Significance
Despite its importance in epithelial development and in breast cancer, the role of EGF receptor (EGFR) signaling and its regulation in the postnatal mammary stroma has remained unclear. Here, we show that sprouty 1 (Spry1) modulates epithelial–stromal interactions by inhibiting EGFR-dependent stromal paracrine signaling and ECM remodeling. Spry1 loss leads to upregulated EGFR signaling and enhanced stromal activities, including ECM remodeling and growth factor production, and ultimately increased epithelial branching. By contrast, EGFR signaling reduction in fibroblasts leads to stunted epithelial branching. We show that EGFR and FGF receptor (FGFR) signaling constitute a critical part of stromal–epithelial interactions during postnatal development and that these signaling pathways are fine-tuned by different members of the Sprouty gene family to ensure normal epithelial morphogenesis of the mammary gland.
Keywords: branching morphogenesis, FGF signaling, EGF signaling, epithelial–stromal interactions, stromal microenvironment
Abstract
The role of the local microenvironment in influencing cell behavior is central to both normal development and cancer formation. Here, we show that sprouty 1 (SPRY1) modulates the microenvironment to enable proper mammary branching morphogenesis. This process occurs through negative regulation of epidermal growth factor receptor (EGFR) signaling in mammary stroma. Loss of SPRY1 resulted in up-regulation of EGFR–extracellular signal–regulated kinase (ERK) signaling in response to amphiregulin and transforming growth factor alpha stimulation. Consequently, stromal paracrine signaling and ECM remodeling is augmented, leading to increased epithelial branching in the mutant gland. By contrast, down-regulation of EGFR–ERK signaling due to gain of Sprouty function in the stroma led to stunted epithelial branching. Taken together, our results show that modulation of stromal paracrine signaling and ECM remodeling by SPRY1 regulates mammary epithelial morphogenesis during postnatal development.
A central theme in developmental and cancer biology research is to understand the mechanisms by which the local microenvironment, or niche, influences fundamental aspects of cell and tissue behavior during organ formation and function (1). The importance of mesenchyme, as the embryonic niche, in determining organ identity and fate of the epithelium was demonstrated long ago by classic embryological studies (2, 3). More recent studies have shown, however, that postnatal stroma, a derivative of embryonic mesenchyme, is essential for maintenance of cell fate and differentiation status in adult life (4, 5). Indeed, abnormal stroma can lead to the formation of a cancer microenvironment, and it plays a causative role during tumor onset and progression (1, 6). As such, understanding postnatal stromal biology and the mechanism by which its deregulation may promote tumorigenesis is of key importance.
The mouse mammary gland is a powerful model for understanding the genetic and cellular basis of stromal biology (7). The observation that epithelial branching of the mammary gland persists for many weeks after birth has made amenable the detection of stromal influences on epithelial invasion and patterning (8, 9). Indeed, postnatal mammary stroma is composed of many cell types, including periductal fibroblasts and white adipocytes, endothelial cells, nerve cells, and a variety of infiltrating immune cells, including macrophages and eosinophils, which play an important role in postnatal branching (10).
Mammary stroma regulates epithelial branching by at least two mechanisms. Stromal cells produce several paracrine factors, including fibroblast growth factors (FGFs) and insulin-like growth factor (IGF) that activate receptor tyrosine kinases (RTK) (11, 12). For example, a reduction in FGF signaling due to loss of FGF10 or its receptor FGF receptor 1/2 (FGFR1/2), or reduced IGF signaling, leads to stunted epithelial branching (13–16). By contrast, excessive FGF signaling due to overactive FGF ligand or receptor causes breast tumorigenesis in mouse models (17–19). In addition, stromal cells also play a major role in remodeling of the ECM, whose unique physical, biochemical, and biomechanical properties make it an essential component for a wide variety of developmental processes including epithelial branching (1, 6). ECM remodeling is highly regulated and its deregulation is a major contributor to tumorigenesis (20).
The postnatal mammary gland stroma is under both systemic hormonal and local control based on epithelial-derived paracrine signaling via, for example, the EGF receptor (EGFR) pathway (21). Whereas a reduction in stromal EGFR signaling due to genetic ablation of the receptor or its ligands, including amphiregulin (AREG) and transforming growth factor alpha (TGFα), leads to deficient epithelial branching (22–24), excessive EGFR signaling causes breast tumorigenesis and is a therapeutic target of human cancer (25, 26). Thus, stromal EGFR signaling is important for epithelial branching of the mammary gland.
Despite its requirement for epithelial branching, the mechanisms by which EGFR signaling functions and is modulated in the mammary gland stroma have remained unclear. Here, we hypothesized that members of the Sprouty family, which encode intracellular RTK signaling inhibitors essential for numerous vertebrate developmental processes (27–30), regulate EGFR signaling in the mammary gland stroma. To test this hypothesis, we determined the mammary gland phenotype in mice lacking sprouty 1 (Spry1) and analyzed the mechanisms by which SPRY1 functions. These studies revealed an important role for SPRY1 in mammary gland stromal–epithelial cross talk.
Results
Spry1 Is Expressed in Mammary Stroma and Basal Epithelium.
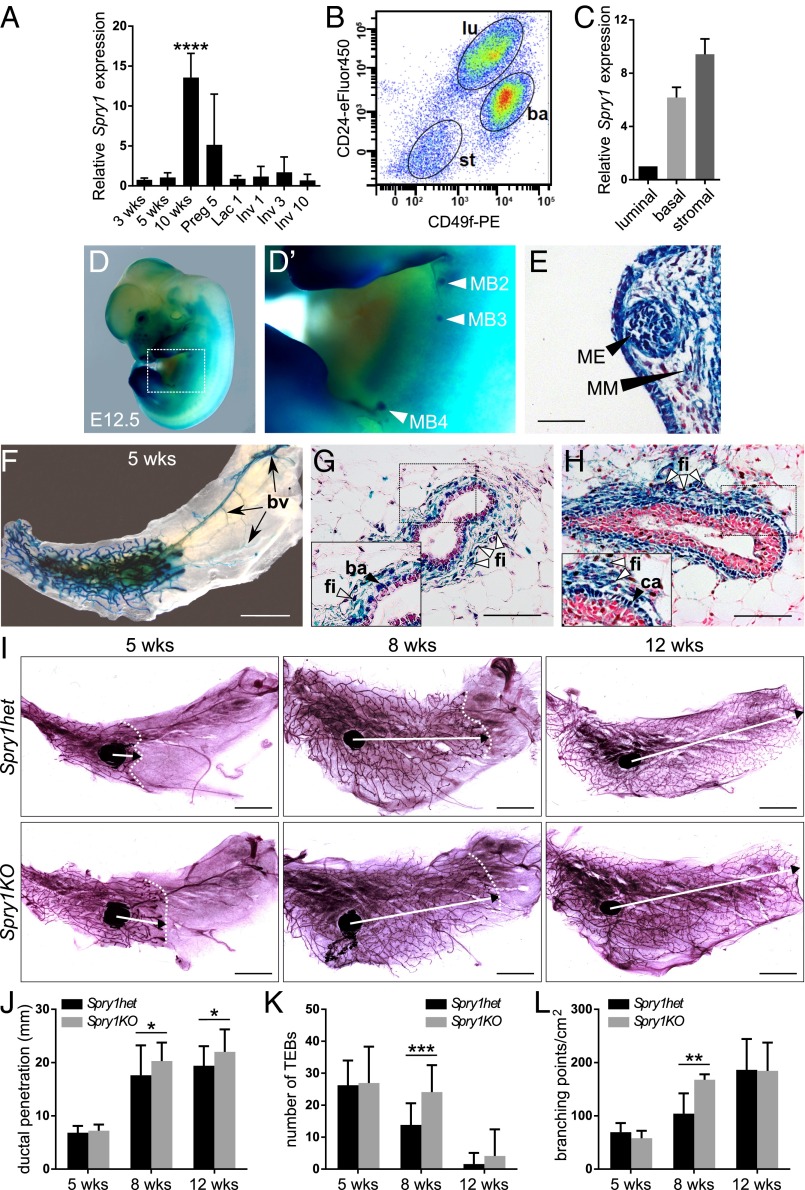
We examined the expression of Spry1 during various stages of postnatal mammary gland development. Spry1 mRNA was readily detected at all of the stages examined by quantitative real-time PCR (qPCR), including during epithelial branching, pregnancy, lactation, and involution (Fig. 1A). We used fluorescence-activated cell sorting (FACS) to sort mammary epithelia based on their expression of CD24 and CD49f cell surface markers (Fig. 1B). Spry1 was highly expressed in basal (CD24medCD49fhi) and stromal (CD24negCD49fneg) cells compared with low levels in luminal cells (CD24hiCD49flow) (Fig. 1C).
Fig. 1.
Epithelial branching is increased in mammary glands lacking Spry1 function. (A–C) Spry1 expression in mammary gland. (A) Relative Spry1 expression during mammary gland development as detected by qPCR and normalized to Actb. RNA was isolated from mammary glands from virgin female mice at 3, 5, and 10 wk of age, and female mice on day 5 of pregnancy (Preg), day 1 of lactation (Lac), and days 1, 3, and 10 of involution (Inv). Graph shows mean ± SD (n = 3). Statistical analysis was performed using one-way ANOVA; ****P < 0.0001. (B) Gating strategy for sorting of mammary gland cell populations by FACS; basal (ba), luminal (lu), and stromal (st) cells. (C) Relative expression of Spry1 in sorted luminal, basal, and stromal cells, normalized to Actb. (D–H) Spry1 expression during embryonic (D and E) and postnatal mammary gland development (F–H) as detected by β-galactosidase (β-gal) staining on Spry1het embryos or tissues. (D and D′) Spry1 expression in mammary bud (MB) #2, #3, and #4 (arrowheads) at E12.5. (E) Cross-section of the MB; ME, mammary epithelium; MM, mammary mesenchyme. Nuclei were counterstained with nuclear fast red. (Scale bar, 50 µm.) (F–H) Spry1 expression in the whole-mount (F) and paraffin sections (G and H) of mammary gland at 5 wk of age as detected by β-gal staining. (F) Spry1 is expressed in the branching epithelial network as well as in the blood vessels (bv; arrows). (Scale bars, 5 mm.) (G, H) Cross sections of β-gal–stained mammary duct (G) and terminal end bud (TEB) (H) revealed expression of Spry1 in fibroblasts (fi), basal cells (ba), and caps cells (ca). Nuclei were counterstained with nuclear fast red. (Scale bars, 100 µm.) (I) Whole mounts of mammary glands from 5-, 8-, and 12-wk-old Spry1het and Spry1KO mice stained with carmine red. Images of parts of the mammary gland were merged in Photoshop (Adobe) to create a full view of the wholemount gland. Arrows indicate the extent of ductal penetration in the fat pad; dotted white line illustrates the epithelial invasion front. (Scale bars, 5 mm.) (J–L) Quantitative comparisons of ductal penetration (J), number of TEBs (K), and branching (L) between Spry1het and Spry1KO glands. Plots show mean ± SD (n = 3–6 litters/age). Statistical analysis was performed using two-way ANOVA; *P < 0.05; **P < 0.01; ***P < 0.001.
To assess the spatiotemporal patterns of Spry1 expression during mammary gland development, we took advantage of Spry1LacZ/+ (Spry1 heterozygous, Spry1het) mice carrying the Spry1LacZ allele, which is a null allele that also reports Spry1 expression due to replacement of essential functional domains by the LacZ gene (31). Spry1 was expressed in many tissues of the developing embryo at embryonic day 12.5 (E12.5) (Fig. 1 D and D′), including both the mammary bud epithelium and mesenchyme (Fig. 1E). In pubertal mammary glands, when epithelial branching is ongoing, we detected Spry1 expression in the branching epithelial network and blood vessels in the distal stroma (Fig. 1F). Histological sections of stained mammary glands confirmed that Spry1 is expressed in periductal fibroblasts (Fig. 1 G and H), cap cells of terminal end buds (Fig. 1H), and the basal ductal epithelium (Fig. 1G), but there was little expression in the luminal epithelium.
Loss of Spry1 Function Leads to Increased Branching Activities in the Mammary Epithelium.
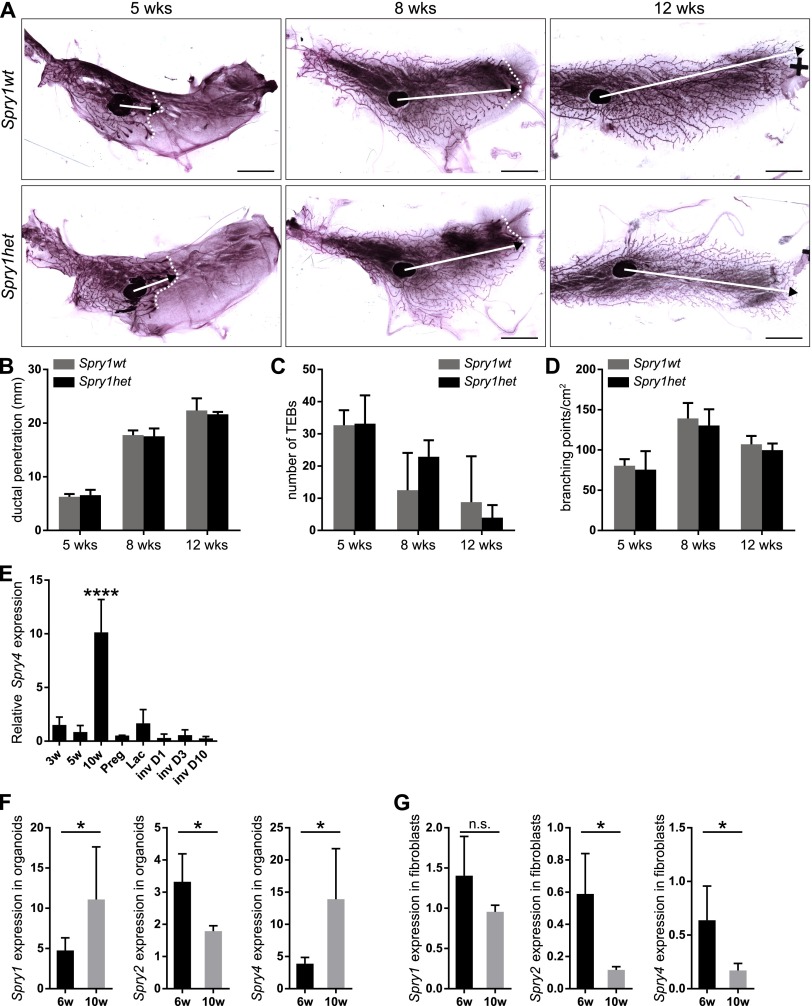
We examined whether the mammary gland develops normally in mice lacking Spry1 function. Compared with mammary glands from pubertal and adult virgin Spry1+/+ [Spry1 wild-type (Spry1wt)] mice, those from Spry1LacZ/+ (Spry1het) mice did not show any significant differences in ductal penetration, terminal end buds (TEBs), or branching (Fig. S1 A–D). These findings were consistent with previously reported haplosufficiency of Sprouty genes in various vertebrate developmental processes (27, 32, 33).
Fig. S1.
Whole-mount analysis reveals no difference between Spry1wt and Spry1het mammary glands. (A) Whole mounts of mammary glands from 5-, 8-, and 12-wk-old Spry1wt and Spry1het mice stained with carmine red. Images of parts of the mammary gland were merged in Photoshop (Adobe) to create a full view of the wholemount gland. Arrows indicate the extent of ductal penetration in the fat pad; dotted white line illustrates the epithelial invasion front. (Scale bars, 5 mm.) (B–D) Quantitative comparisons of ductal penetration (B), number of terminal end buds (TEBs) (C), and branching (D) between Spry1wt and Spry1het glands. Plots show mean ± SD (n = 3–5 litters/age). Statistical analysis was performed using two-way ANOVA and revealed no significant difference between Spry1wt and Spry1het mammary gland. (E–G) Expression of Sprouty genes in mammary gland. (E) Relative Spry4 expression during mammary gland development. RNA was isolated from mammary glands from virgin female mice at 3, 5, and 10 wk of age, and female mice on day 5 of pregnancy (Preg), day 1 of lactation (Lac), and days 1, 3, and 10 of involution (Inv). Graph shows mean ± SD (n = 3). Statistical analysis was performed using one-way ANOVA; ****P < 0.0001. (F and G) Relative expression of Sprouty genes in mammary epithelial organoids (F) and fibroblasts (G) from 6- and 10-wk-old mice. Graphs show mean ± SD (n = 3–7). Statistical analysis was performed using unpaired t test; *P < 0.05; n.s., not significant.
However, when we compared mammary glands from pubertal and adult virgin Spry1het and Spry1LacZ/LacZ [Spry1 knockout (Spry1KO)] female mice, we found significant differences in epithelial morphogenesis. In mice at 8 and 12 wk of age, the epithelium of the KO animals reached further into the fat pad (Fig. 1 I and J). Moreover, at 8 wk of age, mammary glands from Spry1KO mice showed increased number of TEBs (Fig. 1K) and increased branching (Fig. 1L).
Interestingly, this branching phenotype was resolved at 12 wk of age. The lack of overall major defects in Spry1KO mammary glands suggests that other family members could compensate for loss of Spry1. Indeed, Spry2 (30) and Spry4 were expressed in the mammary gland (Fig. S1 E–G), although Spry3 was not detectable. The spatiotemporal pattern of Spry4 expression was most similar to Spry1, suggesting that Spry4 may be redundant for Spry1.
These results reveal that Spry1 negatively regulates epithelial invasion and branching in pubertal mammary gland. However, our data suggest that the loss of Spry1 can be overcome by other Spry proteins later in development.
Spry1 Loss in the Mammary Epithelium Does Not Affect Mammary Branching.
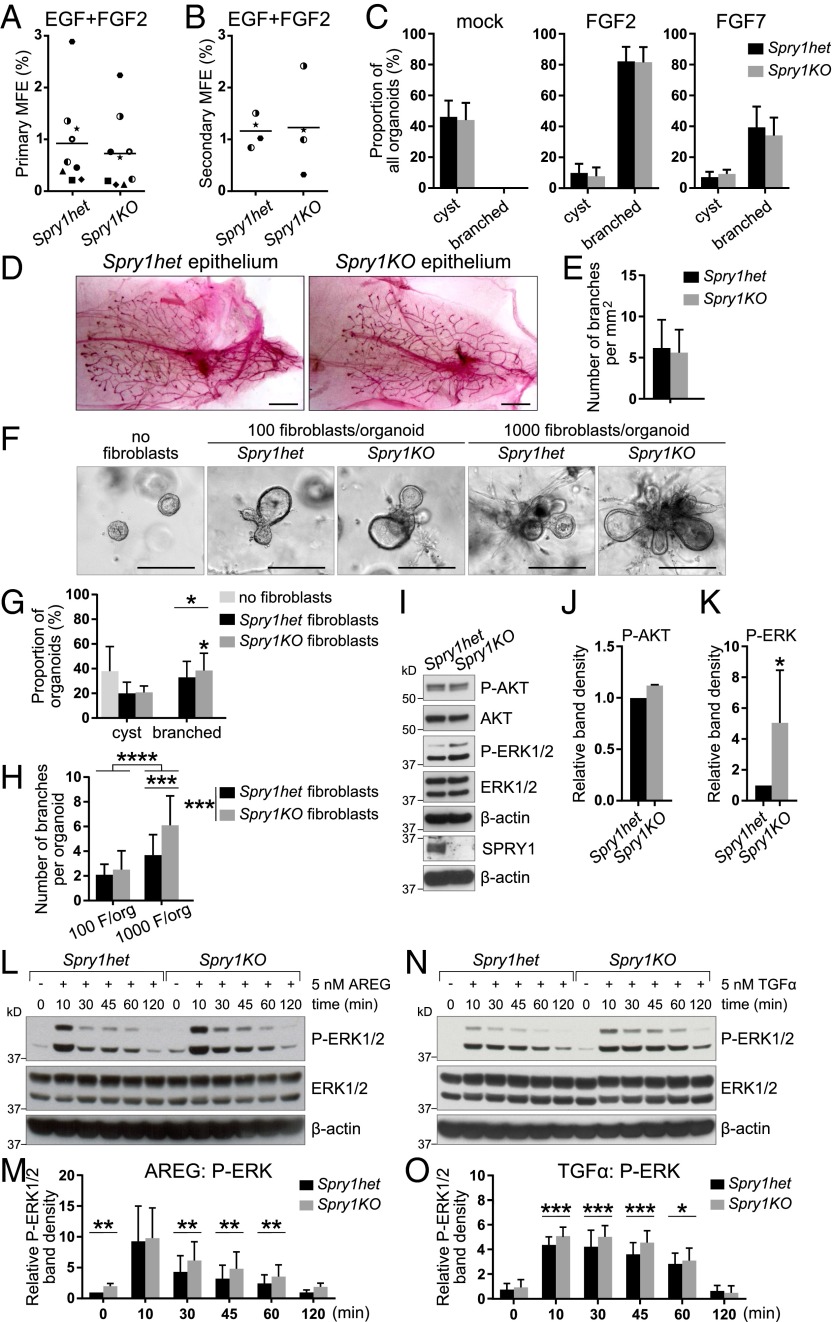
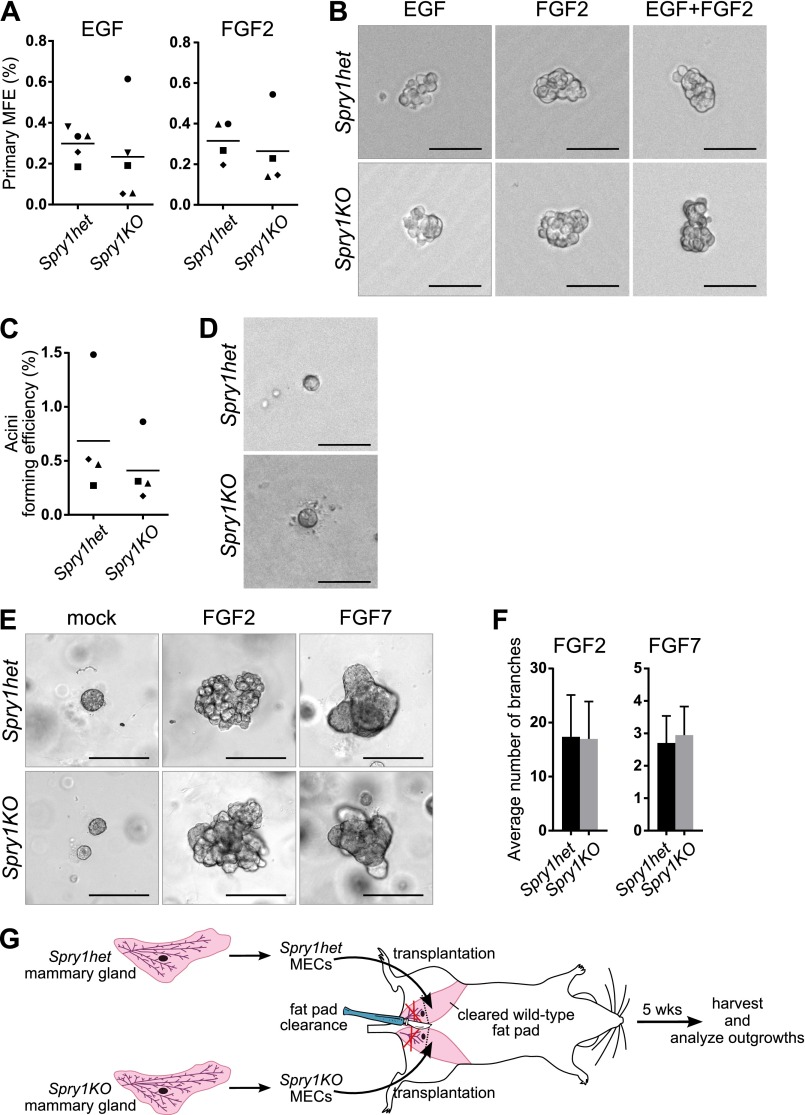
Our observation that Spry1 is expressed in both the epithelium and the stroma of the mammary gland indicated that the branching phenotype of Spry1KO mice could result from defects in either compartment. Considering its expression in the basal layer, which is rich in adult stem cells (34), we asked whether Spry1 is essential for mammary stem cell biology and thus epithelial expansion during branching morphogenesis. We used the mammosphere assay (35), which measures progenitor/stem cell potential of mammary epithelial cells based on their ability to survive and form mammospheric colonies in suspension (primary mammosphere assay) and to self-renew (secondary mammosphere assay), with two commonly used growth factors, EGF and/or FGF2. We observed no statistically significant difference in mammosphere forming efficiency between Spry1het and Spry1KO cells in all cases (Fig. 2 A and B, and Fig. S2 A and B).
Fig. 2.
Spry1-null fibroblasts are sensitized to AREG/TGFα–ERK signaling and show elevated activities in promoting epithelial branching in vitro. (A and B) Plots depicting primary (A) and secondary (B) mammosphere-forming efficiency (MFE) of Spry1het and Spry1KO basal cells. Data points show paired values, and lines indicate mean (n = 4 or 9). Statistical analysis was performed using paired t test and revealed no significant difference in MFE between Spry1het and Spry1KO basal cells. (C) In vitro branching assay. Spry1het and Spry1KO organoids were cultured in Matrigel in basal organoid medium without any growth factor or supplemented with FGF2 or FGF7. Graphs show frequencies of formation of cysts and branched organoids, presented as mean ± SEM (n = 4). Statistical analysis was performed using paired t test and revealed no significant difference in organoid morphogenesis between Spry1het and Spry1KO basal cells. (D and E) Transplantation of Spry1het and Spry1KO MECs into wild-type fat pads. (D) Representative photographs of whole mounts harvested 5 wk after transplantation and stained with carmine red. (Scale bars, 2.5 mm.) (E) Quantification of epithelial branching. The plot shows mean ± SEM (n = 7 per genotype). Statistical analysis was performed using paired t test and revealed no significant difference in epithelial outgrowths between Spry1het and Spry1KO basal cells. (F–H) In vitro coculture branching assay, in which Spry1het or Spry1KO fibroblasts were cocultured with wild-type organoids [at the ratio of 100 or 1,000 fibroblasts (F) per organoid] in Matrigel in basal organoid medium without addition of growth factors. (F) Representative photographs of organoids in cocultures. (G) The graph shows frequencies of formation of cysts and branched organoids, presented as mean ± SEM (n = 4). Statistical analysis was performed using paired t test; *P < 0.05. (H) The graph shows number of branches per branched organoid, presented as mean ± SD (n = 13–22). Statistical analysis was performed using two-way ANOVA with multiple comparisons; ***P < 0.001; ****P < 0.0001. (I–K) Western blot analysis of the phosphorylation status of AKT and ERK1/2 in Spry1het and Spry1KO fibroblasts cultured in normal medium with serum. (I) P-AKT (Ser473), AKT, P-ERK1/2 (Thr202/Tyr204), ERK, and β-actin signals were detected on a single blot. (J and K) Quantitative comparison of AKT (J) and ERK1/2 (K) phosphorylation. The plots show P-AKT (Ser473) band density, normalized to total AKT (J), and P-ERK1/2 (Thr202/Tyr204) band density, normalized to total ERK1/2 (K), as mean ± SD (n = 3). Statistical analysis was performed using paired t test; *P < 0.05. (L–O) Western blot analysis of phosphorylation status of ERK1/2 in Spry1het and Spry1KO fibroblasts in response to AREG (L and M) or TGFα stimulation (N and O). Fibroblasts were serum-starved, treated with 5 nM AREG or TGFα for the indicated time, and lysed, and on a single blot, P-ERK1/2 (Thr202/Tyr204), ERK, and β-actin signals were detected. (M and O) Quantitative comparison of ERK1/2 phosphorylation, normalized to total ERK1/2. Data are mean ± SD (n = 3–5). Statistical analysis was performed using two-way ANOVA; *P < 0.05; **P < 0.01; ***P < 0.001.
Fig. S2.
Loss of Spry1 function in the epithelium is not the cause of branching defects in mutant mice. (A) Mammosphere-forming efficiency (MFE) of Spry1het and Spry1KO basal cells cultured in mammosphere medium supplemented with EGF or FGF2. Data points show paired values, and the lines indicate mean (n = 4–5). Statistical analysis was performed using paired t test and revealed no significant difference in MFE between Spry1het and Spry1KO basal cells. (B) Representative photographs of mammospheres. (C) Plot depicting acini-forming efficiency of Spry1het and Spry1KO basal cells. Data points show paired values, and lines indicate mean (n = 4). Statistical analysis was performed using paired t test and revealed no significant difference between Spry1het and Spry1KO basal cells. (D) Representative photographs of Spry1het and Spry1KO acini. (E and F) In vitro branching assay. (E) Representative photographs of Spry1het and Spry1KO organoids cultured in Matrigel in basal organoid medium without any growth factor or supplemented with FGF2 or FGF7. (Scale bars, 200 µm.) (F) Plots show average number of branches per branching organoid ± SD (n = 4). Statistical analysis was performed using paired t test and revealed no significant difference between Spry1het and Spry1KO organoids. (G) Diagram depicting the procedure of transplantation of Spry1het and Spry1KO MECs into wild-type fat pad.
Similarly, using the acinar assay (36), which measures the ability of mammary epithelial progenitor/stem cells to form acinus-like colonies in 3D Matrigel, we observed no statistically significant difference between Spry1het and Spry1KO basal cells in acinus-forming efficiency (Fig. S2 C and D). Together, these results suggest that Spry1 loss in basal cells is unlikely to have a major effect on mammary stem cell biology and to be the cause of the branching phenotype observed in Spry1KO mice.
Another possibility is that Spry1-null epithelium undergoes branching more readily than normal during postpubertal development. To examine this possibility, we performed 3D in vitro culture studies in which mammary epithelium forms branches in response to growth factor treatment (30). Mammary epithelium did not branch in the absence of growth factors but was able to form branched structures in the presence of FGF2 or FGF7 (Fig. S2E). We found no significant differences between Spry1het organoids and Spry1KO organoids in their responses to FGF2 or FGF7 in terms of proportion of branched organoids (Fig. 2C) or number of branches per organoid (Fig. S2F). These data suggest that Spry1-null epithelium is not defective and responds to branching signals similarly to control epithelium.
To test whether loss of epithelial Spry1 affects mammary branching in vivo, we transplanted 10,000 mammary epithelial cells (MECs) from Spry1het and Spry1KO mice into “cleared” fat pads lacking the endogenous epithelial network of the #4 mammary gland in 3-wk-old nude mice (Fig. S2G). Five weeks after surgery, we harvested transplants and stained them with carmine red. We found that MECs derived from both Spry1het and Spry1KO mice formed epithelial networks, but no significant difference was detected in epithelial branching (Fig. 2 D and E).
These results show that the loss of Spry1 in the epithelium does not affect mammary gland branching and is unlikely to be the cause of accelerated branching in Spry1KO mice.
Loss of Spry1 in Stromal Cells Promotes Increased Epithelial Branching in Vitro.
To determine whether Spry1-null stroma promotes increased epithelial branching, we developed an in vitro assay in which wild-type organoids were cocultured with fibroblasts harvested from Spry1het and Spry1KO mice. Mammary organoids did not form branched structures when cultured in basal medium alone (Fig. 2F). However, when they were cocultured with fibroblasts, mammary organoids formed branches in 4–5 d (Fig. 2F), with more branches forming when more fibroblasts and organoids were present.
When cocultured with Spry1KO fibroblasts, the organoids branched more readily and formed more branches than with Spry1het fibroblasts (Fig. 2 G and H). This observation is consistent with the in vivo phenotype and suggests that Spry1-null stroma promotes epithelial branching more strongly than controls.
These results indicate that the ductal phenotype arising from the loss of Spry1 arises from defects within the stroma, rather than within the epithelium.
In Mammary Fibroblasts, Spry1 Is a Negative Regulator of EGFR-ERK Signaling in Response to AREG and TGFα.
Sprouty genes regulate RTK signaling in various developmental contexts (37, 38). Therefore, we used Western blotting to examine the phosphorylation and activation status of protein kinase B (AKT) and extracellular signal-regulated kinase (ERK), key components of the two main branches downstream of RTK signaling. We found that AKT activation (phospho-Ser473) was the same in Spry1het and Spry1KO fibroblasts (Fig. 2 I and J). In contrast, ERK phosphorylation at Thr202/Tyr204 was up-regulated in Spry1KO fibroblasts compared with Spry1het fibroblasts (Fig. 2 I and K). These data indicate that Spry1 loss leads to up-regulation of RTK signaling through the ERK arm of the pathway in mammary fibroblasts.
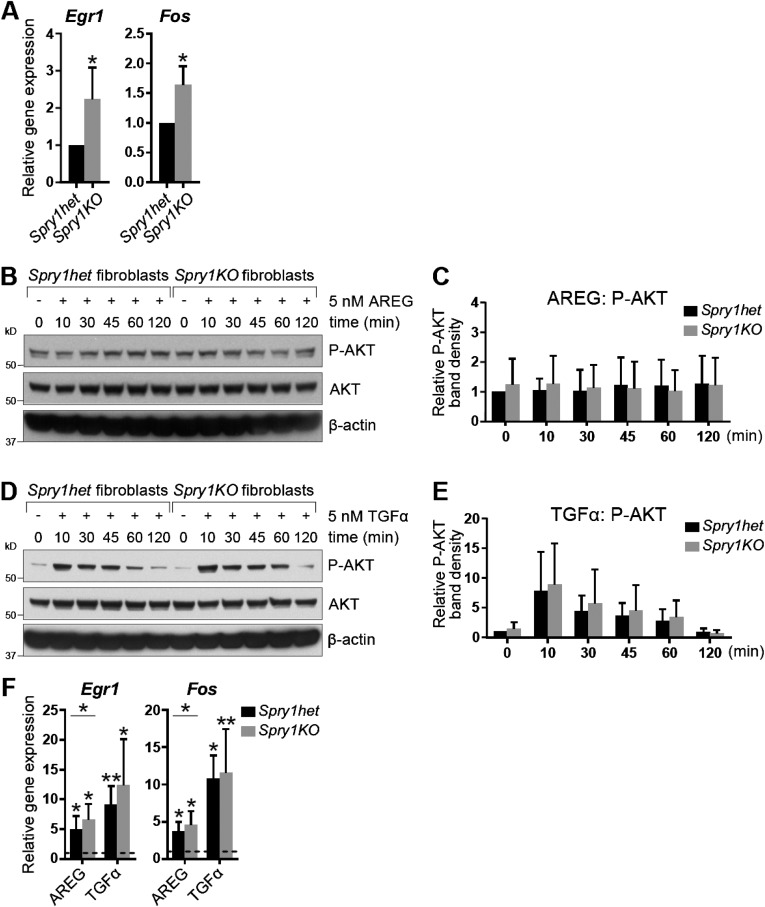
Next, we set out to determine whether Spry1 regulates signaling via EGFR, an RTK that mediates epithelium-to-stroma communication during mammary branching (24). Interestingly, we found increased expression of candidate EGFR-responsive genes Egr1 and Fos (39, 40) in Spry1KO fibroblasts (Fig. S3A). Moreover, treatment of Spry1het and Spry1KO fibroblasts with EGFR ligands, including AREG (Fig. 2 L and M) and TGFα (Fig. 2 N and O), led to an increase of the strength and duration of ERK activation in Spry1KO fibroblasts compared with Spry1het fibroblasts. By contrast, we observed no significant difference in AKT activation between Spry1het and Spry1KO fibroblasts in response to these two growth factors (Fig. S3 B–E). Furthermore, expression of both Egr1 and Fos was up-regulated more strongly in Spry1KO fibroblasts than in Spry1het fibroblasts in response to AREG or TGFα treatment (Fig. S3F), suggesting that Spry1KO fibroblasts are sensitized to AREG/TGFα–EGFR–ERK signaling.
Fig. S3.
Spry1KO fibroblasts show increased expression of EGFR-responsive genes in response to EGFR ligand stimulation. (A) Relative expression of target genes of EGFR signaling, Fos and Egr1, in serum-starved Spry1het and Spry1KO fibroblasts, normalized to Actb. The plots show mean ± SD. Statistical analysis was performed using paired t test; *P < 0.05. (B–E) Western blot analysis of phosphorylation status of AKT in Spry1het and Spry1KO fibroblasts in response to AREG (B and C) or TGFα stimulation (D and E). Fibroblasts were serum-starved, treated with 5 nM AREG or TGFα for the indicated time, and lysed, and on a single blot, P-AKT (Ser473), AKT, and β-actin signals were detected. (C and E) Quantitative comparison of AKT phosphorylation, normalized to total AKT. Data are mean ± SD (n = 3–5). Statistical analysis was performed using two-way ANOVA and revealed no significant difference in P-AKT between Spry1het and Spry1KO fibroblasts. (F) Relative expression of Fos and Egr1 in Spry1het and Spry1KO fibroblasts in response to AREG or TGFα stimulation, normalized to Actb and compared with their expression in mock-treated cells (indicated by dotted line). The plots show mean ± SD. Statistical analysis was performed using two-way ANOVA; *P < 0.05; **P < 0.01.
These results show that, when Spry1 is deleted in mammary fibroblasts, there is increased EGFR signaling and more epithelial branching. Thus, fibroblast Spry1 normally functions to negatively regulate AREG/TGFα–EGFR–ERK signaling.
Mammary Stromal Fibroblasts Lacking Spry1 Promote Epithelial Expansion and Migration in a Paracrine Fashion.
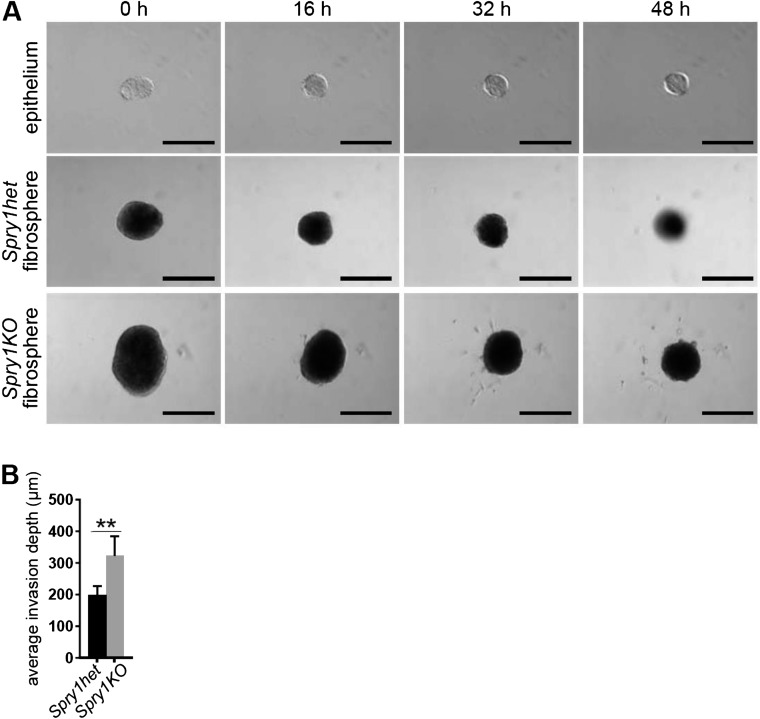
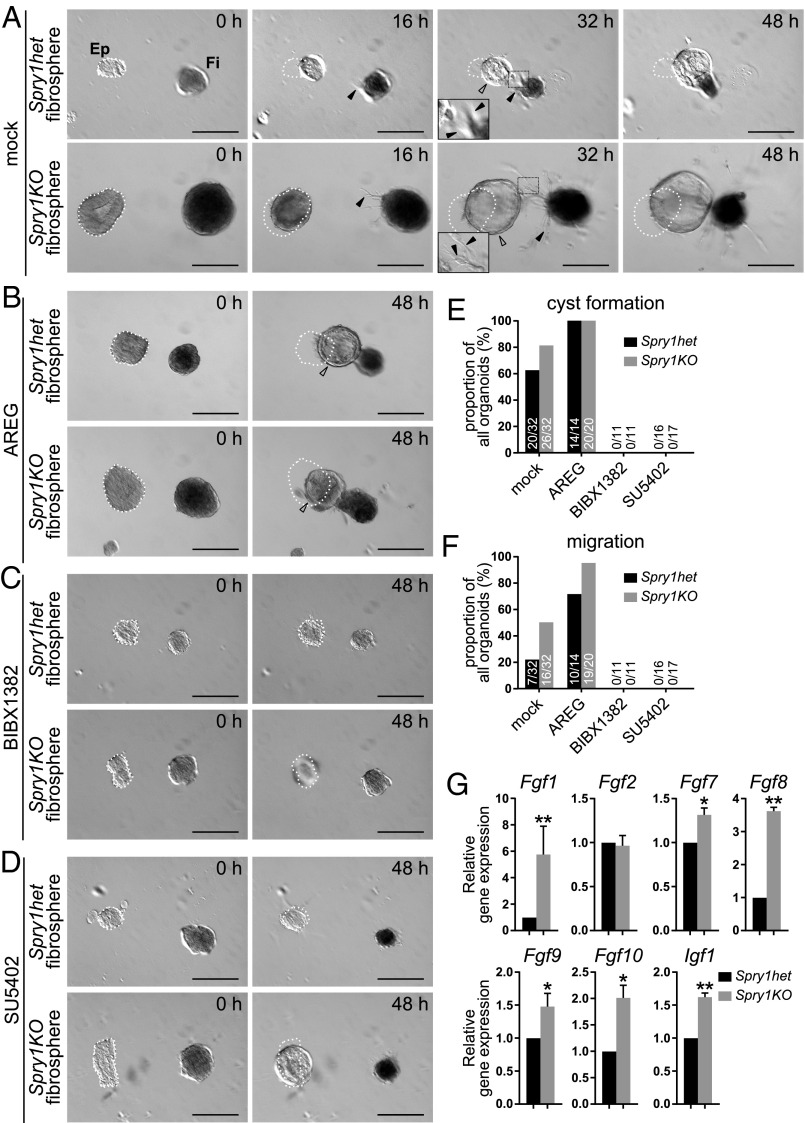
One mechanism by which Spry1-null fibroblasts might promote epithelial branching more strongly than normal is by increasing paracrine signaling to the epithelium. To examine this possibility, we developed an in vitro system in which aggregated fibroblasts, or “fibrospheres,” were cultured with organoids in close proximity (∼100 μm). When cultured alone in basal medium without growth factors, organoids and fibrospheres gradually shrank over the course of 2 d (Fig. S4). However, when cultured together, we observed mutual interactions between organoids and fibrospheres. Specifically, in the presence of Spry1het fibrospheres, a majority of organoids underwent cyst formation, and some of these organoids also migrated toward the fibrospheres (Fig. 3 A, E, and F, and Movie S1). Epithelial cyst formation and collective migration occurred at a higher frequency in the presence of Spry1KO fibrospheres (Fig. 3 A, E, and F and Movie S2). Under both experimental conditions, we observed extensions composed of fibroblasts protruding from fibrospheres, often in the direction of organoid epithelium (Fig. 3A).
Fig. S4.
Epithelial and stromal cell behavior when cultured separately. (A) Time course of in vitro cultures of wild-type epithelium or Spry1het and Spry1KO fibrospheres cultured in basal medium (without any growth factor stimulation). (Scale bars, 200 µm.) (B) Quantification of organotypic invasion assay. The plot shows average invasion depth of MCF7-ras cells per section. Values shown are the mean ± SEM (n = 5). Statistical analysis was performed using paired t test; **P < 0.01.
Fig. 3.
Mammary fibroblasts lacking Spry1 promote epithelial morphogenesis more strongly than normal in a paracrine fashion. (A–D) Time course of in vitro cocultures of wild-type epithelium (Ep; Left) with Spry1het or Spry1KO fibrospheres (Fi; Right) in the absence (A) or presence (B) of AREG stimulation, EGFR inhibitor BIBX1382 (C), or FGFR inhibitor SU5402 (D). (A) The epithelium formed a cyst (hollow arrowhead) and underwent directional collective migration toward fibrospheres, whereas fibrospheres formed spiky extensions made of fibroblasts (filled arrowheads), often in the direction of the epithelium. The images were recorded using time-lapse microscopy over 48 h, and the respective movies can be seen in Movies S1 and S2. (B–D) The ability of the epithelium to undergo cyst formation and collective migration was increased by AREG addition (B) and inhibited by EGFR inhibitor BIBX1382 (C) or FGFR inhibitor SU5402 (D). (Scale bars, 200 µm.) White dotted line indicates the original position of organoids at time 0 h. (E and F) Quantification of epithelial response to Spry1het and Spry1KO fibrospheres. Plots show proportions, as well as actual numbers of organoids, that underwent cyst formation (E) or migration (F) out of total number of organoids per condition. (G) Relative expression of candidate growth factor genes in serum-starved Spry1het and Spry1KO fibroblasts, normalized to Actb. Data shown are mean ± SEM (n = 3–4). Statistical analysis was performed using paired t test; *P < 0.05; **P < 0.01. Refer to Table S1 for primer sequences.
The ability of fibrospheres to promote epithelial cyst formation and collective migration was increased by addition of AREG into the coculture (Fig. 3 B, E, and F), whereas it was inhibited by the highly selective EGFR inhibitor BIBX1382 (41) (Fig. 3C). These results suggest that the ability of fibrospheres to promote cyst formation and migration is positively regulated by EGFR signaling. Moreover, fibrosphere-induced epithelial response was rapid and was evident within 24 h (Fig. 3A and Movies S1 and S2), unlike the assays in which organoids formed branches after several days. The results thus suggest that fibrosphere influence on the epithelium is based on paracrine growth factors.
FGF ligands play distinct roles in epithelial morphogenesis of the mammary gland and, when delivered locally on heparin beads rather than uniformly in the medium, can trigger cyst formation and/or collective migration similar to fibrospheres (42). Thus, likely candidates by which fibrospheres influence the epithelium are FGF ligands, which are major epithelial branching signals produced in the mammary gland stroma. Indeed, the addition of FGFR signaling inhibitor SU5402 at 20 µM, the concentration at which it selectively inhibits FGFR but not insulin receptor or PDGFR (43), effectively abrogated fibrosphere-induced epithelial morphogenesis (Fig. 3 D–F), suggesting that fibroblast-derived signals are, at least in part, ligands of the FGF family. Accordingly, we hypothesized that, if FGFs are indeed responsible for the Spry1 loss-of-function phenotype, then FGF production would be higher in Spry1KO fibroblasts than in Spry1het fibroblasts. To test this hypothesis, we examined mRNA expression of several Fgfs that are expressed in the mammary gland stroma (42). As predicted, some of these Fgfs, including Fgf1, Fgf7, Fgf8, Fgf9, and Fgf10 were more highly expressed in Spry1KO fibroblasts than in Spry1het fibroblasts (Fig. 3G). Interestingly, Igf1, another growth factor that promotes mammary gland development (14), was also up-regulated at the mRNA level in Spry1KO fibroblasts (Fig. 3G).
These results reveal that stromal fibroblasts can promote epithelial branching by providing growth factors, particularly those of the FGF family. In the Spry1KO fibroblasts, paracrine production is higher than in Spry1het cells, leading to an increase in ductal branching.
Loss of Spry1 Increases Collagen Remodeling and Promotes Epithelial Invasion.
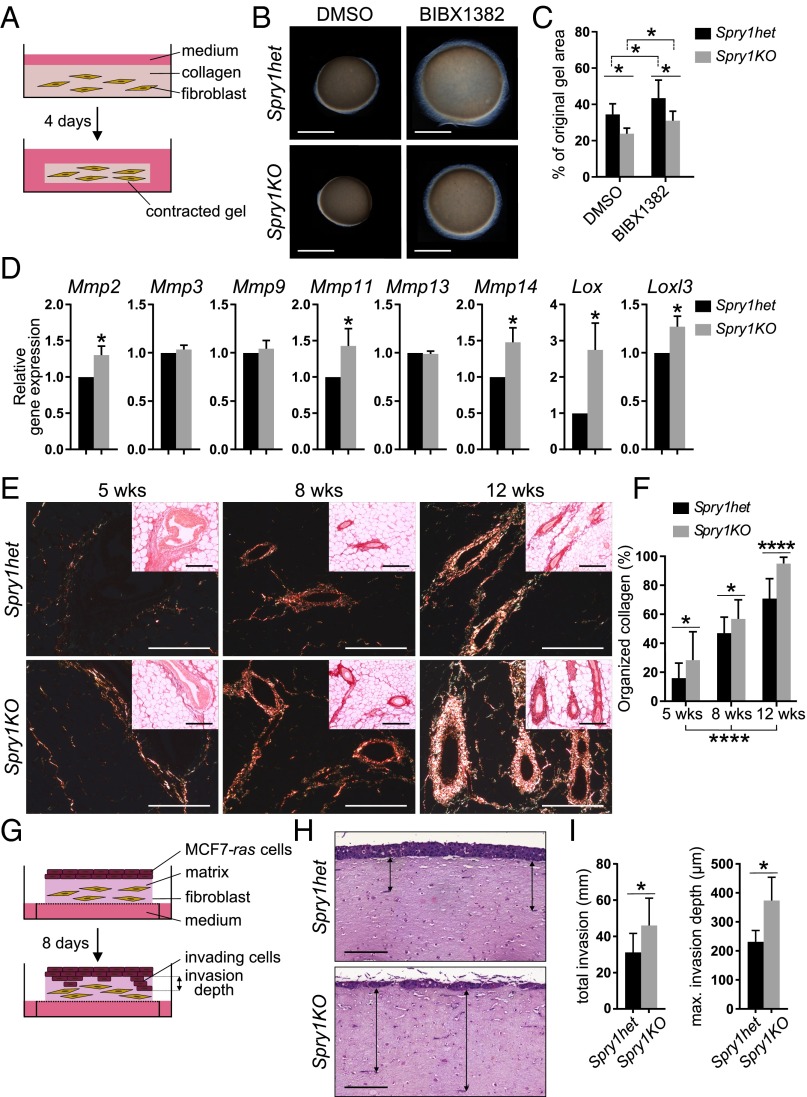
Another mechanism by which Spry1KO stromal fibroblasts might promote epithelial branching more strongly than normal is by modulating ECM remodeling, which is essential for epithelial behavior, including invasion and branching morphogenesis (1, 20). To test this possibility, we used a collagen contraction assay, in which remodeling of collagen matrix leads to contraction, to compare the ECM remodeling abilities between Spry1het fibroblasts and Spry1KO fibroblasts (Fig. 4A). We found that Spry1KO fibroblasts contracted collagen matrix more strongly than Spry1het fibroblasts (Fig. 4 B and C). Moreover, treatment with the EGFR inhibitor BIBX1382 attenuated collagen contraction abilities of Spry1het and Spry1KO fibroblasts (Fig. 4 B and C), suggesting that EGFR signaling regulates collagen remodeling in mammary fibroblasts.
Fig. 4.
Spry1KO fibroblasts show increased ECM-remodeling activities and promote epithelial invasion more strongly than Spry1het fibroblasts. (A–C) Collagen contraction assay. (A) Diagram depicting the experimental procedure. (B) Photographs of a representative experiment. (C) Graph shows size change of collagen gels, expressed as percentage of the original area ± SEM (n = 7). Statistical analysis was performed using two-way ANOVA; *P < 0.05. (D) Expression of candidate ECM-modifying genes in Spry1het and Spry1KO fibroblasts cultured with serum, normalized to Actb. Values shown are the mean ± SEM (n = 3). Statistical analysis was performed using paired t test; *P < 0.05. Refer to Table S1 for primer sequences. (E and F) Collagen deposition in Spry1het and Spry1KO mammary glands during development as detected by picrosirius red staining on paraffin sections. (E) Bundles of organized collagen were visualized under polarized light; Insets show images in bright field, which visualizes total collagen. (F) Quantification of organized collagen relative to the total amount of collagen. Values shown are the mean ± SD (n = 9–16 per genotype). Statistical analysis was performed using two-way ANOVA; *P < 0.05; ****P < 0.0001. (Scale bars, 200 µm.) (G–I) Organotypic invasion assays. (G) Diagram depicting the experimental procedure. (H) Representative images of hematoxylin- and eosin-stained cross-sections from the organotypic gels. Arrows show examples of MCF7-ras cell invasion depths. (Scale bars, 200 µm.) (I) Quantification of organotypic invasion assay. The plots show pooled invasion depth or average maximum invasion depth of MCF7-ras cells per section. Values shown are the mean ± SEM (n = 5). Statistical analysis was performed using paired t test; *P < 0.05.
Using qPCR, we next examined expression of several ECM-remodeling genes, including matrix metalloproteinases (Mmps) and lysyl oxidases (Lox, Lox3) in stromal fibroblasts. Compared with Spry1het fibroblasts, we found that expression of Mmp2, Mmp11, Mmp14, Lox, and Lox3 was up-regulated in Spry1KO fibroblasts (Fig. 4D). These data suggest that Spry1-null fibroblasts promote epithelial branching in part by up-regulating expression of ECM-remodeling enzymes.
We also examined collagen deposition during mammary development by staining sections with Picrosirius red. The amount of organized collagen progressively increased with age (Fig. 4 E and F), and there was a higher amount of organized collagen in Spry1KO glands than in Spry1het glands (Fig. 4F). Thus, the ECM becomes altered in Spry1KO mice.
Next, we investigated whether increased collagen-remodeling activity of Spry1KO fibroblasts could promote epithelial cell invasion. To this end, we used an organotypic invasion assay in which fibroblast-mediated ECM remodeling activity, rather than paracrine signaling, is the primary factor affecting epithelial/cancer cell invasion (44). Because primary MECs do not invade in this experimental setting, we used a stable breast cancer cells line MCF7-ras, which invades depending on fibroblast-mediated ECM remodeling. We measured matrix invasion by the MCF7-ras cells, which had been seeded on top of a dense collagen/Matrigel matrix embedded with either Spry1het or Spry1KO fibroblasts, 8 d after the coculture (Fig. 4G). We found that MCF7-ras cells invaded significantly deeper into the matrix remodeled by Spry1KO fibroblasts than by Spry1het fibroblasts (Fig. 4 H and I and Fig. S4B), indicating that Spry1KO fibroblasts are more effective at promoting epithelial invasion.
These results indicate that Spry1KO fibroblasts contribute to epithelial branching more strongly than normal, by up-regulating ECM remodeling. This upregulation of matrix remodeling is in part due to an increased expression of ECM-remodeling enzymes.
Reduction of Stromal EGFR Signaling Leads to Decreased Epithelial Branching in Vivo.
A prediction that follows from our finding that up-regulation of EGFR signaling due to Spry1 loss in stromal fibroblasts causes accelerated epithelial branching, is that reduction of EGFR signaling, for example, by gain of Spry1 function in the mammary stroma, will lead to stunted epithelial branching.
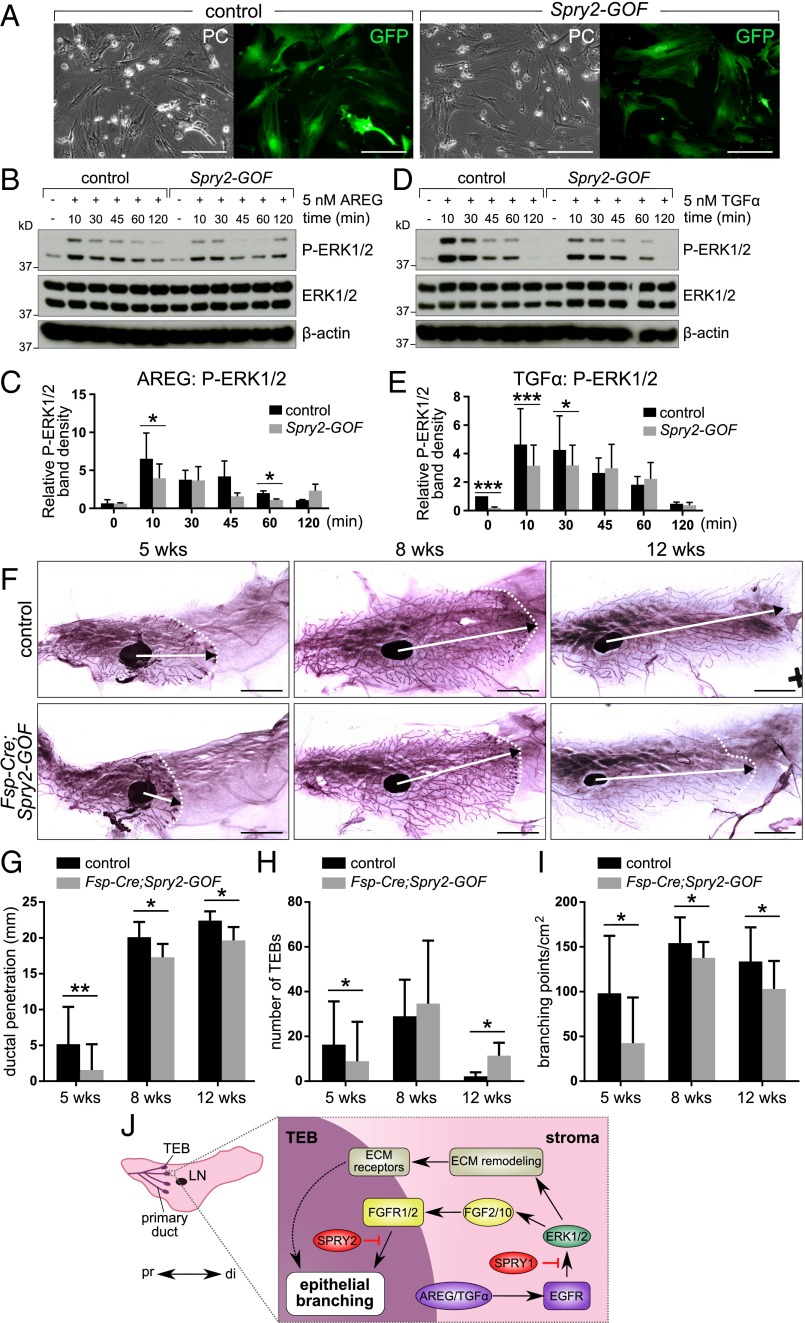
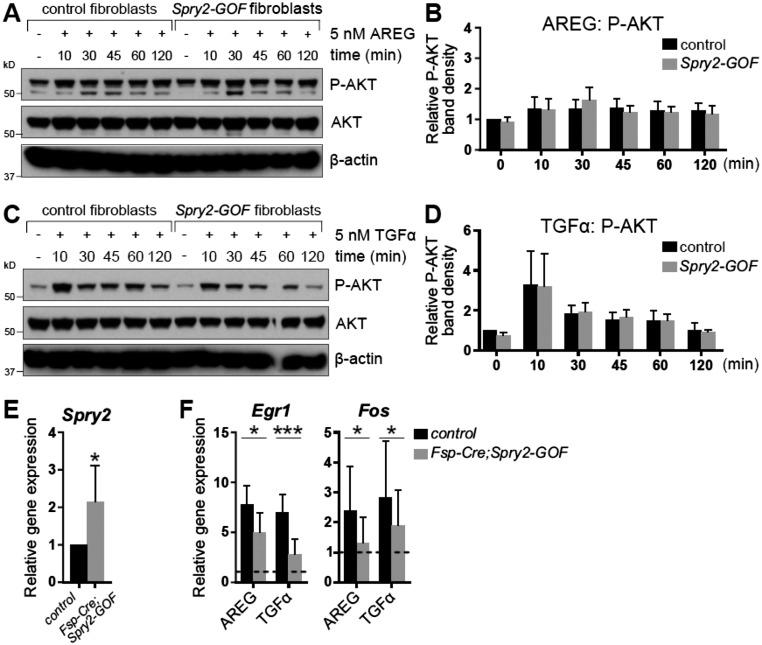
SPRY1 and SPRY2 are biochemically similar and play redundant roles in the same cellular context (33, 37, 45). We therefore exploited a mouse line carrying a conditional Spry2 gain-of-function allele (Spry2-GOF) (30, 46). We first determined whether Cre-mediated recombination could activate Spry2-GOF and, if so, whether Spry2-GOF could reduce EGFR signaling in the mammary stroma. To this end, we exposed fibroblasts from control and Spry2-GOF mice to adenovirus (Ad)-Cre-GFP, resulting in a high proportion expressing GFP (Fig. 5A). We then examined the responses of Ad-Cre-GFP–infected control and Spry2-GOF fibroblasts to AREG and TGFα treatment by Western blot analysis. As expected, AKT activation was unaffected by Spry2 gain of function (Fig. S5 A–D). By contrast, ERK activation in Spry2-GOF fibroblasts was reduced compared with control fibroblasts (Fig. 5 B–E). Thus, ectopic expression of Spry2 dampens AREG/TGFα-ERK signaling, validating the use of the Spry2-GOF mouse line to reduce EGFR signaling in stromal fibroblasts.
Fig. 5.
Ectopic Sprouty expression dampens AREG/TGFα-ERK activation in stromal fibroblasts. (A) Photographs of control and Spry2-GOF fibroblasts transfected with Ad-Cre-GFP; Left, phase contrast (PC) images; Right, fluorescence (GFP) images. (Scale bars, 100 µm.) (B–E) Western blot analysis of ERK1/2 phosphorylation status in control and Spry2-GOF fibroblasts in response to AREG (B and C) or TGFα stimulation (D and E). Fibroblasts were serum-starved, treated with 5 nM AREG or TGFα for the indicated durations, and lysed, and on a single blot, P-ERK1/2 (Thr202/Tyr204), ERK, and β-actin signals were detected. (C and E) Quantitative comparison of ERK1/2 phosphorylation, normalized to total ERK1/2, respectively. Data are mean ± SD (n = 2/3). Statistical analysis was performed using two-way ANOVA; *P < 0.05; **P < 0.01, ***P < 0.001. (F–H) Carmine-stained whole-mount mammary glands from control and Fsp-Cre;Spry2-GOF mice at 5 and 8 wk of age. Arrows indicate the extent of ductal penetration in the fat pad; dotted lines indicate the epithelial invasion front. (G–I) Quantitative comparisons of ductal penetration (G), number of terminal end buds (TEBs) (H), and branching (I) between control and Fsp-Cre;Spry2-GOF glands. The plots show mean ± SD (n = 3–12 per genotype); *P < 0.05; **P < 0.01. (Scale bars, 5 mm.) (J) Model diagram depicting the mechanism by which stromal EGFR signaling and epithelial FGFR signaling is regulated by Sprouty genes during mammary gland epithelial branching. Terminal end buds (TEBs) develop at the onset of puberty (3 wk after birth) at the distal tip of each primary duct, which invades the stroma in a proximal (pr)-to-distal (di) direction. Epithelial EGF ligands, including AREG and TGFα, activate EGFR signaling that is essential for FGF paracrine production and ECM remodeling. Levels of EGFR signaling are fine-tuned by stromal SPRY1 function so that an appropriate amount of FGFs and ECM-remodeling enzymes are produced during branching. By contrast, the interpretation of FGF activation by their receptors FGFR1/2 on the epithelium is modulated by SPRY2. As such, ductal elongation, collective migration, and other cell behavior essential for epithelial branching are orchestrated for the formation of the mammary gland.
Fig. S5.
AKT phosphorylation is not affected by ectopic expression of Spry2 in mammary fibroblasts. (A–D) Western blot analysis of phosphorylation status of AKT in control and Spry2-GOF fibroblasts in response to AREG (A and B) or TGFα stimulation (C and D). Fibroblasts were serum-starved, treated with 5 nM AREG or TGFα for the indicated durations, and lysed, and on a single blot, P-AKT (Ser473), AKT, and β-actin signals were detected. (B and D) Quantitative comparison of AKT phosphorylation, normalized to total AKT. Data are mean ± SD (n = 2–3). Statistical analysis was performed using two-way ANOVA and revealed no significant difference in P-AKT between control and Spry2-GOF fibroblasts. (E) Relative expression of Spry2 in control and Fsp-Cre;Spry2-GOF fibroblasts, normalized to Actb. The plot shows mean ± SD. Statistical analysis was performed using paired t test; *P < 0.05. (F) Relative expression of Egr1 and Fos in control and Fsp-Cre;Spry2-GOF fibroblasts in response to AREG or TGFα stimulation, normalized to Actb and compared with their expression in mock-treated cells (indicated by dotted line). The plots show mean ± SD. Statistical analysis was performed using two-way ANOVA; *P < 0.05; ***P < 0.001.
We crossed Spry2-GOF mice with Fsp-Cre mice [which have Cre activity in mammary fibroblasts (47)] to generate Fsp-Cre;Spry2-GOF female progeny, in which Spry2-GOF was activated in the mammary stroma (Fig. S5E) and there was down-regulated EGFR signaling (Fig. S5F). We found that, when the mammary glands of Fsp-Cre;Spry2-GOF mice were compared with those of controls at 5, 8, or 12 wk of age, ductal penetration and branch formation were reduced (Fig. 5 F–I).
These results confirm that reducing EGFR signaling in stromal fibroblasts leads to decreased epithelial branching in vivo.
Discussion
In this study, we have gained insights into the nature of epithelial–stromal interactions in mammary gland development. We show that Spry1 modulates epithelial–stromal interactions by negatively regulating EGFR-dependent stromal paracrine signaling and ECM remodeling (Fig. 5J). Loss of Spry1 leads to up-regulated EGFR signaling and enhanced stromal activities, including ECM remodeling and growth factor production, which in turn leads to increased epithelial branching. By contrast, down-regulation of EGFR signaling in fibroblasts leads to stunted epithelial branching. Taken together with our findings of the significance of FGFR signaling within the epithelium (15, 16, 42), the results show that EGFR and FGFR signaling constitute a critical part of the molecular basis of stromal–epithelial interactions during postnatal development, and their modulation by Sprouty genes is essential for mammary gland formation and homeostasis.
Spry1 Is Haplosufficient for Normal Mammary Gland Development.
In agreement with Spry1 haplosufficiency in development of kidney, urinary tract, and external genitalia (27, 33), our study revealed that Spry1 is haplosufficient during mammary development because we did not observe any difference in mammary branching between Spry1wt and Spry1het glands. The relatively minor phenotype of Spry1KO mammary glands suggests that loss of Spry1 might be compensated by other stromal Sprouty genes. Indeed, we found that Spry4 shows a similar expression pattern as Spry1 in the developing mammary gland, suggesting that it may play a redundant function as Spry1. Future studies of other Sprouty genes in the mammary gland stroma, alone or in combination with Spry1, should address this question regarding whether they play a similar or distinct role during mammary gland development.
It remains unclear why Spry1 expression remains relatively stable throughout postpubertal development of the mammary gland but is greatly up-regulated at the 10-wk stage, when branching morphogenesis has all but finished in the adult female mice. One intriguing explanation for this observation is that, consistent with it being a negative regulator of mammary epithelial branching, Spry1 may be up-regulated by an increased production of certain growth factor(s) to function as a critical component to halt the epithelial movement when the mammary tree has already been built and “unwanted”/dysregulated epithelial invasiveness beyond this point could harm the integrity and homeostasis of the functional mammary gland as in the case of breast cancer development.
Stromal EGFR Signaling Promotes Epithelial Branching by Regulating ECM Remodeling.
Although it is well established that EGFR expression in the mammary gland stroma is required for epithelial branching, the mechanism by which EGFR signaling functions in the stroma has remained largely unclear. Our data show that stromal fibroblasts provide an essential component of the microenvironment for epithelial morphogenesis by promoting EGFR-dependent ECM remodeling via regulation of MMPs and other ECM remodeling enzymes including Lox genes. Our data showing up-regulation of Mmp2, Mmp11, Mmp14, Lox, and Lox3 are consistent with a previous report on regulation of Mmp expression and collagen contraction by EGFR–ERK signaling in mouse embryonic fibroblasts (48). Importantly, several Mmps, including Mmp2, Mmp3, and Mmp14, have been shown to regulate mammary epithelial branching in vivo (49, 50).
Consistent with increased Mmp and Lox gene expression, we found that organized collagen deposition was higher in Spry1KO mammary glands than in Spry1het glands during development. Although it remains unclear whether the increased deposition of organized collagen directly causes increased epithelial branching in Spry1KO mammary glands, emerging evidence now shows that stiffness of the ECM, including collagen, plays an important role in regulating epithelial behavior (reviewed in refs. 6, 20, and 51). Indeed, MCF7-ras cells invaded significantly deeper in the ECM that had been remodeled by Spry1KO fibroblasts than by Spry1het fibroblasts. These results thus suggest that epithelial invasion, an important aspect of vertebrate epithelial branching, is increased in the absence of Spry1.
It is interesting that organized collagen deposition progressively increased with time during normal development. These results are consistent with an increase in collagen bundles and ECM stiffness in the stromal microenvironment in mouse tumor models (6). Taken together, the increased epithelial branching observed in Spry1KO mice is most likely a direct result of up-regulation of EGFR–ERK signaling in mutant stroma due to Spry1 loss.
EGFR and FGFR Signaling Pathways Underlie Stromal–Epithelial Interactions During Postnatal Mammary Gland Development.
In the presence of fibrospheres, mammary epithelium underwent rapid cyst formation and collective migration, suggesting that fibroblast influence is likely to be paracrine in nature. Interestingly, similar epithelial responses could be triggered by local delivery, as in the case of fibrospheres, of FGF ligands (42). The data thus point to FGFs as candidate mediators of the epithelial responses. Indeed, the addition of the FGFR inhibitor SU5402 abolished cyst formation and epithelial migration, suggesting that stroma-derived paracrine factors can, at least in part, be ascribed to ligands of the FGF family. This notion was confirmed by gene expression analysis based on qPCR, which showed that several stromal Fgfs were more strongly expressed in Spry1KO fibroblasts than in Spry1het fibroblasts.
Thus, the increased epithelial branching in Spry1KO gland is caused by overproduction of FGF ligands and possibly other paracrine factors, including IGF1, due to EGFR signaling up-regulation in the mutant stroma. This conclusion is consistent with the observation that increased epithelial branching can also directly result from FGF signaling up-regulation in the absence of its inhibitor SPRY2 in the epithelium (30). By contrast, EGFR signaling down-regulation due to gain of Sprouty function in the stromal fibroblasts led to retardation of epithelial branching. Indeed, epithelial branching is severely retarded or absent in the mammary stroma lacking Egfr function (24) or in the mammary epithelium lacking Fgfr (15, 16, 42).
Taken together, a reciprocal cross-talk mechanism exists in the developing mammary gland to regulate epithelial morphogenesis (Fig. 5J). On the one hand, epithelium-derived EGF family ligands, including AREG and TGFα, are essential for epithelial branching by targeting the mammary stroma. On the other hand, upon activation by AREG and TGFα, EGFR signaling promotes epithelial branching by a dual mechanism: indirectly by enhancing ECM remodeling and subsequently epithelial invasion, and directly by producing paracrine factors, including FGF ligands that target mammary epithelium via FGFR1 and FGFR2 signaling. Importantly, our data do not exclude the possibility that Spry1 may regulate other RTKs, including IGFR, FGFR, or PDGFR in fibroblasts. Future studies will address whether these RTKs function in the mammary gland stroma and, if so, whether they are regulated by Spry1.
Reciprocal cross talk between epithelium and mesenchyme is a central theme in developmental biology, and this type of interaction is responsible for the specification, differentiation, and morphogenesis of many vertebrate organs during embryonic development (7). Our studies and others show that the mammary stroma, a derivative of embryonic mammary mesenchyme, engages in a similar cross talk with the epithelium during postnatal development, and perhaps in tissue homeostasis. A key aspect of future studies will be to understand how deregulation of stromal–epithelial cross talk may contribute to tumor onset and progression in the breast.
A Model of Sprouty Gene Function in Epithelial Branching of the Mammary Gland.
Our working model is that SPRY1 suppresses overactive epithelial branching by inhibiting EGFR-dependent paracrine signaling and ECM remodeling in the mammary gland stroma (Fig. 5J). In the absence of SPRY1, epithelial branching is increased because paracrine signaling and ECM remodeling in the stromal microenvironment is augmented. By contrast, SPRY2 suppresses precocious epithelial branching by inhibiting FGFR signaling in the epithelium (30). In the absence of SPRY2, epithelial branching is increased because the epithelium is more sensitized to FGF stimulation, and undergoes epithelial expansion, collective migration, and other essential aspects of epithelial branching more readily than normal.
It is interesting that different members of the Sprouty gene family are deployed in different tissues to modulate the reciprocal signaling between epithelium and stroma, thereby ensuring that overactive epithelial branching is prevented. Whether SPRY1 functions as an extrinsic “brake,” or SPRY2 acts as an intrinsic one in epithelial branching, they appear to perform the same function. The use of different Sprouty family members to fine-tune signaling between epithelium and mesenchyme reinforces the robustness of complex biological systems so that they can better withstand fluctuations of essential intrinsic or extrinsic factors during development and homeostasis.
Materials and Methods
Mouse Strains and Surgery.
Immunologically deficient female nude (nu/nu) mice were from Charles River Laboratories. CD-1 mice were obtained from institutional mouse colony (University of Manchester). Mice carrying the Fsp-Cre allele (47) were purchased from The Jackson Laboratory. Mice carrying the Spry1LacZ (31) allele and Spry2-GOF mice (46) were provided by M. Albert Basson, Department of Developmental and Stem Cell Biology, King's College of London, United Kingdom. Because Sprouty genes are haplosufficient for various vertebrate developmental processes (27, 32, 33), we adopted a mating scheme wherein a Spry1LacZ/LacZ male mouse was crossed with a Spry1LacZ/+ female mouse so that Spry1het control and Spry1KO mutant female mice were generated at a 1:1 ratio and this was in compliance with the United Kingdom’s 3Rs policy. Mice were housed and maintained according to the Animal (Scientific Procedures) Act, 1986 (UK), project licence PPL 40/9865, and approved by the University of Manchester Ethical Review Process Committee and by ShanghaiTech University's Institutional Animal Care and Use Committee (IACUC# 2015SHT0006), and were genotyped according to methods in the publications describing establishment of the mouse lines.
Isolation and Culture of Primary Mammary Organoids and Fibroblasts.
Primary mammary organoids were isolated and cultured as previously described (30). Briefly, mouse mammary glands were finely chopped, and mince was digested in collagenase buffer [with 0.2% collagenase (Sigma) and 0.2% trypsin (Life Technologies)]. Samples were centrifuged, washed several times, and treated with DNase I. Then they were subjected to five rounds of differential centrifugation (a short-pulse centrifugation at 450 × g), during which supernatant was collected and pellets were resuspended in DMEM/F12 (Invitrogen) for another round of differential centrifugation. The pellet containing mammary organoids was either plated in Matrigel (Corning) and overlaid with basal medium (DMEM/F12, 10 μg/mL insulin, 5.5 μg/mL transferrin, 6.7 ng/mL selenium, 100 U/mL penicillin, and 100 μg/mL streptomycin), supplemented with 2.5 nM FGF2 or FGF7 if needed (for in vitro epithelial branching assay), or the organoids were further trypsinized to obtain a single-celled MEC solution (for FACS). Supernatant fractions were pooled and centrifuged, and pellets were resuspended in Fibroblast medium [DMEM (Invitrogen) with 10% (vol/vol) FBS (Sigma), 10 μg/mL insulin, 5.5 μg/mL transferrin, 6.7 ng/mL selenium, 100 U/mL penicillin, and 100 μg/mL streptomycin] and seeded on a cell culture dish. After 30 min, when fibroblasts had already adhered to the dish whereas other cellular types remained in suspension, medium was aspirated and cell culture dish was washed twice with PBS and fresh Fibroblast medium was added. To prepare fibrospheres, fibroblasts were aggregated in low-adhesion plates overnight.
See SI Materials and Methods for additional details and more methods, including mammosphere and acinar assay, RNA isolation and qPCR (Table S1), mammary gland staining, in vitro epithelial branching and migration assays, Western blotting, collagen contraction and organotypic invasion assays, and statistical analyses.
Table S1.
Sequences of primers used for qPCR
| Gene name | Forward sequence, 5′ → 3′ | Reverse sequence, 5′ → 3′ |
| Actb | ggctgtattcccctccatcg | ccagttggtaacaatgccatgt |
| Eef1g | ttcctgccggcaaggttcca | tgccgcctctggcgtacttc |
| Egr1 | agcgaacaaccctatgagcac | tcgtttggctgggataactcg |
| Fgf1 | ggacaccgaagggcttttat | gcatgcttcttggaggtgtaa |
| Fgf2 | cggctctactgcaagaacg | tgcttggagttgtagtttgacg |
| Fgf7 | aagggacccaggagatgaag | actgccacggtcctgattt |
| Fgf8 | caggtcctggccaacaag | ggtctccacaatgagcttcg |
| Fgf9 | actgcaggactggatttcatttag | ccaggcccactgctatactg |
| Fgf10 | cgggaccaagaatgaagact | aacaactccgatttccactga |
| Fos | aagggaacggaataagatggc | caacgcagacttctcatcttcaa |
| Igf1 | tcggcctcatagtacccact | acgacatgatgtgtatctttattgc |
| Lox | tcttctgctgcgtgacaacc | gagaaaccagcttggaaccag |
| Loxl3 | ctactgctgctacactgtctgt | gaccttcatagggctttctagga |
| Mmp2 | taacctggatgccgtcgt | ttcaggtaataagcacccttgaa |
| Mmp3 | ttgttctttgatgcagtcagc | gatttgcgccaaaagtgc |
| Mmp9 | acgacatagacggcatcca | gctgtggttcagttgtggtg |
| Mmp11 | tggatgcagcttttgaggat | aggactggcttctcaccatc |
| Mmp13 | gccagaacttcccaaccat | tcagagcccagaattttctcc |
| Mmp14 | gagaacttcgtgttgcctga | ctttgtgggtgaccctgact |
| Spry1 | taggtcagatcgggtcatcc | gtggggtcctctttcaagg |
| Spry2 | gagaggggttggtgcaaag | ctccatcaggtcttggcagt |
SI Materials and Methods
Cell Culture.
Preparation of primary mammary organoids and fibroblasts.
Mouse mammary glands were finely chopped, and the mince was digested in collagenase buffer [0.2% collagenase (Sigma) and 0.2% trypsin (Life Technologies) in DMEM/F12 supplemented with 5% (vol/vol) FBS, 50 µg/mL gentamicin, 5 µg/mL insulin (all from Sigma)] for 30 min at 37 °C. Samples were centrifuged, and pellets were washed with DMEM/F12 and treated with DNase I (2 U/μL) for 5 min. After washing with DMEM/F12, pellets were resuspended in DMEM/F12 and subjected to a short-pulse centrifugation at 450 × g (differential centrifugation). Supernatant was collected, and pellets were resuspended in DMEM/F12 for another round of differential centrifugation. After five rounds of differential centrifugation, pellets containing mammary organoids were plated in Matrigel (Corning) and overlaid with basal medium (DMEM/F12, 10 μg/mL insulin, 5.5 μg/mL transferrin, 6.7 ng/mL selenium, 100 U/mL penicillin, and 100 μg/mL streptomycin). Supernatant fractions were pooled and centrifuged, and pellets were resuspended in DMEM (Invitrogen) with 10% (vol/vol) FBS (Sigma), 10 μg/mL insulin, 5.5 μg/mL transferrin, 6.7 ng/mL selenium, 100 U/mL penicillin, and 100 μg/mL streptomycin (fibroblast medium), and seeded on a cell culture dish. After 30 min, when fibroblasts had already adhered to the dish whereas other cellular types remained in suspension, medium was aspirated and cell culture dish was washed twice with PBS before adding fresh medium. Fibroblast cultures were allowed to grow until they reached ∼80% confluence; then they were subcultured. Only early-passage fibroblasts (up to passage number 5) were used in experiments.
Transfection.
Fibroblasts were resuspended in the fibroblast medium and infected overnight with Adenovirus-Cre-GFP (green fluorescent protein) at a multiplicity of infection of ∼25 particles per cell. Fibroblasts were washed twice with PBS and supplied with fresh fibroblast medium.
Mammosphere and Acinar Assays.
Freshly isolated mammary organoids were trypsinized to obtain single-cell suspension, stained with antibodies against CD24 (FITC), CD49f (PE), and CD45 (PeCy7; eBioscience) and with 7-AAD (BD Biosciences), and subjected to FACS using a FACSAriaII (BD Biosciences) to sort for different cell populations. For mammosphere assay, sorted cells were seeded on low-attachment plates [plates treated with poly(2-hydroxyethyl methacrylate) (Sigma)] in phenol red-free DMEM/F12, supplemented with B-27, 100 μg/mL penicillin, 100 U/mL streptomycin, 20 ng/mL EGF, and/or 20 ng/mL FGF2, and cultured for 7 d. For secondary mammosphere assay, primary mammospheres were collected, trypsinized to obtain single-cell suspension, and seeded on low-attachment plates at the same density and in the same culture medium as primary mammospheres. For acinar assay, sorted cells were plated in Matrigel and cultured in DMEM/F12 with 10% (vol/vol) heat-inactivated FBS (Sigma), penicillin, streptomycin, 5 ng/mL EGF, 5 µg/mL insulin, and 1 µg/mL hydrocortisone for 14 d. Mammosphere (or acinus)-forming efficiency was calculated using the following equation: 100 × [number of mammospheres (or acini, respectively) formed/number of cells seeded].
RNA Isolation and qPCR.
RNA was isolated using the RNAqueous kit (Life Technologies) or RNeasy Mini kit (Qiagen). cDNA was prepared using TaqMan Reverse Transcription Reagents (Life Technologies). Real-time qPCR was performed on 5 ng of cDNA using SYBR Green PCR Master Mix (Life Technologies) and 0.45 µM gene-specific primers in a final volume of 11 µL using the ABI 7900HT or QuantStudio 12K Flex Real-Time PCR System (Life Technologies). Sequences of the oligonucleotides used are described in Table S1. Gene expression data were normalized to housekeeping genes, β-actin (Actb) or eukaryotic translation elongation factor 1γ (Eef1g).
Mammary Gland Staining and Histology.
Carmine staining.
Inguinal (#4) mammary glands were harvested and mounted on glass slides. After overnight fixation in Carnoy’s fixative at 4 °C, they were hydrated and stained in Carmine alum stain for 4 h. Stained mammary glands were dehydrated and cleared in Histoclear before they were photographed using a Leica M205 stereoscope fitted with Leica DFC345 FX camera. Photographs were processed and analyzed using GIMP and ImageJ (NIH). Ductal length was measured as the distance between the center of the lymph node and the tip of the epithelial branch that invaded most distally into the fat pad. Measurements were normalized to average ductal length in Spry1het glands to reduce batch-to-batch variations. Branch number was counted as the number of branch tips distal to the lymph node and normalized to ductal length.
β-Galactosidase staining.
The #4 mammary glands were dissected, spread on glass, and fixed with 4% (wt/vol) paraformaldehyde (PFA) in PBS for 30 min. Embryos were dissected from pregnant mice and fixed with ice-cold 4% PFA for 15 min. Subsequently, samples were washed with PBS and stained for 6–14 h with the β-gal staining kit (Roche) at 37 °C according to manufacturer’s instructions.
Picrosirius red staining.
Mammary glands were fixed with 4% PFA, embedded in paraffin, and sectioned. Sections were dewaxed, hydrated, stained with picrosirius red (0.1% picrosirius red in saturated picric acid), dehydrated, mounted, and analyzed using a Leica DM RXP microscope fitted with a polarizer and Infinity X camera. Imaging parameters were kept constant across all samples. Photographs were processed and analyzed using ImageJ.
In Vitro Epithelial Branching and Migration Assays.
Freshly extracted mammary organoids were plated in Matrigel and cultured in basal medium containing growth factors of interest (2.5 nM FGF2 or FGF7; Sigma) for 10 d; culture medium was replaced with fresh medium every 3 d. For organoid–fibroblast cocultures, organoids from CD1 mice were mixed with fibroblasts at the ratio of 100 or 1,000 fibroblasts per organoid, plated in Matrigel, and cultured in the basal medium (supplemented with 5 nM AREG or FGF2 when necessary) for 5 d. Alternatively, fibroblasts were aggregated in low-adhesion plates overnight to form fibrospheres, which were juxtaposed with organoids in Matrigel at a distance of ∼100 µm using a tungsten needle and cultured in basal medium, with or without the inhibitors SU5402 (20 µM; Sigma), or BIBX1382 (10 µM; R&D), for 48 h at 37 °C.
Imaging was conducted using a Zeiss Observer Z1 and an AxioCam MRM camera. Images were collected at 15-min intervals with exposure times of around 400 ms and were played at 15 frames per second. A Solent environmental chamber was used to maintain the culture condition with CO2 at 5% and temperature at 37 °C.
Western Blotting.
Cells were incubated in serum-reduced fibroblast medium (0.2% FBS) for 24 h and then treated with growth factors for the specified time. After treatment, cells were washed twice in ice-cold PBS and lysed in the RIPA buffer (150 mM NaCl, 1.0% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris, pH 8.0) supplied with proteinase and phosphatase inhibitors (10 mM β-glycerophosphate, 5 mM NaF, 1 mM Na3VO4, 1 mM DTT, 0.5 mM phenylmethanesulfonylfluoride, 2 µg/mL aprotinin, and 10 µg/mL leupeptin). Protein lysates were homogenized by sonication (Microson XL2000; QSonica) and cleared by centrifugation, and protein concentration was measured using the Bradford reagent. Denatured, reduced samples were resolved on 10% (vol/vol) NuPAGE Bis-Tris gels (Life Technologies) and blotted onto nitrocellulose membranes. Membranes were blocked with 5% (wt/vol) nonfat milk in PBS with 0.05% Tween 20 (Sigma; blocking buffer) and incubated with primary antibodies diluted in blocking buffer overnight at 4 °C. After washing in PBS with 0.05% Tween 20, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. Signal was developed using the ECL substrates (SuperSignal West Pico/Dura, Thermo Scientific; or Clarity Western ECL Substrate, Bio-Rad) and exposed on X-ray films (Kodak), which were then scanned and band density was analyzed using ImageJ. Samples comparing Spry1het and Spry1KO lysates were resolved on a single gel for each experiment. Phosphorylated and total proteins and actin were analyzed on a single blot. Primary antibodies used were β-actin (Sigma); p-AKT 1/2/3 (Ser473), p-ERK 1/2 (Thr202/Tyr204), Spry1 (Santa Cruz); and AKT, p44/p42 MAPK (ERK1/2) (Cell Signaling). Secondary antibodies used were as follows: goat anti-mouse antibody (DAKO) and goat anti-rabbit antibody (Santa Cruz Biotechnology).
Collagen Contraction Assays.
Collagen gels were prepared by combining 12.5 vol of collagen type I (3.9 mg/mL; Corning) with 2.5 vol of collagen reconstitution buffer (5× MEM, 20 µg/mL NaHCO3, 0.1 M Hepes), 1 vol of 0.22 M NaOH, 3.1 vol of FBS, and 3.1 vol of fibroblasts in DMEM (2 × 105 fibroblasts per milliliter), yielding a final concentration of collagen of 2.2 mg/mL. Equal volumes of the gel–fibroblast mixture were plated in 24-well plates and incubated at 37 °C for 1 h before fibroblast medium (with the addition of 10 µM BIBX1382, as indicated) was added on the top. After 4 d of culture, samples were fixed in 4% PFA, photographed using a SteREO LumarV12 stereoscope, and analyzed using ImageJ.
Organotypic Invasion Assay.
Fibroblasts (106 per milliliter) were embedded in a mixture of collagen I and Matrigel (Corning). Final collagen concentration was ∼3.6 mg/mL, and Matrigel concentration was ∼2.2 mg/mL. Equal volumes of gel–fibroblast mixture were plated in 24-well plates and incubated at 37 °C for 1 h before fibroblast medium was added on the top. On the following day, 5 × 105 MCF7-ras cells were seeded on top of the gels. After 24 h, breast cancer cells were covered with a thin layer of gel. The gels were then mounted on a metal bridge and fed from underneath using DMEM supplemented with 10% (vol/vol) FBS, 10 µg/mL insulin, 5.5 µg/mL transferrin, 6.7 ng/mL selenium, 5 nM EGF, 100 U/mL penicillin, and 100 µg/mL streptomycin. Medium was changed daily, and samples were cultured for 8 d before they were fixed in 4% PFA. Samples were then sectioned in paraffin and stained with hematoxylin and eosin. Images were taken using Pannoramic 250 Flash scanning microscope and analyzed using Pannoramic Viewer software (3DHISTECH). Invasion depths were measured as the distance from the lower surface of the noninvasive cell layer to the deepest edge of the invading cells in 10 sections per gel (all invading cells were measured) and assessed as total invasion in a gel, average invasion depth, or average maximum invasion depth per gel section.
Statistical Analyses.
Statistical analyses were performed using Prism (GraphPad Software). Mean values and SD or SEM are shown in graphs that were generated from several repeats (n) of biological experiments. P values were obtained from t tests with paired or unpaired samples or from ANOVA, with significance set at P < 0.05. Graphs show symbols describing it as *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Supplementary Material
Acknowledgments
We thank Urszula Polanska for MCF7-ras cells and Xin Guo for performing fibrosphere assays. We thank Gary Freeman for critical reading of the manuscript. This work was supported by the project “Employment of Best Young Scientists for International Cooperation Empowerment” (CZ.1.07/2.3.00/30.0037), cofinanced by the European Social Fund and the state budget of the Czech Republic (GJ16-20031Y) (Czech Science Foundation); funds from the Faculty of Medicine, Masaryk University to junior research (to Z.K.); grants from Breast Cancer Now (to R.B.C. and P.L.); National Institutes of Health (NIH) Grant R03 HD060807 and a start-up grant from ShanghaiTech University (to P.L.); and NIH Grants R01 CA057621 (to Z.W.) and R01 DE021420 (to O.D.K.). The Wellcome Trust Center for Cell-Matrix Research, University of Manchester, is supported by core funding from the Wellcome Trust (088785/Z/09/Z).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1611532113/-/DCSupplemental.
References
- 1.Lu P, Weaver VM, Werb Z. The extracellular matrix: A dynamic niche in cancer progression. J Cell Biol. 2012;196(4):395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Propper A, Gomot L. Control of chick epidermis differentiation by rabbit mammary mesenchyme. Experientia. 1973;29(12):1543–1544. doi: 10.1007/BF01943907. [DOI] [PubMed] [Google Scholar]
- 3.Propper A, Gomot L. [Tissue interactions during organogenesis of the mammary gland in the rabbit embryo.] C R Acad Sci Hebd Seances Acad Sci D. 1967;264(22):2573–2575. French. [PubMed] [Google Scholar]
- 4.Booth BW, et al. The mammary microenvironment alters the differentiation repertoire of neural stem cells. Proc Natl Acad Sci USA. 2008;105(39):14891–14896. doi: 10.1073/pnas.0803214105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulanger CA, Mack DL, Booth BW, Smith GH. Interaction with the mammary microenvironment redirects spermatogenic cell fate in vivo. Proc Natl Acad Sci USA. 2007;104(10):3871–3876. doi: 10.1073/pnas.0611637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egeblad M, Rasch MG, Weaver VM. Dynamic interplay between the collagen scaffold and tumor evolution. Curr Opin Cell Biol. 2010;22(5):697–706. doi: 10.1016/j.ceb.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howard BA, Lu P. Stromal regulation of embryonic and postnatal mammary epithelial development and differentiation. Semin Cell Dev Biol. 2014;25-26:43–51. doi: 10.1016/j.semcdb.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Watson CJ, Khaled WT. Mammary development in the embryo and adult: A journey of morphogenesis and commitment. Development. 2008;135(6):995–1003. doi: 10.1242/dev.005439. [DOI] [PubMed] [Google Scholar]
- 9.Macias H, Hinck L. Mammary gland development. Wiley Interdiscip Rev Dev Biol. 2012;1(4):533–557. doi: 10.1002/wdev.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu P, Sternlicht MD, Werb Z. Comparative mechanisms of branching morphogenesis in diverse systems. J Mammary Gland Biol Neoplasia. 2006;11(3-4):213–228. doi: 10.1007/sl0911-006-9027-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim EJ, Jung HS, Lu P. Pleiotropic functions of fibroblast growth factor signaling in embryonic mammary gland development. J Mammary Gland Biol Neoplasia. 2013;18(2):139–142. doi: 10.1007/s10911-013-9278-4. [DOI] [PubMed] [Google Scholar]
- 12.Lu P, Werb Z. Patterning mechanisms of branched organs. Science. 2008;322(5907):1506–1509. doi: 10.1126/science.1162783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richards RG, Klotz DM, Walker MP, Diaugustine RP. Mammary gland branching morphogenesis is diminished in mice with a deficiency of insulin-like growth factor-I (IGF-I), but not in mice with a liver-specific deletion of IGF-I. Endocrinology. 2004;145(7):3106–3110. doi: 10.1210/en.2003-1112. [DOI] [PubMed] [Google Scholar]
- 14.Bonnette SG, Hadsell DL. Targeted disruption of the IGF-I receptor gene decreases cellular proliferation in mammary terminal end buds. Endocrinology. 2001;142(11):4937–4945. doi: 10.1210/endo.142.11.8500. [DOI] [PubMed] [Google Scholar]
- 15.Pond AC, et al. Fibroblast growth factor receptor signaling is essential for normal mammary gland development and stem cell function. Stem Cells. 2013;31(1):178–189. doi: 10.1002/stem.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu P, Ewald AJ, Martin GR, Werb Z. Genetic mosaic analysis reveals FGF receptor 2 function in terminal end buds during mammary gland branching morphogenesis. Dev Biol. 2008;321(1):77–87. doi: 10.1016/j.ydbio.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peters G, Brookes S, Smith R, Dickson C. Tumorigenesis by mouse mammary tumor virus: Evidence for a common region for provirus integration in mammary tumors. Cell. 1983;33(2):369–377. doi: 10.1016/0092-8674(83)90418-x. [DOI] [PubMed] [Google Scholar]
- 18.Xian W, Schwertfeger KL, Rosen JM. Distinct roles of fibroblast growth factor receptor 1 and 2 in regulating cell survival and epithelial-mesenchymal transition. Mol Endocrinol. 2007;21(4):987–1000. doi: 10.1210/me.2006-0518. [DOI] [PubMed] [Google Scholar]
- 19.Xian W, Schwertfeger KL, Vargo-Gogola T, Rosen JM. Pleiotropic effects of FGFR1 on cell proliferation, survival, and migration in a 3D mammary epithelial cell model. J Cell Biol. 2005;171(4):663–673. doi: 10.1083/jcb.200505098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell. 2010;141(1):52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sternlicht MD, Kouros-Mehr H, Lu P, Werb Z. Hormonal and local control of mammary branching morphogenesis. Differentiation. 2006;74(7):365–381. doi: 10.1111/j.1432-0436.2006.00105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luetteke NC, et al. Targeted inactivation of the EGF and amphiregulin genes reveals distinct roles for EGF receptor ligands in mouse mammary gland development. Development. 1999;126(12):2739–2750. doi: 10.1242/dev.126.12.2739. [DOI] [PubMed] [Google Scholar]
- 23.Ciarloni L, Mallepell S, Brisken C. Amphiregulin is an essential mediator of estrogen receptor alpha function in mammary gland development. Proc Natl Acad Sci USA. 2007;104(13):5455–5460. doi: 10.1073/pnas.0611647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sternlicht MD, et al. Mammary ductal morphogenesis requires paracrine activation of stromal EGFR via ADAM17-dependent shedding of epithelial amphiregulin. Development. 2005;132(17):3923–3933. doi: 10.1242/dev.01966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stern DF. Tyrosine kinase signalling in breast cancer: ErbB family receptor tyrosine kinases. Breast Cancer Res. 2000;2(3):176–183. doi: 10.1186/bcr51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Issa A, et al. Combinatorial targeting of FGF and ErbB receptors blocks growth and metastatic spread of breast cancer models. Breast Cancer Res. 2013;15(1):R8. doi: 10.1186/bcr3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basson MA, et al. Sprouty1 is a critical regulator of GDNF/RET-mediated kidney induction. Dev Cell. 2005;8(2):229–239. doi: 10.1016/j.devcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Klein OD, et al. An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors. Development. 2008;135(2):377–385. doi: 10.1242/dev.015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panagiotaki N, Dajas-Bailador F, Amaya E, Papalopulu N, Dorey K. Characterisation of a new regulator of BDNF signalling, Sprouty3, involved in axonal morphogenesis in vivo. Development. 2010;137(23):4005–4015. doi: 10.1242/dev.053173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Qiao G, Lu P. Modulation of fibroblast growth factor signaling is essential for mammary epithelial morphogenesis. PLoS One. 2014;9(4):e92735. doi: 10.1371/journal.pone.0092735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thum T, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456(7224):980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 32.Shim K, Minowada G, Coling DE, Martin GR. Sprouty2, a mouse deafness gene, regulates cell fate decisions in the auditory sensory epithelium by antagonizing FGF signaling. Dev Cell. 2005;8(4):553–564. doi: 10.1016/j.devcel.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 33.Ching ST, Cunha GR, Baskin LS, Basson MA, Klein OD. Coordinated activity of Spry1 and Spry2 is required for normal development of the external genitalia. Dev Biol. 2014;386(1):1–11. doi: 10.1016/j.ydbio.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shackleton M, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439(7072):84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 35.Shaw FL, et al. A detailed mammosphere assay protocol for the quantification of breast stem cell activity. J Mammary Gland Biol Neoplasia. 2012;17(2):111–117. doi: 10.1007/s10911-012-9255-3. [DOI] [PubMed] [Google Scholar]
- 36.Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4(4):359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dikic I, Giordano S. Negative receptor signalling. Curr Opin Cell Biol. 2003;15(2):128–135. doi: 10.1016/s0955-0674(03)00004-8. [DOI] [PubMed] [Google Scholar]
- 38.Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA. sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell. 1998;92(2):253–263. doi: 10.1016/s0092-8674(00)80919-8. [DOI] [PubMed] [Google Scholar]
- 39.Arora S, et al. Egr1 regulates the coordinated expression of numerous EGF receptor target genes as identified by ChIP-on-chip. Genome Biol. 2008;9(11):R166. doi: 10.1186/gb-2008-9-11-r166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tarcic G, et al. EGR1 and the ERK-ERF axis drive mammary cell migration in response to EGF. FASEB J. 2012;26(4):1582–1592. doi: 10.1096/fj.11-194654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solca FF, et al. Inhibition of epidermal growth factor receptor activity by two pyrimidopyrimidine derivatives. J Pharmacol Exp Ther. 2004;311(2):502–509. doi: 10.1124/jpet.104.069138. [DOI] [PubMed] [Google Scholar]
- 42.Zhang X, et al. FGF ligands of the postnatal mammary stroma regulate distinct aspects of epithelial morphogenesis. Development. 2014;141(17):3352–3362. doi: 10.1242/dev.106732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohammadi M, et al. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science. 1997;276(5314):955–960. doi: 10.1126/science.276.5314.955. [DOI] [PubMed] [Google Scholar]
- 44.Gaggioli C, et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol. 2007;9(12):1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 45.Casaletto JB, McClatchey AI. Spatial regulation of receptor tyrosine kinases in development and cancer. Nat Rev Cancer. 2012;12(6):387–400. doi: 10.1038/nrc3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calmont A, et al. An FGF response pathway that mediates hepatic gene induction in embryonic endoderm cells. Dev Cell. 2006;11(3):339–348. doi: 10.1016/j.devcel.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 47.Bhowmick NA, et al. TGF-beta signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science. 2004;303(5659):848–851. doi: 10.1126/science.1090922. [DOI] [PubMed] [Google Scholar]
- 48.Kajanne R, et al. EGF-R regulates MMP function in fibroblasts through MAPK and AP-1 pathways. J Cell Physiol. 2007;212(2):489–497. doi: 10.1002/jcp.21041. [DOI] [PubMed] [Google Scholar]
- 49.Wiseman BS, et al. Site-specific inductive and inhibitory activities of MMP-2 and MMP-3 orchestrate mammary gland branching morphogenesis. J Cell Biol. 2003;162(6):1123–1133. doi: 10.1083/jcb.200302090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alcaraz J, et al. Collective epithelial cell invasion overcomes mechanical barriers of collagenous extracellular matrix by a narrow tube-like geometry and MMP14-dependent local softening. Integr Biol (Camb) 2011;3(12):1153–1166. doi: 10.1039/c1ib00073j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu P, Takai K, Weaver VM, Werb Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb Perspect Biol. 2011;3(12):a005058. doi: 10.1101/cshperspect.a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.