Significance
Image-based approaches to single-cell transcriptomics offer the ability to quantify not only the copy number of RNAs within cells but also the intracellular RNA location and the spatial organization of cells within cultures or tissues. Here we report advances in multiplexed error-robust fluorescence in situ hybridization (MERFISH) that increase the measurement throughput by two orders of magnitude and allow gene expression profiling of ∼40,000 human cells in a single 18-h measurement. This drastic increase in throughput should facilitate the identification and study of rare populations of cells as well as the characterization of transcriptionally distinct cell types within large tissue regions.
Keywords: single-cell analysis, fluorescence, in situ hybridization, transcriptomics, multiplexed imaging
Abstract
Image-based approaches to single-cell transcriptomics, in which RNA species are identified and counted in situ via imaging, have emerged as a powerful complement to single-cell methods based on RNA sequencing of dissociated cells. These image-based approaches naturally preserve the native spatial context of RNAs within a cell and the organization of cells within tissue, which are important for addressing many biological questions. However, the throughput of these image-based approaches is relatively low. Here we report advances that lead to a drastic increase in the measurement throughput of multiplexed error-robust fluorescence in situ hybridization (MERFISH), an image-based approach to single-cell transcriptomics. In MERFISH, RNAs are identified via a combinatorial labeling approach that encodes RNA species with error-robust barcodes followed by sequential rounds of single-molecule fluorescence in situ hybridization (smFISH) to read out these barcodes. Here we increase the throughput of MERFISH by two orders of magnitude through a combination of improvements, including using chemical cleavage instead of photobleaching to remove fluorescent signals between consecutive rounds of smFISH imaging, increasing the imaging field of view, and using multicolor imaging. With these improvements, we performed RNA profiling in more than 100,000 human cells, with as many as 40,000 cells measured in a single 18-h measurement. This throughput should substantially extend the range of biological questions that can be addressed by MERFISH.
Single-cell transcriptomics, powered by next-generation RNA sequencing (RNA-seq), has transformed many aspects of cellular and tissue-scale biology (1–3). This capability has allowed researchers to address exciting questions ranging from the response of single immune cells to antigen (4–6) to the number of transcriptionally distinct cell types and the cellular heterogeneity within complex tissues (7–13). Recent advances in the automated handling of individual cells and the sequencing library preparation for these cells have substantially increased the number of cells that can be routinely characterized with these approaches; notably, state-of-the-art droplet-based RNA-seq approaches provide the ability to quantify the transcriptome of tens of thousands or more cells (14, 15). This throughput allows rare populations of cells to be characterized and transcriptionally distinct cell types within sizable tissue blocks to be mapped.
However, in most approaches to single-cell transcriptomics, cells are dissociated from tissues, and RNAs are extracted from cells; as a result, the native spatial context of these RNAs is lost. However, this spatial information is important for a complete understanding of many biological behaviors (16). For example, the spatial organization of individual cell types within most tissues is crucial to how tissue function or dysfunction arises from the behavior of individual cells. Likewise, the intracellular spatial organization of RNAs is a powerful form of posttranscriptional regulation; thus, it is often important to know not only how many RNA copies are present within a cell but also where they are located within that cell (17). Addressing questions such as these requires spatially resolved approaches to single-cell transcriptomics (16).
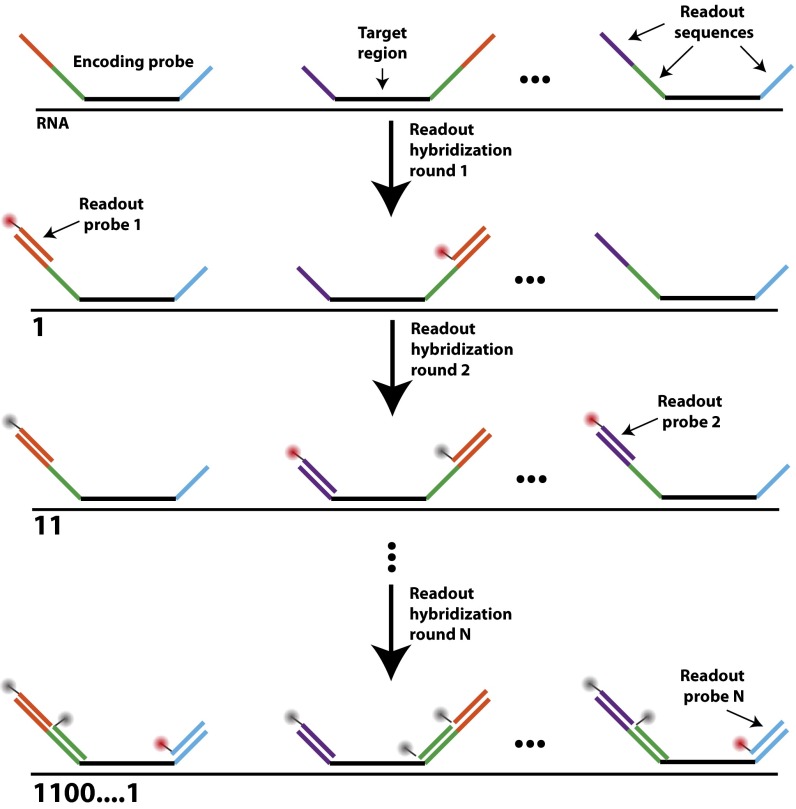
Recently we introduced an image-based approach to spatially resolved, single-cell transcriptomics, multiplexed error-robust fluorescence in situ hybridization (MERFISH) (18). In this approach, RNAs are identified via single-molecule FISH (smFISH) (19, 20), as opposed to alternative in situ methods using sequencing (21, 22). MERFISH uses error-robust barcoding schemes to encode RNA species and reads out these barcodes with sequential rounds of smFISH measurements (Fig. 1A). In our previous implementation of MERFISH (18), RNAs were encoded with binary barcodes and hybridized with complex sets of oligonucleotide probes termed “encoding probes” (Fig. S1). Each encoding probe contains a targeting sequence that binds a given cellular RNA and multiple readout sequences. The collection of readout sequences associated with a cellular RNA corresponds to the barcode of that RNA species. These barcodes then are read out through a series of smFISH measurements; in each round, the sample is stained with a readout probe complementary to one of the readout sequences, the sample is imaged, and the fluorescence signal is extinguished via photobleaching. This process then is repeated with a different readout probe, and the specific on/off pattern of fluorescence observed across multiple smFISH rounds defines the binary barcode (“1”: readout probe bound, “0” readout probe not bound) used to identify each RNA. We use error-robust barcodes that allow measurement errors to be identified and, in some cases, corrected to ensure high-accuracy MERFISH measurements (18). Using this approach we have previously demonstrated the ability to image 140 RNA species with an 80% detection efficiency using 16 rounds of smFISH imaging with an encoding scheme capable of detecting and correcting errors and to image 1,000 RNA species with a 30% detection efficiency with an encoding scheme capable of detecting but not correcting errors (18). In both cases, we were able to quantify the copy number and spatial distribution of these RNAs within ∼100 human fibroblast cells in a single ∼18-h measurement. However, for many biological questions, such as the study of rare populations of cells or the survey of sizeable volumes of tissues, it is highly desirable to increase the throughput of MERFISH so that many more cells can be measured.
Fig. 1.
Approaches to improve the measurement throughput of MERFISH. (A) Simplified schematic of a MERFISH readout protocol. Target RNAs are stained with encoding probes that contain a barcode comprising a combination of readout sequences unique to each RNA species. The barcode then is identified through successive rounds of smFISH, each with a readout probe complementary to one readout sequence. A registered stack of smFISH images for each sample produces an ensemble of fluorescence spots with on/off patterns that define binary barcodes (“1” represents fluorescent signal on, and “0” represents fluorescent signal off) which allow individual RNA species to be identified. A more detailed hybridization and imaging procedure is shown in Fig. S1. (B) The time required to perform a MERFISH experiment for a given sample area for the published protocol (18, 23) that uses photobleaching to remove smFISH signal (red line), a modified protocol without photobleaching (purple line), a modified protocol without photobleaching and a larger FOV (green line), and a modified protocol without photobleaching, a large FOV, and two-color imaging (blue line).
Fig. S1.
Diagram of the hybridization and imaging procedure with encoding and readout probes. Encoding probes are first hybridized to each cellular RNA. Each encoding probe contains a 30-nt target region (black) that binds to the target RNA and three 20-nt readout sequences (purple, green, blue, or orange). The specific choice of readout sequences for a given RNA determines the barcode that will be used to identify it. During each readout hybridization, one readout probe complementary to a given readout sequence (depicted in orange for the first hybridization round) conjugated to a dye (red circle) is hybridized to the sample. The sample is imaged, and the fluorescence signal is eliminated (as indicated by the gray circles). This process is repeated, with a different readout probe hybridized in each of the N rounds of readout hybridization. If the readout probe in a specific round of hybridization is bound to the RNA, we assign “1” to the corresponding bit of the binary barcode of the RNA. Otherwise, a value “0” is assigned to the bit.
Here we present an improved MERFISH method that drastically increases the throughput of this technique, simplifies several aspects of this protocol, and increases the measurement accuracy. With these improvements, we demonstrated the ability to perform spatially resolved gene expression profiling of ∼40,000 cultured human osteosarcoma (U-2 OS) cells in a single 18-h experiment. As a simple illustration of the benefits of this increased throughput, we characterized 130 genes in ∼100,000 cells, identified a subpopulation of cells undergoing DNA replication or cell division, and characterized both the expression profile and the spatial distribution of the cells that comprised this subpopulation.
Results
Increasing the Throughput of MERFISH Measurements.
The total time required for a MERFISH measurement can be divided into an area-dependent time that scales with the total imaged area and an area-independent time that does not. The area-dependent time includes the time required to position, focus, and image each field of view (FOV). In addition, because of the high illumination intensity required to photobleach the fluorescence signals between consecutive rounds of smFISH, each FOV must be photobleached individually; thus, this time is also a part of this area-dependent time. The area-independent time includes buffer-exchange times and incubation times required for sample staining and thus scales with the number of rounds of smFISH that must be performed. Fig. 1B illustrates the scaling of the duration of a MERFISH measurement with the imaged area (red line). For 16 rounds of hybridization and imaging, the total area-independent time amounts to several hours; however, this area-independent time is exceeded by the area-dependent time when the imaged sample area is larger than ∼1 mm2.
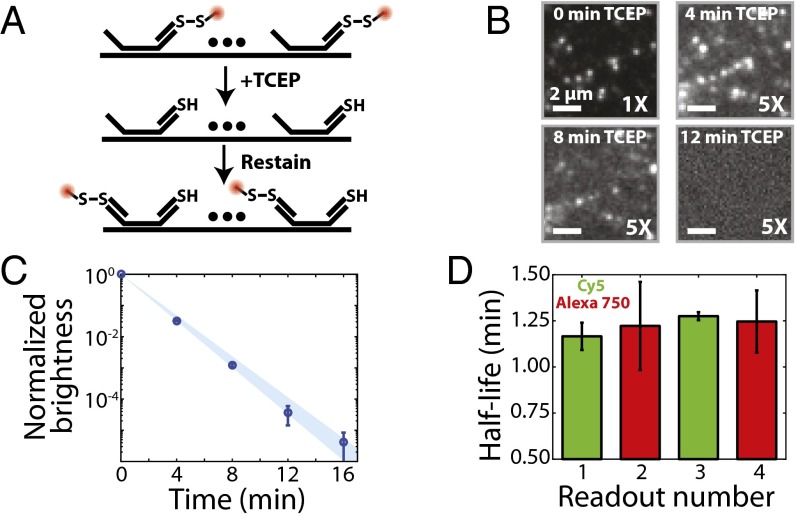
To improve the throughput of MERFISH, we first sought to decrease the area-dependent time. In our previously published MERFISH protocols (18, 23), imaging an FOV of ∼40 × 40 µm required only 0.1 s, but photobleaching of this same FOV required a significantly longer exposure, ∼3 s. Thus, we devised a scheme in which the smFISH signal from the entire sample could be extinguished simultaneously by chemical reaction instead of photobleaching. Specifically, we reasoned that fluorescent dyes conjugated to readout probes via a disulfide linkage could be cleaved from these probes rapidly with a mild reducing agent such as Tris(2-carboxyethyl)phosphine (TCEP) (Fig. 2A).
Fig. 2.
Reductive cleavage of disulfide-linked fluorophores removes the fluorescent signal efficiently. (A) Schematic diagram of the use of TCEP to extinguish the fluorescence signal via cleavage of a disulfide bond linking a fluorescent dye to a readout probe. (B) Images of a region of a human fibroblast (IMR-90) stained with an encoding probe for the FLNA RNA and a readout probe linked to Cy5 via a disulfide bond as a function of time exposed to 50 mM TCEP. Each panel represents the same portion of an FOV. (Scale bars: 2 µm.) Except for the upper left panel, the contrast has been increased fivefold to illustrate better the fluorescent signal remaining in the sample after TCEP treatment. (C) The average brightness of readout probe 1 bound to encoding probes targeting FLNA (normalized to the brightness before TCEP exposure) as a function of the total time of exposure to 50 mM TCEP. Error bars represent SEM (n provided in Fig. S2B), and the blue region represents the 95% confidence interval for a fit to an exponential decay. (D) The measured half-life for the average brightness when exposed to 50 mM TCEP for four readout probes (1–4), each with a different sequence and linked to either Cy5 (green) or Alexa750 (red). Error bars represent the 95% confidence interval for the fit to an exponential decay shown in C for readout probe 1 and in Fig. S2A for readout probes 2–4.
To test this approach, we hybridized encoding probes containing readout sequences to the filamin A (FLNA) mRNA in human lung fibroblast (IMR-90) cells and then stained this sample with a readout probe that was conjugated to a Cy5 dye via a disulfide bond. As expected, the sample exhibited bright fluorescent spots representing individual molecules of the FLNA mRNA, and these fluorescent spots reduced in brightness and eventually disappeared upon treatment with 50 mM TCEP (Fig. 2B). When averaged across thousands of RNAs, the brightness of these spots decayed exponentially (Fig. 2C) with a half-life of 1.17 ± 0.07 min (95% confidence interval). This half-life did not depend on the sequence of the readout probe or the dye to which it was conjugated (Fig. 2D). After ∼15 min of TCEP treatment, the average brightness of each RNA spot and the number of detected RNA spots were reduced by 105-fold and 104-fold, respectively (Fig. 2C and Fig. S2 A and B). Furthermore, the TCEP treatment did not inhibit the ability of the next round of readout probes to bind to the sample (Fig. S2C). Our calculation shows that the use of this chemical approach to remove fluorescence signals between successive rounds of smFISH should reduce measurement time and increase throughput substantially (Fig. 1B, purple line).
Fig. S2.
TCEP cleavage efficiently extinguishes the fluorescence signal from readout probes for different readout sequences and fluorophores. (A) The average brightness of all smFISH spots observed for labeled FLNA mRNAs in human fibroblast (IMR-90) cells as a function of the total time of exposure to cleavage buffer (50 mM TCEP in 2× SSC) for four different readout sequences (blue, green, cyan, and red) and two different fluorophores (Cy5 was conjugated to readouts 1 and 4, and Alexa750 was conjugated to readouts 2 and 3). The readout sequences are provided in Table S1. The brightness values are normalized to the values observed before TCEP treatment (time 0). (B) The fraction of smFISH spots that have a brightness greater than half the brightness determined for a single dye (either Cy5 or Alexa750) as a function of the total exposure time to TCEP cleavage buffer. The colors indicate the readout and dye combinations depicted in A. (C) Representative images of the FLNA mRNA stained with a readout probe corresponding to the first bit (Top), treated with TCEP cleavage buffer for 16 min (Middle), and restained with a readout probe corresponding to the second bit (Bottom). The error bars in A represent SEM based on the number of RNA spots observed at each time point. The numbers of RNA spots observed before TCEP treatment (time 0) were 19,696, 17,644, 20,156, 17,415 for readout probes 1, 2, 3, and 4, respectively. The number of spots determined at all other time points is specified by the survival fraction in B. Missing data points indicate times at which no spots were visible in the sample. (Scale bars: 2 µm.)
Next, we reasoned that, without the requirement for high illumination intensities needed for efficient photobleaching, it should be possible to decrease the area-dependent time further by expanding the size of the imaging FOV. To explore this idea, we designed and constructed a microscope that uses a 2,048 × 2,048 pixel, scientific complementary metal-oxide semiconductor (sCMOS) camera in combination with a high numerical aperture (NA = 1.3) and a high-magnification (60×) silicone oil objective (SI Materials and Methods). We used a silicone oil objective because we found that it had less field curvature than comparable oil immersion 60× objectives. With this optical configuration, we could image an FOV of 223 × 223 µm, an area∼25-fold larger than our previously reported FOV, with an exposure time of 0.5 s. This increase in the size of the FOV should further increase imaging speed, and hence measurement throughput, substantially (Fig. 1B, green line).
As a third step to reduce measurement time, we used multicolor imaging. Specifically, we stained the sample simultaneously with two readout probes per hybridization round, each probe conjugated to one of two spectrally distinct dyes, and used two-color imaging to reduce the number of imaging rounds by half, thereby cutting the area-independent time required to stain, wash, and extinguish signals (Fig. 1B, blue line). We used Cy5 and Alexa750 dyes because of the low cellular autofluorescence observed in the red and near-infrared spectral ranges. In total, the use of reductive cleavage to extinguish fluorescence signal between successive imaging rounds in combination with the increase in the FOV area and the use of two-color imaging should dramatically reduce the time required to perform MERFISH for a given area and increase the sample area that can be measured in a given time (Fig. 1B, blue line vs. red line).
Improving the Performance of MERFISH Measurements.
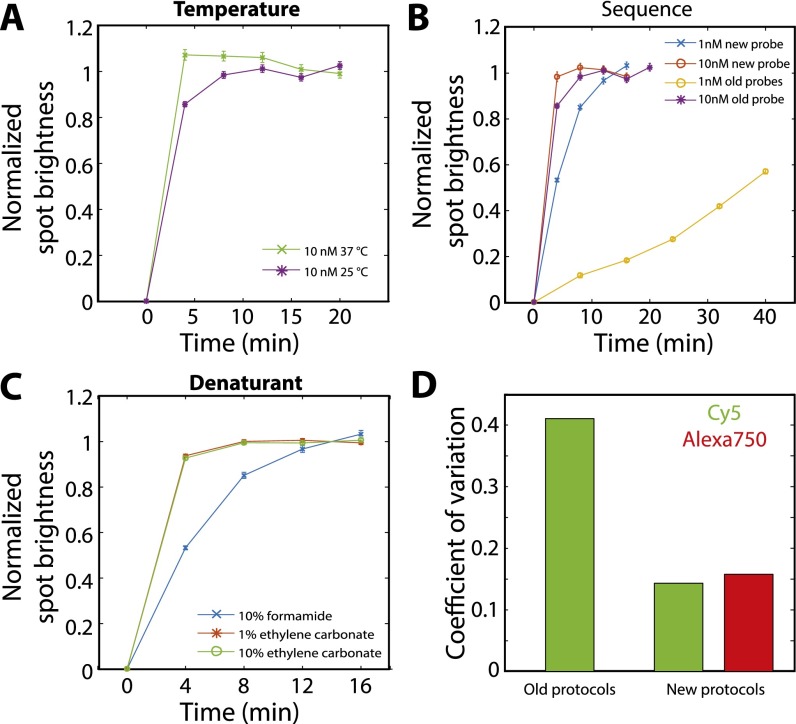
We also made a series of protocol changes aimed at simplifying measurement procedures and improving the robustness of the measurement. First, we found that readout probes can bind to encoding probes at room temperature with rates similar to those observed at 37 °C (Fig. S3A). Room-temperature hybridization avoids any variation in measurement results associated with nonuniform sample heating. Second, we shortened readout probes from 30 to 20 nt, allowing us to include more readout sequences on each encoding probe without increasing the total length of the probe. This modification allows us either to increase the brightness of signals from single mRNA molecules by preserving the number of encoding probes per RNA or to achieve the same signal brightness with fewer encoding probes per RNA, allowing shorter RNAs to be targeted. Third, we created readout probes that bind to readout sequences with rates comparable to those of our previous probes but at 10-fold lower concentrations. Specifically, we exploited the published observation (24) that oligonucleotide sequences that contain only three of the four nucleotides have significantly less secondary structure than sequences that use all four nucleotides and thus have faster hybridization rates (Fig. S3B). Fourth, we replaced the toxic RNA denaturing agent formamide used in the readout hybridization and wash buffers with nontoxic ethylene carbonate (25); we found that this substitution also moderately increased the rate of readout hybridization (Fig. S3C).
Fig. S3.
Characterization of the hybridization properties of different readout probes and different hybridization conditions. (A) The average normalized smFISH spot brightness for FLNA molecules labeled first with encoding probes and then with readout probes vs. the total time the sample is exposed to 10 nM of readout probes at 37 °C (green crosses) or at room temperature (25 °C; purple stars). The sequence of the readout probe is CGCAACGCTTGGGACGGTTCCAATCGGATC, which is one of our previously published readout probe sequences. The hybridization buffer is our previously published, formamide-based hybridization buffer (18, 23). (B) The average normalized smFISH spot brightness as in A but with the sample stained with 10 nM of a previously published 30-nt four-letter readout probe (purple stars; reproduced from A), 10 nM of a 20-nt three-letter readout probe (ATCCTCCTTCAATACATCCC) that does not contain G (red circles), 1 nM of the previously published 30-nt four-letter readout probe (orange circles), or 1 nM of a 20-nt three-letter readout probe (blue crosses). Hybridization was conducted at room temperature in the formamide-based buffer. (C) The average normalized smFISH spot brightness as in A for 1 nM of a 20-nt three-letter readout probe hybridized at room temperature but using different buffers: a hybridization buffer containing 10% formamide as described previously (18, 23) (blue crosses, reproduced from B), a hybridization buffer in which formamide was replaced with 1% (vol/vol) ethylene carbonate (red stars), or a hybridization buffer with 10% (vol/vol) ethylene carbonate (green circles). (D) The coefficient of variation (the SD divided by the mean) for the average brightness of smFISH spots across all rounds of imaging in the 16-bit MERFISH experiment conducted with the previously published 30-nt readout probes and formamide-based hybridization protocol (18, 23) (old protocols) and with the readout protocols published here (new protocols; 20-nt 3-letter readout sequence and an ethylene-carbonate–based hybridization protocol). Error bars in A–C represent SEM across all measured RNA spots; more than 10,000 RNA spots were measured for each data point.
We also found that these modified readout probes and readout hybridization protocols improved MERFISH performance by reducing the variance in staining quality among different rounds of readout hybridization as compared with our previous protocols (Fig. S3D). Of the multiple changes made above, the modified readout sequences likely account for the majority of this improvement, because we previously have observed that some of the variability across different readout staining rounds (Fig. S3D) can be attributed to sequence variations, presumably resulting from unanticipated secondary structures. By design, such secondary structures should be far less likely with the modified readout sequences that use only three of the four nucleotides (24). We anticipate these improvements will increase the accuracy of our MERFISH measurements because lower-quality (or varying-quality) readout hybridizations can result in dim fluorescence signals in some imaging rounds and increase the rate at which readout errors are made.
An Image Analysis Algorithm to Handle High-Throughput MERFISH Data.
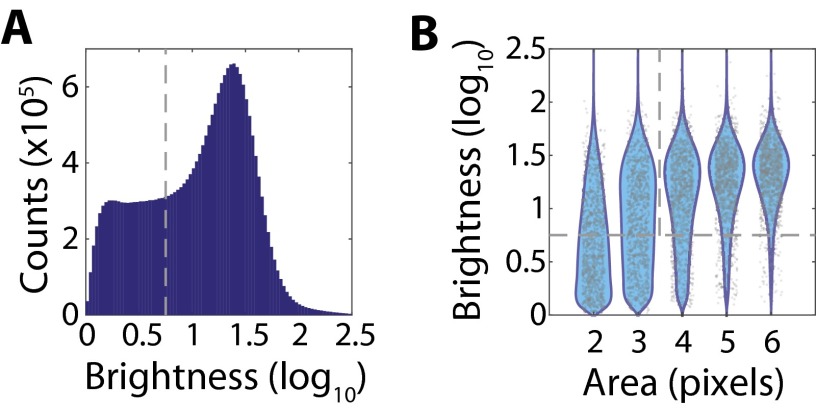
In parallel, we anticipated that our previous computational methods for MERFISH data analysis (18, 23), which typically required several hours to a day to analyze a single MERFISH dataset, would not be adequate for analyzing the two orders of magnitude higher data volume generated per experiment. Thus, we developed an analysis pipeline capable of handling this drastic increase in imaging throughput (SI Materials and Methods). The major advance in this pipeline is the adoption of a pixel-based decoding approach, as opposed to a spot-finding approach, to reduce computation time. Briefly, images of the same FOV from different imaging rounds are registered using images of fiducial beads collected in each round. These images are high-pass filtered to remove background and are deconvolved to sharpen and better resolve closely positioned spots. Previously we observed that signals from the same RNA often varied in position from round to round by ∼100 nm (18). Thus, to connect signals from one round to another more accurately, we applied a low-pass filter with a kernel size of 100-nm radius. The intensities of each pixel across all 16 rounds of images then were used to form a 16-dimensional vector, which we normalized to unit amplitude. This vector then was compared with the set of unit vectors defined by all valid barcodes. The pixel was assigned to a given barcode if the Euclidean distance between its normalized intensity vector and the closest barcode vector was less than the distance defined by a single-bit error. Contiguous sets of pixels that matched to the same barcode were combined to form a single detected RNA. Background pixels mistakenly matched to a barcode were identified and removed based on their low brightness and small number of contiguous pixels matched to the same barcode (Fig. S4). With this pipeline, analysis of large MERFISH datasets (∼40 mm2 with ∼40,000 human cells) can be completed in 2–3 d using multiple cores on a computer cluster.
Fig. S4.
Thresholding of RNA signals based on area and brightness. (A) Histogram of the log10 brightness for all observed single-RNA-molecule signals from the data presented in Fig. 3. The gray dashed line defines the brightness threshold used to discard dim single-molecule signals that likely represent background rather than real RNA signals. (B) Scatter plot of the observed log10 brightness for single-molecule signals with a given area (gray markers), i.e., the number of contiguous pixels assigned to the same RNA molecule, with the associated probability distributions (cyan). For clarity, only 1,000 randomly selected single-molecule signals are plotted for each area. Note that single-molecule signals with smaller areas also tend to be low brightness. The gray dashed lines represent the cuts applied to separate spurious background signals from foreground RNA signals, i.e., a brightness greater than 100.75 and an area of four pixels or larger.
High-Throughput MERFISH Measurements of Tens of Thousands of Cells.
To demonstrate the substantial increase in imaging throughput made possible by the above advances, we measured 130 RNAs in cultured U-2 OS cells with a previously published 16-bit, modified Hamming distance-4 (MHD4) encoding scheme (18). In this encoding scheme, all barcodes used are separated by a Hamming distance of at least 4, and hence at least four bits must be read incorrectly to change one valid barcode to another. Therefore, every single-bit error produces a barcode uniquely close to a single valid barcode, allowing such errors to be detected and corrected. Two-bit errors also can be detected but are not correctable because the resulting barcode is no longer uniquely close to a single valid barcode. To account further for the fact that it is more likely to miss a hybridization event (1-to-0 error) than to misidentify a background spot as an RNA (0-to-1 error) in smFISH measurements, our MHD4 code contains a constant and relatively low number (four) of “1” bits. This 16-bit MHD4 encoding scheme includes 140 distinct barcodes in total (18). We assigned 130 of these barcodes to different RNA species, leaving 10 barcodes unused to serve as blanks (not corresponding to any RNA) for misidentification controls.
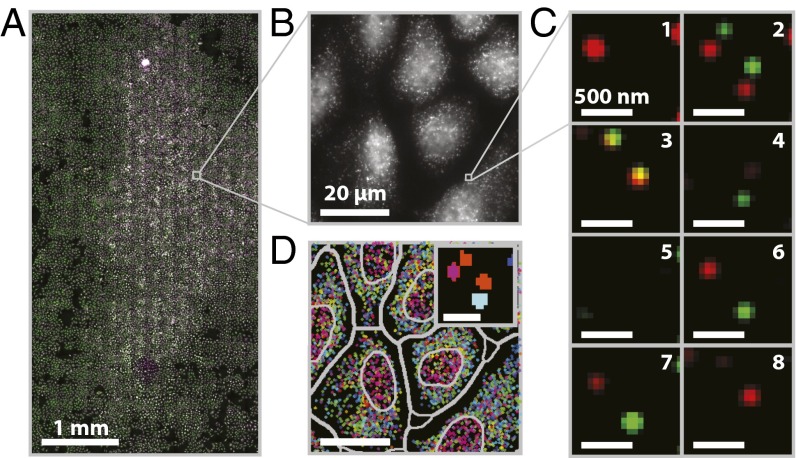
Fig. 3A illustrates one such measurement over an area of 3.2 × 6.2 mm. The cells were fixed, permeabilized, and labeled with encoding probes to 130 RNA species. We then performed eight rounds of hybridization, imaging, and TCEP cleavage with 16 different readout probes; each round of imaging used two readout probes conjugated to Cy5 and Alexa750, respectively. Single-molecule spots were clearly observed across the entire imaged area in both Cy5 and Alexa750 channels in each round of smFISH staining and imaging (Fig. 3 B and C). The identities of individual RNA molecules then were decoded via the algorithm described above (Fig. 3D). To assign RNAs to individual cells, we used DAPI to identify cell nuclei and the local density of RNAs to define cellular boundaries (SI Materials and Methods). In total, Fig. 3 contains 15,181 cells. Among these, 12,607 segmented cells satisfied our conservative criteria for cell morphology designed to eliminate segmentation errors (SI Materials and Methods), and these properly segmented cells contained 9.7 million identified RNA molecules.
Fig. 3.
A MERFISH measurement of an ∼20 mm2 sample area (∼15,000 cells). (A) Mosaic image of a 3.2 × 6.2 mm region of cultured U-2 OS cells stained with DAPI (purple), encoding probes for 130 RNAs and a Cy5-labeled readout probe (green). (Scale bar: 1 mm.) (B) Image of the Cy5 channel in the first round of readout hybridization for the small portion of the field in A marked by the gray square. (Scale bar: 20 µm.) (C) Two-color images of the smFISH stains for all eight rounds of hybridization and imaging for the small portion of the field in B marked by the gray square after the application of a high-pass filter to remove background, deconvolution to tighten spots, and a low-pass filter to connect spots in different images more accurately (SI Materials and Methods). Green, red, and orange represent the Cy5 channel, the Alexa750 channel, and the overlay between the two, respectively. (Scale bars: 500 nm.) (D) The decoded barcodes for the region shown in B. Spots represent individual molecules color-coded based on their RNA species identities (barcodes). Both the nuclear boundaries and the boundaries used to assign RNAs to individual cells are depicted (gray). (Scale bar: 20 µm.) (Inset) An image of the barcode assignment (indicated by color) for each pixel in the images shown in C. (Scale bar: 500 nm.)
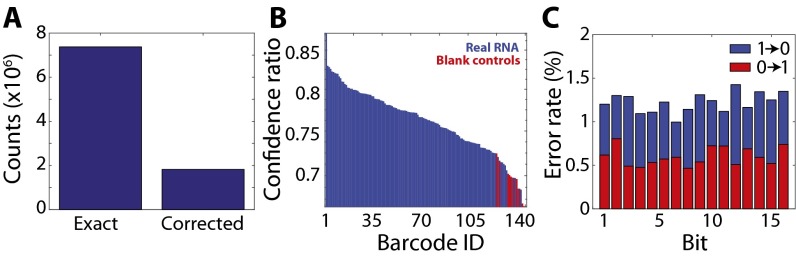
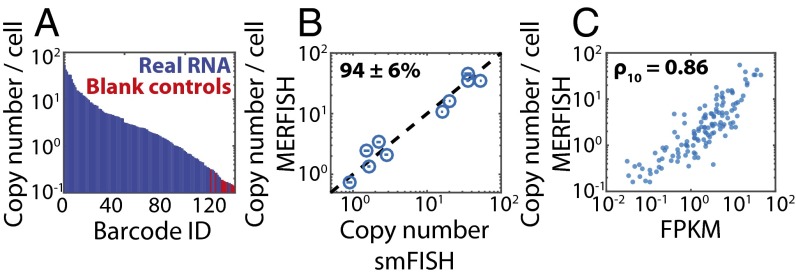
To determine the RNA decoding quality, we considered two types of errors for each RNA species. First, some RNAs can be misidentified as the wrong species, leading to a nonzero misidentification rate. Second, some RNAs can be missed, leading to a non-100% calling rate. To assess these errors, we first examined the fraction of decoded RNAs that required error correction (Fig. S5A). In our previous published MERFISH experiments using the same 16-bit MHD4 code, we observed that ∼60% of all decoded RNAs required error correction (18). By contrast, with the protocols described here, only ∼20% of RNAs required correction. Lower levels of error correction would suggest a lower level of misidentification and a higher calling rate. To test the level of misidentification, we examined the number of times that the blank barcodes were counted. Indeed, these barcodes were counted relatively infrequently: 120 of the 130 (92%) RNA species were counted more frequently than the most abundant blank barcode (Fig. 4A). In addition, we used an alternative metric, the confidence ratio, to assess the misidentification rate further. As previously defined (18), the confidence ratio for each measured barcode was determined as the number of RNA molecules exactly matching this barcode over the total number of exact matches and matches with single-bit errors for this barcode. We have previously shown that blank barcodes tend to have lower confidence ratio values than RNA-encoding barcodes (18). Indeed, here we found that 95% of the 130 RNA species had a confidence ratio higher than the maximum confidence ratio observed for the blank barcodes (Fig. S5B). Next, to examine the calling rate of these measurements, we first used the frequency with which errors were corrected at each bit to determine the average per-bit error rate, as described previously (18). Previously we observed an average 1-to-0 error rate of ∼10% and an average 0-to-1 error rate of ∼4% (18). By contrast, the MERFISH protocol described here produced substantially lower per-bit error rates, namely, a 1-to-0 error rate of ∼1% and a 0-to-1 error rate of ∼0.5% (Fig. S5C). With these per-bit error rates we would predict a very high calling rate of ∼99%. To assess the calling rate experimentally, we determined the copy numbers of 10 different RNAs using conventional smFISH and compared them with our MERFISH results. We found that the average copy number per cell for these 10 RNAs determined with MERFISH correlated strongly with the values determined via smFISH (Fig. 4B). Moreover, the average ratio of copy numbers between the MERFISH and smFISH measurements was 0.94 ± 0.06 (SEM; n = 10), consistent with the high calling rate estimated from our observed per-bit error rates. Together, these metrics indicate a moderately lower misidentification error rate and higher calling rate than obtained with our previous lower-throughput MERFISH measurements (18).
Fig. S5.
Additional metrics to evaluate the performance of MERFISH measurements. (A) The total number of RNAs decoded without (Exact) and with (Corrected) error correction. (B) The confidence ratio for all barcodes representing real RNAs (blue) and the blank controls (red) sorted from largest to smallest value. The confidence ratio for any given gene (or barcode) is defined as the ratio between the number of exact matches to this barcode and the total number of exact matches to this barcode plus matches with single-bit errors. Of the 130 barcodes encoding real RNAs, 123 have a confidence ratio larger than that of the largest confidence ratio of the blank barcodes. (C) The error rate (the fraction of measured barcodes that contain a given bit flip) for each bit. Both 1-to-0 error rates (blue) and 0-to-1 error rates (red) are shown for each bit. The data presented in this figure represent the error properties of the dataset presented in Fig. 3 and are representative of those observed for all other datasets.
Fig. 4.
Performance of the high-throughput MERFISH measurements. (A) The average RNA copy numbers per cell measured in Fig. 3 sorted from largest to smallest abundance. Barcodes assigned to real RNAs are marked in blue, and those not assigned to RNAs, i.e., blank controls, are marked in red. (B) The average RNA copy numbers per cell determined via MERFISH vs. that determined via conventional smFISH for 10 of the 130 RNAs. The dashed line represents equality. The average ratio of counts determined by MERFISH to that determined by smFISH indicates a calling rate (mean ± SEM) of 94 ± 6% (n = 10). Plotted error bars represent the SEM across the number of measured cells (>300 cells) for each gene measured via smFISH. (C) The average RNA copy number per cell determined by MERFISH vs. the abundance as determined by bulk sequencing. The Pearson correlation coefficient between the log10 values (ρ10) is 0.86 with a P value of 6 × 10−39. FPKM, fragments per kilobase per million reads.
We further compared the average copy number per cell determined by MERFISH with that determined from published bulk RNA-seq for U-2 OS cells (26). The values determined by MERFISH correlated with those determined from RNA-seq with a high Pearson correlation coefficient for the logarithmic abundances (ρ10 = 0.86) (Fig. 4C).
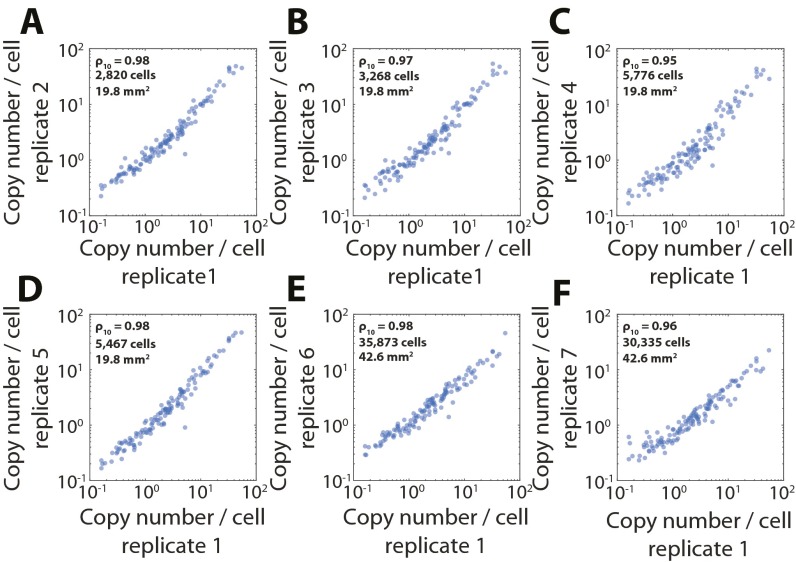
Finally, to demonstrate the reproducibility of these high-throughput measurements, we performed MERFISH measurements for a range of confluencies of cells and for two different sample areas, ∼20 mm2 and ∼40 mm2. Fig. S6 shows that the average RNA copy number determined by each of these measurements correlated strongly with those determined by the measurement presented in Fig. 3 (ρ10 ≥ 0.95). Across all seven measurements we observed an average calling rate of 90% ± 10% (SEM across seven replicate measurements) by comparison with smFISH results. In total, we measured 105,966 cells with 87,632 cells segmented. The largest of these datasets contained 39,523 cells (35,873 segmented) in an area of 40 mm2 measured in less than 18 h. This throughput represents a 250-fold increase in the sample area imaged in a single 18-h measurement relative to previously published throughputs (18) and, because the U-2 OS cells used here are smaller than the IMR-90 cells used previously, a nearly 400-fold increase in the number of measured cells.
Fig. S6.
Reproducibility of high-throughput MERFISH measurements. (A) The average RNA copy number per cell for a replicate MERFISH measurement vs. that shown in Fig. 3. ρ10 represents the Pearson correlation coefficient between the log10 copy numbers. (B–F) As in A but for five additional MERFISH measurements. The strong correlation between these values shows the high reproducibility of MERFISH measurements. The number of segmented cells and the total imaged area are also listed for replicates 2–7. The number of segmented cells and the total imaged area for replicate 1 is described in the main text. The P values for all Pearson correlation coefficients are less than 1 × 10−71.
Characterization of a Subpopulation of Cells.
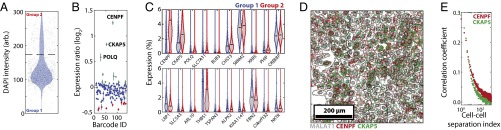
One advantage of the significantly enhanced throughput is the ability to image potentially rare or transient subpopulations of cells with sufficient statistics to characterize the properties of such subpopulations. As a simple illustration of this ability, we identified a subpopulation of cells undergoing DNA replication or cell division in the three datasets collected at the highest confluency (total 78,815 cells). To identify this subpopulation, we determined the distribution of DAPI signal intensity observed in individual cells (Fig. 5A). A local minimum in this distribution divided the cells into two groups: group 1 cells contained lower DAPI levels, and group 2 cells contained roughly twice the DAPI signal of group 1 cells, suggesting that group 2 contained cells undergoing DNA replication or cell division. Group 2 represented a relatively small population of ∼20% of the measured cells; nonetheless, because of the large number of cells measured, this population contained 16,036 cells. To identify how the transcriptional profile of these 130 genes differed between group 2 and group 1, we determined for each gene a fractional expression level defined as the copy number of this RNA divided by the total copy number of all 130 RNAs detected in the cell. Fig. 5B displays the ratio of this fractional expression level in group 1 vs. group 2 cells for each gene, showing that some genes were up-regulated and some were down-regulated in group 2 cells. The large number of cells measured here allowed us to distinguish even small changes in expression levels with confidence. Fig. 5C plots the observed distribution of expression levels for both groups for the 10 most up-regulated (Fig. 5C, Upper) and 10 most down-regulated (Fig. 5C, Lower) genes. The most up-regulated genes included the centromere-binding protein CENPF, the spindle-binding protein CKAP5, the DNA polymerase POLQ, and the mitotic checkpoint protein BUB2, supporting the association of group 2 with cells undergoing DNA replication or cell division. Interestingly, the expression of these genes, in particular CKAP5 and CENPF, also could be used to identify this subpopulation of cells without the DAPI signal information. The set of the most down-regulated genes included thrombosin (THBS1), fibrillin (FBN2), and tetraspanin (TSPAN3) as well as other genes involved in cell–cell interactions and adhesion. We speculate that the differential regulation of these proteins might facilitate the disruption and reformation of cell–cell interactions that must occur during cell division.
Fig. 5.
Characterizing the expression differences of a subpopulation of cells undergoing DNA replication or cell division. (A) Violin plot of the distribution of total DAPI intensity for individual cells. The dashed line defines the intensity threshold (based on a local minimum) used to group cells with low DAPI signal into group 1 and cells with high DAPI signal into group 2. Gray dots indicate the values for individual cells, and the blue-shaded area represents the probability distributions. For clarity, only 1,000 randomly selected cells are displayed. (B) The log2 ratio of the mean fractional expression level of each RNA species in group 2 relative to that of group 1. The fractional expression level for an RNA species is defined as the copy number of that RNA divided by the total copy number of all 130 RNAs detected in the cell. The mean and SEM are computed across three biologic replicates. Green and red markers indicate genes further examined in C. (C) Violin plots of the distribution of expression levels for individual genes within group 1 (blue) or group 2 (red) for the 10 genes with the largest magnitude of up-regulation (Upper; marked green in B) or the 10 genes with the largest magnitude of down-regulation (Lower; marked red in B) in group 2 relative to group 1. The solid black lines represent the mean, and the colored curves represent the probability distributions. The gray dots represent the expression levels for 1,000 randomly selected cells. (D) A small region of one dataset showing the location of MALAT1 (gray), CENPF (red), and CKAP5 (green). The gray lines represent the boundaries of cells (segmented based on the density profile of all 130 measured RNAs). Note that MALAT1 clearly defines the nucleus. (E) The Pearson correlation coefficient for the relative expression of CENPF (red) or CKAP5 (green) observed between pairs of cells separated by various distances. The cell–cell separation index is defined as 1 for any given cell and its nearest neighbor, 2 for any given cell and its second closest neighbor, and so forth. These correlations were calculated for all cells within each of the three datasets and then were averaged across these datasets.
Finally, to illustrate the power of a spatially resolved measurement, we investigated the spatial distribution of the group 2 cells. To probe this organization, we examined the copy numbers of CKAP5 and CENPF, the two RNAs most up-regulated in group 2 cells (Fig. 5D). As expected, we found that the expression levels of these RNAs were highly correlated and varied significantly among cells. Moreover, Fig. 5 D and E reveals that neighboring cells tended to express a similar level of these RNAs. Such spatial correlations could have been caused by a variety of potential mechanisms, e.g., neighboring cells likely share a common progenitor, resulting in an apparent synchronization of their cell cycles, or there may have been local cues that promoted or repressed cell division. The ability to reveal these cellular-scale spatial organizations directly is one of the benefits of an image-based approach to single-cell transcriptomics.
SI Materials and Methods
Encoding Probe Design.
Each encoding probe comprises two priming regions, multiple readout sequences, and a target region. To design the target regions, we developed a computational pipeline that requires significantly less computational cost than the methods we previously used (18, 23, 34). The central challenge in targeting region design was determining the optimal specificity of the probes, e.g., how narrow or broad the range of local GC content or melting temperature (TM) should be. The specificity should be as high as possible to promote specific binding while maintaining conditions permissive enough to produce ample target regions for all desired genes. Our previous computational approach was inefficient, in part because basic calculations of specificity properties, e.g., GC content or TM, were reperformed whenever a new set of stringency conditions was probed. To address this issue, we developed an alternative approach in which we precalculated properties of the transcriptome that allow the rapid calculation of specificity parameters, namely, local GC content, TM, relative abundance of potential off-target binding partners, and relative specificity of a given probe to a given isoform of a gene. This approach reduced the time required to design probes for a given set of stringency conditions from days to minutes.
Specifically, using the human transcriptome from the genome build hg38 (ensembl.org/Homo_sapiens/Info/Index), we calculated the local GC content and the local nearest neighbor thermodynamic properties (entropy and enthalpy) using the parameters defined previously by SantaLucia et al. (35). From the predetermined local nearest neighbor thermodynamic properties, the TM for any length of probe could be computed rapidly, assuming a monovalent salt concentration of 300 mM (the concentration of NaCl in the encoding and hybridization readout buffers) and a probe concentration of 5 nM. In addition, we created a series of look-up tables that allowed us to calculate rapidly a penalty for off-target binding for each potential target region. First, we created a look-up table for each gene comprising all the unique 17-nt sequences present in all of the isoforms of that gene with the penalty associated with each sequence defined as the sum of the abundance of each isoform determined from RNA-seq (see the section RNA-Seq below) in which that sequence appeared. When the same 17-nt sequence appeared multiple times in the same isoform, each appearance contributed to this penalty. We termed these tables “isoform penalty tables.” Second, we created a look-up table comprising the same penalty terms but for the entire transcriptome. We termed this table the “transcriptome penalty table.” For both types of penalty tables, we chose 17-nt homology sequences to balance the desire to eliminate short regions of homology with the dramatic increase in the frequency of such regions in the transcriptome as this length is decreased. Finally, because some noncoding RNAs (ncRNAs) are far more abundant than coding RNAs and thus could contribute more significantly to background, we calculated an additional penalty table corresponding to the number of times all unique 15-nt sequences appear in the set of human rRNA and tRNAs as well as the human mitochondrial rRNAs and tRNAs (GRCh38, ncRNA: ftp://ftp.ensembl.org/pub/release-84/fasta/homo_sapiens/ncrna/); we termed this table the “ncRNA penalty table.” We decreased the homology length for off-target binding to these ncRNAs to increase the stringency of selection against partial homology against these highly abundant RNAs.
Using the isoform and transcriptome penalty tables, we calculated two quantities for each 17-nt region in each transcript—an isoform-specificity index and a gene-specificity index. The isoform-specificity index for each 17-nt region within every transcript was calculated by dividing the measured abundance of the given isoform (i.e., the abundance of the correct target) by the isoform-specific penalty for that sequence as determined by the isoform penalty table for that gene (i.e., the sum of the abundance of the correct target plus all potential off-targets in other isoforms). This value varies between 0 and 1 and can be roughly thought of as the fraction of probes that contain the given 17-nt sequence that would bind to this given isoform out of those that could bind to any isoform derived from that gene. This quantity is most likely an underestimate of that fraction because it is unlikely that a probe would bind to a 17-nt region of homology with the same affinity as to the full-length (30-nt) target of the probe. The gene-specificity index for each 17-nt sequence in a given transcript was calculated by dividing the penalty associated with this 17-nt sequence derived from the specific isoform penalty table for that gene (i.e., the abundance of that 17-nt sequence in all isoforms of that gene) by the penalty for that sequence derived from the transcriptome penalty table (i.e., the abundance of that sequence in all transcripts, which includes all isoforms of the target gene as well as all other transcripts). Again, this quantity varies between 0 and 1 and can be roughly thought of as the fraction of a potential probe containing a given 17-nt region that would bind to any of the isoforms of a given gene as opposed to any other member of the transcriptome. The specificity indices for individual target regions, which were 30-nt long, were derived by averaging the isoform and gene specificity indices for all 17-nt sequences within each potential target region.
Using the GC and thermodynamic annotations in conjunction with these specificity indices, we calculated the GC content, TM, and isoform- and gene-specificity indices for all possible 30-nt target regions as well as the frequency of homology regions in ncRNAs and then chose a subset of target regions based on the desired ranges for each of these quantities. From these chosen target regions, we identified nonoverlapping regions starting with the first valid target region at the 5′ end of each isoform. Computationally, construction of the penalty tables and GC and thermodynamic annotations for the transcriptome was slow, requiring a few hours on a desktop computer running in parallel on multiple cores. However, once these annotations were computed, construction of target regions for a given range of stringencies, e.g., TM, GC, specificity index ranges, and so forth, required only ∼5–10 min. Thus, we were able to screen a wide range of stringency ranges and identify the set of parameters that provided the narrowest, most stringent conditions on the target regions while still producing enough target regions for the desired set of transcripts. For the library reported here, the target regions used were designed with a GC range of 43–63%, a TM range of 66–76 °C, an isoform-specificity index range of 75–100%, an gene-specificity index range of 75–100%, and no regions of homology longer than 15 nt to human rRNAs or tRNAs or to mitochondrial rRNAs and tRNAs (calculated using the ncRNA penalty table). The used target regions are provided as part of the encoding probes in Dataset S1. All calculations were performed in MATLAB with custom functions and scripts which are available upon request.
The 20-nt, three-letter readout sequences were designed by generating a random set of sequences with the per-base probability of 25% for A, 25% for T, and 50% for G. Sequences generated in this fashion can vary in their nucleotide content. To eliminate outlier sequences, we kept only sequences with a GC content between 40 and 50%. In addition, sequences with internal stretches of G longer than 3 nt were removed to eliminate the presence of G-quadruplets, which can form secondary structures that inhibit synthesis and binding. To remove the possibility of significant cross-binding between these readout sequences, we used a published algorithm (36) to identify a subset of these sequences with no cross-homology regions longer than 11 contiguous bases. We then used BLAST (33) to identify and eliminate sequences with contiguous homology regions longer than 11 nt to the human transcriptome. From the readout sequences satisfying the above requirements, 16 were selected. The corresponding readout probes, i.e., the reverse complement of these readout sequences, are provided in Table S1.
Table S1.
Readout probe sequences
| Bit | Readout probe name | Sequence | Dye |
| 1 | RS0015 | ATCCTCCTTCAATACATCCC | Cy5 |
| 2 | RS0083 | ACACTACCACCATTTCCTAT | Alexa750 |
| 3 | RS0095 | ACTCCACTACTACTCACTCT | Alexa750 |
| 4 | RS0109 | ACCCTCTAACTTCCATCACA | Cy5 |
| 5 | RS0175 | ACCACAACCCATTCCTTTCA | Cy5 |
| 6 | RS0237 | TTTCTACCACTAATCAACCC | Alexa750 |
| 7 | RS0247 | ACCCTTTACAAACACACCCT | Cy5 |
| 8 | RS0255 | TCCTATTCTCAACCTAACCT | Alexa750 |
| 9 | RS0307 | TATCCTTCAATCCCTCCACA | Alexa750 |
| 10 | RS0332 | ACATTACACCTCATTCTCCC | Cy5 |
| 11 | RS0343 | TTTACTCCCTACACCTCCAA | Cy5 |
| 12 | RS0384 | TTCTCCCTCTATCAACTCTA | Alexa750 |
| 13 | RS0406 | ACCCTTACTACTACATCATC | Cy5 |
| 14 | RS0451 | TCCTAACAACCAACTACTCC | Alexa750 |
| 15 | RS0468 | TCTATCATTACCCTCCTCCT | Alexa750 |
| 16 | RS0548 | TATTCACCTTACAAACCCTC | Cy5 |
The dye was attached to each readout probe via a disulfide bond at the 5′ end of the listed probe sequences.
To construct the library of encoding probes, we first selected a set of target RNAs (130 genes) drawn from the human transcriptome. We chose 85 of the genes used in our previous 140-gene MERFISH library (18) and then selected the remaining 45 genes at random from those expressed in the range of 10−1 to 103 FPKM. We assigned to each RNA a unique barcode drawn from the same 16-bit MHD4 code that we used previously (18). This code, which has a Hamming distance of 4 and a constant Hamming weight (i.e., the number of “1”’ bits per barcode) of 4, contains 140 barcodes. We randomly assigned one of the 140 barcodes to each of the 130 RNA species and left the remaining 10 as blank controls. Ninety-two putative encoding probes were created for each gene; each of the encoding probes contained a target region that was randomly selected from the target regions of the gene and three readout sequences that were randomly selected from the four readout sequences associated with the gene. These readout sequences were concatenated with the target regions in one of two randomly selected configurations, i.e., with one or two readout sequences at the 5′ end of the target region and with the remaining sequences at the 3′ end of the target region. Additional adenosine nucleotide spacers were added between readout sequences and target regions to prevent terminal G triplets in the readout sequences and to prevent target regions from combining with Gs from adjacent sequences to form G quadruplets. Priming regions for the amplification of these probes were designed by randomly generating a set of 20-nt sequences, selecting those with a TM in the range of 70–72 °C, a GC content in the range of 50–65%, no contiguous region of four or more of the same base, and no region of self-complementarity longer than 6 nt. A final set of orthogonal primers then was designed as described previously (36) with the requirement that there be no region of cross-homology longer than 8 nt. Two primers were drawn at random from these sequences and added to the 5′ and 3′ of each encoding probe. Finally, this set of putative probes was screened for any additional homology to human rRNA or tRNA or to mitochondrial rRNA or tRNA, using the same approach and parameters used in the design of the target regions. These sequences (Dataset S1) then were ordered from CustomArray. The code for the construction of this library was written in MATLAB and is available upon request.
Encoding Probe Construction.
Encoding probes were constructed via a high-yield, enzymatic amplification protocol published previously (18, 23), with a few notable differences to account for the use of the three-letter readout sequences. Briefly, we created in vitro transcription templates from the complex oligo pool using limited-cycle PCR and then amplified RNA from these templates using a high-yield in vitro transcription kit (E2050S; New England Biolabs). To account for the disproportionate use of C in these sequences, we added additional CTP (R0451; Thermo Fisher) to bring the final concentration to 16.7 mM; the concentrations of ATP, GTP, and UTP were each 10 mM. We then transcribed ssDNA probes from these RNA templates via reverse transcription (Maxima RT H) (EP0752; Thermo Fisher). To address the additional requirement for G in this reaction, we doubled the concentration of all dNTPs to 3 mM. The RNA template was removed via alkaline hydrolysis, the sample was neutralized with 1 N HCl, and the DNA probes were purified through phenol-chloroform extraction and two rounds of ethanol precipitation with ammonium acetate. The final probes were resuspended in RNase-free water and stored at −20 °C.
Readout Probe Construction.
Readout probes complementary to the readout sequences on the encoding probes and conjugated to the desired dye via a disulfide linkage were synthesized and purified by Bio-synthesis, Inc. Lyophilized probes were resuspended immediately in Tris-EDTA (TE) buffer, pH 8 (AM9849; Thermo Fisher) to a concentration of 100 µM to prevent degradation of the fluorophore linkage (observed only for the Alexa750-linked readout probes) and were stored at −20 °C. To reduce the number of freeze–thaw cycles experienced by these probes, 1-µM aliquots were made in TE buffer and stored at −20 °C.
High-Throughput Imaging Platform.
Samples were imaged on a custom-built, high-throughput imaging platform. Briefly, the system was constructed around an Olympus IX71 microscopy body. Illumination was provided at 754, 647, 561, and 405 nm with solid-state lasers (DL100/BoosTA, Toptica; F-04306-113, MBP Communications; GCL-150561, CrystaLaser; Cube 405, Coherent). These laser lines were used to excite Alexa750- and Cy5-labeled readout probes, orange fiducial beads, and DAPI, respectively. The 647-, 561-, and 405-nm lasers were collimated with custom three-lens, 0.4×-to-3× zoom systems, combined via a series of long-pass filters (z561bcm-xr, Chroma; LM01-503, Semrock; BLP01-405R, Semrock), and then coupled into a single-mode fiber (S405-XP; Thorlabs) to purify each mode. The output of this fiber and that of the single-mode fiber-coupled, 754-nm laser were each collimated with a 60-mm achromat and combined with a long-pass filter (Semrock; FF735-Di02). The sizes of these collimated beams were adjusted to 6.2 mm using a pair of custom three-lens, 0.5×-to-2× zoom systems (one for the output of the 754-nm laser fiber and the other for all other beams). The Gaussian distribution of these beams was then converted to a round, flat-top distribution using a refractive beam shaper (piShaper 6_6; AdlOptica). This distribution was focused onto a pair of galvanometer mirrors (GVS201; Thorlabs) and then relayed to the back-focal plane of a 300-mm achromat, which focused this illumination onto the back-focal plane of a 60×, Plan Apo, 1.3 NA, silicone oil objective (UPLSAPO 60XS2; Olympus). The fluorescence emission from the sample was separated from the laser illumination using a penta-band dichroic (zy405/488/561/647/752RP-UF1; Chroma). Stray laser light was further removed with two copies of a custom notch filter (ZET405/488/561/647-656/752m; Chroma), and the fluorescent signal was imaged with a sCMOS camera (Zyla 4.2; Andor). The sample was positioned with a motorized microscope stage (SCAN IM 112 × 74; Marzhauser), and the focus was maintained via a custom-built autofocus system that uses an objective nanopositioner (NanoF200; Mad City Labs) to maintain the position of a reflected IR laser (LP980-SF15; Thorlabs) on an inexpensive CMOS camera (uc480; Thorlabs). The sCMOS camera pixel size, 109.2 nm, was calibrated by imaging fields of fluorescent beads moved in defined increments with the motorized stage.
The sample coverslip was housed in a flow chamber (FCS2; Bioptechs), and the flow through this chamber was controlled via a home-built fluidics system composed of three computer-controlled eight-way valves (MVP and HVXM 8-5; Hamilton) and a computer-controlled peristaltic pump (MINIPULS 3; Gilson) as described previously (18, 23). The entire system was fully automated, so that imaging and fluid handling were performed for the entire experiment without user intervention, using home-built software that is available upon request.
Encoding Probe Staining.
U-2 OS cells (ATCC) were cultured with Eagle’s Minimum Essential Medium (30-2003; ATCC) containing 10% (vol/vol) FBS (10437; Thermo Fisher). Cells were plated on 40-mm-diameter, no.1.5 coverslips (0420-0323-2; Bioptechs) at 300,000 cells per coverslip and were incubated in Petri dishes at 37 °C with 5% CO2 for 48–72 h. Cells were fixed, permeabilized, and stained with encoding probes as described previously (18, 23). Briefly, cells were fixed for 20 min in 4% paraformaldehyde (15714; Electron Microscopy Sciences) in 1× PBS at room temperature, washed three times with 1× PBS, permeabilized for 10 min with 0.5% (vol/vol) Triton X-100 (T8787; Sigma) in 1× PBS at room temperature, and washed three times with 1× PBS. Permeabilized cells were incubated for 5 min in encoding wash buffer comprising 2× SSC (AM9763; Ambion), 30% (vol/vol) formamide (AM9342; Ambion), and 2 mM vanadyl ribonucleoside complex (VRC) (S1402S; New England Biolabs). Then 30 μL of ∼200 μM encoding probes (the final concentration was titrated for each probe batch) in encoding hybridization buffer was added to a glass microscope slide (22-265446; Fisher Scientific) and was covered with a cell-containing coverslip. Samples then were incubated in a humid chamber inside a hybridization oven at 37 °C for 36–48 h. Encoding hybridization buffer is composed of encoding wash buffer supplemented with 0.1% (wt/vol) yeast tRNA (15401-011; Life Technologies), 1% (vol/vol) murine RNase inhibitor (M0314L; New England Biolabs), and 10% (wt/vol) dextran sulfate (D8906-50G; Sigma). Cells then were washed with encoding wash buffer and were incubated at 47 °C for 30 min; this washing was repeated once. The cells were stained with DAPI (D1306; Thermo Fisher) during the second washing by adding 10 μg/mL DAPI to the encoding wash buffer. The sample then was incubated for 10 min with a 1:200,000 dilution of 0.1-μm-diameter carboxylate-modified orange fluorescent beads (F-8800; Life Technologies). The bead solution was aspirated, and the sample was postfixed with 4% (vol/vol) paraformaldehyde in 1× PBS at room temperature for 10 min. The sample was washed three times with 2× SSC and was either imaged immediately or stored for no longer than 24 h at 4 °C in 2× SSC containing 0.1% (vol/vol) murine RNase inhibitor. All solutions were prepared as RNase-free. The beads were used as fiducial markers to align images obtained from successive rounds of hybridization.
IMR-90 cells (ATCC) were prepared using the protocols as described above for the U-2 OS cells but without the DAPI staining.
MERFISH Imaging.
Each readout hybridization mixture contained 1 nM each of two different readout probes, one conjugated to Cy5 and the other to Alexa750 via a disulfide bond, in readout hybridization buffer comprising 2× SSC, 1% (vol/vol) ethylene carbonate (E26258; Sigma-Aldrich), 10% (wt/vol) dextran sulfate, and 2 mM VRC. The sample chamber was initially flushed with 2 mL of this readout hybridization mixture over the span of 5 min to exchange buffers fully. Then an additional 2 mL of this readout mixture was flowed continuously across the sample for an additional 6 min. This total incubation time was several times that required to saturate binding (Fig. S3) to reduce round-to-round and experiment-to-experiment variation in hybridization. The sample then was washed by flowing 2 mL of readout wash buffer [2× SSC, 10% (vol/vol) ethylene carbonate, and 2 mM VRC complex] for 9 min. Then 2 mL of imaging buffer comprising 2× SSC, 50 mM Tris⋅HCl (pH 8), 10% (wt/vol) glucose, 2 mM Trolox (238813; Sigma-Aldrich), 0.5 mg/mL glucose oxidase (G2133; Sigma-Aldrich), 40 μg/mL catalase (C30; Sigma-Aldrich), and 50 units/mL murine RNase inhibitor was flowed across the sample for 4 min. Flow was stopped, and ∼500 to ∼1,000 FOVs were imaged. Because the imaging buffer is sensitive to oxygen, it was stored under a layer of mineral oil (330779; Sigma-Aldrich) throughout the measurement (18, 23). Stock solutions of 50% (vol/vol) ethylene carbonate were made by melting solid ethylene carbonate in a water bath at 65 °C and followed by dilution to 50% (vol/vol) with RNase-free water.
After imaging, the fluorescence of the readout probes was extinguished via reductive cleavage using TCEP. Two microliters of cleavage buffer comprising 2× SSC and 50 mM TCEP hydrochloride (646547; Sigma) was flowed across the sample for 4 min, the flow speed was reduced to 0.1 mL/min, and the sample was incubated in this continuous flow for 15 min. After cleavage, the chamber was flushed with 2 mL of 2× SSC for 4 min to remove the risk of premature cleavage of the probes within the subsequent hybridization buffer. All buffers were freshly prepared for each experiment.
The above hybridization, imaging, and chemical cleavage process was repeated eight times with the 488-nm and 405-nm channels imaged in conjunction with the first round of readout imaging. A complete MERFISH measurement of ∼500 FOVs covering 19.8 mm2 required 12 h, and measurement of ∼1,000 FOVs covering 40.8 mm2 required 18 h.
smFISH.
smFISH stains were prepared at 1-µM probe concentrations following the procedures described above in the Encoding Probe Staining section. Probes for smFISH on U-2 OS cells were designed using the same target regions selected for MERFISH measurements and were synthesized conjugated to Quasar760 (Stellaris; Biosearch Technologies). Probes for smFISH on IMR-90 cells (ATCC) were generated using the target regions published previously for the FLNA mRNA (18) conjugated to multiple readout sequences, drawn either from the previously published sequences (18, 23) or from those provided in Table S1. These probes were synthesized by Biosearch Technologies.
Image Registration and Decoding.
Registration of images of the same FOV in different rounds of hybridization was performed as described previously (18, 23). Briefly, the centroids of individual beads within the fiducial bead images collected for each FOV in each round of imaging were found using the multi-emitter fitting routine 3D-DAOSTORM (37) and were used to align images of the different rounds of hybridization using an affine transformation (nonreflective similarity) that corrected for translation, rotation, and a uniform coordinate scaling. In practice, we found that only an X and Y translation was required and that calculated transforms that contained a nonzero rotation or a nonuniform scaling were indicative of a rare registration failure. These affine transformations then were used to warp each image to the same coordinate system using linear interpolation. We found no systematic offsets between the centroid of spots in the Cy5 and Alexa750 channels and thus did not perform any additional chromatic warping.
Warped images were saved as tiff stacks, one per FOV, with the frames in the order of the bits in the barcodes that they represented. These stacks then were preprocessed to remove background and to resolve overlapping fluorescent spots better. Specifically, we used a high-pass filter comprised of a Gaussian filter with a kernel size of three pixels to remove background. This kernel was slightly larger than the point-spread function (PSF) of the system so as not to remove regions of partially overlapping RNA signals. These high-pass–filtered images were then deconvolved using Lucy–Richardson deconvolution and the estimated PSF of the system (two pixels). In practice, we found it unnecessary to modify this kernel for the slight difference in PSF size between the Cy5 and Alexa750 channels. We then low-pass filtered these images using a Gaussian kernel with a width of one pixel (∼100 nm). We found that this low-pass filter improved the quality of decoding, a result consistent with our previous observation that the spot centroids for the same RNA varied in position by ∼100 nm in different imaging rounds, possibly because of the finite cellular volume occupied by each RNA (18).
To decode these images, we first recognized a few geometric properties of this problem. First, each set of 16 normalized intensity values observed for each pixel in each FOV represents a vector in 16-dimensional space; we term this vector a “pixel vector”; second, the 140 barcodes of our 16-bit MHD4 code represent preferred directions in this space; and, finally, the set of all 16 single-bit errors generated from any of the 140 barcodes define a unique volume within this space, containing the set of all possible deviations from these barcodes that correspond to a single-bit error (or less in the case of an analog signal). The central premise of our decoding approach was that pixel vectors that fell within the volume defined by all single-bit errors from a given barcode should be associated with that barcode (we term this volume “Hamming-sphere 1,” HS1). To identify all pixel vectors that fell within one of the 140 different HS1s, we first mapped each pixel vector to the 16-dimensional unit sphere by dividing it by its magnitude, i.e., the L2-norm. All barcodes were mapped to the unit sphere in a similar fashion. Because all barcodes shared the same Hamming weight, the HS1 for each barcode was defined by the same distance; in this case the maximum Euclidean distance between a barcode and all single-bit errors was 0.5176. Thus, occupancy in this volume could be calculated simply by determining the nearest barcode for each pixel vector and thresholding on the distance to that barcode. Any pixel vector with a distance to the nearest barcode larger than 0.5176 was left unassigned.
Because this decoding approach was conducted on individual pixels and because we observed that the signal from RNAs spreads across multiple pixels, we combined adjacent pixels assigned to the same barcode into a single putative RNA. We then calculated various properties of this RNA, including its magnitude-weighted centroid, the area (in pixels) it covered, the average magnitude across all pixels, and the average pixel vector across all combined pixels. Because this approach assigned barcodes within HS1 to a given barcode, it can be thought of as applying error correction. To determine whether error correction was applied to a given RNA, we computed the distance between the average 16-dimensional pixel vector for the set of pixels associated with each RNA and a set of normalized barcodes including the barcode to which it was assigned and all barcodes generated from single-bit errors. If the nearest barcode to the average pixel vector for an RNA was one of the single-bit error barcodes, we considered the RNA decoded with error correction applied. The specific single-bit-error barcode to which it was closest defined the bit at which the error occurred. These quantities then were used to calculate the total number of RNAs decoded with or without error correction (Fig. S5A), the confidence ratio associated with the counts of each barcode (Fig. S5B), and the error rate for each type of error (1-to-0 or 0-to-1) at each bit (Fig. S5C).
In this decoding approach, differences in brightness in different imaging rounds and color channels will lead to different weights for the “1” values in each of the different bits, and these different weights, in turn, can lead to increased per-bit error rates. To remove this source of error, we developed a two-step approach to remove these brightness differences between imaging rounds. First, we applied a crude normalization by setting the 90% quantile of pixel intensities for each imaging round to 1. Second, we used this normalization to decode RNAs from 100 randomly selected FOVs as described above and then used the observed pixel vectors from these decoded RNAs to refine this normalization. Specifically, we selected all RNAs for which the barcodes read “1” in the first bit and averaged the first component of their pixel vectors (calculated as described above). This calculation produced an average intensity for a “1” in the first bit. In a similar way we then calculated this average intensity quantity for all other bits. We then calculated an overall average value by taking the average of this average intensity quantity for each bit, and we calculated the deviation from this average for each bit. We then renormalized the brightness of each imaging round based on the deviation observed for the average intensity of the corresponding bit to the overall average. Because this renormalization step will change the quality of the decoding, we iterated this second step. In practice, we found that 10 iterations were sufficient to remove any substantial variation in the intensity of “1” values in each bit.
The majority of computations were run on the Odyssey cluster supported by the Faculty of Arts and Sciences Division of Science, Research Computing Group at Harvard University. By using 32 cores and 64 GB of RAM, the complete analysis of a MERFISH dataset could be completed in 3 d. Some datasets were analyzed on a desktop server that contained two 10-core Intel Xeon E5-2680 2.8 GHz CPUs and 256 GB of RAM. Data were stored on a Synology DiskStation connected directly to this server via a high-speed switch. With this configuration, it was possible to analyze a single MERFISH dataset in 2 d using only 10 of the available cores and no more than 64 GB of RAM. This increase in speed relative to that of the Odyssey computing cluster resulted from the increased read/write speed provided by the high-speed connection between the data storage system and the server.
Cell Segmentation.
Our cell-segmentation approach in associating individual barcodes with individual cells exploited the observation that the density of RNAs dropped significantly at the edges of cells. Specifically, to generate the cell boundaries, we first created composite mosaic images in which a single FOV was flanked by its eight surrounding neighbors. We then normalized the DAPI signal within this composite to the maximum observed value, thresholded this signal, and defined the cell nuclei as contiguous sets of pixels above this threshold. We then calculated the local density of barcodes throughout each composite image by binning decoded barcodes into 2 × 2 µm bins. These binned images then were smoothed with a Gaussian filter equal in width to the bin size and resized to the original pixel size via bicubic interpolation. Segmentation boundaries for individual cells were calculated using the watershed algorithm, using an inverted image of the barcode density (so that regions of low density formed natural watershed boundaries) and with the cell nuclear regions set to zero to ensure that each watershed region contained a cell nucleus. Even though we included the eight flanking FOVs for the computation, only cells with the centroid of the nucleus within the center FOV were kept. This approach produced some segmentation errors on a small fraction of cells, which were identified and removed via a series of thresholds. First, improperly segmented cells were identified by a threshold on the total effective cytoplasm area of 3,000 µm2 and by removing cells with segmented boundaries that shared more than 10 µm of that boundary with the edge of an FOV. Second, multiple overlapping nuclei were identified via a combination of thresholds on the total nucleus size and on the ratio of the area covered by the nuclei boundary to that of a convex hull defined by the same boundary. We set the threshold of the nucleus size to 1,000 µm2 (nuclei with sizes larger than this value are considered to arise from multiple overlapping nuclei). We set the threshold of the ratio to 1.06 (nuclei with a ratio larger than this value are considered to arise from multiple overlapping nuclei because the boundaries of overlapping nuclei have regions of concavity that tend to increase this ratio). Finally, this algorithm occasionally produced two boundaries for the same cell if the nucleus happened to be shared between two FOVs. These cells were identified by finding cells for which 97% of the boundary was contained within the boundary of another cell. On average, roughly 98% of the identified cells passed the cellular boundary thresholds, and ∼90% of the resulting cells passed thresholds associated with the nuclear segmentation thresholds.
RNA-Seq.
All three replicates of RNA-seq data for U-2 OS (Gene Expression Omnibus accession no. GSM1231610) (26) were downloaded as sra files, converted to fastq files, and analyzed using the human transcriptome (hg38), the indices provided by Illumina’s iGenomes (support.illumina.com/sequencing/sequencing_software/igenome.html), Bowtie 2.2.1, TopHat 2.0.11, and Cufflinks 2.2.1 (38). The reported FPKM represents the average derived from these three replicates.
Discussion
Image-based approaches to single-cell RNA profiling, which identify RNAs via multiplexed smFISH (18, 27–31) or in situ sequencing (21, 22), can directly provide the native spatial context of individual RNAs both within cells and within the context of the culture or tissue. Recently we introduced MERFISH, which uses massively multiplexed smFISH to perform spatially resolved RNA profiling of single cells at the transcriptomic scale (18). However, the measurement throughput of these image-based approaches (i.e., the number of measured cells) has been relatively limited. Here we describe several advances in the MERFISH method that increase the throughput of this approach by two orders of magnitude: We profiled 130 RNAs across 40 mm2 of sample containing as many as 39,000 human cells in only 18 h. In total, we performed such measurements in ∼100,000 cells, generating a dataset comparable in size to those published using droplet-based single-cell sequencing approaches (14, 15). Previously, using a very similar experimental procedure but different encoding schemes, we have shown that MERFISH can be used to measure ∼1,000 RNA species in individual cells (18). Thus we anticipate that this increase in throughput could be applied to the measurement of thousands of RNAs with MERFISH.
This substantial increase in throughput should extend the range of questions that can be addressed via MERFISH. For example, we demonstrate here the ability to identify a subpopulation of cells and to use the sizeable number of cells within this subpopulation to quantify the potentially small differences in their gene-expression profiles with statistical significance. We also envision that the increase in imaging throughput reported here will be instrumental in applying MERFISH to the de novo identification of cell types in sizable volumes of tissues. Finally, we anticipate that with further optimization of the hybridization protocol, utilization of faster fluorescence signal removal protocols, incorporation of more colors per imaging round, and additional improvements in camera, optics, and light sources to increase the FOV area and reduce the imaging time further, it will be possible to increase the throughput of MERFISH further and to characterize millions of individual cells in their native culture and tissue contexts. Given that the MERFISH experimental setup is, at its core, a simple fluorescence microscope with a sensitive camera in combination with an automated fluid handling system composed of commercially available components and controlled by open-source software (18, 23), we anticipate that this technique can be readily adopted by many laboratories.
Materials and Methods
Detailed protocols for all methods used in this work can be found in SI Materials and Methods. All software is available upon request.
Human U-S OS cells (American Type Culture Collection, ATCC) or human fibroblasts (IMR-90; ATCC) were fixed, permeabilized, stained with encoding probes, and coated with fiducial beads as described previously (18, 23). MERFISH imaging was done on a custom, high-throughput imaging platform built around an Olympus IX71 body, a 60× silicone oil 1.3 NA Plan Apochromat objective (UPLSAPO 60XS2; Olympus), and an sCMOS camera (Zyla 4.2; Andor). Automated fluid handling and sequential staining with readout probes were performed as described previously (18, 23) with the notable exception that the readout hybridization and wash buffers contained ethylene carbonate (E26258; Sigma-Aldrich) instead of formamide.
We created the high-diversity encoding probes by adopting and modifying the Oligopaint approach (32) with a high-yield enzymatic amplification protocol (18, 23) and a high-speed probe-design algorithm. The targeting regions of encoding probes were designed using the human transcriptome (hg38) sequences downloaded from Ensembl, published RNA abundances (26), and a custom probe-design algorithm and computational pipeline that selects target regions based on a narrow range of melting temperature (66–76 °C), GC content (43–63%), and a series of penalties associated with the presence of short homology regions within alternative isoforms of the same gene, all other genes, and abundant noncoding RNAs. A library of template oligonucleotides for making encoding probes was ordered from CustomArray (Dataset S1). Encoding probes were amplified from this library via a high-yield protocol as described previously (18, 23) with minor adjustments to nucleotide concentrations.
We generated the sequences of readout probes randomly, with an A/T probability of 25% and a C probability of 50%, and probes with significant homology to the human transcriptome, as determined via BLAST (33), were removed. Readout probes (Table S1) were purchased from Bio-Synthesis, Inc.
Supplementary Material
Acknowledgments
We thank Alistair Boettiger, Yaron Sigal, George Emanuel, Bryan Harada, Pallav Kosuri, and Tian Lu for helpful discussions. This work was supported in part by the Howard Hughes Medical Institute (HHMI). X.Z. is an HHMI investigator.
Footnotes
Conflict of interest statement: X.Z., J.R.M., and K.H.C. are inventors on a patent applied for by Harvard University that covers the MERFISH method.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612826113/-/DCSupplemental.
References
- 1.Sandberg R. Entering the era of single-cell transcriptomics in biology and medicine. Nat Methods. 2014;11(1):22–24. doi: 10.1038/nmeth.2764. [DOI] [PubMed] [Google Scholar]
- 2.Eberwine J, Sul J-Y, Bartfai T, Kim J. The promise of single-cell sequencing. Nat Methods. 2014;11(1):25–27. doi: 10.1038/nmeth.2769. [DOI] [PubMed] [Google Scholar]
- 3.Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet. 2013;14(9):618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 4.Shalek AK, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498(7453):236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shalek AK, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510(7505):363–369. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343(6172):776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Treutlein B, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509(7500):371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hashimshony T, Feder M, Levin M, Hall BK, Yanai I. Spatiotemporal transcriptomics reveals the evolutionary history of the endoderm germ layer. Nature. 2015;519(7542):219–222. doi: 10.1038/nature13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Achim K, et al. High-throughput spatial mapping of single-cell RNA-seq data to tissue of origin. Nat Biotechnol. 2015;33(5):503–509. doi: 10.1038/nbt.3209. [DOI] [PubMed] [Google Scholar]
- 11.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33(5):495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeisel A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347(6226):1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 13.Petropoulos S, et al. Single-Cell RNA-Seq Reveals Lineage and X Chromosome Dynamics in Human Preimplantation Embryos. Cell. 2016;165(4):1012–1026. doi: 10.1016/j.cell.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein AM, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161(5):1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161(5):1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crosetto N, Bienko M, van Oudenaarden A. Spatially resolved transcriptomics and beyond. Nat Rev Genet. 2015;16(1):57–66. doi: 10.1038/nrg3832. [DOI] [PubMed] [Google Scholar]
- 17.Buxbaum AR, Haimovich G, Singer RH. In the right place at the right time: Visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol. 2015;16(2):95–109. doi: 10.1038/nrm3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348(6233):aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280(5363):585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 20.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5(10):877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ke R, et al. In situ sequencing for RNA analysis in preserved tissue and cells. Nat Methods. 2013;10(9):857–860. doi: 10.1038/nmeth.2563. [DOI] [PubMed] [Google Scholar]
- 22.Lee JH, et al. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343(6177):1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moffitt JR, Zhuang X. RNA imaging with multiplexed error-robust fluorescence in situ hybridization (MERFISH) Methods Enzymol. 2016;572:1–49. doi: 10.1016/bs.mie.2016.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z, Revyakin A, Grimm JB, Lavis LD, Tjian R. Single-molecule tracking of the transcription cycle by sub-second RNA detection. eLife. 2014;3:e01775. doi: 10.7554/eLife.01775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matthiesen SH, Hansen CM. Fast and non-toxic in situ hybridization without blocking of repetitive sequences. PLoS One. 2012;7(7):e40675. doi: 10.1371/journal.pone.0040675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walz S, et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014;511(7510):483–487. doi: 10.1038/nature13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levsky JM, Shenoy SM, Pezo RC, Singer RH. Single-cell gene expression profiling. Science. 2002;297(5582):836–840. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- 28.Lubeck E, Cai L. Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nat Methods. 2012;9(7):743–748. doi: 10.1038/nmeth.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levesque MJ, Raj A. Single-chromosome transcriptional profiling reveals chromosomal gene expression regulation. Nat Methods. 2013;10(3):246–248. doi: 10.1038/nmeth.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jakt LM, Moriwaki S, Nishikawa S. A continuum of transcriptional identities visualized by combinatorial fluorescent in situ hybridization. Development. 2013;140(1):216–225. doi: 10.1242/dev.086975. [DOI] [PubMed] [Google Scholar]
- 31.Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat Methods. 2014;11(4):360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beliveau BJ, et al. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc Natl Acad Sci USA. 2012;109(52):21301–21306. doi: 10.1073/pnas.1213818110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Camacho C, et al. BLAST+: Architecture and applications. BMC Bioinformatics. 2009;10(1):421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rouillard J-M, Zuker M, Gulari E. OligoArray 2.0: Design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 2003;31(12):3057–3062. doi: 10.1093/nar/gkg426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.SantaLucia J, Jr, Hicks D. The thermodynamics of DNA structural motifs. Annu Rev Biophys Biomol Struct. 2004;33(1):415–440. doi: 10.1146/annurev.biophys.32.110601.141800. [DOI] [PubMed] [Google Scholar]
- 36.Xu Q, Schlabach MR, Hannon GJ, Elledge SJ. Design of 240,000 orthogonal 25mer DNA barcode probes. Proc Natl Acad Sci USA. 2009;106(7):2289–2294. doi: 10.1073/pnas.0812506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Babcock H, Sigal YM, Zhuang X. A high-density 3D localization algorithm for stochastic optical reconstruction microscopy. Opt Nanoscopy. 2012;1(6):6. doi: 10.1186/2192-2853-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.