Abstract
Prevention is an essential component of cancer eradication. Next-generation sequencing of cancer genomes and epigenomes has defined large numbers of driver mutations and molecular subgroups, leading to therapeutic advances. By comparison, there is a relative paucity of such knowledge in premalignant neoplasia, which inherently limits the potential to develop precision prevention strategies. Studies on the interplay between germ-line and somatic events have elucidated genetic processes underlying premalignant progression and preventive targets. Emerging data hint at the immune system’s ability to intercept premalignancy and prevent cancer. Genetically engineered mouse models have identified mechanisms by which genetic drivers and other somatic alterations recruit inflammatory cells and induce changes in normal cells to create and interact with the premalignant tumor microenvironment to promote oncogenesis and immune evasion. These studies are currently limited to only a few lesion types and patients. In this Perspective, we advocate a large-scale collaborative effort to systematically map the biology of premalignancy and the surrounding cellular response. By bringing together scientists from diverse disciplines (e.g., biochemistry, omics, and computational biology; microbiology, immunology, and medical genetics; engineering, imaging, and synthetic chemistry; and implementation science), we can drive a concerted effort focused on cancer vaccines to reprogram the immune response to prevent, detect, and reject premalignancy. Lynch syndrome, clonal hematopoiesis, and cervical intraepithelial neoplasia which also serve as models for inherited syndromes, blood, and viral premalignancies, are ideal scenarios in which to launch this initiative.
Keywords: premalignancy, biology, vaccines, cancer prevention, immune oncology
Cancer development is a complex process influenced by inherited variation in germ-line DNA and acquired somatic alterations. The stepwise accumulation of genetic changes leads to oncogenic transformation (1), and also co-opts neighboring normal cells (e.g., neuronal and vascular) to support tumor development and progression (1, 2). The immune system recognizes transformed cells, and avoiding immune elimination is now an accepted hallmark of cancer (2). Large-scale somatic sequencing initiatives, such as The Cancer Genome Atlas (TCGA), in parallel with large genome-wide association studies (GWAS) of germ-line variants have analyzed an increasing array of cancers (3). However, there remain some notable lacunae in our understanding of the biology of premalignancy and cancer development, including the roles of the immune system. Although cancers are increasingly being defined by alterations in genetic, epigenetic, and signaling networks, premalignant lesions [with few exceptions (4, 5)] are still largely identified only through morphological criteria.
In this Perspective, we discuss the influence and interactions of omic and cellular [e.g., tumor microenvironment (TME) and microbiome] events on the development and progression of premalignancy (1, 2, 4, 6). There is an unprecedented opportunity in single-cell next-generation sequencing (NGS), computational biology, high-throughput functional screens, and preclinical models (7, 8) to achieve an integrated understanding of premalignant biology and cancer risk to drive immune-based prevention.
Colorectal Adenoma-Carcinoma Model
Even though the seminal multistep genetic model of human carcinogenesis was defined in the colorectal adenoma-carcinoma sequence nearly three decades ago (9), it is unfortunate that NGS of only 25 sporadic colorectal adenomas have been reported to date (10, 11). This number contrasts radically with the plethora of genomic information (12–14) generated by major initiatives at multiple levels for colorectal carcinomas (CRC). Most reported molecular analyses of colorectal adenomas have interrogated only a limited number of genes or restricted-region assessments of copy number, rendering a narrow view of the biology of premalignancy. New technologies, including human organoids with CRISPR/Cas9-based gene editing, are being applied to this model (15). NGS studies of minute tissue specimens with isolated reports of small numbers of premalignant lesion types, such as Barrett’s esophagus (16), ductal and lobular carcinoma in situ (DCIS, LCIS) (4, 17), serous tubal intraepithelial carcinoma (18), pancreatic intraepithelial neoplasia (PanIN) (19), monoclonal gammopathy of unknown significance (MGUS) (20), and high-count monoclonal B-cell lymphocytosis (MBL) (21), with colorectal adenomas being the most salient example.
Knowledge on the genomic annotation of intestinal carcinogenesis has come mainly from the study of premalignant lesions in hereditary CRC syndromes, which are thought to recapitulate the two major pathways of genomic instability. Familial adenomatous polyposis (FAP), caused by germ-line adenomatous polyposis coli (APC) mutations, is a molecular model for 85% of sporadic CRC characterized by Wnt alterations and chromosomal instability (22). Recent whole-exome sequencing characterized the genomic landscape of early adenoma tissue in FAP, which confirmed and extended the proposed “Big Bang” theory of CRC development (11), identifying >200 somatic hits in 25 adenomas, clonal selection, and a mutational load similar to that of stage I CRC (23). An estimated 25% of the mutational load (all passenger mutations) was present in adjacent, apparently normal mucosa (field effect). This study (23) and others (24) in FAP have provided a catalog of the somatic variation cooperating with APC in early colorectal carcinogenesis and indicated that a substantial proportion of the genomic variation present in CRCs is acquired in the earliest at-risk tissues. Understanding FAP biology has led to breakthrough combinatorial chemoprevention for this devastating syndrome (25).
Lynch syndrome (LS), caused by germ-line defects in the DNA mismatch repair (MMR) system, is a model for 15% of sporadic CRCs characterized by microsatellite instability (MSI). The absence of proficient MMR surveying DNA for these errors generates an exponential accumulation of frameshift (FS) mutations at microsatellite tracts, thus increasing mutation rate by several orders of magnitude and accelerating oncogenesis (26) (see LS as a Model for Hypermutability and Immune-Based Prevention, below). Chromosomal instability, defective DNA repair, and APOBEC (apolipoprotein B mRNA-editing enzyme; discussed in Expanding the Scope of Immune Prevention to BRCA1/2 and APOBEC-Associated Neoplasia) are major drivers of oncogenesis and clonal diversity/heterogeneity in other hereditary and sporadic cancers (27). The role of the microbiome in CRC risk is discussed below.
Germ-Line–Somatic Landscape
Colorectal neoplasia also provides examples of germ-line effects on somatic events and phenotype. The location and mechanism (point mutation versus deletion) of germ-line APC inactivation in FAP determines the somatic second hit in APC and amount of β-catenin optimal to promote intestinal carcinogenesis (“‘just right’ model of APC”) (28). Germ-line 5′ APC mutations in FAP affect interactions with wild-type APC, allowing residual Wnt activity (29). Germ-line biallelic MUTYH causes G:T transversions due to base excision repair defects (24). Further, the APC I1307K (c.3920T > A) polymorphism, linked to CRC risk (30), generates a hypermutable, mononucleotide repeat (A8) that impairs replication fidelity, forming a mutational hotspot facilitating biallelic inactivation of APC.
Germ-line mutations of the transcription factor GATA2 confer monocytopenia, atypical mycobacterial infections, and a propensity to develop preleukemia (myelodysplastic syndrome; MDS) or acute myeloid leukemia (AML). GATA2 mutation carriers that develop myeloid malignancies harbor somatic ASXL1 mutations (and monosomy 7) at rates much greater than expected by chance, suggesting a functional or epistatic interaction between these events in myeloid-lineage cells (31). Other examples of hereditary mutated transcription factors that predispose to hematologic neoplasia include mutations in CEBPA, RUNX1, ETV6, and PAX5 (32). The culprit germ-line variants are typically heterozygous and may have dominant-negative activity against the remaining germ-line allele. Cooperating somatic mutations, often including mutation of the remaining wild-type allele, are clearly required and can be identified during periods of clonally skewed hematopoiesis that precede transformation (32).
The development of myeloproliferative neoplasms (MPN) can involve a JAK2 haplotype (termed 46/1) that is highly associated with the acquisition of a somatic JAK2 mutation in MPN patients. Strikingly, the somatic JAK2 mutation associated with the 46/1 haplotype occurs on the cis (vs. trans) allele more often than predicted by chance, suggesting a local interaction (33). However, this mutational predisposition effect is not limited to the nearby JAK2 gene. Patients with mutations of another MPN gene, MPL, are also more likely to carry the 46/1 variant (34).
Integrated analysis of germ-line and somatic variants is also beginning to inform precision prevention. Large-scale sequencing of over 4,000 tumors (12 cancer types) from the TCGA found rare germ-line truncations in 114 cancer-susceptibility-associated genes, ranging in frequency from 4% (AML) to 19% (ovarian cancer) (35). Of the 1% of lung cancer patients with somatic EGFR T790M resistance mutations at diagnosis, most actually carry germ-line EGFR T790M mutations. These families appear to have a different biology of lung neoplasia (slow-growing lung nodules) and so may be good candidates for lung cancer screening and precision chemoprevention with T790M inhibitors (36). Finally, repurposed NGS of “control” blood from large TCGA and GWAS cohorts identified clonal hematopoiesis as a new premalignant state, characterized by age-related myeloid malignancy driver mutations (mostly in DNMT3A, TET2, and ASXL1) (37, 38). The vast majority of these individuals harbored a single-driver mutation. Patients with idiopathic cytopenias of undetermined significance (ICUS) were noted to have higher rates (∼40%) of clonal hematopoiesis and possibly transformation to MDS/AML (39, 40). Once germ-line–somatic relationships have been mapped, an atlas of shared and distinct oncogenic events can be analyzed for targetability.
Harnessing the Immune System for Cancer Prevention
There exists a fascinating duality regarding the immune system’s role in oncogenesis, the depths of which remain incompletely understood (41, 42). It is well known that inappropriate immune responses (as seen in chronic inflammatory conditions) are strongly associated with high risks of developing cancer (e.g., CRC in ulcerative colitis; 43). Immunosurveillance/immunity, however, is thought to be a critical mechanism for inhibiting cancer development and progression, as evidenced by the success of immune checkpoint inhibitors [e.g., programmed cell death protein 1 (PD-1) antibodies], which have been a game-changer for a number of patients, producing deep and durable clinical responses in a variety of malignancies, particularly high-mutational burden cancers (13, 44, 45). In parallel, the incredible efficacy of human papillomavirus (HPV) vaccines has shown the great promise for using the immune system for cancer prevention (46).
Vaccines to prevent cervical intraepithelial neoplasia (CIN) are standard practice (46) and work best when given to healthy individuals before they are exposed to HPV, so as to induce neutralizing antibodies against viral proteins while the cervical tissue is normal, without an immunosuppressive TME. Viral E6 and E7 proteins are well-understood oncogenic drivers, and CIN is relatively indolent and directly accessible by routine screening. Therapeutic vaccine studies targeting E6/E7 antigens in CIN2/3, including results from single-cell T-cell receptor sequencing, suggest that inducing efficient trafficking of functional effector T-cells to the epithelial disease site is critical to eliminate both the disease and virus (47). HPV E6 induces APOBEC3B, which in turn mutates chromosomal DNA and most likely contributes to precancer development (48). HPV also induces tumor-associated stromal fibroblasts (49), and E6 inactivates p53, which induces PD-L1 and cervical Tregs, causing immune evasion (47). HPV16 integration into the PD-L1 3′UTR enhances PD-L1 expression (50).
Mouse data have clearly shown that tumors can escape immune recognition by losing their antigenicity in a process termed “immunoediting” (51, 52). Furthermore, knockout mice lacking an adaptive immune system have dramatically increased rates of tumor (e.g., intestinal adenoma and adenocarcinoma) formation compared with wild-type mice (53). In humans, severe combined immunodeficiency (SCID) is similarly characterized by fundamental defects in adaptive immunity, although the associated risk of cancer (mostly lymphomas) is modest, possibly because SCID is almost universally fatal by age 2 (without stem cell transplant) as a result of infections (54). Other forms of inherited immunodeficiency—such as common variable immunodeficiency, X-linked hyper-IgM syndrome, Bloom syndrome, and ataxia telangiectasia—have been linked to increased risks of cancers, predominantly lymphomas but also a wide spectrum of solid malignancies, MDS and AML (54). Similar findings were reported in acquired immunosuppression, including people with HIV/AIDS and solid organ transplant recipients (55). The mechanisms underlying such cancer risks in immunodeficient patients are not well understood, given the complex and overlapping functions/components of innate and adaptive immunity, which may partially compensate for specific immune defects. Such gaps in knowledge further indicate the need to fully map the biology of premalignancy.
The Premalignant Antigenic Repertoire and Microenvironment.
The premalignant antigenic repertoire/vaccine targets can include driver mutations and nonmutated self-proteins that are expressed at abnormal levels. It is unknown what determines immunogenicity, although it is more complex than simply the category of antigen (Fig. 1). Posttranslational modifications, such as glycosylation, can have complex, poorly understood effects on immune response (56) and evasion (57). Tumor-specific mutant epitopes (called neoantigens) may be important factors for understanding and determining the specificity of an immunotherapy (58). This theory was confirmed experimentally in mice using genomics and bioinformatics to predict those cancer-specific mutations that function as neoantigens and demonstrate their effective use in cancer vaccines (59). Of note, vaccines against immunogenic tumor mutations in mice can be as effective as immune checkpoint blockade (60). This approach has led to considerable interest in the cancer epitope (mutation) landscape and has supported the potential to generate novel immunogenic neoepitopes. In CRC, whole-exome sequencing has been implemented to computationally predict the neoantigenic repertoire from archival specimens (61). A large-scale initiative, including high-throughput mass spectroscopy and single-cell proteomics (7), and rigorous clinical characterization and follow-up will be essential to define immunogenicity of premalignant antigens.
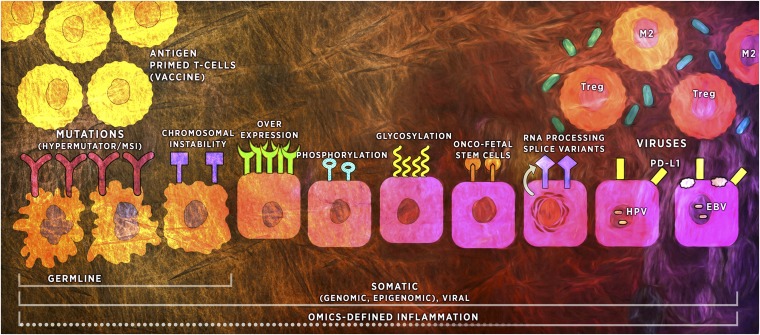
Fig. 1.
The immunogenic repertoire of premalignancy. The horizontal lines at the bottom represent the layers of factors that can stimulate immunity, among them germ-line and somatic alterations and their complex dynamic interplay with the inflammatory TME (Upper Right). The upper half of the figure depicts the progressively immunosuppressive TME from left to right. The epithelial cells (middle row) illustrate two pathways of genomic instability on the left (irregular cell borders)—MSI and chromosomal instability—which can be inherited or acquired (see Colorectal Adenoma-Carcinoma Model). Inherited and acquired MSI-H lesions are highly immunogenic. The somatic cell alterations in the middle include complex posttranslational modifications (e.g., glycosylation), onco-fetal, and splice variants, important parts of the immunogenic repertoire, but their order in terms of cancer risk or immunogenicity is unclear. The cells on the far right middle row are virally infected cells, which have similar TME issues as the nonviral premalignancies. Vaccine-primed T-cells (Upper Left), capable of generating type I Th and CD8+ cells, could overcome early TME changes to eradicate cells in the transformation process.
The first prevention example of a cancer vaccine targeting a driver mutation in premalignancy involved Kras in a pancreas genetically engineered mouse model (GEMM). Kras mutations are the earliest genetic drivers in human pancreas neoplasia, present in both early- and late-stage PanINs. Early Kras-mutated neoplastic cells secrete cytokines (e.g., IL-6), VEGF, and GM-CSF, which recruit Tregs, myeloid-derived suppressor cells (MDSCs), adipocytes and neutrophils, macrophage PI3Kγ, and chemokines (e.g., CXCL13), which recruit B-cells leading to a progressively immunosuppressive TME and immune escape (42, 62, 63). Kras-p53-Cre pancreatic GEMM were immunized with a Listeria vector encoded with the KrasG12D mutation and were found to generate CD8+ T-cells specific for the Kras mutation (64). Kras vaccine combined with cyclophosphamide Treg depletion significantly slowed the progression of early (but not late) PanIN, compared with control mice. These and other mouse-model data show the potential of driver mutation-specific vaccination to prevent premalignant progression (56) and underscore immune evasion mechanisms. Serious concerns with checkpoint inhibitors in the prevention setting include potentially severe immune adverse effects and a dearth of long-term safety data. Modulators of ten-eleven translocation (TET) proteins (65) and other epigenetic regulators (e.g., deliverable forms of miRNAs) and metformin given during vaccination could reprogram early immunosuppressive cell populations. Metformin (a safe FDA-approved oral diabetes agent) can increase CD8+ cells, reduce T-cell exhaustion, reprogram macrophages and stellate cells (to reduce desmoplasia) in pancreatic neoplasia (66), and can alter T-cell metabolism to generate long-lived immune memory (67, 68).
The influence of the premalignant TME is also well illustrated in DCIS: integrated DNA- and RNA-seq of high-grade DCIS identified high rates of p53 pathway inactivation and a molecular subclass of lesions characterized by a highly proliferative, basal-like phenotype with genomic signatures of activated Treg cells and checkpoint complexes indicative of a tumor-associated immunosuppressive phenotype (4). Suppressed immunity (e.g., high Tregs and CD8+ HLA-DR-neg T-cells) correlated with progression from normal to DCIS to invasive ductal carcinoma. PD-L1+ tumor-infiltrating lymphocytes are most prominent in triple-negative DCIS and microinvasive cancer (69).
There is increasing evidence that somatically mutated MDS cells can alter their TME to provide a clonal growth advantage. Examples include activation of inflammatory molecules, s100a8 and s100a9, and induction of TP53 in mouse models. Similarly, MDS cells with various types of somatic mutations can activate inflammasome-mediated pathways that increase MDSC bone marrow number (70). Alteration to stromal cells may promote clonal hematopoiesis (e.g., mice carrying a Dicer1 deletion in osteoblasts developed clonally derived leukemias) (71).
The presence of a robust adaptive T-cell immune response, evident either in tumor or peripheral blood from patients with certain cancers, has been associated with improved survival (72). Antibody response to vaccines is also important and can enhance T-cell immunity (73). Naturally occurring cytotoxic T-cell responses to tumor antigens can be detected in one-third of healthy people without cancer (74). Precancer-specific natural immune surveillance also exists and can prevent the development of cancer (56, 75). For example, MGUS patients can mount a T-cell immune response to SOX2, a transcription factor critical for self-renewal in stem cells, which is associated with reduced risk of progression to multiple myeloma (MM), supporting the potential for a vaccine to boost SOX2-specific immunity (76).
Host–microbiome interactions are important in premalignancy, adding to TME complexity (6). Studies in GEMMs have found that APC loss disrupts the intestinal epithelial barrier, facilitating invasion of microbes and microbial nucleic acids that activate adenoma-associated macrophages to produce IL-23, which then stimulates IL-17 production by T-cells, accelerating adenoma development and progression (77). Bacterial translocation can activate Toll-like receptors that can up-regulate other inflammatory elements. These barrier defects drive innate immunosuppressive TME, and adenoma proliferation (e.g., F. nucleatum in the TME can inhibit NK-cell cytotoxicity producing bacteria-dependent immune evasion) (43, 78). Metagenome study found different taxa in adenomas compared to carcinomas and healthy controls (79). Gut microbiota may explain the provocative link between MMR-deficiency and CRC (80). The interplay between the microbiome, virome, autophagy, inflammatory bowel disease, GWAS, and the immune system is also under active study in CRC development and prevention (81, 82).
LS as a Model for Hypermutability and Immune-Based Prevention.
Somatic hypermutation can arise through diverse mechanisms. As described above, MSI is a form of hypermutability in which DNA MMR defects lead to genome-wide accumulation of FS mutations within predictable nucleotide repeat loci (microsatellites). MSI-related FS mutations drive tumorigenesis by occurring within microsatellite loci that lead to inactivation of tumor suppressors enriched for genes functionally involved in immune regulation (e.g., TGFBR2 and BAX) in both CRCs and adenomas (83). When they occur in coding regions, such mutations generate FS-mutation-derived peptides (FSP), which function as highly immunogenic neoantigens and cause specific CD8+ T-cell responses and neoplastic infiltrates. The “hotspot” nature of these MSI-related FS mutations leads to FSPs with predictable sequences, suggesting that multivalent vaccine development targeting specific, expected T-cell epitopes may be an effective prevention strategy for MSI-induced neoplasia (83). The feasibility of this approach was demonstrated by using a panel of FSPs expected to be generated by MSI-induced FS mutations at a specific hotspot locus within MSH3; engineered CD8+ T-cells from a healthy volunteer specifically targeted these FSP-lysed, HLA-matched, high-level MSI (MSI-H) CRC cell lines (84).
MSI may represent a unique form of hypermutability, expressing high amounts of neoantigens, which up-regulate inhibitory molecules (e.g., PD-L1) to counterbalance the infiltrating immune cells; this is distinct from overall mutational load, in that it renders tumors very susceptible to immune-based destruction. There has been major progress in using PD-1 inhibitors to treat advanced cancers with MSI-H and MMR deficiency (MMR-D), such as those that arise in LS or sporadic MSI-H CRC (44). LS patients are at very high risk of CRC and endometrial cancers, and recent data suggest that this classic LS-cancer spectrum is wider than traditionally appreciated.
The recognition in healthy (screened) LS carriers of MSI-H/MMR-D in preinvasive, normal-appearing tissues and circulating FSP-specific T-cells (85) suggests that immune surveillance mechanisms may help reduce MSI-H tumor development. Histologically normal but MSI-H/MMR-D intestinal crypt foci in LS carriers harbor MSI-related FS mutations, which may be a key source of these FSP-specific T-cells (86). There are conflicting data as to whether the size of LS adenomas correlates with MSI-H/MMR-D, although this may reflect technical aspects of MSI and MMR testing rather than actual adenoma biology (87). NGS can likely address this limitation, because mutational burden appears to be a reliable surrogate for MSI-H status (8).
In LS-associated MSI-H CRCs and adenomas, immune evasion can occur by MHC I loss as a result of β2-microglobulin mutations (a mechanism distinct from sporadic MSI-H CRCs) (88). Additionally, there is evidence of an immune-suppressive TME (increased density of FOXP3+ Tregs) in normal mucosa adjacent (but not distant) to CRC in LS patients with wild-type β2-microglobulin (89).
Children who are homozygous for germ-line LS mutations have biallelic MMR deficiency (BMMR-D) (90) and present a compelling scenario for vaccine-prevention. BMMR-D confers a devastating phenotype of pediatric lymphomas, brain tumors, and intestinal cancers (91). In stark contrast to other pediatric cancers, which classically display few somatic mutations (1), BMMR-D–associated cancers have an “ultrahypermutated” phenotype (90) because of acquisition of somatic FS mutations in the proof-reading domains of the DNA polymerases POLE or POLD1 and have mutational loads exceeding those in adult MSI-H CRC. BMMR-D cancers may be particularly responsive to PD-1 inhibitors (92).
Expanding the Scope of Immune Prevention to BRCA1/2- and APOBEC-Associated Neoplasia.
LS represents an ideal proof-of-principle for using immune-based prevention, relevant to other hereditary cancers. This is particularly important as NGS data continue to expand the spectrum of cancers linked to various germ-line mutations, including BRCA1/2 (93). Germ-line BRCA1/2 mutations induce defects in homologous recombination (HR)-based DNA repair and confer markedly increased risks of cancers of the breast, ovaries/fallopian tubes, pancreas, prostate, and melanoma, although NGS germ-line testing suggest that they may also be linked to cancers more classically LS-associated (CRC and endometrial cancers) (93). Somatic mutational patterns found in HR deficient BRCA1/2-associated breast cancers and BRCA2-mutated prostate cancers demonstrate predictable “signatures” of somatic mutations (94, 95), suggesting the plausibility of creating vaccines to target specific hotspot neoantigens. Similarly, BRCA1/2-associated ovarian cancers (96) have been shown to exhibit an increased effector lymphocytic reaction (which likely first develop in serous tubal intraepithelial carcinoma) (97) and high numbers of immunogenic mutations.
A growing array of data are examining the role that loss of wild-type BRCA1/2 function plays in the development and progression of breast, fallopian tube, pancreatic, and prostatic premalignant lesions from individuals with germ-line BRCA1/2 mutations, including data suggesting that BRCA1 haploinsufficiency promotes genomic instability in nonneoplastic breast epithelium before loss of the wild-type allele (98). Nonneoplastic breast epithelium from BRCA1 mutation carriers have gene-expression profiles similar to luminal progenitor cells (which differs from the basal features of most BRCA1 breast cancers) (99). Further efforts toward characterizing BRCA1/2-associated premalignancy are vital to developing preventive strategies for these high-risk patients. Poly ADP ribose polymerase (PARP) inhibitors, compelling precision therapy of certain BRCA1/2-associated cancers, have been shown to delay mammary tumor development in BRCA1-deficient mice (100). Exciting data suggest that the RANKL/RANK pathway has an integral role in breast oncogenesis in germ-line BRCA1 mutation carriers. Interference with this pathway produced significant preventive activity, including pharmacologic RANK-ligand inhibition (e.g., denosumab) in BRCA1-mutant breast organoids and Brca1-deficient and mutation-driven mouse models (101, 102). Denosumab is an FDA-approved agent for bone loss with an established safety profile and could be repurposed for prevention trials for healthy mutation carriers.
NGS and biochemical characterization have identified key roles of APOBEC3 (A3) enzymes in inducing a hypermutated phenotype as part of innate immunity. A3 induction is a critical early event in HPV-related neoplasia (see above). A3 can be induced by IFN-α, IFN-γ, and other inflammatory cytokines (103), although the induction mechanism (104) in nonviral cancers is unclear and the timing varies by site and etiology (27, 103). A3A and A3B have intrinsic preference for deaminating cytosine residues in TCA and TCG trinucleotide contexts, and it is thus assumed that A3B-mediated neoplasia will be characterized by A3B-catalyzed mutational hotspots (e.g., generating PIK3CA driver mutations at helical domain hotspots E542K and E545K) (27) that could be used as part of a vaccine. A common germ-line APOBEC3A/3B chimeric deletion polymorphism (ΔA3B) has been associated with risk of breast, liver, and certain other cancers (105–108). Paradoxically, this ΔA3B deletion leads to increased A3A activity as a result of increased stability of the chimeric APOBEC3A/B mRNA (109). This increased A3A activity is thought to underlie the associated modest breast cancer risk, because cancers associated with these ΔA3B polymorphisms have A3 mutation signatures (distinct from those seen in HR-deficient or MMR-D breast cancers) that correlate with germ-line copy number (105) and seemingly higher penetrance of hypermutability and immune activation (106, 108). Study of the regulation of APOBEC3 in neoplasia will be critical, including ADAR1 oncogenic effects linking RNA editing to an innate inflammatory TME and potential suppression of hypermutation and immunity (110). Furthermore, the ΔA3B polymorphism is highly prevalent in certain populations (37% East Asians, 58% Native Americans, >90% Pacific Islanders) (111), suggesting that vaccines targeting A3-related neoantigens could have an important public health impact for preventing both ΔA3B- and viral-associated cancers (27, 103, 107, 109), the former possibly providing a roadmap to investigate preventive approaches for other germ-line polymorphisms linked to cancer risk. Increasing evidence from GWAS suggest a substantial germ-line effect in adult “sporadic” tumors (112), and suggest that most loci identified in cancer patients are present in the precursor (e.g., DCIS) and can influence chemoprevention (113, 114).
Summary and Next Steps
A new national investment in cancer—driven by the Vice President’s Cancer Moonshot Initiative that includes the NIH, academia, Food and Drug Administration, private foundations, philanthropic partners, and industry—includes prevention and cancer vaccines (115, cancer.gov/research/key-initiatives/moonshot-cancer-initiative/blue-ribbon-panel/blue-ribbon-panel-report-2016.pdf). A large fund infusion from The American Recovery and Reinvestment Act of 2009 advanced the TCGA from a small pilot program of three cancer types to the tremendous resource it has become (116). A similar opportunity exists in the realm of premalignant biology: a new prospective initiative in this setting could leverage and expand TCGA, GWAS, and related model infrastructures for systematic specimen collection, processing, storage, analyses to bioinformatics and data sharing (116, 117).
This initiative will require collaborations across diverse disciplines. For example, the oncogenic mechanisms of IDH mutations [discovered through broad sequencing (118)] remained unclear until modern metabolomic profiling (119) detected the novel “oncometabolite” 2-hydroxyglutarate, which inhibits TETs and other enzymes that are important in certain premalignancies (120), thereby identifying a completely novel method of oncogenesis and turning a genetic discovery into a drug and vaccine target. There is also a need for: (i) better preclinical models, e.g., CRISPR/Cas9 engineered immunosuppressive mouse strains (121), immune organoids (122), and new model organisms (e.g., zebrafish) providing insight into early premalignant biology, and the role of epigenetic reprogramming in transformation (123), (ii) discern site-specific patterns and timing of driver mutations and genomic instability in neoplastic progression (124) and specific acquired mutations predictive of immune resistance in premalignancy (88) and cancer (125), (iii) imaging immune responses (e.g., NK- and T-cell subtype trafficking) and TME composition to optimize priming and boosting regimens (126), and (iv) new single cell and computational methods to understand the increasingly complex cellular (e.g., adipocyte, myocyte interplay) compartment and tissue microenvironment (e.g., aging fibroblast effects on adaptive immunity) from which the malignancy arises (7, 127, 128).
The development of effective prevention will not be easy (1), but the potential public health benefits are extensive as can be illustrated by the case of cervical cancer, for which screening and HPV vaccination offer the potential to eradicate this disease, whereas recent progress in treating advanced disease included 2- to 3-mo improvements in median survival (129). Cancer vaccines have been studied extensively in thousands of people for many decades and have a very favorable safety profile setting the stage for prevention testing (56). HPV vaccine research supporting one- or two-dose regimens may apply to other cancer prevention vaccines and would greatly improve costs and adherence (46).
The initial phase of this initiative should include LS, clonal hematopoiesis/ICUS, and CIN—an inherited syndrome, blood, and viral premalignancy, respectively—which also serve as models for related disorders. The rationale for cancer-prevention vaccines in healthy LS carriers is particularly compelling: early immune surveillance, reduced MHC loss, predictable FSP patterns, high cancer risk, and young, immunocompetent probands who require serial cancer screening (85). Potential vaccine benefit could extend to LS-associated cancers beyond just CRC, because MSI-H has been found in a wide spectrum of preinvasive LS neoplasia (e.g., 130). This approach would also facilitate vaccine-based prevention for sporadic MSI-H carcinogenesis, which is implicated in subsets of many cancers (131). Outside of the colorectum and stomach, however, little is known about MSI in sporadic premalignancy. Sporadic MSI-H CRCs that arise from sessile serrated adenomas demonstrate FS mutations at the same hotspot microsatellite loci as in LS CRC. Although LS awareness is increasing the use of universal tumor testing of CRC (and now endometrial) specimens for MSI-H/MMR-D (132) and access to NGS germ-line testing, implementation is a major challenge. The estimated prevalence of LS in the general United States population is 1:280 (1.1 million). Colonoscopies are highly effective at reducing CRC risk in LS patients, but strategies for preventing other LS-associated cancers are limited (132). Existing infrastructure includes the international Colon Cancer Family Registry (133). Next steps could include web-based patient recruitment, successfully developed for other related hereditary cancer efforts (e.g., the PROMPT registry, promptstudy.info) and statewide LS registries. These registries would facilitate rapid, large-scale, systematic collection of data and tissue samples from LS and serve as a model that could be expanded to other inherited cancers, such as pancreatic cancer risk/precursors (e.g., identified by germline CDKN2A mutations) to drive vaccine prevention (134) and early detection (135). GWAS (and other) modifiers of high-penetrance mutation effects on risk, biology, precursors, and sites are also needed (136).
The timing is ideal to include clonal hematopoiesis in the initial phase of this initiative (37–40). First, it is important to leverage existing efforts of the National MDS Study (https://thenationalmdsstudy.net/), which will now include a longitudinal biobanking cohort of 500 patients with ICUS. Similarly, the MDS/AML CONNECT Registry sponsored by Celgene will follow 200 ICUS patients over time. Second, there is an opportunity to partner with the Leukemia and Lymphoma Society, patient advocacy groups, and commercial hematopathology laboratories to rapidly identify thousands of potential patients for focused longitudinal studies. Third, innovative prevention, including immune approaches, which have shown promise in MDS and AML, need to be developed for patients at highest risk of malignant transformation (to minimize over diagnosis). Patients with clonal hematopoiesis can harbor small clones for long periods of time (39, 40), and provocatively can account for “therapy-related” MDS/AML (137). Drugs targeting the inflammasome and innate immune responses implicated in remodeling the microenvironment to favor clonal expansion and vaccines against clonal antigens (138) are potential approaches. Analogous approaches can be adopted for MGUS and MBL. Solid-tumor incidence is three- to fourfold higher in MBL and CLL patients vs. healthy controls, likely due to defects in immune surveillance, which could dampen cancer vaccine response (139). Lenalidomide is in clinical trial to improve vaccine response in MBL via its beneficial T-cell effects (NCT02309515). Focusing on premalignancies of the blood has several advantages, including the ease of repeatedly acquiring neoplastic cells to study their clonal evolution over time, and although slightly more invasive, repeated access to the bone marrow to study changes in the cellular microenvironment is also safe and feasible within the scope of research study. Furthermore, study of MPNs (120) provide the only direct data that somatic mutation order (JAK2 and TET2) can greatly influence disease features.
Finally, expanding the development of vaccines for HPV-related neoplasia is a major global need (46). CIN provides an invaluable model for developing these vaccines, for example: targeting E6/7 and/or A3B (to prevent other HPV-related cancers), including routine screening for longitudinal follow-up; and nonviral vaccines, which share premalignant biology (e.g., p53, A3B), T-cell trafficking, TME features (e.g., PD-L1, Tregs), and mechanisms of immune evasion. Epstein–Barr virus vaccine development has been more challenging than HPV in part because of complex virion surface and viral antigen expression patterns (115, 140). Analogous to the TCGA pan-cancer analyses (117), it will be important to combine premalignancy omic and immune TME data from multiple sites, etiologies, and types to understand molecular alterations, timing, and interactions to target common and distinct events that drive oncogenesis across different lesions. The central theme of this initiative, to elucidate premalignant biology, requires collaborations across diverse disciplines, and leveraging other related initiatives, including the Global Human Vaccines Project, which brings tremendous expertise from infectious diseases and immunology to immune oncology, focused on decoding immune response, evasion, and immunogenicity (141).
Prevention research has produced encouraging results (25, 135, 142–144), in some cases possibly due to previously unrecognized immune effects (5, 67, 145–147). To move this field from isolated examples of progress to near elimination of all cancers will take a radically different focus and approach to premalignant disease and cancer prevention. For example, an imperative of cancer vaccines is the induction of long-term memory T-cell responses (68), overcoming a major limitation of chemoprevention. Fulfilling this vision will require a concerted effort across different initiatives and disciplines, the defining theme of the concept of Convergence Research (www.convergencerevolution.net/2016-report). We will need large-scale, systematic, integrated NGS with multiple omics and immuno-informatic platforms and clinically annotated longitudinal follow-up to lay the foundation of an effective framework for more precise early detection (e.g., liquid biopsy) and prevention and to develop cancer vaccines that reprogram the immune response at the earliest stages to durably reject tumor development. Providing adequate resources and developing multidisciplinary teams of expert prevention-focused scientists is the roadmap to success.
Acknowledgments
The authors thank Nikki Lytle and Leona Flores for editorial assistance with this article. S.M.L. was supported for this work by National Cancer Institute Grant P30-CA023100-29.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Hoadley KA, et al. Cancer Genome Atlas Research Network Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158(4):929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abba MC, et al. A molecular portrait of high-grade ductal carcinoma in situ. Cancer Res. 2015;75(18):3980–3990. doi: 10.1158/0008-5472.CAN-15-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.William WN, Jr, et al. Erlotinib and the risk of oral cancer: The Erlotinib Prevention of Oral Cancer (EPOC) Randomized Clinical Trial. JAMA Oncol. 2016;2(2):209–216. doi: 10.1001/jamaoncol.2015.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garrett WS. Cancer and the microbiota. Science. 2015;348(6230):80–86. doi: 10.1126/science.aaa4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heath JR, Ribas A, Mischel PS. Single-cell analysis tools for drug discovery and development. Nat Rev Drug Discov. 2016;15(3):204–216. doi: 10.1038/nrd.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stadler ZK, et al. Reliable detection of mismatch repair deficiency in colorectal cancers using mutational load in next-generation sequencing panels. J Clin Oncol. 2016;34(18):2141–2147. doi: 10.1200/JCO.2015.65.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogelstein B, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319(9):525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 10.Nikolaev SI, et al. A single-nucleotide substitution mutator phenotype revealed by exome sequencing of human colon adenomas. Cancer Res. 2012;72(23):6279–6289. doi: 10.1158/0008-5472.CAN-12-3869. [DOI] [PubMed] [Google Scholar]
- 11.Sottoriva A, et al. A Big Bang model of human colorectal tumor growth. Nat Genet. 2015;47(3):209–216. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang B, et al. NCI CPTAC Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513(7518):382–387. doi: 10.1038/nature13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drost J, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521(7550):43–47. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- 16.Stachler MD, et al. Paired exome analysis of Barrett’s esophagus and adenocarcinoma. Nat Genet. 2015;47(9):1047–1055. doi: 10.1038/ng.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakr RA, et al. Targeted capture massively parallel sequencing analysis of LCIS and invasive lobular cancer: Repertoire of somatic genetic alterations and clonal relationships. Mol Oncol. 2016;10(2):360–370. doi: 10.1016/j.molonc.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDaniel AS, et al. Next-generation sequencing of tubal intraepithelial carcinomas. JAMA Oncol. 2015;1(8):1128–1132. doi: 10.1001/jamaoncol.2015.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy SJ, et al. Genetic alterations associated with progression from pancreatic intraepithelial neoplasia to invasive pancreatic tumor. Gastroenterology. 2013;145(5):1098–1109. doi: 10.1053/j.gastro.2013.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker BA, et al. Intraclonal heterogeneity is a critical early event in the development of myeloma and precedes the development of clinical symptoms. Leukemia. 2014;28(2):384–390. doi: 10.1038/leu.2013.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrio S, et al. Genomic characterization of high-count MBL cases indicates that early detection of driver mutations and subclonal expansion are predictors of adverse clinical outcome. Leukemia. July 29, 2016 doi: 10.1038/leu.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jasperson KW, Tuohy TM, Neklason DW, Burt RW. Hereditary and familial colon cancer. Gastroenterology. 2010;138(6):2044–2058. doi: 10.1053/j.gastro.2010.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borras E, et al. Genomic landscape of colorectal mucosa and adenomas. Cancer Prev Res (Phila) 2016;9(6):417–427. doi: 10.1158/1940-6207.CAPR-16-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rashid M, et al. Adenoma development in familial adenomatous polyposis and MUTYH-associated polyposis: Somatic landscape and driver genes. J Pathol. 2016;238(1):98–108. doi: 10.1002/path.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samadder NJ, et al. Effect of Sulindac and Erlotinib vs placebo on duodenal neoplasia in familial adenomatous polyposis: A randomized clinical trial. JAMA. 2016;315(12):1266–1275. doi: 10.1001/jama.2016.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vilar E, Tabernero J. Molecular dissection of microsatellite instable colorectal cancer. Cancer Discov. 2013;3(5):502–511. doi: 10.1158/2159-8290.CD-12-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swanton C, McGranahan N, Starrett GJ, Harris RS. APOBEC enzymes: Mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov. 2015;5(7):704–712. doi: 10.1158/2159-8290.CD-15-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Albuquerque C, et al. The ‘just-right’ signaling model: APC somatic mutations are selected based on a specific level of activation of the beta-catenin signaling cascade. Hum Mol Genet. 2002;11(13):1549–1560. doi: 10.1093/hmg/11.13.1549. [DOI] [PubMed] [Google Scholar]
- 29.Knudsen AL, Bisgaard ML, Bulow S. Attenuated familial adenomatous polyposis (AFAP). A review of the literature. Fam Cancer. 2003;2(1):43–55. doi: 10.1023/a:1023286520725. [DOI] [PubMed] [Google Scholar]
- 30.Laken SJ, et al. Familial colorectal cancer in Ashkenazim due to a hypermutable tract in APC. Nat Genet. 1997;17(1):79–83. doi: 10.1038/ng0997-79. [DOI] [PubMed] [Google Scholar]
- 31.Micol JB, Abdel-Wahab O. Collaborating constitutive and somatic genetic events in myeloid malignancies: ASXL1 mutations in patients with germline GATA2 mutations. Haematologica. 2014;99(2):201–203. doi: 10.3324/haematol.2013.101303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Churpek JE, et al. Genomic analysis of germ line and somatic variants in familial myelodysplasia/acute myeloid leukemia. Blood. 2015;126(22):2484–2490. doi: 10.1182/blood-2015-04-641100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kilpivaara O, et al. A germline JAK2 SNP is associated with predisposition to the development of JAK2(V617F)-positive myeloproliferative neoplasms. Nat Genet. 2009;41(4):455–459. doi: 10.1038/ng.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pietra D, et al. JAK2 GGCC haplotype in MPL mutated myeloproliferative neoplasms. Am J Hematol. 2012;87(7):746–747. doi: 10.1002/ajh.23229. [DOI] [PubMed] [Google Scholar]
- 35.Lu C, et al. Patterns and functional implications of rare germline variants across 12 cancer types. Nat Commun. 2015;6:10086. doi: 10.1038/ncomms10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oxnard GR, et al. Screening for germline EGFR T790M mutations through lung cancer genotyping. J Thorac Oncol. 2012;7(6):1049–1052. doi: 10.1097/JTO.0b013e318250ed9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaiswal S, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie M, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwok B, et al. MDS-associated somatic mutations and clonal hematopoiesis are common in idiopathic cytopenias of undetermined significance. Blood. 2015;126(21):2355–2361. doi: 10.1182/blood-2015-08-667063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steensma DP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shalapour S, Karin M. Immunity, inflammation, and cancer: An eternal fight between good and evil. J Clin Invest. 2015;125(9):3347–3355. doi: 10.1172/JCI80007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaneda MM, et al. Macrophage PI3Kγ drives pancreatic ductal adenocarcinoma progression. Cancer Discov. May 13, 2016;6(8):870–885. doi: 10.1158/2159-8290.CD-15-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.West NR, McCuaig S, Franchini F, Powrie F. Emerging cytokine networks in colorectal cancer. Nat Rev Immunol. 2015;15(10):615–629. doi: 10.1038/nri3896. [DOI] [PubMed] [Google Scholar]
- 44.Le DT, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell. 2015;27(4):450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bailey HH, et al. American Society of Clinical Oncology Statement: Human papillomavirus vaccination for cancer prevention. J Clin Oncol. 2016;34(15):1803–1812. doi: 10.1200/JCO.2016.67.2014. [DOI] [PubMed] [Google Scholar]
- 47.Trimble CL. HPV infection-associated cancers: Next-generation technology for diagnosis and treatment. Cancer Immunol Res. 2014;2(10):937–942. doi: 10.1158/2326-6066.CIR-14-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan K, et al. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat Genet. 2015;47(9):1067–1072. doi: 10.1038/ng.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.den Boon JA, et al. Molecular transitions from papillomavirus infection to cervical precancer and cancer: Role of stromal estrogen receptor signaling. Proc Natl Acad Sci USA. 2015;112(25):E3255–E3264. doi: 10.1073/pnas.1509322112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kataoka K, et al. Aberrant PD-L1 expression through 3′-UTR disruption in multiple cancers. Nature. 2016;534(7607):402–406. doi: 10.1038/nature18294. [DOI] [PubMed] [Google Scholar]
- 51.DuPage M, Jacks T. Genetically engineered mouse models of cancer reveal new insights about the antitumor immune response. Curr Opin Immunol. 2013;25(2):192–199. doi: 10.1016/j.coi.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teng MW, Galon J, Fridman WH, Smyth MJ. From mice to humans: Developments in cancer immunoediting. J Clin Invest. 2015;125(9):3338–3346. doi: 10.1172/JCI80004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shankaran V, et al. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410(6832):1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 54.Salavoura K, Kolialexi A, Tsangaris G, Mavrou A. Development of cancer in patients with primary immunodeficiencies. Anticancer Res. 2008;28(2B):1263–1269. [PubMed] [Google Scholar]
- 55.Grulich AE, van Leeuwen MT, Falster MO, Vajdic CM. Incidence of cancers in people with HIV/AIDS compared with immunosuppressed transplant recipients: A meta-analysis. Lancet. 2007;370(9581):59–67. doi: 10.1016/S0140-6736(07)61050-2. [DOI] [PubMed] [Google Scholar]
- 56.Finn OJ, Beatty PL. Cancer immunoprevention. Curr Opin Immunol. 2016;39:52–58. doi: 10.1016/j.coi.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nardy AF, Freire-de-Lima L, Freire-de-Lima CG, Morrot A. The sweet side of immune evasion: Role of glycans in the mechanisms of cancer progression. Front Oncol. 2016;6:54. doi: 10.3389/fonc.2016.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Segal NH, et al. Epitope landscape in breast and colorectal cancer. Cancer Res. 2008;68(3):889–892. doi: 10.1158/0008-5472.CAN-07-3095. [DOI] [PubMed] [Google Scholar]
- 59.Castle JC, et al. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012;72(5):1081–1091. doi: 10.1158/0008-5472.CAN-11-3722. [DOI] [PubMed] [Google Scholar]
- 60.Yadav M, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515(7528):572–576. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 61.Giannakis M, et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Reports. 2016;15:1–9. doi: 10.1016/j.celrep.2016.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Phan VT, et al. Oncogenic RAS pathway activation promotes resistance to anti-VEGF therapy through G-CSF--induced neutrophil recruitment. Proc Natl Acad Sci USA. 2013;110(15):6079–6084. doi: 10.1073/pnas.1303302110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roghanian A, Fraser C, Kleyman M, Chen J. B cells promote pancreatic tumorigenesis. Cancer Discov. 2016;6(3):230–232. doi: 10.1158/2159-8290.CD-16-0100. [DOI] [PubMed] [Google Scholar]
- 64.Keenan BP, et al. A Listeria vaccine and depletion of T-regulatory cells activate immunity against early stage pancreatic intraepithelial neoplasms and prolong survival of mice. Gastroenterology. 2014;146(7):1784–1794. doi: 10.1053/j.gastro.2014.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blaschke K, et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500(7461):222–226. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Incio J, et al. Metformin reduces desmoplasia in pancreatic cancer by reprogramming stellate cells and tumor-associated macrophages. PLoS One. 2015;10(12):e0141392. doi: 10.1371/journal.pone.0141392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eikawa S, et al. Immune-mediated antitumor effect by type 2 diabetes drug, metformin. Proc Natl Acad Sci USA. 2015;112(6):1809–1814. doi: 10.1073/pnas.1417636112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pearce EL, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460(7251):103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thompson E, et al. The immune microenvironment of breast ductal carcinoma in situ. Mod Pathol. 2016;29(3):249–258. doi: 10.1038/modpathol.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schneider RK, et al. Rps14 haploinsufficiency causes a block in erythroid differentiation mediated by S100A8 and S100A9. Nat Med. 2016;22(3):288–297. doi: 10.1038/nm.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Raaijmakers MH, et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature. 2010;464(7290):852–857. doi: 10.1038/nature08851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: Impact on clinical outcome. Nat Rev Cancer. 2012;12(4):298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 73.Cavallo F, De Giovanni C, Nanni P, Forni G, Lollini PL. The immune hallmarks of cancer. Cancer Immunol Immunother. 2011;60(3):319–326. doi: 10.1007/s00262-010-0968-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lutz M, et al. Boost and loss of immune responses against tumor-associated antigens in the course of pregnancy as a model for allogeneic immunotherapy. Blood. 2015;125(2):261–272. doi: 10.1182/blood-2014-09-601302. [DOI] [PubMed] [Google Scholar]
- 75.Domchek SM, et al. Telomerase-specific T-cell immunity in breast cancer: effect of vaccination on tumor immunosurveillance. Cancer Res. 2007;67(21):10546–10555. doi: 10.1158/0008-5472.CAN-07-2765. [DOI] [PubMed] [Google Scholar]
- 76.Dhodapkar MV, et al. Prospective analysis of antigen-specific immunity, stem-cell antigens, and immune checkpoints in monoclonal gammopathy. Blood. 2015;126(22):2475–2478. doi: 10.1182/blood-2015-03-632919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grivennikov SI, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012;491(7423):254–258. doi: 10.1038/nature11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gur C, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–55. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Feng Q, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015;6:6528. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- 80.Belcheva A, et al. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell. 2014;158(2):288–299. doi: 10.1016/j.cell.2014.04.051. [DOI] [PubMed] [Google Scholar]
- 81.Levy J, et al. Intestinal inhibition of Atg7 prevents tumour initiation through a microbiome-influenced immune response and suppresses tumour growth. Nat Cell Biol. 2015;17(8):1062–1073. doi: 10.1038/ncb3206. [DOI] [PubMed] [Google Scholar]
- 82.Yang JY, et al. Enteric viruses ameliorate gut inflammation via toll-like receptor 3 and toll-like receptor 7-mediated interferon-B production. Immunity. 2016;44(4):889–900. doi: 10.1016/j.immuni.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 83.Saeterdal I, et al. Frameshift-mutation-derived peptides as tumor-specific antigens in inherited and spontaneous colorectal cancer. Proc Natl Acad Sci USA. 2001;98(23):13255–13260. doi: 10.1073/pnas.231326898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Garbe Y, Maletzki C, Linnebacher M. An MSI tumor specific frameshift mutation in a coding microsatellite of MSH3 encodes for HLA-A0201-restricted CD8+ cytotoxic T cell epitopes. PLoS One. 2011;6(11):e26517. doi: 10.1371/journal.pone.0026517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schwitalle Y, et al. Immune response against frameshift-induced neopeptides in HNPCC patients and healthy HNPCC mutation carriers. Gastroenterology. 2008;134(4):988–997. doi: 10.1053/j.gastro.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 86.Staffa L, et al. Mismatch repair-deficient crypt foci in Lynch syndrome--molecular alterations and association with clinical parameters. PLoS One. 2015;10(3):e0121980. doi: 10.1371/journal.pone.0121980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yurgelun MB, et al. Microsatellite instability and DNA mismatch repair protein deficiency in Lynch syndrome colorectal polyps. Cancer Prev Res (Phila) 2012;5(4):574–582. doi: 10.1158/1940-6207.CAPR-11-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kloor M, et al. Beta2-microglobulin mutations in microsatellite unstable colorectal tumors. Int J Cancer. 2007;121(2):454–458. doi: 10.1002/ijc.22691. [DOI] [PubMed] [Google Scholar]
- 89.Echterdiek F, et al. Low density of FOXP3-positive T cells in normal colonic mucosa is related to the presence of beta2-microglobulin mutations in Lynch syndrome-associated colorectal cancer. OncoImmunology. 2015;5(2):e1075692. doi: 10.1080/2162402X.2015.1075692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shlien A, et al. Biallelic Mismatch Repair Deficiency Consortium Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat Genet. 2015;47(3):257–262. doi: 10.1038/ng.3202. [DOI] [PubMed] [Google Scholar]
- 91.Aronson M, et al. Gastrointestinal findings in the largest series of patients with hereditary biallelic mismatch repair deficiency syndrome: Report from the International Consortium. Am J Gastroenterol. 2016;111(2):275–284. doi: 10.1038/ajg.2015.392. [DOI] [PubMed] [Google Scholar]
- 92.Bouffet E, et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J Clin Oncol. 2016;34(19):2206–2211. doi: 10.1200/JCO.2016.66.6552. [DOI] [PubMed] [Google Scholar]
- 93.Yurgelun MB, et al. Identification of a variety of mutations in cancer predisposition genes in patients with suspected Lynch syndrome. Gastroenterology. 2015;149(3):604–613. doi: 10.1053/j.gastro.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nik-Zainal S, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534(7605):47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Decker B, et al. Biallelic BRCA2 mutations shape the somatic mutational landscape of aggressive prostate tumors. Am J Hum Genet. 2016;98(5):818–829. doi: 10.1016/j.ajhg.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Strickland KC, et al. Association and prognostic significance of BRCA1/2-mutation status with neoantigen load, number of tumor-infiltrating lymphocytes and expression of PD-1/PD-L1 in high grade serous ovarian cancer. Oncotarget. 2016;7(12):13587–13598. doi: 10.18632/oncotarget.7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.George SH, Milea A, Shaw PA. Proliferation in the normal FTE is a hallmark of the follicular phase, not BRCA mutation status. Clin Cancer Res. 2012;18(22):6199–6207. doi: 10.1158/1078-0432.CCR-12-2155. [DOI] [PubMed] [Google Scholar]
- 98.Sedic M, et al. Haploinsufficiency for BRCA1 leads to cell-type-specific genomic instability and premature senescence. Nat Commun. 2015;6:7505. doi: 10.1038/ncomms8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lim E, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15(8):907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 100.To C, et al. The PARP inhibitors, veliparib and olaparib, are effective chemopreventive agents for delaying mammary tumor development in BRCA1-deficient mice. Cancer Prev Res (Phila) 2014;7(7):698–707. doi: 10.1158/1940-6207.CAPR-14-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nolan E, et al. RANK ligand as a potential target for breast cancer prevention in BRCA1-mutation carriers. Nat Med. 2016;22(8):933–939. doi: 10.1038/nm.4118. [DOI] [PubMed] [Google Scholar]
- 102.Sigl V, et al. RANKL/RANK control Brca1 mutation-driven mammary tumors. Cell Res. 2016;26(7):761–774. doi: 10.1038/cr.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Knisbacher BA, Gerber D, Levanon EY. DNA editing by APOBECs: A genomic preserver and transformer. Trends Genet. 2016;32(1):16–28. doi: 10.1016/j.tig.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 104.Leonard B, et al. The PKC/NF-κB signaling pathway induces APOBEC3B expression in multiple human cancers. Cancer Res. 2015;75(21):4538–4547. doi: 10.1158/0008-5472.CAN-15-2171-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nik-Zainal S, et al. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat Genet. 2014;46(5):487–491. doi: 10.1038/ng.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wen WX, et al. Germline APOBEC3B deletion is associated with breast cancer risk in an Asian multi-ethnic cohort and with immune cell presentation. Breast Cancer Res. 2016;18(1):56. doi: 10.1186/s13058-016-0717-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang T, et al. Evidence of associations of APOBEC3B gene deletion with susceptibility to persistent HBV infection and hepatocellular carcinoma. Hum Mol Genet. 2013;22(6):1262–1269. doi: 10.1093/hmg/dds513. [DOI] [PubMed] [Google Scholar]
- 108.Cescon DW, Haibe-Kains B, Mak TW. APOBEC3B expression in breast cancer reflects cellular proliferation, while a deletion polymorphism is associated with immune activation. Proc Natl Acad Sci USA. 2015;112(9):2841–2846. doi: 10.1073/pnas.1424869112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Caval V, Suspène R, Shapira M, Vartanian JP, Wain-Hobson S. A prevalent cancer susceptibility APOBEC3A hybrid allele bearing APOBEC3B 3'UTR enhances chromosomal DNA damage. Nat Commun. 2014;5:5129. doi: 10.1038/ncomms6129. [DOI] [PubMed] [Google Scholar]
- 110.O'Connell MA, Mannion NM, Keegan LP. The epitranscriptome and innate immunity. PLoS Genet. 2015;11(12):e1005687. doi: 10.1371/journal.pgen.1005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kidd JM, Newman TL, Tuzun E, Kaul R, Eichler EE. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007;3(4):e63. doi: 10.1371/journal.pgen.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lu Y, et al. Most common 'sporadic' cancers have a significant germline genetic component. Hum Mol Genet. 2014;23(22):6112–6118. doi: 10.1093/hmg/ddu312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nan H, et al. Association of aspirin and NSAID use with risk of colorectal cancer according to genetic variants. JAMA. 2015;313(11):1133–1142. doi: 10.1001/jama.2015.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Petridis C, et al. Genetic predisposition to ductal carcinoma in situ of the breast. Breast Cancer Res. 2016;18(1):22. doi: 10.1186/s13058-016-0675-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lowy DR, Collins FS. Aiming high—Changing the trajectory for cancer. N Engl J Med. 2016;374(20):1901–1904. doi: 10.1056/NEJMp1600894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Giordano TJ. The cancer genome atlas research network: A sight to behold. Endocr Pathol. 2014;25(4):362–365. doi: 10.1007/s12022-014-9345-4. [DOI] [PubMed] [Google Scholar]
- 117.Weinstein JN, et al. Cancer Genome Atlas Research Network The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Losman JA, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339(6127):1621–1625. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ortmann CA, et al. Effect of mutation order on myeloproliferative neoplasms. N Engl J Med. 2015;372(7):601–612. doi: 10.1056/NEJMoa1412098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhou J, et al. One-step generation of different immunodeficient mice with multiple gene modifications by CRISPR/Cas9 mediated genome engineering. Int J Biochem Cell Biol. 2014;46:49–55. doi: 10.1016/j.biocel.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 122.Zumwalde NA, et al. Analysis of immune cells from human mammary ductal epithelial organoids reveals Vdelta2+ T cells that efficiently target breast carcinoma cells in the presence of bisphosphonate. Cancer Prev Res (Phila) 2016;9(4):305–316. doi: 10.1158/1940-6207.CAPR-15-0370-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kaufman CK, et al. A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science. 2016;351(6272):aad2197. doi: 10.1126/science.aad2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kato S, Lippman SM, Flaherty KT, Kurzrock R. The conundrum of genetic “drivers” in benign conditions. J Natl Cancer Inst. 2016 doi: 10.1093/jnci/djw036. 108(8).pii:djw036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zaretsky JM, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;37(9):819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rashidian M, et al. Noninvasive imaging of immune responses. Proc Natl Acad Sci USA. 2015;112(19):6146–6151. doi: 10.1073/pnas.1502609112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wellbrock C. Melanoma and the microenvironment–age matters. N Engl J Med. 2016;375(7):696–698. doi: 10.1056/NEJMcibr1606907. [DOI] [PubMed] [Google Scholar]
- 128.Lucia A, Ramirez M. Muscling in on cancer. N Engl J Med. 2016;375(9):892–894. doi: 10.1056/NEJMcibr1606456. [DOI] [PubMed] [Google Scholar]
- 129.Tewari KS, et al. Improved survival with bevacizumab in advanced cervical cancer. N Engl J Med. 2014;370(8):734–743. doi: 10.1056/NEJMoa1309748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Everett JN, et al. Screening for germline mismatch repair mutations following diagnosis of sebaceous neoplasm. JAMA Dermatol. 2014;150(12):1315–1321. doi: 10.1001/jamadermatol.2014.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dudley JC, Lin MT, Le DT, Eshleman JR. Microsatellite instability as a biomarker for PD-1 blockade. Clin Cancer Res. 2016;22(4):813–820. doi: 10.1158/1078-0432.CCR-15-1678. [DOI] [PubMed] [Google Scholar]
- 132.Syngal S, et al. American College of Gastroenterology ACG clinical guideline: Genetic testing and management of hereditary gastrointestinal cancer syndromes. Am J Gastroenterol. 2015;110(2):223–262, quiz 263. doi: 10.1038/ajg.2014.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Newcomb PA, et al. Colon Cancer Family Registry Colon Cancer Family Registry: An international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev. 2007;16(11):2331–2343. doi: 10.1158/1055-9965.EPI-07-0648. [DOI] [PubMed] [Google Scholar]
- 134.Beatty PL, et al. Immunobiology and immunosurveillance in patients with intraductal papillary mucinous neoplasms (IPMNs), premalignant precursors of pancreatic adenocarcinomas. Cancer Immunol Immunother. 2016;65(7):771–778. doi: 10.1007/s00262-016-1838-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Vasen H, et al. Benefit of surveillance for pancreatic cancer in high-risk individuals: Outcome of long-term prospective follow-up studies from three European expert centers. J Clin Oncol. 2016;34(17):2010–2019. doi: 10.1200/JCO.2015.64.0730. [DOI] [PubMed] [Google Scholar]
- 136.Milne RL, Antoniou A. 2016. Modifiers of breast and ovarian cancer risks for BRCA1 and BRCA2 mutation carriers Endocr Relat Cancer. pii:ERC-16-0277.
- 137.Wong TN, et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature. 2015;518(7540):552–555. doi: 10.1038/nature13968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Di Stasi A, Jimenez AM, Minagawa K, Al-Obaidi M, Rezvani K. Review of the results of WT1 peptide vaccination strategies for myelodysplastic syndromes and acute myeloid leukemia from nine different studies. Front Immunol. 2015;6:36. doi: 10.3389/fimmu.2015.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Solomon BM, et al. Risk of non-hematologic cancer in individuals with high-count monoclonal B-cell lymphocytosis. Leukemia. 2016;20(2):331–336. doi: 10.1038/leu.2015.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mesri EA, Feitelson MA, Munger K. Human viral oncogenesis: A cancer hallmarks analysis. Cell Host Microbe. 2014;15(3):266–282. doi: 10.1016/j.chom.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Romero P, et al. The human vaccines project: A roadmap for cancer vaccine development. Sci Transl Med. 2016;8(334):334ps9. doi: 10.1126/scitranslmed.aaf0685. [DOI] [PubMed] [Google Scholar]
- 142.Imperiale TF, Ransohoff DF, Itzkowitz SH. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287–1297. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 143.Nair S, et al. Clonal immunoglobulin against lysolipids in the origin of myeloma. N Engl J Med. 2016;374(6):555–561. doi: 10.1056/NEJMoa1508808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Silvestri GA, et al. AEGIS Study Team A bronchial genomic classifier for the diagnostic evaluation of lung cancer. N Engl J Med. 2015;373(3):243–251. doi: 10.1056/NEJMoa1504601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Im JS, et al. Immune-modulation by epidermal growth factor receptor inhibitors: implication on anti-tumor immunity in lung cancer. PLoS One. 2016;11(7):e0160004. doi: 10.1371/journal.pone.0160004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Corriden R, et al. Tamoxifen augments the innate immune function of neutrophils through modulation of intracellular ceramide. Nat Commun. 2015 doi: 10.1038/ncomms9369. 13;6:8369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Forslund K, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528(7581):262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]