Significance
Numerous reports indicate that amyloid-β peptide (Aβ) oligomers, considered the pathogenic molecular form of Aβ in Alzheimer´s disease (AD), exert their neurotoxicity within the membrane. Therefore, it is critical to characterize them in such an environment. Here, we worked with two major Aβ variants and handled them as if they were membrane proteins. By doing so, we found that the Aβ variant most strongly linked to AD assembled into stable Aβ oligomers that adopted a specific structure and incorporated into membranes as pores, a feature linked to neurotoxicity. Having access to pore-forming Aβ oligomers with such a specific structure offers unique opportunities to fully characterize them and establish their involvement in AD.
Keywords: Alzheimer’s disease, amyloid-β peptide, membrane pore, oligomer
Abstract
The formation of amyloid-β peptide (Aβ) oligomers at the cellular membrane is considered to be a crucial process underlying neurotoxicity in Alzheimer’s disease (AD). Therefore, it is critical to characterize the oligomers that form within a membrane environment. To contribute to this characterization, we have applied strategies widely used to examine the structure of membrane proteins to study the two major Aβ variants, Aβ40 and Aβ42. Accordingly, various types of detergent micelles were extensively screened to identify one that preserved the properties of Aβ in lipid environments—namely the formation of oligomers that function as pores. Remarkably, under the optimized detergent micelle conditions, Aβ40 and Aβ42 showed different behavior. Aβ40 aggregated into amyloid fibrils, whereas Aβ42 assembled into oligomers that inserted into lipid bilayers as well-defined pores and adopted a specific structure with characteristics of a β-barrel arrangement that we named β-barrel pore-forming Aβ42 oligomers (βPFOsAβ42). Because Aβ42, relative to Aβ40, has a more prominent role in AD, the higher propensity of Aβ42 to form βPFOs constitutes an indication of their relevance in AD. Moreover, because βPFOsAβ42 adopt a specific structure, this property offers an unprecedented opportunity for testing a hypothesis regarding the involvement of βPFOs and, more generally, membrane-associated Aβ oligomers in AD.
One of the main pathological features of Alzheimer′s disease (AD) is the extracellular accumulation of the amyloid-beta peptide (Aβ) as fibrillar amyloid plaques (1). Aβ is obtained from the transmembrane amyloid precursor protein (APP) by consecutive action of the enzymes β-secretase and γ-secretase. The cleavage of γ-secretase occurs sequentially, giving rise to amphipathic Aβ peptides of lengths ranging from 38 to 43 residues (Aβ38 to Aβ43) (2). Because amyloid plaques are detected extracellularly, it is generally considered that, after APP processing, Aβ variants are fully released into the extracellular media. It is for this reason that researchers have focused on the study of Aβ in a solution environment (3–5). However, several of the most likely mechanisms of Aβ neurotoxicity are associated with the cell membrane, including interactions with membrane receptors (6), induction of membrane bilayer disorder (7), and generation of amyloid pores or channel-like structures (8–12). Therefore, apart from studying Aβ in solution, it is crucial to examine this peptide in a membrane environment.
As in all membrane-associated research, it is of paramount importance to use a suitable biomimetic membrane environment. In this context, previous studies devoted to Aβ in a membrane environment have used either liposomes (8–12) or detergent micelles (13–15). The study of Aβ reconstituted in liposomes has been mainly through electrical recording using planar lipid bilayers. Pioneering research by Arispe et al. revealed that this peptide forms pores across phospholipid bilayer membranes and that these pores show spontaneous transitions between defined levels of conductance (8, 9). This work led to the “amyloid pore hypothesis,” which proposes the formation of Aβ pores at the membrane as a key process in the neurotoxicity observed in AD. Subsequent research by other groups led to somewhat conflicting results due to the diversity of the Aβ pores reported (10–12). Such diversity has prevented the identification of specific features that define Aβ pore structure and conductivity properties, thus preventing confirmation of the amyloid pore hypothesis.
Detergent micelles are commonly used to solubilize membrane proteins for structural research. Indeed, their small size—compared with that of other biomimetic membrane environments—enables the application of well-established solution NMR techniques, thus allowing the obtention of high-resolution structural information for the membrane protein under study (16). However, unlike lipid bilayers, detergent micelles are spheroid and vary in shape and size depending on the chemical structure of the detergent. Therefore, when using detergents to study membrane protein structure and function, several types of detergent micelles must be extensively screened to find one that preserves the native structure and function of the membrane protein under study (16, 17). Such screening has not been performed when studying Aβ in the presence of detergent micelles. Instead, multiple detergents and conditions have been used, leading to diverse and contradictory results ranging from Aβ being monomeric and adopting an α-helical structure (13) to Aβ forming oligomers rich in β-sheet structure (14, 15). Moreover, the links between the Aβ species formed in the presence of detergent micelles and those formed in lipid environments has not been established.
In the present paper, we contribute to establishing such a link. We report on micelle conditions to prepare specific Aβ oligomers with the same function as that observed for Aβ oligomers formed in a lipid environment—namely pore formation in lipid bilayers. To develop such conditions, established strategies used to examine the structure and function of membrane proteins and their complexes were applied to study the two major Aβ variants, Aβ40 and Aβ42 (consisting of 40 and 42 residues, respectively). Remarkably, under the optimized micelle conditions, Aβ42 assembles into oligomers that insert into lipid bilayers as well-defined pores and adopt a specific structure with characteristics of a β-barrel arrangement. On the basis of these observations, we have named this Aβ42 oligomer preparation β-barrel Pore-Forming Aβ42 Oligomers (βPFOsAβ42). Notably, βPFOsAβ42 maintain their structural integrity in a lipid environment provided by bicelles.
Results
Screening Conditions for Aβ Oligomer Formation in the Presence of Detergent Micelles.
To study Aβ oligomer formation in a membrane environment, we used detergent micelles, one of the most commonly used biomimetic membrane environments to examine the structure of membrane proteins and their complexes (18, 19). However, detergent micelles have been reported to compete with the factors that stabilize protein–protein interactions (20). Accordingly, we envisioned that a large excess of free micelles could act as a “hydrophobic sink,” in which Aβ subunits could disperse, leading to the disruption of Aβ oligomers. Taking this idea into account, we used the ratio of the Aβ concentration ([Aβ]) to the micelle concentration ([M]) as a defined variable. We maintained [Aβ] constant at 150 μM and worked under two [M] conditions: one in which [M] was higher than [Aβ], at an [Aβ]/[M] ratio of 1:4.7, referred to as high micelle conditions, and the other in which [M] was lower than [Aβ], at an [Aβ]/[M] ratio of 1:0.5, referred to as low micelle conditions. Using this rationale, we examined 150-μM samples of Aβ40 and Aβ42 reconstituted in octyl glucoside (OG), dodecylmaltoside, decylmaltoside (DM), 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC), lauryldimethylamine-N-oxide (LDAO), and dodecylphosphocholine (DPC) under both high and low micelle conditions. Moreover, because Aβ is highly prone to aggregate into amyloid fibrils, we analyzed samples after immediate reconstitution and after incubation for 24 h at 37 °C. Inspired by the work with membrane proteins, we assessed the formation and stability of Aβ oligomers in the distinct detergent micelle conditions by using size exclusion chromatography (SEC). As a control, we used Aβ40 reconstituted in SDS micelles at an [Aβ]/[MSDS] ratio of 1:4.7, conditions under which Aβ40 has been reported to be monomeric and to adopt an α-helical structure (13). Indeed, SEC of this sample revealed a single peak eluting at 34.7 mL (SI Appendix, Fig. S1), which we assigned to monomeric Aβ. Thus, our criterion to assess oligomer formation in the samples under study was based on the observation of peaks eluting earlier than 34.7 mL.
We detected distinct oligomerization behavior between Aβ40 and Aβ42 in all of the conditions tested (SI Appendix, Fig. S2). After immediate reconstitution, Aβ40 samples eluted mainly at the same volume as the Aβ40 SDS control, thereby indicating that they were monomeric. Only a small peak consistent with oligomer formation was observed for some of the samples. After the incubation period, only Aβ40 samples reconstituted under high OG, DHPC, and DPC micelle conditions eluted as monomers. The rest of the Aβ40 samples showed hardly any signal in SEC, thereby suggesting that Aβ40 assembled into aggregates that were too large to pass through the filter applied before SEC analysis. In contrast, after immediate reconstitution, Aβ42 samples eluted both as monomers and oligomers, the latter at different elution volumes, thereby suggesting the formation of a heterogeneous population of oligomers. After the incubation period, all of the Aβ42 samples except those incubated in DPC either eluted in the void volume or showed hardly any signal, thus pointing to the formation of larger aggregates. Remarkably, only Aβ42 samples incubated in DPC eluted as a major symmetric peak, consistent with the formation of a homogeneous population of Aβ42 oligomers (SI Appendix, Fig. S2F). Indeed, calibration of the SEC column with globular protein standards indicated that the Aβ42 oligomer–micelle complex had a mass of approximately 60 kDa (SI Appendix, Fig. S3).
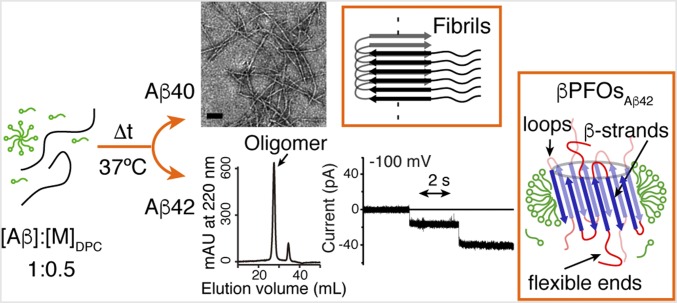
Having observed such remarkable evolution for the Aβ42 oligomer samples formed under DPC micelle conditions (Fig. 1 and SI Appendix, Fig. S2F), we considered it appropriate to further characterize the behavior of Aβ40 under the same conditions (Fig. 1). Aβ40 samples reconstituted under high DPC micelle conditions eluted like the monomeric SDS control before and after incubation, thereby indicating that Aβ40 remained monomeric throughout the incubation period. In contrast, Aβ40 samples reconstituted under low DPC micelle conditions eluted like the monomeric SDS control before the incubation period, but hardly any signal was detected in SEC after completion of the incubation. These results reveal that initially Aβ40 was monomeric and that it assembled into large aggregates with time. To learn more about the morphology of these aggregates, we used transmission electron microscopy (TEM), which showed the presence of abundant amyloid fibrils (Fig. 1B). In summary, under high DPC micelle conditions and after 24 h incubation at 37 °C, Aβ40 remained monomeric and Aβ42 assembled into size homogeneous oligomers (Fig. 1). In contrast, under low DPC micelle conditions and after 24 h incubation at 37 °C, Aβ40 aggregated into amyloid fibrils whereas Aβ42 continued to assemble into size homogeneous oligomers (Fig. 1). Because the latter condition mimicked two possible extreme scenarios for Aβ in the brain of AD patients—evolution into amyloid fibrils, as observed for Aβ40, and assembly into membrane-associated oligomers, as observed for Aβ42—we chose it as the conditions to pursue further studies.
Fig. 1.
Evolution of Aβ40 and Aβ42 samples prepared under low and high DPC micelle conditions. Schematics of four micelles and one micelle, shown in green, represent high and low micelle conditions, respectively. Samples were prepared at 150 μM Aβ under low and high DPC micelles at pH 7.4 and analyzed immediately after being reconstituted (t = 0 h) and after 24 h incubation at 37 °C. SEC chromatograms (A) and electron micrographs (B) at the indicated times. In SEC chromatograms, the orange and purple lines correspond to the elution volumes of the monomeric Aβ40 control (SI Appendix, Fig. S1A) and the Aβ oligomers formed under the studied conditions, respectively. The width of the purple line represents the size distribution of the Aβ oligomers formed under each of the studied conditions. (Scale bars, 100 nm.)
The Aβ42 Oligomers Stabilized by DPC Micelles Insert into Lipid Bilayers as Well-Defined Pores.
Next, we continued to apply established strategies in the field of membrane proteins and tested whether the Aβ42 oligomers formed under the chosen conditions preserved the same pore functionality as observed for Aβ oligomers formed in lipid environments (8–12). To this end, we used electrical recordings with planar lipid bilayers. Samples corresponding to Aβ40 prepared under low DPC micelle conditions, to Aβ42 and Aβ40 monomers, and to empty micelles were also analyzed. Neither the Aβ40 nor the empty micelle samples showed any activity in lipid bilayers. The addition of Aβ42 monomers to the cis side of a planar lipid bilayer induced fast, transient, and heterogeneous ionic current events from approximately −10 to approximately −40 pA at −200 mV (SI Appendix, Fig. S4A). This “spiky” behavior has been reported for Aβ and attributed to the formation of a highly heterogeneous population of Aβ pores (10, 12). The addition of Aβ42 oligomers prepared under low DPC micelle conditions often led to transient disruptions of bilayer conductance (SI Appendix, Fig. S4B), similar to those observed when Aβ42 monomers were allowed to interact with the lipid bilayer (SI Appendix, Fig. S4A). However, 5–15 min after the addition of the Aβ42 oligomers to the chamber, step-wise changes in bilayer conductance were observed (Fig. 2A), behavior typical of the incorporation of individual proteins that form nanopores in membrane bilayers (21). As these oligomers induced various types of nanopore-like behavior, we grouped the responses into three classes, denoted type 1, 2, and 3 on the basis of the signal observed (Fig. 2 B–D). Type 1, observed in approximately 17% of the experiments (n = 105), was characterized by fast and noisy transitions with undefined open pore conductance values (current levels ranging from −40 to −100 pA at −100 mV) (Fig. 2B). Type 2, observed in approximately 48% of the experiments, showed a reasonably well-defined open pore conductance (current level approximately −20 pA at −100 mV) accompanied by significant, rapidly fluctuating noise [root-mean-square (rms) noise > ∼4 pA at −100 mV applying a 2-kHz Bessel filtering] (Fig. 2C). Finally, type 3, which was observed in 35% of the experiments, indicated the presence of a well-defined open pore with no current fluctuations (current level −20 pA at −100 mV) (Fig. 2D). These nanopore-like currents typically appeared to be irreversible, although opening and closing events were occasionally observed. Type 2 and type 3 conductance showed an average open pore current of −19.2 ± 3.5 pA at −100 mV (SI Appendix, Fig. S4 C and D). Moreover, assuming that the internal surface of the Aβ42 oligomeric pores does not affect their conductivity and considering that the pore length is 3 nm, which corresponds to the hydrophobic length of a lipid bilayer, type 2 and 3 pores are consistent with a cylinder with an inner diameter of approximately 0.7 nm (22). All together, these results revealed that Aβ42 oligomers prepared under low DPC micelle conditions have the capacity to form pores in lipid bilayers, the same as Aβ in lipid environments (8–12).
Fig. 2.
Aβ42 oligomers incorporate into lipid bilayers as well-defined pores. Aβ42 oligomers (150-μM Aβ42 concentration) were prepared under low DPC micelles at pH 7.4 and incubated for 24 h at 37 °C (referred to as βPFOsAβ42). (A) Multiple βPFOAβ42 pore insertions. Typical current traces for type 1 (B), type 2 (C), and type 3 (D) βPFOAβ42 pores. Electrical recordings were carried out on diphytanoyl-sn-glycero-3-phosphocholine planar lipid bilayers at the indicated applied potentials.
The Pore-Forming Aβ42 Oligomers Adopt a Specific Structure with Characteristics of a β-Barrel Arrangement.
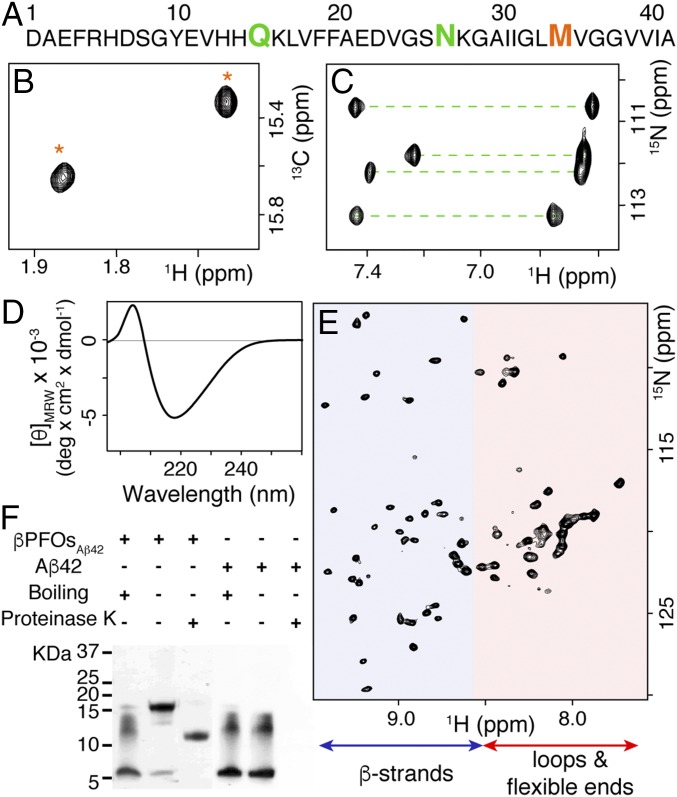
Having confirmed the pore-forming capacity of the Aβ42 oligomers, we proceeded with their structural characterization. Although initially we prepared the oligomer sample at pH 7.4, it was found to have the same structure while being more stable when prepared at pH 9.0 (SI Appendix, Fig. S5). Therefore, we conducted all structural characterization at pH 9.0. Initially, we characterized the Aβ42 oligomers by using carbon-13 incorporation into the methyl group of the methionine side chain (Met-[13CH3]). These methyl groups are highly dynamic and thus have longer relaxation times than those of most hydrogen and carbon atoms in the protein. Moreover, because the sequence of Aβ42 contains a single methionine at residue 35 (Fig. 3A), Met35-[13CH3] Aβ42 labeling offers the additional advantage of spectral simplification. 1H-13C HMQC spectra of the Met35-[13CH3] Aβ42 oligomer sample led to the observation of two sharp and dispersed peaks (Fig. 3B), indicating that the Met-35 side chain perceives two well-defined structural environments. To obtain further information on the number of environments at different sites of the Aβ42 peptide sequence, we measured the 1H-15N HSQC spectra of the 2H,15N Aβ42 oligomer sample and focused on the region of the spectra characteristic of the side-chain amides of Gln and Asn residues. Residues 15 and 27 in the Aβ42 sequence are Gln and Asn, respectively (Fig. 3A). Each side chain amide of Asn and Gln results in two peaks with identical 15N but different 1H chemical shifts. We observed eight peaks, consistent with Asn and Gln chains also perceiving two well-defined environments (Fig. 3C).
Fig. 3.
Pore-forming Aβ42 oligomers adopt a specific β-sheet structure with characteristics of a β-barrel arrangement. Aβ42 oligomers were prepared under low DPC micelles at pH 9.0 and incubated for 24 h at 37 °C (referred to as βPFOsAβ42). (A) Aβ42 sequence highlighting residues Gln-15, Asn-27, and Met-35. (B) 1H-13C HMQC NMR spectrum of Met35-[13CH3] βPFOsAβ42 (1.2-mM Aβ42 concentration). (C) Region of 1H-15N HSQC NMR spectra characteristic of the side chain amides of Gln and Asn residues measured on a 2H,15N βPFOAβ42 sample (1.2 mM Aβ42 concentration). (D) Far-UV CD characterization of βPFOsAβ42 (150-μM Aβ42 concentration). (E) 1H-15N TROSY-HSQC NMR spectrum of 2H,15N βPFOsAβ42 (1.2-mM Aβ42 concentration). The peaks clustered in the random coil region (region shown in red) would be attributable to the loops and flexible ends, whereas the downfield-shifted resonances would correspond to the β-strands of the β-barrel (region shown in blue). (F) SDS/PAGE analysis of monomeric Aβ42 and βPFOsAβ42 samples with and without boiling them and before and after incubation with proteinase K.
Next, to learn about the average secondary structure adopted by the Aβ42 oligomer, we analyzed it by circular dichroism (CD). CD spectra revealed a minimum at 218 nm, indicating that the oligomer adopted a β-sheet structure (Fig. 3D). Further evidence of this type of secondary structure was obtained by analyzing the fingerprint region of 1H-15N TROSY-HSQC spectrum of the 2H,15N Aβ42 oligomer sample (Fig. 3E). The spectrum showed that signals appeared in two differentiated regions—a set of approximately 27 peaks, clustered in the random coil region and a set of 37 downfield-shifted, the latter characteristic of a protein assembly dominated by β-sheet secondary structure. Aβ42 has 42 residues, and NMR experiments using Gln-15, Asn-27, and Met-35 side chains as probes indicated that this peptide adopted two distinct environments (Fig. 3 B and C). Therefore, we expected to detect 82 peaks in the 1H-15N TROSY-HSQC, but only detected 64. This result may be explained because of peak overlap and the fact that at pH 9.0, backbone amides of residues comprising disordered regions of the oligomer undergo fast exchange with the solvent, a process that results in signal loss. To further characterize the two distinct regions of the 1H-15N TROSY-HSQC spectrum, we carried out hydrogen/deuterium exchange (HDX) experiments, which revealed that peaks clustered in the random coil region readily exchanged with solvent deuterons, whereas those in the downfield-shifted region were resistant to exchange (SI Appendix, Fig. S6). All together, these results indicated that the structure of Aβ42 oligomers comprise flexible/disordered regions, and β-strands. Distinct structural arrangements could give rise to the 1H-15N TROSY-HSQC and CD spectra observed. However, given that Aβ42 oligomers form well-defined pores in lipid bilayers (Fig. 2), the most likely structural arrangement of these oligomers is that of a β-barrel structure. In this context, the set of sharp peaks clustered in the random coil region of the spectrum would be attributable to the loops and flexible ends of the β-barrel, whereas the set of downfield-shifted resonances would correspond to the β-strands of the β-barrel (Fig. 3E) (18, 23).
To find additional evidence for a β-barrel arrangement, we analyzed Aβ42 oligomers by means of experiments used to characterize membrane proteins that adopt such a structure. First, even before the first β-barrel membrane protein structure was solved, SDS/PAGE analysis revealed that β-barrel membrane proteins share a characteristic, namely that, when SDS is added, nonboiled samples retain their structure whereas boiled ones lose it (24, 25). Consequently, SDS/PAGE analysis of the nonboiled Aβ42 oligomer sample led to a major band at 18 kDa, which was assigned to the folded structure of the oligomer (Fig. 3F). Upon boiling the sample, the 18-kDa band became a 5-kDa band and a smear ranging from 11 to 14 kDa appeared, an SDS/PAGE pattern characteristic of monomeric Aβ42, thereby indicating that the oligomer had been disrupted (Fig. 3F). Second, another characteristic of β-barrel membrane proteins is that proteases generate polypeptide fragments within the solvent-accessible flexible regions while leaving the β-barrel intact (23). Consistent with this finding, SDS/PAGE analysis of the nonboiled Aβ42 oligomer sample previously incubated with proteinase K revealed a band at 11 kDa (Fig. 3F). To further confirm the effect of the proteases on the structure of the oligomer at atomic level, we measured a 1H-15N HSQC spectrum of an oligomer sample prepared with 15N Aβ42 and incubated with proteinase K. The spectrum revealed that the resonances assigned to residues comprising loops and flexible ends shifted 15N downfield and 1H upfield within the fingerprint region, consistent with the generation of small peptides. In contrast, resonances assigned to residues comprising the β-strands remained intact (SI Appendix, Fig. S7). Taken together, the structural features of the oligomer (Fig. 3) and their pore-forming capacity (Fig. 2) are consistent with a β-barrel structural arrangement. On the basis of these findings, we named this oligomer preparation βPFOsAβ42.
βPFOsAβ42 Structural Integrity Is Maintained in Lipid Environments.
To establish that the structure of βPFOsAβ42 formed in micelles is maintained when incorporated into lipids, we reconstituted the Aβ42 oligomer into bicelles. Because βPFOsAβ42 form under DPC, reconstitution of the oligomers into 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC)/DPC bicelles would be extremely advantageous because it would not require additional detergent exchange steps. However, the DMPC/DPC bicelle system had only previously established for the preparation of large bicelles useful for solid-state NMR measurements (26). Therefore, we first confirmed formation of isotropic small DMPC/DPC bicelles, with molar ratio q = [DMPC]:[DPC] = 0.33, appropriate for solution NMR studies (SI Appendix, Fig. S8). Afterward, we reconstituted βPFOsAβ42 in these bicelles. To confirm oligomer incorporation, we performed 1D [15N,1H]-TROSY for rotational correlation times (TRACT) experiments (27) on βPFOAβ42 samples reconstituted in DPC micelles and DMPC/DPC bicelles (SI Appendix, Fig. S9). The overall correlation time (τc) of βPFOsAβ42 at 37 °C was 22.4 and 30.5 ns, respectively. These values are consistent with previously reported τc for OmpX, an 18-kDa β-barrel protein reconstituted in DHPC micelles (τc = 21–24 ns measured at 30 °C) (27) and DMPC/DHPC bicelles (τc = 35 ns measured with q = 0.5, at 30 °C) (28) and are in complete agreement with the fact that bicelles are larger than micelles.
To assess the number of environments detected for Met-35, Asn-27, and Gln-15 side chains of the oligomer reconstituted in bicelles, we measured 1H-13C HMQC (Fig. 4A) and 1H-15N HSQC (Fig. 4B) on appropriately labeled βPFOAβ42-bicelle samples. Spectra (Fig. 4 A and B) revealed the same number of peaks as those detected in spectra for the micelle samples (Fig. 3 B and C), thereby indicating that the Aβ42 subunits comprising the oligomer reconstituted in bicelles also perceived two environments. Next, we measured a 1H-15N TROSY-HSQC spectrum of 2H, 15N βPFOsAβ42. As expected because of the larger τc measured for βPFOsAβ42 reconstituted in bicelles, the peaks were broader (Fig. 4C) than those detected for the micelle sample (Fig. 3E). However, despite peak broadening, the bicelle spectrum showed good chemical shift dispersion and exhibited approximately the same number of peaks as their respective spectrum counterpart measured in micelles (compare Fig. 3E to Fig. 4C). Indeed, most peaks assigned to the β-strands of the β-barrel in the micelle sample were also detected, with similar chemical shifts, in the bicelle spectrum. In addition, limited proteolysis experiments carried out on the βPFOAβ42 sample reconstituted in bicelles and analyzed by SDS/PAGE further supported a β-barrel arrangement (SI Appendix, Fig. S10). Furthermore, characterization of the bicelle sample using electrical recording with planar lipid bilayers led to the observation of pores with slightly higher average conductance (SI Appendix, Fig. S11) than that observed for the micelle sample (SI Appendix, Fig. S4) but leading to the same type of pores (Fig. 4 D–F) as observed for the micelle sample (Fig. 2). All together these experiments provide strong evidence that the structural integrity of βPFOsAβ42 is preserved in a lipid environment.
Fig. 4.
βPFOsAβ42 maintain their structural integrity in a lipid environment provided by DMPC/DPC bicelles. βPFOsAβ42 were prepared at a 1.2-mM Aβ42 concentration. (A) 1H-13C HMQC NMR spectrum of Met35-[13CH3] βPFOsAβ42 reconstituted in DMPC/DPC bicelles. (B) Region of 1H-15N HSQC NMR spectra characteristic of the side chain amides of Gln and Asn residues measured on a 2H,15N βPFOsAβ42 sample reconstituted in DMPC/DPC bicelles. (C) 1H-15N TROSY-HSQC NMR spectrum of 2H,15N βPFOsAβ42 reconstituted in DMPC/DPC bicelles. Color coding is the same as that detailed in the legend of Fig. 3E. βPFOsAβ42 reconstituted in DMPC/DPC bicelles incorporate into diphytanoyl-sn-glycero-3-phosphocholine planar lipid bilayers as type 1 (D), type 2 (E), and type 3 (F) pores.
Discussion
This study represents a significant advance in our understanding of the role of Aβ oligomers in AD, particularly in a membrane environment. Our work reveals that by handling Aβ as a membrane protein, Aβ variants that differ in hydrophobicity and play a distinct role in the disease show completely different behavior. Under our optimized DPC micelle conditions, the least hydrophobic and most abundantly produced Aβ40 variant aggregated into amyloid fibrils (Fig. 5). In contrast, the more hydrophobic Aβ42 variant, most strongly linked to the etiology of AD, assembled into specific pore-forming β-barrel oligomers, βPFOsAβ42 (Fig. 5). Because Aβ42, relative to Aβ40, has a more prominent role in AD, the higher propensity of Aβ42 to form βPFOs constitutes an indication of their relevance in AD. Notably, because βPFOsAβ42 adopt a specific structure (Fig. 3) that is maintained when the oligomer is reconstituted in a lipid environment (Fig. 4), this study offers the basis for testing a hypothesis regarding the involvement of βPFOs in AD.
Fig. 5.
Evolution of Aβ40 and Aβ42 under our optimized low DPC micelle conditions. After incubation for 24 h at 37 °C, Aβ40 aggregates into amyloid fibrils (Upper), whereas Aβ42 assembles into specific pore-forming β-barrel oligomers, βPFOsAβ42 (Lower). (Scale bar, 100 nm.)
The workflow carried out to conduct this research followed that used to handle membrane proteins. In this regard, various types of detergent micelles were extensively screened to identify the one that preserved the properties of Aβ in lipid environments, namely the capacity to form oligomers and function as pores (8–12). Incubation at 37 °C for 24 h under low DPC micelle conditions led to the formation of a homogenous population of Aβ42 oligomers, as assessed by SEC (Fig. 1A and SI Appendix, Fig. S2F), with the capacity to form pores with defined conductance in lipid bilayers, as tested by electrical recordings using planar lipid bilayers (Fig. 2). Upon selection of optimized micelle conditions, we used specific labels and appropriate NMR experiments to establish the number of environments perceived by the Aβ42 subunits comprising the oligomer (Fig. 3 A–C). The observation of two main environments indicated two possible scenarios, namely the formation of two symmetric oligomers or a asymmetric oligomer with the subunits adopting two distinct structural environments. Although further studies are required to distinguish between these two possibilities, the observation of a single peak by SEC (Fig. 1A) and a single band by SDS/PAGE analysis without boiling the sample (Fig. 3F) is consistent with the latter notion. Next, we determined the structural features of βPFOsAβ42 by CD (Fig. 3D), 1H-15N TROSY-HSQC experiments (Fig. 3E), HDX experiments (SI Appendix, Fig. S6), SDS/PAGE analysis (Fig. 3F), and limited proteolysis (Fig. 3F and SI Appendix, Fig. S7). These experiments were consistent with βPFOsAβ42 adopting a specific structure with characteristics of a β-barrel arrangement.
This structural arrangement has already been proposed for Aβ oligomers formed in solution, on the basis of solvent accessibility measurements derived from top-down HDX mass spectrometry experiments (29) and sequence compatibility with a cylindrin model (30). The latter study reported that the inside of the barrel is filled with packed side chains. This feature would prevent pore formation and would therefore be inconsistent with the properties of βPFOsAβ42. Indeed, structural differences between cylindrin-type oligomers and βPFOsAβ42 are expected because they are formed under different environments, the former in solution and the latter in a biomimetic membrane environment. Aβ oligomers that adopt β-barrel structures and form pores in membranes have also been proposed. Guy and coworkers used modeling approaches to construct a 36 Aβ42 subunit β-barrel oligomer (31). In contrast to the proposed structure for βPFOsAβ42, this model does not encompass flexible/disordered regions. Madhu, Maiti, and coworkers used surface enhanced Raman spectroscopy and solid-state NMR to study membrane-attached Aβ40 oligomers. They detected the presence of a β-turn, flanked by a β-sheet. The orientation of the backbone H-bonds forming the β-sheet was found to be compatible with the notion that Aβ40 membrane oligomers adopt a β-barrel structure (32). Lal and coworkers characterized the structural features of Aβ42 reconstituted in lipids using atomic force microscopy (AFM) imaging (11). They reported donut-shaped structures and oligomeric walls protruding 1 nm above the embedding lipid bilayer surface. The donut shape and the protrusion would be consistent, respectively, with the capacity of βPFOsAβ42 to form pores in lipid bilayers and the presence of flexible/disordered regions outside the micelle within the βPFOAβ42 structure. Using this AFM imaging data, Nussinov and coworkers carried out molecular dynamics simulations to derive atomic models for the structure of β-barrel pore-forming Aβ oligomers (33). This work led to pore structures with inner pore diameters slightly larger (1.7–2.5 nm) than that estimated for βPFOAβ42 (0.7 nm). These models were derived by using shorter Aβ sequences, ranging from residues 9 to 42 or from 17 to 42, and assuming, without any direct 3D structural data, that each Aβ subunit within the β-barrel adopts the same structure as that of Aβ in the fibril. This assumption leads to a β-barrel formed by double β-sheets (33) instead of a single circular β-sheet, as described for transmembrane β-barrel proteins (18, 23). The properties of βPFOsAβ42 are such that they are amenable to studies designed to obtain their 3D atomic structure, thus providing a unique opportunity to obtain a 3D structure of a pore-forming Aβ oligomer.
Pore formation can lead to membrane leakage, which can ultimately cause a depletion of cellular energy stores, neuronal dysfunction, and neuronal death. The amyloid pore hypothesis was proposed for Aβ more than two decades ago (8, 9). Although there is significant evidence supporting this hypothesis, the diversity of Aβ pores reported (10–12) and the lack of specific structural properties characterizing them have impeded confirmation or rejection of this hypothesis. βPFOsAβ42 adopt a specific structure and form well-defined pores, thereby offering a unique opportunity to establish the relevance of pore formation in the context of AD, for example by producing antibodies that specifically recognize this form of Aβ. Such antibodies could then be used to validate βPFOAβ42 structures in relevant AD models. In addition, because, as mentioned, the properties of βPFOsAβ42 are amenable to 3D atomic structure studies, upon βPFOAβ42 validation, the 3D structure could be used to develop new therapeutic agents. Therefore, the βPFOAβ42 species whose formation and functional characteristics are described in this paper have the potential to become viable targets through which to search for new types of molecular agents designed to fight AD.
Materials and Methods
SI Appendix, Materials and Methods provides detailed protocols for the following: preparation of monomeric Aβ, reconstitution of Aβ in different detergent micelles, and preparation of Aβ40 monomeric in the presence of SDS micelles and of βPFOsAβ42. It also provides a description of how the electrical recordings with planar lipid bilayers and NMR studies were carried out, as well as how SEC, TEM, CD, SDS/PAGE, and limited proteolysis experiments were performed.
Supplementary Material
Acknowledgments
We thank Prof. C. M. Dobson, Dr. J. García, Prof. E. Giralt, and Prof. M. Pons for helpful discussions; and the Protein Expression Core Facility at Institute for Research in Biomedicine (IRB) Barcelona for technical support. IRB Barcelona is the recipient of a Severo Ochoa Award of Excellence from Ministerio de Economía y Competitividad (MINECO) (Government of Spain). This work was supported by MINECO-Fondo Europeo de Desarrollo Regional (FEDER) Formación de Personal Investigador (FPI) Program Grants SAF2012-35226 and SAF2015-68789 (to N.C.); Fundació La Marató de TV3 Program Grant 20140730/31 (to N.C. and G.M.). M.N.-P. acknowledges the Spanish Government program for a predoctoral fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605104113/-/DCSupplemental.
References
- 1.Masters CL, Selkoe DJ. Biochemistry of amyloid β-protein and amyloid deposits in Alzheimer disease. Cold Spring Harb Perspect Med. 2012;2(6):a006262. doi: 10.1101/cshperspect.a006262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chávez-Gutiérrez L, et al. The mechanism of γ-Secretase dysfunction in familial Alzheimer disease. EMBO J. 2012;31(10):2261–2274. doi: 10.1038/emboj.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambert MP, et al. Diffusible, nonfibrillar ligands derived from Abeta1-42 are potent central nervous system neurotoxins. Proc Natl Acad Sci USA. 1998;95(11):6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barghorn S, et al. Globular amyloid beta-peptide oligomer—A homogenous and stable neuropathological protein in Alzheimer’s disease. J Neurochem. 2005;95(3):834–847. doi: 10.1111/j.1471-4159.2005.03407.x. [DOI] [PubMed] [Google Scholar]
- 5.Chimon S, et al. Evidence of fibril-like β-sheet structures in a neurotoxic amyloid intermediate of Alzheimer’s β-amyloid. Nat Struct Mol Biol. 2007;14(12):1157–1164. doi: 10.1038/nsmb1345. [DOI] [PubMed] [Google Scholar]
- 6.Laurén J, Gimbel DA, Nygaard HB, Gilbert JW, Strittmatter SM. Cellular prion protein mediates impairment of synaptic plasticity by amyloid-beta oligomers. Nature. 2009;457(7233):1128–1132. doi: 10.1038/nature07761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kayed R, et al. Permeabilization of lipid bilayers is a common conformation-dependent activity of soluble amyloid oligomers in protein misfolding diseases. J Biol Chem. 2004;279(45):46363–46366. doi: 10.1074/jbc.C400260200. [DOI] [PubMed] [Google Scholar]
- 8.Arispe N, Rojas E, Pollard HB. Alzheimer disease amyloid beta protein forms calcium channels in bilayer membranes: Blockade by tromethamine and aluminum. Proc Natl Acad Sci USA. 1993;90(2):567–571. doi: 10.1073/pnas.90.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arispe N, Pollard HB, Rojas E. Zn2+ interaction with Alzheimer amyloid beta protein calcium channels. Proc Natl Acad Sci USA. 1996;93(4):1710–1715. doi: 10.1073/pnas.93.4.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirakura Y, Lin MC, Kagan BL. Alzheimer amyloid abeta1-42 channels: Effects of solvent, pH, and Congo Red. J Neurosci Res. 1999;57(4):458–466. [PubMed] [Google Scholar]
- 11.Lin H, Bhatia R, Lal R. Amyloid β protein forms ion channels: Implications for Alzheimer’s disease pathophysiology. FASEB J. 2001;15(13):2433–2444. doi: 10.1096/fj.01-0377com. [DOI] [PubMed] [Google Scholar]
- 12.Kourie JI, Henry CL, Farrelly P. Diversity of amyloid beta protein fragment [1-40]-formed channels. Cell Mol Neurobiol. 2001;21(3):255–284. doi: 10.1023/a:1010995121153. [DOI] [PubMed] [Google Scholar]
- 13.Shao H, Jao S, Ma K, Zagorski MG. Solution structures of micelle-bound amyloid β-(1-40) and β-(1-42) peptides of Alzheimer’s disease. J Mol Biol. 1999;285(2):755–773. doi: 10.1006/jmbi.1998.2348. [DOI] [PubMed] [Google Scholar]
- 14.Mandal PK, Pettegrew JW. Alzheimer’s disease: Soluble oligomeric Abeta(1-40) peptide in membrane mimic environment from solution NMR and circular dichroism studies. Neurochem Res. 2004;29(12):2267–2272. doi: 10.1007/s11064-004-7035-1. [DOI] [PubMed] [Google Scholar]
- 15.Yu L, et al. Structural characterization of a soluble amyloid β-peptide oligomer. Biochemistry. 2009;48(9):1870–1877. doi: 10.1021/bi802046n. [DOI] [PubMed] [Google Scholar]
- 16.Sanders CR, Sönnichsen F. Solution NMR of membrane proteins: Practice and challenges. Magn Reson Chem. 2006;44(Spec No) S1:S24–S40. doi: 10.1002/mrc.1816. [DOI] [PubMed] [Google Scholar]
- 17.Columbus L, et al. Mixing and matching detergents for membrane protein NMR structure determination. J Am Chem Soc. 2009;131(21):7320–7326. doi: 10.1021/ja808776j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiller S, et al. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science. 2008;321(5893):1206–1210. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett PJ, et al. The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol. Science. 2012;336(6085):1168–1171. doi: 10.1126/science.1219988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popot J-L. Amphipols, nanodiscs, and fluorinated surfactants: Three nonconventional approaches to studying membrane proteins in aqueous solutions. Annu Rev Biochem. 2010;79(1):737–775. doi: 10.1146/annurev.biochem.052208.114057. [DOI] [PubMed] [Google Scholar]
- 21.Soskine M, Biesemans A, De Maeyer M, Maglia G. Tuning the size and properties of ClyA nanopores assisted by directed evolution. J Am Chem Soc. 2013;135(36):13456–13463. doi: 10.1021/ja4053398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kowalczyk SW, Grosberg AY, Rabin Y, Dekker C. Modeling the conductance and DNA blockade of solid-state nanopores. Nanotechnology. 2011;22(31):315101. doi: 10.1088/0957-4484/22/31/315101. [DOI] [PubMed] [Google Scholar]
- 23.Fox DA, Columbus L. Solution NMR resonance assignment strategies for β-barrel membrane proteins. Protein Sci. 2013;22(8):1133–1140. doi: 10.1002/pro.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burgess NK, Dao TP, Stanley AM, Fleming KG. β-barrel proteins that reside in the Escherichia coli outer membrane in vivo demonstrate varied folding behavior in vitro. J Biol Chem. 2008;283(39):26748–26758. doi: 10.1074/jbc.M802754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otzen DE, Andersen KK. Folding of outer membrane proteins. Arch Biochem Biophys. 2013;531(1-2):34–43. doi: 10.1016/j.abb.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 26.Nolandt OV, Walther TH, Grage SL, Ulrich AS. Magnetically oriented dodecylphosphocholine bicelles for solid-state NMR structure analysis. BBA - Biomembranes. 2012;1818(5):1142–1147. doi: 10.1016/j.bbamem.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 27.Lee D, Hilty C, Wider G, Wüthrich K. Effective rotational correlation times of proteins from NMR relaxation interference. J Magn Reson. 2006;178(1):72–76. doi: 10.1016/j.jmr.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 28.Lee D, et al. Bilayer in small bicelles revealed by lipid-protein interactions using NMR spectroscopy. J Am Chem Soc. 2008;130(42):13822–13823. doi: 10.1021/ja803686p. [DOI] [PubMed] [Google Scholar]
- 29.Pan J, Han J, Borchers CH, Konermann L. Structure and dynamics of small soluble Aβ(1-40) oligomers studied by top-down hydrogen exchange mass spectrometry. Biochemistry. 2012;51(17):3694–3703. doi: 10.1021/bi3002049. [DOI] [PubMed] [Google Scholar]
- 30.Laganowsky A, et al. Atomic view of a toxic amyloid small oligomer. Science. 2012;335(6073):1228–1231. doi: 10.1126/science.1213151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shafrir Y, Durell S, Arispe N, Guy HR. Models of membrane-bound Alzheimer’s Abeta peptide assemblies. Proteins. 2010;78(16):3473–3487. doi: 10.1002/prot.22853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhowmik D, et al. Cell-membrane-mimicking lipid-coated nanoparticles confer Raman enhancement to membrane proteins and reveal membrane-attached amyloid-β conformation. ACS Nano. 2015;9(9):9070–9077. doi: 10.1021/acsnano.5b03175. [DOI] [PubMed] [Google Scholar]
- 33.Jang H, et al. Disordered amyloidogenic peptides may insert into the membrane and assemble into common cyclic structural motifs. Chem Soc Rev. 2014;43(19):6750–6764. doi: 10.1039/c3cs60459d. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.