Significance
RIG-I-like receptors (RLRs) belong to a family of DExD/H box RNA helicases and comprise three members: RIG-I, MDA5, and LGP2. The function of RLRs as key cytoplasmic sensors of pathogen-associated molecular patterns has been subjected to numerous pathogenic challenges and undergone dynamic evolution, making it an excellent example for studying innate immunity evolution. We characterized the loss of RIG-I and found that tMDA5 acted as the pattern-recognition receptor responsible for sensing Sendai virus infection in the tree shrew, with an involvement of tLGP2 and tMITA. The positively selective sites in tMDA5 were crucial for the tMDA5-mediated antiviral signaling pathway. Our results provided insights into the evolutionary adaptation and functional diversity of antiviral activity in vertebrates.
Keywords: RIG-I, MDA5, tree shrew, positive selection, functional replacement
Abstract
The function of the RIG-I-like receptors (RLRs; including RIG-I, MDA5, and LGP2) as key cytoplasmic sensors of viral pathogen-associated molecular patterns (PAMPs) has been subjected to numerous pathogenic challenges and has undergone a dynamic evolution. We found evolutionary evidence that RIG-I was lost in the Chinese tree shrew lineage. Along with the loss of RIG-I, both MDA5 (tMDA5) and LGP2 (tLGP2) have undergone strong positive selection in the tree shrew. tMDA5 or tMDA5/tLGP2 could sense Sendai virus (an RNA virus posed as a RIG-I agonist) for inducing type I IFN, although conventional RIG-I and MDA5 were thought to recognize distinct RNA structures and viruses. tMDA5 interacted with adaptor tMITA (STINGTMEM173/ERIS), which was reported to bind only with RIG-I. The positively selected sites in tMDA5 endowed the substitute function for the lost RIG-I. These findings provided insights into the adaptation and functional diversity of innate antiviral activity in vertebrates.
In persistent struggle between host and virus, the evolution of the innate immune system is a pivotal turning point. When virus enters and replicates inside the host cells, host innate immunity, as the first line of immune system to defense against pathogen infection, is quickly motivated. Host proteins, such as the germline-encoded pattern-recognition receptors (PRRs), which interact directly with viral protein, are subjected to molecular arms races (1). Cytosolic RIG-I-like receptors (RLRs), encompassing RIG-I (retinoic acid-inducible gene I, also known as DDX58) (2), MDA5 (melanoma differentiation factor 5, also known as IFIH1) (3), and LGP2 (laboratory of genetics and physiology 2, also known as DHX58) (4), which act as PRRs detecting viral PAMPs, mainly sense viral RNA in the cytosol (5) and trigger a series of signal cascades that lead to the production of type I IFN and various cytokines (6–8). These proteins bear the imprint of the long-term evolutionary arms race against viral RNA and other molecules (9). RIG-I and MDA5 share similar signaling features and structural homology: with two N-terminal caspase-recruitment domains (CARDs), a DExD/H box RNA helicase domain, and a C-terminal repressor domain (RD), whereas LGP2 lacks a CARD (2, 4, 10). The role of RIG-I and MDA5 in response to virus stimulation is not redundant: MDA5 specifically recognizes Picornaviruses, such as the encephalomyocarditis virus (EMCV), and RIG-I recognizes a wide variety of RNA viruses belonging to the Paramyxovirus and Rhabdovirus families (11). In contrast to MDA5 and RIG-I, the role of LGP2 in cytosolic RNA sensing remains controversial. Some reports suggested that LGP2 is essential for the production of type I IFN in response to several RIG-I– and MDA5-dependent viruses (10), whereas others described LGP2 as a negative regulator of the RIG-I signaling (12).
Evolutionary studies have painted an intricate picture of how RLRs have arisen and functionally diversified based on the CARDs of RIG-I and MDA5 (9, 13–15), which are essential in triggering the IFN response (2). It has been suggested that the CARD domains have been gained by RIG-I and MDA5 in two separate events: the first domain being acquired before the duplication that developed RIG-I and MDA5 and the second domain gained after their divergence (13). Moreover, the two CARDs found at separate loci in the sea anemone Nematostellavectensis suggested that these CARDs might have occurred in these loci after the divergence of the chordates (14). A recent study showed that RLR and MAVS CARDs diversified in early deuterostomes, probably through a series of tandem, partial-gene duplication events (15). In contrast to the CARDs, the divergence of RIG-I, MDA5, and LGP2 have remained unresolved (16). Zou et al. (14) demonstrated that RIG-I diverged in the early deuterostomes, with LGP2 and MDA5 diverging later in the vertebrates, whereas Sarkar et al. (13) showed that LGP2 diverged in the early chordates, followed by the divergence of RIG-I and MDA5 in the tetrapods. Recently, Mukherjee et al. showed that the RLR-based immune system might arise with the emergence of multicellularity (9). Although MDA5 and LGP2 homologs were found in many teleost fishes, clear RIG-I homologs have only been identified in salmon and carp (17). RIG-I was absent in the chicken genome, although MDA5 and LGP2 were both present (18, 19). The loss of RIG-I might affect the first line of defense at the lung epithelial cells during influenza infection in chickens. Therefore, it was not surprising that chickens suffer severely from avian influenza virus (AIV) infection compared with ducks (18). No mammalian species has been found to have a defective RIG-I until recently when we noted the absence of RIG-I in the Chinese tree shrew (Tupaia belangeri chinensis) genome (20). An understanding of the biological effects of the loss of RIG-I in the tree shrew will undoubtedly offer insights into the origin and development of the innate immunity in mammals.
In this study, we performed an evolutionary analysis and a functional characterization of the loss of RIG-I in the Chinese tree shrew. We confirmed that the absence of RIG-I in the tree shrew lineage, which accompanying with the presence of a positive selection signal on the other two RLR members, the tree shrew MDA5 (tMDA5) and LGP2 (tLGP2). Functional assays have shown that tMDA5 or tMDA5 combined with tLGP2 (tMDA5/tLGP2) could sense Sendai virus (SeV), which is a RIG-I agonist. tMDA5 could also interact with adaptor tMITA (21–23), which was reported to specifically bind with RIG-I. These results have suggested tMDA5 can replace or at least partially replace the function of RIG-I to sense RNA virus and enhance signaling via interaction with tMITA.
Results
RIG-I Is Absent in the Chinese Tree Shrew.
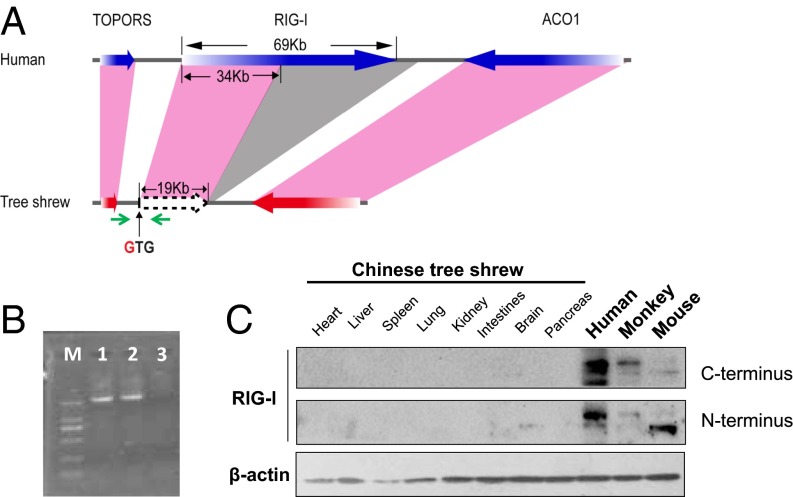
An inspection of the Chinese tree shrew genome (20) revealed the RIG-I gene has been severely damaged so that functional RIG-I protein cannot be made (Fig. 1A). We identified the mutational change affecting the start codon of RIG-I, with the expected methionine-codon being replaced by a valine-codon (Fig. S1), in all 52 Chinese tree shrews collected from different geographic regions (Fig. S1). The RIG-I transcript was missing in tree shrew, but was expressed in both Malayan flying lemur (which has a close affinity to tree shrew) (24) and human cells (Fig. 1B). The RIG-I protein could be detected in human, monkey, and mouse, but not in tree shrew tissues (Fig. 1C). These results confirmed the loss of RIG-I in the tree shrew and showed this event occurred after the divergence of tree shrew from these other species.
Fig. 1.
RIG-I is absent in the Chinese tree shrew. (A) Diagram illustrating the structure of the tree shrew RIG-I genomic region. Color codes: pink shading, homologous genes from human to tree shrew; gray shading, tree shrew RIG-I deficient region relative to human alignment. The green arrows indicated the primers for amplifying the fragment containing the predictive start codon of the RIG-I gene. (B) PCR amplification of the RIG-I gene fragment using the cDNAs of human HeLa (lane 1), Malayan flying lemur (lane 2) cells, and Chinese tree shrew primary renal cells (lane 3). Lane M refers to DNA ladder. (C) Western blot showing the RIG-I protein in human, monkey, mouse, and Chinese tree shrew tissues. RIG-I antibodies recognized the C terminus and the N terminus of RIG-I, respectively.
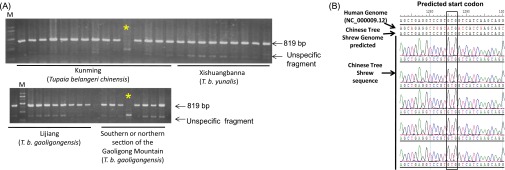
Fig. S1.
RIG-I is absent in the Chinese tree shrew. (A) The PCR products derived from the genomic DNA of the Chinese tree shrew covering the RIG-I gene region have a length of 819 bp. We analyzed a total of 52 Chinese tree shrew individuals that were collected from Kunming (T. b. chinensis), Xishuangbanna (T. b. yunalis), Lijiang, and Southern and northern sections of the Gaoligong Mountain (T. b. gaoligongensis). Each lane refers to one animal and the PCR products of 41 individuals are presented here. The two lanes marked by * mean failure of PCR amplification in those two individuals, and reamplification of the original DNAs yielded the correct bands. Lane M refers to DNA ladder. (B) Sequencing electropherograms of different Chinese tree shrew individuals showing the change of the start codon from ATG to GTG.
A Substitute for the Lost RIG-I Acts as the Ligand of RNA Viruses and 5′ Triphosphate RNA.
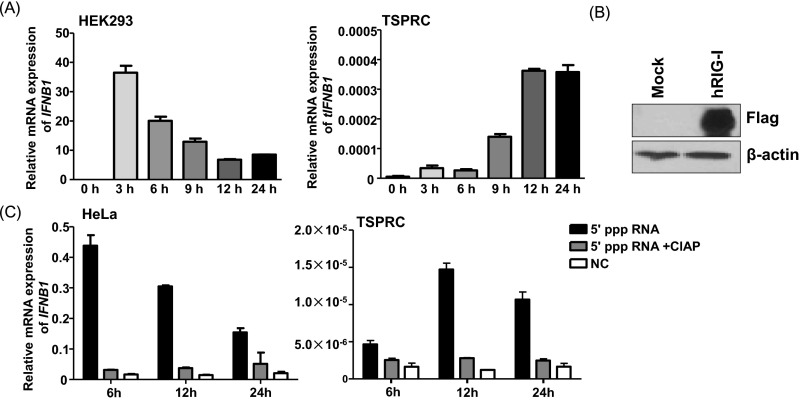
The absence of RIG-I in tree shrew has led us to hypothesize that the functioning of its antiviral innate immune system would be affected. Indeed, we observed different time-dependent expression patterns of IFNB1 mRNA in tree shrew primary renal cells (TSPRCs) and HEK293 cells in response to Newcastle disease virus (NDV) infection (Fig. S2A). We further assessed the tIFNB1 mRNA levels in TSPRCs challenged by a set of negative-sense RNA viruses, including SeV, AIV and VSV (vesicular stomatitis virus), and found that these viruses also significantly induced the tIFNB1 mRNA expression (Fig. 2A). When TSPRCs were overexpressed with human RIG-I (hRIG-I; Fig. S2B), followed by infection of RNA viruses, the tIFNB1 mRNA level was significantly up-regulated earlier than in the mocked cells (Fig. 2A). This observation suggested that loss of RIG-I in tree shrew had not suppressed, but only delayed the tIFNB1 mRNA expression, and overexpression of hRIG-I had the compensatory effect. Evidently, there is an alternative sensor that recognizes RNA viruses and induces an antiviral response in TSPRCs. It is well established that RIG-I mediates antiviral response to RNAs bearing 5′ triphosphate RNA (5′ ppp RNA) (25); we therefore transfected 5′ ppp dsRNA into HeLa cells and TSPRCs. Owing to the presence of RIG-I in HeLa cells, the 5′ ppp dsRNA quickly induced the IFNB1 mRNA expression and reached a high level at 6 h (Fig. 2B). In contrast, the 5′ ppp RNA had a delayed stimulation, with a slight effect at 6 h, significantly increased the tIFNB1 mRNA levels at 12 h in TSPRCs (Fig. 2B and Fig. S2C). Similar results were obtained when SeV viral RNA (vRNA) was transfected in these cells. The SeV vRNA induced IFNB1 mRNA to a high level in HeLa cells at 12 h but had a slow up-regulation effect in TSPRCs (Fig. 2C). However, when SeV vRNAs were treated with CIAP (to ablate the 5′ ppp ends), it had a weak effect on the IFNB1 mRNA levels in both cells (Fig. 2C). These results suggested that human and tree shrew cells are different in sensing SeV vRNA, and there is a functional substitute for the loss of RIG-I in sensing these RNAs in the Chinese tree shrew.
Fig. S2.
HEK293 cells and TSPRCs have a different pattern in response to the NDV infection. (A) Different patterns of IFNB1 mRNA expression in HEK293 and TSPRCs. The TSPRCs or HEK293 cells (1 × 105/well) were grown in 12-well plate overnight and were infected with NDV (Newcastle disease virus; MOI = 10) for the indicated time before the harvest for total RNA isolation. Real-time qPCR was used for measuring the IFNB1 mRNA level. (B) Overexpression of human RIG-I (hRIG-I) in TSPRCs. The TSPRCs (1 × 105/well) were grown in a 12-well plate overnight and were transfected with Flag-tagged hRIG-I expression vector (hRIG-I, 1 μg) or empty vector (Mock, 1 μg). Cells were harvested at 48 h after transfection, and cell lysates were analyzed by immunoblot using the anti-Flag antibody. Blot for β-actin was used as a control. (C) The 5′ ppp RNA up-regulates the IFNB1 mRNA expression in HeLa and TSPRCs with a different time-dependent pattern; 5′ ppp RNA (100 ng/mL) and 5′ ppp RNA treated with CIAP (100 ng/mL) were transfected into 1 × 105 cells for the indicated times, respectively. The IFNB1 mRNA expression levels in cells transfected with 5′ ppp RNA, 5′ ppp RNA treated with CIAP, and nontransfected cells (NCs) were measured by qRT-PCR. Bars represent mean ± SEM. All experiments were repeated for three times, with similar results.
Fig. 2.
A functional substitute for the lost RIG-I as the ligand of RNA viruses and 5′ triphosphate RNA (5′ ppp RNA) in TSPRCs. (A) Overexpression of human RIG-I (hRIG-I) can advance tIFNB1 mRNA expression in response to different viral infections in TSPRCs. Cells (1 × 105) were transfected with hRIG-I expression vector or empty vector (1 μg) for 24 h and then were infected by viruses for indicated times. The 5′ ppp RNA (B) and SeV vRNAs (C) up-regulate IFNB1 mRNA expression in HeLa and TSPRCs with a different time-dependent pattern; 5′ ppp RNA (100 ng/mL), 5′ ppp RNA control (100 ng/mL), SeV RNAs (100 ng/mL), and CIAP-treated SeV RNAs (100 ng/mL) were transfected into 1 × 105 cells for the indicated times, respectively. The IFNB1 mRNA expression was measured by qRT-PCR. Experiments were performed in duplicate. Data are representative of three independent experiments.
Tree Shrew Cells Sensed SeV Through tMDA5, with an Involvement of tLGP2.
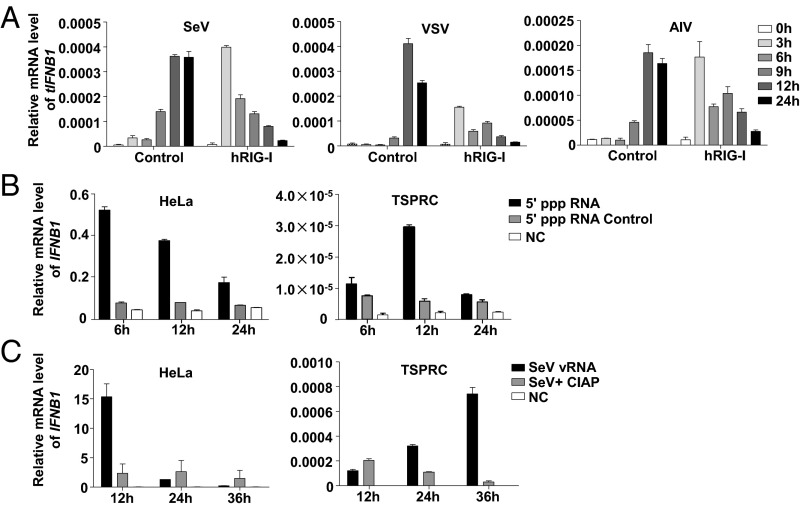
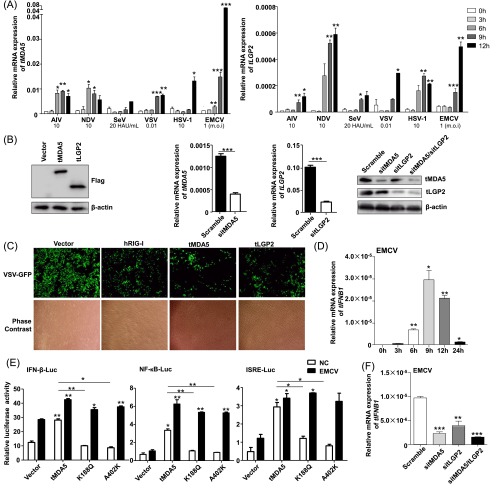
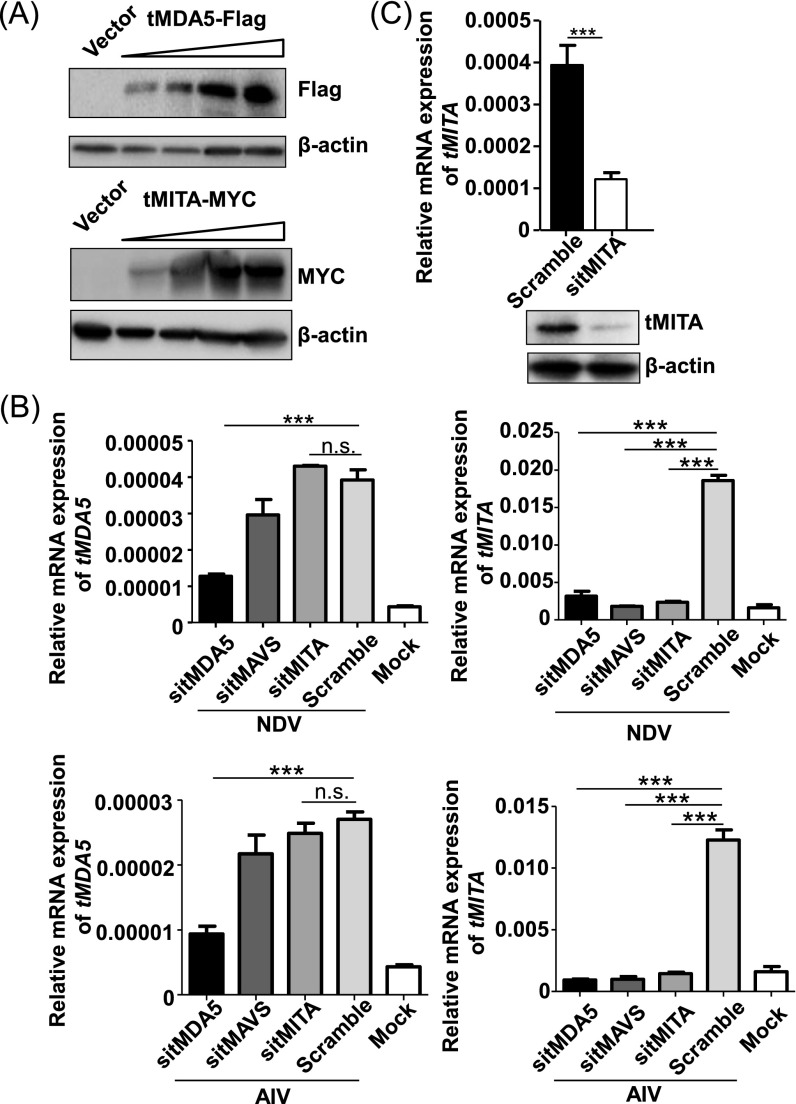
We next sought to identify the cytosolic recognition elements in TSPRCs, which could replace the RIG-I function as the ligand of virus RNA. We found that the two members of RLRs, tMDA5 and tLGP2, had a significantly increased mRNA expression after AIV, SeV, VSV, NDV, EMCV, or HSV-1 (herpes simplex virus-1) infection for 6 h or even later (Fig. S3A). Overexpression of tMDA5 and tLGP2 (Fig. S3B) significantly potentiated the virus-triggered activation of tIFN-β promoter luciferase (IFN-β-Luc) reporter activity (Fig. 3A), whereas knockdown of tMDA5 and tLGP2 (Fig. S3B) displayed the opposite effect (Fig. 3B). Moreover, as indicated by the diminished GFP expression, overexpression of tMDA5 and tLGP2 inhibited GFP-tagged VSV replication, with a comparable effect to that of hRIG-I overexpression (Fig. 3C and Fig. S3C). In addition, tIFNB1 mRNA expression increased in TSPRCs on EMCV infection (Fig. S3D). Overexpression of tMDA5 markedly activated the EMCV-induced IFN-β-Luc, NF-κB-Luc, and ISRE-Luc reporters (Fig. S3E). Knockdown of tMDA5 and/or tLGP2 significantly inhibited EMCV-induced tIFNB1 mRNA expression (Fig. S3F). These observations indicated that the function of tMDA5 and tLGP2 was homologous to that of the other mammals and had an obvious antiviral activity.
Fig. S3.
The function of tMDA5 and tLGP2 in response to RNA virus infections in TSPRCs. (A) Up-regulation of the tMDA5 and tLGP2 mRNA expression levels after challenging with different viruses. TSPRCs (1 × 105/well) were grown in a 12-well plate overnight and were harvested after infection with different viruses at the indicated times for total RNA isolation. The tMDA5 and tLGP2 mRNA expression levels (relative to β-actin) were quantified using qRT-PCR. Statistical difference was performed between the uninfected (0 h) and infected cells at the indicated time points. (B) Overexpression and knockdown of tMDA5 and tLGP2 in TSPRCs. The TSPRCs (1 × 105/well) were grown in a 12-well plate overnight and were transfected with Flag-tagged tMDA5 expression vector, Flag-tagged tLGP2 expression vector or siRNA (sitMDA5, sitLGP2, and sitMDA5/sitLGP2; 50 nM each), and were harvested at 36 h after transfection. Cell lysates were analyzed by immunoblot using the anti-Flag antibody. The endogenous tMDA5 and tLGP2 mRNA and protein expression levels were analyzed by using qRT-PCR and immunoblot (using anti-MDA5 antibody and anti-LGP2 antibody), respectively. (C) Overexpression of tMDA5 or tLGP2 inhibited VSV-GFP replication. The TSPRCs (1 × 105/well) were grown in a 12-well plate overnight and were transfected with the empty vector (pCMV-3Tag-8 vector/Vector; 1 μg) or indicated expression vector (hRIG-I, tMDA5, tLGP2; each 1 μg) for 12 h, followed by infection with VSV-GFP (MOI = 0.01) for 12 h. Cells were photographed under an Olympus microscopy (original magnification, ×10). (D) Up-regulation of the tIFNB1 mRNA expression in response to EMCV (MOI = 1) infection at the indicated times. (E) Effects of overexpression of tMDA5, tMDA5 K188Q, and tMDA5 A402K on the IFN-β-Luc, NF-κB-Luc, and ISRE-Luc reporters upon EMCV infection. The TSPRCs (1 × 104/well) were grown in a 24-well plate overnight and were transfected with the indicated expression vector or empty vector (400 ng), together with the respective luciferase vector (100 ng), TK (10 ng, as an inner control) for 36 h, followed by infection with EMCV (MOI = 1) for 12 h before the harvest for luciferase assay. Statistical difference was performed between cells transfected with vector and tMDA5 or between cells transfected with vector and tMDA5 mutant, unless otherwise marked. (F) Knockdown of tLGP2 (sitLGP2), tMDA5 (sitMDA5), and tLGP2/tMDA5 (sitMDA5/sitLGP2) caused an inhibition of EMCV-induced up-regulation of the tIFNB1 mRNA expression. Statistical difference was performed between the scramble cells and cells transfected with siRNA. Bars represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, Student t test. All experiments were repeated for three times, with similar results.
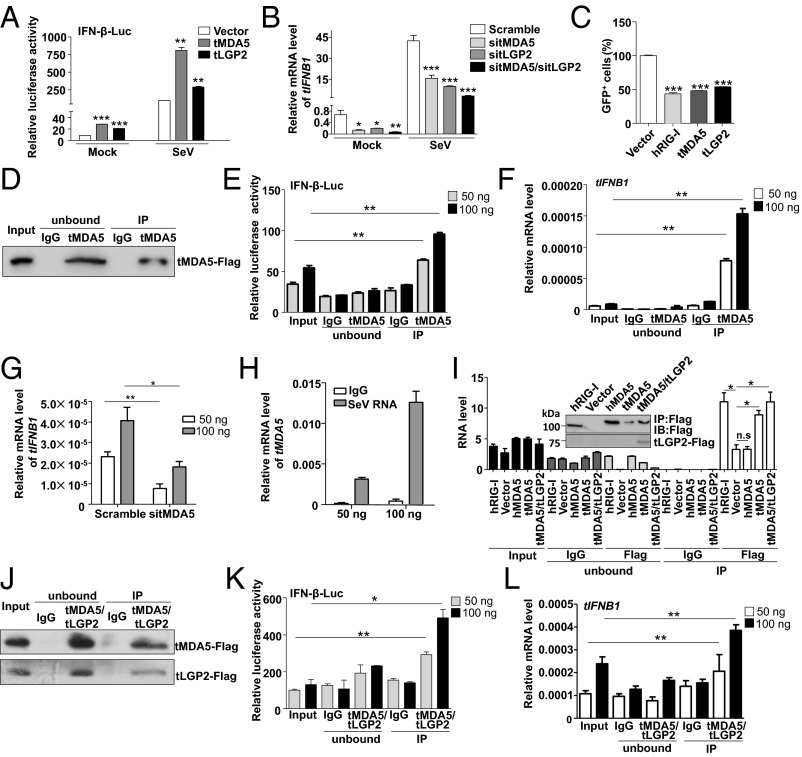
Fig. 3.
tMDA5 or tMDA5/tLGP2 can sense SeV. (A) tLGP2 or tMDA5 activates the IFN-β-Luc reporter. TSPRCs (1 × 104) were transfected with IFN-β-Luc (100 ng), TK (10 ng), and expression vector for tLGP2 or tMDA5 (400 ng) for 36 h, and then were infected with SeV (20 HAU/mL) for 12 h before the luciferase assay. (B) Knockdown of tLGP2, tMDA5, or tLGP2/tMDA5 inhibits virus-induced tIFNB1 mRNA expression. TSPRCs (1 × 105) were transfected with siRNA negative control (Scramble, 50 nM) or indicated siRNA (50 nM) for 24 h, followed by SeV infection for 12 h. (C) Overexpression of tLGP2 or tMDA5 inhibits VSV replication. TSPRCs (1 × 105) were transfected with equal amount of empty vector, hRIG-I, tLGP2, or tMDA5 expression vector (1 μg) for 12 h, followed by VSV-GFP (MOI = 0.01) infection for 12 h. Proportion of the GFP-positive cells was quantified by flow cytometry. (D–F) tMDA5 pull-down captures agonistic RNA from SeV-infected cells. About 1 × 108 TSPRCs were transfected with 30 µg FLAG-tagged tMDA5 expression vector and were cultured for 24 h and then infected with SeV for 16 h, followed by immunoprecipitation assays. Precipitation efficiency was verified by immunoblotting with an anti-Flag antibody (D). The RNAs from SeV-infected Flag-tMDA5-overexpressing TSPRCs (input), RNAs associated with tMDA5 or IgG immunoprecipitation (IP), or RNAs remaining after tMDA5 or IgG precipitations (unbound) were tested for the ability to stimulate the IFN-β-Luc activity in HEK293 cells (E) and to induce tIFNB1 mRNA in TSPRCs (F). (G) Effect of the tMDA5-associated SeV RNAs was dependent on tMDA5. TSPRCs (1 × 105) were transfected with the indicated siRNA (50 nM) for 24 h and then transfected with the indicated amount of SeV RNAs for 6 h for measuring tIFNB1 mRNA levels by qRT-PCR. (H) Up-regulation of tMDA5 mRNA level by tMDA5-associated immunoprecipitated SeV RNAs in a dose-dependent manner. TSPRCs (1 × 105) were transfected with the indicated amount of SeV RNAs for 6 h. (I) Quantification of viral RNA bound by the Flag-tagged proteins from SeV-infected TSPRCs. (Top) Immunoblots showing Flag-tagged hRIG-I, hMDA5, tMDA5, and tMDA5/tLGP2 in IP. (Bottom) SeV RNA level was measured by the strand-specific qRT-PCR in vector, hRIG-I, hMDA5, tMDA5, tMDA5-tLGP2, and IgG immunoprecipitates. (J–L) tLGP2 enhances the ability to sense SeV by tMDA5 in TSPRCs. The procedures were similar to D–F. Data are representative of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, Student t test. Bars represent mean ± SEM.
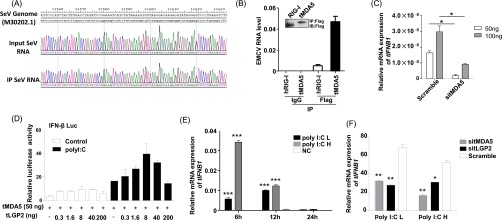
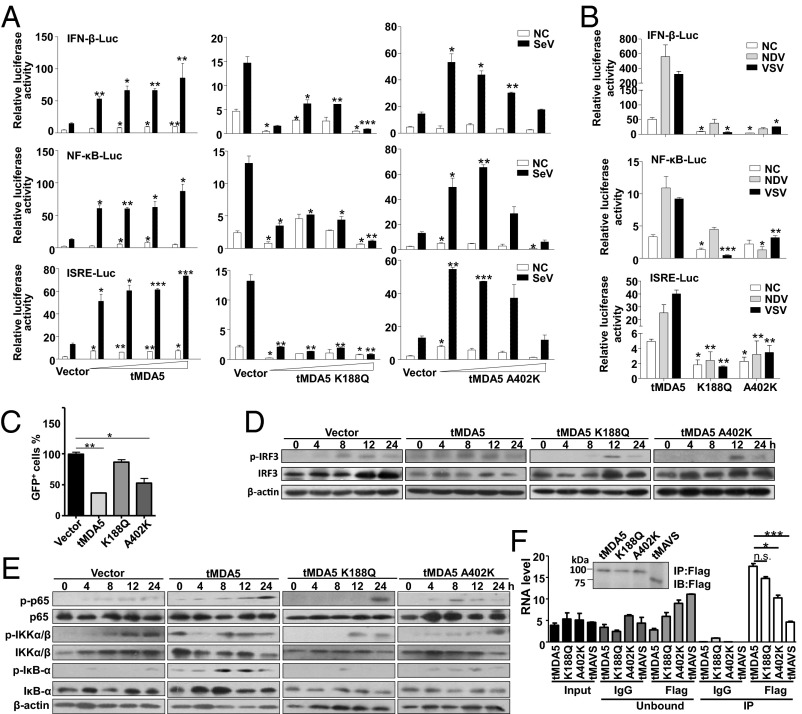
To confirm that tMDA5 was involved in the IFN-β response to SeV and acted as the RIG-I functional substitute, we used previously reported immunoprecipitation (IP) method (26, 27) to isolate tMDA5-associated RNA from SeV-infected cells. We first immunoprecipitated tMDA5 from SeV-infected TSPRCs transiently overexpressing a Flag-tagged tMDA5 protein (Fig. 3D), and then we extracted RNA from the precipitates and analyzed its stimulatory activity on the induction of IFN-β (Fig. 3 E and F). Notably, RNA associated with the tMDA5 precipitates (which contained SeV-derived vRNA; Fig. S4A), but not with the control (IgG) precipitates, stimulated the IFN-β-Luc reporter in HEK293 cells (Fig. 3E) and induced tIFNB1mRNA expression in TSPRCs (Fig. 3F). Moreover, knockdown of tMDA5 inhibited the immunoprecipitated SeV RNA-induced activation of the tIFNB1 mRNA expression in TSPRCs (Fig. 3G). Immunoprecipitated SeV RNA induced tMDA5 mRNA expression in a dose-dependent manner in TSPRCs (Fig. 3H). In human cells, RIG-I specifically bound the defective interfering (DI) particle during SeV infection (27). We used the strand-specific quantitative RT-PCR (qRT-PCR) with primers for SeV DI particle to validate the RNA associated with tMDA5 precipitates. We confirmed that tMDA5, but not human MDA5 (hMDA5), bound SeV RNA (Fig. 3I). tMDA5 had a weaker ability to bind SeV RNA than hRIG-I (Fig. 3I). In addition, we also found that tMDA5 had a higher binding affinity to the L region of EMCV compared with hRIG-I (Fig. S4B), and EMCV RNA was sufficient to trigger the MDA5-dependent IFN response (Fig. S4C). These results indicated that tMDA5 maintained its original function to recognize EMCV and had evolved an additional function for the loss of RIG-I.
Fig. S4.
RNA immunoprecipitation assay showed that the tMDA5- or tMDA5/tLGP2-associated RNAs are SeV RNAs. (A) Sequencing electrophoregrams showing the MDA5- or tMDA5/tLGP2-associated RNA as the SeV RNA. The tMDA5 or tMDA5/tLGP2 immunoprecipitates were extracted for RNA and were reversely transcribed to cDNA, followed by PCR amplification and sequencing. The sequencing results matched to the SeV genome fragment (GenBank accession no. M30202.1). Input Sev RNA: viral RNA isolated from whole cell lysate infected with SeV. IP SeV RNA: viral RNA isolated from the immunoprecipitated protein-RNA complexes. (B) Quantification of the amount of EMCV RNA in hRIG-I, tMDA5, and IgG immunoprecipitates by using the strand-specific RT-PCR. (Top) Immunoblot showing Flag-tagged hRIG-I and tMDA5 in IP. (Bottom) The EMCV RNA levels were calculated by comparison with a dilution curve of cDNA prepared from the EMCV-infected cells. (C) Up-regulation of the tIFNB1 mRNA expression by the tMDA5-associated EMCV RNAs was dependent on tMDA5. TSPRCs (1 × 105) were first transfected with sitMDA5 (50 nM) and siRNA control (Scramble, 50 nM) for 24 h and then transfected with the indicated amount EMCV RNAs for 9 h before the harvest. The relative tIFNB1 mRNA levels were measured by using qRT-PCR. (D) Effects of the overexpression of the tLGP2 and tMDA5 on the induction of the IFN-β-Luc reporter in TSPRCs. Cells (1 × 104 cells per well) were grown in 24-well plate overnight and were transfected with the IFN-β-Luc reporter vector (100 ng) and TK (10 ng, as an inner control). The tMDA5 expression vector was transfected at a constant quantity of 50 ng plasmid per well, whereas the amount of tLGP2 was titrated. After 24 h, cells were transfected with low-molecular-weight poly I:C (1 μg/mL) or without low-molecular-weight poly I:C (Control) for 12 h before the harvest for luciferase assay. (E) Both high-molecular-weight poly I:C (poly I:C H) and low molecular weight poly I:C (poly I:C L) up-regulated the tIFNB1 mRNA levels in TSPRCs. The TSPRCs (1 × 105) were transfected with poly I:C H (100 ng/mL) or poly I:C L (100 ng/mL) or without transfection (NC) and were harvested at the indicated time points after transfection. (F) Knockdown of tLGP2 (sitLGP2) or tMDA5 (sitMDA5) caused an inhibition of poly I:C-induced tIFNB1 mRNA expression. The TSPRCs (1 × 105/well) were grown in 12-well plate overnight and were transfected with the siRNA negative control (Scramble, 50 nM) or indicated siRNA (50 nM) for 36 h, followed by transfection with poly I:C H or poly I:C L (100 ng/mL) for 12 h, respectively, before the harvest. The relative tIFNB1 mRNA levels were measured using qRT-PCR. *P < 0.05, **P < 0.01, ***P < 0.001, relative to the NC or Scramble, Student t test. Bars represent mean ± SEM. All experiments were repeated for three times with similar results.
The LGP2 can assist MDA5-RNA interactions and leads to enhanced MDA5-mediated antiviral signaling (26, 28). Inclusion of tLGP2 significantly increased the tMDA5-dsRNA interaction at a lower concentration, but inhibited the tMDA5 signaling at a high level (Fig. S4D) (28). Transfection with low- or high-molecular-weight poly I:C up-regulated the tIFNB1 mRNA expression (Fig. S4E), whereas tMDA5 and tLGP2 knockdown abolished this induction effect (Fig. S4F). These results suggested that TSPRCs sensed poly I:C through tMDA5, with a modification effect from tLGP2. We cotransfected tLGP2 and tMDA5 to immunoprecipitate tMDA5/tLGP2-associated RNA and analyzed the stimulatory activity of the precipitated RNAs. Consistent with the above results, tMDA5/tLGP2-associated RNAs could induce the IFN-β-Luc activity and tIFNB1 mRNA expression (Fig. 3 J–L) and had a slightly higher binding affinity than tMDA5 alone (Fig. 3I), indicating that tLGP2 was able to synergize with tMDA5 to render cells to be more sensitive to SeV infection, finally leading to an enhanced tMDA5-mediated antiviral signaling.
tMDA5 Interacted with tMITA.
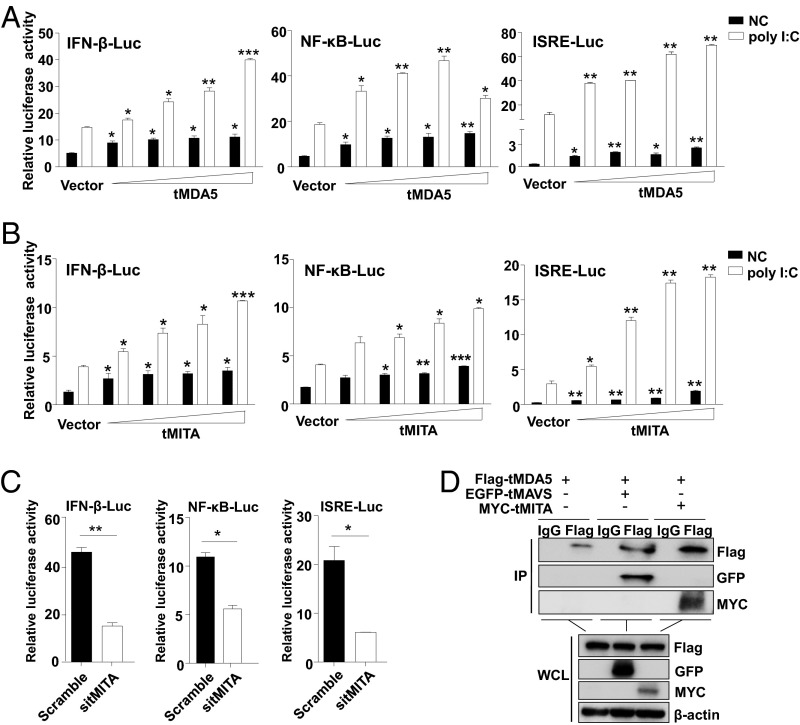
MITA preferentially modulated the RIG-I-, rather than the MDA5-signaling (21, 22). MITA was unable to mediate the signaling triggered by high-molecular-weight poly I:C (poly I:C H), which was known to be sensed by MDA5 (Fig. S4F) (23, 29). To examine whether tMDA5 could replace RIG-I to interact with tMITA, we overexpressed tMDA5 and tMITA in TSPRCs separately and investigated the activation effect of the IFN-β-Luc, NF-κB-Luc, and ISRE-Luc reporters in response to poly I:C. Both tMDA5 and tMITA could induce the IFN-β-Luc, NF-κB-Luc, and ISRE-Luc activities in a dose-dependent manner in response to poly I:C H (Fig. 4 A and B and Fig. S5A), and tMITA was a downstream mediator of the tMDA5 signaling (Fig. S5B). Knockdown of endogenous tMITA (by sitMITA; Fig. S5C) inhibited the activation of the IFN-β-Luc, NF-κB-Luc, and ISRE-Luc by poly I:C H (Fig. 4C). These results suggested that tMITA mediated the tMDA5-dependent signaling triggered by poly I:C H. Coimmunoprecipitation showed that tMDA5 could interact with tMITA in SeV-infected TSPRCs (Fig. 4D). Taken together, tMDA5 can bind to tMITA in the absence of RIG-I and mediate the corresponding signaling in tree shrew.
Fig. 4.
tMDA5 interacts with tMITA in TSPRCs. Overexpression tMDA5 (A) and tMITA (B) activate the IFN-β-Luc, NF-κB-Luc, and ISRE-Luc reporters in a dose-dependent manner. Cells (1 × 104) were transfected with the respective reporter vector (100 ng), TK (10 ng), and increased amount (0, 3.2, 16, 80, and 400 ng) of tMDA5 or tMITA expression vector (with empty vector to reach a total amount of 400 ng) for 36 h and then were transfected with poly I:C (1 μg/mL) for 12 h. (C) Knockdown of tMITA causes an inhibition of poly I:C-induced IFN-β-Luc, NF-κB-Luc, and ISRE-Luc activities. (D) tMDA5 immunoprecipitates with tMITA. Cells (1 × 107) were cotransfected with MYC-tMITA (10 μg) and Flag-tMDA5 (10 μg) expression vectors for 36 h and then were infected with SeV for 16 h, followed by immunoprecipitation (IP) with anti-Flag, anti-MYC, or mouse IgG (control). The EGFP-tagged tMAVS was used as a positive control. Data are representative of three independent experiments.*P < 0.05, **P < 0.01, ***P < 0.001, Student t test. Bars represent mean ± SEM.
Fig. S5.
Knockdown of tMDA5 affects the mRNA expression of tMITA in TSPRCs on RNA virus infection. (A) Overexpression of Flag-tagged tMDA5 and MYC-tagged tMITA in TSPRCs. Cells (1 × 104 cells per well) were grown in 24-well plate overnight and were transfected with different amount of expression vectors (3.2, 16, 80, and 400 ng; with empty vector to reach a total amount of 400 ng). Cells were harvested at 48 h after transfection. Cell lysates were analyzed by immunoblot using the indicated anti-Flag antibody or anti-MYC antibody, respectively. The blot refers to the condition described in Fig. 4 A and B. (B) Effects of knockdown of endogenous tMDA5 (sitMDA5), tMAVS (sitMAVS), or tMITA (sitMITA) on mRNA expression levels of tMDA5 and tMITA in TSPRCs in response to RNA virus infections. TSPRCs (1 × 105 cells per well) were grown in 12-well plate overnight and were transfected with the indicated siRNA (50 nM) for 36 h, followed by infection with NDV (MOI = 10) or AIV (MOI = 1) for 12 h before the harvest. Knockdown of tMAVS by sitMAVS was used as a positive control. Knockdown of tMDA5 prevented the virus-induced tMITA mRNA expression, whereas knockdown of tMITA did not affect the tMDA5 mRNA expression in response to NDV or AIV infection, suggesting that tMITA was a downstream mediator of the tMDA5 signaling. (C) Knockdown of endogenous tMITA in TSPRCs. Cells (1 × 104 cells per well) were grown in 24-well plate overnight and were transfected with siRNA (sitMITA or Scramble, each 50 nM) for 48 h before the harvest. The endogenous tMITA mRNA levels were analyzed by using qRT-PCR. *P < 0.05, **P < 0.01, ***P < 0.001, Student t test. Bars represent mean ± SEM. All experiments were repeated for three times, with similar results.
MDA5 and LGP2 Underwent Positive Selection in the Tree Shrew Lineage.
To understand the evolutionary dynamics and selective pressure on the RLR genes in the tree shrew due to the loss of RIG-I, we used the branch models and the branch-site models based on the maximum-likelihood method implemented in the phylogenetic analysis by maximum likelihood (PAML) package (30) to calculate the average nonsynonymous substitution/synonymous substitution rate (dN/dS, also known as ω) for MDA5 and LGP2. We first tested with the branch models, which were based on the model M0 with same ω for all branches and the model M2 with different ω on selected branches, and then we compared M0 and M2 by the likelihood ratio test (31). As shown in Table S1, tMDA5 and tLGP2 underwent a positive selection (tMDA5, P = 0.032; tLGP2, P = 0.032). The positive selections were further evaluated by using the branch-site models (32) implemented in PAML (30), which is powerful for detecting episodic positive selection and for generating biological hypotheses for mutation and functional analyses (33). Similar positive selection signature was detected in tMDA5 (P = 0.007), whereas no significant sites were found for tLGP2 (P > 0.05) (Table S1). In detail, the branch-site models analysis detected two positively selected residues (Lys188 and Ala402) in the tMDA5 (Table S1).
Table S1.
Analysis of positive selection for the Chinese tree shrew MDA5 and LGP2 genes
| Foreground | lnL (null) | np1 | lnL (alternative) | np2 | 2ΔlnL | P value | PSSs* (BEB analysis) | Parameters |
| Branch analysis | ||||||||
| Tree shrew MDA5 (M0 vs. M2) | −9,394.433345 | 12 | −9,392.138604 | 13 | 4.589482 | 0.032 | / | M0: All branches have the same ω0 = 0.22283 |
| M2: The tree shrew branch has ω2= 0.30320, other branches have ω1= 0.20895 | ||||||||
| Tree shrew LGP2 (M0 vs. M2) | −6,660.077474 | 12 | −6,657.787937 | 13 | 4.579074 | 0.032 | / | M0: All branches have the same ω0 = 0.17109 |
| M2: The tree shrew branch has ω2= 0.24227, other branches have ω1 = 0.15777 | ||||||||
| Branch-site analysis | ||||||||
| Tree shrew MDA5 | −9,433.609553 | 14 | −9,429.916706 | 15 | 7.385694 | 0.007 | 188 Q 0.732, 402 K 0.800 | p0 = 0.77887 p1 = 0.21476 p2a = 0.00499 p2b = 0.00138 ω0 = 0.06695 ω1 = 1.00000 ω2= 957.32532 |
| Tree shrew LGP2 | −6,626.184812 | 14 | −6,626.184812 | 15 | 0 | 1 | 95 R 0.602, 318 T 0.677, 513 V 0.616, 683 L 0.793 | p0 = 0.70426 p1 = 0.19775 p2a = 0.07651 p2b = 0.02148 ω0= 0.05436 ω1 = 1.00000 ω2 = 1.00000 |
BEB analysis, Bayes Empirical Bayes analysis (44); lnL, log-likelihood value; np, number of parameters; 2ΔlnL, twice the difference of ln(likelihood) values (2ΔlnL) between the two models compared; PSSs, positively selected sites; A P value < 0.05 is marked in bold.
The amino acid positions are called based on the tree shrew MDA5.
The Positively Selected Sites in tMDA5 Endowed the Substitute Function for the Lost RIG-I.
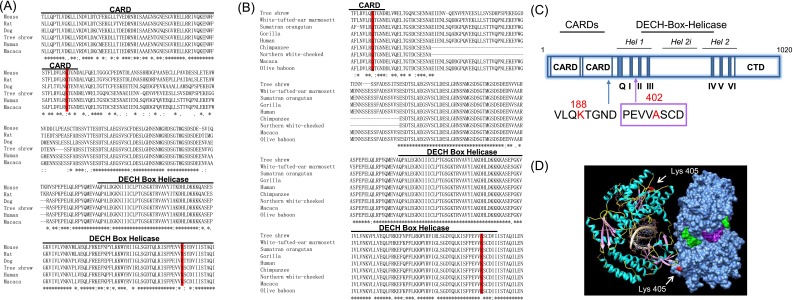
To functionally characterize the potential selection effect on tMDA5, we focused on the two positively selected sites (PSSs) Lys188 and Ala402 in tMDA5, which were highly conserved in the primates (Table S1 and Fig. S6 A and B). Structural modeling showed that the equivalent positions of the PSSs in tMDA5 are located in critical domains: the Lys188 is located in the second CARD near the helicase domain, which is essential for coupling to the downstream signaling adaptors (34); the Ala402 is located in the DECH box helicase domain (35) (Fig. S6 A and C). We located the PSSs based on the crystal structures of hMDA5 (Protein Data Bank ID code 4GL2) and found that Ala402 was adjacent to regions involved in dsRNA binding (35) (Fig. S6D). Alteration of basic amino acid lysine to neutral alanine at this position might have changed the affinity between dsRNA and tMDA5. Evidently, the in silico prediction analysis demonstrated that these PSSs were probably crucial for tMDA5-mediated antiviral signaling pathway.
Fig. S6.
Evolutionary conservation of the positively selected sites in the tree shrew MDA5 and its impact on the predicted protein structure. (A) Sequence alignment of the second CARD domain of the MDA5 and DECH Box Helicase domain in six mammalian species [human, rat, mouse, dog, Chinese tree shrew, macaque; sequence data were retrieved from Ensemble (asia.ensembl.org/index.html)]. PSSs are marked in red. (B) Sequence alignment of the second CARD domain of the MDA5 and the DECH Box Helicase domain in eight primates (white-tufted-ear marmoset, Sumatran orangutan, gorilla, human, chimpanzee, northern white-cheeked gibbon, macaque, olive baboon) and the Chinese tree shrew. (C) Diagram illustrating domain structure of the tree shrew MDA5. The tMDA5 protein is composed of three critical domains: (1) tandem CARDs at its N-terminal, which are essential for interacting with MAVS to mediate the downstream signaling; (2) a central DECH-box helicase domain that encompasses conserved helicase subdomains, Hel1 (surrounding helicase motifs Q, I, II, and III), Hel2 (surrounding helicase motifs IV, V, and VI), and the helicase insert domain Hel2i; and (3) a C-terminal domain (CTD) for auto-regulation and RNA terminus recognition. (D) Structure of human MDA5 binding to dsRNA. The positively selected site (Ala-402; shown in red) in tree shrew was equivalent to Lys-405 in hMDA5.
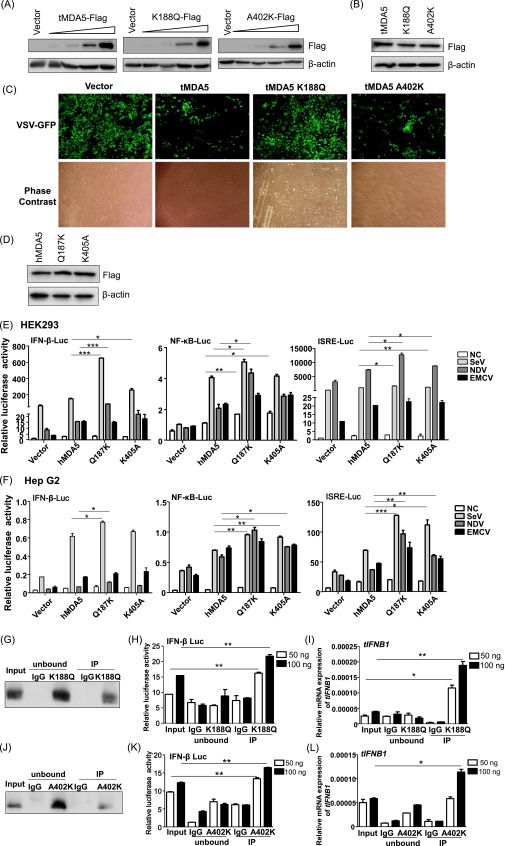
To examine the potential function of the two PSSs in tMDA5, we swapped the Lys188 and Ala402 in tMDA5 back to the evolutionarily conserved and primitive glutamine (K188Q) and lysine (A402K), respectively (Fig. S6). tMDA5 induced the IFN-β-Luc, NF-κB-Luc, and ISRE-Luc activities in a dose-dependent manner by Mock or SeV induction, whereas tMDA5 K188Q robustly inhibited these luciferase activities. tMDA5 A402K only activated these reporters at a low concentration but decreased their activities at a high concentration on SeV infection (Fig. 5A and Fig. S7A). We speculated that this pattern was caused by different regulation effects of heterodimer of endogenous tMDA5 and exogenous tMDA5 A402K and homodimer of exogenous tMDA5 A402K in TSPRCs. Similarly, tMDA5 K188Q and tMDA5 A402K, but not tMDA5, had no apparent ability to activate the luciferase reporters in response to NDV and VSV infections (Fig. 5B and Fig. S7B). tMDA5 and tMDA5 A402K, but not tMDA5 K188Q, inhibited VSV replication, albeit with different capabilities (Fig. 5C and Fig. S7C). To further confirm the critical role of the PSSs of tMDA5, we performed a gain-of-function analysis by introducing mutations at the equivalent positions in hMDA5. We found that hMDA5 Q187K and K405A significantly potentiated the IFN-β-Luc, NF-κB-Luc, and ISRE-Luc activities induced by SeV and NDV in HEK293 and HepG2 cells, but no effect was observed for EMCV infection (Fig. S7 D–F). Thus, the role of hMDA5 mutants was consistent with that of tMDA5 (Fig. 5 A and B and Fig. S3E), suggesting that the two PSSs in MDA5 endowed tMDA5 a substitute function for the lost RIG-I. Note that both hMDA5 mutants have not been found in the general human populations (>60,000 individuals; SI Materials and Methods), suggesting both sites were extremely conserved.
Fig. 5.
Antiviral responses in tree shrew cells overexpressing tMDA5 and its mutants. (A) Different stimulatory effects of tMDA5, tMDA5 K188Q, and tMDA5 A402K on the activation of the IFN-β-Luc, NF-κB-Luc, and ISRE-Luc reporters in a dose-dependent manner. The procedures are same to Fig. 4A, except for SeV infection for 12 h. (B) Effects of overexpression of tMDA5, tMDA5 K188Q, and tMDA5 A402K on IFN-β-Luc, NF-κB-Luc, and ISRE-Luc reporters on NDV and VSV-GFP infections. Cells were transfected as A, except with 400 ng expression vector and different virus infections (NDV, MOI = 10; VSV-GFP, MOI = 0.01). (C) Different inhibition effects of tMDA5 and its mutants on VSV-GFP replication. (D and E) Effects of tMDA5 and its mutants on activation of phospho-IRF3, phospho-p65, phospho-IKKα/β, and phospho-IκBα. (F) Quantification of viral RNA bound by the Flag-tagged tMDA5 and its mutants from SeV-infected TSPRCs. The procedure and labels are the same as Fig. 3I. Data are representative of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, Student t test. Bars represent mean ± SEM.
Fig. S7.
Antiviral activities of MDA5 and its mutants in TSPRCs and human cells. (A and B) Overexpression of tMDA5 and its mutants, tMDA5 K188Q and tMDA5 A402K. TSPRCs (1 × 104 cells per well) were grown in a 24-well plate overnight and were transfected with different amounts of expression vector (section A: 3.2, 16, 80, and 400 ng/well; section B: 400 ng/well) or empty vector (400 ng/well) for 36 h. Cell lysates were analyzed by immunoblot using the anti-Flag antibody. Sections A and B refer to the conditions described in Fig. 5 A and B, respectively. (C) Overexpression of tMDA5 and its mutants had different abilities to inhibit vesicular stomatitis virus (VSV) replication. TSPRCs (1 × 105 cells per well) were grown in a 12-well plate overnight and were transfected with the empty vector (pCMV-3Tag-8 vector, 1 μg) or indicated expression vector (1 μg) for 12 h, followed by infection with VSV-GFP (MOI = 0.01) for 12 h. Cells were photographed under an Olympus microscopy (original magnification, ×10). Data are representative of three independent experiments. (D) Overexpression of Flag-tagged hMDA5, hMDA5 Q187K, and hMDA5 K405A in the HEK293 cells. Cells (1 × 105 cells per well) were grown in 12-well plate overnight and were transfected for 48 h before the harvest. Cell lysates were analyzed by immunoblot using the anti-FLAG antibody. (E and F) Overexpression of hMDA5, hMDA5 Q187K, and hMDA5 K405A on the IFN-β-Luc, NF-κB-Luc, and ISRE-Luc reporter activities upon infection with different RNA viruses in HEK293 cells (E) and Hep G2 cells (F). Cells (1 × 104 cells per well) were grown in a 24-well plate overnight and were transfected with indicated expression vector and luciferase reporter for 24 h, followed by infection with SeV (20 HAU/mL), NDV (MOI = 10) or EMCV (MOI = 1) for 12 h before the harvest for luciferase assay. (G–I) tMDA5 K188Q RNA-IP assay. TSPRCs (1 × 108 cells) were grown in a 150-mm culture dish overnight and were transfected with Flag-tagged mutant tMDA5 K188Q overexpression vector (30 μg) for 24 h, and then cells were infected with SeV (20 HAU/mL) for 16 h. The subsequent procedure was similar to that of Fig. 3 D–F. Precipitation was verified by immunoblot with the anti-Flag antibody (G). RNAs from the SeV-infected TSPRCs overexpressing the Flag-tagged tMDA5 K188Q (Input), RNAs associated with tMDA5 K188Q or IgG (control) immunoprecipitates (IP), or RNAs remaining after tMDA5 K188Q or IgG immunoprecipitations (unbound) were tested for the abilities to stimulate the IFN-β-Luc in HEK293 cells (H) or to induce tIFNB1 mRNA (normalized to the β-actin) in TSPRCs (I). Same assays were performed for mutant tMDA5 K402A (J–L). *P < 0.05, **P < 0.01, ***P < 0.001, Student t test. Bars represent mean ± SEM. All experiments were repeated for three times, with similar results.
We further assessed the antiviral activity of tMDA5 and its mutants by analyzing phosphorylation of the transcription factor IRF3 and NF-κB signaling. Infection with SeV induced phosphor-IRF3 in TSPRCs overexpressing tMDA5 and it mutants, but tMDA5 K188Q and tMDA5 A402K activated the phosphor-IRF3 at a later time point (12 h) (Fig. 5D). Concordantly, tMDA5 K188Q and tMDA5 A402K affected the ability of SeV to induce phospho-p65, phospho-IKKα/β, and phospho-IκBα, with a different pattern compared with tMDA5 (Fig. 5E). The RNA IP assay showed that tMDA5 A402K had a weaker ability to sense SeV RNA than tMDA5 and tMDA5 K188Q (Fig. 5F and Fig. S7 G–L). These results were consistent with the structure prediction that the PSSs K188 and A402 were located in different functional domains (Fig. S6) and affected the SeV-induced IRF3 and NF-κB activation.
Discussion
The RLR proteins play a key role in the innate immune system response against viral infection by recognizing viral RNA molecules (36, 37). RLRs are highly conserved during vertebrate evolution (13, 14), and loss of RIG-I in mammalian species is extremely rare (20). The loss of any RLR member might be expected to have a great impact on immune response. In this study, we characterized the consequences resulting from the loss of RIG-I in the tree shrew and provided direct functional evidence for the diversification of the RLR members in this species due to the inability to produce RIG-I. Accompanying this change, we observed a positive selection signal on tMDA5 and tLGP2. We found that tMDA5 alone or tMDA5/tLGP2 could replace RIG-I in sensing RNA viruses and trigger IFN production. This replacement might be enhanced by the interaction of tMDA5 with tMITA, which was proved to specifically interact with RIG-I to cascade the antiviral signaling (22). The functional divergence between tMDA5 and its mutants indicated that the compensatory effect of tMDA5 might be driven by natural selection. Loss of RIG-I has resulted in a functional replacement by MDA5 or MDA5/LGP2 as a cytosolic RNA sensor to trigger IFN production.
Although the available data suggested that loss of RIG-I has led to MDA5 functional alteration, most likely via the PSSs in tMDA5, it raised an interesting question: the time order of RIG-I loss and the positive selection on MDA5—which one is earlier in the tree shrew? A straightforward explanation is that tMDA5 first mutated and evolved by positive selection to gain a new function to achieve the ability to sense the viruses that were initially recognized by RIG-I, and then the redundant RIG-I gradually eroded in the genome. This assumption would be compatible with the pattern for overlapped recognition of same viruses by both MDA5 and RIG-I (38). The RIG-I is essential for preferentially recognizes RNAs bearing 5′ ppp ends in vertebrates, which serve to define a nonself RNA PAMP (25, 36, 39). It was unlikely that tree shrew accidentally lost RIG-I, because the sudden loss of RIG-I would render this species be more susceptible to virus infections (11, 18, 40). Moreover, RLRs and their downstream signaling molecules could be found in the earliest animals, suggesting for an ancient origin of the RLRs (15). However, it remains a challenge to attribute the selection patterns of RLRs to virus-mediated natural selection in the tree shrew.
Besides MDA5, LGP2 acts as a regulator of RLR-mediated antiviral signaling and was reported to play apparently conflicting roles in different studies (10, 41). In contrast to negative regulation in the RIG-I-mediated signaling pathway (4, 42, 43), tree shrew cells lacking tLGP2 exhibited a decreased IFNB1 mRNA expression in response to RNA virus infections (Fig. 3B). Moreover, tLGP2 could synergize with tMDA5 to sense SeV infection (Fig. 3I). This pattern is consistent with the observation that silencing endogenous chLGP2 reduced chIFN-β mRNA expression induced by AIV, suggested that chLGP2 had a positive role in antiviral signaling (19). Taken both observations in the tree shrew and chicken together, we speculated that LGP2 had a positive role in regulating the RLR signaling pathway upon the RIG-I loss, and this role was endowed by the evolution. One unresolved question arising from this study is that whether other immune genes would undergo similar effect upon the RIG-I loss, as we would anticipate a cascade event for the loss of this important factor.
In short, we uncovered a previously unknown evolutionary signal in response to RIG-I loss in the tree shrew. The loss of RIG-I was accompanied by a functional substitute with MDA5 involving LGP2, which underwent positive selection. Our study provides an example that will assist our understanding of the functional evolution and conservation of the innate immune system in vertebrates.
Materials and Methods
SI Materials and Methods and Table S2 contains complete related information in this study. All of the experimental procedures were performed according to the guidelines approved by the Institutional Animal Care and Use Committee of Kunming Institute of Zoology.
Table S2.
Primers and vectors used in this study
| Primer | Sequence (5′-3′) | Restriction endonuclease | Application and vector |
| tLGP2-qF | GTCTCCCAAGAACCTCTG | Analytical real-time qRT-PCR | |
| tLGP2-qR | GGACCACTTTTTGGCTTG | ||
| tMDA5-qF | TTACAGGGCTCAACCATC | Analytical qRT-PCR | |
| tMDA5-qR | CATGCTTGACCACATTTG | ||
| tLGP2-5′RACE | GCCGTGTCCTCCGCAGTTTGTGCTCTAGGT | 5′ RACE | |
| tLGP2-5′RACE | GTTGTAGGTGGTGTCCTTGTGCGTGTGGTG | 5′ RACE nested | |
| tLGP2-3′RACE | GGGCCAGTCAGAGCGTGTACTCGTTTGT | 3′ RACE | |
| tLGP2-3′RACE | CTGCGGCGGGAGCTGACCAATGAG | 3′ RACE nested | |
| tMDA5-5′RACE | GCACAGGCTCCACCTGGATATATGT | 5′ RACE | |
| tMDA5-3′RACE | CAAGCATGGGGAACGATGATGGTGC | 3′ RACE | |
| tLGP2-BamH-F | CGGGATCCATGGAGCTGCGGCCCTAC | BamHI | PCR for constructing tLGP2-3Tag-8 using pCMV-3Tag-8 vector |
| tLGP2-Xho-R | CCGCTCGAGGTCCCCGGAGAGGTTGG | XhoI | |
| tMDA5-BamH-F | CGGGATCCATGTCGAATGGGCATTCCT | BamHI | PCR for constructing tMDA5-3Tag-8 using pCMV-3Tag-8 vector |
| tMDA5-Xho-R | CCGCTCGAGATCCTCATCACTAAACAAAC | XhoI | |
| tMITA-BspE-F | CGGTCCGGAATGCCCCACTCCAGCCTGC | BspEI | PCR for constructing tMITA-FL-myc using pCS-myc-N vector |
| tMITA-Xho-R2 | CCGCTCGAGTCAGAAGACATCCGTGCGG | XhoI | |
| tMDA5-K188Q-F | TTGAGGGTTCTTCAACAAACAGGAAACGAT | PCR for constructing mutant tMDA5 K188Q using pCMV-3Tag-8 vector | |
| tMDA5-K188Q-R | ATCGTTTCCTGTTTGTTGAAGAACCCTCAA | ||
| tMDA5-A402K-F | GAAGTTGTTAAATCCTGTGATGTTATCATC | PCR for constructing mutant tMDA5 A402K using pCMV-3Tag-8 vector | |
| tMDA5-A402K-R | GATGATAACATCACAGGATTTAACAACTTC | ||
| hMDA5-Q187K-F | ATGTTCTTCGTAAAACAGGAAACAATGAAC | PCR for constructing mutant hMDA5 Q187K based in hMDA5 expression vector | |
| hMDA5-Q187K-R | GTTCATTGTTTCCTGTTTTACGAAGAACAT | ||
| hMDA5-K405A-F | GAAGTTGTCGCGTCCTGTGATATTATTATC | PCR for constructing mutant hMDA5 K405A based in hMDA5 expression vector | |
| hMDA5-K405A-R | GATAATAATATCACAGGACGCGACAACTTC | ||
| tIFN-β-pro.-Kpn-F1 | GGGGTACCTTTGCTTTCCTTTGCTTTG | KpnI | PCR for constructing pGL3-tIFN-β-promoter using pGL3-Basic vector |
| tIFN-β-pro.-Nhe-R | GGAATTCGCTAGCCCTTCCTCCATGGGCATGG | NheI | |
| 6964-fwd1 | CCCCAGCYRCCGCTAGTTGCA | PCR for amplify the tRIG-I genomic region which contains the start codon ATG | |
| 8733-low1 | AAGGCTCCTCAAACTCTG | ||
| RIG-I F | CAGACAAGGAGAACTGGCC | PCR amplification to capture the RIG-I gene using human, flying lemur and tree shrew cDNAs as the template | |
| RIG-I R2 | CTGGAATCTCAAATGTCTTG |
Restriction endonuclease sites introduced by PCR are underlined.
SI Materials and Methods
Reagent, Cell Lines, Viruses, and Experimental Animals.
Reagent.
High-molecular-weight poly(I:C) (HMW, 1.5–8 kb, Catalog # tlrl-piclv), low-molecular-weight poly(I:C) (LMW, 0.2–1 kb, Catalog # tlrl-picwlv), 5′ ppp RNA (Catalog # tlrl-3prna), and 5′ ppp RNA control (Catalog # tlrl-3prnac) were purchased from InvivoGen. The following antibodies were used in this study: mouse monoclonal anti-Flag (Abmart; M20008), mouse anti-c-Myc (9E11) (Life Technologies; MA1-16637), mouse monoclonal anti-GFP (EnoGene; E12-152), rabbit monoclonal anti-IRF3 (Cell Signal Technology; 4302), rabbit monoclonal anti-phospho-IRF3 (Ser396) (Cell Signal Technology; 4947), rabbit polyclonal anti-MDA5 (Merck Millipore; ABF210), rabbit polyclonal anti-LGP2 (IBL; 29030), rabbit polyclonal anti-MITA (LifeSpan BioSciences; LS-B7237), and mouse monoclonal anti β-actin (EnoGene; E1C602-2). Probing of the endogenous phospho-NF-κB, phospho-p65 (Ser536), p65, phospho-IKKα/β (Ser176/180), IKKα/β, phospho-IκBα (Ser32), and IκBα was done using the NF-κB pathway sample kit (Cell Signal Technology; 9936). Rabbit monoclonal anti-RIG-I (D14G6) (Cell Signal Technology; 3743), rabbit polyclonal anti-RIG-I (H-300) (Santa Cruz Biotechology; sc-98911), peroxidase-conjugated anti-mouse antibody (KPL; 474-1806), and peroxidase-conjugated anti-rabbit antibody (KPL; 074-1506) were used in this study.
Cells, viruses, and virus infections.
HEK293, HeLa, and Hep G2 cells were introduced from Kunming Cell Bank, Kunming Institute of Zoology (KIZ), Chinese Academy of Sciences (CAS). In brief, cells were grown in DMEM (Gibco-BRL; 11965-092) supplemented with 10% (vol/vol) FBS (Gibco-BRL; 10099-141) and 1× penicillin/streptomycin (Gibco-BRL; 10378016) at 37 °C in 5% CO2. Primary renal cells were established from the Chinese tree shrews with age range from 1 to 4 months as follows: tree shrew was killed, and a pair of kidneys was dissected. The intact kidney was minced into small pieces (about 1 mm3) in cold PBS, and the pieces were transferred into a 50-mL sterile plastic tube containing a 1 mg/mL DNase (Sigma; AMPD1-1KT) and 5 mg/mL collagenase type IV (Invitrogen; 17104019) solution for 30 min in a 37 °C water bath. After digestion, the solution was filtered through a 200-mesh sieve to remove tissue pieces. The TSPRCs were suspended and washed three times with cold PBS. Finally, cells were resuspended and cultured at a density of 2 × 106 cells/mL in high-glucose DMEM supplemented with 10% FBS and 1× penicillin/streptomycin at 37 °C in 5% CO2 until confluent. Cells were then passaged for the indicated experiments.
A cell line of Malayan flying lemur (Galeopterus variegatus) was retrieved from the Kunming Cell Bank, KIZ, CAS, to extract total RNA for the detection of RIG-I transcript in this species.
Sendai virus (SeV) and Vesicular stomatitis virus (tagged by GFP; VSV-GFP) were kind gifts from Xinwen Chen, Wuhan Institute of Virology, CAS, Yunnan, China. Herpes simplex virus-1 (HSV-1) was introduced from Jumin Zhou’s laboratory at KIZ, Yunnan, China. Avian influenza virus (AIV) and Newcastle disease virus (NDV) were obtained from the China Institute of Veterinary Drug Control, Beijing. Encephalomyocarditis virus (EMCV) was a kind gift from Hanchun Yang, China Agricultural University, Beijing. SeV, AIV, and NDV were propagated by injecting 10-d-old specific pathogen-free embryonated chicken egg with stocks of virus that were diluted in PBS. The 50% tissue culture infective dose (TCID50) of VSV was determined by green fluorescence, and the titers of HSV-1, AIV, NDV, and EMCV were determined by cytopathic effect. The titers were calculated by the Reed-Muench method. The titer of SeV was determined by hemagglutination test on chicken erythrocytes.
For virus infections, TSPRCs were incubated with NDV [multiplicity of infection (MOI) = 10], AIV (MOI = 1), VSV-GFP (MOI = 0.01), HSV-1 (MOI = 10), SeV [20 hemagglutinating units (HAU)/mL], or EMCV (MOI = 1), respectively, for 1 h in DMEM without FBS, and then cells were rinsed and cultured in fresh medium containing 1% FBS for the indicated times before the harvest.
Experimental animals.
Chinese tree shrews (Tupaia balangeri chinensis) were purchased from the experimental animal core facility of the Kunming Institute of Zoology, CAS. After being lethally anesthetized by diethyl ether, tissue samples were taken from the kidney, heart, liver, spleen, lung, intestine, pancreas, and brain. The tissue samples were rapidly dissected and frozen in liquid nitrogen. Monkey and mouse brain tissues and human paracancerous normal tissues were preserved in our laboratory. Genomic DNA samples of 52 Chinese tree shrews collected from different parts of Yunnan Province [including Kunming (Tupaia balangeri chinensis), Xishuangbanna (T. b. yunalis), Lijiang, and southern and northern sections of the Gaoligong Mountain (T. b. gaoligongensis)] were provided by Xuelong Jiang, Kunming Institute of Zoology, CAS, Yunnan, China.
All of the experimental procedures were performed according to the guidelines approved by the Institutional Animal Care and Use Committee of the Kunming Institute of Zoology.
RNA Expression, Transfection, Western Blot, Immunoprecipitation, and Luciferase Reporter Assay.
Total RNA extraction and qRT-PCR.
Total RNA was extracted from primary renal cells and different tissues of the Chinese tree shrew by using the RNAsimple Total RNA Kit (TIANGEN; DP419) according to the manufacturer’s instructions. The A260/A280 ratio of total RNA was measured on a NanoDrop biophotometer (Thermo Fisher Scientific), and only these RNA samples with a value of 1.8–2.0 were used for subsequent reverse transcription. We also evaluated the quality and integrity of RNA samples based on the 28S and 18S rRNA bands on a 1% agarose gel. Total RNA (1 μg) was used to synthesize cDNA by using the oligo-dT18 primer and M-MLV reverse transcriptase (Promega; M1701). qRT-PCR was performed using the SYBR green Premix Ex Taq II (TaKaRa; RR820L) supplemented with gene-specific primers (Table S2) on a MyIQ2 Two-Color Real-Time PCR Detection system (Bio-Rad Laboratories). In brief, a volume of 20 μL containing 0.4 μM of each forward and reverse primer, 1 μL cDNA product, and 10 μL 2× SYBR green Premix Ex Taq II were used for the qRT-PCR reaction. The tree shrew housekeeping gene β-actin was used as the reference gene for normalization. The cycling condition consisted of an initial denaturation cycle for 3 min at 95 °C, 35 cycles of 30 s at 94 °C, 40 s at 55 °C, and a final extension step at 72 °C for 15 s. To verify no nonspecific amplification, following the completion of qRT-PCR, melting curve analysis was performed. The melting protocol consisted of heating from 55 °C to 95 °C at a rate of 0.5 °C per step, and each step was held for 1 s for data acquisition. Standard curves were generated using 10−3–10−10 dilution series of PCR product for each of the indicated genes and β-actin gene, respectively.
Plasmid construction.
For epitope-tagged tree shrew MDA5 (tMDA5) and LGP2 (tLGP2) constructs, the coding region of each gene was amplified using the gene-specific primer pairs in Table S2. PCR product was purified with the TIANquick Midi Purification Kit (TIANGEN; DP204) and was cloned into Flag-tagged pCMV-3Tag-8 with BamHI and XhoI. We generated expression vectors for tMDA5 mutants, p.K188Q (tMDA5 K188Q) and p.A402K (tMDA5 A402K), by using the multisite directed mutagenesis (Stratagene; 200518). Expression vector of human MDA5 (hMDA5) and human IFN-β promoter luciferase reporter were kind gifts from Bin Li, Institute Pasteur of Shanghai, CAS, Yunnan, China. We generated hMDA5 mutants, p.Q187K (hMDA5 Q187K) and p.K405A (hMDA5 K405A), using the same procedure for making tMDA5 mutants. All constructs were verified by direct sequencing.
Luciferase reporter assay.
HEK293, HepG2, and TSPRC cells (1 × 104) were transfected with 0.1 μg of a luciferase reporter vector [IFN-β-Luc: pGL3-tIFN-β-promoter (Table S2) or human IFN-β-promoter luciferase reporter; NF-κB-Luc: pNFκB-TA-Luc (Clontech; 631912); or ISRE-Luc: ISRE cis-reporter (Stratagene; 219092)], together with 0.01 μg pRL-SV40-Renilla (TK; Promega; as an internal control), the indicated amount (400 ng) of an empty vector (Vector), and/or the indicated expression vector by using the Lipofectamine 2000 (Invitrogen; 11668-027). After transfection for 36 h, the transfected cells were left untreated or infected with SeV (20 HAU/mL), VSV-GFP (MOI = 0.01), NDV (MOI = 10), and EMCV (MOI = 1) for 12 h. Cells were lysed, and luciferase activity was measured by using the Dual-Luciferase Reporter Assay System (Promega; E1960) on Infinite M1000 Pro multimode microplate reader (Tecan; 30064852).
Western blot and immunoprecipitation.
HEK293 cells and TSPRCs were transfected with the indicated expression vectors using X-tremeGENE HP DNA Transfection Reagent (Roche; 06366546001). Cells were lysed on ice in RIPA lysis buffer (Beyotime Institute of Biotechnology; P0013). After centrifugation at 12,000 × g at 4 °C for 5 min. Protein concentration was determined using the BCA protein assay kit (Beyotime Institute of Biotechnology; P0012). BSA (Beyotime Institute of Biotechnology; P0007) was used as a protein standard. Cell lysates (30 μg total protein per sample) were separated by electrophoresis on a 12% (vol/vol) SDS-polyacrylamide gel and transferred to PVDF membranes (Roche Diagnostics; IPVH00010) using the standard procedures. The membranes were soaked in the blocking buffer [5% (wt/vol) skimmed milk or 5% (wt/vol) BSA in TBST (Tris-buffered saline; Cell Signaling Technology; #9997) with Tween 20 (0.1%; Sigma; P1379)] at room temperature for 2 h. The membranes were then incubated with the indicated primary antibodies overnight at 4 °C: antibody for Flag (1:5,000), MYC (1:5,000), tIRF3 (1:1,000), phospho-IRF3 (Ser396) (1:1,000), tMDA5 (1:1,000), tLGP2 (1:2,000), and tβ-actin (1:10,000). To probe the endogenous phospho-NF-κB, p65 (Ser536), p65, phospho-IKKα/β (Ser176/180), IKKα/β, phospho-IκBα (Ser32), and IκBα, membranes were incubated overnight at 4 °C with the NF-κB pathway sample kit (1:1,000). After washing with 1× TBST three times (5 min each), membranes were incubated with TBST-conjugated anti-mouse or anti-rabbit secondary antibody (depends on the primary antibody; 1:10,000; KPL) for 1 h at room temperature. After another round of three washes with TBST, the proteins on the membrane were detected by using enhanced chemiluminescence (ECL) reagents (Millipore; WBKLS0500). ImageJ (NIH) was used to evaluate the densitometry.
For the detection of the RIG-I protein in tree shrew and other species, different tree shrew tissues (including heart, liver, spleen, lung, kidney, intestine, brain, and pancreas tissues), monkey and mouse brain tissues, and human paracancerous normal tissues were used. Protein samples were separated and transferred to PVDF membranes by using the above-described procedure. Membranes were incubated with rabbit anti-RIG-I (D14G6) (1:1,000; Cell Signal Technology; against Lys652 at the C terminus of human RIG-I) and rabbit anti-RIG-I (H-300) (1:1,000; Santa Cruz Biotechology; against aa 1–281 at the N terminus of human RIG-I), respectively, to detect the presence and expression of endogenous RIG-I.
For immunoprecipitation, appropriate antibodies were incubated with protein G-agarose beads (Life Technologies; 15920010) for 1 h. At the same time, 1 × 107 cells were lysed in the RIPA lysis buffer (Beyotime Institute of Biotechnology; P0013) on ice for 1 h, followed by a centrifugation at 12,000 × g for 10 min. Lysates were immunoprecipitated with previous bead-antibody complex at 4 °C overnight, followed by washing with wash buffer (RIPA lysis buffer; Beyotime Institute of Biotechnology; P0013) four times using DynaMag-Spin (Life Technologies; 12320D). The precipitated complexes were resuspended in SDS sample buffer for Western blot analysis.
RNA immunoprecipitation.
The RNA immunoprecipitation assays were performed by the procedure described in a previous study by Deddouche and coworkers (26). Briefly, about 1 × 108 TSPRCs were transfected with 30 µg FLAG-tagged tMDA5 expression vector by using the X-tremeGENE HP DNA Transfection Reagent (Roche; 06366546001) following the manufacturer’s instructions. Transfected cells were cultured for 24 h in growth medium and then infected with SeV (20 HAU/mL) for 16 h. Cells were subsequently washed and lysed in the lysis buffer [10 mM Tris, pH 7.4, 2.5 mM MgCl2, 200 mM NaCl, 0.5% Nonidet P-40, 1× protease inhibitor mixture (Millipore; 539131), and 0.5 U/mL RNasin (Promega; N2111)] (26). In brief, a small fraction of the input was collected for protein and RNA extraction. Five micrograms of anti-Flag (Abmart; M20008) or mouse IgG isotype (Beyotime Institute of Biotechnology; A7028) control antibody was added to 500 μL lysate and incubated on a rotating shaker for 1 h at 4 °C; 300 μL washed Gamma Bind Plus Sepharose Beads (GE Healthcare Bioscience AB; 10004D) were then added to the mixture for another 1 h at 4 °C. The beads were then precipitated by centrifugation and washed four times with 1 mL lysis buffer. The beads were then split into two samples for protein or RNA extraction. Proteins were extracted from protein–RNA complexes by boiling the beads for 10 min in SDS sample buffer for Western blot analysis. RNAs were extracted from protein–RNA complexes by using TRIzol (Invitrogen; 15596018).
For strand-specific detection of the SeV RNA, we first reverse transcribed the SeV RNA using the M-MLV reverse transcriptase (Promega) in the presence of a primer (SeV R15360: 5′-accagacaagagtttaagagatatg-3′) specific for the SeV defective-interfering (DI) particles (27). Next, qRT-PCR was performed in presence of the SYBR green Premix Ex Taq II (TaKaRa; RR820L) by using the primer pair SeV F15160: 5′-tgttcggggccaggcaaaat-3′/SeV R15247: 5′-gttctgcacgatagggacta-3′. For strand-specific detection of the EMCV RNA, we used a primer specific for the L antisense (5′-ggccgtcatggtggcgaataagcgcactctctcacttttga-3′) reported by Dedeouche et al. (26). The quantitative PCR procedure (EMCV common F: 5′-aataaatcataaggccgtcatggtggcgaataa-3′/L reverse primer: 5′-aataaatcataatcgaaaacgacttccatgtct-3′) (26) was same to the above described one. The amount of SeV or EMCV RNAs was analyzed by the strand-specific qRT-PCR. The RNA levels were calculated by comparison with a dilution curve of cDNA prepared from the SeV- or EMCV-infected cells.
Positive selection analysis and structural modeling.
The MDA5 and LGP2 sequences of human, macaque, Chinese tree shrew, rat, mouse, and dog were retrieved from the Ensembl (asia.ensembl.org/index.html.html) database to test for possible selective pressure in the Chinese tree shrew lineage by using the CODEML program implemented in the PAML4 package (30).
We used the following models to test the selective pressure: (i) a one-ratio model, which assumes all branches have an identical ω0 value, with ω as the ratio of nonsynonymous to synonymous substitution rate; (ii) a two-ratio model, which allows different ω values between the foreground branch (ω2) and background branch (ω1); and (iii) a branch-site model with fixed foreground branch ω2 = 1 or nonfixed foreground branch ω2, issued to determine whether the gene has undergone positive selection on a foreground branch. Finally, the likelihood ratio test (LRT) was performed: (i) the two-ratio model vs. the one-ratio model were compared with the test for whether the foreground branch ω ratio has a significant difference; (ii) branch-site model vs. branch-site model with fixed ω2 = 1 (test 2 of branch-site model) were compared with a test for whether a proportion of sites in the sequence provides a statistically significant support for ω > 1 on the foreground branch.
The branch model and the branch-site model are two likelihood ratio tests to detect positive selection (31–33). The branch-site test aims to detect episodic Darwinian selection along prespecified branches on a tree that affects only a few codons in a protein-coding gene, with selection measured by the nonsynonymous/synonymous rate ratio (ω = dN/dS) and positive selection indicated by ω > 1 (33). The branch models allow the ω ratio to vary among branches in the phylogeny and are useful for detecting positive selection acting on particular lineages. The branch-site models had improved power in detecting positive selection than the branch models.
A protein 3D structure for MDA5 (Protein Data Bank ID code 4GL2) (35) was derived from the Protein Data Bank (www.ncbi.nlm.nih.gov/protein). Sites were mapped into structures using the PyMOL (The PyMOL Molecular Graphics System, Version 1.5.0.2; Schrödinger).
Data mining of the human MDA5 nonsynonymous variants.
We checked the human Exome Aggregation Consortium (ExAC) database (exac.broadinstitute.org), to identify any (rare) nonsynonymous variants/mutations that occurred at the 187th and 405th sites in human MDA5. The ExAC is a collection of exome sequencing data from a variety of large-scale sequencing projects. The data set contains 60,706 unrelated individuals and serves as a useful reference data set of population genetics. All of the raw data from these projects have been reprocessed through the same pipeline and jointly variant-called to increase consistency across projects. We found no variation at both sites in 60,706 individuals, suggesting that these two residues were extremely conserved in human due to possible constrained function.
Statistical analysis.
Differences of relative mRNA and luciferase activity levels between groups with different treatments were conducted by the Student t test using the PRISM software (GraphPad Software). Data were represented as mean ± SEM. In all cases, P < 0.05 was considered statistically significant. All statistical analyses were two-tailed, with a 95% CI.
Acknowledgments
We thank Ian Logan for language editing and Dr. Xuelong Jiang for sharing the tree shrew DNA samples. This study was supported by National Natural Science Foundation of China Grant U1402224.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. H.S.M. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604939113/-/DCSupplemental.
References
- 1.Duggal NK, Emerman M. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat Rev Immunol. 2012;12(10):687–695. doi: 10.1038/nri3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 3.Andrejeva J, et al. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci USA. 2004;101(49):17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoneyama M, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175(5):2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 5.Baccala R, et al. Sensors of the innate immune system: Their mode of action. Nat Rev Rheumatol. 2009;5(8):448–456. doi: 10.1038/nrrheum.2009.136. [DOI] [PubMed] [Google Scholar]
- 6.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 7.Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- 8.Ronald PC, Beutler B. Plant and animal sensors of conserved microbial signatures. Science. 2010;330(6007):1061–1064. doi: 10.1126/science.1189468. [DOI] [PubMed] [Google Scholar]
- 9.Mukherjee K, Korithoski B, Kolaczkowski B. Ancient origins of vertebrate-specific innate antiviral immunity. Mol Biol Evol. 2014;31(1):140–153. doi: 10.1093/molbev/mst184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Satoh T, et al. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci USA. 2010;107(4):1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441(7089):101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 12.Bruns AM, Horvath CM. Activation of RIG-I-like receptor signal transduction. Crit Rev Biochem Mol Biol. 2012;47(2):194–206. doi: 10.3109/10409238.2011.630974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarkar D, Desalle R, Fisher PB. Evolution of MDA-5/RIG-I-dependent innate immunity: Independent evolution by domain grafting. Proc Natl Acad Sci USA. 2008;105(44):17040–17045. doi: 10.1073/pnas.0804956105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou J, Chang M, Nie P, Secombes CJ. Origin and evolution of the RIG-I like RNA helicase gene family. BMC Evol Biol. 2009;9(1):85. doi: 10.1186/1471-2148-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korithoski B, et al. Evolution of a novel antiviral immune-signaling interaction by partial-gene duplication. PLoS One. 2015;10(9):e0137276. doi: 10.1371/journal.pone.0137276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis SH, Obbard DJ. Recent insights into the evolution of innate viral sensing in animals. Curr Opin Microbiol. 2014;20(0):170–175. doi: 10.1016/j.mib.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biacchesi S, et al. Mitochondrial antiviral signaling protein plays a major role in induction of the fish innate immune response against RNA and DNA viruses. J Virol. 2009;83(16):7815–7827. doi: 10.1128/JVI.00404-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barber MRW, Aldridge JR, Jr, Webster RG, Magor KE. Association of RIG-I with innate immunity of ducks to influenza. Proc Natl Acad Sci USA. 2010;107(13):5913–5918. doi: 10.1073/pnas.1001755107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liniger M, Summerfield A, Zimmer G, McCullough KC, Ruggli N. Chicken cells sense influenza A virus infection through MDA5 and CARDIF signaling involving LGP2. J Virol. 2012;86(2):705–717. doi: 10.1128/JVI.00742-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan Y, et al. Genome of the Chinese tree shrew. Nat Commun. 2013;4:1426. doi: 10.1038/ncomms2416. [DOI] [PubMed] [Google Scholar]
- 21.Zhong B, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29(4):538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455(7213):674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun W, et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci USA. 2009;106(21):8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy WJ, et al. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 2001;294(5550):2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- 25.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314(5801):994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 26.Deddouche S, et al. Identification of an LGP2-associated MDA5 agonist in picornavirus-infected cells. eLife. 2014;3:e01535. doi: 10.7554/eLife.01535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baum A, Sachidanandam R, García-Sastre A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci USA. 2010;107(37):16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bruns AM, Leser GP, Lamb RA, Horvath CM. The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5-RNA interaction and filament assembly. Mol Cell. 2014;55(5):771–781. doi: 10.1016/j.molcel.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abe T, Barber GN. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-κB activation through TBK1. J Virol. 2014;88(10):5328–5341. doi: 10.1128/JVI.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen R, Yang Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148(3):929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22(12):2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 33.Yang Z, dos Reis M. Statistical properties of the branch-site test of positive selection. Mol Biol Evol. 2011;28(3):1217–1228. doi: 10.1093/molbev/msq303. [DOI] [PubMed] [Google Scholar]
- 34.Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38(5):855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu B, et al. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152(1-2):276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 36.Yoo JS, Kato H, Fujita T. Sensing viral invasion by RIG-I like receptors. Curr Opin Microbiol. 2014;20:131–138. doi: 10.1016/j.mib.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 37.Yoneyama M, Onomoto K, Jogi M, Akaboshi T, Fujita T. Viral RNA detection by RIG-I-like receptors. Curr Opin Immunol. 2015;32:48–53. doi: 10.1016/j.coi.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 38.Schoggins JW, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472(7344):481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34(5):680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoneyama M, Fujita T. Recognition of viral nucleic acids in innate immunity. Rev Med Virol. 2010;20(1):4–22. doi: 10.1002/rmv.633. [DOI] [PubMed] [Google Scholar]
- 41.Rodriguez KR, Bruns AM, Horvath CM. MDA5 and LGP2: Accomplices and antagonists of antiviral signal transduction. J Virol. 2014;88(15):8194–8200. doi: 10.1128/JVI.00640-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saito T, et al. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci USA. 2007;104(2):582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rothenfusser S, et al. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol. 2005;175(8):5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 44.Yang Z, Wong WS, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22(4):1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]