Significance
Splice-switching oligonucleotide (SSO) treatment in spinal muscular atrophy (SMA) has quickly become a clinical reality, but without an effective delivery system, the practicalities of delivering SSO therapy efficiently might preclude its widespread use. Our peptide-conjugated SSOs are being designed for clinical trials for the treatment of Duchenne muscular dystrophy. Here, we report advanced phosphorodiamidate oligomer (PMO) internalizing peptide (Pip) peptides that effectively deliver SSOs bodywide and at doses an order-of-magnitude lower than required by naked SSOs in a mouse model of SMA. Furthermore, our peptide-SSO is able to deliver to the CNS of adult mice. This study thus presents an oligonucleotide showing activity in the CNS following a systemic route with peptide delivery.
Keywords: spinal muscular atrophy, survival motor neuron, antisense oligonucleotide, splice switching oligonucleotide, cell-penetrating peptide
Abstract
The development of antisense oligonucleotide therapy is an important advance in the identification of corrective therapy for neuromuscular diseases, such as spinal muscular atrophy (SMA). Because of difficulties of delivering single-stranded oligonucleotides to the CNS, current approaches have been restricted to using invasive intrathecal single-stranded oligonucleotide delivery. Here, we report an advanced peptide-oligonucleotide, Pip6a-morpholino phosphorodiamidate oligomer (PMO), which demonstrates potent efficacy in both the CNS and peripheral tissues in severe SMA mice following systemic administration. SMA results from reduced levels of the ubiquitously expressed survival motor neuron (SMN) protein because of loss-of-function mutations in the SMN1 gene. Therapeutic splice-switching oligonucleotides (SSOs) modulate exon 7 splicing of the nearly identical SMN2 gene to generate functional SMN protein. Pip6a-PMO yields SMN expression at high efficiency in peripheral and CNS tissues, resulting in profound phenotypic correction at doses an order-of-magnitude lower than required by standard naked SSOs. Survival is dramatically extended from 12 d to a mean of 456 d, with improvement in neuromuscular junction morphology, down-regulation of transcripts related to programmed cell death in the spinal cord, and normalization of circulating insulin-like growth factor 1. The potent systemic efficacy of Pip6a-PMO, targeting both peripheral as well as CNS tissues, demonstrates the high clinical potential of peptide-PMO therapy for SMA.
Spinal muscular atrophy (SMA), a leading genetic cause of infant mortality primarily due to lower motor neuron degeneration and progressive muscle weakness, results from loss of the ubiquitous survival motor neuron 1 gene (SMN1) (1, 2). Humans have a second nearly identical copy, SMN2, that differs from SMN1 by a crucial nucleotide transition within exon 7 leading to the predominant generation of an alternative exon 7-excluded transcript and only marginally functional protein (3–7). SMN2 therefore fails to compensate for loss of SMN1 unless sufficient copies are present to generate functional levels of full-length SMN protein (2).
A rational, gene therapy-based approach for SMA uses single-stranded antisense splice-switching oligonucleotides (SSOs) to enhance SMN2 pre-mRNA exon 7 inclusion via steric block of splice regulatory pre-mRNA elements (8). Targeting the intron splice silencer N1 (ISS-N1) site within intron 7, by deletion or SSO-mediated splice switching, improves exon 7 inclusion (9, 10). ISS-N1–targeted SSOs used to treat presymptomatic severely affected neonatal SMA mice, via systemic or intracerebroventricular administration, extend survival from 10 to >100 d (11, 12). Although SSO targeting to the CNS is essential, there is also evidence for a peripheral role for the SMN in SMA (13–24).
Although SSO therapy is currently at an advanced stage of development and one of the most promising approaches for SMA, a major challenge is efficient delivery. The current generation of SSOs does not cross the blood–brain barrier and must be administered by repeated intrathecal injection. Although this mode of administration appears to be safe, nearly a third of treated patients experience the typical side effects associated with lumbar puncture, and patients who develop scoliosis, which frequently occurs in SMA, pose additional challenges associated with lumbar puncture (25). To improve delivery of neutrally charged SSOs dramatically, we have developed an advanced phosphorodiamidate oligomer (PMO) internalizing peptide (Pip) peptide delivery technology. Pip peptides are covalently conjugated (26, 27) and capable of SSO delivery to a variety of adult tissues, including liver, kidney, skeletal muscle, diaphragm, and heart (26, 28–30). Here, we report that a highly active peptide, Pip6a, directly conjugated to a morpholino PMO permits highly efficient systemic delivery, which enhances bodywide SMN expression, including in brain and spinal cord; rescues the phenotype; and dramatically prolongs the life span of severe SMA mice. These data demonstrate powerful SMA disease modification by peptide-PMO therapy, a benefit that could be extended to many other neurodegenerative disorders (8, 31, 32).
Results
Systemic Pip6a-PMO Treatment of Severe SMA Mice Rescues the Disease Phenotype and Enhances Survival.
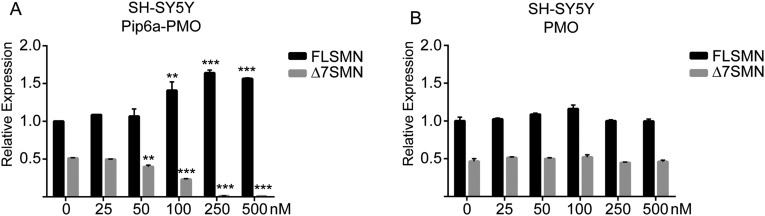
Pip6a, our lead delivery peptide (28, 29), was directly conjugated to a 20-mer PMO sequence targeting the ISS-N1 element of SMN2 intron 7 (12). We evaluated the efficacy of Pip6a-PMOs in vitro using the neuroblastoma cell line SH-SY5Y, which resulted in a significant dose-dependent increase of full-length SMN2 (FLSMN2) expression (Fig. S1A). By contrast, PMO alone failed to increase FLSMN2 expression even at the highest dose of 500 nM (Fig. S1B). Pip6a is thus able to improve cellular uptake of PMO by neurons.
Fig. S1.
Improved uptake of Pip6a-PMO over naked PMO in cultured neuronal cell line SH-SY5Y. Twenty-five nanomolar to 500 nM Pip6a-PMO (A) or naked PMO (B) was added to neuroblastoma SH-SY5Y cells cultured in full media without a transfection reagent. Twenty-four hours postadministration, cells were harvested and RNA was extracted for quantitative RT-PCR analysis of SMN2 transcripts. FLSMN2 and transcripts lacking exon 7 (Δ7SMN2) transcripts were normalized to “total SMN2” transcripts represented by exons 2a and 2b. Untreated FLSMN2 expression in untreated cells (0 nM) was set to 1 and used for comparison. The Δ7SMN2 transcripts were observed at an expression level of 0.5 ± 0.015 relative to FLSMN2 transcripts in untreated cells. (A) Significant reduction in expression was observed in Δ7SMN2 transcripts at 50 nM (0.4 ± 0.04), 100 nM (0.23 ± 0.01), 250 nM (0.013 ± 0.003), and 500 nM (0.01 ± 0.0) concentrations. FLSMN2 expression was significantly increased at 100 nM (1.4 ± 0.2), 250 nM (1.64 ± 0.07), and 500 nM (1.56 ± 0.02) concentrations of Pip6a-PMO. (B) Naked PMO showed no activity in treated cells. Data are represented as mean ± SEM (**P ≤ 0.005; ***P ≤ 0.0005 by Student’s t test relative to untreated expression).
For in vivo studies, we used the Taiwanese severe SMA model (33). Pups were treated at postnatal day 0 (PND0) with a single dose of 10 μg/g of PMO or Pip6a-PMO via the facial vein. Survival was recorded until the humane end point was reached. Untreated and saline-treated SMA mice survived a median of 12 d, as is typical of this model (34) (Table S1). PMO-treated mice showed no change in survival, whereas Pip6a-PMO–treated mice survived a median of 167 d (Fig. 1A and Table S1).
Table S1.
Treatment and survival
| Treatment group | n | Facial vein (μg/g) | Median survival (d) | Mean survival ± SD (d) | Age range (d) | Log-rank (Mantel–Cox) P value | |
| PND0 | PND2 | ||||||
| Single administration | |||||||
| Untreated | 9 | 12 | 11.9 ± 2.0 | 7–14 | |||
| Saline | 4 | 11 | 11.0 ± 1.2 | 10–12 | N.S. | ||
| PMO | 20 | 10 | 11.5 | 12.0 ± 3.1 | 9–22 | N.S. | |
| Pip6a-PMO scrambled | 4 | 10 | 15.5 | 15.5 ± 2.4 | 13–18 | 0.0164 | |
| Pip6a-PMO | 5 | 1 | 10 | 10.4 ± 0.9 | 10–12 | N.S. | |
| Pip6a-PMO | 11 | 2.5 | 33 | 50.6 ± 36.3 | 15–194 | ≤0.0001 | |
| Pip6a-PMO | 11 | 5 | 55 | 103.6 ± 76.8 | 42–232 | ≤0.0001 | |
| Pip6a-PMO | 9 | 10 | 167 | 196.4 ± 114.9 | 79–428 | ≤0.0001 | |
| Double administration | |||||||
| PMO | 7 | 10 | 10 | 54 | 40.1 ± 27.8 | 9–70 | 0.0041 |
| Pip6a-PMO scrambled | 3 | 10 | 10 | 14 | 14.7 ± 4.0 | 11–19 | N.S. |
| Pip6a-PMO | 5 | 5 | 5 | 283 | 283 ± 55.1 | 209–339 | ≤0.0001 |
| Pip6a-PMO | 5 | 10 | 10 | 457 | 436.2 ± 93.4 | 321–557 | ≤0.0001 |
N.S., not significant.
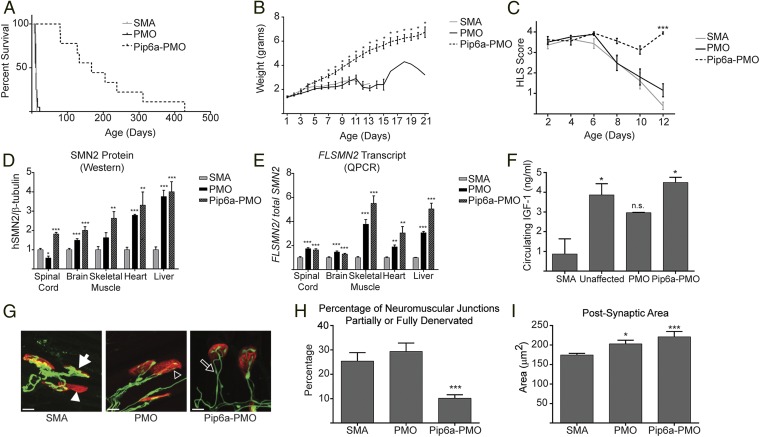
Fig. 1.
Direct comparison of PMO versus Pip6a-PMO administration in severe SMA mice. (A) Survival curves for SMA untreated (n = 9) pups and for SMA pups administered 10 μg/g PMO (n = 20) or 10 μg/g of Pip6a-PMO (n = 9). Pip6a-PMO treatment significantly enhanced survival (mean of 196.4 ± 114.9, median of 167 d) (P ≤ 0.0001, log-rank Mantel–Cox test). (B) Weights of treated and untreated severe SMA pups plotted as mean ± SEM. Pip6a-PMO–treated pups weighed significantly more than SMA untreated pups starting from PND7 (*P ≤ 0.005, Student’s t test). (C) Hind-limb suspension score plotted as mean ± SEM (***P = 3.55 × 10−13, Fisher’s tests with significance after Bonferroni correction). (D) Protein in tissues analyzed by Western blots for human SMN and mouse β-tubulin. SMN expression of untreated SMA pups was normalized to 1 for comparative expression. (E) RNA expression of FLSMN transcript analyzed by quantitative RT-PCR related to “total SMN” transcripts, represented by exon 2a and 2b amplicons. The expression of FLSMN in each untreated SMA tissue was normalized to 1 for comparative expression. (F) Circulating serum levels of IGF1 determined by murine IGF1 ELISA plotted as mean ± SEM. Statistical significance was determined by one-way ANOVA with multiple comparisons (*P ≤ 0.01). n.s., not significant. (G) Representative confocal images of NMJs at PND12. Several defects of the NMJs were observed in SMA untreated and PMO-treated pups, including neurofilament accumulation (white arrow), denervated endplates (white arrowhead), and reduced terminal arborization (open arrowhead). Also observed in all treated and untreated pups, although not quantified, were multiinnervated endplates, indicative of normally developing NMJs (open arrow). Postsynaptic endplates were stained in red (α-bungarotoxin) and neurons were stained in green in neurofilament (2H3) and synaptic vesicles (SV2). (Scale bars, 10 μm.) (H) Percentage of NMJs scored as partially or fully denervated. (I) Postsynaptic areas were quantified using ImageJ. In D, E, H, and I, data are represented as mean ± SEM (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001 by Student’s t test in comparison to untreated SMA mice).
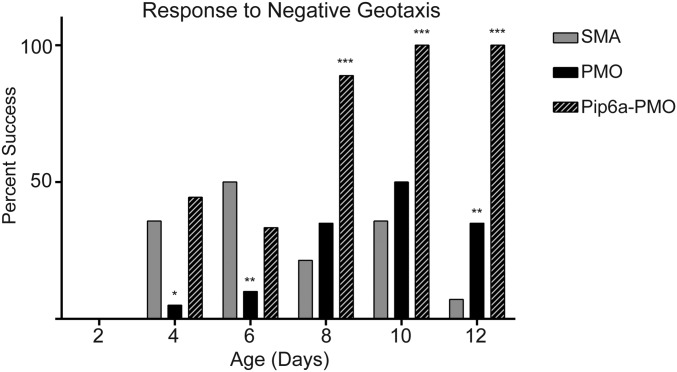
Pip6a-PMO–treated pups were significantly heavier than their untreated or PMO-treated littermates from day 7 (Fig. 1B). Movement and coordination were tested via the natural response to negative geotaxis. A greater percentage of Pip6a-PMO–treated pups were able to turn upright by day 8 compared with either untreated or PMO-treated pups (Fig. S2). Muscle strength was tested by the hind-limb suspension tube test scoring system (35). Pups treated with 10 μg/g of PMO were not significantly different from untreated pups, whereas Pip6a-PMO–treated pups were significantly stronger by day 12 (Fig. 1C).
Fig. S2.
Movement and coordination were tested by placing the pups head down on a 30° inclined plane and scoring their natural response to turn themselves upright. The percentage of pups able to turn themselves upright in reaction to negative geotaxis was greatly enhanced in Pip6a-PMO–treated pups over PMO-treated and untreated pups (*P ≤ 0.01; **P ≤ 0.001; ***P ≤ 0.0001 by binomial test).
Although only Pip6a-PMO–treated pups showed an improved phenotype, both PMO- and Pip6a-PMO–treated pups demonstrated significantly increased FLSMN mRNA and SMN protein expression in all tissues tested (Fig. 1 D and E). Insulin-like growth factor 1 (IGF1) is a neurotrophic factor reduced in severe SMA models (11, 36). Both Pip6a-PMO and PMO treatment rescued IGF1 expression at PND7, with Pip6a-PMO restoring IGF1 to levels found in unaffected littermates (Fig. 1F). Impaired maturation of neuromuscular junctions (NMJs) is a hallmark of SMA, notable by small endplate area and complexity, denervation, poor terminal arborization, and neurofilament accumulation at the presynaptic nerve terminal (37–41). We therefore studied the morphology of NMJs in the transversus abdominis (TVA), one of the most severely affected muscles, between untreated and treated SMA pups at PND12 (42). Here, we observed the strongest difference between treatment groups. Pups treated with Pip6a-PMO had far fewer denervated or partially denervated NMJs, indicative of poor end-terminal arborization, in comparison to PMO-treated or untreated SMA pups (Fig. 1 G and H). Although significantly greater than in untreated SMA pups, both treatments resulted in similar restoration of postsynaptic motor endplate areas (Fig. 1I). In this side-by-side comparison using a low dose of compounds, Pip6a-PMO was clearly better able to correct the SMA phenotype over naked PMO-treated mice.
Pip6a-PMO Treatment Demonstrates Dose-Dependent Phenotypic Rescue and Enhanced Survival.
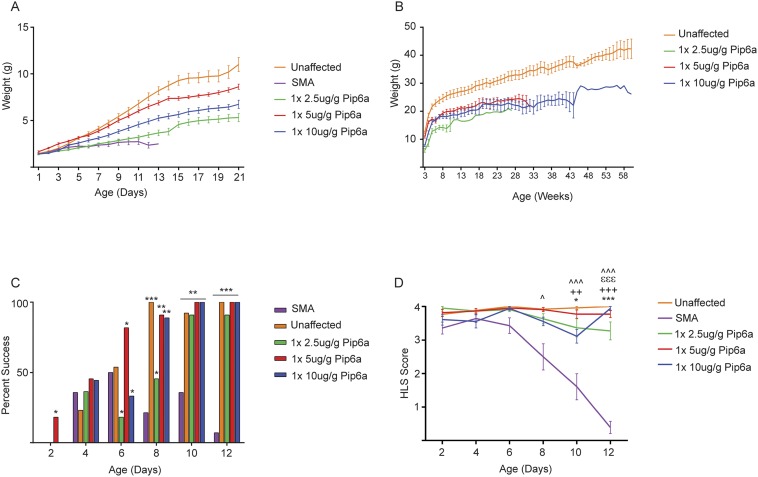
As established above, pups treated with a single dose of 10 μg/g of Pip6a-PMO survived a median of 167 d. We next evaluated lower doses of Pip6a-PMO, demonstrating that survival of treated pups was dose-dependent. Untreated mice survived a median of 12 d, whereas a single dose of 5 μg/g of Pip6a-PMO improved median survival to 55 d (Fig. 2A and Table S1). Furthermore, Pip6a-PMO–treated animals receiving a single 2.5-μg/g dose survived a median of 33 d, whereas no increase in survival was recorded for pups treated with a 1-μg/g dose (Table S1). Reduced doses of 2.5 and 5 μg/g of Pip6a-PMO still improved weight, muscle strength, movement, and coordination over untreated SMA pups (Fig. S3). Delayed onset of peripheral ear and tail necrosis, a characteristic pathological finding in this model, was also found to be dose-dependent (Table S2).
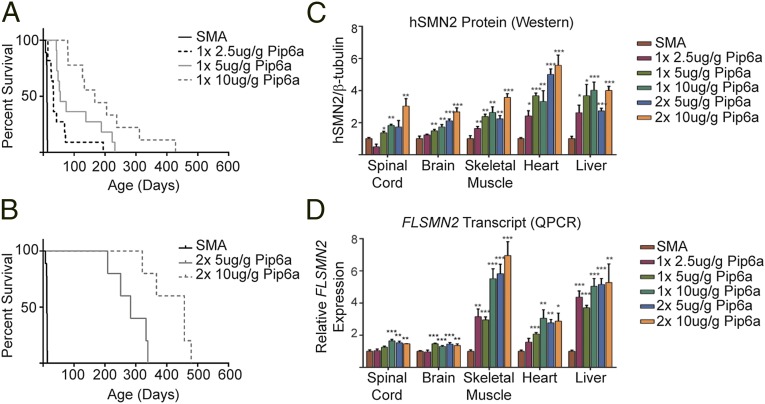
Fig. 2.
Dose-dependent Pip6a-PMO restoration of phenotype. (A) Survival curves for untreated severe SMA mice (n = 9) and for pups treated with a single dose (PND0) of 2.5 μg/g of Pip6a-PMO (n = 11), 5 μg/g of Pip6a-PMO (n = 11), or 10 μg/g of Pip6a-PMO (n = 9). All administrations of Pip6a-PMO resulted in statistically significant improvement of survival over untreated SMA mice (P ≤ 0.0001, log-rank Mantel–Cox text). (B) Survival curves for untreated severe SMA mice (n = 9) and two doses (PND0/2) of 5 μg/g (n = 5) and 10 μg/g of Pip6a-PMO (n = 5). Both administrations with Pip6a-PMO resulted in statistically significant improvement of survival over untreated SMA mice (P ≤ 0.0001, log-rank Mantel–Cox test). (C) Protein expression of SMN in tissues following single PND0 (1×) or double PND0 and PND2 (2×) administration of Pip6a-PMO. SMN protein was analyzed with Western blots for human SMN and mouse β-tubulin. SMN expression of untreated SMA pups was normalized to 1 for comparative expression. (D) FLSMN2 transcript levels were measured by qPCR. Expression of FLSMN2 in each untreated SMA tissue was normalized to 1 for comparative expression. Samples are plotted as mean ± SEM, and statistical significance compared with untreated SMA mice was determined by Student’s t test (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).
Fig. S3.
Improved phenotype after single administration of 2.5, 5, or 10 μg/g of Pip6a-PMO in severe SMA mice. (A) Preweaning weights after administration of Pip6a-PMO are significantly greater than preweaning weights in untreated SMA pups by day 12. Weights were taken daily from PND0 until weaning at PND21 and plotted as mean ± SEM. (B) Weekly weights plotted as mean ± SEM. All pups administered Pip6a-PMO maintained similar weights. (C) Percentage of pups administered a single dose of Pip6a-PMO able to turn themselves upright in reaction to negative geotaxis, similar to their unaffected littermates (*P ≤ 0.05; **P ≤ 0.0001; ***P ≤ 1 × 10−6 determined by binomial test). (D) Hind-limb suspension (HLS) score given to treated, untreated SMA, and unaffected littermates for single administrations (*P ≤ 1 × 10−4; **P ≤ 1 × 10−6; ***P ≤ 1 × 10−8 by Fisher’s tests with significance after Bonferroni correction). Statistical comparisons are shown between SMA untreated and 2.5-μg/g Pip6a-PMO–treated (*), SMA untreated and 5-μg/g Pip6a-PMO–treated (+), SMA untreated and 10-μg/g Pip6a-PMO–treated (ε), and SMA and unaffected (^) littermates.
Table S2.
Age at necrosis
| Treatment group | Facial vein (μg/g) | Mean age at necrosis ± SD (d) | ||||
| PND0 | PND2 | Tail | Ear | Nose | Feet | |
| Single administration | ||||||
| Untreated | NA | NA | NA | NA | ||
| PMO | 10 | NA | NA | NA | NA | |
| Pip6a-PMO scrambled | 10 | NA | NA | NA | NA | |
| Pip6a-PMO | 2.5 | 47 ± 3.8 | 49 ± 5.5 | 59 ± 0.0 | 75 ± 18.4 | |
| Pip6a-PMO | 5 | 49 ± 6.5 | 41 ± 4.9 | 70 ± 7.1 | 111 ± 22.5 | |
| Pip6a-PMO | 10 | 70 ± 9.7 | 47 ± 10 | 71 ± 11 | 193 ± 54.0 | |
| Double administration | ||||||
| PMO | 10 | 10 | 49 (n = 1) | 50 (n = 1) | ||
| Pip6a-PMO scrambled | 10 | 10 | NA | NA | NA | NA |
| Pip6a-PMO | 5 | 5 | 65 ± 11.8 | 50 ± 5.7 | 84 ± 21.4 | 175 (n = 1) |
| Pip6a-PMO | 10 | 10 | 67 ± 0.0 | 55 ± 6.7 | 104 ± 14.8 | 296 ± 67.3 |
NA, not applicable.
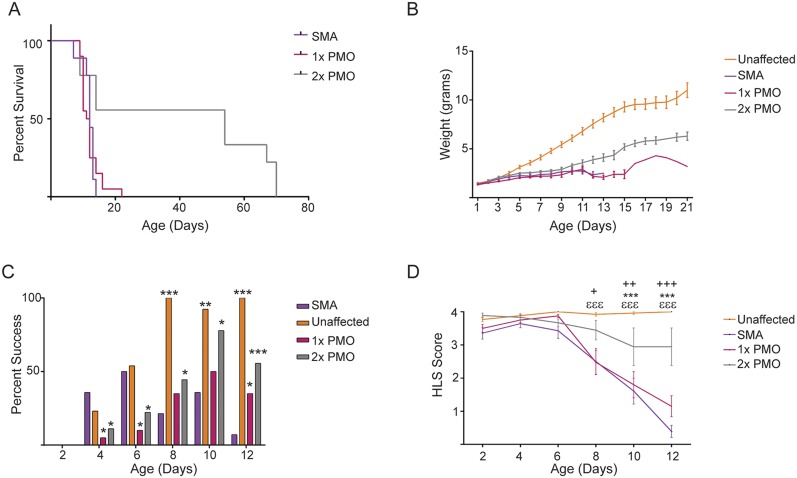
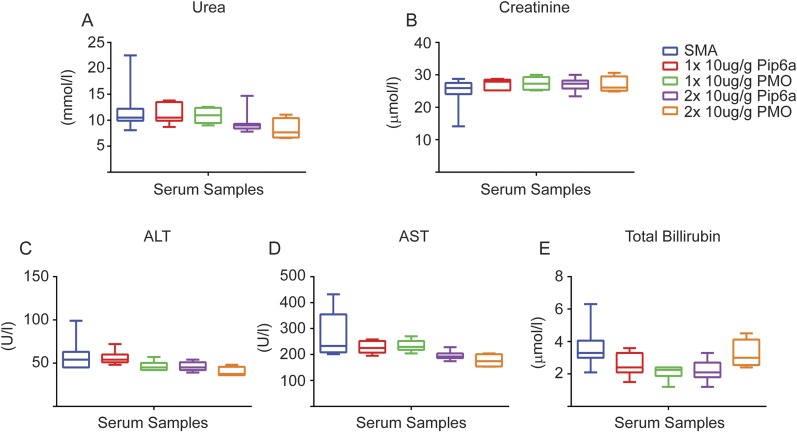
Several seminal studies have reported that using two doses of SSOs confers stronger phenotypic improvements than single doses (11, 43, 44). We therefore treated severe SMA pups at PND0 and PND2 with our most effective doses of Pip6a-PMO (5 and 10 μg/g), as well as 10 μg/g of Pip6a-PMO scrambled and 10 μg/g of naked PMO. Two doses of scrambled Pip6a-PMO showed no improvement on survival (Table S1). Median survival of pups treated with two doses of naked PMO was improved over single-dose treatment (median survival of 54 d and 11.5 d, respectively) (Table S1). The second dose of naked PMO also moderately improved weight and motor function (Fig. S4). By contrast, all mice treated with two doses of 5 or 10 μg/g of Pip6a-PMO survived at least 200 d, with median survivals of 283 and 457 d, respectively, the greatest life span extension published for any SSO-based therapy to date (Fig. 2B and Table S1). Importantly, doubling the doses did not generate a toxic response in these pups. Indeed, analysis of serum markers for liver and kidney toxicity revealed no differences between treated and untreated SMA pups 7 d postadministration (Fig. S5).
Fig. S4.
Early phenotype in SMA severe pups administered one or two doses of 10 μg/g of PMO. (A) Survival curve between untreated SMA pups (n = 9) and pups treated with one (n = 20) or two (n = 7) doses of 10 μg/g of PMO. The two-dose treatment with PMO resulted in a statistically significant improvement of survival over untreated pups (P ≤ 0.005, log-rank Mantel–Cox test). (B) Preweaning weights of treated and untreated pups. Early improvement in phenotype was measured by the percentage of pups able to turn themselves upright in reaction to negative geotaxis (C) (*P ≤ 0.05; **P ≤ 0.0001; ***P ≤ 1 × 10−6 by binomial test) and scoring hind-limb strength from PND2 to PND12 (D) (+P ≤ 1 × 10−4, ++P ≤ 1 × 10−6, +++P ≤ 1 × 10−8, ***P ≤ 1 × 10−8, εεεP ≤ 1 × 10−8, determined by Fisher's tests with significance after Bonferroni correction). Statistical comparisons between SMA untreated and 1 × 10-μg/g PMO-treated (*), SMA untreated and 2 × 10-μg/g PMO-treated (+), and SMA untreated and unaffected (ε) littermates.
Fig. S5.
(A–E) Serum analysis for markers of kidney toxicity (urea and creatinine) as well as markers for liver toxicity [alanine transaminase (ALT), aspartate aminotransferase (AST), and total bilirubin] were analyzed in pups administered one (PND0) or two (PND0/2) doses of 10 μg/g of Pip6a-PMO or PMO. Serum was extracted at PND7. Biochemical analysis revealed no difference between treated and untreated groups.
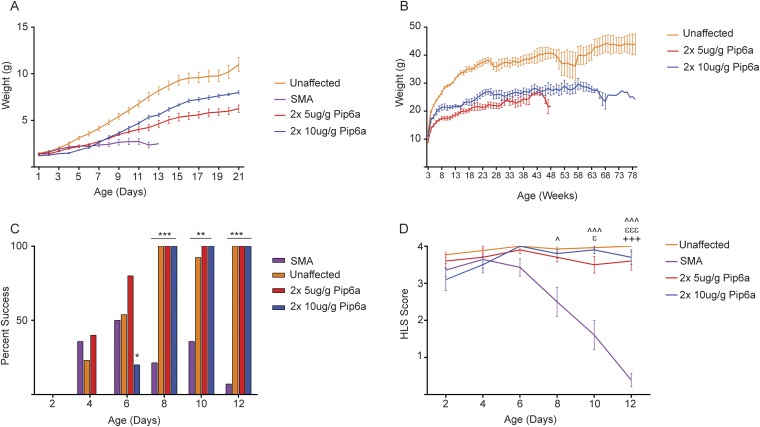
Early assessment of phenotypes after two doses of 5 and 10 μg/g of Pip6a-PMO in SMA pups showed an overall improvement over untreated pups (Fig. S6). Pre- and postweaning weights for Pip6a-PMO–treated animals were greater than for untreated groups, yet never reaching the pre- and postweaning weights of unaffected littermates (Fig. S6 A and B). Response to negative geotaxis was similar between unaffected littermates and Pip6a-PMO–treated pups, with the majority of treated pups completing the task by day 8. A much smaller percentage of untreated SMA pups were able to complete the task, even by day 12 (Fig. S6C). Hind-limb strength of Pip6a-PMO–treated pups was also similar to unaffected littermates (Fig. S6D).
Fig. S6.
Improved phenotype after two doses of 5 or 10 μg/g of Pip6a-PMO in severe SMA mice. (A) Preweaning weights after administration of Pip6a-PMO are greater than preweaning weights of untreated SMA pups by day 12. Weights were taken daily from PND0 until weaning at PND21 and plotted as mean ± SEM. (B) Postweaning weekly weights plotted as mean ± SEM. All pups administered Pip6a-PMO maintained similar weights. (C) Percentage of pups administered two doses of Pip6a-PMO able to turn themselves upright in reaction to negative geotaxis, similar to their unaffected littermates (*P ≤ 0.05; **P ≤ 0.0001; ***P ≤ 1 × 10−6 by binomial test). (D) Hind-limb suspension score given to treated, untreated SMA, and unaffected littermates for single-dose administration (^P ≤ 1 × 10−4, ^^^P ≤ 1 × 10−8, εP ≤ 1 × 10−4, εεεP ≤ 1 × 10−8, +++P ≤ 1 × 10−8 determined by Fisher's tests with significance after Bonferroni correction). Statistical comparisons between (+) SMA untreated and 2 × 5-μg/g Pip6a-PMO–treated, (ε) SMA untreated and 2 × 10-μg/g Pip6a-PMO–treated, and (^) SMA and unaffected littermates.
To elucidate molecular efficacy, SMN mRNA expression and protein expression were determined in treated tissues (Fig. 2 C and D). Spinal cord, brain, lower limb skeletal muscle, heart, and liver were harvested at PND7, and FLSMN2 transcripts were analyzed by quantitative PCR (qPCR), normalized to total SMN2 expression, whereas total SMN protein expression was determined by Western blot. A dose-dependent increase of FLSMN2 mRNA as well as SMN protein was observed in nearly all tissues of mice treated with Pip6a-PMO. These results demonstrate that the efficacy of Pip6a-PMO is dose-dependent and improves with multiple administrations.
Pip6a-PMO Improves NMJ Structure and Innervation.
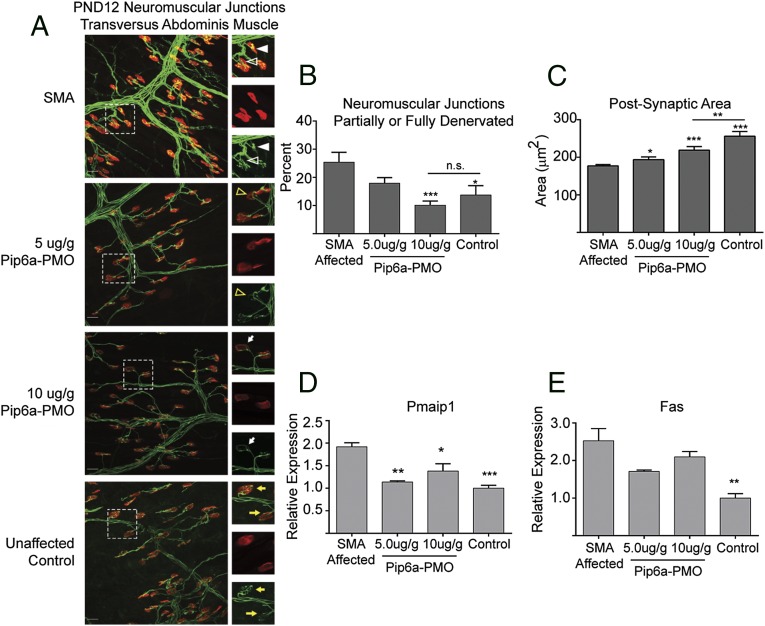
We previously described rescue of NMJ pathology in pups treated with 10 μg/g of Pip6a-PMO. To understand further the dose-dependent improvement on phenotype of Pip6a-PMO–treated animals, we analyzed the innervation and endplate area of NMJs from the TVA of pups treated with the reduced dose of 5 μg/g compared with 10 μg/g of Pip6a-PMO (Fig. 3). Severe SMA mice have greater numbers of partially or fully denervated NMJs (45). A single dose of 10 μg/g rescued NMJs to normal levels of innervation, and NMJs from pups treated with 5 μg/g of Pip6a-PMO also showed an improvement over untreated pups (Fig. 3B). Similarly, postsynaptic endplate areas were significantly greater in both treatment groups compared with SMA-affected mice (Fig. 3C). The extent of NMJ phenotypic correction therefore nicely correlates with life span extension and Pip6a-PMO concentration.
Fig. 3.
Rescue of SMA pathology in Pip6a-PMO–administered SMA pups. (A) Representative confocal images of NMJs from untreated SMA pups, unaffected controls, and pups administered 5 μg/g or 10 μg/g of Pip6a-PMO PND0. Postsynaptic endplates were stained in red (α-bungarotoxin) and neurons were stained in green in neurofilament (2H3) and synaptic vesicles (SV2). (Scale bar, 25 μm.) Reduced innervation (open white triangle) and accumulation of neurofilament (filled white triangle) were observed in SMA-untreated pups. Partial innervation (open yellow triangle) and poor arborization (white arrow) were visible in Pip6a-PMO–treated pups. The optimal complexity of terminal arborizations and fully innervated NMJs is observable in unaffected controls (yellow arrows). (B) NMJs were scored for denervation, either partial or full, and plotted as the percentage of total NMJs analyzed. (C) Postsynaptic areas were quantified using ImageJ. Quantitative levels of markers for apoptosis are shown: Pmaip1 (D) and Fas (E) relative transcripts in spinal cord of PND7-treated and untreated mice, normalized to mGapdh transcript levels. In B–E, data are represented as mean ± SEM (*P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.0005 by Student’s t test in comparison to untreated SMA mice).
Loss of motor neurons in the spinal cord appears to be a late feature of SMA mouse models, and there are no universally accepted methods for quantifying motor neuron loss, hindering consistency across different studies. The health of neurons in the spinal cord was thus demonstrated by measuring two transcripts for apoptosis, TNF receptor superfamily member 6 (fas) and phorbol-12-myristate-13-acetate-induced protein 1 (Pmaip1), which are dysregulated in the spinal cord of SMA mice (46). At PND7, spinal cords from unaffected littermates expressed significantly lower transcript levels of fas and Pmaip1 than spinal cords from SMA-affected pups (Fig. 3 D and E). Treatment with a single dose of 5 or 10 μg/g of Pip6a-PMO reduced their transcript levels, indicating that early treatment protects against programmed cell death within the spinal cord.
Pip6a Delivers SSO to the CNS in Adult Mice.
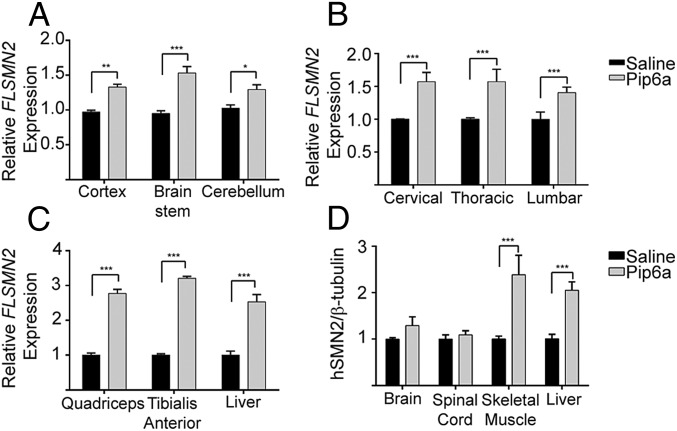
Although we have achieved systemic delivery in early postnatal pups, targeting the CNS of patients at all ages will be critical for a successful SMA therapy. To test this objective, we chose to use adult mice carrying the human SMN2 transgene (Smn1tm1Hung/WT;SMN2tg/tg). These mice are indistinguishable from wild-type animals. At 7.5 wk of age, mice were treated twice by tail vein administration 2 d apart with 18 μg/g of Pip6a-PMO or saline. Seven days postadministration, tissues of the CNS and relevant peripheral tissues were harvested and SMN expression was assessed. Transcript levels of FLSMN2 relative to total SMN2 transcript levels were significantly increased in all parts of the brain and spinal cord, as well as in skeletal muscle and liver (Fig. 4 A–C). SMN protein levels were augmented in skeletal muscles and liver only (Fig. 4D). Suboptimal sensitivity in the protein detection method may have limited the observation of increased levels in CNS tissues, which are predicted to be lower than the levels observed in peripheral tissues. This study demonstrates that our peptide delivery of antisense oligonucleotides causing changes in transcripts within the adult CNS.
Fig. 4.
Pip6a-PMO administration in unaffected adult mice harboring the human SMN2 allele (Smn1tm1Hung/WT;SMN2tg/tg). (A–C) Quantitative RT-PCR from tissues of mice administered 18 mg/kg, i.v. 2 d apart. Tissues from brain (A), spinal cord (B), and peripheral skeletal muscles and liver (C) were harvested 7 d postadministration. In all tissues, FLSMN2 expression was significantly increased over saline-treated mice. (D) Protein expression of SMN in tissues. Separate tissues of spinal cord and skeletal muscles were combined. SMN protein was analyzed with Western blots for human SMN and mouse β-tubulin. Data are represented as mean ± SEM (*P ≤ 0.005; **P ≤ 0.001; ***P ≤ 0.0001 by Student’s t test in comparison to saline-treated mice).
Discussion
In developing an effective SMA therapy, it is important to consider the CNS and peripheral requirements for SMN restoration. Multiple studies in SMA mouse models have generated conflicting data regarding the importance of systemic versus CNS-only treatment (11, 12, 43, 44, 47, 48). The single most important pathological finding for SMA is the loss of lower motor neurons, and denervation at the NMJ is the earliest pathological change in SMA mice (37–41). However, reduced SMN expression has also been observed to cause skeletal muscle (13, 14, 16) and vascular system (19, 20, 49) defects in SMA mice, and SMN is involved in general cellular functions, including small nuclear ribonucleic protein (snRNP) biogenesis (13, 22–24) and glucose metabolism (17, 18, 21). Thus, although delivery to the CNS is primordial for SMA therapy, combined targeting of both the CNS and the periphery has the potential of being the optimal approach. We have shown that Pip6a-conjugated PMO heralds a potent therapeutic option combining the genetic precision of SSOs with the systemic delivery efficacy of a small molecule. Systemic Pip6a-PMO treatment demonstrates ultrahigh potency, with only a single 10-μg/g dose at PND0 yielding a median survival of 167 d in severe SMA pups and the longest lived mouse surviving 428 d, which is far beyond the greatest extension recorded for naked PMO (12) (Fig. 2A and Table S1). This dramatic extension in survival is associated with increased SMN expression in brain, spinal cord, and all examined peripheral tissues; rescued levels of circulating IGF1; reduced NMJ denervation; and decreased expression of apoptotic markers in the spinal cord (Figs. 1 and 3). Furthermore, we were able to enhance median survival to over 450 d with two 10-μg/g doses (Fig. 2B and Table S1). Significantly enhanced survival in severe SMA models has been reported using systemic administration of SSOs targeting the ISS-N1 site. The best survival data published to date include reports by Hua et al. (11), who reported a median survival of 248 d, and by Zhou et al. (44), who reported a median survival of 261 d. However, the former administered two doses of 160 mg/kg and the latter used a single dose of 40 mg/kg. Pip6a delivery allows us to reduce the dose required for maximum survival by at least fourfold, an important step for generating clinically relevant compounds.
Current clinical trials (Ionis Pharmaceuticals) use intrathecal delivery of a 2'-O-methoxyethyl (2'-MOE) SSO, primarily targeting SMN expression in the CNS. In an open-label phase 2 study, infants with SMA were treated with SSO targeting ISS-N1 (Nusinerson, IONIS-SMNRX), which modestly improved their median event-free age (age of permanent ventilation or death) (50). More encouragingly, treated infants showed improvement in muscle function and motor milestones. In a separate phase 2 study in children with SMA, the same SSO resulted in improved mobility [6-min walk test) (51), upper limb mobility test (52), and Hammersmith functional motor scale-expanded function (53)] compared with pretreatment functions. F. Hoffman-La Roche in Switzerland, PTC Therapeutics, and the SMA Foundation have partnered to develop a small-molecule drug (RG7800) capable of positively influencing exon 7 splicing systemically. This trial, termed Moonfish, has been suspended because of safety concerns in treated animals. However, Roche has a new similarly acting drug (RG7916) for which they have initiated a phase 1 study to investigate safety, tolerability, pharmacokinetics, and pharmacodynamics in healthy volunteers. A related trial with a Novartis-sponsored orally active small molecule that enhances SMN2 splicing, elevates full-length SMN protein, and extends survival in a severe SMA mouse model is also underway, sponsored by Novartis. The molecular mechanism of action of this drug is via stabilization of the transient double-stranded RNA structure formed by the SMN2 pre-mRNA and U1 snRNP complex (54). It will be of great interest to compare results of the CNS restoration of SMN protein (intrathecal SSO) with the outcome of the combined peripheral and CNS restoration of SMN protein level, because these trials could provide the greatest indication in patients of the need for a systemic and/or CNS treatment.
Although a previous study observed brain delivery of antisense oligonucleotides with tagged dyes (55), we report the bona fide activity of a CNS-peptide–delivered SSO. We have demonstrated here the potent impact of peptide delivery on PMO SSO therapy. In addition to SMA, other diseases for antisense oligonucleotide therapies include ALS (56, 57), Huntington’s disease (58, 59), and Parkinson’s disease (60, 61), which all ideally require both systemic and CNS SSO delivery. Future work will focus on extending further the clinical applications of Pip-PMOs.
Materials and Methods
Synthesis of Peptide-PMO Conjugates.
Pip6a Ac-(RXRRBRRXRYQFLIRXRBRXRB)-COOH was synthesized and conjugated to PMO as described previously (28). The PMO sequence targeting ISS-N1 intron 7 (-10-27) (5′-ATTCACTTTCATAATGCTGG-3′) and PMO scrambled (5′-ATTGTCTATCAAAATCCTGC-3′) was purchased from Gene Tools LLC.
In Vitro Study.
Human neuroblastoma SH-SY5Y cells were treated using 25, 50, 100, 250, and 500 nM oligonucleotides in 500 μL of full-serum DMEM and analyzed for FLSMN2 mRNA expression 24 h later.
Animal Models.
Experiments were carried out in the Biomedical Sciences Unit, University of Oxford, according to procedures authorized by the UK Home Office. SMA-like mouse strain FVB.Cg-Smn1tm1HungTg(SMN2)2Hung/J was generated and maintained as previously described (33, 34). The intravenous injections were performed by single (PND0) or double (PND0 and PND2) administration via the facial vein. Doses of 2.5 μg/g (0.25 nmol/g), 5.0 μg/g (0.5 nmol/g), and 10 μg/g (1 nmol/g) of Pip6a-PMO and 10 μg/g of PMO (1.48 nmol/g) were diluted in 0.9% saline and given at a volume of 5 μL/g of body weight. Treated mice were given mash and jelly as food supplementation. Tail vein administration in adult mice was performed in unaffected mice (Smn1tm1Hung/WT;SMN2tg/tg) at 7.5 wk of age. Two doses of 18 μg/g of Pip6a-PMO or saline were given 2 d apart. Tissues were harvested 7 d postadministration.
Measuring Early Disease Phenotype.
Weights were taken daily, and overall health was assessed. Following the humane end point, mice were euthanized according to the approved procedure of rising CO2. Negative geotaxis was performed as previously described (35). Statistical significance was determined by a binomial test. The hind-limb suspension tube test was performed as described in procedures for preclinical studies by Translational Research in Europe – Assessment & Treatment of Neuromuscular Diseases (35). For the purposes of reproducibility, factors such as leg pulls and time were left out of the analysis. Statistical significance was determined by Fisher’s test with Bonferroni correction in SPSS.
NMJ Immunohistochemistry.
Animals treated with Pip6a-PMO were harvested 12 d postadministration (PND12). The TVA muscle was isolated, and immunohistochemistry was performed on NMJs as previously described (42, 62). NMJs were imaged using an Olympus FV1000 confocal microscope. Images were taken with a 40× objective as a z-stack. Only NMJs with fully observable postsynapses were used for denervation counts. More than 300 NMJs were analyzed for full or partial denervation per group, which was observed by the colocalization of presynaptic neurons with postsynaptic acetylcholine receptors. The area of postsynaptic motor endplates was measured using ImageJ (NIH) as previously published (63).
qPCR.
RNA extraction from harvested tissues was carried out using TRIzol reagent (Invitrogen), and cDNA was generated using an ABI High Capacity cDNA Reverse Transcription Kit (Invitrogen) following the manufacturer’s instructions. A qPCR reaction using Power SYBR Green Master Mix (Life Technologies) was performed and analyzed on an Applied Biosystems StepOnePlusTM real-time PCR system (Life Technologies). FLSMN2 and total SMN2 transcripts were amplified using gene-specific primers (Table S3). Fas, Pmaip1, and Gapdh were amplified using an Integrated DNA Technologies-derived probe and primer set (Table S3).
Table S3.
Probe and primers used for quantitative real-time PCR
| mRNA | Forward primer (5′ to 3′) | Reverse primer (5′′ to 3′) | Probe (5′ to 3′) |
| FLSMN2 | GCTTTGGGAAGTATGTTAATTTCA | CTATGCCAGCATTTCTCCTTAATT | SYBR Green |
| Total SMN2 | GCGATGATTCTGACATTTGG | GGAAGCTGCAGTATTCTTCT | SYBR Green |
| Fas | ACCAGACTTCTACTGCGATTC | GTTTGTATTGCTGGTTGCTGT | CCTCGTGTGAACATGGAACCCTTGA |
| Pmaip1 | CCGGACATAACTGTGGTTCT | GCACACTCGTCCTTCAAGT | TCATCCTGCTCTTTTGCGACTTCCC |
| Gapdh | AATGGTGAAGGTCGGTGTG | GTGGAGTCATACTGGAACATGTAG | TGCAAATGGCAGCCCTGGTG |
Protein Extraction and Western Blot.
Protein was harvested from ∼300 mg of tissue homogenized into radioimmunoprecipitation buffer with complete mini-proteinase inhibitors (Roche). Protein (30–40 µg) was probed for human SMN protein using anti-SMN, clone SMN-KH monoclonal IgG1 (Millipore), and mouse β-tubulin utilizing anti–β-actin monoclonal IgG2a (Sigma) and secondary antibody IRDye 800CW goat anti-mouse IgG (LI-COR Biosciences). Membranes were imaged on a LI-COR Odyssey FC imager and analyzed with Image StudioTM software (LI-COR Biosciences).
IGF1 ELISA.
Concentration of serum IGF1 was determined using a Murine IGF1 ELISA kit (900-K170; Peprotech) following the manufacturer’s instructions.
Clinical Biochemistry.
Serum samples from mice administered Pip6a-PMO or PMO at PND0 and PND0/2 were extracted from the jugular vein upon harvest at PND7, and analysis of toxicity biomarkers was performed by a clinical pathology laboratory (Mary Lyon Centre, Medical Research Council, Harwell, UK).
Acknowledgments
The spinal muscular atrophy (SMA)-like mouse strain was obtained through a material transfer agreement. Work in the laboratory of M.J.G. was supported by Medical Research Council (MRC) Program U105178803. F.S. is a recipient of a C. J. Martin Fellowship from the Australian National Health and Medical Research Council and also acknowledges support by the Bethlehem Griffith Research Foundation (BGRF1501). Work in the laboratory of M.J.A.W. was supported by the MRC Program G0900887 and SMA Europe. F.M. is supported by the National Institute for Health Research Biomedical Research Centre at Great Ormond Street Hospital for Children National Health Service Foundation Trust and University College London. M.B. is an SMA Trust Career Development Fellow.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605731113/-/DCSupplemental.
References
- 1.Lefebvre S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80(1):155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 2.Wirth B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA) Hum Mutat. 2000;15(3):228–237. doi: 10.1002/(SICI)1098-1004(200003)15:3<228::AID-HUMU3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 3.Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet. 2002;30(4):377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 4.Kashima T, Manley JL. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet. 2003;34(4):460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 5.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci USA. 1999;96(11):6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monani UR, et al. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999;8(7):1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 7.Le TT, et al. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet. 2005;14(6):845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 8.Hammond SM, Wood MJ. Genetic therapies for RNA mis-splicing diseases. Trends Genet. 2011;27(5):196–205. doi: 10.1016/j.tig.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Singh NK, Singh NN, Androphy EJ, Singh RN. Splicing of a critical exon of human survival motor neuron is regulated by a unique silencer element located in the last intron. Mol Cell Biol. 2006;26(4):1333–1346. doi: 10.1128/MCB.26.4.1333-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hua Y, Vickers TA, Baker BF, Bennett CF, Krainer AR. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007;5(4):e73. doi: 10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hua Y, et al. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478(7367):123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porensky PN, et al. A single administration of morpholino antisense oligomer rescues spinal muscular atrophy in mouse. Hum Mol Genet. 2012;21(7):1625–1638. doi: 10.1093/hmg/ddr600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braun S, Croizat B, Lagrange MC, Warter JM, Poindron P. Constitutive muscular abnormalities in culture in spinal muscular atrophy. Lancet. 1995;345(8951):694–695. doi: 10.1016/s0140-6736(95)90869-2. [DOI] [PubMed] [Google Scholar]
- 14.Rajendra TK, et al. A Drosophila melanogaster model of spinal muscular atrophy reveals a function for SMN in striated muscle. J Cell Biol. 2007;176(6):831–841. doi: 10.1083/jcb.200610053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bogdanik LP, et al. Systemic, postsymptomatic antisense oligonucleotide rescues motor unit maturation delay in a new mouse model for type II/III spinal muscular atrophy. Proc Natl Acad Sci USA. 2015;112(43):E5863–E5872. doi: 10.1073/pnas.1509758112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayhurst M, Wagner AK, Cerletti M, Wagers AJ, Rubin LL. A cell-autonomous defect in skeletal muscle satellite cells expressing low levels of survival of motor neuron protein. Dev Biol. 2012;368(2):323–334. doi: 10.1016/j.ydbio.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowerman M, et al. Glucose metabolism and pancreatic defects in spinal muscular atrophy. Ann Neurol. 2012;72(2):256–268. doi: 10.1002/ana.23582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowerman M, et al. Defects in pancreatic development and glucose metabolism in SMN-depleted mice independent of canonical spinal muscular atrophy neuromuscular pathology. Hum Mol Genet. 2014;23(13):3432–3444. doi: 10.1093/hmg/ddu052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Araujo Ap, Araujo M, Swoboda KJ. Vascular perfusion abnormalities in infants with spinal muscular atrophy. J Pediatr. 2009;155(2):292–294. doi: 10.1016/j.jpeds.2009.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rudnik-Schöneborn S, et al. Digital necroses and vascular thrombosis in severe spinal muscular atrophy. Muscle Nerve. 2010;42(1):144–147. doi: 10.1002/mus.21654. [DOI] [PubMed] [Google Scholar]
- 21.Davis RH, Miller EA, Zhang RZ, Swoboda KJ. Responses to fasting and glucose loading in a cohort of well children with spinal muscular atrophy type II. J Pediatr. 2015;167(6):1362–1368.e1. doi: 10.1016/j.jpeds.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Q, Fischer U, Wang F, Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90(6):1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- 23.Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90(6):1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 24.Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298(5599):1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- 25.Haché M, et al. Intrathecal injections in children with spinal muscular atrophy: Nusinersen clinical trial experience. J Child Neurol. 2016;31(7):899–906. doi: 10.1177/0883073815627882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jearawiriyapaisarn N, et al. Sustained dystrophin expression induced by peptide-conjugated morpholino oligomers in the muscles of mdx mice. Mol Ther. 2008;16(9):1624–1629. doi: 10.1038/mt.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lebleu B, et al. Cell penetrating peptide conjugates of steric block oligonucleotides. Adv Drug Deliv Rev. 2008;60(4-5):517–529. doi: 10.1016/j.addr.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Betts C, et al. Pip6-PMO, a new generation of peptide-oligonucleotide conjugates with improved cardiac exon skipping activity for DMD treatment. Mol Ther Nucleic Acids. 2012;1(8):e38. doi: 10.1038/mtna.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Betts CA, et al. Implications for cardiac function following rescue of the dystrophic diaphragm in a mouse model of Duchenne muscular dystrophy. Sci Rep. 2015;5:11632. doi: 10.1038/srep11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin H, et al. Pip5 transduction peptides direct high efficiency oligonucleotide-mediated dystrophin exon skipping in heart and phenotypic correction in mdx mice. Mol Ther. 2011;19(7):1295–1303. doi: 10.1038/mt.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osorio FG, et al. Splicing-directed therapy in a new mouse model of human accelerated aging. Sci Transl Med. 2011;3(106):106ra107. doi: 10.1126/scitranslmed.3002847. [DOI] [PubMed] [Google Scholar]
- 32.Disterer P, et al. Development of therapeutic splice-switching oligonucleotides. Hum Gene Ther. 2014;25(7):587–598. doi: 10.1089/hum.2013.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsieh-Li HM, et al. A mouse model for spinal muscular atrophy. Nat Genet. 2000;24(1):66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- 34.Gogliotti RG, Hammond SM, Lutz C, Didonato CJ. Molecular and phenotypic reassessment of an infrequently used mouse model for spinal muscular atrophy. Biochem Biophys Res Commun. 2010;391(1):517–522. doi: 10.1016/j.bbrc.2009.11.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.El-Khodor BF, et al. Identification of a battery of tests for drug candidate evaluation in the SMNDelta7 neonate model of spinal muscular atrophy. Exp Neurol. 2008;212(1):29–43. doi: 10.1016/j.expneurol.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 36.Murdocca M, et al. IPLEX administration improves motor neuron survival and ameliorates motor functions in a severe mouse model of spinal muscular atrophy. Mol Med. 2012;18:1076–1085. doi: 10.2119/molmed.2012.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cifuentes-Diaz C, et al. Neurofilament accumulation at the motor endplate and lack of axonal sprouting in a spinal muscular atrophy mouse model. Hum Mol Genet. 2002;11(12):1439–1447. doi: 10.1093/hmg/11.12.1439. [DOI] [PubMed] [Google Scholar]
- 38.Kariya S, et al. Reduced SMN protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy. Hum Mol Genet. 2008;17(16):2552–2569. doi: 10.1093/hmg/ddn156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kong L, et al. Impaired synaptic vesicle release and immaturity of neuromuscular junctions in spinal muscular atrophy mice. J Neurosci. 2009;29(3):842–851. doi: 10.1523/JNEUROSCI.4434-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGovern VL, Gavrilina TO, Beattie CE, Burghes AH. Embryonic motor axon development in the severe SMA mouse. Hum Mol Genet. 2008;17(18):2900–2909. doi: 10.1093/hmg/ddn189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin TL, et al. Selective neuromuscular denervation in Taiwanese severe SMA mouse can be reversed by morpholino antisense oligonucleotides. PLoS One. 2016;11(4):e0154723. doi: 10.1371/journal.pone.0154723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murray LM, et al. Selective vulnerability of motor neurons and dissociation of pre- and post-synaptic pathology at the neuromuscular junction in mouse models of spinal muscular atrophy. Hum Mol Genet. 2008;17(7):949–962. doi: 10.1093/hmg/ddm367. [DOI] [PubMed] [Google Scholar]
- 43.Nizzardo M, et al. Effect of combined systemic and local morpholino treatment on the spinal muscular atrophy Δ7 mouse model phenotype. Clin Therapeutics. 2014;36(3):340–356.e5. doi: 10.1016/j.clinthera.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 44.Zhou H, et al. Repeated low doses of morpholino antisense oligomer: An intermediate mouse model of spinal muscular atrophy to explore the window of therapeutic response. Hum Mol Genet. 2015;24(22):6265–6277. doi: 10.1093/hmg/ddv329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riessland M, et al. SAHA ameliorates the SMA phenotype in two mouse models for spinal muscular atrophy. Hum Mol Genet. 2010;19(8):1492–1506. doi: 10.1093/hmg/ddq023. [DOI] [PubMed] [Google Scholar]
- 46.Murray LM, Beauvais A, Gibeault S, Courtney NL, Kothary R. Transcriptional profiling of differentially vulnerable motor neurons at pre-symptomatic stage in the Smn (2b/-) mouse model of spinal muscular atrophy. Acta Neuropathol Commun. 2015;3:55. doi: 10.1186/s40478-015-0231-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hua Y, et al. Motor neuron cell-nonautonomous rescue of spinal muscular atrophy phenotypes in mild and severe transgenic mouse models. Genes Dev. 2015;29(3):288–297. doi: 10.1101/gad.256644.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou H, et al. A novel morpholino oligomer targeting ISS-N1 improves rescue of severe spinal muscular atrophy transgenic mice. Hum Gene Ther. 2013;24(3):331–342. doi: 10.1089/hum.2012.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Somers E, et al. Vascular defects and spinal cord hypoxia in spinal muscular atrophy. Ann Neurol. 2016;79(2):217–230. doi: 10.1002/ana.24549. [DOI] [PubMed] [Google Scholar]
- 50.Walke DW, Blackley A. 2014 Isis Pharmaceuticals reports data from ISIS-SMN Rx phase 2 studies in infants and children with spinal muscular atrophy. Available at ir.ionispharma.com/phoenix.zhtml?c=222170&p=irol-newsArticle&ID=1976144. Accessed August 26, 2016.
- 51.Montes J, et al. Weakness and fatigue in diverse neuromuscular diseases. J Child Neurol. 2013;28(10):1277–1283. doi: 10.1177/0883073813493663. [DOI] [PubMed] [Google Scholar]
- 52.Mazzone E, et al. Assessing upper limb function in nonambulant SMA patients: Development of a new module. Neuromuscul Disord. 2011;21(6):406–412. doi: 10.1016/j.nmd.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 53.O’Hagen JM, et al. An expanded version of the Hammersmith Functional Motor Scale for SMA II and III patients. Neuromuscul Disord. 2007;17(9-10):693–697. doi: 10.1016/j.nmd.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 54.Palacino J, et al. SMN2 splice modulators enhance U1-pre-mRNA association and rescue SMA mice. Nat Chem Biol. 2015;11(7):511–517. doi: 10.1038/nchembio.1837. [DOI] [PubMed] [Google Scholar]
- 55.Du L, et al. Arginine-rich cell-penetrating peptide dramatically enhances AMO-mediated ATM aberrant splicing correction and enables delivery to brain and cerebellum. Hum Mol Genet. 2011;20(16):3151–3160. doi: 10.1093/hmg/ddr217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lagier-Tourenne C, et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci USA. 2013;110(47):E4530–E4539. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith RA, et al. Antisense oligonucleotide therapy for neurodegenerative disease. J Clin Invest. 2006;116(8):2290–2296. doi: 10.1172/JCI25424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Skotte NH, et al. Allele-specific suppression of mutant huntingtin using antisense oligonucleotides: Providing a therapeutic option for all Huntington disease patients. PLoS One. 2014;9(9):e107434. doi: 10.1371/journal.pone.0107434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Southwell AL, et al. In vivo evaluation of candidate allele-specific mutant huntingtin gene silencing antisense oligonucleotides. Mol Ther. 2014;22(12):2093–2106. doi: 10.1038/mt.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kalbfuss B, Mabon SA, Misteli T. Correction of alternative splicing of tau in frontotemporal dementia and parkinsonism linked to chromosome 17. J Biol Chem. 2001;276(46):42986–42993. doi: 10.1074/jbc.M105113200. [DOI] [PubMed] [Google Scholar]
- 61.Monti B, et al. Alpha-synuclein protects cerebellar granule neurons against 6-hydroxydopamine-induced death. J Neurochem. 2007;103(2):518–530. doi: 10.1111/j.1471-4159.2007.04778.x. [DOI] [PubMed] [Google Scholar]
- 62.Sleigh JN, Grice SJ, Burgess RW, Talbot K, Cader MZ. Neuromuscular junction maturation defects precede impaired lower motor neuron connectivity in Charcot-Marie-Tooth type 2D mice. Hum Mol Genet. 2014;23(10):2639–2650. doi: 10.1093/hmg/ddt659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sleigh JN, Burgess RW, Gillingwater TH, Cader MZ. Morphological analysis of neuromuscular junction development and degeneration in rodent lumbrical muscles. J Neurosci Methods. 2014;227:159–165. doi: 10.1016/j.jneumeth.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]