Plants rely on the levels and concentration gradients of the hormone auxin as crucial information cues to trigger and modulate almost every aspect of their life cycle, from establishing embryo polarity to promoting phototropic and gravitropic responses (1). Thus, it is critical for plants to finely control the levels of the bioactive form of this hormone, both at spatial and temporal levels. Local production, transport, conjugation, storage, and catabolism are all well-known processes participating in the dynamic regulation of auxin homeostasis (2). It is, however, unknown whether or how these different auxin homeostasis mechanisms talk to each other. In the past 30 y, the identification of the genes coding for the key components of the auxin biosynthetic, transport, and conjugation machineries has proven instrumental in assessing the contribution of each of these processes to concrete developmental pathways, as well as to specific plant responses to environmental changes. Although much less is known about the ways the predominant auxin, indole-3-acetic acid (IAA), is catabolized, classical labeling and metabolite quantification experiments indicate that the oxidation of IAA into 2-oxindole-3-acetic acid (oxIAA) is one of the most prevalent mechanisms to inactivate this hormone (3). Identifying the specific enzymes catalyzing this reaction in vivo has proved to be more challenging than anticipated, as different plant peroxidases and oxygenases, the original suspects for catalyzing this reaction, were found not to play a significant physiological role in the production of oxIAA (3). The recent identification of a rice 2-oxoglutarate-dependent-Fe(II) dioxygenase, DIOXYGENASE FOR AUXIN OXIDATION (DAO), originally linked to male fertility in rice and capable of oxidizing IAA into oxIAA (1–4), has opened new avenues for addressing the physiological significance of auxin catabolism in plant growth and development. In PNAS, Zhang et al. (5), Porco et al. (6), and Mellor et al. (7) identify and characterize the Arabidopsis orthologs of the rice DAO gene, AtDAO1 and AtDAO2, not only demonstrating that this important auxin catabolism pathway is well conserved in monocots and dicots but also expanding the list of physiological processes in which this IAA oxidation plays potential regulatory roles. Even more importantly, an intricate regulatory network comprising the production, conjugation, and catabolism of auxin was experimentally uncovered (5–7), and some of its complexity was mathematically described (7).
As stated above, precise control of local concentrations of IAA is at the core of many developmental programs and plant responses to the environment. Intuitively, this fine-tuning of auxin levels could be accomplished by altering the production, transport, storage, and degradation of this hormone. Auxin transport, with its well-known net of influx and efflux transporters, has long been recognized as a central component of this refined mechanism to alter local levels of auxin (8). With the identification of the genes involved in auxin biosynthesis and their exquisite spatial–temporal regulation, a new layer of regulation in the dynamic control of local auxin levels emerged (9). Finally, although storage and degradation are processes both known to contribute to the auxin homeostasis, one of the most prevalent IAA inactivation pathways, its oxidation to oxIAA, has remained elusive (10). The recent characterization of a male-sterile mutant in rice resulted in the identification of a candidate enzyme, DAO, capable of catalyzing the conversion of IAA into oxIAA (4). Phylogenetic approaches were then used to find the putative functional orthologs of this gene, AtDAO1 and AtDAO2, in the Arabidopsis genome (5, 6). Interestingly, a related dioxygenase gene, ARRO-1, has been previously described in apples and has been shown to be inducible by auxin in apple seedlings’ roots (11). By identifying homologous genes in Arabidopsis, a large battery of genetic tools and phenotypic information became immediately available, facilitating the thorough functional characterization of what was then shown to be the key enzymes of one of the predominant auxin catabolic pathways in plants. A simple search of the existing large collections of gene-indexed mutants in Arabidopsis, followed by the quantitative analysis of the expression of the genes of interest in the candidate T-DNA lines, resulted in the identification of both gain- and loss-of-function alleles for AtDAO1, as well as of a hypomorphic mutant allele for AtDAO2. The metabolic and morphological analysis of these mutants, combined with in vitro enzymatic assays, confirmed the central role of this dioxygenase in the oxidation of IAA into oxIAA (5, 6). Detailed phenotypic analysis indicated that the oxidation of IAA plays regulatory roles in a variety of developmental processes, including root hair elongation, lateral root formation, rosette size, and fertility (5, 6). Although dao1 dao2 double-knockout mutants are not yet available and will likely take time to generate given the physical proximity of the two genes in the genome, the phenotypes observed in the single dao1 loss-of-function mutant were relatively mild and variable dependent on the growth conditions (5, 6). Although this mild phenotype could be explained by the redundant function of an active AtDAO2 in the dao1 knockout, the low expression levels of AtDAO2, together with the dramatic reduction in the levels of oxIAA in the single dao1 mutant, point toward a different explanation. Very revealing in this regard was the fact that no significant changes in the levels of IAA were observed in the dao1 knockouts despite the twofold or even greater reduction in the levels of oxIAA in the mutants, at least in the tissues and developmental stages tested (5–7). Taken together, these results suggested that either the oxidation of IAA plays only a minor physiological role in modulating the levels of bioactive auxin or, most likely, plants possess redundant mechanisms to catabolize IAA. In fact, the latter possibility is supported by the observation that the IAA conjugates, IAA-Glu and IAA-Asp, the products of another major route of IAA inactivation, accumulate to much higher levels in the dao1 knockout mutant than in wild-type plants (5–7), whereas a gain-of-function allele of dao1 shows reduced levels of these conjugates (6). Consistent with this observation, an increase in the transcript level of GH3.3, one of the key enzymes in the conjugation of IAA to amino acids, in the dao1 loss-of-function mutant could be observed (5–7). This idea of the functional redundancy between the DAO- and the GH3-mediated auxin inactivation pathways was further supported by metabolite feeding experiments in which labeled auxin precursors were much more rapidly incorporated into the IAA conjugates, IAA-Asp and IAA-Glu, in the dao1 knockout mutant than in the wild-type controls (6).
With the key genes of the oxIAA pathway at hand and strong experimental evidence for the existence of a compensatory mechanism between the two main IAA catabolic pathways, the question arose as to how these two pathways influence each other’s activities. The first clue to answering this question came from the observation by Porco et al. (6) that AtDAO1 was auxin inducible, as it has been previously shown for the apple AtDAO1 ortholog, ARRO-1a (11), and for members of the auxin-conjugating enzyme family in Arabidopsis, GH3 (7, 12). Thus, one could imagine a simple mechanistic model in which a decrease in the rate of IAA oxidation due to a disruption in DAO1 in Arabidopsis would result in a transient accumulation of IAA. This increase in IAA will then trigger an increase in GH3 transcription, leading to more GH3 protein and a subsequent increase in IAA conjugation, thus bringing the levels of active IAA back to “normal.” To better evaluate the role of these interactions, Mellor et al. (7) developed mathematical models that could explain how a decrease in the levels of IAA oxidation in the dao1 knockout mutant could trigger not only the observed decrease in the levels of oxIAA but also the parallel dramatic increase in the levels of IAA-Asp and IAA-Glu detected experimentally (5–7). Interestingly, even when a 10-fold increase in the GH3 activity in the dao1 knockout was considered, the original mathematical model only predicted minor increases in the levels of IAA conjugates. The failure of the original model suggested the existence of additional factors or regulatory interactions, not yet accounted for in the model, that were playing a central role in auxin homeostasis in planta. Another possible mechanism that could be behind the large increase in IAA-Asp and IAA-Glu in the dao1 knockout, which was not originally considered, is an increase in the rates of IAA biosynthesis. In fact, when even a minor increase in auxin production was introduced, the mathematical model now predicted large increases in IAA conjugates, resulting in a much better match between the experimentally observed and model-predicted levels of IAA catabolites (7). Consistent with the increase in IAA biosynthesis needed for the mathematical model to accurately reflect the observed changes in auxin catabolites, an increase in the IAA precursor indole-3-pyruvic acid (IPyA) was detected in the dao1 loss-of-function mutant (6, 7). Although additional experiments will be required to further demonstrate that the DAO1 disruption indeed results in an increase in the metabolic flux through the IPyA pathway in planta and to determine the exact molecular mechanism responsible for such increase, the computational model was successful at explaining in mathematical terms the interactions between production and two different IAA catabolic pathways. Integral to this model was the experimentally observed differential regulation of the two catabolic pathways by IAA. Thus, the GH3-dependent conjugation pathway is rapidly activated by high levels of IAA, whereas the DAO oxidation pathway is much slower to respond to IAA, but can be modulated by even minor changes in the levels of this hormone (6, 7, 12) (Fig. 1).
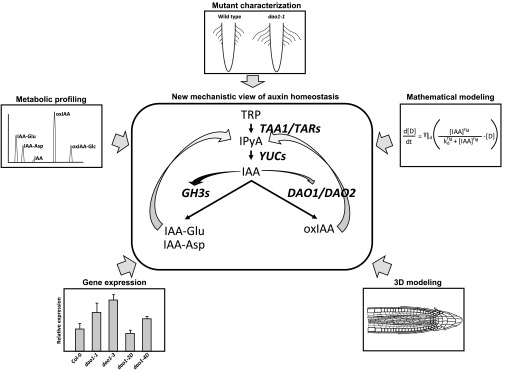
Fig. 1.
Schematic representation of the main routes of auxin metabolism and the new interactions found between anabolic and catabolic pathways. In the central panel, the indole-3-pyruvic acid (IPyA) route of auxin biosynthesis from tryptophan (TRP) to indole-3-acetic acid (IAA) is depicted, as well as the two main routes of auxin inactivation, the conjugation of IAA to glutamate (IAA-Glu) and aspartate (IAA-Asp), and the oxidation of IAA to 2-oxindole-3-acetic acid (oxIAA). The genes coding for the enzymes involved in the depicted enzymatic conversions are shown in bold italic letters. Straight arrows indicate the conversion of a precursor into a product. Curved arrows indicate regulatory interactions. The arrows converging on IPyA mark the feedback on IAA biosynthesis by a yet-undetermined molecular mechanism. The curved arrows originating from IAA signify the quick and strong stimulatory effect of high levels of IAA on GH3 activity (dark curved arrow) or a slow and weak induction of DAO1 by IAA (gray curved arrow) via transcriptional up-regulation of the respective genes. The peripheral panels depict some of the strategies used in the studies by Zhang et al. (5), Porco et al. (6), and Mellor et al. (7) to uncover the new mechanisms of auxin homeostasis.
The identification of the genes responsible for key aspects of auxin biosynthesis, transport, conjugation, and now oxidative catabolism has opened the possibility not only to examine how each of these processes responds to specific perturbations but also to begin to infer potential interactions among these auxin homeostasis pathways and to map these interactions in 3D models. Moving toward such comprehensive 3D models of auxin metabolism, Mellor et al. (7) integrated their mathematical model discussed above with their previously developed 3D model of auxin transport. Interestingly, this 3D model made the prediction of a greater effect of the dao1 loss-of-function mutation on the levels of IAA specifically in the outer cell layers of the root. The observation by both Zhang et al. (5) and Porco et al. (6) of a robust increase in the root hair length in dao1 knockouts and a corresponding reduction in the length of dao1 gain-of-function mutant root hairs is consistent with the model prediction and provides an illustrative example of the potential power of such 3D models.
The future expansion of the current 3D models of auxin homeostasis would likely benefit from incorporating additional high-resolution spatial and temporal expression information of genes responsible not only for auxin transport but also for biosynthesis, conjugation, and catabolism, as well as from equivalent high-resolution data on metabolite levels and fluxes. With the genes for the major auxin pathways at hand and the constant improvement of quantitative methods that capture gene expression and metabolite levels with cellular resolution, a systems understanding of auxin metabolism should become a reality in the foreseeable future.
Acknowledgments
This work was supported by National Science Foundation Grant IOS 1444561 (to J.M.A. and A.N.S.).
Footnotes
References
- 1.Vanneste S, Friml J. Auxin: A trigger for change in plant development. Cell. 2009;136(6):1005–1016. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Ljung K. Auxin metabolism and homeostasis during plant development. Development. 2013;140(5):943–950. doi: 10.1242/dev.086363. [DOI] [PubMed] [Google Scholar]
- 3.Pencík A, et al. Regulation of auxin homeostasis and gradients in Arabidopsis roots through the formation of the indole-3-acetic acid catabolite 2-oxindole-3-acetic acid. Plant Cell. 2013;25(10):3858–3870. doi: 10.1105/tpc.113.114421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao Z, et al. A role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev Cell. 2013;27(1):113–122. doi: 10.1016/j.devcel.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Zhang J, et al. DAO1 catalyzes temporal and tissue-specific oxidative inactivation of auxin in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2016;113:11010–11015. doi: 10.1073/pnas.1604769113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porco S, et al. Dioxygenase-encoding AtDAO1 gene controls IAA oxidation and homeostasis in Arabidopsis. Proc Natl Acad Sci USA. 2016;113:11016–11021. doi: 10.1073/pnas.1604375113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mellor N, et al. Dynamic regulation of auxin oxidase and conjugating enzymes AtDAO1 and GH3 modulates auxin homeostasis. Proc Natl Acad Sci USA. 2016;113:11022–11027. doi: 10.1073/pnas.1604458113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peer WA, Blakeslee JJ, Yang H, Murphy AS. Seven things we think we know about auxin transport. Mol Plant. 2011;4(3):487–504. doi: 10.1093/mp/ssr034. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Y. Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol. 2010;61:49–64. doi: 10.1146/annurev-arplant-042809-112308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korasick DA, Enders TA, Strader LC. Auxin biosynthesis and storage forms. J Exp Bot. 2013;64(9):2541–2555. doi: 10.1093/jxb/ert080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butler ED, Gallagher TF. Characterization of auxin-induced ARRO-1 expression in the primary root of Malus domestica. J Exp Bot. 2000;51(351):1765–1766. doi: 10.1093/jexbot/51.351.1765. [DOI] [PubMed] [Google Scholar]
- 12.Park JE, et al. GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J Biol Chem. 2007;282(13):10036–10046. doi: 10.1074/jbc.M610524200. [DOI] [PubMed] [Google Scholar]