Significance
Neuroimaging has largely focused on two goals: mapping associations between neuroanatomical features and phenotypes and building individual-level prediction models. This paper presents a complementary analytic strategy called morphometricity that aims to measure the neuroanatomical signatures of different phenotypes. Inspired by prior work on heritability, we define morphometricity as the proportion of phenotypic variation that can be explained by brain morphology (e.g., as captured by structural brain MRI). In the dawning era of large-scale datasets comprising traits across a broad phenotypic spectrum, morphometricity will be critical in prioritizing and characterizing behavioral, cognitive, and clinical phenotypes based on their neuroanatomical signatures. Furthermore, the proposed framework will be significant in dissecting the functional, morphological, and molecular underpinnings of different traits.
Keywords: neuroimaging, brain morphology, statistical association
Abstract
Complex physiological and behavioral traits, including neurological and psychiatric disorders, often associate with distributed anatomical variation. This paper introduces a global metric, called morphometricity, as a measure of the anatomical signature of different traits. Morphometricity is defined as the proportion of phenotypic variation that can be explained by macroscopic brain morphology. We estimate morphometricity via a linear mixed-effects model that uses an anatomical similarity matrix computed based on measurements derived from structural brain MRI scans. We examined over 3,800 unique MRI scans from nine large-scale studies to estimate the morphometricity of a range of phenotypes, including clinical diagnoses such as Alzheimer’s disease, and nonclinical traits such as measures of cognition. Our results demonstrate that morphometricity can provide novel insights about the neuroanatomical correlates of a diverse set of traits, revealing associations that might not be detectable through traditional statistical techniques.
The structural, functional, and molecular properties of the brain support numerous traits spanning the behavioral, cognitive, and clinical spectra. Neuroanatomical features are in turn influenced by factors such as age, sex, training, and genetics (1–4). Neuroimaging allows us to characterize these bidirectional associations by revealing variation in brain structure and function across individuals. Conventional methods that we use to probe these associations aim to anatomically map effects, build prediction models, or test hypotheses. However, we do not have a standard technique to measure and compare the often spatially distributed and complex patterns of neuroanatomical correlates of different phenotypes. Here we present a metric called morphometricity that offers this capability.
To date, structural neuroimaging studies have primarily relied on three classes of analytic approaches. The first strategy is hypothesis-driven and uses a regression model to examine associations between behavioral traits or clinical conditions and a small number of a priori image-derived measurements, such as those restricted to an anatomical region of interest (ROI) (5). The ROI-based approach provides useful insights about the underlying biology and can be efficient in limited-sample-size scenarios but is restricted to the tested hypothesis. The second approach is exploratory and aims to compute maps of associations by conducting brain-wide tests (6), as exemplified in voxel-based morphometry (7) or vertex-wise cortical thickness analysis (see, e.g., ref. 8). Such massive univariate analyses can offer a comprehensive picture of the underlying anatomical associations, yet they can also be inefficient in revealing subtle, multivariate patterns of association, because each anatomical location is typically examined in isolation and the burden of multiple testing correction can constrain statistical power. The third class includes multivariate techniques such as canonical correlation analysis (9), partial least squares (10), Bayesian inference algorithms (11), or other machine learning methods (12, 13). These studies are focused on either discovering the multivariate patterns of association or demonstrating individual-level prediction capabilities, but the biological interpretation of trained multivariate models can be challenging (14). Furthermore, these methods often suffer from high computational demand and can be sensitive to implementation details, such as the choice of learning rule, optimization algorithm, and local optima in training.
We present morphometricity analysis as an approach to examine the global statistical association between brain morphology and observable traits. Inspired by prior work on trait heritability in population and statistical genetics (15, 16), we define morphometricity as the proportion of phenotypic variation that can be explained by brain morphology (e.g., as captured by measurements derived from structural brain MRI scans). Unlike ROI-based or massive univariate association tests, morphometricity analysis is not concerned with specific anatomical structures or the precise anatomic localization of effects. In contrast to the application of machine learning to population data, the primary aim of morphometricity analysis is not to maximize individual-level prediction accuracy but to examine and quantify statistical associations. The proposed strategy thus affords a unique perspective on the biological underpinnings of different phenotypes and allows us to compare and contrast imaging modalities, types of anatomical measurements, and processing streams.
Morphometricity is grounded in linear mixed effects (LME) modeling, a classical statistical framework that was recently used in population genetics to quantify the heritability of a trait using genome-wide genetic variants (17–19). The model relates the variation in brain morphology computed from brain-wide, MRI-derived measurements to the variation in observable traits and can be fitted using well-established, robust computational tools. In our implementation, we use FreeSurfer (20), a freely available, widely used, and extensively validated brain MRI analysis software package, to automatically process structural MRI scans and obtain a vector of volumetric measurements across subcortical structures and cortical thickness measurements across the entire cortical mantle, which constitute a comprehensive description of the structural neuroanatomy.
We applied the morphometricity analysis to over 3,800 unique brain MRI scans from nine large-scale studies and computed the morphometricity of clinical conditions including Alzheimer’s disease, attention-deficit hyperactivity disorder (ADHD), schizophrenia, autism spectrum disorder, and Parkinson’s disease and nonclinical traits including sex, age, intelligence, education level, and an array of cognitive measures. Our results demonstrate that morphometricity analysis promises to offer a unique perspective on the relationship between brain anatomy and behavioral, cognitive, and physiological traits.
Results
Overview of the Model.
The proposed morphometricity analysis is based on the following LME model:
| [1] |
where is an -dimensional column vector of a quantitative phenotype with being the sample size (i.e., number of subjects), is the (design) matrix of confounding variables (sometimes called covariates or nuisance variables) such as age and sex, is the (fixed effect) coefficient vector, is an -dimensional random effect vector drawn from a zero-mean multivariate Gaussian distribution with a covariance matrix that is equal to the scaled anatomic similarity matrix (ASM) , and the elements of the noise vector are assumed to be drawn from independent and zero-mean Gaussian distributions with homogeneous variance . The ASM encodes global morphological resemblance between pairs of individuals in the sample and in principle can be any nonnegative definite matrix with its diagonal elements constrained to be equal to 1 on average, and can thus be interpreted as the total variance captured by the ASM. In this paper, we considered two types of intuitive and widely used metrics that quantify the similarity of volumetric and cortical thickness measurements extracted from structural brain MRI scans between pairs of individuals: (i) a linear similarity metric (i.e., inner product between normalized imaging measurements) and (ii) a nonlinear Gaussian-type similarity metric (Methods). Using a model selection approach, we found that the Gaussian similarity metric provided consistently better description of the data across the traits we studied (Methods and Table S1). Therefore, all reported morphometricity estimates were based on the Gaussian metric.
Table S1.
AIC and BIC values (Eq. 6) for the primary morphometricity analyses conducted with the linear (Eq. 4) and Gaussian (Eq. 5) kernels
| AIC (× 104) | BIC (× 104) | |||
| Disease/trait (study) | Gaussian | Linear | Gaussian | Linear |
| Alzheimer’s (ADNI) | −1.508 | −0.309 | −1.362 | −0.163 |
| ADHD | −2.040 | −0.655 | −1.826 | −0.441 |
| Schizophrenia (MCIC) | −0.772 | −0.216 | −0.716 | −0.160 |
| Autism (ABIDE) | −2.022 | −0.637 | −1.804 | −0.419 |
| Parkinson’s | −0.522 | −0.194 | −0.491 | −0.163 |
| Age (GSP) | −2.967 | −0.276 | −2.433 | 0.258 |
| IQ (ADHD 200) | −0.521 | −0.107 | −0.474 | −0.059 |
| Sex (GSP) | −3.559 | −0.992 | −3.024 | −0.457 |
| Education (PPMI) | −0.512 | −0.188 | −0.477 | −0.153 |
Formally, we define morphometricity based on the LME model of Eq. 1 as
| [2] |
where is the phenotypic variance. Morphometricity is thus the proportion of phenotypic variation that can be explained by brain morphology, the variation of which is captured by the ASM. Estimates of morphometricity can be computed by plugging the restricted maximum likelihood (ReML) estimates (21, 22) of the variance components, and , into Eq. 2.
We extend this definition to the case-control design, that is, binary disease traits (affected vs. unaffected), using the classical liability-threshold model, which is widely used in population and statistical genetic studies (23, 24). The model assumes that the underlying disease liability (which is a quantitative variable) follows a Gaussian distribution and individuals are cases (affected) if their liability exceeds a threshold. The morphometricity estimate for a disease trait on the observed scale, obtained by fitting the model of Eq. 1 to the binary phenotype data, can be easily transformed to the liability scale (24, 25):
| [3] |
where is the morphometricity on the liability scale, is the prevalence of the disease in the general population (i.e., the proportion of the population having the disease), is the prevalence of the disease in the study sample, is the liability threshold, is the standard Gaussian cumulative distribution function, and is the standard Gaussian density function. In real disease studies, cases are often considerably oversampled relative to their population prevalence (known as nonrandom ascertainment), in which case is larger than . Transforming morphometricity estimates from the observed scale to the liability scale makes them independent of population and sample prevalence and thus comparable across different diseases.
Overview of the Data.
We analyzed brain MRI scans and trait data from over 3,800 unique individuals spanning nine large-scale studies: the Harvard/Massachusetts General Hospital Brain Genomic Superstruct Project (GSP) (26), the Human Connectome Project (HCP) (27), the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (28), the Attention-Deficit Hyperactivity Disorder (ADHD 200) sample (29), the Open Access Series of Imaging Studies (OASIS) cross-sectional sample (30), the Center for Biomedical Research Excellence (COBRE) schizophrenia sample (31), the MIND Clinical Imaging Consortium (MCIC) schizophrenia sample (32), the Autism Brain Imaging Data Exchange (ABIDE) (33), and the Parkinson Progression Marker Initiative (PPMI) (34). See Methods for further details on the datasets. The traits of interest were grouped into three categories: clinical diagnoses, general nonclinical traits, and measures of cognition.
Morphometricity of Clinical Diagnoses.
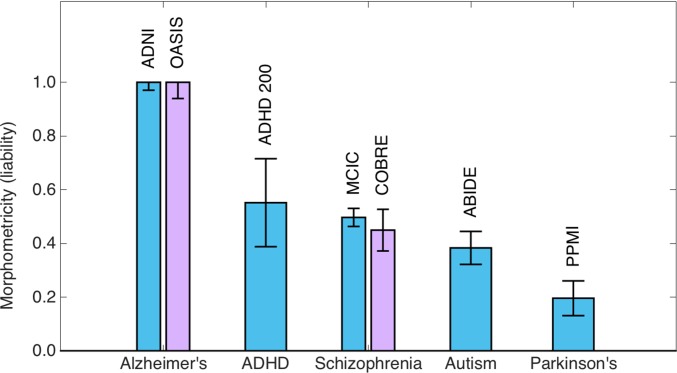
The clinical traits we examined included Alzheimer’s disease, ADHD, schizophrenia, autism spectrum disorder, and Parkinson’s disease. Table 1 lists the characteristics of the corresponding samples, along with morphometricity estimates (on the liability scale) and assumed population prevalence values (35–41). Fig. 1 shows the estimated morphometricity values on the liability scale. Our analyses revealed that Alzheimer’s disease is substantially morphometric (with a 95% confidence interval of [0.94–1.00]), suggesting that this clinical condition is associated with a substantial anatomical signature. However, ADHD, schizophrenia, and autism showed moderate morphometricity values, all greater than 0.35. Finally, we found that Parkinson’s disease was modestly morphometric, with an estimated liability-scale value of 0.20. All examined clinical conditions were statistically significantly associated with whole-brain macroscopic morphology, that is, the estimated morphometricity values were significantly larger than zero (all P < 0.005). Table S2 lists point estimates of morphometricity and their SEs computed via jackknife resampling (42). These results are in strong agreement with the parametric estimates in Table 1.
Table 1.
Sample characteristics and morphometricity estimates for analyzed disease traits
| Disease | Case N | Case age (mean ± SE) | Case females, % | Control N | Control age (mean ± SE) | Control females, % | Study name | Assumed prevalence, % | Morphometricity (liability scale) | SE |
| Alzheimer’s | 154 | 74.6 ± 7.6 | 46.8 | 219 | 75.9 ± 5 | 48.4 | ADNI | 13 | 1.00*** | 0.03 |
| ADHD | 122 | 11.6 ± 3.2 | 75.4 | 384 | 11.8 ± 2.9 | 62.2 | ADHD | 1 | 0.55** | 0.16 |
| Schizophrenia | 92 | 33 ± 11.2 | 75.0 | 85 | 32.8 ± 11.7 | 67.1 | MCIC | 1 | 0.50*** | 0.03 |
| Autism | 209 | 17.6 ± 7.9 | 14.4 | 305 | 17.3 ± 7.2 | 18.4 | ABIDE | 1 | 0.38*** | 0.06 |
| Parkinson's | 376 | 61.4 ± 9.7 | 35.1 | 152 | 60.4 ± 11.4 | 47.8 | PPMI | 0.2 | 0.20* | 0.06 |
*P < 5e-3, **P < 5e-4, ***P < 5e-5.
Fig. 1.
Morphometricity estimates of various diseases (on the liability scale) computed using the Gaussian ASM of Eq. 5. Each bar is annotated with study names used to compute these estimates. For Alzheimer’s disease and schizophrenia, we had independent samples used to compute replication estimates (purple bars). Error bars indicate SE of the estimates.
Table S2.
Parametric and jackknife estimates of morphometricity and SEs
| Morphometricity | SE | |||
| Disease/trait (study) | Parametric | Jackknife | Parametric | Jackknife |
| Alzheimer’s (ADNI) | 1.00 | 1.00 | 0.03 | 0.03 |
| ADHD | 0.55 | 0.55 | 0.16 | 0.17 |
| Schizophrenia (MCIC) | 0.50 | 0.50 | 0.03 | 0.03 |
| Autism (ABIDE) | 0.38 | 0.38 | 0.06 | 0.06 |
| Parkinson’s | 0.20 | 0.20 | 0.06 | 0.06 |
| Age (GSP) | 1.00 | 1.00 | 0.01 | 0.01 |
| IQ (ADHD 200) | 0.95 | 0.95 | 0.05 | 0.06 |
| Sex (GSP) | 0.93 | 0.93 | 0.02 | 0.02 |
| Education (PPMI) | 0.81 | 0.81 | 0.08 | 0.07 |
Morphometricity estimates for disease traits are on the liability scale.
We had access to two independent samples that allowed us to replicate our morphometricity estimates of Alzheimer’s disease (OASIS) and schizophrenia (COBRE). We observed that there was strong agreement between the estimates from independent samples (Fig. 1). Table S3 provides further data about these replication analyses.
Table S3.
Independent replication on nonoverlapping data: Sample characteristics and morphometricity estimates for analyzed disease traits
| Disease | Case N | Case age (mean ± SE) | Case females, % | Control N | Control age (mean ± SE) | Control females, % | Study name | Assumed prevalance, % | Morphometricity (liability scale) | SE |
| Alzheimer's | 100 | 76.8 ± 7.1 | 59.0 | 98 | 75.9 ± 9.0 | 73.4 | OASIS | 13 | 1.00 | 0.06 |
| Schizophrenia | 56 | 35.8 ± 12.4 | 16.1 | 73 | 35.7 ± 11.6 | 31.5 | COBRE | 1 | 0.45 | 0.08 |
Morphometricity of General, Nonclinical Traits.
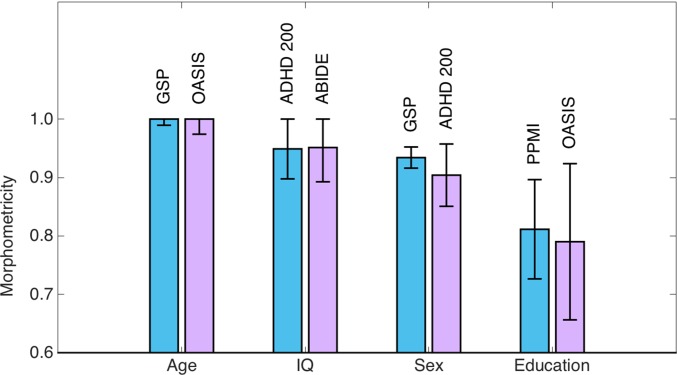
The nonclinical traits we examined were age, general intelligence (IQ), sex, and education level. Table 2 lists the characteristics of the samples used in the primary analyses, along with the estimates of morphometricity. These results revealed that all of the examined general traits are significantly and substantially morphometric (Fig. 2); all morphometricity point estimates were greater than 0.8, and all P values were <1e-8. Table S2 lists morphometricity estimates and their SEs computed via jackknife resampling. These results and the parametric estimates in Table 2 are virtually identical.
Table 2.
Sample characteristics and morphometricity estimates for the analyzed general nonclinical traits
| Trait name | Sample size | Age (mean ± SE) (minimum–maximum) | Females, % | Study name | Trait (mean ± SE) (minimum–maximum) | Morhopmetricity estimate | SE |
| Age | 1,073 | 22 ± 5.8 (18–81) | 56 | GSP | N/A | 1.00 | 0.01 |
| IQ | 155 | 11.3 ± 2.8 (7.2–17.7) | 60 | ADHD | 110.9 ± 14.4 (73–144) | 0.95 | 0.05 |
| Sex | 1,074 | 25.3 ± 13.7 (18–84) | 50 | GSP | N/A | 0.93 | 0.02 |
| Education | 152 | 60.4 ± 11.4 (30–82) | 34.8 | PPMI | 16.1 ± 2.8 (9–24) | 0.81 | 0.08 |
N/A, not applicable.
Fig. 2.
Morphometricity estimates of general nonclinical traits computed using the Gaussian ASM of Eq. 5. IQ denotes general intelligence. Each bar is annotated with study names used to compute these estimates. Blue bars correspond to results from the primary analyses, whereas purple bars correspond to independent replication analyses. Error bars indicate SE of the estimates.
We had access to independent replication samples for all of the general nonclinical traits we examined. It can be seen in Fig. 2 that the replication analyses revealed remarkably consistent morphometricity estimates. Table S4 provides further data about these replication analyses.
Table S4.
Independent replication on nonoverlapping data: Sample characteristics and morphometricity estimates for analyzed general nonclinical traits
| Trait name | Sample size | Age (mean ± SE) (minimum–maximum) | Females, % | Study name | Trait (mean ± SE) (minimum–maximum) | Morhopmetricity estimate | SE |
| Age | 124 | 68.0 ± 13.7 (33–94) | 72.6 | OASIS | N/A | 1.00 | 0.03 |
| IQ | 108 | 17.5 ± 8.0 (10.5–56.2) | 14.8 | ABIDE | 109.33 ± 11.0 (84–133) | 0.95 | 0.06 |
| Sex | 298 | 11.3 ± 2.9 (7.17–21.74) | 50 | ADHD | N/A | 0.90 | 0.05 |
| Education | 124 | 68.0 ± 13.7 (33–94) | 72.6 | OASIS | 3.5 ± 1.2 (1–5) | 0.79 | 0.13 |
N/A, not applicable.
Contrasting with ROI-Based Association Analyses.
The most common analytic strategy in today’s neuroimaging studies involves examining associations between traits and measurements from ROIs. Our goal in this analysis was to contrast the proposed whole-brain morphometricity analysis with such ROI-based techniques. We restricted our analysis to the six phenotypes (age, IQ, sex, education, Alzheimer’s disease, and schizophrenia), for which we had two independent datasets. We then used one of the samples (the replication sample in the analyses above) for the discovery of the most significantly associated ROI with the trait and the other sample (the primary sample in the analyses above) to quantify the strength and magnitude of association. Table S5 lists the structures that exhibited the strongest association with the traits in the discovery analysis.
Table S5.
ROI-based morphometricity estimates for the traits with two independent samples
| Trait name | ROI-based morphometricity | SE | P value | Most-associated ROI |
| Age | 0.82 | 0.03 | <1e-15 | Third ventricle |
| IQ | 0.87 | 0.05 | <1e-15 | Right lateral ventricle |
| Sex | 0.44 | 0.07 | 1.9e-9 | Right thalamus |
| Education | 0.00 | 0.06 | 1.00 | Right hippocampus |
| Alzheimer’s | 0.66 | 0.16 | 2.6e-5 | Right hippocampus |
| Schizophrenia | 0.06 | 0.05 | 0.22 | Right inferior lateral ventricle |
Most-associated ROIs were identified on independent sample. For disease traits (Alzheimer’s and schizophrenia) we list morphometricity estimates on the liability scale.
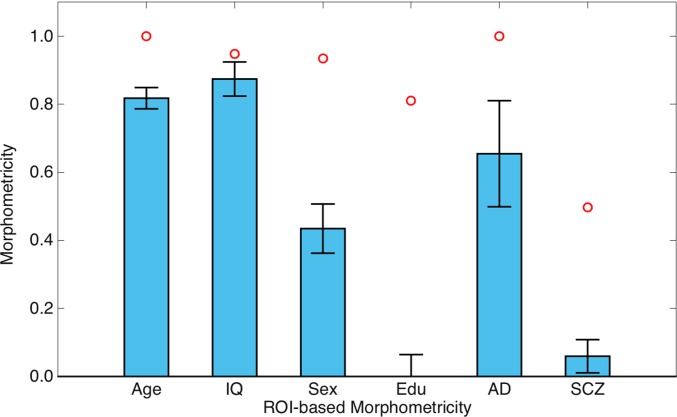
Fig. 3 visualizes the magnitude of association between the ROI-based measurements and phenotypic variation, assessed using the same LME modeling framework of whole-brain morphometricity. Here, we replaced the global ASM with one computed based on ROI measurements (see Methods for further details). It can be seen that the proportion of variance explained by ROI-based measurements was consistently lower than whole-brain morphometricity estimates (with general intelligence exhibiting the smallest discrepancy). Most notably, for education and schizophrenia, ROI-based associations were much weaker (both in magnitude and statistical significance) than whole-brain associations (Fig. 3 and Table S5). In fact, education and schizophrenia did not exhibit a statistically significant correlation with individual ROI-based measurements, whereas whole-brain morphometricity analyses revealed significant associations.
Fig. 3.
ROI-based morphometricity estimates of general nonclinical traits (age, intelligence, sex, and education), Alzheimer’s disease (AD), and schizophrenia (SCZ). For AD and SCZ, morphometricity estimates have been transformed to the liability scale. Red circles denote whole-brain morphometricity estimates for each trait. Error bars indicate SE of the estimates.
Morphometricity Analysis of Cognitive Measures.
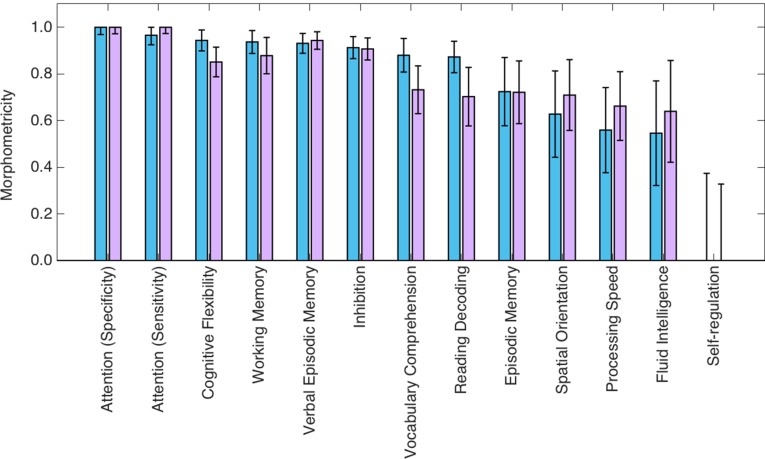
We used the most recent release of the HCP data (downloaded on December 15, 2015) to compute morphometricity estimates for an array of cognitive measures. Our primary analysis relied on 190 nontwin subjects of non-Hispanic European ancestry (28.9 ± 3.8 y of age, 47.3% female) drawn from separate families (i.e., there were no siblings in this sample). Fig. 4 shows the morphometricity estimates computed for variables that measure sustained attention, nonverbal and verbal episodic memory, working memory, executive function, delay discounting, language (vocabulary comprehension and reading decoding), spatial orientation, processing speed, fluid intelligence, and self-regulation (impulsivity). We conducted a secondary (replication) analysis on 208 non-Hispanic white twins (29.4 ± 3.2 y of age, 61.4% female), with one twin drawn from each family, and thus there were no siblings in this replication sample. Fig. 4 shows the results obtained from this secondary analysis as well.
Fig. 4.
Morphometricity estimates of various measures of cognition computed on data from the HCP and using the Gaussian ASM. Blue and purple bars correspond to primary and secondary analyses, respectively. Error bars indicate SE of the estimates.
Our results demonstrated that all examined variables, except for the measure of self-regulation, are statistically significantly morphometric. We note further that there was an important amount of variability in the degree of morphometricity across cognitive measures. This variation was remarkably consistent between the primary and secondary analyses. Measures of attention, cognitive flexibility, working memory, verbal episodic memory, and inhibition were substantially morphometric (with estimates greater than 0.80 in both primary and secondary analyses). Measures of language, nonverbal episodic memory, spatial orientation, processing speed, and fluid intelligence were moderately morphometric (with point estimates greater than 0.50).
Discussion
Morphometricity: A Metric to Quantify Whole-Brain Associations with a Trait.
In this paper, we introduced a technique for analyzing the neuroanatomical underpinnings of various clinical, physiological, and behavioral traits using large-scale neuroimaging data. In contrast with association testing techniques widely used in today’s neuroimaging studies, our approach does not focus on a priori ROIs or conduct independent (massive univariate) interrogations at each candidate region or voxel. Instead, morphometricity is a global quantification of the whole-brain anatomical signature of a trait.
Although the proposed approach is intimately related to image-based multivariate prediction performance, there are two characteristics of morphometricity that make it different from the application of machine learning. First, the metric does not require cross-validation, which is often the technique used in machine learning to gauge prediction accuracy. Cross-validation is usually computationally demanding and relies on the unbiased setting of model parameters (which might be achieved via a nested cross-validation strategy) and repeated and balanced partitioning of data into train and test sets. In contrast, the proposed LME-based approach exploits the entire dataset to fit the model and estimate the unknown variance component parameters, and in turn morphometricity, in an unbiased fashion. Second, morphometricity is a classical statistical measure of explained variance and is therefore familiar to interpret.
Morphometricity has many parallels to heritability in genetics (15, 16). Both concepts, statistical in nature, are about phenotypic variation and the proportion of variance explained. Thus, the interpretation has to be carried out within a probabilistic framework, is limited by the studied population, depends on the technique used to quantify the trait, and can be confounded by unmeasured variables acting through unknown mechanisms. The biases due to confounds such as (cryptic) relatedness between subjects or population admixture are well-studied in heritability analysis. As we elaborate below, morphometricity alone cannot be used to infer causal relationships but has to be followed up with further studies that will home in on potential mechanisms. The core difference between morphometricity and heritability is the direction of association. In heritability, this direction is known and fixed, because there is no known biological mechanism that would allow the phenotype to alter the genotype. In morphometricity, however, the directionality can go either way and has to be dissected with further biological studies.
Traits Can Be Morphometric to Different Degrees.
Virtually all traits we examined in this study were significantly morphometric. However, our analyses also revealed interesting variation in the whole-brain anatomical signature of different traits. Certain phenotypes, such as Alzheimer’s disease, age, and (maybe surprisingly) general intelligence (IQ), were substantially morphometric (with estimates exceeding 0.90), whereas other measures, such as nonverbal episodic memory, spatial orientation, processing speed, and fluid intelligence exhibited moderate morphometricity. Furthermore, the psychiatric disorders we examined (schizophrenia, ADHD, and autism) were all moderately morphometric, unequivocally pointing to a neuroanatomical substrate for these clinical conditions. The proposed morphometricity analysis is the first coherent framework that enables us to directly quantify and compare the morphological signatures of such diverse sets of traits.
The traits we presented in this study have been examined extensively in prior structural neuroimaging studies to reveal morphological correlates. Whereas many of these studies relied on regional or voxel-level association tests that are conducted at each location independently, there is growing evidence that multiple brain regions are implicated in complex, multivariate relationships with many common phenotypes. For example, patterns of atrophy in neurodegenerative conditions such as Alzheimer’s disease have been shown to spread through large-scale, distributed brain networks that can be circumscribed based on resting-state activity (43). Furthermore, developmental mechanisms such as neuronal migration, synapse formation, myelination, and synaptic pruning follow predictable and robust spatiotemporal patterns that are likely associated with behavioral traits such as intelligence and disrupted in psychiatric disorders such as schizophrenia (44, 45). Our data from ROI-based analyses support the premise of analyzing brain-wide patterns, rather than isolated regions, for associations between neuroanatomical features and behavioral or clinical phenotypes.
Morphometricity Estimates Are Consistent Across Independent Samples.
For most of the traits we examined in this study we replicated our analyses on independent data. All point estimates fell within 95% confidence intervals of the estimates computed on the corresponding independent data. These results suggested that the presented morphometricity estimates are consistent across different samples. We present these results with a cautionary note, however. As we emphasized above, morphometricity is a statistical metric that depends on the studied population and the measurement of the trait. Thus, variations in the patient composition, for example, or changes in diagnostic criteria will inevitably lead to different estimates of morphometricity. In our replication analyses, these factors seemed to play only a minor role.
Whole-Brain Morphometricity Analyses Can Be More Powerful Than ROI-Based Analyses.
We present morphometricity analysis as an alternative to the classical region-based interrogation conducted in neuroimaging, which is often focused on discovering or characterizing biomarkers and mapping biological effects. The central challenge in region-based approaches is that we need to either confine our analyses to a priori ROIs or exhaust statistical power by probing a large number of candidate regions. In our experiments, we conducted a direct comparison between whole-brain morphometricity analysis and an ROI-based approach. To identify trait-specific ROIs, a discovery analysis was run on independent samples of each trait. The associations between the identified ROIs and traits were then tested in nonoverlapping samples. Identifying the most associated ROI and estimating the magnitude of association in independent samples avoided the issue of circular analysis (46, 47) and produced unbiased morphometricity estimates for individual ROIs. Our results demonstrated that whole-brain morphology consistently explained more of the phenotypic variation than single ROIs. Furthermore, morphometricity analysis could reveal associations that were not detectable when focused on isolated regions. For example, education and schizophrenia were found to be not significantly associated with volumetric/thickness measurements of any of the individual ROIs, yet both traits were moderately and significantly morphometric in whole-brain analyses, which indicates that they may have spatially distributed neuroanatomical signatures that cannot be captured by individual ROIs. In addition, whole-brain morphometricity analysis offers the capability of capturing interactions between brain regions and thus can be more powerful than analyzing each ROI independently.
Potential Limitations and Drawbacks of Morphometricity.
Morphometricity is a statistical metric and assumes a particular, linear model of the relationship between variables. One critical component of the model is the ASM, which captures the covariance structure of the random effect that accounts for the morphological variation in the sample. In this work, we considered a linear metric and a nonlinear Gaussian-type metric to quantify the similarity of volumetric/thickness measurements between pairs of subjects and used a model selection technique to find the metric that better describes the data. Our analyses suggest that the Gaussian metric is consistently better than the linear metric across the traits we studied and captures a significant portion of relevant intersubject variation under different conditions. However, we have not attempted to exhaustively explore other types of similarity metrics, which may emphasize different aspects of the data and produce different results. Alternatively, the ASM can be built using a bottom-up approach and expressed as a combination of elementary matrices, and the parameters of this combination can be treated as unknown variables. By increasing the unknowns in the model, however, this approach will likely reduce statistical power.
Another important point to consider is that ASM should reflect brain-wide global morphology and cannot be optimized for a specific trait. This latter observation is critical to be able to objectively compare morphometricity estimates across different traits. Our global definition of ASM and morphometricity, however, constrains the interpretation of the results. Certain traits with very dramatic yet focal effects might not yield large morphometricity estimates, because the proposed model is insensitive to localized effects.
Finally, as in any association testing framework, the interpretation of morphometricity analysis results should be done very carefully and consider confounding mechanisms. This is well understood in the context of heritability, where nongenetic (e.g., dietary, cultural, and socioeconomic) influences that vary across racial groups or families can confound genetic analyses. Similar drawbacks apply to morphometricity. For example, siblings might have similar brain morphologies and phenotypic expressions, yet there might not be any causal link between brain morphology and the phenotype. Therefore, we recommend running morphometricity analyses on a set of unrelated subjects. Furthermore, we advise constraining the analysis (if possible) to a homogeneous sample of uniform ancestry and explicitly controlling for other potential confounding factors, such as age, sex, and scan site.
Potential Uses and Extensions of Morphometricity.
Morphometricity analysis can be used to prioritize imaging modalities, acquisition parameters, and processing pipelines. For example, there are a growing number of software packages that allow us to automatically extract numerous structural measurements from brain images. Different constructions of feature vectors and different metrics that quantify the similarity of imaging features between individuals will result in distinct ASMs, which can be compared with the model selection framework used in this study. Therefore, morphometricity analysis offers a way to quantitatively and objectively identify the imaging features and intersubject similarity metric that best describe the trait-relevant aspects of whole-brain morphology.
Alternatively, one can imagine estimating the functional, structural, connectomic, and molecular signatures of a trait within a single statistical model, where each of these components is represented with a random effect and the corresponding similarity matrix computed from a relevant modality. Similarly, one can partition the phenotypic variation into contributions from, for instance, cortical and subcortical features, or different large-scale brain networks. This strategy might offer novel insights about the neural correlates of certain phenotypes by integrating multiple modalities and/or modeling spatial heterogeneity in a unified analytic framework. This novel perspective might also allow us to quantify the complementary information contained in different imaging modalities and spatial locations.
As longitudinal imaging studies continue to grow, it will be interesting to extend morphometricity to examine the relationships between temporal dynamics in brain morphology (e.g., global atrophy rates) and clinical or behavioral traits. We envision using the LME strategy to model longitudinal data (48, 49), and we will define the morphometricity of longitudinal changes in phenotypes within this framework.
Finally, the proposed framework can also offer a novel perspective on examining relationships between different phenotypes. We plan to extend morphometricity, which is essentially the degree of association between global brain morphology and a phenotype, to quantify the “morphological correlation” between phenotypes. This will be analogous to genetic correlation analysis (50), which quantifies the genetic overlap between traits. We believe that morphological correlation will be an invaluable tool to examine the complex biological relationships between the various dimensions of human behavior and will inform basic and translational research into exploring and redefining the landscape of brain diseases.
Methods
The Imaging Measurements.
In this study we used the extensively studied FreeSurfer-derived measurements to describe the whole-brain morphology. The imaging measurements included volumes of noncortical structures (51) (left and right cerebral white matter, lateral ventricle, inferior lateral ventricle, cerebellum white matter, cerebellum cortex, thalamus proper, caudate, putamen, pallidum, hippocampus, amygdala, and the third and fourth ventricles) and thickness measurements of cortical regions (52) (left and right superior frontal, rostral middle frontal, caudal middle frontal, pars opercularis, pars triangularis, pars orbitalis, lateral orbitofrontal, medial orbitofrontal, precentral, paracentral, frontal pole, superior parietal, inferior parietal, supra marginal, post central, precuneus, superior temporal, middle temporal, inferior temporal, banks of the superior temporal sulcus, fusiform, transverse temporal, entorhinal, temporal pole, parahippocampal, lateral occipital, lingual, cuneus, pericalcarine, rostral anterior frontal, caudal anterior frontal, posterior parietal, isthmus parietal, and insula).
The Anatomical Similarity Matrix.
The ASM plays a central role in the proposed morphometricity analysis. The ASM is an symmetric matrix, where is the number of subjects in the analyzed sample. Entries in the ASM quantify the pairwise global similarity between the brain morphologies of two individuals. In principle, the ASM can be any nonnegative definite matrix with its diagonal elements constrained to be equal to 1 on average. In this study, we considered two widely used similarity metrics (linear and Gaussian) to construct the ASM.
Assume that denotes the -th imaging measurements from subject , is the total number of measurements, and is the sample SD of the -th measurement. The first ASM we considered uses a linear kernel. Thus, the similarity is measured as the linear correlation between pairs of imaging vectors and the -th entry is computed as
| [4] |
This is equivalent to modeling the random effect in Eq. 1 as a linear combination of the imaging features: , where is an matrix comprising the standardized imaging measurements [i.e., the -th entry of is ], and is an random vector distributed as (i.e., each imaging measurement is associated with an independent and normally distributed effect size). The covariance of can then be computed as , that is, is the ASM with its entries explicitly stated in Eq. 4.
The second ASM we considered was computed using a Gaussian kernel on standardized imaging features, with the -th entry defined as
| [5] |
Note that each ASM definition corresponds to different models of trait-relevant variation. For example, the Gaussian kernel can capture nonlinear and multivariate associations between brain morphology and traits. One can further decide to weigh different features differently, based on, for example, some a priori information about trait relevance. Each of these ASM choices will correspond to a particular semiparametric regression model, where the morphological association is captured with a function that belongs to a specific space of functions induced by the used kernel (53). Below, we describe an empirical strategy to choose the most appropriate ASM model from a selection of candidates.
In the presented study, for a given similarity metric, we computed an ASM for the cortical thickness measurements and an ASM for the head-size-normalized volumes of noncortical structures (i.e., divided by total intracranial volume estimates). The global ASM was then computed as the average of the cortical and noncortical ASMs.
Model Selection.
To select the ASM that can best describe the data, we used a model selection technique derived for LME models and proposed in ref. 53. Specifically, for a given ASM , if we denote as the ReML estimate of the covariance of , , and define , it can be shown that , that is, the trace of the matrix , is a measure of model complexity. Liu et al. (53) thus proposed the following Akaike Information Criterion (AIC) and Bayesian Information Criterion (BIC) in the LME modeling framework:
| [6] |
where is the residual sum of squares of the LME model. Both AIC and BIC reward the goodness of fit (the first term) of the model and penalize complex models (the second term) to avoid overfitting. BIC has a larger penalty term than AIC for large and thus favors simpler models. We selected the ASM that gave smaller AIC/BIC values.
The Analysis Pipeline.
We first ran FreeSurfer on all structural brain MRI scans available in each of the analyzed studies and dropped the subjects that FreeSurfer failed to complete successfully. Next, we conducted automatic quality control on the measurements computed by FreeSurfer by identifying outliers; if >25% of the used morphometric variables exhibited values that were more than two SDs away from the population mean, we deemed that subject an outlier and discarded it. For the remaining subjects, we computed pairwise similarity measures based on the linear or Gaussian kernel described above. If there were two subjects that exhibited a cortical and noncortical similarity measure greater than 0.95, we dropped one of those subjects, accounting for the possibility that this was a duplicate case or a closely related individual. Finally, we included sex and age as covariates in all our analyses (unless sex or age was the trait of interest, in which case that variable was not included). We further introduced dummy variables that indicated site IDs when analyzing multisite data [except for ADNI, where there were a large number of sites, yet the imaging parameters were carefully calibrated across sites (28)].
Given individual-level data and the ASM, we fit the model of Eq. 1 to estimate the variance component parameters via the ReML algorithm (21, 22). In MATLAB, we implemented an efficient Fisher scoring method to iteratively maximize the restricted likelihood of the model. The SE of the variance component estimates can be derived using the inverse of the Fisher information matrix when the algorithm converges. Empirically we confirmed the parametric estimates using jackknife resampling (Table S2). Significance of the morphometricity estimate was obtained via a likelihood ratio test, comparing LME models with and without the random effects. Because the null hypothesis () lies on the boundary of the parameter space, the likelihood ratio test statistic follows a half–half mixture of (a χ2 distribution with all probability mass at zero) and (a χ2 distribution with one degree of freedom) (54). Implementations of the developed morphometricity tools are available at people.csail.mit.edu/msabuncu/morphometricity.
ROI-Based Morphometricity Analysis.
To compare the proposed whole-brain morphometricity analysis to more conventional ROI-based analyses, we implemented an ROI-based adaptation of Eq. 1. Here, instead of computing the ASM using an array of variables that span the brain, we computed ROI-based ASMs only using a single ROI biomarker: the variable that had the strongest association with the trait of interest. To identify the ROI biomarker of each trait, we conducted an independent discovery analysis on a nonoverlapping sample, examining the association between each of the candidate imaging variables and the trait in a regression analysis while appropriately controlling for age, sex, and site. The imaging variable that exhibited the smallest P value was then identified as the ROI biomarker for the trait of interest and used in the morphometricity analysis. We note that for the ROI-discovery analyses we used the replication (secondary) samples of the whole-brain morphometricity analyses. This way, we computed the ROI-based morphometricity results using the primary samples.
The Data.
All of the data analyzed in this study have been made publicly available and are described in detail in prior publications (28–34, 55). As explained in these references, the data collection efforts have been approved by pertinent institutional committees and all subjects have received appropriate informed consent. We obtained access to these data following the relevant procedures outlined in corresponding data dissemination websites.
We used baseline brain MRI scans (T1-weighted acquired on 1.5T machines), clinical diagnosis, and demographic variables from phase 1 of the ADNI (28). In the ADHD 200 sample (29), cases were defined as those with evidence of nontypical development and an ADHD-Combined diagnosis, as per the published phenotypic key (fcon_1000.projects.nitrc.org/indi/adhd200/general/ADHD-200_PhenotypicKey.pdf). In the cross-sectional OASIS sample (30), subjects (of 60 y or older) with a clinical dementia rating (CDR) greater than 0 were classified as having dementia. Elderly subjects with a 0 CDR were classified as healthy controls. For the 20 control subjects with repeat scans, we only used the data from the first imaging session. In the COBRE sample (31), schizophrenia subjects were identified according to the phenotype file (fcon_1000.projects.nitrc.org/indi/retro/cobre.html). The MCIC data were compiled from a shared repository of multisite brain imaging data collected for the clinical investigation of schizophrenia (32). The ABIDE analyses (33) were conducted on subjects who were older than 10 y, and cases were defined as those having a nonzero diagnostic group entry in the phenotype table (fcon_1000.projects.nitrc.org/indi/abide/). In the PPMI analyses (34), cases were determined to be those diagnosed with Parkinson’s disease at baseline and controls were those who were clinically healthy and not prodromal, again at baseline (www.ppmi-info.org/access-data-specimens/download-data/).
All general trait analyses were conducted on healthy control samples (see criteria of relevant study). Both the OASIS and GSP samples cover a substantial portion of the adult life span and thus were used to estimate the morphometricity of age. All GSP analyses were constrained to unrelated, healthy controls of non-Hispanic European ancestry, with high-quality structural brain MRI scans acquired on a 12-channel coil (26). In the general intelligence (IQ) analyses, we used the Wechsler Abbreviated Scale of Intelligence as the phenotype (both in the ADHD 200 and ABIDE samples). For the morphometricity analysis of sex, we created subsamples that were gender-balanced (50% female) and age-matched between sexes. In the PPMI data, education was measured in years (minimum 9 and maximum 24), whereas in the OASIS sample, education levels were encoded as 1: less than high school graduate, 2: high school graduate, 3: some college, 4: college graduate, and 5: beyond college.
In the cognitive measure analyses, we used the demographic and behavioral measures reported in the “open access” and “restricted” subject information spreadsheets available from the HCP database website (www.humanconnectome.org/data). The HCP collected a range of well-validated and reliable behavioral measures, including those from the NIH Toolbox Assessment of Neurological and Behavioral Function, and several additional measures to assess domains not covered by the NIH Toolbox. For more information on the rationale behind the development of the behavioral batteries used in HCP, see ref. 55.
All MRI scans from ADNI, OASIS, ADHD 200, MCIC, COBRE, PPMI, and HCP were processed with FreeSurfer version 5.3. The GSP MRI scans were processed with FreeSurfer version 4.5.
Acknowledgments
We thank the 20 individual investigators at Harvard and Massachusetts General Hospital (MGH) who generously contributed data to the overall project. Support for this research was provided in part by National Institute for Biomedical Imaging and Bioengineering Grants P41EB015896, R01EB006758, R21EB018907, and R01EB019956; National Institute on Aging Grants 5R01AG008122 and R01AG016495; and National Institute for Neurological Disorders and Stroke Grants R01NS0525851, R21NS072652, R01NS070963, R01NS083534, and 5U01NS086625 and was made possible by the resources provided by Shared Instrumentation Grants 1S10RR023401, 1S10RR019307, and 1S10RR023043. Additional support was provided by the NIH Blueprint for Neuroscience Research Grant 5U01-MH093765, part of the multiinstitutional Human Connectome Project. Data were provided in part by the Brain Genomics Superstruct Project of Harvard University and MGH, with support from the Center for Brain Science Neuroinformatics Research Group, Athinoula A. Martinos Center for Biomedical Imaging, Center for Human Genetic Research, and Stanley Center for Psychiatric Research. Data used in preparation of this article were also obtained from the ADNI database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf. This research was also funded in part by NIH Grants R01NS083534, R01NS070963, R41AG052246, 1K25EB013649-01, and 1R21AG050122-01A1 (to M.R.S.); K01MH099232 (to A.J.H.); K24MH094614 and R01MH101486 (to J.W.S.); and an MGH ECOR Tosteson Postdoctoral Fellowship Award (to T.G.). J.W.S. is a Tepper Family MGH Research Scholar.
Footnotes
Conflict of interest statement: B.F. has a financial interest in CorticoMetrics, a company whose medical pursuits focus on brain imaging and measurement technologies. His interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
Data used in the preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu/). As such, the investigators within the ADNI contributed to the design and implementation of ADNI or provided data but did not participate in analysis or writing of this report. (A complete listing of ADNI investigators is available at https://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.)
This article is a PNAS Direct Submission. M.D. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604378113/-/DCSupplemental.
Contributor Information
Collaborators: Michael W. Weiner, Paul Aisen, Michael Weiner, Paul Aisen, Ronald Petersen, Clifford R. Jack, Jr., William Jagust, John Q. Trojanowki, Arthur W. Toga, Laurel Beckett, Robert C. Green, Andrew J. Saykin, John Morris, Leslie M. Shaw, Zaven Khachaturian, Greg Sorensen, Maria Carrillo, Lew Kuller, Marc Raichle, Steven Paul, Peter Davies, Howard Fillit, Franz Hefti, David Holtzman, M. Marcel Mesulam, William Potter, Peter Snyder, Adam Schwartz, Robert C. Green, Tom Montine, Ronald Petersen, Paul Aisen, Ronald G. Thomas, Michael Donohue, Sarah Walter, Devon Gessert, Tamie Sather, Gus Jiminez, Archana B. Balasubramanian, Jennifer Mason, Iris Sim, Laurel Beckett, Danielle Harvey, Michael Donohue, Clifford R. Jack, Jr., Matthew Bernstein, Nick Fox, Paul Thompson, Norbert Schuff, Charles DeCArli, Bret Borowski, Jeff Gunter, Matt Senjem, Prashanthi Vemuri, David Jones, Kejal Kantarci, Chad Ward, William Jagust, Robert A. Koeppe, Norm Foster, Eric M. Reiman, Kewei Chen, Chet Mathis, Susan Landau, John C. Morris, Nigel J. Cairns, Erin Franklin, Lisa Taylor-Reinwald, Leslie M. Shaw, John Q. Trojanowki, Virginia Lee, Magdalena Korecka, Michal Figurski, Arthur W. Toga, Karen Crawford, Scott Neu, Andrew J. Saykin, Tatiana M. Foroud, Steven Potkin, Li Shen, Kelley Faber, Sungeun Kim, Kwangsik Nho, Michael W. Weiner, Lean Thal, Leon Thal, Neil Buckholtz, Michael W. Weiner, Peter J. Snyder, William Potter, Steven Paul, Marilyn Albert, Richard Frank, Zaven Khachaturian, John Hsiao, Jeffrey Kaye, Joseph Quinn, Lisa Silbert, Betty Lind, Raina Carter, Sara Dolen, Lon S. Schneider, Sonia Pawluczyk, Mauricio Becerra, Liberty Teodoro, Bryan M. Spann, James Brewer, Helen Vanderswag, Adam Fleisher, Judith L. Heidebrink, Joanne L. Lord, Ronald Petersen, Sara S. Mason, Colleen S. Albers, David Knopman, Kris Johnson, Rachelle S. Doody, Javier Villanueva-Meyer, Valory Pavlik, Victoria Shibley, Munir Chowdhury, Susan Rountree, Mimi Dang, Yaakov Stern, Lawrence S. Honig, Karen L. Bell, Beau Ances, John C. Morris, Maria Carroll, Mary L. Creech, Erin Franklin, Mark A. Mintun, Stacy Schneider, Angela Oliver, Daniel Marson, David Geldmacher, Marissa Natelson Love, Randall Griffith, David Clark, John Brockington, Erik Roberson, Hillel Grossman, Effie Mitsis, Raj C. Shah, Leyla deToledo-Morrell, Ranjan Duara, Maria T. Greig-Custo, Warren Barker, Marilyn Albert, Chiadi Onyike, Daniel D’Agostino, II, Stephanie Kielb, Martin Sadowski, Mohammed O. Sheikh, Anaztasia Ulysse, Mrunalini Gaikwad, P. Murali Doraiswamy, Jeffrey R. Petrella, Salvador Borges-Neto, Terence Z. Wong, Edward Coleman, Steven E. Arnold, Jason H. Karlawish, David A. Wolk, Christopher M. Clark, Charles D. Smith, Greg Jicha, Peter Hardy, Partha Sinha, Elizabeth Oates, Gary Conrad, Oscar L. Lopez, MaryAnn Oakley, Donna M. Simpson, Anton P. Porsteinsson, Bonnie S. Goldstein, Kim Martin, Kelly M. Makino, M. Saleem Ismail, Connie Brand, Steven G. Potkin, Adrian Preda, Dana Nguyen, Kyle Womack, Dana Mathews, Mary Quiceno, Allan I. Levey, James J. Lah, Janet S. Cellar, Jeffrey M. Burns, Russell H. Swerdlow, William M. Brooks, Liana Apostolova, Kathleen Tingus, Ellen Woo, Daniel H.S. Silverman, Po H. Lu, George Bartzokis, Neill R Graff-Radford, Francine Parfitt, Kim Poki-Walker, Martin R. Farlow, Ann Marie Hake, Brandy R. Matthews, Jared R. Brosch, Scott Herring, Christopher H. van Dyck, Richard E. Carson, Martha G. MacAvoy, Pradeep Varma, Howard Chertkow, Howard Bergman, Chris Hosein, Sandra Black, Bojana Stefanovic, Curtis Caldwell, Ging-Yuek Robin Hsiung, Benita Mudge, Vesna Sossi, Howard Feldman, Michele Assaly, Elizabeth Finger, Stephen Pasternack, Irina Rachisky, Dick Trost, Andrew Kertesz, Charles Bernick, Donna Munic, Marek-Marsel Mesulam, Emily Rogalski, Kristine Lipowski, Sandra Weintraub, Borna Bonakdarpour, Diana Kerwin, Chuang-Kuo Wu, Nancy Johnson, Carl Sadowsky, Teresa Villena, Raymond Scott Turner, Kathleen Johnson, Brigid Reynolds, Reisa A. Sperling, Keith A. Johnson, Gad Marshall, Jerome Yesavage, Joy L. Taylor, Barton Lane, Allyson Rosen, Jared Tinklenberg, Marwan N. Sabbagh, Christine M. Belden, Sandra A. Jacobson, Sherye A. Sirrel, Neil Kowall, Ronald Killiany, Andrew E. Budson, Alexander Norbash, Patricia Lynn Johnson, Thomas O. Obisesan, Saba Wolday, Joanne Allard, Alan Lerner, Paula Ogrocki, Curtis Tatsuoka, Parianne Fatica, Evan Fletcher, Pauline Maillard, John Olichney, Charles DeCarli, Owen Carmichael, Smita Kittur, Michael Borrie, T-Y Lee, Dr Rob Bartha, Sterling Johnson, Sanjay Asthana, Cynthia M. Carlsson, Steven G. Potkin, Adrian Preda, Dana Nguyen, Pierre Tariot, Anna Burke, Ann Marie Milliken, Nadira Trncic, Adam Fleisher, Stephanie Reeder, Vernice Bates, Horacio Capote, Michelle Rainka, Douglas W. Scharre, Maria Kataki, Brendan Kelley, Earl A. Zimmerman, Dzintra Celmins, Alice D. Brown, Godfrey D. Pearlson, Karen Blank, Karen Anderson, Laura A. Flashman, Marc Seltzer, Mary L. Hynes, Robert B. Santulli, Kaycee M. Sink, Leslie Gordineer, Jeff D. Williamson, Pradeep Garg, Franklin Watkins, Brian R. Ott, Geoffrey Tremont, Lori A. Daiello, Stephen Salloway, Paul Malloy, Stephen Correia, Howard J. Rosen, Bruce L. Miller, David Perry, Jacobo Mintzer, Kenneth Spicer, David Bachman, Elizabeth Finger, Stephen Pasternak, Irina Rachinsky, John Rogers, Andrew Kertesz, Dick Drost, Nunzio Pomara, Raymundo Hernando, Antero Sarrael, Susan K. Schultz, Karen Ekstam Smith, Hristina Koleva, Ki Won Nam, Hyungsub Shim, Norman Relkin, Gloria Chiang, Michael Lin, Lisa Ravdin, Amanda Smith, Balebail Ashok Raj, Kristin Fargher, Michael W. Weiner, Paul Aisen, Michael Weiner, Paul Aisen, Ronald Petersen, Robert C. Green, Danielle Harvey, Clifford R. Jack, Jr., William Jagust, John C. Morris, Andrew J. Saykin, Leslie M. Shaw, Arthur W. Toga, John Q. Trojanowki, Thomas Neylan, Jordan Grafman, Robert C. Green, Tom Montine, Michael Weiner, Ronald Petersen, Paul Aisen, Ronald G. Thomas, Michael Donohue, Devon Gessert, Tamie Sather, Melissa Davis, Rosemary Morrison, Gus Jiminez, Thomas Neylan, Jacqueline Hayes, Shannon Finley, Danielle Harvey, Michael Donohue, Clifford R. Jack, Jr., Matthew Bernstein, Bret Borowski, Jeff Gunter, Matt Senjem, Kejal Kantarci, Chad Ward, William Jagust, Robert A. Koeppe, Norm Foster, Eric M. Reiman, Kewei Chen, Susan Landau, John C. Morris, Nigel J. Cairns, Erin Householder, Leslie M. Shaw, John Q. Trojanowki, Virginia Lee, Magdalena Korecka, Michal Figurski, Arthur W. Toga, Karen Crawford, Scott Neu, Andrew J. Saykin, Tatiana M. Foroud, Steven Potkin, Li Shen, Kelley Faber, Sungeun Kim, Kwangsik Nho, Michael W. Weiner, Karl Friedl, Lon S. Schneider, Sonia Pawluczyk, Mauricio Becerra, James Brewer, Helen Vanderswag, Yaakov Stern, Lawrence S. Honig, Karen L. Bell, Debra Fleischman, Konstantinos Arfanakis, Raj C. Shah, Ranjan Duara, Daniel Varon, Maria T Greig, P. Murali Doraiswamy, Jeffrey R. Petrella, Olga James, Anton P. Porsteinsson, Bonnie Goldstein, Kimberly S. Martin, Steven G. Potkin, Adrian Preda, Dana Nguyen, Jacobo Mintzer, Dino Massoglia, Olga Brawman-Mintzer, Carl Sadowsky, Walter Martinez, Teresa Villena, William Jagust, Susan Landau, Howard Rosen, David Perry, Raymond Scott Turner, Kelly Behan, Brigid Reynolds, Reisa A. Sperling, Keith A. Johnson, Gad Marshall, Marwan N. Sabbagh, Sandra A. Jacobson, Sherye A. Sirrel, Thomas O. Obisesan, Saba Wolday, Joanne Allard, Sterling C. Johnson, J. Jay Fruehling, Sandra Harding, Elaine R. Peskind, Eric C. Petrie, Gail Li, Jerome A. Yesavage, Joy L. Taylor, Ansgar J. Furst, Steven Chao, Norman Relkin, Gloria Chiang, and Lisa Ravdin
References
- 1.Smith CD, Chebrolu H, Wekstein DR, Schmitt FA, Markesbery WR. Age and gender effects on human brain anatomy: A voxel-based morphometric study in healthy elderly. Neurobiol Aging. 2007;28(7):1075–1087. doi: 10.1016/j.neurobiolaging.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 2.Draganski B, et al. Temporal and spatial dynamics of brain structure changes during extensive learning. J Neurosci. 2006;26(23):6314–6317. doi: 10.1523/JNEUROSCI.4628-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson PM, et al. Genetic influences on brain structure. Nat Neurosci. 2001;4(12):1253–1258. doi: 10.1038/nn758. [DOI] [PubMed] [Google Scholar]
- 4.Thompson PM, Ge T, Glahn DC, Jahanshad N, Nichols TE. Genetics of the connectome. Neuroimage. 2013;80:475–488. doi: 10.1016/j.neuroimage.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poldrack RA. Region of interest analysis for fMRI. Soc Cogn Affect Neurosci. 2007;2(1):67–70. doi: 10.1093/scan/nsm006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Penny WD, Friston KJ, Ashburner JT, Kiebel SJ, Nichols TE, editors. Statistical Parametric Mapping: The Analysis of Functional Brain Images. Academic; New York: 2011. [Google Scholar]
- 7.Ashburner J, Friston KJ. Voxel-based morphometry--the methods. Neuroimage. 2000;11(6 Pt 1):805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 8.Salat DH, et al. Thinning of the cerebral cortex in aging. Cereb Cortex. 2004;14(7):721–730. doi: 10.1093/cercor/bhh032. [DOI] [PubMed] [Google Scholar]
- 9.Avants BB, Cook PA, Ungar L, Gee JC, Grossman M. Dementia induces correlated reductions in white matter integrity and cortical thickness: A multivariate neuroimaging study with sparse canonical correlation analysis. Neuroimage. 2010;50(3):1004–1016. doi: 10.1016/j.neuroimage.2010.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McIntosh AR, Lobaugh NJ. Partial least squares analysis of neuroimaging data: Applications and advances. Neuroimage. 2004;23(Suppl 1):S250–S263. doi: 10.1016/j.neuroimage.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 11.Friston K, et al. Bayesian decoding of brain images. Neuroimage. 2008;39(1):181–205. doi: 10.1016/j.neuroimage.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 12.Pereira F, Mitchell T, Botvinick M. Machine learning classifiers and fMRI: A tutorial overview. Neuroimage. 2009;45(1) Suppl:S199–S209. doi: 10.1016/j.neuroimage.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabuncu MR, Konukoglu E. Alzheimer’s Disease Neuroimaging Initiative Clinical prediction from structural brain MRI scans: A large-scale empirical study. Neuroinformatics. 2015;13(1):31–46. doi: 10.1007/s12021-014-9238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haufe S, et al. On the interpretation of weight vectors of linear models in multivariate neuroimaging. Neuroimage. 2014;87:96–110. doi: 10.1016/j.neuroimage.2013.10.067. [DOI] [PubMed] [Google Scholar]
- 15.Wray N, Visscher P. Estimating trait heritability. Nature Education. 2008;1(1):29. [Google Scholar]
- 16.Visscher PM, Hill WG, Wray NR. Heritability in the genomics era--Concepts and misconceptions. Nat Rev Genet. 2008;9(4):255–266. doi: 10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- 17.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42(7):565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ge T, et al. Massively expedited genome-wide heritability analysis (MEGHA) Proc Natl Acad Sci USA. 2015;112(8):2479–2484. doi: 10.1073/pnas.1415603112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischl B. FreeSurfer. Neuroimage. 2012;62(2):774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patterson HD, Thompson R. Recovery of inter-block information when block sizes are unequal. Biometrika. 1971;58(3):545–554. [Google Scholar]
- 22.Harville DA. Maximum likelihood approaches to variance component estimation and to related problems. J Am Stat Assoc. 1977;72(358):320–338. [Google Scholar]
- 23.Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Vol 1 Sinauer; Sunderland, MA: 1998. [Google Scholar]
- 24.Dempster ER, Lerner IM. Heritability of threshold characters. Genetics. 1950;35(2):212–236. doi: 10.1093/genetics/35.2.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88(3):294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holmes AJ, et al. Brain Genomics Superstruct Project initial data release with structural, functional, and behavioral measures. Sci Data. 2015;2:150031. doi: 10.1038/sdata.2015.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Essen DC, et al. WU-Minn HCP Consortium The WU-Minn human connectome project: An overview. Neuroimage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jack CR, Jr, et al. The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27(4):685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milham MP, Fair D, Mennes M, Mostofsky SH. HD-200 Consortium The ADHD-200 consortium: A model to advance the translational potential of neuroimaging in clinical neuroscience. Front Syst Neurosci. 2012;6:62. doi: 10.3389/fnsys.2012.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcus DS, et al. Open Access Series of Imaging Studies (OASIS): Cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. J Cogn Neurosci. 2007;19(9):1498–1507. doi: 10.1162/jocn.2007.19.9.1498. [DOI] [PubMed] [Google Scholar]
- 31. The Mind Research Network (2011) COBRE. Available at fcon_1000.projects.nitrc.org/indi/retro/cobre.html. Accessed November 16, 2012.
- 32.Gollub RL, et al. The MCIC collection: A shared repository of multi-modal, multi-site brain image data from a clinical investigation of schizophrenia. Neuroinformatics. 2013;11(3):367–388. doi: 10.1007/s12021-013-9184-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Martino A, et al. The autism brain imaging data exchange: Towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol Psychiatry. 2014;19(6):659–667. doi: 10.1038/mp.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marek K, et al. Parkinson Progression Marker Initiative The Parkinson progression marker initiative (PPMI) Prog Neurobiol. 2011;95(4):629–635. doi: 10.1016/j.pneurobio.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alzheimer’s Association 2014 Alzheimer’s disease facts and figures. Alzheimers Dement. 2014;10(2):e47–e92. doi: 10.1016/j.jalz.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 36.Hebert LE, Scherr PA, Bienias JL, Bennett DA, Evans DA. Alzheimer disease in the US population: Prevalence estimates using the 2000 census. Arch Neurol. 2003;60(8):1119–1122. doi: 10.1001/archneur.60.8.1119. [DOI] [PubMed] [Google Scholar]
- 37.Polanczyk G, de Lima MS, Horta BL, Biederman J, Rohde LA. The worldwide prevalence of ADHD: A systematic review and metaregression analysis. Am J Psychiatry. 2007;164(6):942–948. doi: 10.1176/ajp.2007.164.6.942. [DOI] [PubMed] [Google Scholar]
- 38.Lee SH, et al. Schizophrenia Psychiatric Genome-Wide Association Study Consortium (PGC-SCZ); International Schizophrenia Consortium (ISC); Molecular Genetics of Schizophrenia Collaboration (MGS) Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44(3):247–250. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simonoff E, et al. Psychiatric disorders in children with autism spectrum disorders: Prevalence, comorbidity, and associated factors in a population-derived sample. J Am Acad Child Adolesc Psychiatry. 2008;47(8):921–929. doi: 10.1097/CHI.0b013e318179964f. [DOI] [PubMed] [Google Scholar]
- 40.Keller MF, et al. International Parkinson’s Disease Genomics Consortium (IPDGC); Wellcome Trust Case Control Consortium 2 (WTCCC2) Using genome-wide complex trait analysis to quantify ‘missing heritability’ in Parkinson’s disease. Hum Mol Genet. 2012;21(22):4996–5009. doi: 10.1093/hmg/dds335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee SH, et al. Cross-Disorder Group of the Psychiatric Genomics Consortium; International Inflammatory Bowel Disease Genetics Consortium (IIBDGC) Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45(9):984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Efron B, Tibshirani RJ. An Introduction to the Bootstrap. CRC; Boca Raton, FL: 1994. [Google Scholar]
- 43.Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. Neurodegenerative diseases target large-scale human brain networks. Neuron. 2009;62(1):42–52. doi: 10.1016/j.neuron.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huttenlocher PR. 1994. Synaptogenesis, synapse elimination, and neural plasticity in human cerebral cortex. Threats to Optimal Development: Integrating Biological, Psychological, and Social Risk Factors, ed Nelson CA (Erlbaum, Hillsdale, NJ), Vol 27, pp 35–54.
- 45.Lewis DA, Levitt P. Schizophrenia as a disorder of neurodevelopment. Annu Rev Neurosci. 2002;25(1):409–432. doi: 10.1146/annurev.neuro.25.112701.142754. [DOI] [PubMed] [Google Scholar]
- 46.Vul E, Harris C, Winkielman P, Pashler H. Puzzlingly high correlations in fMRI studies of emotion, personality, and social cognition. Perspect Psychol Sci. 2009;4(3):274–290. doi: 10.1111/j.1745-6924.2009.01125.x. [DOI] [PubMed] [Google Scholar]
- 47.Kriegeskorte N, Simmons WK, Bellgowan PS, Baker CI. Circular analysis in systems neuroscience: The dangers of double dipping. Nat Neurosci. 2009;12(5):535–540. doi: 10.1038/nn.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bernal-Rusiel JL, Greve DN, Reuter M, Fischl B, Sabuncu MR. Alzheimer’s Disease Neuroimaging Initiative Statistical analysis of longitudinal neuroimage data with Linear Mixed Effects models. Neuroimage. 2013;66:249–260. doi: 10.1016/j.neuroimage.2012.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bernal-Rusiel JL, Reuter M, Greve DN, Fischl B, Sabuncu MR. Alzheimer’s Disease Neuroimaging Initiative Spatiotemporal linear mixed effects modeling for the mass-univariate analysis of longitudinal neuroimage data. Neuroimage. 2013;81:358–370. doi: 10.1016/j.neuroimage.2013.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28(19):2540–2542. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fischl B, et al. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 52.Desikan RS, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31(3):968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 53.Liu D, Lin X, Ghosh D. Semiparametric regression of multidimensional genetic pathway data: Least-squares kernel machines and linear mixed models. Biometrics. 2007;63(4):1079–1088. doi: 10.1111/j.1541-0420.2007.00799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Molenberghs G, Verbeke G. Likelihood ratio, score, and Wald tests in a constrained parameter space. Am Stat. 2007;61(1):22–27. [Google Scholar]
- 55.Barch DM, et al. WU-Minn HCP Consortium Function in the human connectome: Task-fMRI and individual differences in behavior. Neuroimage. 2013;80:169–189. doi: 10.1016/j.neuroimage.2013.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]