Significance
HdeA and HdeB are essential chaperones for many Gram-negative enteric bacteria to survive acid stress. How they cooperate to protect a broad range of client proteins from acid denaturation while avoiding nonspecific binding remains elusive. Here we developed a comparative proteomic strategy combining genetically encoded releasable photocross-linking with 2D difference gel electrophoresis, which allows an unbiased side-by-side comparison of the entire client pools from these two acid-activated chaperones in Escherichia coli. Our results reveal the existence of client specificities between these chaperones, and this pH-regulated client specificity mechanism demonstrates their seemingly paradoxical specific but promiscuous client recognition. Our comparative proteomic strategy provides a powerful platform for profiling protein–protein interactions under harsh conditions and for elucidating the otherwise elusive mode of action within native cellular contexts.
Keywords: comparative proteomics, conditionally disordered chaperones, bacteria acid resistance, 2D-DIGE, photocross-linking
Abstract
HdeA and HdeB constitute the essential chaperone system that functions in the unique periplasmic space of Gram-negative enteric bacteria to confer acid resistance. How this two-chaperone machinery cooperates to protect a broad range of client proteins from acid denaturation while avoiding nonspecific binding during bacterial passage through the highly acidic human stomach remains unclear. We have developed a comparative proteomic strategy that combines the genetically encoded releasable protein photocross-linker with 2D difference gel electrophoresis, which allows an unbiased side-by-side comparison of the entire client pools from these two acid-activated chaperones in Escherichia coli. Our results reveal distinct client specificities between HdeA and HdeB in vivo that are determined mainly by their different responses to pH stimulus. The intracellular acidity serves as an environmental cue to determine the folding status of both chaperones and their clients, enabling specific chaperone–client binding and release under defined pH conditions. This cooperative and synergistic mode of action provides an efficient, economical, flexible, and finely tuned protein quality control strategy for coping with acid stress.
The highly acidic human stomach (pH <3) is a natural barrier against many enteric bacterial pathogens, which must survive through this hostile environment to establish infection in intestine (1–3). Thus, these gut-resident pathogens, such as Escherichia coli and Shigella, have evolved extensive protection mechanism to cope with this extreme acidity (1, 2). In the cytosolic space, at least four well-studied acid-resistance systems have been developed to keep the internal pH above a dangerous level, including the oxidative (AR1), glutamate-dependent (AR2), arginine-dependent (AR3), and lysine-dependent (AR4) acid-resistance systems (4). In contrast, proteins residing in the ATP-deprived periplasmic space are more vulnerable to acid attack, owing to the highly permeable nature of the outer membrane of these Gram-negative bacteria.
Two homologous acid stress chaperones, HdeA (5) and HdeB (6), have been found to preserve proteome homeostasis in periplasm, the pH of which can drop to the same level as the extracellular gastric acid environment (pH < 3). Noteworthy, the HdeA and HdeB genes are regulated by a single promoter (7) and are well-conserved in various serotypes and pathotypes of E. coli, Shigella flexneri, and Brucella abortus (2). This hdeAB operon is highly expressed in response to acid stress (8), whereas disruption of these genes renders such bacteria extremely vulnerable to acid (5, 9, 10). Determining how these two acid chaperones cooperate to protect a broad range of periplasmic client proteins from acid denaturation during bacterial passage through the human stomach is central to understanding their acid-resistance mechanism.
Both HdeA and HdeB are well-folded “inactive” dimers at neutral pH that on acid stress become partially unfolded monomers with chaperoning capability (11, 12). This stress-specific unfolding mechanism enables the exposure of these monomers’ hydrophobic surfaces to interact with an array of client proteins in a promiscuous fashion (13–15); however, this unique disorder-triggered interaction between chaperones and clients makes direct characterization of the recognition mechanism very challenging, especially within native cellular contexts. HdeA has been recognized as the major acid chaperone, whereas HdeB has a smaller hydrophobic surface with weaker chaperoning activity in vitro (6). In particular, although HdeB has been suggested to have certain functions together with HdeA at pH 3, no distinct HdeB clients have been identified to date. Therefore, the physiological roles of HdeB, as well as the in vivo functional redundancy between HdeA and HdeB, remain to be clarified.
To examine whether HdeA and HdeB have distinct in vivo chaperoning roles and client profiles in facilitating bacterial acid resistance, in the present study we compared their client profiles within the entire periplasmic proteome in E. coli, in an effort to identify and differentiate the functional specificities in these two seemingly redundant acid chaperones.
Comparative proteomics has shown unprecedented potential for illustrating proteome changes in response to various altered physiological and/or pathological conditions (16, 17). Methods for globally profiling and comparing these condition-specific client pools from two different proteins are limited, however (18–21), owing mainly to the lack of strategies for capturing and comparing native client pools without interference from their distinct “bait” proteins. Genetically encoded protein photocross-linkers offer a powerful platform for capturing native clients of a given protein under living conditions (22). Despite the rapidly increasing use of gel-free technologies, 2D electrophoresis coupled with tandem mass spectrometry remains a powerful, versatile, and straightforward procedure for proteome analysis after sample preparation (16, 17, 23). The 2D difference gel electrophoresis (2D-DIGE) technique, in which two protein samples are separately labeled with different fluorescent dyes and then coelectrophoresed on the same 2D gel, has expanded its application in comparative proteomics (23).
Based on these considerations, we decided to combine the protein photocross-linking and 2D-DIGE techniques to provide comparative profiling of the client pools from two closely related bait proteins, such as HdeA and HdeB. Conventional protein photocross-linking strategies are not suitable for such unbiased comparisons among multiple chaperones, because the captured clients remain bound to their respective chaperones after photocross-linking, which alters their migration behaviors on gels and thus interferes with the comparison. For example, the pIs (isoelectric points) of these complexes are difficult to predict owing to such factors as the partially buried protein–protein interaction interface, which may cause unpredictable variations of the net charge for each client when the chaperone is bound, thus generating biased results. Therefore, releasing these bait chaperones before 2D-DIGE is essential for unbiased side-by-side comparisons of the pure client pools.
To address this challenge, we have developed a new strategy, termed CAPP-DIGE, which couples the cleavage after protein photocross-linking (CAPP) method with 2D-DIGE for comparative proteomic analysis. This strategy allows direct visual comparison of the unbiased native client pools between HdeA and HdeB in E. coli at the whole proteome level, which reveals unique client profiles between these two chaperones under acid stress conditions. Furthermore, by monitoring the pH-dependent chaperone activation, client unfolding, and chaperone–client interactions during both the acid stress and acid recovery processes, we revealed the pH-regulated distinct client interaction profiles of these two chaperones. This pH-regulated two-chaperone system avoids potential nonspecific binding and ensures efficient and economical protein quality control under acid stress.
Results
CAPP-DIGE Reveals Unique in Vivo Client Profiles for HdeA and HdeB.
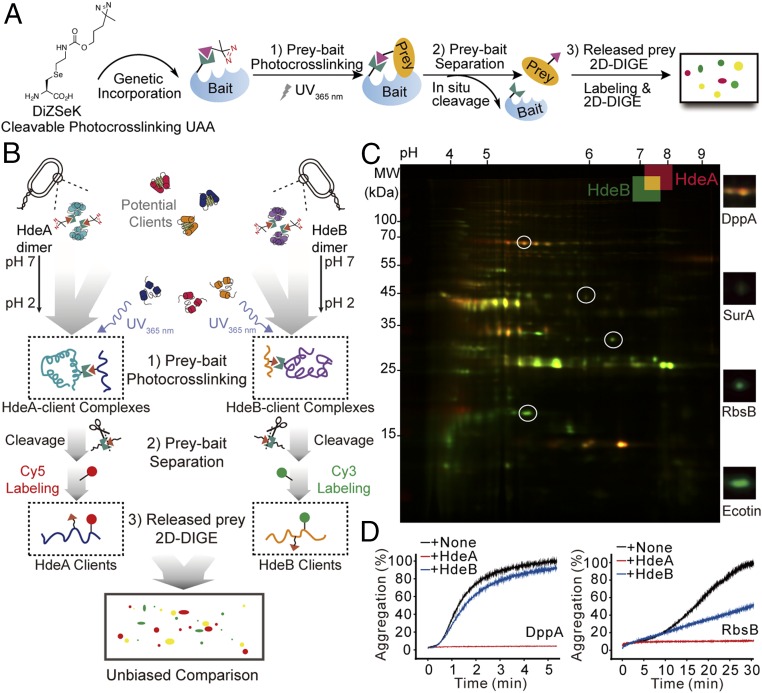
To perform side-by-side comparisons of in vivo clients of HdeA and HdeB in the entire proteome, we used our recently developed CAPP strategy that permitted the efficient separation of the clients from their interacting chaperones (i.e., HdeA or HdeB) after photocross-linking. This cleavable photocross-linker, DiZSeK (24), bears a C-Se moiety that can undergo oxidative cleavage, allowing subsequent release of the captured client pools from their respective chaperones for unbiased 2D-DIGE analysis (Fig. 1 A and B). We applied this newly developed CAPP-DIGE protocol to HdeA and HdeB according to the scheme shown in Fig. 1B. In brief, using a previously evolved pyrrolysyl-tRNA synthetase-tRNA pair (22), DiZSeK was site-specifically incorporated at residues located at the dimer interface of HdeA (HdeA-35DiZSeK) (5, 25) and HdeB (HdeB-24DiZSeK) (12). E. coli cells expressing HdeA-35DiZSeK or HdeB-24DiZSeK were grown at 30 °C for 12 h and treated at pH 2.3 for 30 min before being subjected to 365-nm UV irradiation. Purified HdeA-client and HdeB-client complexes were oxidatively cleaved (8 mM H2O2 for 2 h), and the released client pools were fluorescent-labeled by Cy5 and Cy3 dyes, respectively. Finally, a 1:1 ratio between the Cy5-labeled client pool from HdeA and the Cy3-labeled client pool from HdeB were combined and separated on 2D-PAGE (Fig. 1B).
Fig. 1.
CAPP-DIGE reveals unique client specificities for HdeA and HdeB in vivo. (A) Schematic overview of the CAPP-DIGE method. (B) Workflow for an unbiased, whole-proteome comparison of client profiles between acid-activated chaperones HdeA and HdeB using a CAPP-DIGE strategy. (C) Profiling and comparison of HdeA and HdeB client pools at pH 2 by CAPP-DIGE. Shown is a representative 2D-DIGE gel, with red spots corresponding to HdeA-specific clients, green spots corresponding to HdeB-specific clients, and yellow spots corresponding to their shared clients. The client specificity patterns were verified (Fig. S1). Close-up views of circled selected spots are shown on the right (Fig. S2 and Table S1). (D) In vitro chaperoning activities of HdeA and HdeB toward DppA and RbsB. Aggregation of client proteins was measured by monitoring light-scattering signals at 340 nm in the presence and absence of HdeA or HdeB.
On the overlaid 2D-DIGE images in Fig. 1C, proteins that bound specifically to either HdeA or HdeB appeared as red spots or green spots, respectively, whereas those bound to both HdeA and HdeB appeared as yellow spots. Comparison of the patterns and fluorescence intensities of the entire HdeA and HdeB client pools showed that the common clients of HdeA and HdeB (Fig. 1C, yellow spots) were in the center of the DIGE gel, whereas HdeA-preferred clients (Fig. 1C, red spots) and HdeB-preferred clients (Fig. 1C, green spots) were located on the top and bottom of the gel, respectively. Further analysis by ImageMaster identified nearly 100 client spots for HdeA and HdeB, among which 80% were common clients (volume ratio < twofold). These unique client patterns on 2D-DIGE gel were reproducible from three independent biological samples.
To further verify the unique client patterns that we observed, we performed a series of control experiments. First, E. coli cells expressing HdeA-35DiZSeK or HdeB-24DiZSeK were exposed to acidic solutions without UV irradiation and obtained no bound proteins (Fig. S1 A and B), which confirmed that these binding proteins formed cross-linked complexes with HdeA or HdeB. Secondly, no cross-linked bands could be detected by Western blot analysis with H2O2 treatment (Fig. S1C), which ensured that these identified proteins were the products from H2O2-mediated cleavage. And finally, reciprocal labeling was performed, and the patterns on 2D-DIGE gels showed no distinction (Fig. S1D), demonstrating that the observed patterns were not related to a labeling preference of Cy5-NHS or Cy3-NHS. Taken together, the findings from our side-by-side comparison reveal partially overlapping client protein profiles of HdeA and HdeB. The high similarity of their client profiles suggests that they may have similar client recognition mechanism, whereas the unique subsets of their client proteins indicate that they have different roles against acid stress.
Fig. S1.
Confirmation of CAPP-DIGE results. (A and B) SDS/PAGE (A) and Western blot (B) analysis of photocross-linking products from E. coli cells expressing HdeA-35DiZSeK or HdeB-24DiZSeK at pH 2 (with and without 15 min of UV irradiation). (C) Western blot analysis of photocross-linking products from HdeA-35DiZSeK or HdeB-24DiZSeK with and without H2O2 treatment. Samples were diluted in 20 mM Tris⋅HCl buffer containing 40 mM imidazole (pH 7.4) after H2O2 treatment and then subjected to Ni-NTA purification. Fractions of flow-through (FT) and elution (eluted by 500 mM imidazole) were collected. (D) Reciprocal labeling of photocross-linked HdeA and HdeB clients with Cy3 and Cy5 dyes on the 2D-DIGE gels shown. (Left) Cy5-HdeA vs. Cy3-HdeB. (Right) Cy3-HdeA vs. Cy5-HdeB.
We next sought to identify the specific in vivo binding proteins of HdeA and HdeB by mass spectrometry (Fig. 1C and Fig. S2A). As shown in Table S1, a total of five and six unique proteins were identified as HdeA- and HdeB-preferred clients, respectively (Fig. S2 B and C), which accounted for ∼10% of the total client pool of HdeA and HdeB. The remaining 47 proteins were identified as their common clients. Among these proteins, the dipeptide transporter protein DppA was a preferred client for HdeA, the ribose transporter protein RbsB and the serine protease inhibitor protein Ecotin were preferred clients for HdeB, and the periplasmic chaperone SurA was a common client for both HdeA and HdeB. We then used light-scattering analysis to monitor the chaperoning activities of HdeA and HdeB toward these identified clients in vitro (Fig. 1D). Consistent with the aforementioned 2D-DIGE results, HdeA prevented DppA aggregation almost completely at pH 2, whereas HdeB had only a slight chaperoning effect. Interestingly, although HdeB showed much greater chaperoning activity on its preferred client RbsB compared with DppA, HdeA still had greater efficiency in preventing RbsB aggregation in vitro. Therefore, the unique client specificity that we identified in vivo was not observed in solution, indicating that the in vivo specificities may result from certain environmental kinetic factors rather than thermodynamic factors, such as the recognition interface. In fact, this observation is consistent with previous reports demonstrating that HdeA is a stronger acid chaperone than HdeB in vitro and that no HdeB-specific client proteins have been reported from the in vitro study. Identifying the molecular mechanism underlying the in vivo specificity of these two chaperones will help elucidate the acid-resistant mechanism in the acid-vulnerable E. coli periplasm.
Fig. S2.
(A) Coomassie blue-stained 2D-DIGE gel for LC-MS/MS identification. (B and C) The preferred binding clients of HdeA (B) and HdeB (C) at pH 2. The preferred clients are circled in green. Images were analyzed by ImageMaster 2D (Amersham).
Table S1.
Clients identified by mass spectrometry
| Number | Name | Molecular weight (kDa) | Function |
| Common client proteins shared by HdeA and HdeB | |||
| 1 | BamA | 113 | Formate dehydrogenase-O major subunit |
| 2 | PtrA* | 107 | Protease |
| 3 | FdoG | 90 | Outer membrane protein assembly factor |
| 4 | BglX | 83 | Periplasmic β-glucosidase |
| 5 | Tsp* | 76 | Tail-specific protease |
| 6 | LpoA | 73 | Penicillin-binding protein activator |
| 7 | CpdB | 71 | 2,3-cyclic-nucleotide 2-phosohodiesterase/3-nucleotidase |
| 8 | PpiD* | 68 | Peptidyl-prolyl cis-trans isomerase D |
| 9 | TreA | 63 | Periplasmic trehalase |
| 10 | YdeN | 63 | Uncharcterized sulfatase |
| 11 | UshA | 60 | Protein ushA |
| 12 | NikA | 59 | Nickel-binding periplasmic protein |
| 13 | YgiS | 61 | Putative binding protein ygiS |
| 14 | SapA | 61 | Peptide transport periplasmic protein sapA |
| 15 | MdoG | 58 | Glucans biosynthesis protein G |
| 16 | YhjJ | 55 | Protein yhjJ |
| 17 | DegP* | 49 | Protease |
| 18 | DadA | 48 | d-amino acid dehydrogenase small subunit |
| 19 | UgpB | 48 | sn-glycerol-3-phosphate–binding periplasmic protein |
| 20 | DegQ* | 47 | Protease |
| 21 | SurA* | 47 | Chaperone |
| 22 | TolB | 46 | Protein tolB |
| 23 | YbhC | 46 | Putative acyl-CoA thioester hydrolase |
| 24 | Agp | 46 | Glucose 1-phosphatase |
| 25 | HflK | 46 | Modulator of FtsH protease HflK |
| 26 | GlpQ | 41 | Glycerophosphoryl diester phosphodiesterase |
| 27 | PotF | 40 | Putrescine-binding periplasmic protein |
| 28 | PotD | 38 | Spermidine/putrescine-binding periplasmic protein |
| 29 | AspG2 | 37 | l-asparaginase |
| 30 | OmpA | 37 | Outer membrane protein A |
| 31 | DgaL | 36 | d-galactose–binding periplasmic protein |
| 32 | AraF | 36 | l-arabinose–binding periplasmic protein |
| 33 | XylF | 36 | d-xylose–binding periplasmic protein |
| 34 | YphF | 35 | ABC transporter periplasmic-binding protein yphF |
| 35 | BamC | 35 | Outer membrane protein assembly factor BamC |
| 36 | GltI | 34 | Glutamate/aspartate periplasmic-binding protein |
| 37 | YtfQ | 34 | ABC transporter periplasmic-binding protein ytfQ |
| 38 | Mdh | 32 | Malate dehydrogenase |
| 39 | FliY | 29 | Cystine-binding periplasmic protein |
| 40 | DsbG* | 28 | Thiol:disulfide interchange protein |
| 41 | ArgT | 28 | Lysine-arginine-ornithine–binding periplasmic protein |
| 42 | HisJ | 28 | Histidine-binding periplasmic protein |
| 43 | GlnH | 27 | Glutamine-binding periplasmic protein |
| 44 | ModA | 27 | Molybdate-binding periplasmic protein |
| 45 | Cat | 26 | Chloramphenicol acetyltransferase |
| 46 | DsbC* | 26 | Thiol:disulfide interchange protein DsbC |
| 47 | LolA | 23 | Outer-membrane lipoprotein carrier protein |
| HdeA preferred client proteins | |||
| 1,2 | DppA | 60 | Periplasmic dipeptide transport protein |
| 1,2 | OppA | 60 | Periplasmic oligopeptide transport protein |
| 3 | MalE | 43 | Maltose-binding periplasmic protein |
| 3 | BamB | 42 | Outer membrane protein assembly factor |
| 4,5 | MetQ | 30 | d-methionine–binding lipoprotein metQ |
| HdeB preferred client proteins | |||
| 1 | RbsB | 31 | d-ribose–binding periplasmic protein |
| 2 | DsbA* | 23 | Thiol:disulfide interchange protein dsbA |
| 3 | YfgM | 22 | UPF0070 protein YfgM |
| 4 | EmtA | 22 | Endo-type membrane-bound lytic murein transglycosylase A |
| 5 | Ecotin | 18 | Serine protease inhibitor Ecotin |
| 6 | OsmE | 12 | Osmotically inducible protein OsmE |
The spot numbers of HdeA and HdeB preferred client proteins are consistent with those noted on the image shown in Fig. S2 B and C, respectively.
Protein quality control factor in periplasmic space.
HdeA and HdeB Have Different Responses to pH-Induced Activation.
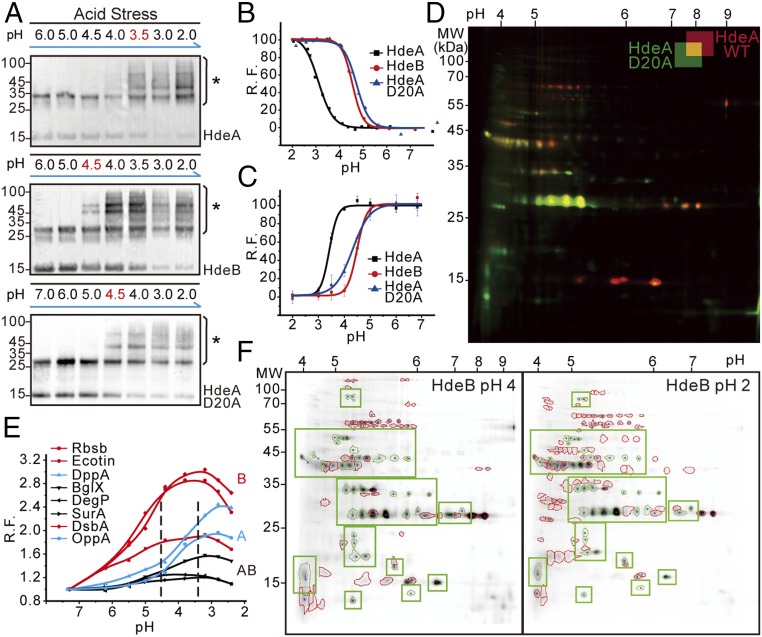
As acid-activated chaperones, HdeA and HdeB undergo pH-triggered unfolding and have a different optimum pH for activation. These results suggest that the chaperoning effects of these two chaperones likely depend on their responses to specific pH conditions. To explore whether pH is a kinetic factor contributing to in vivo client specificity, we systematically varied the environmental acidity of E. coli cells expressing the photocross-linker-containing HdeA or HdeB variants and performed photocross-linking experiments under each pH value between neutral pH and pH 2 (Fig. 2A). The crosslinked chaperone-client complexes on Western blot gels indicate that HdeB starts to bind clients at pH ≤4.5, whereas HdeA does not bind to clients until the pH drops to ≤3.5. Notably, these pH-dependent client-binding features of HdeA and HdeB are highly similar to their respective pH-dependent unfolding processes as monitored by a bis-ANS assay (Fig. 2B). Site-specific introduction of an environment-sensitive dye, 4-DMN, to residues located at HdeA and HdeB dimer interfaces allowed us to monitor the more local hydrophobic-to-hydrophilic transition on pH-induced dimer disruption (26) (Fig. 2C and Fig. S3 A and B). When the environmental pH dropped from 7 to 2, these dimeric chaperones gradually became monomers and exposed their hydrophobic dimer interfaces in a pH-dependent fashion. Notably, the fluorescence signals on these 4-DMN–bearing HdeA and HdeB variants decreased markedly during acidification, confirming that the pH-dependent dimer exposure of HdeA and HdeB is highly consistent with their global conformational changes that we observed (Fig. S3 C and D). Taken together, these results reveal that both HdeA and HdeB are able to couple their pH-triggered unfolding processes with client binding during acid stress, and that they have different responses to pH-induced activation.
Fig. 2.
pH-regulated unfolding and activation determine client specificities of HdeA and HdeB during acid stress. (A) pH-dependent photocross-linking revealing the in vivo chaperoning pH windows of HdeA, HdeA-D20A mutant, and HdeB on acidification. An asterisk denotes cross-linked complexes. (B) pH-dependent global conformational changes of HdeA, HdeA-D20A, and HdeB detected by bis-ANS. R.F., relative bis-ANS fluorescence. (C) pH-dependent unfolding of HdeA, HdeA-D20A, and HdeB detected by the site-specifically labeled environment-sensitive fluorophore 4-DMN (Fig. S3). (D) CAPP-DIGE analysis of client pools of HdeA and HdeA-D20A mutant at pH 2. Shown is a representative 2D-DIGE gel, with red spots corresponding to HdeA-specific clients, green spots corresponding to HdeA-D20A–specific clients, and yellow spots corresponding to their shared clients. (E) pH-dependent global conformational changes of the client proteins detected by bis-ANS and recorded by fluorometry. (F) Comparison of the binding patterns in the entire client pool of HdeB at pH 4 (Left) and pH 2 (Right), with overlapped clients circled in green.
Fig. S3.
Site-specific 4-DMN labeling of HdeA and HdeB for monitoring local conformational changes. (A) The molecular structure of alk-4-DMN. (B) Fluorescence gel and corresponding Coomassie blue-stained SDS/PAGE of 4-DMN–labeled proteins. Purified azide-containing proteins at specific sites were labeled with alk-4-DMN by Cu(I)-catalyzed azide-alkyne cycloaddition reaction (CuAAC). A WT, HdeA-WT; A35, HdeA-35LAPK; A35 (D20A), HdeA-D20A/35LAPK; B24, HdeB-24LAPK. (C) Schematic model for the conformational changes of HdeA and HdeB with the corresponding fluorescence change at pH 7 and pH 2. (D) The fluorescence intensity of 4-DMN labeled proteins measured at pH 7 (black line) and pH 2 (red line).
pH-Regulated Client Binding Ensures Client Specificity.
Our in vivo results suggest that the different responses to pH stimulus observed on HdeA and HdeB may affect their client binding, which in turn may dictate their client specificities. Because D20 is an acid-sensing residue on HdeA that has been previously shown to affect HdeA’s acid-dependent chaperoning effects (11), we investigated the pH-dependent in vivo client binding and conformational changes of the HdeA-D20A variant (Fig. 2 A–C). Interestingly, the pH-dependent behaviors of HdeA-D20A were quite different from those of wild-type HdeA (WT-HdeA) but very similar to those of HdeB. We next compared the client profiles of the HdeA-D20A mutant and WT-HdeA at pH 2 (Fig. 2D). CAPP-DIGE analysis showed that WT-HdeA and HdeA-D20A had different client profiles, whereas the client-binding pattern of HdeA-D20A was highly similar to that of HdeB, as analyzed by ImageMaster (Fig. S4). Because WT-HdeA and HdeA-D20A share the same hydrophobic regions for client binding, the altered client specificity should result from their different responses to pH stimulus.
Fig. S4.
Comparison of client-binding patterns of HdeA-D20A and HdeB at pH 2. Green spots represent the preferred binding clients of HdeA-D20A (A) and HdeB (B) over clients bound to HdeA. Images were analyzed using ImageMaster 2D (Amersham).
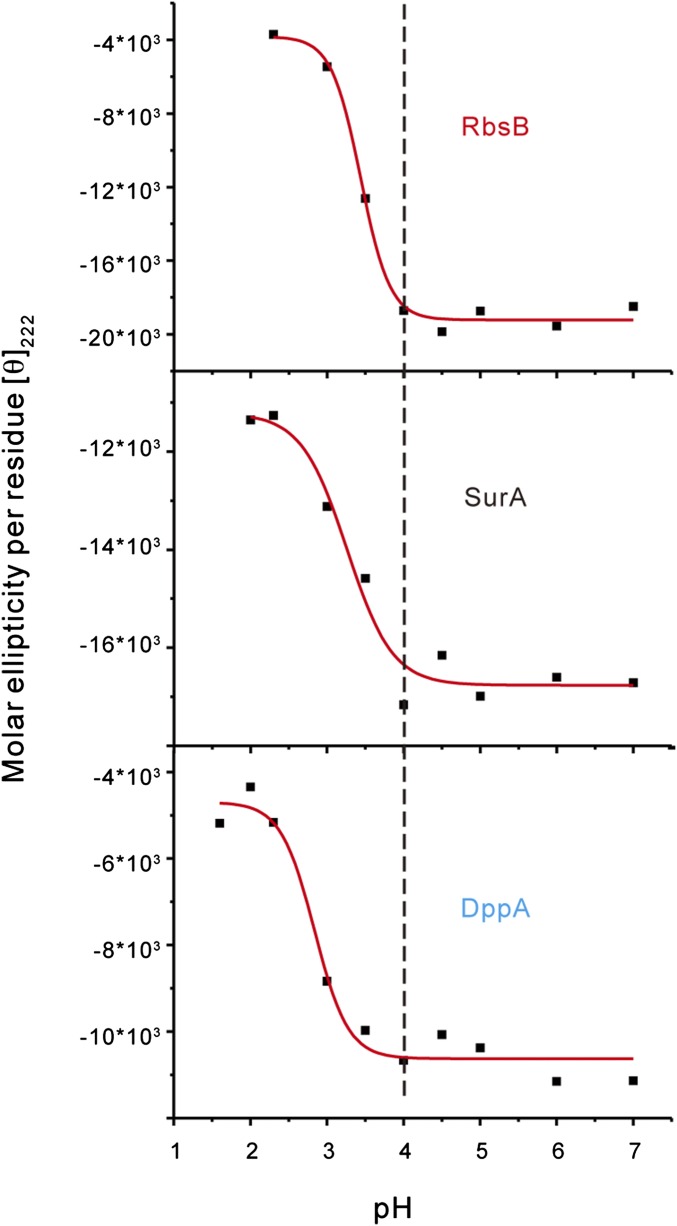
To further confirm that the observed client specificities result from pH-dependent client binding, we used CAPP-DIGE to compare the client binding profiles of HdeB between pH 4 and pH 2 (Fig. 2F). A detailed comparison demonstrated that HdeB’s clients at pH 4 were a subset of its entire clients at pH 2. In particular, some HdeB-specific clients that we identified at pH 2 were found to interact with HdeB at pH 4. This observation confirms our speculation that the in vivo binding specificity of HdeB toward certain proteins is due to its early activation at a higher pH than the dimer-to-monomer transition pH for HdeA (e.g., pH 3). We also used the bis-ANS assay and far-UV circular dichroism (CD) spectra to monitor the pH-dependent hydrophobic surface exposure and structure changes of their client proteins (Fig. 2E). We found that the hydrophobic surfaces of HdeB-preferred clients, such as RbsB, Ecotin, and DsbA, were all significantly exposed when the pH dropped to 4.5, whereas the hydrophobic surfaces of HdeA-preferred clients, such as DppA and OppA, would not be exposed until the pH dropped to <3.5. In contrast, the changes in hydrophobic surfaces from HdeA and HdeB common clients, such as BglX, SurA, and DegP, were much smaller during acidification. The pH-dependent far-UV CD spectra also suggested that the HdeB-preferred clients were more acid-sensitive than HdeA-preferred clients (Fig. S5). Through pH-mediated unfolding of HdeA, HdeB, and client proteins, specific interactions between these chaperones and their clients were established at each given pH during this acidification process, ultimately leading to the different client profiles of these two acid chaperones.
Fig. S5.
Secondary structure changes of different client proteins at various pH conditions. The conformational changes were monitored by CD ellipticity at 222 nm. Conditions for the CD experiments were as described in SI Methods.
pH-Regulated Client Release During Neutralization.
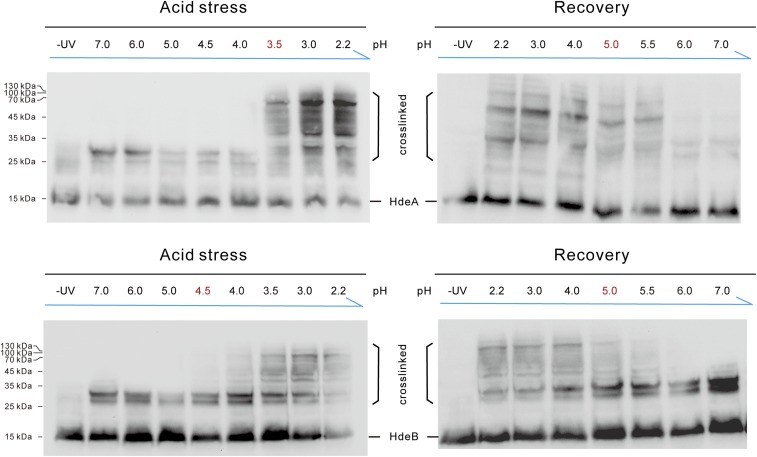
After analyzing the client-binding mechanism on acidification, we next investigated the client release process of these chaperones during neutralization. We conducted pH-dependent photocross-linking of HdeA and HdeB by systematically shifting the acidity from pH 2 to neutral pH (Fig. 3A). In brief, we treated E. coli cells expressing HdeA-35DiZPK or HdeB-24DiZPK at pH 2.3 for 30 min and then restored the pH to different values for another 30 min before exposing the cells to 365-nm UV irradiation. Surprisingly, our Western blot analysis showed that the clients remained bound to their respective chaperones until the pH value was restored to >5. This finding is in direct contrast to the photocross-linking results during acidification, with the client-binding events occurring after pH 4 for HdeB and after pH 3 for HdeA. Therefore, although the pH-induced unfolding and refolding processes for purified HdeA and HdeB proteins are “symmetric” as shown on the bis-ANS assay in vitro (Fig. 3B), the “acid-protection windows” for these chaperones are asymmetric in vivo. HdeA and HdeB remain bound to their clients at a higher pH value during the neutralization process than during the acidification process (see, for example, Fig. S8A). This is also consistent with the previously demonstrated “slow-release” mechanism for HdeA-assisted client refolding in vitro assays (27). The unique asymmetric pH working window was also confirmed in the enteropathogenic E. coli O127:H6 strain, indicating that the mode of action of this acid chaperone system is conserved in both pathogenic and nonpathogenic E. coli strains (Fig. S6).
Fig. 3.
pH-regulated client release from HdeA and HdeB during acid recovery. (A) pH-dependent photocross-linking revealing the in vivo chaperoning pH windows of HdeA and HdeB on neutralization. (B) The pH symmetric unfolding and refolding of HdeA and HdeB monitored by bis-ANS fluorescence. (C) Both HdeA and HdeB release DegP at around pH 4 and release SurA at around pH 5, as monitored by Western blot analysis during acid recovery. (D) Rescue of DegP protease activity in the presence and absence of HdeA during acid recovery. Data are the mean ± SD of three independent experiments.
Fig. S8.
(A) A schematic view of the chaperoning pH windows of HdeA and HdeB during the entire acid stress and recovery processes. The pH-regulated client release mechanism facilitates the refolding of client proteins on neutralization. Furthermore, in addition to their previously demonstrated roles in preventing client aggregation, HdeA and HdeB also aid in the clearance of misfolded clients by rescuing the protease activity of DegP at an acidic pH. (B) Schematic model demonstrating how HdeA and HdeB reconcile client specificities and promiscuities via pH-regulated chaperone–client interactions during acid stress. pH-regulated client binding during acid stress ensures specific but promiscuous client recognition and successfully avoids unwanted interactions, whereas pH-regulated client release during acid recovery allows clients to search for folding-competent states that maximize folding efficiency while suppressing client aggregation.
Fig. S6.
pH-regulated client binding and release from HdeA and HdeB in enteropathogenic E. coli O127:H6 strain.
Among the client proteins identified by CAPP-DIGE, we found many important protein quality control (PQC) factors, including three main protein folding catalysts (DsbA, DsbC, and DsbG), two proline isomerases (SurA and PpiD), and four proteases (DegP, DegQ, Tsp, and PtrA), suggesting that a network of periplasmic PQC factors may be protected by HdeA and HdeB under acid stress (28). These results also indicate a potential linkage between the HdeA/B acid chaperone machinery to the broader proteostasis systems (e.g., DegP) in E. coli (Table S1). By applying Western blot analysis with the antibodies against DegP or SurA, two previously identified periplasm PQC factors protected by HdeA during acid stress (22), we found that DegP was released from both HdeA and HdeB when pH was restored to >4, whereas SurA was not released until pH was restored to >5 (Fig. 3C). These results were further verified by FRET analysis between fluorescent-labeled PQC clients and HdeA or HdeB (Fig. S7). Taken together, our observations indicate that different clients are released at different pH values during the neutralization process, and that the same client is released at the same pH condition regardless of whether it binds to HdeA or to HdeB. Therefore, the client release processes of HdeA and HdeB are also regulated by the environmental pH. Such a pH-induced client release mechanism means that the released clients are able to gain their folding-competent states during acid recovery, thus greatly decreasing the aggregation-prone species.
Fig. S7.
Interactions between HdeA/B and their clients as measured by FRET analysis during the acid-recovery process. HdeA and HdeB were labeled with bimane, and DegP and SurA were labeled with Cy3. FRET signals between HdeA and DegP (A), HdeA and SurA (B), HdeB and DegP (C), and HdeB and SurA (D) were recorded at differently recovered pH conditions after initial acid stress (pH 2) for 30 min.
We have previously shown that HdeA-protected DegP and SurA can regain chaperoning activities during acid recovery and can further assist in the refolding of additional clients (22). Because DegP has chaperone–protease dual functions (29), we assessed whether its protease activity could be rescued as well. To do so, we incubated DegP and its fluorescent-labeled protease substrate β-casein in the presence and absence of HdeA at pH 2 for 30 min, then adjusted the pH back to 5 to monitor the fluorescence change (Fig. 3D). Compared with its protease activity at pH 7, DegP resumed 70% of its activity at pH 5 in the presence of HdeA, but only 35% of its protease activity at the same pH without HdeA. Therefore, HdeA may help DegP regain most of its correct folding at pH 5, as indicated by the significantly rescued protease activity. Notably, the presence of a functional protease DegP at an acidic pH may play essential roles in eliminating aggregation-prone proteins that fail to fall into the proper folding pathways during acid recovery. Indeed, in addition to preventing client aggregation and aiding their refolding by acid chaperones, clearance of the misfolded species that would otherwise form lethal aggregates adds another layer of protein quality control within the E. coli cell envelope (Fig. S8A). The functional role of protease activity in maintaining proteostasis during acid stress merits further investigation.
Distinct Roles of HdeB in Supporting E. coli Acid Resistance.
Because HdeB starts its client binding at pH >4, whereas HdeA is not engaged in client binding until pH <3.5, we next assessed the physiological functions of HdeB at pH 4. SDS/PAGE analysis of the insoluble pellet fraction of E. coli periplasmic extracts incubated with HdeA or HdeB revealed lower amounts of proteins in the insoluble fractions in the presence of HdeB, but not in the presence of HdeA (Fig. 4A). In addition, we performed the acid susceptibility assay at pH 4 on E. coli WT strain, ΔhdeAB, and ΔhdeB strains, as well as these mutant strains complemented with plasmids expressing HdeA or HdeB (Fig. 4B). Interestingly, when complemented with HdeB, both ΔhdeAB and ΔhdeB mutant strains displayed similar survival rates as the WT strain. In contrast, even though it had a similar activation pH as HdeB, the HdeA-D20A variant afforded no protection to the ΔhdeAB strain at pH 4. Therefore, whereas HdeA is the major acid chaperone under extreme acidity, HdeB confers E. coli acid resistance under mild acid stress conditions (e.g., pH 4). Moreover, overexpression of the HdeA-D20A/D51A double mutant, a constitutively active and monomeric HdeA variant at all pH conditions (11), resulted in severe cell death at pH 4 (Fig. 4B). We reason that this might be due to the nonspecific binding of various periplasmic proteins by this “super-hydrophobic” HdeA variant under neutral conditions. To test this hypothesis, we conducted bis-ANS fluorescence assay to measure the hydrophobic surface areas of these proteins (i.e., HdeB, HdeA-D20A, and HdeA-D20A/D51A) at different pH values, which showed that these variants indeed displayed much larger hydrophobic surfaces than HdeB under both neutral and acidic conditions (Fig. 4C). Because both HdeA and HdeB undergo pH-dependent graduate opening of hydrophobic regions for client binding, the exposure of unwanted extra hydrophobic surfaces near neutral pH might lead to nonspecific protein binding and result in cell death.
Fig. 4.
Distinct roles of HdeB in supporting E. coli acid resistance. (A) SDS/PAGE analysis of the aggregated pellet of periplasmic extracts incubated with HdeA or HdeB at pH 4. (B) The survival rates of acid-stressed (pH 4) E. coli WT strain K12 BW25113 with and without the empty pBAD vector (WT+pBAD, WT), the hdeAB KO strain, the hdeB KO strain, and the KO strains complemented with the following plasmids: HdeA, pBAD-HdeA; HdeB, pBAD-HdeB; A-D20A/D51A, pBAD-HdeA-D20A/D51A; A-D20A, pBAD-HdeA-D20A; and A-D51A, pBAD-HdeA-D51A. (C) Bis-ANS fluorescence of HdeB, HdeA-D20A, and HdeA-D20A/D51A at neutral and acidic pH. (D) The synergistic effects of HdeA and HdeB in preventing the aggregation of SurA at pH 2, as detected by SDS/PAGE analysis of supernatant and pellet samples. (E) HdeA and HdeB synergistically suppress SurA aggregation at pH 2, as monitored by light scattering at 340 nm.
Finally, because the synergistic chaperoning effects of HdeA and HdeB have been tested only on whole cell extracts and some model proteins (6), we examined the synergistic effects of these two acid-activated chaperones on a physiological client SurA previously identified by our MS study. The aggregation behavior of SurA with and without equal amounts of HdeA or HdeB at pH 2 (SurA:chaperone ratio 1:1, n/n) was first monitored by analysis of 15,000 × g supernatants and pellets (Fig. 4D). In the absence of chaperones, only a tiny amount of SurA remained in the soluble fraction. In contrast, both HdeA and HdeB increased the soluble fraction of SurA, whereas the combination of HdeA and HdeB was more efficient than the same amount of HdeA or HdeB alone. To more precisely examine the synergistic effects, we used light scattering analysis to monitor SurA aggregation at pH 2 (Fig. 4E). In agreement with the gel-based assay, the combination of 3 μM HdeA and 3 μM HdeB suppressed SurA aggregation at pH 2 more efficiently than the presence of 6 μM HdeA or 6 μM HdeB alone, further confirming the synergistic effect between these two chaperones.
Discussion
The HdeA/HdeB chaperone system in enteric bacterial periplasm efficiently supports their acid-resistance capabilities, which is essential to ensure the pathogenic dosage needed to cause infections. Because this two-chaperone machinery is highly conserved in many gut-resident pathogens, revealing this mode of action would help elucidate the acid-resistance mechanism during host–pathogen interactions. HdeA and HdeB are typical members of a growing group of so-called “conditionally disordered chaperones” that use stress-induced unfolding to trigger chaperone–client interactions and chaperoning activation. The disorder-based interactions enable them to protect a broad array of client proteins with the same interacting interface (30, 31); however, these disordered interfaces should be under tight control to avoid toxic nonspecific interactions, including self-aggregation (32).
Both the client promiscuity and specificity of chaperones are essential for maintaining proteostasis during acid stress. Understanding how the disorder-triggered interactions can reconcile specific but promiscuous client recognition remains highly desirable yet challenging, particularly within an intracellular context. Our comparative proteomics-based CAPP-DIGE strategy for systematically comparing the chaperone–client interactions between two homologous acid-activated chaperones HdeA and HdeB demonstrates that stimulative conditions, such as pH, can serve as an environmental cue to regulate specific binding and release events between disordered chaperones and their clients. The pH-dependent photocross-linking strategy allowed us to observe the pH-dependent activation and inactivation of HdeA/B in vivo. As illustrated by the schematic model shown in Fig. S8B, all E. coli periplasmic proteins are gradually unfolded on acidification from neutral to pH 2, and HdeB becomes active at pH 4.5 during this process to bind those unfolded proteins above this pH value. Meanwhile, although it has a stronger chaperoning activity, HdeA is not activated until the pH drops further to <3.5, and thus interacts only with those proteins that are unfolded under more acidic conditions. This pH-regulated client binding ensures that whereas specific chaperone–client binding occurs at each given pH condition, these chaperones can bind to a broad spectrum of clients during the entire acidification process.
This strategy ensures that diverse clients can be protected by the same set of chaperones, whereas unwanted nonspecific binding can be effectively avoided. Similarly, during acid recovery, clients are released from their respective chaperones in a pH-regulated manner. The stable chaperone–client complexes permit most of these aggregation prone clients to reach folding-competent states before being released from their respective chaperones. This pH-regulated client release strategy allows chaperones to maximally suppress the aggregation of fragile clients. This cooperative network between HdeA and HdeB provides an efficient, economical, flexible, and finely tuned protein quality control system to cope with acid stress.
A number of stress-specific chaperones that specifically activate their chaperone functions through stress-induced conformational rearrangements, unfolding or changes in oligomerization states have been identified (30, 31). The stress conditions also may contribute to the tight control of their client profiles, thereby avoiding nonspecific binding. Our work provides a facile and generally applicable approach for investigating the distinct protein–protein interaction profiles among such seemingly redundant homologous chaperones. In particular, our strategy offers valuable insight into the seemingly paradoxical protein disorder-mediated specific but promiscuous client recognition. Beyond HdeA and HdeB, our unbiased whole-proteome CAPP-DIGE strategy provides a powerful platform for profiling protein–protein interactions under dynamic conditions and uncovering the otherwise elusive mode of action within a native cellular context.
Methods
The cultures expressing DiZSeK-incorporated HdeA or HdeB (SI Methods) were treated at pH 2.3 for 30 min and then subjected to UV light (365 nm) for 15 min using a Hoefer UVC 500 UV cross-linker installed with 365-nm UV lamps (Amersham Biosciences) at a distance of 3 cm on ice. Cells were then lysed by sonication in buffer A (20 mM Tris⋅HCl and 0.5 M NaCl, pH 7.4), and the cross-linked products were purified in a Ni-NTA column (GE Healthcare). The purified products were desalted in PBS buffer (pH 8.0) and then treated with 0.5% SDS at 95 °C for 20 min. Denatured products were treated with 8 mM H2O2 at 30 °C for 2 h, diluted into buffer B (20 mM Tris⋅HCl, 0.5 M NaCl and 40 mM imidazole, pH 7.4), and then purified in an Ni-NTA column. The flow-through solution was collected, concentrated, and dialyzed in PBS (pH 7.4). The pools of clients thus obtained were subjected to standard 2D-DIGE protocols (23). Images were scanned with a Typhoon FLA 9500 laser scanner (GE Healthcare) and analyzed using ImageMaster 2D (Amersham).
SI Methods
pH-Dependent Photocross-Linking of DiZPK-Incorporated HdeA or HdeB.
Aliquots (1 mL) of the cultures expressing DiZPK-incorporated HdeA or HdeB were transferred to 24-well plates and treated at different pH for 30 min. For photocross-linking during the acid-recovery process, the pH of the aliquots was adjusted back to 7, followed by treatment for another 30 min before subjection to photocross-linking. The cells were harvested by centrifugation at 13,000 × g for 2 min and then analyzed by Tricine SDS/PAGE and Western blot analysis.
Expression and Incorporation of DiZPK/DiZSeK into HdeA or HdeB.
The HdeA or HdeB expression plasmid carrying an in-frame amber mutation was transformed into Escherichia coli strain JWK3478 with the plasmid pSUPAR-Mb-DiZPK-RS (22), which allowed the incorporation of DiZSeK or DiZPK into a desired site on HdeA or HdeB. The transformed bacteria cells were grown at 30 °C in LB medium supplemented with 0.2 mM DiZSeK or DiZPK to the midlog phase, at which point the expression of amber-mutated HdeA or HdeB proteins was induced by the addition of 0.4% arabinose, followed by incubation at 30 °C for another 12 h.
Fluorescence Labeling and FRET Measurements.
To monitor the interaction between HdeA or HdeB and their client proteins, we engineered the mutant HdeA-S27C and HdeB-A20C, which introduce a unique reactive cysteine near the dimer interface. HdeA-S27C and HdeB-A20C were labeled with monobromobimane as recommended by Molecular Probes and a previous report (14). HdeA-S27C or HdeB-A20C were incubated with 10-fold molar excess of monobromobimane dye in 50 mM Tris buffer and 50 mM NaCl (pH 8) for 1 h at 25 °C. Excess dyes were removed with desalting columns. The labeling efficiency was determined spectrophotometrically by comparing the absorbance of bimane (ε380 = 5,000 M−1 cm−1; ε280 = 3,600 M−1 cm−1) to the absorbance of unlabeled HdeA (ε280 = 11,200 M−1 cm−1). The labeling efficiency of both proteins was between 0.8 and 0.9. The client proteins SurA and DegP were labeled with Cy3-NHS as recommended by Molecular Probes. These fluorescent-labeled HdeA or HdeB and SurA or DegP were incubated at pH 2 in the buffer containing 10 mM citrate and 150 mM NaCl for 30 min. The solutions were then recovered to different pH by addition of 0.6 M NaOH and incubated for another 30 min before FRET measurements. Emission spectra were recorded from 400 to 700 nm by excitation at 390 nm.
Light-Scattering Analysis.
Client proteins were diluted into incubation buffer [8 mM H3PO4, 150 mM KCl, and 150 mM (NH4)2SO4, pH 2 or pH 4] to a final concentration of 5 M in the absence or presence of HdeA, HdeA variants, or HdeB. Aggregation was monitored by observing light-scattering signals at 340 nm using a Cary Eclipse fluorescence spectrophotometer.
Bis-ANS Assay.
Binding of 100 μM bis-ANS with 5 μM HdeA, HdeA variants, or HdeB was measured by monitoring the fluorescence increase of bis-ANS in buffer containing 10 mM citrate and 150 mM NaCl, which was titrated from pH 7 to pH 2 by the stepwise addition of 0.6 M HCl or from approximately pH 2 to pH 7 by the stepwise addition of 0.6 M NaOH. Samples were excited at 385 nm, and the emissions were recorded between 410 and 650 nm using a Cary Eclipse fluorescence spectrophotometer. For the pH window comparison, bis-ANS fluorescence was normalized to the signals observed at neutral pH (relative bis-ANS fluorescence of 0) and low pH (relative bis-ANS fluorescence of 100). For the acid-sensitivity comparison of client proteins, bis-ANS fluorescence was normalized to the proteins’ signals observed at neutral pH (relative bis-ANS fluorescence of 1, deduction of the background without proteins).
DMN Assay of LAPK-Incorporated HdeA or HdeB.
HdeA or HdeB (1–2 mg/mL) containing LAPK was reacted at room temperature for 1 h with alk-4-DMN (0.5 mM) in the presence of Cu(I)/ligand (100 μM) and sodium ascorbate (2 mM) in the sodium phosphate buffer (pH 7.4). The reaction mixture was diluted five times with PBS and concentrated (to 5×) using a 3,000-molecular weight cutoff centrifuge filter, repeated three times to remove all of the unreacted dyes and Cu(I) ligand. The alk-4-DMN–labeled proteins were then diluted into citrate buffer with different pH values at a concentration of 5 μM. Samples were excited at 480 nm, and emission was monitored at 550 nm.
DegP Protease Activity Assessment.
Equal amounts of DegP (50 μg) and resorufin-labeled casein (Roche) were incubated in reaction buffer [8 mM H3PO4, 150 mM KCl, and 150 mM (NH4)2SO4, pH 2.5] at 37 °C for 30 min. The solution was subsequently adjusted to pH 5 by the stepwise addition of 1 M NaOH for another 1 h of incubation. The reaction was stopped by adding 120 μL 5% (wt/vol) trichloroacetic acid, and the solution was incubated for 10 min at 37 °C. After a 5-min centrifugation, supernatants were diluted in assay buffer (0.5 M Tris⋅HCl, pH 8.8), and the fluorescent signals were immediately recorded by a 96-well microplate reader (BioTek). Relative protease activity was calculated as (FrpH 5 − FpH 2)/FpH 7. DegP protease activity at neutral pH was measured following a standard protocol (33).
Chaperone Activity Assay.
Periplasmic extracts or client proteins were incubated in buffer A [8 mM H3PO4, 150 mM KCl, and 150 mM (NH4)2SO4, pH 2.5] for 60 min in the presence and absence of HdeA or HdeB at acidic pH conditions. The appearance of the proteins in the 16,000 × g pellet or supernatant was monitored by SDS/PAGE.
Acid-Resistance Assay.
E. coli ΔhdeAB and ΔhdeB mutant strains, derived from the parent strain BW25113 [rrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-l ] (22), were used for the acid-resistance assay. Plasmids pBAD (expressing HdeA or its mutants), pBAD (expressing HdeB), or pBAD (empty vector control) were transformed into the mutant strains or BW25113 strain for complementary or control experiments. The assay was performed as described previously (34). In brief, these strains were grown in LB at 37 °C in the presence of 0.05% arabinose and the required antibiotics on a shaker (150 rpm) to stationary phase (16 h), followed by dilution to ∼106 CFU/mL and incubation for 2 h in acidified LB (pH 4, acidified with HCl). Control experiments were conducted at pH 7. After incubation, the cultures were neutralized and spot-titered onto LB plates containing the required antibiotics. Each strain was tested in at least four independent experiments. The survival rate was calculated by comparing the viable cells of the acid-treated group with those of the control group. The expressions of HdeA and HdeB and their variants were confirmed by Western blot analysis with the antibody of HdeA or HdeB.
CD.
CD spectroscopy measurements were performed with a J-815 CD spectrometer (JASCO) using quartz cuvettes with a path length of 0.1 cm. Spectra were recorded and signals at 222 nm were monitored for 100 μg/mL client proteins in citrate buffer with different pH values.
Acknowledgments
We thank Prof. Zengyi Chang for providing the antibodies for Escherichia coli HdeA, HdeB, SurA, and DegP; Rong Meng and Wen Zhou for assisting with protein identification by LC-MS/MS analysis; and Maiyun Yang for providing unnatural amino acids LAPK and alk-4-DMN. This work was supported by the National Key Research and Development Program (2016YFA0501500) and the National Natural Science Foundation of China (21225206, 21432002, and 21521003).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606360113/-/DCSupplemental.
References
- 1.Foster JW. Escherichia coli acid resistance: Tales of an amateur acidophile. Nat Rev Microbiol. 2004;2(11):898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- 2.Hong W, Wu YE, Fu X, Chang Z. Chaperone-dependent mechanisms for acid resistance in enteric bacteria. Trends Microbiol. 2012;20(7):328–335. doi: 10.1016/j.tim.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Hingorani KS, Gierasch LM. How bacteria survive an acid trip. Proc Natl Acad Sci USA. 2013;110(14):5279–5280. doi: 10.1073/pnas.1303297110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao B, Houry WA. Acid stress response in enteropathogenic gammaproteobacteria: An aptitude for survival. Biochem Cell Biol. 2010;88(2):301–314. doi: 10.1139/o09-182. [DOI] [PubMed] [Google Scholar]
- 5.Gajiwala KS, Burley SK. HDEA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria. J Mol Biol. 2000;295(3):605–612. doi: 10.1006/jmbi.1999.3347. [DOI] [PubMed] [Google Scholar]
- 6.Kern R, Malki A, Abdallah J, Tagourti J, Richarme G. Escherichia coli HdeB is an acid stress chaperone. J Bacteriol. 2007;189(2):603–610. doi: 10.1128/JB.01522-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mates AK, Sayed AK, Foster JW. Products of the Escherichia coli acid fitness island attenuate metabolite stress at extremely low pH and mediate a cell density-dependent acid resistance. J Bacteriol. 2007;189(7):2759–2768. doi: 10.1128/JB.01490-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tucker DL, Tucker N, Conway T. Gene expression profiling of the pH response in Escherichia coli. J Bacteriol. 2002;184(23):6551–6558. doi: 10.1128/JB.184.23.6551-6558.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waterman SR, Small PLC. Identification of sigma S-dependent genes associated with the stationary-phase acid-resistance phenotype of Shigella flexneri. Mol Microbiol. 1996;21(5):925–940. doi: 10.1046/j.1365-2958.1996.00058.x. [DOI] [PubMed] [Google Scholar]
- 10.Valderas MW, et al. Role of HdeA in acid resistance and virulence in Brucella abortus 2308. Vet Microbiol. 2005;107(3-4):307–312. doi: 10.1016/j.vetmic.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 11.Foit L, George JS, Zhang BW, Brooks CL, 3rd, Bardwell JC. Chaperone activation by unfolding. Proc Natl Acad Sci USA. 2013;110(14):E1254–E1262. doi: 10.1073/pnas.1222458110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang W, et al. Salt bridges regulate both dimer formation and monomeric flexibility in HdeB and may have a role in periplasmic chaperone function. J Mol Biol. 2012;415(3):538–546. doi: 10.1016/j.jmb.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu YE, Hong W, Liu C, Zhang L, Chang Z. Conserved amphiphilic feature is essential for periplasmic chaperone HdeA to support acid resistance in enteric bacteria. Biochem J. 2008;412(2):389–397. doi: 10.1042/BJ20071682. [DOI] [PubMed] [Google Scholar]
- 14.Tapley TL, et al. Structural plasticity of an acid-activated chaperone allows promiscuous substrate binding. Proc Natl Acad Sci USA. 2009;106(14):5557–5562. doi: 10.1073/pnas.0811811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dahl JU, et al. HdeB functions as an acid-protective chaperone in bacteria. J Biol Chem. 2015;290(1):65–75. doi: 10.1074/jbc.M114.612986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minden J. Comparative proteomics and difference gel electrophoresis. Biotechniques. 2007;43(6) doi: 10.2144/000112653. 739741, 743 passim. [DOI] [PubMed] [Google Scholar]
- 17.Jiang Y, Wang X. Comparative mitochondrial proteomics: Perspective in human diseases. J Hematol Oncol. 2012;5(1):11. doi: 10.1186/1756-8722-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deuerling E, et al. Trigger Factor and DnaK possess overlapping substrate pools and binding specificities. Mol Microbiol. 2003;47(5):1317–1328. doi: 10.1046/j.1365-2958.2003.03370.x. [DOI] [PubMed] [Google Scholar]
- 19.Winter J, Linke K, Jatzek A, Jakob U. Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol Cell. 2005;17(3):381–392. doi: 10.1016/j.molcel.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 20.Hirtreiter AM, et al. Differential substrate specificity of group I and group II chaperonins in the archaeon Methanosarcina mazei. Mol Microbiol. 2009;74(5):1152–1168. doi: 10.1111/j.1365-2958.2009.06924.x. [DOI] [PubMed] [Google Scholar]
- 21.Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature. 1999;400(6745):693–696. doi: 10.1038/23301. [DOI] [PubMed] [Google Scholar]
- 22.Zhang M, et al. A genetically incorporated crosslinker reveals chaperone cooperation in acid resistance. Nat Chem Biol. 2011;7(10):671–677. doi: 10.1038/nchembio.644. [DOI] [PubMed] [Google Scholar]
- 23.Tannu NS, Hemby SE. Two-dimensional fluorescence difference gel electrophoresis for comparative proteomics profiling. Nat Protoc. 2006;1(4):1732–1742. doi: 10.1038/nprot.2006.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin S, et al. Genetically encoded cleavable protein photo-cross-linker. J Am Chem Soc. 2014;136(34):11860–11863. doi: 10.1021/ja504371w. [DOI] [PubMed] [Google Scholar]
- 25.Yang F, Gustafson KR, Boyd MR, Wlodawer A. Crystal structure of Escherichia coli HdeA. Nat Struct Biol. 1998;5(9):763–764. doi: 10.1038/1796. [DOI] [PubMed] [Google Scholar]
- 26.Yang M, et al. Converting a solvatochromic fluorophore into a protein-based pH indicator for extreme acidity. Angew Chem Int Ed Engl. 2012;51(31):7674–7679. doi: 10.1002/anie.201204029. [DOI] [PubMed] [Google Scholar]
- 27.Tapley TL, Franzmann TM, Chakraborty S, Jakob U, Bardwell JC. Protein refolding by pH-triggered chaperone binding and release. Proc Natl Acad Sci USA. 2010;107(3):1071–1076. doi: 10.1073/pnas.0911610107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weski J, Ehrmann M. Genetic analysis of 15 protein folding factors and proteases of the Escherichia coli cell envelope. J Bacteriol. 2012;194(12):3225–3233. doi: 10.1128/JB.00221-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krojer T, et al. Structural basis for the regulated protease and chaperone function of DegP. Nature. 2008;453(7197):885–890. doi: 10.1038/nature07004. [DOI] [PubMed] [Google Scholar]
- 30.Jakob U, Kriwacki R, Uversky VN. Conditionally and transiently disordered proteins: Awakening cryptic disorder to regulate protein function. Chem Rev. 2014;114(13):6779–6805. doi: 10.1021/cr400459c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bardwell JC, Jakob U. Conditional disorder in chaperone action. Trends Biochem Sci. 2012;37(12):517–525. doi: 10.1016/j.tibs.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gsponer J, Futschik ME, Teichmann SA, Babu MM. Tight regulation of unstructured proteins: From transcript synthesis to protein degradation. Science. 2008;322(5906):1365–1368. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schickaneder E, Hosel W, Vondereltz H, Geuss U. Casein-resorufin, a new substrate for a highly sensitive protease assay. Fresenius Z Anal Chem. 1988;330(4-5):360. [Google Scholar]
- 34.Carter MQ, et al. Evolutionary silence of the acid chaperone protein HdeB in enterohemorrhagic Escherichia coli O157:H7. Appl Environ Microbiol. 2012;78(4):1004–1014. doi: 10.1128/AEM.07033-11. [DOI] [PMC free article] [PubMed] [Google Scholar]