of electrons from d-LDH during d-lactate oxidation, we measured d-LDH activity in isolated mitochondria using antimycin A. Antimycin A binds to the Qi site of complex III, thereby inhibiting the electron transfer from cytochrome b to CYTc (Campo et al., 1992). We found no significant difference in d-LDH activity in the presence or absence of antimycin A (Supplemental Fig. S7A). This indicates that ubiquinone and its associated dehydrogenase systems (Schertl and Braun, 2014) do not serve as electron acceptors and are upstream of the entry site of d-LDH-derived electrons into the electron transport chain. To support these biochemical data, we tested the effect of d-lactate on the growth of plants in which complex III was inhibited by myxothiazol. Myxothiazol binds to the quinone-binding site and blocks electron transfer from ubiquinol to cytochrome b (von Jagow et al., 1984). At the concentration tested (20 μm), myxothiazol did not affect the sensitivity of wild-type plants to d-lactate (Supplemental Fig. S7, B and C), suggesting that inhibition of complex III does not affect d-lactate oxidation. To test if myxothiazol effectively blocks electron transport through complex III under these conditions, we included in the assay an antisense AOX1a line in which the capacity of the alternative respiratory pathway is decreased (Umbach et al., 2005). These plants cannot divert electrons to the alternative pathway upon inhibition of complex III and are then sensitive to myxothiazol (Supplemental Fig. S7, B and C). The aox1a seedlings were not affected in d-lactate oxidation, while dldh mutants showed sensitivity to d-lactate and not to myxothiazol (Supplemental Fig. S7, B and C). This provides in vivo evidence that the activity of complex III is not required for d-lactate oxidation in Arabidopsis. Both biochemical and physiological data indicate that electrons from d-lactate oxidation enter the respiratory chain downstream of complex III.
To test the hypothesis that CYTc acts as such an entry point in vivo, we expanded our combinatorial reverse genetic and chemical biological approach. Knockdown and knockout T-DNA insertion mutants in both CYTC genes (cytc-1b for CYTC-1 and cytc-2a and cytc-2b for CYTC-2; Welchen et al., 2012; for schematic representation, see Supplemental Fig. S9) were grown on plates with supplementation of different amounts of d-lactate or MGO. Double mutants (cytc-1b2a and cytc-1b2b), which possess severely reduced amounts of CYTc (Welchen et al., 2012), showed impaired growth in the presence of both compounds similar to that observed with the dldh mutant plants (Fig. 4, A, rows 1, 4, and 5, and B). The simultaneous analysis of the CYTc single mutants indicated a predominant role of CYTC-1 in MGO detoxification, since mutants lacking a fully functional CYTc-1 (cytc-1b) were more strongly affected than mutants lacking CYTc-2 (cytc-2b), which behaved similarly to wild-type plants (Fig. 4A, rows 1, 6, and 7). The double cytc mutants were even more sensitive than the single mutants in CYTC-1, indicating that CYTc-2 also participates in the detoxification of d-lactate and MGO in vivo, albeit with a smaller contribution.
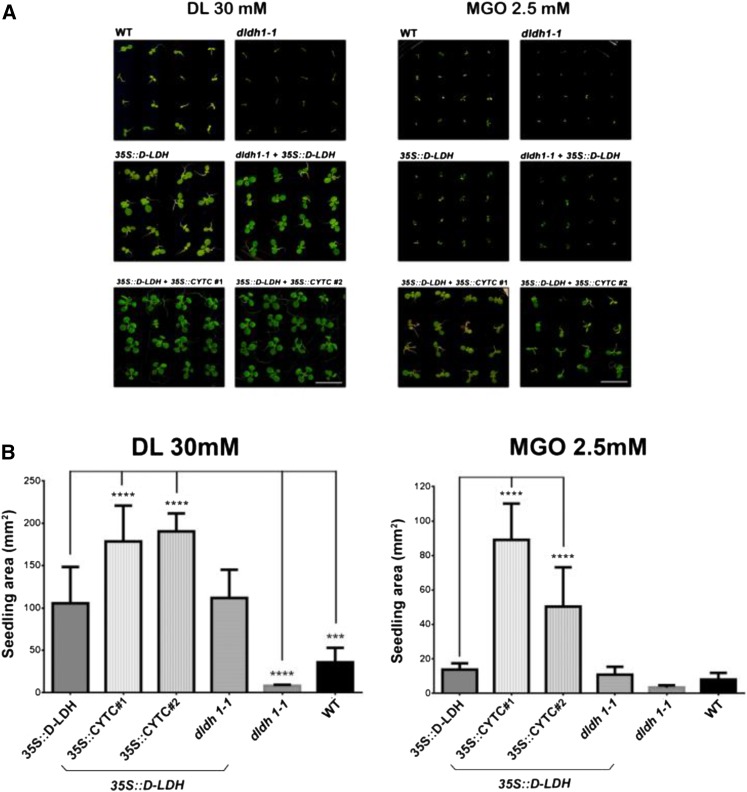
Figure 6.
Combined overexpression of d-LDH and CYTc allows plant development at very high d-lactate and MGO concentrations. Seedlings of the different genotypes were grown on plates containing either 30 mm d-lactate (DL) or 2.5 mm MGO. 35S::d-LDH + 35S::CYTC 1 and 2 represent two different lines obtained after transformation of a d-LDH-overexpressing line with a construct expressing CYTC-1 driven by the CaMV35S promoter (for a schematic representation of the transgenic lines used in this experiment, see Supplemental Fig. S9). A, Representative images of plants 10 d after germination. Bars = 10 mm. B, Area of seedlings of the different genotypes at 10 d after germination grown under different conditions. The results are expressed as means ± sd of 100 seedlings from three different experiments. Data were analyzed by one-way ANOVA and Dunnett’s test. Asterisks indicate significant differences from 35S::d-LDH plants (***, P < 0.001; and ****, P < 0.0001). WT, Wild type.
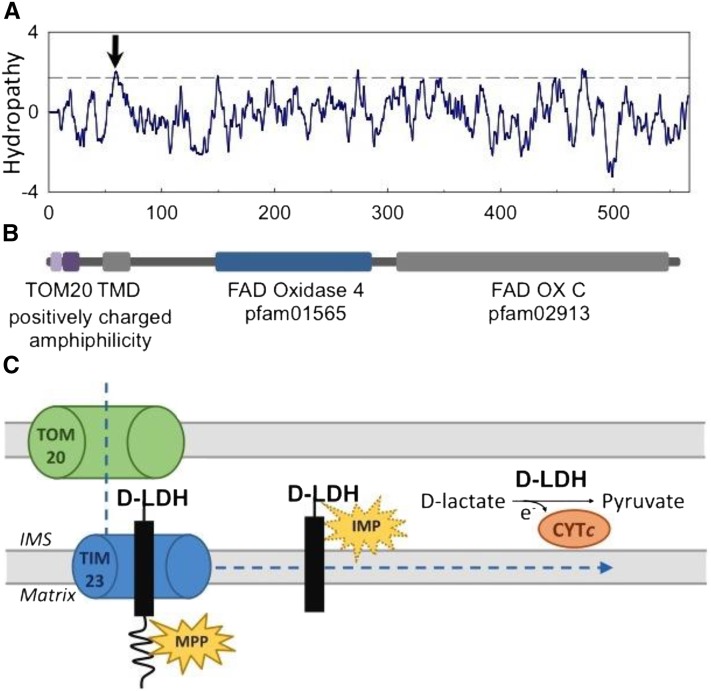
Figure 7.
A, Hydropathy plot of the 567-amino acid-long sequence of d-LDH. Window size was nine amino acids; the dashed line indicates the threshold for possible transmembrane domains (TMD). The hydropathy plot supports the predicted transmembrane domain between amino acids 48 and 68 predicted by TMpred (arrow). B, Structural and functional domain features of d-LDH. C, Predicted stop-transfer mechanism of mitochondrial d-LDH import and its mode of action. In the model, d-LDH is recognized via TOM20 and guided through the OMM into the IMS. The mitochondrial processing peptidase (MPP) cleaves the positively charged amphiphilic region to yield a membrane-anchored IMS protein. d-LDH may be further processed by an IMS protease (IMP) to be released as a soluble IMS protein. In both scenarios, d-LDH converts d-lactate to pyruvate, passing electrons to CYTc.
We hypothesized that the sensitivity of the CYTc mutants may be due to decreased d-LDH protein amount or activity. To test this, we generated lines that express d-LDH driven by the CaMV35S promoter (35S::d-LDH) in the single cytc-1b or cytc-1b2a and cytc-1b2b double mutant backgrounds. d-LDH expression did not influence the sensitivity of the cytc mutants to d-lactate and MGO. The plants remained more sensitive to both compounds than wild-type plants (Fig. 5, A, rows 5–7, and B). Immunoblot analysis using an anti-d-LDH antibody confirmed that the increased sensitivity of cytc mutants was not the result of a lower amount of d-LDH protein (Supplemental Fig. S8A). Hence, impairment of d-LDH activity in the cytc mutant backgrounds appears to result from limited electron flux downstream of d-LDH, which is consistent with the hypothesis that CYTc is the in vivo acceptor of those electrons.
To further evaluate the role of CYTc in d-lactate and MGO degradation, we generated plants that express CYTC-1 driven by the CaMV35S promoter (35S::CYTC) in different backgrounds. These plants showed an increase in CYTc protein amount, as evidenced by immunoblot analysis (Supplemental Fig. S8B). Increased expression of CYTC-1 in either the wild-type or cytc background led to increased tolerance to toxic concentrations of d-lactate and to a lesser extent also to MGO (Fig. 5, C, rows 1–5, and D). In contrast, CYTC-1 overexpressed in the dldh background did not change the sensitivity of dldh plants to either compound, as the plants remained more sensitive than wild-type plants (Fig. 5, C, rows 6 and 7, and D). These results indicate that CYTc, as an electron acceptor of the d-LDH reaction, can limit the capacity of the system to oxidize d-lactate and provide in vivo evidence for the participation of d-LDH and CYTc in the same detoxification pathway.
Combined Overexpression of d-LDH and CYTc Increases the Tolerance to d-Lactate and MGO
Since constitutive expression of either d-LDH or CYTc caused increased tolerance to high concentrations of d-lactate and MGO in the wild-type background, we hypothesized that expressing both proteins simultaneously might further increase the tolerance, allowing the plants to cope with even higher d-lactate and MGO concentrations. To test this hypothesis, we expressed d-LDH in the 35S::CYTC background (Supplemental Fig. S8C) and analyzed seedling development on solid medium supplemented with very high d-lactate (30 mm) and MGO (2.5 mm) concentrations. Quantification of seedling area under different growth conditions (Fig. 6) indicates that the double overexpressors were less affected than plants expressing d-LDH alone, suggesting that the tolerance to high d-lactate and MGO concentrations could be boosted.
DISCUSSION
d-LDH Localizes to the IMS and Does Not Form Part of Respiratory Supercomplexes
Using a combination of in silico, biochemical, physiological, and imaging analyses, we here shed light on enzymatic and structural properties of d-LDH. We confirm that higher plant mitochondria possess a protein that corresponds to d-LDH that uses only d-lactate and not glycolate as a substrate in vivo (Fig. 3).
d-LDH does not form part of OXPHOS supercomplexes (Fig. 3) and is localized to the IMS and/or the IMM (Figs. 1 and 2). Anchoring in the inner membrane with exposure to the IMS appears particularly likely. In silico analysis indicates that d-LDH possesses a transmembrane α-helix span from amino acids 48 to 68 (TMpred; Hofmann and Stoffel, 1993) after the N-terminal targeting signal (Fig. 7A). The new developed and trained prediction program MitoFates (Fukasawa et al., 2015) clearly identifies a TOM20 recognition motif with good confidence (amino acids 3–7) followed by an amphiphilic region (amino acids 11–20) in the Arabidopsis d-LDH. The prediction of the canonical cleavage site (at Gln-22 in the recognition motif RQLHS) for the matrix-located mitochondrial processing peptidase in context with a predicted transmembrane domain (high hydrophobic region: amino acids 48–68; Fig. 7B) is consistent with a stop-transfer mechanism, leading to an import arrest of d-LDH at the inner membrane. In this scenario, the premature d-LDH protein is translocated through the outer membrane via TOM20 and further guided to TIM23, where it comes to an import arrest at the transmembrane domain. The matrix-exposed signal peptide is cleaved by the mitochondrial processing peptidase, and the protein is released into the IMM. The stop-transfer sequence might be further cleaved by the inner membrane protease, releasing the soluble d-LDH into the IMS (Fig. 7C). Protein anchoring in the IMM with exposure to the IMS side, or in the outer membrane with exposure to the IMS, or just localization as a soluble IMS protein, cannot be ruled out. When considering the mechanisms of the mitochondrial IMS protein import machinery, however, anchoring in the outer membrane with exposure to the IMS is unlikely (Hewitt et al., 2014). An IMS-oriented active center allows d-LDH to interact with CYTc to donate the electrons produced during the oxidation of d-lactate to pyruvate.
Similarly, in yeast and bacteria, the oxidation of d-lactate occurs through a d-LDH coupled to CYTc (Lodi and Ferrero, 1993; Lodi et al., 1994) and CYTc-2 (Horikiri et al., 2004), respectively. Although d-LDH from different species are phylogenetically related (Cristescu and Egbosimba, 2009), current data indicate that there are fundamental differences in their submitochondrial localization and electron acceptors. While in yeast, d-LDH is anchored in the IMM, with the bulk of the protein residing as a folded domain in the IMS and possessing high acceptor specificity for CYTc (Pallotta et al., 2004), mammalian d-LDH has been found to be a membrane-bound matrix-oriented enzyme and to transfer electrons to complex III through ubiquinone (de Bari et al., 2002). Our results indicate a situation in Arabidopsis that is similar to that in yeast.
CYTc Is the Electron Acceptor of the d-LDH Reaction
Our previous in vitro studies with the recombinant isolated d-LDH protein suggested that CYTc might be the electron acceptor of the d-LDH reaction (Engqvist et al., 2009). This was confirmed and refined in this work by the fact that the inhibition of complex III by antimycin A does not interfere with d-LDH activity in isolated mitochondria and that myxothiazol does not affect the sensitivity of plants to d-lactate in vivo (Supplemental Fig. S7). In addition, the analysis of CYTc knockdown and knockout mutants and of overexpression plants clearly indicates that CYTc is involved in d-lactate and MGO detoxification in vivo, since sensitivity to these compounds is affected by changes in CYTc levels (Figs. 4 and 5). Furthermore, the fact that CYTc action requires an active d-LDH suggests that CYTc detoxifies these compounds, acting in the same pathway as d-LDH. The data obtained in this work collectively and consistently indicate that CYTc receives the electrons of d-lactate oxidation and delivers them to the electron transport chain, which ultimately reduces oxygen. This is also supported by the fact that increased expression of d-LDH does not improve the tolerance of CYTc-deficient plants to d-lactate and MGO. It is noteworthy that increasing the expression levels of either CYTc or d-LDH increases the tolerance of plants to both compounds, suggesting that both proteins can limit the detoxification process (depending on the expression level of the other). Even increased tolerance was observed when both proteins were overexpressed simultaneously (Fig. 6). This agrees with a model in which CYTc acts as a direct acceptor of electrons from d-LDH. Our observation that d-LDH is located in the mitochondrial IMS is consistent with this model. Similarly, in yeast and bacteria, the oxidation of d-lactate occurs through a d-LDH coupled to CYTc (Lodi and Ferrero, 1993; Lodi et al., 1994) and CYTc-2 (Horikiri et al., 2004), respectively.
Our observation exemplifies that, in addition to its canonical role in electron transfer from complex III to complex IV during respiration, CYTc also participates in other electron transfer reactions in plants. As such, CYTc acts as an electron sink for different oxidation reactions that take place in the mitochondrial IMS or the cytosol (Schertl and Braun, 2014; Welchen and Gonzalez, 2016) CYTc is thus a flexible platform to integrate reductant inputs from different metabolic pathways, including the detoxification of cytosolic MGO and d-lactate.
MATERIALS AND METHODS
Plant Lines
The following plant lines were used in this work: Arabidopsis (Arabidopsis thaliana) wild-type ecotype Columbia-0 (Col-0), single T-DNA mutants dldh1-1 (Salk_001490; Engqvist et al., 2009), dldh1-2 (Salk_026859; Engqvist et al., 2009), cytc-1b (Salk_143142), cytc-2a (Salk_037790), and cytc-2b (Salk_029663), and double mutants in CYTc genes (cytc-1b2a and cytc-1b2b; Welchen et al., 2012; Supplemental Fig. S8). To validate the effectiveness of myxothiazol inhibition, an antisense AOX1a line (Nottingham Arabidopsis Stock Centre stock no. N6599; Umbach et al., 2005) was used. Lines expressing d-LDH driven by the CaMV35S promoter (35S::d-LDH) were produced by transforming plants with the pGWB2-d-LDH construct (Wienstroer et al., 2012; Supplemental Fig. S8). To express CYTC-1 driven by the CaMV35S promoter (Supplemental Fig. S8), a 515-bp SalI/EcoRI cDNA fragment comprising the entire open reading frame and additional 5′ and 3′ untranslated regions of CYTC-1 (At1g22840) was amplified using specific oligonucleotides (5′-GGCGTCGACTCTCTTGTTGTTCCTGATC-3′ and 5′-GGCGAATTCCAAATTCCCACCAAGAAC-3′) and cloned into pENTR-3C (Thermo Fisher Scientific). This fragment, together with its flanking recombination sites, was amplified by PCR and transferred to the destination binary vector pAUL1 (Lyska et al., 2013) using the Gateway cloning system (Thermo Fisher Scientific).
Transformed plants were obtained by floral dip using Agrobacterium tumefaciens GV3103 followed by selection on plates containing 50 mg L−1 BASTA (glufosinate ammonium) or 50 mg L−1 hygromycin for the CYTc or d-LDH construct, respectively.
Plant Growth Conditions
Plants were grown on soil pots of 10 cm height at 22°C to 24°C under a long-day (16 h of light/8 h of darkness) photoperiod and a light intensity of approximately 100 µmol m−2 s−1. Alternatively, Arabidopsis seeds were surface sterilized in a solution containing 70% (v/v) ethanol and 0.1% (w/v) SDS for 5 min, washed in distilled water, and sown in petri dishes containing 0.5× Murashige and Skoog basal medium (M5519; Sigma) and 1% (w/v) agar (pH 7.4). Plates were cold stratified at 4°C for 2 d and transferred to a growth chamber under long-day conditions. The toxicity of MGO and d-lactate was assayed in a range of concentrations (0.1–5 mm MGO and 0.5–40 mm d-lactate) on agar plates containing Murashige and Skoog medium (pH 7.4).
Arabidopsis Suspension Culture Cultivation and Isolation of Mitochondria
Arabidopsis Col-0 suspension cells were cultivated as described by Sunderhaus et al. (2006). Isolation of mitochondria from leaves was carried out according to Keech et al. (2005) using leaves of 4-week-old plants grown under long-day conditions (16 h of light/8 h of dark). Mitochondria isolation from suspension cells was performed according to Werhahn et al. (2001) with slight modifications. Briefly, cells were disrupted in a Waring blender and in the presence of 450 mm Suc, 15 mm MOPS, 1.5 mm EGTA, 0.6% (w/v) polyvinylpyrrolidone, 0.2% (w/v) bovine serum albumin, and 0.2 mm phenylmethylsulfonyl fluoride (PMSF), pH 7.4, with KOH). The suspension was supplemented with 2-mercaptoethanol at a final concentration of 14.3 mm and centrifuged twice for 5 min at 2,700g and once for 10 min at 8,300g to remove cell debris. Mitochondria were pelleted by centrifugation at 17,000g for 10 min, and the pellet was resuspended in 3 to 5 mL of washing buffer (300 mm Suc, 10 mm MOPS, 1 mm EGTA, and 0.2 mm PMSF, pH 7.2, with KOH). The suspension was homogenized using a Potter-Elvehjem homogenizer (two strokes), transferred to a Percoll gradient, and centrifuged for 45 min at 70,000g. The Percoll gradient consisted of 40%, 23%, and 18% Percoll in 0.3 m Suc and 10 mm MOPS, pH 7.2, with KOH. Mitochondria were enriched at the 23%-40% interphase. Mitochondria from the interphase were washed two times with resuspension buffer (400 mm mannitol, 10 mm Tricine, 1 mm EGTA, and 0.2 mm PMSF, pH 7.2, with KOH) and pelleted by centrifugation at 14,300g for 10 min. The pellet was frozen in liquid nitrogen and stored at −80°C.
BN-PAGE and in Gel Activity Assay
Mitochondria were centrifuged for 10 min at 14,300g and resuspended in 100 μL of digitonin solubilization buffer (30 mm HEPES, pH 7.4, 150 mm potassium acetate, 10% [v/v] glycerol, and 50 mg mL−1 digitonin). After incubation on ice for 15 min, samples were centrifuged at 18,000g for 10 min to remove insoluble material, and the supernatant was supplemented with 5 μL of Coomassie Blue solution (5% [w/v] Coomassie Brilliant Blue G250 in 750 mm aminocaproic acid). Samples were loaded on a one-dimensional BN-PAGE device (Wittig et al., 2006). Gradient gels (4.5%–16% [w/v] acrylamide) were used for the separation of mitochondrial proteins. The gels were run for 45 min at 100 V and afterward for 16 h at 15 mA, maximum 500 V. The Coomassie Blue-containing cathode buffer was replaced by a cathode buffer without Coomassie Blue at half completion of each run to reduce background staining.
d-LDH activity was analyzed in gel using d-lactate or glycolate as a substrate as described previously (Engqvist et al., 2009). d-LDH activity becomes visible as purple bands. The activity staining was stopped by washing the gel with water. To improve visualization and reduce the background, the gel was transferred into destaining solution (40% [v/v] methanol and 10% [v/v] acetic acid) overnight. The gels were scanned on a transmission scanner (PowerLook III; UMAX).
d-LDH Activity Assay
d-LDH activity was measured using extracts of isolated mitochondria at 25°C using an Epoch Microplate Spectrophotometer (Biotek) in a total volume of 300 µL. Kinetic values were corrected by values obtained in parallel control experiments without substrate or added protein. Protein quantification was carried out using the Pierce Coomassie (Bradford) Protein Assay (Thermo Scientific). The reaction mixture contained 50 mm Tris-HCl, pH 8.4, 60 µm CYTc, 1 mm KCN, and 50 mm d-lactate. Complex III was inhibited by the addition of 20 µm antimycin A. The reaction was started by adding 5 µg of mitochondria extract. The reduction of CYTc was monitored at 550 nm (E = 19 mm−1 cm−1).
Western-Blot Analysis
For western-blot analysis, mitochondrial crude protein extracts were separated by 10% Tricine SDS-PAGE and transferred to Hybond-ECL (GE Healthcare). Blots were incubated with polyclonal rabbit antibodies raised against Arabidopsis d-LDH (Eurogentec) at a dilution of 1:100 and against COX2 (Agrisera; AS04 053A) at a dilution of 1:1,000 or with purified mouse antibody against CYTc (BD Pharmingen; 556433) diluted 1:1,000. Reactions were developed with secondary anti-rabbit (Thermo Scientific; 31462) or anti-mouse (Thermo Scientific; 31450) immunoglobulin conjugated with horseradish peroxidase using the SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Oxygen Consumption Measurements
Oxygen consumption was measured according to Welchen et al. (2012). Plants were kept in darkness for 40 min, and then 100 to 150 mg of aerial parts was transferred to 800 µL of reaction buffer (300 mm mannitol, 1% [w/v] bovine serum albumin, 10 mm potassium phosphate, pH 7.2, 10 mm KCl, and 5 mm MgCl2). Measurements were made at 25°C using a Clark-type oxygen electrode (Hansatech). The cyanide-sensitive pathway was inhibited by the addition of 1 mm KCN.
Mass Spectrometry
Tryptic digestion of proteins and mass spectrometry were performed as described previously (Klodmann et al., 2010). Protein identifications were based on the MASCOT search algorithm using the Arabidopsis protein database, The Arabidopsis Information Resource release 10 (www.arabidopsis.org).
Confocal Imaging and Protease Protection Analysis
The coding sequence of d-LDH (At5g06580) was amplified from Arabidopsis Col-0 cDNA (using the following primer pair: 5′-TCTCCTACGTTGTGAAATCCAC-3′ and 5′-AGAAACAGTATGAGGCCAAACG-3′) and prepared for Gateway cloning using primer pair 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGCTTTCGCTTCAAAATTCGC-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTGAAACATACATGAGGAGGAATTAAC-3′. The amplicon was inserted into pDONR207 to generate an entry vector. For the generation of a C-terminal GFP fusion, the insert was transferred into the destination vector pSS01 (Brach et al., 2009), which contains the sequence for roGFP2 downstream of and in frame with the Gateway cassette. For the C-terminal YFP fusion, pSS01 destination vector and cpYFP sequence (Schwarzländer et al., 2011) amplified with the primer pair 5′-TGCCTAGGTACAACAGCGACAACGTCTATATC-3′ and 5′-TCTTAATTAATTAGGTACCGTTGTACTCCAGCT-3′ were digested with AvrII and PacI and subsequently ligated into the linearized pSS01 destination vector with cpYFP sequence downstream of and in frame with the Gateway cassette. Transient expression in wild tobacco (Nicotiana benthamiana) leaf epidermis, generation of stable Arabidopsis lines, MitoTracker Orange staining, confocal imaging of GFP and MitoTracker Orange, and quantitative pixel analysis were performed as described before (Wagner et al., 2015) using the same controls, mito:GFP (Logan and Leaver, 2000) and NMT1:GFP (Logan and Knight, 2003) for soluble matrix and outer mitochondrial membrane localization, respectively. For protease protection analysis, a d-LDH:cpYFP line was used and a mito:cpYFP line (Schwarzländer et al., 2011) as well as mito:GFP (Logan and Leaver, 2000) served as controls for soluble matrix localization.
For mitochondrial isolation, Arabidopsis seedlings were hydroponically grown for 2 weeks as described previously (Schwarzländer et al., 2011). Freshly purified mitochondria from each line were checked for OMM integrity by CYTc latency assays and subjected to treatments on ice for 2 h each with thermolysin at 66.7 mg g−1 total mitochondrial protein (Sigma; 9073-78-3) and 0.1% (v/v) Triton X-100. Reactions were stopped by the addition of Laemmli sample buffer and boiling at 95°C for 6 min. Amounts of 24 µg (NMT1:GFP), 12 µg (d-LDH:YFP), and 6 µg (mito:YFP and mito:GFP) of total mitochondrial protein were separated by 12% SDS-PAGE and blotted onto a polyvinylidene difluoride membrane. The membrane was blocked and probed with GFP Tag Antibody (Thermo; A-6455). Goat anti-rabbit poly-horseradish peroxidase secondary antibody (Thermo; 32260) was used for chemiluminescent detection with Pierce ECL Western Blotting Substrate (Thermo; 32109).
Statistical Analysis of Seedling Area
The area of seedlings of the different genotypes grown under different conditions was calculated from images of at least three different experiments using ImageJ 1.48v public software (https://imagej.nih.gov/ij/). Data were analyzed by one-way ANOVA, and the mean values were compared by Dunnett’s multiple comparison test. Statistical analysis was performed using InfoStat for Windows (http://www.infostat.com.ar).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At5g06580 and At1g22840.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Transient expression of d-LDH:GFP in N. benthamiana leaves.
Supplemental Figure S2. Protease protection analysis by GFP immunodetection after thermolysin treatment of intact, purified mitochondria expressing C-terminal fusions of NMT1, d-LDH, as well as the Nicotiana plumbaginifolia β-ATPase mitochondrial presequence with YFP or GFP in the absence and presence of Triton X-100 to disrupt mitochondrial membranes.
Supplemental Figure S3. d-LDH activity in protein extracts of isolated mitochondria.
Supplemental Figure S4. d-LDH identification using GelMap.
Supplemental Figure S5. Respiration is not altered in dldh mutant plants.
Supplemental Figure S6. d-LDH deficiency does not affect the growth of plants under standard conditions.
Supplemental Figure S7. Effect of complex III inhibition on d-LDH activity and plant growth in the presence of d-lactate.
Supplemental Figure S8. Western-blot analysis of d-LDH and CYTc protein levels in plants with different backgrounds.
Supplemental Figure S9. Schematic representation of the plant lines used in this study.
Acknowledgments
We thank David C. Logan (Université d’Angers) for providing the mito:GFP and NMT1:GFP lines and Natanael Mansilla and Manuel Franco (Instituto de Agrobiotecnología del Litoral) for help in the genetic experiments using different Arabidopsis lines.
Glossary
- MGO
methylglyoxal
- IMS
intermembrane space
- CaMV35S
cauliflower mosaic virus 35S
- OMM
outer mitochondrial membrane
- IMM
inner mitochondrial membrane
- BN
blue native
- Col-0
Columbia-0
- PMSF
phenylmethylsulfonyl fluoride
Footnotes
This work was supported by the Deutsche Akademische Austauschdienst and the Ministerio de Ciencia, Tecnología, e Innovación Productiva (Argentina) through a cooperative research grant to D.H.G. and V.G.M.; the Deutsche Forschungsgemeinschaft (grant no. FOR 1186 to V.G.M. and H.-P.B. and Emmy-Noether program grant no. SCHW1719/1–1, Research Training Group grant no. GRK2064, and grant no. SCHW1719/5–1 to M.S., as part of the package PAK918, jointly with V.G.M.), the Agencia Nacional de Promoción Científica y Tecnológica (Argentina), the Consejo Nacional de Investigaciones Científicas y Técnicas (Argentina), and the Universidad Nacional del Litoral (to E.W. and D.H.G.).
References
- Atlante A, de Bari L, Valenti D, Pizzuto R, Paventi G, Passarella S (2005) Transport and metabolism of D-lactate in Jerusalem artichoke mitochondria. Biochim Biophys Acta 1708: 13–22 [DOI] [PubMed] [Google Scholar]
- Brach T, Soyk S, Müller C, Hinz G, Hell R, Brandizzi F, Meyer AJ (2009) Non-invasive topology analysis of membrane proteins in the secretory pathway. Plant J 57: 534–541 [DOI] [PubMed] [Google Scholar]
- Braun HP, Schmitz UK (1992) Affinity purification of cytochrome c reductase from potato mitochondria. Eur J Biochem 208: 761–767 [DOI] [PubMed] [Google Scholar]
- Campo ML, Kinnally KW, Tedeschi H (1992) The effect of antimycin A on mouse liver inner mitochondrial membrane channel activity. J Biol Chem 267: 8123–8127 [PubMed] [Google Scholar]
- Chen M, Thelen JJ (2010) The plastid isoform of triose phosphate isomerase is required for the postgerminative transition from heterotrophic to autotrophic growth in Arabidopsis. Plant Cell 22: 77–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristescu ME, Egbosimba EE (2009) Evolutionary history of D-lactate dehydrogenases: a phylogenomic perspective on functional diversity in the FAD binding oxidoreductase/transferase type 4 family. J Mol Evol 69: 276–287 [DOI] [PubMed] [Google Scholar]
- de Bari L, Atlante A, Guaragnella N, Principato G, Passarella S (2002) d-Lactate transport and metabolism in rat liver mitochondria. Biochem J 365: 391–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist M, Drincovich MF, Flügge UI, Maurino VG (2009) Two D-2-hydroxy-acid dehydrogenases in Arabidopsis thaliana with catalytic capacities to participate in the last reactions of the methylglyoxal and beta-oxidation pathways. J Biol Chem 284: 25026–25037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa Y, Tsuji J, Fu SC, Tomii K, Horton P, Imai K (2015) MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol Cell Proteomics 14: 1113–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt VL, Gabriel K, Traven A (2014) The ins and outs of the intermembrane space: diverse mechanisms and evolutionary rewiring of mitochondrial protein import routes. Biochim Biophys Acta 1840: 1246–1253 [DOI] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W (1993) TMbase: a database of membrane spanning proteins segments. Biol Chem Hoppe Seyler 374: 166 [Google Scholar]
- Hoque TS, Uraji M, Ye W, Hossain MA, Nakamura Y, Murata Y (2012) Methylglyoxal-induced stomatal closure accompanied by peroxidase-mediated ROS production in Arabidopsis. J Plant Physiol 169: 979–986 [DOI] [PubMed] [Google Scholar]
- Horikiri S, Aizawa Y, Kai T, Amachi S, Shinoyama H, Fujii T (2004) Electron acquisition system constructed from an NAD-independent D-lactate dehydrogenase and cytochrome c2 in Rhodopseudomonas palustris No. 7. Biosci Biotechnol Biochem 68: 516–522 [DOI] [PubMed] [Google Scholar]
- Kalapos MP. (1999) Methylglyoxal in living organisms: chemistry, biochemistry, toxicology and biological implications. Toxicol Lett 110: 145–175 [DOI] [PubMed] [Google Scholar]
- Keech O, Dizengremel P, Gardestrom P (2005) Preparation of leaf mitochondria from Arabidopsis thaliana. Physiol Plant 124: 403–409 [Google Scholar]
- Klodmann J, Senkler M, Rode C, Braun HP (2011) Defining the protein complex proteome of plant mitochondria. Plant Physiol 157: 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klodmann J, Sunderhaus S, Nimtz M, Jänsch L, Braun HP (2010) Internal architecture of mitochondrial complex I from Arabidopsis thaliana. Plant Cell 22: 797–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon K, Choi D, Hyun JK, Jung HS, Baek K, Park C (2013) Novel glyoxalases from Arabidopsis thaliana. FEBS J 280: 3328–3339 [DOI] [PubMed] [Google Scholar]
- Lodi T, Ferrero I (1993) Isolation of the DLD gene of Saccharomyces cerevisiae encoding the mitochondrial enzyme D-lactate ferricytochrome c oxidoreductase. Mol Gen Genet 238: 315–324 [DOI] [PubMed] [Google Scholar]
- Lodi T, O’Connor D, Goffrini P, Ferrero I (1994) Carbon catabolite repression in Kluyveromyces lactis: isolation and characterization of the KIDLD gene encoding the mitochondrial enzyme D-lactate ferricytochrome c oxidoreductase. Mol Gen Genet 244: 622–629 [DOI] [PubMed] [Google Scholar]
- Logan DC, Knight MR (2003) Mitochondrial and cytosolic calcium dynamics are differentially regulated in plants. Plant Physiol 133: 21–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan DC, Leaver CJ (2000) Mitochondria-targeted GFP highlights the heterogeneity of mitochondrial shape, size and movement within living plant cells. J Exp Bot 51: 865–871 [PubMed] [Google Scholar]
- Lyska D, Engelmann K, Meierhoff K, Westhoff P (2013) pAUL: a Gateway-based vector system for adaptive expression and flexible tagging of proteins in Arabidopsis. PLoS ONE 8: e53787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurino VG, Engqvist MK (2015) 2-Hydroxy acids in plant metabolism. The Arabidopsis Book 13: e0182, doi/10.1199/tab.0182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra K, Banerjee AB, Ray S, Ray M (1995) Glyoxalase III from Escherichia coli: a single novel enzyme for the conversion of methylglyoxal into D-lactate without reduced glutathione. Biochem J 305: 999–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallotta ML, Valenti D, Iacovino M, Passarella S (2004) Two separate pathways for D-lactate oxidation by Saccharomyces cerevisiae mitochondria which differ in energy production and carrier involvement. Biochim Biophys Acta 1608: 104–113 [DOI] [PubMed] [Google Scholar]
- Phillips SA, Thornalley PJ (1993) The formation of methylglyoxal from triose phosphates: investigation using a specific assay for methylglyoxal. Eur J Biochem 212: 101–105 [DOI] [PubMed] [Google Scholar]
- Rode C, Senkler M, Klodmann J, Winkelmann T, Braun HP (2011) GelMap: a novel software tool for building and presenting proteome reference maps. J Proteomics 74: 2214–2219 [DOI] [PubMed] [Google Scholar]
- Saito R, Yamamoto H, Makino A, Sugimoto T, Miyake C (2011) Methylglyoxal functions as Hill oxidant and stimulates the photoreduction of O2 at photosystem I: a symptom of plant diabetes. Plant Cell Environ 34: 1454–1464 [DOI] [PubMed] [Google Scholar]
- Salvato F, Havelund JF, Chen M, Rao RS, Rogowska-Wrzesinska A, Jensen ON, Gang DR, Thelen JJ, Møller IM (2014) The potato tuber mitochondrial proteome. Plant Physiol 164: 637–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertl P, Braun HP (2014) Respiratory electron transfer pathways in plant mitochondria. Front Plant Sci 5: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzländer M, Logan DC, Fricker MD, Sweetlove LJ (2011) The circularly permuted yellow fluorescent protein cpYFP that has been used as a superoxide probe is highly responsive to pH but not superoxide in mitochondria: implications for the existence of superoxide ‘flashes.’ Biochem J 437: 381–387 [DOI] [PubMed] [Google Scholar]
- Senkler M, Braun HP (2012) Functional annotation of 2D protein maps: the GelMap portal. Front Plant Sci 3: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subedi KP, Choi D, Kim I, Min B, Park C (2011) Hsp31 of Escherichia coli K-12 is glyoxalase III. Mol Microbiol 81: 926–936 [DOI] [PubMed] [Google Scholar]
- Sunderhaus S, Dudkina NV, Jänsch L, Klodmann J, Heinemeyer J, Perales M, Zabaleta E, Boekema EJ, Braun HP (2006) Carbonic anhydrase subunits form a matrix-exposed domain attached to the membrane arm of mitochondrial complex I in plants. J Biol Chem 281: 6482–6488 [DOI] [PubMed] [Google Scholar]
- Thornalley PJ. (2003) Glyoxalase I: structure, function and a critical role in the enzymatic defence against glycation. Biochem Soc Trans 31: 1343–1348 [DOI] [PubMed] [Google Scholar]
- Umbach AL, Fiorani F, Siedow JN (2005) Characterization of transformed Arabidopsis with altered alternative oxidase levels and analysis of effects on reactive oxygen species in tissue. Plant Physiol 139: 1806–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vögtle FN, Burkhart JM, Rao S, Gerbeth C, Hinrichs J, Martinou JC, Chacinska A, Sickmann A, Zahedi RP, Meisinger C (2012) Intermembrane space proteome of yeast mitochondria. Mol Cell Proteomics 11: 1840–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Jagow G, Ljungdahl PO, Graf P, Ohnishi T, Trumpower BL (1984) An inhibitor of mitochondrial respiration which binds to cytochrome b and displaces quinone from the iron-sulfur protein of the cytochrome bc1 complex. J Biol Chem 259: 6318–6326 [PubMed] [Google Scholar]
- Wagner S, Behera S, De Bortoli S, Logan DC, Fuchs P, Carraretto L, Teardo E, Cendron L, Nietzel T, Füßl M, et al. (2015) The EF-hand Ca2+ binding protein MICU choreographs mitochondrial Ca2+ dynamics in Arabidopsis. Plant Cell 27: 3190–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchen E, Gonzalez DH (2016) Cytochrome c, a hub linking energy, redox, stress and signaling pathways in mitochondria and other cell compartments. Physiol Plant 157: 310–321 [DOI] [PubMed] [Google Scholar]
- Welchen E, Hildebrandt TM, Lewejohann D, Gonzalez DH, Braun HP (2012) Lack of cytochrome c in Arabidopsis decreases stability of complex IV and modifies redox metabolism without affecting complexes I and III. Biochim Biophys Acta 1817: 990–1001 [DOI] [PubMed] [Google Scholar]
- Werhahn W, Niemeyer A, Jänsch L, Kruft V, Schmitz UK, Braun H (2001) Purification and characterization of the preprotein translocase of the outer mitochondrial membrane from Arabidopsis: identification of multiple forms of TOM20. Plant Physiol 125: 943–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienstroer J, Engqvist MK, Kunz HH, Flügge UI, Maurino VG (2012) d-Lactate dehydrogenase as a marker gene allows positive selection of transgenic plants. FEBS Lett 586: 36–40 [DOI] [PubMed] [Google Scholar]
- Wittig I, Braun HP, Schägger H (2006) Blue native PAGE. Nat Protoc 1: 418–428 [DOI] [PubMed] [Google Scholar]
- Yadav SK, Singla-Pareek SL, Ray M, Reddy MK, Sopory SK (2005) Methylglyoxal levels in plants under salinity stress are dependent on glyoxalase I and glutathione. Biochem Biophys Res Commun 337: 61–67 [DOI] [PubMed] [Google Scholar]