Significance
Although auxin oxidation has long been known to be the primary mechanism of auxin catabolism and Arabidopsis seedlings have 10–100 more 2-oxindole 3-acetic acid compared with other auxin catabolic products, the enzymes that constitutively catalyze this process remained unknown. This work fills the gap by identifying and characterizing the Arabidopsis proteins DIOXYGENASE FOR AUXIN OXIDATION 1 (DAO1) and DAO2, which catalyze auxin oxidation under normal growth conditions and shows that this activity has a physiological function in planta. The protein localization and phenotypes of the loss/gain of function mutants support that DAO1 is the primary constitutive mechanism of auxin catabolism in Arabidopsis and that the temporal- and tissue-specific oxidative inactivation of auxin by DAO adjusts indole-3-acetic acid levels throughout the life of the plant to optimize growth and development.
Keywords: auxin homeostasis, auxin oxidation, auxin oxidase, lateral roots, flowers
Abstract
Tight homeostatic regulation of the phytohormone auxin [indole-3-acetic acid (IAA)] is essential to plant growth. Auxin biosynthetic pathways and the processes that inactivate auxin by conjugation to amino acids and sugars have been thoroughly characterized. However, the enzyme that catalyzes oxidation of IAA to its primary catabolite 2-oxindole-3-acetic acid (oxIAA) remains uncharacterized. Here, we show that DIOXYGENASE FOR AUXIN OXIDATION 1 (DAO1) catalyzes formation of oxIAA in vitro and in vivo and that this mechanism regulates auxin homeostasis and plant growth. Null dao1-1 mutants contain 95% less oxIAA compared with wild type, and complementation of dao1 restores wild-type oxIAA levels, indicating that DAO1 is the primary IAA oxidase in seedlings. Furthermore, dao1 loss of function plants have altered morphology, including larger cotyledons, increased lateral root density, delayed sepal opening, elongated pistils, and reduced fertility in the primary inflorescence stem. These phenotypes are tightly correlated with DAO1 spatiotemporal expression patterns as shown by DAO1pro:β-glucuronidase (GUS) activity and DAO1pro:YFP-DAO1 signals, and transformation with DAO1pro:YFP-DAO1 complemented the mutant phenotypes. The dominant dao1-2D mutant has increased oxIAA levels and decreased stature with shorter leaves and inflorescence stems, thus supporting DAO1 IAA oxidase function in vivo. A second isoform, DAO2, is very weakly expressed in seedling root apices. Together, these data confirm that IAA oxidation by DAO1 is the principal auxin catabolic process in Arabidopsis and that localized IAA oxidation plays a role in plant morphogenesis.
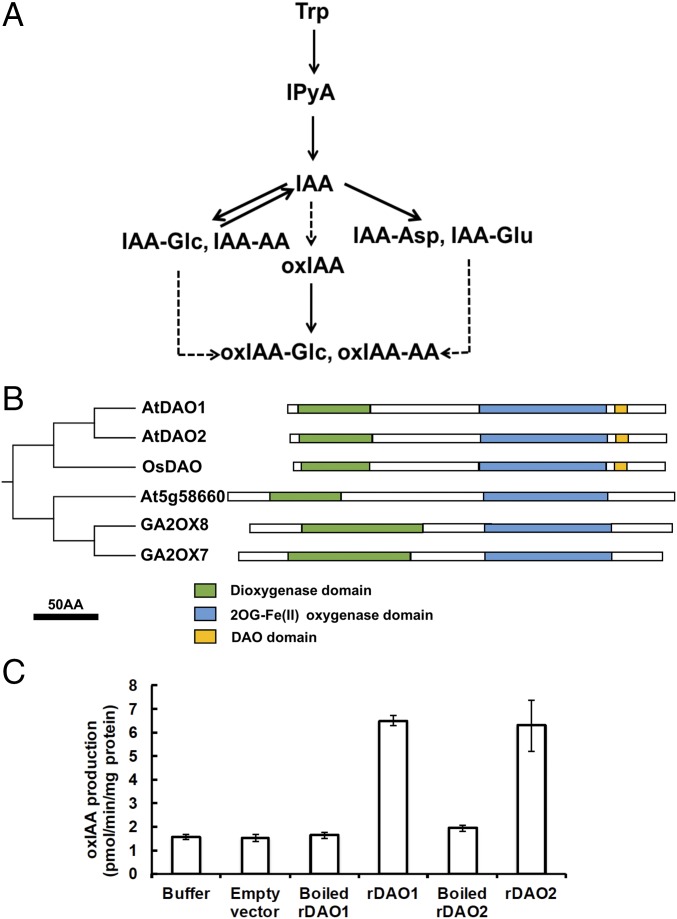
The phytohormone auxin [indole-3-acetic acid (IAA)] regulates plant architecture, tropic growth, and multiple other aspects of plant development. The spatiotemporal distribution of auxin is regulated at the cellular level by control of biosynthesis, transport, reversible conjugation, compartmentalization, and catabolism. Loss of any one of these mechanisms is often balanced by compensatory homeostatic responses but can also produce morphological or physiological changes. The enzymes that catalyze auxin biosynthesis via the indole-3-pyruvic acid (IPyA) pathway (1, 2) and the UDP-glucose transferases (3–5) and amido synthetases that inactivate IAA by conjugation to small molecules (6, 7) are well-characterized (Fig. 1A). Most IAA conjugation is reversible and functions in compartmentation or storage (7–9). Exceptions are the formation of the irreversible catabolites indole-3-acetic acid-aspartic acid (IAA-Asp) and indole-3-acetic acid-glutamic acid (IAA-Glu) in discrete plant tissues (10–12).
Fig. 1.
Identification and characterization of 2-oxindole acetic acid synthetases in A. thaliana. (A) Simplified auxin metabolic pathway via IPyA. IAA-AA, indole-3-acetic acid-amino acid conjugate; IAA-Glc, indole-3-acetic acid-glucose; oxIAA-AA, 2-oxindole-3-acetic acid-amino acid conjugate; oxIAA-Glc, 2-oxindole-3-acetic acid-glucose; Trp, tryptophan. (B) Pictogram of conserved domains among OsDAO, AtDAO1, AtDAO2, and the nearest clade of a subfamily of 2-oxoglutarate oxygenases in Arabidopsis. (C) Enzyme assays of rDAO (rDAO1 and rDAO2) with buffer control, empty vector control, and heat-inactivated (boiled) enzyme control with IAA as a substrate. Data are means ± SD of at least independent 10 experiments.
Oxidation of IAA has long been known to be the primary mechanism of auxin catabolism (13). Arabidopsis seedlings contain 10–100 times more 2-oxindole-3-acetic acid (oxIAA) than the major IAA conjugates IAA-Glu and IAA-Asp (10, 14, 15), but the enzymes catalyzing oxIAA formation have been elusive. In part, this has been because of early implication of peroxidases and oxygenases in auxin oxidative processes (13, 16–18). However, reexamination by liquid chromatography-mass spectrometry (LC-MS) of the reaction products from previously published horseradish and grape leaf peroxidases as well as two Arabidopsis peroxidases showed that these reactions produce the decarboxylation products indole-3-carbinol, 3-methylene-2-oxindole, and 3-hydroxymethyl-2-oxindole present in low levels in plants (19) but not oxIAA (20). In Zea mays, oxIAA was reported to be produced by an oxygen-requiring soluble enzyme and a more active detergent-soluble, stable component (17). A correlation of cellular oxIAA and IAA levels (15, 20) and with expression of a gene with similarity to a rice gene named DIOXYGENASE FOR AUXIN OXIDATION (DAO) (21, 22) and sequence homology to apple ADVENTITIOUS ROOTING RELATED OXYGENASE 1 (23) suggested that similar genes in Arabidopsis, DAO1 and DAO2, encode the enzymes that produce oxIAA (24, 25). Here, we show that DAO1 and DAO2 catalyze oxIAA formation and that DAO1 is the major IAA oxidase in Arabidopsis. The temporal- and tissue-specific oxidative inactivation of auxin by DAO fine tunes IAA levels throughout the life of the plant to maintain optimal growth and development.
Phylogenetic Analysis of Arabidopsis 2-Oxoglutarate and Fe(II)-Dependent Oxygenase Superfamily
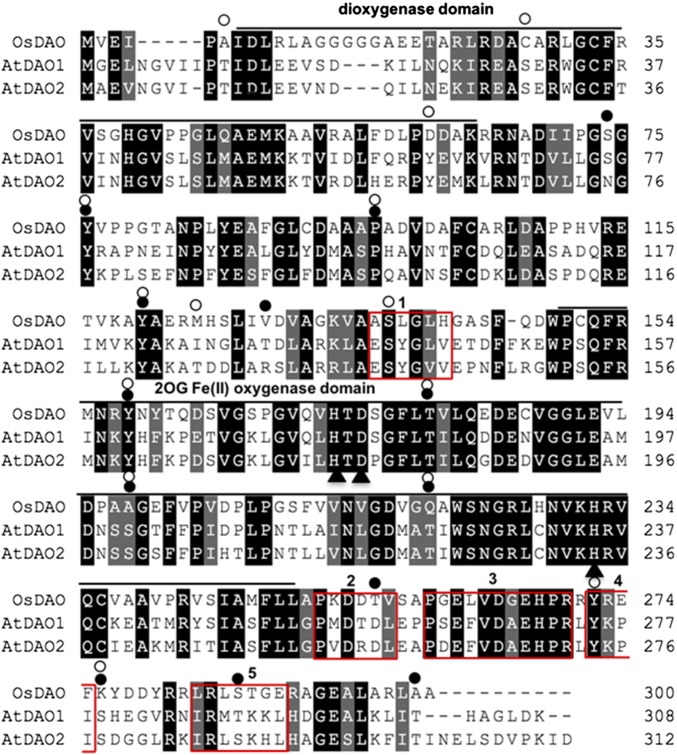
Previous work from our laboratory suggested that oxIAA formation was catalyzed by a member of the 2-oxoglutarate and Fe(II)-dependent [2OG Fe(II)] oxygenase superfamily (20). AtDAO1 (At1g14130) and AtDAO2 (At1g14120) in Arabidopsis thaliana were identified as homologs of rice OsDAO (Os04g0475600) (Fig. S1); all members of this superfamily are characterized by the conserved dioxygenase and 2OG Fe(II) oxygenase domains (Fig. 1B and Fig. S1). Another conserved sequence P(S/D/G)E(F/L)VD(A/G)EHPR without any known motifs was identified among the rice and Arabidopsis DAO proteins and examined further (Fig. 1B and Fig. S1). Sequence alignment with Viridiplantae (taxid: 33090) suggests that this motif is specific to proteins in a subset of the 2OG Fe(II) oxygenase superfamily annotated as DAO and DAO like (Table S1). Motif analysis (26, 27) also showed μ-Adaptor Protein and actin binding motifs (Fig. S1), 12 putative serine/threonine phosphorylation sites in DAO1, and 15 sites in DAO2.
Fig. S1.
Protein sequence alignment of AtDAO1, AtDAO2, and OsDAO with Clustalx2.1. Dark gray shading indicates conserved residues, light gray shading indicates residues with similar properties, - indicates a gap inserted to maximize the alignment. Bars, oxygenase or dioxygenase domains; black circles, putative phosphorylation sites in AtDAO2; triangles, Fe binding residues; white circles, putative phosphorylation sites in AtDAO1. Red boxes numbered 1–5 indicate putative motifs: 1, μ-Adaptor Protein motif; 2, CK2 phosphorylation site; 3, DAO motif; 4, μ-Adaptor Protein motif; and 5, actin cleft binding motif. AtDAO1 (DAO1) has 71.7% similarity and 45% identity and AtDAO2 (DAO2) has 71.9% similarity and 43% identity to OsDAO, and DAO1 and DAO2 show 72% identity to each other.
Table S1.
Viridiplantae (taxid: 33090) BLAST analyses of a degenerate DAO motif PSELVDAEHPR
| GenInfo Identifier (GI) no. | Sequence | % Identity |
| >gi|460374964|ref|XP_004233278.1|:259–269 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Solanum lycopersicum]* | PSELVDAEHPR | 100 |
| >gi|674243627|gb|KFK36392.1|:216–226 hypothetical protein AALP_AA4G118300 [Arabis alpina] | PSELVDADHPR | 90.909 |
| >gi|661890588|emb|CDP06085.1|:258–268 unnamed protein product [Coffea canephora] | PSELVDSEHPR | 90.909 |
| >gi|698552242|ref|XP_009769574.1|:214–224 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like isoform X2 [Nicotiana sylvestris]* | PPELVDAEHPR | 90.909 |
| >gi|723676585|ref|XP_004233915.2|:214–224 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Solanum lycopersicum]* | PPELVDAEHPR | 90.909 |
| >gi|698552236|ref|XP_009769572.1|:259–269 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like isoform X1 [Nicotiana sylvestris]* | PPELVDAEHPR | 90.909 |
| >gi|697104708|ref|XP_009606157.1|:259–269 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like isoform X2 [Nicotiana tomentosiformis]* | PPELVDAEHPR | 90.909 |
| >gi|565398913|ref|XP_006365008.1|:259–269 PREDICTED: probable 2-oxoglutarate–dependent dioxygenase AOP1-like [Solanum tuberosum] | PPELVDAEHPR | 90.909 |
| >gi|565398911|ref|XP_006365007.1|:259–269 PREDICTED: probable 2-oxoglutarate–dependent dioxygenase AOP1-like [Solanum tuberosum] | PPELVDAEHPR | 90.909 |
| >gi|697104706|ref|XP_009606156.1|:266–276 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like isoform X1 [Nicotiana tomentosiformis]* | PPELVDAEHPR | 90.909 |
| >gi|674907065|emb|CDY26052.1|:24–34 BnaC06g05720D [Brassica napus] | PEELVDAEHPR | 90.909 |
| >gi|703112634|ref|XP_010100168.1|:264–274 Gibberellin 3-beta-dioxygenase 4 [Morus notabilis] | PSELVDDEHPR | 90.909 |
| >gi|674951431|emb|CDX81613.1|:134–144 BnaC08g39880D [Brassica napus] | PSEFVDAEHPR | 90.909 |
| >gi|922508960|ref|XP_013590817.1|:247–257 PREDICTED: LOW-QUALITY PROTEIN: probable 2-oxoglutarate–dependent dioxygenase AOP1.2 [Brassica oleracea var. oleracea] | PEELVDAEHPR | 90.909 |
| >gi|923832425|ref|XP_013698039.1|:262–272 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Brassica napus]* | PSEFVDAEHPR | 90.909 |
| >gi|923818163|ref|XP_013693229.1|:262–272 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Brassica napus]* | PSEFVDAEHPR | 90.909 |
| >gi|685316161|ref|XP_009148694.1|:262–272 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Brassica rapa]* | PSEFVDAEHPR | 90.909 |
| >gi|674883357|emb|CDY49110.1|:262–272 BnaA06g08960D [Brassica napus] | PSEFVDAEHPR | 90.909 |
| >gi|923874821|ref|XP_013710739.1|:263–273 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Brassica napus]* | PSEFVDAEHPR | 90.909 |
| >gi|923825585|ref|XP_013695754.1|:263–273 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Brassica napus]* | PSEFVDAEHPR | 90.909 |
| >gi|923763987|ref|XP_013677797.1|:262–272 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Brassica napus]* | PSEFVDAEHPR | 90.909 |
| >gi|923719998|ref|XP_013664686.1|:263–273 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Brassica napus]* | PSEFVDAEHPR | 90.909 |
| >gi|923693597|ref|XP_013657373.1|:262–272 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Brassica napus]* | PSEFVDAEHPR | 90.909 |
| >gi|922545847|ref|XP_013600713.1|:263–273 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Brassica oleracea var. oleracea]* | PSEFVDAEHPR | 90.909 |
| >gi|922491820|ref|XP_013585242.1|:263–273 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Brassica oleracea var. oleracea]* | PSEFVDAEHPR | 90.909 |
| >gi|685350016|ref|XP_009110557.1|:262–272 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Brassica rapa]* | PSEFVDAEHPR | 90.909 |
| >gi|685316165|ref|XP_009148696.1|:263–273 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Brassica rapa]* | PSEFVDAEHPR | 90.909 |
| >gi|923915368|ref|XP_013727221.1|:263–273 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Brassica napus]* | PSEFVDAEHPR | 90.909 |
| >gi|674883356|emb|CDY49109.1|:263–273 BnaA06g08970D [Brassica napus] | PSEFVDAEHPR | 90.909 |
| >gi|702482924|ref|XP_010033632.1|:260–270 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Eucalyptus grandis]* | PEELVDAEHPR | 90.909 |
| >gi|674899181|emb|CDY33774.1|:264–274 BnaA09g45800D [Brassica napus] | PSEFVDAEHPR | 90.909 |
| >gi|727432894|ref|XP_010495771.1|:262–272 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Camelina sativa]* | PSEFVDAEHPR | 90.909 |
| >gi|565492802|ref|XP_006304040.1|:262–272 hypothetical protein CARUB_v10009839mg [Capsella rubella] | PSEFVDAEHPR | 90.909 |
| >gi|297849816|ref|XP_002892789.1|:262–272 hypothetical protein ARALYDRAFT_471571 [Arabidopsis lyrata subsp. lyrata] | PSEFVDAEHPR | 90.909 |
| >gi|15223096|ref|NP_172865.1|:263–273 2OG Fe(II) oxygenase-like protein [A. thaliana] | PSEFVDAEHPR | 90.909 |
| >gi|923874876|ref|XP_013710756.1|:280–290 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Brassica napus]* | PSEFVDAEHPR | 90.909 |
| >gi|923755136|ref|XP_013675537.1|:280–290 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Brassica napus]* | PSEFVDAEHPR | 90.909 |
| >gi|923719992|ref|XP_013664684.1|:280–290 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Brassica napus]* | PSEFVDAEHPR | 90.909 |
| >gi|922545843|ref|XP_013600711.1|:280–290 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Brassica oleracea var. oleracea]* | PSEFVDAEHPR | 90.909 |
| >gi|685367957|ref|XP_009117936.1|:280–290 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Brassica rapa]* | PSEFVDAEHPR | 90.909 |
| >gi|685314120|ref|XP_009147659.1|:279–289 PREDICTED: probable 2-oxoglutarate–dependent dioxygenase AOP1 [Brassica rapa] | PEELVDAEHPR | 90.909 |
| >gi|685316163|ref|XP_009148695.1|:282–292 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Brassica rapa]* | PSEFVDAEHPR | 90.909 |
| >gi|674936830|emb|CDX96865.1|:326–336 BnaA08g24080D [Brassica napus] | PSEFVDAEHPR | 90.909 |
| >gi|674949734|emb|CDX84043.1|:395–405 BnaC08g16020D [Brassica napus] | PSEFVDAEHPR | 90.909 |
| >gi|695051445|ref|XP_009413747.1|:336–346 PREDICTED: gibberellin 20 oxidase 1-D [Musa acuminata subsp. malaccensis] | PAELVDADHPR | 81.818 |
| >gi|296088490|emb|CBI37481.3|:183–193 unnamed protein product [Vitis vinifera] | PPELVDSEHPR | 81.818 |
| >gi|118484910|gb|ABK94321.1|:213–223 unknown [Populus trichocarpa] | PPELVDSEHPR | 81.818 |
| >gi|147781479|emb|CAN69444.1|:256–266 hypothetical protein VITISV_016474 [Vitis vinifera] | PPELVDSEHPR | 81.818 |
| >gi|743920686|ref|XP_011004396.1|:255–265 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Populus euphratica]* | PPELVDSEHPR | 81.818 |
| >gi|224056727|ref|XP_002298993.1|:255–265 adventitious rooting related oxygenase family protein [Populus trichocarpa] | PPELVDSEHPR | 81.818 |
| >gi|566195567|ref|XP_002317640.2|:255–265 adventitious rooting related oxygenase family protein [Populus trichocarpa] | PPELVDSEHPR | 81.818 |
| >gi|225431637|ref|XP_002263124.1|:256–266 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Vitis vinifera]* | PPELVDSEHPR | 81.818 |
| >gi|590711065|ref|XP_007049001.1|:257–267 2OG Fe(II) oxygenase superfamily protein [Theobroma cacao] | PPELVDSEHPR | 81.818 |
| >gi|743887366|ref|XP_011038125.1|:258–268 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Populus euphratica]* | PPELVDSEHPR | 81.818 |
| >gi|460374960|ref|XP_004233276.1|:259–269 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO [Solanum lycopersicum]* | PPELVNAEHPR | 81.818 |
| >gi|694388066|ref|XP_009369758.1|:265–275 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Pyrus x bretschneideri]* | PPELVDSEHPR | 81.818 |
| >gi|694388061|ref|XP_009369756.1|:265–275 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Pyrus x bretschneideri]* | PPELVDSEHPR | 81.818 |
| >gi|694388042|ref|XP_009369750.1|:265–275 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Pyrus x bretschneideri]* | PPELVDSEHPR | 81.818 |
| >gi|658054610|ref|XP_008363059.1|:265–275 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Malus domestica]* | PPELVDSEHPR | 81.818 |
| >gi|657947856|ref|XP_008392065.1|:265–275 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Malus domestica]* | PPELVDSEHPR | 81.818 |
| >gi|695080829|ref|XP_009387900.1|:562–572 PREDICTED: gibberellin 20 oxidase 1-d like [Musa acuminata subsp. malaccensis] | PPELVDADHPR | 81.818 |
| >gi|565495888|ref|XP_006305583.1|:179–189 hypothetical protein CARUB_v10010246mg [Capsella rubella] | PEEMVDAEHPR | 81.818 |
| >gi|125590728|gb|EAZ31078.1|:212–222 hypothetical protein OsJ_15175 [Oryza sativa Japonica Group] | PGELVDGEHPR | 81.818 |
| >gi|641842951|gb|KDO61853.1|:257–267 hypothetical protein CISIN_1g036868mg [Citrus sinensis] | PAEFVDAEHPR | 81.818 |
| >gi|567922574|ref|XP_006453293.1|:257–267 hypothetical protein CICLE_v10009037mg [Citrus clementina] | PAEFVDAEHPR | 81.818 |
| >gi|567922576|ref|XP_006453294.1|:258–268 hypothetical protein CICLE_v10009037mg [Citrus clementina] | PAEFVDAEHPR | 81.818 |
| >gi|115458950|ref|NP_001053075.1|:260–270 Os04g0475600 [Oryza sativa Japonica Group]† | PGELVDGEHPR | 81.818 |
| >gi|922545717|ref|XP_013600645.1|:262–272 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Brassica oleracea var. oleracea]* | PSEFVNAEHPR | 81.818 |
| >gi|567132038|ref|XP_006392866.1|:279–289 hypothetical protein EUTSA_v10011658mg [Eutrema salsugineum] | PEEMVDAEHPR | 81.818 |
| >gi|747076935|ref|XP_011085561.1|:292–302 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Sesamum indicum]* | PPELVDVEHPR | 81.818 |
| >gi|567151883|ref|XP_006417054.1|:262–272 hypothetical protein EUTSA_v10008310mg [Eutrema salsugineum] | PSEFVDAQHPR | 81.818 |
| >gi|674878202|emb|CDY53799.1|:247–257 BnaA06g01840D [Brassica napus] | PEELVDPEHPR | 81.818 |
| >gi|16118978|gb|AAL14687.1|:264–274 2-oxoglutarate–dependent dioxygenase [Arabidopsis halleri] | PEELVDEEHPR | 81.818 |
| >gi|922449065|ref|XP_013627893.1|:255–265 PREDICTED: probable 2-oxoglutarate–dependent dioxygenase AOP1.2 [Brassica oleracea var. oleracea] | PDEIVDAEHPR | 81.818 |
| >gi|727565328|ref|XP_010455020.1|:281–291 PREDICTED: probable 2-oxoglutarate–dependent dioxygenase AOP1.2 [Camelina sativa] | PEELVDEEHPR | 81.818 |
| >gi|727439110|ref|XP_010500908.1|:257–267 PREDICTED: probable 2-oxoglutarate–dependent dioxygenase AOP1 isoform X3 [Camelina sativa] | PEELVDEEHPR | 81.818 |
| >gi|923915384|ref|XP_013727228.1|:262–272 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Brassica napus]* | PSEFVDVEHPR | 81.818 |
| >gi|731339915|ref|XP_010681112.1|:261–271 PREDICTED: 2-oxoglutarate–dependent dioxygenase DAO-like [Beta vulgaris subsp. vulgaris]* | PSEFVDVEHPR | 81.818 |
| >gi|674893502|emb|CDY39340.1|:263–273 BnaC05g10390D [Brassica napus] | PSEFVDVEHPR | 81.818 |
| >gi|674238139|gb|KFK30904.1|:274–284 hypothetical protein AALP_AA6G041200 [Arabis alpina] | PEELVDEEHPR | 81.818 |
| >gi|727439108|ref|XP_010500907.1|:275–285 PREDICTED: probable 2-oxoglutarate–dependent dioxygenase AOP1 isoform X2 [Camelina sativa] | PEELVDEEHPR | 81.818 |
| >gi|685314118|ref|XP_009147658.1|:275–285 PREDICTED: probable 2-oxoglutarate–dependent dioxygenase AOP1 [Brassica rapa] | PDELVDEEHPR | 81.818 |
| >gi|922508768|ref|XP_013590757.1|:275–285 PREDICTED: probable 2-oxoglutarate–dependent dioxygenase AOP1 [Brassica oleracea var. oleracea] | PDELVDEEHPR | 81.818 |
| >gi|15219099|ref|NP_175691.1|:275–285 putative 2-oxoglutarate–dependent dioxygenase [A. thaliana] | PDELVDEEHPR | 81.818 |
| >gi|567132034|ref|XP_006392865.1|:275–285 hypothetical protein EUTSA_v10011667mg [Eutrema salsugineum] | PDELVDEEHPR | 81.818 |
| >gi|565495504|ref|XP_006305391.1|:275–285 hypothetical protein CARUB_v10009783mg [Capsella rubella] | PDELVDEEHPR | 81.818 |
| >gi|727439106|ref|XP_010500906.1|:276–286 PREDICTED: probable 2-oxoglutarate–dependent dioxygenase AOP1 isoform X1 [Camelina sativa] | PEELVDEEHPR | 81.818 |
| >gi|470110454|ref|XP_004291498.1|:273–283 PREDICTED: probable 2-oxoglutarate–dependent dioxygenase AOP1 [Fragaria vesca subsp. vesca] | PKELVDEEHPR | 81.818 |
| >gi|16118889|gb|AAL14645.1|AF417857_1:276–286 AOP1 [Arabidopsis lyrata] | PEELVDEEHPR | 81.818 |
| >gi|674238138|gb|KFK30903.1|:277–287 hypothetical protein AALP_AA6G041100 [Arabis alpina] | PEELVDEEHPR | 81.818 |
| >gi|922563880|ref|XP_013610213.1|:279–289 PREDICTED: probable 2-oxoglutarate–dependent dioxygenase AOP1 [Brassica oleracea var. oleracea] | PEELVDEEHPR | 81.818 |
| >gi|674238136|gb|KFK30901.1|:279–289 hypothetical protein AALP_AA6G040900 [Arabis alpina] | PEELVDEEHPR | 81.818 |
| >gi|923684279|ref|XP_013654615.1|:280–290 PREDICTED: probable 2-oxoglutarate–dependent dioxygenase AOP1 [Brassica napus] | PDEIVDAEHPR | 81.818 |
| >gi|685340431|ref|XP_009107019.1|:280–290 PREDICTED: probable 2-oxoglutarate–dependent dioxygenase AOP1 [Brassica rapa] | PDEIVDAEHPR | 81.818 |
| >gi|674238137|gb|KFK30902.1|:280–290 hypothetical protein AALP_AA6G041000 [Arabis alpina] | PEELVDEEHPR | 81.818 |
| >gi|565463434|ref|XP_006289716.1|:283–293 hypothetical protein CARUB_v10003282mg [Capsella rubella] | PDELVDEEHPR | 81.818 |
| >gi|727427883|ref|XP_010467243.1|:289–299 PREDICTED: probable 2-oxoglutarate–dependent dioxygenase AOP1 [Camelina sativa] | PEELVDEEHPR | 81.818 |
| >gi|923804578|ref|XP_013688552.1|:279–289 PREDICTED: probable 2-oxoglutarate–dependent dioxygenase AOP1 [Brassica napus] | PEELVDEEHPR | 81.818 |
| >gi|116788351|gb|ABK24846.1|:308–318 unknown [Picea sitchensis] | PPELVDNEHPR | 81.818 |
| >gi|646283286|gb|AIB53819.1|:252–262 2-oxoacid–dependent dioxygenase [Paeonia suffruticosa] | PEELVDAENPR | 81.818 |
Sequences in GenBank annotated as DAO or DAO-like.
Rice DAO was identified in the search.
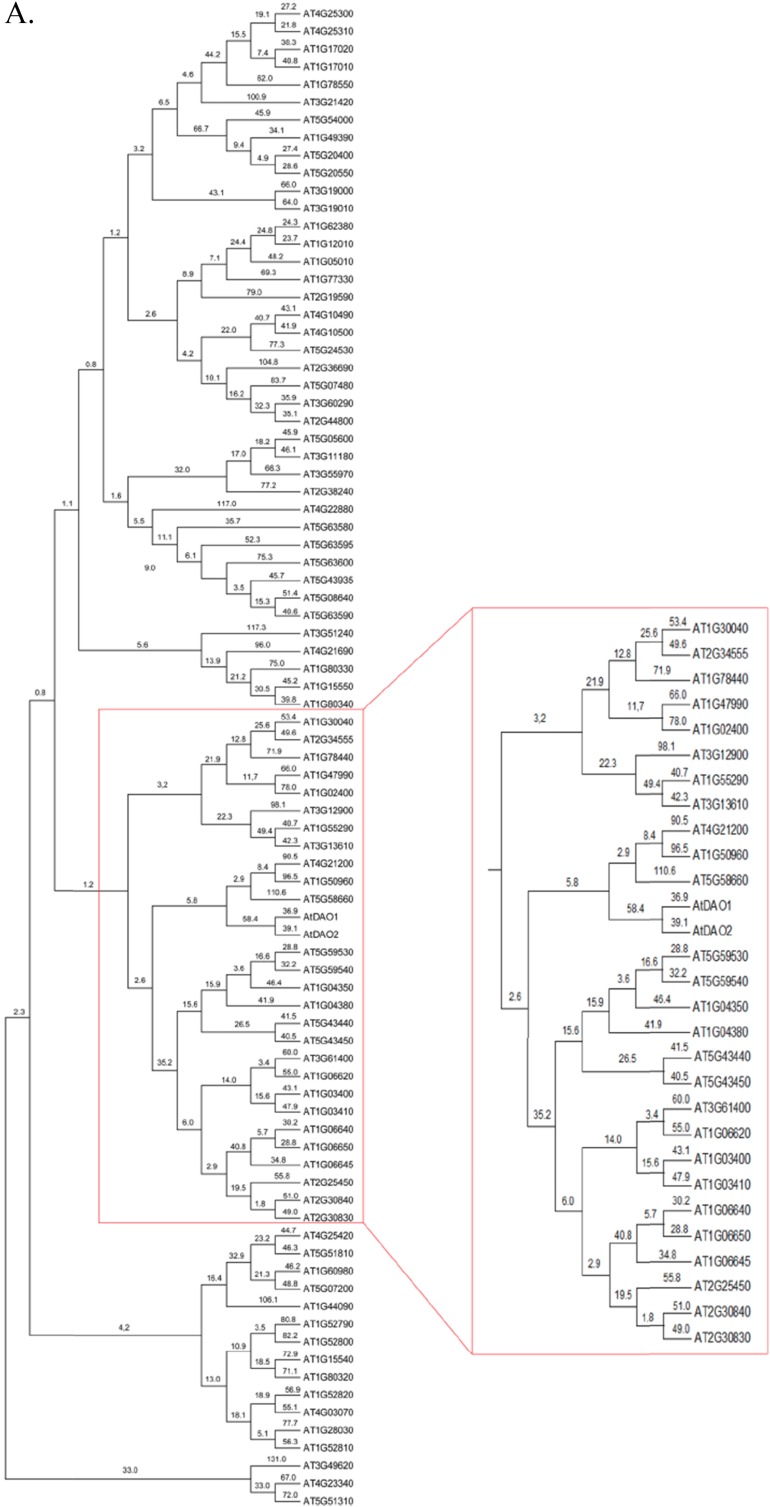
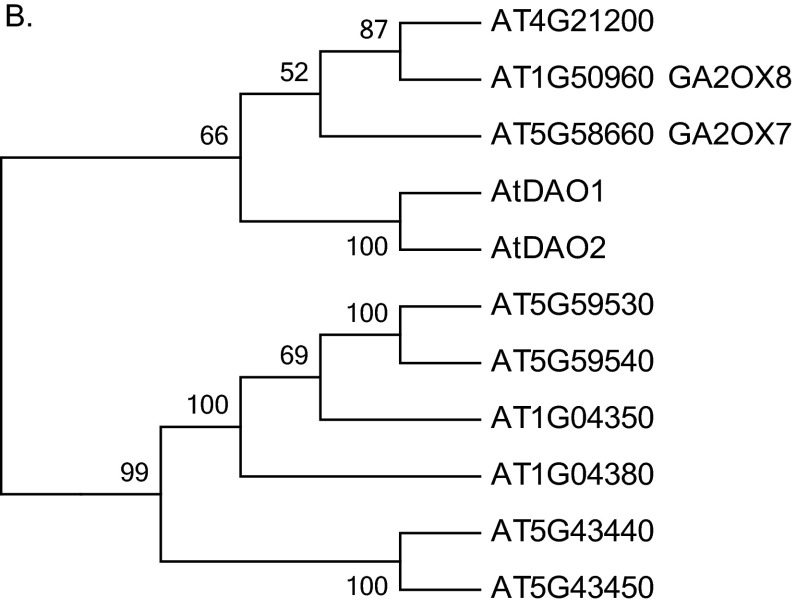
The majority of the proteins in the 2OG Fe(II) oxygenase superfamily have not been characterized, and a phylogenetic analysis of the superfamily in Arabidopsis was undertaken to identify any nearest neighbors to DAO1 and DAO2 (Fig. S2) (24). The nearest neighbors are AT5G58660, an uncharacterized gene, AT1G50960 (GA2OX7), and AT4G21200 (GA2OX8), all of which lack the DAO motif (Fig. 1B). Similarly, Porco et al. (24) report that DAO1/2 are in a separate clade closest to GAOX7/8.
Fig. S2.
Phylogenetic analysis of 2OG Fe(II) oxygenase superfamily in A. thaliana. The evolutionary history was inferred using the neighbor-joining method in MEGA 6.0. The bootstrap consensus tree inferred from 10,000 replicates is taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The evolutionary distances were computed using the number of differences method and are shown in the units. A shows the entire tree, B is a zoom view of A.
Recombinant DAO Can Catalyze the Formation of oxIAA
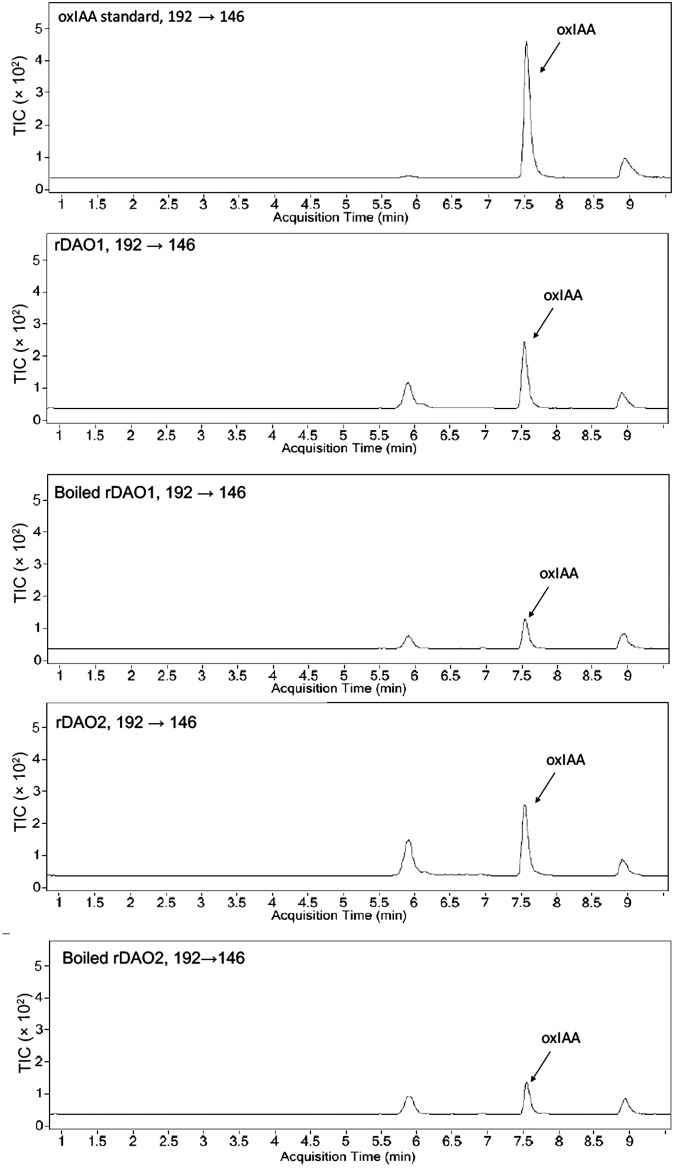
To test the hypothesis that DAO1 and/or DAO2 catalyze the oxidation of IAA into oxIAA, enzyme assays were conducted with recombinant DIOXYGENASE FOR AUXIN OXIDATION (rDAO; 36 kDa) (Fig. S3 and Table S2) and IAA as a substrate. Ultra performance liquid chromatography (UPLC) and LC-MS/MS were used to analyze the products from in vitro enzyme assays, wherein IAA was incubated with purified rDAO1 or rDAO2. In both cases, one major peak with the same retention time and mass transitions as the oxIAA standard was observed (Fig. S4), with low background levels of oxIAA (presumably derived from nonenzymatic conversion of IAA) observed in buffer only, empty vector, and inactivated (boiled) enzyme controls (Fig. 1C). The calculated enzymatic activity of rDAO1 was 6.5 ± 0.2 pmol oxIAA−1 min−1 mg protein, and rDAO2 was 6.3 ± 1.1 pmol oxIAA−1 min−1 mg protein. These rates are consistent with the IAA oxidation rate previously observed in planta, which is between 10 and 40 nM−1 h (28), and indirectly support the hypothesis that DAO1 and DAO2 function in IAA oxidation in planta.
Fig. S3.
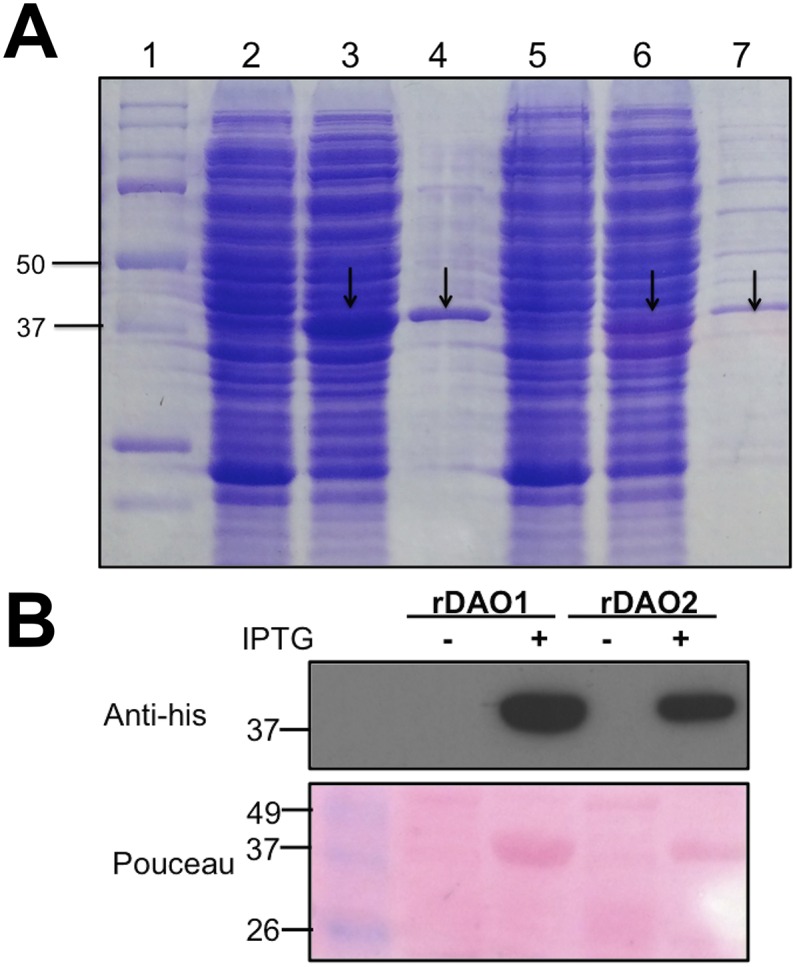
Heterologous expression and purification of rDAO1 and rDAO2 in E. coli. (A) SDS/PAGE analysis of rDAO1 and rDAO2 expression after IPTG induction. Lane 1, protein marker (Bio-Rad); lane 2, cell lysate of noninduced E. coli harboring rDAO1 construct; lane 3, cell lysate of IPTG-induced E. coli harboring rDAO1 construct; lane 4, purified rDAO1 from IPTG-induced E. coli; lane 5, cell lysate of noninduced E. coli harboring rDAO2 construct; lane 6, cell lysate of IPTG-induced E. coli harboring rDAO2 construct; and lane 7, purified rDAO2 from IPTG-induced E. coli. (B, Upper) Western blot analysis of noninduced (−) and purified rDAO from IPTG-induced (+) E. coli harboring either rDAO1 or rDAO2 construct as indicated. (B, Lower) Ponceau staining of the gel.
Table S2.
Primer list
| Primer | Sequence (5′-3′) |
| For heterogeneous expression of recombinant DAO1 and DAO2 | |
| EcoRI-DAO1-F | GAATTCGATGGAAGCTT GTTTGAGATG |
| Xho1 -DAO1-R | CTCGAGGAGTTCTGAAACCAAGATCCG |
| EcoRI-DAO2-F | GAATTCG ATGGCGGAAG TAAATGGAGT |
| Xho1 -DAO2-R | CTCGAG ATCTATCTTTGGGACGTCA |
| For genotyping of T-DNA insertion lines | |
| Salk_093162-F | TTCCCCACGGAATTAAGGTAC |
| Salk_093162-R | CAAGTCCATTGATAGCCTTCG |
| CS816250-F | GGTTTGGAGATAAGGCTCAGG |
| CS816250-R | CGCCTCGTTCTTATTAGGACC |
| Salk_209298-F | TCCGTTTAGTTCCCCCATATC |
| Salk_209298-R | AAAAGTCGTGATGTTTGGCAC |
| Salk_082522-F | GGTCTGAATTGGACAATGACG |
| Salk_082522-R | CTTGAGAAGTAGCATCAGTTTCTG |
| For qRT-PCR | |
| RTDAO1-F | ATCCGTTGCAAGTCCATTGA |
| RTDAO1-R | GTTACAGAGCTCCAAACGAAA |
| RTDAO2-F | AAAATTGGGCTCTACCACTCC |
| RTDAO2-R | TTGTGATAAACTCGACGCCTC |
| RTGH3.3-F | ACAATTCCGCTCCACAGTTC |
| RTGH3.3-R | ACGAGTTCCTTGCTCTCCAA |
| RTGH3.4 -F | CGTTGGAGATACGTGTGGTG |
| RTGH3.4-R | GCAGTTTCATGATCGGTGTG |
| RTGH3.5-F | GTCTTCGAGGACTGCTGCTT |
| RTGH3.5-R | ATGTCCCTGGCTCAACAATC |
| RTGH3.6-F | CCTTGTTCCGTTTGATGCTT |
| RTGH3.6-R | CGTGTTACCGTTCAAGCAGA |
| For making constructs of DAO1pro:YFP: DAO1, DAO2pro:YFP: DAO2, DAO1pro:GUS | |
| DAO1pro-F-SAC1 | GAGCTCTCGTAGATTTCCGGCGAAGTT |
| DAO1pro-R-spe1 | ACTAGTATCTTCAATGGAGAGGTTAAC |
| DAO2pro-F-SAC1 | GAGCTCCCTTACTATGAAGCATTAGGTCT |
| DAO2pro-R-spe1 | ACTAGTTATCTTCAATGAAAAGGCTT |
| CACC-DAO1-F | CACCATGGGGGAACTAAACGGAGTC |
| DAO1-R | TCATTTATCTAGTCCTGCATGG |
| CACC-DAO2-F | CACCATGGCGGAAGTAAATGGAGT |
| DAO2-R | TTAATCTATCTTTGGGACGTCAC |
| CACC-GUS -F | CACCATGTTACGTCCTGTAGAAACCC |
| GUS-R | TCATTGTTTGCCTCCCTGCT |
Fig. S4.
LC-MS/MS analysis of the genuine oxIAA standard and the in vitro product of rDAO1 and rDAO2. Total ion counts (TICs) show a product peak (m/z 192→146) at 7.5 min, which is identical to oxIAA standard.
DAO1 Mutants Show Altered Auxin Metabolites and Auxin-Related Phenotypes
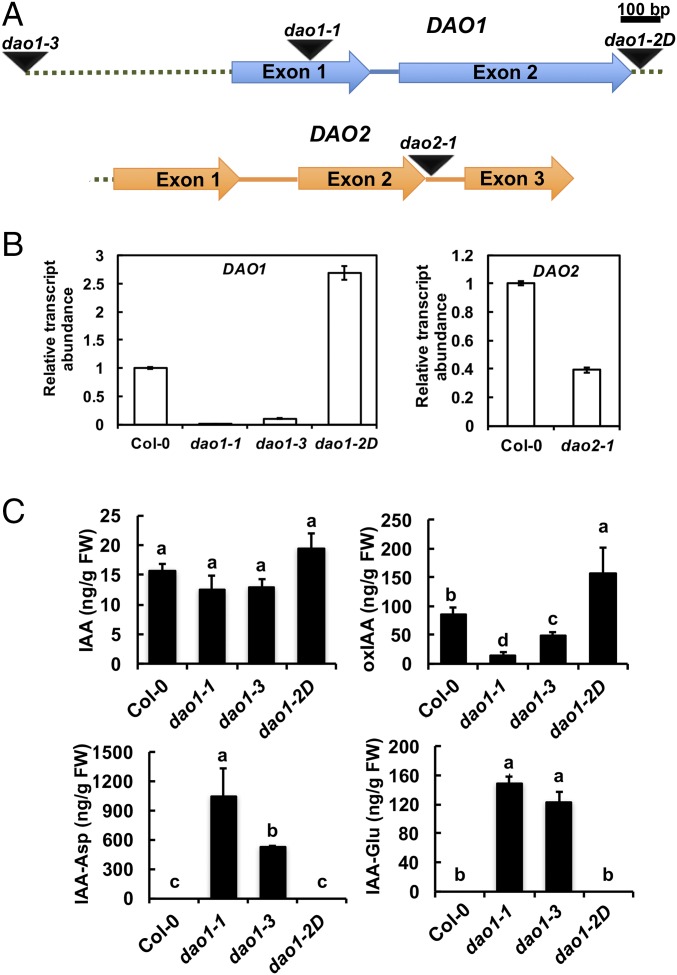
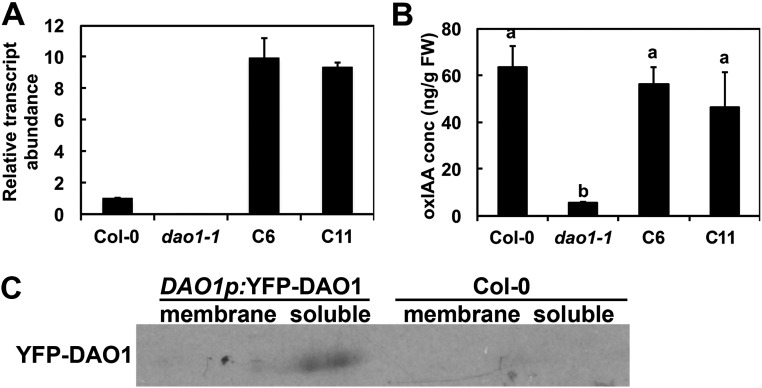
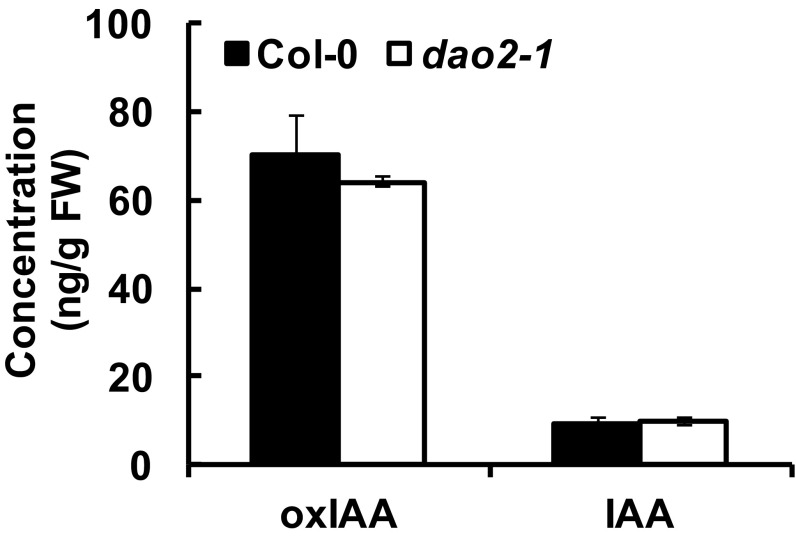
DAO1 and DAO2 IAA oxidation activity was investigated in planta using reverse genetics. DAO1 and DAO2 are located in tandem on chromosome 1, with 1,091 bp between the stop codon of DAO1 and the start codon of DAO2. Transfer-DNA (T-DNA) insertional mutants for DAO1 and DAO2 were obtained (29) and verified (Fig. 2 A and B). Quantitative real-time PCR (qPCR) showed that DAO1 transcripts were not detected in dao1-1 (first exon insertion), were reduced to 10% of the wild type in dao1-3 (promoter insertion), and increased 1.7-fold in dao1-2D (3′ UTR insertion) compared with the wild type (Fig. 2 A and B). qPCR showed that DAO2 expression was 61% less in dao2-1 than the wild type (Fig. 2B). Seven-day-old mutant seedlings were analyzed for altered auxin metabolism, and dao1-1 and dao1-3 mutants exhibited 94.3% and 41.6% decreases in oxIAA production, respectively, whereas dao1-2D gain of function seedlings showed an 83% increase in oxIAA levels compared with the wild type [Columbia-0 (Col-0); P < 0.001] (Fig. 2C). Furthermore, the oxIAA levels in dao1-1 were restored to wild type by the expression of native promoter-driven YFP-DAO1 (dao1-1c6 and dao1-1c11) (Fig. S5), although transcript abundance was greater than wild type. However, oxIAA accumulation in whole seedlings of the dao2-1 knockdown line was not different from wild type (P > 0.05) (Fig. S6). Together, these data indicate that DAO1 is the primary IAA oxidase during seedling establishment.
Fig. 2.
Identification of DAO1 and DAO2 T-DNA insertion lines. (A) Gene organization and schematic representation of T-DNA insertion lines of DAO1 (At1G14130) and DAO2 (At1G14120). Arrows, exons; dotted line, 1,091-bp interval between DAO1 stop codon and DAO2 start codon; intervening lines, introns; triangles, T-DNA sites. (B) Transcript abundance of DAO1 and DAO2 in mutants compared with Col-0. (C) IAA and IAA metabolite concentrations in 7-d-old Col-0 (wild type) and mutant seedlings. Data are means ± SD of at least four biological repeats. Different letters indicate statistical significance (ANOVA; P < 0.05). FW, fresh weight.
Fig. S5.
DAO1pro:YFP-DAO1 complemented oxIAA levels in dao1-1. Comparison among Col-0, dao1-1, and DAO1 complemented lines C6 and C11. (A) Relative transcript abundance of DAO1 in 7-d-old seedlings. (B) oxIAA levels in 7-d-old whole seedlings. FW, fresh weight. (C) Western blot with anti-YFP antibody shows that YFP-DAO1 is in soluble fraction in the DAO1pro: YFP-DAO1 line.
Fig. S6.
LC-MS analysis of oxIAA and IAA concentrations in 7-d-old Col-0 and the DAO2 knockdown dao2-1 mutant seedlings. Data represent means ± SD of four replicates. No significant difference (Student’s t test). FW, fresh weight.
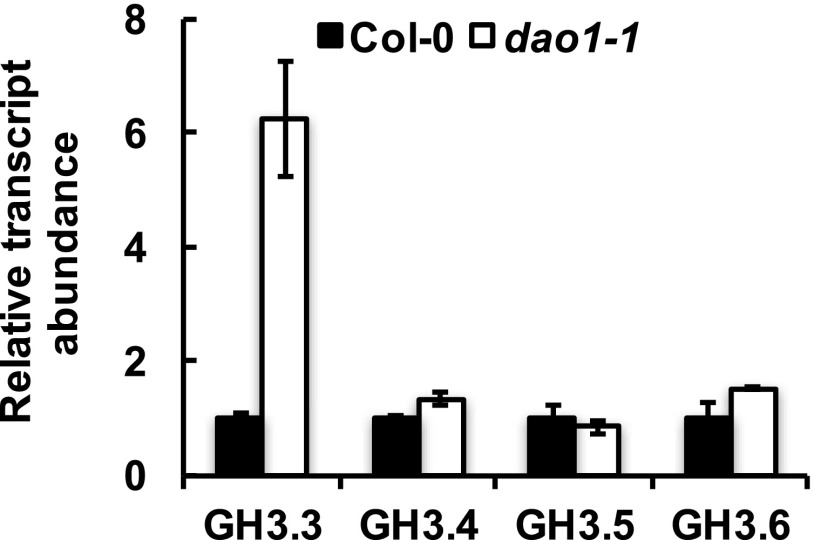
Although the amount of free IAA was not significantly altered in dao1-1 or dao1-3 (P > 0.05) (Fig. 2C), two of the major IAA-amino acid conjugates, IAA-Asp and IAA-Glu, were substantially increased in dao1-1 and dao1-3 compared with wild type (Fig. 2C), consistent with the levels reported in the work by Porco et al. (24). The accumulation of IAA-Asp and IAA-Glu was noteworthy, because levels of these metabolites were below the limit of detection in wild type. Furthermore, Gretchen Hagen3.3 (GH3.3), which can synthesize IAA-Asp, was up-regulated in dao1 loss of function mutants (Fig. S7) as noted by Porco et al. (24) and predicted by Mellor et al. (25). These data suggest that the IAA-amino acid conjugation pathway is induced to compensate for loss of DAO-mediated attenuation of auxin signaling.
Fig. S7.
Transcript abundance of GH3 genes in Col-0 and the dao1-1 loss of function mutant. RNA was extracted from 7-d-old seedlings of the wild type (Col-0) and dao1-1, and quantitative RT-PCR was conducted with ACTIN2 as an internal standard. Data represent means ± SD of six biological replicates.
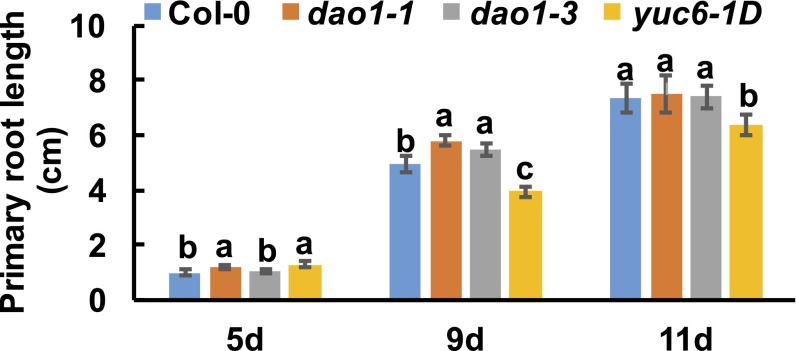
However, the increased irreversible IAA conjugation was apparently not sufficient to complement the loss of IAA oxidase activity, because both dao1-1 and dao1-3 showed altered morphological phenotypes (Figs. 3 and 4) similar to those reported in the work by Porco et al. (24). Those morphological phenotypes were reminiscent of those observed in yuc6-1D (kidari), in which auxin levels are slightly elevated, and the expression domain of the YUCCA6 auxin biosynthetic gene, normally restricted to flowers, carpels, pollen, and the root tip (30, 31), is expanded into other vegetative organs (32, 33). When dao1-1, dao1-3, and yuc6-1D were compared with wild type, significant (P < 0.001) increases in lateral root density (+86%, 47%, and 85%, respectively) were observed (Fig. 3 A and B). Primary roots in young dao1-1 and dao1-3 seedlings were initially longer than the wild type, with root hair phenotypes similar to those reported by Porco et al. (24), but primary roots were not different in length compared with wild type at 11 d (Fig. S8). Root hairs in yuc6-1D were also elongated, but primary roots were 13% shorter than the wild type (P < 0.05).
Fig. 3.
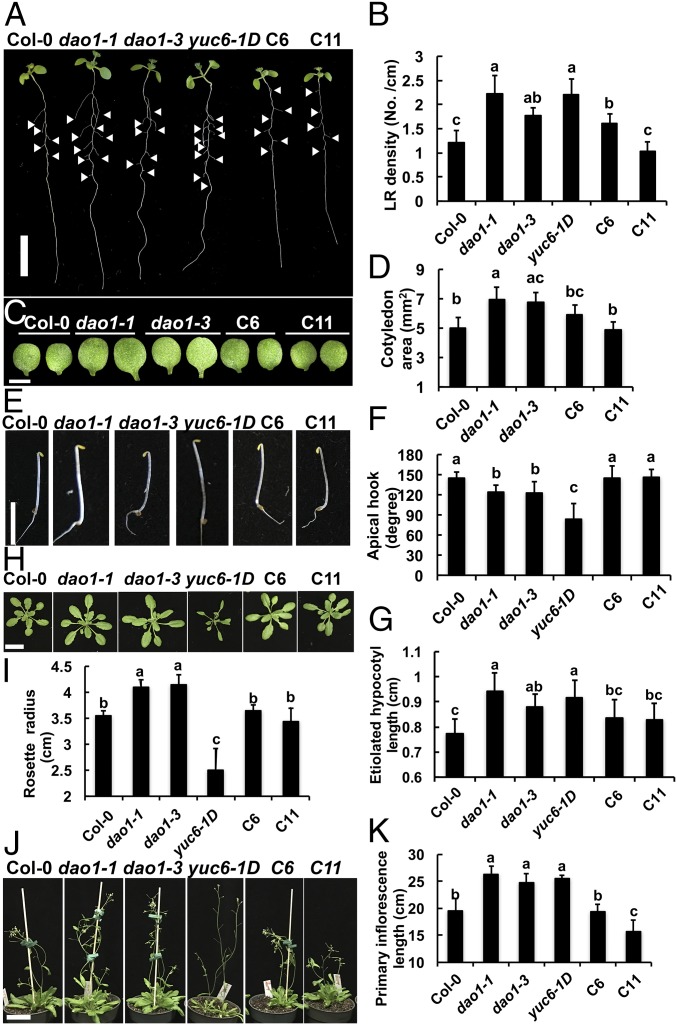
DAO1 loss of function plants show altered phenotypes and dao1 complementation. Phenotypes of Col-0, dao1-1, dao1-3, yuc6-1D, and dao1-1 complemented lines (C6 and C11). (A) Nine-day-old light-grown seedlings. (B) Quantification of lateral root (LR) density in 9-d-old seedlings. (C and D) Cotyledons from 7-d-old seedlings and quantification of cotyledon area. (E) Three-day-old etiolated seedlings. (F and G) Quantification of hypocotyl length and apical hook angle of 3-d-old etiolated seedlings. (H) Rosette leaves at bolting. (I) Rosette radii at bolting. (J) Thirty-two–day-old plants. (K) Primary inflorescence length of 32-d-old plants. Data are means ± SD (n ≥ 10). Different letters indicate statistical significance (ANOVA; P < 0.05). (Scale bars: A and H, 1 cm; C, 0.5 cm; J, 5 cm.)
Fig. 4.
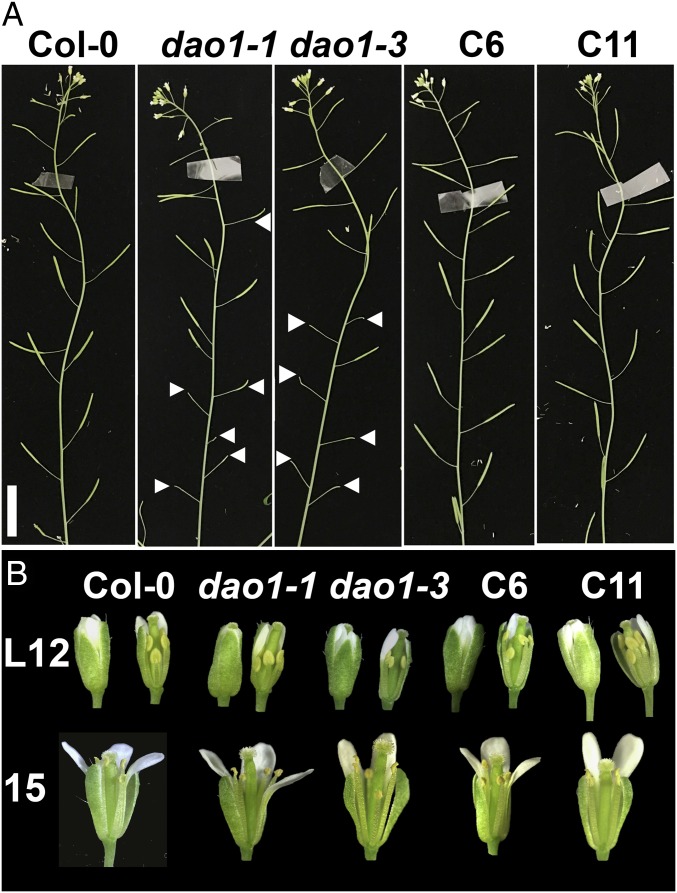
Comparison of fertility and floral phenotypes of dao1 mutants and complementation lines. (A) Primary inflorescences from 36-d-old Col-0, dao1-1, and complemented lines (C6 and C11). (B) Floral anatomy at late stage 12 (L12) and stage 15; L12 (Left) shows intact flowers, and in L12 (Right) and 15, sepals and petals were removed to show the pistil.
Fig. S8.
Root elongation of dao1 mutants. Primary root lengths of 5-, 9-, and 11-d-old dao1 mutants, yuc6-1D, and Col-0 were measured by ImageJ. Data represent means ± SD (n ≥ 10). Letters indicate statistical significance (Student’s t test).
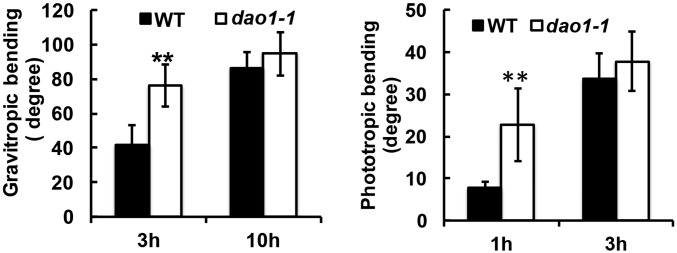
Both dao1-1 and dao1-3 had planar and significantly larger cotyledons than the wild type (38% and 35%, respectively; P < 0.05) (Fig. 3 C and D). In 3-d-old etiolated seedlings, the hypocotyls of dao1-1, dao1-3, and yuc6-1D seedlings were 22%, 14%, and 19% longer than wild type, respectively (P < 0.01) (Fig. 3 E and F), with 14.5%, 15.5%, and 40% reductions in the apical hook angle in 3-d-old etiolated seedlings, respectively (P < 0.001) (Fig. 3G). Tropic responses were also altered in dao1-1, and root gravitropic bending and phototropic bending in postphotomorphogenic seedlings were faster in dao1-1 than wild type during the initial stages of the tropic response (P < 0.01 and P < 0.03, respectively) (Fig. S9 and Movies S1 and S2).
Fig. S9.
Tropism analyses of Col-0 and dao1-1. Gravitropic bending assay: 5-d-old seedlings grown on plates were rotated 90°, and images were taken at 3 and 10 h. Phototropism assay: 3-d-old postphotomorphogenic seedlings grown on sand slurry were treated with unilateral blue light, and images were taken every 5 min (n ≥ 10). Asterisks indicate statistical significance (Student’s t test). **P < 0.01.
Rosette radii in dao1-1 and dao1-3 were larger than the wild type (15.5% and 17%, respectively; P < 0.01) (Fig. 3 H and I) and contrasted strongly with the narrow and curling rosette leaves in yuc6-1D. Primary inflorescences were 35% longer in dao1-1 and 27% longer in dao1-3 than the wild type at 32 d (P < 0.001) (Fig. 3 J and K), similar to those in yuc6-1D. In contrast, the dominant mutant dao1-2D had smaller and rounder rosette leaves and shorter inflorescences compared with the wild type (P < 0.001) (Fig. S10). All of the loss of function phenotypes were restored to wild type in dao1-1c6 and dao1-1c11 YFP-DAO1 expression lines (Fig. 3). The knockdown mutant dao2-1 had no obvious phenotypes (Fig. S11). Differences in the phenotypes reported here and in the work by Porco et al. (24) can be attributed to light intensity. Here, plants were grown under light conditions that result in lower basal IAA levels (100 μmol m−2 s−1), which seems to accentuate some of the observed phenotypes.
Fig. S10.
dao1-2D mature plant phenotypes. (A) Col-0 and dao1-2D before bolting. (B) Col-0 and dao1-2D at 38 d after sowing. (C) Quantitation of rosette radii and primary inflorescence height in Col-0 and dao1-2D (n ≥ 6). Asterisks indicate statistical significance (Student’s t test). ***P < 0.001.
Fig. S11.
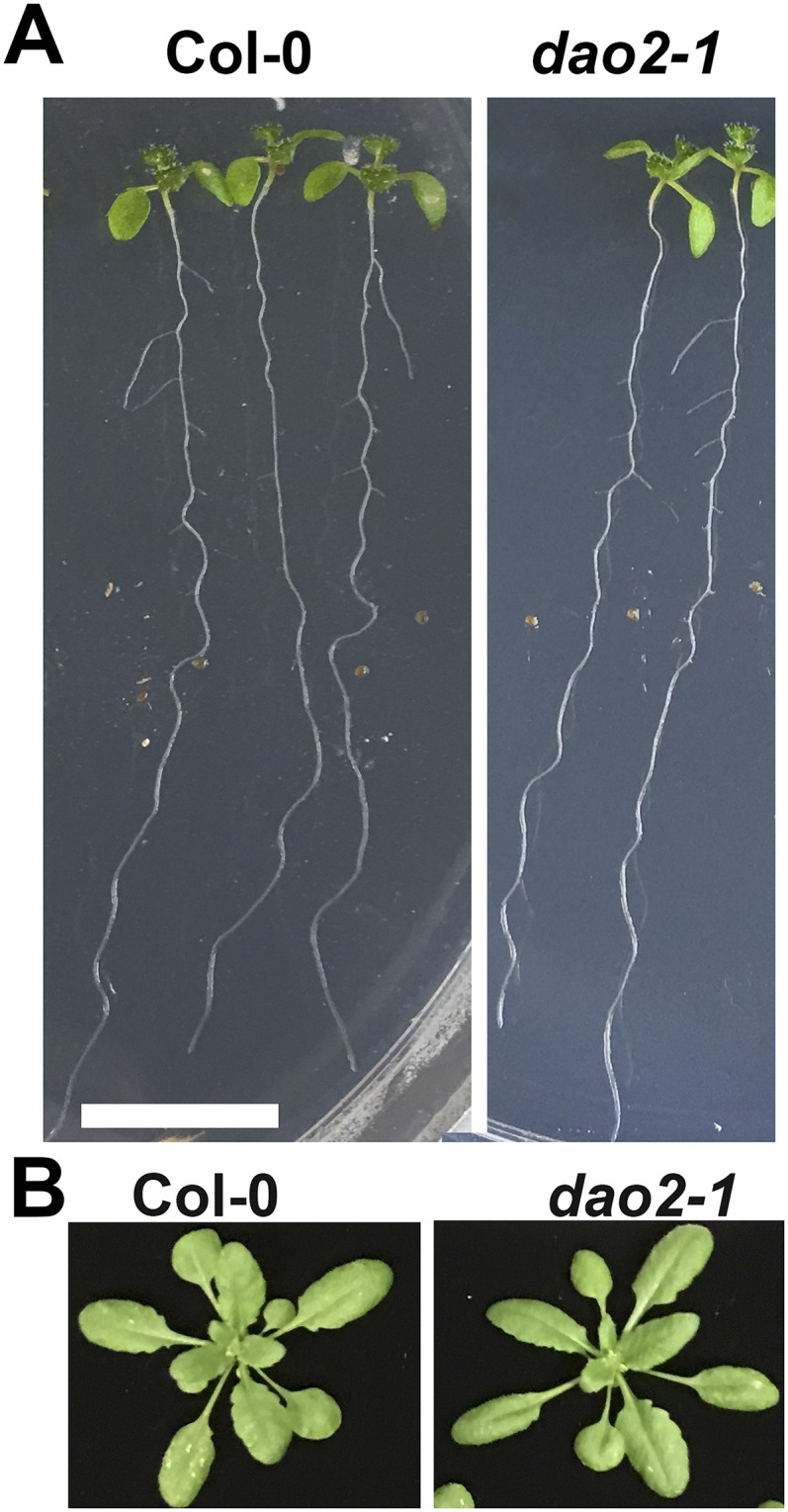
Phenotypes of dao2-1. (A) Eight-day-old Col-0 and dao2-1 seedlings grown on plates. (B) Col-0 and dao2-1 rosettes 20 d after sowing on soil.
As also reported by Porco et al. (24), dao1 loss of function mutants had small siliques with incomplete filling in an irregular pattern in the primary inflorescence (Fig. 4A). Closer investigation revealed that sepal opening was delayed at floral stage 13 in dao1-1 mutants (Fig. 4B). Furthermore, at floral stage 15, the pistils in dao1-1 and dao1-3 were elongated compared with the wild type, resulting in variable pollination. These phenotypes are strong in primary inflorescences at the beginning of bolting but weaken later, especially in axillary inflorescences. These results are consistent with the concentration dependence of auxin metabolism levels during floral development and tissue specificity of DAO1 function. Together, these results support the hypothesis that DAO1 functions as an IAA oxidase in vivo and regulates plant morphogenesis.
DAO1 Is Expressed Throughout the Plant, and DAO2 Is Expressed in the Root Cap
The spatial and temporal expression patterns of DAO1 and DAO2 were examined to determine when and where DAO activity was required to maintain auxin homeostasis during growth and development. Microarray and transcriptomic analyses showed that Arabidopsis DAO1 is expressed in reproductive organs (33, 34). DAO1 is also expressed in the cortical, endodermal, and pericycle cells of the root distal elongation zone and maturation zone, the root columella, and atrichoblasts (35, 36). DAO2 is expressed weakly in the root columella and procambium of the root maturation zone (35, 36). Transcriptomic data also indicate that DAO2 expression in the root is several hundred fold lower than DAO1 expression in the roots of Arabidopsis seedlings (36). These expression data are consistent with results of the accumulation profiles of IAA metabolites in dao1-1 and dao2-1 in seedlings (Fig. 2C and Fig. S6) (24, 25) but also suggest that some posttranslational regulation of DAO is likely.
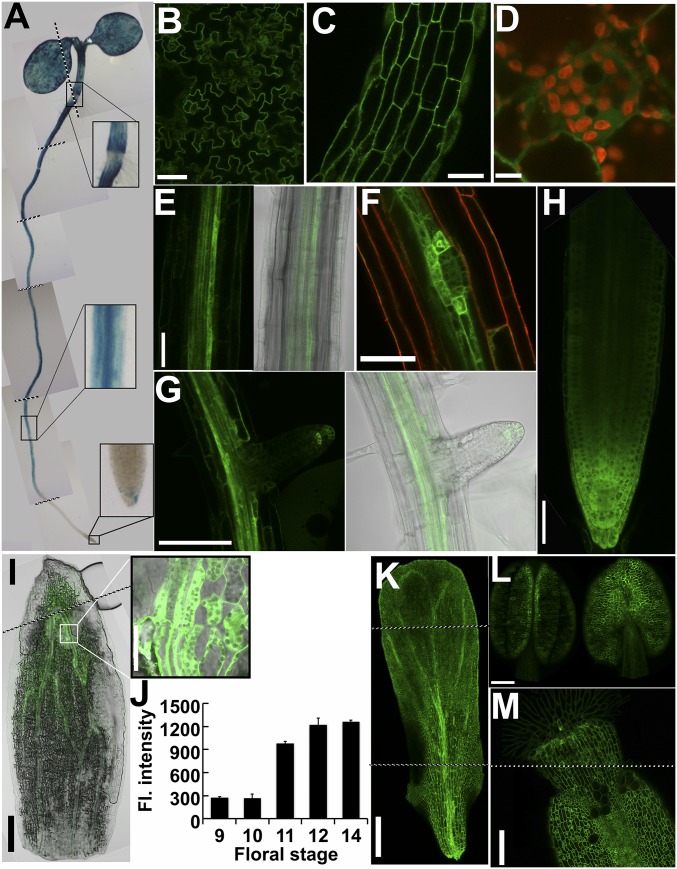
DAO1 and DAO2 spatiotemporal expression patterns were analyzed using DAO native promoter-driven reporter and functional DAO fluorescent protein fusion constructs [DAO1pro:β-glucuronidase (GUS), DAO1pro:YFP-DAO1, and DAO2pro:YFP-DAO2]. Strong and constitutive DAO1pro:GUS activity was observed at the cotyledon margins and ubiquitously in the epidermis and vascular tissue in 7-d-old seedlings (Fig. 5A). These results are consistent with the larger cotyledons and leaves observed in dao1-1 and dao1-3 (Fig. 3 C and D) and the smaller leaves in dao1-2D, indicating that localized auxin oxidation mediated by DAO1 is involved in controlling cotyledon and leaf size. GUS activity had a distinct delineation at the root–shoot transition zone, where the adjacent root cells had little or no activity, whereas the adjacent shoot epidermal cells showed strong staining. These patterns are consistent with the florescence signals of DAO1pro:YFP-DAO1 in dao1-1 seedlings. Confocal microscopy (Fig. 5 B–G and autofluorescence controls in Fig. S12) of 7-d-old seedlings showed cytosolic DAO1pro:YFP-DAO1 signals in the pavement cells of cotyledons, the epidermis, and the pericycle in the root and in the root cap. Porco et al. (24) also showed soluble DAO1-GFP signals in the root tip, pericycle, and trichoblasts. The YFP-DAO1 signal was much stronger in the pericycle cells flanking the developing lateral root primordia, and this pattern was maintained throughout the stages of lateral root development. However, no DAO1 signals were detectable in the tips of lateral roots until they emerged from the primary root (Fig. 5 F and G). The localization of DAO1 at the organ borders was similar to that observed at the root–shoot junction (Fig. 5A) and suggests a role in organ delineation.
Fig. 5.
Spatial and temporal expression pattern of DAO1. (A) DAO1pro:GUS activity in 7-d-old seedling. The boxes show magnified regions as indicated. (B–E and G) DAO1pro:YFP-DAO1 signals in 5-d-old seedling. (B) Cotyledon, (C) hypocotyl, and (D) subcellular localization of DAO1 in hypocotyl epidermis. Red indicates chloroplast autofluorescence. (E) Upper root. (F) Upper root with a lateral root primordium in a 9-d-old seedling. Red indicates FM4-64, a plasma membrane dye. (G) Lateral root in the 9-d-old seedling. (H) Primary root tip. (I–M) DAO1pro:YFP-DAO1 signals in floral stage 12. (I) Sepal. (J) Florescence intensity of DAO1pro:YFP-DAO1 in the upper sepal during flower development. Data are means ± SD. (K) Petal, (L) stamen, and (M) stigma. [Scale bars: B, C, E–H, and I (Inset), 0.5 mm; D, 0.01 mm; I and L, 0.2 mm; K and M, 0.1 mm.]
Fig. S12.
Autofluorescence controls of wild-type plants corresponding to confocal images shown in Fig. 5. (A–D) Five-day-old seedlings: (A) cotyledon, (B) shoot apex, (C) upper root, and (D) root tip. (E) Confocal image of apical hook of 3-d-old etiolated seedling. (F–I) Reproductive tissues: (F) sepal, (G) petal, (H) stamen, and (I) stigma. F–H are from one flower at stage 12, and I is from one flower at stage 13. All of the images were taken under the same settings as the corresponding images in Fig. 5. (Scale bars: 0.05 mm.)
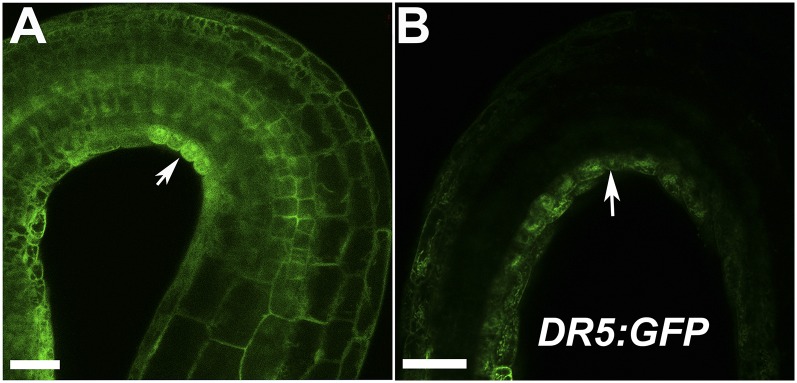
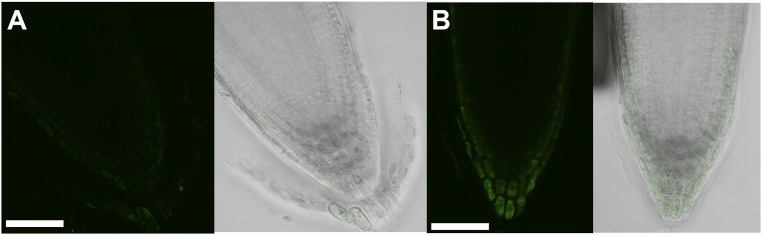
Coinciding with DAO1 signals in the root cap, the DR5:GUS indirect auxin reporter construct in the dao1-1 mutant showed enhanced activity in the root cap compared with the wild type (Fig. S13), a phenotype predicted by the modeling of auxin distribution in dao1 loss of function mutants (25). These data support the hypothesis that localized IAA oxidation plays an important role in the regulation of auxin signaling in the root cap. Furthermore, in etiolated seedlings, YFP-DAO1 signals were greater on the inside of the apical hook, where DR5:GFP signals were also strong (Fig. S14), suggesting that DAO1 functions in apical hook maintenance. In contrast, analyses of YFP-DAO2 patterns showed that DAO2 signals were only weakly detected in the root cap of seedlings (Fig. S15 with autofluorescence controls), consistent with the low transcript levels of DAO2 detected in the root and other tissues (36) (arexdb.org).
Fig. S13.
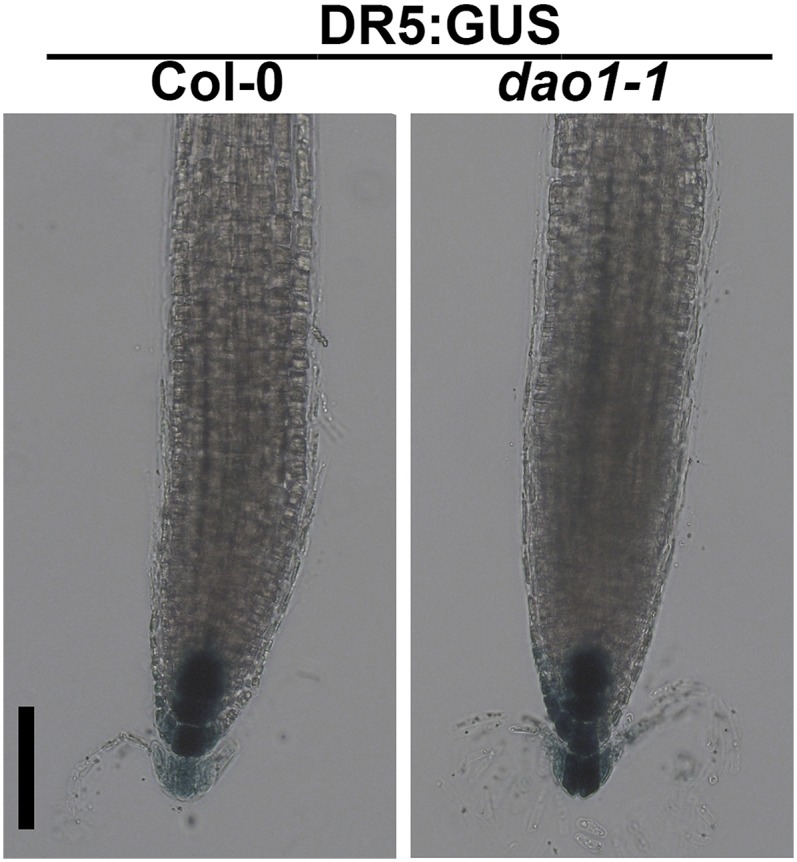
DR5:GUS expression in 7-d Col-0 and dao1-1. Five-day-old seedlings were stained for GUS activity. (Scale bar: 100 µm.)
Fig. S14.
DAO1 is asymmetrically localized inside of the apical hook. (A) DAO1pro:YFP-DAO1 signal in apical hook of 3-d-old etiolated seedlings. (B) DR5:GFP signal in apical hook of 3-d-old etiolated seedlings. (Scale bars: 0.05 mm.)
Fig. S15.
DAO2pro:YFP-DAO2 expression in the root cap. (A) Autofluorescence control in Col-0. (B) DAO2pro:YFP-DAO2 signal in root tip of T2 seedlings (second generation of transformed lines). YFP signals were not observed in T3 seedlings (third generation of transformed lines). (Scale bars: 0.05 mm.)
In floral organs, YFP-DAO1 signals were strongest in the sepal and petal vasculature as well as in the epidermis of all four whorls (Fig. 5 I–M). DAO1 was strongly expressed near the sepal apex, especially in the bundle sheath cells and adjacent mesophyll cells just before flower opening (Fig. 3 J and K and autofluorescence controls in Fig. S12). This result is consistent with microarray data showing that DAO1 expression in the sepals is highest at floral stage 15 (34). Before bud opening, the sepal apex is curved inward around the other floral organs to maintain a closed flower that protects the developing reproductive organs (37). Auxin regulation of sepal opening was suggested by high expression of the rate-limiting auxin biosynthetic gene YUCCA4 at the sepal apex of young flowers (30). The delayed flower opening observed in dao1-1 (Fig. 4A) supports the notion that IAA oxidation in sepal bundle sheath cells plays an important role in controlling the timing of sepal opening. LC-MS determinations showed that auxin levels are high in the sepals of very young flowers (42 ± 3.4 pg mg−1) and reduced (6.7 ± 2.9 pg mg−1) by the time of bud opening, indicating that YUCCA4 and DAO1 expressions are coordinated to fine tune auxin levels during sepal opening. This finding is also consistent with a defect in lemma and palea opening associated with DAO loss of function in rice (21). Strong DAO1 signals in the carpel (Fig. 5 L and M) correlated with the elongated carpel observed and reduced fertility in dao1-1 and dao1-3 (Fig. 4 A and B). However, no severe defects in fertility were observed in dao1-1.
Discussion
In contrast to observations in rice, where dao mutants were shown to have defects limited to anther dehiscence, pollen growth, and seed development (21), the data presented here suggest that temporal- and tissue-specific degradation of auxin by DAO1 plays an important role in plant growth and development, including regulation of lateral root density, inflorescence height, and leaf and floral organ size, with subsequent effects on fertility (Figs. 3 and 4). Strong and uniform DAO1 localization in cotyledons correlates with the increased cotyledon size in dao1 loss of function alleles, indicating that localized oxidation of auxin is involved in regulating cotyledon size. The strong localization of DAO1 in the pericycle (Fig. 5E) and the border cells flanking the lateral root primordia but not in lateral root primordia (Fig. 5F) indicate that DAO1 functions in the process that generates the localized auxin maxima involved in initiating lateral root formation.
Recently, IAA derived from the root cap was found to be involved in the prepatterning of lateral root formation (38). Xuan et al. (38) reported that IAA oscillation amplitudes in the root cap determined the prebranch sites of the lateral root. However, the role of auxin oxidation was not addressed in that study. Both DAO1 and DAO2 are expressed in root cap, and dao1 mutants show increased lateral root density (Figs. 3A and 5 A and G and Fig. S11), supporting the hypothesis that auxin oxidation in the root cap plays an important role in lateral root initiation. Our data indicate that localized auxin oxidation by both DAO1 and DAO2 in the root cap is involved in the regulation of lateral root density, consistent with the results presented by Xuan et al. (38). The oscillations that induce lateral root development (38) also seem to have a modulating effect on circadian rhythms, and both oxIAA production and DAO expression levels have been shown to follow circadian patterns (22, 25). However, DAO2 null and dao1 dao2 double mutants are still needed to fully characterize the role of auxin oxidation in lateral root development. Unfortunately, the tight linkage of DAO1 and DAO2 prevents ready analysis of dao1 dao2 double mutants, and so far, we have not been able to stably inactivate both DAO1 and DAO2 with CRISPR-Cas9 or amiRNA (artificial microRNA).
Arabidopsis is self-fertile, and before anthesis, the stamens extend above the stigma at floral stage 14 (39). This process requires highly coordinated filament and pistil elongation, which is achieved by differentially regulated auxin levels in the respective organs (30, 40). The abcb1 abcb19 double auxin transporter mutant shows reduced fertility as a consequence of impaired filament elongation (41). Similarly, our results show that loss of IAA oxidation results in an extended growth phase of the pistil (40) (Fig. 4). This finding suggests that auxin homeostasis is synergistically regulated in stamens and pistils for subsequent pollination. Our study indicates that spatiotemporal auxin oxidation by DAO1 plays an important role in coordination of filament and pistil elongation by reducing active auxin levels in the carpel and thus, restraining pistil elongation.
Data presented here and in the work by Porco et al. (24) show that GH3 gene expression increases as a result of loss of DAO function. Porco et al. (24) also show a decrease in the auxin precursor indole pyruvate in dao1, which suggests either a down-regulation of auxin biosynthetic genes expression or posttranslational regulation of the enzymes. The low substrate specificities (Kcats) of auxin biosynthetic enzymes (30) and membrane localization of YUCCA isoforms suggest the possibility of functional, membrane-associated metabalons (42, 43). The slow kinetics of the DAOs are more similar to those of the auxin biosynthetic enzymes than to the those observed with GH3s. Accordingly, we hypothesize that DAO functions in tandem with the auxin biosynthetic machinery and may be recruited to the metabolon to fine tune auxin levels. Our data indicate that precise cooperation between IAA biosynthesis and IAA oxidation allows dynamic regulation of IAA levels that is likely required for flower development and lateral root formation.
These results presented here and in the works by Porco et al. (24) and Mellor et al. (25) also show that the irreversible major IAA conjugates, IAA-Glu and IAA-Asp, occur at higher levels in dao1 loss of function alleles than in wild type (Fig. 2C). So why did plants evolve two different pathways to catabolize auxin and terminate auxin signaling? A functional comparison suggests that Asp conjugation is a rapid response, whereas auxin oxidation via DAO represents a constitutive mechanism to slowly remove auxin signals. The GH3 family genes that encode the amino acid transferases are expressed at very low levels under normal growth conditions but are stimulated by application of exogenous auxin [GH3.1, 2, 3, 4, 6; Gene Expression Omnibus (GEO) accession no. GSM9969], blue light (GH3.10) (44), or jasmonic acid treatment (GH3.9) (45). GH3 genes are among the fastest to respond to exogenous auxin, with transcript levels increasing more than 10 times within 1 h (GEO accession no. GSM9969). Most GH3 KO mutants (6) are hypersensitive to long-term exogenous IAA treatment. In contrast, DAO1 has a constitutively high expression level in most tissues under normal conditions. In addition, GH3s have faster enzyme kinetics compared with DAOs from in vitro enzyme assays (6) (Fig. 1). Recombinant GH3.6 conjugates Asp to IAA at 244 nmol IAA-Asp min−1 mg−1 protein (6), whereas rDAO1 oxidizes IAA at 5 pmol oxIAA−1 min−1 mg protein (Fig. 1C), more than 10,000 times slower than GH3.6 (6). Therefore, DAO1 seems to function spatiotemporally to terminate active auxin constitutively during development in contrast to GH3, which responds quickly to environmental factors that increase cellular IAA levels. This finding also explains why increased irreversible auxin conjugates in dao1-1 could not compensate for the altered morphological phenotypes (24). Taken together, our results show that DAO1 regulates IAA homeostasis in a manner different from IAA conjugation and is functionally important in plant growth and development.
More details are in SI Materials and Methods.
SI Materials and Methods
Plant Material.
All of the Arabidopsis lines used in this research are in Col-0 background; dao1-1 (Salk_093162), dao1-3 (Salk_082522), dao1-2D (CS816250), and dao2-1 (Salk_205223) were obtained from Arabidopsis Biological Resource Center (ABRC). Plants were grown as previously described (20). yuc6-1D, DR5:GUS, and DR5:GFP have been described (32, 46, 47). Briefly, seeds were surface sterilized and planted on 1/4 Murashige and Skoog medium (RPI Corp.) containing 0.5% sucrose and 0.8% agar, pH 5.5. Seeds were stratified at 4 °C for 3 d, and then placed in growth chambers at 22 °C for 24 h at 100 µmol m−2 s−1 light, except as indicated for specific treatments. Plants on soil were grown in growth chambers at 22 °C at 100 µmol m−2 s−1 light (16 h light/8 h darkness).
Cloning and Protein Expression of AtDAO1 and AtDAO2.
Plasmids with AtDAO1 (At1g14130; U50705) and AtDAO2 (At1g14120; C105263) cDNA were obtained from the ABRC (The Ohio State University). The cDNA fragments were subcloned into pET23B+ (Novagen) between the EcoRI and XhoI using the restriction sites introduced by the PCR primers (Table S2) and sequenced. The plasmid was transformed into Escherichia coli BL21 (DE3) plysE, and protein expression was optimized. In the preparative procedure, E. coli containing the plasmid was grown in shaker flasks (250 rpm at 37 °C) containing Luria Broth supplemented with 15 μg/mL ampicillin to late exponential phase (A600 ∼ 0.8). Recombinant protein production was induced by the addition of isopropyl-d-thiogalactopyrano-side (IPTG) to a final concentration of 0.4 mM. Cells were grown for another 4 h at 27 °C, and then harvested. Cell paste was stored at −80 °C. Thawed cell pellets were suspended in lysis buffer containing 20 mM PBS, pH 7.4, 10% (vol/vol) glycerol, 0.02 mg/mL DNase, 1 mM MgCl2, 1 mM PMSF, 20 mM imidazole, and 500 mM NaCl. Unbroken cells, cell debris, and precipitated nuclei were then removed by centrifugation at 14,000 × g at 4 °C for 15 min. The supernatant was run through this spin trap column (GE), washed with buffer A (20 mM imidazole, 500 mM NaCl, 20 mM PBS, pH 7.4), and eluted with buffer B (buffer A plus 250 mM imidazole). The eluate was concentrated with Millipore Amicon 30-kb Membrane by centrifugation at 10,000 × g at 4 °C for 10 min.
SDS/PAGE and Western Blot Analyses.
SDS loading buffer (48) was added to the cell pellet or purified enzyme and boiled for 5 min. Samples were centrifuged at 12,000 × g for 5 min. Then, supernatant was loaded on a 12% (wt/vol) SDS/PAGE gel (48). After running the gels, the gels were either blotted or stained with Coomassie blue. For Western blot analyses, proteins were transferred onto nitrocellulose by wet electroblotting. For detection of recombinant protein, a mouse monoclonal 6× HIS antibody (Abcam) and an anti-mouse antibody conjugated to peroxidase (Sigma) were used at 1:1,000 and 1:5,000 dilutions, respectively. Blots were developed using the HyGLO Quick Spray, and chemiluminescence emitted from the filter was exposed to a Kodak film for 10–30 s and then, developed.
Enzyme Assays.
The reaction mixtures contained 40 mM PBS, pH 7.4, 5 mM 2-oxoglutarate, and 0.5 mM Fe(SO4)2 in a final volume of 500 µL; 0.002 mg/mL IAA and 6 µg boiled or unboiled enzyme were added to the mixtures and incubated in the dark at 30 °C for 1 h. Then, 2 vol acetone mixed with 0.001 mg/mL indole-3-propionic acid as a standard was added to the sample and placed at −20 °C overnight to precipitate protein. After centrifugation at 14,000 × g for 10 min, the supernatants were dried with a speed vacuum and dissolved with 50 µL 10% (vol/vol) methanol and 0.3% acetic acid; 20-µL samples were introduced by an autosampler onto a C18 column (EC 150/3; 3 μm Nucleodur; Macherey Nagel) on a high-pressure LC system (1260 infinity; Agilent). The mobile phase was delivered at a flow rate of 0.3 mL/min with an initial mixture of 34% (vol/vol) methanol in water (with 0.3% acetic acid in both solvents) for 5 min followed by a 20-min linear gradient to 90% (vol/vol) methanol. A diode array detector (DAD) (1260 infinity; Agilent) was set up to detect the effluence absorbance at 200, 254, 280, and 325 nm. Boiled or unboiled rDAO1 and rDAO2 were incubated with 0.2, 1, 2, and 5 µmol/L IAA for 30 min, and oxIAA production was quantified by UPLC. Each data point is the average of at least six repeats.
LC-MS/MS Analyses.
For enzyme assays, samples were injected into an Agilent 1260 Infinity LC System, and compounds were separated using an Agilent Poroshell 120EC-C18 (3.5 × 50 mm; 2.7 µm) Column and an acidified water:methanol buffer system (solvent A: 0.1% acetate, 5% (vol/vol) MeOH in water; solvent B: 0.1% acetate in methanol). Gradient conditions were as follows: 1 min 0–10% (vol/vol) B, 5 min 10–60% (vol/vol) B, 5 min 60–100% (vol/vol) B, hold at 100% (vol/vol) B for 3 min, and then back to 0% B in 2 min. Eluted compounds were further separated and quantified using an Agilent 6460 Triple Quadrupole Dual Mass Spectrometer equipped with an electrospray ionization source. Compounds were quantified in positive ion mode, and MS/MS settings were as described (49) and conducted by Agilent G6460.
For plant tissue, Arabidopsis seedling samples were collected, weighed, and chilled in liquid nitrogen before storing in −80 °C. Samples were ground in liquid nitrogen, and 1 mL 50 mM sodium-phosphate buffer, pH 7.0 (contains 1% diethyldithiocarbamic acid was immediately added into each tube afterward; 25 ng d5-IAA (part 0311532; OlChemIm) and 25 ng d3-tryptophan (part d-7419; CDN Isotopes) were added into each tube as internal standards. Samples were vortexed, extracted for 20 min at 4 °C on a laboratory nutator, and finally, centrifuged at 12,000 x g for 15 min at 4 °C. The pH value of supernatant was adjusted to 3 using 1 N HCl, and the supernatants were purified using an HLB Column [column conditioned with 1 mL methanol (LC-MS/MS grade; part A456-1; Fisher Scientific) followed with 1 mL water and 0.5 mL 50 mM Na-Phosphate buffer, pH 2.7; after loading the sample, the column was washed with 2 mL 5% (vol/vol) methanol and finally, eluted with 2 mL 80% (vol/vol) methanol]. The eluted samples were dried under nitrogen gas, redissolved with 500 µL methanol, and filtered through 4-mm 0.2-µm PTFE Filters (part 03–391-4E; Fisher Scientific); 1 µL each sample was injected for LC-MS/MS analyses.
RNA Extraction and Transcription Analysis.
Total RNA was extracted from designated tissues using the RNeasy Plant Mini Kit (Zymo Research). After treatment with DNaseI (Invitrogen), 2 μg total RNA was used for the synthesis of the first-strand cDNA using the thermoscript RT-PCR system and oligo(dT) as primers (Invitrogen). qPCR was performed using SYBR Green on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). PCR was performed in 96-well optical reaction plates heated for 5 min to 95 °C followed by 40 cycles of denaturation for 10 s at 95 °C and annealing–extension for 30 s at 60 °C.
The gene-specific primers used to detect the transcripts were as follows: RTDAO1-F: ATCCGTTGCAAGTCCATTGA; RTDAO1-R: GTTACAGAGCTCCAAACGAAA; RTDAO2-F: AAAATTGGGCTCTACCACTCC; RTDAO2-R: TTGTGATAAACTCGACGCCTC; RTGH3.3-F: ACAATTCCGCTCCACAGTTC; and RTGH3.3-R: ACGAGTTCCTTGCTCTCCAA. Expression levels were normalized to actin-2 using the following primers: AtACT2- F: ACACTGTGCCAATCTACGAGGGTT; and AtACT2- R: ACAATTTCCCGCTCTGCTGTTGTG. All quantitative RT-PCR experiments were performed in triplicate, and the values presented represent means ± SE.
Generation of Transgenic Lines.
Gateway vector pUBNYFP-dest (50) was digested with SacI and SpeI followed by ligation with the DAO1 promoter (1.8 kb) or the DAO2 promoter (2.0 kb), respectively, into the vector replacing the UBQ10 promoter. DAO1 and DAO2 genomic coding sequences were PCR amplified and ligated into modified pUBNYFP vector to construct DAO1pro:YFP-DAO1 and DAO2pro:YFP-DAO2 using the Gateway System (Invitrogen). Transformation of Arabidopsis thaliana with Agrobacterium tumefasciens (GV3101) was performed as described previously (51). All complementation experiments were performed with two independent homozygous T3 lines.
Gravitropism Assay.
Seedlings were grown vertically on plates as above for 4 d. After reorienting the plates by 90°, the root tip position was marked every 3 h over a 24-h time period. The angles of curvature were measured using the ImageJ (52), and the data were analyzed by Microsoft Excel. Averages and SDs were calculated from 50 seedlings.
Phototropism Assay.
Seeds were placed on 50–70 mesh sand (catalog no. 274739; Sigma) with water, covered with a clear vented lid, and light treated for 12 h. Subsequently, seeds were placed in the dark until 2 d old or about 0.4 cm in height. Seedlings then undergo light treatment at 65 μmol m−2 s−1 light (Philips f32t8/tl741; Philips) at 24 °C for 12 h to undergo photomorphogenesis and then are returned to dark for 12 h. The vented lid was removed, and seedlings are placed in front of a 450-nm 100 LED light source (Rothner Laser Technic) between 0.9 and 0.6 μM m−2 s−1 at 24 °C. Video was captured using an MTI ccd72 (DAGE-MTI) Camera, and the image was snapped every 5 min. The bending angles were measured using ImageJ.
GUS Staining.
GUS staining was as described in the work by Geisler et al. (53). Briefly, seedlings or adult plant tissues were incubated in GUS substrate for 12 h at 37 °C and then, destained in 70% ethanol before imaging.
Microsomal Membrane Preparations.
Microsomal preparations of 7-d-old Arabidopsis seedlings were prepared and assayed for purity as described previously (54), with the exception that 500 μM benzamide and 500 μM benzamidine were added to the original homogenization buffer. Prepared membranes were stored in liquid nitrogen. Membrane proteins were detergent solubilized by incubation with gentle shaking at 4 °C for 30 min in a buffer consisting of 0.1% (wt/vol) Brij 35, 0.05% (wt/vol) CHAPS, 10 mM BisTris propane-MES, pH 7.8, 250 mm Suc, 20% (wt/vol) glycerol, and 1 mm DTT followed by centrifugation at 100,000 × g for 30 min.
Microscopy.
Protein localizations in Arabidopsis seedlings were visualized using an LSM 710 Laser Spectral Scanning Confocal Microscope (Zeiss; www.zeiss.com) with either a 20× lens or a 40× water immersion lens and pixel dwell time of 0.01 ms. The master gain was always set to less than 893, with a digital gain of 1.5. For YFP acquisition, 514-nm (5%) excitation and 519- to 560-nm emission were used. For GFP, 488-nm (5%) excitation and 493- to 598-nm emission were used. Quantification of florescence intensity was analyzed using ZEN Lite 2012. Briefly, representative images from three sepals in each floral stage were selected for analysis. Six cells from each image were selected in each image. The florescence intensity of at least six bundle sheath cells in the upper sepal from each stage was quantified with the histogram analyses tool in the ZEN Lite 2012 software (n = 3). All of the images were taken under the same conditions. All images were processed with ZEN Lite 2012 (Zeiss; www.zeiss.com) and Photoshop (Adobe; www.adobe.com).
Sequence and Phylogenetic Analysis.
tBLASTn was used to identify homologous genes of OsDAO (Os04g0475600) in A. thaliana (www.ncbi.nlm.nih.gov/). Protein sequences of the 2OG Fe(II) oxygenase super family in A. thaliana were obtained from the Arabidopsis Information Resource (v10; https://www.arabidopsis.org). The sequences were aligned using the CustalW 2.0 (55). The alignment file was used to generate an unrooted tree with MEGA 6.0 (56) applying the neighbor-joining method, with 10,000 bootstrap replications and handling gaps with pairwise deletion.
Supplementary Material
Acknowledgments
We thank Candace Pritchard for assistance with the phototropism assays; Sarah Turner and Jongmi Park for assistance with enzyme assays, RNA extractions, and growing plants; Dr. Angus Murphy for providing the auxin quantitation data in the sepals, helpful discussions, and critically reading the manuscript; and Dr. Jeong-Im Kim for the yuc6-1D seeds and discussion of the phenotypes. This work was supported by the Maryland Agricultural Research Station, the University of Maryland College of Agriculture and Natural Resources, and the Ohio Agricultural Research and Development Center SEEDS Grant Program, The Ohio State University, and Coordenação de Aperfeiçoamento de Pessoa de Nível Superior (Capes), Brazil.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 10742.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604769113/-/DCSupplemental.
References
- 1.Mashiguchi K, et al. The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA. 2011;108(45):18512–18517. doi: 10.1073/pnas.1108434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stepanova AN, et al. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell. 2011;23(11):3961–3973. doi: 10.1105/tpc.111.088047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson RG, et al. Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. J Biol Chem. 2001;276(6):4350–4356. doi: 10.1074/jbc.M006185200. [DOI] [PubMed] [Google Scholar]
- 4.Jackson RG, et al. Over-expression of an Arabidopsis gene encoding a glucosyltransferase of indole-3-acetic acid: Phenotypic characterisation of transgenic lines. Plant J. 2002;32(4):573–583. doi: 10.1046/j.1365-313x.2002.01445.x. [DOI] [PubMed] [Google Scholar]
- 5.Tognetti VB, et al. Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell. 2010;22(8):2660–2679. doi: 10.1105/tpc.109.071316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Staswick PE, et al. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell. 2005;17(2):616–627. doi: 10.1105/tpc.104.026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ludwig-Müller J. Auxin conjugates: Their role for plant development and in the evolution of land plants. J Exp Bot. 2011;62(6):1757–1773. doi: 10.1093/jxb/erq412. [DOI] [PubMed] [Google Scholar]
- 8.Korasick DA, Enders TA, Strader LC. Auxin biosynthesis and storage forms. J Exp Bot. 2013;64(9):2541–2555. doi: 10.1093/jxb/ert080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishimaru K, et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat Genet. 2013;45(6):707–711. doi: 10.1038/ng.2612. [DOI] [PubMed] [Google Scholar]
- 10.Östin A, Kowalyczk M, Bhalerao RP, Sandberg G. Metabolism of indole-3-acetic acid in Arabidopsis. Plant Physiol. 1998;118(1):285–296. doi: 10.1104/pp.118.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LeClere S, Tellez R, Rampey RA, Matsuda SPT, Bartel B. Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J Biol Chem. 2002;277(23):20446–20452. doi: 10.1074/jbc.M111955200. [DOI] [PubMed] [Google Scholar]
- 12.Rampey RA, et al. A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 2004;135(2):978–988. doi: 10.1104/pp.104.039677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ray PM. Destruction of auxin. Annu Rev Plant Physiol. 1958;9(1):81–118. [Google Scholar]
- 14.Kowalczyk M, Sandberg G. Quantitative analysis of indole-3-acetic acid metabolites in Arabidopsis. Plant Physiol. 2001;127(4):1845–1853. [PMC free article] [PubMed] [Google Scholar]
- 15.Pencík A, et al. Regulation of auxin homeostasis and gradients in Arabidopsis roots through the formation of the indole-3-acetic acid catabolite 2-oxindole-3-acetic acid. Plant Cell. 2013;25(10):3858–3870. doi: 10.1105/tpc.113.114421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandberg G, Ernstsen A. Dynamics of indole-3-acetic acid during germination of Picea abies seeds. Tree Physiol. 1987;3(2):185–192. doi: 10.1093/treephys/3.2.185. [DOI] [PubMed] [Google Scholar]
- 17.Reinecke DM, Bandurski RS. Oxidation of indole-3-acetic acid to oxindole-3-acetic acid by an enzyme preparation from Zea mays. Plant Physiol. 1988;86:868–872. doi: 10.1104/pp.86.3.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu T, Dryhurst G. Electrochemical and peroxidase O2-mediated oxidation of indole-3-acetic acid at physiological pH. J Electroanal Chem. 1997;432(1-2):7–18. [Google Scholar]
- 19.Normanly J. Auxin metabolism. Physiol Plant. 1997;100(3):431–442. [Google Scholar]
- 20.Peer WA, Cheng Y, Murphy AS. Evidence of oxidative attenuation of auxin signalling. J Exp Bot. 2013;64(9):2629–2639. doi: 10.1093/jxb/ert152. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Z, et al. A role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev Cell. 2013;27(1):113–122. doi: 10.1016/j.devcel.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 22.Voß U, et al. The circadian clock rephases during lateral root organ initiation in Arabidopsis thaliana. Nat Commun. 2015;6:7641. doi: 10.1038/ncomms8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butler ED, Gallagher TF. Characterization of auxin-induced ARRO-1 expression in the primary root of Malus domestica. J Exp Bot. 2000;51(351):1765–1766. doi: 10.1093/jexbot/51.351.1765. [DOI] [PubMed] [Google Scholar]
- 24.Porco S, et al. Dioxygenase-encoding AtDAO1 gene controls IAA oxidation and homeostasis in Arabidopsis. Proc Natl Acad Sci USA. 2016;113:11016–11021. doi: 10.1073/pnas.1604375113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mellor N, et al. Dynamic regulation of auxin oxidase and conjugating enzymes AtDAO1 and GH3 modulates auxin homeostasis. Proc Natl Acad Sci USA. 2016;113:11022–11027. doi: 10.1073/pnas.1604458113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dinkel H, et al. ELM 2016--data update and new functionality of the eukaryotic linear motif resource. Nucleic Acids Res. 2016;44(D1):D294–D300. doi: 10.1093/nar/gkv1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Durek P, et al. PhosPhAt: The Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res. 2010;38(Database issue):D828–D834. doi: 10.1093/nar/gkp810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kramer EM, Ackelsberg EM. Auxin metabolism rates and implications for plant development. Front Plant Sci. 2015;6(March):150. doi: 10.3389/fpls.2015.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301(5633):653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 30.Cheng Y, Dai X, Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006;20(13):1790–1799. doi: 10.1101/gad.1415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JI, et al. yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol. 2007;145(3):722–735. doi: 10.1104/pp.107.104935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim JI, et al. YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. J Exp Bot. 2011;62(11):3981–3992. doi: 10.1093/jxb/err094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swanson R, Clark T, Preuss D. Expression profiling of Arabidopsis stigma tissue identifies stigma-specific genes. Sex Plant Reprod. 2005;18(4):163–171. [Google Scholar]
- 34.Schmid M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37(5):501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 35.Winter D, et al. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007;2(8):e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318(5851):801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 37.Takeda S, et al. Physical interaction of floral organs controls petal morphogenesis in Arabidopsis. Plant Physiol. 2013;161(3):1242–1250. doi: 10.1104/pp.112.212084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xuan W, et al. Root cap-derived auxin pre-patterns the longitudinal axis of the Arabidopsis root. Curr Biol. 2015;25(10):1381–1388. doi: 10.1016/j.cub.2015.03.046. [DOI] [PubMed] [Google Scholar]
- 39.Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2(8):755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tashiro S, Tian CE, Watahiki MK, Yamamoto KT. Changes in growth kinetics of stamen filaments cause inefficient pollination in massugu2, an auxin insensitive, dominant mutant of Arabidopsis thaliana. Physiol Plant. 2009;137(2):175–187. doi: 10.1111/j.1399-3054.2009.01271.x. [DOI] [PubMed] [Google Scholar]
- 41.Titapiwatanakun B, et al. ABCB19/PGP19 stabilises PIN1 in membrane microdomains in Arabidopsis. Plant J. 2009;57(1):27–44. doi: 10.1111/j.1365-313X.2008.03668.x. [DOI] [PubMed] [Google Scholar]
- 42.Müller A, Weiler EW. IAA-synthase, an enzyme complex from Arabidopsis thaliana catalyzing the formation of indole-3-acetic acid from (S)-tryptophan. Biol Chem. 2000;381(8):679–686. doi: 10.1515/BC.2000.088. [DOI] [PubMed] [Google Scholar]
- 43.Kriechbaumer V, Botchway SW, Hawes C. Localization and interactions between Arabidopsis auxin biosynthetic enzymes in the TAA/YUC-dependent pathway. J Exp Bot. 2016;67(14):4195–4207. doi: 10.1093/jxb/erw195. [DOI] [PubMed] [Google Scholar]
- 44.Takase T, et al. ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J. 2004;37(4):471–483. doi: 10.1046/j.1365-313x.2003.01973.x. [DOI] [PubMed] [Google Scholar]
- 45.Khan S, Stone JM. Arabidopsis thaliana GH3.9 in auxin and jasmonate cross talk. Plant Signal Behav. 2007;2(6):483–485. doi: 10.4161/psb.2.6.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ. Composite structure of auxin response elements. Plant Cell. 1995;7(10):1611–1623. doi: 10.1105/tpc.7.10.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ottenschläger I, et al. Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc Natl Acad Sci USA. 2003;100(5):2987–2991. doi: 10.1073/pnas.0437936100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 49.Novák O, et al. Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J. 2012;72(3):523–536. doi: 10.1111/j.1365-313X.2012.05085.x. [DOI] [PubMed] [Google Scholar]
- 50.Grefen C, et al. A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 2010;64(2):355–365. doi: 10.1111/j.1365-313X.2010.04322.x. [DOI] [PubMed] [Google Scholar]
- 51.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 52.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Geisler M, et al. Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 2005;44(2):179–194. doi: 10.1111/j.1365-313X.2005.02519.x. [DOI] [PubMed] [Google Scholar]
- 54.Murphy A, Taiz L. Localization and characterization of soluble and plasma membrane aminopeptidase activities in Arabidopsis seedlings. Plant Physiol Biochem. 1999;37(6):431–443. [Google Scholar]
- 55.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 56.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.